Abstract
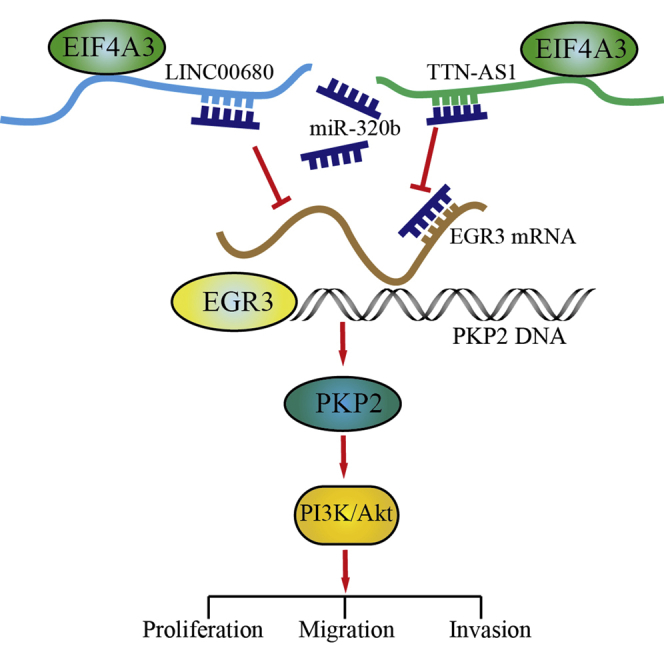
Glioblastomas are the most common and malignant intracranial tumors with a low survival rate. Dysregulation of long non-coding RNAs and RNA-binding protein causes various diseases, including cancers. However, the function of LINC00680 and TTN-AS1 in the progression of glioblastomas is still elusive. In this study, we detected that LINC00680 and TTN-AS1 were upregulated in glioblastoma cells. RNA-binding protein EIF4A3 could prolong the half-life of LINC00680 and TTN-AS1. Knockdown of EIF4A3, LINC00680, and TTN-AS1 impaired proliferation, migration, and invasion and inhibited the growth of tumor in vivo and promoted apoptosis of glioblastoma cells. miR-320b was proven to be a target of LINC00680 and TTN-AS1. They interacted with miR-320b as competing endogenous RNAs, which resulted in the reduction of binding between transcriptional factor EGR3 (early growth response 3) mRNA and miR-320b. The accumulation of EGR3 promoted expression of plakophilin (PKP)2, which could activate the epidermal growth factor receptor (EFGR) pathway, leading to the malignant biological behaviors of glioblastoma cells. In summary, LINC00680 and TTN-AS1 promoted glioblastoma cell malignant biological behaviors via the miR-320b/EGR3/PKP2 axis by being stabilized by EIF4A3, which may provide a novel strategy for glioblastoma therapy.
Keywords: LINC00680, TTN-AS1, EIF4A3, miR-320b, EGR3, glioblastoma
Graphical Abstract
Introduction
Gliomas are the most common primary brain tumors and represent about 80% of malignant brain tumors, and about 50% of the patients present the most lethal form, glioblastoma.1 Despite the major advances in therapy for gliomas, the overall 5-year survival of patients with glioblastomas is less than 5%.2 The invasive growth of the tumor cells and resistance to the induction of cell death make it difficult for systemic therapy.3 Thus, it is urgent to identify an effective molecular therapy for glioblastomas.
RNA-binding proteins (RBPs) play an important role in many cellular processes. RBPs conduct their functions by interacting with different kinds of target RNAs and forming ribonucleoprotein (RNP) complexes. Dysregulation of RBPs is involved in the progression of different diseases, cancers included. For instance, HuR could bind to LINC00324 to increase the stability of FAM83B mRNA and promote the proliferation of gastric cancer cells.4 EIF4A3 is one of the three core components of the exon junction complex (EJC), which is critical for RNA splicing, location, and degradation. The EJC is vital in brain development, neuronal outgrowth, and neuronal activity.5 However, the role of EIF4A3 in the development of glioblastomas is still elusive.
Long non-coding RNAs (lncRNAs) are a group of RNAs with a molecular weight of >200 bases in length that generally do not code for proteins. lncRNAs have shown to be involved in modulation of many aspects of the tumor biological process. SNHG-12 and NEAT1, which were expressed aberrantly in glioma tissues and cell lines, regulate the biological behaviors of glioma cells.6,7 LINC00680 might be a protective biomarker and an independent prognostic indicator of soft-tissue sarcoma.8 TTN-AS1 promotes esophageal squamous cell carcinoma cell proliferation and invasion.9 The clinical significance of lncRNA LINC00680 and TTN-AS1 in glioblastomas remains unclear.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs of 18–24 nt, acting as modulators in post-transcriptional and translational gene expression. miRNAs can bind to Argonaute (Ago) protein and form an RNA-induced silencing complex (RISC), in which miRNAs are able to recognize and bind to the 3′ UTR (untranslated region) of target mRNA so that target mRNAs can be cleaved with perfect complementarity or translational repression with imperfect complementarity.10 Upregulation of miR-320b could promote apoptosis and inhibit proliferation, migration, and invasion of glioma cells.11 However, its mechanism is indistinct.
EGR3 (early growth response 3) is a member of the zinc finger transcription factor family. This family contains a three-zinc finger motif that can bind to a 9-bp consensus sequence of GC-rich consensus DNA, the EGR response element.12 EGR3 has diverse functions on tumor biological behavior.13,14 EGR3 is highly expressed in glioblastoma.15 However, the function of EGR3 on biological behavior and its mechanism in glioblastomas have not been stated.
Plakophilin (PKP)2 is a member of the armadillo-like protein subfamily that has another three members, PKP1, PKP3, and PKP4. PKP proteins localize to cell desmosomes, and its mutations are related to arrhythmogenic cardiomyopathy.16 PKP2 could directly activate epidermal growth factor receptor (EGFR) signaling.17 It has been reported that PKP2 is upregulated and correlated to the progression of gliomas. Knockdown of PKP2 could inhibit proliferation, migration, and invasion of glioma cells.18 However, its mechanism is still unclear.
In this study, we investigated the expression of EIF4A3, LINC00680, TTN-AS1, miR-320b, and EGR3 in glioma tissues and cells. Furthermore, we examined the role of EIF4A3, LINC00680, TTN-AS1, miR-320b, EGR3, and PKP2 in the regulation of glioma malignant progression and their interactions. These results may provide a potential strategy for glioblastoma therapy.
Results
EIF4A3 Promoted Malignant Biological Behaviors of Glioblastoma Cells by Prolonging the Half-Life of LINC00680 and TTN-AS1
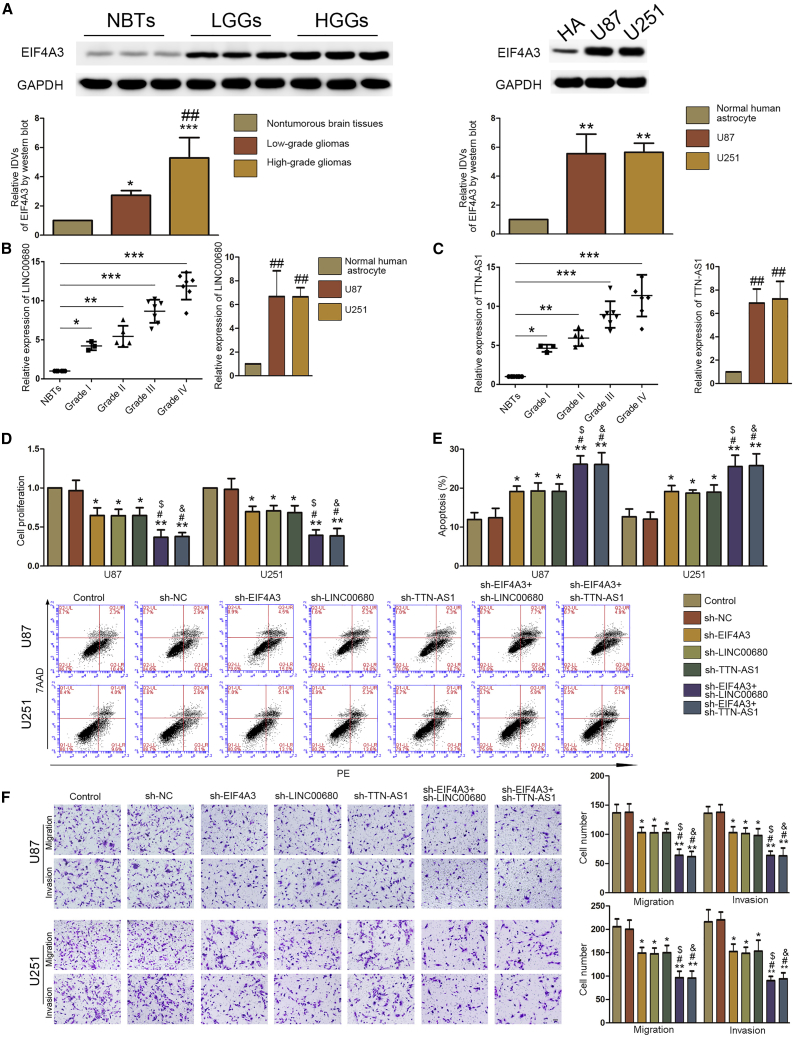
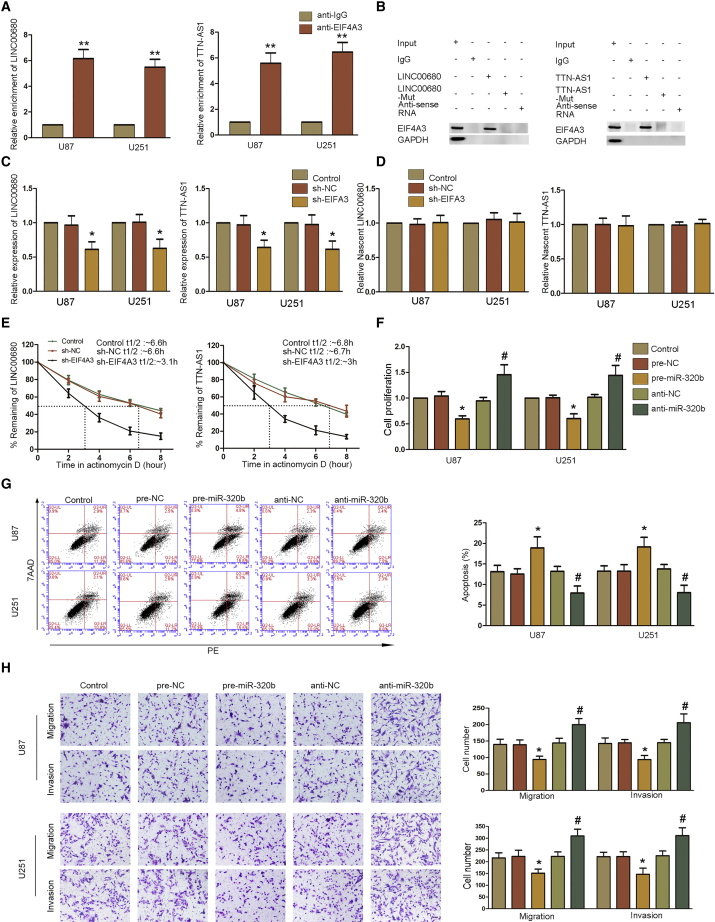
