Significance
DNA methylation is an important chromatin modification that helps delineate heterochromatin and transcriptional silencing. Mutations that disrupt robust targeting of DNA methylation to heterochromatin result in ectopic DNA methylation on genes that can alter transcription. By examining variation in DNA methylation among hundreds of natural accessions of the model plant Arabidopsis thaliana, we found evidence that robust targeting of DNA methylation to heterochromatin is a trait that varies within this species. Plant genotypes that had lower levels of methylation in heterochromatin had more genes with DNA methylation, and these genes were prone to transcriptional silencing. These results reveal that epigenetic alleles can arise as a byproduct of maintaining methylation of heterochromatin-associated DNA.
Keywords: DNA methylation, heterochromatin, gene body DNA methylation, epiallele, epigenetics
Abstract
In plants and mammals, DNA methylation plays a critical role in transcriptional silencing by delineating heterochromatin from transcriptionally active euchromatin. A homeostatic balance between heterochromatin and euchromatin is essential to genomic stability. This is evident in many diseases and mutants for heterochromatin maintenance, which are characterized by global losses of DNA methylation coupled with localized ectopic gains of DNA methylation that alter transcription. Furthermore, we have shown that genome-wide methylation patterns in Arabidopsis thaliana are highly stable over generations, with the exception of rare epialleles. However, the extent to which natural variation in the robustness of targeting DNA methylation to heterochromatin exists, and the phenotypic consequences of such variation, remain to be fully explored. Here we describe the finding that heterochromatin and genic DNA methylation are highly variable among 725 A. thaliana accessions. We found that genic DNA methylation is inversely correlated with that in heterochromatin, suggesting that certain methylation pathway(s) may be redirected to genes upon the loss of heterochromatin. This redistribution likely involves a feedback loop involving the DNA methyltransferase, CHROMOMETHYLASE 3 (CMT3), H3K9me2, and histone turnover, as highly expressed, long genes with a high density of CMT3-preferred CWG sites are more likely to be methylated. Importantly, although the presence of CG methylation in genes alone may not affect transcription, genes containing CG methylation are more likely to become methylated at non-CG sites and silenced. These findings are consistent with the hypothesis that natural variation in DNA methylation homeostasis may underlie the evolution of epialleles that alter phenotypes.
Heterochromatin is abundant in eukaryotic genomes, and it is important for the transcriptional silencing of repeats and transposons (TEs), as well as for centromere function (1). Heterochromatin is composed of more tightly condensed chromatin compared to euchromatin. At the molecular level, heterochromatin is typically demarcated by Histone H3 lysine 9 methylation (H3K9me), and there are a variety of enzymes that “read” this covalent histone modification to help establish and maintain heterochromatin (2–5). In plant and mammalian genomes, heterochromatin is associated with an additional chromatin modification, DNA cytosine methylation, which, in plants, is established in a feedback loop with H3K9 methylation (5–13). Loss of maintenance of heterochromatin leads to genome instability, resulting in numerous phenotypic consequences. In humans, disease progression in many cancer types is characterized by global losses of DNA methylation, accompanied by ectopic gains of DNA methylation in CpG islands that may silence transcription of tumor suppressors (14). Similarly, in the model plant, Arabidopsis thaliana, mutations in the nucleosome remodeler, DECREASE IN DNA METHYLATION 1 (DDM1), result in a global decrease in DNA methylation in heterochromatin and a redistribution of DNA methylation to genes that results in pleiotropic developmental defects (15, 16). Therefore, it is critical to maintain chromatin homeostasis, or proper targeting of chromatin modifications that demarcate heterochromatin from euchromatin, through both mitotic and meiotic cell divisions. Consistent with this, methylated regions were found to be faithfully propagated genome-wide over multiple generations in A. thaliana with 99.998% accuracy (17). Yet, despite this demonstrated stability in an individual accession of A. thaliana, DNA methylation has been widely characterized both within and between plant species and found to be highly variable in genome-wide levels and distribution (18–20). This variation is due, at least in part, to genetic variation in genes encoding the machinery responsible for the maintenance and targeting of DNA methylation (19–24). As a whole, however, epigenetic diversity may be more generally conceptualized as consequence of natural, population-level variation in chromatin homeostasis, the extent, causes, and phenotypic consequences of which remain to be fully explored.
Most angiosperm (flowering) plants studied to date encode multiple functionally distinct DNA methyltransferase enzymes that combine methylate cytosines in all sequence contexts, including CG, CHG, and CHH (H = A, C, or T) (8, 9, 20, 23). Methylation in all contexts is characteristic of silenced repeats in constitutive heterochromatin (25, 26). Protein-coding genes located in euchromatin can be characterized by DNA methylation as well, although the patterns of methylation can vary. Many constitutively expressed genes in flowering plant genomes are characterized by strictly CG context methylation, which is referred to as gene body methylation (gbM) (27). Genes can also be characterized by TE-like methylation (teM) that occurs in multiple sequence contexts, and, when teM is also associated with H3K9me2, teM genes are generally transcriptionally silent. Finally, many genes in flowering plant genomes are essentially devoid of DNA methylation and thus referred to as unmethylated (UM; reviewed in ref. 28).
From an evolutionary standpoint, gbM has garnered much interest, as it is a feature of not just flowering plants, but many animal species as well, suggesting it may represent an ancestral state conserved over long periods of evolutionary time (29–32). Curiously, it is absent from fungal genomes that contain cytosine DNA methylation (33). In general, although there is accumulation of methylation variation at the level of individual cytosines within genes, gbM is faithfully inherited over generational timescales (17, 34, 35). Furthermore, among flowering plant species, gbM is often found on conserved orthologous genes between species (20, 24, 36–41). GbM is also consistently associated with genes with characteristic features including constitutive, but moderate, expression levels and reduced expression variance between species relative to UM genes (21, 25, 42–44). The correlations of gbM with higher but less variable levels of expression have been interpreted as evidence that gbM is functionally implicated in gene expression and maintained in populations by natural selection (29, 42, 45). However, numerous studies of experimentally induced and natural losses of gbM have found no evidence for expression changes directly associated with the loss of gbM, suggesting that the correlation between gbM and expression is not a functional relationship (19, 20, 22, 34, 37, 40, 43, 44). Also, although the presence of gbM on some conserved orthologous genes between species is well established, there is wide variation in the percentage of orthologous genes characterized by gbM, both within and between species, with species differences ranging from ∼0 to 60% of all genes characterized by gbM in a given species (20).
Mechanistic studies that have identified the molecular processes that underlie the establishment of gbM have provided evidence for an alternative hypothesis for the conservation of gbM, which does not invoke a conserved, universal function (22, 23, 46–51). The establishment of gbM requires the activity of the DNA methyltransferase CHROMOMETHYLASE 3 (CMT3) (22, 24, 28, 46–48). CMT3 preferentially methylates cytosines in the CHG sequence context (with a preference for CWG relative to CCG; W = A or T) in a self-reinforcing feedback loop with H3K9me2 to maintain constitutive heterochromatin (6, 12, 52–55). CMT3 physically binds H3K9me2 to activate methyltransferase activity on nearby cytosines, whereas histone methyltransferases (HMTs) can bind DNA methylation to methylate nearby histones (10, 12, 13, 52, 56). In addition to heterochromatin, evidence suggests that CMT3 and HMTs are localized to transcribed genes with gbM in euchromatin, where they may transiently establish DNA methylation and H3K9me2 that is characteristic of heterochromatin (46, 51). This activity can recruit additional methyltransferases that methylate cytosines in CG and CHH contexts as well (46, 51). However, heterochromatin and transcriptional silencing are not ultimately established at these genic loci due to the activity of the H3K9 de-methylase, INCREASED BONSAI METHYLATION 1 (IBM1), which removes H3K9me2 in a cotranscriptional process (51, 57, 58). IBM1 activity disrupts the feedback loop between CMT3 and H3K9me2, and DNA methylation in all sequence contexts is lost passively following DNA replication except at CG sites, which is characteristic of gbM (46). CG methylation is maintained due to the preferential activity of the CG maintenance methyltransferase METHYLTRANSFERASE 1 (MET1) for hemimethylated CG sites, following DNA replication, to which it is recruited to methylate the complementary strand (59–62).
