Abstract
The antibiotic 2,4-diacetylphoroglucinol (2,4-DAPG), produced by the Gram-negative rod-shaped bacterium Pseudomonas fluorescens 2P24, is active against various soil-borne bacterial and fungal pathogens that cause plant diseases. Biosynthesis of 2,4-DAPG is controlled by regulating expression of the phlACBD operon at the post-transcriptional level. The phlG gene is located between the phlF and phlH genes, upstream of the phlACBD biosynthetic operon. Herein, we cloned the phlG gene, generated a phlG deletion mutant, and investigated its regulatory role in 2,4-DAPG biosynthesis. The results showed that deletion of phlG had no effect on the biosynthesis of 2,4-DAPG, but it affected conversion of 2,4-DAPG to its precursor monoacetylphloroglucinol (MAPG). The global regulatory factor encoded by gacS positively regulated expression of phlG, while rsmE negatively regulated its expression. Deleting phlG did not alter the ability of the bacterium to colonise plants or promote plant growth. These results suggest that phlG collaborates with other factors to regulate production of the antibiotic 2,4-DAPG in P. fluorescens 2P24.
Subject terms: Antimicrobials, Applied microbiology
Introduction
The antibiotic 2,4-diacetylphloroglucinol (DAPG) is produced by several Pseudomonas sp., including 2P24, CHA0, Pf-5, and YGJ3, and it plays a key role in inhibiting the growth of pathogenic microorganisms surrounding the plant rhizosphere1–4. As a phenolic secondary metabolite, 2,4-DAPG from some bacteria above has shown the capacity to control various plant pathogens. For example, P. fluorescens CHA0 protects plants against tobacco black root rot, P. fluorescens F113 protects sugar beet against Pythium damping-off, and P. fluorescens 2P24 protects against tomato bacterial wilt and wheat take-all diseases5–11. To further improve its potential applications, chemically synthesised 2,4-DAPG analogues have been developed and tested against plant diseases, and MP4, one of analogue of 2,4-DAPG, exhibited particularly potent antifungal activity, with inhibition rates of 84% and 63% against Penicillium. digitatum and Penicillium. italicum, respectively, and lower toxicity toward human cells compared with a fungicide widely used to treat harvested citrus fruit12. Such discoveries may assist the utilisation of DAPG analogues as novel biological fungicides for controlling plant diseases.
In P. fluorescens, the DAPG locus contains the four biosynthetic genes phlACBD that together produce 2,4-DAPG. PhlA, PhlC and PhlB are required for transacetylation of the monoacetylphloroglucinol (MAPG) precursor to generate DAPG13,14, and PhlD is critical for the biosynthesis of MAPG. In P. fluorescens 2P24, multiple factors in the GacS/GacA two-component system are involved in the biosynthesis of 2,4-DAPG during the late exponential and stationary phases2,15,16. The small RNA-binding proteins RsmA and RsmE, the resistance-nodulation-division efflux pump EmhABC, and the sigma factors RpoD, RpoN and RpoS are also associated with 2,4-DAPG biosynthesis1,17–21. In addition, PsrA is also a regulator of a sigma factor and involves in 2,4-DAPG biosynthesis PsrA negatively regulates phlA expression via either direct binding to an operator in the PhlA promoter region, or post-transcriptionally by affecting RpoS and RsmA expression. Inactivation of PsrA leads to a significant increase in 2,4-DAPG biosynthesis.
The phlG gene is also present in the DAPG biosynthetic locus of P. fluorescens 2P24, located between phlF and phlH. The PhlG protein is a DAPG hydrolase that may also modulate DAPG production10. The three-dimensional structure of PhlG revealed that the enzyme converts DAPG into MAPG by cleaving the carbon-carbon bond that attaches the acetyl group to the phenolic ring, hence PhlG differs functionally from classical α/β-hydrolases in terms of catalytic mechanism and substrate specificity10. Expression of PhlG is controlled by the pathway-specific regulators PhlF and PhlH, and phlH and PhlG impose negative feedback regulation on 2,4-DAPG biosynthesis21.
