Abstract
Gut microbial communities (microbiomes) profoundly shape the ecology and evolution of multicellular life. Interactions between host and microbiome appear to be reciprocal, and ecological theory is now being applied to better understand how hosts and their microbiome influence each other. However, some ecological processes that underlie reciprocal host–microbiome interactions may be obscured by the current convention of highly controlled transplantation experiments. Although these approaches have yielded invaluable insights, there is a need for a broader array of approaches to fully understand host–microbiome reciprocity. Using a directed review, we surveyed the breadth of ecological reality in the current literature on gut microbiome transplants with non-human recipients. For 55 studies, we categorized nine key experimental conditions that impact the ecological reality (EcoReality) of the transplant, including host taxon match and donor environment. Using these categories, we rated the EcoReality of each transplant. Encouragingly, the breadth of EcoReality has increased over time, but some components of EcoReality are still relatively unexplored, including recipient host environment and microbiome state. The conceptual framework we develop here maps the landscape of possible EcoReality to highlight where fundamental ecological processes can be considered in future transplant experiments.
Keywords: conservation, ecological adaptation, ecophysiology, holobiont, metacommunity, ecosystem on a leash
1. A quest for ecological reality
We shall not cease from exploration
And the end of all our exploring
Will be to arrive where we started
And know the place for the first time.
T.S. Eliot - Little Gidding [1, section 5].
Far from passive passengers, resident microbial communities (microbiomes) are integral to the basic biological functioning of multicellular life. This revelation, ushered in by advances in sequencing and computing technology, is grounded in a growing understanding that microbiomes profoundly shape their host's biology, influencing factors such as immunity [2], adiposity [3], thermogenesis [4], hormonal regulation [5], physiological development [6], memory [7] and behaviour [8]. To date, highly controlled experiments with laboratory rodent microbiomes have provided foundational and indispensable knowledge on host–microbiome interactions. Furthermore, these initial experiments have set the stage for integrative contributions by comparative animal physiologists, ecologists and evolutionary biologists to fill knowledge gaps in our understanding of host–microbiome evolution and the interactions which underlie these partnerships [9].
Recently, researchers have started to appreciate the intertwining nature of host–microbiome interactions. Evidence is mounting that hosts can shape the composition of their microbiome community [10], and that microbiomes can influence their host's behaviour [8] and physiology [5]. Based on differing cases of how host and microbiome might interact, Foster et al. [11] proposed four distinct models: (i) ‘host control', in which the host unilaterally governs the composition of its microbiome; (ii) ‘symbiont control’, in which the microbiome shapes the host phenotype; (iii) ‘open ecosystem', in which the host and microbiome do not interact; and (iv) ‘ecosystem on a leash', in which the host influences the microbiome by selecting upon microbial function rather than for specific microbial taxa. These connections can be so intimate that some researchers [12,13] proposed that a host and its associated microorganisms are a single biological entity—termed the ‘holobiont'—on which selection acts, challenging notions of organismal individuality. Using this holobiont perspective, Alberdi et al. [14] posited that the microbial component of the holobiont, with its greater mutability compared to the host genome, may be an important mechanism facilitating host adaptation to rapid environmental change. Therefore, understanding the interplay between the host and the microbiome is crucial for addressing both fundamental and applied questions about the microbiome.
Host–microbiome interactions are shaped by ecological and evolutionary processes [15,16]. Because host–microbiome interactions are potentially reciprocal, these processes act on three levels: the assembly and dynamics of the microbiome, the influence of the host on the microbiome and the influence of the microbiome on the host. Microbiome assembly is governed by a variety of factors including environmental filtering, priority effects, random sampling and dispersal limitation [16,17]. The within-microbiome community dynamics are influenced by new invasions, competition, mutualisms and other interactions [15]. A host's actions can also shape their associated microbiomes. For example, the host's social behaviour can impact microbial dispersal [18]. Conversely, the dynamics of the microbiome can impact the host; the change in microbiome community composition leading to Clostridium difficile colonization and pathogenicity is a classic example in humans [19]. Evolutionary processes also occur in tandem with all the ecological processes mentioned previously because of the short timescales associated with microbial turnover relative to microbial evolutionary rates [17]. Consequently, considering the ecological processes that underlie host–microbiome interactions is critical for making sense of the reciprocity between the host and its microbiome.
The most convincing evidence for host–microbiome interactions has been gleaned through microbiome transplantation studies. In these studies, researchers experimentally translocate microbial species or communities from donor hosts or external substrates to recipient hosts. Highly controlled transplantation studies have been and will continue to be invaluable to experimentally probe the host–microbiome relationship. However, there is a trade-off: highly controlled experiments isolate mechanisms of interest, but they cannot simultaneously capture the full suite of ecological processes (drift, dispersal, competition, etc.) that influence reciprocal host–microbiome interactions in nature. For example, the use of germ-free recipients may preclude competition between introduced and resident microbes [20], and isolated laboratory conditions may limit the potential for microbial dispersal from influencing the composition of the resulting microbiome [21]. How researchers weigh this trade-off depends on the research question of interest. If a researcher's goal is to understand the effect of specific microbes on host physiology, or to develop applications for human health and domestic animal production, controlled experimental conditions are preferred. By contrast, when examining the role and consequences of reciprocal host–microbiome interactions in ecological and evolutionary contexts (e.g. fitness effects, intergenerational microbial transmission, speciation, species persistence, etc.), ecological complexity needs to be considered [22]. Therefore, a comparison of highly controlled transplants and ecologically realistic (which we term EcoReality, see box 1 for a full definition) transplants that match what the host plus its microbiome would experience in a wild ecosystem is required. The trade-offs of laboratory approaches and the need for comparison to studies that use ecologically realistic conditions have long been recognized by comparative animal physiologists [23], though to date there does not seem to have been a similar recognition in microbiome research. Specifically, the breadth of EcoReality in microbiome transplant studies has not been examined, meaning such an evaluation remains an exciting potential avenue for future work.
