Abstract
Cells succumbing to stress via regulated cell death (RCD) can initiate an adaptive immune response associated with immunological memory, provided they display sufficient antigenicity and adjuvanticity. Moreover, multiple intracellular and microenvironmental features determine the propensity of RCD to drive adaptive immunity. Here, we provide an updated operational definition of immunogenic cell death (ICD), discuss the key factors that dictate the ability of dying cells to drive an adaptive immune response, summarize experimental assays that are currently available for the assessment of ICD in vitro and in vivo, and formulate guidelines for their interpretation.
Keywords: oncology, immunology, molecular biology
Introduction
Regulated cell death (RCD), a form of cellular demise that is governed by a genetically encoded molecular machinery,1 2 has long been considered as an immunologically silent or even tolerogenic event.3 At least in part, this widely accepted view originated from the highly tolerogenic nature of programmed cell death (PCD), the physiological variant of RCD that contributes to postembryonic development and adult tissue turnover.1 4 However, it has now become clear that, at least under specific circumstances, stress-induced RCD can drive an inflammatory response that may culminate with the activation of cytotoxic T lymphocyte (CTL)-driven adaptive immunity coupled with the establishment of long-term immunological memory. Such a functionally unique form of stress-driven RCD is now usually referred to as immunogenic cell death (ICD).5
Cellular stressors that are associated with ICD encompass (but are not limited to): (1) obligate intracellular pathogens including multiple bacterial and viral species6–8; (2) therapeutic oncolytic viruses9–16; (3) various molecules with oncolytic potential17–19; (4) conventional chemotherapeutics such as numerous anthracyclines (ie, doxorubicin, epirubicin, idarubicin and mitoxantrone), some (but importantly not all) DNA-damaging agents (ie, cyclophosphamide and oxaliplatin, but not cisplatin), poly-A-ribose polymerase (PARP) inhibitors, mitotic poisons (ie, docetaxel and patupilone) and proteasomal inhibitors (ie, bortezomib and carfilzomib)20–25; (5) epigenetic modifiers including DNA methyltransferase, histone deacetylase (HDAC) and bromodomain inhibitors26–30; (6) targeted anticancer agents such as the tyrosine kinase inhibitor crizotinib, the epidermal growth factor receptor (EGFR)-specific monoclonal antibody cetuximab, the cyclin-dependent kinase (CDK) inhibitor dinaciclib and the Bruton tyrosine kinase (BTK) inhibitor ibrutinib31–33; (7) other chemicals including the ubiquitin-specific peptidase inhibitor spautin-1, the antibiotic bleomycin, the protein phosphatase-2A inhibitor LB-100, the Chinese herbal medicine component shikonin and capsaicin34–38 and (8) numerous physical interventions, encompassing various forms of ionizing radiation, extracorporeal photochemotherapy, hypericin-based photodynamic therapy (PDT), near‐infrared photoimmunotherapy, high hydrostatic pressure, severe cytotoxic heat shock, nanopulse stimulation and electrohyperthermia.39–49 Importantly, dose and administration schedules have a major impact on the ability of many of these agents to initiate productive ICD.50–52
The aforementioned ICD inducers have been instrumental not only for identifying the molecular machinery that underlies the immunogenicity of some variants of RCD,5 but also for elucidating the pathophysiological and therapeutic implications of the process.53 Indeed, the ability of ICD to initiate adaptive immunity not only is critical for the optimal eradication of infectious pathogens,54 but also influences the cancer-immunity cycle by tipping the balance toward antitumor immunity.55 Consistent with this notion, both pathogens and progressing tumors harness strategies that enable immunoevasion by avoiding ICD induction.5 Moreover, accumulating clinical evidence demonstrates that numerous ICD inducers commonly employed in the management of cancer patients synergize with immunotherapy with immune checkpoint blockers (ICBs), as long as they do not compromise immunostimulatory signals or the activity of tumor-infiltrating lymphocytes.56 57
The morphological features displayed by dying cells and the molecular mechanisms that are mechanistically responsible for the cellular demise do not necessarily correlate with the immunogenicity of RCD.58 Thus, while specific instances of caspase 3 (CASP3)-dependent apoptosis and mixed lineage kinase domain-like pseudokinase (MLKL)-dependent necroptosis initiate adaptive immunity in certain experimental settings,20 59 60 RCD accompanied by CASP3 or MLKL activation is not necessarily immunogenic.61–63 Moreover, while the perception of RCD as immunogenic has been etiologically attributed to the emission of specific signals from dying cells (see Definition of immunogenic cell death), the presence of such signals is not necessarily predictive of the ability of dying cells to drive adaptive immunity in vivo.34 64 Altogether, these observations highlight the importance of defining standardized experimental settings that enable the assessment of ICD in the context of a robust conceptual framework for the interpretation of results. Here, we provide a general overview of the factors that underpin the immunogenicity or RCD, and attempt to provide such a framework by formulating guidelines for the definition, detection and interpretation of ICD.
Definition of ICD
The Nomenclature Committee on Cell Death has recently defined ICD as ‘a form of RCD that is sufficient to activate an adaptive immune response in immunocompetent syngeneic hosts’,1 which properly reflects the two major components of ICD as a process, that is, the cellular component and the host component. Importantly, the latter does not refer to potential defects of the host that prevent the initiation of adaptive immunity (eg, HLA mismatch, systemic immunodeficiency), but to features intrinsic to dying cells that render them immunogenic only in specific hosts. Indeed, the ability of RCD to drive adaptive immunity depends on two major parameters, neither of which is ultimately intrinsic to dying cells: antigenicity and adjuvanticity.
Antigenicity is conferred by the expression and presentation of antigens that fail to induce clonal deletion in the context of central tolerance in a specific host, implying that the host contains naïve T cell clones that can recognize such antigens.65 66 Thus, healthy cells are limited in their ability to drive ICD, as their antigens are typically expressed by the thymic epithelium during T cell development. As an exception, some naïve T cell clones expressing self-reactive low-affinity T cell receptors (TCRs) escape thymic selection, implying that such antigens may support ICD in the context of peripheral tolerance breakdown (see Sources of ICD antigenicity). Conversely, infected cells, as well as malignant cells, display sufficient antigenicity to drive immune responses, as they express a panel of antigenic epitopes for which naïve T cell clones are generally available. These antigenic determinants include neoepitopes that are highly immunogenic as they are not covered by central tolerance as well as (non-mutated) epitopes that may be immunogenic due to gaps in central tolerance and/or incomplete peripheral tolerance.67 Adjuvanticity is provided by the spatiotemporally coordinated release or exposure of danger signals that are necessary for the recruitment and maturation of antigen-presenting cells (APCs), which are cumulatively referred to as damage-associated molecular patterns (DAMPs).68 69 Although most (if not all) cells contain DAMPs in levels that are sufficient to drive robust APC stimulation, the kinetics and intensity of their release are dictated by intracellular responses driven by the initiating stressor.70–72 This may explain why some cytotoxic agents can drive ICD while others are unable to, despite their similar RCD-inducing capability.23 73
Microenvironmental conditions also have a dramatic influence on the propensity of infected or neoplastic cells undergoing a potentially immunogenic variant of RCD to initiate adaptive immunity and/or be susceptible to CTL-dependent lysis, thus impacting both the priming and the effector phase of the immunological response. As an example, mouse cancer cells irradiated in vitro can be successfully employed to immunize immunocompetent syngeneic mice against a subsequent challenge with living cells of the same type, demonstrating the elicitation of immunological memory.74 Conversely, in therapeutic settings, the immunosuppressive microenvironment that characterizes a majority of tumors can considerably limit ICD-driven immunity.75 76 Thus, irradiating a neoplastic lesion established in immunocompetent, syngeneic mice generally fails to generate a CTL-dependent immune response of sufficient strength to eradicate a distant, non-irradiated lesion, unless additional immunostimulatory molecules are provided.77–79
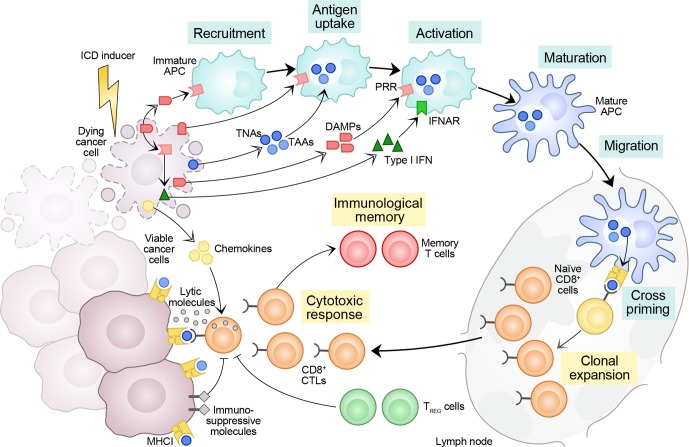
Taken together, these observations suggest that while the adjuvanticity of RCD depends on dying cells and the capability of the initiating stressor to elicit danger signaling, its immunogenicity ultimately depends on dying cells and the host, which also determines the propensity of dying cells to drive adaptive immunity at the microenvironmental level (figure 1).
Figure 1.
Major factors dictating the immunogenicity of cell death. Cells undergoing regulated cell death (RCD) in response to stress can prime an adaptive immune response specific for dead cell-associated antigens provided that (1) those antigens are not perfectly covered by central tolerance, and (2) dying cells emit a panel of immunostimulatory damage-associated molecular patterns (DAMPs) and cytokines that, when delivered according to a precise spatiotemporal pattern, support the recruitment, phagocytic activity and maturation of antigen-presenting cells (APCs), de facto enabling them to engulf antigenic material, migrate to lymph nodes and prime a cytotoxic T lymphocyte (CTL)-dependent immune response. As they express tumor neoantigens (TNAs, which are not covered by central tolerance) and/or tumor-associated antigens (TAAs, for which central tolerance is leaky), cancer cells can undergo bona fide immunogenic cell death (ICD) in response to select stimuli, including (but not limited to) some chemotherapeutic agents commonly employed in the clinic, as well as radiation therapy. However, the TME is generally characterized by an immunosuppressive profile that may prevent either the initiation or the execution of ICD-driven anticancer immunity. Thus, the ultimate ability of RCD to drive adaptive immunity does not depend only on the initiating stimulus and the dying cell, but also on features that are intrinsic to the host. IFNAR, interferon-alpha/beta receptor; PRR, pattern recognition receptor; TREG, regulatory T; TME, tumor microenvironment.
Sources of ICD antigenicity
Infection by pathogenic microbes is an obvious source of antigenic determinants, as microbial proteins are not covered by central tolerance and hence their epitopes are highly antigenic.80 81 Together with the ability of conserved microbial products cumulatively referred to as microbe-associated molecular patterns (MAMPs) to deliver potent immunostimulatory signals, this explains why RCD driven by intracellular pathogens is highly immunogenic.82–84 The same generally does not apply to healthy syngeneic cells, as thymic and peripheral tolerance result in the deletion or functional inactivation of self-reactive T cell clones from the mature host T cell repertoire,85 although some naïve T cell clones expressing self-reactive low-affinity TCRs can escape thymic selection and hence drive (especially in the context of lost peripheral tolerance) autoimmune reactions.86
There are at least two exceptions to this principle, which may underlie the ability of healthy cells to undergo ICD. First, the genome of normal cells contains a significant number of endogenous retroviruses, which are generally latent (ie, not transcribed) in physiological conditions.87 88 In response to some cellular stressors, however, endogenous retroviruses can become activated and/or retroviral genes can be expressed, resulting in the synthesis of potentially antigenic proteins.89 Second, antigenic determinants can be generated by enzymatic or non-enzymatic post-translational modifications (PTMs) that alter protein structure, encompassing (but perhaps not limited to) phosphorylation, acetylation, glycosylation, citrullination, nitration/nitrosylation, glycation, oxidation and ubiquitination.90 Moreover, the antigenic peptide repertoire can be boosted by alterations in the activation of reticular aminopeptidases such as endoplasmic reticulum aminopeptidase 1 (ERAP1) and ERAP2.91 Importantly, the signal transduction cascades that regulate enzymatic PTMs are sensitive to a variety of microenvironmental signals, and not necessarily activated in a similar manner in the periphery and the thymic epithelium during clonal T cell selection. This implies that some PTM-containing epitopes may not be covered by central tolerance. Along similar lines, microenvironmental conditions that impose non-enzymatic PTMs (eg, an oxidative extracellular milieu) are common at sites of inflammation, ischemia or malignant progression, but not in the thymus.92 As a note, such stressful conditions can also lead to the generation of antigenic peptides derived from ‘cryptic’ translation (ie, from untranslated mRNAs).93 In support of the ability of healthy cells to drive ICD, at least in some settings, PTM-dependent epitopes have been attributed pathogenic value in some autoimmune disorders including diabetes and rheumatoid arthritis.94 95
The majority of human tumors are not driven by active viral infections. Nonetheless, like pathogen-infected dying cells, malignant cells can display a high antigenicity, largely reflecting the elevated mutational rate that frequently accompanies malignant transformation and disease progression in the context of immunoevasion.96 97 In a fraction of cases, such mutations prime immune responses as they affect coding regions of the genome. This is the case of non-synonymous point mutations (ie, mutations altering the amino acid sequence) as well as frameshift mutations caused by small insertions and deletions (indels) in proteins that are expressed and properly processed by the antigen presentation machinery, culminating in the exposure of tumor neoantigens (TNAs). TNAs exposed on the surface of malignant cells may have poor structural homology to self epitopes, hence partially resemble microbial epitopes and efficiently prime de novo immune responses.75 98–101 Some self antigens expressed by cancer cells can also initiate antitumor immunity. Because they are not unique to neoplastic tissues, but are also expressed by healthy or immunoprivileged tissues, such antigens have been designated tumor-associated antigens (TAAs). TAAs that have been shown to drive anticancer immunity, especially in the setting of therapeutic anticancer vaccination,102 103 encompass: (1) tissue differentiation antigens such as CD19, CD20, premelanosome protein (PMEL, best known as gp100), and melan-A (MLANA, best known as MART-1), as well as (2) ectopically expressed proteins such as carcinoembryonic antigens (CEAs), cancer/testis antigens, as well as multiple members of the MAGE and SSX protein families.104–106 Central tolerance against these antigens is leaky (implying that naïve T cell clone that express low-affinity TCRs are available) and peripheral tolerance can be overcome in the context of robust adjuvanticity.106–108 Thus, although TAAs are generally weaker at eliciting anticancer immunity as compared with TNAs,109 they can be relevant for ICD-driven immunity in tumors with low TNA load. Of note, the harsh conditions that characterize the tumor microenvironment (TME) and the extensive rewiring of signal transduction that characterizes malignant cells suggest that PTMs may play a predominant role in determining the antigenicity of cancer cells, a possibility that remains largely unexplored.110
Several factors influence the antigenicity of tumors evolving in immunocompetent, syngeneic hosts. First, the mutational burden (and thus the potential to generate TNAs) is heterogeneous across and within tumors, ranging from ~1 mutation/Mb in hematologic malignancies to >10 mutations/Mb in solid tumors with a hypermutant phenotype.111–113 Mutational burden and the TNA landscape also evolve over space (ie, in distinct tumor areas) and time (ie, at distinct stages of malignant progression) under the pressure of ongoing immunity and in response to increased genomic instability as well as chemotherapeutic or radiotherapeutic interventions,97 114 115 resulting in tumors or tumor areas with distinct antigenicity and hence differential ability to drive adaptive immune responses on RCD.116 That said, while mutational burden has been associated with the sensitivity of multiple tumors to ICBs117–119 and tumors with extensive immune infiltration (which often are genetically unstable) are characterized by a transcriptional signature of ICD,120 formal demonstration that mutational burden also influences the antineoplastic effects of ICD-based therapeutic regimens in the clinic is lacking. Preclinical evidence suggests that even cells with reduced mutational load can drive adaptive immunity,121 although the choice of the experimental model is expected to have a major role in this context (see Detection of ICD in cancer). Second, the antigenicity of malignant cells is directly related to antigen presentation, implying that genetic and epigenetic defects that compromise it can be beneficial for cancer cells.122 123 These defects, which are common in tumors with high mutational burden and robust T cell infiltration,124 125 include: (1) antigen loss and subclonal evolution, that is, the preferential expansion of cancer cell clones that do not express an antigen subjected to active immunity114 and (2) impaired antigen presentation as a consequence of mutations, deregulated expression, or structural alterations of key components of the antigen-presenting machinery including MHC Class I molecules, beta-2-microglobulin (B2M), transporter 1, ATP binding cassette subfamily B member (TAP1), TAP2 and proteasomal subunits.125 126
Of note, while the majority of ICD inducers are believed to have little impact on antigenicity and to operate by driving the correct spatiotemporal emission of DAMPs in the context of cell death (see Sources of ICD adjuvanticity), at least some ICD-triggering regimens may also boost antigenicity. This applies to potentially mutagenic agents, and to interventions that drive the reactivation of endogenous retroviruses and/or induce the expression of mutated genes or TAAs, such as CDK4/CDK6 inhibitors,127 ionizing radiation,79 128 DNA damage response (DDR) inhibitors,129 130 oncolytic viruses131 or HDAC inhibitors and other epigenetic regulators.132–134 Irrespective of this possibility, the ability of RCD to drive adaptive immunity intimately depends on the antigenicity of dying cells with respect to the availability and the functional status of the mature TCR repertoire of the host.
