Bacterial cells regulate their own chromosomal DNA synthesis and cell division depending on the growth conditions, producing more DNA when growing in nutritionally rich media than in poor media (i.e., human gut versus water reservoir). The accumulation of the nucleotide analog (p)ppGpp is usually viewed as serving to warn cells of impending peril due to otherwise lethal sources of stress, which stops growth and inhibits DNA, RNA, and protein synthesis. This work importantly finds that small physiological changes in (p)ppGpp basal levels associated with slow balanced exponential growth incrementally inhibit the intricate process of initiation of chromosomal DNA synthesis. Without (p)ppGpp, initiations mimic the high rates present during fast growth. Here, we report that the effect of (p)ppGpp may be due to the regulation of the expression of gyrase, an important enzyme for the replication of DNA that is a current target of several antibiotics.
KEYWORDS: chromosome initiation, DNA replication, Escherichia coli, growth rate, ppGpp
ABSTRACT
The initiation of Escherichia coli chromosomal DNA replication starts with the oligomerization of the DnaA protein at repeat sequences within the origin (ori) region. The amount of ori DNA per cell directly correlates with the growth rate. During fast growth, the cell generation time is shorter than the time required for complete DNA replication; therefore, overlapping rounds of chromosome replication are required. Under these circumstances, the ori region DNA abundance exceeds the DNA abundance in the termination (ter) region. Here, high ori/ter ratios are found to persist in (p)ppGpp-deficient [(p)ppGpp0] cells over a wide range of balanced exponential growth rates determined by medium composition. Evidently, (p)ppGpp is necessary to maintain the usual correlation of slow DNA replication initiation with a low growth rate. Conversely, ori/ter ratios are lowered when cell growth is slowed by incrementally increasing even low constitutive basal levels of (p)ppGpp without stress, as if (p)ppGpp alone is sufficient for this response. There are several previous reports of (p)ppGpp inhibition of chromosomal DNA synthesis initiation that occurs with very high levels of (p)ppGpp that stop growth, as during the stringent starvation response or during serine hydroxamate treatment. This work suggests that low physiological levels of (p)ppGpp have significant functions in growing cells without stress through a mechanism involving negative supercoiling, which is likely mediated by (p)ppGpp regulation of DNA gyrase.
INTRODUCTION
Among bacterial second messengers, the (p)ppGpp nucleotides are often thought to be key regulators of many of the cellular physiological responses of growing cells to severe nutritional starvation or physical stress. In part, this is because stress can provoke a dramatic burst of (p)ppGpp that soon reaches levels approximating those of GTP and can have severe effects on physiology. These major adjustments to very high levels of (p)ppGpp are usually viewed as ensuring bacterial survival, whether in free-living environments or for pathogens trying to survive in hosts.
Both pppGpp and ppGpp are analogs of GTP and GDP that accumulate in response to nutritional and physical stress throughout the bacterial kingdom and in plant chloroplasts (1–3). Early observations of (p)ppGpp in Escherichia coli suggested a regulatory role in RNA control during the stringent response to amino acid starvation (4, 5) and documented both the inhibition and activation of gene expression by (p)ppGpp (6, 7). The accumulation of (p)ppGpp provokes a still-growing list of intricate adjustments of gene expression, metabolism, and cell physiology that favor survival (8).
Several genetic studies on E. coli provided further insights into (p)ppGpp functions. First, it was found that the complete absence of (p)ppGpp [(p)ppGpp0], occasioned by the deletion of genes that encode both (p)ppGpp synthetases in E. coli (relA and spoT), results in multiple amino acid requirements (9–11). The addition of 9 amino acids (DEILVFHST), known as the Σ set, allows balanced exponential growth of (p)ppGpp0 strains (12). Second, a series of constitutive suppressor mutants of the single (p)ppGpp hydrolase (SpoT) have been isolated, which display incremental elevations of (p)ppGpp basal levels that are associated with a progressive decrease of balanced growth rates despite the absence of stress (13). Finally, spontaneous mutants that suppress the amino acid auxotrophy of (p)ppGpp0 strains are found to occur in the RNA polymerase RpoB, RpoC, and RpoD subunits (14, 15). These mutants display regulatory patterns that mimic those of (p)ppGpp but in the absence of (p)ppGpp (10). This can be taken to implicate regulatory functions exerted by (p)ppGpp that operate through effects on transcription. A large number of (p)ppGpp-mediated regulatory effects occur at the level of promoter activation or inhibition (16), but many regulatory interactions also occur through the binding of (p)ppGpp to specific proteins (17).
Classical experiments revealed a fundamental relationship: the DNA, RNA, and protein contents of E. coli cells were found to covary as a function of the rates of balanced exponential growth; this regulatory relationship came to be known as growth rate control (18, 19). The notion that (p)ppGpp is the key determinant of the cellular content of RNA was first based on measurements correlating the fraction of total transcripts devoted to rRNA synthesis with the abundance of (p)ppGpp at different balanced growth rates (20, 21). More recently, the (p)ppGpp0 state was shown to result in constitutive, high RNA/protein and RNA/DNA ratios independent of balanced growth rates, while artificially increasing (p)ppGpp basal levels without stress mimics growth rate control (12). This behavior is taken to suggest that (p)ppGpp is both necessary and sufficient for the regulation of the cellular contents of RNA and protein as a function of the growth rate. The mechanism by which this occurs was subsequently rigorously attributed to (p)ppGpp inhibition of rRNA promoters since constitutive overexpression of apparently normal, mature rRNA could be documented at all growth rates (12).
Incremental elevations of predominantly either ppGpp or pppGpp basal levels without stress have been achieved by the progressive induction in E. coli of heterologous (streptococcal) ppGpp synthetase fragments combined with genetic manipulation of GppA (guanosine pentaphosphatase), which mediates the conversion of pppGpp to ppGpp (22). This revealed that even modest elevations of ppGpp basal levels are sufficient to slow growth, with ppGpp found to be a more potent regulator than pppGpp (22). This difference in potency was retained for several other in vitro and cellular assays of ppGpp regulation, including rRNA promoter (rrnB P1) activity (22).
Our primary concern here is to explore the role of (p)ppGpp as a possible regulator of chromosomal DNA synthesis initiation as a function of balanced growth rates in E. coli. During balanced growth, the amount of DNA and the number of replication origins per cell have been shown to directly correlate with the growth rate (20, 23, 24). There are clear indications that (p)ppGpp can inhibit DNA replication elongation by binding DNA primase (25–28), but our focus is on the initiation of DNA synthesis.
Replication of the E. coli chromosome initiates from a single replication origin, oriC, and proceeds bidirectionally around the circular chromosome until the two replication forks meet and terminate at the terminus region (ter). During rapid growth in rich media, the time required to complete chromosomal replication is longer than the time for cell division. Additional rounds of replication are then necessarily initiated at the ori region before the previous round is completed, leading to overlapping rounds of chromosome replication (29). The abundance of DNA at the oriC region can be normalized to the abundance of ter region DNA to give an ori/ter ratio, which provides a quantitative estimate of initiation activity in growing cells during even small changes in balanced growth rates.
The initial step of chromosomal DNA replication involves the ATP-dependent oligomerization of DnaA that binds at repeated sites within oriC. This is followed by transient unwinding of the DNA, loading of the DnaB helicase onto the exposed single strands, binding of primase, and, finally, assembly of the replisome. To avoid triggering excessive premature replication forks, hydrolysis of the ATP moiety in DnaA-ATP and sequestration of both the dnaA promoter and oriC occur. Changes in the balance of any of these regulatory steps can affect overlapping replication cycles or multifork replication (for reviews, see references 30 to 33). Nevertheless, while a modest 2-fold inhibition of dnaA transcription by (p)ppGpp has been repeatedly observed (16, 34), this 2-fold change of DnaA levels has been argued as not being enough to affect ori/ter ratios (35, 36).
A fundamental question is how DNA synthesis is regulated as a function of varying balanced exponential growth. Population-averaged measurements of bacterial mass and DNA replication initiation frequencies over a range of growth rates resulted in the long-standing hypothesis that the bacteria initiate replication once they reach a constant, critical mass (37). In this study, we compare ori/ter ratios for wild-type (WT) and (p)ppGpp0 cells as a function of balanced growth rates. With wild-type cells, we verify a monotonic decrease in the rate of DNA initiation that occurs as cells grow more slowly, which is accompanied by a proportionate decrease in cell mass at lower growth rates. In striking contrast, cells lacking (p)ppGpp maintain high frequencies of DNA synthesis initiation regardless of whether balanced growth rates are high or low. These persistent high rates of DNA initiation independent of the growth rate in (p)ppGpp0 cells are accompanied by a constant cell mass, estimated as dry weight.