Box 1. Key terms and definitions.
| term | definition |
|---|---|
| transplant instance | a transplant of a microbial strain or community from its native host or substrate to a different host population. A given study can involve multiple transplant instances, which are delineated based on non-substitutability of host populations or of transplant parameters |
| experimental conditions | a decision or step in a transplant instance where there is the potential for variation in ecological reality |
| level of EcoReality | the degree to which an experimental condition matches the conditions that a host–microbiome interaction would experience in a wild ecosystem. Each experimental condition possesses its own intrinsic EcoReality. Each transplant instance can also be assigned an EcoReality score |
Here, we probe the current EcoReality of microbiome transplantation studies. Our work here is not unlike Hanage's [24] questioning of the reality and applicability of biomedical microbiome studies. By taking advantage of the recent explosion of studies conducting microbiome transplants, we evaluated whether the current microbiome transplant literature limits opportunities for ecological processes to influence study outcomes. We investigated two key questions: (i) how EcoReal are the experimental conditions in the current microbiome transplant literature? and (ii) does the literature currently cover the full potential range of EcoReality? Using long-established ecological concepts, we categorized microbiome transplantations into different experimental conditions which can impact the EcoReality of the transplant (figure 1 and box 2). Using this framework, we scored the EcoReality of microbiome transplant studies that used non-human recipients. We show that, overall, the breadth of EcoReality of the present microbiome transplant literature has increased over time. However, EcoReality has been constrained by hosts bred and kept in laboratory conditions, and with transplants into germ-free recipient hosts. Importantly, we provide a conceptual framework, illustrated in figure 1, to help broaden the range of EcoReality in transplant experiments and to facilitate comparisons between transplants of varying EcoReality.
Figure 1.
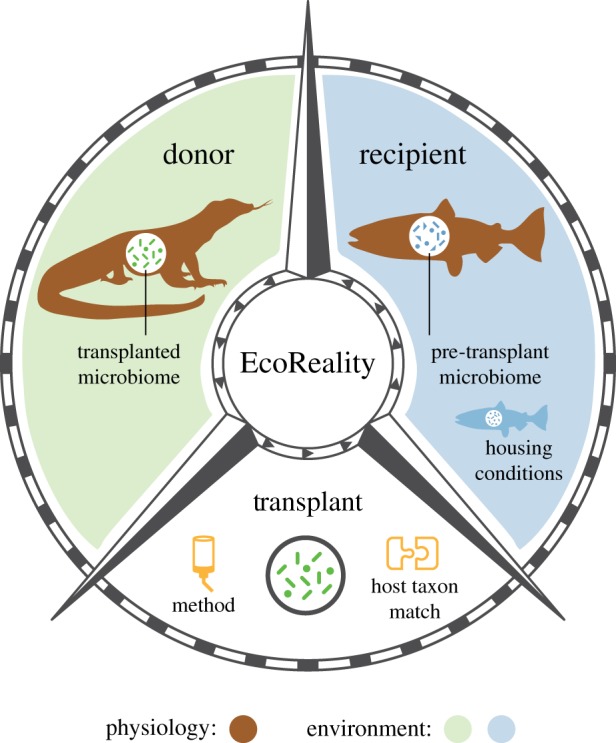
Conceptual framework of all the experimental conditions in a microbial transplant where EcoReality can vary. See box 2 for explanations for each experimental condition. (Online version in colour.)
Box 2. Ecological reasoning for each experimental condition within a transplant.
| experimental condition | reasoning: ecological theory + application to microbiome |
|---|---|
| taxon match | organisms can become locally adapted [25]. Local adaptation of a microbial species to a given host may mean it is not adapted to hetero-specific hosts and will perform poorly after transplantation [26] |
| donor and recipient environment | during community assembly, the local environment acts as a filter, incorporating species from a wider species pool [21]. From the microbiome's perspective, the host's physiology and the external environment are one intertwined environment. Therefore, the external environment can affect microbiome dynamics in two ways: indirectly through impacting the host physiology [27], and directly through the wider microbial species pool that the host and its microbiome has access to |
| donor and recipient physiology | the local environment acts as a filter in community assembly [21]. For this experimental condition, we define physiology as physiological states that would occur regardless of the external environmental context (e.g. gene knock-out, disease-state). We differentiate intrinsic physiology from mutable host physiological responses to the external environmental context. Although these indirect environmental effects acting through host physiology are relevant [27], they are captured by the ‘environment’ experimental condition. A host's physiology is the de facto environment of inhabitant microbes, and changes or dysregulation in the host may disrupt associations between host and the microbes that persist under homeostatic physiological conditions |
| transplanted microbiome | the interactions within an invading community, including predation or mutualism, can impact whether colonisation is successful or not [20]. Thus, a full community microbiome transplantation may differ significantly from the transplantation of a single microbe monoculture at artificially high densities |
| transplant method | species have different dispersal abilities [27] and local environments filter species from the wider species pool [28]. Active transplantations may circumvent differing dispersal abilities of microbial species and may undermine host filtering of the microbial community. Furthermore, active transplant methods can stress the host thereby changing host physiology and disrupting endogenous microbial communities [29] |
| recipient pre-transplant microbiome | high species diversity in a community is predicted to reduce niche opportunities and to increase invasion resistance [20]. Germ-free or antibiotic perturbed recipients are likely to have lower invasion resistance than recipients with intact microbiomes |
| housing conditions | dispersal between patches is an integral ecological process which can maintain stable populations or can rescue extirpated populations [21,30]. Recipient host cohabitation allows for further transmissions of the microbiome. |
2. Lay of the land
(a). Literature search
We conducted a directed review of the existing literature on gut microbiome transplants, finishing on 26 October 2018. We conducted our literature search in three stages. First, to gauge the extent of the current literature, we did a preliminary search of gut microbiome transplant studies using both Google Scholar and Web of Science (University of Guelph subscription). Based on this preliminary search, we conducted a more methodical search using both Google Scholar and Web of Science. Search terms can be found in the electronic supplementary material. We then sought additional publications through searching the citations of papers already collected using the Web of Science citations tool. We retained only those studies that conducted at least one gut microbiome transplant into a non-human recipient organism. To ensure our findings were generalizable to ecological and evolutionary frameworks across a broad range of taxa and ecosystems, we excluded studies focused on a single human disease, C. difficile.
