Abstract
BACKGROUND.
Prostate-specific membrane antigen (PSMA) is a validated target for the imaging and therapy of prostate cancer. Here, we report the detailed characterization of four novel murine monoclonal antibodies (mAbs) recognizing human PSMA as well as PSMA orthologs from different species.
METHODS.
Performance of purified mAbs was assayed using a comprehensive panel of in vitro experimental setups including Western blotting, immunofluorescence, immunohistochemistry, ELISA, flow cytometry, and surface-plasmon resonance. Furthermore, a mouse xenograft model of prostate cancer was used to compare the suitability of the mAbs for in vivo applications.
RESULTS.
All mAbs demonstrate high specificity for PSMA as documented by the lack of cross-reactivity to unrelated human proteins. The 3F11 and 1A11 mAbs bind linear epitopes spanning residues 226–243 and 271–288 of human PSMA, respectively. 3F11 is also suitable for the detection of PSMA orthologs from mouse, pig, dog, and rat in experimental setups where the denatured form of PSMA is used. 5D3 and 5B1 mAbs recognize distinct surfaceexposed conformational epitopes and are useful for targeting PSMA in its native conformation. Most importantly, using a mouse xenograft model of prostate cancer we show that both the intact 5D3 and its Fab fragment are suitable for in vivo imaging.
CONCLUSIONS.
With apparent affinities of 0.14 and 1.2 nM as determined by ELISA and flow cytometry, respectively, 5D3 has approximately 10-fold higher affinity for PSMA than the clinically validated mAb J591 and, therefore, is a prime candidate for the development of next-generation theranostics to target PSMA.
Keywords: monoclonal antibody, glutamate carboxypeptidase II, NAALADase, in vivo imaging, prostate cancer
INTRODUCTION
Prostate carcinoma (PCa) is by far the most common non-cutaneous malignancy in men and the second cause of cancer-related deaths, accounting for 9% of all male cancer-related fatalities in the US in 2015 [1]. A recently published comprehensive validation of immunohistochemical biomarkers of PCa emphasized prostate-specific membrane antigen (PSMA) as one of only four independent prognostic markers for prostate-specific antigen relapse following radical prostatectomy [2]. PSMA, also known as glutamate carboxypeptidase II (GCPII), is a membrane-bound metallopeptidase with an expression pattern restricted mainly to the healthy prostate secretory-acinar epithelium and the plasma membrane of epithelial PCa. Dysplastic and neoplastic transformation of the prostate tissue is accompanied by substantial increase in PSMA levels, with the most prominent expression observed in high-grade, metastatic, and castration-resistant disease [3]. Apart from PCa tissue, PSMA was also found in the neovasculature of a variety of solid tumors, but not physiological healthy vasculature [4,5], aside from within granulation tissue, secretory endometrium, and keloid scars [6]. As a result of a fairly restricted PSMA expression pattern, bioactive molecules targeting PSMA associated with either PCa or tumor neovasculature provide excellent therapeutic opportunities and offer versatile diagnostic tools for the detection of various solid cancers [7–9].
Small-molecule ligands comprise the most prominent class of PSMA-specific reagents. For biomedical applications (in particular, PCa imaging and therapy) the inhibitor molecules are functionalized with a suitable tracer such as a radionuclide, fluorescent dye, magnetic resonance (MR) contrast agent, or a toxin [8–10]. Within the last 10 years, urea-based compounds have become most prominent in the field, and numerous clinical trials are ongoing to validate their use in patients with PCa and other cancers [11–13]. Small molecules offer distinct advantages such as high affinity, very rapid clearance, and ease of synthesis and formulation. On the other hand, potential caveats especially for therapeutic applications might include promiscuous binding to glutamate carboxypeptidase 3 (GCP3; a human paralog of PSMA with high structural similarity) [14], nephrotoxicity and pronounced accumulation to lacrimal and salivary glands [15].
Macromolecular reagents, most notably monoclonal antibodies (mAbs), offer a viable alternative to small-molecule PSMA ligands for imaging and therapy [16–19]. Consequently, several mAbs as well as their conjugates and recombinant fragments are being evaluated in a variety of experimental and preclinical models. At present, J591 and 7E11 (including their conjugates) are the only two anti-PSMA antibodies that have been developed beyond phase I clinical trials, with the 111In-labeled 7E11/CYT-356 (ProstaScint®) constituting the only mAb approved by the FDA for PCa imaging. However, ProstaScint® recognizes an intracellular epitope of PSMA and, therefore, it primarily binds to necrotic cells. Accordingly, ProstaScint® displays compromised sensitivity and is not suitable for live cell staining, including the imaging of tumor neovasculature [20]. These limitations were mitigated by the development of second generation mAbs that recognize extracellular epitopes of human PSMA, most notably J591. The murine mAb J591 was described and characterized in 1997 by Liu et al. [21] and, currently, is the most advanced second generation mAb. Various conjugates of J591 (or its humanized form) have been prepared and characterized as potential diagnostic and therapeutic agents and are subject to late-stage clinical trials [21–25].
Nevertheless, there is still an unmet need for the development of new mAbs against this tumor target. Drawbacks of existing anti-PSMA antibodies include limited commercial availability, poorly defined epitopes, and lack of data on cross-reactivity toward GCPII paralogs and orthologs. Here, we report the identification and detailed characterization of four novel PSMA-specific mAbs and provide a direct comparison with J591, currently the “gold standard” in this field.
METHODS
Cell Lines
HEK293T/17 cells obtained from the American Type Culture Collection were grown in DMEM medium (Sigma–Aldrich, Steinheim, Germany) supplemented with 10% v/v fetal bovine serum (FBS; Gibco, Life Technologies, Carlsbad, CA). PC-3 and DU145 cells, also from the American Type Culture Collection, LNCaP cells, kindly provided by Z. Hodny (IMG, Prague, Czech Republic), and CW22Rv1 cells, a gift from R. Lapidus (UMBC, Baltimore, MD), were all grown in RPMI 1640 medium (Sigma–Aldrich) with 10% v/v FBS. All cell lines were kept under humidified 5% CO2 atmosphere at 37°C. Stable overexpression of human PSMA in HEK293T/17 cells was realized by transfection of pcDNA4/V5-His A vector (Invitrogen, Carlsbad, CA) carrying the cDNA for full-length human PSMA (FOLH1; NCBI Reference sequence: NM_004476.1). Following transfection using jetPRIME (Polyplus-transfection, Illkirch, France), cells were selected in the presence of Zeocin (25 μg/ml; InvivoGen, San Diego). Stable transfectants were isolated by repeated cloning of single cell progeny.
Proteins Used in This Study
rhPSMA.
Purification of the extracellular part of human PSMA (rhPSMA; denoted rhGCPII in the original paper, residues 44–750) was described in detail elsewhere [26]. Briefly, the recombinant protein was expressed in Schneider S2 cells and purified by ion-exchange chromatography (Q and SP Sepharose FF), affinity chromatography on Lentil-Lectin Sepharose, and size-exclusion chromatography (SEC) on a Superdex 200 column with 20 mM Tris–HCl, 150 mM NaCl, pH 8.0 as mobile phase (all resins/columns from GE Healthcare Bio-Sciences, Uppsala, Sweden). Purified rhPSMA was concentrated and stored at −80°C until further use.
Avi-PSMA.
The extracellular part of human PSMA comprising an N-terminal Avi-tag (Avi-PSMA) was prepared as previously described [27]. Briefly, the recombinant protein was expressed in Schneider S2 cells stably transfected with E. coli biotin protein ligase localized to the endoplasmic reticulum. Avi-PSMA was purified from the cell culture supernatant by affinity chromatography using Streptavidin Mutein Matrix (Roche, Basel, Switzerland) and eluted with 2 mM D-biotin. Pooled fractions were concentrated and loaded onto a Superdex 200 column equilibrated with 20 mM Tris–HCl, 150 mM NaCl, pH 8.0 as mobile phase. Purified Avi-PSMA was aliquoted, shock-frozen in liquid nitrogen and stored at −80°C until further use.
Mouse GCPII, rat GCPII, pig GCPII, human GCP3, human NAALADase L, mouse GCP3.
The recombinant proteins were a kind gift from J. Konvalinka, IOCB, Prague, and their cloning, expression, and purification was described elsewhere [28,29].