RESULTS
(p)ppGpp is necessary for inhibition of DNA synthesis initiation.
To measure DNA synthesis initiation, ratios of the E. coli origin (ori) DNA and terminus region (ter) DNA were determined by quantitative PCR (qPCR). Different growth rates (μ) for E. coli strain MG1655 and its derivative (p)ppGpp0 strain (CF10237) were achieved using the same media as the ones previously reported (12) (growth rates and medium conditions are shown in Table S1 in the supplemental material). Figure 1 shows that the known direct correlation between growth rate and chromosomal DNA synthesis initiation (ori/ter ratio) for WT cells is abolished in (p)ppGpp0 cells. During fast growth in rich media, such as LB (μ = 2.3 h−1), multifork initiation of WT chromosomal replication results in a high ori/ter ratio of 4.3 ± 0.35. The same cells growing slowly in poor media (μ = 0.52 h−1) show low ori/ter ratios approaching 1 (1.7 ± 0.05). In fast-growing (p)ppGpp0 cells in LB (μ = 1.94 h−1), the ori/ter ratio is 4.2 ± 0.23, similar to the value of 4.3 for the WT. The lowest growth rate that we can achieve for the (p)ppGpp0 strain is 0.47 h−1 in minimal medium necessarily supplemented with the Σ set of amino acids. As mentioned above, this supplement is needed because these strains are auxotrophs for multiple amino acids. The ori/ter ratios for slow-growing (p)ppGpp0 strains (3.6 ± 0.21) are higher than that for the WT (1.7 ± 0.05). Figure 1 reveals that the linear dependence of DNA synthesis initiation on the medium-imposed growth rate observed for the WT strain is essentially abolished in the (p)ppGpp0 strain.
FIG 1.
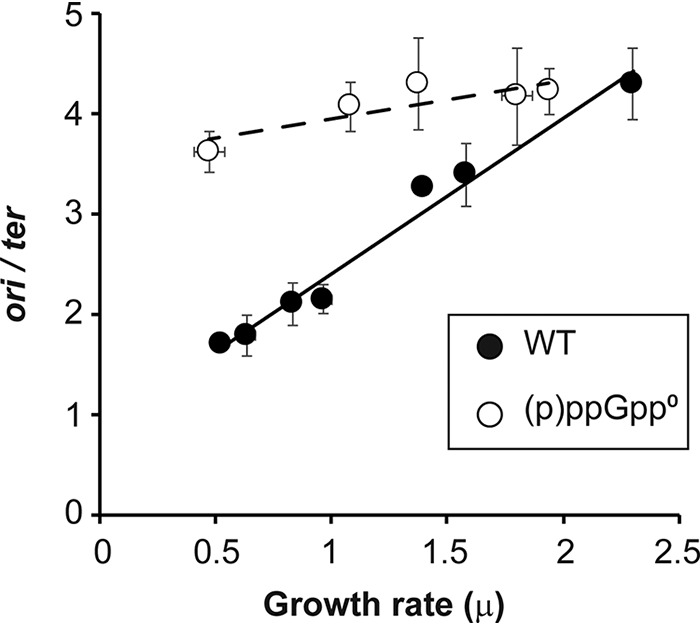
Ratios of ori/ter at different growth rates in the presence or absence of (p)ppGpp. Strains MG1655 (WT) and CF10237 [(p)ppGpp0] were grown in different media in order to achieve different balanced growth rates (see Table S1 in the supplemental material), and ori/ter ratios were measured by qPCR. Error bars represent the SD from 2 biological replicates and 3 technical replicates. The P values were calculated by comparing the values to the highest growth rate (LB) for each strain. All samples of the WT strain are significantly different, with a P value of <0.01, while the (p)ppGpp0 samples are not significantly different (P value of >0.01).
Growth rates (μ) and standard deviations (STDV) measured for MG1655 and (p)ppGpp0 strains in different media. Download Table S1, PDF file, 0.1 MB (139.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The levels of (p)ppGpp increase not only in poor media but also in the early stationary phase. At the same time, stationary phase is known for the lack of cellular replication; therefore, ratios of ori/ter were measured in WT and (p)ppGpp0 cells grown in LB at exponential and stationary phases (Fig. S1). At stationary phase, there is a decrease of the ori/ter ratio up to 1 in WT strains, but it stays elevated (up to 4) in (p)ppGpp0 strains. These data seem consistent with a regulatory role for (p)ppGpp as being necessary to inhibit the initiation of DNA replication. The persistence of high ori/ter ratios in slow-growing (p)ppGpp0 cells could be due to higher levels of ori, lower levels of ter if termination is somehow impaired, or both. Next-generation sequencing (NGS) has been used by others to detect aberrant fork dynamics or inappropriate replication stalling or blockage near the ter site (38). When performing NGS on WT genomes grown in rich medium, higher reads per million of the ori region are observed than for the ter region (Fig. 2). This difference is consistent with multifork replication. The same WT strain grown in poor medium resulted in values for reads per million that remained constant over the entire genome. In the (p)ppGpp0 strain, multifork replication seems independent of the growth rate, which reinforces the qPCR data shown in Fig. 1. The ter region shows neither these anomalies nor inappropriate stalling. The effects of (p)ppGpp on ori/ter ratios seem to be preferentially exerted on the ori region, as expected if (p)ppGpp might inhibit initiation.
FIG 2.
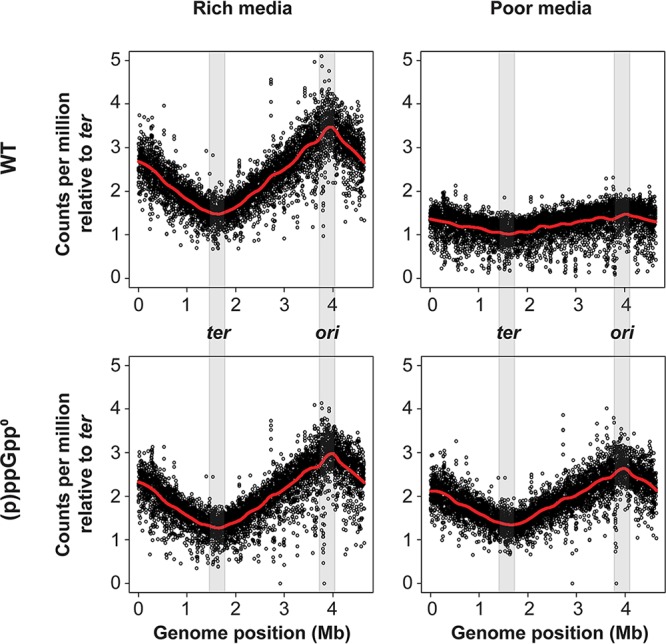
Next-generation sequencing (NGS) profiles for WT and (p)ppGpp-deficient strains. Strains were grown in rich medium (LB) or poor medium (M9 with the Σ set and 0.2% glucose). The total read count was corrected per million reads and normalized to the number of ter region reads. Both the ori and ter regions are shaded in gray. Locally estimated scatterplot smoothing (LOESS) regression curves are shown in red.
Ratios of ori/ter at exponential and stationary phases in the presence or absence of (p)ppGpp. Strains MG1655 (WT) and CF10237 [(p)ppGpp0] were grown in LB up to exponential phase (A600 of 0.2) and stationary phase (A600 of 1.5), and ori/ter ratios were measured by qPCR. Error bars represent the SD from 2 biological replicates and 3 technical replicates. Values significantly different with a P value of <0.01 are marked with *, while those not significantly different (P value of >0.01) are marked with ns (not significant). Download FIG S1, TIF file, 0.4 MB (406.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(p)ppGpp is sufficient for inhibition of DNA synthesis initiation.
It has been previously observed that (p)ppGpp abundance controls balanced growth rates, in the sense that artificially elevating (p)ppGpp levels per se provokes slower growth without stress, even in rich media (12). This behavior led us to ask if artificially elevated (p)ppGpp levels are also sufficient to inhibit the initiation of DNA replication. Therefore, ori/ter ratios were compared between the WT and the two hydrolase mutants (spoT202 and spoT203), with constitutive 4-fold and 8-fold increases of (p)ppGpp basal levels above those of the wild type, respectively (13). Figure 3A reveals that the mutant-mediated elevations of basal (p)ppGpp levels in LB decrease both growth rates and ori/ter ratios. Moreover, comparison of the data in Fig. 1 and Fig. 3A reveals that the relationship of ori/ter ratios and growth rates when (p)ppGpp levels are manipulated with the spoT alleles is similar to the correlation found when growth rates are determined by medium composition, consistent with the notion that WT growth rates are responding to (p)ppGpp basal levels, even in rich media.
FIG 3.
