Abstract
In this work, we describe a microfluidic three-dimensional (3D) chondrocyte culture mimicking in vivo articular chondrocyte morphology, cell distribution, metabolism, and gene expression. This has been accomplished by establishing a physiologic nutrient diffusion gradient across the simulated matrix, while geometric design constraints of the microchambers drive native-like cellular behavior. Primary equine chondrocytes remained viable for the extended culture time of 3 weeks and maintained the low metabolic activity and high Sox9, aggrecan, and Col2 expression typical of articular chondrocytes. Our microfluidic 3D chondrocyte microtissues were further exposed to inflammatory cytokines to establish an animal-free, in vitro osteoarthritis model. Results of our study indicate that our microtissue model emulates the basic characteristics of native cartilage and responds to biochemical injury, thus providing a new foundation for exploration of osteoarthritis pathophysiology in both human and veterinary patients.
Keywords: Organ-on-a-Chip, Cartilage, Primary cells, Tissue-on-a-Chip
1. Introduction
Articular cartilage is exposed to high loads and stresses during joint loading and has limited regenerative capacity. Hence, damage to articular cartilage commonly leads to osteoarthritis, the most common joint disease, which affects 10–12% of the adult population [1]. Despite the high incidence of osteoarthritis, currently no disease-modifying treatments are available, motivating large-scale research efforts. Animal models including sheep, goats, mice, rabbits, dogs, and horses are validated for cartilage research. Among these, the horse, which also suffers from naturally occurring osteoarthritis, provides the closest approximation to human articular cartilage and subchondral bone thicknesses [2]. However, animal trials are expensive and controversial because of ethical considerations, creating the demand for alternative, ethically responsible, and economic research methods. Among others [3], one promising alternative is organ-on-a-chip technology, where the recreation of physiological conditions promotes the formation of tissue-like structures on a microchip platform [4], [5]. Another benefit of organ-on-a-chip technology is the inherent flexibility in design, which allows for relevant concentration gradients and shear force conditions [6], both vital to cartilage physiology and function. For instance, an integrated microfluidic device was used to investigate the effects of growth factor gradients on morphology and proliferation of rabbit articular chondrocytes embedded in Matrigel [7]. A microphysiological cartilage-on-a-chip model that applied strain-controlled compression to three-dimensional (3D) articular cartilage microtissues demonstrated that a 30% confined compression recapitulates the mechanical factors involved in osteoarthritis pathogenesis [8]. Another example integrated a microfluidic base in a multichamber bioreactor containing human bone marrow stem cell (hBMSC)–derived constructs to create tissue-specific microenvironments in which chondral and osseous tissues develop and mature [9]. Despite these recent advances, existing microfluidic technologies are either not scalable and/or do not fully recapitulate the physiological cartilage niche in terms of its in vivo physical dimensions, nutrient availability, and time-dependent diffusion gradients inside the matrix, as well as shear forces. It is also important to note that articular cartilage is an avascular and relatively acellular tissue with the single-cell population, chondrocytes, only being nourished by the synovial fluid through diffusion. The chondrocytes contribute significantly to the stability and integrity of the tissue by synthesizing a strong extracellular matrix (ECM) mainly consisting of collagen type II and proteoglycans [10]. In addition, the functional role of articular cartilage is reflected by its unique organization into three different zones including (a) the superficial zone toward the synovial cavity of the joint, where chondrocytes show a flattened morphology [11]; (b) the middle zone with chondrocytes exhibiting a more typical, spherical morphology surrounded by more randomly arranged collagen fibers and high amounts of aggrecan, a glucosaminoglycan (GAG) typical for articular cartilage; and (c) the deep zone that has the lowest cell density but high contents of aggrecan and collagen fibrils [12].
To combine the need for device scalability, ease of use, and proper tissue functionality, the premise of our work was to recreate an approximate 1:1 articular cartilage model in a single step by inducing chondrocyte redifferentiation that promotes spontaneous self-organization into cartilaginous tissue structures such as the superficial zone [13]. This is achieved by loading hydrogel-embedded primary (equine) chondrocytes into a 3-mm-long and 1-mm-high semicircular tissue chamber designed to establish defined, unidirectional, and reproducible nutrient concentration gradients across the in vitro tissue constructs. Additional regular media exchanges provide cyclic shear stress on chondrocytes residing at the hydrogel to media interface, thus mimicking joint movement resulting in fresh nutrient supply. In this work, we show that our microfluidic 3D chondrocyte culture-on-a-chip array features a number of relevant cartilage characteristics including structural organization, cellular morphologies, and gene expression patterns, as well as limited proliferative capacity and low metabolic activity of resident chondrocytes. Furthermore, we demonstrate that on-chip–established equine cartilage microtissues strongly react to biochemical injury using inflammatory mediators and also respond to triamcinolone steroid treatment, a well-known veterinary therapy option.