Hybridomas
Murine mAbs were prepared by immunizing BALB/c mice with purified rhPSMA using a standard protocol [30]. Briefly, two 12-week old female mice were injected subcutaneously with 50 μg rhPSMA in 100 μl PBS mixed with 100 μl of Complete Freund’s Adjuvant (Sigma–Aldrich). Three subcutaneous booster injections (50 μg rhPSMA in 100 μl PBS + 100 μl Incomplete Freund’s Adjuvant) were applied in weekly intervals. The final intraperitoneal booster of 100 μg rhPSMA in 200 μl PBS was administered approximately 1 month later. Three days after that, mice were sacrificed and spleen-derived immune cells were fused with SP2/0Ag14 myeloma cells using 50% w/v polyethylene glycol 1,450 solution (Sigma–Aldrich) [30]. Positive clones were selected by ELISA with rhPSMA as target antigen, then, mAb-producing cells were re-cloned by dilution into Opti-Clone Hybridoma Cloning Factor (MP Biomedicals, Santa Ana, CA) to isolate a single cell colony, which was expanded and stored in liquid nitrogen.
mAb Expression and Purification
A starter culture of the hybridoma was expanded in RPMI 1640 supplemented with 10% v/v FBS, ribonucleosides (Gibco), penicillin, and streptomycin (PAA, Pasching, Austria).
mAb production was performed in a spinner cultivation system in serum-free RPMI 1640 at 37°C and 95% humidity under 5% CO2 atmosphere. The production was carried on for 10 days while the culture was additionally spiked twice with a new aliquot of growing hybridoma cells. Cell culture supernatants were harvested by centrifugation at 2,701g for 10 min and concentrated to approximately 1/10 of the original volume using tangential flow filtration (TFF; Millipore, Mosheim, France). mAbs were then purified by affinity chromatography on HiTrap rProtein A Sepharose (GE Healthcare Bio-Sciences). To this end, the concentrated supernatants were loaded onto the column equilibrated in PBS (equilibration buffer), followed by washing with 10 volumes of the equilibration buffer, and the captured mAbs were eluted with 100 mM Na-citrate, pH 5.0. The eluate was immediately neutralized by addition of 1/10 volume of 1 M Tris–HCl, pH 8.0, concentrated and subjected to SEC on a Superdex 200 column with PBS as mobile phase. Purified mAbs were concentrated to approximately 5 mg/ml (concentration determined by A280) and stored at 4°C until further use. Purified J591 and GCP-04 mAbs were obtained from Dr. Bander and Dr. Konvalinka, respectively [26,31].
Isotyping
Individual mAbs were isotyped using the Rapid ELISA Mouse mAb Isotyping Kit (Pierce, Thermo Fisher Scientific, Rockford, IL) according to the manufacturer’s protocol. Briefly, 50 μl of the tested sample (250 ng/ml of purified mAb in PBS) was added to all eight wells of a strip, which are pre-coated with different class- or subclass-specific capture antibodies. Then, 50 μl of the Goat Anti-Mouse IgG + IgA + IgM HRP Conjugate was added to each well and incubated for 1 hr at room temperature. Following extensive washing, 75 μl of the supplied tetramethylbenzidine (TMB) substrate was added to each well and the signal was quantified via absorbance measurement at 450 nm (CLARIOstar, BMG Labtech, Ortenberg, Germany).
Fab Fragment Preparation
Fab fragments of the individual mAbs were prepared using the Mouse IgG1 Fab and F(ab′)2 Preparation Kit (Pierce) according to the manufacturer’s protocol. Briefly, 5 mg of a given mAb was incubated with 100 μl of Immobilized Ficin in the digestion buffer supplemented with 25 mM cysteine in a final volume of 300 μl for 5 hr at 37°C. The digested mAb was separated from the Ficin resin by centrifugation and the Fab fragment was recovered and separated from undigested mAb and Fc fragment by affinity chromatography using an immobilized Protein A resin in a spin column format. Finally, the Fab fragment was concentrated, loaded onto the Superdex 200 column with PBS as mobile phase and fractions containing the purified Fab were pooled, concentrated, shock-frozen in liquid nitrogen, and stored at −80°C until further use.
Immunoprecipitation
Purified mAb (8 μg in 400 μl TBS/T; Tris buffered saline/0,1% v/v Tween-20) was added to 25 μl of settled Protein G Dynabeads (Life Technologies) and the mixture was incubated on a rotator for 20 min at room temperature. The beads were then washed three times with 0.3 ml TBS/T, mixed with 100 μl of rhPSMA in TBS (8 μg total) and the mixture was incubated on a rotator for 20 min at room temperature. Following magnetic separation, supernatant containing free rhPSMA was discarded and the beads were washed three times with TBS/T. Finally, captured proteins were eluted from the beads with 50 μl 50 mM glycine-HCl, pH 2.8, for 3 min at room temperature. The eluate was immediately neutralized with 1/10 volume of 1 M Tris–HCl, pH 8.0, and analyzed by SDS–PAGE followed by Coomassie Brilliant Blue G-250 staining.
Western Blotting
Samples of cell lysates, cell culture supernatants, and purified proteins were resolved by reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE; Supplementary Figs. S1 and S2). Following SDS–PAGE, the proteins were electroblotted onto a PVDF membrane using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad Laboratories, Hercules, CA). The membrane was blocked with 5% w/v non-fat dry milk/TBS (blocking buffer) for 1 hr and subsequently incubated overnight with 5 μg/ml of mAb in the blocking buffer. Membranes were washed three times with TBS/T and incubated for 2 hr with HRP-conjugated goat anti-mouse antibody (1 mg/ml; Bio-Rad) in blocking buffer at 1:10,000 dilution. Finally, blots were washed in TBS/T and developed with Luminata Forte chemiluminescence substrate (Merck, Millipore) according to the manufacturer’s protocol. Chemiluminescence signals were visualized using the ImageQuant LAS4000 Imaging System (GE Healthcare Bio-Sciences).
Epitope Mapping
A set of 18-mer peptides, in total 83 peptides with 9-residue overlaps, covering the complete sequence of human PSMA was custom-made by PepScan (Lelystad, The Netherlands). A biotin tag was attached to the N-terminus of each peptide via a 6-aminohexanoic acid linker. MaxiSorp microtiter plates (Nunc, Thermo Fisher Scientific) were coated with 50 μl of 5 μg/ml strepavidin in TBS buffer overnight at 4°C. The coating solution was discarded and plates were blocked with 200 μl of 0.5% w/v BSA for 2 hr at room temperature. Subsequently, 50 μl of the peptide solution (2 μM in PBS + 0.5 mg/ml BSA) was added to individual wells and incubated for 1 hr at room temperature. Excess peptide was washed away and plates were treated with 50 μl mAb solution in PBS (2 μg/ml) for 2 hr at RT. Finally, the plates were washed three times in PBS/0.05% v/v Tween-20 and remaining bound mAb was detected after 1 hr incubation with an goat anti-mouse secondary antibody conjugated to horseradish peroxidase (Bio-Rad) diluted 1:10,000 in PBS. Signals were developed with 0.5 mg/ml o-phenylenediamine dihydrochloride (OPD) + 0.015% v/v hydrogen peroxide in 0.05 M phosphate-citrate buffer, pH 5, and monitored via absorbance measurement at 492 nm (CLARIOstar).
Native ELISA
ELISA experiments were carried out in white 384-well MaxiSorp plates at 25°C. Plates were coated with 20 μl of streptavidin solution (5 μg/ml in 100 mM Na-borate, pH 9.5) for 1 hr. The coating solution was discarded, plates were washed twice with TBS and blocked with 80 μl of 1% w/v BSA for 1 hr. Following washing steps (3× TBS/0.05% v/v Tween-20), 20 μl of Avi-PSMA (0.4 nM in TBS/0.1% v/v Tween-20) was added to each well and incubated for 1 hr. Plates were washed again with TBS/T and probed with a twofold serial dilution of the mAb (20 μl, starting concentration 50 nM in TBS/0.1% v/v Tween-20). Following washing steps, bound mAb was detected after 1 hr incubation with a goat anti-mouse secondary antibody conjugated to horseradish peroxidase (Bio-Rad) diluted 1:50,000 in TBS/T. Signals were developed using the Luminata Forte ELISA chemiluminescence substrate (Merck) according to the manufacturer’s protocol. Baseline-corrected data were analyzed with Prism 5 software (GraphPad, San Diego, CA).
Surface Plasmon Resonance Real-Time Affinity Measurements
Surface plasmon resonance (SPR) spectroscopy was performed on a BIAcore 2000 instrument (BIAcore, GE Healthcare Bio-Sciences). The Mouse Antibody Capture Kit (GE Healthcare Bio-Sciences) was used to immobilize ~6,000 resonance units (ΔRU) of anti-mouse IgG on a measuring and a reference channel of a CM5 sensorchip using the amine coupling kit (both GE Healthcare Bio-Sciences). The mAbs 5B1, 5D3, and J591 were diluted to 1 μg/ml in HEPES buffered saline (HBS; 10 mM HEPES-NaOH, 150 mM NaCl, pH 7.4) with 0.005% v/v Tween-20 (HBS/T0.005) and applied to the sensorchip to reach ~Δ80 RU immobilized antibody (5B1: Δ85 RU; 5D3: Δ75 RU; J591: Δ80–85 RU). A dilution series from 128 to 1 nM rhPSMA was prepared in HBS/T0.005 and applied to the sensorchip in the same buffer. Complex formation was monitored for 240 sec at a flow rate of 25 μl/min, then dissociation was followed for 4,000 sec. Regeneration of the sensorchip was achieved by applying up to four injections of glycine-HCl, pH 1.7. The sensorgrams were corrected by double subtraction of the corresponding signals measured for the in-line reference channel and an averaged baseline determined from three buffer blank injections [32]. Kinetic parameters were determined by data fitting using BIAevaluation software version 4.1 (BIAcore).
Immunofluorescence Microscopy
Cells grown on coverslips coated by 0.1% w/v gelatin from porcine skin were fixed, washed with PBS and incubated with each mAb (20 μg/ml) overnight at 4°C. Three fixation protocols were used: (i) 4% w/v formaldehyde/PBS for 15 min followed by permeabilization with 0.1% v/v Triton X-100/PBS for 15 min, (ii) methanol cooled to −20°C for 10 min, and (iii) 5% v/v acetic acid diluted in 95% v/v ethanol cooled to −20°C for 10 min. After washing with PBS/0.05% v/v Tween-20 (PBS/T) slides were incubated for 1 hr at RT with the goat anti-mouse secondary antibody conjugated with Alexa Fluor 488 (5 μg/ml in PBS/T; Life Technologies). Following a washing step with PBS/T, processed slides were treated with 4′, 6-diamidino-2-phenylindole (DAPI; 1 μg/ml) for 5 min. Finally, cells were mounted in VectaShield medium (Vector Laboratories, Burlingame, CA). Fluorescence signal was visualized under the confocal microscope TCS SP8 (Leica Microsystems, Wetzlar, Germany) using the immersion oil objective with 63× magnification. Digital scans were processed with Adobe Photoshop software (Adobe Systems, San Jose, CA).