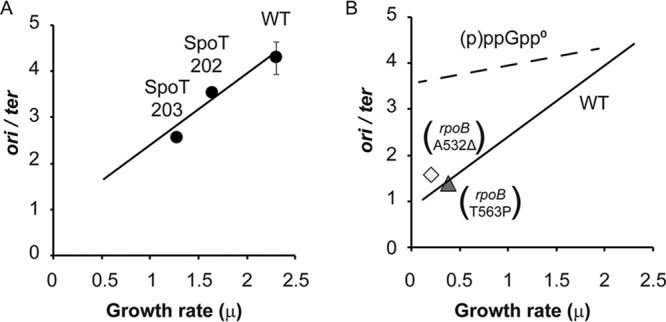
(p)ppGpp is sufficient to inhibit initiation of DNA replication, probably involving gene expression. (A) Strains with increased (p)ppGpp basal levels due to (p)ppGpp hydrolase defects (MG1655 [WT], CF17960 [SpoT202], and CF17961 [SpoT203]) were grown in LB, and ori/ter ratios were determined. The line shows the relation between the ori/ter ratio and the growth rate observed for the WT growing in different media (Fig. 1). A chi-square goodness-of-fit test was used to determine that the samples follow the WT pattern with a 98.1% probability. (B) Strains lacking (p)ppGpp but containing RNA polymerase phenocopy mutants that mimic the presence of (p)ppGpp. Mutant alleles rpoB-A532Δ (CF11760) and rpoB-T563P (CF11768) suppress the amino acid requirements of (p)ppGpp0 hosts and allow growth in M9 glucose minimal medium. Under these conditions, the ori/ter ratios are lowered to approximate values found for a slow-growing WT strain. The solid line reflects the relation between ori/ter ratios and growth rates for the WT strain. The dashed line reflects the relation for (p)ppGpp0 strains without rpoB mutations, varying balanced growth rates when grown in different media, as shown in Fig. 1. A chi-square goodness-of-fit test was used to determine that the samples follow the WT pattern with a 93.3% probability. Error bars represent the SD from 2 biological replicates and 3 technical replicates.
While ppGpp and pppGpp are known to inhibit balanced growth rates, manipulations that lead to the accumulation (arabinose inducible) of predominately one or the other reveal that ppGpp is a more potent inhibitor of the growth rate than pppGpp (22). The effects of the preferential accumulation of each analog were measured on ori/ter ratios with increasing levels of arabinose, as shown in Fig. S2. As previously observed (22), ppGpp has a more potent ability to reduce the growth rate for a given concentration of arabinose than pppGpp. Both nucleotides inhibit DNA replication initiation in proportion to the resulting growth rate.
Both ppGpp and pppGpp affect growth rates and ori/ter ratios. The results confirm the higher potency of ppGpp than of pppGpp for growth rate inhibition and extend this relationship to include a similar higher potency for inhibition of ori/ter ratios. Strains CF16762 (ppGpp) and CF16760 (pppGpp) were grown in M9 medium containing 0.4% glycerol plus all the amino acids with increasing levels of arabinose (0, 0.005%, 0.01%, and 0.02%). The black line, taken from Fig. 1, represents the correlation between ori/ter ratios and growth rates observed for WT growth in different media. A goodness-of-fit (chi-square) test was used to determine that the samples follow the WT pattern shown in Fig. 1, with 99.2% and 99.7% probabilities for ppGpp and pppGpp samples, respectively (error bars represent the SD from 2 biological replicates and 3 technical replicates). Download FIG S2, TIF file, 0.6 MB (585.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Inhibitory effects of (p)ppGpp on initiation of DNA replication are also found with RNA polymerase suppressor mutants in the absence of (p)ppGpp.
Spontaneous RNA polymerase mutants can be isolated that allow the growth of (p)ppGpp0 cells in minimal media, suppressing the (p)ppGpp0 multiple-amino-acid requirements (15). Suppression can be viewed as altering the conformation of RNA polymerase to phenocopy the regulatory effects of (p)ppGpp on the transcription of amino acid biosynthetic pathway genes (10). Two such mutants (rpoB-A523Δ and rpoB-T563P), first found by Jin and Gross (39), were employed previously in (p)ppGpp0 hosts to document their effects on the regulation of rRNA content as a function of balanced growth rates (12). Evidently, these two rpoB alleles also lower ori/ter ratios in the (p)ppGpp0 strain grown on minimal medium (Fig. 3B) and therefore also suppress the high ori/ter ratios characteristic of the (p)ppGpp0 strain with wild-type RNA polymerase (Fig. 1). Since these same RNA polymerase mutant suppressor alleles can also mimic positive transcription regulation by (p)ppGpp, this phenocopy suggests that (p)ppGpp also acts by altering RNA polymerase gene expression rather than binding directly to components of the replisome as seen with other proteins (17).
The high rates of initiation of chromosomal DNA replication in the absence of (p)ppGpp are not an indirect consequence of topological perturbations due to (p)ppGpp inhibition of rrn operon transcription transmitted to the ori region.
Recently, it has been proposed that the mechanism of (p)ppGpp inhibition of DNA initiation is primarily an effect of (p)ppGpp inhibition of the transcription of the seven E. coli rrn operons, which indirectly increases oriC superhelical density so as to retard the strand separation required for the initiation of replication by DnaA (36). In these experiments, induction of (p)ppGpp to high levels was achieved either by adding serine hydroxamate (SHX) to inhibit serinyl-tRNA charging or by inducing relA expression on a multicopy plasmid. We attempted to test one aspect of this hypothesis, namely, the prediction that active rrn operons, in particular the rrnC operon that is a close neighbor of oriC, must be present on the same chromosome as the ori locus for (p)ppGpp regulation to occur. We employed a strain that has all seven rrn loci deleted, including their (p)ppGpp-sensitive promoters. The strain is viable only because of constitutively expressed genes carried on two compatible multicopy plasmids. A single rrnB operon is present on one plasmid, and several rrn intragenic tRNA genes are present on the other. We also constructed a (p)ppGpp0 derivative of this strain with deletions of the spoT and relA genes and confirmed its inability to grow on minimal media.
Figure 4A shows that when the source of all rRNA transcription is a plasmid rather than the host chromosome (referred as Δ7 rrn), both WT and (p)ppGpp0 strains retain the relationship between growth rates and ori/ter ratios seen in the strains containing the ribosomal operons in the chromosome (Fig. 1). This provides a strong argument that regulation by (p)ppGpp does not require one or more ribosomal operons to be present in cis with oriC.
FIG 4.
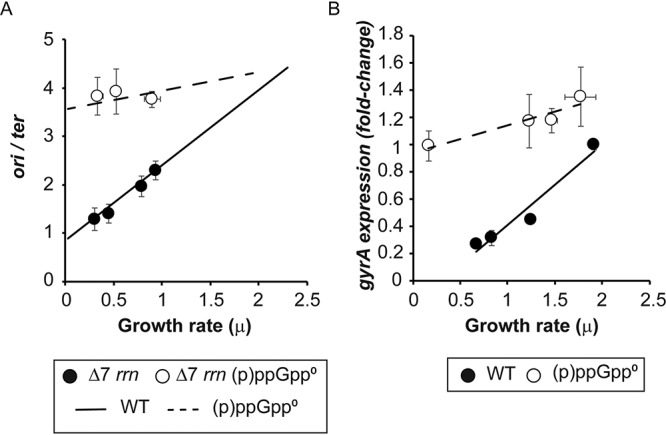
(p)ppGpp effects on DNA initiation are not due to rrn operons but possibly are due to changes in gyrase expression. (A) ori/ter ratios of the Δ7 rrn strain and its (p)ppGpp0 mutant plotted against growth rates. The lines represent the relation between ori/ter ratios and growth rates for the WT and (p)ppGpp0 strains shown in Fig. 1. A chi-square goodness-of-fit test was used to determine that the samples follow the pattern shown in Fig. 1 with a 99% probability. (B) Influence of growth rate on relative gyrA gene expression for WT and (p)ppGpp-deficient strains. Strains MG1655 and CF10237 were grown in the following media: LB, medium A plus 0.2% glucose and all amino acids, and M9 medium plus 0.2% glucose and the Σ set. The WT strain was also grown in M9 medium plus glucose, and the (p)ppGpp strain was grown in M9 medium plus 0.2% glucose and all amino acids. The expression level of the dnaA gene was determined by RT-PCR. The values plotted are normalized to the value for the fastest-growing WT strain. The P values were calculated by comparing the values to the highest growth rate (LB) for each strain. All samples of the WT strain are significantly different, with a P value of <0.01, while the (p)ppGpp0 samples are not significantly different (P value of >0.01). Error bars represent the SD from 2 biological replicates and 3 technical replicates.
(p)ppGpp inhibits gene expression of DNA gyrase and topoisomerase IV.