(b). Literature evaluation
For each study that met our criteria, we determined the number of transplant instances, which we defined as the transfer of a microbial strain or community from its native host or substrate to a different host population (see box 1). We used transplant instances as our unit of focus because many studies contained multiple transplant instances which sometimes differed substantially in EcoReality (e.g. [31]). For studies that had sequential transplants (i.e. transplant from donor to a first recipient, which then was the donor for a second recipient, e.g. [31]), we used only the first phase of the transplant experiment.
We identified nine key experimental conditions in a transplant where variation in EcoReality might substantially affect the outcome of the experiment: host taxon match, donor environment, donor physiology, transplanted microbiome, transplant method, recipient pre-transplant microbiome, recipient environment, recipient physiology and recipient housing conditions (figure 1 and box 2). Each experimental condition was given an ordinal data scale (see the electronic supplementary material, table S1) based on the range of observed and possible levels for that condition, with one always representing the lowest level of EcoReality. Our goal was to maintain a similar resolution for each highly dimensional experimental condition within our framework. For example, taxon match could have included more levels to capture phylogenetic distance, geographical distance and feeding relationships between host and recipient [32]. However given the variation in scale, generalization across host taxa would have then been difficult. The levels in each experimental condition were based on likely conditions found in the wild. For example, with respect to the transplanted microbiome experimental condition, a single bacterial strain at high densities entering a host in the wild is less likely than invasion by mixed communities. For each transplant instance, we characterized the level of EcoReality in each of the nine experimental conditions. To ensure consistent evaluation methods, EcoReality scores for each transplant instance were determined independently by two co-authors (separate pairs of co-authors randomized per paper). The co-author pairs then compared their scores and agreed upon the final transplant EcoReality scores.
To determine the overall standardized EcoReality score of a transplant instance, we divided each score by its corresponding maximum potential EcoReality score and then added the scaled scores for each experimental condition. Thus all experimental conditions were equally weighted in the overall calculation of standardized EcoReality.
We separated laboratory rodents from other animals in our results for each experimental condition because the ecology, physiology and genetics of laboratory-reared, inbred rodent models are heavily modified from wild-type rodents and other wild animals in ways that may affect our understanding of reciprocal host–microbiome interactions (for example [23,33]).
(c). Literature EcoReality patterns
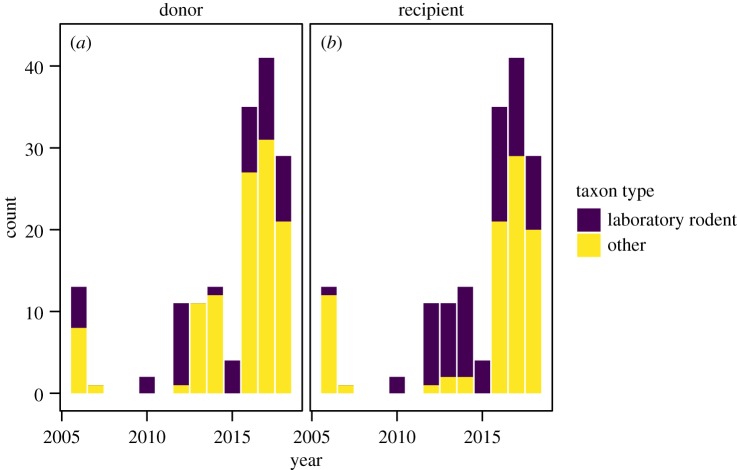
Our literature search returned 55 articles that met our criteria for inclusion. These articles ranged from having 1 to 13 transplant instances with an average of 2.91 transplant instances per article and a total of 160 from all articles. There was a clear shift over time in the number of articles using microbiome transplants. Notably, there were 20 articles in the first 10 years of our search period in comparison to almost 40 articles during 2015–2018 (electronic supplementary material, figure S1). This increase coincided with a switch from mainly laboratory rodent studies to a more diverse group of donor hosts (figure 2a, around 2013), and later also to more diverse recipient hosts (figure 2b, around 2016).
Figure 2.
Number of transplant instances over time where the donor or recipient animal was either a laboratory rodent (mouse or rat) or another animal. (Online version in colour.)
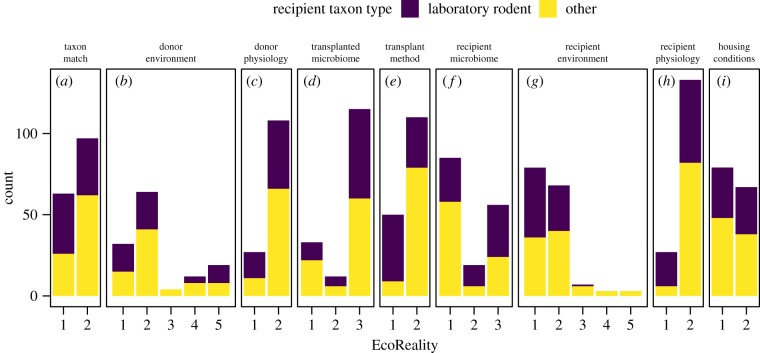
The transplant conditions donor and recipient physiology had the highest EcoReality with average scores of 1.8 out of 2 (figure 3c,h). Taxon match (score 1.6 out of 2, figure 3a), transplanted microbiome (score 2.5 out of 3, figure 3d), transplant method (score 1.7 out of 2, figure 3e) and housing condition (score 1.5 out of 2, figure 3i) were moderately EcoReal. Donor environment (score 2.4 out of 5, figure 3b), recipient environment (score 1.6 out of 5, figure 3g) and recipient microbiome (score 1.8 out of 3, figure 3f) had the lowest EcoReality. Breaking EcoReality into recipient laboratory rodents and other animals, we see that active transplant methods (score of 1) were used more for laboratory rodents, and passive transplant methods (score of 2) were used more for other animals (figure 3e). Interestingly, there were fewer transplants with germ-free recipient laboratory rodents than germ-free recipient other animals (score of 1) (figure 3f). This pattern was driven by bees (19 out of 85 transplant instances from five articles) and zebrafish (14 out of 85 transplant instances from two articles). Overall, most transplants were performed with matching (score of 2, figure 3a) wild-type, non-diseased (score of 2, figure 3c,h) donor and recipient hosts using passive transplant methods (score of 2, figure 3e) of whole microbial communities (score of 3, figure 3d) and with a mixture of individual (score of 1, figure 3i) and cohousing (score of 2, figure 3i) of recipient hosts. However, transplants were mostly in sterile or normal laboratory conditions (score of 1 and 2, figure 3b,g) with germ-free recipient hosts (score of 1, figure 3f).
