Abstract
The cell culture-based retrotransposition reporter assay has been (and is) an essential tool for the study of vertebrate Long INterspersed Elements (LINEs). Developed more than 20 years ago, this assay has been instrumental in characterizing the role of LINE-encoded proteins in retrotransposition, understanding how ribonucleoprotein particles are formed, how host factors regulate LINE mobilization, etc. Moreover, variations of the conventional assay have been developed to investigate the biology of other currently active human retrotransposons, such as Alu and SVA. Here, we describe a protocol that allows combination of the conventional cell culture-based LINE-1 retrotransposition reporter assay with short interfering RNAs (siRNAs) and microRNA (miRNAs) mimics or inhibitors, which has allowed us to uncover specific miRNAs and host factors that regulate retrotransposition. The protocol described here is highly reproducible, quantitative, robust and flexible, and allows the study of several small RNA classes and various retrotransposons. To illustrate its utility, here we show that siRNAs to Fanconi anaemia proteins (FANC-A and FANC-C) and an inhibitor of miRNA-20 upregulate and downregulate human L1 retrotransposition, respectively.
This article is part of a discussion meeting issue ‘Crossroads between transposons and gene regulation’.
Keywords: cell culture-based retrotransposition reporter assay, LINE-1, siRNAs, miRNAs, miR-20, Fanconi anaemia
1. Background
Long INterspersed Element class 1 retrotransposons (LINE-1s or L1s) are the only autonomous active transposable elements in the human genome, and constitute approximately 17% of its mass [1]. Although the majority of the more than 500 000 genomic L1 copies are no longer active owing to 5′ truncation and/or the presence of inactivating mutations/internal rearrangements, it has been estimated that a reference human genome contains 80–100 retrotransposition competent or active L1s (RC-L1s) [2,3]. Recent studies have demonstrated that ongoing LINE-1 retrotransposition can target the human genome in a random manner [4,5], implying that new L1 insertions can impact our genome through a myriad of mechanisms (reviewed in [6–8]). As a result, retrotransposition events mediated by RC-L1s have been associated with a diverse group of more than 100 genetic disorders in man [9].
Human RC-L1s are 6 kb in length, and, from 5′ to 3′, an active element contains a 900 bp long 5′ untranslated region (UTR) with sense and antisense promoter activities [10,11], two non-overlapping open reading frames (L1-ORF1 and L1-ORF2) and a short 3′UTR ending in a poly(A) tail [6–8] (figure 1). L1-ORF1 codes for a protein with sequence-independent RNA binding and nucleic acid chaperone activities, while L1-ORF2 codes for a protein with endonuclease (EN) and reverse transcriptase (RT) activities (reviewed in [6,7,13]). L1 retrotransposition starts with the transcription of a full-length L1 mRNA using the internal sense promoter [11]; in the cytoplasm, L1-ORF1p and L1-ORF2p are translated [14,15], and both proteins bind back in cis to their encoding RNA [16], forming a RiboNucleoprotein Particle (L1-RNP, a retrotransposition intermediate [17,18]). The L1-RNP can access the nucleus without cell division [19,20], although in dividing cells retrotransposition peaks during the S phase [21]. In the nucleus, a new L1 insertion is generated by a mechanism known as target-primed reverse transcription (TPRT [22,23], reviewed in [6,7,13]), which requires the enzymatic activities encoded by the L1-RNP but also host factors. RC-L1s are also responsible for the mobilization of Alu and SVA RNAs in trans [24–26], and even cellular mRNAs to generate processed pseudogenes [27]. In summary, L1 activity has generated at least a third of the human genome during evolution, and RC- L1s, Alus and SVAs continue to impact our genome.
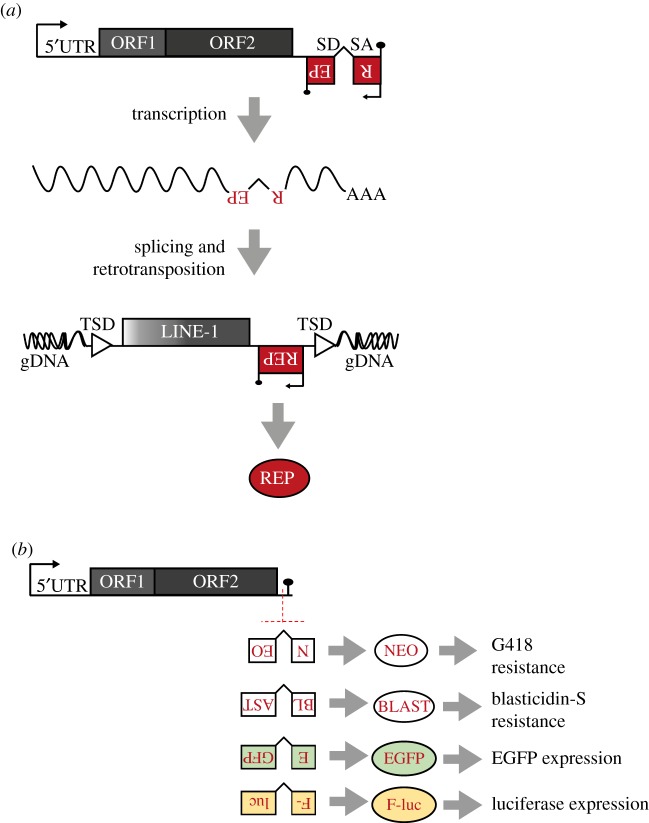
Figure 1.
Rationale of the retrotransposition reporter assay. (a) The structure of a human RC-L1 is shown. In the conventional retrotransposition reporter assay, an active L1 is tagged with a retrotransposition indicator cassette in its 3′UTR (red boxes labelled with backward REP), which is normally cloned at nucleotide 5982 with respect to L1.3 (accession number L19088.1 [12], a reference L1 used in most L1 biology studies). With this configuration, only transcripts generated from the L1 promoter can undergo a round of retrotransposition and activate expression of the reporter (red oval labelled REP). Further details can be found in the main text. Black arrows and lollipops indicate promoters and polyadenylation signals, respectively. TSD, target-site duplications flanking the new insertion (white triangles). (b) Schematic of the different retrotransposition reporter assays used to study LINE-1 biology. Below a scheme of an active human L1, are shown the structures of the four most commonly used L1 retrotransposition indicator cassettes: mneoI, mblastI, megfpI and mlucI. Also shown on the right is the reporter activated by each indicator cassette.
Given its mutagenic potential, retrotransposition needs to be controlled, allowing the evolutionary success of L1 over evolution without decreasing the fitness of species. Over the years, dozens of host factors regulating retrotransposition have been identified (recently reviewed in [6,9,13,28,29]). Epigenetic repression of L1 expression by DNA-methylation of the L1 CpG island is a well-known restriction mechanism [30–32], although additional cellular host factors that regulate other steps of the L1 retrotransposition cycle have also been characterized ([28,33], reviewed in [13,29]). Notably, the conventional L1-retrotransposition reporter assay developed by Moran and colleagues in 1996 ([34], reviewed in [35,36]) was instrumental in most of these studies, revealing the complexity of the ‘L1 regulome’. The L1-retrotransposition reporter assay exploits the rationale developed by Boeke and colleagues in 1985 to demonstrate that the mobilization of a long terminal repeat (LTR)-containing retrotransposon from yeast, Ty1, occurs through an intermediate RNA [37]. Briefly, in the L1 retrotransposition reporter assay, the 3′UTR of an active L1 is tagged with a retrotransposition indicator cassette oriented antisense with respect to L1 transcription (figure 1a). The retrotransposition indicator cassette consists of a reporter gene containing its own promoter and polyadenylation signal, but the reporter is interrupted by an intron that is located in the same transcriptional orientation as the L1 (i.e. antisense to the reporter, figure 1a). With this configuration, only transcripts arising from the L1 promoter could remove the intron by canonical splicing. If the resulting spliced RNA undergoes a round of retrotransposition, expression of the reporter gene will be activated upon L1 insertion, giving rise to a functional reporter product (figure 1a). The first version of the L1 retrotransposition reporter assay exploited a retrotransposition indicator cassette developed by Dixie Mager's laboratory, termed mneoI [34,38,39]; the mneoI cassette consists of the neomycin phosphotransferase gene, containing its own promoter (SV40) and polyadenylation sequence, and confers resistance against neomycin/G418 to cultured cells upon retrotransposition (figure 1b). The development of this assay has been instrumental in uncovering many aspects of L1 biology, identifying and characterize active LINEs in genomes, identifying how retrotransposition is regulated, etc. A wider view of all the implications and achievements facilitated by the retrotransposition reporter assay can be found in [7,35,36]. Over time, different reporter genes have been created to study LINE biology, including enhanced green fluorescent protein (EGFP, [40]), the blasticidin S-resistance gene [41,42] and luciferase [43] (figure 1b); similarly, modifications of the original retrotransposition assay have been developed to study the mobilization of Alu [44] and SVA retrotransposons [25,26], and to demonstrate pseudogene formation [16,27]. In sum, the cell culture-based L1 retrotransposition reporter assay represents an essential tool for most laboratories working on LINE biology.