Since supercoiling is clearly involved in (p)ppGpp effects on DNA replication initiation (36) but evidently not due to cis-dependent transmission of (p)ppGpp inhibition of rrn transcription, we decided to ask if the expression of factors responsible for increasing or decreasing supercoiling might be affected by (p)ppGpp (Fig. S3). While (p)ppGpp seems to only mildly affect the gene expression of topoisomerase I (topA), it strongly represses the expression of gyrase (gyrA) and topoisomerase IV (parC) during stationary phase. Therefore, we asked if variations in the expression of gyrA at different growth rates respond to (p)ppGpp (Fig. 4B). In the WT strain, gyrA expression decreases as a function of the growth rate, but in the absence of (p)ppGpp, it remains quite constant, which is similar to observations for ori/ter ratios (Fig. 1). The observed gyrA expression levels correlate with ori/ter ratios (Fig. S4) in the presence or absence of (p)ppGpp (correlation coefficients of 0.97 and 0.94, respectively). Although DNA replication initiation is a highly regulated process involving many factors, our data can be taken to suggest that (p)ppGpp could affect ori/ter ratios by controlling gyrase gene expression.
Effect of (p)ppGpp on gene expression of factors that control global supercoiling. Strains MG1655 (WT) and CF10237 [(p)ppGpp0] were grown in LB up to exponential phase (A600 of 0.2) and stationary phase (A600 of 1.5), and the expression levels of topA (A), gyrA (B), and parC (C) were measured by RT-qPCR. Error bars represent the SD from 2 biological replicates and 3 technical replicates. The values plotted are normalized to the value for the WT strain in exponential phase. Values significantly different with a P value of <0.01 are marked with *, while those not significantly different (P value of >0.01) are marked with ns. Download FIG S3, TIF file, 0.8 MB (876.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Correlation between gyrA expression (data from Fig. 4B) and ori/ter ratios. Correlation coefficients are 0.975 for the WT and 0.938 for the (p)ppGpp0 strain. Download FIG S4, TIF file, 0.4 MB (381.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cell mass is also affected by (p)ppGpp.
A strong correlation exists between cell mass and growth rate, which led to the hypothesis that cells require the accumulation of a critical mass before they divide (37). We asked if (p)ppGpp might alter the cell mass, estimated as dry weight, again comparing WT and (p)ppGpp0 strains, while varying the growth rate as in Fig. 1. The data in Fig. 5A reaffirm the correlation between cell mass and growth rate in wild-type cells. In contrast, for the (p)ppGpp0 strain, the cell mass becomes independent of the growth rate (Fig. 5A). Instead, the cell mass of the (p)ppGpp0 strain remains constant at about half of the range displayed by WT cells. If cell mass estimates are plotted against the corresponding ori/ter ratios instead of growth rates, as in Fig. 5B, then a linear correlation is observed for the WT strain (correlation coefficient of 0.98), indicating a three-way correlation. In the absence of (p)ppGpp, the correlation between the cell mass and the ori/ter ratio disappears (correlation coefficient of 0.29).
FIG 5.
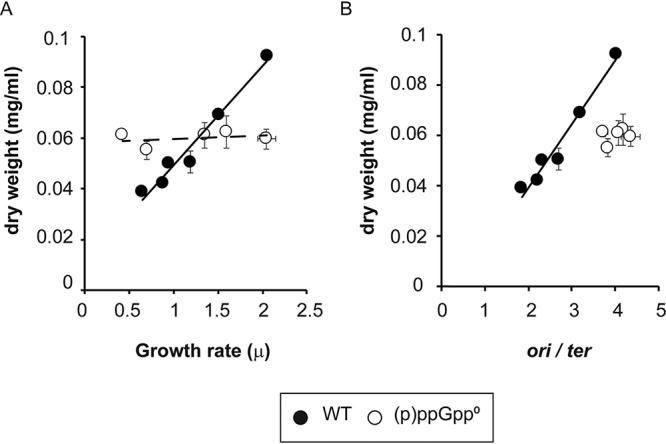
(p)ppGpp deficiency alters the normal dependence of cell mass on growth rate. (A) Measurements of the cellular dry-weight estimates of mass as a function of growth rate variations achieved with different media (see Table S1 in the supplemental material. The wild-type strain is MG1655, and the (p)ppGpp0 strain is CF10237. The P values were calculated by comparing the values to the highest growth rate (LB) for each strain. All samples of the WT strain are significantly different, with a P value of <0.01, while the (p)ppGpp0 samples are not significantly different (P value of >0.01). (B) Correlation between cell mass and ori/ter ratios. Correlation coefficients are 0.983 for the WT and 0.288 for the (p)ppGpp0 strain. Error bars represent the SD from 2 biological replicates.
A similar behavior is suggested by observations of effects of growth rate on cell size in (p)ppGpp0 strains (Fig. S5), despite considerable variability in the absence of (p)ppGpp. It is noteworthy that (p)ppGpp0 cells show cell division defects accompanied by abnormal filamentation (7, 9), as shown in Fig. S5C. While there is evidently a reduction of cell size that is dependent on the growth rate in the WT strain, for the (p)ppGpp0 strain, no statistically significant differences are found (Fig. S5A).
(p)ppGpp deficiency seems to alter the normal dependence of cell volume on growth rate. (A) Plot of cellular volume versus growth rate variations achieved with the different media listed in Table S1 in the supplemental material. The wild-type strain is MG1655 (solid symbols), and the (p)ppGpp0 strain is CF10237. The P values were calculated by comparing the values to the highest growth rate (LB) for each strain. All samples of the WT strain are significantly different, with a P value of <0.01, while the (p)ppGpp0 samples are not significantly different (P value of >0.01). (B) Correlation between cell volume and ori/ter ratios. Correlation coefficients are 0.976 for the WT and −0.051 for the (p)ppGpp0 strain. Error bars represent the SD from 2 biological replicates. (C) Microscopy image of (p)ppGpp0 strains in LB showing deficiencies in segregation and filamentation. Download FIG S5, TIF file, 2.6 MB (2.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The distribution of chromosomes per cell depends on growth rate but it is not affected by (p)ppGpp.
The number of chromosomes per cell was also measured by flow cytometry (Table 1; Fig. S6) for WT and (p)ppGpp-deficient strains comparing growth on rich medium (LB plus Glu) with growth on poor medium (M9 medium plus the Σ set and glucose). A set of standards was included to relate the number of chromosomes to fluorescence intensity (Fig. S6, inset). This approach yields several puzzling observations.
TABLE 1.
Number of chromosomes per cell of WT and (p)ppGpp0 strains grown in rich and poor media measured by flow cytometrya
| Strain | No. of chromosomes | % of cells with no. of chromosomes |
|
|---|---|---|---|
| Rich medium | Poor medium | ||
| WT | 1 | 0.13 | |
| 2 | 17.35 | ||
| 4 | 0.83 | 73.40 | |
| 8 | 84.15 | 9.00 | |
| 16 | 14.45 | 0.07 | |
| (p)ppGpp0 | 1 | 5.36 | |
| 2 | 18.40 | ||
| 4 | 0.15 | 45.25 | |
| 8 | 27.30 | 29.65 | |
| 16 | 70.15 | 1.36 | |
Data are presented as percentages of cells from Fig. S6 in the supplemental material. The average numbers of chromosomes per cell in rich medium and poor medium were 9.07 and 4.02, respectively, for the WT and 13.55 and 4.82, respectively, for the (p)ppGpp0 strain.
The number of chromosomes per cell is growth rate dependent and independent of (p)ppGpp. Shown are flow cytometry profiles for WT and (p)ppGpp-deficient strains grown in rich (LB plus 0.2% glucose) or poor (M9 medium with the Σ set plus 0.2% glucose) medium. The inset shows standards used to associate the intensity peaks with the number of chromosomes. The light gray vertical bars identify the chromosomal number per cell. The numerical distribution of these data is shown in Table 1. Download FIG S6, TIF file, 1.5 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
During fast growth in rich medium, up to 84% of WT cells have 8 chromosomes, while up to 70% of (p)ppGpp0 cells have 16 chromosomes (Table 1; Fig. S6). This occurs even though the ori/ter ratios during fast growth are nearly the same (Fig. 1).
For slow growth, although (p)ppGpp0 cells have more chromosomes per cell than do WT cells [29% of (p)ppGpp0 cells have 8 chromosomes, compared to 9% of WT cells], most of the cells of both strains [73% of WT and 45% of (p)ppGpp0 cells] have 4 chromosomes (Table 1; Fig. S6). In the presence or absence of (p)ppGpp, cells growing in rich medium have a higher average number of chromosomes per cell than when growing in poor medium [9.07 versus 4.02 for the WT and 13.55 versus 4.82 for the (p)ppGpp0 strain]. This suggests that the number of chromosomes per cell depends on the growth rate, independent of the presence or absence of (p)ppGpp. (p)ppGpp0 cells have similar ori/ter ratios in rich and poor media (Fig. 1), but they have different numbers of chromosomes per cell (Table 1), suggesting differences in the rate of synthesis of DNA.