First, we determined the expression of EIF4A3 in glioma tissues and cells by western blot. EIF4A3 was significantly upregulated in glioma tissues and cells (U87, U251, and BTIC [brain tumor-initiating cell]) (Figure 1A; Figure S2B). We found that EIF4A3 was highly expressed in glioma tissues in the Oncomine database, and lower expression of EIF4A3 presented a longer survival in the GEPIA database (Figures S1G and S1I). By searching bioinformatics with Starbase v2.0 software, we found that there were putative binding sites between EIF4A3 and LINC00680 or TTN-AS1 (Figures S1A and S1B). Then, the expression of LINC00680 and TTN-AS1 in glioma tissues and cells (U87, U251, and BTIC) was detected using qRT-PCR. As shown in Figures 1B, 1C, and S2C, LINC00680 and TTN-AS1 were significantly upregulated in glioma tissues and cells. For further study, we constructed cell lines of short hairpin RNA (shRNA)-induced (sh-)EIF4A3, sh-LINC00680, sh-TTN-AS1, sh-EIF4A3+sh-LINC00680, and sh-EIF4A3+sh-TTNA-AS1. Compared to sh-negative control (NC) cells, knockdown of EIF4A3, LINC00680, or TTN-AS1 could significantly decrease the ability of proliferation, migration, and invasion and promote apoptosis of glioblastoma cells. Meanwhile, co-silencing of EIF4A3 and LINC00680 or co-silencing of EIF4A3 and TTN-AS1 could significantly inhibit proliferation, migration, and invasion and promote apoptosis of glioblastoma cells compared to sh-EIF4A3, sh-LINC00680, and sh-TTN-AS1 (Figures 1D–1F; Figures S2D–S2F). To verify the interaction of EIF4A3 and LINC00680 or TTN-AS1, we performed RNA immunoprecipitation (RIP) and RNA pull-down assays. LINC00680 and TTN-AS1 were significantly enriched in anti-EIF4A3 compared to anti-immunoglobulin G (IgG) (Figure 2A). RNA pull-down verified that EIF4A3 could bind to LINC00680 and TTN-AS1 (Figure 2B). The expression of LINC00680 and TTN-AS1 in sh-EIF4A3 cells was significantly downregulated compared to sh-NC cells (Figure 2C; Figure S2G). A nascent RNA capture assay showed that there was no significant difference between sh-EIF4A3 cells and sh-NC cells (Figure 2D). However, the half-life of LINC00680 and TTN-AS1 was shortened in sh-EIF4A3 cells compared to sh-NC cells (Figure 2E). All of these results proved that EIF4A3 could bind to LINC00680 and TTN-AS1 and help stabilize them to promote malignant biological behaviors of glioblastoma cells.
Figure 1.
EIF4A3, LINC00680, and TTN-AS1 Served as Oncogenes in Glioblastoma Cells
(A) Western blot was used to detect expression of EIF4A3 in glioma tissues (left) and cells (right). Data are presented as the mean ± SD (n = 8, NBTs; n = 3, grade I; n = 5, grade II; n = 7, grade III; n = 7, grade IV; n = 3 for each cell line). Left: *p < 0.05 versus nontumorous brain tissues; ***p < 0.001 versus nontumorous brain tissues; ##p < 0.01 versus low-grade glioma tissues. Right: **p < 0.01 versus normal human astrocytes. (B) Expression of LINC00680 in glioma tissues (left) and cells (right) was detected by real-time PCR. Data are presented as the mean ± SD (n = 8, NBTs; n = 3, grade I; n = 5, grade II; n = 7, grade III; n = 7, grade IV; n = 3 for each cell line). *p < 0.05 versus NBTs group; **p < 0.01 versus NBTs group; ***p < 0.001 versus NBTs group; ##p < 0.01 versus normal human astrocytes group. (C) Expression of TTN-AS1 in glioma tissues (left) and cells (right) was detected by real-time qPCR. Data are presented as the mean ± SD (n = 8, NBTs; n = 3, grade I; n = 5, grade II; n = 7, grade III; n = 7, grade IV). *p < 0.01 versus NBTs group; ***p < 0.001 versus NBTs group; ##p < 0.01 versus normal human astrocytes group. (D) A CCK-8 assay was conducted to investigate the effect of EIF4A3, LINC00680, and TTN-AS1 on proliferation in U87 and U251 cells. (E) Flow cytometry analysis of EIF4A3, LINC00680, and TTN-AS1 knockdown in U87 and U251 cells. (F) Transwell assays were used to measure the effect of EIF4A3, LINC00680, and TTN-AS1 on cell migration and invasion in U87 and U251 cells. Representative images and statistical plots are presented. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group (empty vector); **p < 0.01 versus sh-NC group; #p < 0.05 versus sh-EIF4A3 group; $p < 0.05 versus sh-LINCC0680 group; &p < 0.05 versus sh-TTN-AS1 group. Scale bar represents 80 μm.
Figure 2.
EIF4A3 Stabilized LINC00680 and TTN-AS1; miR-320b Promoted Malignant Biological Behaviors of Glioblastoma Cells
(A) A RIP assay showed that LINC00680 (left) and TTN-AS1 (right) could bind with EIF4A3. Real-time PCR was used to measure enrichment of LINC00680 and TTN-AS1. Data represent mean ± SD (n = 3 in each group). **p < 0.01 versus anti-IgG group. (B) RNA pull-down assay indicated EIF4A3 binding with LINC00680 (left) and TTN-AS1 (right). (C) Expression of LINC00680 (left) and TTN-AS1 (right) was detected in U87 and U251 cells with EIF4A3 knockdown by PCR. Data represent mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group. (D) A nascent RNA capture assay showed no significant difference between the sh-EIF4A3 and sh-NC group in the expression of LINC006780 and TTN-AS1. (E) Relative levels of LINC00680 (left) and TTN-AS1 (right) after U87 and U251 cells with EIF4A3 knockdown being treated in actinomycin D for different periods were measured by PCR. (F) CCK-8 assay was conducted to investigate the effect of miR-320b on proliferation in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus pre-NC group; #p < 0.05 versus anti-NC group. (G) Flow cytometry analysis effect of miR-320b on apoptosis in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus pre-NC group; #p < 0.05 versus anti-NC group. (H) Transwell assays were used to measure the effect of miR-320b on cell migration and invasion in U87 and U251 cells. Representative images and statistical plots are presented. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus pre-NC group; #p < 0.05 versus anti-NC group. Scale bar represents 80 μm.
miR-320b Inhibited Proliferation, Migration, and Invasion of Glioblastoma Cells by Targeting LINC00680 and TTN-AS1
The expression of miR-320b was downregulated in glioblastoma cells and tissues.11 We predicted via bioinformatics software LncBase that there were binding sites between miR-320b and LINC00680 or TTN-AS1 (Figures S1C and S1D). The effects of miR-320b on biological behaviors of glioblastoma cells are shown in Figures 2F–2H and S2H–S2J. As shown in Figures 2F and S2H, knockdown of miR-320b could promote proliferation of glioblastoma cells, and overexpression of miR-320b could inhibit proliferation of glioblastoma cells. Knockdown of miR-320b could decrease apoptosis of glioblastoma cells, and overexpression of miR-320b could increase apoptosis of glioblastoma cells (Figure 2G; Figure S2I). Knockdown of miR-320b could promote migration and invasion of glioblastoma cells. Overexpression of miR-320b had the opposite results (Figure 2H; Figure S2J).