2. Results and discussion
2.1. Organ-on-a-chip specifications and design characterization
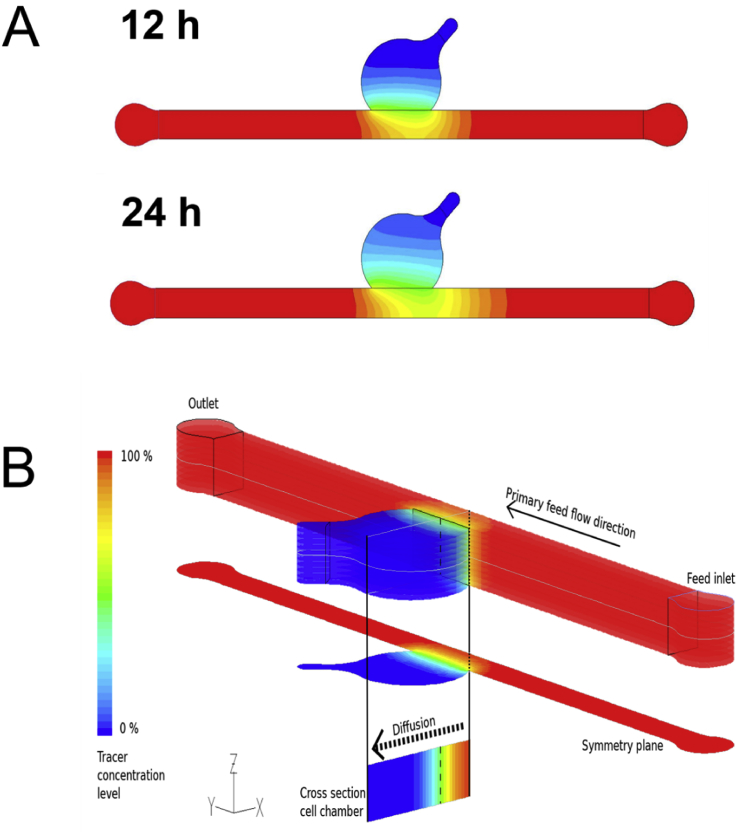
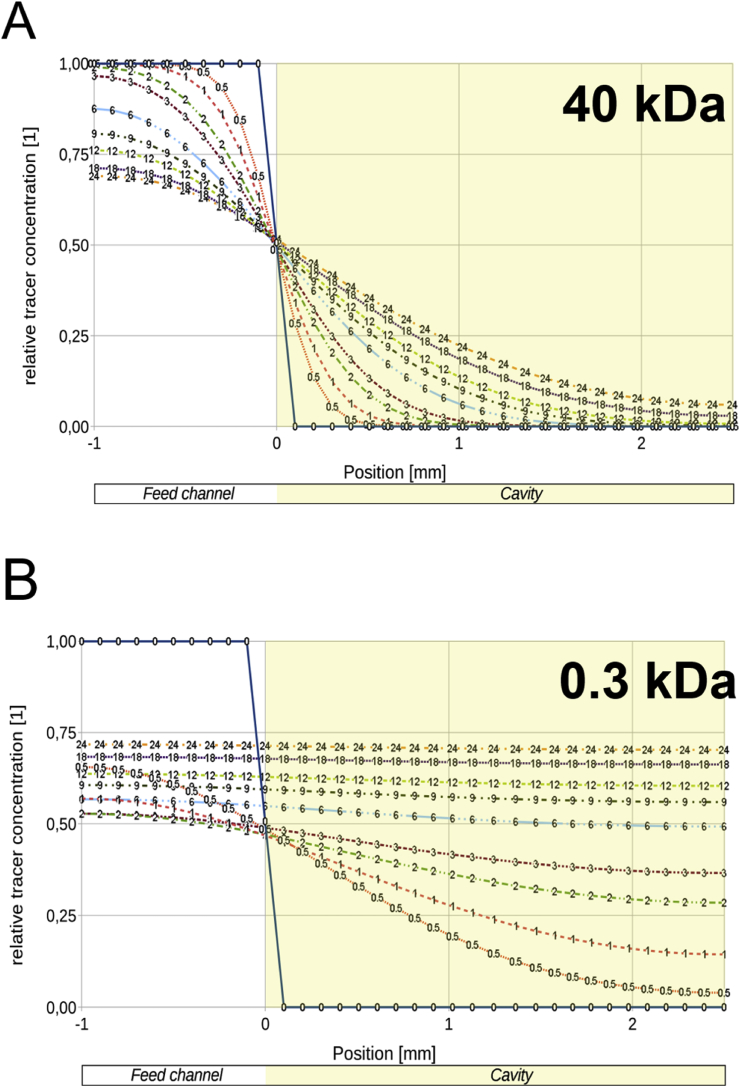
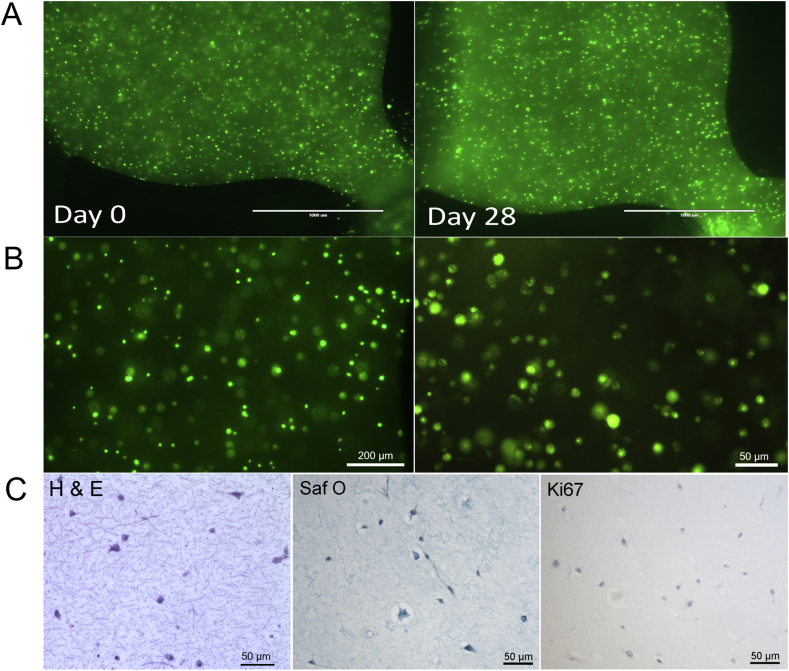
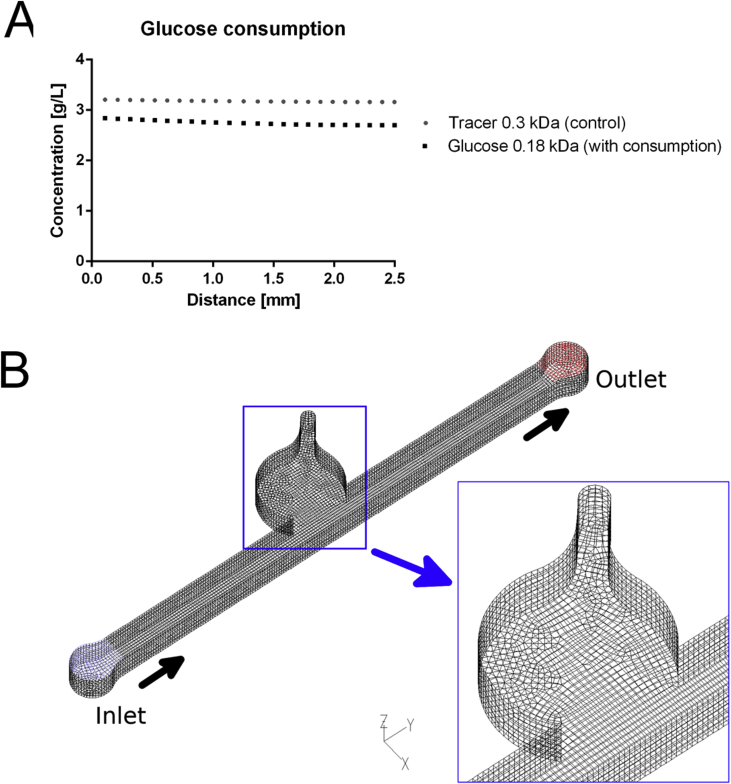
Owing to articular cartilage's avascular nature, nutrient transport in native tissue is mainly accomplished by joint movement and subsequent diffusion from the synovial cavity into the cartilage tissue [3]. In other words, nutrient gradients are present across the cartilage thickness which drive cellular behavior [14] and need to be accounted for in the design of the biological niche on chip. Fig. 1A shows the optimized tissue chamber geometry emulating the in vivo cartilage thickness of the equine stifle joint featuring a 3-mm-wide and 1-mm-high semispherical cultivation chamber adjacent to a rectangular medium supply channel. It is important to highlight that although simple, the present device configuration supports the main tissue requirements including (i) formation of nutrient gradients across the tissue construct without perfusion, (ii) application of periodic shear stress and fresh nutrient supply to the chondrocytes residing at the matrix to fluid interface, and (iii) reliable loading of cell-laden hydrogel to establish reproducible 3D environments for chondrocyte redifferentiation, cultivation, and maturation (Fig. 1). Using our simple chamber design, high cellular viability of chondrocytes was maintained over a period of 28 days as shown by live cell staining in Fig. 1B and S1A. Staining primary chondrocytes with 5-chloromethylfluorescein diacetate (CMFDA Cell Tracker Green™ Dye), a cytoplasmic dye that only persists in viable cells [15], also allowed for morphology assessment inside the turbid fibrin hydrogel. Chondrocyte morphologies are distinct and change from elongated at the superficial layer and round in the middle and deep layers, as shown in Fig. 1C and E. In addition, sectioning and histological evaluation as well as gene expression analysis are accomplished after extracting the complete 3D chondrocyte constructs (Fig. 1D) from the microarray. For instance, Fig. 1D shows hematoxylin and eosin (H&E) staining of the superficial layer, indicating a clear alignment along the medium channel featuring elongated cells. Elongation is most likely caused by medium exchange resulting in a nutrient-rich zone at the biointerface, which promotes cell migration and proliferation from the deeper zones of the hydrogel. Cell migration from the middle and deeper zones strongly points at the