CONSPECTUS:
Oxidative post-translational modifications (OxiPTMs) of cysteine residues are the molecular foundation of thiol-based redox regulation that modulates physiological events such as cell proliferation, differentiation, and migration and, when dysregulated, can lead to biomolecule damage and cell death. Common OxiPTMs of cysteine thiols (─SH) include reversible modifications such as S-sulfenylation (─SOH), S-glutathionylation (─SSG), disulfide formation (─SSR), S-nitrosylation (─SNO), and S-sulfhydration (─SSH) as well as more biologically stable modifications like S-sulfinylation (─SO2H) and S-sulfonylation (─SO3H). In the past decade, our laboratory has developed first-in-class chemistry-based tools and proteomic methods to advance the field of thiol-based redox biology and oxidative stress. In this Account, we take the reader through the historical aspects of probe development and application in our laboratory, highlighting key advances in our understanding of sulfur chemistry, in the test tube and in living systems.
Offering superior resolution, throughput, accuracy, and reproducibility, mass spectrometry (MS)-based proteomics coupled to chemoselective “activity-based” small-molecule probes is the most rigorous technique for global mapping of cysteine OxiPTMs. Herein, we describe the evolution of this field from indirect detection to state-of-the-art site-centric quantitative chemoproteomic approaches that enable mapping of physiological and pathological changes in cysteine oxidation. These methods enable protein and site-level identification, mechanistic studies, mapping fold-changes, and modification stoichiometry. In particular, this Account focuses on activity-based methods for profiling S-sulfenylation, S-sulfinylation, and S-sulfhydration with an eye toward new reactions and methodologies developed in our group as well as their applications that have shed new light on fundamental processes of redox biology. Among several classes of sulfenic acid probes, dimedone-based C-nucleophiles possess superior chemical selectivity and compatibility with tandem MS. Cell-permeable dimedone derivatives with a bioconjugation handle are capable of detecting of S-sulfenylation in living cells. In-depth screening of a C-nucleophile library has yielded several entities with significantly enhanced reactivity over dimedone while maintaining selectivity, and reversible linear C-nucleophiles that enable controlled target release. C-Nucleophiles have also been implemented in tag-switch methods to detect S-sulfhydration. Most recently, activity-based detection of protein S-sulfinylation with electrophilic nitrogen species (ENS), such as C-nitroso compounds and electron deficient diazines, offers significant advantages in simplicity-of-use and target specificity compared to label-free methods.
When feasible, the rich information provided by site-centric quantitative proteomics should not be tainted by oxidation artifacts from cell lysis. Therefore, chemoselective probes that function in a native environment with low cytotoxicity, good cell-permeability, and competitive kinetics are desired in modern redox chemoproteomics approaches. As our understanding of sulfur chemistry and redox signaling evolves, newly discovered cysteine OxiPTMs in microorganisms, plants, cells, tissues, and disease models should innovatively promote mechanistic and therapeutic research.
Graphical Abstract
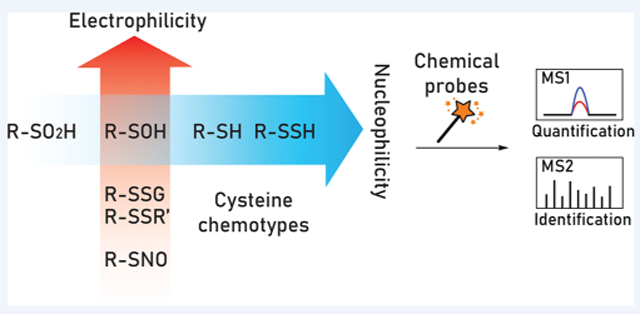
OXIDATIVE POST-TRANSLATIONAL MODIFICATION OF CYSTEINE
Post-translational modification (PTM) exponentially expands the chemical repertoire available to proteins beyond the 20 standard amino acids. The human proteome contains more than 200 types of PTMs that modulate protein function.1 Among these, oxidative post-translational modifications (OxiPTMs) of cysteine residues have emerged as a fundamental mechanism in thiol-based redox regulation and signaling in physiological processes such as cell proliferation, differentiation, and migration.2 The sulfur atom of cysteine can exist in various oxidation states from −2 to +6. Two electron oxidation of cysteine thiol (─SH) by the reactive oxygen species (ROS) hydrogen peroxide (H2O2) produces cysteine sulfenic acid (─SOH), which may be stabilized by the protein microenvironment or form a disulfide with an adjacent cysteine residue (─SSR) or mixed disulfide with glutathione (─SSG). Likewise, S-nitrosothiols (─SNO) and persulfides (─SSH) are generated from reactive nitrogen/sulfur species. The aforementioned cysteine OxiPTMs can be biologically reduced back to their thiol form with cellular enzymes like glutaredoxin (Grx) or thioredoxin (Trx), as a key mechanism in maintaining redox homeostasis.3 On the other hand, higher oxidation states of cysteine, like cysteine sulfinic acid (─SO2H) and sulfonic acid (─SO3H), are more stable and can accumulate over time during oxidative stress.4 The distinct reactivity of each cysteine OxiPTM provides the chemical basis for differential redox regulation of protein function. Cysteine oxidation also has a profound impact on PTMs that require a reduced thiol for modifications such as palmitoylation and drug pharmacology with covalent thiol-reactive inhibitors.5,6 Protein cysteines can exhibit a range of reactivity, dictated by factors such as pKa, electrostatic interactions, and solvent exposure.7 The reaction rate between cysteine thiolates and H2O2 can vary substantially: antioxidant enzyme peroxiredoxins (Prxs) are hyperactive (105−108 M−1·s−1), followed by redox-sensor proteins like GAPDH and OxyR (103−105 M−1·s−1), while the bulk of the cysteinome reacts slowly (~10 M−1·s−1).8,9
A wealth of cysteine OxiPTMs in microorganisms, plants, animal tissues, and disease models await discovery. To this end, proteome-wide profiling of cysteine oxidation is an essential molecular technique to elucidate redox-regulated signaling pathways and offer new insights into pathological states involving oxidative stress, such as cancer, diabetes, and neurodegenerative and cardiovascular diseases.10 The rapidly growing area of chemical proteomics or chemoproteomics has been fueled by advances in instrumentation and bioinformatics as well as small-molecule probes that enable tagging, isolation, identification, and quantification.11-13 In the past decade, researchers have developed MS-based techniques to identify low pKa or “reactive cysteines” as well as cysteine OxiPTMs with detailed information including the site, type, dynamic fold-change, and extent of modification. Herein, we spotlight recent developments in profiling cysteine reactivity and oxidation with emphasis on site-centric quantitative approaches.
ORTHOGONAL REACTIVITY
Pioneered by Bertozzi and co-workers, bioorthogonal chemistry enables researchers to investigate native biomolecules under complex biological settings.14 Understanding the reactivity profile of each cysteine “chemotype” is key to designing an effective activity-based proteomic workflow. To achieve orthogonality, two types of approaches are commonly employed: (1) blocking undesired forms of cysteine (e.g., thiols), reducing cysteine OxiPTMs, followed by trapping nascent thiols with detectable tags (“tag-switch” method); and (2) probing distinct classes of cysteine OxiPTMs with reagents that are chemically selective. In the experimental design, all biologically relevant forms of cysteine must be taken into consideration. Summarized in Table 1, cysteine OxiPTMs can be categorized on the basis of sulfur charge density. Electronrich species or nucleophiles (when deprotonated) include cysteine thiols (─SH), persulfides (─SSH), and sulfinic acids (─SO2H). Electron-deficient species or electrophiles include disulfides (─SSR), S-glutathione adducts (─SSG), and S-nitrosothiols (─SNO). Some species, like sulfenic acids (─SOH), are unique in that they exhibit both nucleophilic and electrophilic reactivity.
Table 1.
Orthogonal Reactivity Chart of Common Cysteine Chemotypes
| cysteine chemotype |
typical behaviora |
thiol-reactive reagents |
unselective reductants |
reducing agents or specific probes |
dimedone-based C-nucleophiles |
strained alkynes/ alkenes |
|---|---|---|---|---|---|---|
| ─SH | Nu | yes | no | no | no | partial |
| ─SSH | Nu | yes | yes | no | no | partial |
| ─SOH | Nu/E | partialb | yes | arsenite,c dimedone | yes | yes |
| ─SNO | weak E | no | yes | ascorbate,c etc.d | no | no |
| ─SSR | weak E | no | yes | no | no | no |
| ─SSG | weak E | no | yes | glutaredoxin | no | no |
| ─SO2H | weak Nu | partial | no | ENSe | no | no |
Nu = Nucleophilic chemotypes; E = electrophilic chemotypes.
Partial reactivity indicates a subset of chemical reagents react with this modification.
Arsenite and ascorbate are not fully selective.
Several chemical probes for ─SNO are not discussed here.
Electrophilic nitrogen species also reacts with thiols, but only forms stable products with sulfinic acids.