Figure 3.
Number of transplant instances in each experimental condition, separated into whether the recipient animal was a laboratory rodent or another animal. The x-axis is the level of EcoReality, with 1 always the lowest EcoReality. The levels are explained in the electronic supplementary material, table S1. (Online version in colour.)
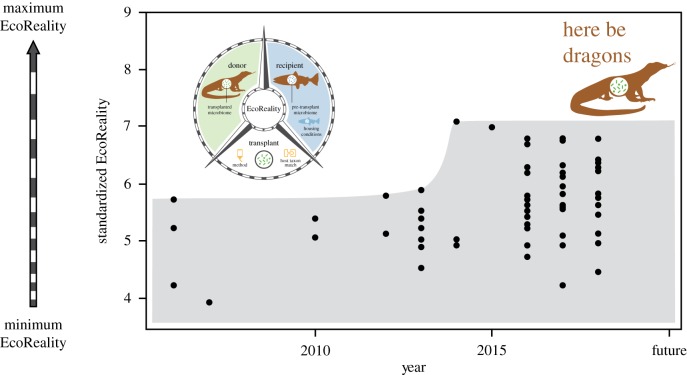
Although the maximal EcoReality score increased, increasing the breadth of EcoReality studied, the maximal EcoReality score was still below the theoretical maximum standardized EcoReality score of nine possible outlined in our framework (figure 4).
Figure 4.
Standardized EcoReality score for each transplant instance. The grey area identifies the zone of EcoReality that has been studied in the literature, and the ‘here be dragons' area is the unexplored zone of EcoReality that is bound at the top by the theoretical maximum standardized EcoReality score of nine. (Online version in colour.)
3. Here be dragons!
The burgeoning field of microbiome research is integrating the traditionally disparate disciplines of ecology, evolution and physiology, which examine distinct but interrelated processes at different scales. Yet, these interrelated processes across scales are inherent in host–microbiome relationships (e.g. [34]), and thus further integration of ecology, evolution and physiology with microbiology will be crucial for unlocking important insights about the interactions between hosts and their microbiome. As microbiome research expands further to include ecological processes that are well established in traditional ecosystems, studies that can capture these processes will be necessary. Here, we expand on the insights from foundational highly controlled experiments that identified key mechanisms in host–microbiome interactions. We surveyed the state of the microbiome transplant literature and identified gaps in how well ecological processes are captured in transplants, what we term as EcoReality (see box 1). Our results are promising; the breadth of EcoReality is increasing over time in transplant experiments (figure 4), but there are still some key gaps in the types of studies conducted on host–microbiome interactions (figure 3). We suggest that a critical step in understanding reciprocal host–microbiome interactions includes explicitly designing a broader array of studies that can evaluate the role of various ecological processes that are known to shape traditional ecological systems.
Our evaluation of the microbiome transplantation literature revealed broadening EcoReality in experimental procedures. Lately, there has been a sharp increase in taxonomic diversity of both donor and recipient hosts (figure 2). Transplants often used passive transplantation methods with wild-type non-diseased donors as well as a mixture of individual and cohabitation housing conditions (figure 3). Finally, the maximal EcoReality score of microbiome transplant studies has increased over time (figure 4). These results are encouraging because they suggest that researchers are building on the initial flurry of highly controlled transplant experiments and designing diverse studies that differ in their degree of EcoReality in several of the categories we examined. Continuing to broaden EcoReality will be essential for understanding the ecological and evolutionary processes at work in reciprocal host–microbiome interactions.
However, our results show that the current literature lacks EcoReality in two key areas: the host environment and the state of the recipient microbiome (figure 3). Although the environment of the donor hosts was on average more EcoReal than the environment of the recipient hosts, the EcoReality of the donor and recipient host's environments was generally low. Most studies that we evaluated used laboratory settings that excluded the chance for hosts to encounter the broader microbial species pool in the environment [18,21]. Laboratory conditions can also either increase or decrease conspecific interactions relative to what would be observed in nature, thus affecting the dispersal of microbes between hosts [18]. Furthermore, laboratory conditions may be obscuring feedbacks between the host and its microbiome that can impact diet and habitat choice [18]. The second key area lacking EcoReality is the state of the recipient microbiome where most recipient hosts were germ-free. Although some animals naturally start out with germ-free gastrointestinal tracts (e.g. newly eclosed worker bees [35]) or do not have a resident microbiome [36], most animal species host substantial microbial communities [37]. Germ-free gastrointestinal tracts may lack key biotic processes such as predation, competition and facilitation, which are important filters in classic ecological communities that act to mediate incoming species [20,38]. Overall, neglecting natural environments and intact recipient microbiomes risks constraining the fundamental processes that impact reciprocal host–microbiome interactions.
Consequently, we advocate for more breadth in EcoReality in microbiome transplant experiments. This breadth includes highly controlled laboratory transplants, which offer critical points of comparison, and provide a focused understanding of particular mechanisms. The wider breadth of EcoReality which we are advocating for requires that we venture into the largely untested realm of highly EcoReal experimental conditions (figure 4), despite the logistical challenges probably associated with wild conditions. There are many ways in which we might venture beyond our present frontier. For example, researchers could use wild-caught animals that are either allowed to roam freely or are housed in outdoor enclosures, or use recipient animals with intact microbiomes rather than germ-free microbiomes (figure 3). We also suggest identifying and addressing the major phylogenetic gaps in the Tree of Life for the donor and recipient host taxa. Overall, we call for a balance of studies dealing with all permutations of EcoReality in each experimental condition. We hope researchers will use and adapt our conceptual framework (figure 1) in their own systems to incorporate EcoReality and, where appropriate, consider how constrained EcoReality may impact their conclusions. Likewise, we encourage researchers to report the methodological details pertaining to each experimental condition we have identified. We hope that our literature evaluation and conceptual framework will stimulate new avenues of collaborative research that will evaluate the role of ecological processes in host–microbiome interactions.