Sources of ICD adjuvanticity
ICD driven by microbial pathogens is associated with arguably the most potent adjuvant signals for mammalian organisms, MAMPs. MAMPs encompass a variety of microbial products that operate both within infected cells and in their microenvironment to drive the recruitment and maturation of APCs, culminating with antigen (cross-)presentation to naïve T cells. MAMPs include microbial nucleic acid species (eg, viral single-stranded or double-stranded RNA or DNA, bacterial CpG-rich DNA) as well as structural components (eg, lipopolysaccharide, peptidoglycans, flagellin), and mostly mediate immunostimulatory effects via pattern recognition receptors (PRRs).82 83 The latter encompass numerous Toll-like receptors (TLRs) expressed on the cell surface and in endosomal compartments,135–138 as well as (1) cyclic GMP-AMP synthase (CGAS), a sensor of cytosolic double-stranded DNA (dsDNA)139; (2) RIG-I-like receptors (RLRs), a group of RNA-specific PRRs named after DExD/H-box helicase 58 (DDX58, best known as RIG-I)140 141; (3) NOD-like receptors (NLRs), a family of PRRs with broad ligand specificity named after a common nucleotide-binding oligomerization domain (NOD)142 143; (4) Z-DNA binding protein 1 (ZBP1), a nucleic acid sensor also known as DAI whose precise mechanism of activation remains unknown144; (5) heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1), a sensor of viral nuclear DNA and N6-methyladenosine-bearing RNAs.145 146 PRRs drive the production of numerous immunostimulatory that are key for pathogen control by the immune system.147
The same PRRs activated by MAMPs are also involved in the adjuvanticity of ICD induced in cancer cells by non-microbial stimuli.68 148 Thus, malignant cells exposed to a potentially immunogenic RCD inducer emit numerous DAMPs and cytokines that have been mechanistically linked to the initiation of adaptive immunity in preclinical models (table 1). These immunostimulatory molecules include (but most likely are not limited to): ATP,149 150 cellular nucleic acids,151 152 the non-histone, nuclear DNA-binding protein high mobility group box 1 (HMGB1),153–156 the member of the annexin superfamily annexin A1 (ANXA1),157 cytokines like type I interferon (IFN), C-C motif chemokine ligand 2 (CCL2), C-X-C motif chemokine ligand 1 (CXCL1) and CXCL10,151 158 159 ER chaperones like calreticulin (CALR), protein disulfide isomerase family A member 3 (PDIA3, also known as ERp57), heat shock protein family A (Hsp70) member 1A (HSPA1A, best known as HSP70), Hsp90 alpha family class A member 1 (HSP90AA1, best known as HSP90),21 22 160 cytosolic components like F-actin,161 and other mitochondrial products like DNA, reactive oxygen species (ROS), cardiolipin and transcription factor A, mitochondrial (TFAM).36 162–164 The major roles of ICD-associated DAMPs and cytokines are to: (1) enable the recruitment of APCs or their precursors to sites of RCD (eg, ATP)149 150 165, (2) spatially guide the interaction between APCs and dying cells (eg, ANXA1)157, (3) favor the phagocytosis of dying cells or their corpses (eg, CALR, ERp57, HSP70, HSP90, F-actin)21 22 160 161, (4) promote the maturation of APCs and their capacity to effect cross-presentation (eg, ATP, HMGB1, type I IFN and TFAM),149 153 166–168 and (5) facilitate the recruitment of T cells (eg, CCL2, CXCL1 and CXCL10).151 158 Of note, some emitted DAMPs are immunosuppressive (eg, adenosine),169 while others can switch to anti-inflammatory depending on the engaged PRR (eg, HMGB1), biochemical modifications such as oxidation (eg, HMGB1) or chronic release (eg, type I IFN).53 70 170 Indeed, while acute, robust inflammatory responses such as those driven by ICD ultimately engage anticancer immunity, indolent, chronic inflammation has consistently been associated with immunoevasion and tumor progression.171 Of note, the emission of DAMPs by dying cells occurs in the context of failing intracellular responses to stress, which relay danger signals to the rest of the organism for the preservation of systemic homeostasis.172 This implies that defects in multiple mechanisms that support cellular adaptation to stress may favor cell death, but at the same time may compromise the ability of dying cells to initiate adaptive immunity as a consequence of limited adjuvanticity.
Table 1.
Major immunostimulatory DAMPs and cytokines mechanistically linked to ICD in cancer
| Factor | Class | Effect | Main receptor(s) | Ref. |
| ANXA1 | Surface protein | Directs APCs to dying cells | FPR1 | 157 |
| ATP | Nucleotide | Promotes the recruitment, maturation and cross-presentation activity of APCs | P2RX7 P2RY2 |
149 150 |
| CALR | ER chaperone | Promotes the uptake of dying cells and type I IFN secretion by APCs | LRP1 | 21 22 160 178 191 |
| CCL2 | Cytokine | Promotes T cell and neutrophil recruitment | CCR2 | 151 159 |
| CXCL1 | Cytokine | Promotes T cell and neutrophil recruitment | CXCR2 | 151 159 |
| CXCL10 | Cytokine | Promotes T cell and neutrophil recruitment | CXCR3 | 151 158 159 |
| Cytosolic RNA | Nucleic acid | Promotes the secretion of type I IFN and other proinflammatory factors | MDA5 RIG-I TLR3 |
152 |
| Cytosolic DNA | Nucleic acid | Promotes the secretion of type I IFN and other proinflammatory factors | AIM2 CGAS ZBP1 |
162 164 |
| ERp57 | ER chaperone | Promotes the uptake of dying cells by APCs | LRP1 (?) | 160 |
| Extracellular DNA | Nucleic acid | Promotes the recruitment and activation of neutrophils | TLR9 | 151 163 |
| F-actin | Cytoskeletal component | Promotes the uptake of dying cells by APCs | CLEC9A | 161 |
| HMGB1 | Nuclear DNA-binding protein | Promotes the maturation and cross-presentation activity of APCs | AGER TLR2 TLR4 |
153 154 |
| HSP70 | ER chaperone | Favors the uptake of dying cells by APCs | LRP1 | 21 |
| HSP90 | ER chaperone | Favors the uptake of dying cells by APCs | LRP1 | 21 |
| TFAM | Transcription factor | Promotes APC maturation and recruitment | AGER | 36 |
| Type I IFN | Cytokine | Promotes APC maturation, cross-presentation, and T cell recruitment | IFNARs | 158 194 195 |
AGER, advanced glycosylation end-product specific receptor; AIM2, absent in melanoma 2; ANXA1, annexin A1; APC, antigen-presenting cell; CALR, calreticulin; CCL, C-C motif chemokine ligand 2; CGAS, cyclic GMP-AMP synthase; CLEC9A, C-type lectin domain containing 9A; CXCL1, C-X-C motif chemokine ligand 1; CXCL10, C-X-C motif chemokine ligand 10; CXCR2, C-X-C motif chemokine receptor 2; CXCR3, C-X-C motif chemokine receptor 3; DAMP, danger-associated molecular pattern; ER, endoplasmic reticulum; FPR1, formyl peptide receptor 1; HMGB1, high mobility group box 1; HSP, heat shock protein; ICD, immunogenic cell death; IFN, interferon; IFNAR, interferon-alpha/beta receptor; LRP1, LDL receptor-related protein 1; P2RY2, purinergic receptor P2Y2; P2X7, purinergic receptor P2X 7; TFAM, transcription factor A, mitochondrial; TLR2, Toll-like receptor 2; TLR3, toll like receptor 3; TLR4, toll like receptor 4; TLR9, toll like receptor 9; ZBP1, Z-DNA binding protein 1.
Macroautophagy (herein referred to as autophagy) is a cytoprotective lysosomal pathway for the degradation of superfluous or potentially dangerous cytosolic material and organelles.173–176 Robust evidence indicates that autophagy is required for the preservation of lysosomal ATP stores in the course of most (but not all) variants of ICD.149 166 As a consequence of cellular blebbing, which is mediated by lysosomal-associated membrane protein 1 (LAMP1) and pannexin 1 (PANX1), ATP is released into the extracellular space.165 177 As an alternative, the ICD-associated release of ATP can occur through anterograde ER-to-Golgi transport.178 Extracellular ATP exerts its adjuvanticity by binding to purinergic receptor P2Y2 (P2RY2), which favors the recruitment of APCs and their precursors, and purinergic receptor P2X7 (P2RX7), which favors their activation and consequent release of the immunostimulatory cytokine interleukin (IL)−1β.149 150 179
The integrated stress response (ISR), a multipronged molecular mechanism for the preservation of cellular homeostasis is required for the exposure of ER chaperones on the cell surface during ICD.46 180–182 In particular, ER stress induced by anthracyclines stimulates the inactivating phosphorylation of eukaryotic translation initiation factor 2 subunit alpha (EIF2S1, best known as eIF2α) by eukaryotic translation initiation factor 2 alpha kinase 3 (EIF2AK3, best known as PERK),183 culminating in the CASP8- and B cell receptor associated protein 31 (BCAP31)-dependent translocation of ER chaperones to the outer leaflet of the plasma membrane.160 178 180 For most ICD inducers, the entire process also relies on anterograde ER-to-Golgi transport mediated by vesicle-associated membrane protein 1 (VAMP1) and synaptosomal-associated protein 25 (SNAP25)53 180 184 and requires the concomitant production of ROS.71 Of note, the ICD-associated exposure of some ER chaperones (notably, CALR) on the cell surface appears to be regulated by C-X-C motif chemokine ligand 8 (CXCL8),184 ER Ca2+ levels,185 as well as CASP2, long non-coding RNAs (eg, ncRNA-RB1 and miR-27a), and plasma membrane integrins, at least in some settings.186–189 Surface-exposed CALR (and other ER chaperons) promotes the uptake of dying cells or their corpses by APCs, at least in some settings on interaction with LDL receptor related protein 1 (LRP1).178 190 Moreover, CALR exposure appears to drive type I IFN secretion by APCs,191 192 which is also expected to contribute to the immunogenicity of RCD.
The mechanisms for the preservation of cellular homeostasis in response to infection are also intimately involved in the adjuvanticity of ICD, even when the latter is not driven by microbes.193 Indeed, multiple nucleic acids of endogenous derivation can be detected by PRRs to initiate danger signalling, generally based on ectopic localization or structural modifications that arise during stress responses.135 Thus, chemotherapy-driven ICD involves the activation of TLR3 by endogenous RNA species, resulting in type I IFN secretion and the consequent initiation of an autocrine loop that culminates with CXCL10 release.158 Along similar lines, cancer cells succumb to ionizing radiation as they produce type I IFN downstream of CGAS signalling driven by cytosolic dsDNA.194 195 By binding to its cognate receptor, type I IFN mediates robust immunostimulatory effects on both APCs and effector cells.196 It also triggers the production of IFN-stimulated genes (ISGs) like CXCL10. CXCL10 then acts as a chemoattractant for T cells and, together with CXCL1 and CCL2, for neutrophils, which (at least in some settings) appear to contribute to the ICD-driven killing of residual cancer cells in an antigen-independent fashion.151 158 Of note, cancer cell-derived nucleic acids can also mediate immunostimulatory effects by driving type I IFN secretion in APCs, generally on the transfer of nucleic acid-containing extracellular microvesicles between these two cellular compartments.195 197
The precise stress responses involved in the release of other ICD-associated DAMPs including ANXA1 and HMGB1 remain to be fully elucidated. Irrespective of this unknown, ANXA1 is known to spatially direct APCs to dying cells on interaction with formyl peptide receptor 1 (FPR1),157 while HMGB1 can mediate robust immunostimulatory functions by signaling via advanced glycosylation end-product specific receptor (AGER, also known as RAGE) and TLR4,153 154 167 198–200 an activity that appears to depend on oxidation status.201 202 Moreover, it has become clear that the molecular machinery involved in ICD-associated DAMP emission exhibits some degree of variation depending on ICD inducer and cell type. Thus, CALR exposure and ATP secretion are required for the full-blown immunogenicity of cancer cells succumbing to hypericin-based PDT, but this can occur independently of eIF2α phosphorylation and autophagy activation.178 203 Likewise, neoplastic cells succumbing to necroptosis-driven ICD release ATP and HMGB1, but CALR is exposed at low levels on the plasma membrane and ISR activation appears to be dispensable.59 204
The critical role of DAMP signaling in the immunogenicity of RCD has been established by a plethora of complementary mechanistic experiments aimed at: (1) the pharmacological or genetic inhibition of the signaling pathways involved in DAMP release (eg, with cancer cells expressing a non-phosphorylatable variant of eIF2α)180; (2) the artificial activation of stress responses responsible for DAMP emission (eg, with ER stressors in neoplastic cells undergoing a variant of RCD otherwise not associated with activation of the ISR)205 206; (3) the knockout or knockdown of DAMP-coding genes (eg, with cancer cells depleted of HMGB1 by RNA interference)23 153; (4) DAMP neutralization/blockade (eg, with cells overexpressing an intracellular enzyme degrading cytosolic dsDNA)194; (5) the exogenous complementation of DAMPs (eg, with recombinant CALR administered to cells undergoing RCD in the absence of CALR exposure)22 180 207 208; (6) the upregulation of antagonistic processes (eg, with cancer cells overexpressing the antiphagocytic molecule CD47)209 210; (7) the blockade of DAMP receptors (eg, with monoclonal antibodies specific for type I IFN receptors)158 and (8) the knockout or knockdown of genes encoding for DAMP receptors in the host (eg, with Fpr1-/- mice).157 Several lines of clinical evidence also suggest that proficient danger signaling is critical for cancer patients to obtain clinical benefits from ICD-inducing therapies.211 These findings generally relate to the prognostic or predictive value of (1) activation of stress responses impinging on DAMP emission in cancer cells (eg, eIF2α phosphorylation in association with high CALR levels in biopsies from patients with non-small cell lung cancer)212; (2) the expression levels of specific DAMPs (eg, HMGB1 levels in biopsies from patients with breast cancer subjected to adjuvant anthracycline-based chemotherapy)213 214; (3) DAMP emission by cancer cells (eg, CALR exposure on blasts in patients with acute myeloid leukemia)215 216; (4) actual danger signaling in the TME (eg, gene signatures of type I IFN signaling in subjects with breast cancer)217; (5) loss-of-function polymorphisms in genes encoding DAMP receptors (eg, P2R× 7, TLR4 and FPR1 polymorphisms in patients with breast carcinoma receiving neoadjuvant anthracyclines)23 150 153 157 and (6) the expression levels of DAMP antagonists (eg, CD47 expression on cancer cells in patients with acute myeloid leukemia, esophageal squamous cell carcinoma and ovarian clear cell carcinoma).218–220 These are only a few examples corroborating the relevance of DAMP signaling for RCD to be sensed as immunogenic in patients.
Microenvironmental factors influencing ICD
Although some tissues respond to pathogenic infection more robustly than others (reflecting the differential abundance of tissue-resident APCs), cells succumbing to microbial infection generally drive adaptive immunity irrespective of anatomical location.221 Conversely, the microenvironment of dying cancer cells is a major determinant of their ability to initiate adaptive immune responses, even in the presence of sufficient antigenicity and adjuvanticity,5 222 and this has major implications for the choice of experimental models for the assessment of ICD in vivo (see In vivo models).
There are several mechanisms whereby the microenvironment of developing tumors can antagonize the initiation or execution of ICD, largely reflecting the ability of various neoplasms to establish peripheral tolerance. So-called ‘cold’ and ‘excluded’ tumors are poorly infiltrated by immune cells including APCs and their precursors at baseline, implying that the likelihood for dying cancer cells and their corpses to be productively processed and drive cross-priming is reduced.223 224 Priming is also limited by coinhibitory receptors expressed by tumor-infiltrating T cells including CTL-associated protein 4 (CTLA4) and hepatitis A virus cellular receptor 2 (HAVCR2, best known as TIM-3), a glycoprotein that binds to HMGB1 as well as the ‘eat me’ signal phosphatidylserine on the surface of dying cells.152 225
Moreover, the activity of APCs that infiltrate malignant lesions is generally inhibited by immunosuppressive cytokines including (but not limited to) IL-10 and transforming growth factor beta 1 (TGFB1).226 227 These bioactive factors are abundantly produced in response to hypoxia and during chronic inflammation, and are robustly associated with immunoevasion and tumor progression.228 IL-10 and TGFB1 are secreted by cancer cells and by immunosuppressive immune cells actively recruited to the TME, such as CD4+CD25+FOXP3+ regulatory T (TREG) cells, M2-polarized tumor-associated macrophages (TAMs), and/or myeloid-derived suppressor cells (MDSCs).229–231 Importantly, these immune cell populations express high levels of ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1, best known as CD39) and 5'-nucleotidase ecto (NT5E, best known as CD73),232–234 two enzymes that cooperate to convert extracellular ATP into adenosine, which also mediates robust immunosuppressive effects.235 Thus, TREG cells, M2-polarized TAMs and MDSCs also have direct ICD antagonizing effects.