It is also noticeable that the total numbers of cells analyzed in (p)ppGpp0 strains are half those of the WT. The reason for this is unknown, but it could be due to cephalexin sensitivity differences. The (p)ppGpp0 strains are twice as sensitive to cephalexin as the WT (40).
(p)ppGpp can be important to maintain a constant initiation mass.
Our data allow comparisons of several cell cycle parameters for WT and (p)ppGpp0 cells grown in rich and poor media. Figure 6A shows that the C period (time required to complete a round of replication) for the WT, as expected, remains similar in both media (40 to 50 min). However, the C period increases nearly 3-fold (43 to 135 min) during slow growth in the absence of (p)ppGpp, as predicted based on the flow cytometry data. For WT cells, the D period (time between the end of replication and cell division) increases in poor medium (42 min compared to 22 min) (Fig. 6B), while the D period for (p)ppGpp0 does not change much. For WT cells, measurements of the initiation mass remain constant and independent of growth (Fig. 6C), as classically described (41). In the absence of (p)ppGpp, the initiation mass increases up to 7-fold (0.006 for slow growth to 0.045 for fast growth) (Fig. 6C).
FIG 6.
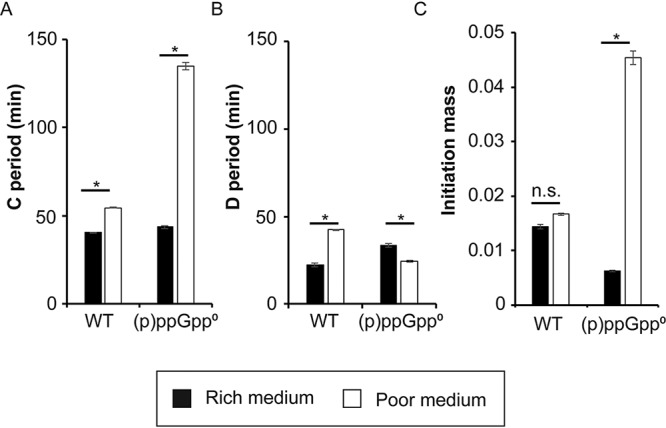
Growth parameters of WT and (p)ppGpp0 strains in rich and poor media. (A and B) The lengths of the C period (A) and the D period (B) were measured as described previously (66). (C) Initiation mass was measured as described previously (67). Values significantly different with a P value of <0.01 are marked with *, while those not significantly different (P value of >0.01) are marked with n.s. (not significant).
DISCUSSION
The exponential rates of Escherichia coli growth determined by media with all components present in excess but utilized with different efficiencies can be predicted to require a myriad of subtle adjustments. Slowly emerging evidence over the past 50 years suggests that (p)ppGpp plays increasingly generalized roles, with subtle, as well as major, adjustments (Fig. 7). Initially, (p)ppGpp was a contested participant linking these adjustments to charged tRNA availability, later to carbon source availability, later still to robust lipid metabolism, and finally to extended rRNA synthesis and ribosome formation. It was appreciated as being critical for controlling the growth rate and rRNA synthesis (20). Later evidence, using methods similar to those described here, linked basal (p)ppGpp levels to the cellular ribosome content (12, 22).
FIG 7.
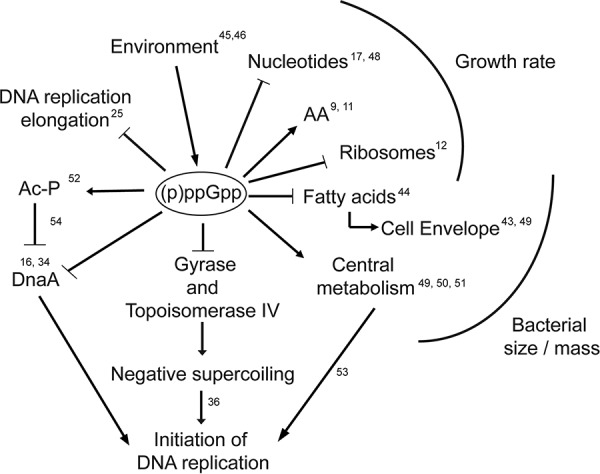
Large array of effects of (p)ppGpp on diverse cellular processes. Numbers correspond to the references shown in the text (9, 11, 12, 16, 17, 25, 34, 36, 43–46, 48–54). Arrows represent positively regulated pathways, while crossed lines are negatively regulated pathways. AA, amino acid biosynthesis; Ac-P, acetyl-phosphate.
The experiments here suggest that (p)ppGpp also plays a key role as a regulator of the initiation of DNA replication (Fig. 1). The regulation of cellular mass as a function of the growth rate can also be dependent on (p)ppGpp, although both phenomena seem to be regulated by different pathways (Fig. 5). As a simplistic model, it can be hypothesized that (p)ppGpp could control cellular size by controlling phospholipid production (42–44), while effects on ori/ter ratios occur through modifying ori region DNA topology (36) through changes in the expression of gyrase (Fig. 4B; see also Fig. S3 in the supplemental material). However, the coordination of chromosomal DNA initiation with the growth rate and cellular mass is a complex process, and it seems likely that the cumulative effects of diverse adjustments might combine to modulate major determinants (Fig. 7).
The RelA and SpoT proteins cooperatively sense environmental changes to increase the (p)ppGpp levels (3, 45, 46), which will limit active ribosome content (12, 22, 47), that are a major contributor to growth rate control. Both the synthesis and uptake of precursors are regulated by (p)ppGpp (9, 11, 17, 48), resulting in the control of DNA, RNA, and protein. Moreover, central metabolism is also controlled by (p)ppGpp (49–52). Mutations in central metabolism genes such as tktB, pgi, and pta change the thermosensitivities of dnaB8, dnaE486, and dnaA46 mutant alleles, respectively (53), but these mutations do not seem to affect DNA replication initiation (Fig. S7A). These mutants show similar ori/ter ratios as those observed in the WT with different rates of balanced growth (Fig. S7A).
The effect of (p)ppGpp on DNA replication initiation is not due to effects on central metabolism or dnaA expression. (A) ori/ter ratios are related to various growth rates obtained with strains deficient in tktB, pgi, or pta and grown in different media. The dashed line represents the correlation between ori/ter ratios and growth rates observed for the WT, as shown in Fig. 1. A chi-square goodness-of-fit test was used to determine that the samples follow the WT pattern shown in Fig. 1, with 88%, 90.2%, and 91.6% probabilities for tktB, pgi, and pta samples, respectively. Error bars represent the SD from 2 biological replicates and 3 technical replicates. (B) Strains MG1655 and CF10237 were grown in the same media as the ones for Fig. 4. The expression level of the dnaA gene was determined by RT-PCR. Error bars represent the SD from 2 biological replicates and 3 technical replicates. The values plotted are normalized to the value for the fastest-growing WT strain. The P values were calculated by comparing the values to the highest growth rate (LB) for each strain. All samples are significantly different, with a P value of <0.01. Download FIG S7, TIF file, 0.6 MB (654.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The question arises as to whether the effects of (p)ppGpp are also exerted on dnaA promoter activity at different growth rates (Fig. S7B). Up to 2-fold increases in dnaA mRNA levels have been reported in the absence of (p)ppGpp (16, 34). Our observations suggest that growth rate control over dnaA expression persists even in the absence of (p)ppGpp (Fig. S7B). It is noteworthy that the acetylation of a DnaA lysine in the ATP binding domain by acetyl-phosphate affects its ability to bind to the origin (54). Since the levels of acetyl-phosphate are proportional to the levels of (p)ppGpp (52), it is possible that (p)ppGpp could change the levels of acetylation of DnaA and affect DNA replication initiation. However, we find that ori/ter ratios at different growth rates are unchanged with a pta mutant deficient for acetyl-phosphate production (Fig. S7A) as well as with an ackA-pta double mutant that abolishes acetyl-phosphate formation. The control of DNA replication initiation by (p)ppGpp seems not to involve the acetylation of DnaA. However, under some other condition, a combination of these effects might contribute to the control of the initiation of DNA replication.
