Significance
Models that use genetic deficiency to infer gene function can be confounded by compensatory actions of coexpressed paralogs. For the MAX-binding proteins, such as MYC, compensation can occur during embryogenesis and hematopoiesis. The present study defines the roles of MYCL and MXD1 in Batf3-dependent dendritic cells. Our results support the prevailing model of antagonism between MYC and MXD family members. We show that MYCL and MXD1 have reciprocal actions that converge on a shared transcriptional program of biosynthesis during dendritic cell maturation. More broadly, we identify a physiological setting in which compensation is insufficient to rescue transcriptional deficiencies in Mycl−/− and Mxd1−/− mice.
Keywords: dendritic cells, transcription factors, maturation, MYC, cancer
Abstract
We previously found that MYCL is required by a Batf3-dependent classical dendritic cell subset (cDC1) for optimal CD8 T cell priming, but the underlying mechanism has remained unclear. The MAX-binding proteins encompass a family of transcription factors with overlapping DNA-binding specificities, conferred by a C-terminal basic helix-loop-helix domain, which mediates heterodimerization. Thus, regulation of transcription by these factors is dependent on divergent N-terminal domains. The MYC family, including MYCL, has actions that are reciprocal to the MXD family, which is mediated through the recruitment of higher-order activator and repressor complexes, respectively. As potent proto-oncogenes, models of MYC family function have been largely derived from their activity at supraphysiological levels in tumor cell lines. MYC and MYCN have been studied extensively, but empirical analysis of MYCL function had been limited due to highly restricted, lineage-specific expression in vivo. Here we observed that Mycl is expressed in immature cDC1s but repressed on maturation, concomitant with Mxd1 induction in mature cDC1s. We hypothesized that MYCL and MXD1 regulate a shared, but reciprocal, transcriptional program during cDC1 maturation. In agreement, immature cDC1s in Mycl−/−-deficient mice exhibited reduced expression of genes that regulate core biosynthetic processes. Mature cDC1s from Mxd1−/− mice exhibited impaired ability to inhibit the transcriptional signature otherwise supported by MYCL. The present study reveals LMYC and MXD1 as regulators of a transcriptional program that is modulated during the maturation of Batf3-dependent cDC1s.
Myc, Mycn, and Mycl compose a highly conserved family of proto-oncogenes that support elevated transcription in transformed cells (1–5). All members of the MYC family are structural partners of MAX, which shares a basic helix-loop-helix (bHLH) domain and confers DNA-binding specificity to the heterodimer (6, 7). The MYC:MAX dimer in turn activates transcription via transcriptional activation domains intrinsic to MYC (8). Analysis of mice deficient in Myc and Mycn revealed that these factors have nonredundant, essential roles in the regulation of embryogenesis (9, 10). However, developmental defects associated with Mycn deficiency can be rescued by expression of the Myc coding sequence from the endogenous Mycn locus (11). Notwithstanding, redundancy is context-dependent, and Myc expression is insufficient to rescue all the cell-intrinsic functions of Mycn, such as during myogenesis and lymphocyte proliferation (11, 12). Despite significant overlap in the functions of MYC, MYCN, and MYCL, distinct enhancer elements at their respective genomic loci enforce a requirement for both Myc and Mycn. This paradigm has since been extended to a number of developmental pathways, including hematopoiesis. Sustained production of lymphocytes, for example, requires early expression of Mycn by hematopoietic stem cells and subsequent transition to Myc after restriction to lymphoid lineages (13–15).
Unlike Myc and Mycn, Mycl is dispensable for development into adulthood (16, 17). However, Mycl is known to retain transcriptional activity and to serve as a functional proto-oncogene in numerous cancer cell lines (3, 18, 19). Following a precise analysis of transcription factors expressed by dendritic cells (DCs), the first hematopoietic lineage that requires normal Mycl expression for its function was identified. DCs, including plasmacytoid DCs (pDCs) and both subsets of classical DCs (cDCs), develop normally in Mycl−/−-deficient mice, but these mice exhibit an impaired capacity to prime CD8 T cells in response to bacterial and viral infections (20). This effect has been attributed specifically to the Batf3-dependent cDC subset, called cDC1, thus providing a model for studying MYCL in a primary cell lineage.
Terminal differentiation of diverse cellular lineages is associated with reduced expression of MYC, coincident with reductions in the rate of growth and proliferation (21). MYC-supported transcription can also be repressed directly by bHLH domain-containing repressors, such as the Mxd family of genes. Like the MYC proteins, MXD proteins dimerize with MAX. As repressors of transcription, dimerization and DNA binding impose reciprocal actions at MYC-regulated loci (22). Surveys of the major hematopoietic lineages have revealed that Mxd1 is expressed primarily by granulocytes, innate lymphoid cells, and mature DCs (23, 24). In addition, recent studies have revealed suppression of Mycl and induction of Mxd1 expression during the transition of cDC1s from the immature state to the mature state (25, 26).
Since the initial report that described Mycl expression in the hematopoietic system (20), more precise surface markers have been defined to distinguish immature and mature cDC1 across tissues and species (25, 27). The present study extends the analysis of Myclgfp/+ mice and reports that expression of Mycl is restricted to immature subsets of DCs. Among cDCs, the immature cDC1 subset expresses the highest level of Mycl across all the tissues examined. Therefore, we set out to examine the impact of Mycl deficiency on immature splenic cDC1s at steady state and during inflammation. We also asked whether MXD1 acts to suppress the transcriptional program supported by MYCL during inflammatory maturation. The results demonstrate that cDC1 cells from Mxd1−/− mice have an impaired capacity to suppress the same core biosynthetic processes that are otherwise supported by Mycl. Therefore, we conclude that Mycl and Mxd1 cooperate to regulate the fitness of cDC1s in vivo through the regulation of a shared transcriptional program of core biosynthetic processes.
Results
Mycl Is Highly Expressed in cDC1s and Is Repressed on Homeostatic Maturation in Vivo.