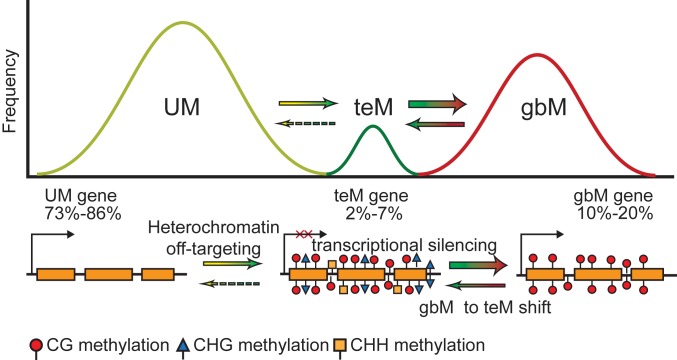
The mechanism of gbM establishment is consistent with the possibility that gbM may be a passive byproduct related to perturbations to or constraints on chromatin homeostasis that promote the transient off-targeting of the heterochromatin machinery to genes. Indeed, variation of levels of gbM within and between species has been correlated with genome-wide levels of CMT3-preferred CHG methylation, and, in A. thaliana, gbM genes have been found on average to be localized closer to dense, pericentromeric heterochromatin relative to UM genes (19, 20, 42). GbM genes also tend to be longer and have a higher frequency of CMT3-preferred CWG context cytosines relative to UM genes, which may increase the probability of CMT3 activity (22, 46). Thus, a model has emerged that posits that gbM is a byproduct resulting from machinery that facilitates heterochromatin formation. The presence of molecular pathways that uncouple DNA methylation from H3K9me2 at PolII transcribed loci, such as IBM1, reduce deleterious consequences of gbM by preventing transcriptional silencing, such that gbM is not eliminated from populations via selection, but rather maintained passively by maintenance methyltransferases. Under this model, the various patterns of DNA methylation that characterize genes (UM, gbM, or teM) are on a continuous spectrum, such that factors that influence the balance between pathways that promote or remove methylation dictate the methylation state of a given gene (Fig. 1) (19, 48, 63).
Fig. 1.
Genic DNA methylation patterns represent a continuum of epigenetic states. In A. thaliana, most genes located within euchromatin lack DNA methylation (UM). Some genes are prone to off-targeting by the heterochromatin machinery, which can result in transposable element-like DNA methylation (teM), characterized by cytosine methylation in all sequence contexts, CG, CHG, and CHH (H = A, C, or T), and transcriptional silencing. To combat the negative consequences of off-targeting of DNA methylation to genes, cells encode pathways that disrupt heterochromatin formation at genes by promoting the loss of non-CG methylation. This allows CG methylation to be maintained passively by maintenance methyltransferases, as CG methylation alone is inconsequential for transcription, which results in the strictly CG methylation pattern characteristic of gene body methylation (gbM). gbM then increases the probability of a gbM-to-teM shift due to feedback loops associated with DNA and histone methylation. The proportion of genes characterized by each methylation state within an individual vary due to factors that influence homeostatic targeting of DNA methylation. Percentages indicate the variation in the proportion of genes classified with each methylation pattern across 725 A. thaliana accessions.
We conducted a comprehensive within-species analysis of gbM in 725 natural A. thaliana accessions derived from across the globe to explore the relationship between the maintenance of heterochromatin DNA methylation and gbM (19). We found that the number of genes characterized by gbM varied widely in these accessions, ranging from ∼9 to 20% of all genes, and that some of the variation in the number of gbM genes likely has a genetic basis. We categorized genes based on the conservation of gbM status within A. thaliana and found that genic features associated with gbM genes relative to UM genes, including expression characteristics, gene length, and CWG frequency, among others, were generally correlated with gbM status conservation. Using machine learning, we created a model that could utilize genic features alone, independent of methylation data, to accurately predict gbM status, with gene length and CWG frequency being the most important predictors. Importantly, we also experimentally demonstrated that genes with conserved gbM status in A. thaliana are preferentially methylated by CMT3 relative to other genes and that CMT3 is further biased toward genes with preexisting gbM. Finally, to explore possible phenotypic implications of gbM, we examined the epiallelic states of gbM genes within these accessions. Although the majority alternate epiallelic state of gbM genes was UM, the most conserved gbM genes were more likely than other gbM genes to exist as polyepialleles in the form of teM in these accessions, and epiallelic shifts to teM were at times associated with transcriptional silencing. All together, we interpret these findings to be consistent with a passive model for the conservation of gbM as a trait across angiosperm species, arising as a byproduct of population-level variation in DNA methylation homeostasis (Fig. 1). We further posit that the evolutionary relevance of gbM derives from the predisposition of gbM loci to epiallelic shifts from gbM to teM, which affects transcription and could lead to phenotypic consequences.
Results
Distribution of Genic DNA Methylation Patterns within A. thaliana.
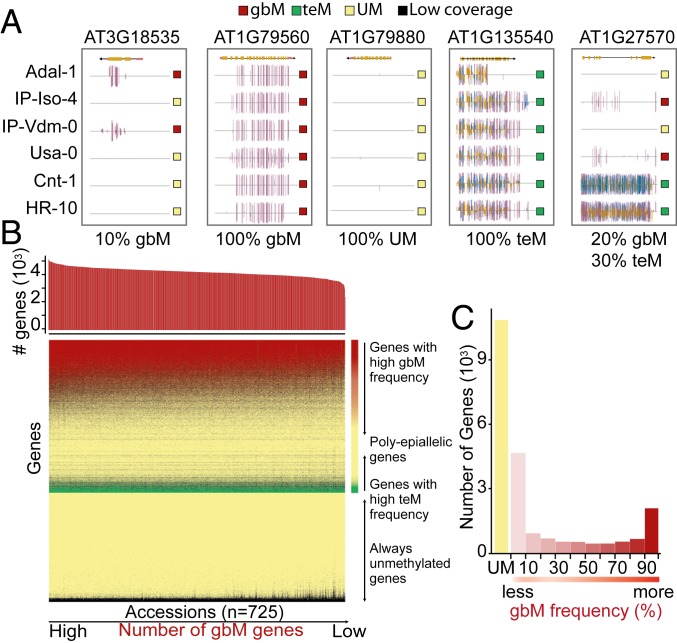
To gain insights into the within-species level variation in gbM, we first classified all genes according to three genic DNA methylation patterns across 725 natural accessions of A. thaliana: (i) gbM (strictly CG context methylation), (ii) teM (TE-like methylation, or methylation in CHH, CHG, or multiple sequence contexts), and (iii) UM (unmethylated; Dataset S1). Methylation patterns were defined based on cytosine methylation in exons only (Materials and Methods). We found that, similar to prior results (19), the methylation pattern of many genes was highly conserved across accessions, whereas a subset of genes showed variability in their methylation pattern within the population (termed polyepialleles; Dataset S1; Fig. 2A shows representative genome browser views of genes with various epiallelic classes). On average, in a given accession, ∼79.2% of genes are classified as UM, ∼3.6% genes are classified as teM, and ∼17.2% are classified as gbM (Fig. 2B and Dataset S2). However, the number of genes in each category in a given accession showed a wide range of variability across accessions. The number of UM genes ranged from 18,488 to 21,605 (68.9 to 80.5% of all genes), the number of teM genes ranged from 593 to 1,668 (2.2 to 6.2% of all genes), and the number of gbM genes ranged from 2,428 to 5,221 (9.1 to 19.5% of all genes; Dataset S2). Thus, although the presence of gbM as a genic methylation pattern is conserved across species (20, 41), there is considerable variation in the proportion of genes classified as gbM within a single species, similar to results found between species (20, 24).
Fig. 2.
Distribution of genic DNA methylation patterns within A. thaliana. (A) Genome browser views demonstrating the three genic DNA methylation patterns and distributions within A. thaliana, represented by six accessions. Genic methylation patterns detected include gene body methylation (gbM), which is restricted to mCG only (denoted by a red square); transposable element-like methylation (teM), where methylation is detected in sequence contexts other than or in addition to mCG (mCG and/or mCHG and/or mCHH; denoted by a green square); and unmethylated genes (UM; denoted by a yellow square). The methylation pattern of a given gene can vary within the 725 A. thaliana accessions, with some genes predominantly UM, whereas others are rarely gbM in some individuals (first panel). Some genes are predominantly gbM (second panel). Some genes are always UM (third panel). Some genes are always teM (fourth panel). Finally, some genes are variable between all methylation patterns (fifth panel). Within the browser views, mCG is denoted by red lines, mCHG by blue lines, and mCHH by yellow lines. (B) Classification of genes based on DNA methylation patterns within A. thaliana. All annotated genes are listed on the y axis of the heat map, ordered by the percentage of accessions in which the genes were classified as a given DNA methylation pattern. Genes were classified as UM (yellow), gbM (red), and teM (green) based on a binominal test. Black indicates genes that were not classified due to low coverage. Accessions are arranged on the x-axis by the number of gbM genes identified, denoted in the histogram at the top. (C) Number of genes classified as gbM within A. thaliana. Genes identified as gbM in at least one accession were categorized into 10 groups based on gbM frequency, or the percentage of accessions a given gene was classified as gbM. The histogram shows the number of genes classified in each category, and UM genes are also shown for comparison.