On the other hand, the discovery of RNA interference [45] and microRNAs (miRNAs, [46]) were instrumental to uncover many aspects of human biology. miRNAs are small non-coding RNAs that function as key regulators of gene expression [47]; miRNAs are transcribed in the nucleus as long pri-miRNAs, undergo two subsequent processing steps by the Microprocessor in the nucleus and by Dicer in the cytoplasm, and are then loaded into AGO proteins to form the effector complex RISC (RNA-induced silencing complex, [48]); similarly, cytoplasmic siRNAs can be loaded into RISC. The RISC complex, guided by the miRNA/siRNA sequence, will bind its target RNA with imperfect or perfect pairing, respectively, and either promote its degradation or decrease its translation (miRNAs, reviewed in [49]), or it will promote degradation of mRNAs (siRNAs [45]). Indeed, while a given siRNA can normally regulate only one mRNA, a single miRNA can regulate a variety of mRNAs [50]. As a result, miRNAs are involved in a myriad of biological processes [47,51,52], including the regulation of LINE-1 retrotransposition (by miR-128 [53]). Interestingly, mammalian LINE-1s contain bidirectional promoters conserved through evolution, and it has been postulated that L1 derived RNAs could generate L1-siRNAs regulating retrotransposition [54].
The inherent specificity of siRNAs to degrade target RNAs with complementary sequences has resulted in useful molecular tools to ablate gene expression in a specific manner, and siRNAs can be routinely used to reduce gene expression of specific genes and study a plethora of biological processes [55]. Consistently, siRNAs are starting to be used as a therapeutic tool for several human disorders [56]. Here, we introduce sRNA/L1 retrotransposition, an optimized L1 retrotransposition reporter protocol that allows: (i) testing of whether a given miRNA can regulate L1 mobilization, and (ii) ablation of expression of specific proteins using siRNAs to test their role on L1 retrotransposition/regulation. The sRNA/L1 retrotransposition protocol is quantitative, sensitive and reproducible. As a proof of concept, we used the sRNA/L1 retrotransposition protocol to explore the role of miR-20 and of Fanconi anaemia mutated proteins on L1 retrotransposition.
2. Methods
(a). Cell culture
All reagents were purchased from Gibco-ThermoFisher unless otherwise indicated. Hela-JVM, HCT116 and U2OS cells were obtained from ATCC and cultured in high-glucose (4.5 g l−1) Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Hyclone), 2 mM Glutamax, 100 U ml−1 penicillin and 50 µg ml−1 streptomycin (Invitrogen). Cells were maintained at 37°C, 5% CO2 and atmospheric O2, and were passaged using 0.05% trypsin-EDTA. The absence of Mycoplasma spp. was confirmed at least once a month using the Lonza-Mycoalert Mycoplasma Detection Kit (at GENYO). Once a year, the identity of the cell lines was confirmed using short tandem repeat (STR) analyses at Lorgen (Granada, Spain).
(b). Plasmids
All plasmids used were purified using the Qiagen Plasmid Midi Kit (Qiagen), and were checked in 1% agarose gels prior to use. Only preparations of highly supercoiled DNAs (greater than 80%) were used for transfection.
(i). pJM101/L1.3
pJM101/L1.3 has been previously described in [12]. It contains a full-length copy of the human L1.3 element (accession number L19088.1 [12]) tagged with the mneoI retrotransposition indicator cassette [39]. It is cloned in pCEP4 (Life Technologies).
(ii). pJJ101/L1.3
pJJ101/L1.3 has been previously described in [57]. It contains a full-length copy of the human L1.3 element [12] tagged with the mblastI retrotransposition indicator cassette [41,42]. It is cloned in pCEP4 (Life Technologies).
(iii). Pu6ineo
Pu6ineo has been previously described in [58]. It contains the neomycin phosphotransferase (NEO) expression cassette from pEGFP-N1 (Clontech) cloned into a modified pBSKS-II(+) (Stratagene) that contains a Pol-III U6 promoter in the multi-cloning site. However, the U6 promoter does not participate or affect expression of the NEO gene.
(iv). pcDNA6.1
pcDNA6.1 was purchased from Invitrogen. It contains an expression cassette for the blasticidin-S deaminase gene.
(c). MicroRNA inhibitors and siRNAs
The miR-20a (accession number MIMAT0000075) inhibitor (antimiR, IH-300491-05), an antimiR negative control (C-, IN-002005-01), ON-TARGET plus SMART-pool siRNAs against XRCC4 (L-004494-00-0010), FANC-A (L-019283-00-0005) and FANC-C (L-016941-00-0005) and their non-targeting (NT) control (D-001810-10-20, ON-TARGETplus Non-targeting Pool) were purchased from Dharmacon. They were dissolved in 1× siRNA Buffer (Thermo) to a working concentration of 20 µM, aliquotted and stored at −80°C.
(d). MicroRNA expression analyses
We used RT-qPCR to quantify expression of miRNAs in cultured cells. Briefly, total RNA was isolated from confluent cultures of HeLa cells using TRIzol (Thermo fisher Scientific), following standard protocols. Next, we used the qScript microRNA cDNA Synthesis Kit (QuantaBio) to polyadenylate 1 µg of total RNAs, which were subsequently converted to cDNAs in a single step, following the manufacturer's instructions. cDNAs were diluted using UltraPure double-distilled (dd)H2O (Invitrogen) and quantitative PCR (qPCR) analyses were performed using a universal primer against poly(A) and miRNA-specific primers (electronic supplementary material, table S1) that allows the specific detection of polyadenylated mature miRNAs and not their precursors (QuantaBio). qPCRs were performed (20 µl, triplicate) using GoTaq qPCR MasterMix (Promega) and 0.15 µM of each primer. The qPCR cycling conditions were: 2 min at 95°C; 40 cycles of 15 s at 95°C, 30 s at 57°C and 30 s at 72°C. A melting curve was recorded to confirm the identity of amplified products. We used the Small Nucleolar RNA C/D Box44 (SNORD44) to normalize miRNA expression.
(e). miRNA-retrotransposition assays
Different concentrations of HeLa cells were plated in six-well tissue culture plates (Corning): 2 × 105 cells well−1 (triplicate) for the retrotransposition reporter assay and RNA extraction (duplicate), and 1 × 105 cells well−1 (triplicate) for the clonability/toxicity assay (i.e. using pU6iNeo or pcDNA6.1, figure 2c). After 18 h, cells were transfected with 1 µg of pJM101/L1.3 or 0.5 µg of pU6iNeo and 40 nM of an antimiR or its negative control (C-), using 4 µl of Dharmafect DUO (Dharmacon) per well and following the manufacturer's instructions. Eight hours post-transfection, cells were fed with fresh medium, and total RNA was extracted 48 h post-transfection using TRIzol (Thermo Fisher Scientific) and following standard protocols. For the clonability and retrotransposition reporter assays, selection with 400 µg ml−1 G418 (Invitrogen) was started 48 h post-transfection, and cells were fed every other day. After 12 days, cells were washed in 1× PBS (Gibco), fixed (using 2 ml well−1 of 2% (v/v) formaldehyde, 0.2% (v/v) glutaraldehyde prepared in 1× PBS), washed with ddH2O, and stained with 0.5% (w/v, prepared in ddH2O) crystal violet. Plates were dried and colonies were counted manually. Toxicity/clonability data was used to normalize changes in L1 retrotransposition, as described [35].