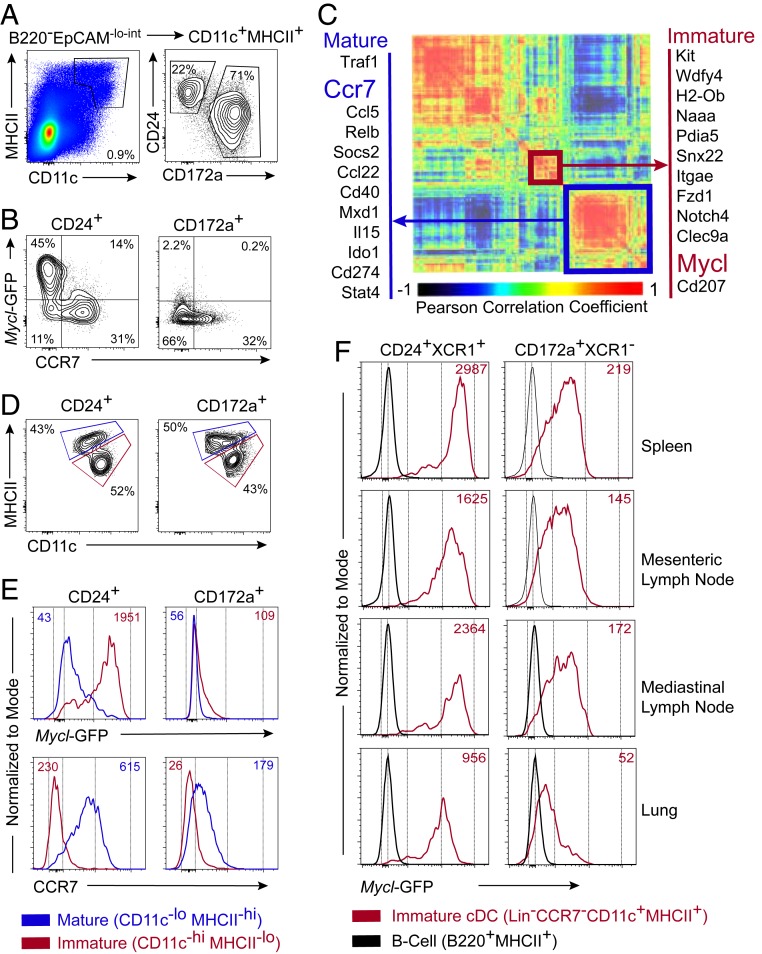
We recently reported that GFP expression driven from the Mycl locus in Myclgfp/+ mice is restricted to DCs, and that Myc is not coexpressed with Mycl (20). Myclgfp/gfp mice do not express a functional Mycl transcript, and thus served as an in vivo model to demonstrate that MYCL is required for optimal CD8 T cell priming in vivo (20). Recent whole-transcriptome analyses of DCs during homeostatic and inflammatory maturation found suppression of Mycl, suggesting that its expression is most likely limited to immature cDCs (25, 26). Thus, we examined Mycl-GFP expression in Myclgfp/+ mice, using CCR7 expression as a canonical marker of maturation status of DCs in peripheral lymphoid organs (28). In the skin-draining lymph nodes (LNs), we show that mature CD11c+MHCII+CCR7+ cDCs are GFP−, confirming that Mycl expression is suppressed on maturation in this tissue (Fig. 1 A and B).
Fig. 1.
Mycl expression is restricted to immature cDC1s in lymphoid organs. (A and B) Flow cytometry of CD24+ cDC1s and CD172a+ cDC2s (A) and expression of Mycl-GFP and CCR7 (B) in skin-draining lymph nodes of Myclgfp/+ mice. (C) Heatmap of Pearson correlation coefficients for expressed genes differentially expressed among DC subsets from expression microarray analysis published by the ImmGen Consortium (23, 24). Modules characteristic of immature and mature phenotypes defined by hierarchical clustering and highlighted in red and blue, respectively. (D and E) Definition of immature/resident and mature/migratory CD24+ cDC1s and CD172a+ cDC2s based on CD11c and MHCII expression (D) and histograms of Mycl-GFP and CCR7 median fluorescence intensity (E) for immature and mature subsets of cDC1s and in skin-draining lymph nodes of Myclgfp/+ mice analyzed by flow cytometry. (F) For respective populations defined in D, histograms of Mycl-GFP median fluorescence intensity were analyzed by flow cytometry. Expression of Mycl-GFP in splenic B cells served as a negative control for GFP. Flow cytometry results are representative of three to five independent experiments each with two or three mice.
We extended these observations across several other cDC subsets in various tissues. We identified a strong positive correlation between Mycl expression and immature DC markers, such as Kit, Wdfy4, Clec9a, Naaa, and Snx22 (Fig. 1C) (23–25, 29–31). Inversely, a strong negative correlation exists between the expression of Mycl and canonical markers of mature cDCs, such as Ccr7, Ido1, CD40, Mxd1, and Cd274. To establish whether this pattern of Mycl expression is conserved across cDCs in peripheral lymphoid organs in vivo, we analyzed GFP expression in Myclgfp/+ mice. The results demonstrate that Mycl expression is restricted to immature cDCs, as defined by CD11chiMHCIIint cells, in the skin-draining LNs, spleen, mesenteric LNs, mediastinal LNs, and lung (Fig. 1 D–F).
MYCL Regulates cDC1 Cell Size and Supports Transcription Broadly.