SITE-CENTRIC QUANTITATIVE PROTEOMICS
Reversible or labile cysteine OxiPTMs are extremely susceptible to perturbation during sample preparation and subsequent analyses. Therefore, chemical labeling is widely recognized in redox proteomics, because oxidation states are “trapped” by chemical tags, which may also enable affinity enrichment for sensitivity improvement. Stable isotope-coded tags are often used in quantitative proteomics, since corresponding peptides have identical ionization ability, and therefore can serve as internal MS standards.15 In addition, tagged peptides must be stable and have a clear fragmentation pattern on tandem MS to reveal modified sites. Such site-centric quantitative approaches not only show the location and multiplicity of modifications, but also provide definitive confirmation by greatly decreasing the rate of false discovery.
Indirect Profiling of Cysteine Oxidation
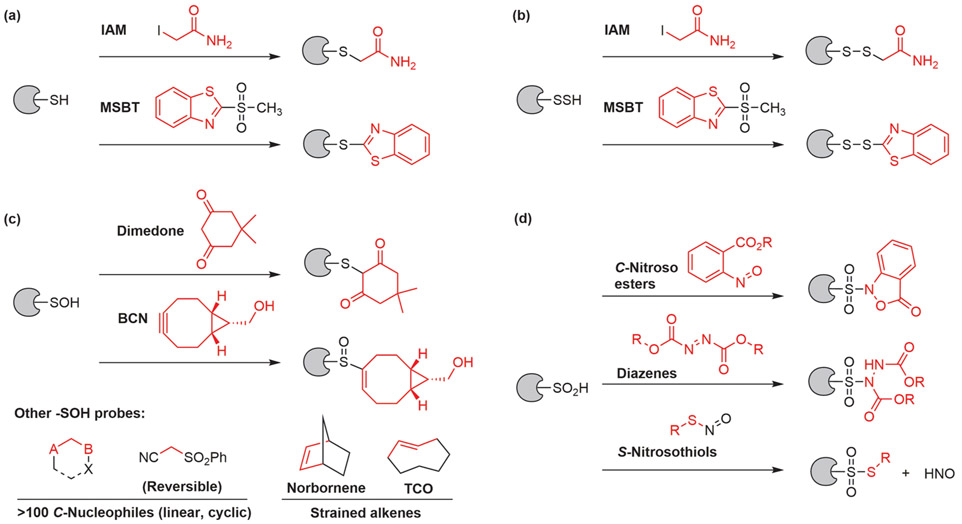
With estimated concentration up to 50 mM, protein-bound cysteine thiols are mostly reduced and constitute the majority (up to 70%) of reduced thiols in cells.16,17 Iodoacetamide (IAM) and its derivatives are among the most frequently used thiol-reactive alkylating agents, but at the high concentrations typically employed they are plagued by off-target modification of basic amino acid residues (e.g., N-terminus, lysine and histidine residue)18,19 and other cysteine OxiPTMs including ─SOH, ─SO2H, and ─SSH.20-22 Several novel reagents with enhanced selectivity, like methylsulfonyl benzothiazole (MSBT),23 do not react with ─SOH, ─SO2H, and ─SNO (but thiol alkylation generates sulfinic acids, which may cross react with other species, including ─SNO)24 (Figure 1a). However, alkylation of persulfides (─SSH) is expected, due to its enhanced nucleophilicity originated from low pKa (CysSSH pKa = 4.3; CysSH pKa = 8.3).25 In fact, MSBT and IAM derivatives have been used in detection of ─SSH (Figure 1b).26,27
Figure 1.
Common chemical reagents that selectively target cysteine oxoforms. (a) Reagents that label free cysteine thiols. (b) Thiol-reactive reagents also label persulfides. (c) Sulfenic acid probes. (d) Sulfinic acid probes.
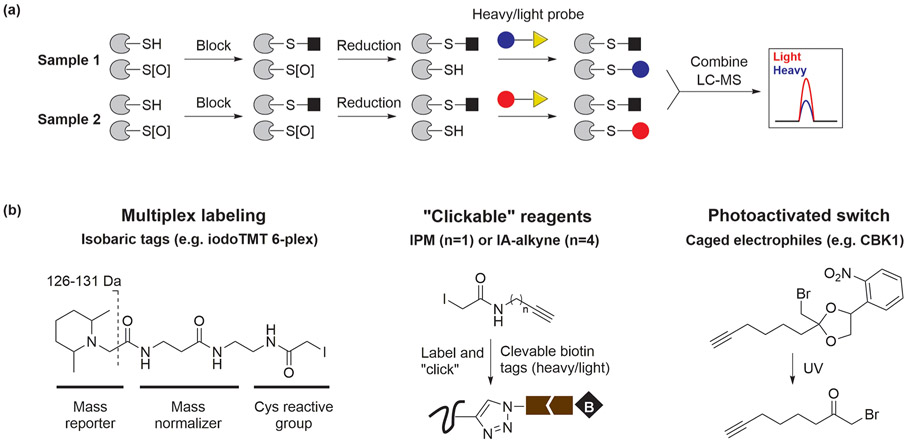
Reversibly oxidized cysteine modifications can be indirectly detected by thiol-blocking, reduction of oxidized species, followed by labeling of nascent thiols (Figure 2a). DTT and TCEP (standard reduction potential = −0.33 and −0.32 V, respectively)28 are powerful reducing agents that are capable of converting reversible cysteine OxiPTMs to thiols, but they do not effectively reduce ─SO2H or ─SO3H. Alternatively, a given OxiPTM may be selectively reduced and tagged for detection. Sodium ascorbate and sodium arsenite were considered as selective reducing agents for ─SNO and ─SOH, respectively.29,30 Likewise, enzymatic reduction of ─SSG by Grxs has been employed to detect S-glutathionylation.31 Nevertheless, extreme caution is advised when interpreting data obtained from these strategies, as incomplete blocking, reduction, or sample degradation can occur during lengthy sample processing workflows, especially for labile modifications. In addition, clear examples of cross-reactivity with other cysteine OxiPTMs indicate that ascorbate and arsenite are far less selective than previously believed.9,29
Figure 2.
(a) Indirect detection of cysteine OxiPTMs with thiol-reactive reagents. (b) Recent advances in thiol-reactivity profiling.
In early studies, cysteine OxiPTMs were typically profiled with isotope-coded affinity tag (ICAT)-based platforms;32,33 however, many recent improvements to this workflow have been made (Figure 2b). Cysteine-reactive isobaric tags have been introduced for multiplexed redox quantitation, reducing run-to-run variance. For example, 6-plex tandem mass tag (TMT) reagents have been used to quantify ─SNO and other reversible cysteine OxiPTMs in three competitive samples.34,35
The application of smaller electrophiles has expanded the coverage of targetable cysteines due to easier access to sterically encumbered sites. Alkynyl iodoacetamide probes IA-alkyne and IPM can be conjugated to isotope-coded reporter groups via copper-catalyzed azide–alkyne cycloaddition (CuAAC) or “click chemistry”. With the IPM probe, over 6500 cysteines in multiple human cell lines were profiled, and most of them responded to H2O2 in a cell-specific manner.36 The Weerapana group has developed cost-effective isotope-coded electrophiles and applied these reagents to identify almost 1000 reversibly oxidized cysteine sites in HeLa lysate.37 Moreover, the inherent cytotoxicity of IA-alkyne (LC50 = 16 μM) can be mitigated, at least in part, by using photocaged electrophiles. Epidermal growth factor (EGF)-induced cysteine oxidation in A431 cells has been analyzed using the photocaged probe, CBK1, which displayed little detectable cytotoxicity below 250 μM and allowed a degree of temporal control over cysteine labeling.38
In combination with other methodologies, researchers aim to integrate quantitative information on relative cysteine oxidation, termed fold-change, with total protein levels. For example, the SILAC-iodoTMT (metabolic labeling), GELSILOX (18O proteolytic labeling), cysTMTRAQ (coupled isobaric labeling), and oxSWATH (integrative chemical labeling)39-42 platforms have been recently reported to comprehensively evaluate the effect of oxidation on cysteine proteomes.