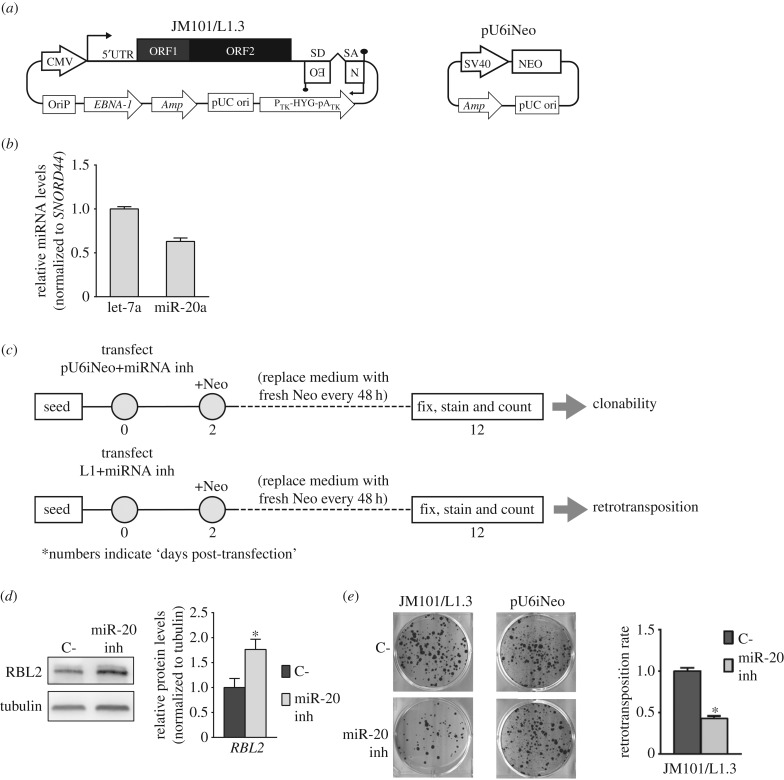
Figure 2.
The miRNA/L1-retrotransposition reporter assay. (a) Scheme of the two plasmids used, following the nomenclature described in figure 1. CMV and SV40, cytomegalovirus and simian virus 40 promoters; OriP and pUC ori denote mammalian and prokaryotic origins of replication; Amp, denotes the gene conferring resistance to ampicillin in bacteria; PTK-HYG-pATK denotes the hygromycin-resistance marker present in pCEP4, driven by the herpes simplex virus thymidine kinase (TK) promoter, and which also uses the polyadenylation signal from this viral gene. (b) The graph indicates the expression level of two miRNAs, let-7 and miR-20a, in HeLa cells. The standard deviation (s.d.) of the assay (triplicate) is indicated. (c) Scheme of the optimized protocol developed to study the role of miRNAs in L1 retrotransposition. Details are included in the panel, and numbers indicate ‘days post-transfection’. Briefly, HeLa cells are co-transfected with antimiR-20 and pU6iNeo (to control for toxicity/changes in clonability), or with antimiR-20 and JM101/L1.3 (to measure changes in retrotransposition), and G418-resistant foci selected during 12 days. In parallel, HeLa cells were also co-transfected with a non-targeting control antimiR and pU6iNeo or JM101/L1.3 (see Methods). (d) AntimiR-20 transfection in HeLa cells can increase the expression of validated miR-20 targets [59], as revealed by Western blot analyses of RBL2 protein expression. In the Western blot, tubulin was used as a loading control. The graph shown on the right depicts the quantification of the Western blot results shown on the left side. (e) Representative retrotransposition (left) and toxicity/clonability (right) results in HeLa cells transfected with antimiR-20a (miR-20 inh) or an internal negative control (C-). The graph on the right side plots the retrotransposition rate upon miR-20a inhibition, relative to C- transfected cells. Toxicity/clonability values were used to normalize retrotransposition data. The s.d. of the assay is indicated. Unpaired two-sided t-test, *p < 0.05.
(f). siRNA-retrotransposition assays
We used a protocol adapted from [60], where we first transfected siRNAs and then co-transfected siRNAs with L1 reporter plasmids. Besides functional validation of siRNAs, it is important to note that some ‘non-targeting control’ siRNAs (NTCs) are known to interfere with L1 retrotransposition, and their use should be avoided [61]. Here, we confirmed that the NTC-siRNA used had no significant effect on L1 retrotransposition (electronic supplementary material, figure S2) For the first siRNA depletion, 4 × 105 cells were plated (duplicate) per well of a six-well tissue culture plate (Corning). After 18 h, cells were transfected with each siRNA (final concentration 40 nM) using DharmaFECT 4 following the manufacturer's instructions (4 µl transfection reagent per transfected well). After 20 h, culture medium was replaced with fresh medium. Forty-eight hours after the first transfection, cells in the two transfected wells were trypsinized, pooled and counted, and these are termed ‘depleted cells step1’. We then reseeded ‘depleted cells step1’ under different plating conditions (see below), and 18 h later cells were co-transfected with each siRNA (40 nM) and: (a) plasmid JJ101/L1.3 (mitomycin C (MMC) and RNA level assays); (b) 0.5 µg well−1 of plasmid pcDNA6.1, in clonability/toxicity assays; or (c) 1 µg well−1 of plasmid JJ101/L1.3 for retrotransposition assays (figure 3b). In the second transfection, we used DharmaFECT DUO, following the manufacturer's instructions and using the same ratio of transfection reagent per well as above (i.e. 4 µl well−1 of a six-well tissue culture plate).
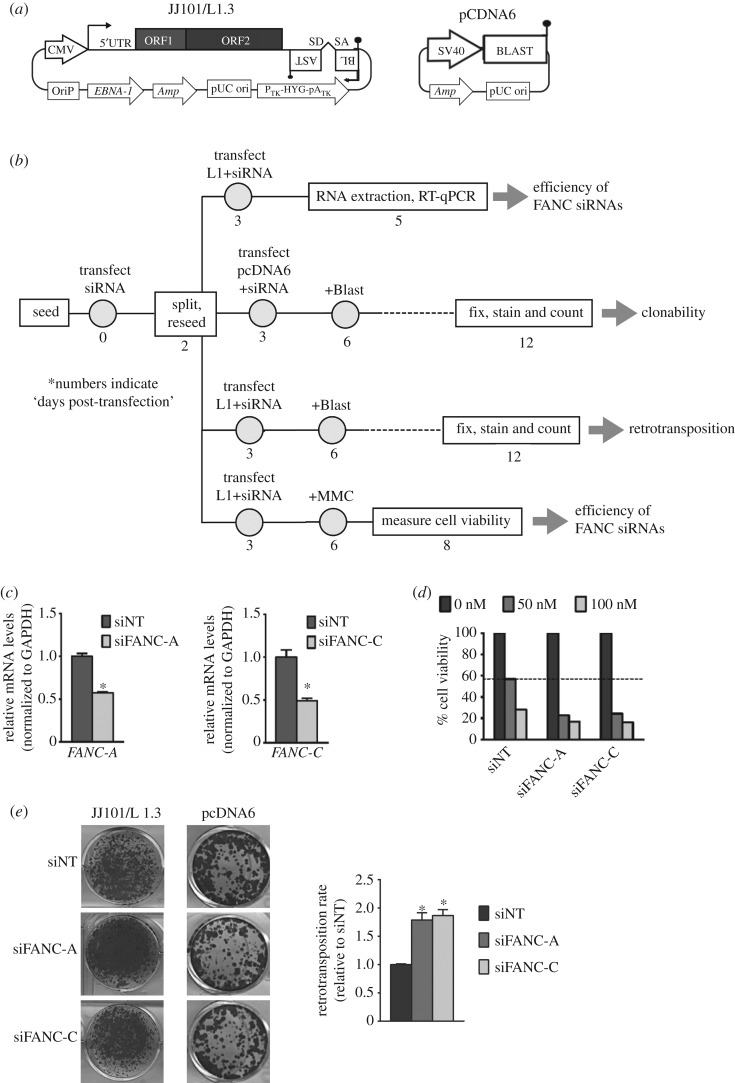
Figure 3.