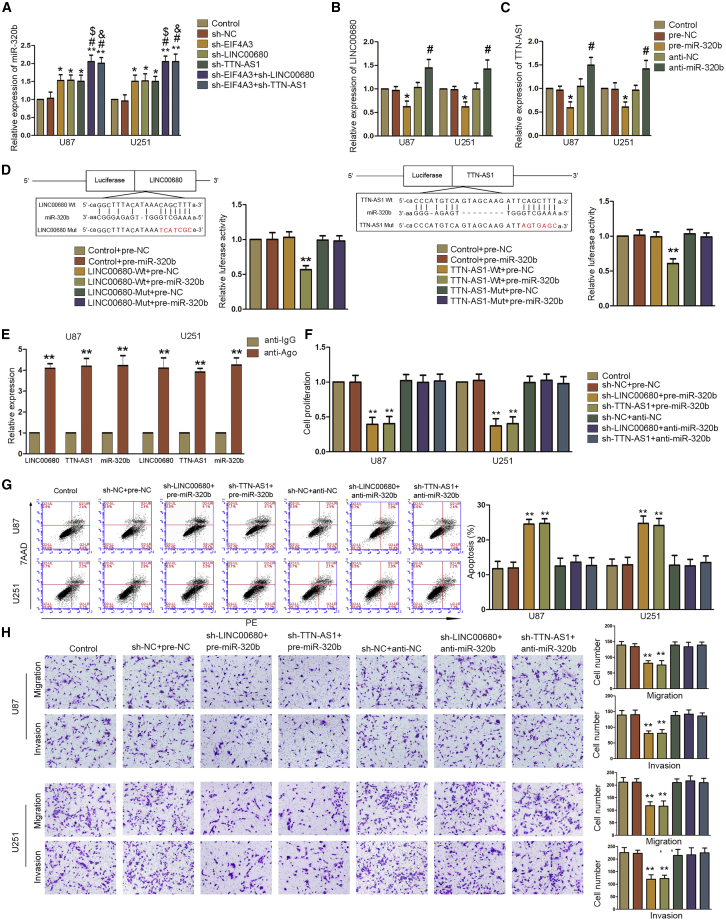
To determine how LINC00680 and TTN-AS1 interact with miR-320b, we detected the expression of miR-320b in different cell lines. Compared to sh-NC cells, silencing EIF4A3, LINC00680, or TTN-AS1 could upregulate the expression of miR-320b. Co-silencing EIF4A3 and LINC00680 or co-silencing EIF4A3 and TTN-AS1 could significantly increase the expression of miR-320b compared to silencing EIF4A3, LINC00680, or TTN-AS1 alone (Figure 3A; Figure S3A). Knockdown or overexpression of miR-320b could upregulate or downregulate LINC00680 and TTN-AS1, respectively (Figures 3B and 3C; Figure S3B). A dual-luciferase reporter assay showed that co-transfection of wild-type (Wt) LINC00680 or TTN-AS1 and miR-320b could reduce fluorescence (Figure 3D). LINC00680, TTN-AS1, and miR-320b were much more enriched in the anti-Ago group than that in the anti-IgG group (Figure 3E). These results suggested that LINC00680 or TTN-AS1 could interact with miR-320b to form RISC. To investigate how LINC00680 or TTN-AS1 and miR-320b influence biological behaviors of glioblastoma cells, we constructed sh-LINC00680+anti-miR-320b, sh-TTN-AS1+anti-miR-320b, sh-LINC00680+pre-miR-320b, sh-TTN-AS1+pre-miR-320b, and their negative control cell lines. As Figures 3F–3H and S3C–S3E show, knockdown of LINC00680 and TTN-AS1 while overexpression of miR-320b could significantly inhibit proliferation, migration, and invasion and increase apoptosis of glioblastoma cells. Nevertheless, knockdown of miR-320b could entirely invert the effect of knockdown of LINC00680 or TTN-AS1 on the biological behaviors of glioblastoma cells.
Figure 3.
LINC00680 and TTN-AS1 Were Targets of miR-320b: Reintroducing miR-320b Impaired Malignant Behavior of Glioblastoma Cells Induced by LINC00680 and TTN-AS1
(A) Real-time PCR analysis for EIF4A3, LINC00680, and TTN-AS1 regulating miR-320b expression in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group; **p < 0.01 versus sh-NC group; #p < 0.05 versus sh-EIF4A3 group; $p < 0.05 versus sh-LINCC0680 group; &p < 0.05 versus sh-TTN-AS1 group. (B) Real-time PCR analysis for miR-320b modulating LINC00680 expression in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus pre-NC group; #p < 0.05 versus anti-NC group. (C) Real-time PCR analysis for miR-320b modulating TTN-AS1 expression in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus pre-NC group; #p < 0.05 versus anti-NC group. (D) Putative binding sites of LINC00680 (left) and TTN-AS1 (right) with miR-320b are indicated. Relative luciferase activity was conducted after cells were transfected with LINC00680/TTN-AS1-3′ UTR-wild-type (Wt) or LINC00680/TTN-AS1-3′ UTR-mutant (Mut). Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus LINC00680/TTN-AS1-3′ UTR-Wt+pre-NC group. (E) miR-320b was identified in the LINC00680-miR-320b and TTN-AS1-miR-320b RISC complex. Enrichment of LINC00680, TTN-AS1, and miR-320b was measured via real-time PCR. Data represent mean ± SD (n = 3 in each group). **p < 0.01 versus anti-IgG group. (F) A CCK-8 assay was conducted to investigate the effect of LINC00680/TTN-AS1 and miR-320b on proliferation in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus sh-NC+pre-NC group. (G) Flow cytometry analysis of LINC00680/TTN-AS1 and miR-320b in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus sh-NC+pre-NC group. (H) Transwell assays were used to measure the effect of LINC00680/TTN-AS1 and miR-320b on cell migration and invasion in U87 and U251 cells. Representative images and statistical plots are presented. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus sh-NC+pre-NC group. Scale bar represents 80 μm.
EGR3 Was a Target of miR-320b and Promoted Proliferation, Migration, and Invasion of Glioblastoma Cells
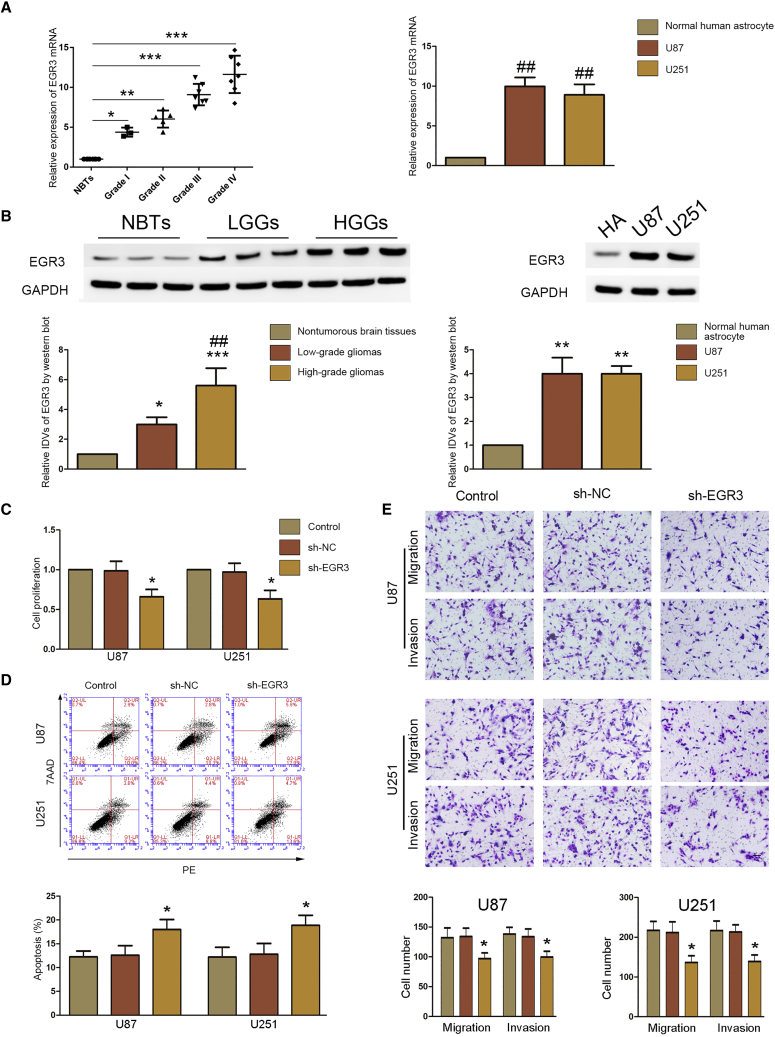
With the help of the bioinformatics software TargetScan, we predicted that EGR3 was the target of miR-320b (Figure S1E). The expression of mRNA and protein of EGR3 was upregulated in glioblastoma cells and tissues (Figure 4A and 4B; Figures S3F and S3G). We found that EGR3 was highly expressed in glioma tissues in the Oncomine database, and lower expression of EGR3 presented a longer survival in database GEPIA (Figures S1G and S1H). A Cell Counting Kit-8 (CCK-8) and transwell assay showed that knockdown of EGR3 could inhibit proliferation, migration, and invasion of glioblastoma cells (Figures 4C and 4E; Figure S3H and S3J). A flow cytometry assay proved that knockdown of EGR3 increased apoptosis of glioblastoma cells (Figure 4D; Figure S3I).
Figure 4.
EGR3 Acts as an Oncogene in Glioma Tissues and Cells
(A) Expression of EGR3 mRNA in glioma tissues (left) and cells (right) was detected by real-time PCR. Data are presented as the mean ± SD (n = 8, NBTs; n = 3, grade I; n = 5, grade II; n = 7, grade III; n = 7, grade IV; n = 3 for each cell line). *p < 0.05 versus NBTs group; **p < 0.01 versus NBTs group; ***p < 0.001 versus NBTs group; ##p < 0.01 versus normal human astrocytes group. (B) western blot was used to detect expression of EGR3 in glioma tissues (left) and cells (right). Data are presented as the mean ± SD (n = 8, NBTs; n = 3, Grade I; n = 5, Grade II; n = 7, Grade III; n = 7, Grade IV; n = 3 each cell line). Left: *p < 0.05 versus nontumorous brain tissues; ***p < 0.001 versus nontumorous brain tissues; ##p < 0.01 versus low-grade glioma tissues. Right: **p < 0.01 versus normal human astrocytes. (C) A CCK-8 assay was conducted to investigate the effect of EGR3 on proliferation in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group. (D) Flow cytometry analysis of EGR3 in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group. (E) Transwell assays were used to measure the effect of EGR3 on cell migration and invasion in U87 and U251 cells. Representative images and statistical plots are presented. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group. Scale bar represents 80 μm.