presence of a nutrient gradient across the 3D hydrogel construct. To gain a deeper understanding of gradient formation across the 20-mg mL-1 fibrin hydrogel construct, a series of CFD-finite volume simulations were performed to portray small (0.3 kDa) and large (40 kDa) molecule diffusion [6], as well as glucose (0.18 kDa) consumption, using a previously reported consumption rate of 1.28 × 10−7 mol/(L·s) [16]. Results of the first computational fluid dynamics (CFD) calculation are shown in Fig. 2A describing the time-dependent formation of a nutrient gradient based on an average protein of 40 kDa after 12 h and 24 h. Interestingly, even after 24 h of cultivation, a defined and stable nutrient gradient is still present across the 3-mm tissue chamber. A 3D representation of the established nutrient gradient is shown in Fig. 2B, further highlighting the diffusion-based nutrient supply in the present device configuration. In other words, the geometric features of the tissue chamber provide optimized conditions to cultivate chondrocyte tissue constructs inside a microfluidic device. In an attempt to investigate the established gradients in more detail, diffusion distance and speed were calculated for a large (40 kDa) protein and small (0.3 kDa) biomolecules to gain a deeper insight into the nutrient distributions within the hydrogel construct. Fig. 3A shows distance-concentration profiles of a 40-kDa protein over time. Results of the simulation demonstrate that at a distance larger than 2 mm from the fluid interface, only 10% of large molecules reach the deeper zone of the chondrocyte construct within 24 h. The constant nutrient limitation beyond the threshold of 2 mm represents similar conditions as found in deep-zone cartilage [17]. While at the biointerface, a constant nutrient supply of at least 50% is present, at a distance of 1 mm, a time-dependent protein gradient is established ranging from 15% to 23% after 12 h and 24 h, respectively. In turn, Fig. 3B shows that small molecules (0.3 kDa) are rapidly distributed across the entire hydrogel chamber after medium exchange, resulting in a 50% and 75% concentration after 6 h and 24 h, respectively. In addition, simulations including the consumption rate of chondrocytes, shown in Fig S2A, confirm constant supply of glucose to all cells within the hydrogel construct. Time-resolved representations of all conditions can be found in the supplementary video 1. Overall, a reproducible and reliable gradient-based nutrient supply offer optimal cell culture conditions for 3D hydrogel-based chondrocytes.