The siRNA/L1-retrotransposition reporter assay. (a) Scheme of the two plasmids used, following the nomenclature described in figure 2. (b) Scheme of the optimized protocol developed to study the role of FANC siRNAs on L1 retrotransposition. Details are included in the panel, and numbers indicate ‘days post-transfection’. Briefly, U2OS cells were first transfected with FANC siRNAs (or non-targeting control siRNAs), and cells grown for 2 days. After 2 days, cells were collected, reseeded, and co-transfected again with FANC siRNAs and the control plasmid pcDNA6.1 (to control for toxicity/clonability), or with plasmid JJ101/L1.3 to test changes in retrotransposition and to conduct siRNA depletion controls (mitomycin C (MMC) and RT-qPCR experiments). To measure the efficiency of siRNAs, we took a double approach and we determined the extent of MMC sensitivity induced by the double siRNA treatment, but we also determined the reduction in each targeted FANC mRNA using RT-qPCR. When exploring changes in L1 retrotransposition, toxicity data were used to normalize data (see Methods). (c) The graphs indicate the overall efficacy of each siRNA depletion, measured as RNA levels of each targeted gene by RT-qPCR. In the graphs, the expression level of each FANC gene upon siNT transfection was arbitrarily assigned as 1. The s.d. of the assay is included, and we used an unpaired two-sided t-test, *p < 0.05. (d) Results from the MMC sensitivity assay. The graph plots the percentage of cell viability detected upon treatment of siRNA-transfected cells with 0 (black bars, assigned 100%), 50 (dark grey) and 100 nM MMC (light grey). (e) Representative retrotransposition (left) and toxicity/clonability (right) results in U2OS cells transfected with the indicated FANC siRNA (A or C) or with a NT control (siNT), used as an internal negative control. The graph on the right side plots the retrotransposition rate upon siRNA treatment (FANC-A and FANC-C, dark and light grey bars, respectively) relative to siNT. The s.d. of the assay is indicated. Unpaired two-sided t-test, *p < 0.05.
In (a), and to determine the efficiency of each siRNA, we analysed mRNA levels of each targeted FANC gene 48 h after the second transfection (efficiency of FANC siRNAs, top, figure 3b); similarly, 72 h after the second siRNA transfection, we analysed the sensitivity to mitomycin C (MMC) induced by each siRNA (efficiency of FANC siRNAs, bottom, figure 3b). For RNA level experiments, we reseeded 1 × 105 ‘depleted cells step1’ (duplicate) per well of a six-well tissue culture plate, and 18 h later cells were transfected with a second dose of each siRNA and 1 µg of plasmid JJ101/L1.3, but using DharmaFECT DUO. Next day, medium was replaced with fresh medium, and 48 h later total RNA was isolated from cells and used to analyse expression levels of each targeted FANC gene (see below). In parallel and for MMC experiments, we reseeded 5 × 103 ‘depleted cells step1’ (triplicate) per well of a 12-well tissue culture plate, and 18 h later cells were transfected with a second dose of each siRNA and 0.5 µg of plasmid JJ101/L1.3, but using DharmaFECT DUO. After 72 h, cells were treated with 0, 50 or 100 nM MMC for 48 h and sensitivity to MMC was determined using a colorimetric method (see below). Untransfected parental cells were also used in MMC assays as an internal negative control.
In (b), to analyse changes in clonability/toxicity induced by the double siRNA transfection (clonability, figure 3b), we reseeded 5 × 103 and 1 × 104 ‘depleted cells step1’ (each in triplicate) per well of a 12-well tissue culture plate; 18 h later, cells were co-transfected with a second dose of each siRNA (40 nM) and 0.5 µg of plasmid pcDNA6.1, using DharmaFECT DUO (2 µl transfection reagent per well). Next day, medium was replaced with fresh medium, and 72 h post-co-transfection we started selection with blasticidin S (blast, 5 µg ml−1, Invitrogen), which was continued for 7 days (changing the medium every other day). Next, cells were washed, fixed and stained as described above. The toxicity results were used to normalize retrotransposition rates upon siRNA transfection.
Finally, in (c), and to analyse changes in L1 retrotransposition, we reseeded 2 × 104 and 1 × 105 ‘depleted cells step1’ (each in triplicate) per well of a six-well tissue culture plate (retrotransposition, figure 3b); 18 h later, cells were co-transfected with a second dose of each siRNA (40 nM) and 1 µg of plasmid JJ101/L1.3, using DharmaFECT DUO (4 µl transfection reagent per well). Next day, medium was replaced with fresh medium, and 72 h post-co-transfection we started selection with blast (5 µg ml−1), which was continued for 7 days (changing the medium every other day). Next, cells were washed, fixed and stained as described above.
The above assays were optimized for amount of: siRNA transfected as a function of cellular toxicity and MMC sensitivity achieved; DharmaFECT reagent used to transfect siRNAs and co-transfect plasmids and siRNAs; time to initiate selection with blast after the second transfection. While optimizing the protocol, we included additional controls to demonstrate that the transfection efficiency of plasmids was constant among samples (i.e. different siRNAs), using the same DNA batch in all transfections. Finally, and when indicated, we used the extent of MMC sensitivity conferred by each siRNA to normalize retrotransposition values. We also tested the reproducibility of these protocols using three cell lines: HeLa, U2OS and HCT116, with very similar results among cell lines. For simplicity, in the main manuscript, we only show results in U2OS cells, while data on HeLa and HCT116 cells are shown in electronic supplementary material, figure S4).
(g). Quantification of siRNA target mRNA expression by RT-qPCR
TRIzol was used to isolate total RNA, following the manufacturer's instructions. We then measured mRNA transcript levels using previously described methods [62]. Briefly, after two cycles of RNase-free DNaseI (Invitrogen) treatment on 1 µg of total RNA, cDNAs were synthesized using a high-capacity cDNA Reverse Transcription Kit (Applied Biosystems). Triplicate samples were next analysed in a StepOne real-time PCR system (Applied Biosystems) using GoTaq qPCR MasterMix (Promega), and serial dilutions of cDNAs (1/5 and 1/10, prepared using UltraPure ddH2O (Invitrogen)). An internal control (no RT added) was performed in all subsequent qPCR reactions. Diluted cDNAs were analysed in 20 µl reaction volumes, using GoTaq qPCR MasterMix (Promega) and 0.15 µM of each primer (Invitrogen). The qPCR cycling conditions were described above, and to normalize expression we used GADPH. Results were analysed using the 2−ΔΔCT method [63].
(h). Western blot analyses of antimir-transfected cells
To test the effect of antimiR-20 on cultured HeLa cells, we measured the expression of a known target of miRNA-20 (RBL2 gene, [59]) by Western blot. Briefly, 2 × 105 HeLa cells were plated per well of a six-well tissue culture plate. After 18 h, cells were co-transfected with 1 µg of JM101/L1.3 and 40 nM of either the miR-20 inhibitor (i.e. antimir-20) or the internal negative control (a non-targeting antimiR) using Dharmafect DUO and the conditions described above. Forty-eight hours after transfection, cells were washed with 1× PBS, trypsinized and pelleted at 200 g for 4 min. To extract proteins, cell pellets were resuspended in 50 µl of RIPA buffer (Sigma) supplemented with 1× Complete EDTA-free Protease Inhibitor cocktail (Roche), 1 mM PMSF (Sigma), 0.25% β-mercaptoethanol (Sigma) and incubated for 10 min on ice. Extracts were then centrifuged (13000 r.p.m. at 4°C for 10 min) and debris-free supernatants were transferred to new tubes. Protein concentration was determined using the Micro BCA Protein Assay Kit (Thermo). Proteins were resolved in 4–15% Mini-PROTEAN TGX Precast Gels (BioRad), and transferred to polyvinylidene difluoride (PVDF) membrane using Trans-Blot Turbo Mini PVDF Transfer Packs (BioRad) and the Trans-Blot Turbo Transfer System (BioRad). The following antibodies were used: a monoclonal rabbit anti-RBL2 (D9T7M, 1 : 1000, Cell Signaling) and a monoclonal mouse anti-tubulin (1 : 1000, Santa Cruz). For chemiluminescence detection, we used anti-rabbit HRP (1 : 2000, Cell Signaling) or anti-mouse HRP (1 : 2000, Cell Signaling), and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo). Images were acquired with an ImageQuant LAS4000 and quantified using ImageJ software.