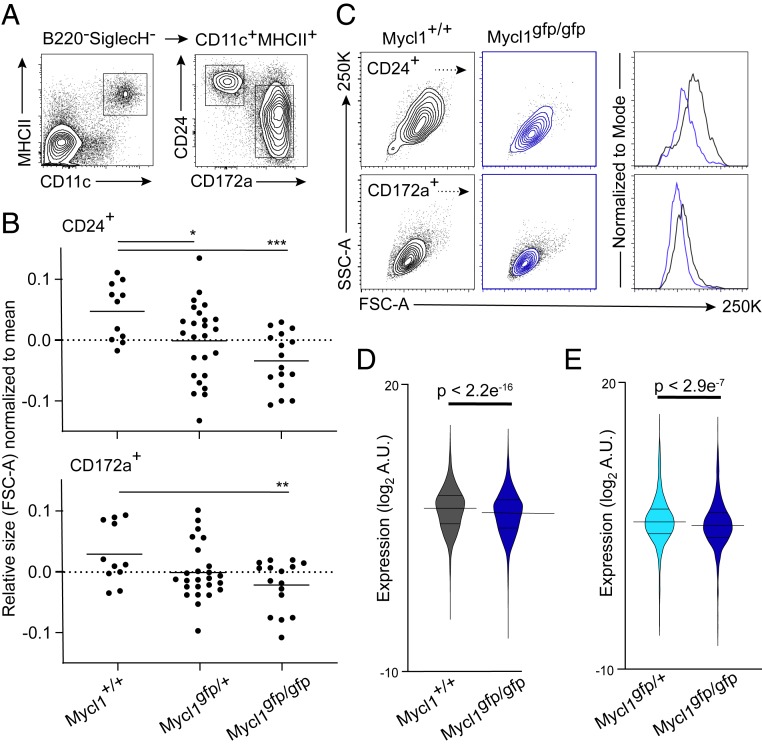
Although MYCL is required for optimal CD8 T cell priming by cDC1s (20), the transcriptional mechanisms for this action of MYCL are not established. A hallmark of MYC deficiency, from insects to mammals, is a reduction in cell size (32–34). Therefore, we asked whether cDCs in MYCL-deficient mice would exhibit a similar phenotype. By flow cytometry, steady-state splenic cDCs in Myclgfp/gfp mice exhibited significantly reduced forward scatter area (FSC-A) compared with Mycl+/+ mice, an indication of reduced cell size (Fig. 2 A–C). Although CD172+ splenic cDC2 cell size is also affected by Mycl deficiency (Fig. 2 B and C), a role for MYCL in the regulation of cDC2 function has not been established. In addition, Mycl expression is highest in immature cDC1s in all the tissues examined (Fig. 1 E and F). Therefore, to define the mechanism by which MYCL supports transcription, we focused our analysis on splenic CD24+ cDC1 cells.
Fig. 2.
Mycl expression supports cDC1 fitness with respect to cell size and global transcription in vivo. (A) Representative flow cytometry analysis of splenic CD24+ cDC1s and CD172a+ cDC2s from Mycl+/+, Myclgfp/+, and Myclgfp/gfp mice. Populations (Right) are pregated on CD11c and MHCII expression (Left). (B) Relative cell size of splenic CD24+ (Top) and CD172a+ (Bottom) cDCs as measured by median fluorescence intensity of the FSC-A. *P < 0.05, Student’s t test between the indicated populations in Myclgfp/gfp and Mycl+/+ mice. Data were pooled and normalized from three independent experiments (n = 11 to 25). (C) Representative 2D histograms (Left) of cell size for cDC subsets defined in A, measured as a function of FSC-A and side scatter area (SSC-A) for the indicated genotypes. Representative histogram (Right) of FSC-A from which the data in B are derived. Colors correspond to the indicated genotypes (Left). (D and E) Violin plots of microarray expression levels of genes with significant signal to noise ratios between Myclgfp/gfp and Mycl+/+ splenic cDC1s (n = 5 per genotype) mice and Myclgfp/gfp and Myclgfp/+ (n = 4 to 5 per genotype), respectively. Results of Welch’s two-sample t test are reported as P values or indicated as *P < 0.05, **P < 0.01, or ***P < 0.005.
To define the transcriptional footprint of MYCL, we used RNA spike-in controls to normalize signal intensity to RNA content (35). This allowed for an examination of the effect of Mycl deficiency on cDC1 transcriptional fitness with more precision than in our initial studies (20). We hypothesized that MYCL, like MYC, might support transcription broadly as a transcriptional activator. This model would predict a uniform reduction in mRNA levels for genes that are supported transcriptionally by MYCL. Normalization of expression microarray signal intensity to RNA spike-in concentration identified a uniform and significant reduction in transcription in splenic cDC1s from Myclgfp/gfp mice relative to Mycl+/+ mice (Fig. 2D). To rule out an effect of GFP expression, an independent experiment demonstrated the same effect when cDC1s were compared between Myclgfp/gfp and Myclgfp/+ mice (Fig. 2E).
MYCL Supports Core Biosynthetic Processes in cDC1s at Homeostasis and during Inflammation.
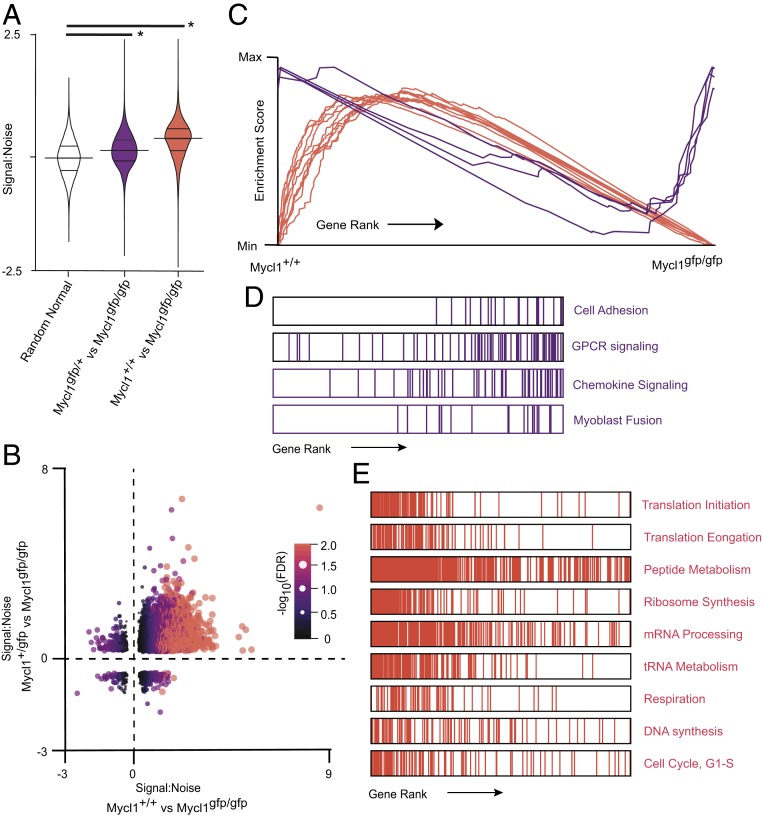
Since MYC is a positive regulator of transcription with broad activity, MYC deficiency in primary cells is characterized by a directional, downward shift in mRNA levels (35). We used gene set enrichment analysis (GSEA) to determine the direction of the effect of Mycl deficiency on the transcription of genes grouped by functional annotations with Gene Ontology (GO) terms (36–39). Signal-to-noise ratios for all expressed genes were calculated, with positive values corresponding to reduced expression in cDC1s from Myclgfp/gfp mice relative to cDC1s from Mycl+/+ or Myclgfp/+ mice. When compared with a random normal distribution of signal-to-noise ratios, the top 1,000 genes, ranked on the basis of statistical significance, were more highly expressed in Mycl+/+ and Myclgfp/+ cDC1s (Fig. 1 A and B). These results demonstrate that LMYC functions to activate transcription in cDC1s at steady state.