To further investigate the function of PhlG in the synthesis and metabolism of 2,4-DAPG in P. fluorescens 2P24, we performed thin-layer chromatography (TLC) and liquid chromatography (LC) assays to analyse 2,4-DAPG degradation, and generated and characterised PhlG mutants. The results confirm that PhlG converts DAPG to MAPG, and rsmE and gacS negatively or positively regulate phlG gene expression.
Results
Deletion of PhlG does not affect the growth of P. fluorescens 2P24
To investigate the influence of the phlG gene on the growth of P. fluorescens 2P24, the PhlG deletion (2P24-ΔG) and complementary line (2P24-G) were generated and transformed into 2P24 and PM901 as shown in Fig. 1A. The bacteria growth of 2P24, 2P24-ΔG and 2P24-G were evaluated at different time points (Fig. 1B). The results showed that the growth rate of the deletion mutant 2P24-ΔG was comparable with that of the WT and complementary strain 2P24-G, suggesting that deletion of phlG did not affect bacterial cell growth.
Figure 1.
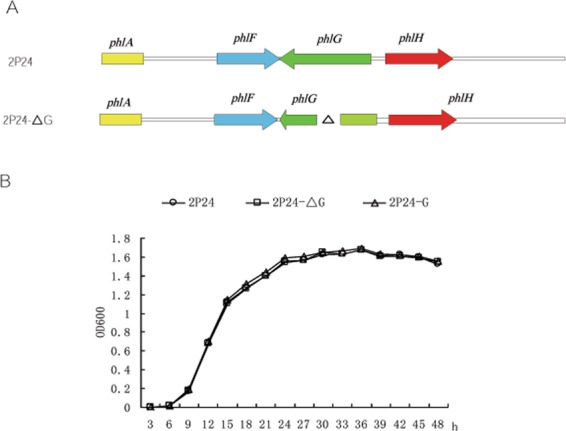
(A) Schematic diagram of the construction of the phlG deletion mutant in Pseudomonas fluorescens 2P24 and PM901. (B) Growth rate of wild-type (WT) P. fluorescens 2P24 and the phlG deletion mutant.
PhlG mediates the conversion of DAPG to MAPG
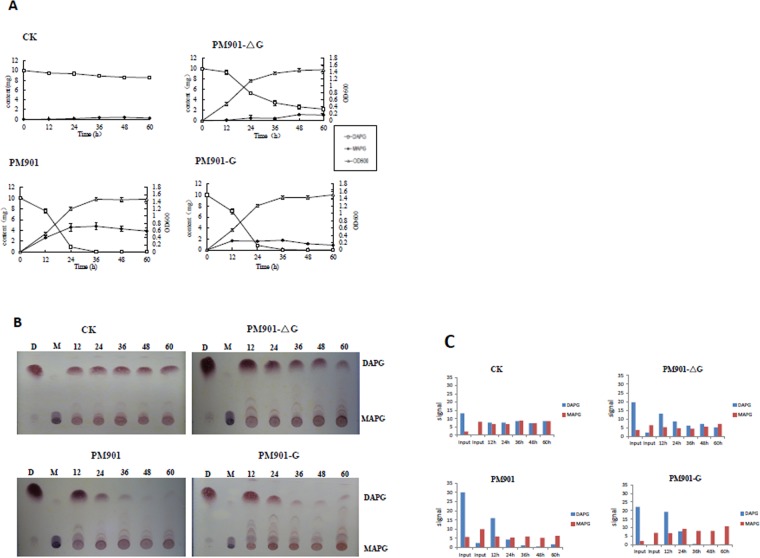
A previous report showed that strain CHA0 degrades the potent antimicrobial agent DAPG to the much less toxic MAPG14. To investigate whether PhlG is also involved in this function in P. fluorescens 2P24, we deleted the phlG gene in strain PM901, a derivative of 2P24 in which phlA is deleted, rendering the strain unable to produce DAPG and MAPG. TLC and LC assays were performed to examine the potential regulatory role of PhlG in 2,4-DAPG biosynthesis. The culture of PM901(the phlA mutant, ΔphlA), PM901-ΔG (the phlA and phlG double mutant, ΔphlAΔphlG) and PM901-G (the phlG complementary strain) were incubated in the presence (100 μM) and absence of DAPG. The results showed that the timing of DAPG reduction and MAPG increase is quite similar. With the PM901 strain, the DAPG concentration rapidly declined and the compound was undetectable after 48 h of incubation (Fig. 2A and Table S2). However, the DAPG concentration was decreased from 10 mg to 2 mg in the culture medium of PM901-ΔG strain lacking PhlG after 60 h incubation. When the PhlG is complemented in PM901-G strain, the accumulation of DAPG is similar to those of PM901, whereas the MAPG concentration increased rapidly from 12 h and was maintained at an elevated level at 60 h.