The redox status of the TME and individual DAMPs or their receptors may also affect the ability of RCD to drive adaptive anticancer immunity. For example, the release of oxidized HMGB1 by cancer cells undergoing pyroptosis, a gasdermin-dependent form of RCD generally associated with inflammasome activation,1 limits anticancer immunity as it favors the expression of coinhibitory ligands.236 In contrast, oxidized mitochondrial DNA favors inflammasome activation and hence the secretion of immunostimulatory factors such as IL-1β in the TME,237 although the actual pathologic relevance of this pathway remains unknown.
Another major mechanism for progressing tumors to evade ICD at the execution phase (ie, the ability of ICD-driven CTLs to mediate cytotoxic effects) relies on immune exhaustion, that is, the establishment of dysfunction in tumor-infiltrating T cells.238–241 Coinhibitory receptors including programmed cell death 1 (PDCD1, also known as PD-1) are major (but not the sole) players in this setting. Indeed, activated CTLs have elevated metabolic demands, and both glucose and amino acids are generally limited in the TME.242 243 Moreover, several immunosuppressive metabolites that are enriched in the TME besides adenosine, such as kynurenine and lactate,244 245 and cytokine-dependent immunosuppression also operate on CTLs.224 246 247 Finally, vascular defects and the dense stromal reaction that characterize some tumors (eg, pancreatic cancer) can constitute a physical barrier to tumor infiltration by CTLs cross-primed in tumor-draining lymph nodes,248 249 de facto hampering the execution phase of ICD. Besides exemplifying the critical importance of local microenvironment for the immunogenicity of RCD, these observations explain why the same cancer cells receiving the same ICD inducers in vitro and in vivo may differ in their ability to initiate adaptive immunity.
Importantly, multiple ICD-inducing therapeutics as well as various therapies that do not promote ICD can mediate immunomodulatory effects on the TME by directly interacting with immune cell populations (rather than with cancer cells).73 Although such immunomodulatory effects are important for the ultimate efficacy of therapy in patients, they are conceptually and mechanistically unrelated to ICD induction, and hence will not be discussed further here.
Detection of ICD in cancer
Over the past two decades, an intensive wave of investigation has unveiled major mechanistic and correlative aspects of ICD as a process culminating in the activation of adaptive immunity against dying cells. Experimental strategies conceived to assess the immunogenicity of RCD encompass: (1) the study of DAMP emission from (and activation of relevant stress responses in) dying cells; (2) biochemical and functional tests to assess the activation of APCs and their ability to mediate cross-priming, in vitro and (3) the ability of dying cells to initiate adaptive immunity in vivo, in immunocompetent syngeneic hosts. Here, we will summarize current methods to assess ICD in oncological settings.
In vitro assays with cancer cells
Robust experimental evidence indicates that the ability of RCD to promote adaptive immunity critically relies on a progressing (but not exhausted) CTL response.121 Presumably, this reflects the need for the availability of one or more TNAs/TAAs above a specific threshold, coupled to the timely delivery of danger signals (which are likely to vary depending on RCD inducer and tumor cell type). Thus, while mouse cancer cells exposed to cardiac glycosides release ICD-associated DAMPs, they are unable to initiate protective anticancer immunity once inoculated in immunocompetent syngeneic hosts as cytotoxicity is limited and tumors develop at the vaccination sites.250 Similarly, repeated freeze-thawing causes rapid cell death coupled to massive TNA/TAA release, but mouse cancer cells subjected to this harsh procedure cannot be used to protect syngeneic immunocompetent hosts against a subsequent challenge with living cells of the same type, most likely due to suboptimal adjuvanticity.20 59 The cytotoxic response driven by ICD inducers has classically been assessed by terminal cell death biomarkers such as plasma membrane permeabilization, as well as by conventional biomarkers of apoptosis, including phosphatidylserine exposure, mitochondrial transmembrane potential dissipation and initiator or effector caspase activation.148 The rationale and principles of these assays have been extensively described by us and others,58 251 252 and hence will not be discussed further here. That said, the existence of ICD variants relying on the necroptotic machinery and/or occurring independent of caspase activation1 59 61 identifies a need for tools to measure ICD induction that can accommodate all potentially relevant RCD pathways.
Intracellular responses to stress mechanistically involved in DAMP release have also been employed as surrogate ICD biomarkers. ISR activation has generally been monitored in its three reticular components by the assessment of eIF2α phosphorylation, X-box binding protein 1 (XBP1) splicing and activating transcription factor 6 (ATF6) nuclear translocation, in some instances along with heat shock protein family A (Hsp70) member 5 (HSPA5, also known as GRP78) upregulation. These biomarkers are detected by immunoblotting, flow cytometry, immunofluorescence microscopy, immunohistochemistry and/or RT-PCR, using dedicated antibodies or probes.33 183 253 ICD-associated PRR activation in cancer cells has been classically measured by immunoblotting with antibodies specific for key phosphorylated transducers, such as phosphorylated IFN regulatory factor 3 (IRF3) downstream of CGAS signaling,254 255 or with transcription-based luminescent reporters.254 256–258 As an alternative approach, the ectopic accumulation of nucleic acids has been monitored, generally by immunofluorescence microscopy with dedicated antibodies or by subcellular fractionation coupled to optional enzymatic digestion of nucleic acids and absorbance-based quantification.158 256 Activation of autophagy in cells exposed to potential ICD inducers has largely been monitored by the concomitant assessment of microtubule associated protein 1 light chain 3 (MAP1LC3, best known as LC3) lipidation and degradation of an autophagic substrate such as sequestosome 1 (SQSTM1, also known as p62) by immunoblotting, in the context of appropriate control conditions.259 Autophagy is indeed a dynamic process and the mere analysis of LC3 lipidation in cells responding to stress conditions cannot be employed to discriminate between autophagy activation and inhibition.259
Cell surface exposure of ER chaperones as a surrogate biomarker of ICD can be determined by (1) flow cytometry in non-permeabilized early apoptotic cells using specific antibodies or dedicated constructs (eg, CALR fused to the HaloTag) combined with vital dyes like 7-aminoactinomycin D (7-AAD), 4′,6-diamidino-2-phenylindole (DAPI) or propidium iodide (PI), which enable the exclusion of dead cells from the analysis41 180 205 260; (2) fluorescence microscopy, in cells fixed with low concentration of paraformaldehyde and then immunostained with antibodies specific for ER chaperones21 32 43 160 261; (3) fluorescence microscopy or video microscopy, in cells constitutively expressing a construct in which specific the ER chaperone of choice is fused to GFP or another fluorescent tag33 205 250 262 and (4) immunoblotting, in cells previously subjected to cell surface protein biotinylation followed by streptavidin-mediated precipitation.160 263
Two main approaches have been harnessed to determine the release of ICD-associated soluble DAMPs and cytokine from dying cells: (1) DAMP/cytokine quantification in culture supernatants and (2) quantification of residual DAMPs/cytokines in the intracellular microenvironment. Of note, while the former approach is universally applicable, the latter cannot be employed for DAMPs and cytokines that are actively synthesized before release, such as type I IFN and CXCL10.148 Thus, extracellular and intracellular ATP can be quantified with commercial luminescence-based assays on the culture supernatants and cell lysates, respectively.149 150 178 264 Extracellular ATP and its degradation products (ADP, AMP and adenosine) can also be quantified by targeted mass spectrometry,149 while its intracellular counterpart can be monitored by flow cytometry or fluorescence microscopy on staining with the ATP-specific dye quinacrine or implementation of ATP-specific fluorescence resonance energy transfer (FRET)-based reporters.165 265 Finally, ANXA1, HMGB1, type I IFN and CXCL10 secretion by cells undergoing RCD have been classically monitored by commercial ELISA or immunoblotting on culture supernatants or cell lysates.32 151 153 157 158 266 Moreover, a fluorescent version of HGMB1 is available that enables the assessment of HMGB1 release by fluorescence microscopy or video microscopy, as a function of residual cell fluorescence.33 42 250 267 Of note, while RT-PCR is commonly employed to monitor type I IFN and CXCL10,158 268 this approach de facto measures PRR signaling, as transcription is not necessarily coupled with translation and secretion.
In vitro assays with immune cells
While intracellular responses to stress and DAMP emission can be equally monitored in mouse and human tumor models, in vivo studies can currently be performed only in the former (see In vivo models). To partially circumvent this issue and enable a functional assessment of the immunogenicity of RCD in both the mouse and human system, the field has borrowed multiple classical assays from immunology. In general, these experiments aim at evaluating whether dying cancer cells can stimulate the ability of APCs to optimally cross-prime CTLs and hence initiate an adaptive immune response.
In particular, human or mouse APCs exposed to human or mouse dying cancer cells, respectively, are often investigated for: (1) their ability to engulf dying cells or their corpses; (2) their maturation status and migratory capacity and (3) their ability to cross-present antigenic material to CTLs. Phagocytosis is often assessed by coculturing APCs or their precursors and dying cancer cells on individual prelabeling of both compartments with distinct non-toxic fluorescent dyes that remain in the cytoplasm, such as carboxyfluorescein succinimidyl ester (CFSE) or PKH26, followed by fluorescence microscopy or flow cytometry.32 269 270 As an alternative, only cancer cells are prelabeled, and phagocytosis is monitored on staining the coculture with monoclonal antibodies specific for the APC of choice.264 266 Moreover, apoptotic cell uptake has been measured by injecting pre-labeled dying mouse cancer cells intravenous, followed by splenocyte isolation and flow cytometry.268 270 The maturation status of APCs has been classically measured by flow cytometry, on staining of cell cocultures with antibodies specific for MHC class II molecules and costimulatory molecules including CD80, CD83 and CD86 (which are all upregulated during maturation).21 43 263 271 As an alternative or complementary approach, functional maturation has been monitored by the detection of cytokines secreted in culture supernatants by APCs acquiring an immunostimulatory phenotype, including (but not limited to) IL-1β, IL-6, IL-12 and IL-23.41 150 263 ELISA and flow cytometry on intracellular staining with dedicated antibodies remain the techniques of choice for the latter strategy. That said, the intracellular assessment of IL-1β requires an antibody directed against the fully mature variant of the protein, as its precursor is not secreted.272 273 Migratory capacity (which reflects the ability of dying cells to secrete chemotactic factors) has been evaluated by transwell assays274 275 or by dedicated microfluidic devices that allow for video microscopy (if cells are prelabeled with fluorescent dyes).157 276
The cross-priming potential of APCs exposed to cancer cells undergoing RCD has most frequently been assessed by coculturing them with syngeneic, naïve T cells, followed by the assessment of: (1) proliferative T cell response, generally by flow cytometry on previous labeling with CFSE271 277; (2) T cell activation status, most often by flow cytometry on staining with monoclonal antibodies specific for surface proteins linked to activation (eg, CD69, LAMP1, PD-1)150 153 278; (3) T cell functional profile, either by flow cytometry on intracellular staining with monoclonal antibodies specific for effector molecules such as IFN-γ, perforin 1 (PRF1) and granzyme B (GZMB) or by quantification of extracellular IFN-γ by ELISPOT.21 41 268 278 Finally, cytotoxic T cell functions on cross-priming can be tested by measuring the lysis of living cancer cells of the same type of those employed for APC pulsing46 279 280 or by analyzing the response to specific TAAs.281 282
In vivo models
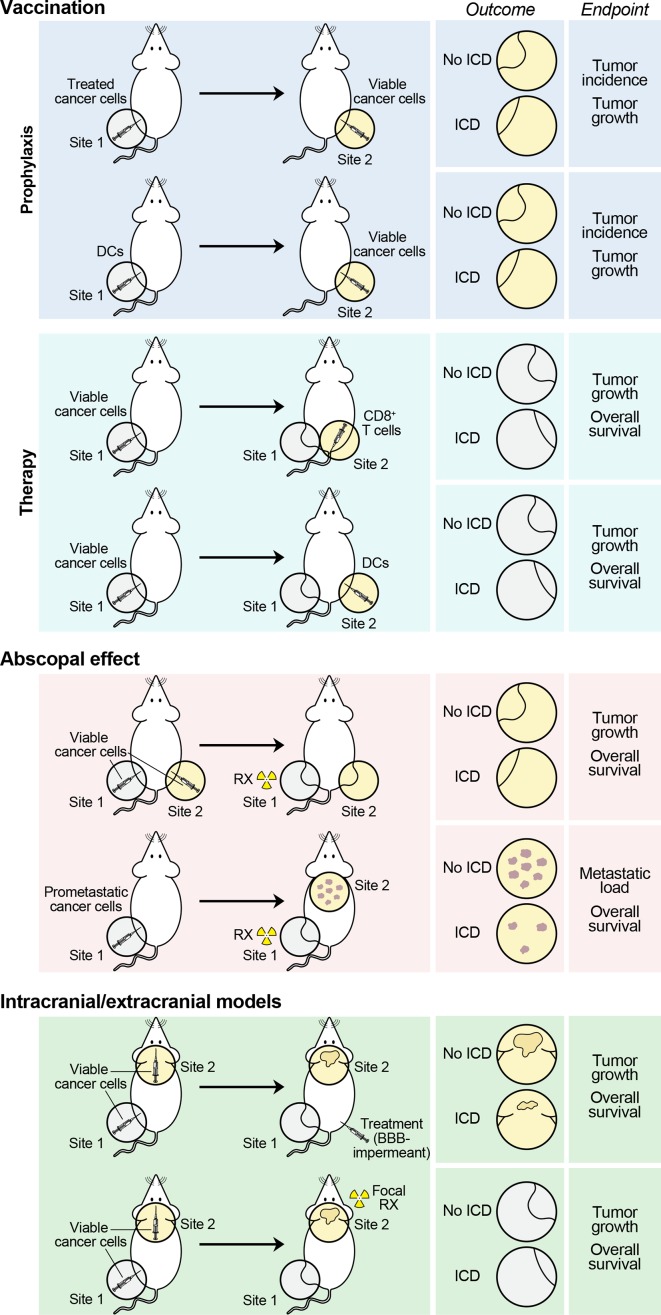
Only a few models are currently available to investigate ICD in vivo (figure 2). The gold standard approach to assess the ability of dying cells to initiate adaptive immunity involves vaccination assays with immunocompetent, syngeneic mice.148 In this context, mouse cancer cells are exposed to a potential ICD inducer in vitro and then administered as a vaccine s.c., in the absence of any immunological adjuvant and on the removal of exogenous chemical entities (if any, such as the ICD inducer itself). One to two weeks later, mice are challenged s.c. with living cancer cells of the same type (at the minimal dose that is 100% effective at generating progressing lesions in naïve mice) and followed over 40–60 days for tumor incidence and growth.20 22 153 178 208 Not only the percentage of tumor-free mice, but also the growth rate of tumors potentially developing despite a vaccine-induced adaptive immune response are usually employed as indicators of (at least some) degree of immunogenicity. Specificity is confirmed by re-challenging tumor-free mice at the end of the experiment with another syngeneic cancer cell line, which is expected to generate progressing neoplastic lesions in 100% of mice. Of note, vaccination can also be performed with APCs exposed to dying cancer cells in vitro or implemented in therapeutic (rather than prophylactic) settings, that is, as a treatment of established tumors.157 283–285 Moreover, CD8+ T cells cross-primed in vitro by APCs exposed to cancer cells undergoing ICD have been employed in adoptive transfer experiments to treat tumors previously established with the living cancer cells of the same type.32 286 Importantly, comparing the efficacy of any RCD inducer against mouse cancer cells growing in immunocompetent, syngeneic versus immunodeficient mice can provide hints on the ability of such intervention to drive ICD (in such case, therapeutic efficacy will be limited in immunodeficient hosts). However, this latter experimental setting is intrinsically unsuitable to discriminate between ICD induction and ICD-unrelated immunostimulation (see Interpretation of ICD).
Figure 2.