Analysis of the functional consequences of Mycl deficiency on transcription revealed a consistent enrichment of GO terms associated with core biosynthetic processes (Fig. 3 C–E) These results support a model in which LMYC enhances the expression of genes that regulate core biological processes. MYC family members amplify transcription at loci in which transcription has already been initiated by increasing the rate of RNA polymerase elongation (40–42). Consistent with this model, Mycl deficiency did not result in the absolute inhibition of transcription from target loci. Rather, cDC1s from mice with at least one copy of Mycl exhibited broad amplification of gene expression (Fig. 3 A–C). Genes that were not amplified by MYCL were grouped by GSEA into poorly supported functional gene sets as a result of lower signal-to-noise ratios (Fig. 3 B–D).
Fig. 3.
Mycl supports a dose-dependent transcriptional program that regulates core biosynthetic processes homeostatically in splenic cDC1s. (A) Violin plots of signal-to-noise ratios for 1000 differentially expressed genes ranked by false discovery rate (FDR). Signal-to-noise ratios are derived from independent expression microarray experiments that compared cDC1s from Myclgfp/gfp and Myclgfp/+ mice with splenic cDC1s from Mycl+/+ mice. Welch two-sample t tests were performed for each independent experiment compared with a random normal distribution of signal-to-noise ratios. *P < 2.2e−16. (B) Scatterplot of signal-to-noise ratios of genes with for the indicated experimental comparisons, with point color and size scaled to -log10(FDR). (C–E) Summary of results of GSEA for annotated GOs. (D and E) Representative examples of enrichment plots and respective rank-ordered gene sets. GSEA results are provided in SI Appendix, Tables S1 and S2.
The cDC1 lineage can provide critical signals required for the innate immune responses in some contexts (43). Thus, we next asked whether MYCL regulates the transcriptional fitness of cDC1s after acute activation. The synthetic analog to pathogen-associated double-stranded RNA, poly(I:C), is sufficient to activate innate immune cells in vivo and to induce inflammatory maturation of DCs (25, 38, 44). We activated splenic cDC1s in vivo using poly(I:C) and analyzed their transcriptional response in Mycl+/+ or Myclgfp/gfp mice after 5 h. After activation, pairwise comparisons of the normalized enrichment scores of enriched gene sets revealed shared transcriptional support by MYCL at steady state and after activation. This is illustrated in Fig. 4A, where the gene sets with the highest enrichment scores are associated with higher expression of constituent genes in Mycl+/+ splenic cDC1s. At the gene level, nearly one-half of the genes that contribute to the core enrichment signature are unique to steady-state or inflammatory conditions (Fig. 4B). Therefore, we asked whether LMYC controls discrete biological processes at steady state and activation. We generated an adjacency matrix of the core enrichment genes from steady-state and activating conditions (Fig. 4B and SI Appendix, Tables S1 and S3). In turn, the matrix was visualized as a network of significantly enriched gene sets and their corresponding core enrichment genes, which were clustered using a prefuse force-directed layout algorithm in Cytoscape (45). Gene sets that shared constituent genes were grouped into core biological processes, which included respiration, regulation of chromatin and transcription, protein processing, and translation (Fig. 4 C–F). For these major biological processes, genes that were unique to steady-state or activating conditions contributed to the enrichment of the same core biological processes (Fig. 4 G and H).
Fig. 4.
The transcriptional program of biosynthesis supported by MYCL is independent of activation status. Results from GSEA of expression microarray data from two independent experiments, in which splenic cDC1s were isolated from Myclgfp/gfp(n = 4 to 5) and Mycl+/+ (n = 5) mice under homeostatic conditions or activation with poly I:C. (A) Scatterplot of normalized enrichment scores of all biological process GO gene sets for the indicated experiments. Each gene set is represented as a colored dot whose color and size are scaled to their respective FDR. (B) Venn diagram of genes classified as part of the core enrichment signature by GSEA at homeostasis and under activation with poly I:C. (C–F) Network analysis of genes that contribute to the core enrichment signature for a given gene set. Individual genes are represented as nodes, which are colored based on their membership to the core-enrichment signatures defined in B. Edges are lines that connect individual genes to the functional gene sets, represented as black nodes, to which they are annotated by GO. Clusters were generated by a prefuse force-directed algorithm in Cytoscape, in which nodes that are connected by one or more edges are positioned closer together and disconnected nodes are drawn farther apart. Gene node color corresponds to whether they are shared or unique to a given experimental condition, as illustrated in B. The clusters of genes are centered around gene set nodes associated broadly with respiration (C), regulation of chromatin and transcription (D), protein processing (E), and translation (F). (G and H) Heat maps of core enrichment genes expressed by splenic cDC1s at homeostasis (G) or after poly I:C activation (H). Genes are grouped based on their inclusion in the indicated functional gene set, which are derived from GSEA results in SI Appendix, Table S3. Normalized expression is scaled by row z-score.
MXD1 Regulates the Repression of MYCL-Supported Transcription in Mature cDC1s.
We also found that Mxd1 expression changes during cDC maturation across various tissues, being induced rather than suppressed (Fig. 1C), in agreement with a recent in vivo study (25). Given the established role of MXD1 as a transcriptional repressor that can antagonize MYC-supported transcription, we next asked whether it also carries out a similar function in mature cDC1s.