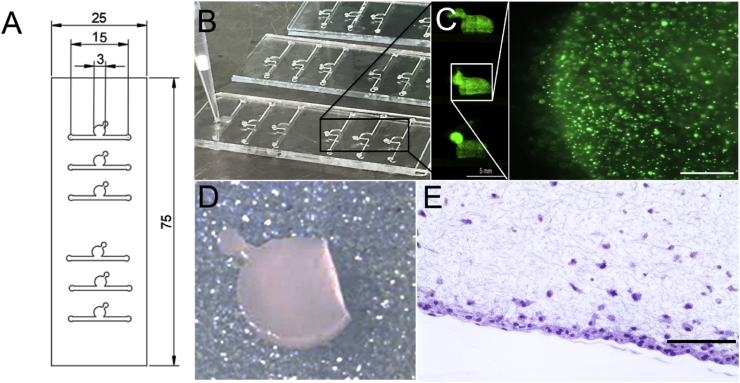
Fig. 1.
(A) CAD design of cartilage-on-a-chip device featuring six individually addressable circular chambers on one object slide, enabling multiplexing of experiments. (B) Photograph of an actual cartilage-on-a-chip device with glass slide as the top layer and PDMS slab with microstructures as the bottom layer showing loading of cell-laden hydrogel in the top chamber using a pipette tip. (C) Lift side showing overview picture of three cell culture chambers and right side showing one individual culture chamber featuring CMFDA-stained primary equine chondrocytes cultivated on-chip for 21 days. Scale bars 5 mm and 500 μm. (D) Intact cell-laden fibrin hydrogel clot released from the device before downstream analyses and (E) histological section of chondrocytes-on-a-chip. Scale bar 100 μm. CMFDA, 5-chloromethylfluorescein diacetate
Fig. 2.
(A) Finite volume (CFD) simulation of diffusion distance of a 40-kDa-sized biomolecule 12 h and 24 h after addition and (B) 3D representation of a 40-kDa-sized biomolecule 12 h after addition, demonstrating the establishment of a diffusion gradient. CFD, computational fluid dynamics.
Fig. 3.
Comparison of diffusion distance and speed of (A) 40-kDa-sized and (B) 0.3-kDa-sized biomolecules into the fibrin hydrogel at various time points up to 24 h. A nutrient gradient is formed for the 40-kDa-sized molecule, stabilizing in the time frame of 12 h–24 h, while the 0.3-kDa-sized molecule diffuses rapidly into the culture chamber and levels out at 75% of feed concentration.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.mtbio.2019.100023.
The following is the supplementary data related to this article:
2.2. Development of gradient-based microfluidic chondrocyte culture
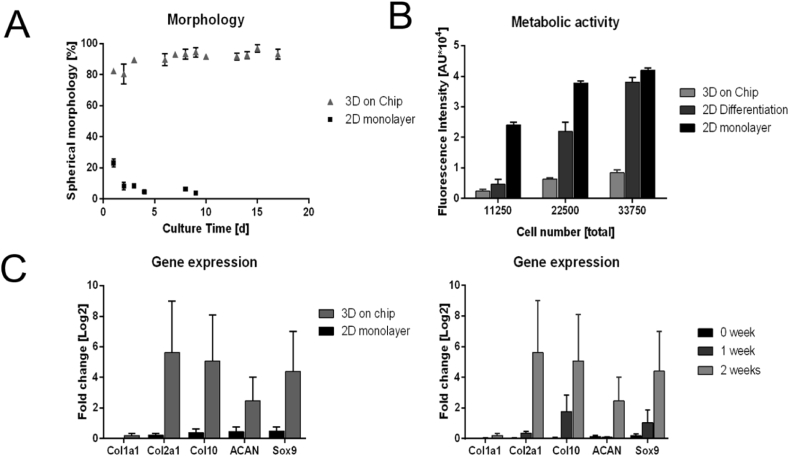
The biological impact of the gradient-based 3D cell culture condition within our microfluidic device was investigated in subsequent experiments to determine (a) cell morphology, (b) cellular metabolism, (c) gene expression, and (d) histology. All microfluidic cultures (3000 cells/mm3) were compared with same-passage, same-donor monolayer chondrocyte cultures (2500 cells/cm2) as controls. Results of our cell morphology experiments are shown in Fig. 4A revealing spherical cell morphologies, a hallmark of articular chondrocytes, which increased from day one to day 8 from 84% to 99% of round cells, remaining at this level throughout prolonged gradient-based 3D culture. In addition to spherical cellular morphologies, low metabolic activities are an important indicator of physiological behavior of mature native articular chondrocytes. Metabolic activity was compared between 2D monolayer and 3D hydrogel primary chondrocyte cultures via Resazurin-based TOX8 Assay. Results in Fig. 4B display lowest metabolic activities by chondrocytes cultivated in 3D hydrogels on chip, while highest metabolic rates are present in chondrocytes in 2D culture. The resulting sevenfold reduction in metabolic activity under gradient-based microfluidic 3D culture conditions highlights the redifferentiated status of the chondrocytes-on-chip. To verify that the redifferentiation seen on chip was due to the 3D gradient-based conditions and not associated with factors in the medium, a comparison of chondrogenic differentiation medium with Ham's F12 was performed in monolayer cultures. As evidenced by the 40% reduction in metabolic rates when chondrocytes were cultured in chondrogenic differentiation medium vs. Ham's F12 medium, an influence toward phenotypic behavior is noted. Further reduction in the metabolic rate was observed under microfluidic 3D culture conditions, demonstrating that the metabolism of on-chip–cultivated 3D chondrocytes resembles the low metabolic activity seen in vivo [18]. Although limited metabolic activity is an indicator of articular chondrocyte behavior, gene expression analysis is needed to substantiate chondrocytic phenotype. To compare expression of cartilage matrix proteins aggrecan (Acan), collagen type 2 (Col2), and a marker for chondrogenic differentiation (Sox9), real-time quantative PCR (RT-qPCR) analysis was performed on the microtissues. Fig. 4C (left panel) highlights the differences