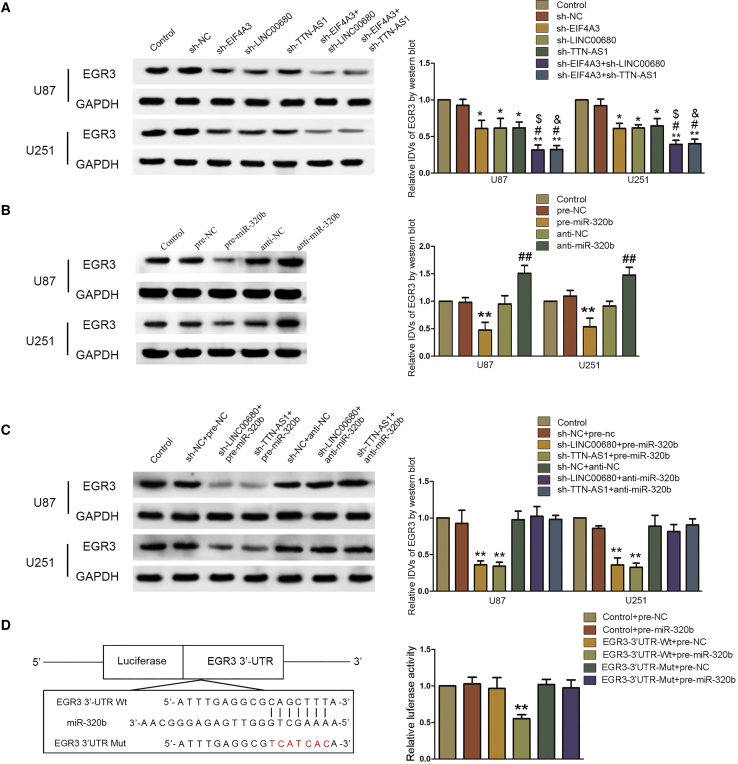
To verify that EGR3 was involved in the EIF4A3/LINC00680, TTN-AS1/miR-320b regulatory axis, we detected the expression of EGR3 in knocked down EIF4A3A, LINC00680 and TTN-AS1 cells. Compared to the sh-NC group, EGR3 was expressed at lower levels in sh-EIF4A3, sh-LINC00680, and sh-TTN-AS1 cells (Figure 5A; Figure S4A). Furthermore, EGR3 was significantly lower expressed in sh-EIF4A3+sh-LINC00680 and sh-EIF4A3+sh-TTN-AS1 cells compared to sh-EIF4A3, sh-LINC00680, and sh-TTN-AS1 cells, respectively (Figure 5A; Figure S4A). However, knockdown or overexpression of miR-320b could upregulate or downregulate the expression of EGR3 (Figure 5B; Figure S4B). Overexpression of miR-320b could strengthen the downregulation of EGR3 caused by knockdown of LINC00680 or TTN-AS1, while knockdown of miR-320b could entirely invert the downregulation of EGR3 caused by knockdown of LINC00680 or TTN-AS1 (Figure 5C; Figure S4C). A dual-luciferase reporter assay indicated that miR-320b could bind to EGR3 (Figure 5D).
Figure 5.
EGR3 Was a Target of miR-320b and Was Regulated by LINC00680, TTN-AS1, and miR-320b
(A) Expression of EGR3 was regulated in U87 and U251 cell lines with knockdown of EIF4A3 and/or LINC00680/TTN-AS1. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group; **p < 0.01 versus sh-NC group; #p < 0.05 versus sh-EIF4A3 group; $p < 0.05 versus sh-LINCC0680 group; &p < 0.05 versus sh-TTN-AS1 group. (B) Expression of EGR3 was regulated in U87 and U251 cell lines with knockdown or overexpression of miR-320b. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus pre-NC group; ##p < 0.01 versus anti-NC group. (C) Expression of EGR3 was regulated by LINC00680/TTN-AS1 and miR-320b. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus sh-NC+pre-NC group. (D) Putative binding sites of miR-320b and the 3′ UTR of EGR3 mRNA are indicated. Relative luciferase activity was conducted after cells were transfected with EGR3-3′ UTR-Wt or EGR3-3′ UTR-Mut. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus EGR3-3′UTR-Wt+pre-NC group.
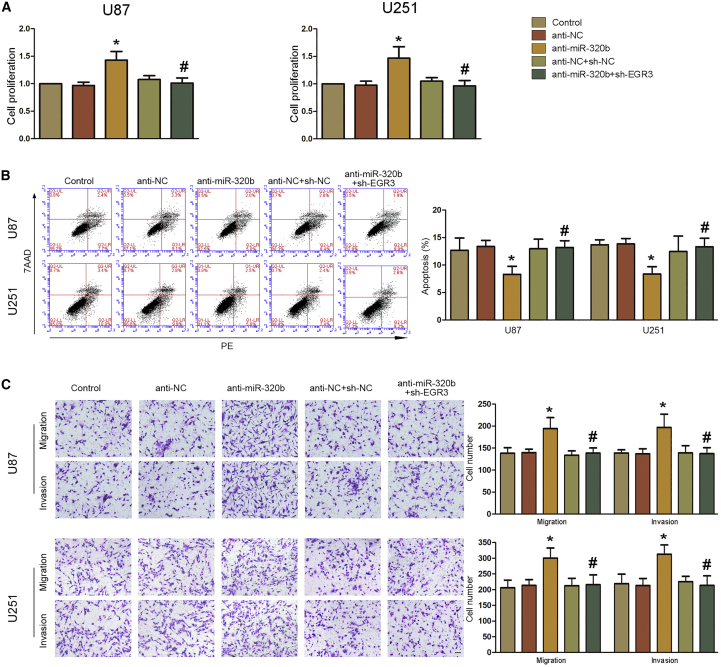
To further determine the effect of miR-320b and EGR3 on biological behaviors of glioblastoma cells, we constructed anti-miR-320b+sh-EGR3 cells and their negative controls. As Figures 6A–6C and S3K–S3M show, knockdown of miR-320b could promote proliferation, migration, and invasion of glioblastoma cells, while this effect could be inverted by knockdown of EGR3. Knockdown of miR-320b could decrease apoptosis of glioblastoma cells, and the effect could be inverted by knockdown of EGR3. These results indicated that EGR3 could entirely invert the effect of miR-320b on biological behaviors of glioblastoma cells.
Figure 6.
EGR3 Could Reverse the Biological Function of miR-320b in Glioblastoma Cells
(A) CCK-8 assays were performed in U87 and U251 cells with the altered expression of miR-320b and EGR3. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.01 versus anti-NC group; #p < 0.05 versus anti-miR-320b group. (B) Flow cytometry analysis of U87 and U251 cells with the altered expression of miR-320b and EGR3. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus anti-NC group; #p < 0.05 versus anti-miR-320b group. (C) Quantification number of migration and invasion cells treated with anti-miR-320b and sh-EGR3. Representative images and statistical plots are presented. Scale bar represents 80 μm. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus anti-NC group; #p < 0.05 versus anti-miR-320b group.
PKP2 Was a Target of EGR3 and Was Involved in EGR3-Induced Regulation of Glioblastoma Cell Malignant Progression
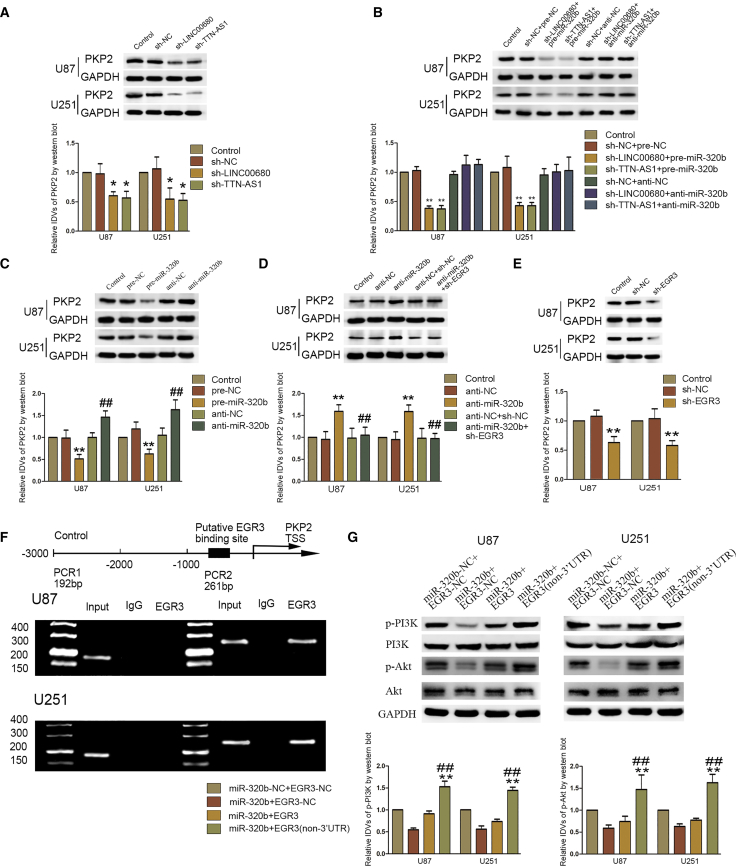
PKP2 was a predicted target of EGR3 via the bioinformatics software Jaspar (Figure S1F). The expression of PKP2 was correlated to grades of glioblastoma, and inhibition of PKP2 could inhibit proliferation, migration, and invasion of glioblastoma cells.18 Knockdown of LINC00680 or TTN-AS1 could significantly reduce the expression of PKP2 in glioblastoma cells (Figure 7A; Figure S4D); however, knockdown of miR-320b could invert this effect, and overexpression of miR-320b could strengthen this effect (Figure 7B; Figure S4E). Knockdown or overexpression of miR-320b could upregulate or downregulate the expression of PKP2, respectively (Figure 7C; Figure S4F). Furthermore, the effect of miR-320b knockdown on the expression of PKP2 could be completely inverted by knockdown of EGR3 (Figure 7D; Figure S4G). Knockdown of EGR3 could downregulate the expression of PKP2 (Figure 7E; Figure S4H). As Figure 7F shows, Chromatin Immunoprecipitation (ChIP) confirmed that EGR3 could bind to the promotor of PKP2.