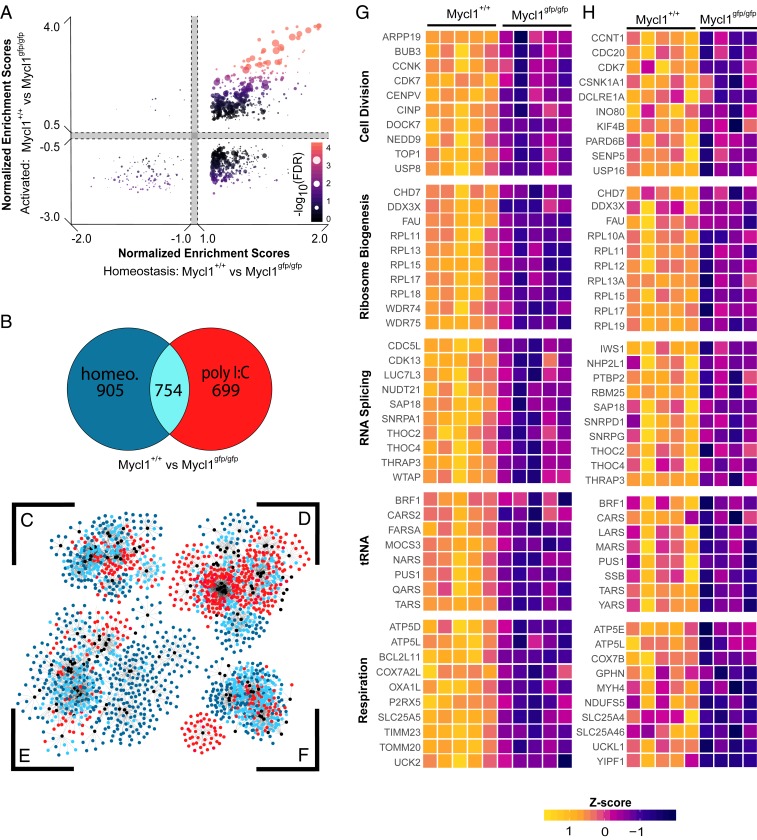
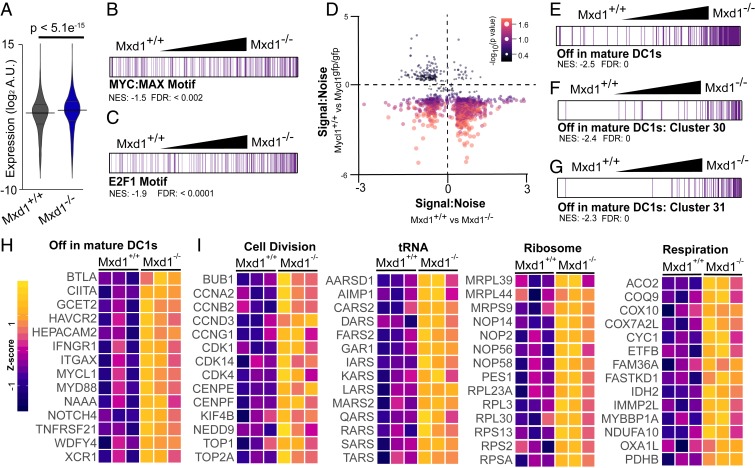
Before the present study, the role of MXD1 in the regulation of DC function or development had not been examined. However, we found that Mxd1−/− mice have phenotypically normal expression of surface markers that identify cDCs in peripheral lymphoid organs, including normal expression of CCR7 by mature CD11cintMHCIIhi cDCs. This allowed us to examine the targets of transcriptional regulation by MXD1 in cDC1s (Fig. 5). Mature splenic cDC1s were isolated from Mxd1+/+ and Mxd1−/− mice after 18 h of activation with poly(I:C) and used for expression microarray analysis. cDC1s from Mxd1−/− mice exhibited significant, uniform increases in the expression levels of genes with signal-to-noise ratios that contributed to the enrichment profile identified by GSEA (Fig. 5A). These results indicate that MXD1 acts to broadly repress transcription in mature cDC1s.
Fig. 5.
Broad transcriptional support of biosynthesis is suppressed by MXD1 during inflammatory maturation. Results of GSEA of expression microarray data of splenic cDC1s isolated from Mxd1+/+ and Mxd1−/− after activation with poly I:C (n = 3). (A) Violin plot of expression levels for genes with signal-to-noise ratio >1. P values reported for Welch two-sample t tests. (B and C) Rank-ordered genes, normalized enrichment scores, and FDR for gene sets defined as having known cis-regulatory elements for the transcription factors MYC:MAX (B) and E2F1 (C) within 4 kb of their transcription start sites. (D) Scatterplot of signal-to-noise ratios of genes enriched in activated cDC1s from experiments described here and in Fig. 4. (E–G) Rank-ordered gene sets as described in B and C for published gene sets related to cDC1 maturation (25). (H and I) Heat maps of gene expression for gene sets of cDC1 maturation markers (H) and core biosynthetic processes (I), which are derived from GSEA results in SI Appendix, Tables S4 to S6. Expression is scaled by row z-score.
Although there is evidence that MXD1 regulates a unique transcriptional program, models of its function have been developed with respect to its activity as a MAX-binding partner that antagonizes MYC-supported transcription (46, 47). We set out to test whether the suppressive activity of MXD1 in mature cDC1s overlaps with the support of transcription by LMYC in immature cDC1s. GSEA results identified elevated expression of genes with promoter-proximal MYC:MAX and E2F1 DNA-binding motifs (Fig. 5 B and C). In addition, a model for reciprocal action of MXD1 and MYCL in activated cDC1s is supported by a large subset of genes with concordance between MXD1-mediated suppression and MYCL-mediated support (Fig. 5D).
Numerous factors have been identified as sufficient to induce maturation, but none has been shown to be absolutely required for maturation at steady state or during immune responses in vivo (26). The precise description of transcriptional changes that occur during cDC1 maturation in vivo can now be used to identify putative regulators of this process. Therefore, we conducted preliminary analyses that can be used to inform future work on the role of MXD1 in cDC1 function. Using curated gene sets related to cDC1 maturation (24), we performed GSEA to investigate whether MXD1 activity correlated with maturation gene sets. In activated cDC1s from Mxd1−/− mice, there was significant enrichment and elevated expression of genes known to be repressed on maturation (Fig. 5E). Functionally, these gene sets corresponded to cell cycle control and DNA replication (Fig. 5 F and G). Finally, GO-based GSEA revealed that MXD1 regulates the transcriptional fitness of cDC1s by repressing transcription associated with biosynthetic processes (Fig. 5 H and I).
