Abstract
The plant-specific RNA Polymerase IV (Pol IV) transcribes heterochromatic regions, including many transposable elements (TEs), with the well-described role of generating 24 nucleotide (nt) small interfering RNAs (siRNAs). These siRNAs target DNA methylation back to TEs to reinforce the boundary between heterochromatin and euchromatin. In the male gametophytic phase of the plant life cycle, pollen, Pol IV switches to generating primarily 21–22 nt siRNAs, but the biogenesis and function of these siRNAs have been enigmatic. In contrast to being pollen-specific, we identified that Pol IV generates these 21–22 nt siRNAs in sporophytic tissues, likely from the same transcripts that are processed into the more abundant 24 nt siRNAs. The 21–22 nt forms are specifically generated by the combined activities of DICER proteins DCL2/DCL4 and can participate in RNA-directed DNA methylation. These 21–22 nt siRNAs are also loaded into ARGONAUTE1 (AGO1), which is known to function in post-transcriptional gene regulation. Like other plant siRNAs and microRNAs incorporated into AGO1, we find a signature of genic mRNA cleavage at the predicted target site of these siRNAs, suggesting that Pol IV-generated 21–22 nt siRNAs may function to regulate gene transcript abundance. Our data provide support for the existing model that in pollen Pol IV functions in gene regulation.
This article is part of a discussion meeting issue ‘Crossroads between transposons and gene regulation’.
Keywords: pollen, small interfering RNA, siRNA, RNA polymerase IV, pol IV, transposable element
1. Background
Transposable elements (TEs) are mobile DNA fragments that cause mutations by inserting into genes and creating chromosomal breaks. To repress their mobility, and therefore limit the number of new mutations, eukaryotes target TE activity at the transcriptional, post-transcriptional and translational levels (reviewed in [1]). A major regulatory mechanism used to repress TEs is small RNAs, which target TE mRNAs for degradation, inhibit the translation of TE protein and can guide de novo chromatin modification of TE loci, resulting in transcriptional silencing. In flowering plants, TE small interfering RNAs (siRNAs) are well studied and fall into two major categories: 21–22 nucleotide (nt) siRNAs generated by RNA Polymerase II (Pol II) and 24 nt siRNAs generated by the plant-specific RNA Polymerase IV (Pol IV) (reviewed in [2]).
Early in plant evolution, the protein subunits of the Pol II holoenzyme duplicated and subfunctionalized into two additional RNA polymerase complexes, Pol IV and Pol V [3]. The biological function of Pol IV is to transcribe heterochromatic regions of plant genomes into non-polyadenylated transcripts that are created for the sole purpose of siRNA generation [4]. Pol IV is guided to heterochromatic target regions of the genome by the mark of histone H3 lysine 9 dimethylation (H3K9me2) [5] and creates short 26–45 nt transcripts that are converted into double-stranded RNA via the RNA-DEPENDENT RNA POLYMERASE 2 protein (RDR2) [6,7]. This double-stranded RNA is then cleaved by DICER-LIKE 3 (DCL3) into predominantly 23–24 nt siRNAs [8]. Pol IV-derived 24 nt siRNAs are incorporated into the ARGONAUTE4 (AGO4) and AGO6 proteins, to guide AGO function in RNA-directed DNA methylation (RdDM) of the target locus [9]. Therefore, Pol IV's overall function in plant biology is to generate the siRNAs necessary to reinforce heterochromatic marks and maintain euchromatin/heterochromatin boundaries [10]. A secondary role is to generate an siRNA defense against any new or active TEs that share sequence homology [11].
Pollen is the male gametophytic generation of flowering plants and contains two sperm cell gametes encapsulated in a larger vegetative cell, which directs the delivery of the sperm cells upon pollination. There is a known broad activation of TE expression in the nucleus of the pollen vegetative cell, resulting in steady-state TE mRNAs in pollen [12,13]. This TE activation occurs simultaneously with abundant TE 21–22 nt siRNA production in the pollen grain [12,14,15]. Other cases of TE transcriptional activation in the sporophytic plant body, for example, in mutants of the TE master chromatin-modifying gene DDM1, are also associated with 21–22 nt siRNA production from Pol II-derived TE mRNAs [16]. These siRNAs were termed epigenetically activated siRNAs (easiRNAs) because they appear only when TEs lose transcriptional repression and produce Pol II-derived mRNAs [15,17]. It was therefore assumed that in pollen, the reactivated TE-generated Pol II mRNAs were the source of pollen easiRNAs [12]. However, a recent publication demonstrated that pol IV mutants fail to generate 21–22 nt TE siRNAs [15]. This suggested a key role of Pol IV beyond the known production of 24 nt siRNAs.
We aimed to determine whether pollen 21–22 nt easiRNAs are actually produced from Pol IV transcripts, or alternatively whether Pol IV is necessary to trigger siRNA production from Pol II transcripts. We found that pollen TE easiRNA production is a product of Pol IV transcription, and this activity of Pol IV is not specific to pollen. We find that in the absence of the more abundant 24 nt siRNAs, Pol IV-derived 21–22 nt siRNAs can participate in RdDM. Like other 21–22 nt siRNAs generated from Pol II, Pol IV 21–22 nt siRNAs are incorporated into AGO1, which is the main effector protein of post-transcriptional gene silencing. Our data suggest that like other siRNAs and microRNAs incorporated into AGO1, Pol IV-dependent 21–22 nt siRNAs may participate in the post-transcriptional targeting of genic mRNAs.