Figure 2.
(A) Requirement of PhlG for the conversion of DAPG to MAPG. Strains PM901, PM901-ΔG (PhlG mutant), and PM901-G (PhlG complemented mutant) were grown at 27 °C in media supplemented with 100 μM DAPG, and bacterial cell growth and the concentrations of DAPG and MAPG were measured at the indicated time periods. Values are means and standard deviations from three independent cultures, and all experiments were performed in triplicate. (B) Degradation of 2,4-DAPG is regulated by PhlG. D, 2,4-DAPG at start of experiment; M, MAPG at start of experiment; measurements were made at 12, 24, 36, 48, and 60 h after inoculation. CK: Degradation profile of 2,4-DAPG without any treatment in 60 h. The original pictures of gels are shown in the supplementary information Fig. S3. (C) The signals of Fig. 3B from the original picture (Fig. S3) were quantified by Quantity ONE software.
Consistent with Fig. 3A, the results of TLC assays showed that DAPG degradation after 24 h was accompanied by the temporary accumulation of MAPG in the PM901 culture medium (Fig. 2B,C). In PM901-ΔG, the accumulation of DAPG is remained at higher level after 24 h and decreased at 60 h. By the contrast, the formation of MAPG increased from 24 h and showed high accumulation at 60 h. When PhlG was complemented in PM901-G, the patterns of DAPG and MAPG were similar to those of PM901. This suggested that PhlG is functional in conversion of DAPG to MAPG in P. fluorescens 2P24.
Figure 3.
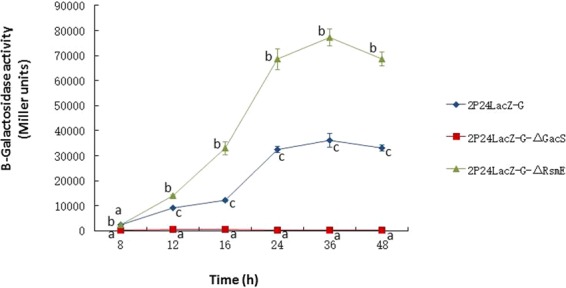
GacS and RsmZ regulate phlG expression in P. fluorescens 2P24.
GacS and RsmZ regulate phlG expression in P. fluorescens 2P24
In the biocontrol strain P. fluorescens CHA0, posttranscriptional repression of GacS/GacA-controlled genes relies highly on the RNA-binding protein RsmE and RsmA21–23. To explore whether DAPG biosynthesis is regulated by the GacS/GacA regulatory cascade through affecting the PhlG expression, gacS and rsmE genes in 2P24-LacZ-G were mutated and β-galactosidase activity was measured. We found that expression of phlG was completely inhibited when gacS gene was deleted, whereas phlG expression was markedly increased following deletion of rsmE (Fig. 3 and Table S3). Taken together, these results indicate that the global regulatory factor gacS positively regulates expression of phlG, while rsmE negatively regulates its expression.
PhlG does not affect root colonization or plant growth in P. fluorescens 2P24
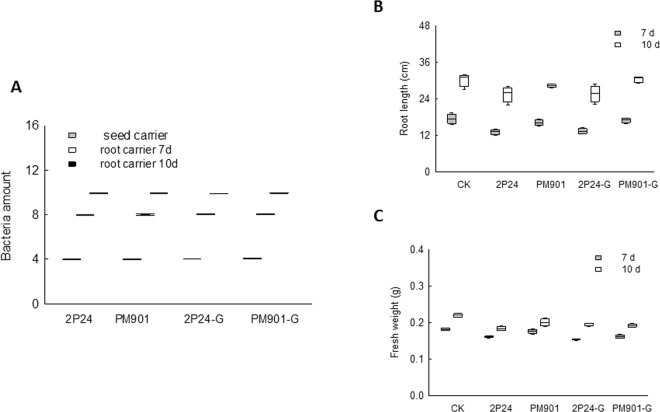
In order to investigate whether the colonisation ability of the 2P24 strain is influenced by phlG, the seed germination rate was determined after soaking seeds in cultures of strains 2P24, PM901, 2P24-ΔG and PM901-ΔG. The number of bacteria present on seeds and roots was measured at 7 d and 10 d post-sowing. As shown in Fig. 4A and Table S4, deletion of phlG did not alter the colonisation capacity. The effects of strains 2P24, PM901, 2P24-ΔG and PM901-ΔG on plant growth were also examined. There was no significant difference in plant height and root length when roots were colonised by 2P24-ΔG, PM901-ΔG and the respective WT strains (Fig. 4B,C and Table S5). This indicates that PhlG does not affect root colonization or plant growth in P. fluorescens 2P24.