Figure 7.
PKP2 Was Regulated by LINC00680, TTN-AS1, miR-320b, and EGR3
(A) Western blot analysis of PKP2 in U87 and U251 cell knockdown of LINC00680 or TTN-AS1. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group. (B) Western blot analysis of PKP2 in U87 and U251 cells regulated by LINC00680/TTN-AS1 and miR-320b. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus sh-NC+pre-NC group. (C) Western blot analysis of PKP2 in U87 and U251 cells regulated by miR-320b. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus pre-NC group; ##p < 0.01 versus anti-NC group. (D) Western blot analysis of PKP2 in U87 and U251 cells regulated by miR-320b and EGR3. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus anti-NC group; ##p < 0.05 versus anti-miR-320b group. (E) Western blot analysis of PKP2 in U87 and U251 cells regulated by EGR3. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus sh-NC group. (F) Putative EGR3 binding sites are indicated. Immunoprecipitated DNA was amplified by PCR. (G) Expression of p-PI3K and p-Akt was regulated by mir-320b and EGR3. Data are presented as the mean ± SD (n = 3 in each group). **p < 0.01 versus miR-320b+EGR3-NC group. ##p < 0.01 versus miR-320b+EGR3 group.
PKP2 may take effect by activating the EGFR pathway. As shown in Figure 7G, miR320b could significantly downregulate expression of phosphorylated (p-)phosphatidylinositol 3-kinase (PI3K) and p-Akt. Binding of miR-320b and the 3′ UTR of EGR3 mRNA could downregulate expression of p-PI3K and p-Akt. However, EGR3 without the 3′ UTR could reverse the effect.
Knockdown of EIF4A3, LINC00680, or TTN-AS1 Could Inhibit Tumor Growth In Vivo
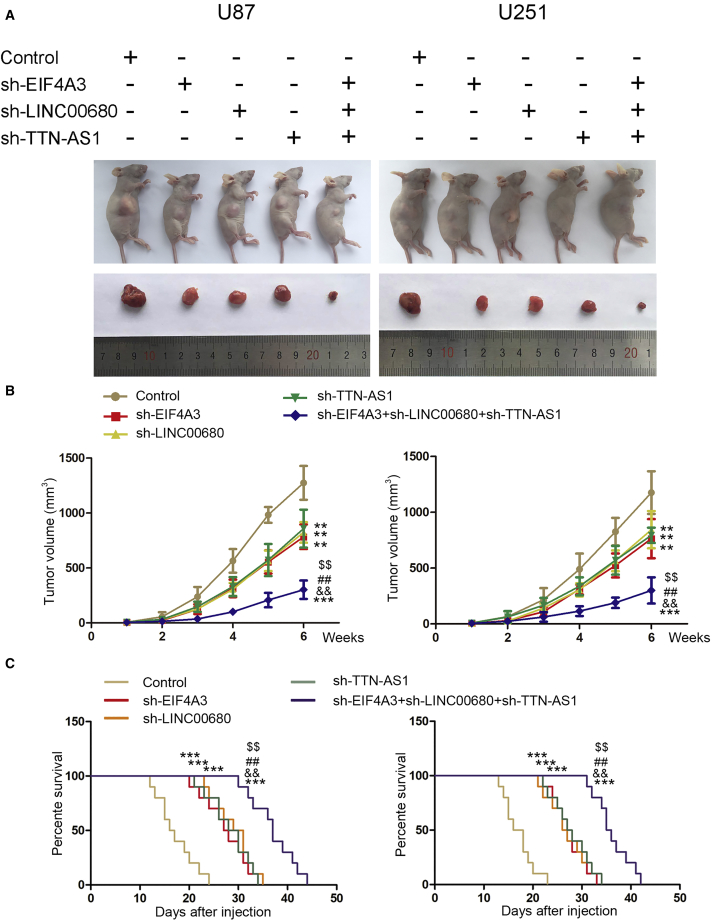
To study the effects of these molecules in vivo, we constructed sh-EIF4A3, sh-LINC00680, sh-TTN-AS1, and sh-EIF4A3+LINC00680+TTN-AS1 cells. Compared to controls, knockdown of EIF4A3, LINC00680, and TTN-AS1 could significantly reduce the volume of tumor and prolong survival of nude mice. sh-EIF4A3+LINC00680+TTN-AS1 could significantly reduce the volume of tumors and prolong the survival of nude mice compared to sh-EIF4A3, sh-LINC00680, and sh-TTN-AS1 (Figures 8A–8C).
Figure 8.
In Vivo Study
(A) The stable expressing cells were used for the in vivo study. The nude mice carrying tumors from respective groups are shown. The sample tumors from respective groups are shown. (B) Tumor volume was calculated every week after injection, and the tumor was excised after 6 weeks. **p < 0.01, ***p < 0.001 versus control group; $$p < 0.01 versus sh-EIF4A3 group; ##p < 0.01 versus sh-LINC00680 group; &&p < 0.01 versus sh-TTN-AS1 group. (C) The survival curves of nude mice with xenografts injected into the right striatum (n = 10). ***p < 0.001 versus control group; $$p < 0.01 versus sh-EIF4A3 group; ##p < 0.01 versus sh-LINC00680 group; &&p < 0.01 versus sh-TTN-AS1 group.
Discussion
In the present study, we have confirmed that EIF4A3, LINC00680, and TTN-AS1 were highly expressed in glioblastoma cells and tissues. EIF4A3 could help prolong the half-life of LINC00680 and TTN-AS1. Knockdown of EIF4A3, LINC00680, or TTN-AS1 inhibited proliferation, migration, and invasion and promoted apoptosis of glioblastoma cells. However, miR-320b had an opposite effect on glioblastoma cells compared to EIF4A3, LINC00680, or TTN-AS1. miR-320b could bind to the 3′ UTR of EGR3 mRNA to hinder the expression of EGR3. Knockdown of LINC00680 and TTN-AS1 could downregulate the expression of EGR3. EGR3 could bind to the promotor of PKP2 and activate the PI3K/Akt pathway. Knockdown EIF4A3, LINC00680, and TTN-AS1 could reduce the growth of xenograft tumor and prolonged the survival of nude mice.
RBPs have been proven to be involved in many aspects of the cell process. Its dysfunction may cause diseases, including cancers.19 EIF4A3 is a core component of the EJC, which stimulates precursor (pre-)mRNA splicing, mRNA export, translation, and degradation.20 EIF4A3 was overexpressed in several kinds of cancers and was closely related to the prognostic index for survival, and thus EIF4A3 was considered as a diagnostic marker or therapeutic target for cancers.21 Inhibition of EIF4A3 could impair the formation and maintenance of stress granules in the cell after stress and change the expression of cell cycle-related transcripts in tumor cells, both of which are important for the survival and progression of tumor cells.22 In this study, EIF4A3 was highly expressed in glioblastoma cells and tissues. Furthermore, the expression in high-grade gliomas was higher than that in low-grade gliomas. As glioma grade increases, the glioma exhibits more invasiveness and less apoptosis. Since EIF4A3 was related to cell cycle and stress in tumor cells, there might be a correlation between its expression and glioma grade. However, more samples are needed for further research. In vivo study showed that silencing EIF4A3 could reduce tumor growth and prolong the survival of nude mice. In addition to our results, the expressions of EIF4A3 and survival from database results demonstrate that EIF4A3 is highly expressed in glioblastoma and that lower expression of EIF4A3 shows longer survival. These results indicate that EIF4A3 may be a diagnostic marker for glioblastomas, but this needs more research. Knockdown of EIF4A3 could inhibit the proliferation, migration, and invasion and promote the apoptosis of glioblastoma cells. These results suggest that EIF4A3 could promote malignant biological behaviors of glioblastoma cells.
lncRNA could regulate gene expression on the post-transcriptional level.23 Dysfunction of lncRNA relates to the numbers of cancers. LINC01121 represses the expression of GLP1R and inhibition of the cyclic AMP (cAMP)/protein kinase A (PKA) signaling pathway, thus inhibiting apoptosis and promoting proliferation, migration, and invasion of pancreatic cancer cells.24 LINC01133 is downregulated in gastric cancer cells. LINC01133 depletion promotes cell proliferation, migration, and the epithelial-mesenchymal transition (EMT) in gastric cancer cells.25 Moreover, a lot of lncRNAs have been identified as diagnostic and prognostic bio-markers for a variety of cancers, including gliomas.26,27 In our study, LINC00680 and TTN-AS1 were highly expressed in glioma tissues, and the expression of them was higher in high-grade gliomas than that in low-grade gliomas. Furthermore, LINC00680 and TTN-AS1 could significantly reduce the volume of tumor and prolong the survival of nude mice. These results indicate that LINC00680 and TTN-AS1 could promote progression of glioblastoma.