Our literature evaluation suggests that we may understand only a small subset of possible reciprocal host–microbiome interactions impacting our ability to assess the conservation potential of the microbiome. Because we are presently probably constraining fundamental ecological and evolutionary processes, host–microbiome studies may be biased towards results that indicate a strong role of the microbiome on the host. Yet researchers have already made strong and general assertions about the role of the microbiome in the biology of the host. Owing to the large effects of the microbiome on its host and its mutability, Alberdi et al. [14] argued that the microbiome could act as an additional axis of ecological adaptation for hosts. If the microbiome does act as an additional axis, conserving microbial diversity and using bioaugmentation tools (probiotic therapy and transplantation of microbiomes) would then be critical tools for animal conservation [39,40]. We caution that experimental protocols which lack EcoReality might lead us to overestimate the capacity for microbiome variation to shape host phenotypes in nature by biasing our understanding of the host–microbiome relationship towards models of symbiont control [11]. We suspect that a full reckoning of the spectrum of EcoReality in microbiome transplant studies will uncover more examples of the ‘ecosystem on a leash' model [11], which posits an important but more limited reciprocity between the host and the ‘ecosystem' of the microbiome. These sorts of nuanced interactions may or may not include the large microbiome effects which underpin the ecological adaptation and conservation arguments above. Thus, we may not yet have the level of understanding about reciprocal host–microbiome interactions that is required to know the role of the microbiome in host adaptation or to confidently inform conservation efforts. Moving forward, we assert that a consideration of EcoReality is required in the design and interpretation of every study that explores how the host–microbiome relationship impacts ecological adaptation.
Microbiome research has undoubtedly fascinated biologists across disciplines, prompting advances in both pure and applied research and raising questions about some of the most fundamental ideas in biology [13]. Yet, the lay of the land in terms of the ecological reality of this rapidly growing research area was unexplored. Our objective here—to survey the breadth of EcoReality in the microbiome transplant literature and identify key areas lacking EcoReality—was not unlike a fact-finding mission expanding the map of our understanding of reciprocal host–microbiome interactions. We recommend a full, extended journey into the wilds to round out the literature's coverage of the landscape of possible EcoReality. Charting all territories, from highly controlled laboratory studies to free-ranging organisms, is necessary to fully comprehend the interplay between microbiomes and their hosts.
Supplementary Material
Acknowledgements
We thank the authors, animals and microbes of the many studies that provided the peaks and valleys that we traversed in our exploration. We thank the three anonymous reviewers for being our north star, guiding us forward. C.J.G.G. thanks his PhD supervisor, Kevin McCann, for his support. Thanks go to KAP Design for designing figures 1 and 4. We thank the attendees of the Evelyn Pielou Discussion Group for inspiring this voyage.
Data accessibility
The supporting information is included in the electronic supplementary material. The supporting information plus the data, and R script for this manuscript can be found on Zenodo/Github [41]. The full list of transplant studies used in this article can be found in the dataset on Zenodo/GitHub [41], and [10,26,31,35,42–92] in the bibliography below.
Authors' contributions
All authors conceived of and produced the directed review. C.J.G.G. wrote the first draft and all authors contributed to editing the manuscript.
Competing interests
We declare we have no competing interests.
Funding
C.J.G.G. was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) CGS-D. K.C. acknowledges the support of NSERC, RGPIN-2018-04399. M.R.S. was supported by an NSERC Vanier CGS.
References
- 1.Eliot T, Eliot T, Hodgson A, Mairet P. 1942. Little Gidding. London, UK: Faber and Faber. [Google Scholar]
- 2.Lee YK, Mazmanian SK. 2010. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768–1773. ( 10.1126/science.1195568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. ( 10.1038/nature05414) [DOI] [PubMed] [Google Scholar]
- 4.Chevalier C, et al. 2015. Gut microbiota orchestrates energy homeostasis during cold. Cell 163, 1360–1374. ( 10.1016/j.cell.2015.11.004) [DOI] [PubMed] [Google Scholar]
- 5.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, Kubo C, Koga Y. 2004. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice: commensal microbiota and stress response. J. Physiol. 558, 263–275. ( 10.1113/jphysiol.2004.063388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sommer F, Bäckhed F. 2013. The gut microbiota: masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. ( 10.1038/nrmicro2974) [DOI] [PubMed] [Google Scholar]
- 7.Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, Macqueen G, Sherman PM. 2011. Bacterial infection causes stress-induced memory dysfunction in mice. Gut 60, 307–317. ( 10.1136/gut.2009.202515) [DOI] [PubMed] [Google Scholar]
- 8.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. 2014. Microbiota is essential for social development in the mouse. Mol. Psychiatry 19, 146–148. ( 10.1038/mp.2013.65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohl KD, Carey HV. 2016. A place for host–microbe symbiosis in the comparative physiologist's toolbox. J. Exp. Biol. 219, 3496–3504. ( 10.1242/jeb.136325) [DOI] [PubMed] [Google Scholar]
- 10.Zhang X-Y, Sukhchuluun G, Bo T-B, Chi Q-S, Yang J-J, Chen B, Zhang L, Wang D-H. 2018. Huddling remodels gut microbiota to reduce energy requirements in a small mammal species during cold exposure Microbiome 6, 103 ( 10.1186/s40168-018-0473-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. 2017. The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. ( 10.1038/nature23292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. ( 10.1038/nrmicro1635) [DOI] [PubMed] [Google Scholar]