Figure 4.
The effect of phlG mutation on the colonization capacity of 2P24 and the wheat growth. (A) phlG gene mutation does not change the colonization capacity of P. fluorescens 2P24. The values represent the number of bacteria carried on roots and seeds. The bacteria carrying on roots and seeds when treated with 2P24, PM901, 2P24-ΔG, PM9014-ΔG was represented by box plot. (B) phlG gene mutation does not promote wheat growth in length. The length of wheat plant when treated with 2P24, PM901, 2P24-ΔG, PM9014-ΔG was shown by box plot. (C) phlG gene mutation does not promote wheat growth in length. The weight of wheat plant when treated with 2P24, PM901, 2P24-ΔG, PM9014-ΔG was represented by box plot. CK: control plant, no treatment.
Discussion
2,4-DAPG is a key secondary metabolite in the biocontrol bacterium Pseudomonas fluorescens that inhibits the growth of pathogenic microorganisms in the plant rhizosphere. Our data showed that PhlG does not affect the antifungal activities of 2P24 against Rhizoctonia solani Kühn, which is different with the role of PhlG in CHA05.
The protein encoded by the phlG gene catalyses the conversion of DAPG into the much less toxic MAPG by cleaving a carbon-carbon bond linking an acetyl group to the phenolic ring24. In this study, we confirmed the function of the product of the phlG gene that is located in the DAPG biosynthetic cluster in P. fluorescens 2P24, which is consistent with that phlG might also be associated with the DAPG biosynthetic locus in P. fluorescens strains Pf-5, Q2-87, F113 and CHA03,8,10,13,14.
Together with a previous report in P. fluorescens CHA010, our results of mutation of the phlG gene and DAPG degradation assays are underling that PhlG probably encodes a hydrolase that converts DAPG to MAPG. This suggests that PhlG-mediated degradation of DAPG is a conserved feature among DAPG-producing pseudomonads. In addition, we also found that deletion of phlG slowed down but did not eliminate degradation of 2,4-DAPG, suggesting other factors involoved in the DAPG degradation in this bacterium. MAPG is a direct precursor of DAPG biosynthesis10,14,25. We therefore propose that MAPG is also a degradation product of DAPG generated by PhlG, although the biological significance of this conversion remains controversial. PhlG appears to act on both the DAPG metabolite itself, and the DAPG biosynthetic operon. For example, accumulation of MAPG was increased in the phlA deletion mutant. By converting DAPG to the much less toxic MAPG, PhlG may help to prevent accumulation of the toxic metabolite, as demonstrated in strains CHA0 and F11310.
In P. fluorescens, the GacS/GacA system positively regulated the transcription of noncoding small RNAs, which further bind with repressor proteins RsmA and RsmE and release them from their target mRNAs22,23. The GacS mutant was used to investigate whether PhlG is regulated by the GacS/GacA system in P. fluorescens, and expression of phlG gene was markedly decreased when gacS was deleted, suggesting that this global regulatory factor positively regulates phlG expression. This further suggests that DAPG biosynthesis is highly dependent on the GacS/GacA regulatory cascade14,16. In P. fluorescens CHA0, the RNA-binding protein RsmA is a key regulatory element in the GacS/GacA signal transduction pathway that acts at the posttranscriptional level. The 64 amino acid polypeptide RsmE is a homolog of RsmA in strain CHA0, and RsmA and RsmE function together to cause maximal repression in the GacS/GacA cascade in this organism21. Deletion of the rsmE gene significantly up-regulated the expression of phlG in P. fluorescens 2P24, suggesting the negative regulation of RsmE on PhlG-mediated DAPG degradation. Additionally, PhlG expression is also negatively regulated by PhlF, a known pathway-specific transcriptional repressor of DAPG gene expression21.