Flow Cytometry
Cells were harvested by treatment with 0.025% w/v Trypsin/0.01% w/v EDTA/PBS for 3 min, washed and incubated with each mAb at a final concentration of 5 μg/ml in a total volume of 20 μl for 30 min at 4°C. In case of titration experiments, the anti-PSMA mAbs 5B1, 5D3, and J591 were used in twofold dilution series spanning the concentration range from 533 nM down to 0.25 pM. Following washing step, cells were incubated with a goat anti-mouse secondary antibody conjugated to Alexa Fluor 647 (4 μg/ml; Life Technologies). In the competition experiment, rhodamine-labeled 5D3 mAb was used at a final concentration of 2.5 μg/ml and non-labeled antibodies were used in the concentration range 2.5–250 μg/ml. Finally, cells were washed and stained with Hoechst 33258 to gate the viable cell population. All incubation and washing steps were done in PBS supplemented with 0.5% w/v gelatin from cold water fish skin. Cell samples were immediately analyzed using the LSRII or LSRFortessa flow cytometer (both from BD Biosciences, San Jose, CA). At least 30,000 viable cells were gated for subsequent analysis with FlowJo software (FlowJo, LLC, Ashland, OR).
Antibody Labeling for In Vivo Imaging
A total of 50 μg of each carrier-free IgG or Fab were labeled by pipetting into a microcentrifuge tube containing 10 μg of IRDye680RD-NHS ester or IRDye800CW-NHS ester (LI-COR Biosciences, Lincoln, NE) dissolved in 2 μl DMSO and complementing the volume to 100 μl by adding 50–70 μl of PBS, pH 7.4. The conjugation reaction was performed for 12 min at ambient temperature before loading the sample on a PBS conditioned Sephadex G-25 size-exclusion column (GE Healthcare Bio-Sciences), which was operated according to the manufacturer’s instructions. Antibody was assayed to contain ≤5% (by fluorescence) unincorporated dye using silica gel HLF in normal phase TLC (Analtech, Newark, DE) developed in 100% v/v acetonitrile + 0.1% v/v TFA, where the Rf of the antibody conjugate is 0 and the Rf of free dye is >0.5. Labeled antibodies were formulated as 30 μg of conjugate in 200 μl of sterile PBS, pH 7.5 immediately prior to injection.
In Vivo Near Infrared Fluorescence (NIRF) Imaging
All animal studies were conducted in full compliance with a protocol approved by the Johns Hopkins University Animal Care and Use Committee. Five young adult male athymic nude mice (Taconic Biosciences, Hudson, NY) were prepared as described previously [33,34] to carry a single subcutaneous xenograft each of PSMA-positive PC-3 PIP cells and PSMA-negative PC-3 flu cells (a gift from Warren B. Heston, the Cleveland Clinic). Near IR dye-labeled intact IgG or 5D3 Fab, as indicated, was injected via the tail vein when tumor xenografts had reached 4–6 mm in diameter. Imaging was performed on a Pearl Impulse imager (LI-COR Biosciences). All images were displayed using the manufacturer’s software and normalized to the same acquisition time to facilitate direct comparison between mice and over time. Image acquisition began 45 min after fluorescent antibody injection and concluded 72 hr post-injection. Following the 72 hr image acquisition, each mouse was euthanized by 3% v/v isoflurane-anesthetized cervical dislocation and dissected to allow imaging of the tumors without attenuation from the skin.
Ex Vivo NIRF Imaging of Tumor Sections
Immediately following euthanasia, both PSMA-positive PC-3 PIP and PSMA-negative PC-3 flu tumors were harvested and frozen over dry ice. The tumors were then sectioned to 20 μm using an HM Microm 550 cryotome (Thermo Fisher Scientific) and annealed to charged glass slides (VWR, Radnor, PA). Slides were allowed to thaw to ambient temperature and dry prior to scanning using a LI-COR Odyssey imager (LI-COR Biosciences). Both 700 and 800 nm emission channels were captured and displayed using the manufacturer’s software.
RESULTS AND DISCUSSION
mAb Preparation and Purification
Four hybridoma cell lines were prepared according to standard protocols using rhPSMA as an immunogen and later as a target for the identification of antibody-producing hybridoma clones. The PSMA-specific mAbs were purified to homogeneity from the hybridoma supernatants by protein A affinity chromatography followed by size exclusion chromatography (Supplementary Fig. S1). The overall yield was 5.2, 5.7, 17.6, and 33.4 mg per liter cell culture supernatant for 1A11, 3F11, 5B1, and 5D3, respectively. Using Rapid ELISA Mouse Antibody Isotyping Kit we determined that all mAbs are of the IgG1/k isotype (Table I).
TABLE I.
Basic Characteristics of the Newly Developed mAbs
| mAb | Isotype | Epitope | Residuesa | Speciesb | Experimental usec |
|---|---|---|---|---|---|
| 1A11 | IgG1/κ | Linear | 271–288 | Human | WB, ICC, IHC, ELISA |
| 3F11 | IgG1/κ | Linear | 226–243 | Human, mouse, rat, pig, dog | WB, ICC, IHC, ELISA |
| 5B1 | IgG1/κ | Conformational | n.d. | Human | IF, ICC, IHC, ELISA, FC, in vivo |
| 5D3 | IgG1/κ | Conformational | n.d. | Human | IF, ICC, IHC, ELISA, FC, in vivo |
Residues of human PSMA recognized by a given mAb.
PSMA orthologs recognized by a given mAb.
WB, Western blotting; ELISA, enzyme-linked immunosorbent assay; IHC, immunohistochemistry; ICC, immunocytochemistry; FC, flow cytometry; IF, immunofluorescence.
Immunoprecipitation and Epitope Mapping
Our initial experiments were aimed to determine whether a given mAb recognizes a linear or a conformational epitope of human PSMA as this information helps in governing subsequent experimental approaches. To this end, we used immunoprecipitation (IP) to identify mAbs recognizing rhPSMA in its native conformation. Individual mAbs were captured on protein-G paramagnetic beads and then incubated with rhPSMA. Following extensive washing, beads were treated with 100 mM glycine-HCl, pH 2.8, and released proteins were analyzed by SDS–PAGE (Supplementary Fig. S2). Results clearly showed that clones 5D3 and 5B1 are able to bind and immunoprecipitate native rhPSMA, while 1A11 and 3F11 did not recognize the fully folded native PSMA ectodomain. The J591 and GCP-04 mAbs, which are known to recognize native and denatured PSMA, respectively, were used as control mAbs [21,26].
To pinpoint epitopes recognized by individual mAbs we used a set of 83 overlapping 18-mer peptides covering the entire sequence of human PSMA. N-terminally biotinylated peptides were immobilized on a streptavidin-coated 96-well plate and probed with a given mAb at 2 μg/ml concentration. Following extensive washing, bound mAbs were detected by anti-mouse secondary antibody conjugated to horseradish peroxidase. The results were in agreement with the immunoprecipitation experiments. First, the absence of any detectable signal for 5B1 and 5D3 suggests that these two mAbs only recognize conformational epitopes present at the PSMA surface and cannot bind to linear 18-mer peptides comprising the library. Conversely, 1A11 and 3F11 specifically bound the PSMA-derived linear peptides spanning residues 271–288 and 226–243, respectively (Table I, Fig. 1A). Of note, within fully folded PSMA the 271–288 amino acid segment is located at the dimer interface and, therefore, not accessible to mAb binding. The 226–243 epitope is partially buried in the three-dimensional structure of the native enzyme and adopts a distinct α-helical fold, again precluding efficient binding of mAb 3F11 to this epitope within the context of native PSMA.
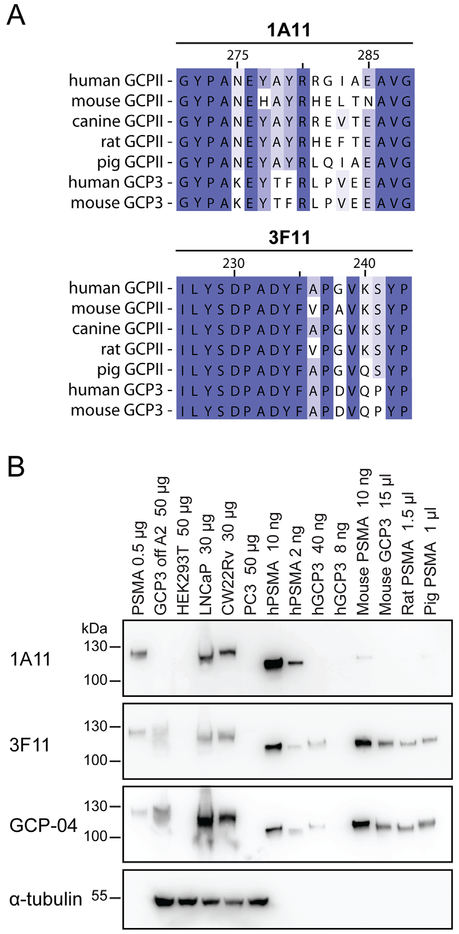
Fig. 1.