The Chemical Odyssey of Sulfenic Acid (─SOH) Detection
A chemical probe is a small-molecule reagent that allows mechanistic and phenotypic studies on its protein targets.43 In the context of redox proteomics, the chemical probe is expected to target a specific class of cysteine OxiPTM among the whole proteome, with minimal cross-reactivity. Because of its dynamic nature and central role in OxiPTM-based redox regulation, the odyssey of trapping cysteine sulfenic acids (─SOH) with nucleophilic compounds began in 1974, when Allison and co-workers introduced the cyclic 1,3-diketone, dimedone, and other nucleophiles that reacted with cysteine sulfenic acid in GAPDH.44 Thereafter, nucleophiles functionalized with biotin, azide, or alkyne reporter groups and other handles for bioorthogonal ligation based upon the dimedone structure have been reported.9 Our laboratory has performed a comprehensive study, wherein a library of ~100 carbon-based nucleophiles (C-nucleophiles) were designed, prepared, and screened, ultimately identifying several entities with exceptional reactivity (up to 3170 M−1·s−1, a more than 300-fold increase over dimedone) while maintaining selectivity.45-47Furthermore, our survey of linear C-nucleophiles showed reversible covalent labeling of sulfenic acids for the first time, enabling target release under defined conditions (Figure 1c).46
Complementary to nucleophilic probes, several strained alkynes and alkene electrophilic reagents have been engineered to capture ─SOH.48-51 Because these reactions are strain-promoted and the alkenes/alkynes are not as electron-deficient as Michael acceptors, thiol–ene/yne side reactions can be suppressed, yet incompletely.49,52 The issue of thiol–ene/yne side reaction is magnified given the much higher abundance of ─SH than ─SOH in cells. Moreover, ─SSH was found to participate in the reaction with strained alkynes under a mechanism analogous to that of ─SOH.53
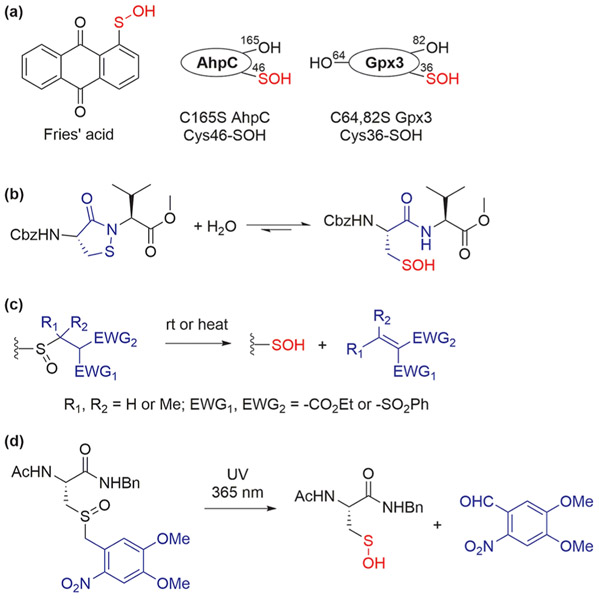
Regardless of the chemistry or probe used for trapping, sulfenic acid models are essential touchstones in reactivity and selectivity studies (Figure 3). A small-molecule anthraquinone-1-sulfenic acid (Fries’ acid) is stabilized via intramolecular hydrogen bonding, but not “bio-representative”.9 In comparison, protein-stabilized sulfenic acids are typically preferred choices, including C165S AhpC and C64,82S Gpx3.45,48 On the other hand, certain electrostatic or steric effects can clearly modulate the rate of probe reactivity with protein ─SOH. Therefore, an alternative model developed by our group consists of a dipeptide cyclic sulfenamide, which is stable in dry, powdered form and organic solvent, but readily hydrolyzes to form sulfenic acid in aqueous solution; this model has been employed with great success to benchmark our extensive library of ─SOH probes and reactive fragments.45-47 Thermal-driven sulfoxide elimination reactions also produce sulfenic acids.9,54 Recently, we have reported an approach for the generation of small-molecule and protein sulfenic acids from photocaged sulfoxide precursors with applications in unnatural amino acid (UAA) incorporation.55
Figure 3.
Small-molecule and protein models used in developing sulfenic acid probes. (a) Stabilized small-molecule and protein sulfenic acids. (b) Hydrolytic equilibrium of cyclic sulfenamide. (c) Generation of sulfenic acids via sulfoxide elimination. (d) Photocaged sulfenic acid.
The Selectivity of Dimedone-Based Probes for Sulfenic Acid (─SOH)
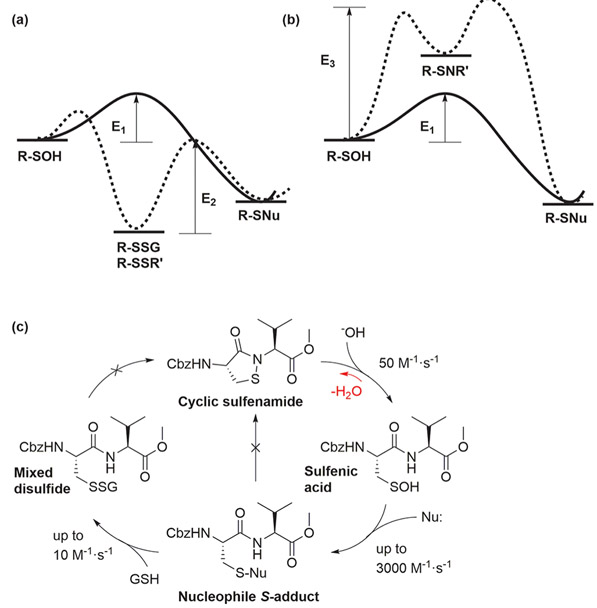
Dimedone-based C-nucleophile probes for ─SOH do not possess intrinsic reactivity with biological nucleophiles, such as the thiol in cysteine, hydroxyl in serine, amino in lysine, or cysteine sulfinate (─SO2−).45 They are also inert toward weak electrophiles, such as the disulfide of a protected cysteine (Z-Cys-OH)2, oxidized glutathione (GSSG), or S-nitrosoglutathione (GSNO) (Figure 4a).45,56 No reaction between dimedone-based C-nucleophiles and protein disulfides (such as those in Trx, Gpx3, and Prx I) has been observed either. It has been noted that the activated methylene in dimedone can undergo Knoevenagel-type condensation with nonprotein bound aldehydes, including pyridoxal, acrolein, and glyceraldehyde.57 From a chemoproteomic standpoint, however, this reaction is a red herring. Indeed, the rate constant for the reaction of dimedone with glyceraldehyde (0.06 M−1·s−1) is approximately 200-fold less than that of ─SOH (~10 M−1·s−1). Also, no reaction was observed between butyraldehyde (a representative model of an aldehyde-modified protein) and dimedone at physiological pH and temperatures up to 100 °C (Gupta, unpublished results).
Figure 4.
Energy profiles of cysteine OxiPTMs. Graphs (a,b) are drawn qualitatively based on reactivity of a Cys-Val dipeptide representing an unperturbed cysteine residue. (a) Dimedone-based nucleophiles readily label ─SOH but do not react with disulfides, indicating E1 < E2. (b) Cyclic sulfenamide (─SNR′) rapidly hydrolyzes to form ─SOH, indicating E1 < E3. (c) Representative rate constants of chemotype transformations of the dipeptide model compound. Data was extracted from ref 45.
Another electrophilic species that could conceivably react with dimedone-based probes is the cyclic sulfenamide, which exists in equilibrium with the corresponding sulfenic acid (Figure 4c).45 Protein tyrosine phosphatase 1B (PTP1B) is the first and best-characterized example of this cysteine OxiPTM, whose crystal structure showed a cyclic sulfenamide linkage in its active site.58 The nitrogen atom on an amide (pKa ~ 17) is less nucleophilic than the nitrogen of an amine, due to the resonance stabilization afforded by the amide carbonyl group. However, in the case of PTP1B, the pKa of the Ser216 amide nitrogen is decreased via extensive hydrogen bonding within the phosphate-binding loop (P-loop), which enables nucleophilic attack on the neighboring sulfur atom of the Cys215─SOH precursor. Although quite interesting, this phenomenon is rare and there are few well-characterized examples.58,59 Additionally, dimedone does not react with PTP1B in its cyclic sulfenamide state.60 Due to the high energy barrier for cyclization, cyclic sulfenamide formation is quite slow.54,61 On this basis, after ─SOH is formed, sulfenamidation would be outcompeted by nucleophilic attack of a thiol or, if present, dimedone-based probes (Figure 4b), in contrast to some recent assertions.62 Similarly, it is energetically unfavorable for weakly electrophilic species such as ─SSR, ─SSG, ─SSH, and ─SNO to form a cyclic sulfenamide and subsequently be tagged with dimedone-based probes.63
Other studies have suggested that dimedone labeled polysulfides (─SnH) or polythiosulfenic acid (─SnOH), but they were performed in vitro under nonphysiological conditions, with 0.2–0.3 mM Na2S2.64,65 Such adducts have never been reported in any proteome-wide experiments, most likely due to their exceedingly rare existence in cells. Given consideration to all aspects above, ─SOH is the only meaningful protein target of C-nucleophile probes in biological systems.
Profiling Protein Sulfenic Acids (─SOH)
Compared to indirect thiol reactivity-based approaches that involve blocking, reduction, and reprobing steps, chemoselective reactions allow direct tagging of a specific cysteine chemotype, providing precise molecular information about the OxiPTM identity and minimizing artifacts stemming from incomplete blocking and reduction, which can be significant in cell lysates and lengthy workflows. In order to capture dynamic cysteine OxiPTMs in living systems, successful activity-based probes should be both selective and kinetically competitive, while maintaining high stability and membrane permeability as well as low cytotoxicity.