Theoretically, mutant 2P24-∆G should be more inhibitory to the bacterial or fungal pathogens than 2P24, because it produces more DAPG. However, it appears that PhlG-mediated DAPG degradation does not affect either the root colonisation or plant growth-promoting activity of P. fluorescens 2P24. In P. fluorescens strains CHA0 and Pf-5, DAPG biosynthesis is negatively affected by pyoluteorin14,26,27, we thus speculate that other unknown factors probably mediate the antifungal activity of P. fluorescens 2P24 in the absence of PhlG. This means PhlG might have no direct effect on the synthesis of 2,4-DAPG; instead, phlG regulates the metabolism of 2,4-DAPG by assisting the conversion of DAPG to MAPG, which indicating that the deletion of phlG may have an indirect effect on the total amount of DAPG in the cell.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are listed in Table S1. E. coli and P. fluorescens were cultured as described previously in28.
Plasmid construction and transformation
To generate the phlG deletion mutant p299ΔphlG, fragments flanking the phlG gene were amplified with four pair of primers using 2P24 genomic DNA as template. Forward primer Ga (5′-GGCGGTACCTACCGGTCAGCATGTG-3′) and reverse primer Gb (5′-CAGGGATCCCTGCGAGCTGGGC-3′) were used to amplify the left flanking sequence of the phlG gene, and the right flanking sequence was amplified with forward primer Gc (5′-GAGGATCCGGTTGAGGTCTTC-3′) and reverse primer Gd (5′-GTCAAGCTTCTGGAGAGACGATCGG-3′). After digestion with the relevant restriction enzymes, PCR fragments Gab and Gcd were cloned into pHSG299 and pHSG399 (TaKaRa), respectively. After digestion of p399ΔGcd with Hind III and BamH I, the excised fragment was ligated into the corresponding plasmid p299ΔphlG, and the two fragments were ligated at the BamH I site, resulting in the p299ΔphlG mutant, which resulted in the sequence of 317 bp (247–567) of PhlG gene (GenBank: DQ083928.1) was deleted (Fig. S1). The construct was verified by diagnostic PCR by perimer pair of G1/G2 and Ga/Gd using 2P24 genomic DNA and plasmid p299△G (Fig. S1A, B). We found that 300 bp fragment from both diagnosis PCR was lost in p299△G.
To generate the complementation plasmid, the coding region of the phlG gene was amplified by PCR with primer pair G1 (5′- ATGAATTCCCGTCATTGTCCCTTTAC -3′) and G2 (5′-ATGTCGACACTTCTGCTGAACGG-3′). The amplified fragment was digested and ligated with plasmid pRK415 to generate recombinant plasmid p415-phlG referenced to Wei et al., 2005 and Wu et al., 2012, which was transformed into 2P24 and PM901, resulting in strains 2P24-G and PM901-G.
Plasmid p299-phlG and 3 kb LacZ gene fragments were digested with BamH I, and ligated to create p299-LacZ-G. The correct plasmid was transferred to the strain 2P24. The homologous recombination mutant 2P24-LacZ-G was screened X-gal-containing LB plates. Using strains 2P24-LacZ-G and 2P24-ΔG as templates, using primer G1: 5′-ATGTCGACACTTCTGCTGAACGG-3′ and G2: 5′-ATGTCGACACTTCTGCTGAACGG-3′ were subjected to PCR examination (Fig. S2C).
To obtain gacS gene from P. fluorescens 22P4, the chromosomal DNA of P. fluorescens 22P4 was digested with Mob l and ligated to pLAFRS with BamH l and Sac I. Then, it was transfected into E. coli DH5a after in vitro packaging to construct a genomic library. According to the published sequence of gacS gene, primers S-H: 5′-TCGGCATCAACCGCATGGC-3′ and S-G: 5′-GTGCCTTCGCGGGTGAACTT-3′ in the conserved region were designed. PCR amplification was performed using the strain 2P24 genome DNA as a template. The resulting fragment (0.54 kb) was verified by sequencing, and the 2P24 genomic library was screened by PCR using this as a marker. The verified PCR containing 3.2 kb fragment and 2.5 kb was shown in Fig. S2D. The resulting positive fragment was sub-cloned and ligated into plasmid pBluescript and verified by sequencing.