Recently, inhibition of DNA replication by high (p)ppGpp levels was hypothesized to be indirectly due to inhibition of rRNA gene transcription that changes the DNA topology neighboring the ori sequences (36). However, we find that (p)ppGpp inhibition of initiation persists even when chromosomal rRNA operons are deleted and instead expressed on a separate plasmid, which persists whether (p)ppGpp is present or absent (Fig. 4A). Inhibition of topoisomerase I seems to bypass the negative effect of (p)ppGpp on DNA replication, suggesting that the (p)ppGpp effect is due to a decrease of negative supercoiling (36). Both gyrase and topoisomerase IV are able to reduce positive supercoiling, at the same time that gyrase introduces negative supercoiling (55). Our data show that (p)ppGpp has no effect on topoisomerase I expression, while it inhibits the expression of gyrase and topoisomerase IV (Fig. S3). At the same time, the levels of gyrase seem to correlate with ori/ter ratio measurements in both the presence and absence of (p)ppGpp (Fig. 4B), suggesting a possible mechanism of action. Further studies are required.
The ori/ter ratios (Fig. 1) show that (p)ppGpp-deficient strains contain multiple replication forks in poor medium, but the numbers of chromosomes per cell (Table 1) are similar to WT numbers. At the same time, C periods of these cells are extremely long (up to 135 min), suggesting that DNA replication elongation has been slowed. This could not be a direct effect, because (p)ppGpp inhibits DNA primase in vitro, and higher elongation rates would be expected; however, in vitro effects seem to be masked in vivo (26). The lower elongation rates could instead result from stalled RNA polymerase complexes that slow DNA replication elongation (56, 57). Since these events are not evident from the NGS experiments (Fig. 2), they are judged to either occur at random widespread sites instead of a small number of specific chromosomal sites or not be severe enough to be detected. Notably, these lower elongation rates seem to be dependent on growth rates but must be independent of (p)ppGpp because they are observed in the absence of (p)ppGpp.
The general growth law was defined under certain constraints, and our study employed the same parameters to compare effects in the presence and absence of (p)ppGpp. This includes sampling at low cell densities (A600 of ≤0.2) during balanced exponential growth, where growth rates are determined only by the ability to use nutrients present in excess. Different media have been used in this study (Table S1), with different compositions: while LB is an undefined medium, A and M9 media differ in their contents of micronutrients. Although small effects might well be attributed to medium differences, growing cells adapt but are never starved, keeping with the classically defined constraints. In minimal media with a carbon source but without amino acids, E. coli can reproduce itself from salts available in the media. In contrast to this classical way of altering the growth rate, quite different regulatory effects can result from starvation for one or more specific nutrients that delay or avoid the severe consequences of starvation, stasis or cell death (58). Protocols for nutrient starvation can entail either abrupt removal, steady-state limitation (chemostat), or exhaustion. Changing of the growth rates by other means, such as temperature, antibiotics, or overexpression of proteins, does not result in the classical correlations (20, 59), for example, the use of chloramphenicol (Cm) to reduce protein synthesis, which blocks the consumption of charged tRNAs and reduces the levels of (p)ppGpp. In a previous study (59), the authors used chloramphenicol to reduce the growth rate, and they observed a constant cell size, mass, and amount of DNA and RNA per cell. However, under those conditions, a drop in the levels of (p)ppGpp is expected, resulting in results similar to the ones observed in (p)ppGpp0 strains. Blocking of serinyl-tRNASer acylation with serine hydroxamate (SHX) forces the accumulation of excessive levels of (p)ppGpp and also disallows normal feedback mechanisms, such as restoration of charged serinyl-tRNA ratios due to protein synthesis inhibition. The physiological consequences can be to activate more severe stress survival regulatory circuits rather than adaptation.
In this study, we have seen that (p)ppGpp is necessary and sufficient to inhibit DNA replication initiation in E. coli, probably due to changes in the expression of gyrase and topoisomerase IV that would alter supercoiling around the origin. As mentioned above, (p)ppGpp is present in all bacteria and plants; therefore, further studies are warranted to determine if it is an evolutionarily conserved mechanism. Also, it will be interesting to determine which other effects associated with (p)ppGpp require changes in the supercoiling state of the chromosome.
MATERIALS AND METHODS
Media and growth conditions.
Strains and plasmids used in this study are listed in Table S2 in the supplemental material. M9 minimal medium was prepared as described previously (12), and minimal A medium was described previously (60). After experiments with (p)ppGpp0 strains, the low frequency of suppressor mutants that had gained the ability to grow without amino acids was verified as being below levels that would alter results (12). The following required antibiotics were added: 20 μg/ml kanamycin (Km), 20 μg/ml chloramphenicol (Cm), and 15 μg/ml tetracycline (Tc).
Bacterial strains and plasmids used in this study. Download Table S2, PDF file, 0.1 MB (126.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ori/ter measurements.
The ratios of ori to ter were measured by qPCR. Two independent cultures (25 ml in 125-ml flasks) for each strain and each growth condition were grown with aeration at 37°C to an A600 of 0.2, 10-ml samples were harvested by centrifugation, and the pellets were frozen. DNA was isolated with TRIzol (Thermo Fisher) as indicated by the manufacturer and fragmented with EcoRI. Traces of RNA were removed with RNase T1. The regions of ori and ter were amplified with SYBR green PCR master mix from Applied Biosystems in a LightCycler 480 instrument using 200 ng of template DNA with the primers listed in Table S3. The method (61) was used to calculate ori/ter ratios, where ΔCT is the threshold cycle (CT) value for the ori region subtracted from the CT value for the ter region. Means and standard deviations (SD) of data from 3 technical replicates for each biological duplicate were calculated (6 values in total).
Oligonucleotides used in this study. Download Table S3, PDF file, 0.01 MB (91.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Next-generation sequencing.
Two independent cultures of each strain were grown under the conditions described above, and cell pellets were frozen. Pellets were resuspended in 1 ml GTE (0.9% glucose, 25 mM Tris-HCl [pH 8.0], 10 mM EDTA) with lysozyme and RNase T1 and incubated for 10 min at 37°C. Next, 0.6% SDS was added, and the cells were incubated at 37°C until cell lysis, usually less than 2 min. The DNA was then purified with phenol-chloroform (62) and quantified with the Qubit double-stranded DNA (dsDNA) BR assay kit, and 250 ng of DNA was used for fragmentation and library preparation with the Nextera DNA library prep kit (Illumina). Libraries were sequenced using the Illumina NextSeq500 system. BCL files were converted to FASTQ format using bcl2fastq conversion software (Illumina) and uploaded to the Galaxy server, where TrimGalore was used to eliminate adaptors. Reads were mapped with Bowtie2 for Illumina (63, 64). The resulting BAM file analyses were performed using SeqMonk software (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/). A set of running 1-kb windows with a step size of 1 kb was generated to analyze the number of reads, which was corrected for the total read count per million reads. Duplicates were counted only once.
Dry-weight measurements.
Two independent cultures for each strain and growth condition were grown as described above but with 250-ml flasks to an A600 of 0.2. Next, 40-ml samples were pelleted, resuspended in 1 ml of phosphate-buffered saline (PBS), and transferred into a previously weighed 1.5-ml microcentrifuge tube. The pellet was quickly washed 3 times with PBS (1 min/wash) and finally dried in a heating block (90°C) for at least 24 h. Tubes were again weighed, the empty-tube weight was subtracted, and values were normalized to the culture volume.
Confocal microscopy and cell size measurements.
Cells were grown at different growth rates as described above, and 500 μl of cultures at an A600 of 0.2 to 0.3 was fixed with 100 μl of fixative solution consisting of 1 ml of 16% paraformaldehyde, 6.25 μl of 8% glutaraldehyde, and 20 μl of 1 M Na2HPO4 buffer (pH 7.4). Samples were incubated for 15 min at room temperature and then for 30 min on ice. After fixation, cells were centrifuged, washed three times in 1 ml of 1× PBS (pH 7.4), and then resuspended in 100 μl of GTE (0.9% glucose, 25 mM Tris-HCl, 10 mM EDTA [pH 8.0]), and the fixed cells were stored overnight at 4°C (43). The fixed cells were stained the next day, first with BODIPY TR dye (Thermo Fisher) for 30 min at 4°C and then with Hoechst dye (Thermo Fisher) for 5 to 7 min at room temperature. Finally, the excess dye was removed by washing with PBS. Stained cells (100 μl) were mixed with 200 μl of melted 1% low-melting-point agarose (liquified at 37°C), and the cells were immobilized by applying the agar to a 35-mm glass-bottom dish. Multifluorescence images were acquired at two different laser wavelengths (405 nm for Hoechst dye and 561 nm for BODIPY TR dye). Imaging was performed with an A64×/oil immersion lens (numerical aperture [NA], 0.55) using a Zeiss LSM 880 confocal microscope. Cell lengths and widths were determined using the MATLAB-based cell segmentation program SuperSegger (65). To define segmentation parameters that would enable the measurement of elongated (p)ppGpp0 cells that are present under some growth conditions, we used the SuperSegger training GUI (graphical user interface) to train the program to a data set of phase-contrast images of (p)ppGpp0 cells cultured in LB. Cells cultured in each medium were then segmented using the “fluorescence segmentation” function and the newly trained segmentation parameters. Segmented images were checked manually, and cells that were incorrectly segmented by the software were removed from the data sets. Cell volumes were calculated assuming shapes of geometrical cylinders with two hemispherical ends for rod-shaped cells. Between 100 and 200 cells were analyzed for cell size for each growth rate.