- 13.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735. ( 10.1111/j.1574-6976.2008.00123.x) [DOI] [PubMed] [Google Scholar]
- 14.Alberdi A, Aizpurua O, Bohmann K, Zepeda-Mendoza ML, Gilbert MTP. 2016. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31, 689–699. ( 10.1016/j.tree.2016.06.008) [DOI] [PubMed] [Google Scholar]
- 15.Koskella B, Hall LJ, Metcalf CJE. 2017. The microbiome beyond the horizon of ecological and evolutionary theory. Nat. Ecol. Evol. 1, 1606–1615. ( 10.1038/s41559-017-0340-2) [DOI] [PubMed] [Google Scholar]
- 16.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. ( 10.1126/science.1224203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. ( 10.1038/nrmicro1978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller ET, Svanbäck R, Bohannan BJM. 2018. Microbiomes as metacommunities: understanding host-associated microbes through metacommunity ecology. Trends Ecol. Evol. 33, 926–935. ( 10.1016/j.tree.2018.09.002) [DOI] [PubMed] [Google Scholar]
- 19.Samarkos M, Mastrogianni E, Kampouropoulou O. 2018. The role of gut microbiota in Clostridium difficile infection. Eur. J. Intern. Med. 50, 28–32. ( 10.1016/j.ejim.2018.02.006) [DOI] [PubMed] [Google Scholar]
- 20.Shea K, Chesson P. 2002. Community ecology theory as a framework for biological invasions. Trends Ecol. Evol. 17, 170–176. ( 10.1016/S0169-5347(02)02495-3) [DOI] [Google Scholar]
- 21.Leibold MA, et al. 2004. The metacommunity concept: a framework for multi-scale community ecology. Ecol. Lett. 7, 601–613. ( 10.1111/j.1461-0248.2004.00608.x) [DOI] [Google Scholar]
- 22.Carrier TJ, Reitzel AM. 2017. The hologenome across environments and the implications of a host-associated microbial repertoire. Front. Microbiol. 8, 802 ( 10.3389/fmicb.2017.00802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman AEM, Edmunds NB, Ferraro S, Heffell Q, Merritt GM, Pakkala JJ, Schilling CR, Schorno S. 2015. Using ecology to inform physiology studies: implications of high population density in the laboratory. Am. J. Physiol. Regul. Integr. Comp. Physiol. 308, R449–R454. ( 10.1152/ajpregu.00328.2014) [DOI] [PubMed] [Google Scholar]
- 24.Hanage WP. 2014. Microbiome science needs a healthy dose of scepticism: to guard against hype, those interpreting research on the body's microscopic communities should ask five questions. Nature 512, 247–248. ( 10.1038/512247a) [DOI] [PubMed] [Google Scholar]
- 25.Kawecki TJ, Ebert D. 2004. Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241. ( 10.1111/j.1461-0248.2004.00684.x) [DOI] [Google Scholar]
- 26.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR. 2016. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 ( 10.1371/journal.pbio.2000225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bie T, et al. 2012. Body size and dispersal mode as key traits determining metacommunity structure of aquatic organisms. Ecol. Lett. 15, 740–747. ( 10.1111/j.1461-0248.2012.01794.x) [DOI] [PubMed] [Google Scholar]
- 28.Chase JM. 2003. Community assembly: when should history matter? Oecologia 136, 489–498. ( 10.1007/s00442-003-1311-7) [DOI] [PubMed] [Google Scholar]
- 29.Allen-Blevins CR, You X, Hinde K, Sela DA. 2017. Handling stress may confound murine gut microbiota studies. PeerJ 5, e2876 ( 10.7717/peerj.2876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zelnik YR, Arnoldi J, Loreau M. 2019. The three regimes of spatial recovery. Ecology 100, e02586 ( 10.1002/ecy.2586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seedorf H, et al. 2014. Bacteria from diverse habitats colonize and compete in the mouse gut Cell 159, 253–266. ( 10.1016/j.cell.2014.09.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moeller AH, Suzuki TA, Lin D, Lacey EA, Wasser SK, Nachman MW. 2017. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc. Natl Acad. Sci. USA 114, 13 768–13 773. ( 10.1073/pnas.1700122114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter J, Armet AM, Finlay BB, Shanahan F. 2020. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 180, 221–232. ( 10.1016/j.cell.2019.12.025) [DOI] [PubMed] [Google Scholar]
- 34.Stothart MR, Palme R, Newman AEM. 2019. It's what's on the inside that counts: stress physiology and the bacterial microbiome of a wild urban mammal. Proc. R. Soc. B 286, 20192111 ( 10.1098/rspb.2019.2111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Näpflin K, Schmid-Hempel P. 2016. Immune response and gut microbial community structure in bumblebees after microbiota transplants Proc. R. Soc. B 283, 20160312 ( 10.1098/rspb.2016.0312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N. 2017. Caterpillars lack a resident gut microbiome. Proc. Natl Acad. Sci. USA 114, 9641–9646. ( 10.1073/pnas.1707186114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell JA, Dubilier N, Rudgers JA. 2014. Nature's microbiome: introduction. Mol. Ecol. 23, 1225–1237. ( 10.1111/mec.12676) [DOI] [PubMed] [Google Scholar]
- 38.Jackson DA, Peres-Neto PR, Olden JD. 2001. What controls who is where in freshwater fish communities: the roles of biotic, abiotic, and spatial factors. Can. J. Fish. Aquat. Sci. 58, 157–170. ( 10.1139/cjfas-58-1-157) [DOI] [Google Scholar]
- 39.Trevelline BK, Fontaine SS, Hartup BK, Kohl KD. 2019. Conservation biology needs a microbial renaissance: a call for the consideration of host-associated microbiota in wildlife management practices. Proc. R. Soc. B 286, 20182448 ( 10.1098/rspb.2018.2448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee A, Cornejo J, Bandopadhyay R. 2020. Emergent climate change impact throughout the world: call for ‘Microbiome Conservation’ before it's too late. Biodivers. Conserv. 29, 345–348. ( 10.1007/s10531-019-01886-6) [DOI] [Google Scholar]
- 41.Greyson-Gaito CJ, Bartley TJ, Cottenie K, Jarvis W, Newman AEM, Stothart MR. 2020. Data, SI & R Script for: Into the wild: microbiome transplant studies need broader ecological reality (version v2.2). Zenodo/Github. See 10.5281/zenodo.2652255. [DOI] [PMC free article] [PubMed]