Recently, the focus on interaction between RBPs and RNAs has shifted from mRNA to non-coding RNA (ncRNA). RBP could regulate splicing, translocation, and stability of RNA on the post-transcriptional level.28, 29, 30 H19 could bind to EIF4A3 and hinder the recruitment of EIF4A3 to the cell-cycle gene mRNA to promote the proliferation of colorectal cancer cells.31 EIF4A3 could bind to circPVRL3 and promote the malignant biological behaviors of gastric cancer cells.32 In our study, we predicted that there were binding sites between EIF4A3 and LINC00680 or TTN-AS1 via Starbase v2.0. RIP and RNA pull-down assays verified that EIF4A3 could bind to LINC00680 and TTN-AS1. Knockdown of EIF4A3 and LINC00680 or TTN-AS1 together could significantly inhibit the proliferation, migration, and invasion and promote apoptosis of glioblastoma cells compared to knockdown of EIF4A3 or LINC00680 or TTN-AS1 alone, which indicated there may be interaction between EIF4A3, LINC00680, and TTN-AS1. In this study, we provide a new insight of EIF4A3 for its function in glioblastoma progression. Nascent RNA and half-life tests showed knockdown of EIF4A3 to shorten the half-life of LINC00680 and TTN-AS1 without increasing their expression. Thus, we assumed that EIF4A3 promotes malignant behaviors of glioblastoma cells by stabilizing LINC00680 and TTN-AS1.
miRNAs were involved in the regulation of tumorigenesis, cell apoptosis, migration, and invasion of cancers.33 miRNAs have drawn much attention in cancer research since they can form RISC with Ago10. miR-320 could inhibit proliferation, migration, and invasion of breast cancer, ovarian cancer, and cholangiocarcinoma cells.34, 35, 36 miR-320b was lowly expressed in glioma cells and could inhibit proliferation, migration, and invasion of glioma cells,11 which was consistent with our results.
lncRNAs, serving as competitive endogenous RNAs (ceRNAs), could function as a miRNA sponge and regulate target gene expression by sharing the same miRNA response elements (MREs).10 NEAT1 acts as a ceRNA to regulate miR-34a expression, repress the miR-34a/SIRT1 axis, and activate the Wnt/β-catenin signaling pathway to promote proliferation and metastasis of colorectal cancer cells.37 We predicted that LINC00680 and TTN-AS1 could bind to miR-320b by using the biological information software LncBase. Dual-luciferase reporter assays confirmed that miR-320b could bind to LINC00680 and TTN-AS1. Knockdown of EIF4A3, LINC00680, and TTN-AS1 could upregulate the expression of miR-320b. However, knockdown or overexpression of miR-320b could upregulate or downregulate LINC00680 and TTN-AS1, respectively. RIP assays verified that LINC00680 or TTN-AS1 and miR-320b were enriched in Ago. These results proved that LINC00680 and TTN-AS1 could bind to miR-320b in Ago to form RISC to regulate malignant biological behaviors of glioblastoma cells.
EGR3 is a zinc-finger transcription factor that contains a highly conserved DNA-binding domain encoding zinc finger protein. EGR3 is a diagnostic and prognostic biomarker of cutaneous squamous cell carcinoma.38 Consistent with a previous study,15 mRNA and protein of EGR3 in our research were highly expressed in glioblastoma tissues and cells. It has been reported that mature brain-derived neurotrophic factor (BDNF) was upregulated in human glioma tissues and that its expression was correlated with the glioma grade.39 Furthermore, BDNF could regulate the expression of EGR3 on both the mRNA and protein levels,40,41 which was consistent with our results that the expression of EGR3 in high-grade glioma was higher than that in low-grade glioma. However, whether the expression of EGR3 was correlated to the grade of glioma needs more research since we have limited tumor tissue. Knockdown of EGR3 could inhibit proliferation, migration, and invasion but promote apoptosis of glioma cells, which indicated that EGR3 facilitates the malignant biological behaviors of glioblastoma cells.
miRNAs target mRNAs of some proteins and regulate their expression by binding to the 3′ UTR of their mRNAs.42 Inhibition of CCNA2 mediated by miR-381-3p modulates the proliferation and EMT progression in bladder cancer cells.43 Overexpression of miR-489 could inhibit HER2-induced tumorigenesis by altering progenitor cell populations and could decrease tumor growth and metastasis via influencing tumor-promoting genes DEK and SHP2.44 As shown in the present study, we found that miR-320b could bind to the 3′ UTR of EGR3 mRNA via a dual-luciferase reporter assay. LINC00680 and TTN-AS1 may serve as ceRNAs of miR-320b, competing with EGR3 mRNA, thus repressing the binding of miR-320b and EGR3 mRNA so as to upregulate expression of EGR3.
With the use of JASPAR, we predicted that PKP2 was one of the targets of EGR3. PKP2 is upregulated in glioma tissues and cells and is related to the progression of gliomas. Knockdown of PKP2 could inhibit the proliferation and migration of glioma cells,18 which is consistent with our results. PKP2 activates the EGFR pathway.17,45 PKP2 is the most widespread in the desmosomes of all proliferative epithelial cells of normal tissues as well as of the tumors, and it was related to metastasis of cancer cells.46 ChIP verified that EGR3 could bind to the promotor of PKP2. Knockdown of EGR3 inhibited the expression of PKP2. Wild-type EGR3 binding to miR-320b could inhibit the expression of p-PI3K and p-Akt, indicating that PKP2 was involved in EGR3-induced regulation of glioblastoma cell malignant progression via the EGFR pathway.
In summary, this study presented that knockdown of LINC00680 and TTN-AS1 could inhibit proliferation, migration, and invasion but promote apoptosis of glioblastoma cells. While EIF4A3 could help stabilize LINC00680 and TTN-AS1, LINC00680 and TTN-AS1 might be ceRNAs of miR-320b, which might result in the accumulation of EGR3 and activation of the EGFR pathway. The interactions of EIF4A3, LINC00680, TTN-AS1, miR-320b, EGR3, and PKP2 were discovered for the first time, and they might be a promising target in glioblastoma therapy.
Materials and Methods
Patients and Clinical Specimens
Glioma tissues and normal brain tissues (NBTs) were collected during surgery in the Department of Neurosurgery of Shengjing Hospital with the approval of the Ethics Committee of Shengjing Hospital of China Medical University. Informed consent was obtained from all the patients. The tumor specimens were classified into four grades according to the 2016 World Health Organization (WHO) classification by two independent neuropathologists. All samples were immediately frozen in liquid nitrogen after resection. NBTs acquired from patients undergoing decompression caused by trauma, hemorrhage, or infarction were used as NCs.
Cell Culture
Human astrocyte (HA) cells were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in RPMI 1640 culture medium (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA). Glioblastoma cell lines (U87 and U251) and human embryonic kidney (HEK) 293 T cells were purchased from the Shanghai Institutes for Biological Sciences Cell Resource Center. BTICs are a derivate from glioblastoma. Tumor tissues were stored in liquid nitrogen after surgery. Tissues were washed in normal saline and cut into pieces about 1 mm3. Then, they were digested in 0.25% trypsin at 37°C for 10 min. After that, the solution was filtered and centrifuged and then suspended in culture solution. Tumor cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/high glucose with 10% FBS. All cells were cultured at 37°C with 5% CO2 in a humidified atmosphere.
RNA Extraction and PCR
Total RNA was extracted from cells and tissues with TRIzol reagent (Life Technologies, Carlsbad, CA, USA). A NanoDrop spectrophotometer (ND-100, Thermo Scientific, USA) was used to detect the concentration of RNA at 260 nm/280 nm absorbance. Expression of mRNA of EIF4A3 and EGR3, LINC00680, and TTN-AS1 was detected by a One-Step SYBR PrimeScript RT-PCR kit (Takara Bio, Japan) in a 7500 Fast RT-PCR system. Expression of miR-320b was detected by a TaqMan miRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) with TaqMan Universal Master Mix II. GAPDH and U6 were used to normalize the expression of mRNA, lncRNA, and miRNA respectively. Expressions were normalized to endogenous controls, and the relative quantification (2−ΔΔCt) method was used for fold-change calculation. Primer sequences of PCR are shown in Table S1.
Cell Transfection
shRNAs against EIF4A3, LINC00680, TTN-AS1, EGR3, EGR3 without the 3′ UTR (EGR3 non-3′ UTR), and their non-target sequences (NCs) were constructed (GenePharama, Shanghai, China). miR-320b agomir, miR-320b antagomir, and their respective NCs were synthesized (GenePharama, Shanghai, China). Glioma cells were seeded in a 24-well plate (Corning, Corning, NY, USA). After incubating for 24 h, the cells reached about 70% confluence. Lipofectamine 3000 reagent and Opti-MEM I (Life Technologies, Waltham, MA, USA) were used according to the manufacturer’s instructions after the cells were incubated to reach about 70% confluence. Stable transfected cells were selected with G418 and puromycin (Sigma-Aldrich, St. Louis, MO, USA). qPCR and western blot were used to evaluate the transfection efficacy. The shRNA concentration was 500 ng/μL, the agomiR-320b concentration was 2 μg/μL, and the antagomiR-320b concentration was 1 μg/μL. Sequences of RNA interference (RNAi) are shown in Table S2.
Western Blot
Total proteins of cells and tissues were extracted by lysing in radioimmunoprecipitation assay buffer (RIPA buffer; Beyotime Institute of Biotechnology, Jiangsu, China) on ice for 30 min and then centrifuged at 17,000 × g for 40 min at 4°C. The concentration of protein was determined by a bicinchoninic acid (BCA) assay kit (Beyotime Institute of Biotechnology). An equal amount of protein was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF) membranes. Tween 20 with Tris-buffered saline (TTBS) with 10% non-fat milk was used to block the membranes at room temperature for 2 h. Then, membranes were incubated in primary antibody of EIF4A3 (1:500, Proteintech, Rosemont, IL, USA), EGR3 (1:300, Santa Cruz Biotechnology, Santa Cruz, TX, USA), PKP2 (1:200, Santa Cruz Biotechnology, Santa Cruz, TX, USA), PI3K, Akt (1:500, Proteintech, Rosemont, IL, USA), and GAPDH (1:5,000, Proteintech, Rosemont, IL, USA) overnight at 4°C followed by horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000, Proteintech, Rosemont, IL, USA) at room temperature for 2 h. The blots were visualized with an enhanced chemiluminescence (ECL) kit (Beyotime Institute of Biotechnology) and scanned by ChemImager 5500 v2.03 software.