Flow cytometry.
Cultures were grown in medium supporting either low or high balanced exponential rates for 4 to 5 generations before sampling at an A600 of 0.15 and treated with 300 μg/ml of rifampin (Sigma-Aldrich) and 10 mg/ml of cephalexin (Sigma-Aldrich) for 4 to 5 h. Samples were then harvested, resuspended in TE buffer (20 mM Tris-HCl [pH 7.5], 1 mM EDTA), fixed in 70% ethanol, and stored at 4°C. For flow cytometry, fixed cells were washed once in 50 mM trisodium citrate, resuspended in the same buffer containing 5 μg/ml RNase (Sigma-Aldrich), and shaken for at least 3 h before staining of DNA with Sytox dye (Thermo Fisher). Samples of bacteria containing 1, 2, 4, and 8 chromosomes were used as internal standards. Flow cytometry analysis was performed using a FACSCalibur flow cytometer (Becton, Dickinson) equipped with a 488-nm argon ion laser. The data obtained from the flow cytometry measurements were processed with FlowJo software from Tree Star, Inc.
Gene expression determination by RT-qPCR.
Expression levels of dnaA, gyrA, topA, and parC were determined by reverse transcription-qPCR (RT-qPCR). Briefly, 2 independent cultures for each strain were grown to an A600 of 0.2, at which time samples were subjected to RNA isolation with TRIzol (Thermo Fisher) as indicated by the manufacturer. The RNA samples were retrotranscribed into cDNA using the high-capacity cDNA reverse transcription kit from Applied Biosystems, and target genes were amplified with SYBR green PCR master mix from Applied Biosystems in a LightCycler 480 instrument. Relative gene expression was determined with the comparative CT method described above (61), with 3 technical replicates (6 values under each experimental condition). The gadA gene was used as an internal control. Primers used in this study are listed in Table S3.
Statistical analysis.
Statistical analysis was carried out with the use of Microsoft Excel for Office 365 MSO. To determine whether two samples are significantly different, Student’s two-tailed t test was used (employed in Fig. 1, Fig. 4B, Fig. 5A, Fig. 6, Fig. S1, Fig. S3, Fig. S5A, and Fig. S7B). To determine if samples follow a described pattern, a goodness-of-fit (chi-square) test was used (employed in Fig. 3, Fig. 4A, Fig. S2, and Fig. S7A). The coefficient of correlation was determined for data in Fig. 5B, Fig. S4, and Fig. S5B.
ACKNOWLEDGMENTS
We thank Dhruba Chattoraj (NCI, NIH) for encouragement during this project, Nathan Blewett (NICHD, NIH) for introducing us to next-generation sequencing, Daniel Castranova (NICHD, NIH) for his help with the confocal microscopy, and Ciaran Condon (Institut de Biologie Physico-Chemique, Paris, France) for strains constructed in the laboratory of Cathy Squires with chromosomal deletions of seven ribosomal operons.
This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), by the Narodowe Centrum Nauki (2016/23/D/NZ1/02601), and by the National Institutes of Health (NIH R35 GM127331).
Footnotes
Citation Fernández-Coll L, Maciag-Dorszynska M, Tailor K, Vadia S, Levin PA, Szalewska-Palasz A, Cashel M. 2020. The absence of (p)ppGpp renders initiation of Escherichia coli chromosomal DNA synthesis independent of growth rates. mBio 11:e03223-19. https://doi.org/10.1128/mBio.03223-19.
REFERENCES
- 1.Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol 14:45–54. doi: 10.1016/j.tim.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson GC, Tenson T, Hauryliuk V. 2011. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irving SE, Corrigan RM. 2018. Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. Microbiology 164:268–276. doi: 10.1099/mic.0.000621. [DOI] [PubMed] [Google Scholar]
- 4.Cashel M, Gallant J. 1969. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 5.Cashel M. 1969. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem 244:3133–3141. [PubMed] [Google Scholar]
- 6.Cashel M, Gentry D, Hernandez V, Vinella D. 1996. The stringent response, p 1458–1489. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 7.Magnusson LU, Farewell A, Nyström T. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol 13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Steinchen W, Bange G. 2016. The magic dance of the alarmones (p)ppGpp. Mol Microbiol 101:531–544. doi: 10.1111/mmi.13412. [DOI] [PubMed] [Google Scholar]
- 9.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 10.Vinella D, Potrykus K, Murphy H, Cashel M. 2012. Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J Bacteriol 194:261–273. doi: 10.1128/JB.06238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traxler MF, Summers SM, Nguyen H-T, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potrykus K, Murphy H, Philippe N, Cashel M. 2011. ppGpp is the major source of growth rate control in E. coli. Environ Microbiol 13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarubbi E, Rudd KE, Cashel M. 1988. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet 213:214–222. doi: 10.1007/bf00339584. [DOI] [PubMed] [Google Scholar]
- 14.Zhou YN, Jin DJ. 1998. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci U S A 95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy H, Cashel M. 2003. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol 371:596–601. doi: 10.1016/S0076-6879(03)71044-1. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez-Vazquez P, Dewey CN, Kitten N, Ross W, Gourse RL. 2019. Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc Natl Acad Sci U S A 116:8310–8319. doi: 10.1073/pnas.1819682116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B, Dai P, Ding D, Del Rosario A, Grant RA, Pentelute BL, Laub MT. 2019. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat Chem Biol 15:141–150. doi: 10.1038/s41589-018-0183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaechter M, Maaløe O, Kjeldgaard NO. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Microbiol 19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 19.Kjeldgaard NO, Maaløe O, Schaechter M. 1958. The transition between different physiological states during balanced growth of Salmonella typhimurium. J Gen Microbiol 19:607–616. doi: 10.1099/00221287-19-3-607. [DOI] [PubMed] [Google Scholar]
- 20.Bremer H, Dennis PP. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p 1553–1569. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 21.Ryals J, Little R, Bremer H. 1982. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol 151:1261–1268. doi: 10.1128/JB.151.3.1261-1268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. 2013. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res 41:6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Churchward G, Estiva E, Bremer H. 1981. Growth rate-dependent control of chromosome replication initiation in Escherichia coli. J Bacteriol 145:1232–1238. doi: 10.1128/JB.145.3.1232-1238.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis PP, Bremer H. 7 October 2008. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- 25.Maciag M, Kochanowska M, Lyzeń R, Wegrzyn G, Szalewska-Pałasz A. 2010. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid 63:61–67. doi: 10.1016/j.plasmid.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Maciąg-Dorszyńska M, Szalewska-Pałasz A, Węgrzyn G. 2013. Different effects of ppGpp on Escherichia coli DNA replication in vivo and in vitro. FEBS Open Bio 3:161–164. doi: 10.1016/j.fob.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rymer RU, Solorio FA, Tehranchi AK, Chu C, Corn JE, Keck JL, Wang JD, Berger JM. 2012. Binding mechanism of metal·NTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure 20:1478–1489. doi: 10.1016/j.str.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JD, Sanders GM, Grossman AD. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper S, Helmstetter CE. 1968. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol 31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 30.Leonard AC, Méchali M. 2013. DNA replication origins. Cold Spring Harb Perspect Biol 5:a010116. doi: 10.1101/cshperspect.a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riber L, Frimodt-Møller J, Charbon G, Løbner-Olesen A. 2016. Multiple DNA binding proteins contribute to timing of chromosome replication in E. coli. Front Mol Biosci 3:29. doi: 10.3389/fmolb.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katayama T, Kasho K, Kawakami H. 2017. The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front Microbiol 8:2496. doi: 10.3389/fmicb.2017.02496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Messer W, Weigel C. 1996. Initiation of chromosome replication, p 1579–1601. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 34.Chiaramello AE, Zyskind JW. 1990. Coupling of DNA replication to growth rate in Escherichia coli: a possible role for guanosine tetraphosphate. J Bacteriol 172:2013–2019. doi: 10.1128/jb.172.4.2013-2019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flåtten I, Fossum-Raunehaug S, Taipale R, Martinsen S, Skarstad K. 2015. The DnaA protein is not the limiting factor for initiation of replication in Escherichia coli. PLoS Genet 11:e1005276. doi: 10.1371/journal.pgen.1005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraemer JA, Sanderlin AG, Laub MT. 2019. The stringent response inhibits DNA replication initiation in E. coli by modulating supercoiling of oriC. mBio 10:e01330-19. doi: 10.1128/mBio.01330-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donachie WD. 1968. Relationship between cell size and time of initiation of DNA replication. Nature 219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 38.Maduike NZ, Tehranchi AK, Wang JD, Kreuzer KN. 2014. Replication of the Escherichia coli chromosome in RNase HI-deficient cells: multiple initiation regions and fork dynamics. Mol Microbiol 91:39–56. doi: 10.1111/mmi.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin DJ, Gross CA. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol 202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 40.Greenway DLA, England RR. 1999. The intrinsic resistance of Escherichia coli to various antimicrobial agents requires ppGpp and σ(s). Lett Appl Microbiol 29:323–326. doi: 10.1046/j.1472-765x.1999.00642.x. [DOI] [PubMed] [Google Scholar]