(i). Sensitivity to mitomycin C assays
To determine the functional efficiency of siRNAs, we analysed mitomycin C (MMC, Sigma) resistance, following the protocol described in [64]. Briefly, double-siRNA-transfected cells and untransfected controls were treated with 0, 50 and 100 nM MMC (which was made from a freshly prepared Streptomyces caespitosus MMC stock solution, Sigma) for 72 h at 37°C. Cells were then washed with 1 × PBS (Gibco) and fixed to plates by adding 0.5 ml well−1 of an aqueous solution containing 10% acetic acid (Sigma) and 10% methanol (Sigma), and incubating plates for 10 min at room temperature. After fixation, plates were washed twice with ddH2O and stained with an aqueous 0.1% (w/v) crystal violet solution for 10 min at room temperature. After staining, plates were washed twice with ddH2O and allowed to dry overnight. On the following day, the crystal violet solution was dissolved from the wells by incubating them for 2 h at room temperature using 250 µl well−1 of 1% (w/v) Sodium dodecyl sulfate (SDS, Sigma) prepared in 100% methanol (Sigma). An aliquot (100 µl) from each well was transferred to a well of a 96-well tissue culture plate and OD595 values were determined using a GloMax-Multi Detection System (Promega). For comparisons, the value obtained with untreated samples was arbitrarily assigned as 100%.
3. Results
(a). The sRNA/L1-retrotransposition reporter assay allows testing on the role of microRNAs in L1 activity
To test the effect of selected miRNAs on L1 retrotransposition, we used RC-L1s tagged with the mneoI retrotransposition indicator cassette [34,38,39], which confers resistance to neomycin(NEO)/G418 upon retrotransposition (plasmid JM101/L1.3, figure 2a, left side). Previously, the mneoI-based L1-retrotransposition reporter assay has been used to test the effect of transiently overexpressed host factors on retrotransposition, using plasmid co-transfection (reviewed in [35]). From these studies, it has become evident that changes in clonability/toxicity induced by the co-transfected host factor need to be taken into account when analysing potential changes in L1 retrotransposition (see for example [58]). Thus, and to control for potential changes in toxicity/clonability induced by miRNA inhibition, we used a plasmid that constitutively expresses the same NEO reporter gene (plasmid pU6iNeo, but lacking the antisense intron, figure 2a); to quantify potential changes in L1 retrotransposition, we considered data from the toxicity/clonability assays. We tested different transfection reagents specifically designed to transfect small RNAs (sRNAs) to cultured cells, as this would allow us to test the role of miRNAs and siRNAs. Among the different reagents used, we reproducibly observed that Dharmafect DUO from Dharmacon generated constant transfection efficiency rates among experiments and consistent results (see Methods). These experiments resulted in an optimized retrotransposition protocol that could be used to potentially analyse the role of any miRNA on L1 mobilization/regulation (see protocol schematic figure 2c).
Notably, using the rna22 prediction software [65], we identified two putative miR-20 binding sites in the consensus sequence of human RC-L1s [2], located in the 5′UTR and 3′UTR (electronic supplementary material, figure S1). miR-20 has been associated with several biological processes, including cancer [66], and we selected this miRNA for further experimentation. RT-qPCR analyses revealed that in HeLa cells, the scale of miR-20 expression is similar to that of other miRNAs previously shown to be highly expressed, such as let-7 (figure 2b) [67]. Thus, we decided to inhibit miR-20 and test any effect on human L1 retrotransposition, using HeLa cells where human L1s are known to retrotranspose at high frequencies [34,38]. To inhibit the action of miRNAs, we used antimiR oligonucleotides, as this loss-of-function approach has been previously shown to work robustly on cultured cells (reviewed in [68]). In these experiments, we used a non-targeting antimiR as an internal negative control (see Methods).
To analyse the effect of antimiR-20 on retrotransposition, we used the protocol shown in the schematic in figure 2c, where we also controlled for potential changes in clonability/toxicity. Briefly, cells transfected with pU6iNeo (figure 2a, right side) could express the NEO gene after transfection, generating G418-resistant foci. However, cells transfected with plasmid JM101/L1.3 (figure 2a, left side) could only activate the NEO gene after a round of retrotransposition, also generating G418-resistant foci. Thus, we used the number of G418-resistant foci generated with the control plasmid pU6iNeo in cells co-transfected with antimiR-20 to normalize changes in L1 retrotransposition. As an internal negative control, cells were also co-transfected with a non-targeting antimiR (see Methods). Notably, Western blot analyses revealed a significant increase in RBL2 protein expression in HeLa cells transfected with antimiR-20a, which is consistent with validated targets for this human miRNA (figure 2d) [59]. However, control assays revealed that antimiR-20 did not elicit toxicity to HeLa cells, at least under our experimental conditions (figure 2e, compare number of G418 resistant foci generated upon pU6iNEO co-transfection with antimiR-20a (labelled ‘miR-20 inh’) or with a non-targeting antimiR negative control (labelled ‘C-’)). Surprisingly, we found that inhibition of miR-20 decreased human L1 retrotransposition by half when compared with the control (figure 2e). We confirmed these results using a second retrotransposition indicator marker (mblastI, data not shown). While further research is required to clarify the molecular mechanisms involved in downregulation of L1 retrotransposition, these data suggest that, contrary to expectations, miR-20 does not act as a negative regulator of L1 retrotransposition (see Discussion).
(b). Using siRNAs and the sRNA/L1-retrotransposition reporter assay
The discovery of interference RNA [45] allowed the development of small interfering RNAs (siRNAs) as tools to transiently knock-down expression of genes of interest. Thus, we next optimized the L1 retrotransposition reporter protocol using siRNAs. Notably, a previous study demonstrated that widely used non-targeting control siRNAs (NTCs) could interfere with L1 retrotransposition [61]. The mechanism of the interference is unknown, and the use of these NTC siRNA controls should be avoided, at least in combination with the L1-retrotransposition reporter assay (see Methods for further details). Here, we used SMART-pool siRNAs, and we specifically tested that the SMART-pool NTC used did not interfere with engineered L1 retrotransposition. To do that, we compared the rate of L1 retrotransposition (using plasmid JJ101/L1.3, figure 3a) on siRNA-NTC and mock-transfected HeLa cells, and we detected very similar levels of L1 retrotransposition (electronic supplementary material, figure S2). As an additional control, we also confirmed that SMART-pool siRNAs to XRCC4 had no effect on L1 retrotransposition (electronic supplementary material, figure S2), consistent with a previous report on mammalian cells [33,42,69]. Although siRNAs are powerful tools, protein depletion induced by siRNAs is transient, and this could confound the interpretation of results using mneoI-tagged RC-L1s, especially considering that G418 selection in cultured cells requires more than 7 days. As blasticidin S has a fast and potent mode of action that induces rapid cell death [70], we reasoned that we could exploit this peculiarity to measure retrotransposition while siRNAs were successfully depleting mRNAs (i.e. score retrotransposition in the first 72 h after siRNA transfection). Thus, we used RC-L1s tagged with mblastI, a retrotransposition indicator cassette that activates the blasticidin S-resistance gene upon retrotransposition (figure 3a, [41,42]). To control for changes in toxicity/clonability, we used plasmid pcDNA6.1, which constitutively expresses the same blast marker after transfection (figure 3a). In the conventional assay using mblastI-tagged L1s, cells are transfected with L1 reporter constructs and cultured for 5 days prior to initiation of blasticidin S selection [41,42] (see scheme in electronic supplementary material, figure S3). However, here we analysed the kinetics of mblastI-tagged L1 retrotransposition in HeLa cells, starting blast selection at 1, 2, 3, 4 or 5 days after transfection (electronic supplementary material, figure S3). Remarkably, we consistently observed that the majority of retrotransposition events accumulated during the first 72 h after transfection, and that the numbers of blast-resistant foci were similar if selection was started 2, 3, 4 or 5 days after transfection (electronic supplementary material, figure S3). Similar data were obtained using U2OS and HCT116 cells. In summary, we started blast selection 3 days after transfection when using siRNAs, (see scheme in figure 3b), ensuring that scored retrotransposition events accumulated while siRNAs were effectively depleting candidate genes.