Current methods to assess ICD in vivo, in oncological settings. Current models to ascertain the ability of dying cancer cells to elicit an adaptive, tumor-specific immune response in vivo invariably rely on mouse neoplasms established in immunocompetent syngeneic hosts. In prophylactic models, mouse cancer cells succumbing in vitro to a potential inducer of immunogenic celldeath (ICD) are used as a vaccine, either as such, or on loading on immature, syngeneic dendritic cells (DCs). The ability of mice to reject (tumor incidence) or control (tumor growth) a rechallenge with living cancer cells of the same type inoculated 1–2 weeks later is monitored as a sign of protective anticancer immunity. In therapeutic settings, mouse tumors developing in immunocompetent syngeneic hosts are treated with autologous DCs preloaded with cancer cells exposed to a potential ICD inducer in vitro (generally in combination with immunological adjuvants), or with autologous CD8+ cytotoxic lymphocytes primed in vitro by the same DCs (generally in combination with IL-2 or other cytokines that support expansion in vivo). Tumor control and mouse survival are monitored as indicators of therapeutic anticancer immunity. In abscopal models, mouse cancer cells are harnessed to generate lesions at distant anatomical sites (either artificially, or exploiting the natural capacity of some cell lines to generate metastases), followed by treatment at only one disease site (generally in the context of otherwise inactive systemic immunostimulation). Tumor control at the non-treated disease site and mouse survival are monitored as signs of systemic anticancer immunity with therapeutic relevance. Finally, in intracranial/extracranial models, mouse cancer cells are employed to generate one intracranial and one extracranial tumor, only one of which receives treatment (generally, a systemic agent that cannot cross the blood–brain barrier [BBB] for extracranial lesion, or radiation therapy for intracranial lesions, in both cases in combination with otherwise inactive immunostimulants). As in abscopal models, tumor control at the non-treated disease site and mouse survival are monitored as indicators of therapeutic anticancer immunity with systemic outreach. In all these models, mice achieving systemic, long-term disease eradication are often rechallenged with cancer cells to monitor durability (with the same cancer cells employed for disease establishment) and specificity (with unrelated, but syngeneic cancer cells). ICD, immunogenic cell death; IL-2, interleukin 2.
Three alternative approaches to assess ICD in vivo, in immunocompetent syngeneic systems, critically rely on the measurement of tumor growth at non-treated disease sites, which implies they can only be implemented with localized therapies (eg, focal ionizing radiation and intratumoral delivery of therapeutic agents that fail to reach active concentrations systemically)79 287–289 or when the non-treated site is biologically inaccessible to treatment but accessible to CTLs (eg, brain metastases in a host receiving chemotherapeutics that do not cross the blood-brain barrier).290 Models of the so-called ‘abscopal response’, that is, the regression of an out-of-field lesion in patients receiving ionizing radiation to a distant disease site,291 have proven highly instrumental in this setting. Usually, immunocompetent mice are grafted with either (1) cancer cells to generate two slightly asynchronous lesions s.c., at anatomically distant sites or (2) metastasis-prone cancer cells to generate a palpable lesion s.c. and metastatic (lung) dissemination.79 288 292 In both scenarios, only one of the subcutaneous lesion(s) receives ionizing radiation (generally in the presence of an immunostimulatory molecule that has no single-agent systemic effect), and the response of non-irradiated lesion, metastatic load and overall survival are monitored as indicators of ICD induction coupled to activation of adaptive immunity with systemic outreach.78 79 287 288 292 293 Mice rejecting irradiated and non-irradiated lesions can be rechallenged 30–40 days after disease eradication with the same cancer cells employed originally to assess the durability of protection, as well as with syngeneic cancer cells of another type to monitor its specificity.49 292 293 Likewise, distinct cell types can be employed for the generation of primary and secondary lesions to verify that the in situ vaccination effect generated by ICD is antigen specific.294 Finally, models of simultaneous intracranial and extracranial disease have been harnessed to monitor the ability of systemic chemotherapeutic agents that are unable to penetrate the blood–brain barrier and focal ionizing radiation to elicit anticancer immunity in the periphery.295 Of note, in these latter models, the presence of extracranial lesions increases the efficacy of immunotherapy against cranial neoplasms by stimulating CTL trafficking,296 although the relevance of this phenomenon for ICD-driven immunity remains to be ascertained.
All these models are amenable to ex vivo studies aimed at: (1) the characterization of DAMPs released by cancer cells responding to stress in situ; (2) the immunological profiling of APCs and CTLs underlying the initiation and execution of anticancer immunity in vivo and (3) the identification of mechanistic vs correlative aspects of ICD elicited in vivo by the stressor(s) of choice (eg, with depletion, blockage or neutralization strategies).79 287 288 293
Interpretation of ICD
Most of the assays that are currently available to estimate the propensity of RCD to drive adaptive immune responses fail to take into proper consideration the complexity that is inherent to ICD, which (as detailed above) ultimately represents a highly contextual process that depends on (1) initiating stimulus, (2) responding cell and (3) the host (figure 1). In this context, the interpretation of ICD-related assays faces a number of challenges that should be always kept under consideration.
Surrogate biomarkers of ICD, such as the accumulation of cytosolic DNA, the exposure of ER chaperones on the cell surface or the release of ATP and/or HMGB1 by dying cells, as well as biomarkers of the stress responses that underlie their emission, have been instrumental for the characterization of the key molecular players of process, and the identification of potential ICD inducers in large screening efforts.183 184 262 However, not all triggers of ICD operate via the same molecular mechanisms, as demonstrated by the ability of hypericin-based PDT to initiate anticancer immunity irrespective of the autophagic proficiency of malignant cells.203 Moreover, the emission of ICD-associated DAMPs according to the correct spatiotemporal pattern is required, but not sufficient, for APCs to initiate CTL-dependent immune responses against dying cells. Thus, cardiac glycosides trigger a multipronged stress response culminating with all major surrogate biomarkers of ICD, and yet cannot establish protective immunity in gold-standard vaccination assays.250 These observations suggest that the ability of any stressor to drive a variant of RCD associated with adaptive immunity cannot be extrapolated by in vitro assays focusing on cancer cells.
Although their scalability to screening applications is limited, immunological assays testing the ability of APCs primed with dying cells to engage in the sequential process leading to cross-priming (which is an absolute requirement for adaptive immunity in this setting) obviously offer a more precise assessment of the immunogenicity of RCD. However, these assays are also inherently limited in that they are unable to assess two major prerequisites for cross-priming: (1) the ability of APCs to physically reach sites of RCD and (2) the existence of naïve T cell clones specific for antigens expressed by dying cells, ultimately calling for validation with in vivo models.
Vaccination assays have been highly instrumental for the demonstration that syngeneic dying cells can drive adaptive immunity in the presence of adequate antigenicity and adjuvanticity.20 22 153 178 208 297 However, the use of tumor-naïve hosts enables a very high degree of sensitivity, which may not necessarily be advantageous if clinical applications are the ultimate goal. In this context, abscopal and intracranial/extracranial models may offer increased pathophysiological relevance, as (with the limitations described above) they mimic established metastatic disease in humans. However, these models are limited in that they can only measure systemic immunity to local therapies or agents that do not penetrate the blood–brain barrier.79 288 Moreover, largely reflecting the clinical scenario, eliciting systemic immunity with disease-eradicating potential in these models is challenging, and often requires the provision of additional immunostimulatory signals that antagonize peripheral tolerance.77–79 Such a limited sensitivity (although clinically relevant) may be detrimental at early stages of discovery when optimal ICD conditions are yet to be determined.
As a group, ICD-relevant in vivo models are affected by multiple issues including the fact that only murine systems can be investigated, and it cannot be excluded that the molecules and cell populations at play in the human setting may be different (at least to some degree).298 299 Moreover, all current models for the detection of ICD in vivo rely on the establishment of primary lesions with murine cancer cell lines, which (1) have been immunoedited and acquired the ability to evade immunosurveillance in their original host (implying that they do not properly recapitulate primary oncogenesis), (2) have a relatively low and homogeneous mutational burden compared with human disease (and hence fail to recapitulate the mutational and antigenic landscape of human tumors), (3) generally do not establish a complex stromal and endothelial architecture as a consequence of their elevated proliferation rate (which differs from human neoplasms) and (4) are implanted by injection, which per se may mediate at least some degree of immunostimulation.300–303 Finally, the need for fully syngeneic settings (to prevent rejection based on HLA mismatch) restricts remarkably the spectrum of cell lines and rodent strains that can be harnessed for this purpose, with a predominance of models based on the C57BL/6 or BALB/c background.304
Current efforts to circumvent, at least in part, these issues involve the use of orthotopic models (which offer improved microenvironmental features), transgene-driven models (which may offer a superior view on early oncogenesis) and carcinogen-driven models (which have superior heterogeneity).305–307 However, not all of these systems are compatible with vaccination and/or abscopal assays, implying that ICD induction can only be imprecisely addressed by complementing in vitro observations with therapeutic efficacy in immunocompetent vs immunodeficient animals. Moreover, considerable efforts are being devoted to the development of humanized mice, which ultimately may enable the assessment of ICD induction in vivo with human cancer cells.308–310 Most often, these models involve the engraftment of functional human immune cells from various sources into highly immunodeficient mice as a means to (partially and temporarily) reconstitute a functional human immune system.304 The major limitations of this approach (which vary in severity depending on the precise experimental protocol) include: (1) the ability of human immune cells to react against their mouse counterparts due to cross-species incompatibility and consequently graft-versus-host disease311 312; (2) the limited ability of (at least some) mouse cytokines to support immune cell reconstitution and function via human cytokine receptors313; (3) the hitherto poorly understood cross-talk between residual components of the mouse immune system (eg, macrophages, granulocytes, endothelial cells) and reconstituted human cells309 and (4) the lack of thymic selection.314 Although refined strategies to circumvent these issues are being investigated, including the use of mice engineered to express human cytokines as well as the coimplantation of thymic fragments,313 315–320 this technology is hitherto immature for the assessment of ICD in the human system.
In summary, the assessment of ICD requires experimental support from a variety of in vitro and in vivo assays that should cumulatively assess the ability of malignant cells undergoing RCD at the natural anatomical location to recruit APCs and stimulate them to initiate adaptive anticancer immunity.
Conclusions and perspectives
Accumulating evidence demonstrates that the initiation of ICD is critical for the elimination of infectious pathogens and stands out as a major therapeutic goal for cancer therapy, especially in consideration of the current clinical success of ICBs.321 322 Indeed, the ability of several agents to drive ICD in oncological settings is hampered by the robust immunosuppressive circuitries established in the TME during the tumor-host coevolution, and ICBs may be instrumental for the inactivation of such circuitries.56 77 79 323 324
There are three major obstacles precluding the full clinical potential of ICD inducers to be realized. First, most of these agents have been developed clinically based on the paradigm of maximum tolerated dose (MTD), and it is now clear that greater cytotoxicity does not necessarily coincide with optimal immunogenicity.194 Moreover, the vast majority of clinically employed ICD inducers have been developed preclinically in immunodeficient models of disease, implying that little is known of their effects on the host immune system.299 302 Second, despite considerable progress over the past two decades, our understanding of RCD-associated DAMP signaling remains limited. In particular, limited attention has been dedicated to the study of immunosuppressive DAMPs, including (but not limited to) phosphatidylserine, prostaglandin E2, and adenosine, especially in the context of ICD.68 325–327 Intriguingly, MAMPs can also be immunosuppressive, and these are important for the establishment of symbiosis.328 The impact of metabolism,329 the gut microbiota330 and the central nervous system331 on the release and activity of DAMPs also remains to be elucidated. Finally, the key role of the host in ICD detection has been mostly studied from an adjuvanticity perspective, that is, linked to the capacity of the host to decode DAMP signaling via PRRs.5 Conversely, little is known about the TCR repertoire of hosts that respond to ICD with robust adaptive immunity versus hosts that do not, and about the impact of environmental and behavioral features (eg, microbiome, dietary habits, stress) on host ICD sensing.
Along with the establishment of humanized rodent models that enable the investigation of ICD in vivo (although with the caveats of a murine microenvironment), we believe that these issues currently stand as the major challenges for the field in the near future. We surmise that the clinical efficacy of numerous agents currently employed for the management of cancer could be remarkably boosted if we acquire the capacity to use them as ICD inducers. Novel technologies are constantly improving our ability to monitor the immunological changes occurring in patients responding to ICD inducers, including alterations in the intratumoral and circulating TCR repertoire.332 Overall, the time is mature to take on the challenge to realize the clinical potential of ICD inducers and improve disease outcome for a variety of patients with cancer.
Acknowledgments
LG is supported by a Breakthrough Level 2 grant from the US Department of Defense (DoD), Breast Cancer Research Program (BRCP) (#BC180476P1), by a startup grant from the Dept. of Radiation Oncology at Weill Cornell Medicine, by industrial collaborations with Lytix and Phosplatin, and by donations from Sotio a.s., Phosplatin, and the Luke Heller TECPR2 Foundation. IV is supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC, IG 2017 grant number 20417) and a startup grant from the Italian Institute for Genomic Medicine (Candiolo, Turin, Italy) and Compagnia di San Paolo (Turin, Italy).
Footnotes
LG, IV and SW contributed equally.
Contributors: LG, IV, SW, AC and FM conceived the article. LG, IV and SW wrote the first version of the manuscript with constructive input from all authors. LG, IV and SW addressed concerns raised by coauthors and reviewers. All authors approved the final version of the article.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing Interests: LG has provided/provides remunerated consulting to AstraZeneca, Boehringer Ingelheim, Inzen and the Luke Heller TECPR2 Foundation, serves on advisory boards for Boehringer Ingelheim and OmniSEQ, has received research support from Lytix Biopharma and Phosplatin, and is an inventor on a patent concerning the use of caspase inhibitors for cancer therapy. TAC is a co-founder of Gritstone Oncology, holds equity in An2H and Gritstone Oncology, has served as an advisor for Bristol-Myers, MedImmune, Squibb, Illumina, Eisai, AstraZeneca, and An2H, has received grant funding from Bristol-Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H, and Eisai, holds ownership of intellectual property on using tumor mutation burden to predict immunotherapy response, with pending patent, which has been licensed to PGDx. GC has received honoraria for consultations or presentations by Roche, Genentech, BMS, AstraZeneca, Sanofi-Aventis, Nextcure and GeneosTx, has received grants, research support or is coinvestigator in clinical trials from/by BMS, Celgene, Boehringer Ingelheim, Roche, Iovance and Kite, has patents in the domain of antibodies and vaccines targeting the tumor vasculature as well as technologies related to T-cell expansion and engineering for T-cell therapy, and holds patents around TEM1 antibodies and receives royalties from the University of Pennsylvania regarding technology licensed to Novartis. SD has received compensation for consultant/advisory services from AstraZeneca, EMD Serono, Lytix Biopharma, and Mersana Therapeutics, and has received research support from Lytix Biopharma and Nanobiotix. ED has received personal fees from Accuray, Amgen, AstraZeneca, Merck Serono, and Roche, and has received grants from AstraZeneca, Boehringer, BMS, Lilly, Merck Serono, MSD, Roche and Servier. DD is an employee and options holder of Calidi Biotherapeutics. USG has served on the advisory boards for AstraZeneca, and BMS, and has received funding for IITs from MSD and AstraZeneca. AH is shareholder in Targovax ASA, and employee and shareholder in TILT Biotherapeutics Ltd. DH is employed by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA. HLK is an employee of Replimune, Inc. OK is a cofounder of Samsara Therapeutics. GK has been holding research contracts with Bayer Healthcare, Genentech, Glaxo Smyth Kline, Institut Mérieux, Kaleido, Lytix Pharma, Nucana, Oncolinx, PharmaMar, Samsara, Sotio and Vasculox, serves on the Board of Directors of the Bristol Myers Squibb Foundation France, and is a scientific co-founder of everImmune and Samsara Therapeutics. SL has acted as non-compensated consultant for AstraZeneca, BMS, Merck, Novartis, Pfizer, Roche-Genentech and Seattle Genetics, and as compensated consultant (via her institution) for Aduro Biotech, and receives research funding from BMS, Eli Lilly, Merck, Novartis, Puma Biotechnology, Pfizer and Roche-Genentech, MTL serves as an advisor for Checkmate, iRepetoire, Nurix, Myst, Instilbio, and Torque. TM is consultant for Leap Therapeutics, Immunos Therapeutics and Pfizer, co-founder of Imvaq therapeutics, has equity in Imvaq therapeutics, reports grants from Bristol-Myers Squibb, Surface Oncology, Kyn Therapeutics, Infinity Pharmaceuticals, Peregrine Pharmeceuticals, Adaptive Biotechnologies, Leap Therapeutics, Aprea, and is inventor on patent applications related to work on oncolytic viral therapy, alphavirus-based vaccines, neo-antigen modeling, CD40, GITR, OX40, PD-1 and CTLA-4. ØR is an employee and shareholder in Lytix Biopharma. MR serves as an advisor for Gelesis, MillBo, Casillo, has research contracts with fKraftHeinz, Gelesis, AlfaSigma and is founder of Postbiotica. JS is a permanent member of the scientific advisory board and owns stocks of Surface Oncology. TY has received research support from Lytix Biopharma and Phosplatin. DZ reports consulting fees from Merck, Synlogic Therapeutics, Tesaro, Trieza Therapeutics, and Western Oncolytics, and is an inventor on a patent concerning the use of Newcastle Disease Virus for cancer therapy. LZ has served on the Board of Directors of Transgene and on advisory boards for EpiVax, Lytix Biopharma, NeoVax, Transgene, Vedanta, has research contracts with BMS, GSK, Incyte, Innovate Bopharma, Kaleido, Pilege, and Transgene, and is founder of everImmune.