The construction of 2P24ΔGacS is referenced to Hailei Wei and Liqun Zhang29. Based on the obtained gacS gene sequence from 2P24 strain, primer S1: 5′-GATAAGCTTGGCAGCACTC-3′ and S3186: 5′- ATGGATCCAGCTTAACCGC-3′ were designed. PCR amplification was carried out using the strain 2P24 genome as a template. The PCR product was digested with the corresponding enzyme and ligated with pBluescript to construct the recombinant plasmid pS-BH. pS-BH was digested with EcoR I and self-ligated to obtain the recombinant vector pS-BEH. The The pS-BEH was digested with Sal l and BamH I, and ligated with the corresponding digested pSR47s to construct a suicide vector (pSR47s△S). The pSR47s△S was transformed into 2P24 by the method of parental hybridization, and the recombinant bacteria were screened by using kanamycin as a marker. The selected recombinant bacteria were cultured at 28°C for 36 h without antibiotic pressure, and continuously transferred to culture for 4 times. The strains which lost the kanamycin resistance were screened, and the second recombinant strain was produced, thereby obtaining the deletion mutant (2P24ΔGacS).
The rsmE gene of P. fluorescens 2P24 was found in the genome sketch of P. fluorescens 2P24 by homology alignment according to the sequence of other rsmE genes of P. fluorescens that have been reported. The left flanking fragment (1.015 kb) was amplified by primers RsmE-2285 (5′-ATC TGCAGAAGGGCCAGTACGGCTC-3′) and RsmE-3330 (5′-ATGGATCC TATGAGTGGGCGTTTCAGCC- 3′) using 2P24 genome DNA as template. After digestion with Pst I and BamH I, the fragment was ligated into the corresponding digested pHSG399 to obtain p399RsmE (L) and confirmed by sequencing. The primers RsmE-3498 and RsmE-4685 were designed for PCR amplification using the 2P24 strain genome as a template. The right flanking fragment (1.108 kb) was amplified with primers RsmE-3498 (5′- ATGGATCC TCACCGCCCCGACAAGCCGC - 3′) and RsmE-4658 (5′- CGGTGCTGTTCGAAATGGTGCG - 3′) using 2P24 genome DNA as template. The same digestion strategy was followed as used in p399RsmE (L) construction. Then, the plasmid p399RsmE (L) was cultured and digested with Pst I. The digested fragment was recovered and then ligated into the corresponding digested pHSG399RsmE(R), and the resulting vector was named pHSG399ΔRsmE.
The pHSG399ΔRsmE was digested with EcoR I-Sac I. The fragment was recovered and then ligated into the corresponding digested pBSKm. The resulting vector was named pBSKmΔRsmE. The vectors containing GacS and RsmE mutant were introduced into the mutant 2P24-LacZ-G by electroporation, and the two-step homologous recombination mutants lacking the GacS and RsmE genes were screened by PCR (Fig. S2E). The β-galactosidase enzyme activity of the mutant strain was simultaneously determined.
Measurement of β-galactosidase activity
The procedure for the construction of reporter fusions and measurement of β-galactosidase activity is described in Wu et al. (2012).
Determination of DAPG and MAPG production by TLC and LC assays
Strains were cultured overnight and adjusted to an absorbance at 600 nm (OD600) of 0.8 with LB. Control (CK) cultures were not mock inoculated. Next, 10 mg of 2,4-DAPG dissolved in methanol was added after 10 h and cultures were incubated at 28 °C with shaking at 150 rpm for 12, 24, 36, 48, or 60 h. Cells were centrifuged at 7000 g for 10 min and the supernatant was acidified to pH 2.0 with 1 M HCl. The organic phase was extracted by rotary evaporation with an equal volume of ethyl acetate, and the dry solid was dissolved in 0.15 mL of methanol. Samples were spotted on the bottom of a silicone plate, dried with a sterilised airflow, and separated by TLC using a solvent system comprised of chloroform:acetone (19:1 v:v). A 1–5% ferric chloride ethanol solution spray was applied for visualised of the developed plate. LC was used to assess 2,4-DAPG and MAPG standards.