Epitope mapping and mAb Western blotting. (Panel A): Alignment of the epitopes on PSMA from different species recognized by the mAbs 1A11 and 3F11 as revealed by peptide scanning. Panel B: Purified ectodomains of human PSMA and several orthologs/paralogs, cell culture supernatants as well as cell lysates were separated by reducing 10% SDS–PAGE, electrotransferred onto a PVDF membrane, and probed with individual mAbs. Lanes: 1. human PSMA-overexpressing HEK293T/17 lysate (0.5 μg); 2. GCP3 overexpressing HEK293T/17 lysate (50 μg); 3. HEK293T/17 lysate (50 μg); 4. LNCaP lysate (30 μg); 5. CW22Rv1 lysate (30 μg); 6. PC-3 lysate (50 μg); 7. human PSMA (10 ng); 8. human PSMA (2 ng); 9. human GCP3 (40 ng); 10. human GCP3 (8 ng); 11. mouse PSMA (10 ng); 12. mouse GCP3 (cell culture supernatant; 15 μl); 13. Rat PSMA (cell culture supernatant; 1.5 μl); 14. pig PSMA (cell culture supernatant; 1 μl).
Combined IP and epitope-mapping data clearly define suitable experimental setups for the individual mAbs to be used (Table I). 1A11 and 3F11 are mostly suitable for techniques dealing with denatured PSMA, including Western blotting, immunocytochemistry, immunohistochemistry, and ELISA under denaturing conditions. On the contrary, 5B1 and 5D3 are ideally suited for IF, ICC, IHC, ELISA, FC, and in vivo experiments where native PSMA prevails.
PSMA Specificity and Species Cross-Reactivity
Although all four mAbs were raised against purified human PSMA we wanted to know whether they (i) cross-react with non-related human proteins; (ii) recognize human glutamate carboxypeptidase 3 (GCP3), a close PSMA homolog with 67% identity at the amino acid level; and (iii) recognize PSMA/GCP3 orthologs from different species. To this end we first employed Western blotting, in which PSMA-expressing cells and purified PSMA orthologs/paralogs were used to probe the specificity of 1A11 and 3F11. The data (Fig. 1B and Supplementary Fig. S3) can be summarized as follows: (i) both 1A11 and 3F11 do not cross-react with non-related human proteins as a panel of human tumor cell lines of different origins (prostate, skin, breast, ovary, kidney, nervous tissue, colon, and lung) was negative for any staining and only a single band with a size corresponding to PSMA was observed in lysates from PSMA-positive LNCaP and CW22Rv1 cells; (ii) 1A11 does not recognize any PSMA paralogs/orthologs whereas 3F11 can be used to detect mouse, rat and pig PSMA, human and mouse GCP3 (Fig. 1B), but not human and mouse NAALADase L (not shown).
Figure 1A shows epitopes of human PSMA recognized by 1A11 and 3F11, together with the alignment with corresponding sequences from PSMA orthologs/paralogs. Although not tested experimentally, this sequence alignment suggests that 3F11 will also recognize canine PSMA, expanding thus the utility of this mAb to yet another experimental animal model. The 3F11 PSMA cross-reactivity with several mammalian species is similar to that of the mAb GCP-04, although the latter is somewhat more sensitive for human PSMA (cf. Fig. 1B). However, as 3F11 and GCP-04 recognize distinct epitopes within the PSMA sequence, they may be used back-to-back to confirm the specificity of staining in human/animal tissues.
We recently determined that epitopes recognized by YPSMA-1 and YPSMA-2, two widely used commercial mAbs, span amino acids 469–486 [29]. Consequently, these mAbs will definitely cross-react with human PSMA-L, an intracellular protein with 98% amino acid sequence identity to human PSMA [35]. The shorter PSMA-L is only found in humans and higher primates, and two sequence homology regions between PSMA-L and PSMA comprise residues 1–442 and 309–750, respectively. Contrary to YPSMA-1 and YPSMA-2, our 3F11 and 1A11 mAbs (as well as GCP-04 and the mAb 3E6 from Dako) will not cross-react with PSMA-L as their epitopes are missing there. Consequently, the new mAbs can be preferably used for IHC staining to ensure high specificity for PSMA in human tissue samples.
Immunofluorescence Microscopy
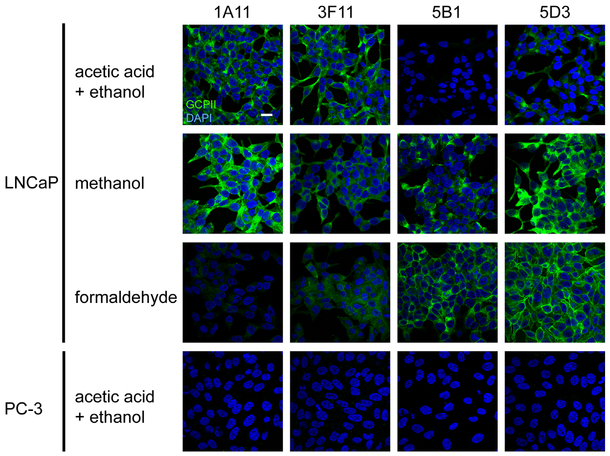
To determine compatibility of individual mAbs with different methods of sample preparation for immunofluorescence microscopy, we evaluated their performance using LNCaP and control PC-3 cells (Fig. 2) treated by denaturing or non-denaturing fixatives.
Fig. 2.
PSMA detection on LNCaP (PSMA-positive) and PC-3 (PSMA-negative) cell lines by immunofluorescence microscopy. Individual cell lines were fixed on glass coverslips using three different fixation protocols and probed with tested mAbs (20 μg/ml), followed by detection with a secondary antibody conjugated with Alexa Fluor 488 (green channel). Under these varying conditions distinct intensities of both cytoplasmic and plasma membrane staining were observed on LNCaP cells. The PSMA-negative PC-3 prostate cell line revealed no staining (cells fixed by acetic acid + ethanol are shown as an example). Nuclei were visualized with DAPI (blue channel); scale bar: 20 μm.
The observed staining pattern is in line with the findings on mAb specificity stated above including their recognition of the denatured (3F11 and 1A11) or native (5B1 and 5D3) antigen. First, the absence of a fluorescence signal in PC-3 cells confirmed high specificity of all mAbs for human PSMA. For 3F11 and 1A11 and LNCaP cells, the strongest signal was observed when a denaturing fixation by ethanol/acetic acid or methanol was used while much weaker labeling was seen when cells were fixed with paraformaldehyde. To the contrary, the staining intensity for 5B1 and 5D3 was highest using the “native” paraformaldehyde fixation, less pronounced in methanol fixation and very weak (negligible) when acetic acid/ethanol mixture was used.
Side-by-side comparison of the mAbs 5B1 and 5D3 with GCP-04 and J591 revealed identical staining pattern and similar staining intensity for the particular type of fixation. Furthermore, we also compared the staining pattern of 5B1 on Tritonpermeabilized and non-permeabilized LNCaP cells. With permeabilized cells both cytoplasmic and plasma-membrane localization of PSMA was visible, while only the cell-surface signal was detected using non-permeabilized cells (Supplementary Fig. S4).
Flow Cytometry
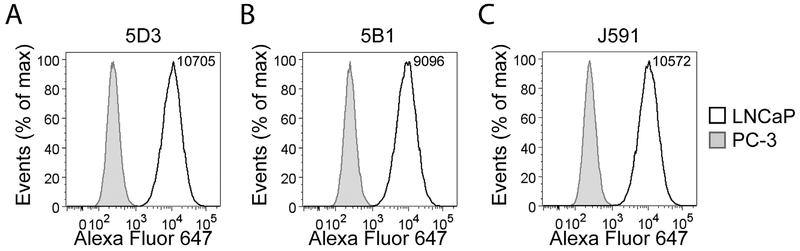
LNCaP and PC-3 cells were used to assess the suitability of mAbs 5D3 and 5B1 for flow cytometry as well as to compare their performance with J591. mAb binding (5 μg/ml) to the PSMA displayed on the surface of live cells was detected via indirect staining with a secondary antibody labeled with Alexa Fluor 647 (Fig. 3). As s1n from the flow cytometry histograms, no staining was observed for PSMA-negative PC-3 cells, confirming the lack of cross-reactivity of all tested mAbs with other human proteins. At the same time, the PSMA-positive LNCaP cell line was specifically labeled by all three mAbs with the median signal intensity highest for 5D3, followed by J591 and 5B1 (Fig. 3). These results were further corroborated using additional PSMA-positive (CW22Rv1) and PSMA-negative (DU-145) cells (Supplementary Fig. S5). Additionally, HEK293T cells transfected with human PSMA, human GCP3 or mouse PSMA were used to test reactivity of the three mAbs toward these proteins. As predicted, all three mAbs stained cells expressing human PSMA, but no staining was observed for HEK293T cells transfected with mouse PSMA and a much weaker signal was detected for human GCP3 (Supplementary Fig. S5).
Fig. 3.
Flow cytometry analysis of PSMA expression on live cells. Specificity and labeling intensity of the mAbs 5D3 (A) and 5B1 (B) were compared to J591 (C) using LNCaP (PSMA-positive) and PC-3 (PSMA-negative) cell lines of prostate origin. Harvested cells were incubated with 5 μg/ml of each mAb and binding detected by a secondary antibody conjugated to Alexa Fluor 647 using an LSRII flow cytometer. A minimum of 30,000 cells were analyzed for each sample using FlowJo software. While staining on PC-3 cells was negative, staining profiles of LNCaP cells suggest comparable performance for all three mAbs tested. According to the indicated median fluorescence intensity, 5D3 revealed the strongest binding affinity toward PSMA on LNCaP cells.