p, li { white-space: pre-wrap; }
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the nomenclature Committee on cell death 2018. Cell Death Differ 2018;25:486–541. 10.1038/s41418-017-0012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tang D, Kang R, Berghe TV, et al. The molecular machinery of regulated cell death. Cell Res 2019;29:347–64. 10.1038/s41422-019-0164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Green DR, Ferguson T, Zitvogel L, et al. Immunogenic and tolerogenic cell death. Nat Rev Immunol 2009;9:353–63. 10.1038/nri2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell 2011;147:742–58. 10.1016/j.cell.2011.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galluzzi L, Buqué A, Kepp O, et al. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 2017;17:97–111. 10.1038/nri.2016.107 [DOI] [PubMed] [Google Scholar]
- 6. Torchinsky MB, Garaude J, Martin AP, et al. Innate immune recognition of infected apoptotic cells directs Th17 cell differentiation. Nature 2009;458:78–82. 10.1038/nature07781 [DOI] [PubMed] [Google Scholar]
- 7. Campisi L, Barbet G, Ding Y, et al. Apoptosis in response to microbial infection induces autoreactive Th17 cells. Nat Immunol 2016;17:1084–92. 10.1038/ni.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nair-Gupta P, Baccarini A, Tung N, et al. Tlr signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell 2014;158:506–21. 10.1016/j.cell.2014.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fend L, Yamazaki T, Remy C, et al. Immune checkpoint blockade, immunogenic chemotherapy or IFN-α blockade boost the local and Abscopal effects of oncolytic virotherapy. Cancer Res 2017;77:4146–57. 10.1158/0008-5472.CAN-16-2165 [DOI] [PubMed] [Google Scholar]
- 10. Koks CA, Garg AD, Ehrhardt M, et al. Newcastle disease virotherapy induces long-term survival and tumor-specific immune memory in orthotopic glioma through the induction of immunogenic cell death. Int J Cancer 2015;136:E313–25. 10.1002/ijc.29202 [DOI] [PubMed] [Google Scholar]
- 11. Donnelly OG, Errington-Mais F, Steele L, et al. Measles virus causes immunogenic cell death in human melanoma. Gene Ther 2013;20:7–15. 10.1038/gt.2011.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown MC, Holl EK, Boczkowski D, et al. Cancer immunotherapy with recombinant poliovirus induces IFN-dominant activation of dendritic cells and tumor antigen–specific CTLs. Sci Transl Med 2017;9:eaan4220 10.1126/scitranslmed.aan4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hirvinen M, Rajecki M, Kapanen M, et al. Immunological effects of a tumor necrosis factor Alpha–Armed oncolytic adenovirus. Hum Gene Ther 2015;26:134–44. 10.1089/hum.2014.069 [DOI] [PubMed] [Google Scholar]
- 14. Siurala M, Bramante S, Vassilev L, et al. Oncolytic adenovirus and doxorubicin-based chemotherapy results in synergistic antitumor activity against soft-tissue sarcoma. Int J Cancer 2015;136:945–54. 10.1002/ijc.29048 [DOI] [PubMed] [Google Scholar]
- 15. Liikanen I, Ahtiainen L, Hirvinen MLM, et al. Oncolytic adenovirus with temozolomide induces autophagy and antitumor immune responses in cancer patients. Molecular Therapy 2013;21:1212–23. 10.1038/mt.2013.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diaconu I, Cerullo V, Hirvinen MLM, et al. Immune response is an important aspect of the antitumor effect produced by a CD40L-encoding oncolytic adenovirus. Cancer Res 2012;72:2327–38. 10.1158/0008-5472.CAN-11-2975 [DOI] [PubMed] [Google Scholar]
- 17. Zhou H, Forveille S, Sauvat A, et al. The oncolytic peptide LTX-315 triggers immunogenic cell death. Cell Death Dis 2016;7:e2134 10.1038/cddis.2016.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Forveille S, Zhou H, Sauvat A, et al. The oncolytic peptide LTX-315 triggers necrotic cell death. Cell Cycle 2015;14:3506–12. 10.1080/15384101.2015.1093710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou H, Sauvat A, Gomes-da-Silva LC, et al. The oncolytic compound LTX-401 targets the Golgi apparatus. Cell Death Differ 2016;23:2031–41. 10.1038/cdd.2016.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Casares N, Pequignot MO, Tesniere A, et al. Caspase-Dependent immunogenicity of doxorubicin-induced tumor cell death. J Exp Med 2005;202:1691–701. 10.1084/jem.20050915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fucikova J, Kralikova P, Fialova A, et al. Human tumor cells killed by anthracyclines induce a tumor-specific immune response. Cancer Res 2011;71:4821–33. 10.1158/0008-5472.CAN-11-0950 [DOI] [PubMed] [Google Scholar]
- 22. Obeid M, Tesniere A, Ghiringhelli F, et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med 2007;13:54–61. 10.1038/nm1523 [DOI] [PubMed] [Google Scholar]
- 23. Tesniere A, Schlemmer F, Boige V, et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene 2010;29:482–91. 10.1038/onc.2009.356 [DOI] [PubMed] [Google Scholar]
- 24. Kopecka J, Salaroglio IC, Righi L, et al. Loss of C/EBP-β lip drives cisplatin resistance in malignant pleural mesothelioma. Lung Cancer 2018;120:34–45. 10.1016/j.lungcan.2018.03.022 [DOI] [PubMed] [Google Scholar]
- 25. Spisek R, Charalambous A, Mazumder A, et al. Bortezomib enhances dendritic cell (DC)–mediated induction of immunity to human myeloma via exposure of cell surface heat shock protein 90 on dying tumor cells: therapeutic implications. Blood 2007;109:4839–45. 10.1182/blood-2006-10-054221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christiansen AJ, West A, Banks K-M, et al. Eradication of solid tumors using histone deacetylase inhibitors combined with immune-stimulating antibodies. Proc Natl Acad Sci U S A 2011;108:4141–6. 10.1073/pnas.1011037108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fragale A, Romagnoli G, Licursi V, et al. Antitumor effects of Epidrug/IFNα combination driven by modulated gene signatures in both colorectal cancer and dendritic cells. Cancer Immunol Res 2017;5:604–16. 10.1158/2326-6066.CIR-17-0080 [DOI] [PubMed] [Google Scholar]
- 28. Riganti C, Lingua MF, Salaroglio IC, et al. Bromodomain inhibition exerts its therapeutic potential in malignant pleural mesothelioma by promoting immunogenic cell death and changing the tumor immune-environment. Oncoimmunology 2018;7:e1398874 10.1080/2162402X.2017.1398874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. West AC, Mattarollo SR, Shortt J, et al. An intact immune system is required for the anticancer activities of histone deacetylase inhibitors. Cancer Res 2013;73:7265–76. 10.1158/0008-5472.CAN-13-0890 [DOI] [PubMed] [Google Scholar]
- 30. Sonnemann J, Greßmann S, Becker S, et al. The histone deacetylase inhibitor vorinostat induces calreticulin exposure in childhood brain tumour cells in vitro. Cancer Chemother Pharmacol 2010;66:611–6. 10.1007/s00280-010-1302-4 [DOI] [PubMed] [Google Scholar]
- 31. Sagiv-Barfi I, Kohrt HE, Burckhardt L, et al. Ibrutinib enhances the antitumor immune response induced by intratumoral injection of a TLR9 ligand in mouse lymphoma. Blood 2015;125:2079–86. 10.1182/blood-2014-08-593137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pozzi C, Cuomo A, Spadoni I, et al. The EGFR-specific antibody cetuximab combined with chemotherapy triggers immunogenic cell death. Nat Med 2016;22:624–31. 10.1038/nm.4078 [DOI] [PubMed] [Google Scholar]
- 33. Liu P, Zhao L, Pol J, et al. Crizotinib-induced immunogenic cell death in non-small cell lung cancer. Nat Commun 2019;10:1486 10.1038/s41467-019-09415-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bugaut H, Bruchard M, Berger H, et al. Bleomycin exerts ambivalent antitumor immune effect by triggering both immunogenic cell death and proliferation of regulatory T cells. PLoS One 2013;8:e65181 10.1371/journal.pone.0065181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen H-M, Wang P-H, Chen S-S, et al. Shikonin induces immunogenic cell death in tumor cells and enhances dendritic cell-based cancer vaccine. Cancer Immunol Immunother 2012;61:1989–2002. 10.1007/s00262-012-1258-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang M, Li C, Zhu S, et al. Tfam is a novel mediator of immunogenic cancer cell death. Oncoimmunology 2018;7:e1431086 10.1080/2162402X.2018.1431086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ho WS, Wang H, Maggio D, et al. Pharmacologic inhibition of protein phosphatase-2A achieves durable immune-mediated antitumor activity when combined with PD-1 blockade. Nat Commun 2018;9:2126 10.1038/s41467-018-04425-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. D’Eliseo D, Manzi L, Velotti F. Capsaicin as an inducer of damage-associated molecular patterns (DAMPs) of immunogenic cell death (ICD) in human bladder cancer cells. Cell Stress and Chaperones 2013;18:801–8. 10.1007/s12192-013-0422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garg AD, Vandenberk L, Koks C, et al. Dendritic cell vaccines based on immunogenic cell death elicit danger signals and T cell-driven rejection of high-grade glioma. Sci Transl Med 2016;328:ra27. [DOI] [PubMed] [Google Scholar]
- 40. Ogawa M, Tomita Y, Nakamura Y, et al. Immunogenic cancer cell death selectively induced by near infrared photoimmunotherapy initiates host tumor immunity. Oncotarget 2017;8:10425–36. 10.18632/oncotarget.14425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fucikova J, Moserova I, Truxova I, et al. High hydrostatic pressure induces immunogenic cell death in human tumor cells. Int. J. Cancer 2014;135:1165–77. 10.1002/ijc.28766 [DOI] [PubMed] [Google Scholar]
- 42. Golden EB, Frances D, Pellicciotta I, et al. Radiation fosters dose-dependent and chemotherapy-induced immunogenic cell death. Oncoimmunology 2014;3:e28518 10.4161/onci.28518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Adkins I, Sadilkova L, Hradilova N, et al. Severe, but not mild heat-shock treatment induces immunogenic cell death in cancer cells. Oncoimmunology 2017;6:e1311433 10.1080/2162402X.2017.1311433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guo S, Jing Y, Burcus NI, et al. Nano-pulse stimulation induces potent immune responses, eradicating local breast cancer while reducing distant metastases. Int J Cancer 2018;142:629–40. 10.1002/ijc.31071 [DOI] [PubMed] [Google Scholar]
- 45. Rubner Y, Muth C, Strnad A, et al. Fractionated radiotherapy is the main stimulus for the induction of cell death and of Hsp70 release of p53 mutated glioblastoma cell lines. Radiat Oncol 2014;9:89 10.1186/1748-717X-9-89 10.1186/1748-717X-9-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gameiro SR, Jammeh ML, Wattenberg MM, et al. Radiation-Induced immunogenic modulation of tumor enhances antigen processing and calreticulin exposure, resulting in enhanced T-cell killing. Oncotarget 2014;5:403–16. 10.18632/oncotarget.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ventura A, Vassall A, Robinson E, et al. Extracorporeal photochemotherapy drives Monocyte-to-Dendritic cell maturation to induce anticancer immunity. Cancer Res 2018;78:4045–58. 10.1158/0008-5472.CAN-18-0171 [DOI] [PubMed] [Google Scholar]
- 48. Weiss EM, Meister S, Janko C, et al. High hydrostatic pressure treatment generates inactivated mammalian tumor cells with immunogeneic features. J Immunotoxicol 2010;7:194–204. 10.3109/15476911003657414 [DOI] [PubMed] [Google Scholar]
- 49. Vancsik T, Kovago C, Kiss E, et al. Modulated electro-hyperthermia induced loco-regional and systemic tumor destruction in colorectal cancer allografts. J Cancer 2018;9:41–53. 10.7150/jca.21520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu J, Waxman DJ. Immunogenic chemotherapy: dose and schedule dependence and combination with immunotherapy. Cancer Lett 2018;419:210–21. 10.1016/j.canlet.2018.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ko EC, Benjamin KT, Formenti SC. Generating antitumor immunity by targeted radiation therapy: role of dose and fractionation. Adv Radiat Oncol 2018;3:486–93. 10.1016/j.adro.2018.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deutsch E, Chargari C, Galluzzi L, et al. Optimising efficacy and reducing toxicity of anticancer radioimmunotherapy. Lancet Oncol 2019;20:e452–63. 10.1016/S1470-2045(19)30171-8 [DOI] [PubMed] [Google Scholar]
- 53. Garg AD, Agostinis P. Cell death and immunity in cancer: from danger signals to mimicry of pathogen defense responses. Immunol Rev 2017;280:126–48. 10.1111/imr.12574 [DOI] [PubMed] [Google Scholar]
- 54. Yatim N, Albert ML. Dying to replicate: the orchestration of the viral life cycle, cell death pathways, and immunity. Immunity 2011;35:478–90. 10.1016/j.immuni.2011.10.010 [DOI] [PubMed] [Google Scholar]
- 55. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. 10.1016/j.immuni.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 56. Pfirschke C, Engblom C, Rickelt S, et al. Immunogenic chemotherapy sensitizes tumors to checkpoint blockade therapy. Immunity 2016;44:343–54. 10.1016/j.immuni.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Voorwerk L, Slagter M, Horlings HM, et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the tonic trial. Nat Med 2019;25:920–8. 10.1038/s41591-019-0432-4 [DOI] [PubMed] [Google Scholar]
- 58. Galluzzi L, Bravo-San Pedro JM, Vitale I, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ 2015;22:58–73. 10.1038/cdd.2014.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aaes TL, Kaczmarek A, Delvaeye T, et al. Vaccination with Necroptotic cancer cells induces efficient anti-tumor immunity. Cell Rep 2016;15:274–87. 10.1016/j.celrep.2016.03.037 [DOI] [PubMed] [Google Scholar]
- 60. Van Hoecke L, Van Lint S, Roose K, et al. Treatment with mRNA coding for the necroptosis mediator MLKL induces antitumor immunity directed against neo-epitopes. Nat Commun 2018;9:3417 10.1038/s41467-018-05979-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang H, Ma Y, Chen G, et al. Contribution of RIP3 and MLKL to immunogenic cell death signaling in cancer chemotherapy. Oncoimmunology 2016;5:e1149673 10.1080/2162402X.2016.1149673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Werthmöller N, Frey B, Wunderlich R, et al. Modulation of radiochemoimmunotherapy-induced B16 melanoma cell death by the pan-caspase inhibitor zVAD-fmk induces anti-tumor immunity in a HMGB1-, nucleotide- and T-cell-dependent manner. Cell Death Dis 2015;6:e1761 10.1038/cddis.2015.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature 2015;517:311–20. 10.1038/nature14191 [DOI] [PubMed] [Google Scholar]
- 64. Menger L, Vacchelli E, Kepp O, et al. Trial Watch: cardiac glycosides and cancer therapy. Oncoimmunology 2013;2:e23082 10.4161/onci.23082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Palucka AK, Coussens LM. The basis of oncoimmunology. Cell 2016;164:1233–47. 10.1016/j.cell.2016.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Han J, Lotze MT. The Adaptome as biomarker for assessing cancer immunity and immunotherapy. Methods Mol Biol 2055;2020:369–97. [DOI] [PubMed] [Google Scholar]
- 67. Goldszmid RS, Dzutsev A, Trinchieri G. Host immune response to infection and cancer: unexpected commonalities. Cell Host Microbe 2014;15:295–305. 10.1016/j.chom.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Garg AD, Galluzzi L, Apetoh L, et al. Molecular and translational classifications of DAMPs in immunogenic cell death. Front Immunol 2015;6:588 10.3389/fimmu.2015.00588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bloy N, Garcia P, Laumont CM, et al. Immunogenic stress and death of cancer cells: contribution of antigenicity vs adjuvanticity to immunosurveillance. Immunol Rev 2017;280:165–74. 10.1111/imr.12582 [DOI] [PubMed] [Google Scholar]
- 70. Krysko DV, Garg AD, Kaczmarek A, et al. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer 2012;12:860–75. 10.1038/nrc3380 [DOI] [PubMed] [Google Scholar]
- 71. Rufo N, Garg AD, Agostinis P. The unfolded protein response in immunogenic cell death and cancer immunotherapy. Trends in Cancer 2017;3:643–58. 10.1016/j.trecan.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 72. Hou W, Zhang Q, Yan Z, et al. Strange attractors: DAMPs and autophagy link tumor cell death and immunity. Cell Death Dis 2013;4:e966 10.1038/cddis.2013.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Galluzzi L, Buqué A, Kepp O, et al. Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell 2015;28:690–714. 10.1016/j.ccell.2015.10.012 [DOI] [PubMed] [Google Scholar]
- 74. Obeid M, Panaretakis T, Joza N, et al. Calreticulin exposure is required for the immunogenicity of γ-irradiation and UVC light-induced apoptosis. Cell Death Differ 2007;14:1848–50. 10.1038/sj.cdd.4402201 [DOI] [PubMed] [Google Scholar]
- 75. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69–74. 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 76. Vesely MD, Kershaw MH, Schreiber RD, et al. Natural innate and adaptive immunity to cancer. Annu Rev Immunol 2011;29:235–71. 10.1146/annurev-immunol-031210-101324 [DOI] [PubMed] [Google Scholar]