We next classified all gbM genes according to their epiallele frequency, or the proportion of the population in which a given gene was classified as gbM. Within the accessions analyzed, more than one third of all coding genes (10,940 of 26,834 total genes) were classified as gbM in at least one accession (Dataset S3). For the remaining analyses, we refer to these genes as gbM genes. Of these gbM genes, approximately one fifth (n = 2,078) were found to be highly conserved and classified as gbM in more than 90% of the accessions analyzed (hereafter referred to as “core” gbM genes; Fig. 2C and Dataset S3). In contrast, approximately half of the gbM genes (n = 4,607) were rare events, as they had a gbM frequency of less than 10% (Fig. 2C and Dataset S3). The remaining gbM genes were fairly evenly distributed across the frequencies between 10 to 90% (Fig. 2C and Dataset S3). In comparison, 10,852 of 26,834 total genes were categorized as UM across accessions and 4,712 of 26,834 total genes were classified as teM genes (Dataset S3). For the remainder of the study, this highly conserved set of unmethylated genes is referred to as the UM genes and utilized for comparison to the gbM genes described here earlier. The teM genes were excluded from further analyses that focused on gbM genes, since the major alternative epiallelic status of gbM genes is UM and not teM (Dataset S4). The remaining genes were not classified due to missing data or low coverage.
Genetic and Epigenomic Features Associated with gbM Gene Number.
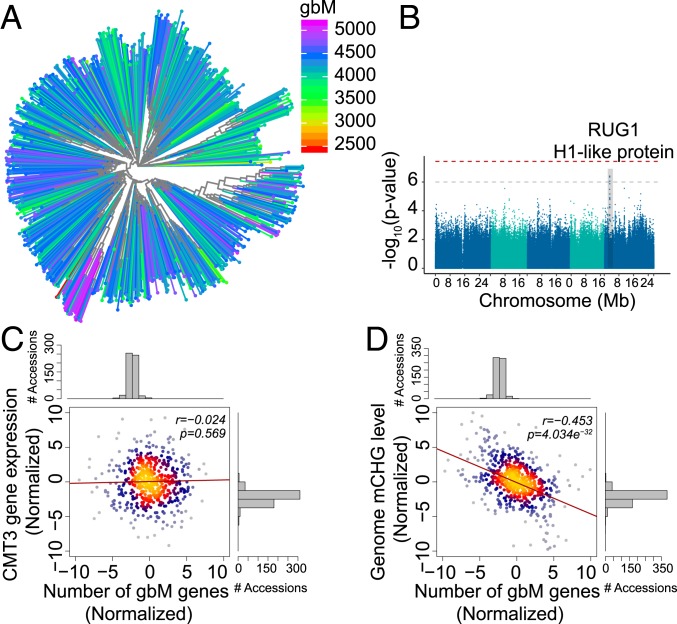
There is a wide variation in the number of gbM genes across accessions (Fig. 2B). Thus, we considered number of gbM genes as a trait and sought to determine whether there was a genetic basis for this variation. First, we performed an SNP-based heritability analysis, which determined that genetic variance explained 10.5% of the variation in gbM gene number. Similarly, mapping the number of gbM genes onto a phylogenetic tree was consistent with gbM gene number being affected by genetic background (Fig. 3A). We next conducted a genome-wide association analysis utilizing gbM gene number as a trait, but found no clear association signal (SI Appendix, Fig. S1). This is likely indicative that gbM gene number is a multifactorial trait, and the influence of any one genetic factor could vary widely depending on the genetic background. To address this possibility, we refined the analysis to a subset of 198 accessions with a similar mCG level (0.22 ± 0.01), since the number of gbM genes is positively correlated with a gene’s mCG level (19). Within this subpopulation, we identified a QTL (18) on chromosome 5, nucleotides 2,811,722 to 2,890,303 (Fig. 3B). The association signal reveals a clear peak that passes the threshold of 1e−6. Candidate genes in the peak region include RUG1 (AT5G08710), a putative regulator of chromosome condensation and an RCC1 family protein (the top SNP is found in the intron, chr5, 2,835,403; −log10 P = 6.40) (64, 65), and a linker histone H1 and H5 family protein (AT5G08780; the top SNP is a missense variant, Lys to Asn, chr5, 2857876; −log10 P = 5.23) (66). Intriguingly, similar, but not identical, genes encoding an RCC1 family protein and a histone H1 family protein were identified in GWAS that considered levels of gbM rather than number of gbM genes (21).
Fig. 3.
Genetic and epigenetic factors associated with gbM gene number. (A) Close genetic relatives have similar gbM gene numbers within A. thaliana. The number of gbM genes, indicated by a color gradient, is projected on a neighbor-joining tree of 725 A. thaliana accessions. (B) Manhattan plots of GWAS of gbM gene number using the CMLM model in the GAPIT package in R. The horizontal gray line indicates a threshold of 1e−6, and the red line indicates genome-wide threshold P = 0.05 with a Bonferroni correction. The candidates in the peak region include RUG1 (AT5G08710), regulator of chromosome condensation RCC1 family protein (the top SNP is found in the intron, chr5, 2,835,403, −log10 P = 6.40), and a linker histone H1 and H5 family protein (AT5G08780; the top SNP is a missense variant from Lys to Asn, chr5, 2857876, −log10 P = 5.23). (C) The correlation between the number of gbM genes and CMT3 gene expression levels. The density of samples is represented by brighter colors on the scatter plot. (D) The correlation between the number of gbM genes and the genome-wide mCHG levels estimated by mapping bisulfite sequenced reads to the Col-0 reference genome.
Because gbM status and the number of CWG sites within a gene are correlated (22), we also considered that genetic variation resulting in changes in CWG frequency could influence gbM gene number across accessions. However, we found no relationship between the total number of CWG sites within genes and the gbM gene number across accessions (SI Appendix, Fig. S2A). Another possible cause of variation in gbM gene number is the expression level of CMT3, which is thought to facilitate the origins of gbM (22, 28, 46). However, we found little correlation between CMT3 expression and gbM gene number across natural accessions of A. thaliana (Fig. 3C). Correspondingly, variation of CMT3 expression also showed little correlation with whole-genome CHG methylation levels (Dataset S5 and SI Appendix, Fig. S2B). These results may be consistent with the possibility that, given a uniform genetic background, altering expression of CMT3 could influence gbM gene number and CHG methylation levels, but there are likely many other factors influencing this trait at the population level.
Finally, we considered the possibility that there may be a relationship between whole-genome CHG methylation and gbM gene number. This was tested using phylogenetically independent contrasts (PICs), a method that removes the phylogenetic relationships of accessions to account for genetic relatedness. Comparing genome-wide CHG levels and gbM gene numbers across accessions revealed that these two traits are negatively correlated (Pearson correlation = 0.453; P = 4.034e−32; Fig. 3D). Importantly, the significance of this correlation remained whether whole-genome CHG methylation levels were estimated from reads mapped to the reference genome or independently of mapping, suggesting that this variation was not significantly influenced by variation in repetitive regions between accessions (SI Appendix, Fig. S2 C and D). Since the majority of CHG methylation co-occurs with H3K9me2 in dense pericentromeric heterochromatin (11), and if we assume that gbM is an indirect readout of CMT3 activity on genes, as is suggested by other studies (22, 28, 46), this result is consistent with the conclusion that there is a tradeoff between CMT3 activity in heterochromatin and genic loci. This tradeoff is not necessarily related to the intrinsic enzymatic activity of CMT3, but rather the robustness of properly targeting CMT3 to heterochromatin.
Genic Features Are Predictive of gbM Status and Correlate with Epiallele Frequency.