2. Results
(a). Pol IV is required for the production of TE 21–22 nt siRNAs
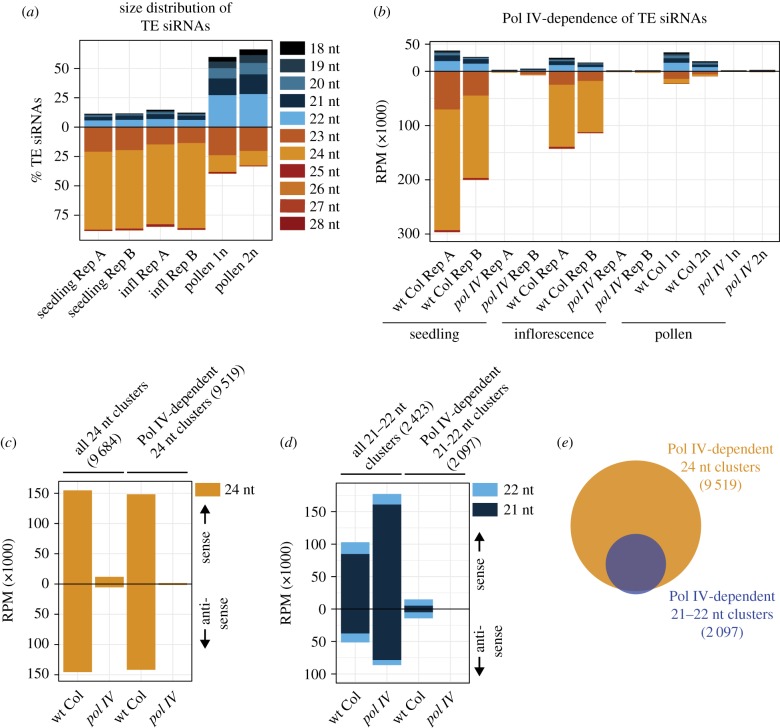
Arabidopsis easiRNAs were discovered in gametophytic pollen and found to be primarily 21–22 nt in length. By contrast, heterochromatic siRNAs are produced during sporophytic stages and are primarily 24 nt in length. To compare the change in siRNA size distribution during development, we analysed small RNAs sequenced from wt Col seedling [18], inflorescence (this study) and pollen [15]. We used diploidized 2n wt Col pollen (derived from 4n wt Col) as a second pollen replicate (see §4). We confirmed that compared to seedling and inflorescence, there is a sharp increase in relative amounts of TE 21–22 nt siRNAs and a corresponding decrease in TE 24 nt siRNAs in pollen (figure 1a). However, we found that the shift in small RNA size relative accumulation of figure 1a was primarily owing to a sharp decrease in TE 24 nt siRNA production in pollen and not an increase in TE 21–22 nt siRNA abundance (figure 1b).
Figure 1.
Pol IV-dependent 21–22 nt siRNAs (a) Per cent of TE siRNAs in different size classes in wild-type Columbia (wt Col) seedlings, inflorescence and pollen tissue. Rep A and Rep B are distinct biological replicates, 1n is normal haploid pollen and 2n is diploid pollen derived from a 4n parent [15]. (b) Reads per million (RPM) counts of TE siRNAs in wt Col and pol IV mutant plants. (c) 24 nt siRNA accumulation in inflorescence from all identified 24 nt siRNA-producing clusters (left) or clusters specifically identified as dependent on Pol IV (right). Above zero are sense siRNAs, and below are antisense. (d) 21–22 nt siRNA accumulation in inflorescence from all identified 21–22 nt siRNA-producing clusters (left) or clusters identified that are dependent on Pol IV (right). (e) Overlap between the regions of the genome where Pol IV produces 24 nt versus 21–22 nt siRNAs.
The recently reported Pol IV-dependence of 21–22 nt siRNAs in pollen [15] was unexpected, since Pol IV had previously only been shown to generate 23–24 nt siRNAs [19] (reviewed in [20]). Therefore, we aimed to determine if Pol IV-dependent TE 21–22 nt siRNA production is specific to pollen or occurs in non-gametophytic tissues. We confirmed that both 21–22 and 24 nt TE siRNAs are dependent on Pol IV in pollen (figure 1b) [15]. We also identified that in the TE-silent sporophytic seedling and inflorescence tissue, Pol IV is responsible for the accumulation of TE 21–22 nt siRNAs (figure 1b). For comparison across different tissue types and sequencing libraries, we normalized the TE siRNA counts by total sequenced small RNAs that match the Arabidopsis genome (figure 1a,b). To confirm that these observations were not biased owing to our specific normalization method, we alternatively normalized TE siRNAs using Pol IV-independent miRNA counts (electronic supplementary material, figure S1). We find that the reduction in the accumulation of TE siRNAs in pol IV mutants is consistent regardless of normalization method. We conclude that Pol IV-dependent TE 21–22 nt siRNAs are not specific to pollen, and a similar mechanism of 21–22 nt siRNA production exists during sporophytic stages.
To take an unbiased approach to investigate all small RNAs (sRNAs) beyond annotated TEs, we identified clusters of 24 nt sRNAs and 21–22 nt sRNAs in wt Col inflorescence (figure 1c,d). As expected, almost all 24 nt sRNAs are lost from the 24 nt clusters in a pol IV mutant (figure 1c). By contrast, global levels of 21–22 nt sRNAs increase in pol IV mutants (figure 1d), which has been previously reported [21]. A majority of these 21–22 nt sRNAs are miRNAs and/or miRNA-induced (tasiRNAs), which are not dependent on Pol IV production. This increased overall level of 21–22 nt sRNAs has likely obscured the fact that Pol IV-dependent 21–22 nt sRNA regions of the genome do exist in wt Col inflorescence (figure 1d), accounting for why they were not discovered earlier. In addition, more than 99% of Pol IV-dependent 21–22 nt clusters overlap with 24 nt clusters (figure 1e). We conclude that there is a genome-wide population of Pol IV-dependent 21–22 nt siRNAs, so far uninvestigated, which are generated from a subset of loci that also produce Pol IV-dependent 24 nt siRNAs.
(b). Pol IV-dependent 21–22 nt siRNAs are produced from Pol IV transcripts
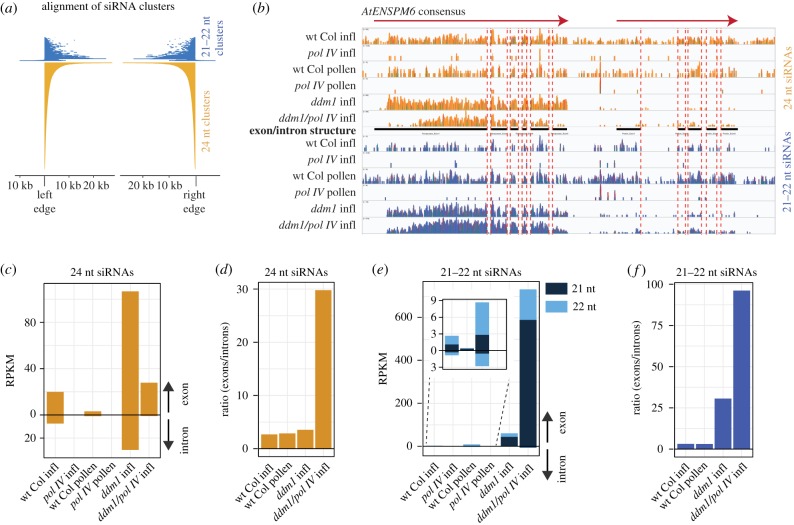
It is well established that 24 nt siRNAs are produced from Pol IV transcripts (reviewed in [20]). We aimed to determine whether 21–22 nt siRNAs are also produced from Pol IV transcripts or are instead produced from Pol II transcripts but somehow dependent on Pol IV. As shown in figure 1e, almost all 21–22 nt clusters overlap with 24 nt siRNAs clusters, suggesting that 21–22 nt siRNAs are produced from a subset of 24 nt clusters and therefore likely from Pol IV transcripts. To investigate the extent of overlap between the two clusters, we positioned each 21–22 nt cluster relative to its corresponding overlapping aligned 24 nt siRNA cluster (figure 2a). We found that most of the 21–22 nt clusters (92%) aligned within the boundaries of 24 nt clusters. Upon investigation of the remaining (8%) 21–22 nt clusters, we found that these loci also shadow 24 nt siRNA loci, but were falsely classified as extending beyond 24 nt clusters owing to bioinformatic artefacts of cluster identification. Therefore, we failed to identify any locus producing Pol IV-dependent 21–22 nt siRNAs that does not also produce 24 nt siRNAs. This observation strongly suggests that Pol IV transcripts that feed into the 24 nt siRNA pathway also produce 21–22 nt siRNAs.