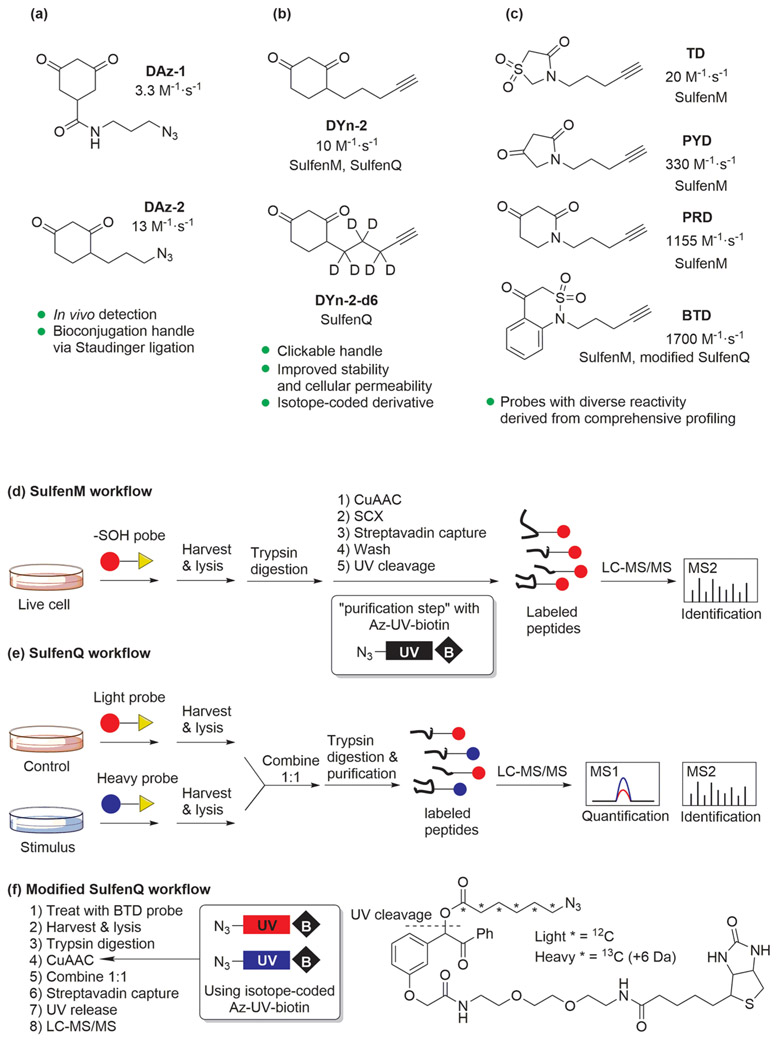
As discussed above, dimedone-based nucleophilic probes show remarkable chemoselectivity with ─SOH. We have also demonstrated that dimedone and related probes are well suited for tandem MS.66 The first dimedone-based probes, reported by the Poole and King groups, were directly conjugated to biotin or fluorescent tags for visualization.9 However, these tools were incompatible with MS and suffered from poor cell permeability. To address this issue, our lab developed azide-functionalized dimedone derivatives, termed DAz-1 and DAz-2, that are compatible with bioconjugation techniques, including Staudinger ligation (Figure 5a). Although both probes are small in size, DAz-2 exhibited better reactivity and membrane permeability due to the replacement of the amide linkage at C-6 with a hydrocarbon linker at C-4. Using DAz-2, we reported the first first survey of S-sulfenylation in living cells, identifying both established redox-sensitive targets and more than 175 new candidates.67 As a continuation of developing ─SOH probes for in situ detection, we replaced the azide functional group on our probes with an alkynyl group, since alkyne reporters offer superior sensitivity and stability in vivo.68 At the same time, we switched from Staudinger ligation chemistry to the more efficient CuAAC reaction. Subsequently, two platforms, termed SulfenM and SulfenQ, were developed in close collaboration with the Liebler group. These methods featured an alkyne-containing probe, DYn-2, to probe ─SOH modifications in human cancer RKO and A431 cells (Figure 5d, e).69 Both strategies enabled precise mapping of sulfenic acid modifications to individual cysteines in complex proteomes. SulfenM was employed to map more than 1000 S-sulfenylation sites from 700 proteins in RKO cells. SulfenQ featured a heavy isotopomer, DYn-2-d6, to quantify the fold-change in ─SOH between samples (Figure 5b). Using the SulfenQ workflow, cells were treated with stimulus (H2O2 or EGF) or vehicle control in the presence of heavy or light DYn-2. Treatment of RKOs with exogenous H2O2 led to detectable changes in S-sulfenylation (heavy/light > 2) in >89% of 360 identified ─SOH sites; similar observations were made with H2O2 treatment of A431 cells. By contrast, fewer than half of ─SOH sites were modulated by stimulation of A431 cells with EGF, demonstrating that growth factor-driven cysteine S-sulfenylation was more target-selective.
Figure 5.
(a) Structures of DAz sulfenic acid probes. (b) Structures of DYn probes. (c) Structures of dimedone-based probes with enhanced reactivity. (d) General workflow of the SulfenM strategy. (e) General workflow of the SulfenQ strategy. (f) Modified workflow of the SulfenQ strategy with the BTD probe.
Although DYn-2 is an effective probe, its moderate reactivity (10 M−1·s−1) poses some inherent limitations for quantitative detection, particularly ─SOH with short cellular lifetimes. To address the demand for kinetically superior probes, we made structural modifications to the dimedone 1,3-diketone scaffold, swapping one or both ketones to other electron withdrawing groups, such as amides, esters, sulfones, sulfonamides, and nitro or cyano groups on a linear or cyclic skeleton.45-47 Four representative cyclic nucleophiles equipped with alkyne handles (PD, PYD, PRD, BTD) showed diverse reactivity with 2–170-fold increased rates relative to DYn-2 (Figure 5c). These five probes were applied in the SulfenM platform, resulting in the discovery of 1283 sulfenylated sites on 761 proteins.70 The most efficient probe, BTD, was also applied in a site-centric quantitative proteomics platform for global detection of ─SOH in cell lysates and living cells. The platform is a modified version of SulfenQ, where paired samples are distinguished by isotope-coded UV-cleavable azido-biotin (Az-UV-biotin) instead of an isotope-coded ─SOH probe (Figure 5f). Due to the enhanced reactivity of BTD relative to DYn-2, the input of lysate material was reduced more than 10-fold (e.g., from 30 mg of protein for DYn-2 to 2 mg of protein for BTD) while increasing the coverage of ─SOH sites (1867 sites for BTD versus 1105 sites for DYn-2) in RKO cells.71
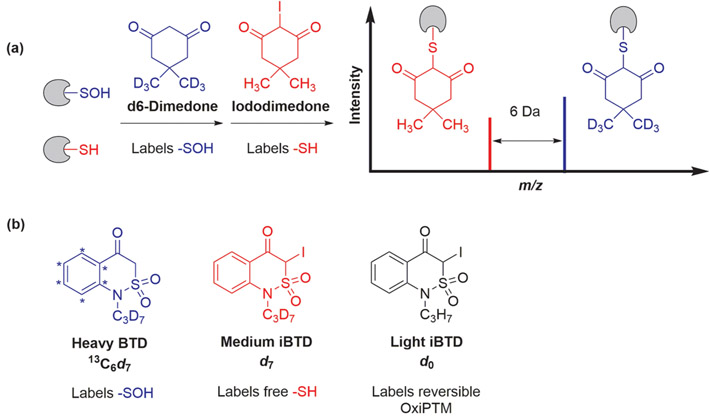
In addition to quantifying relative fold-changes in cysteine oxidation, another important goal is to measure the stoichiometry of ─SOH modification. By installing an iodo group on dimedone at C-2, we created a new thiol-reactive alkylating agent, yielding the same reaction product as sulfenic acid labeling by dimedone. Isotope-coded dimedone and iododimedone (ICDID) reagents were subsequently prepared such that the two reagents were separable by 6 Da, which enabled absolute quantification of the fraction ─SOH at a given cysteine (Figure 6a).72 In one proof-of-concept study, oxidation of Cys149 and Cys244 of GAPDH were profiled simultaneously. Importantly, only redox-sensitive Cys149 exhibited an increase in the fraction of sulfenic acid modification in response to H2O2. In an elegant modification of this chemical strategy, Guengerich and co-workers prepared BTD and iodo-BTD derivatives 13C6d7-pBTD, d7-ipBTD, and d0-ipBTD to quantify sulfenic acids, free thiols, and reversible OxiPTMs in complex proteomes (Figure 6b).73 A relatively low number (~600) of sulfenylated peptides can be attributed to the lack of an enrichment handle.
Figure 6.
(a) Basic principle of the ICDID strategy of sulfenic acid quantification. (b) BTD-derived probes used in the quantification of cysteine oxidation.
Although other chemical probes for sulfenic acids have been reported, few of them have actually been used to globally quantify sulfenomes using MS platforms. Fox and co-workers recently reported the SAM-TCO probe, a strained trans-cyclooctene with an axial hydroxyl group proposed to facilitate the reaction via an intramolecular attack. Additionally, a bioorthogonal tetrazine-alkene reaction was employed to quench excess SAM-TCO, which may help to minimize artifacts that can result from cell lysis.51
To date, site-specific mapping of protein S-sulfenylation in the aforementioned chemoproteomic studies has illuminated many novel thiol-based regulatory mechanisms. For example, application of DYn-2 in A431 cells revealed S-sulfenylation of epidermal growth factor receptor (EGFR) at a critical cysteine adjacent to its active site, Cys797. Further biochemical and cellular studies in our laboratory demonstrated that S-sulfenylation of this site augments EGFR tyrosine kinase activity, thus creating a positive feedback loop with the enzymatic source of EGF-induced generation of H2O2 and NADPH oxidase (Nox).6,74 In another example, we demonstrated that S-sulfenylation of SIRT6 Cys18 leads to intermolecular disulfide formation with the transcription factor, HIF1A, disclosing a new mechanism for regulating the expression of multiple glycolytic genes.69 A recent study of S-sulfenylation with BTD in Arabidopsis thaliana mapped more than 1000 protein targets including AtMAPK4, an orthologue to human MAPK1, which is a known redox switch.75 Finally, application of BTD in mouse livers revealed the redox control of circadian pacemaker CLOCK via a reversibly oxidized cysteine.76
In addition to the discovery of new mechanisms of thiol-based redox regulation, quantitative site-specific mapping of S-sulfenylation has applications in drug design. Profiling with structurally and chemically distinct cyclic C-nucleophiles (DYn-2, PD, PYD, PRD, and BTD) identified > 1280 S-sulfenylated cysteines present in “druggable” proteins and orphan targets, revealing both disparate reactivity profiles and target preferences.70 Among the unique ligand–protein interactions, we identified a planar pyrrolidinedione nucleophile, PYD, that reacted preferentially with protein tyrosine phosphatases (PTPs). This study shows that fragment-based covalent ligand discovery with C-nucleophiles can be applied to generate an expansive view of the ligandable “redoxome” with significant implications for covalent inhibitor pharmacology.