Discussion
In the present study, we examined the impacts of Mycl and Mxd1 deficiency on global transcription as a function of cDC1 maturation state. Genetic analyses have demonstrated that Myc and Mycn are necessary for the development of hematopoietic and nonhematopoietic lineages during embryogenesis and adulthood (9, 10, 48, 49). However, models of transcriptional regulation by MYC family members are based largely on empirical studies of MYC function. All MYC family members have the ability to function as proto-oncogenes (2, 3, 5, 18, 19, 50–52). Mediated by interactions with MAX, their functions can be redundant when amplified to supraphysiological levels in transformed cells (11, 53). Analysis of transformed cell lines has identified that a core transcriptional program is conserved across MYC family members, but MYC-, MYCN-, and MYCL-specific signatures have also been reported (11, 54). For example, MYCN is required during neurogenesis, and MYCN-amplified cancers are enriched with a gene signature associated with neuronal function (10, 16). Likewise, both MYC and MYCN are necessary for hematopoiesis and immune responses, and thus MYC- and MYCN-amplified cancers are enriched with a gene signature associated with cytokines and immune responses (13, 48).
Initial examination of the Myclgfp/+ mutant mouse model demonstrated that GFP+ hematopoietic cells in vivo were restricted to the DC lineage, including cDC1, cDC2, and pDC subsets (20). More recent whole-transcriptome datasets suggest that Mycl expression is regulated differentially between tissues and among DC subsets, however (Fig. 1C) (23–26). We found that expression of Mycl is an order of magnitude higher in cDC1s than in cDC2s (Fig. 1 E and F). Therefore, in the present study we focused on the role of LMYC in the transcriptional fitness specifically of cDC1s, which correspond to a single developmental lineage of Batf3- and Irf8-dependent cDCs present in lymphoid and nonlymphoid tissues (55, 56). Our analysis revealed that maturation is associated with suppression of Mycl expression across tissues (Fig. 1 B–E).
Immature cDC1s are present in all major lymphoid organs and are the predominant population among cDC1s in the spleen. Therefore, we focused on splenic cDC1s at steady state and during acute inflammation to determine the impact of Mycl deficiency on the cDC1 fitness. Examination of cDC1 cell size and mRNA content revealed that both were reduced in Myclgfp/gfp mice (Fig. 2 A–E). This phenomenon is also associated with MYC deficiency in cancer and activated lymphocytes, suggesting that the core function of MYCL in cDC1 cells is conserved with other MYC family members (32, 34).
Detailed expression microarray analysis using GSEA revealed that MYCL supports transcription broadly in cDC1s at steady state (Fig. 3). We found that MYCL regulates the transcription of genes associated with core biosynthetic processes, such as nucleic acid and peptide biosynthesis (Fig. 3 C–E). Independent of activation status, MYCL activated transcription uniformly, manifested as a uniform reduction in mRNA signal intensity in cDC1s from Myclgfp/gfp (Figs. 3 A and B and 4) (44). A major fraction of MYCL-supported genes was unique to cDC1s isolated from steady-state and activating conditions (Fig. 4B); however, GSEA revealed that the broader functions of these genes converged on the regulation of the biosynthetic processes (Fig. 4 A and C–F). The mechanism by which MYCL-regulated gene expression is sensitive to activation status was not examined here, but in vivo analysis revealed that cDC1 maturation coincides with suppression of Mycl expression (Fig. 1 C–F).
In other contexts, such as lymphocyte activation and MYC amplification, MYC is known to function as an analog regulator of gene expression, where the level of MYC protein expression positively correlates with transcription and binding to dose-dependent enhancer elements (34, 42, 57). Given the ability of poly(I:C) to induce cDC1 maturation in vivo, it is possible that MYCL levels vary between activating and steady-state conditions (25). Additional work is needed to identify the DNA-binding sites of MYCL in cDC1s in immature and activated cDC1s. To inform future investigations, the evidence presented here supports a role for MYCL in the support of transcription of genes that regulate core biosynthetic processes and thus the fitness of cDC1s.
Proteins structurally similar to MYC, such as MXD1, can dimerize with MAX and represses transcription at MYC-regulated loci (22, 46, 58). Generation of an MXD1-deficient mouse model demonstrated a role for MXD proteins in granulocyte cell cycle exit, and Mxd1 expression is associated with terminal differentiation of a number of cellular lineages (59–63). cDC1 maturation is marked by the induction of Mxd1 and concomitant repression of Mycl (Fig. 1C) (23–26). Induction of cDC1 maturation in vivo with poly(I:C) revealed broad inhibition of transcription by MXD1, demonstrated by significantly enhanced mRNA signal intensities in cDC1s from Mxd1−/− mice (Fig. 5A). The repressive activity of MXD1 correlated significantly with genes enriched with promoter-adjacent MYC:MAX and E2F DNA-binding motifs (Fig. 5 B and C). Qualitatively, a large fraction of genes repressed by MXD1 are otherwise supported by MYCL in immature cDC1s (Fig. 5D). Further studies will need to determine whether MYCL and MXD1 execute their reciprocal actions on the same loci. With respect to the independent actions of MXD1 during cDC1 maturation, genes that are normally repressed in mature cDC1s had elevated expression in Mxd1−/− mice (Fig. 5 E–H). Therefore, MXD1 broadly represses the transcription of genes that regulate biosynthesis as a function of cDC1 maturation.
It is widely accepted that DC maturation correlates with the capacity of DCs to effectively regulate immune responses (28). The present study reveals mechanisms by which MYCL and MXD1 regulate cellular fitness in a primary cell lineage. We demonstrate that MYCL and MXD1 have overlapping but reciprocal actions that regulate biological processes associated with cDC1 maturation. This contributes to the growing body of evidence supporting the evolutionary conservation of structure and function of MAX-binding transcription factors across species and cell types (46, 64).
Materials and Methods
Mice.
The generation of Myclgfp/gfp mice has been described previously, and the strain used in this study, B6.129S6(C)-Mycltm1.1Kmm/J, is available publicly from The Jackson Laboratory (20). Mice were maintained in a pathogen-free animal facility and experiments conducted in accordance with institutional guidelines and protocols established by the Animal Studies Committee at Washington University in St. Louis. The generation of Mxd1−/− mice has been described previously (73). Mice were provided by R. Eisenman, Fred Hutchinson Cancer Research Center, and maintained as described above at Washington University in St. Louis. Mice aged 8 to 12 wk were used for all experiments.
Antibodies and Flow Cytometry.