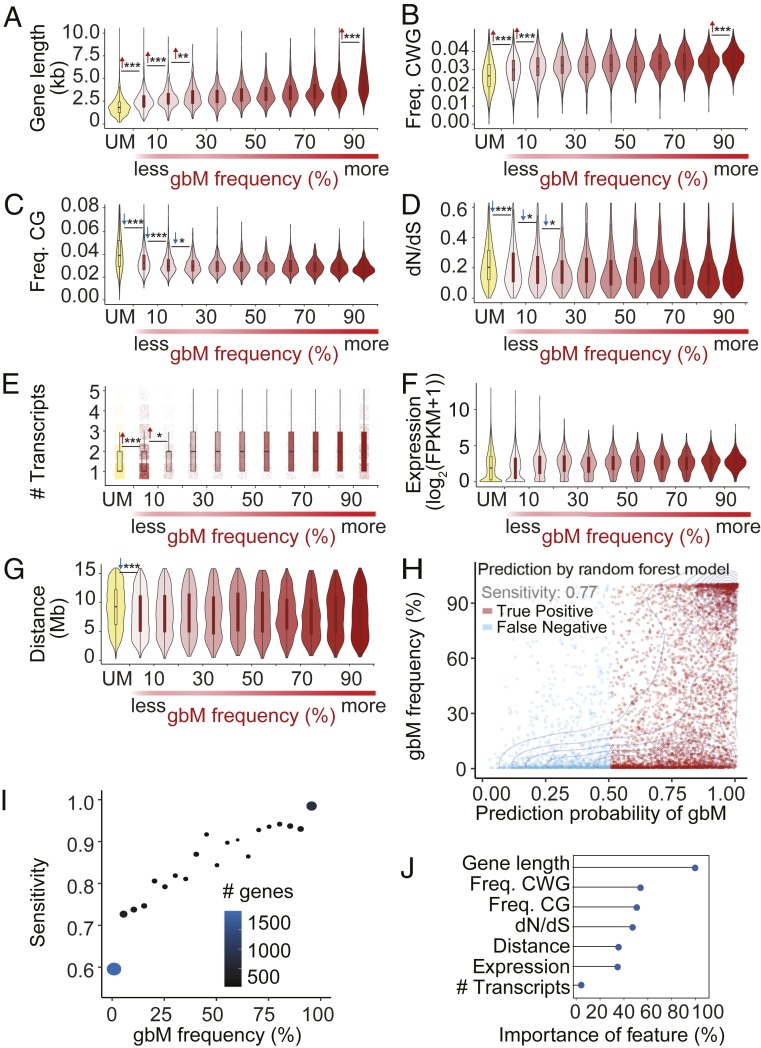
gbM genes are generally distinguished from UM genes by being characterized as having longer gene lengths; more moderate, but on average higher, expression levels; a greater number of associated transcripts; lower substitution rates (dN/dS); a higher frequency of CMT3-preferred CWG (W = A or T) context cytosines; and a lower frequency of MET1-preferred CG context cytosines (22, 41, 57, 67). Thus, we next sought to determine the relationship between these features and the epiallele frequency of gbM genes in A. thaliana and to identify the extent to which these genomic features were predictive of gbM status. The gbM genes were divided into 10 groups based on gbM frequency within A. thaliana. Next, pairwise comparisons between adjacent groups based on their gbM frequencies were completed for each genic feature, with gbM loci with the lowest gbM frequency (0 to 0.1 in the population) compared to UM genes (Fig. 4 A–F and Dataset S6). The results revealed that all genic features show general trends relative to gbM frequency. This was most pronounced with gene length and CWG frequency, which demonstrated the most significant differences in pairwise comparisons, with higher gbM frequencies associated with higher values for each feature (Fig. 4 A and B). For CG frequency, dN/dS, and transcript number, significant differences between pairwise comparisons were limited to genes with lower epiallele frequencies and UM genes (Fig. 4 C–E). Finally, for gene expression levels, no significant differences were detected between pairwise comparisons, but a general trend toward more moderate and slightly higher expression was evident as gbM frequency increased (Fig. 4F).
Fig. 4.
Genic features are predictive of gbM status and correlate with gbM frequency. (A–G) Association of seven gene features across the gbM frequency categories: (A) gene length, (B) frequency of CWGs relative to gene length, (C) frequency of CGs relative to gene length, (D) dN/dS rate, (E) transcript number, (F) gene expression level, and (G) chromosomal location relative to the centromere. The significance of differences between neighboring groups was calculated using a Welch’s two-sample t test for most features with normal distributions except for the number of transcripts category, which was tested using a Chi-square goodness-of-fit test. Significance and direction are indicated. (H) Predictability of gbM based on seven gene features. The scatter plot shows the prediction probability of a gene being classified as gbM on the x-axis based on a random forest prediction model and each gene’s empirically determined gbM frequency in A. thaliana on the y axis. Red dots show gbM genes that were successfully predicted as gbM, and blue dots show gbM genes that failed to be predicted as gbM. (I) Plot of gbM genes divided into 20 groups in 5% intervals based on their gbM frequency (x-axis) relative to the sensitivity of the random forest prediction model (the proportion of successfully predicted gbM genes; y axis) shows that genes that are classified as gbM in a higher percentage of individuals in the population are more likely to be successfully predicted as gbM based on gene features alone. Dot size and color are proportional to the number of genes in each group. (J) Importance ranking of gene features in predicting gbM status shows that gene length and frequency of CWG context cytosines are the most predictive features. ***P < 0.001, **P < 0.01, *P < 0.05.
Recent work has demonstrated that the establishment of gbM is initiated by CMT3 (22, 46), which mainly methylates cytosines in dense, pericentromeric heterochromatin (11, 13). We therefore hypothesized that location on the chromosome relative to pericentromeric heterochromatin may be related to a gene’s gbM status, as a correlation was noted previously by using methylation data from microarrays (42). We first compared the distance to the centromere of gbM genes relative to UM genes using seven A. thaliana accessions, including the reference accession, Col-0 (note that gene positions were based on the reference genome, but methylation status was identified individually in each accession). Results demonstrated that, on all or nearly all five chromosomes in each accession, gbM genes were significantly closer, on average, than UM genes to the centromere (SI Appendix, Fig. S3 and Dataset S7). A notable exception was chromosome 3, in which gbM genes were found to be significantly closer to the centromere than UM genes in only two of the seven accessions (SI Appendix, Fig. S3 and Dataset S7). Next, we examined the relationship between the distance to the centromere and the epiallele frequency of gbM genes. Similar to other genic features, a significant difference in distance to the centromere was found in the pairwise comparison between gbM genes with the lowest gbM frequency and UM genes, yet no additional significant differences were detected between the additional gbM frequency groups (Fig. 4G). These results further demonstrate that gbM is correlated with multiple genic features, which are also related to the gbM frequency of a locus in A. thaliana.
We next sought to determine whether genic properties could be utilized to predict the gbM status of a gene. To do so, we combined the 10,940 gbM genes identified in A. thaliana with the 10,852 UM genes and randomly divided the genes into two equal groups that maintain the same proportion of gbM genes and UM genes. We utilized one group as a training data set to build a machine learning binary prediction model based on the seven genic features described here earlier, which we then used to predict the gbM vs. UM status of the remaining test group (SI Appendix, Fig. S4A). We evaluated machine learning prediction models trained using 12 different machine learning algorithms. The random forest algorithm was found to have the highest prediction accuracy by cross-validation of the training data set (SI Appendix, Fig. S4B) and was chosen to predict a gene’s methylation status in the test group.
For all genes in the test group, the methylation status of ∼79% of the genes could be accurately predicted, which is greater than the 50% accuracy that would be expected by chance (Fig. 4H). The sensitivity of the model was also high, with ∼77% of all gbM genes correctly identified (Fig. 4H). The sensitivity of the model increased with increasing gbM frequency, and genes with a gbM frequency of more than 90% could be predicted with ∼97% accuracy (Fig. 4I and Dataset S8). In contrast, for genes with less than 10% gbM frequency, the prediction accuracy was ∼59%, and there was much more uncertainty associated with the prediction (the probability of random prediction for these genes was 50.2%; Fig. 4I and Dataset S8). Within the model, the various genic features differed in their importance for predicting gbM status (Materials and Methods describes how importance was determined). Most important was gene length, followed by CWG and CG frequency, which showed similar levels of importance (Fig. 4J). Next, in order of importance, were dN/dS, relative distance to the centromere, expression level, and, finally, transcript number was the least important factor (Fig. 4J). Collectively, these results show that several genomic features are correlated with gbM and frequency of gbM in A. thaliana and can be used to accurately predict the gbM status of a given gene.
Genes with High gbM Frequencies Are Preferentially Methylated by CMT3.
Given the role of CMT3 in the establishment and maintenance of gbM (22, 28, 46), we hypothesized that the genic features associated with gbM status and gbM frequency would also be predictive of CMT3 targeting. To test this hypothesis, we utilized data from previously described experimental systems that promote persistent CMT3 targeting to genes (15, 46) to determine whether CMT3 is preferentially targeted to genes with high gbM frequencies and associated genic features.