As candidate factors, we decided to test the effect of siRNAs on Fanconi anaemia proteins, as a previous CRISPR/Cas9-based genome-wide study identified several of these genes as L1 regulators [33]. Briefly, Fanconi anaemia (FANC) is a rare recessive genetic disorder characterized by genomic instability resulting from the accumulation of unrepaired interstrand crosslinks (ICLs) (reviewed in [71–73]). More than 20 FANC genes have been discovered in humans [74], and the FANC pathway is essential for the DNA damage response; as a result, patients' cells exhibit characteristic sensitivity to DNA crosslinking agents such as mitomycin C (MMC), and are prone to chromosome breakage [71–73]. To validate the siRNA/L1-retrotransposition reporter assay, we selected two commonly mutated FANC genes, FANC-A and FANC-C, mutated in approximately 65% and approximately 14% of all known FANC cases [75].
In the siRNA/L1-retrotransposition reporter assay, we used ON-TARGET plus SMART-pool siRNAs to avoid off-target effects, and to increase the effectiveness of the siRNA-mediated depletion, we used a double siRNA transfection protocol as previously described [60] (figure 3b). Additionally, we used a different cell line, U2OS osteosarcoma cells, where human RC-L1s are also known to retrotranspose at high frequencies [17]. In the assay, U2OS cells were transfected with each FANC siRNA; 48 h later, siRNA-depleted cells were pooled and reseeded, and this population of FANC-depleted cells was used to analyse: (i) the overall efficiency of the siRNA depletion (using MMC sensitivity assays and RT-qPCR); (ii) changes in toxicity/clonability induced by siRNAs, using co-transfection with the control plasmid pcDNA6.1 (figure 3a, right side); and (iii) changes in L1 retrotransposition induced by the FANC siRNAs (see schematic in figure 3b), using the JJ101/L1.3 vector (figure 3a, left side). As we used two rounds of siRNA-mediated depletion, in the second co-transfection we co-transfected either the mblastI-tagged RC-L1 vector (JJ101/L1.3) or the toxicity/clonability control plasmid pcDNA6.1, again with each siRNA (figure 3a,b). To measure the efficiency of the double siRNA-mediated depletion, we extracted total RNA 48 h after the second transfection, and RT-qPCR analyses revealed that both siRNAs reduced expression of each FANC gene more than 50% (figure 3c). To analyse whether this reduction in FANC mRNAs could induce a FANC phenotype in U2OS cells, we determined the sensitivity to MMC induced by each siRNA. Notably, we determined that the double siRNA transfection increased the sensitivity to MMC of U2OS cells when compared with siNT-transfected cells and untransfected controls (figure 3d).
Next, we analysed whether the double siRNA-mediated depletion had any effect on L1 retrotransposition rates, using the protocol optimized above where blast selection is started 72 h after the second transfection (day 6 in scheme shown in figure 3b). In parallel, we also tested if siRNAs induced toxicity/changes in clonability, using plasmid pcDNA6.1, and we used the number of blast-resistant foci generated upon transfection with the toxicity/clonability control plasmid (pcDNA6.1) to correct changes in retrotransposition (figure 3b). Notably, we reproducibly detected that both FANC siRNAs led to elevated levels of L1 retrotransposition, without significant changes in toxicity/clonability (average increase twofold, grey bars, figure 3e). Using this optimized protocol, we also analysed the effect of FANC-A and FANC-C siRNAs on HeLa and HCT116 cells, with similar results (electronic supplementary material, figures S2 and S4). Notably, our results are in agreement with those from Liu and colleagues [33], who used a completely different approach to knock-out FANC genes using CRISPR/Cas9 and a different cell line (K562 cells). In summary, these data further suggest that FANC members act as regulators of human RC-L1 retrotransposition in cultured cells.
4. Discussion
The development of the engineered L1 retrotransposition reporter assay more than 20 years ago [34,38] significantly improved our capability to study how active retrotransposons impact genomes and how they are regulated. Indeed, this assay is still the gold standard in the field, because of its simplicity, robustness and sensitivity, and the list of processes uncovered using the retrotransposition reporter assay is long and continues to grow (recently reviewed in [35,36]). To illustrate its sensitivity, it is worth noting that the retrotransposition reporter assay was successfully used to uncover the mechanism of L1-ORF2p translation [14], which is translated at an incredibly low level, perhaps as little as one molecule of L1-ORF2p per L1 transcript. Here, we developed minor modifications of this robust assay to analyse the role of small RNAs on L1 biology, and to exploit siRNAs to further understand how host factors interact with L1-RNPs and can regulate their activity.
Work from several labs including ours has demonstrated that small RNAs (sRNAs) are major components of the L1 ‘regulome’ [33,54,60]. Among sRNAs, miR-128 was recently shown to repress L1 retrotransposition by binding to its mRNA [53]. Another class of sRNAs often associated with L1 retrotransposons are Piwi interacting RNAs (piRNAs) and endosiRNAs, which seem to have a critical role to control retrotransposons in the germline [76–79]. Besides sRNAs, many factors associated with RNA metabolism, such as Dicer, the Microprocessor, MOV10, small nuclear RNAs and TUTases, are also known members of the L1 ‘regulome’ [28,29,33,60,80–82]. Thus, developing protocols that allow testing of the role of small RNAs on L1 retrotransposition would ultimately increase our knowledge on the L1 ‘regulome’ at a mechanistic level. In this study, we identified two potential binding sites for miR-20 in the consensus sequence of human RC-L1s, and as this miRNA is expressed in HeLa cells (figure 2b), we used the optimized protocol to test whether miR-20 inhibition would impact retrotransposition. Unexpectedly, we reproducibly observed that antimiR-20 reduced L1 retrotransposition rates in cultured cells. Thus, we speculate that if miR-20 binds the 3′UTR of RC-L1s, it does not act as a canonical miRNA, which would reduce transcript levels and/or translation of L1-encoded proteins. While this might seem paradoxical, it is possible that miR-20 might regulate expression of L1 repressors [33], explaining why miR-20 inhibition would result in increased levels of L1 repressors and thus reduced L1 retrotransposition, acting through an in trans mechanism. Interestingly, a previous study validated a small number of human miRNA-20 targets (n = 7, see electronic supplementary material, table S2); furthermore, we found a second group of human genes (n = 15, electronic supplementary material, table S2) potentially regulated by miRNA-20, which were identified by three independent softwares (microrna.org, targetScan and miRdb.org). Intriguingly, 21 of 22 genes potentially regulated by miR-20 were found to behave as L1 repressors in a recent LINE-1 retrotransposition genome-wide CRISPR/Cas9 screening [33] (electronic supplementary material, table S2). Alternatively, miR-20 binding to the 5′UTR of RC-L1s in cis could positively regulate L1 retrotransposition, using a mechanism discovered with other sRNAs that when binding 5′UTRs facilitate gene expression/translation rather than inhibiting targeted RNAs [83–85]. Either way, these data revealed that some miRNAs could behave as L1 activators within the ‘L1 regulome’, and we currently continue to research the unexpected molecular mechanism of regulation elicited by miR-20 on L1 retrotransposition.