Profiling Protein Sulfinic Acids (─SO2H)
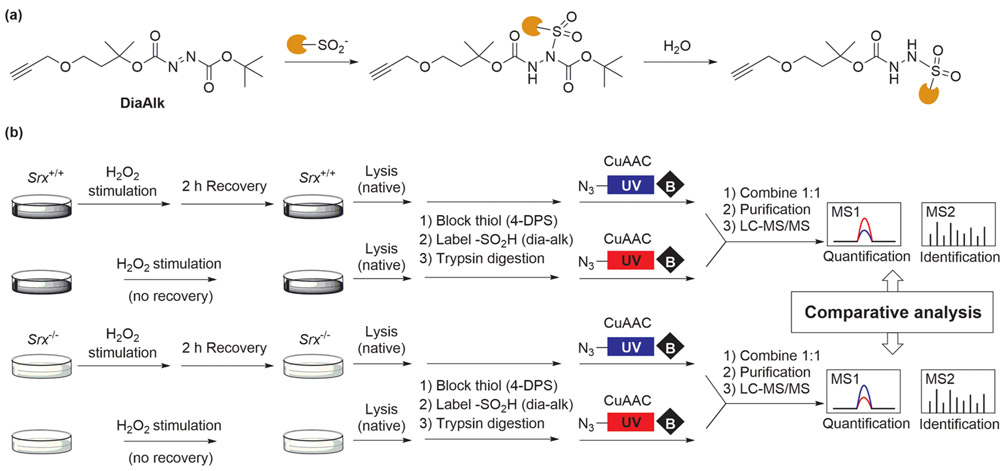
Protein S-sulfinylation was traditionally detected by antibodies or label-free MS. These methods suffer from limited affinity and specificity, or lengthy preparation steps. On the other hand, electrophilic nitrogen species (ENS) only form stable adducts with ─SO2H and can therefore serve as chemoselective probes for activity-based detection of S-sulfinylation (Figure 1d). Our group has designed two ENS-based ─SO2H probes: one compound is a C-nitroso benzoic ester named NO-Bio and the other probe, DiaAlk, is based upon an electron deficient diazine scaffold.77-79 The latter has superior sensitivity and compatibility with MS, facilitating site-centric quantitative analyses of protein sulfinic acids (Figure 7a). Biotinylated S-nitrosothiols have been used to detect ─SO2H,24 but side reactions generate an aggressive electrophilic oxidant nitroxyl (HNO) that converts thiols to sulfinic acids.79
Figure 7.
(a) DiaAlk reacts with sulfinic acids followed by hydrolysis to form a stable MS-compatible product. (b) Discovery of SRX substrates by a chemical proteomics approach. With a recovery period, putative SRX substrates showed a lower recovery/control (blue/red) ratio in wild type cells, and a higher ratio in SRX knockout cells.
Most recently, in close collaboration with the Yang laboratory, we have developed a site-centric proteomic approach to profile cysteine sulfinic acids. Application of DiaAlk uncovered 387 ─SO2H sites from native lysates of A549 and HeLa cells, hundreds of which were previously unknown. Interestingly, S-sulfinylation levels under acute H2O2 exposure remained relatively static at the majority of sites, in contrast to the dynamic changes observed in the S-sulfenylome, suggesting that sulfinic acid modification likely takes place on a different time scale, with fewer biological pathways for repair. To investigate these observations in greater detail, we probed S-sulfinylome repair in a cysteine sulfinic acid reductase sulfiredoxin (SRX) knockout model. Comparative study of Srx+/+ and Srx−/− mouse embryonic fibroblasts (MEFs) revealed more than 50 potential substrates of SRX, with subsequent biochemical validation of several new targets (Figure 7b). Our findings expand the substrate scope of SRX significantly beyond Prx isoforms 1–4, with new implications for the role of SRX in oxidative stress-associated diseases and drug development programs.79
Profiling Protein Persulfides (─SSH)
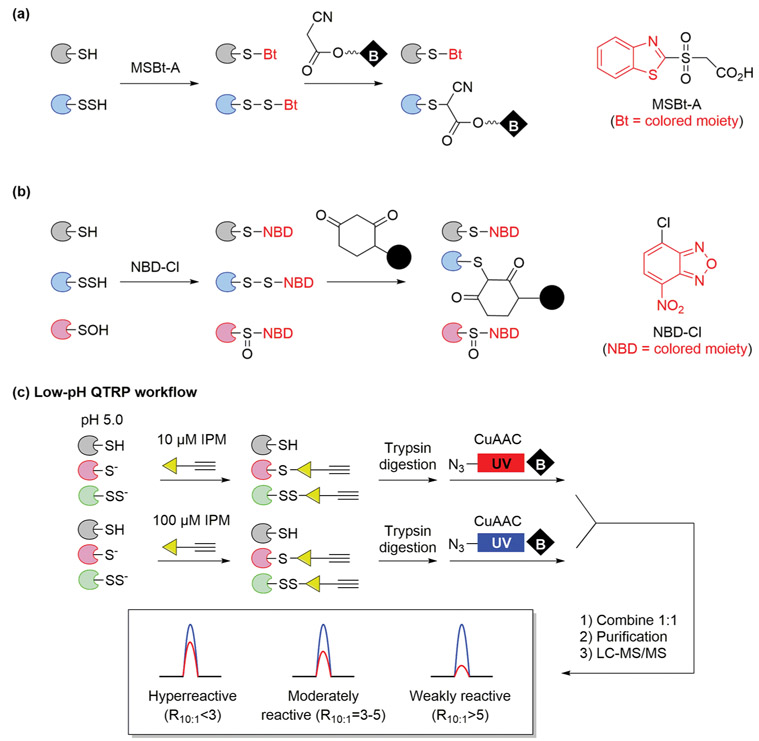
Protein persulfidation (S-sulfhydration) generates persulfides (─SSH) that were detectable by alkylating agents. Although there are limited reports from small-molecule models, possibly as a result of low yield from product decomposition,80 our lab has convincingly demonstrated that a variety of persulfide models, including protein persulfide Gpx3─SSH, readily react with most thiol-reactive agents.22 However, they are often accompanied by issues in selectivity. An alkylation/tag-switch approach was introduced as a reliable method to detect persulfides, where the disulfide products of ─SSH alkylation are substituted by a nucleophilic probe and alkylated ─SH are irreversibly modified. For example, thiols and persulfides were alkylated with MSBt-A, and only the latter subsequently reacted with a cyanoacetate nucleophile with a biotin reporter (Figure 8a).26 Recently, the Filipovic group used NBD-Cl and a dimedone derivative to detect protein persulfides. An evolutionary-conserved correlation between aging and a decay in persulfidation levels was observed, suggesting a protective role of ─SSH against irreversible cysteine oxidation (Figure 8b).81 A direct method to simultaneously profile thiol and persulfide proteomes has also been reported.27 In this method, termed low-pH quantitative thiol reactivity profiling (QTRP), the alkylation by IPM is performed at pH 5.0, because persulfides (pKa of Cys─SSH = 4.3) are expected to maintain high reactivity, whereas more abundant free thiols (pKa of Cys─SH = 8.3) are mostly protonated and less reactive (Figure 8c).
Figure 8.
(a) Persulfide detection by MSBt-A alkylation and cyanoacetate tag-switch. (b) Persulfide detection by NBD-Cl alkylation and dimedone derivative tag-switch. (c) Direct thiol and persulfide activity profiling with the low-pH QTRP workflow.
CONCLUSIONS AND OUTLOOK
LC-MS platforms featuring chemoselective probes (Table 2) offer compelling advantages on resolution, throughput, accuracy, and reproducibility. At the same time, our evolving understanding of cysteine OxiPTMs presents new challenges in detection. For example, reagents believed to be thiol-selective were found to react with ─SOH, ─SO2H, and ─SSH species, which are of increasing biological relevance. Certain cysteine OxiPTMs exhibit dual reactivity (e.g., ─SOH is both nucleophilic and electrophilic), diverse reactivity (e.g., ─SSR), or similar reactivity to other OxiPTMs (e.g., ─SH and ─SSH). Therefore, it is critical to conduct a comprehensive survey of cross-reactivity while keeping the biological relevance and abundance of cysteine OxiPTMs in mind. More importantly, caution is advised against extrapolating the reactivity of an entire cysteine chemotype on the basis of studies with compounds that are not biorepresentative, as such models which may not represent the true reaction profiles as in complex proteomes.