Cell Proliferation Assay
Cells were seeded into wells of a 96-well plate, with 2,000 cells in each well. After cells were cultured in the incubator for 24 h, 20 μL of CCK-8 (Beyotime Institute of Biotechnology, Jiangsu, China) was added to each well and then the plate was displaced at 37°C for another 2 h. The absorbance value of each well was measured spectrophotometrically at 450 nm on a SpectraMax M5 microplate reader (Molecular Devices, USA).
Cell Migration and Invasion Assays
A transwell chamber (Costar, Corning, NY, USA) was used to evaluate the migration and invasion ability of cells. Cells were resuspended in serum-free medium and loaded onto the upper chamber with a density of 2 × 105 cells per chamber for the cell migration assay. For the cell invasion assay, the upper chamber was pre-coated with 50 ng/μL Matrigel solution (BD Biosciences, Franklin Lakes, NJ, USA). After incubation for 24 h, the chamber was fixed with methanol and glacial acetic acid at the ratio of 3:1 and stained with 20% Giemsa stain. Cells were observed and counted using an inverted microscope within five randomly chosen fields, and the average number of cells was calculated.
Cell Apoptosis Assay
The cell apoptosis rate was assessed by annexin V-phycoerythrin (PE)/7-aminoactinomycin D (7AAD) staining (SouthernBiotech, Birmingham, AL, USA) according to the instructions of the manufacturer. After washing with 4°C PBS twice, cells were collected and stained, and then samples were detected by flow cytometry (FACScan, BD Biosciences) and apoptotic fractions were investigated by CellQuest 3.0 software.
Dual-Luciferase Reporter Assay
The sequence of the putative binding site was substituted by LINC00680-mutant (Mut), TTN-AS1-Mut, and EGR3-Mut to mutate the putative binding site of LINC00680, TTN-AS1, or EGR3 and miR-320b. HEK293 cells were seeded in a 96-well plate, and after the cells reached 50%–70% confluence, then they were co-transfected with LINC00680-Wt (or LINC00680-Mut), TTN-AS1 (or TTN-AS1-Mut), or EGR3-Wt (or EGR3-Mut) and miR-320b or miR-320b-NC plasmids. The luciferase activities were detected by a Dual-Luciferase reporter assay kit (Promega) 48 h after transfection.
ChIP Assay
A SimpleChIP enzymatic ChIP kit (Cell Signaling Technology) was used to perform a ChIP assay. Glioma cells were cross-linked with 1% formaldehyde for 10 min and terminated by glycine. 2% lysates were used as an input control, and the remainder was incubated with normal rabbit IgG or anti-EGR3 antibody. DNA crosslinks were reversed by NaCl and proteinase K and then purified and amplified by PCR. ChIP primer sequences are shown in Table S3.
RIP Assay
RIP assays were conducted with an EZ-Magna RIP kit (Millipore, Billerica, MA, USA). Glioblastoma cells were lysed by a complete RNA lysis buffer according to the manufacturer’s protocol. Magnetic beads conjugated with human anti-Argonaute2 (Ago2) antibody (Millipore) or normal mouse IgG (Millipore) as a negative control were incubated beforehand. Samples were incubated with proteinase K buffer at 4°C overnight, and then immunoprecipitated RNA was isolated, purified, and analyzed by qRT-PCR.
RNA Pull-Down Assay
In brief, LINC00680 and TTN-AS1 transcripts were transcribed by using T7 RNA polymerase (Ambion), and then by using the RNeasy Plus mini kit (QIAGEN) and treatment with DNase I (QIAGEN). Purified RNAs were biotin labeled with a biotin RNA labeling mix (Ambion). Cell lysates were mixed and incubated with biotinylated RNAs. Streptavidin agarose beads were added to each binding reaction and incubated at room temperature for 1 h. Afterward, the beads were washed and the retrieved protein was analyzed by western blot.
RNA Stability Measurement
Transfected cells (sh-NC, sh-EIF4A3) were seeded in culture dishes with 2 μg/mL actinomycin D (ActD, NobleRyder, China) to block de novo RNA synthesis. Total RNA was extracted every 2 h. The expression of LINC00680 and TTN-AS1 was measured by qRT-PCR. The half-life of LINC00680 and TTN-AS1 was determined at the time when they reached the 50% of RNA before adding actinomycin D.
Nascent RNA Capture Assay
A nascent RNA capture assay was performed using a Click-iT nascent RNA capture kit (Invitrogen) following the manufacturer’s protocol. Cells were seeded in 5-ethynyl uridine (EU). Total RNA was extracted at this time. EU-labeled RNA was biotinylated in a Click-iT reaction buffer and then collected using streptavidin magnetic beads.
Tumor Xenograft Models
Female BALB/c nude mice (4–5 weeks of age; Vital River Laboratory Animal Technology, Beijing, China) were used for xenograft models in accordance with a protocol approved by the Administrate Panel on Laboratory Animal Care of China Medical University. Nude mice were divided into five groups: control, sh-EIF4A3, sh-LINC00680, sh-TTN-AS1, and sh-EIF4A3+LINC00680+TTN-AS1. Cell transfection and selection were described as before. 3 × 105 cells were subcutaneously injected in the right flanks of the mice. Tumor size was calculated as (length × width2)/2 and recorded every week. Mice were sacrificed 42 days after injection. The number of surviving nude mice was recorded and survival analysis was performed using a Kaplan-Meier survival curve in the orthotopic inoculation experiment.
Statistical Analysis
Data are presented as mean ± SD from three independent experiments. All statistical analyses were performed by Graphpad Prism 5.0 (GraphPad, La Jolla, CA, USA) with a Student’s t test or one-way analysis of variance; survival data were analyzed by a Kaplan-Meier survival curve and log-rank test. p < 0.05 indicated statistical significance.
Author Contributions
Y.L. contributed to the experiment design, manuscript draft, and data analysis. W.T. designed the experiments, performed the experiments, and wrote the manuscript. D.W. and L.S. contributed to the experiment implementation, manuscript draft, and data analysis. J.Z., X.L., and Y.X. conceived or designed the experiments. X.R., C.Y., and J.M. performed the experiments. L.L. and Z.L. analyzed the data. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of China (81872073, 81672511, 81872503, and 81573010), Liaoning Science and Technology Plan Project (nos. 2017225020 and 2015225007), Project of Key Laboratory of Neuro-oncology in Liaoning Province (112-2400017005), a special developmental project guided by the central government of Liaoning Province (no. 2017011553-301), and by the outstanding scientific fund of Shengjing Hospital (no. 201304).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.043.