To assess overlaps in epitopes recognized by 5D3, 5B1, and J591 we carried out competition experiments using flow cytometry. To this end, rhodamine-labeled 5D3 and 5B1 were mixed with unlabeled “competing” mAbs in different molar ratios (1:1, 1:10, and 1:100) and binding of the mixtures to the surface of LNCaP cells was evaluated using flow cytometry. We did not observe any competition between rhodamine-labeled 5B1 and the two remaining mAbs, thus pointing toward spatially distinct epitopes. On the other hand, we clearly observed competition between 5D3 and J591 (and self-competition of 5D3 used as a positive control; Supplementary Fig. S6). This finding suggests that 5D3 and J591 share spatially fully or partially overlapping epitopes.
Affinity Determined by ELISA, Surface-Plasmon Resonance, and Flow Cytometry
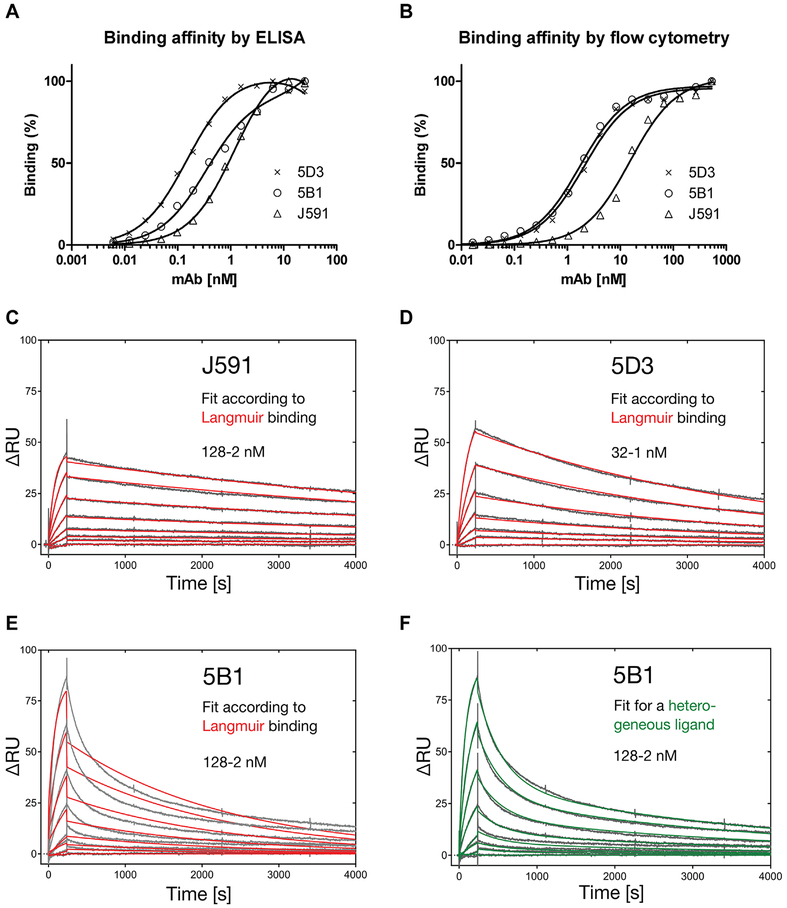
To determine affinity of mAbs 5D3 and 5B1 in comparison with J591 for PSMA we employed three complementary experimental setups including ELISA, surface-plasmon resonance, and flow cytometry. The results are summarized in Table II and Figure 4. First, we used dilution series of 5B1, 5D3, and J591 processed by ELISA under native conditions to both verify the practical applicability of 5B1 and 5D3 in this experimental setup and determine binding affinities. Resulting signals were fitted using GraphPad (one site total binding model), and normalized binding curves are shown in Figure 4A. Dissociation constants (KD) for 5B1, 5D3, and J591 were determined as 0.26, 0.14, and 1.12 nM, respectively (Table II). Notably, despite its lower affinity, J591 yielded approximately 1.5-fold and 3-fold stronger signal under saturating conditions than 5D3 and 5B1, respectively (data not shown). These observations are in line with findings reported in our recent publication where J591 consistently yielded the strongest signal in native ELISA from all mAbs tested [29], but the mechanism underlying this observation is unknown at present.
TABLE II.
Affinities of Individual mAbs as Determined by ELISA, Flow Cytometry, and Surface-Plasmon Resonance
| SPR | ||||||
|---|---|---|---|---|---|---|
| mAb | ELISA [nM] | FC [nM] | kon [105·(M·s)−1] | koff [10−4·s−1] | KD [nM] | χ2 for Langmuir fit |
| 5B1 | 0.26 ± 0.07 | 1.8 ± 0.13 | n.d.a | n.d.a | n.d.a | 5.86a |
| 5D3 | 0.14 ± 0.01 | 2.1 ± 0.10 | 2.36 ± 0.003 | 2.56 ± 0.002 | 1.08 ± 0.002 | 0.40 |
| J591 | 1.12 ± 0.10 | 15.2 ± 1.3 | 1.02 ± 0.0008 | 1.23 ± 0.002 | 1.21 ± 0.002 | 0.29 |
| (1.12 ± 0.06)b | (0.84 ± 0.15)b | (0.75 ±0.14)b | ||||
The binding mode of the 5B1 antibody did not show a simple 1:1 association/dissociation according to the Langmuir model as indicated by the high χ2 value.
Kinetic data obtained with an inverted SPR setup by [1] for J591 are given in brackets.
Fig. 4.
Affinity of the mAbs 5B1 and 5D3 for PSMA determined by ELISA, flow cytometry and surface-plasmon resonance. Panel A: For direct ELISA a 384-well MaxiSorp plate was coated with streptavidin and charged with N-terminally biotinylated Avi-PSMA. mAbs were applied in twofold dilution series and binding was detected by a secondary antibody conjugated to horseradish peroxidase. The resulting chemiluminescence signal was plotted against the mAb concentration and data were analyzed by curve fitting with GraphPad. Panel B: LNCaP cells were incubated with twofold dilution series of tested mAbs and binding was detected by a secondary antibody conjugated to Alexa Fluor 647 using a LSRII flow cytometer. Median values of fluorescence signals were plotted against the mAb concentration and data were analyzed by curve fitting with GraphPad. Panels C–F: Real-time SPR measurements for individual mAbs were made on a BIAcore 2000 instrument. A CM5 sensorchip was amino-coupled with anti-murine Fc-specific capture antibody and ~Δ80 RU of the respective mAb was immobilized. Application of rhPSMA in a dilution series resulted in sensograms which were fitted to a Langmuir 1:1 binding model for J591 (C) and 5D3 (D). In contrast, 5B1 clearly showed biphasic dissociation, not in agreement with the Langmuir model (E) but suggesting a heterogeneous mAb species (F).
In a complementary approach, affinities of 5B1, 5D3, and J591 were determined by flow cytometry analysis of dilution series of all three mAbs (533 nM–0.25 pM) applied to LNCaP cells. Results were fitted using GraphPad (one site total binding model), and normalized binding curves are shown in Figure 4B while calculated affinity constants are listed in Table II. Overall, these data are consistent with those from ELISA measurements showing that under this experimental setup our novel mAbs have approximately eightfold higher affinity for PSMA than J591.
Finally, to precisely determine kinetic and thermodynamic binding constants of individual mAbs we performed surface-plasmon resonance (SPR) real-time analyses on a BIAcore instrument. Here, a sensorchip surface was coated with an anti-mouse IgG capture antibody, which was subsequently used to immobilize the respective mAb. The sensograms obtained by applying recombinant rhPSMA were fitted to a 1:1 binding model according to Langmuir, which resulted in dissociation constants of 1.08 nM for 5D3 and 1.21 nM for J591 (Fig. 4C, D). In line with the ELISA and FC measurements, these results reveal a KD in the low single-digit nanomolar range, with the best affinity for mAb 5D3. Previous affinity measurements for J591 in an inverted setup with Avi-PSMA immobilized on the sensorchip via NeutrAvidin indicated 1.6-fold higher affinity (750 pM) [29].
In contrast, the fit of the 5B1 sensograms to a Langmuir binding model (Fig. 4E) resulted in a strong discrepancy, causing an elevated Chi2 (χ2) of 5.9 compared to 0.4 and 0.3 for the 5D3 and J591 antibodies, respectively. Hence, the raw data measured for 5B1 were fitted to different binding models including (i) bivalent analyte (χ2 3.7), (ii) 1:1 binding with mass transfer (χ2 4.1), (iii) two state reaction (χ2 χ 46.1), and (iv) heterogeneous ligand (χ2 0.75). Thus, the biphasic dissociation observed for the 5B1sample could point to an inhomogeneous antibody pool secreted by the hybridoma clone, such as the differential N-glycosylation or other post-translational modifications in the V-regions. However, additional experiments including 5B1 gene cloning/sequencing, separation of the putative mAb mixture by anion- or cation-exchange chromatography as well as isoelectric focusing did not confirm the existence of different antibody species (data not shown). This issue thus warrants further studies.