Cells were prepared for staining and analysis at 4 °C in PBS with 0.5% BSA and 2 mM EDTA (MACS buffer). Antibodies used in this study were manufactured by BD Biosciences, Tonbo Biosciences, BioLegend, and Thermo Fisher Scientific. Anti-CD16/32 was used as an Fc-block (2.4G2). The following antibodies were used for staining and depletion: CD11c (N418), MHCII (M5/114/15/2), CD24 (M1/69), CD172a (P84), XCR1 (ZET), B220 (RA3-6B2), SiglecH (eBio440C), Ly6G (IA8), CCR7 (4B12), CD3e (145-2C11), CD19 (1D3), NK1.1 (PK136), and TER-119. Cells were analyzed and sorted on a FACSAria Fusion flow cytometer (BD Biosciences). Data were analyzed using FlowJo software.
Relative cell size was quantified as a function of forward scatter area (FSC-A). Normalization across three independent experiments was performed by dividing FSC-A of each sample by the mean FSC-A within the same experiment and genotype, followed by –log2 transformation. Welch’s t test was performed on the normalized sample means and variance.
Cell Activation, Isolation, and Preparation.
Spleens, skin-draining lymph nodes, mesenteric lymph nodes, mediastinal lymph nodes, lungs, and spleens were dissociated mechanically and enzymatically with 250 μg/mL collagenase B (Roche) and 30 U/mL DNase I (Sigma-Aldrich) with gentle agitation at 37 °C for 45 min in up to 5 mL of Iscove's modified Dulbecco's medium. Red blood cells were lysed with hypotonic buffer containing NH4Cl and KHCO3. Before analysis, cell suspensions were filtered through 70-μm mesh. For microarray experiments, maturation was induced by activation with i.p. injection of 100 μg poly(I:C). For Myclgfp/gfp and Mxd1−/− experiments, mice were euthanized at 5 h and 18 h, respectively. CD24+ cDC1s were purified from the spleen by FACS after labeling with biotinylated anti-CD3e, CD19, NK1.1, TER-119, and Ly6G and depletion by negative selection with MojoSort streptavidin microbeads (BioLegend). Lineage-positive cells were also excluded by staining with Qdot 605 streptavidin conjugate (Thermo Fisher Scientific). Cells were sorted into microcentrifuge tubes containing MACS buffer at 4 °C.
Expression Microarray Analysis.
Cells were lysed in cell lysis buffer provided with the RNeasy Micro Kit (Qiagen). The volume of lysis buffer used for each sample was normalized to cell number as estimated by FACS. An equal volume of External RNA Controls Consortium (ERCC) RNA spike-in mix diluted in lysis buffer (Thermo Fisher Scientific) was added to each sample for downstream normalization of expression microarray signal intensities (79). Total RNA was isolated and genomic DNA digested as described in the manufacturer’s protocol. RNA was amplified with the GeneChip WT Pico Kit (Applied Biosystems) in accordance with the manufacturer’s protocol and hybridized to GeneChip Mouse Gene 1.0 ST microarrays (Affymetrix). Data were processed by robust microarray average (RMA) summarization. Normalization across samples with the 4pl curve fitting function of GraphPad Prism. Standard curves of RNA concentration were generated for each sample by plotting known ERCC concentrations to expression of microarray signal intensities. Experimental signal intensities were estimated in turn by interpolation. Where indicated, statistical analyses are described in the figure legends. Microarray data presented in Fig. 1B are publicly available from the Immunological Genome Project (21, 22) and were analyzed by RMA summarization and quantile normalization with ArrayStar (DNASTAR). Gene sets used in Fig. 5 E–G have been published previously (24).
GSEA and Network Analysis.
Expression microarray data were analyzed by GSEA (36). Probe set signal intensities were normalized to spike-in concentrations and used as the input dataset. Phenotype labels were assigned according to sample genotype. In Figs. 3 to 5, biological process GO-based curated gene sets from MSigDB were used for enrichment analysis (37–39, 65). For Fig. 5, GSEA was also performed with curated gene sets of transcription factor targets and recently published gene sets associated with cDC1 maturation (25). Network analysis and visualization of GSEA results were done using core packages available through Cytoscape, R, R Studio, ggplot2, and tidyverse (66–68). Clustering of genes and gene sets in Fig. 4 C–F was performed in Cytoscape using a prefuse force-directed layout algorithm, as explained in the figure legend (45).
Data Availability Statement.
Original expression microarray data are available at NCBI GEO (accession no. GSE141492) (69).
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (K.M.M.), the National Science Foundation (Graduate Research Fellowship DGE-1143954, to D.A.A.), and the National Cancer Institute at the National Institutes of Health (R35 CA231989, to R.N.E. and F31 CA228240, to D.A.A.).
Footnotes
The authors declare no competing interest.
Data deposition: Data have been deposited at the National Center for Biotechnology Information (NCBI) (BioProject: PRJNA593609), and can be downloaded from the Gene Expression Omnibus (GEO) (accession no. GSE141492).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915060117/-/DCSupplemental.