Table 2.
List of Chemical Probes Discussed in This Account
| name of probe | targets | notes | refs |
|---|---|---|---|
| DYn-2, BTD | ─SOH | BTD is ~170× faster than DYn-2 | 69-71,74 |
| ICDID, iTROC reagents | ─SH, ─SOH | for stoichiometric analysis | 72,73 |
| BCN, Norb-bio, SAM-TCO | ─SOH | may react with Cys─SH, ─SSH | 48-51 |
| NO-bio, DiaAlk | ─SO2H | DiaAlk is MS-compatible | 77-79 |
| IPM | ─SH, ─SSH | operates at low pH | 27,36 |
| MSBT, NBD-Cl | ─SH, ─SSH | tag-switch with C-nucleophiles | 26,81 |
In order to profile cysteine OxiPTMs with high fidelity, many efforts have been made in recent years including but not limited to the following: (1) Direct “activity-based” sensing of OxiPTMs with chemoselective probes to minimize various sources of artifacts generated by tag-switch methods. (2) In situ detection with smaller, cell permeable, nontoxic probes, which makes discovery in live cells or tissues possible. (3) Temporal control of labeling with probes that can be switched on/off. This decreases the cytotoxicity of the probes and can reduce labeling after cell lysis. (4) Site-centric profiling, which is significantly more stringent than protein level discovery and provides mechanistic information. (5) Multiplex labeling that allows multiple samples or modifications to be analyzed simultaneously, reducing the cost and run-to-run variations.
Novel discovery of proteomic cysteine oxidation fuels the future development in the fields of redox signaling and disease mechanism. Moreover, complementary to covalent inhibitors that target free cysteine thiols, redox-based covalent inhibitors targeting cysteine OxiPTMs may be developed. For example, we have shown that sulfenylation of EGFR not only enhances its activity but also masks its reactivity toward thiol-targeting covalent inhibitors like afatinib.6,74 Proteome-wide ligand discovery on cysteine OxiPTMs is ongoing and is expected to reveal additional targets. Another interesting direction is organelle-specific profiling of cysteine oxidation. As one of the major source of ROS, mitochondria are an intriguing host of cysteine OxiPTMs. Along these lines, the Furdui and King laboratories have reported ─SOH probes conjugated with mitochondria targeting vectors, such as fluorescent coumarin and rhodamine moieties, or triphenylphosphonium groups, which serve as proof-of-concept for localized profiling of OxiPTMs.82,83 Last but not least, direct probing of protein disulfides remains an extremely challenging but vital mission. To date, protein disulfides are detected by indirect tag-switch methods, hampering our ability to profile disulfide reactivity and assign disulfide pairs. Although a sophisticated MS-based technique to characterize disulfide linkages in IgG2λ antibodies has been reported,84 going forward, a reaction-based approach to map disulfides at the proteome level is still in great demand. This is an active area of research in our group, as we currently explore several promising chemical strategies; these findings are forthcoming and will be reported in due course.
ACKNOWLEDGMENTS
The authors acknowledge the National Institutes of Health (R01GM102187, R01GM087638, R01CA227849) for funding. We thank J. Yang (Beijing Proteome Research Center) for helpful advice for the manuscript.
Biography
Yunlong Shi received his B.S. in Chemistry from Zhejiang University, where he conducted research on organometallic chemistry and bioengineering. He earned his Ph.D. degree in Organic Chemistry at North Carolina State University in the laboratory of Prof. Joshua G. Pierce, where he focused on the synthesis and medicinal chemistry of natural products. He is currently a postdoctoral associate in the laboratory of Prof. Kate S. Carroll, working on chemical biology and designing novel probes for proteomics and therapy.
Kate S. Carroll is an Associate Professor with tenure in the Department of Chemistry at The Scripps Research Institute in Jupiter, Florida. She received her B.A. degree in Biochemistry from Mills College in 1996 and Ph.D. in Biochemistry from Stanford University in 2003. Her postdoctoral work was completed at the University of California, Berkeley, where she was a Damon Runyon-Walter Winchell Chancer Fund Fellow with Prof. Carolyn Bertozzi. She was an Assistant Professor at the University of Michigan until 2010, when she joined the Chemistry faculty at Scripps. Her research interests span the disciplines of chemistry and biology with an emphasis on studies of sulfur biochemistry pertinent to human disease. Her lab focuses on the development of chemical probes to study redox modifications of cysteine thiols, profiling changes in protein oxidation associated with disease, and exploiting this information for development of diagnostic and therapeutic approaches.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Deribe YL; Pawson T; Dikic I Post-Translational Modifications in Signal Integration. Nat. Struct. Mol. Biol 2010, 17, 666–672. [DOI] [PubMed] [Google Scholar]
- (2).Paulsen CE; Carroll KS Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev 2013, 113, 4633–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ray PD; Huang B-W; Tsuji Y Reactive Oxygen Species (ROS) Homeostasis and Redox Regulation in Cellular Signaling. Cell. Signalling 2012, 24, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Dickinson BC; Chang CJ Chemistry and Biology of Reactive Oxygen Species in Signaling or Stress Responses. Nat. Chem. Biol 2011, 7, 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ho GPH; Selvakumar B; Mukai J; Hester LD; Wang Y; Gogos JA; Snyder SH S-Nitrosylation and S-Palmitoylation Reciprocally Regulate Synaptic Targeting of PSD-95. Neuron 2011, 71 (1), 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Truong TH; Ung PM-U; Palde PB; Paulsen CE; Schlessinger A; Carroll KS Molecular Basis for Redox Activation of Epidermal Growth Factor Receptor Kinase. Cell Chem. Biol 2016, 23, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wible RS; Sutter TR Soft Cysteine Signaling Network: The Functional Significance of Cysteine in Protein Function and the Soft Acids/Bases Thiol Chemistry That Facilitates Cysteine Modification. Chem. Res. Toxicol 2017, 30, 729–762. [DOI] [PubMed] [Google Scholar]
- (8).D’Autréaux B; Toledano MB ROS as Signalling Molecules: Mechanisms That Generate Specificity in ROS Homeostasis. Nat. Rev. Mol. Cell Biol 2007, 8, 813–824. [DOI] [PubMed] [Google Scholar]
- (9).Gupta V; Carroll KS Sulfenic Acid Chemistry, Detection and Cellular Lifetime. Biochim. Biophys. Acta, Gen. Subj 2014, 1840, 847875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Valko M; Leibfritz D; Moncol J; Cronin MTD; Mazur M; Telser J Free Radicals and Antioxidants in Normal Physiological Functions and Human Disease. Int. J. Biochem. Cell Biol 2007, 39, 44–84. [DOI] [PubMed] [Google Scholar]
- (11).Bantscheff M; Lemeer S; Savitski MM; Kuster B Quantitative Mass Spectrometry in Proteomics: Critical Review Update from 2007 to the Present. Anal. Bioanal Chem 2012, 404, 939–965. [DOI] [PubMed] [Google Scholar]
- (12).Cravatt BF; Simon GM; Yates III JR The Biological Impact of Mass-Spectrometry-Based Proteomics. Nature 2007, 450, 991–1000. [DOI] [PubMed] [Google Scholar]
- (13).Wright MH; Sieber SA Chemical Proteomics Approaches for Identifying the Cellular Targets of Natural Products. Nat. Prod. Rep 2016, 33, 681–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Sletten EM; Bertozzi CR Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem., Int. Ed 2009, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gygi SP; Rist B; Gerber SA; Turecek F; Gelb MH; Aebersold R Quantitative Analysis of Complex Protein Mixtures Using Isotope-Coded Affinity Tags. Nat. Biotechnol 1999, 17, 994–999. [DOI] [PubMed] [Google Scholar]