In Vivo/Ex Vivo NIRF Imaging
Finally, as a proof-of-concept experiment for our future endeavors focusing on the development of imaging and therapeutic reagents, we used an established mouse model implanted with paired PC-3 based xenograft lines that are isogenic except for PSMA expression [36] to (i) assess the suitability of 5D3, the best performing mAb, as well as its Fab fragment for in vivo imaging; and (ii) to directly compare the performance of our mAbs to J591 and GCPII-04, two commercially available mAbs. Each whole IgG antibody (5B1, 3F11, 5D3, GCPII-04, and 1A11) was fluorescently labeled with IRDye680RD to allow for co-injection and direct comparison with the gold standard for immunoimaging of PSMA, J591 [37–40], which was labeled with the orthogonal IRDye800CW.
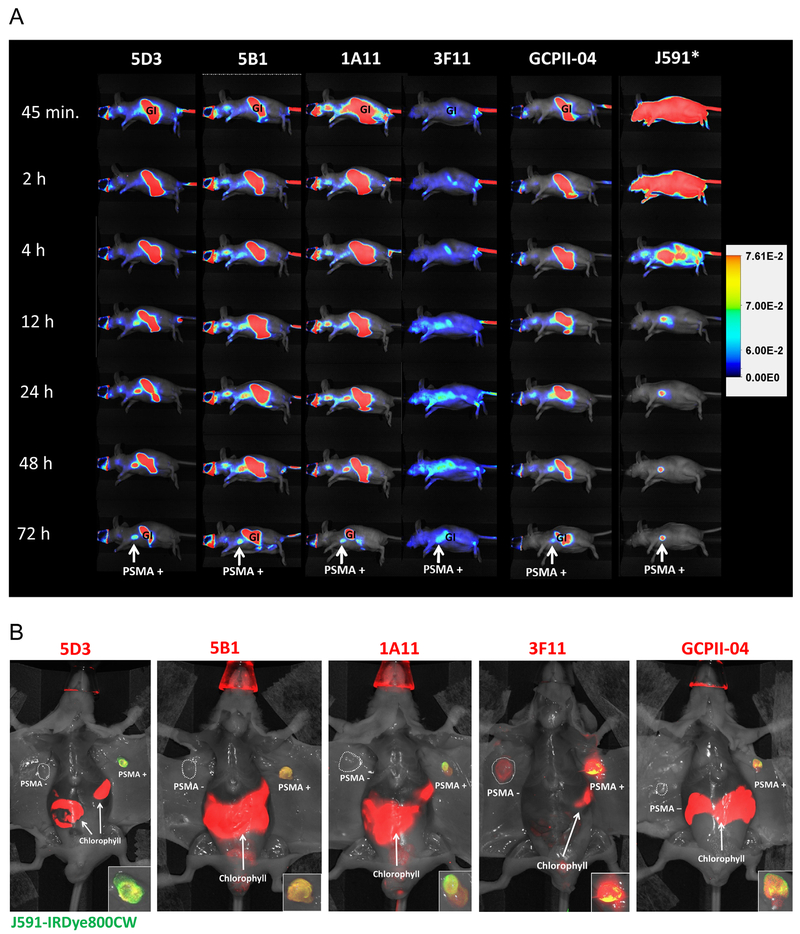
As shown in Figure 5A, all antibodies accumulated within the PSMA-positive PC-3 tumor and the highest signal-to-noise occurred around 72 hr post-injection for all clones with the exception of J591, which yielded tumor-specific images 48 hr post-injection. Notably, clone 3F11 displayed an overall low uptake in comparison with all other clones.
Fig. 5.
Pharmacokinetics of new anti-PSMA mAbs compared with J591 and ex vivo NIRF imaging showing tumor specificity. Panel A: Representative mice, each bearing a PSMA-positive and PSMA-negative (not depicted in views shown) xenograft, was co-injected with the indicated IRDye680RD-labeled mAb (designated in the top row) and J591-IRDye800CW. Images collected at various times post-injection at the 710 nm peak emission (except for J591, asterisk) were normalized to the same exposure time. All mAbs were bound to the PSMA-positive tumor, as detectable 12 hr post-injection, and were cleared from non-target sites by 72 hr. 3F11 displayed particularly low uptake overall while 5D3 and 5B1 both showed high tumor uptake with 5D3 displaying the highest tumor signal as well as extensive non-target tissue clearance by 48 hr, similar to J591. Autofluorescence due to dietary chlorophyll was observed across the gastrointestinal (GI) tract. As J591 was imaged at 800 nm autofluorescence was not detected in this case. Panel B: Each mouse shown is from panel A after the 72 hr data point. The ventral skin was removed to reduce attenuation and reveal both tumors. J591-IRDye800CW is displayed in green; hence, yellow denotes co-localization of J591 uptake (green) with the indicated mAb (red) tested in each mouse. Panels at the lower right show an enlargement of the PSMA-positive tumor. Notably, all antibodies tested except 3F11 displayed specificity for the PSMA-positive PC-3 PIP tumor versus the antigen-negative tumor. 5D3 and 5B1 both showed mostly yellow/orange co-localization with J591 while 1A11 showed more heterogeneous tumor uptake. The mAb GCPII-04, which binds to a cytoplasmic epitope of PSMA, also displayed a more heterogeneous binding pattern compared with J591.
The specificity of each antibody was further assessed by opening the ventral skin following euthanasia 72 hr post-injection such that both tumors could easily be imaged without attenuation from skin. Figure 5B shows mice with the indicated IRDye680RD-labeled mAb in red overlaid with IRDye800CW-labeled J591 in green where yellow indicates co-localization. Again, all mAbs were taken up by the PSMA-expressing tumor whereas, notably, 3F11 was also accumulated by the PSMA-negative tumor. Additionally, 3F11 and 1A11 both displayed non-homogenous co-localization with J591, suggesting reduced specificity for native PSMA. Both 5D3 and 5B1 displayed uptake in the PSMA-positive tumor only and co-localized with J591. Likewise, GCPII-04 displayed PSMA-specific tumor uptake as well as homogenous distribution and co-localized with J591 in vivo, even though GCPII-04 recognizes denatured PSMA [26,29].
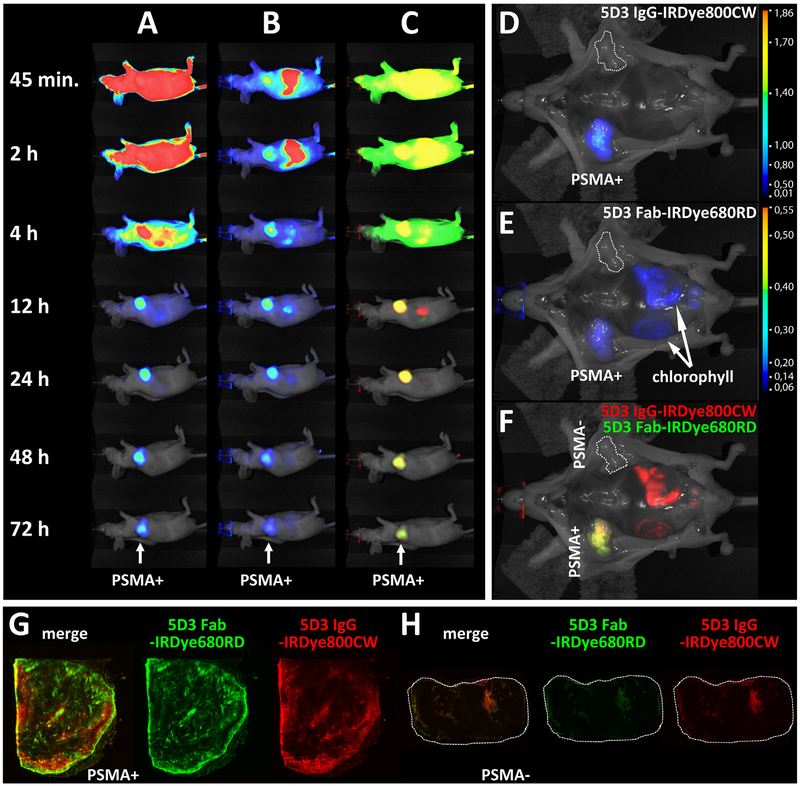
In the interest of developing a theranostic agent suitable for clinical translation, a Fab fragment of 5D3 was prepared and evaluated for its in vivo pharmacokinetics and tumor specificity alongside the parental IgG. Figure 6 shows the pharmacokinetic distribution of 5D3 IgG (panel A), Fab (panel B), and their overlay (panel C), where IgG is displayed in red and the Fab is indicated green. Both the IgG and Fab accumulated in the PSMA-positive tumor although the Fab was observed in the tumor as early as 45 min post-injection, while with the IgG the tumor was only observed starting 4 hr post-injection. The best signal-to-noise for Fab and IgG was achieved at 4 and 24 hr post-injection, respectively. Twelve hours post-injection the degree of observed overlap between IgG and Fab uptake was nearly 100% (C). Ex vivo data reveal homogenous uptake in the PSMA-positive tumor by both IgG and Fab and confirm their high specificity for PSMA (Fig. 5D–F).
Fig. 6.