Figure 2.
Pol IV-dependent 21–22 nt siRNAs are produced from Pol IV transcripts. (a) Alignment of the 21–22 nt siRNA clusters to the ordered alignment of the 24 nt siRNA clusters. Very few 21–22 nt clusters extend beyond the boundaries of the 24 nt clusters, and inspection of these found no confirmable region of the genome that generates Pol IV-dependent 21–22 nt siRNAs but not 24 nt siRNAs in inflorescence tissue. (b) Alignment of siRNAs from various genotypes and tissues to the AtENSPM6 TE consensus sequence, which encodes two transcripts (red arrows). Black bars indicate protein-coding exons. Red dashed lines indicate transcript intron boundaries. Inflorescence is abbreviated as infl. (c) Reads per kilobase per million (RPKM) counts of intron- and exon-aligning 24 nt siRNAs for AtENSPM6. (d) Ratio of exon to intron small RNA abundance for 24 nt siRNAs. (e) RPKM counts of intron- and exon-aligning 21–22 nt siRNAs for AtENSPM6. Inset shows the low-level accumulation when TEs are not transcriptionally activated in the ddm1 mutant. Inset shows the low-level accumulation when TEs are not transcriptionally activated in the ddm1 mutant. (f) Ratio of exon to intron small RNA abundance for 21–22 nt siRNAs.
To further investigate the origin of Pol IV-dependent siRNAs, we used the exon–intron structure of transcripts that produce siRNAs. Pol II-transcribed RNAs are efficiently co-transcriptionally spliced and are therefore cleaved into siRNAs matching only exons (figure 2b–f), but the exon–intron distribution of Pol IV siRNAs has not been investigated. We focused on the consensus sequence of the AtENSPM6 family of TEs and its annotated exon–intron structure in the GIRI Repbase [22]. We aligned both 21–22 and 24 nt siRNAs from six key plant lines to this consensus sequence (figure 2b) and counted the abundance from exons and introns (figure 2c,e). We also calculated the ratio of exonic/intronic siRNAs (figure 2d,f) and aimed to use the relative bias in exonic/intronic ratio as a signature for determining the polymerase origin of siRNAs.
We found that when TEs are transcribed by Pol II while Pol IV is not present (ddm1 pol IV double mutant), siRNAs have a high exon/intron bias for both 24 nt and 21–22 nt siRNAs (figure 2b–f). In this double mutant, when Pol II is the only polymerase generating TE siRNAs, the level of exon reads outweighs intron reads 30× for 24 nt siRNAs, and 95× for 21–22 nt siRNAs (figure 2d,f). When a functional Pol IV protein is present, in the ddm1 single mutant (both Pol IV and Pol II active at TEs), the bias of exon/intron siRNAs is severely reduced, suggesting that Pol IV-derived siRNAs are produced from intronic regions as well (figure 2c–f). This conclusion is supported by 24 nt siRNA production in TE-silent wt Col inflorescence, in which TE siRNAs are known to be produced from Pol IV transcripts (Pol IV active, but Pol II inactive at TEs), and therefore have a low exon/intron ratio bias in wt Col inflorescence (figure 2c,d). Using these observations as controls, we investigated the exon/intron bias of 21–22 nt siRNAs in wt Col. We found that in inflorescence and pollen, the Pol IV-dependent 21–22 nt siRNAs have a low bias of exon/intron siRNAs (figure 2e,f), therefore demonstrating that unspliced and likely Pol IV transcripts produce 21–22 nt siRNAs in TE-silent wt Col inflorescence and in TE-active pollen. Together with the observation that 21–22 nt siRNAs are completely lost in pol IV (figure 2c,e), we conclude that Pol IV (and not Pol II) produces the Pol IV-dependent 21–22 nt siRNAs.
(c). Pol IV-dependent 21–22 nt siRNAs are produced by DCL2 and DCL4
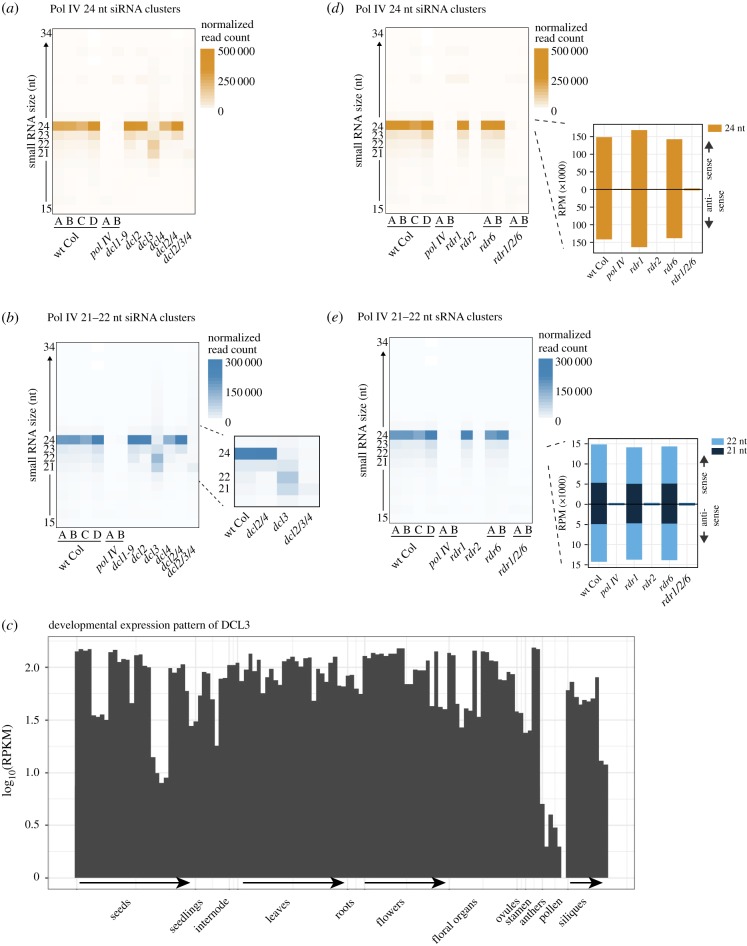
To address whether the Pol IV-derived 21–22 nt siRNAs are non-specific degradation products produced from Pol IV transcripts or full-length 24 nt siRNAs, we compared siRNA accumulation in wt Col and DCL protein family mutants. As expected, we observed the complete loss of 24 nt siRNAs in the dcl3 mutant (figure 3a) [8]. DCL family proteins have known redundancies [25], so when DCL3 is absent, DCL2 and DCL4 substitute and process Pol IV transcripts into 21–22 nt siRNAs (figure 3a,b), confirming that DCL2 and DCL4 have the ability to process Pol IV transcripts [8]. When specifically focused on Pol IV 21–22 nt clusters (figure 3b), we observe a class of Pol IV-dependent 21–22 nt siRNAs in wt Col that are dependent on DCL2 and DCL4 for their production (inset, figure 3b). This demonstrates that even in wt Col sporophytic tissue, Pol IV generates 21–22 nt siRNAs that are not random degradation products of Pol IV transcripts or full-length 24 nt siRNAs but rather are specific cleavage products of DCL2 and DCL4. We conclude that Pol IV transcripts are acted upon by DCL2, DCL3 and DCL4, with a strong bias towards DCL3 and production of 24 nt siRNAs in sporophytic tissues.