in gene expression between 3D and 2D chondrocyte cultures after a period of two weeks in culture. All relevant chondrogenic markers including Col2 and Acan are upregulated within the 3D hydrogel-based chondrocyte constructs. In addition, expression of Col1 is downregulated, thus pointing at the establishment of a stable chondrogenic phenotype. Fig. 4C (right panel) demonstrates time-dependent upregulation in 3D hydrogel chondrocyte constructs of Sox9, Acan, and Col2 chondrogenesis markers, supporting the earlier observations of successful redifferentiation of chondrocytes-on-chip. To further investigate whether discernible tissue remodeling also took place on chip in the presence of gradient-based cultivation conditions, a series of histological investigations were performed. After 3 weeks of culture, 3D chondrocyte microtissues were fixed in formalin, harvested, and subsequently analyzed after sectioning under H&E, Safranin O, and Ki67 staining to visualize chondrocyte organization and morphology, proteoglycan deposition, and cell proliferation. Histology results (shown in S1B) revealed the absence of cell proliferation and little to no appreciable proteoglycan secretion. Although some indication of the formation of cartilage-relevant extracellular matrix was present, the cultivation period of 3 weeks was not sufficient for substantial accumulation of proteoglycans in the matrix. Overall, these results support the redifferentiation capacity of primary chondrocytes inside our microfluidic device including (a) spherical morphologies, (b) low metabolic activity, (c) cartilage-related phenotype, and (d) limited proliferation.
Fig. 4.
(A) Comparison of cell morphologies between 3D chondrocyte cultures and monolayer cultures. (B) Metabolic activities of 2D chondrocytes in the presence of Ham's F12 medium chondrogenic differentiation medium as well as 3D chondrocytes. (C) Gene expression patterns of 3D chondrocyte cultures compared with monolayer cultivation after 2 weeks of cultivation (left) and changes in gene expression during cultivation (right).
2.3. Evidence of osteoarthritis-mimicking response to inflammation and treatment
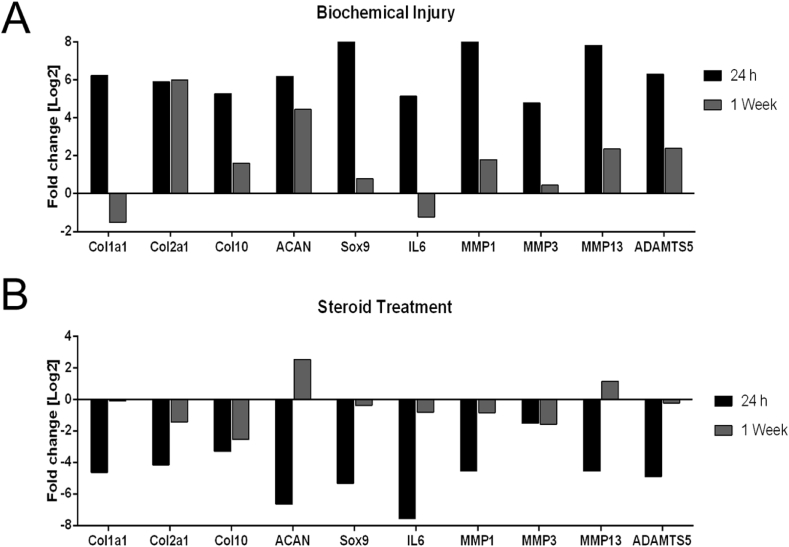
In a final set of experiments, established chip-based 3D chondrocyte constructs were exposed to an inflammatory environment to induce a biochemical injury associated with osteoarthritis. Inflammation of the microtissue was incited after 1 week of culture via biochemical stimulation using 50 pg/mL of tumor necrosis factor (TNF)-α and interleukin (IL)-1β [19], and cellular responses were analyzed by RT-qPCR. A comparison between healthy control and biochemically injured microtissues is shown in Fig. 5A. After 24 h of stimulation, all inflammatory mediators aggrecanase (ADAMTS5), interleukin (IL-6), matrixmetalloproteinases (MMP-1, MMP-3, and MMP-13), as well as ColX, were significantly upregulated (black bars). These results point at an inflammatory phenotype and beginning hypertrophic differentiation in stimulated cultures, thus supporting the onset of an osteoarthritic microenvironment. However, two weeks after biochemical stimulation (gray bars), a number of inflammatory mediators including ADAMTS5, IL-6, MMP-1, MMP-3, and MMP-13, as well as ColX, were downregulated, indicating subsiding inflammatory processes and a cellular rescue. Verification of the microtissue as a potential disease model was conducted by simulating a current clinical osteoarthritis therapy, intra-articular injection of the corticosteroid triamcinolone. Here, inflammation of our microtissues was treated using 60 μg/mL of triamcinolone [20] to mitigate the inflammatory effects of TNF-α and IL-1β. After 24 h of stimulation with TNF-α and IL-1β in the presence of the corticosteroid triamcinolone, a substantial decrease in expression of inflammatory markers was revealed, as shown in Fig. 5B (black bars). In addition, decreased expression of ADAMTS5, IL-6, MMP-1 and MMP-3, as well as ColX, at 1 week after treatment was observed (gray bars). The fact that discernible effects after treatment were demonstrated further indicates the suitability of our microfluidic 3D chondrocyte culture as a simple disease model for osteoarthritis.
Fig. 5.