- 42.Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection Cell 127, 423–433. ( 10.1016/j.cell.2006.08.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang X, Hua X, Yang Q, Ding D, Che C, Cui L, Jia W, Bucheli P, Zhao L. 2007. Inter-species transplantation of gut microbiota from human to pigs. ISME J. 1, 156–162. ( 10.1038/ismej.2007.23) [DOI] [PubMed] [Google Scholar]
- 44.Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, Guigo R, Knight R, Guarner F. 2010. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 20, 1411–1419. ( 10.1101/gr.107987.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bindels LB, et al. 2012. Restoring specific Lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model. PLoS ONE 7, e37971 ( 10.1371/journal.pone.0037971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bindels LB, et al. 2016. Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia. ISME J. 10, 1456–1470. ( 10.1038/ismej.2015.209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koch H, Schmid-Hempel P. 2012. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol. Lett. 15, 1095–1103. ( 10.1111/j.1461-0248.2012.01831.x) [DOI] [PubMed] [Google Scholar]
- 48.Upadhyay V, et al. 2012. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nat. Immunol. 13, 947–953. ( 10.1038/ni.2403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toh MC, Goodyear M, Daigneault M, Allen-Vercoe E, Van Raay TJ. 2013. Colonizing the embryonic zebrafish gut with anaerobic bacteria derived from the human gastrointestinal tract. Zebrafish 10, 194–198. ( 10.1089/zeb.2012.0814) [DOI] [PubMed] [Google Scholar]
- 50.Ridaura VK, et al. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 ( 10.1126/science.1241214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohl KD, Weiss RB, Cox J, Dale C, Denise Dearing M. 2014. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol. Lett. 17, 1238–1246. ( 10.1111/ele.12329) [DOI] [PubMed] [Google Scholar]
- 52.Li M, et al. 2015. Fecal microbiota transplantation and bacterial consortium transplantation have comparable effects on the re-establishment of mucosal barrier function in mice with intestinal dysbiosis. Front. Microbiol. 6, 692 ( 10.3389/fmicb.2015.00692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruce-Keller AJ, Salbaum JM, Luo M, Blanchard E, Taylor CM, Welsh DA, Berthoud H-R. 2015. Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biol. Psychiatry. 77, 607–615. ( 10.1016/j.biopsych.2014.07.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Luccia B, et al. 2015. Rescue of fructose-induced metabolic syndrome by antibiotics or faecal transplantation in a rat model of obesity. PLoS ONE 10, e0134893 ( 10.1371/journal.pone.0134893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hensley-McBain T, et al. 2016. Effects of fecal microbial transplantation on microbiome and immunity in simian immunodeficiency virus-infected macaques. J. Virol. 90, 4981–4989. ( 10.1128/JVI.00099-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Twitchell EL, et al. 2016. Modeling human enteric dysbiosis and rotavirus immunity in gnotobiotic pigs. Gut Pathog. 8, 51 ( 10.1186/s13099-016-0136-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon MY, et al. 2016. A single gene of a commensal microbe affects host susceptibility to enteric infection. Nat. Commun. 7, 11606 ( 10.1038/ncomms11606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelly JR, et al. 2016. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 82, 109–118. ( 10.1016/j.jpsychires.2016.07.019) [DOI] [PubMed] [Google Scholar]
- 59.Tian Z, Liu J, Liao M, Li W, Zou J, Han X, Kuang M, Shen W, Li H. 2016. Beneficial effects of fecal microbiota transplantation on ulcerative colitis in mice. Dig. Dis. Sci. 61, 2262–2271. ( 10.1007/s10620-016-4060-2) [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, et al. 2016. Probiotic Lactobacillus casei Zhang reduces pro-inflammatory cytokine production and hepatic inflammation in a rat model of acute liver failure. Eur. J. Nutr. 55, 821–831. ( 10.1007/s00394-015-0904-3) [DOI] [PubMed] [Google Scholar]
- 61.Yan H, et al. 2016. Gut microbiota can transfer fiber characteristics and lipid metabolic profiles of skeletal muscle from pigs to germ-free mice. Sci. Rep. 6, 31786 ( 10.1038/srep31786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mikaelyan A, Thompson CL, Hofer MJ, Brune A. 2016. Deterministic assembly of complex bacterial communities in guts of germ-free cockroaches. Appl. Environ. Microbiol. 82, 1256–1263. ( 10.1128/AEM.03700-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwarz RS, Moran NA, Evans JD. 2016. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl Acad. Sci. USA 113, 9345–9350. ( 10.1073/pnas.1606631113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohl KD, Stengel A, Dearing MD. 2016. Inoculation of tannin-degrading bacteria into novel hosts increases performance on tannin-rich diets: tannin-degrading bacteria aid hosts consuming tannins. Environ. Microbiol. 18, 1720–1729. ( 10.1111/1462-2920.12841) [DOI] [PubMed] [Google Scholar]
- 65.Sonowal R, et al. 2017. Indoles from commensal bacteria extend healthspan. Proc. Natl Acad. Sci. USA 114, E7506–E7515. ( 10.1073/pnas.1706464114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller AW, Dale C, Dearing MD. 2017. The induction of oxalate metabolism in vivo is more effective with functional microbial communities than with functional microbial species. mSystems 2, e00088-17 ( 10.1128/mSystems.00088-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmidt V, Gomez-Chiarri M, Roy C, Smith K, Amaral-Zettler L. 2017. Subtle microbiome manipulation using probiotics reduces antibiotic-associated mortality in fish. mSystems 2, e00133-17 ( 10.1128/mSystems.00133-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warne RW, Kirschman L, Zeglin L. 2017. Manipulation of gut microbiota reveals shifting community structure shaped by host developmental windows in amphibian larvae. Integr. Comp. Biol. 57, 786–794. ( 10.1093/icb/icx100) [DOI] [PubMed] [Google Scholar]