Figure 3.
Pathway for Pol IV-dependent siRNA production. (a,b,d,e) Heatmap of read counts of 15–34 nt small RNAs in inflorescence from Pol IV-dependent clusters in various mutant combinations. 24 nt siRNAs clusters are used in (a,d) and 21–22 nt siRNA clusters are used in (b,e). (a,b) uses DCL family mutants. dcl2/3/4 refers to the dcl2/dcl3/dcl4 triple mutant. (d,e) uses RDR family mutants, and rdr1/2/6 refers to the rdr1/rdr2/rdr6 triple mutant. Insets of (d,e) show accumulation of 24 nt and 21–22 nt siRNAs, respectively, with abundance averaged across replicates. (c) Developmental expression pattern of DCL3 protein according to ref. [23] and analysed in the eFP-RNA-Seq Browser [24].
The wt pollen siRNA profile of relatively equal amounts of 21, 22 and 24 nt Pol IV-derived siRNAs (figure 1b) could be produced by a partial absence or dysfunction of the DCL3 protein, whereby more Pol IV transcripts are processed by DCL2 and DCL4. We examined DCL3 expression in pollen and found a lack of expression specifically in this tissue (figure 3c). This suggests that DCL3 protein may be simply lacking or reduced in wt pollen; however, mRNAs of many siRNA-generating proteins are reduced or absent in wt pollen (including the largest subunit of Pol IV itself (NRPD1), electronic supplementary material, figure S3). There is a poor correlation between steady-state mRNA levels and protein abundance (reviewed in [26]) and further analysis is needed to confirm if the reduction in DCL3 activity is responsible for the pollen-specific TE siRNA size distribution.
We next aimed to identify a DCL mutant combination that removes all siRNA production from Pol IV transcripts. The overall abundance of siRNAs is reduced in dcl2/3/4 triple mutants; however, 21 nt siRNAs are still detected (figure 3a,b). To investigate the source of 21 nt siRNAs in dcl2/3/4, which was assumed to be DCL1, we interrogated a seedling tissue dataset that includes mutations in all four DCL family proteins (dcl1/2/3/4) [18]. Using this dataset, we first confirmed that the Pol IV-dependent siRNA-producing clusters identified in inflorescence also produce Pol IV-dependent siRNAs in seedlings (electronic supplementary material, figure S2). Second, the loss of 24 nt siRNAs and increase in relative abundance of 21 nt siRNAs in dcl2/3/4 at the Pol IV-dependent siRNA clusters is also observed in seedlings. We found that these 21 nt siRNAs are produced from Pol IV transcripts, since these siRNAs are lost in pol IV dcl2/3/4 (compared to dcl2/3/4). However, we found that all siRNAs are not lost in dcl1/2/3/4 quadruple mutants (electronic supplementary material, figure S2A,B), which therefore must be owing to the DICER-independent pathway of Pol IV small RNA production [18].
In figure 1, we found a relatively equal distribution of Pol IV-dependent 21–22 nt siRNAs between sense and antisense strands (figure 1d), suggesting that like 24 nt siRNAs (figure 1c), 21–22 nt siRNAs are produced from double-stranded Pol IV transcripts. To further elucidate the pathway of siRNA biogenesis, we investigated the production of siRNAs in plants mutated for RDR family proteins. We found that both 21–22 and 24 nt siRNAs are dependent on RDR2 and are unperturbed in either rdr1 or rdr6 single mutants (figure 3d,e). Therefore, Pol IV-derived 21–22 nt siRNA production is distinct from Pol II-derived TE siRNA production (in ddm1 mutants), which requires RDR6 [16]. We conclude that in sporophytic tissues, Pol IV/RDR2 generates double-stranded TE transcripts that are primarily cleaved into 24 nt siRNAs by DCL3 but are also cleaved by DCL2/DCL4 into low levels of 21–22 nt siRNAs.