To unveil the numerous molecular mechanisms involved in the regulation of active retrotransposons, several studies have successfully exploited ‘gain of function’ approaches, where host factors are overexpressed in the retrotransposition reporter assay, increasing our knowledge of the L1 ‘regulome’ [35,36]. Similarly, ‘loss of function’ approaches for L1 regulators will help to ultimately define molecular mechanisms operating in the context of the L1 ‘regulome’. The development of CRISPR/Cas9 genome editing tools represents a revolution in genomics, and these resources will be further used to better understand the L1 ‘regulome’ in the future, either as ‘loss of function’ tools [33], or to remove transposable elements from selected genomic loci [86]. While CRISPR/Cas9 systems are simple and very efficient at generating knock-out models, this strategy is likely to fail with essential genes. However, as siRNAs are not completely effective in removing targeted RNAs, generating hypomorph phenotypes, siRNAs could be an ideal alternative to CRISPR/Cas9 for essential genes. Consistently, another inherent advantage of siRNA-based experiments is that experiments are done in pools of cells, not in many isolated clones, as would occur in genome-wide CRISPR/Cas9-based screenings. Thus, following this rationale we speculate that the development of reproducible siRNA/L1-retrotransposition reporter protocols will be useful to study the role of essential genes that belong to the L1 ‘regulome’. As a proof of principle, we used here the optimized siRNA/L1-retrotransposition reporter protocol to demonstrate that FANC proteins can indeed regulate L1 retrotransposition, which is consistent with two previous reports [33,87], although we know very little about the molecular mechanism of this regulation. Considering the large variety of siRNAs commercially available, the protocol developed here could be used in many different scenarios, with the ultimate goal of increasing our knowledge on L1 biology and the different molecular mechanisms participating in the regulation of L1 activity.
Supplementary Material
Acknowledgements
We acknowledge members of our laboratories for helpful discussions. We also acknowledge Dr John V. Moran (University of Michigan, USA) for critical input and advice.
Data accessibility
This article has no additional data.
Authors' contributions
P.T.-R., S.M., J.L.G.-P. and S.R.H. conceived, designed and interpreted the experiments. P.T.-R., S.M., B.T. and L.S. performed the experiments and data analyses. J.L.G.-P. and S.R.H. supervised the project. P.T-R. and J.L.G.-P. wrote the manuscript, with comments provided by all authors.
Competing interests
None of the authors declares any conflict of interest.
Funding
J.L.G.-P. acknowledges funding from the European Research Council (ERC-Consolidator ERC-STG-2012-309433), MINECO-FEDER (SAF2017-89745-R), the Howard Hughes Medical Institute (IECS-55007420), the Wellcome Trust-University of Edinburgh Institutional Strategic Support Fund (ISFF2) and a private donation to his laboratory from Ms Francisca Serrano (Trading y Bolsa para Torpes, Granada, Spain). S.M. was supported by the Government of Andalucia (FEDER-P08-CTS-03678). S.R.H. and P.T.-R. are funded by the Ramon y Cajal programme (RYC-2016-21395) and by MINECO-FEDER (SAF2015-71589-P). This paper is part of the thesis project of P.T.-R., within the Biomedicine Program of the University of Granada, Spain.
References
- 1.Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. ( 10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 2.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA 100, 5280–5285. ( 10.1073/pnas.0831042100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. 2010. LINE-1 retrotransposition activity in human genomes. Cell 141, 1159–1170. ( 10.1016/j.cell.2010.05.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flasch DA, Macia A, Sanchez L, Ljungman M, Heras SR, Garcia-Perez JL, Wilson TE, Moran JV. 2019. Genome-wide de novo L1 retrotransposition connects endonuclease activity with replication. Cell 177, 837–851. ( 10.1016/j.cell.2019.02.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sultana T, et al. 2019. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol. Cell 74, 555–570. ( 10.1016/j.molcel.2019.02.036) [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Perez JL, Widmann TJ, Adams IR. 2016. The impact of transposable elements on mammalian development. Development 143, 4101–4114. ( 10.1242/dev.132639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. 2015. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol. Spectrum 3, MDNA3–0061–2014 ( 10.1128/microbiolspec.MDNA3-0061-2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumann GG, Fuchs NV, Tristan-Ramos P, Sebe A, Ivics Z, Heras SR. 2019. The impact of transposable element activity on therapeutically relevant human stem cells. Mob. DNA 10, 9 ( 10.1186/s13100-019-0151-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancks DC, Kazazian HH Jr. 2016. Roles for retrotransposon insertions in human disease. Mob. DNA 7, 9 ( 10.1186/s13100-016-0065-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speek M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21, 1973–1985. ( 10.1128/MCB.21.6.1973-1985.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swergold GD. 1990. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 10, 6718–6729. ( 10.1128/MCB.10.12.6718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH Jr. 1997. Many human L1 elements are capable of retrotransposition. Nat. Genet. 16, 37–43. ( 10.1038/ng0597-37) [DOI] [PubMed] [Google Scholar]
- 13.Goodier JL. 2016. Restricting retrotransposons: a review. Mob. DNA 7, 16 ( 10.1186/s13100-016-0070-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. 2006. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 20, 210–224. ( 10.1101/gad.1380406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, Merrick WC, Shatsky IN. 2007. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol. Cell. Biol. 27, 4685–4697. ( 10.1128/MCB.02138-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21, 1429–1439. ( 10.1128/MCB.21.4.1429-1439.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doucet AJ, et al. 2010. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 6, e1001150 ( 10.1371/journal.pgen.1001150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulpa DA, Moran JV. 2005. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 14, 3237–3248. ( 10.1093/hmg/ddi354) [DOI] [PubMed] [Google Scholar]
- 19.Kubo S, Seleme Md C, Soifer HS, Perez JL, Moran JV, Kazazian HH Jr, Kasahara N. 2006. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl Acad. Sci. USA 103, 8036–8041. ( 10.1073/pnas.0601954103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macia A. et al. 2017. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res. 27, 335–348. ( 10.1101/gr.206805.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mita P. et al. 2018. LINE-1 protein localization and functional dynamics during the cell cycle. eLife 7, e30058 ( 10.7554/eLife.30058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72, 595–605. ( 10.1016/0092-8674(93)90078-5) [DOI] [PubMed] [Google Scholar]
- 23.Cost GJ, Feng Q, Jacquier A, Boeke JD. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21, 5899–5910. ( 10.1093/emboj/cdf592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48. ( 10.1038/ng1223) [DOI] [PubMed] [Google Scholar]
- 25.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH Jr. 2011. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 20, 3386–3400. ( 10.1093/hmg/ddr245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raiz J, et al. 2012. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40, 1666–1683. ( 10.1093/nar/gkr863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esnault C, Maestre J, Heidmann T. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367. ( 10.1038/74184) [DOI] [PubMed] [Google Scholar]
- 28.Heras SR, Macias S, Caceres JF, Garcia-Perez JL. 2014. Control of mammalian retrotransposons by cellular RNA processing activities. Mob. Genet. Elem. 4, e28439 ( 10.4161/mge.28439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pizarro JG, Cristofari G. 2016. Post-transcriptional control of LINE-1 retrotransposition by cellular host factors in somatic cells. Front. Cell Dev. Biol. 4, 14 ( 10.3389/fcell.2016.00014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez-Luque FJ. et al. 2019. LINE-1 evasion of epigenetic repression in humans. Mol. Cell 75, 590–604. ( 10.1016/j.molcel.2019.05.024 [DOI] [PubMed] [Google Scholar]
- 31.Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340. ( 10.1016/S0168-9525(97)01181-5) [DOI] [PubMed] [Google Scholar]
- 32.Bourc'his D, Bestor TH. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3 L. Nature 431, 96–99. ( 10.1038/nature02886) [DOI] [PubMed] [Google Scholar]
- 33.Liu N, Lee CH, Swigut T, Grow E, Gu B, Bassik MC, Wysocka J. 2018. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553, 228–232. ( 10.1038/nature25179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927. ( 10.1016/S0092-8674(00)81998-4) [DOI] [PubMed] [Google Scholar]