- 77. Golden EB, Demaria S, Schiff PB, et al. An abscopal response to radiation and ipilimumab in a patient with metastatic non-small cell lung cancer. Cancer Immunol Res 2013;1:365–72. 10.1158/2326-6066.CIR-13-0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dewan MZ, Galloway AE, Kawashima N, et al. Fractionated but not single-dose radiotherapy induces an immune-mediated abscopal effect when combined with anti-CTLA-4 antibody. Clinical Cancer Research 2009;15:5379–88. 10.1158/1078-0432.CCR-09-0265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Twyman-Saint Victor C, Rech AJ, Maity A, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 2015;520:373–7. 10.1038/nature14292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Braciale TJ, Hahn YS. Immunity to viruses. Immunol Rev 2013;255:5–12. 10.1111/imr.12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dorhoi A, Kaufmann SHE. Fine-Tuning of T cell responses during infection. Curr Opin Immunol 2009;21:367–77. 10.1016/j.coi.2009.07.004 [DOI] [PubMed] [Google Scholar]
- 82. Broz P, Monack DM. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol 2013;13:551–65. 10.1038/nri3479 [DOI] [PubMed] [Google Scholar]
- 83. Cao X. Self-Regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat Rev Immunol 2016;16:35–50. 10.1038/nri.2015.8 [DOI] [PubMed] [Google Scholar]
- 84. Tang D, Kang R, Coyne CB, et al. Pamps and DAMPs: signal 0s that Spur autophagy and immunity. Immunol Rev 2012;249:158–75. 10.1111/j.1600-065X.2012.01146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Klein L, Kyewski B, Allen PM, et al. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol 2014;14:377–91. 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Waldmann H. Tolerance: an overview and perspectives. Nat Rev Nephrol 2010;6:569–76. 10.1038/nrneph.2010.108 [DOI] [PubMed] [Google Scholar]
- 87. Ishak CA, Classon M, De Carvalho DD. Deregulation of retroelements as an emerging therapeutic opportunity in cancer. Trends Cancer 2018;4:583–97. 10.1016/j.trecan.2018.05.008 [DOI] [PubMed] [Google Scholar]
- 88. Jones PA, Ohtani H, Chakravarthy A, et al. Epigenetic therapy in immune-oncology. Nat Rev Cancer 2019;19:151–61. 10.1038/s41568-019-0109-9 [DOI] [PubMed] [Google Scholar]
- 89. Smith CC, Beckermann KE, Bortone DS, et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest 2018;128:4804–20. 10.1172/JCI121476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Raposo B, Merky P, Lundqvist C, et al. T cells specific for post-translational modifications escape intrathymic tolerance induction. Nat Commun 2018;9:353 10.1038/s41467-017-02763-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zervoudi E, Saridakis E, Birtley JR, et al. Rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses. Proc Natl Acad Sci U S A 2013;110:19890–5. 10.1073/pnas.1309781110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Doyle HA, Mamula MJ. Autoantigenesis: the evolution of protein modifications in autoimmune disease. Curr Opin Immunol 2012;24:112–8. 10.1016/j.coi.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Starck SR, Shastri N. Nowhere to hide: unconventional translation yields cryptic peptides for immune surveillance. Immunol Rev 2016;272:8–16. 10.1111/imr.12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Scally SW, Petersen J, Law SC, et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med 2013;210:2569–82. 10.1084/jem.20131241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. van Lummel M, Duinkerken G, van Veelen PA, et al. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes 2014;63:237–47. 10.2337/db12-1214 [DOI] [PubMed] [Google Scholar]
- 96. Greaves M. Evolutionary determinants of cancer. Cancer Discov 2015;5:806–20. 10.1158/2159-8290.CD-15-0439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 98. Schumacher TN, Hacohen N. Neoantigens encoded in the cancer genome. Curr Opin Immunol 2016;41:98–103. 10.1016/j.coi.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 99. Balachandran VP, Łuksza M, Zhao JN, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017;551:512–6. 10.1038/nature24462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kanaseki T, Tokita S, Torigoe T. Proteogenomic discovery of cancer antigens: neoantigens and beyond. Pathol Int 2019;69:511–8. 10.1111/pin.12841 [DOI] [PubMed] [Google Scholar]
- 101. Schumacher TN, Scheper W, Kvistborg P. Cancer neoantigens. Annu Rev Immunol 2019;37:173–200. 10.1146/annurev-immunol-042617-053402 [DOI] [PubMed] [Google Scholar]
- 102. Sprooten J, Ceusters J, Coosemans A, et al. Trial Watch: dendritic cell vaccination for cancer immunotherapy. Oncoimmunology 2019;8:e1638212 10.1080/2162402X.2019.1638212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bezu L, Kepp O, Cerrato G, et al. Trial Watch: peptide-based vaccines in anticancer therapy. Oncoimmunology 2018;7:e1511506 10.1080/2162402X.2018.1511506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kirkin AF, Dzhandzhugazyan KN, Guldberg P, et al. Adoptive cancer immunotherapy using DNA-demethylated T helper cells as antigen-presenting cells. Nat Commun 2018;9:785 10.1038/s41467-018-03217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Simpson AJG, Caballero OL, Jungbluth A, et al. Cancer/Testis antigens, gametogenesis and cancer. Nat Rev Cancer 2005;5:615–25. 10.1038/nrc1669 [DOI] [PubMed] [Google Scholar]
- 106. Coulie PG, Van den Eynde BJ, van der Bruggen P, et al. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 2014;14:135–46. 10.1038/nrc3670 [DOI] [PubMed] [Google Scholar]
- 107. Schuster H, Peper JK, Bösmüller H-C, et al. The immunopeptidomic landscape of ovarian carcinomas. Proc Natl Acad Sci U S A 2017;114:E9942–51. 10.1073/pnas.1707658114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gilboa E. The makings of a tumor rejection antigen. Immunity 1999;11:263–70. 10.1016/S1074-7613(00)80101-6 [DOI] [PubMed] [Google Scholar]
- 109. Stone JD, Harris DT, Kranz DM. Tcr affinity for p/MHC formed by tumor antigens that are self-proteins: impact on efficacy and toxicity. Curr Opin Immunol 2015;33:16–22. 10.1016/j.coi.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Malaker SA, Penny SA, Steadman LG, et al. Identification of glycopeptides as posttranslationally modified neoantigens in leukemia. Cancer Immunol Res 2017;5:376–84. 10.1158/2326-6066.CIR-16-0280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Campbell BB, Light N, Fabrizio D, et al. Comprehensive analysis of hypermutation in human cancer. Cell 2017;171:1042–56. 10.1016/j.cell.2017.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. McGranahan N, Furness AJS, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463–9. 10.1126/science.aaf1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 2017;171:934–49. 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Vitale I, Sistigu A, Manic G, et al. Mutational and antigenic landscape in tumor progression and cancer immunotherapy. Trends Cell Biol 2019;29:396–416. 10.1016/j.tcb.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 117. Hellmann MD, Ciuleanu T-E, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018;378:2093–104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hellmann MD, Callahan MK, Awad MM, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell 2018;33:853–61. 10.1016/j.ccell.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mandal R, Samstein RM, Lee K-W, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti–PD-1 immunotherapy response. Science 2019;364:485–91. 10.1126/science.aau0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Turan T, Kannan D, Patel M, et al. Immune oncology, immune responsiveness and the theory of everything. J Immunother Cancer 2018;6:50 10.1186/s40425-018-0355-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kroemer G, Galluzzi L, Kepp O, et al. Immunogenic cell death in cancer therapy. Annu Rev Immunol 2013;31:51–72. 10.1146/annurev-immunol-032712-100008 [DOI] [PubMed] [Google Scholar]
- 122. Garrido F, Aptsiauri N, Doorduijn EM, et al. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr Opin Immunol 2016;39:44–51. 10.1016/j.coi.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. O’Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol 2019;16:151–67. 10.1038/s41571-018-0142-8 [DOI] [PubMed] [Google Scholar]
- 124. McGranahan N, Rosenthal R, Hiley CT, et al. Allele-Specific HLA loss and immune escape in lung cancer evolution. Cell 2017;171:1259–71. 10.1016/j.cell.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Grasso CS, Giannakis M, Wells DK, et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov 2018;8:730–49. 10.1158/2159-8290.CD-17-1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Goel S, DeCristo MJ, Watt AC, et al. Cdk4/6 inhibition triggers anti-tumour immunity. Nature 2017;548:471–5. 10.1038/nature23465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lhuillier C, Rudqvist N-P, Elemento O, et al. Radiation therapy and anti-tumor immunity: exposing immunogenic mutations to the immune system. Genome Med 2019;11:40 10.1186/s13073-019-0653-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Chabanon RM, Muirhead G, Krastev DB, et al. Parp inhibition enhances tumor cell–intrinsic immunity in ERCC1-deficient non–small cell lung cancer. J Clin Invest 2019;129:1211–28. 10.1172/JCI123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Dillon MT, Bergerhoff KF, Pedersen M, et al. Atr inhibition potentiates the radiation-induced inflammatory tumor microenvironment. Clin Cancer Res 2019;25:3392–403. 10.1158/1078-0432.CCR-18-1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Woller N, Gürlevik E, Fleischmann-Mundt B, et al. Viral infection of tumors overcomes resistance to PD-1-immunotherapy by broadening Neoantigenome-directed T-cell responses. Molecular Therapy 2015;23:1630–40. 10.1038/mt.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Briere D, Sudhakar N, Woods DM, et al. The class I/IV HDAC inhibitor mocetinostat increases tumor antigen presentation, decreases immune suppressive cell types and augments checkpoint inhibitor therapy. Cancer Immunol Immunother 2018;67:381–92. 10.1007/s00262-017-2091-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Segovia C, San José-Enériz E, Munera-Maravilla E, et al. Inhibition of a G9a/DNMT network triggers immune-mediated bladder cancer regression. Nat Med 2019;25:1073–81. 10.1038/s41591-019-0499-y [DOI] [PubMed] [Google Scholar]
- 134. Weber J, Salgaller M, Samid D, et al. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2'-deoxycytidine. Cancer Res 1994;54:1766–71. [PubMed] [Google Scholar]
- 135. Vanpouille-Box C, Hoffmann JA, Galluzzi L. Pharmacological modulation of nucleic acid sensors — therapeutic potential and persisting obstacles. Nat Rev Drug Discov 2019;18:845–67. 10.1038/s41573-019-0043-2 [DOI] [PubMed] [Google Scholar]
- 136. Gay NJ, Symmons MF, Gangloff M, et al. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol 2014;14:546–58. 10.1038/nri3713 [DOI] [PubMed] [Google Scholar]
- 137. Kawai T, Akira S. Toll-Like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011;34:637–50. 10.1016/j.immuni.2011.05.006 [DOI] [PubMed] [Google Scholar]
- 138. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010;11:373–84. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 139. Ablasser A, Chen ZJ. cGAS in action: expanding roles in immunity and inflammation. Science 2019;363:eaat8657 10.1126/science.aat8657 [DOI] [PubMed] [Google Scholar]
- 140. Chow KT, Gale M, Loo Y-M. Rig-I and other RNA sensors in antiviral immunity. Annu Rev Immunol 2018;36:667–94. 10.1146/annurev-immunol-042617-053309 [DOI] [PubMed] [Google Scholar]
- 141. Galluzzi L, Vanpouille-Box C, Bakhoum SF, et al. Snapshot: cGAS-STING signaling. Cell 2018;173:276–276.e1. 10.1016/j.cell.2018.03.015 [DOI] [PubMed] [Google Scholar]
- 142. Motta V, Soares F, Sun T, et al. Nod-Like receptors: versatile cytosolic sentinels. Physiol Rev 2015;95:149–78. 10.1152/physrev.00009.2014 [DOI] [PubMed] [Google Scholar]
- 143. Kersse K, Bertrand MJM, Lamkanfi M, et al. Nod-Like receptors and the innate immune system: coping with danger, damage and death. Cytokine Growth Factor Rev 2011;22:257–76. 10.1016/j.cytogfr.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 144. Kuriakose T, Kanneganti T-D. Zbp1: innate sensor regulating cell death and inflammation. Trends Immunol 2018;39:123–34. 10.1016/j.it.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Alarcón CR, Goodarzi H, Lee H, et al. Hnrnpa2B1 is a mediator of m6A-Dependent nuclear RNA processing events. Cell 2015;162:1299–308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wang L, Wen M, Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 2019;365:eaav0758 10.1126/science.aav0758 [DOI] [PubMed] [Google Scholar]
- 147. McNab F, Mayer-Barber K, Sher A, et al. Type I interferons in infectious disease. Nat Rev Immunol 2015;15:87–103. 10.1038/nri3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kepp O, Senovilla L, Vitale I, et al. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology 2014;3:e955691 10.4161/21624011.2014.955691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Michaud M, Martins I, Sukkurwala AQ, et al. Autophagy-Dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 2011;334:1573–7. 10.1126/science.1208347 [DOI] [PubMed] [Google Scholar]
- 150. Ghiringhelli F, Apetoh L, Tesniere A, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1β–dependent adaptive immunity against tumors. Nat Med 2009;15:1170–8. 10.1038/nm.2028 [DOI] [PubMed] [Google Scholar]
- 151. Garg AD, Vandenberk L, Fang S, et al. Pathogen response-like recruitment and activation of neutrophils by sterile immunogenic dying cells drives neutrophil-mediated residual cell killing. Cell Death Differ 2017;24:832–43. 10.1038/cdd.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Chiba S, Baghdadi M, Akiba H, et al. Tumor-Infiltrating DCs suppress nucleic acid–mediated innate immune responses through interactions between the receptor Tim-3 and the alarmin HMGB1. Nat Immunol 2012;13:832–42. 10.1038/ni.2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Apetoh L, Ghiringhelli F, Tesniere A, et al. Toll-Like receptor 4–dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med 2007;13:1050–9. 10.1038/nm1622 [DOI] [PubMed] [Google Scholar]
- 154. Yang H, Hreggvidsdottir HS, Palmblad K, et al. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc Natl Acad Sci U S A 2010;107:11942–7. 10.1073/pnas.1003893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Soloff AC, Lotze MT. A peaceful death orchestrates immune balance in a chaotic environment. Proc Natl Acad Sci U S A 2019;116:22901–3. 10.1073/pnas.1916211116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Gorgulho CM, Romagnoli GG, Bharthi R, et al. Johnny on the Spot-Chronic inflammation is driven by HMGB1. Front Immunol 2019;10:1561 10.3389/fimmu.2019.01561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Vacchelli E, Ma Y, Baracco EE, et al. Chemotherapy-Induced antitumor immunity requires formyl peptide receptor 1. Science 2015;350:972–8. 10.1126/science.aad0779 [DOI] [PubMed] [Google Scholar]
- 158. Sistigu A, Yamazaki T, Vacchelli E, et al. Cancer cell–autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med 2014;20:1301–9. 10.1038/nm.3708 [DOI] [PubMed] [Google Scholar]
- 159. Krombach J, Hennel R, Brix N, et al. Priming anti-tumor immunity by radiotherapy: dying tumor cell-derived DAMPs trigger endothelial cell activation and recruitment of myeloid cells. Oncoimmunology 2019;8:e1523097 10.1080/2162402X.2018.1523097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Panaretakis T, Joza N, Modjtahedi N, et al. The co-translocation of ERp57 and calreticulin determines the immunogenicity of cell death. Cell Death Differ 2008;15:1499–509. 10.1038/cdd.2008.67 [DOI] [PubMed] [Google Scholar]
- 161. Ahrens S, Zelenay S, Sancho D, et al. F-Actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity 2012;36:635–45. 10.1016/j.immuni.2012.03.008 [DOI] [PubMed] [Google Scholar]
- 162. Krysko DV, Agostinis P, Krysko O, et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol 2011;32:157–64. 10.1016/j.it.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 163. Zhang Q, Raoof M, Chen Y, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010;464:104–7. 10.1038/nature08780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Rodriguez-Ruiz ME, Buqué A, Hensler M, et al. Apoptotic caspases inhibit abscopal responses to radiation and identify a new prognostic biomarker for breast cancer patients. Oncoimmunology 2019;8:e1655964 10.1080/2162402X.2019.1655964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Martins I, Wang Y, Michaud M, et al. Molecular mechanisms of ATP secretion during immunogenic cell death. Cell Death Differ 2014;21:79–91. 10.1038/cdd.2013.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Martins I, Michaud M, Sukkurwala AQ, et al. Premortem autophagy determines the immunogenicity of chemotherapy-induced cancer cell death. Autophagy 2012;8:413–5. 10.4161/auto.19009 [DOI] [PubMed] [Google Scholar]
- 167. Apetoh L, Ghiringhelli F, Tesniere A, et al. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunol Rev 2007;220:47–59. 10.1111/j.1600-065X.2007.00573.x [DOI] [PubMed] [Google Scholar]
- 168. Yang D, Han Z, Oppenheim JJ. Alarmins and immunity. Immunol Rev 2017;280:41–56. 10.1111/imr.12577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Di Virgilio F, Sarti AC, Falzoni S, et al. Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat Rev Cancer 2018;18:601–18. 10.1038/s41568-018-0037-0 [DOI] [PubMed] [Google Scholar]
- 170. Vanpouille-Box C, Demaria S, Formenti SC, et al. Cytosolic DNA sensing in organismal tumor control. Cancer Cell 2018;34:361–78. 10.1016/j.ccell.2018.05.013 [DOI] [PubMed] [Google Scholar]