(d). Pol IV 21–22 nt siRNAs can target RNA-directed DNA methylation
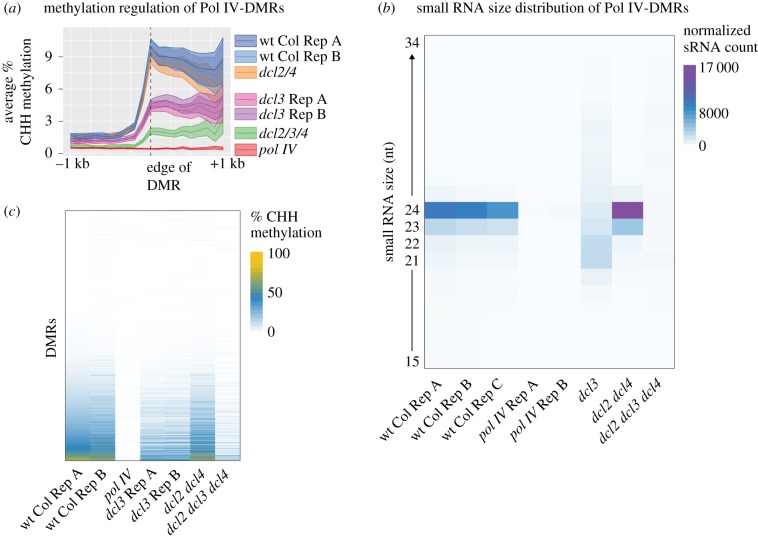
Pol IV-derived 24 nt siRNAs have well-established roles in guiding RdDM [9]. To determine if Pol IV-derived 21–22 nt siRNAs can function in RdDM, we used MethylC-seq to assay genome-wide DNA methylation in a series of DCL family single, double and triple mutants. We identified differentially methylated regions (DMRs) in pol IV mutant plants and aligned CHH context DNA methylation (H = A, C or T) at their edge (figure 4a). Asymmetric CHH methylation, particularly at Pol IV-DMRs, is a hallmark of the RdDM pathway [27]. Importantly, the methylation level of the dcl3 single mutant is not as low as the pol IV mutant (figure 4a), demonstrating that Pol IV-dependent methylation can function through other DCL proteins. In addition, the methylation level in dcl3 is not as low as in as the dcl2/3/4 triple mutant, demonstrating that specifically DCL2 and DCL4 have a function in targeting DNA methylation (figure 4a). SiRNAs from these same regions show the increased abundance of 21–22 nt siRNAs in the dcl3 mutant (figure 4b). These siRNAs must participate in RdDM, as they are lost in dcl2/3/4 (figure 4b), resulting in reduced methylation (figure 4a). Figure 4c shows that the loss of methylation in figure 4a is not the product of just a few loci. Together, these data demonstrate that in the absence of DCL3 and 24 nt siRNA production, Pol IV-generated 21–22 nt siRNAs can participate in RdDM.
Figure 4.
Pol IV-dependent 21–22 nt siRNAs can cause DNA methylation. (a) Metaplot of average per cent CHH context methylation of Pol IV-DMRs in wt Col and different mutant combinations. (b) Small RNA size distribution of the Pol IV-DMRs shown in (a). (c) Heatmap of CHH DNA methylation at Pol IV-DMRs where each row represents an individual DMR. DMRs are sorted by their methylation level in wt Col Rep A.
(e). Pol IV 21–22 nt siRNAs may target gene transcripts
Given the size of Pol IV-dependent 21–22 nt siRNAs, and the known function of tasiRNAs, microRNAs and TE siRNAs of this size to act in post-transcriptional gene silencing (PTGS) of genic mRNAs, we wondered if Pol IV-dependent 21–22 nt siRNAs played a similar role. If Pol IV-derived 21–22 nt siRNAs target genic transcripts for post-transcriptional regulation, then there are four predictions to be tested.
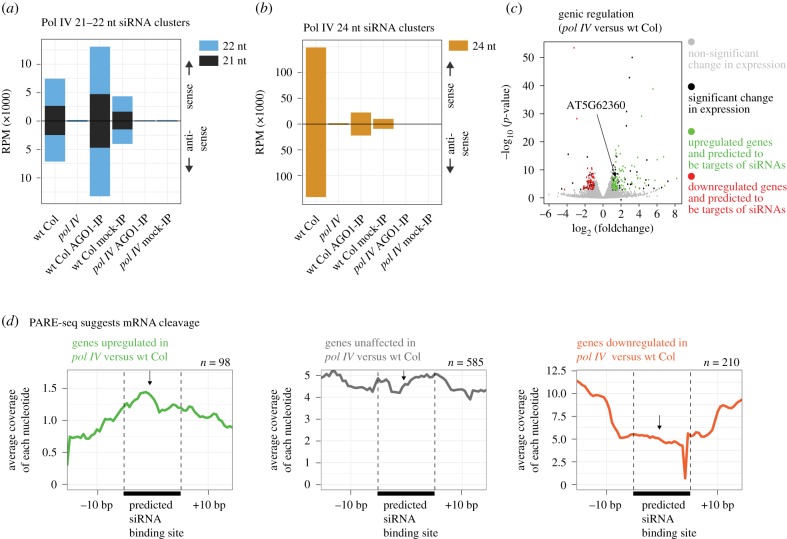
First, these Pol IV-derived 21–22 nt siRNAs would be incorporated into the genic mRNA-regulating AGO protein, AGO1. Pol II-derived 21–22 nt siRNAs are known to cleave mRNA transcripts of genes in trans through the effector protein AGO1 [28]. We investigated whether Pol IV-derived siRNAs are incorporated into AGO1. We sequenced siRNAs from AGO1 immuno-precipitations (IPs) in both wt Col and pol IV mutants. As a control, we also sequenced siRNAs from no-antibody controls (mock IPs) in both wt Col and pol IV. Positive and negative controls that ensure that our IP sRNA-seq experiment worked as expected are shown in electronic supplementary material, figure S4. We found that like Pol II-derived 21–22 nt microRNAs, tasiRNAs and some 21–22 nt TE siRNAs [29], Pol IV-derived 21–22 nt siRNAs are enriched in AGO1 (figure 5a). As expected, these AGO1-incorporated 21–22 nt siRNAs are completely lost in pol IV mutants. As a control, we confirmed that Pol IV-derived 24 nt siRNAs are not strongly enriched in AGO1 (figure 5b). AGO1 incorporation suggests that Pol IV siRNAs could act to target the known activity of AGO1 for mRNA transcript cleavage and translational inhibition.
Figure 5.
A potential role for Pol IV-dependent 21–22 nt siRNAs in post-transcriptional gene silencing. (a) AGO1 incorporation of Pol IV-dependent 21–22 nt siRNAs in wt Col and pol IV mutants. Mock IPs did not include a primary antibody. (b) AGO1 incorporation of Pol IV-dependent 24 nt siRNAs in wt Col and pol IV mutants. (c) Genic expression changes as evidenced by RNA-seq comparing wt Col and pol IV mutants. Black dots represent significant changes and grey dots represent non-significant changes. Green and red dots represent significantly upregulated and downregulated genes, respectively, for genes predicted to be targets of Pol IV siRNAs. (d) Metaplot showing accumulation of PARE sequencing reads aligned to predicted target sites of Pol IV-dependent 21–22 nt siRNAs that show AGO1 incorporation. Genes identified as upregulated (green), unaffected (grey) and downregulated (red) in (c) are aligned separately (left, centre and right). The black arrow marks the predicted target cleavage site by Pol IV-dependent 21–22 nt siRNAs. The black bar at the bottom shows the predicted siRNA binding site used to align multiple genes. n denotes the number of transcripts analysed towards the metaplot.
The second prediction states that for these Pol IV-derived 21–22 nt siRNAs, we could computationally identify target mRNAs and their predicted cleavage sites, although these programmes identify false positives at a high rate. We identified 49 683 proposed target sites of the Pol IV-dependent 21–22 nt siRNAs that are enriched in AGO1 (from figure 5a). This includes 23 668 distinct transcript models encompassing 18 167 total genes. We performed this analysis to inform our experiments below; however, the presence of small RNA target sites by itself provides no direct evidence of function.