In vivo pharmacokinetics and ex vivo high resolution NIRF imaging (tumor sections) of the 5D3 IgG and Fab fragment. Panels A–F: A mouse was co-injected with the IgG-IRDye800CW (A) and the Fab-IRDye680RD (B) with overlay shown in (C), where IgG is displayed in green and the Fab is displayed in red. In (A), high tumor contrast is achieved 12 hr post-injection and by 24 hr the whole-mouse background uptake is also low. In (B), high tumor contrast is achieved already 2 hr post-injection [gastrointestinal (GI) signal is chlorophyll] and continues till 48 hr post-injection. The overlay in row (C) shows a high degree of co-localization from 12 hr onwards. Panels (D, E) show the 72 hr uptake without skin of the IgG and Fab, respectively. Both are selective for the PSMA-positive tumor at 72 hr. Panel (F) reveals largely yellow co-localization of both antibody formats within the PSMA-positive tumor. Panels G, H: PSMA-positive PC-3 PIP and PSMA-negative PC-3 flu tumors were harvested following imaging, sectioned and scanned to detect the high-resolution distribution of IgG-IRDye800CW and Fab-IRDye680RD within the tumors. Row (G) shows a section of the PSMA-positive PC-3 PIP tumor where green depicts IgG, red depicts Fab and yellow shows co-localized IgG and Fab in the leftmost section. Row (H) shows the same investigation for sections of PSMA-negative PC-3 flu tumor (dotted lines). In the PSMA-positive section, IgG uptake (green) appears more confined to the tumor rim and small focal regions near the rim while the Fab (red) appears to display a wider uptake pattern both away from the rim and within tumor.
>Both tumors were sectioned after imaging and then the sections were imaged at high (≥25 μm) resolution to determine whether the 5D3 Fab exhibits broader uptake within the tumor sections relative to IgG distribution. Panels G and H show representative sections of the PSMA-positive and PSMA-negative tumors and the distribution of each immunoglobulin format. The IgG (red) was taken up by the PSMA-positive tumor (panel G) primarily along the rim with focal uptake scattered throughout the interior. The Fab (green) was also primarily distributed along the rim with less scattered uptake in the interior. The overlay (leftmost section) demonstrates more Fab along the rim than IgG, suggesting greater perfusion allows for increased binding to epithelial PSMA. The PSMA-negative tumor section (panel H) revealed very little uptake by either IgG or Fab and showed just a small amount of uptake around the tumor rim as well as a small region in the interior, which might reflect blood flow and pooling [41].
In conclusion, the 5D3 IgG provided high tumor contrast between 12 and 24 hr post-injection (depending on which IRDye was used) and exhibited homogenous uptake consistent with that demonstrated by J591 in the PSMA-positive tumor (Fig. 5). There was no observable mAb uptake in the PSMA-negative tumor. Likewise, the Fab fragment of 5D3 also displayed PSMA-specific tumor uptake and provided high tumor contrast as early as 2 hr post-injection, while maintaining high contrast throughout 24 hr, after which the signal within tumor began to wane (Fig. 6B). Whole mount tumor sections taken at 72 hr post-injection revealed more intense distribution of the Fab fragment within the tumor compared with the IgG, also radiating further from the tumor rim than the IgG (Fig. 6G). The faster pharmacokinetics observed with the 5D3 Fab coupled with its greater penetration into the tumor make it an attractive option for both diagnostic imaging and radioimmunotherapy, which benefit from fast binding kinetics and widespread epithelial target binding [42–44].
The mAb 5B1 also displayed favorable pharmacokinetics but exhibited some non-target background uptake in cervical lymph nodes throughout the first 72 hr, making it less beneficial than 5D3. The mAb 1A11 showed selective PSMA-positive tumor uptake but also suffered from non-target tissue uptake through the initial 48 hr post-injection while mAb 3F11 displayed a large extent of non-specific uptake throughout the mouse, including uptake in the PSMA-negative tumor (Fig. 5). This likely reflects the finding that the 3F11 clone also recognizes murine forms of GCPII, which are present in many tissues including blood [28]. The mAb GCPII-04 was described before and has been included here for comparison due to its known affinity and specificity for denatured PSMA [26,29]. GCPII-04 also displayed high-contrast though lower intensity of PSMA-specific tumor uptake. Comparison of intratumor accumulation with J591 at 72 hr revealed GCPII-04 uptake in regions of the tumor where J591 binding was largely absent (Fig. 5B, tumor inset), suggesting the binding of GCPII-04 to necrotic, denatured tumor tissue, similar to the in vivo binding mechanism of the mAb 7E11 (ProstaScint™) [45].
CONCLUSIONS
In summary, we prepared and fully characterized four novel PSMA-specific monoclonal antibodies. In addition to human PSMA, the mAb 3F11 recognizes orthologs from rat, mouse, pig, and dog, thus expanding its utility for several animal models. Most importantly, mAb 5D3 shows high affinity and specificity for PSMA and is suitable for the in vivo imaging of PSMA-positive tumors, therefore, providing a starting point for future theranostic applications.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. J. Peknicova (Institute of Biotechnology, Prague, Czech Republic) for hybridoma preparation and Dr. J. Konvalinka (Institute of Organic Chemistry and Biochemistry, Prague, Czech Republic) for samples of PSMA othologs/paralogs. C.B. acknowledges the support from the Czech Science Foundation (grant No 301/12/1513). This publication is supported by the project BIOCEV (CZ.1.05/1.1.00/02.0109) from the ERDF, RVO 86652036, and the Deutsche Forschungsgemeinschaft, Germany, in frame of the Collaborative Research Centre SFB 824. Further support was also provided by CA13765 and CA18228.
Grant sponsor: Czech Science Foundation; Grant number: 301/12/1513; Grant sponsor: BIOCEV; Grant number: CZ.1.05/1.1.00/02.0109; Grant sponsor: ERDF, RVO; Grant number: 86652036; Grant sponsor: Deutsche Forschungsgemeinschaft, Collaborative Research Centre SFB 824; Grant sponsor: CA13765; Grant sponsor: CA18228; Grant sponsor: MEYS/BIOCEV-FAR; Grant number: LQ1604.
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2015;65(1):5–29. [DOI] [PubMed] [Google Scholar]
- 2.Huber F, Montani M, Sulser T, Jaggi R, Wild P, Moch H, Gevensleben H, Schmid M, Wyder S, Kristiansen G. Comprehensive validation of published immunohistochemical prognostic biomarkers of prostate cancer—What has gone wrong? A blueprint for the way forward in biomarker studies. Br J Cancer 2015;112(1):140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bostwick DG, Pacelli A, Blute M, Roche P, Murphy GP. Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma: A study of 184 cases. Cancer 1998;82(11):2256–2261. [DOI] [PubMed] [Google Scholar]
- 4.Chang SS, O’Keefe DS, Bacich DJ, Reuter VE, Heston WD, Gaudin PB. Prostate-specific membrane antigen is produced in tumor-associated neovasculature. Clin Cancer Res 1999;5(10): 2674–2681. [PubMed] [Google Scholar]
- 5.Wernicke AG, Kim S, Liu H, Bander NH, Pirog EC. Prostate-specific membrane antigen (PSMA) expression in the neovasculature of gynecologic malignancies: Implications for PSMA-targeted therapy. Appl Immunohistochem Mol Morphol 2016, doi: 10.1097/PAI.0000000000000297 [DOI] [PubMed] [Google Scholar]
- 6.Gordon IO, Tretiakova MS, Noffsinger AE, Hart J, Reuter VE, Al-Ahmadie HA. Prostate-specific membrane antigen expression in regeneration and repair. Mod Pathol 2008;21(12): 1421–1427. [DOI] [PubMed] [Google Scholar]
- 7.Barinka C, Rojas C, Slusher B, Pomper M. Glutamate carboxypeptidase II in diagnosis and treatment of neurologic disorders and prostate cancer. Curr Med Chem 2012;19(6):856–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foss CA, Mease RC, Cho SY, Kim HJ, Pomper MG. GCPII imaging and cancer. Curr Med Chem 2012;19(9):1346–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiess AP, Banerjee SR, Mease RC, Rowe SP, Rao A, Foss CA, Chen Y, Yang X, Cho SY, Nimmagadda S, Pomper MG. Prostate-specific membrane antigen as a target for cancer imaging and therapy. Q J Nucl Med Mol Imaging 2015;59(3): 241–268. [PMC free article] [PubMed] [Google Scholar]
- 10.Sacha P, Knedlik T, Schimer J, Tykvart J, Parolek J, Navratil V, Dvorakova P, Sedlak F, Ulbrich K, Strohalm J, Majer P, Subr V, Konvalinka J. iBodies: Modular synthetic antibody mimetics based on hydrophilic polymers decorated with functional moieties. Angew Chem Int Ed Engl 2016;55(7):2356–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haberkorn U, Eder M, Kopka K, Babich JW, Eisenhut M. New strategies in prostate cancer: Prostate-specific membrane antigen (PSMA) ligands for diagnosis and therapy. Clin Cancer Res 2016;22(1):9–15. [DOI] [PubMed] [Google Scholar]
- 12.Kratochwil C, Giesel FL, Stefanova M, Benesova M, Bronzel M, Afshar-Oromieh A, Mier W, Eder M, Kopka K, Haberkorn U. PSMA-targeted radionuclide therapy of metastatic castration-resistant prostate cancer with 177Lu-labeled PSMA-617. J Nucl Med 2016;57(8):1170–1176. [DOI] [PubMed] [Google Scholar]