- (16).Requejo R; Hurd TR; Costa NJ; Murphy MP Cysteine Residues Exposed on Protein Surfaces Are the Dominant Intramitochondrial Thiol and May Protect against Oxidative Damage. FEBS J 2010, 277, 1465–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hansen RE; Roth D; Winther JR Quantifying the Global Cellular Thiol-disulfide Status. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Suttapitugsakul S; Xiao H; Smeekens J; Wu R Evaluation and Optimization of Reduction and Alkylation Methods to Maximize Peptide Identification with MS-Based Proteomics. Mol. BioSyst 2017, 13, 2574–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Müller T; Winter D Systematic Evaluation of Protein Reduction and Alkylation Reveals Massive Unspecific Side Effects by Iodine-Containing Reagents. Mol. Cell. Proteomics 2017, 16, 1173–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Reisz JA; Bechtold E; King SB; Poole LB; Furdui CM Thiol-Blocking Electrophiles Interfere with Labeling and Detection of Protein Sulfenic Acids. FEBS J 2013, 280, 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kuo Y-H; Konopko AM; Borotto NB; Majmudar JD; Haynes SE; Martin BR Profiling Protein S-Sulfination with Maleimide-Linked Probes. ChemBioChem 2017, 18, 2028–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Pan J; Carroll KS Persulfide Reactivity in the Detection of Protein S-Sulfhydration. ACS Chem. Biol 2013, 8, 1110–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang D; Devarie-Baez NO; Li Q; Lancaster JR; Xian M Methylsulfonyl Benzothiazole (MSBT): A Selective Protein Thiol Blocking Reagent. Org. Lett 2012, 14, 3396–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Majmudar JD; Konopko AM; Labby KJ; Tom CTMB; Crellin JE; Prakash A; Martin BR Harnessing Redox Cross-Reactivity To Profile Distinct Cysteine Modifications. J. Am. Chem. Soc 2016, 138, 1852–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Cuevasanta E; Lange M; Bonanata J; Coitmo EL; Ferrer- Sueta G; Filipovic MR; Alvarez B Reaction of Hydrogen Sulfide with Disulfide and Sulfenic Acid to Form the Strongly Nucleophilic Persulfide. J. Biol. Chem 2015, 290, 26866–26880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhang D; Macinkovic I; Devarie-Baez NO; Pan J; Park C-M; Carroll KS; Filipovic MR; Xian M. Detection of Protein S-Sulfhydration by a Tag-Switch Technique. Angew. Chem., Int. Ed 2014, 53, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Fu L; Liu K; He J; Tian C; Yu X; Yang J Direct Proteomic Mapping of Cysteine Persulfidation. Antioxid. Redox Signaling 2019, DOI: 10.1089/ars.2019.7777. [DOI] [PubMed] [Google Scholar]
- (28).Sanjeeva SK; Korrapati S; Nair CB; Rao PVS; Pullela PK; Vijayalakshmi U; Siva R Hydrophobic Interactions in Donor- Disulphide-Acceptor (DSSA) Probes Looking Beyond Fluorescence Resonance Energy Transfer Theory. J. Fluoresc 2014, 24, 1297–1306. [DOI] [PubMed] [Google Scholar]
- (29).Wang H; Xian M Chemical Methods to Detect S-Nitrosation. Curr. Opin. Chem. Biol 2011, 15, 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Saurin AT; Neubert H; Brennan JP; Eaton P Widespread Sulfenic Acid Formation in Tissues in Response to Hydrogen Peroxide. Proc. Natl. Acad. Sci U. S. A 2004, 101, 17982–17987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Loi VV; Rossius M; Antelmann H Redox Regulation by Reversible Protein S-Thiolation in Bacteria. Front. Microbiol 2015, 6, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Leichert LI; Gehrke F; Gudiseva HV; Blackwell T; Ilbert M; Walker AK; Strahler JR; Andrews PC; Jakob U Quantifying Changes in the Thiol Redox Proteome upon Oxidative Stress in Vivo. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 8197–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Go Y-M; Park H; Koval M; Orr M; Reed M; Liang Y; Smith D; Pohl J; Jones DP A Key Role for Mitochondria in Endothelial Signaling by Plasma Cysteine/Cystine Redox Potential. Free Radical Biol. Med 2010, 48 (2), 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Pan K-T; Chen Y-Y; Pu T-H; Chao Y-S; Yang C-Y; Bomgarden RD; Rogers JC; Meng T-C; Khoo K-H Mass Spectrometry-Based Quantitative Proteomics for Dissecting Multiplexed Redox Cysteine Modifications in Nitric Oxide-Protected Cardiomyocyte Under Hypoxia. Antioxid. Redox Signaling 2014, 20, 1365–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Murray CI; Uhrigshardt H; O’Meally RN; Cole RN; Van Eyk JE Identification and Quantification of S-Nitrosylation by Cysteine Reactive Tandem Mass Tag Switch Assay. Mol. Cell. Proteomics 2012, 11, M111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Fu L; Liu K; Sun M; Tian C; Sun R; Morales Betanzos C; Tallman KA; Porter NA; Yang Y; Guo D; Liebler DC; Yang J Systematic and Quantitative Assessment of Hydrogen Peroxide Reactivity With Cysteines Across Human Proteomes. Mol. Cell. Proteomics 2017, 16, 1815–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Abo M; Li C; Weerapana E Isotopically-Labeled Iodoacetamide-Alkyne Probes for Quantitative Cysteine-Reactivity Profiling. Mol. Pharmaceutics 2018, 15, 743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Abo M; Weerapana E A Caged Electrophilic Probe for Global Analysis of Cysteine Reactivity in Living Cells. J. Am. Chem. Soc 2015, 137, 7087–7090. [DOI] [PubMed] [Google Scholar]
- (39).Vajrychova M; Salovska B; Pimkova K; Fabrik I; Tambor V; Kondelova A; Bartek J; Hodny Z Quantification of Cellular Protein and Redox Imbalance Using SILAC-IodoTMT Methodology. Redox Biol. 2019, 24, 101227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Martínez-Acedo P; Núñez E; Gómez FJS; Moreno M; Ramos E; Izquierdo-Álvarez A; Miró-Casas E; Mesa R; Rodriguez P; Martínez-Ruiz A; Dorado DG; Lamas S; Vázquez J A Novel Strategy for Global Analysis of the Dynamic Thiol Redox Proteome. Mol. Cell. Proteomics 2012, 11, 800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Parker J; Balmant K; Zhu F; Zhu N; Chen S CysTMTRAQ—An Integrative Method for Unbiased Thiol-Based Redox Proteomics. Mol. Cell. Proteomics 2015, 14, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Anjo SI; Melo MN; Loureiro LR; Sabala L; Castanheira P; Grãos M; Manadas B OxSWATH: An Integrative Method for a Comprehensive Redox-Centered Analysis Combined with a Generic Differential Proteomics Screening. Redox Biol. 2019, 22, 101130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Arrowsmith CH; Audia JE; Austin C; Baell J; Bennett J; Blagg J; Bountra C; Brennan PE; Brown PJ; Bunnage ME; Buser-Doepner C; Campbell RM; Carter AJ; Cohen P; Copeland RA; Cravatt B; Dahlin JL; Dhanak D; Edwards AM; Frederiksen M; Frye SV; Gray N; Grimshaw CE; Hepworth D; Howe T; Huber KVM; Jin J; Knapp S; Kotz JD ; Kruger RG; Lowe D; Mader MM; Marsden B; Mueller-Fahrnow A; Müller S; O’Hagan RC; Overington JP; Owen DR; Rosenberg SH; Ross R; Roth B; Schapira M; Schreiber SL; Shoichet B; Sundström M; Superti-Furga G; Taunton J; Toledo-Sherman L; Walpole C; Walters MA; Willson TM; Workman P; Young RN; Zuercher WJ The Promise and Peril of Chemical Probes. Nat. Chem. Biol 2015, 11, 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Benitez LV; Allison WS The Inactivation of the Acyl Phosphatase Activity Catalyzed by the Sulfenic Acid Form of Glyceraldehyde 3-Phosphate Dehydrogenase by Dimedone and Olefins. J. Biol. Chem 1974, 249, 6234–6243. [PubMed] [Google Scholar]
- (45).Gupta V; Carroll KS Profiling the Reactivity of Cyclic C-Nucleophiles towards Electrophilic Sulfur in Cysteine Sulfenic Acid. Chem. Sci 2016, 7, 400–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Gupta V; Carroll KS Rational Design of Reversible and Irreversible Cysteine Sulfenic Acid-Targeted Linear C-Nucleophiles. Chem. Commun 2016, 52, 3414–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Gupta V; Paritala H; Carroll KS Reactivity, Selectivity, and Stability in Sulfenic Acid Detection: A Comparative Study of Nucleophilic and Electrophilic Probes. Bioconjugate Chem 2016, 27, 1411–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Poole TH; Reisz JA; Zhao W; Poole LB; Furdui CM; King SB Strained Cycloalkynes as New Protein Sulfenic Acid Traps. J. Am. Chem. Soc 2014, 136, 6167–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Alcock LJ; Farrell KD; Akol MT; Jones GH; Tierney MM; Kramer HB; Pukala TL; Bernardes GJL; Perkins MV; Chalker JM Norbornene Probes for the Study of Cysteine Oxidation. Tetrahedron 2018, 74, 1220–1228. [Google Scholar]
- (50).Alcock LJ; Oliveira BL; Deery MJ; Pukala TL; Perkins MV; Bernardes GJL; Chalker JM Norbornene Probes for the Detection of Cysteine Sulfenic Acid in Cells. ACS Chem. Biol 2019, 14, 594–598. [DOI] [PubMed] [Google Scholar]
- (51).Scinto SL; Ekanayake O; Seneviratne U; Pigga JE; Boyd SJ; Taylor MT; Liu J; Am Ende CW; Rozovsky S; Fox JM Dual-Reactivity Trans-Cyclooctenol Probes for Sulfenylation in Live Cells Enable Temporal Control via Bioorthogonal Quenching. J. Am. Chem. Soc 2019, 141, 10932–10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).van Geel R; Pruijn GJM; van Delft FL; Boelens WC Preventing Thiol-Yne Addition Improves the Specificity of Strain- Promoted Azide-Alkyne Cycloaddition. Bioconjugate Chem. 2012, 23, 392–398. [DOI] [PubMed] [Google Scholar]