The third prediction states that for at least a subset of the predicted target genes, their mRNA levels would increase in pol IV mutant plants, when the targeting siRNAs are not generated. We used publicly available mRNA-seq expression data [30] to identify genes that have increased steady-state transcripts in pol IV mutants compared to wt Col. Assuming that the steady-state transcript levels of the Pol IV siRNA target genes will increase in pol IV mutants, we overlapped the two sets of genes and found 117 genes with both a predicted target site for a Pol IV-dependent 21–22 nt siRNA and the expected increase in transcript levels in the pol IV mutant (green, figure 5c). We compared the fraction of genes with predicted target sites in the upregulated set (green, figure 5c) and unaffected set (grey, figure 5c) and did not find an enrichment of target sites in upregulated genes (data not shown). However, the increase in mRNA levels can be observed only for a subset of genes because of (a) the false positives identified by the mRNA targeted prediction algorithm and (b) AGO1-incorporated siRNAs could potentially cause translational repression instead of mRNA cleavage. Similar to the second prediction, expression data could not provide direct evidence of Pol IV-dependent mRNA degradation. Therefore, we aimed to further investigate the siRNA–mRNA interaction using a parallel analysis of RNA ends (PARE) sequencing (see below).
The fourth prediction states that for genes that increase in steady-state mRNA abundance in pol IV mutants, their mRNA cleavage products could be detected specifically around the predicted siRNA target sites. To determine if the Pol IV-dependent 21–22 nt siRNAs could cause cleavage of the target mRNA akin to a Pol II-derived siRNA or microRNA in AGO1, we analysed publicly available PARE mRNA cleavage data from wt Col inflorescence [31]. We defined three sets of genes to investigate, one with increased transcripts in pol IV (green, figure 5c), a control with unaffected transcripts (grey, figure 5c) and a second control with decreased transcript abundance in pol IV (red, figure 5c). We aligned the target transcripts by the predicted Pol IV-dependent 21–22 nt siRNA target site and mapped the PARE sequences to these transcripts. We expected there to be increased signature of mRNA cleavage and thus PARE reads around the predicted cleavage site of the target genes. For the transcripts with the increased steady-state levels in pol IV mutants (green, figure 5c), we indeed observed increased coverage of PARE sequences at the predicted siRNA binding site compared to flanking regions (green, figure 5d). By contrast, we found no such change in coverage for the control gene set with unchanged transcript levels in pol IV (grey, figure 5d) or decreased transcript levels in pol IV (red, figure 5d). The combined data of AGO1 incorporation and cleavage transcripts at the predicted target site support a model that Pol IV-dependent 21–22 nt siRNAs may function in gene regulation.
3. Discussion
We began our investigation of Pol IV-dependent 21–22 nt siRNAs based on a single publication that observed an anti-dogmatic siRNA accumulation pattern in pollen [15]. Our work has confirmed the Pol IV-dependence of many TE 21–22 nt siRNAs. These siRNAs differ from other TE 21–22 nt siRNAs that require Pol II transcription, such as in ddm1 mutants [16]. In addition, we find that the Pol IV-dependent 21–22 nt siRNAs are direct products of unspliced Pol IV transcripts and are produced when TEs are transcriptionally silent (wt Col inflorescence) and transcriptionally activated (wt Col pollen). The 21–22 nt siRNAs are generated from the same regions with simultaneous overlapping production of 24 nt siRNAs, suggesting that the same Pol IV/RDR2 transcripts that are acted upon by DCL3 to generate 24 nt siRNAs are also processed by DCL2/DCL4 to generate 21–22 nt siRNAs.
A significant remaining question is why the size ratio of Pol IV-derived siRNAs is heavily skewed towards the production of 21–22 nt siRNAs in pollen. Our analysis shows that the number of 21–22 nt siRNAs is not greatly increased in pollen, but rather the amount of 24 nt siRNAs is drastically reduced in pollen, skewing the ratio of 21–22 versus 24 nt siRNAs (figure 1b). The tissue-specific reduction of 24 nt siRNAs in pollen, while still retaining the Pol IV-dependent 21–22 nt siRNAs, is similar to a dcl3 mutant in sporophytic tissue (figure 3a,b), suggesting that a lack of DCL3 activity in pollen could result in the observed pattern. We observe a lack of DCL3 mRNA expression in pollen; however, several components required for the biogenesis of these siRNAs (such as the largest subunit of Pol IV) also fail to accumulate, necessitating future research. In addition, in pollen, there is TE transcriptional activation [12,13], but the resulting Pol II transcripts are not responsible for generating the TE 21–22 nt siRNAs observed in pollen, and therefore the term epigenetically activated siRNA (easiRNA) is not suitable for pollen siRNAs.
In sporophytic tissue, Pol IV transcripts generate high levels of 24 nt siRNAs and low levels of 21–22 nt siRNAs. We investigated the biological function of these Pol IV-derived 21–22 nt siRNAs and found that in the absence of DCL3 and 24 nt siRNAs, 21–22 nt siRNAs generated by Pol IV/RDR2/DCL2/DCL4 can participate in RdDM. Additionally, these 21–22 nt siRNAs may participate in the post-transcriptional regulation of genic mRNAs. We found evidence that these 21–22 nt siRNAs are loaded into AGO1 and at the predicted mRNA target sites, increased transcript cleavage was detected. However, further research is needed to conclusively demonstrate that these 21–22 nt siRNAs can direct AGO1 to post-transcriptionally cleave genic mRNAs. Nonetheless, we conclude that these siRNAs may be RNAi potent and can in theory target complementary invading TEs and quickly initiate an RNAi defense. A question remains regarding what would limit AGO1 activity and PTGS if Pol IV can generate gene-regulating 21–22 nt siRNAs? However, the levels of these siRNAs are only a fraction of known highly abundant microRNAs and tasiRNAs. Therefore, even though mRNA cleavage can be detected, this may not be occurring on enough mRNA molecules to have a phenotypic consequence.
Although it is not understood why there is a shift in the size distribution of Pol IV siRNAs in pollen, the functional consequence of this shift is clear. Unlike 24 nt siRNAs, 21–22 nt siRNAs are incorporated into AGO1, which participates in gene regulation. Therefore, the function of Pol IV in pollen is likely to shift towards gene regulation (function of 21–22 nt siRNAs). Here we show evidence that Pol IV-derived 21–22 nt siRNAs may participate in post-transcriptional regulation. We speculate that this function may be connected to establishing hybridization barriers. Pol IV mutants fail to establish the triploid block, which ensures that the maternal : paternal ratio of genetic contribution is 2 : 1 in the early endosperm [15]. Pollen Pol IV-derived 21–22 nt siRNAs associate with gene expression changes [15], and TE siRNAs in pollen (of unknown biogenesis) are important regulators of imprinting through the post-transcriptional targeting of the genes UBP1b and PEG2 [32]. We propose that TE regions of the genome contribute towards diverse gene regulation via Pol IV-derived 21–22 nt siRNAs specifically in pollen and the early seed, including imprinting [33] and hybridization barriers [14,15].