Root colonisation test and wheat growth assay
Wheat seeds of uniform size without visible wounds were selected and used to investigate the in vivo biological activities of 2P24-ΔG and PM901-ΔG. Wild-type (WT) 2P24 and PM901 strains were included as controls. Strains 2P24, PM901, 2P24-ΔG and PM901-ΔG were separately inoculated into 5 mL of LB medium and incubated at 28 °C with shaking at 150 rpm for 24 h. When the culture concentration reached an OD600 value of 0.056 or 5×107 colony forming units (cfu)/mL, wheat seeds were soaked inside the culture for overnight. The wheat seeds were planted into a pot containing 30 g mixture of meteorite and soil. Three seeds per pot and 3 pots per treatment were prepared. At 7 days and 10 days, soil was gently removed from roots by shaking, and roots were placed in test tubes containing 10 mL of sterile water and agitated at 150 rpm for 10 min. The number of bacteria in the test tube was calculated, and the root length and plant fresh weight were determined. All tests were repeated for three times.
Statistical analysis
GraphPad Prism software version 5.01 (Graphpad Software, Inc.) was used for analysis of variance, followed by multiple comparisons using one way ANOVA, and p < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
We thank Drs. Gail P. Ferguson, Martin Schuster, Zhaoqing Luo and Rui Zhou for plasmids and reagents, and Mr. Zhenhua Guo for technical support. This work was supported by the National Natural Science Foundation of China (30860166, 31572049), Research projects of agricultural public welfare industry in China(201503109) and The National Key Research and Development Program of China (2017YFD0201101).
Author contributions
Conceived and designed the experiments: Hong-you Zhou and Li-qun Zhang; Performed the experiments: Ning Lyu, Xiao-gang Wu; Analyzed the data: Ming-min Zhao, Ning Lyu and Hong-you Zhou; Contributed reagents/materials/analysis tools: Ning Lyu, Dong Wang,Yuan-zheng Zhao; Wrote the paper: Ming-min Zhao and Hong-you Zhou. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ming-Min Zhao and Ning Lyu.
Supplementary information
is available for this paper at 10.1038/s41598-020-60555-9.
References
- 1.Tian T, Wu XG, Duan HM, Zhang LQ. The resistance-nodulation-division efflux pump EmhABC influences the production of 2,4-diacetylphloroglucinol in Pseudomonas fluorescens 2P24. Microbiology. 2010;156:39–48. doi: 10.1099/mic.0.031161-0. [DOI] [PubMed] [Google Scholar]
- 2.Haas D, Keel C. Regulation of antibiotic production in rootcolonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 2003;41:117–153. doi: 10.1146/annurev.phyto.41.052002.095656. [DOI] [PubMed] [Google Scholar]
- 3.Paulsen IT, et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 2005;23(7):873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saitou H, Watanabe M, Maruyama K. Molecular and catalytic properties of 2,4-diacetylphloroglucinol hydrolase (PhlG) from Pseudomonas sp. YGJ3. Biosci. Biotechnol. Biochem. 2012;76(6):1239–1241. doi: 10.1271/bbb.120054. [DOI] [PubMed] [Google Scholar]
- 5.Keel C, et al. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 1996;62:552–563. doi: 10.1128/AEM.62.2.552-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raaijmakers JM, Weller DM. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol. Plant-Microbe Interact. 1998;11:144–152. doi: 10.1094/MPMI.1998.11.2.144. [DOI] [Google Scholar]
- 7.Sharifi-Tehrani A, Zala M, Natsch A, Moenne-Loccoz Y, Defago G. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol- producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur. J. Plant. Pathol. 1998;104:631–643. doi: 10.1023/A:1008672104562. [DOI] [Google Scholar]
- 8.Delany I, et al. Regulation of production of the antifungal metabolite 2,4-diacetylphloroglucinol in Pseudomonas fluorescens F113: genetic analysis of PhlF as a transcriptional repressor. Microbiology. 2000;146:537–546. doi: 10.1099/00221287-146-2-537. [DOI] [PubMed] [Google Scholar]
- 9.De Souza JT, Weller DM, Raaijmakers JM. Frequency, diversity, and activity of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. In Dutch take-all decline soils. Phytopathology. 2003;93:54–63. doi: 10.1094/PHYTO.2003.93.1.54. [DOI] [PubMed] [Google Scholar]