- 171. Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol 2018;18:309–24. 10.1038/nri.2017.142 [DOI] [PubMed] [Google Scholar]
- 172. Galluzzi L, Yamazaki T, Kroemer G. Linking cellular stress responses to systemic homeostasis. Nat Rev Mol Cell Biol 2018;19:731–45. 10.1038/s41580-018-0068-0 [DOI] [PubMed] [Google Scholar]
- 173. Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. Embo J 2017;36:1811–36. 10.15252/embj.201796697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Sicari D, Igbaria A, Chevet E. Control of protein homeostasis in the early secretory pathway: current status and challenges. Cells 2019;8:1347 10.3390/cells8111347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Long M, McWilliams TG. Monitoring autophagy in cancer: from bench to bedside. Semin Cancer Biol 2019. 10.1016/j.semcancer.2019.05.016 [DOI] [PubMed] [Google Scholar]
- 176. Juste YR, Cuervo AM. Analysis of chaperone-mediated autophagy. Methods Mol Biol 1880;2019:703–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Boyd-Tressler A, Penuela S, Laird DW, et al. Chemotherapeutic drugs induce ATP release via caspase-gated pannexin-1 channels and a caspase/pannexin-1-independent mechanism. J. Biol. Chem. 2014;289:27246–63. 10.1074/jbc.M114.590240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Garg AD, Krysko DV, Verfaillie T, et al. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. Embo J 2012;31:1062–79. 10.1038/emboj.2011.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Elliott MR, Chekeni FB, Trampont PC, et al. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 2009;461:282–6. 10.1038/nature08296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Panaretakis T, Kepp O, Brockmeier U, et al. Mechanisms of pre-apoptotic calreticulin exposure in immunogenic cell death. Embo J 2009;28:578–90. 10.1038/emboj.2009.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Senovilla L, Vitale I, Martins I, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science 2012;337:1678–84. 10.1126/science.1224922 [DOI] [PubMed] [Google Scholar]
- 182. Truxova I, Kasikova L, Salek C, et al. Calreticulin exposure on malignant blasts correlates with improved natural killer cell-mediated cytotoxicity in acute myeloid leukemia patients. Haematologica 2019:haematol.2019.223933 10.3324/haematol.2019.223933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Bezu L, Sauvat A, Humeau J, et al. eIF2α phosphorylation is pathognomonic for immunogenic cell death. Cell Death Differ 2018;25:1375–93. 10.1038/s41418-017-0044-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Sukkurwala AQ, Martins I, Wang Y, et al. Immunogenic calreticulin exposure occurs through a phylogenetically conserved stress pathway involving the chemokine CXCL8. Cell Death Differ 2014;21:59–68. 10.1038/cdd.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Tufi R, Panaretakis T, Bianchi K, et al. Reduction of endoplasmic reticulum Ca2+ levels favors plasma membrane surface exposure of calreticulin. Cell Death Differ 2008;15:274–82. 10.1038/sj.cdd.4402275 [DOI] [PubMed] [Google Scholar]
- 186. Moserova I, Truxova I, Garg AD, et al. Caspase-2 and oxidative stress underlie the immunogenic potential of high hydrostatic pressure-induced cancer cell death. Oncoimmunology 2017;6:e1258505 10.1080/2162402X.2016.1258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Musahl A-S, Huang X, Rusakiewicz S, et al. A long non-coding RNA links calreticulin-mediated immunogenic cell removal to Rb1 transcription. Oncogene 2015;34:5046–54. 10.1038/onc.2014.424 [DOI] [PubMed] [Google Scholar]
- 188. Colangelo T, Polcaro G, Ziccardi P, et al. The miR-27a-calreticulin axis affects drug-induced immunogenic cell death in human colorectal cancer cells. Cell Death Dis 2016;7:e2108 10.1038/cddis.2016.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Liu C-C, Leclair P, Pedari F, et al. Integrins and ERp57 coordinate to regulate cell surface calreticulin in immunogenic cell death. Front Oncol 2019;9:411 10.3389/fonc.2019.00411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Gardai SJ, McPhillips KA, Frasch SC, et al. Cell-Surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 2005;123:321–34. 10.1016/j.cell.2005.08.032 [DOI] [PubMed] [Google Scholar]
- 191. Chen X, Fosco D, Kline DE, et al. Calreticulin promotes immunity and type I interferon-dependent survival in mice with acute myeloid leukemia. Oncoimmunology 2017;6:e1278332 10.1080/2162402X.2016.1278332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Fucikova J, Kasikova L, Truxova I, et al. Relevance of the chaperone-like protein calreticulin for the biological behavior and clinical outcome of cancer. Immunol Lett 2018;193:25–34. 10.1016/j.imlet.2017.11.006 [DOI] [PubMed] [Google Scholar]
- 193. Radogna F, Diederich M. Stress-Induced cellular responses in immunogenic cell death: implications for cancer immunotherapy. Biochem Pharmacol 2018;153:12–23. 10.1016/j.bcp.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 194. Vanpouille-Box C, Alard A, Aryankalayil MJ, et al. Dna exonuclease TREX1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun 2017;8:15618 10.1038/ncomms15618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Deng L, Liang H, Xu M, et al. Sting-Dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 2014;41:843–52. 10.1016/j.immuni.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Zitvogel L, Galluzzi L, Kepp O, et al. Type I interferons in anticancer immunity. Nat Rev Immunol 2015;15:405–14. 10.1038/nri3845 [DOI] [PubMed] [Google Scholar]
- 197. Diamond JM, Vanpouille-Box C, Spada S, et al. Exosomes shuttle TREX1-Sensitive IFN-Stimulatory dsDNA from irradiated cancer cells to DCs. Cancer Immunol Res 2018;6:910–20. 10.1158/2326-6066.CIR-17-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Kang R, Tang D, Schapiro NE, et al. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene 2014;33:567–77. 10.1038/onc.2012.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Kang R, Chen R, Xie M, et al. The receptor for advanced glycation end products activates the AIM2 inflammasome in acute pancreatitis. J.i. 2016;196:4331–7. 10.4049/jimmunol.1502340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Boone BA, Orlichenko L, Schapiro NE, et al. The receptor for advanced glycation end products (RAGE) enhances autophagy and neutrophil extracellular traps in pancreatic cancer. Cancer Gene Ther 2015;22:326–34. 10.1038/cgt.2015.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Bianchi ME, Crippa MP, Manfredi AA, et al. High-Mobility group box 1 protein orchestrates responses to tissue damage via inflammation, innate and adaptive immunity, and tissue repair. Immunol Rev 2017;280:74–82. 10.1111/imr.12601 [DOI] [PubMed] [Google Scholar]
- 202. Venereau E, Casalgrandi M, Schiraldi M, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med 2012;209:1519–28. 10.1084/jem.20120189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Garg AD, Dudek AM, Ferreira GB, et al. Ros-Induced autophagy in cancer cells assists in evasion from determinants of immunogenic cell death. Autophagy 2013;9:1292–307. 10.4161/auto.25399 [DOI] [PubMed] [Google Scholar]
- 204. Yatim N, Jusforgues-Saklani H, Orozco S, et al. RIPK1 and NF- B signaling in dying cells determines cross-priming of CD8+ T cells. Science 2015;350:328–34. 10.1126/science.aad0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Martins I, Kepp O, Schlemmer F, et al. Restoration of the immunogenicity of cisplatin-induced cancer cell death by endoplasmic reticulum stress. Oncogene 2011;30:1147–58. 10.1038/onc.2010.500 [DOI] [PubMed] [Google Scholar]
- 206. Aranda F, Bloy N, Pesquet J, et al. Immune-dependent antineoplastic effects of cisplatin plus pyridoxine in non-small-cell lung cancer. Oncogene 2015;34:3053–62. 10.1038/onc.2014.234 [DOI] [PubMed] [Google Scholar]
- 207. Garg AD, Elsen S, Krysko DV, et al. Resistance to anticancer vaccination effect is controlled by a cancer cell-autonomous phenotype that disrupts immunogenic phagocytic removal. Oncotarget 2015;6:26841–60. 10.18632/oncotarget.4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Dudek-Peri AM, Ferreira GB, Muchowicz A, et al. Antitumor immunity triggered by melphalan is potentiated by melanoma cell surface-associated calreticulin. Cancer Res 2015;75:1603–14. 10.1158/0008-5472.CAN-14-2089 [DOI] [PubMed] [Google Scholar]
- 209. Chao MP, Jaiswal S, Weissman-Tsukamoto R, et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci Transl Med 2010;2:63ra94 10.1126/scitranslmed.3001375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Romano E, Rufo N, Korf H, et al. Bnip3 modulates the interface between B16-F10 melanoma cells and immune cells. Oncotarget 2018;9:17631–44. 10.18632/oncotarget.24815 10.18632/oncotarget.24815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Fucikova J, Moserova I, Urbanova L, et al. Prognostic and predictive value of DAMPs and DAMP-Associated processes in cancer. Front Immunol 2015;6:402 10.3389/fimmu.2015.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Fucikova J, Becht E, Iribarren K, et al. Calreticulin expression in human Non–Small cell lung cancers correlates with increased accumulation of antitumor immune cells and favorable prognosis. Cancer Res 2016;76:1746–56. 10.1158/0008-5472.CAN-15-1142 [DOI] [PubMed] [Google Scholar]
- 213. Ladoire S, Penault-Llorca F, Senovilla L, et al. Combined evaluation of LC3B puncta and HMGB1 expression predicts residual risk of relapse after adjuvant chemotherapy in breast cancer. Autophagy 2015;11:1878–90. 10.1080/15548627.2015.1082022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Yamazaki T, Hannani D, Poirier-Colame V, et al. Defective immunogenic cell death of HMGB1-deficient tumors: compensatory therapy with TLR4 agonists. Cell Death Differ 2014;21:69–78. 10.1038/cdd.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Fucikova J, Truxova I, Hensler M, et al. Calreticulin exposure by malignant blasts correlates with robust anticancer immunity and improved clinical outcome in AML patients. Blood 2016;128:3113–24. 10.1182/blood-2016-08-731737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Wemeau M, Kepp O, Tesnière A, et al. Calreticulin exposure on malignant blasts predicts a cellular anticancer immune response in patients with acute myeloid leukemia. Cell Death Dis 2010;1:e104 10.1038/cddis.2010.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Bidwell BN, Slaney CY, Withana NP, et al. Silencing of IRF7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med 2012;18:1224–31. 10.1038/nm.2830 [DOI] [PubMed] [Google Scholar]
- 218. Suzuki S, Yokobori T, Tanaka N, et al. Cd47 expression regulated by the miR-133a tumor suppressor is a novel prognostic marker in esophageal squamous cell carcinoma. Oncol Rep 2012;28:465–72. 10.3892/or.2012.1831 [DOI] [PubMed] [Google Scholar]
- 219. Majeti R, Chao MP, Alizadeh AA, et al. Cd47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 2009;138:286–99. 10.1016/j.cell.2009.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Wang H, Tan M, Zhang S, et al. Expression and significance of CD44, CD47 and c-Met in ovarian clear cell carcinoma. Int J Mol Sci 2015;16:3391–404. 10.3390/ijms16023391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol 2015;16:343–53. 10.1038/ni.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Fridman WH, Zitvogel L, Sautès–Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–34. 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- 223. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 224. Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Sabatos-Peyton CA, Nevin J, Brock A, et al. Blockade of Tim-3 binding to phosphatidylserine and CEACAM1 is a shared feature of anti-Tim-3 antibodies that have functional efficacy. Oncoimmunology 2018;7:e1385690 10.1080/2162402X.2017.1385690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Mittal SK, Roche PA. Suppression of antigen presentation by IL-10. Curr Opin Immunol 2015;34:22–7. 10.1016/j.coi.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Pickup M, Novitskiy S, Moses HL. The roles of TGFβ in the tumour microenvironment. Nat Rev Cancer 2013;13:788–99. 10.1038/nrc3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Shalapour S, Karin M. Pas de deux: control of anti-tumor immunity by cancer-associated inflammation. Immunity 2019;51:15–26. 10.1016/j.immuni.2019.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res 2017;27:109–18. 10.1038/cr.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Wellenstein MD, de Visser KE. Cancer-Cell-Intrinsic mechanisms shaping the tumor immune landscape. Immunity 2018;48:399–416. 10.1016/j.immuni.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 231. Vitale I, Manic G, Coussens LM, et al. Macrophages and metabolism in the tumor microenvironment. Cell Metab 2019;30:36–50. 10.1016/j.cmet.2019.06.001 [DOI] [PubMed] [Google Scholar]
- 232. Montalbán del Barrio I, Penski C, Schlahsa L, et al. Adenosine-generating ovarian cancer cells attract myeloid cells which differentiate into adenosine-generating tumor associated macrophages – a self-amplifying, CD39- and CD73-dependent mechanism for tumor immune escape. J Immunother Cancer 2016;4:49 10.1186/s40425-016-0154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. d’Almeida SM, Kauffenstein G, Roy C, et al. The ecto-ATPDase CD39 is involved in the acquisition of the immunoregulatory phenotype by M-CSF-macrophages and ovarian cancer tumor-associated macrophages: regulatory role of IL-27. Oncoimmunology 2016;5:e1178025 10.1080/2162402X.2016.1178025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Mandapathil M, Hilldorfer B, Szczepanski MJ, et al. Generation and accumulation of immunosuppressive adenosine by human CD4+CD25highFOXP3+ regulatory T cells. J Biol Chem 2010;285:7176–86. 10.1074/jbc.M109.047423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Vijayan D, Young A, Teng MWL, et al. Targeting immunosuppressive adenosine in cancer. Nat Rev Cancer 2017;17:709–24. 10.1038/nrc.2017.86 [DOI] [PubMed] [Google Scholar]
- 236. Li C, Zhang Y, Cheng X, et al. Pink1 and PARK2 suppress pancreatic tumorigenesis through control of mitochondrial iron-mediated Immunometabolism. Dev Cell 2018;46:441–55. 10.1016/j.devcel.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Shimada K, Crother TR, Karlin J, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 2012;36:401–14. 10.1016/j.immuni.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. McLane LM, Abdel-Hakeem MS, Wherry EJ. Cd8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol 2019;37:457–95. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- 239. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486–99. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Munn DH, Bronte V. Immune suppressive mechanisms in the tumor microenvironment. Curr Opin Immunol 2016;39:1–6. 10.1016/j.coi.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Ribas A. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov 2015;5:915–9. 10.1158/2159-8290.CD-15-0563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Chang C-H, Qiu J, O'Sullivan D, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 2015;162:1229–41. 10.1016/j.cell.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Bantug GR, Galluzzi L, Kroemer G, et al. The spectrum of T cell metabolism in health and disease. Nat Rev Immunol 2018;18:19–34. 10.1038/nri.2017.99 [DOI] [PubMed] [Google Scholar]
- 244. Labadie BW, Bao R, Luke JJ. Reimagining IDO pathway inhibition in cancer immunotherapy via downstream focus on the Tryptophan–Kynurenine–Aryl hydrocarbon axis. Clin Cancer Res 2019;25:1462–71. 10.1158/1078-0432.CCR-18-2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Colegio OR, Chu N-Q, Szabo AL, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014;513:559–63. 10.1038/nature13490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Voron T, Colussi O, Marcheteau E, et al. Vegf-A modulates expression of inhibitory checkpoints on CD8+ T cells in tumors. J Exp Med 2015;212:139–48. 10.1084/jem.20140559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Flavell RA, Sanjabi S, Wrzesinski SH, et al. The polarization of immune cells in the tumour environment by TGFβ. Nat Rev Immunol 2010;10:554–67. 10.1038/nri2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Yamauchi M, Barker TH, Gibbons DL, et al. The fibrotic tumor stroma. J Clin Invest 2018;128:16–25. 10.1172/JCI93554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Schaaf MB, Garg AD, Agostinis P. Defining the role of the tumor vasculature in antitumor immunity and immunotherapy. Cell Death Dis 2018;9:115 10.1038/s41419-017-0061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Menger L, Vacchelli E, Adjemian S, et al. Cardiac glycosides exert anticancer effects by inducing immunogenic cell death. Sci Transl Med 2012;143:ra99. [DOI] [PubMed] [Google Scholar]
- 251. Kepp O, Galluzzi L, Lipinski M, et al. Cell death assays for drug discovery. Nat Rev Drug Discov 2011;10:221–37. 10.1038/nrd3373 [DOI] [PubMed] [Google Scholar]