4. Material and methods
(a). Plants and materials
All plants used for small RNA sequencing as part of this study were grown in growth chambers with standard conditions: 22°C temperature and 16 h light. Stage 1–12 inflorescence tissue was used for RNA isolation. nrpd1a-3 (pol IV), dcl1-9, dcl2-1, dcl3-1, dcl4-2, rdr1-1, rdr2-1, rdr6-15 alleles were used. Wt Col and pol IV have standard 1n pollen samples and 2n pollen generated using the osd1 mutation [15], which were used as replicates in this study.
(b). AGO1 immunoprecipitation
0.5 g inflorescence tissue per sample was ground with liquid nitrogen and homogenized in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 1% IGEPAL, 0.5 mM DTT, 1 mM PMSF and 1× GoldBio protease inhibitor) for 15 min. Lysates pre-cleared for 15 min with 50 µl goat anti-rabbit magnetic beads (NEB). Pre-cleared lysates were then incubated with either goat anti-rabbit magnetic beads only (mock IP) or beads plus 5 µg anti-AGO1 primary antibody (Agrisera) (AGO1 IP). IPs were performed at 4°C for 2 h with end-over-end rotation. Beads were then washed three times for 5 min in wash buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.5 mM DTT). RNA was extracted directly from washed beads using TRIzol reagent, and small RNA libraries were constructed as described below directly from this RNA.
(c). Small RNA sequencing
Total RNA was extracted with phenol chloroform method using TRIzol reagent (Thermo Fisher Scientific). Small RNAs were enriched using miRVana miRNA isolation kit (Thermo Fisher Scientific). The TrueSeq Small RNA Library Preparation Kit (Illumina) was used to make sequencing libraries for total or IP-enriched small RNAs. Multiplexed libraries were sequenced on a HiSeq2500 (Illumina) at the University of Delaware DNA Sequencing and Genotyping Center.
(d). Small RNA processing
Adapter TGGAATTCTCGGGTGCCAAGG was removed from demultiplexed libraries using fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). These sRNAs were mapped to the genome using bowtie 1.2.2 (-v 0) to determine the number of total genome matching reads, which was used to normalize sRNA counts [34]. sRNA Workbench [35] was used to filter out low complexity reads, t/rRNA reads and retain 18–28 nt reads that match the Arabidopsis TAIR10 genome. ShortStack 3.8.5 [36] was used to map the sRNAs to the genome using the parameters --nohp --mmap f --bowtie_m all. Bowtie 1.2.2 was used by ShortStack. For the digital Northern in figure 3c,d, the size limit of 18–28 nt was not applied to allow the visualization of longer RNAs.
(e). Cluster identification
ShortStack 3.8.5 was used to identify clusters of 24 nt and 21–22 nt sRNAs. All small RNA sequencing data used in this study were individually mapped to the Arabidopsis genome using ShortStack [36]. These mapped files were filtered to retain either only 24 nt reads or 21–22 nt reads. All the samples were then merged to create two merged mapped files, one each for 24 nt and 21–22 nt reads. These merged mapped files were used as input for ShortStack to identify clusters with the default parameters except for mincov (set to 10) and pad (set to 50). The identified clusters were then filtered for Arabidopsis miRNA loci from miRBase 22.1 [37]. The miRNA filtered cluster list was filtered for Pol IV-dependent clusters with the criterion that average accumulation of reads was at least two-fold reduced in pol IV compared to wt Col.
(f). Whole-genome DNA methylation analyses
We used inflorescence tissue to isolate DNA and perform MethylC-sequencing as previously described [38]. Statistics of the sequenced reads are shown in electronic supplementary material, table S1. We identified DMRs using default parameters of the methylpy program [39] available in github (https://github.com/yupenghe/methylpy). DMRs were aligned by their edge and CHH methylation was calculated across the region in bins of 50 nt size and averaged across DMRs.
(g). RNA sequencing data analyses
mRNA sequencing data from GSE99691 [30] was reprocessed. Adapters were removed and the sequences were mapped to the genome using STAR 2.6.0c (parameters: –outMultimapperOrder Random –outSAMtype BAM SortedByCoordinate –outFilterMultimapNmax 50 –outFilterMatchNmin 30 –alignSJoverhangMin 3) [40]. Summarize Overlaps from GenomicFeatures [41] was used to count the abundance of genic transcripts using annotation from JGI v11, Arabidopsis v167 TAIR10. DESeq2 [42] was used for differential expression analysis.
(h). Target prediction
A list of candidate sRNAs was prepared and used for target prediction. All 21–22 nt sRNAs enriched in wt Col IP samples (at least five raw counts in each of the two replicate of IP samples and more than twofold accumulation over mock IP) and from Pol IV-dependent 21–22 nt clusters were labelled as candidate small RNAs. These siRNAs were further filtered for loss of accumulation in pol IV AGO1 IP samples (more than twofold reduction in pol IV AGO1 IP). These 440 sRNAs were used in psRNA [43] for target prediction with default parameters.
(i). PARE data analyses
Raw data from GSM1263708 [31] were reprocessed. Adapter sequence was removed and reads with the length of less than 12 nt were discarded. The sequences were mapped to Arabidopsis transcripts (JGI v11, TAIR10, v167 transcripts) using bowtie 1.2.2 and the parameters: -v 0, -a. Bedtools v.2.25.0 [44] was used to count the accumulation of these degradome sequences on transcripts. This count was normalized by the total transcripts investigated in the gene set.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The authors thank Saima Shahid for her advice on bioinformatic approaches.
Data accessibility
All the sRNA sequences generated for this study have been deposited to GEO with the accession number GSE133618. Publicly available small RNA-seq (GSE41755, GSE74398, GSE57191, GSE118705, GSE84122, GSE79780), RNA-seq (GSE99691) and PARE-seq (GSM1263708) were used.
Authors' contributions
K.P. and R.K.S. designed the research. K.P. and A.D.M. performed the research. K.P. analysed the results. K.P. and R.K.S. wrote the article.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by grant MCB-1608392 to R.K.S from the U.S. National Science Foundation.