- 10.Bottiglieri M, Keel C. Characterization of PhlG, a hydrolase that specifically degrades the antifungal compound 2,4-diacetylphloroglucinol in the biocontrol agent Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 2006;72:418–427. doi: 10.1128/AEM.72.1.418-427.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weller DM, et al. Thomashow LS. Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant. Biol. 2007;9(1):4–20. doi: 10.1055/s-2006-924473. [DOI] [PubMed] [Google Scholar]
- 12.Gong L, et al. Novel synthesized 2, 4-DAPG analogues: antifungal activity, mechanism and toxicology. Sci. Rep. 2016;26(6):32266. doi: 10.1038/srep32266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bangera MG, Thomashow LS. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2–87. J. Bacteriol. 1999;181:3155–3163. doi: 10.1128/JB.181.10.3155-3163.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnider-Keel U, et al. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 2000;182(5):1215–1225. doi: 10.1128/JB.182.5.1215-1225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas D, Defago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 16.Zuber S, et al. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 2003;16:634–644. doi: 10.1094/MPMI.2003.16.7.634. [DOI] [PubMed] [Google Scholar]
- 17.Sarniguet A, Kraus J, Henkels MD, Muehlchen AM, Loper JE. The sigma factorδS affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc. Natl Acad. Sci. 1995;92:12255–12259. doi: 10.1073/pnas.92.26.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnider U, et al. Amplification of the house-keeping sigma factor in Pseudomonas fluorescens CHA0 enhances antibiotic production and improves biocontrol abilities. J. Bacteriol. 1995;177:5387–5392. doi: 10.1128/JB.177.18.5387-5392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pechy-Tarr M, et al. RpoN (sigma54) controls production of antifungal compounds and biocontrol activity in Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 2005;18:260–272. doi: 10.1094/MPMI-18-0260. [DOI] [PubMed] [Google Scholar]
- 20.Heeb S, Valverde C, Gigot-Bonnefoy C, Haas D. Role of the stress sigma factor RpoS in GacA/RsmA-controlled secondary metabolism and resistance to oxidative stress in Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 2005;243:251–258. doi: 10.1016/j.femsle.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Reimmann C, Valverde C, Kay E, Haas D. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 2005;187(1):276–85. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valverde C, Heeb S, Keel C, Haas D. RsmY. A small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbio. 2003;50(4):1361–1379. doi: 10.1046/j.1365-2958.2003.03774.x. [DOI] [PubMed] [Google Scholar]
- 23.Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl Acad. Sci. 2005;102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He YX, et al. Crystal structure and computational analyses provide insights into the catalytic mechanism of 2,4-diacetylphloroglucinol hydrolase PhlG from Pseudomonas fluorescens. J. Biol. Chem. 2010;285(7):4603–4611. doi: 10.1074/jbc.M109.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanisich VA, Holloway BW. A mutant sex factor of Pseudomonas aeruginosa. Genet. Res. 1972;19:91–108. doi: 10.1017/S0016672300014294. [DOI] [PubMed] [Google Scholar]
- 26.Baehler E, Bottiglieri M, Pechy-Tarr M, Maurhofer M, Keel C. Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 2005;99:24–38. doi: 10.1111/j.1365-2672.2005.02597.x. [DOI] [PubMed] [Google Scholar]
- 27.Brodhagen M, Henkels MD, Loper JE. Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 2004;70:1758–1766. doi: 10.1128/AEM.70.3.1758-1766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Liu J, Zhang W, Zhang L. Multiple-level regulation of 2,4-diacetylphloroglucinol production by the sigma regulator PsrA in Pseudomonas fluorescens 2P24. PLoS One. 2012;7(11):e50149. doi: 10.1371/journal.pone.0050149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hailei Wei & Liqun Zhang. Cloning and functional characterization of the gacS gene of the biocontrol strain Pseuodomonas fluorescens 2P24. Acta Microbiologica Sinica, 45(3), 10.13343/j. cnki. wsxb.2005.03.011”. (2005). [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.