(A) Gene expression levels of biochemically inflamed microtissues after 1 week of culture and 24-h exposure with 50 pg/mL of TNF-α and IL-1β at day 7 and 2 weeks after stimulation. (B) Gene expression levels of biochemically inflamed microtissues in the presence of 60 μg/mL of triamcinolone, an inflammation inhibiting steroid. TNF-α, tumor necrosis factor-α; IL-1β, interleukin-1β.
3. Conclusion
In conclusion, the nutrient gradient–based microfluidic 3D cell cultivation method provides an improved in vitro environment for primary chondrocytes that fosters (a) redifferentiation including chondrocytic morphology and phenotype, (b) biochemical osteoarthritis-like inflammatory responses, and (c) the ability to evaluate osteoarthritic treatment options. In addition, in contrast to previous reports of engineered cartilage that separately assembled the distinctly different zones of cartilage [21] or used biphasic constructs [22], in the present study, chondrocytes were seeded in a homogenous distribution in fibrin gel and migrated to the gel-medium interface, approximating the superficial zone of cartilage. The observed increased cell density of the resident chondrocytes at the medium-hydrogel interface and their orientation along the microchannel points at a shear force induced cellular orientation similar to native cartilage. Some limitations in the present study remain and include the number of biological replicates (e.g. 4 horses used in the study) and difficulties in obtaining physiologically relevant cell densities of primary cells in low passage to avoid cellular senescence and dedifferentiation. Furthermore, the lack of biomechanical stimulation and absence of corresponding synovial fluid constitute additional limitations not yet addressed in the current work. However, we postulate that the presented microfluid-based chondrocyte culture technique may offer a new platform for future investigations in the behavior of differentiated chondrocytes and subsequently how the cells respond differently in pathophysiological states.
4. Methods
4.1. Microfluidic 3D hydrogel chondrocyte culture-on-a-chip
4.1.1. Microfluidic device and microfabrication
The device consists of two layers, a glass top layer with inlets and a polydimethylsiloxane (PDMS) bottom layer with the microfluidic structures. The device comprises a cell chamber of 3 mm diameter, with a chamber volume of 7.5 μL, a medium channel of 21.5 mm length × 1.0 mm width, and a chamber height of 1 mm. One chip hosts six of these chamber-medium channel complexes. The PDMS bottom can be opened to release intact cell-laden fibrin matrices for histological analysis. Molds for soft lithography of PDMS were designed using AutoCAD software and manufactured by stereolithography (i.materialise). The soft lithography mold was cleaned using 99% isopropanol and dried at 70 °C. PDMS (Sylgard® 184 Silicone Elastomer Kit; Down Corning) polymer was then mixed in a 1:10 ratio of curing agent and base, distributed evenly on the surface of the mold and polymerized at 70 °C for 1 h. Inlets on the glass cover slides were drilled using a 1-mm spheroid diamond drill bit to form the top layer. Before plasma activation, both layers were again cleaned with isopropanol and dried at 70 °C. After drying, substrates were plasma activated for 45 s each using a handheld corona plasma discharge system to create excess hydroxyl groups on both surfaces and ensure stable adhesive bonding. The two layers were then aligned with one another, and gentle pressure was applied before overnight incubation. All devices were sterilized with 70% ethanol and subsequently baked overnight at 70 °C before cell loading.
4.1.2. Isolation of primary equine chondrocytes
Primary chondrocytes were isolated with written owner consent and under strict sterile technique from the knee joints of 4 horses (4 biological replicates) euthanized for reasons unrelated to osteoarthritis and unrelated to this study. After harvest, the cartilage was minced into small pieces (1 mm3), digested in a 1-mg mL-1 collagenase (Type I; Sigma-Aldrich, St. Louis, MO, USA) solution for 6–8 h under constant stirring at 37 °C and filtered through a cell strainer (100 μm; Greiner Bio-One). The obtained chondrocytes were washed twice in Dulbecco's phosphate buffered saline (DPBS + Ca/Mg) and centrifuged at 400g for 5 min. The resulting cell pellet was resuspended in standard culture medium consisting of Ham's (low glucose, with l-Glutamine, [Lonza]), 10% fetal calf serum (FCS) [low endotoxin; Sigma-Aldrich], 1% Pen/Strep [100×; Sigma-Aldrich], and 1% amphotericin B [250 μg/ml; Biochrom]). Chondrocytes were seeded in polystyrene tissue culture flasks (Sarstedt) and cultured in a humidified atmosphere at 37 °C and 5% CO2. The following day, the cells were washed to remove non-adherent cells and tissue debris. Medium was changed twice weekly. Passaging was performed upon colony confluence for the first passage and later upon 80–90% confluence by trypsinization (trypsin 0.05%, Ethylenediaminetetraacetic acid (EDTA) 0.02% [Biochrom]). Chondrocytes were frozen and stored in liquid nitrogen until further processing.
4.1.3. Microfluidic device loading
Primary equine chondrocytes were used for cultivation in passage two or passage three. Immediately before loading, cells were washed twice with phosphate-buffered saline (PBS) and stained for 45 min using 1 μM cytoplasmic Cell Tracker Green™ CMFDA Dye (Thermo Fisher Scientific, Waltham, MA, USA) in pure Ham's F12 medium per the manufacturer's instructions. After staining, the cells were washed twice with PBS, detached using 0.25% trypsin-EDTA solution, and centrifuged at 450 rpm for 5 min. Cell concentration and viability were determined via Trypan blue exclusion using Countess™ automated cell counter (Invitrogen). Microfluidic devices were subsequently loaded with chondrocytes encapsulated in fibrin hydrogel (Tisseel Fibrinkleber, Baxter, Germany/Austria. https://www.baxter.com/healthcare-professionals/surgical-care/tisseel-fibrin-sealant-surgical-care), yielding final concentrations of 3 × 106 cells/mL, 20 mg mL-1 of fibrinogen, and 5 U/mL of thrombin. The fibrin hydrogel was polymerized at 37 °C in a cell culture incubator before adding Ham's F12 complete chondrocyte medium into the medium channel and sealing the microfluidic inlets with clear, self-adhesive foil (Polyolefin StarSeal Xtra-Clear; StarLab). Medium in the microfluidic devices was exchanged by manual pipetting every other day. Chondrocyte-on-chips were harvested for further analysis on day 7 and 21. All experiments and assays were carried out in 3 technical replicates/biological replicates.