References
- 1.Bourque G, et al. 2018. Ten things you should know about transposable elements. Genome Biol. 19, 199 ( 10.1186/s13059-018-1577-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borges F, Martienssen RA. 2015. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. ( 10.1038/nrm4085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Y, et al. 2015. Ancient origin and recent innovations of RNA polymerase IV and V. Mol. Biol. Evol. 32, 1788–1799. ( 10.1093/molbev/msv060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJM. 2005. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 37, 761–765. ( 10.1038/ng1580) [DOI] [PubMed] [Google Scholar]
- 5.Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AMS, Strahl BD, Patel DJ, Jacobsen SE. 2013. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389. ( 10.1038/nature12178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhai J, et al. 2015. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163, 445–455. ( 10.1016/j.cell.2015.09.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blevins T, Podicheti R, Mishra V, Marasco M, Tang H, Pikaard CS. 2015. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. Elife 4, e09591 ( 10.7554/eLife.09591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, E104 ( 10.1371/journal.pbio.0020104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. 2010. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22, 321–334. ( 10.1105/tpc.109.072199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, et al. 2015. RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl Acad. Sci. USA 112, 14 728–14 733. ( 10.1073/pnas.1514680112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fultz D, Slotkin RK. 2017. Exogenous transposable elements circumvent identity-based silencing permitting the dissection of expression-dependent silencing. Plant Cell 29, 360–376. ( 10.1105/tpc.16.00718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijó JA, Martienssen RA. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472. ( 10.1016/j.cell.2008.12.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He S, Vickers M, Zhang J, Feng X. 2019. Natural depletion of H1 in sex cells causes DNA demethylation, heterochromatin decondensation and transposon activation. Elife 8, 974 ( 10.7554/eLife.42530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borges F, Parent JS, van Ex F, Wolff P, Martínez G, Köhler C, Martienssen RA. 2018. Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 50, 186–192. ( 10.1038/s41588-017-0032-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez G, Wolff P, Wang Z, Moreno-Romero J, Santos-González J, Conze LL, DeFraia C, Slotkin RK, Köhler C. 2018. Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat. Genet. 50, 193–198. ( 10.1038/s41588-017-0033-4) [DOI] [PubMed] [Google Scholar]
- 16.McCue AD, Nuthikattu S, Reeder SH, Slotkin RK. 2012. Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 8, e1002474 ( 10.1371/journal.pgen.1002474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calarco JP, Martienssen RA. 2011. Genome reprogramming and small interfering RNA in the Arabidopsis germline. Curr. Opin. Genet. Dev. 21, 134–139. ( 10.1016/j.gde.2011.01.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye R, et al. 2016. A dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol. Cell 61, 222–235. ( 10.1016/j.molcel.2015.11.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daxinger L, Kanno T, Bucher E, van der Winden J, Naumann U, Matzke AJM, Matzke M. 2009. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J. 28, 48–57. ( 10.1038/emboj.2008.260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Lang Z, Zhu J-K. 2018. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 6, 597 ( 10.1038/s41580-018-0016-z) [DOI] [PubMed] [Google Scholar]
- 21.Nobuta K, et al. 2008. Distinct size distribution of endogeneous siRNAs in maize: evidence from deep sequencing in the mop1–1 mutant. Proc. Natl Acad. Sci. USA 105, 14 958–14 963. ( 10.1073/pnas.0808066105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mobile DNA 6, 11 ( 10.1186/s13100-015-0041-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA. 2016. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88, 1058–1070. ( 10.1111/tpj.13312) [DOI] [PubMed] [Google Scholar]
- 24.Sullivan A, et al. 2019. An ‘eFP-Seq Browser’ for visualizing and exploring RNA sequencing data. Plant J. 100, 641–654. ( 10.1111/tpj.14468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15, 1494–1500. ( 10.1016/j.cub.2005.07.024) [DOI] [PubMed] [Google Scholar]
- 26.Vélez-Bermúdez IC, Schmidt W. 2014. The conundrum of discordant protein and mRNA expression. Are plants special? Front. Plant Sci. 5, 619 ( 10.3389/fpls.2014.00619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE. 2003. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13, 2212–2217. ( 10.1016/j.cub.2003.11.052) [DOI] [PubMed] [Google Scholar]
- 28.Mallory A, Vaucheret H. 2010. Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22, 3879–3889. ( 10.1105/tpc.110.080671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCue AD, Nuthikattu S, Slotkin RK. 2013. Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol. 10, 1379–1395. ( 10.4161/rna.25555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou M, Palanca AMS, Law JA. 2018. Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat. Genet. 14, 100 ( 10.1038/s41588-018-0115-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creasey KM, Zhai J, Borges F, Van Ex F, Regulski M, Meyers BC, Martienssen RA. 2014. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508, 411–415. ( 10.1038/nature13069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G, Jiang H, Del Toro de León G, Martinez G, Köhler C. 2018. Sequestration of a transposon-derived siRNA by a target mimic imprinted gene induces postzygotic reproductive isolation in Arabidopsis. Dev. Cell 46, 696–705. ( 10.1016/j.devcel.2018.07.014) [DOI] [PubMed] [Google Scholar]
- 33.Satyaki PRV, Gehring M. 2019. Paternally acting canonical RNA-directed DNA methylation pathway genes sensitize Arabidopsis endosperm to paternal genome dosage. Plant Cell 31, 1563–1578. ( 10.1105/tpc.19.00047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 ( 10.1186/gb-2009-10-3-r25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stocks MB, Mohorianu I, Beckers M, Paicu C, Moxon S, Thody J, Dalmay T, Moulton V. 2018. The UEA sRNA Workbench (version 4.4): a comprehensive suite of tools for analyzing miRNAs and sRNAs. Bioinformatics 34, 3382–3384. ( 10.1093/bioinformatics/bty338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Axtell MJ. 2013. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19, 740–751. ( 10.1261/rna.035279.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozomara A, Birgaoanu M, Griffiths-Jones S. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. ( 10.1093/nar/gky1141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panda K, Ji L, Neumann DA, Daron J, Schmitz RJ, Slotkin RK. 2016. Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17, 170 ( 10.1186/s13059-016-1032-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schultz MD, et al. 2015. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–216. ( 10.1038/nature14465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. ( 10.1093/bioinformatics/bts635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 ( 10.1371/journal.pcbi.1003118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai X, Zhuang Z, Zhao PX. 2018. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54. ( 10.1093/nar/gky316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinlan AR. 2014. BEDTools: the Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–11.12.34. ( 10.1002/0471250953.bi1112s47) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the sRNA sequences generated for this study have been deposited to GEO with the accession number GSE133618. Publicly available small RNA-seq (GSE41755, GSE74398, GSE57191, GSE118705, GSE84122, GSE79780), RNA-seq (GSE99691) and PARE-seq (GSM1263708) were used.