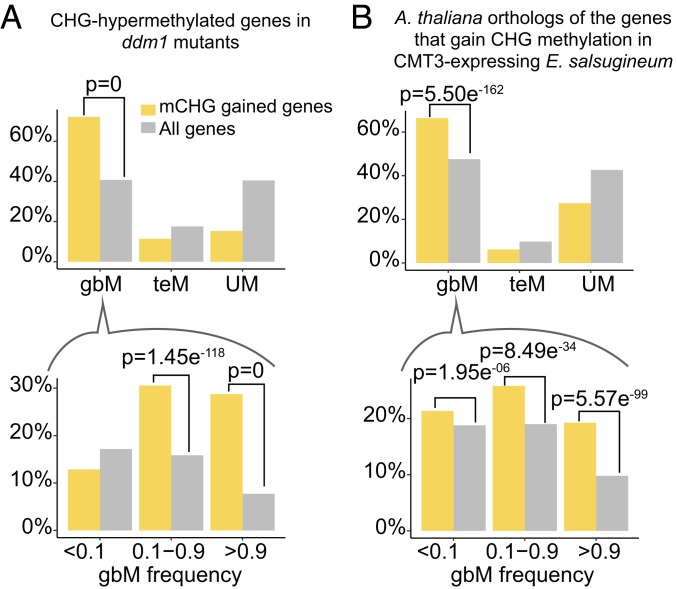
The first experimental system we examined was A. thaliana ddm1 mutants (15). DDM1 is a nucleosome remodeler, and it is required for DNA methylation on heterochromatin (68–70). Mutation of ddm1 results in a hypomethylation of heterochromatin, and a previous study discovered that propagation of ddm1 mutants for nine generations resulted in a genome-wide redistribution of DNA methylation to euchromatin (15). These results support a model whereby a reduction in the ability to maintain heterochromatin can lead to widespread ectopic off-targeting to genes of the heterochromatin maintenance machinery. This mutant phenotype is reminiscent of our finding of a negative correlation between genome-wide CHG methylation levels (a proxy for heterochromatin) and gbM gene numbers, which could be a symptom of variation in the ability to maintain homeostatic targeting of DNA methylation across accessions. Therefore, we used these ddm1 mutant data to test the hypothesis that disruption to maintenance of heterochromatin could lead to preferential CMT3-mediated ectopic CHG methylation of high-frequency gbM genes. We observed the gradual accumulation of CHG methylation on 3,372 genes over nine generations (Materials and Methods). Enrichment tests of these 3,372 genes showed that 2,435 genes are classified as gbM genes (P = 1.27 × 10−342, Fisher’s exact test; Fig. 5A and Dataset S9). Importantly, these genes are specifically enriched for genes with high gbM frequencies in the population (>0.9; P = 2.78 × 10−360, Fisher’s exact test; Fig. 5A and Dataset S9), suggesting that genic features predictive of gbM status are also predictive of CMT3 targeting.
Fig. 5.
Genes with high gbM frequencies are preferentially methylated by CMT3. (A) Distribution of CHG-hypermethylated genes in ninth-generation ddm1 mutants. (Top) Most of the CHG-gain genes in ddm1 mutants are classified as gbM genes in A. thaliana. (Bottom) Among gbM genes, CHG-gain genes are specifically enriched for genes with high gbM frequencies in the population. (B) Distribution of A. thaliana orthologs of the genes that gain CHG methylation following heterologous expression of CMT3 in E. salsugineum, which is a species that has naturally lost CMT3 and gbM. The top and bottom images show that CHG-gain genes in the E. salsugineum system are enriched for gbM genes, specifically high-frequency gbM genes, as well.
An important caveat of the ddm1 system is that gbM genes that gained CHG methylation possessed preexisting gbM, which could influence CMT3 targeting via a self-reinforcing feedback loop between cytosine methylation and H3K9me2 (12), as H3K9 methyltransferases are recruited to targets via their SRA-domain, which binds cytosine methylation (10, 13). Therefore, we also examined a second experimental system, Eutrema salsugineum, which is an angiosperm species that has naturally lost the gene encoding CMT3 and gbM, and thus provides a model to examine the targeting to CMT3 to genes independent of preexisting gbM (22, 46). Heterologous expression of CMT3 in E. salsugineum results in CHG methylation on a subset of genes, and it was previously shown that this subset of genes was significantly enriched in orthologs of gbM genes in A. thaliana, Col-0 accession (46). We further expanded this analysis and identified 4,076 A. thaliana orthologs of the genes that gain CHG methylation in CMT3-expressing E. salsugineum and examined their gbM status across all 725 A. thaliana accessions (Dataset S10). Of these, 2,706 were classified as gbM in at least one A. thaliana accession, which is significantly more than expected by chance (P = 5.50 × 10−162, Fisher’s exact test; Fig. 5B and Dataset S9). These gbM loci were also specifically enriched in the core gbM genes (P = 5.57 × 10−99, Fisher’s exact test; Fig. 5B and Dataset S9). Thus, even without prior gbM, CMT3 preferentially methylates orthologs of gbM loci in A. thaliana, with a significant enrichment found at the core gbM genes, again suggesting that there are intrinsic features of these genes that promote CMT3 targeting.
GbM Genes Are More Susceptible to Epiallelic Shifts That Lead to Transcriptional Silencing.
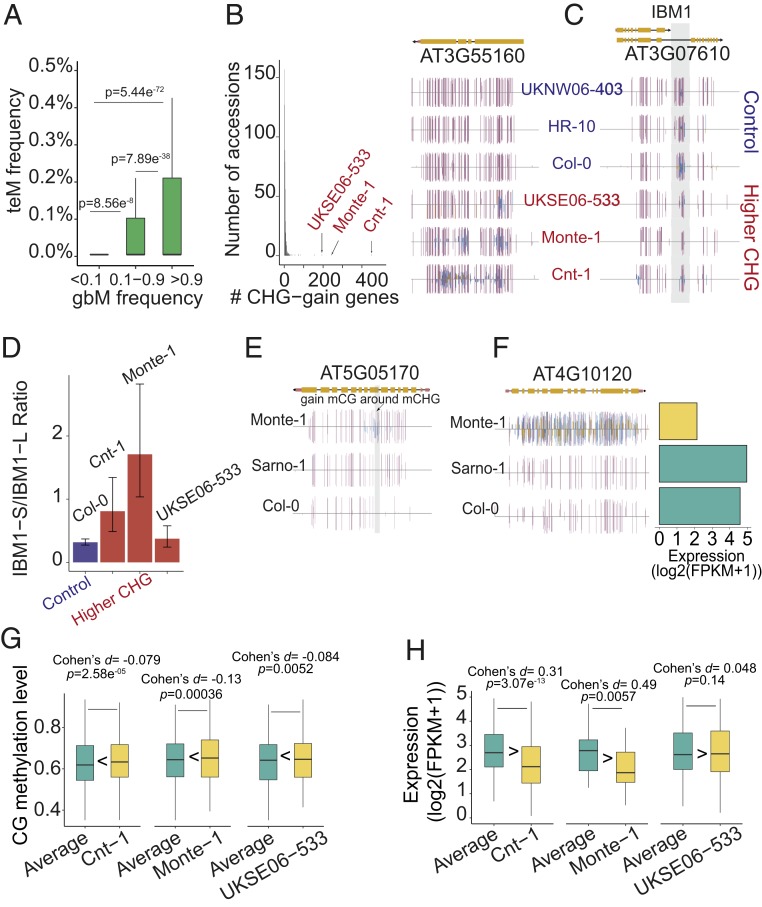
Similar to proposals by others, we hypothesized that gbM loci and teM loci might represent a continuous spectrum of chromatin states (Fig. 1) (19, 48) resulting from the balance between factors that promote or disrupt DNA methylation homeostasis (63). Factors that push this balance to one end of the spectrum or the other may therefore explain the formation of some naturally occurring epialleles, such as those previously described to be dependent on CMT3 (71–73). We reasoned that the core gbM genes, which are preferentially methylated by CMT3, may be more prone to transitions to teM status characteristic of heterochromatin, which is promoted by CMT3. Indeed, genes with the highest gbM frequency showed a significantly higher teM frequency than genes with lower gbM frequencies (Fig. 6A). In addition, the number of gbM genes and teM genes are positively correlated in the population (SI Appendix, Fig. S5A), further supporting the conclusion that gbM and teM status are on a continuous spectrum.
Fig. 6.
GbM genes are more susceptible to epiallelic shifts that lead to transcriptional silencing. (A) Box plots show the distribution of teM epialleles for gbM genes in different gbM frequency categories. Genes with the highest gbM frequency showed a significantly higher teM frequency compared to genes with lower gbM frequencies. Wilcoxon rank-sum test was used for comparisons between groups. (B) (Left) Frequency distribution of the number of CHG-gain genes within the core gbM genes (genes with ≥90% gbM frequency within the population) in accessions. The top three accessions (Cnt-1, Monte-1, and UKSE06-533) with highest CHG-gain genes are indicated with their name and an arrow. (Right) Genome browser view of a representative core gbM gene possessing high CHG methylation in Cnt-1, Monte-1, and UKSE06-533 relative to other accessions shown that do not have these patterns of methylation (UKNW06-403 and HR-10, most closely related to Cnt-1 on the phylogenetic tree) in addition to the reference accession, Col-0, as controls. (C) A genome browser view (Left) shows that accessions with high CHG methylation within core gbM genes show reduced methylation on the longest intron of IBM1, which is known to be important for proper splicing of IBM1 transcripts. Functional IBM1 is required to prevent CHG accumulation at gbM loci. (D) A qRT-PCR result shows that the three highest CHG-gain accessions tend to have a higher proportion of short IBM1 transcripts compared to functional, full-length transcripts that encode a functional protein. (E) A genome browser view shows a representative core gbM gene with additional CG methylation (highlighted with gray background) around CHG methylation in Monte-1 relative to a control (Sarno-1 is most closely related to Monte-1 on the phylogenetic tree). (F) A genome browser view shows a representative core gbM gene possessing teM status in Monte-1 and lower expression level relative to other accessions. (G) In the first panel, the distribution of CG methylation levels of genes that exist as teM genes in Cnt-1 but are otherwise categorized as gbM genes in a majority of accessions (shown in yellow bar) were compared with their pairs in a control (green bar), which were the averaged values across all accessions. Significance of the comparison was given by Wilcoxon signed-rank test, and effect size of the difference is shown as Cohen’s d. The same analysis for Monte-1 and UKSE06-533 are shown in the second and third panels, respectively. (H) In the first panel, the distribution of expression levels of genes that exist as teM genes in Cnt-1 were compared with their pairs in a control group, which were the averaged values across all accessions. The same analysis for Monte-1 and UKSE06-533 are shown in the second and third panels, respectively.