- 13.Rowe SP, Macura KJ, Mena E, Blackford AL, Nadal R, Antonarakis ES, Eisenberger M, Carducci M, Fan H, Dannals RF, Chen Y, Mease RC, Szabo Z, Pomper MG, Cho SY. PSMA-based [(18)F]DCFPyL PET/CT is superior to conventional imaging for lesion detection in patients with metastatic prostate cancer. Mol Imaging Biol 2016;18(3):411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hlouchova K, Barinka C, Konvalinka J, Lubkowski J. Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III. FEBS J 2009;276(16): 4448–4462. [DOI] [PubMed] [Google Scholar]
- 15.Hohberg M, Eschner W, Schmidt M, Dietlein M, Kobe C, Fischer T, Drzezga A, Wild M. Lacrimal glands may represent organs at risk for radionuclide therapy of prostate cancer with [(177)Lu] DKFZ-PSMA-617. Mol Imaging Biol 2016;18(3):437–445. [DOI] [PubMed] [Google Scholar]
- 16.Barinka C, Ptacek J, Richter A, Novakova Z, Morath V, Skerra A. Selection and characterization of Anticalins targeting human prostate-specific membrane antigen (PSMA). Protein Eng Des Sel 2016;29(3):105–115. [DOI] [PubMed] [Google Scholar]
- 17.Dassie JP, Hernandez LI, Thomas GS, Long ME, Rockey WM, Howell CA, Chen Y, Hernandez FJ, Liu XY, Wilson ME, Allen LA, Vaena DA, Meyerholz DK, Giangrande PH. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol Ther 2014;22(11): 1910–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu C, Bandekar A, Sempkowski M, Banerjee SR, Pomper MG, Bruchertseifer F, Morgenstern A, Sofou S. Nanoconjugation of PSMA-targeting ligands enhances perinuclear localization and improves efficacy of delivered alpha-particle emitters against tumor endothelial analogues. Mol Cancer Ther 2016; 15(1):106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiehr S, Buhler P, Gierschner D, Wolf P, Rolle AM, Kesenheimer C, Pichler BJ, Elsasser-Beile U. Pharmacokinetics and PET imaging properties of two recombinant anti-PSMA antibody fragments in comparison to their parental antibody. Prostate 2014;74(7):743–755. [DOI] [PubMed] [Google Scholar]
- 20.Ellis RJ, Kaminsky DA, Zhou EH, Fu P, Chen WD, Brelin A, Faulhaber PF, Bodner D. Ten-year outcomes: The clinical utility of single photon emission computed tomography/computed tomography capromab pendetide (Prostascint) in a cohort diagnosed with localized prostate cancer. Int J Radiat Oncol Biol Phys 2011;81(1):29–34. [DOI] [PubMed] [Google Scholar]
- 21.Liu H, Moy P, Kim S, Xia Y, Rajasekaran A, Navarro V, Knudsen B, Bander NH. Monoclonal antibodies to the extracellular domain of prostate-specific membrane antigen also react with tumor vascular endothelium. Cancer Res 1997;57(17): 3629–3634. [PubMed] [Google Scholar]
- 22.Holland JP, Divilov V, Bander NH, Smith-Jones PM, Larson SM, Lewis JS. 89Zr-DFO-J591 for immunoPET of prostate-specific membrane antigen expression in vivo. J Nucl Med 2010;51(8): 1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kampmeier F, Williams JD, Maher J, Mullen GE, Blower PJ. Design and preclinical evaluation of a 99mTc-labelled diabody of mAb J591 for SPECT imaging of prostate-specific membrane antigen (PSMA). EJNMMI Res 2014;4(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tagawa ST, Akhtar NH, Nikolopoulou A, Kaur G, Robinson B, Kahn R, Vallabhajosula S, Goldsmith SJ, Nanus DM, Bander NH. Bone marrow recovery and subsequent chemotherapy following radiolabeled anti-prostate-specific membrane antigen monoclonal antibody j591 in men with metastatic castration-resistant prostate cancer. Front Oncol 2013;3:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tagawa ST, Milowsky MI, Morris M, Vallabhajosula S, Christos P, Akhtar NH, Osborne J, Goldsmith SJ, Larson S, Taskar NP, Scher HI, Bander NH, Nanus DM. Phase II study of Lutetium-177-labeled anti-prostate-specific membrane antigen monoclonal antibody J591 for metastatic castration-resistant prostate cancer. Clin Cancer Res 2013;19(18):5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barinka C, Mlcochova P, Sacha P, Hilgert I, Majer P, Slusher BS, Horejsi V, Konvalinka J. Amino acids at the N- and C-termini of human glutamate carboxypeptidase II are required for enzymatic activity and proper folding. Eur J Biochem 2004;271(13): 2782–2790. [DOI] [PubMed] [Google Scholar]
- 27.Tykvart J, Sacha P, Barinka C, Knedlik T, Starkova J, Lubkowski J, Konvalinka J. Efficient and versatile one-step affinity purification of in vivo biotinylated proteins: Expression, characterization and structure analysis of recombinant human glutamate carboxypeptidase II. Protein Expr Purif 2012;82(1):106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rovenska M, Hlouchova K, Sacha P, Mlcochova P, Horak V, Zamecnik J, Barinka C, Konvalinka J. Tissue expression and enzymologic characterization of human prostate specific membrane antigen and its rat and pig orthologs. Prostate 2008;68(2): 171–182. [DOI] [PubMed] [Google Scholar]
- 29.Tykvart J, Navratil V, Sedlak F, Corey E, Colombatti M, Fracasso G, Koukolik F, Barinka C, Sacha P, Konvalinka J. Comparative analysis of monoclonal antibodies against prostate-specific membrane antigen (PSMA). Prostate 2014;74(16):1674–1690. [DOI] [PubMed] [Google Scholar]
- 30.Peknicova J, Capkova J, Cechova D, Sulcova B. Preparation and characterization of a monoclonal antibody against boar acrosin. Folia Biol (Praha) 1986;32(4):282–285. [PubMed] [Google Scholar]
- 31.Smith-Jones PM, Vallabahajosula S, Goldsmith SJ, Navarro V, Hunter CJ, Bastidas D, Bander NH. In vitro characterization of radiolabeled monoclonal antibodies specific for the extracellular domain of prostate-specific membrane antigen. Cancer Res 2000;60(18):5237–5243. [PubMed] [Google Scholar]
- 32.Myszka DG. Improving biosensor analysis. J Mol Recognit 1999;12(5):279–284. [DOI] [PubMed] [Google Scholar]
- 33.Banerjee SR, Ngen EJ, Rotz MW, Kakkad S, Lisok A, Pracitto R, Pullambhatla M, Chen Z, Shah T, Artemov D, Meade TJ, Bhujwalla ZM, Pomper MG. Synthesis and evaluation of Gd(III)—Based magnetic resonance contrast agents for molecular imaging of prostate-specific membrane antigen. Angew Chem Int Ed Engl 2015;54(37):10778–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, Mease RC, Pullambhatla M, Lisok A, Chen Y, Foss CA, Wang Y, Shallal H, Edelman H, Hoye AT, Attardo G, Nimmagadda S, Pomper MG. [(18)F]Fluorobenzoyllysinepentanedioic acid carbamates: New scaffolds for positron emission tomography (PET) imaging of prostate-specific membrane antigen (PSMA). J Med Chem 2016;59(1):206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Keefe DS, Bacich DJ, Heston WD. Comparative analysis of prostate-specific membrane antigen (PSMA) versus a prostate-specific membrane antigen-like gene. Prostate 2004;58(2):200–210. [DOI] [PubMed] [Google Scholar]
- 36.Kiess AP, Minn I, Chen Y, Hobbs R, Sgouros G, Mease RC, Pullambhatla M, Shen CJ, Foss CA, Pomper MG. Auger radiopharmaceutical therapy targeting prostate-specific membrane antigen. J Nucl Med 2015;56(9):1401–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouchelouche K, Capala J, Oehr P. Positron emission tomography/computed tomography and radioimmunotherapy of prostate cancer. Curr Opin Oncol 2009;21(5):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milowsky MI, Nanus DM, Kostakoglu L, Sheehan CE, Vallabhajosula S, Goldsmith SJ, Ross JS, Bander NH. Vascular targeted therapy with anti-prostate-specific membrane antigen monoclonal antibody J591 in advanced solid tumors. J Clin Oncol 2007;25(5):540–547. [DOI] [PubMed] [Google Scholar]
- 39.Nakajima T, Mitsunaga M, Bander NH, Heston WD, Choyke PL, Kobayashi H. Targeted, activatable, in vivo fluorescence imaging of prostate-specific membrane antigen (PSMA) positive tumors using the quenched humanized J591 antibody-indocyanine green (ICG) conjugate. Bioconjug Chem 2011;22(8): 1700–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborne JR, Akhtar NH, Vallabhajosula S, Anand A, Deh K, Tagawa ST. Prostate-specific membrane antigen-based imaging. Urol Oncol 2013;31(2):144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng QS, Goh V, Milner J, Padhani AR, Saunders MI, Hoskin PJ. Acute tumor vascular effects following fractionated radiotherapy in human lung cancer: In vivo whole tumor assessment using volumetric perfusion computed tomography. Int J Radiat Oncol Biol Phys 2007;67(2):417–424. [DOI] [PubMed] [Google Scholar]
- 42.Larson SM, Carrasquillo JA, Cheung NK, Press OW. Radioimmunotherapy of human tumours. Nat Rev Cancer 2015;15(6): 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sofou S Radionuclide carriers for targeting of cancer. Int J Nanomedicine 2008;3(2):181–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wittrup KD, Thurber GM, Schmidt MM, Rhoden JJ. Practical theoretic guidance for the design of tumor-targeting agents. Methods Enzymol 2012;503:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Troyer JK, Beckett ML, Wright GL Jr. Location of prostate-specific membrane antigen in the LNCaP prostate carcinoma cell line. Prostate 1997;30(4):232–242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.