- 35.Kopera HC, Larson PA, Moldovan JB, Richardson SR, Liu Y, Moran JV. 2016. LINE-1 cultured cell retrotransposition assay. Methods Mol. Biol. 1400, 139–156. ( 10.1007/978-1-4939-3372-3_10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rangwala SH, Kazazian HH. 2009. The L1 retrotransposition assay: a retrospective and toolkit. Methods 49, 219–226. ( 10.1016/j.ymeth.2009.04.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. 1985. Ty elements transpose through an RNA intermediate. Cell 40, 491–500. ( 10.1016/0092-8674(85)90197-7) [DOI] [PubMed] [Google Scholar]
- 38.Feng Q, Moran JV, Kazazian HH Jr, Boeke JD. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916. ( 10.1016/S0092-8674(00)81997-2) [DOI] [PubMed] [Google Scholar]
- 39.Freeman JD, Goodchild NL, Mager DL. 1994. A modified indicator gene for selection of retrotransposition events in mammalian cells. Biotechniques 17, 48–49. [PubMed] [Google Scholar]
- 40.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH Jr. 2000. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28, 1418–1423. ( 10.1093/nar/28.6.1418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodier JL, Zhang L, Vetter MR, Kazazian HH Jr. 2007. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27, 6469–6483. ( 10.1128/MCB.00332-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31, 159–165. ( 10.1038/ng898) [DOI] [PubMed] [Google Scholar]
- 43.Xie Y, Rosser JM, Thompson TL, Boeke JD, An W. 2011. Characterization of L1 retrotransposition with high-throughput dual-luciferase assays. Nucleic Acids Res. 39, e16 ( 10.1093/nar/gkq1076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esnault C, Casella JF, Heidmann T. 2002. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 30, e49 ( 10.1093/nar/30.11.e49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. ( 10.1038/35888) [DOI] [PubMed] [Google Scholar]
- 46.Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lini-14. Cell 75, 843–854. ( 10.1016/0092-8674(93)90529-y) [DOI] [PubMed] [Google Scholar]
- 47.Bartel DP. 2018. Metazoan microRNAs. Cell 173, 20–51. ( 10.1016/j.cell.2018.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139. ( 10.1038/nrm2632) [DOI] [PubMed] [Google Scholar]
- 49.Fabian MR, Sonenberg N, Filipowicz W. 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. ( 10.1146/annurev-biochem-060308-103103) [DOI] [PubMed] [Google Scholar]
- 50.Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655. ( 10.1016/j.cell.2009.01.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lujambio A, Lowe SW. 2012. The microcosmos of cancer. Nature 482, 347–355. ( 10.1038/nature10888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soifer HS, Rossi JJ, Saetrom P. 2007. MicroRNAs in disease and potential therapeutic applications. Mol. Ther. 15, 2070–2079. ( 10.1038/sj.mt.6300311) [DOI] [PubMed] [Google Scholar]
- 53.Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ, Pedersen IM. 2015. miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat. Struct. Mol. Biol. 22, 824–831. ( 10.1038/nsmb.3090) [DOI] [PubMed] [Google Scholar]
- 54.Yang N, Kazazian HH Jr. 2006. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13, 763–771. ( 10.1038/nsmb1141) [DOI] [PubMed] [Google Scholar]
- 55.Dorsett Y, Tuschl T. 2004. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 3, 318–329. ( 10.1038/nrd1345) [DOI] [PubMed] [Google Scholar]
- 56.Wittrup A, Lieberman J. 2015. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 16, 543–552. ( 10.1038/nrg3978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kopera HC, Moldovan JB, Morrish TA, Garcia-Perez JL, Moran JV. 2011. Similarities between LINE-1 reverse transcriptase and telomerase. Proc. Natl Acad. Sci. USA 108, 20 345–20 350. ( 10.1073/pnas.1100275108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. 2014. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. eLife 3, e02008 ( 10.7554/eLife.02008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trompeter HI, Abbad H, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, Schira J, Muller HW, Wernet P. 2011. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS ONE 6, e16138 ( 10.1371/journal.pone.0016138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Caceres JF. 2013. The Microprocessor controls the activity of mammalian retrotransposons. Nat. Struct. Mol. Biol. 20, 1173–1181. ( 10.1038/nsmb.2658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cook PR, Tabor GT. 2016. Deciphering fact from artifact when using reporter assays to investigate the roles of host factors on L1 retrotransposition. Mob. DNA 7, 23 ( 10.1186/s13100-016-0079-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benitez-Guijarro M, et al. 2018. RNase H2, mutated in Aicardi-Goutières syndrome, promotes LINE-1 retrotransposition. EMBO J. 37, e98506 ( 10.15252/embj.201798506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 64.Howlett NG, Harney JA, Rego MA, Kolling FWT, Glover TW. 2009. Functional interaction between the Fanconi anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. J. Biol. Chem. 284, 28 935–28 942. ( 10.1074/jbc.M109.016352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. 2006. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217. ( 10.1016/j.cell.2006.07.031) [DOI] [PubMed] [Google Scholar]
- 66.Zhu GF. et al. 2019. Mir20a/106a-WTX axis regulates RhoGDIa/CDC42 signaling and colon cancer progression. Nat. Commun. 10, 112 ( 10.1038/s41467-018-07998-x) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. 2009. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 27, 549–555. ( 10.1038/nbt.1543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. 2012. Inhibition of microRNA function by antimiR oligonucleotides. Silence 3, 1 ( 10.1186/1758-907X-3-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. 2007. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 446, 208–212. ( 10.1038/nature05560) [DOI] [PubMed] [Google Scholar]
- 70.Izumi M, Miyazawa H, Kamakura T, Yamaguchi I, Endo T, Hanaoka F. 1991. Blasticidin S-resistance gene (bsr): a novel selectable marker for mammalian cells. Exp. Cell Res. 197, 229–233. ( 10.1016/0014-4827(91)90427-V) [DOI] [PubMed] [Google Scholar]
- 71.Ceccaldi R, Sarangi P, D'Andrea AD. 2016. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 17, 337–349. ( 10.1038/nrm.2016.48) [DOI] [PubMed] [Google Scholar]
- 72.Cheung RS, Taniguchi T. 2017. Recent insights into the molecular basis of Fanconi anemia: genes, modifiers, and drivers. Int. J. Hematol. 106, 335–344. ( 10.1007/s12185-017-2283-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moldovan GL, D'Andrea AD. 2009. How the Fanconi anemia pathway guards the genome. Annu. Rev. Genet. 43, 223–249. ( 10.1146/annurev-genet-102108-134222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nebert DW, Dong H, Bruford EA, Thompson DC, Joenje H, Vasiliou V. 2016. Letter to the editor for “Update of the human and mouse Fanconi anemia genes”. Hum. Genomics 10, 25 ( 10.1186/s40246-016-0081-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morgan NV, Tipping AJ, Joenje H, Mathew CG. 1999. High frequency of large intragenic deletions in the Fanconi anemia group A gene. Am. J. Hum. Genet. 65, 1330–1341. ( 10.1086/302627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aravin AA, Hannon GJ, Brennecke J. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764. ( 10.1126/science.1146484) [DOI] [PubMed] [Google Scholar]
- 77.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799. ( 10.1016/j.molcel.2008.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marchetto MC, et al. 2013. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503, 525–529. ( 10.1038/nature12686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20, 1732–1743. ( 10.1101/gad.1425706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goodier JL, Cheung LE, Kazazian HH Jr. 2012. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 8, e1002941 ( 10.1371/journal.pgen.1002941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Warkocki Z, Krawczyk PS, Adamska D, Bijata K, Garcia-Perez JL, Dziembowski A. 2018. Uridylation by TUT4/7 restricts retrotransposition of human LINE-1s. Cell 174, 1537–1548. ( 10.1016/j.cell.2018.07.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. 2007. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 17, 602–611. ( 10.1101/gr.5870107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vasudevan S, Tong Y, Steitz JA. 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934. ( 10.1126/science.1149460) [DOI] [PubMed] [Google Scholar]
- 84.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27, 3300–3310. ( 10.1038/emboj.2008.244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. 2006. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl Acad. Sci. USA 103, 17 337–17 342. ( 10.1073/pnas.0607015103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087. ( 10.1126/science.aad5497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bregnard C, Guerra J, Dejardin S, Passalacqua F, Benkirane M, Laguette N. 2016. Upregulated LINE-1 activity in the Fanconi anemia cancer susceptibility syndrome leads to spontaneous pro-inflammatory cytokine production. EBioMedicine 8, 184–194. ( 10.1016/j.ebiom.2016.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.