We next sought to more carefully examine examples of genes that exist as a teM gene in certain accessions but are otherwise categorized as a gbM gene in the majority of accessions (>0.9, or the core gbM genes), as these could provide clues to mechanisms underlying polyepiallelic states. We identified several accessions that possessed greater than 100 teM genes that were otherwise classified as core gbM genes across all accessions (Dataset S11). We focused on the three accessions with the highest number of transitions from core gbM to teM methylation states, Cnt-1, Monte-1, and UKSE06-533 (Fig. 6B, SI Appendix, Fig. S5B, and Dataset S11), and sought to identify possible causative factors for these epimutations. Previous studies have demonstrated that mutations in the H3K9me2 demethylase, IBM1, results in gbM-to-teM transitions that affect transcription (51). Functional expression of IBM1 requires the presence of intronic methylation, the removal of which promotes expression of a truncated, nonfunctional transcript (74). Interestingly, IBM1 intronic methylation in Cnt-1, Monte-1, and UKSE06-533 is reduced compared to Col-0, which we predicted may preclude proper accumulation of functional IBM1 transcripts in these lines (Fig. 6C). By conducting qRT-PCR to detect truncated (nonfunctional) vs. full-length (functional) IBM1 transcripts in these accessions, we found that the ratio of nonfunctional vs. functional transcripts was higher in Cnt-1 and Monte-1 compared to Col-0 (Fig. 6D). The ratio was also slightly higher in UKSE06-533, although not significantly higher. Also consistent with loss of IBM1 function in Cnt-1 and Monte-1, genes gaining CHG methylation also gained methylation in additional contexts, including CG, and demonstrated reduced expression (Fig. 6 E–H). Importantly, Cnt-1 and Monte-1 are located on different branches of the phylogenetic tree (SI Appendix, Fig. S5C), suggesting that this reduction of methylation in the intron of IBM1 happened independently in multiple lineages. Although alternative mechanisms are possible, especially in the case of UKSE06-533, these results suggest that the ectopic, non-CG DNA hypermethylation on genes in these accessions may be caused by abnormal processing of IBM1 transcripts, which promotes epiallelic transitions from gbM to teM preferentially on core gbM genes.
Discussion
The comprehensive analysis of gbM in A. thaliana revealed gbM to be a highly variable trait in this species. We found that, within the accessions tested, 10,940 of 26,834 total genes (∼41% of coding genes) were classified as gbM in at least one accession, yet only ∼2,078 genes (∼8% of coding genes) were found to be characterized as gbM in the majority (>90%) of the accessions. These data are consistent with other within- and between-species comparisons of the conservation of gbM on orthologous genes, which also found a wide range of variation (19–21, 36, 40, 43). We interpret these results to be consistent with the hypothesis that gbM is the result of a passive process related to the variation in the robustness of the maintenance of heterochromatin. Overall, our analyses suggest that the establishment and maintenance of gbM is likely a multifactorial trait and is likely the sum result of genetic and/or environmental factors that influence DNA methylation homeostasis, including but not limited to the following.
First is the activity and subgenomic targeting accuracy of DNA methyltransferases and other enzymes involved in establishing heterochromatin-associated modifications exclusively to sequences that require heterochromatinization. Second is the efficiency with which cells can sequester genomic regions targeted for silencing into higher-order structures associated with dense heterochromatin that distinguish them from transcriptionally active regions. Third are genic features that expose genes to targeting by the heterochromatin machinery, including especially gene length and frequency of CMT3-preferred CWG context cytosines, but also expression levels and proximity to TEs and pericentromeric heterochromatin. Finally, fourth is the efficiency of the cotranscriptional machinery, including IBM1, that removes H3K9me2 from genes.
The interplay between these factors dictates the spectrum of loci that are gbM and may have increased susceptibility to epiallelic shifts to teM, which can alter expression and thus have phenotypic consequences (Fig. 1). Under this model, what are the evolutionary implications of gbM as a trait? As gbM loci are generally housekeeping genes without a consensus molecular function, most alterations to transcription would be expected to have negative consequences. In support of this, A. thaliana mutant backgrounds (e.g., ibm1 mutants) that result in gbM-to-teM shifts have altered transcriptional states associated with pleiotropic developmental defects (51, 58). Yet, it is also clear that polyepialleles are present within this species (Fig. 5B) (19, 71, 72, 75–79). In well-studied examples of genes that exist as polyepialleles, polyepiallelic states have been found to be associated with genomic rearrangements that position genes closer to TEs or with genes that have undergone duplications (19, 71, 72, 79). In the case of HISN6 (a histidine biosynthesis gene) and TAD3 (a transfer RNA deaminase), which are essential genes that have undergone duplications in some accessions, the duplication event is associated with CMT3-dependent methylation and silencing of one of the paralogs (71, 72). In this case, silencing of one paralog may be beneficial in correcting for gene dosage. However, these epialleles may also contribute to speciation, as they have been demonstrated to contribute to hybrid incompatibility in crosses to accessions where the gene is not duplicated and inheritance of the silent copy is lethal (71, 72). Thus, there are clear precedents for polyepialleles potentially affecting fitness. However, it is also important to consider that it is currently unclear whether an epiallele can act as a substrate for natural selection and change frequencies in populations over time, as epialleles are also prone to reversions from silent to expressed states (71, 72, 80, 81). Given the current data, it is parsimonious to conclude that, as a whole, gbM, and the associated susceptibility to epiallelic switching associated with silencing, is a mildly deleterious condition present more or less in populations due to trade-offs with selective pressures related to the maintenance of DNA methylation homeostasis and associated constraints to this process imposed by environmental factors and genome architecture. Thus, variation in the robustness of targeting heterochromatin DNA methylation is likely a symptom of the broader phenomenon of population-level variation of chromatin homeostasis, which may contribute to a diverse array of epigenetic phenomena, including epigenetic drivers of disease.
Materials and Methods
Data Acquisition.
Methylomes, transcriptomes, and genomic variants (SNPs) of 725 accessions used in this analysis were obtained from published datasets of the Arabidopsis 1001 Genomes database (http://signal.salk.edu/1001.php) (19, 82) and reanalyzed.
The methylomes of the ninth generation of ddm1 mutants, including four lines of ddm1 mutants that were independently self-pollinated eight times (9G ddm1) (15), were obtained and reanalyzed. The list of CHG-gain genes that showed a minimum 5% increase in CHG methylation in E. salsugineum AtCMT3-L2–expressing lines relative to wild type was obtained from ref. 46, and ortholog gene pairs between E. salsugineum and A. thaliana were those identified in ref. 20.
Methylome Mapping.
WGBS data were processed using “single-end-pipeline” function from Methylpy as described in ref. 83. Quality filtering and adapter trimming were performed using cutadapt v1.9.dev1 (84). Qualified reads were aligned to the A. thaliana TAIR10 reference genome (68) (downloaded from https://phytozome.jgi.doe.gov) using bowtie 2.2.4 (80). Only uniquely aligned and nonclonal reads were retained. Chloroplast DNA (which is fully unmethylated) was used as a control to calculate the sodium bisulfite reaction nonconversion rate of unmodified cytosines. A binomial test was used to determine the methylation status of cytosines with a minimum coverage of three reads.
RNA-Seq Mapping.
Quality filtering and adapter trimming were performed using Trimmomatic v0.33 with default parameters (85). Qualified reads were aligned to the A. thaliana TAIR10 reference genome using HISAT2 v2.0.5 (86). Gene expression (FPKM) values were computed using StringTie v1.3.3b (87). Genes with zero FPKM values among all investigated accessions were removed from expression analyses.
Gene Methylation Status Classification.
The coding genes for each of 725 accessions were classified as gbM, teM, or UM by applying a binomial test to the number of methylated sites in a gene, similar to the methods described in ref. 67. Further details are described in SI Appendix, Supplementary Methods.
Gene Features Preparation.