Supplemental Information
References
- 1.Ostrom Q.T., Bauchet L., Davis F.G., Deltour I., Fisher J.L., Langer C.E., Pekmezci M., Schwartzbaum J.A., Turner M.C., Walsh K.M. The epidemiology of glioma in adults: a “state of the science” review. Neuro-oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sant M., Minicozzi P., Lagorio S., Børge Johannesen T., Marcos-Gragera R., Francisci S., EUROCARE Working Group Survival of European patients with central nervous system tumors. Int. J. Cancer. 2012;131:173–185. doi: 10.1002/ijc.26335. [DOI] [PubMed] [Google Scholar]
- 3.Guichet P.O., Masliantsev K., Tachon G., Petropoulos C., Godet J., Larrieu D., Milin S., Wager M., Karayan-Tapon L. Fatal correlation between YAP1 expression and glioma aggressiveness: clinical and molecular evidence. J. Pathol. 2018;246:205–216. doi: 10.1002/path.5133. [DOI] [PubMed] [Google Scholar]
- 4.Zou Z., Ma T., He X., Zhou J., Ma H., Xie M., Liu Y., Lu D., Di S., Zhang Z. Long intergenic non-coding RNA 00324 promotes gastric cancer cell proliferation via binding with HuR and stabilizing FAM83B expression. Cell Death Dis. 2018;9:717. doi: 10.1038/s41419-018-0758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMahon J.J., Miller E.E., Silver D.L. The exon junction complex in neural development and neurodevelopmental disease. Int. J. Dev. Neurosci. 2016;55:117–123. doi: 10.1016/j.ijdevneu.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J., Cai H., Zheng J., Liu X., Liu Y., Ma J., Que Z., Gong W., Gao Y., Tao W., Xue Y. Long non-coding RNA NEAT1 regulates permeability of the blood-tumor barrier via miR-181d-5p-mediated expression changes in ZO-1, occludin, and claudin-5. Biochim Biophys Acta Mol Basis Dis. 2017;1863:2240–2254. doi: 10.1016/j.bbadis.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Liu X., Zheng J., Xue Y., Qu C., Chen J., Wang Z., Li Z., Zhang L., Liu Y. Inhibition of TDP43-mediated SNHG12-miR-195-SOX5 feedback loop impeded malignant biological behaviors of glioma cells. Mol. Ther. Nucleic Acids. 2018;10:142–158. doi: 10.1016/j.omtn.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He R.Q., Wei Q.J., Tang R.X., Chen W.J., Yang X., Peng Z.G., Hu X.H., Ma J., Chen G. Prediction of clinical outcome and survival in soft-tissue sarcoma using a ten-lncRNA signature. Oncotarget. 2017;8:80336–80347. doi: 10.18632/oncotarget.18165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin C., Zhang S., Wang Y., Wang Y., Nice E., Guo C., Zhang E., Yu L., Li M., Liu C. Functional role of a novel long noncoding RNA TTN-AS1 in esophageal squamous cell carcinoma progression and metastasis. Clin. Cancer Res. 2018;24:486–498. doi: 10.1158/1078-0432.CCR-17-1851. [DOI] [PubMed] [Google Scholar]
- 10.Sheu-Gruttadauria J., MacRae I.J. Structural foundations of RNA silencing by Argonaute. J. Mol. Biol. 2017;429:2619–2639. doi: 10.1016/j.jmb.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lv Q.L., Du H., Liu Y.L., Huang Y.T., Wang G.H., Zhang X., Chen S.H., Zhou H.H. Low expression of microRNA-320b correlates with tumorigenesis and unfavorable prognosis in glioma. Oncol. Rep. 2017;38:959–966. doi: 10.3892/or.2017.5762. [DOI] [PubMed] [Google Scholar]
- 12.O’Donovan K.J., Tourtellotte W.G., Millbrandt J., Baraban J.M. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22:167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z.D., Qu F.Y., Chen Y.Y., Ran Z.S., Liu H.Y., Zhang H.D. Involvement of microRNA-718, a new regulator of EGR3, in regulation of malignant phenotype of HCC cells. J. Zhejiang Univ. Sci. B. 2017;18:27–36. doi: 10.1631/jzus.B1600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baron V.T., Pio R., Jia Z., Mercola D. Early growth response 3 regulates genes of inflammation and directly activates IL6 and IL8 expression in prostate cancer. Br. J. Cancer. 2015;112:755–764. doi: 10.1038/bjc.2014.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salotti J., Sakchaisri K., Tourtellotte W.G., Johnson P.F. An Arf-Egr-C/EBPβ pathway linked to Ras-induced senescence and cancer. Mol. Cell. Biol. 2015;35:866–883. doi: 10.1128/MCB.01489-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alcalde M., Campuzano O., Sarquella-Brugada G., Arbelo E., Allegue C., Partemi S., Iglesias A., Oliva A., Brugada J., Brugada R. Clinical interpretation of genetic variants in arrhythmogenic right ventricular cardiomyopathy. Clin. Res. Cardiol. 2015;104:288–303. doi: 10.1007/s00392-014-0794-z. [DOI] [PubMed] [Google Scholar]
- 17.Kazlauskas A. Plakophilin-2 promotes activation of epidermal growth factor receptor. Mol. Cell. Biol. 2014;34:3778–3779. doi: 10.1128/MCB.00968-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D., Qian Y., Liu X., Yu H., Zhao N., Wu Z. Up-regulation of plakophilin-2 is correlated with the progression of glioma. Neuropathology. 2017;37:207–216. doi: 10.1111/neup.12363. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z.L., Li B., Luo Y.X., Lin Q., Liu S.R., Zhang X.Q., Zhou H., Yang J.H., Qu L.H. Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep. 2018;22:286–298. doi: 10.1016/j.celrep.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Boehm V., Gehring N.H. Exon junction complexes: supervising the gene expression assembly line. Trends Genet. 2016;32:724–735. doi: 10.1016/j.tig.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y., Zhang J., Cai J., Liang R., Chen G., Qin G., Han X., Yuan C., Liu Z., Li Y. Systematic Analysis of Gene Expression Alteration and Co-Expression Network of Eukaryotic Initiation Factor 4A-3 in Cancer. J. Cancer. 2018;9:4568–4577. doi: 10.7150/jca.27655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazloomian A., Araki S., Ohori M., El-Naggar A.M., Yap D., Bashashati A., Nakao S., Sorensen P.H., Nakanishi A., Shah S. Pharmacological systems analysis defines EIF4A3 functions in cell-cycle and RNA stress granule formation. Commun. Biol. 2019;2:165. doi: 10.1038/s42003-019-0391-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melissari M.T., Grote P. Roles for long non-coding RNAs in physiology and disease. Pflugers Arch. 2016;468:945–958. doi: 10.1007/s00424-016-1804-y. [DOI] [PubMed] [Google Scholar]
- 24.Qian Y.G., Ye Z., Chen H.Y., Lv Z., Zhang A.B., Fan L., Zhou J., Zheng S.S., Wang W.L. LINC01121 inhibits cell apoptosis while facilitating proliferation, migration, and invasion though negative regulation of the Camp/PKA signaling pathway via GLP1R. Cell. Physiol. Biochem. 2018;47:1007–1024. doi: 10.1159/000490167. [DOI] [PubMed] [Google Scholar]
- 25.Yang X.Z., Cheng T.T., He Q.J., Lei Z.Y., Chi J., Tang Z., Liao Q.X., Zhang H., Zeng L.S., Cui S.Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol. Cancer. 2018;17:126. doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reon B.J., Anaya J., Zhang Y., Mandell J., Purow B., Abounader R., Dutta A. Expression of lncRNAs in low-grade gliomas and glioblastoma multiforme: an in silico analysis. PLoS Med. 2016;13:e1002192. doi: 10.1371/journal.pmed.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiran M., Chatrath A., Tang X., Keenan D.M., Dutta A. A prognostic signature for lower grade gliomas based on expression of long non-coding RNAs. Mol. Neurobiol. 2019;56:4786–4798. doi: 10.1007/s12035-018-1416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishore S., Luber S., Zavolan M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief. Funct. Genomics. 2010;9:391–404. doi: 10.1093/bfgp/elq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukong K.E., Chang K.W., Khandjian E.W., Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Singh R. RNA-protein interactions that regulate pre-mRNA splicing. Gene Expr. 2002;10:79–92. [PMC free article] [PubMed] [Google Scholar]
- 31.Han D., Gao X., Wang M., Qiao Y., Xu Y., Yang J., Dong N., He J., Sun Q., Lv G. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159–22173. doi: 10.18632/oncotarget.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun H.D., Xu Z.P., Sun Z.Q., Zhu B., Wang Q., Zhou J., Jin H., Zhao A., Tang W.W., Cao X.F. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci. Rep. 2018;8:10111. doi: 10.1038/s41598-018-27837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farazi T.A., Hoell J.I., Morozov P., Tuschl T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C., Duan P., Wang J., Lu X., Cheng J. miR-320 inhibited ovarian cancer oncogenicity via targeting TWIST1 expression. Am. J. Transl. Res. 2017;9:3705–3713. [PMC free article] [PubMed] [Google Scholar]
- 35.Bai J.W., Wang X., Zhang Y.F., Yao G.D., Liu H. MicroRNA-320 inhibits cell proliferation and invasion in breast cancer cells by targeting SOX4. Oncol. Lett. 2017;14:7145–7152. doi: 10.3892/ol.2017.7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu H., Jiang X., Zhou X., Dong X., Xie K., Yang C., Jiang H., Sun X., Lu J. Neuropilin-1 regulated by miR-320 contributes to the growth and metastasis of cholangiocarcinoma cells. Liver Int. 2018;38:125–135. doi: 10.1111/liv.13495. [DOI] [PubMed] [Google Scholar]
- 37.Luo Y., Chen J.J., Lv Q., Qin J., Huang Y.Z., Yu M.H., Zhong M. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/β-catenin signaling pathway. Cancer Lett. 2019;440–441:11–22. doi: 10.1016/j.canlet.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Wei W., Chen Y., Xu J., Zhou Y., Bai X., Yang M., Zhu J. Identification of biomarker for cutaneous squamous cell carcinoma using microarray data analysis. J. Cancer. 2018;9:400–406. doi: 10.7150/jca.21381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong J., Zhou L.I., Lim Y., Yang M., Zhu Y.H., Li Z.W., Fu D.L., Zhou X.F. Mature brain-derived neurotrophic factor and its receptor TrkB are upregulated in human glioma tissues. Oncol. Lett. 2015;10:223–227. doi: 10.3892/ol.2015.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts D.S., Hu Y., Lund I.V., Brooks-Kayal A.R., Russek S.J. Brain-derived neurotrophic factor (BDNF)-induced synthesis of early growth response factor 3 (Egr3) controls the levels of type A GABA receptor α4 subunits in hippocampal neurons. J. Biol. Chem. 2006;281:29431–29435. doi: 10.1074/jbc.C600167200. [DOI] [PubMed] [Google Scholar]
- 41.Pfaffenseller B., Kapczinski F., Gallitano A.L., Klamt F. EGR3 immediate early gene and the brain-derived neurotrophic factor in bipolar disorder. Front. Behav. Neurosci. 2018;12:15. doi: 10.3389/fnbeh.2018.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 43.Li J., Ying Y., Xie H., Jin K., Yan H., Wang S., Xu M., Xu X., Wang X., Yang K. Dual regulatory role of CCNA2 in modulating CDK6 and MET-mediated cell-cycle pathway and EMT progression is blocked by miR-381-3p in bladder cancer. FASEB J. 2019;33:1374–1388. doi: 10.1096/fj.201800667R. [DOI] [PubMed] [Google Scholar]
- 44.Patel Y., Soni M., Awgulewitsch A., Kern M.J., Liu S., Shah N., Singh U.P., Chen H. Overexpression of miR-489 derails mammary hierarchy structure and inhibits HER2/neu-induced tumorigenesis. Oncogene. 2019;38:445–453. doi: 10.1038/s41388-018-0439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arimoto K., Burkart C., Yan M., Ran D., Weng S., Zhang D.E. Plakophilin-2 promotes tumor development by enhancing ligand-dependent and -independent epidermal growth factor receptor dimerization and activation. Mol. Cell. Biol. 2014;34:3843–3854. doi: 10.1128/MCB.00758-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rickelt S. Plakophilin-2: a cell-cell adhesion plaque molecule of selective and fundamental importance in cardiac functions and tumor cell growth. Cell Tissue Res. 2012;348:281–294. doi: 10.1007/s00441-011-1314-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.