4.1.4. Finite volume simulation
A multipurpose finite volume CFD code (e.g. Ansys Fluent 6.3) was used for analyzing diffusion distance and velocity into the hydrogel. The geometry consisting of the hydrogel cavity and the feed channel was split into about 26400 hexahedral control volumes (Supplementary Fig. 2B). Second-order or higher order discretization was selected for all flow variables (momentum, mass) and for the species equations. To ensure physically correct transient solutions, the time step size was selected to guarantee a maximum Courant number Co < 1 within the flow domain over the whole run time of up to 24 h physical time. Wall boundaries were treated as ideally smooth, no-slip boundary conditions were selected for all surfaces. Stagnant flow was assumed in the flow channel – for numerical stability, a small non-zero flow velocity had to be applied (mass flow inlet with plug flow velocity profile, maximum Reynolds number Re << 10-8→ laminar; diffusion-based Peclet number Pe < 0.1 → diffusion dominated transport inside the hydrogel zone). The outlet was set to pressure outlet at a standard pressure of p = 1 atm (101325 Pa). The hydrogel regions were approximated as porous zones with constant porosities of ε = 0.99 and isotropic viscous resistances of R = 6.67.10–12 1/m2. The flow was considered isothermal; no temperature or energy field was solved. For simplicity, Newtonian fluid behavior was assumed with a constant dynamic viscosity and constant density. As the concentrations of the dissolved species in the fluid are low, the properties of the solvent, water, have been used for the simulation (ρ = 998 kg/m³, η = 0.001003 Pa s). The diffusion coefficients for the tracer components have been determined previously [6] or based on literature values (generic tracers with 0.3 kDa–4.10−10 m [2]/s and 40 kDa–1.10−11 m [2]/s, and glucose with 0.18 kDa 4.10−10 m [2]/s), assuming a dilute solution. Tracers in the flow channel (reservoir zone) were added with constant concentration at the initial time T = 0 s; at the same time, a constant zero tracer saturation of the hydrogel zone was set as starting values for the simulation runs. For glucose, the actual concentration levels from experiments were used (1.35 g/l initial concentration in the hydrogel; 4.5 g/l in the feed channel). Simulations were carried out on cae.zserv.tuwien.ac.at.
4.1.5. Live cell imaging and on-chip morphology assessment
Chondrocytes cultivated in fibrin hydrogels and in conventional cell culture were imaged via brightfield, phase contrast, and fluorescent microscopy throughout the entire culture period of 21 days using an EVOS cell imaging system (Thermo Fisher Scientific). Cell viability was assessed by means of Cell Tracker Green™ CMFDA Dye, cell morphology was recorded, and the cells cultured in Ham's F12 chondrocyte medium were counted manually under fluorescent microscopy every day in multiple microfluidic chambers and culture flasks to compare the morphological redifferentiation process of the chondrocytes during the culture period [23]. Chondrocytes were considered redifferentiated when they displayed a round, spherical morphology and fibroblastic when their length was twice their width. If the morphology differed from these specifications, the differentiation status was listed as not assigned [24].
4.1.6. Metabolic activity
A Resazurin-based in vitro toxicology assay (TOX8; Sigma-Aldrich) was performed to determine differences in metabolic activity between microfluidic and monolayer cultures. To study the metabolic changes during chondrogenic differentiation, cells were cultured with the standard cultivation method using Ham's F12 or StemPro™ Chondrogenesis Differentiation Kit (Gibco). Cells were cultured in either monolayer culture or on a microfluidic device for 6 days. To standardize the fluid volume and cell number between the different culture methods, 11250 cells, 22500 cells, or 33750 cells were transferred into a 24-well plate either in continued monolayer culture or embedded in fibrin gel (microfluidic device group). For the metabolic assay, 40 μL of TOX8 reagent (Sigma-Aldrich) was added to 400 μL of medium and incubated for 8 h at 37 °C inside a cell culture incubator. The analysis was performed by aliquoting 100 μL of supernatant per technical triplicate in a flat-bottom 96-well plate and measured fluorometrically at a wavelength of 590 nm with excitation at 560 nm.
4.1.7. Gene expression analysis
Isolation of RNA
Fibrin hydrogel clots were released from the microfluidic device using a scalpel and tweezers before homogenization using a Biopulverizer (Biospec, USA). In detail, the BioPulverizer was cooled thoroughly by submerging in liquid nitrogen for 1 min. Each fibrin hydrogel to be analyzed was placed in a shallow container of the Biopulverizer and pulverized for 1 min by blowing the hammer to the pestle. Powdered contents were transferred to an Eppendorf tube. Total RNA was extracted from both 3D and 2D cultures using peqGOLD Total RNA isolation Kit (Peqlab) per the provided protocol.
4.1.7.2. Quantitative PCR
One nanogram of RNA of each sample was used for quantitative PCR (qPCR) reaction. RevTrans QPCR One-Step EvaGreen kit (Bio&Sell, Germany) was used first for cDNA synthesis and subsequently for qPCR reaction. The reaction mixtures were incubated for 15 min at 50 °C for cDNA generation, followed by qPCR reaction; 95 °C for 5 min, 95 °C for 15 s, 55 °C for 20 s, and 72 °C for 30 s. For each gene, a reaction mixture with water instead of the total RNA template was run at the same time as a PCR negative control. The transcript data were analyzed using the Agilent AriaMx 1.1 software (Agilent Technologies, USA). The transcript level of genes of interest was normalized to the transcriptional level of GAPDH and represented as a relative transcript level relative to GAPDH.