- 69.Ericsson AC, Personett AR, Turner G, Dorfmeyer RA, Franklin CL. 2017. Variable colonization after reciprocal fecal microbiota transfer between mice with low and high richness microbiota. Front. Microbiol. 8, 196 ( 10.3389/fmicb.2017.00196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ekmekciu I, et al. 2017. Immune responses to broad-spectrum antibiotic treatment and fecal microbiota transplantation in mice. Front. Immunol. 8, 397 ( 10.3389/fimmu.2017.00397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith P, Willemsen D, Popkes M, Metge F, Gandiwa E, Reichard M, Valenzano DR. 2017. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. Elife 6, e27014 ( 10.7554/eLife.27014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.von Klitzing E, Ekmekciu I, Kühl AA, Bereswill S, Heimesaat MM. 2017. Intestinal, extra-intestinal and systemic sequelae of Toxoplasma gondii induced acute ileitis in mice harboring a human gut microbiota. PLoS ONE 12, e0176144 ( 10.1371/journal.pone.0176144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Macke E, Callens M, De Meester L, Decaestecker E. 2017. Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat. Commun. 8, 1608 ( 10.1038/s41467-017-01714-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berasategui A, Salem H, Paetz C, Santoro M, Gershenzon J, Kaltenpoth M, Schmidt A. 2017. Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol. Ecol. 26, 4099–4110. ( 10.1111/mec.14186) [DOI] [PubMed] [Google Scholar]
- 75.Kwong WK, Mancenido AL, Moran NA. 2017. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. open sci. 4, 170003 ( 10.1098/rsos.170003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alberoni D, Baffoni L, Gaggìa F, Ryan PM, Murphy K, Ross PR, Stanton C, Di Gioia D. 2018. Impact of beneficial bacteria supplementation on the gut microbiota, colony development and productivity of Apis mellifera L. Benef. Microbes. 9, 269–278. ( 10.3920/BM2017.0061) [DOI] [PubMed] [Google Scholar]
- 77.Billiet A, Meeus I, Cnockaert M, Vandamme P, Van Oystaeyen A, Wäckers F, Smagghe G. 2017. Effect of oral administration of lactic acid bacteria on colony performance and gut microbiota in indoor-reared bumblebees (Bombus terrestris). Apidologie 48, 41–50. ( 10.1007/s13592-016-0447-5) [DOI] [Google Scholar]
- 78.Erkosar B, Kolly S, van der Meer JR, Kawecki TJ. 2017. Adaptation to chronic nutritional stress leads to reduced dependence on microbiota in Drosophila melanogaster. mBio 8, e01496-17 ( 10.1128/mBio.01496-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moghadam NN, Thorshauge PM, Kristensen TN, de Jonge N, Bahrndorff S, Kjeldal H, Nielsen JL. 2018. Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly (Austin) 12, 1–12. ( 10.1080/19336934.2017.1394558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi J, et al. 2017. Intestinal microbiota contributes to colonic epithelial changes in simulated microgravity mouse model. FASEB J. 31, 3695–3709. ( 10.1096/fj.201700034R) [DOI] [PubMed] [Google Scholar]
- 81.Ji S, et al. 2018. Ecological restoration of antibiotic-disturbed gastrointestinal microbiota in foregut and hindgut of cows. Front. Cell. Infect. Microbiol. 8, 79 ( 10.3389/fcimb.2018.00079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schroeder BO, et al. 2018. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe 23, 27–40. ( 10.1016/j.chom.2017.11.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Näpflin K, Schmid-Hempel P. 2018. Host effects on microbiota community assembly. J. Anim. Ecol. 87, 331–340. ( 10.1111/1365-2656.12768) [DOI] [PubMed] [Google Scholar]
- 84.Mockler BK, Kwong WK, Moran NA, Koch H. 2018. Microbiome structure influences infection by the parasite Crithidia bombi in bumble bees. Appl. Environ. Microbiol. 84, e02335-17 ( 10.1128/AEM.02335-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu L, Geng S, Li Y, Cheng S, Fu X, Yue X, Han X. 2018. Exogenous fecal microbiota transplantation from local adult pigs to crossbred newborn piglets. Front. Microbiol. 8, 2663 ( 10.3389/fmicb.2017.02663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Siegerstetter S-C, Petri RM, Magowan E, Lawlor PG, Zebeli Q, O'Connell NE, Dudley EG. 2018. Fecal microbiota transplant from highly feed-efficient donors shows little effect on age-related changes in feed-efficiency-associated fecal microbiota from chickens. Appl. Environ. Microbiol. 84, e02330-17 ( 10.1128/AEM.02330-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bidu C, et al. 2018. The transplantation of ω3 PUFA-altered gut microbiota of fat-1 mice to wild-type littermates prevents obesity and associated metabolic disorders. Diabetes 67, 1512–1523. ( 10.2337/db17-1488) [DOI] [PubMed] [Google Scholar]
- 88.Wrzosek L, et al. 2018. Transplantation of human microbiota into conventional mice durably reshapes the gut microbiota. Sci. Rep. 8, 6854 ( 10.1038/s41598-018-25300-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee H, Lee Y, Kim J, An J, Lee S, Kong H, Song Y, Lee C-K, Kim K. 2018. Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes 9, 155–165. ( 10.1080/19490976.2017.1405209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moeller AH, Suzuki TA, Phifer-Rixey M, Nachman MW. 2018. Transmission modes of the mammalian gut microbiota. Science 362, 453–457. ( 10.1126/science.aat7164) [DOI] [PubMed] [Google Scholar]
- 91.Sommer F, Ståhlman M, Ilkayeva O, Arnemo JM, Kindberg J, Josefsson J, Newgard CB, Fröbert O, Bäckhed F. 2016. The gut microbiota modulates energy metabolism in the hibernating brown bear Ursus arctos. Cell Rep. 14, 1655–1661. ( 10.1016/j.celrep.2016.01.026) [DOI] [PubMed] [Google Scholar]
- 92.Miller AW, Oakeson KF, Dale C, Dearing MD. 2016. Microbial community transplant results in increased and long-term oxalate degradation Microb. Ecol. 72, 470–478. ( 10.1007/s00248-016-0800-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Greyson-Gaito CJ, Bartley TJ, Cottenie K, Jarvis W, Newman AEM, Stothart MR. 2020. Data, SI & R Script for: Into the wild: microbiome transplant studies need broader ecological reality (version v2.2). Zenodo/Github. See 10.5281/zenodo.2652255. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The supporting information is included in the electronic supplementary material. The supporting information plus the data, and R script for this manuscript can be found on Zenodo/Github [41]. The full list of transplant studies used in this article can be found in the dataset on Zenodo/GitHub [41], and [10,26,31,35,42–92] in the bibliography below.