Gene length, exon length, exon number, and number of transcripts for all coding genes were obtained directly from the A. thaliana reference protein-coding gene annotation file in the GFF format (TAIR11) (88). Calculation procedures for determining CWG and CG frequencies, distance to the pericentromere, expression levels, and substitution rates are described in SI Appendix, Supplementary Methods. Differences in gene features between UM genes and genes with varied gbM epiallele frequencies (the proportion of gbM accessions) was assessed using the Welch two-sample t test statistic in R, except for transcript number, which was tested using a χ2 goodness-of-fit test, as this was a discrete datatype.
Prediction Model for Gene’s Methylation Status.
A total of 10 features (Dataset S6) that had shown different distributions between gbM and UM genes from previous studies (41, 67) and our preliminary analysis were selected to train a prediction model of a gene’s methylation status. Machine learning algorithm training, prediction, and evaluation are described in detail in SI Appendix, Supplementary Methods.
Additional Genome-Wide Analyses.
A neighbor-joining tree of 725 accessions was constructed by using MEGA7 (89) using SNPs (82) from coding sequences. SNPs were filtered using a minor allele frequency cutoff of 0.05 and a minimum data integrity cutoff of 0.5, and then the VCF-formatted SNPs were converted into sequence alignments readable by MEGA7.
The number of CWG sites in the CDS was calculated by replacing the reference sequence with each accession’s genotype, and then the number of CWG sites were tabulated based on the replaced CDS for each accession.
Reference mapping-based genome-wide percent methylation was calculated by dividing the total number of aligned methylated reads to the genome by the total number of methylated plus unmethylated reads. To evaluate the accuracy of this estimate, we compared percent methylation values of 17 accessions that possess publicly available reference genomes (90, 91). We observed that the methylation levels estimated by mapping sequence reads to each accession’s own assembly is generally higher than that estimated from mapping to the reference (SI Appendix, Fig. S2C).
To address the possibility of sequence variation among accessions, reads-based genome methylation levels were also calculated by using a nonreference-based DNA methylation predictive model, FASTmC (92). This method estimates methylation levels directly from WGBS reads. Methylation levels estimated in this way generally show a higher value than the value estimated by alignments to their own assembly, but this method also reveals more variation (SI Appendix, Fig. S2C). Because of computational efficiency, this method was based on random sampling on a subset of 10,000 reads, which may include sampling bias. Regardless, this method used to estimate genome CHG methylation levels also shows a significant negative correlation with the number of gbM genes within each accession (SI Appendix, Fig. S2D).
Correlations between number of gbM genes, methylation levels, number of CWG sites, and CMT3 gene expression were performed using R, and the data were corrected for phylogenetic signals among accessions using a phylogenetically independent contrasts (PICs) method in APE (93). Because extremely low pair-wise distances (little genetic variance) between accessions will generate outliers after PIC correction, a cutoff of 0.01 pair-wise distance was applied to prune clades with almost no genetic differences, which resulted in a final set of 620 accessions that were used in the correlation tests. This same set of individuals was also used for GWAS, where the number of gbM genes was used as the phenotype. GWAS analysis was performed with a compressed mixed linear model (94) implemented in the GAPIT package (95) of R.
SNP-based heritability (h2) of the trait is the proportion of the total variance (σ2e + σ2a) explained by the genetic variance (σ2a), and σ2e is the residual variance. Both σ2e and σ2a were estimated by maximum likelihood method in the compressed mixed linear model during GWAS analysis.
Genome-wide SNPs that satisfied a minor allele frequency of 0.05 were used for association studies, and the genome-wide threshold was modified using the Bonferroni method. If a significant SNP, which passed a threshold of 1e−5, lied within 10 kb of another significant SNP, they were combined into a block. Using this block as a starting point, all other significant SNPs within 10 kb of either end of the block were further combined into the block. The procedure was repeated until no significant SNPs could be found within 10 kb of the block ends. These blocks were referred to as QTLs (18). Genes that located within the QTL were identified as candidate genes of the trait.
Characterizing Genes That Are Predisposed to mCHG.
The methylomes of ninth-generation ddm1 mutants (15) were mapped to reference genome using Methylpy (83), and the methylation status of each gene was determined using a binomial test as described earlier. Genes that had reads mapping to at least 20 CHGs, an mCHG q-value < 0.05 in ninth-generation ddm1 mutants, and mCHG q-value > 0.05 in wild type were identified as mCHG-gain genes. The remaining genes were classified as genes that do not gain mCHG. All coding genes of A. thaliana were also categorized into gbM, teM, and UM based on their methylation status in A. thaliana population as defined earlier. Then, a Fisher’s exact test was used to examine the significance of the association between the gene’s mCHG-gain status and their methylation categories. Similarly, all gbM genes defined from the A. thaliana population were categorized into different groups with varied gbM epiallele frequency. Those gbM genes were also classified into two groups based on whether they gain mCHG in ninth-generation ddm1 mutants. A Fisher’s exact test was used to examine the significance of the association between the gene’s mCHG-gain status and groups with varied gbM epiallele frequency.
The genes that gain CHG methylation in E. salsugineum AtCMT3-L2–expressing lines were obtained from published data (46). A. thaliana orthologs of the genes in E. salsugineum were classified into two classes based on whether they gain mCHG in CMT3-expressing E. salsugineum. Those A. thaliana orthologs were also categorized into gbM, teM, and UM based on their methylation status in A. thaliana population (Dataset S10). Then, a Fisher’s exact test was used to examine the significance of the association between the gene’s mCHG-gain status and their methylation categories. The significance of the association between the gene’s mCHG-gain status and groups with varied gbM epiallele frequency were also examined as in ddm1 mutants.
q-RT PCR.
RNA was extracted from cauline leaves of individual plants using TRIzol according to the manufacturer’s instructions. Synthesis of cDNA was completed using SuperScript III with random hexamers (Invitrogen) according to the manufacturer’s instructions. Real-time qRT-PCR was conducted using LightCycler 480 SYBR green master mix in a Light Cycler 480 instrument (Roche). Primers used to detect short and long IBM1 transcripts are those published in ref. 74. Relative expression of the short (nonfunctional) to full-length (functional) IBM1 transcripts was calculated using the double delta threshold cycle (Ct) method (96). Average Ct values were calculated from three technical replicates.
Analysis of Gene Expression and CG Methylation Levels in Cnt-1, Monte-1, and UKSE06-533.
For gene expression analysis in Cnt-1, the Cnt-1 library and FPKM values were averaged across all accessions to create a “control library” and used for further comparisons. Only genes with an FPKM >0 in both libraries were retained. Then, Cnt-1 and control libraries were collectively normalized using quantile normalization method to remove global variation across samples (97). Then, the expression level of genes that exist as teM in Cnt-1 but are otherwise categorized as a gbM gene in a majority of accessions (>0.9 or the core gbM genes) were compared with their gene pairs in the control library. A Wilcoxon signed-rank test was used for comparisons between pairs of genes. In addition to statistical significance, Cohen’s d was used to assess effect size of differences in gene expression. The same procedure was applied to Monte-1 and UKSE06-533.
For mCG level analysis in Cnt-1, the mCG level of core gbM genes in Cnt-1 and the averaged mCG levels across all accessions were normalized using the quantile normalization method (97). Then, among those core gbM genes, genes that exist as a teM gene in Cnt-1 were compared with their gene pairs in the control group. Statistical analysis was the same as that in the expression comparison. The same procedure was applied to Monte-1 and UKSE06-533.
Data Availability.
All sequencing data used in this study were previously published and are available under NCBI GEO accession numbers (GSE43857, GSE80744) and DDBJ Sequence Read Archive (DRA002551). Code used in processing and analysis of these data can be found at GitHub (https://github.com/schmitzlab/Natural_variation_in_DNA_methylation_homeostasis_and_the_emergence_of_epialleles).
Supplementary Material
Acknowledgments
This study was supported by the National Science Foundation (grant MCB-1856143) and the National Institutes of Health (grant R01GM134682) to R.J.S. R.J.S. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts. J.M.W. is a National Plant Genome Initiative Postdoctoral Fellow supported by the NSF (fellowship IOS-1811694). We thank Joseph Gage and Edward S. Buckler for their technical support on machine learning analysis and Nathan Springer and Frank Johannes for valuable feedback on the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Code used in processing and analysis of these data can be found at GitHub (https://github.com/schmitzlab/Natural_variation_in_DNA_methylation_homeostasis_and_the_emergence_of_epialleles).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918172117/-/DCSupplemental.