4.1.8. Histology
Histological sections of cell-laden fibrin hydrogels cultured inside the microfluidic device were performed to analyze cell morphology and distribution. After cultivation, the hydrogels were fixed overnight using 4% buffered formalin, released from the microfluidic device using a scalpel and tweezers, and kept in histology cassettes in 70% ethanol until embedding. The hydrogels were dehydrated, embedded in paraffin using a Shandon Tissue Excelsior (Thermo Fisher Scientific) and cut into 2-μm slices. The sections were mounted onto glass slides using dibutylphthalate polystyrene xylene (DPX) (Sigma-Aldrich) and stained with hematoxylin (Richard Allan Scientific, Waltham, MA, USA), eosin (Carl Roth, Karlsruhe, Germany), and Safranin O/Fast Green (counterstain). Immunohistochemical staining for Ki67 (Cell Signaling, clone 8D5, mouse mc, dilution 1:400) was performed after 30 min of antigen retrieval in a steamer with citrate buffer at pH 6.
4.1.9. Biochemical injury and steroid treatment
Chip-based 3D chondrocyte constructs were exposed to an inflammatory environment using 50 pg/mL of TNF-α and IL-1β (Sigma-Aldrich), as previously described [19]. Chondrocyte cultures were subjected to biochemical stimulation after one week in culture for a time of 24 h, after which chondrocyte constructs were either released from the device for RT-qPCR or cultivated in standard medium for another six days before analysis. For steroid treatment, 60 μg/mL of triamcinolone [20] was added simultaneously with inflammatory factors.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2019.100023.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Fig. S1.
(A) Live staining of chondrocytes-on-chip directly after loading (left) and after 28 of cultivation (right), confirming high viabilities throughout the cultivation time. (B) CMFDA-stained chondrocytes in the cultivation chamber exhibiting spherical morphology. (B) Histological staining of chondrocyte-on-chip cultures with H&E (left), Safranin O (middle), and Ki67 (right). CMFDA, 5-chloromethylfluorescein diacetate .
Fig. S2.
(A) Levels of the tracer molecule compared with glucose levels including glucose consumption after 24 h and (B) representative image showing mesh quality used in in silico experiments .
References
- 1.Hunter D.J. Nat. Rev. Rheumatol. 2014;10(7):437. doi: 10.1038/nrrheum.2014.44. [DOI] [PubMed] [Google Scholar]
- 2.Frisbie D.D. Vet. Comp. Orthop. Traumatol. 2006;19(3):142. [PubMed] [Google Scholar]
- 3.Camarero-Espinosa S. Biomater Sci. 2016;4(5):734. doi: 10.1039/c6bm00068a. [DOI] [PubMed] [Google Scholar]
- 4.Sticker D. Lab Chip. 2015;15(24):4542. doi: 10.1039/c5lc01028d. [DOI] [PubMed] [Google Scholar]
- 5.Rothbauer M. Lab Chip. 2018;18(2):249. doi: 10.1039/c7lc00815e. [DOI] [PubMed] [Google Scholar]
- 6.Bachmann B. Biomicrofluidics. 2018;12(4) doi: 10.1063/1.5027054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y. Exp. Ther. Med. 2017;14(3):2657. doi: 10.3892/etm.2017.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Occhetta P. Nat. Biomed. Eng. 2019;3(7):545. doi: 10.1038/s41551-019-0406-3. [DOI] [PubMed] [Google Scholar]
- 9.Lin H. Mol. Pharm. 2014;11(7):2203. doi: 10.1021/mp500136b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhosale A.M., Richardson J.B. Br. Med. Bull. 2008;87:77. doi: 10.1093/bmb/ldn025. [DOI] [PubMed] [Google Scholar]
- 11.Poole A.R. Clin. Orthop. Relat. Res. 2001:S26. doi: 10.1097/00003086-200110001-00004. 391 Suppl. [DOI] [PubMed] [Google Scholar]
- 12.Sophia Fox A.J. Sport Health. 2009;1(6):461. doi: 10.1177/1941738109350438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H. Connect. Tissue Res. 2014;55(5–6):339. doi: 10.3109/03008207.2014.949698. [DOI] [PubMed] [Google Scholar]
- 14.Maroudas A. J. Anat. 1976;122(Pt 2):335. [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson I. Histochem. J. 1998;30(3):123. doi: 10.1023/a:1003287101868. [DOI] [PubMed] [Google Scholar]
- 16.Spitters T.W.G.M. Tissue Eng. A. 2014;20(23–24):3270. doi: 10.1089/ten.tea.2014.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson A., Gu W. Curr. Rheumatol. Rev. 2009;5(1):40. doi: 10.2174/157339709787315320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aigner T. Arthritis Rheum. 2001;44(6):1304. doi: 10.1002/1529-0131(200106)44:6<1304::AID-ART222>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 19.Sun L. Biomaterials. 2011;32(24):5581. doi: 10.1016/j.biomaterials.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dechant J.E. Equine Vet. J. 2003;35(5):444. doi: 10.2746/042516403775600479. [DOI] [PubMed] [Google Scholar]
- 21.Schuurman W. J. Tissue Eng. Regenerat. Med. 2016;10(4):315. doi: 10.1002/term.1726. [DOI] [PubMed] [Google Scholar]
- 22.Paetzold H. Eur. Cells Mater. 2012;23:209. doi: 10.22203/ecm.v023a16. [DOI] [PubMed] [Google Scholar]
- 23.Dorotka R. Biomaterials. 2005;26(17):3617. doi: 10.1016/j.biomaterials.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 24.Stockwell R.A. J. Anat. 1967;101(Pt 4):753. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.