- (53).Galardon E; Padovani D Reactivity of Persulfides Toward Strained Bicyclo[6.1.0]Nonyne Derivatives: Relevance to Chemical Tagging of Proteins. Bioconjugate Chem. 2015, 26, 1013–1016. [DOI] [PubMed] [Google Scholar]
- (54).Sivaramakrishnan S; Keerthi K; Gates KS A Chemical Model for Redox Regulation of Protein Tyrosine Phosphatase 1B (PTP1B) Activity. J. Am. Chem. Soc 2005, 127, 10830–10831. [DOI] [PubMed] [Google Scholar]
- (55).Pan J; Carroll KS Light-Mediated Sulfenic Acid Generation from Photocaged Cysteine Sulfoxide. Org. Lett 2015, 17, 6014–6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Klomsiri C; Nelson KJ; Bechtold E; Soito L; Johnson LC; Lowther WT; Ryu S-E; King SB; Furdui CM; Poole LB Use of Dimedone-Based Chemical Probes for Sulfenic Acid Detection: Evaluation of Conditions Affecting Probe Incorporation into Redox-Sensitive Proteins In Thiol Redox Transitions in Cell Signaling, Part A: Chemistry and Biochemistry of Low Molecular Weight and Protein Thiols; Cadenas E, Packer L, Eds.; Academic Press, 2010; Vol. 473, pp 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Tom CTMB; Crellin JE; Motiwala HF; Stone MB; Davda D; Walker W; Kuo Y-H; Hernandez JL; Labby KJ; Gomez-Rodriguez L; Jenkins PM; Veatch SL; Martin BR Chemoselective Ratiometric Imaging of Protein S-Sulfenylation. Chem. Commun 2017, 53, 7385–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Salmeen A; Andersen JN; Myers MP; Meng T-C; Hinks JA; Tonks NK; Barford D Redox Regulation of Protein Tyrosine Phosphatase 1B Involves a Sulphenyl-Amide Intermediate. Nature 2003, 423, 769–773. [DOI] [PubMed] [Google Scholar]
- (59).Lee J-W; Soonsanga S; Helmann JD A Complex Thiolate Switch Regulates the Bacillus Subtilis Organic Peroxide Sensor OhrR. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 8743–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Tsutsumi R; Harizanova J; Stockert R; Schroder K; Bastiaens PIH; Neel BG Assay to Visualize Specific Protein Oxidation Reveals Spatio-Temporal Regulation of SHP2. Nat. Commun 2017, 8, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Sarma BK; Mugesh G Redox Regulation of Protein Tyrosine Phosphatase 1B (PTP1B): A Biomimetic Study on the Unexpected Formation of a Sulfenyl Amide Intermediate. J. Am. Chem. Soc 2007, 129, 8872–8881. [DOI] [PubMed] [Google Scholar]
- (62).Forman HJ; Davies MJ; Krämer AC; Miotto G; Zaccarin M; Zhang H; Ursini F Protein Cysteine Oxidation in Redox Signaling: Caveats on Sulfenic Acid Detection and Quantification. Arch. Biochem. Biophys 2017, 617, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Stöcker S; Van Laer K; Mijuskovic A; Dick TP The Conundrum of Hydrogen Peroxide Signaling and the Emerging Role of Peroxiredoxins as Redox Relay Hubs. Antioxid. Redox Signaling 2018, 28, 558–573. [DOI] [PubMed] [Google Scholar]
- (64).Heppner DE; Hristova M; Ida T; Mijuskovic A; Dustin CM; Bogdándi V; Fukuto JM; Dick TP; Nagy P; Li J; Akaike T; van der Vliet A Cysteine Perthiosulfenic Acid (Cys-SSOH): A Novel Intermediate in Thiol-Based Redox Signaling? Redox Biol. 2018, 14, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Bogdándi V; Ida T; Sutton TR; Bianco C; Ditrói T; Koster G; Henthorn HA; Minnion M; Toscano JP; van der Vliet A; Pluth MD; Feelisch M; Fukuto JM; Akaike T; Nagy P Speciation of Reactive Sulfur Species and Their Reactions with Alkylating Agents: Do We Have Any Clue about What Is Present inside the Cell? Br. J. Pharmacol 2019, 176, 646–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Martínez-Acedo P; Gupta V; Carroll KS Proteomic Analysis of Peptides Tagged with Dimedone and Related Probes. J. Mass Spectrom 2014, 49, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Leonard SE; Reddie KG; Carroll KS Mining the Thiol Proteome for Sulfenic Acid Modifications Reveals New Targets for Oxidation in Cells. ACS Chem. Biol 2009, 4, 783–799. [DOI] [PubMed] [Google Scholar]
- (68).Charron G; Zhang MM; Yount JS; Wilson J; Raghavan AS; Shamir E; Hang HC Robust Fluorescent Detection of Protein Fatty-Acylation with Chemical Reporters. J. Am. Chem. Soc 2009, 131, 4967–4975. [DOI] [PubMed] [Google Scholar]
- (69).Yang J; Gupta V; Carroll KS; Liebler DC Site-Specific Mapping and Quantification of Protein S-Sulphenylation in Cells. Nat. Commun 2014, 5, 4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Gupta V; Yang J; Liebler DC; Carroll KS Diverse Redoxome Reactivity Profiles of Carbon Nucleophiles. J. Am. Chem. Soc 2017, 139, 5588–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Fu L; Liu K; Ferreira RB; Carroll KS; Yang J Proteome-Wide Analysis of Cysteine S-Sulfenylation Using a Benzothiazine-Based Probe. Curr. Protoc. Protein Sci 2019, 95, No. e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Seo YH; Carroll KS Quantification of Protein Sulfenic Acid Modifications Using Isotope-Coded Dimedone and Iododimedone. Angew. Chem., Int. Ed 2011, 50, 1342–1345. [DOI] [PubMed] [Google Scholar]
- (73).Albertolle ME; Glass SM; Trefts E; Guengerich FP Isotopic Tagging of Oxidized and Reduced Cysteines (ITORC) for Detecting and Quantifying Sulfenic Acids, Disulfides, and Free Thiols in Cells. J. Biol. Chem 2019, 294, 6522–6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Paulsen CE; Truong TH; Garcia FJ; Homann A; Gupta V; Leonard SE; Carroll KS Peroxide-Dependent Sulfenylation of the EGFR Catalytic Site Enhances Kinase Activity. Nat. Chem. Biol 2012, 8, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Huang J; Willems P; Wei B; Tian C; Ferreira RB; Bodra N; Martinez Gache SA; Wahni K; Liu K; Vertommen D; Gevaert K; Carroll KS; Van Montagu M; Yang J; Van Breusegem F; Messens J Mining for Protein S-Sulfenylation in Arabidopsis Uncovers Redox-Sensitive Sites. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 21256–21261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Pei J-F; Li X-K; Li W-Q; Gao Q; Zhang Y; Wang X-M; Fu J-Q; Cui S-S; Qu J-H; Zhao X; Hao D-L; Ju D; Liu N; Carroll KS; Yang J; Zhang EE; Cao J-M; Chen H-Z; Liu D-P. Diurnal Oscillations of Endogenous H2O2 Sustained by P66Shc Regulate Circadian Clocks. Nat. Cell Biol 2019, 21, 1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Lo Conte M; Carroll KS Chemoselective Ligation of Sulfinic Acids with Aryl-Nitroso Compounds. Angew. Chem., Int. Ed 2012, 51, 6502–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Lo Conte M; Lin J; Wilson MA; Carroll KS A Chemical Approach for the Detection of Protein Sulfinylation. ACS Chem. Biol 2015, 10, 1825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Akter S; Fu L; Jung Y; Conte M. Lo; Lawson JR; Lowther WT; Sun R; Liu K; Yang J; Carroll KS Chemical Proteomics Reveals New Targets of Cysteine Sulfinic Acid Reductase. Nat. Chem. Biol 2018, 14, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Park C-M; Weerasinghe L; Day JJ; Fukuto JM; Xian M Persulfides: Current Knowledge and Challenges in Chemistry and Chemical Biology. Mol. BioSyst 2015, 11, 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Zivanovic J; Kouroussis E; Kohl JB; Adhikari B; Bursac B; Schott-Roux S; Petrovic D; Miljkovic JL; Thomas-Lopez D; Jung Y; Miler M; Mitchell S; Milosevic V; Gomes JE; Benhar M; Gonzales-Zorn B; Ivanovic-Burmazovic I; Torregrossa R; Mitchell JR; Whiteman M; Schwarz G; Snyder SH; Paul BD; Carroll KS; Filipovic MR Selective Persulfide Detection Reveals Evolutionarily Conserved Antiaging Effects of S-Sulfhydration. Cell Metab 2019, 30, 1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Holmila RJ; Vance SA; Chen X; Wu H; Shukla K; Bharadwaj MS; Mims J; Wary Z; Marrs G; Singh R; Molina AJ; Poole LB; King SB; Furdui CM Mitochondria-Targeted Probes for Imaging Protein Sulfenylation. Sci. Rep 2018, 8, 6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Li Z; Forshaw TE; Holmila RJ; Vance SA; Wu H; Poole LB; Furdui CM; King SB Triphenylphosphonium- Derived Protein Sulfenic Acid Trapping Agents: Synthesis, Reactivity, and Effect on Mitochondrial Function. Chem. Res. Toxicol 2019, 32, 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Guan X; Zhang L; Wypych J Direct Mass Spectrometric Characterization of Disulfide Linkages. MAbs 2018, 10, 572–582. [DOI] [PMC free article] [PubMed] [Google Scholar]