References
- 1.Elgin S. C., Heterochromatin and gene regulation in Drosophila. Curr. Opin. Genet. Dev. 6, 193–202 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T., Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Bannister A. J., et al. , Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I., Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Lindroth A. M., et al. , Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23, 4286–4296 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindroth A. M., et al. , Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292, 2077–2080 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Jackson J. P., Lindroth A. M., Cao X., Jacobsen S. E., Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Zhang H., Lang Z., Zhu J. K., Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Law J. A., Jacobsen S. E., Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du J., et al. , Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol. Cell 55, 495–504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du J., et al. , Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151, 167–180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du J., Johnson L. M., Jacobsen S. E., Patel D. J., DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson L. M., et al. , The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17, 379–384 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flavahan W. A., Gaskell E., Bernstein B. E., Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito T., et al. , Genome-wide negative feedback drives transgenerational DNA methylation dynamics in Arabidopsis. PLoS Genet. 11, e1005154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakutani T., Jeddeloh J. A., Flowers S. K., Munakata K., Richards E. J., Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. U.S.A. 93, 12406–12411 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmeister B. T., Lee K., Rohr N. A., Hall D. W., Schmitz R. J., Stable inheritance of DNA methylation allows creation of epigenotype maps and the study of epiallele inheritance patterns in the absence of genetic variation. Genome Biol. 18, 155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz R. J., et al. , Patterns of population epigenomic diversity. Nature 495, 193–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakatsu T., et al. ; 1001 Genomes Consortium , Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166, 492–505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niederhuth C. E., et al. , Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17, 194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubin M. J., et al. , DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 4, e05255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bewick A. J., et al. , On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 113, 9111–9116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bewick A. J., et al. , The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 18, 65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiefer C., et al. , Interspecies association mapping links reduced CG to TG substitution rates to the loss of gene-body methylation. Nat. Plants 5, 846–855 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Cokus S. J., et al. , Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lister R., et al. , Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran R. K., et al. , DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr. Biol. 15, 154–159 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Bewick A. J., Schmitz R. J., Gene body DNA methylation in plants. Curr. Opin. Plant Biol. 36, 103–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zilberman D., An evolutionary case for functional gene body methylation in plants and animals. Genome Biol. 18, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zemach A., McDaniel I. E., Silva P., Zilberman D., Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Feng S., et al. , Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U.S.A. 107, 8689–8694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bewick A. J., Vogel K. J., Moore A. J., Schmitz R. J., Evolution of DNA methylation across insects. Mol. Biol. Evol. 34, 654–665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bewick A. J., et al. , Diversity of cytosine methylation across the fungal tree of life. Nat. Ecol. Evol. 3, 479–490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picard C. L., Gehring M., Proximal methylation features associated with nonrandom changes in gene body methylation. Genome Biol. 18, 73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Graaf A., et al. , Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc. Natl. Acad. Sci. U.S.A. 112, 6676–6681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seymour D. K., Gaut B. S., Phylogenetic shifts in gene body methylation correlate with gene expression and reflect trait conservation. Mol. Biol. Evol., msz195 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Bewick A. J., Zhang Y. W., Wendte J. M., Zhang X. Y., Schmitz R. J., Evolutionary and experimental loss of gene body methylation and its consequence to gene expression. G3 (Bethesda) 9, 2441–2445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takuno S., Seymour D. K., Gaut B. S., The evolutionary dynamics of orthologs that shift in gene body methylation between arabidopsis species. Mol. Biol. Evol. 34, 1479–1491 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Takuno S., Ran J. H., Gaut B. S., Evolutionary patterns of genic DNA methylation vary across land plants. Nat. Plants 2, 15222 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Seymour D. K., Koenig D., Hagmann J., Becker C., Weigel D., Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 10, e1004785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takuno S., Gaut B. S., Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc. Natl. Acad. Sci. U.S.A. 110, 1797–1802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zilberman D., Gehring M., Tran R. K., Ballinger T., Henikoff S., Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39, 61–69 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Vaughn M. W., et al. , Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5, e174 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X., et al. , Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126, 1189–1201 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Muyle A., Gaut B. S., Loss of gene body methylation in Eutrema salsugineum is associated with reduced gene expression. Mol. Biol. Evol. 36, 155–158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wendte J. M., et al. , Epimutations are associated with CHROMOMETHYLASE 3-induced de novo DNA methylation. eLife 8, e47891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wendte J. M., Schmitz R. J., Specifications of targeting heterochromatin modifications in plants. Mol. Plant 11, 381–387 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Inagaki S., Kakutani T., What triggers differential DNA methylation of genes and TEs: Contribution of body methylation? Cold Spring Harb. Symp. Quant. Biol. 77, 155–160 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Teixeira F. K., Colot V., Gene body DNA methylation in plants: A means to an end or an end to a means? EMBO J. 28, 997–998 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reinders J., et al. , Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23, 939–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saze H., Shiraishi A., Miura A., Kakutani T., Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319, 462–465 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Stoddard C. I., et al. , A nucleosome bridging mechanism for activation of a maintenance DNA methyltransferase. Mol. Cell 73, 73–83 e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gouil Q., Baulcombe D. C., DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 12, e1006526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papa C. M., Springer N. M., Muszynski M. G., Meeley R., Kaeppler S. M., Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13, 1919–1928 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartee L., Malagnac F., Bender J., Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15, 1753–1758 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X., et al. , Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 115, E8793–E8802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inagaki S., et al. , Autocatalytic differentiation of epigenetic modifications within the Arabidopsis genome. EMBO J. 29, 3496–3506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miura A., et al. , An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 28, 1078–1086 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woo H. R., Richards E. J., Natural variation in DNA methylation in ribosomal RNA genes of Arabidopsis thaliana. BMC Plant Biol. 8, 92 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woo H. R., Pontes O., Pikaard C. S., Richards E. J., VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21, 267–277 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ronemus M. J., Galbiati M., Ticknor C., Chen J., Dellaporta S. L., Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273, 654–657 (1996). [DOI] [PubMed] [Google Scholar]
- 62.Finnegan E. J., Peacock W. J., Dennis E. S., Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. U.S.A. 93, 8449–8454 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams B. P., Gehring M., Stable transgenerational epigenetic inheritance requires a DNA methylation-sensing circuit. Nat. Commun. 8, 2124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tabata S., et al. ; Kazusa DNA Research Institute; Cold Spring Harbor and Washington University in St Louis Sequencing Consortium; European Union Arabidopsis Genome Sequencing Consortium , Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408, 823–826 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Carazo-Salas R. E., et al. , Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Hergeth S. P., Schneider R., The H1 linker histones: Multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 16, 1439–1453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takuno S., Gaut B. S., Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol. Biol. Evol. 29, 219–227 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Berardini T. Z., et al. , The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeddeloh J. A., Stokes T. L., Richards E. J., Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97 (1999). [DOI] [PubMed] [Google Scholar]
- 70.Vongs A., Kakutani T., Martienssen R. A., Richards E. J., Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928 (1993). [DOI] [PubMed] [Google Scholar]
- 71.Blevins T., Wang J., Pflieger D., Pontvianne F., Pikaard C. S., Hybrid incompatibility caused by an epiallele. Proc. Natl. Acad. Sci. U.S.A. 114, 3702–3707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agorio A., et al. , An arabidopsis natural epiallele maintained by a feed-forward silencing loop between histone and DNA. PLoS Genet. 13, e1006551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen W., et al. , Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening. Sci. Rep. 5, 9192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rigal M., Kevei Z., Pélissier T., Mathieu O., DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J. 31, 2981–2993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Silveira A. B., et al. , Extensive natural epigenetic variation at a de novo originated gene. PLoS Genet. 9, e1003437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Durand S., Bouché N., Perez Strand E., Loudet O., Camilleri C., Rapid establishment of genetic incompatibility through natural epigenetic variation. Curr. Biol. 22, 326–331 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Luff B., Pawlowski L., Bender J., An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell 3, 505–511 (1999). [DOI] [PubMed] [Google Scholar]
- 78.Cubas P., Vincent C., Coen E., An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157–161 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Bender J., Fink G. R., Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83, 725–734 (1995). [DOI] [PubMed] [Google Scholar]
- 80.Quadrana L., et al. , Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 5, 3027 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Manning K., et al. , A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38, 948–952 (2006). [DOI] [PubMed] [Google Scholar]
- 82.Alonso-Blanco C., et al. ; 1001 Genomes Consortium , 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166, 481–491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schultz M. D., et al. , Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 85.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L., Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cheng C. Y., et al. , Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zapata L., et al. , Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. U.S.A. 113, E4052–E4060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gan X., et al. , Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477, 419–423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bewick A. J., et al. , FASTmC: A suite of predictive models for nonreference-based estimations of DNA methylation. G3 (Bethesda) 6, 447–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 94.Zhang Z., et al. , Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42, 355–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lipka A. E., et al. , GAPIT: Genome association and prediction integrated tool. Bioinformatics 28, 2397–2399 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 97.Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P., A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data used in this study were previously published and are available under NCBI GEO accession numbers (GSE43857, GSE80744) and DDBJ Sequence Read Archive (DRA002551). Code used in processing and analysis of these data can be found at GitHub (https://github.com/schmitzlab/Natural_variation_in_DNA_methylation_homeostasis_and_the_emergence_of_epialleles).