- 252. Galluzzi L, Aaronson SA, Abrams J, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ 2009;16:1093–107. 10.1038/cdd.2009.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. van Schadewijk A, van’t Wout EFA, Stolk J, et al. A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell Stress and Chaperones 2012;17:275–9. 10.1007/s12192-011-0306-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Lam AR, Le Bert N, Ho SSW, et al. Rae1 ligands for the NKG2D receptor are regulated by STING-dependent DNA sensor pathways in lymphoma. Cancer Res 2014;74:2193–203. 10.1158/0008-5472.CAN-13-1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Duewell P, Beller E, Kirchleitner SV, et al. Targeted activation of melanoma differentiation-associated protein 5 (MDA5) for immunotherapy of pancreatic carcinoma. Oncoimmunology 2015;4:e1029698 10.1080/2162402X.2015.1029698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Shen YJ, Le Bert N, Chitre AA, et al. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep 2015;11:460–73. 10.1016/j.celrep.2015.03.041 [DOI] [PubMed] [Google Scholar]
- 257. Diner EJ, Burdette DL, Wilson SC, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human sting. Cell Rep 2013;3:1355–61. 10.1016/j.celrep.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Seo GJ, Kim C, Shin W-J, et al. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat Commun 2018;9:613 10.1038/s41467-018-02936-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12:1–222. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Teo ZL, Versaci S, Dushyanthen S, et al. Combined Cdk4/6 and PI3Kα inhibition is synergistic and immunogenic in triple-negative breast cancer. Cancer Res 2017;77:6340–52. 10.1158/0008-5472.CAN-17-2210 [DOI] [PubMed] [Google Scholar]
- 261. Lu J, Liu X, Liao Y-P, et al. Breast cancer chemo-immunotherapy through liposomal delivery of an immunogenic cell death stimulus plus interference in the IDO-1 pathway. ACS Nano 2018;12:11041–61. 10.1021/acsnano.8b05189 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 262. Sukkurwala AQ, Adjemian S, Senovilla L, et al. Screening of novel immunogenic cell death inducers within the NCI mechanistic diversity set. Oncoimmunology 2014;3:e28473 10.4161/onci.28473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Garg AD, Krysko DV, Vandenabeele P, et al. Hypericin-Based photodynamic therapy induces surface exposure of damage-associated molecular patterns like Hsp70 and calreticulin. Cancer Immunol Immunother 2012;61:215–21. 10.1007/s00262-011-1184-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Hossain DMS, Javaid S, Cai M, et al. Dinaciclib induces immunogenic cell death and enhances anti-PD1–mediated tumor suppression. J Clin Invest 2018;128:644–54. 10.1172/JCI94586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Imamura H, Huynh Nhat KP, Togawa H, et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci U S A 2009;106:15651–6. 10.1073/pnas.0904764106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Melis MHM, Simpson KL, Dovedi SJ, et al. Sustained tumour eradication after induced caspase-3 activation and synchronous tumour apoptosis requires an intact host immune response. Cell Death Differ 2013;20:765–73. 10.1038/cdd.2013.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Martins I, Kepp O, Menger L, et al. Fluorescent biosensors for the detection of HMGB1 release. Methods Mol Biol 2013;1004:43–56. 10.1007/978-1-62703-383-1_4 [DOI] [PubMed] [Google Scholar]
- 268. Schiavoni G, Sistigu A, Valentini M, et al. Cyclophosphamide synergizes with type I interferons through systemic dendritic cell reactivation and induction of immunogenic tumor apoptosis. Cancer Res 2011;71:768–78. 10.1158/0008-5472.CAN-10-2788 [DOI] [PubMed] [Google Scholar]
- 269. Schildkopf P, Frey B, Ott OJ, et al. Radiation combined with hyperthermia induces Hsp70-dependent maturation of dendritic cells and release of pro-inflammatory cytokines by dendritic cells and macrophages. Radiother Oncol 2011;101:109–15. 10.1016/j.radonc.2011.05.056 [DOI] [PubMed] [Google Scholar]
- 270. Lorenzi S, Mattei F, Sistigu A, et al. Type I IFNs control antigen retention and survival of CD8α(+) dendritic cells after uptake of tumor apoptotic cells leading to cross-priming. J Immunol 2011;186:5142–50. 10.4049/jimmunol.1004163 [DOI] [PubMed] [Google Scholar]
- 271. Kulzer L, Rubner Y, Deloch L, et al. Norm- and hypo-fractionated radiotherapy is capable of activating human dendritic cells. J Immunotoxicol 2014;11:328–36. 10.3109/1547691X.2014.880533 [DOI] [PubMed] [Google Scholar]
- 272. Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev 2018;281:8–27. 10.1111/imr.12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Mantovani A, Dinarello CA, Molgora M, et al. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity 2019;50:778–95. 10.1016/j.immuni.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Mattei F, Schiavoni G, Sestili P, et al. Irf-8 controls melanoma progression by regulating the cross talk between cancer and immune cells within the tumor microenvironment. Neoplasia 2012;14:1223–43. 10.1593/neo.121444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Sagwal SK, Pasqual-Melo G, Bodnar Y, et al. Combination of chemotherapy and physical plasma elicits melanoma cell death via upregulation of SLC22A16. Cell Death Dis 2018;9:1179 10.1038/s41419-018-1221-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Parlato S, De Ninno A, Molfetta R, et al. 3D microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Sci Rep 2017;7:1093 10.1038/s41598-017-01013-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Nam G-H, Lee EJ, Kim YK, et al. Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer. Nat Commun 2018;9:2165 10.1038/s41467-018-04607-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Ma Y, Aymeric L, Locher C, et al. Contribution of IL-17–producing γδ T cells to the efficacy of anticancer chemotherapy. J Exp Med 2011;208:491–503. 10.1084/jem.20100269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Malamas AS, Gameiro SR, Knudson KM, et al. Sublethal exposure to alpha radiation (223Ra dichloride) enhances various carcinomas' sensitivity to lysis by antigen-specific cytotoxic T lymphocytes through calreticulin-mediated immunogenic modulation. Oncotarget 2016;7:86937–47. 10.18632/oncotarget.13520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Duewell P, Steger A, Lohr H, et al. Rig-I-Like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8+ T cells. Cell Death Differ 2014;21:1825–37. 10.1038/cdd.2014.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Prestwich RJ, Errington F, Ilett EJ, et al. Tumor infection by oncolytic reovirus primes adaptive antitumor immunity. Clinical Cancer Research 2008;14:7358–66. 10.1158/1078-0432.CCR-08-0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Müller LME, Holmes M, Michael JL, et al. Plasmacytoid dendritic cells orchestrate innate and adaptive anti-tumor immunity induced by oncolytic coxsackievirus A21. J Immunother Cancer 2019;7:164 10.1186/s40425-019-0632-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Ma Y, Adjemian S, Mattarollo SR, et al. Anticancer chemotherapy-induced intratumoral recruitment and differentiation of antigen-presenting cells. Immunity 2013;38:729–41. 10.1016/j.immuni.2013.03.003 [DOI] [PubMed] [Google Scholar]
- 284. Wculek SK, Amores-Iniesta J, Conde-Garrosa R, et al. Effective cancer immunotherapy by natural mouse conventional type-1 dendritic cells bearing dead tumor antigen. J Immunother Cancer 2019;7:100 10.1186/s40425-019-0565-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Bauer C, Bauernfeind F, Sterzik A, et al. Dendritic cell-based vaccination combined with gemcitabine increases survival in a murine pancreatic carcinoma model. Gut 2007;56:1275–82. 10.1136/gut.2006.108621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Lu X, Ding Z-C, Cao Y, et al. Alkylating Agent Melphalan Augments the Efficacy of Adoptive Immunotherapy Using Tumor-Specific CD4 + T Cells. J.i. 2015;194:2011–21. 10.4049/jimmunol.1401894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Rodriguez-Ruiz ME, Rodriguez I, Garasa S, et al. Abscopal effects of radiotherapy are enhanced by combined immunostimulatory mAbs and are dependent on CD8 T cells and Crosspriming. Cancer Res 2016;76:5994–6005. 10.1158/0008-5472.CAN-16-0549 [DOI] [PubMed] [Google Scholar]
- 288. Vanpouille-Box C, Diamond JM, Pilones KA, et al. Tgfβ is a master regulator of radiation therapy-induced antitumor immunity. Cancer Res 2015;75:2232–42. 10.1158/0008-5472.CAN-14-3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Hartmann J, Wölfelschneider J, Stache C, et al. Novel technique for high-precision stereotactic irradiation of mouse brains. Strahlenther Onkol 2016;192:806–14. 10.1007/s00066-016-1014-8 [DOI] [PubMed] [Google Scholar]
- 290. Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell 2017;31:326–41. 10.1016/j.ccell.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Ngwa W, Irabor OC, Schoenfeld JD, et al. Using immunotherapy to boost the abscopal effect. Nat Rev Cancer 2018;18:313–22. 10.1038/nrc.2018.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Demaria S, Kawashima N, Yang AM, et al. Immune-Mediated inhibition of metastases after treatment with local radiation and CTLA-4 blockade in a mouse model of breast cancer. Clin Cancer Res 2005;11:728–34. [PubMed] [Google Scholar]
- 293. Rodríguez-Ruiz ME, Rodríguez I, Mayorga L, et al. Tgfβ blockade enhances radiotherapy Abscopal efficacy effects in combination with anti-PD1 and Anti-CD137 immunostimulatory monoclonal antibodies. Mol Cancer Ther 2019;18:621–31. 10.1158/1535-7163.MCT-18-0558 [DOI] [PubMed] [Google Scholar]
- 294. Zamarin D, Holmgaard RB, Subudhi SK, et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med 2014;226:ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Singh M, Savage N, Singh SK. In vivo murine models of brain metastasis. Methods Mol Biol 1869;2019:231–8. [DOI] [PubMed] [Google Scholar]
- 296. Taggart D, Andreou T, Scott KJ, et al. Anti–PD-1/anti–CTLA-4 efficacy in melanoma brain metastases depends on extracranial disease and augmentation of CD8 + T cell trafficking. Proc Natl Acad Sci U S A 2018;115:E1540–9. 10.1073/pnas.1714089115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Seitz C, Rückert M, Deloch L, et al. Tumor cell-based vaccine generated with high hydrostatic pressure synergizes with radiotherapy by generating a favorable anti-tumor immune microenvironment. Front Oncol 2019;9:805 10.3389/fonc.2019.00805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Zschaler J, Schlorke D, Arnhold J. Differences in innate immune response between man and mouse. Crit Rev Immunol 2014;34:433–54. [PubMed] [Google Scholar]
- 299. Buqué A, Galluzzi L. Modeling tumor immunology and immunotherapy in mice. Trends in Cancer 2018;4:599–601. 10.1016/j.trecan.2018.07.003 [DOI] [PubMed] [Google Scholar]
- 300. Bonnotte B, Gough M, Phan V, et al. Intradermal injection, as opposed to subcutaneous injection, enhances immunogenicity and suppresses tumorigenicity of tumor cells. Cancer Res 2003;63:2145–9. [PubMed] [Google Scholar]
- 301. Santi A, Kugeratski FG, Zanivan S. Cancer associated fibroblasts: the architects of stroma remodeling. Proteomics 2018;18:e1700167 10.1002/pmic.201700167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Zitvogel L, Pitt JM, Daillère R, et al. Mouse models in oncoimmunology. Nat Rev Cancer 2016;16:759–73. 10.1038/nrc.2016.91 [DOI] [PubMed] [Google Scholar]
- 303. Gengenbacher N, Singhal M, Augustin HG. Preclinical mouse solid tumour models: status quo, challenges and perspectives. Nat Rev Cancer 2017;17:751–65. 10.1038/nrc.2017.92 [DOI] [PubMed] [Google Scholar]
- 304. Olson B, Li Y, Lin Y, et al. Mouse models for cancer immunotherapy research. Cancer Discov 2018;8:1358–65. 10.1158/2159-8290.CD-18-0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Kemp CJ. Animal models of chemical carcinogenesis: driving breakthroughs in cancer research for 100 years: table 1. Cold Spring Harb Protoc 2015;2015:pdb.top069906–74. 10.1101/pdb.top069906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Kersten K, Visser KE, Miltenburg MH, et al. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med 2017;9:137–53. 10.15252/emmm.201606857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Galuschka C, Proynova R, Roth B, et al. Models in translational oncology: a public resource database for preclinical cancer research. Cancer Res 2017;77:2557–63. 10.1158/0008-5472.CAN-16-3099 [DOI] [PubMed] [Google Scholar]
- 308. Walsh NC, Kenney LL, Jangalwe S, et al. Humanized mouse models of clinical disease. Annu Rev Pathol 2017;12:187–215. 10.1146/annurev-pathol-052016-100332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Shultz LD, Goodwin N, Ishikawa F, et al. Human cancer growth and therapy in immunodeficient mouse models. Cold Spring Harb Protoc 2014;2014:pdb.top073585–708. 10.1101/pdb.top073585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Shultz LD, Brehm MA, Garcia-Martinez JV, et al. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol 2012;12:786–98. 10.1038/nri3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Ali N, Flutter B, Sanchez Rodriguez R, et al. Xenogeneic graft-versus-host-disease in NOD-scid IL-2Rγnull mice display a T-Effector memory phenotype. PLoS One 2012;7:e44219 10.1371/journal.pone.0044219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Mosier DE, Gulizia RJ, Baird SM, et al. Transfer of a functional human immune system to mice with severe combined immunodeficiency. Nature 1988;335:256–9. 10.1038/335256a0 [DOI] [PubMed] [Google Scholar]
- 313. Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol 2007;7:118–30. 10.1038/nri2017 [DOI] [PubMed] [Google Scholar]
- 314. Halkias J, Yen B, Taylor KT, et al. Conserved and divergent aspects of human T-cell development and migration in humanized mice. Immunol Cell Biol 2015;93:716–26. 10.1038/icb.2015.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Saito Y, Ellegast JM, Rafiei A, et al. Peripheral blood CD34+ cells efficiently engraft human cytokine knock-in mice. Blood 2016;128:1829–33. 10.1182/blood-2015-10-676452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Rongvaux A, Willinger T, Martinek J, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol 2014;32:364–72. 10.1038/nbt.2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Melkus MW, Estes JD, Padgett-Thomas A, et al. Humanized mice Mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med 2006;12:1316–22. 10.1038/nm1431 [DOI] [PubMed] [Google Scholar]
- 318. Lan P, Tonomura N, Shimizu A, et al. Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood 2006;108:487–92. 10.1182/blood-2005-11-4388 [DOI] [PubMed] [Google Scholar]
- 319. Billerbeck E, Barry WT, Mu K, et al. Development of human CD4+Foxp3+ regulatory T cells in human stem cell factor–, granulocyte-macrophage colony-stimulating factor–, and interleukin-3–expressing NOD-SCID IL2Rγnull humanized mice. Blood 2011;117:3076–86. 10.1182/blood-2010-08-301507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Ito R, Takahashi T, Katano I, et al. Establishment of a human allergy model using human IL-3/GM-CSF–Transgenic NOG mice. J.i. 2013;191:2890–9. [DOI] [PubMed] [Google Scholar]
- 321. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350–5. 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Zappasodi R, Merghoub T, Wolchok JD. Emerging concepts for immune checkpoint Blockade-Based combination therapies. Cancer Cell 2018;33:581–98. 10.1016/j.ccell.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Postow MA, Callahan MK, Barker CA, et al. Immunologic correlates of the Abscopal effect in a patient with melanoma. N Engl J Med 2012;366:925–31. 10.1056/NEJMoa1112824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Grimaldi AM, Simeone E, Giannarelli D, et al. Abscopal effects of radiotherapy on advanced melanoma patients who progressed after ipilimumab immunotherapy. Oncoimmunology 2014;3:e28780 10.4161/onci.28780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Cekic C, Linden J. Purinergic regulation of the immune system. Nat Rev Immunol 2016;16:177–92. 10.1038/nri.2016.4 [DOI] [PubMed] [Google Scholar]
- 326. Hangai S, Ao T, Kimura Y, et al. Pge2 induced in and released by dying cells functions as an inhibitory dAMP. Proc Natl Acad Sci U S A 2016;113:3844–9. 10.1073/pnas.1602023113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Bondanza A, Zimmermann Valérie S., Rovere-Querini P, et al. Inhibition of phosphatidylserine recognition heightens the immunogenicity of irradiated lymphoma cells in vivo. J Exp Med 2004;200:1157–65. 10.1084/jem.20040327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol 2013;14:668–75. 10.1038/ni.2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Green DR, Galluzzi L, Kroemer G. Cell biology. metabolic control of cell death. Science 2014;345:1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Gopalakrishnan V, Helmink BA, Spencer CN, et al. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 2018;33:570–80. 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 2017;20:145–55. 10.1038/nn.4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Formenti SC, Rudqvist N-P, Golden E, et al. Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nat Med 2018;24:1845–51. 10.1038/s41591-018-0232-2 [DOI] [PMC free article] [PubMed] [Google Scholar]