References
- 1.Brodeur G. M., Seeger R. C., Schwab M., Varmus H. E., Bishop J. M., Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224, 1121–1124 (1984). [DOI] [PubMed] [Google Scholar]
- 2.Lee W. H., Murphree A. L., Benedict W. F., Expression and amplification of the N-myc gene in primary retinoblastoma. Nature 309, 458–460 (1984). [DOI] [PubMed] [Google Scholar]
- 3.Nau M. M., et al. , L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature 318, 69–73 (1985). [DOI] [PubMed] [Google Scholar]
- 4.Payne G. S., Bishop J. M., Varmus H. E., Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature 295, 209–214 (1982). [DOI] [PubMed] [Google Scholar]
- 5.Schwab M., et al. , Enhanced expression of the human gene N-myc consequent to amplification of DNA may contribute to malignant progression of neuroblastoma. Proc. Natl. Acad. Sci. U.S.A. 81, 4940–4944 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwood E. M., Lüscher B., Eisenman R. N., Myc and Max associate in vivo. Genes Dev. 6, 71–80 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Prendergast G. C., Lawe D., Ziff E. B., Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell 65, 395–407 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Eisenman R. N., Deconstructing myc. Genes Dev. 15, 2023–2030 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Davis A. C., Wims M., Spotts G. D., Hann S. R., Bradley A., A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7, 671–682 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Moens C. B., Auerbach A. B., Conlon R. A., Joyner A. L., Rossant J., A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes Dev. 6, 691–704 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Malynn B. A., et al. , N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 14, 1390–1399 (2000). [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz-Martín N., Sierra R., Schimmang T., Villa Del Campo C., Torres M., Myc is dispensable for cardiomyocyte development but rescues Mycn-deficient hearts through functional replacement and cell competition. Development 146, dev170753 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Bahr C., et al. , A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553, 515–520 (2018). [DOI] [PubMed] [Google Scholar]
- 14.King B., et al. , The ubiquitin ligase Huwe1 regulates the maintenance and lymphoid commitment of hematopoietic stem cells. Nat. Immunol. 17, 1312–1321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reavie L., et al. , Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat. Immunol. 11, 207–215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernard O., Drago J., Sheng H., L-myc and N-myc influence lineage determination in the central nervous system. Neuron 9, 1217–1224 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Morgenbesser S. D., et al. , Contrasting roles for c-Myc and L-Myc in the regulation of cellular growth and differentiation in vivo. EMBO J. 14, 743–756 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirvonen H., et al. , Expression of L-myc and N-myc proto-oncogenes in human leukemias and leukemia cell lines. Blood 78, 3012–3020 (1991). [PubMed] [Google Scholar]
- 19.Möröy T., et al. , High frequency of myelomonocytic tumors in aging E mu L-myc transgenic mice. J. Exp. Med. 175, 313–322 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kc W., et al. , L-Myc expression by dendritic cells is required for optimal T-cell priming. Nature 507, 243–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll P. A., Freie B. W., Mathsyaraja H., Eisenman R. N., The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 12, 412–425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayer D. E., Kretzner L., Eisenman R. N., Mad: A heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72, 211–222 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Heng T. S., Painter M. W.; Immunological Genome Project Consortium , The Immunological Genome Project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Jojic V., et al. ; Immunological Genome Project Consortium , Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 14, 633–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ardouin L., et al. , Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity 45, 305–318 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Manh T. P., Alexandre Y., Baranek T., Crozat K., Dalod M., Plasmacytoid, conventional, and monocyte-derived dendritic cells undergo a profound and convergent genetic reprogramming during their maturation. Eur. J. Immunol. 43, 1706–1715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guilliams M., et al. , Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity 45, 669–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson D. A., III, Murphy K. M., Briseno C. G., Development, diversity, and function of dendritic cells in mouse and human. Cold Spring Harb. Perspect. Biol. 10, a028613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brahler S., et al. , Opposing roles of dendritic cell subsets in experimental GN. J. Am. Soc. Nephrol. 29, 138–154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schraml B. U., et al. , Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell 154, 843–858 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Theisen D. J., et al. , WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science 362, 694–699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iritani B. M., Eisenman R. N., c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. U.S.A. 96, 13180–13185 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston L. A., Prober D. A., Edgar B. A., Eisenman R. N., Gallant P., Drosophila myc regulates cellular growth during development. Cell 98, 779–790 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin C. Y., et al. , Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovén J., et al. , Revisiting global gene expression analysis. Cell 151, 476–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashburner M., et al. ; The Gene Ontology Consortium , Gene Ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium G. O.; The Gene Ontology Consortium , The Gene Ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahl P. B., et al. , c-Myc regulates transcriptional pause release. Cell 141, 432–445 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin C. H., Jackson A. L., Guo J., Linsley P. S., Eisenman R. N., Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 28, 3157–3170 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nie Z., et al. , c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mashayekhi M., et al. , CD8α(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 35, 249–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pantel A., et al. , Direct type I IFN but not MDA5/TLR3 activation of dendritic cells is required for maturation and metabolic shift to glycolysis after poly IC stimulation. PLoS Biol. 12, e1001759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su G., Morris J. H., Demchak B., Bader G. D., Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics 47, 1–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conacci-Sorrell M., McFerrin L., Eisenman R. N., An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 4, a014357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen N., et al. , Divergence in DNA specificity among paralogous transcription factors contributes to their differential in vivo binding. Cell Syst. 6, 470–483.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laurenti E., et al. , Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3, 611–624 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knoepfler P. S., et al. , Myc influences global chromatin structure. EMBO J. 25, 2723–2734 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birrer M. J., et al. , L-myc cooperates with ras to transform primary rat embryo fibroblasts. Mol. Cell. Biol. 8, 2668–2673 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins S., Groudine M., Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature 298, 679–681 (1982). [DOI] [PubMed] [Google Scholar]
- 52.Möröy T., et al. , IgH enhancer deregulated expression of L-myc: Abnormal T lymphocyte development and T cell lymphomagenesis. EMBO J. 9, 3659–3666 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mathsyaraja H., et al. , Max deletion destabilizes MYC protein and abrogates Eµ-Myc lymphomagenesis. Genes Dev. 33, 1252–1264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaub F. X., et al. ; Cancer Genome Atlas Network , Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst. 6, 282–300.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edelson B. T., et al. , Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hildner K., et al. , Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeid R., et al. , Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 50, 515–523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hurlin P. J., et al. , Mad3 and Mad4: Novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 14, 5646–5659 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foley K. P., et al. , Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 17, 774–785 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McArthur G. A., et al. , MAD1 and p27(KIP1) cooperate to promote terminal differentiation of granulocytes and to inhibit Myc expression and cyclin E-CDK2 activity. Mol. Cell. Biol. 22, 3014–3023 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chin L., et al. , Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc. Natl. Acad. Sci. U.S.A. 92, 8488–8492 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Västrik I., et al. , (1995) Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J. Cell Biol. 128, 1197–1208 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu S., et al. , TGF-beta enforces senescence in Myc-transformed hematopoietic tumor cells through induction of Mad1 and repression of Myc activity. Exp. Cell Res. 315, 3099–3111 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Zirin J., et al. , Interspecies analysis of MYC targets identifies tRNA synthetases as mediators of growth and survival in MYC-overexpressing cells. Proc. Natl. Acad. Sci. U.S.A. 116, 14614–14619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liberzon A., et al. , Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2017). [Google Scholar]
- 67.Wickham H., ggplot2: Elegant graphics for data analysis (Springer, New York, 2009). [Google Scholar]
- 68.Wickham H., et al. , Welcome to the tidyverse. J. Open Source Software 4, 1686 (2019). [Google Scholar]
- 69.Anderson D. A., Murphy K. M., The MYCL and MXD1 transcription factors regulate the fitness of murine dendritic cells. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141492. Deposited 5 December 2019. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original expression microarray data are available at NCBI GEO (accession no. GSE141492) (69).