- 41.Si F, Li D, Cox SE, Sauls JT, Azizi O, Sou C, Schwartz AB, Erickstad MJ, Jun Y, Li X, Jun S. 2017. Invariance of initiation mass and predictability of cell size in Escherichia coli. Curr Biol 27:1278–1287. doi: 10.1016/j.cub.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Z, Davis RM, Kishony R, Kahne D, Ruiz N. 2012. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc Natl Acad Sci U S A 109:E2561–E2568. doi: 10.1073/pnas.1209742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vadia S, Tse JL, Lucena R, Yang Z, Kellogg DR, Wang JD, Levin PA. 2017. Fatty acid availability sets cell envelope capacity and dictates microbial cell size. Curr Biol 27:1757–1767. doi: 10.1016/j.cub.2017.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.My L, Rekoske B, Lemke JJ, Viala JP, Gourse RL, Bouveret E. 2013. Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp. J Bacteriol 195:3784–3795. doi: 10.1128/JB.00384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 46.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li SH-J, Li Z, Park JO, King CG, Rabinowitz JD, Wingreen NS, Gitai Z. 2018. Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat Microbiol 3:939–947. doi: 10.1038/s41564-018-0199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winslow RM, Lazzarini RA. 1969. Amino acid regulation of the rates of synthesis and chain elongation of ribonucleic acid in Escherichia coli. J Biol Chem 244:3387–3392. [PubMed] [Google Scholar]
- 49.Roghanian M, Semsey S, Løbner-Olesen A, Jalalvand F. 2019. (p)ppGpp-mediated stress response induced by defects in outer membrane biogenesis and ATP production promotes survival in Escherichia coli. Sci Rep 9:2934. doi: 10.1038/s41598-019-39371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Traxler MF, Chang D-E, Conway T. 2006. Guanosine 3′,5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc Natl Acad Sci U S A 103:2374–2379. doi: 10.1073/pnas.0510995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harinarayanan R, Murphy H, Cashel M. 2008. Synthetic growth phenotypes of Escherichia coli lacking ppGpp and transketolase A (tktA) are due to ppGpp-mediated transcriptional regulation of tktB. Mol Microbiol 69:882–894. doi: 10.1111/j.1365-2958.2008.06317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernández-Coll L, Cashel M. 2018. Contributions of SpoT hydrolase, SpoT synthetase, and RelA synthetase to carbon source diauxic growth transitions in Escherichia coli. Front Microbiol 9:1802. doi: 10.3389/fmicb.2018.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maciąg M, Nowicki D, Janniere L, Szalewska-Pałasz A, Węgrzyn G. 2011. Genetic response to metabolic fluctuations: correlation between central carbon metabolism and DNA replication in Escherichia coli. Microb Cell Fact 10:19. doi: 10.1186/1475-2859-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q, Zhou A, Li SS, Ni J, Tao J, Lu J, Wan B, Li SS, Zhang J, Zhao S, Zhao G-P, Shao F, Yao Y-F. 2016. Reversible lysine acetylation is involved in DNA replication initiation by regulating activities of initiator DnaA in Escherichia coli. Sci Rep 6:30837. doi: 10.1038/srep30837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashley RE, Dittmore A, McPherson SA, Turnbough CL, Neuman KC, Osheroff N. 2017. Activities of gyrase and topoisomerase IV on positively supercoiled DNA. Nucleic Acids Res 45:9611–9624. doi: 10.1093/nar/gkx649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. 2005. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell 19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Rasouly A, Pani B, Nudler E. 2017. A magic spot in genome maintenance. Trends Genet 33:58–67. doi: 10.1016/j.tig.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potrykus K, Cashel M. 2018. Growth at best and worst of times. Nat Microbiol 3:862–863. doi: 10.1038/s41564-018-0207-6. [DOI] [PubMed] [Google Scholar]
- 59.Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, Hwa T. 2015. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller JH. 1972. Experiments in molecular genetics, 3rd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 61.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 62.Maniatis T, Sambrook J, Fritsch EF. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 63.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stylianidou S, Brennan C, Nissen SB, Kuwada NJ, Wiggins PA. 2016. SuperSegger: robust image segmentation, analysis and lineage tracking of bacterial cells. Mol Microbiol 102:690–700. doi: 10.1111/mmi.13486. [DOI] [PubMed] [Google Scholar]
- 66.Hill NS, Kadoya R, Chattoraj DK, Levin PA. 2012. Cell size and the initiation of DNA replication in bacteria. PLoS Genet 8:e1002549. doi: 10.1371/journal.pgen.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wold S, Skarstad K, Steen HB, Stokke T, Boye E. 1994. The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J 13:2097–2102. doi: 10.1002/j.1460-2075.1994.tb06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth rates (μ) and standard deviations (STDV) measured for MG1655 and (p)ppGpp0 strains in different media. Download Table S1, PDF file, 0.1 MB (139.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Ratios of ori/ter at exponential and stationary phases in the presence or absence of (p)ppGpp. Strains MG1655 (WT) and CF10237 [(p)ppGpp0] were grown in LB up to exponential phase (A600 of 0.2) and stationary phase (A600 of 1.5), and ori/ter ratios were measured by qPCR. Error bars represent the SD from 2 biological replicates and 3 technical replicates. Values significantly different with a P value of <0.01 are marked with *, while those not significantly different (P value of >0.01) are marked with ns (not significant). Download FIG S1, TIF file, 0.4 MB (406.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Both ppGpp and pppGpp affect growth rates and ori/ter ratios. The results confirm the higher potency of ppGpp than of pppGpp for growth rate inhibition and extend this relationship to include a similar higher potency for inhibition of ori/ter ratios. Strains CF16762 (ppGpp) and CF16760 (pppGpp) were grown in M9 medium containing 0.4% glycerol plus all the amino acids with increasing levels of arabinose (0, 0.005%, 0.01%, and 0.02%). The black line, taken from Fig. 1, represents the correlation between ori/ter ratios and growth rates observed for WT growth in different media. A goodness-of-fit (chi-square) test was used to determine that the samples follow the WT pattern shown in Fig. 1, with 99.2% and 99.7% probabilities for ppGpp and pppGpp samples, respectively (error bars represent the SD from 2 biological replicates and 3 technical replicates). Download FIG S2, TIF file, 0.6 MB (585.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of (p)ppGpp on gene expression of factors that control global supercoiling. Strains MG1655 (WT) and CF10237 [(p)ppGpp0] were grown in LB up to exponential phase (A600 of 0.2) and stationary phase (A600 of 1.5), and the expression levels of topA (A), gyrA (B), and parC (C) were measured by RT-qPCR. Error bars represent the SD from 2 biological replicates and 3 technical replicates. The values plotted are normalized to the value for the WT strain in exponential phase. Values significantly different with a P value of <0.01 are marked with *, while those not significantly different (P value of >0.01) are marked with ns. Download FIG S3, TIF file, 0.8 MB (876.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Correlation between gyrA expression (data from Fig. 4B) and ori/ter ratios. Correlation coefficients are 0.975 for the WT and 0.938 for the (p)ppGpp0 strain. Download FIG S4, TIF file, 0.4 MB (381.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(p)ppGpp deficiency seems to alter the normal dependence of cell volume on growth rate. (A) Plot of cellular volume versus growth rate variations achieved with the different media listed in Table S1 in the supplemental material. The wild-type strain is MG1655 (solid symbols), and the (p)ppGpp0 strain is CF10237. The P values were calculated by comparing the values to the highest growth rate (LB) for each strain. All samples of the WT strain are significantly different, with a P value of <0.01, while the (p)ppGpp0 samples are not significantly different (P value of >0.01). (B) Correlation between cell volume and ori/ter ratios. Correlation coefficients are 0.976 for the WT and −0.051 for the (p)ppGpp0 strain. Error bars represent the SD from 2 biological replicates. (C) Microscopy image of (p)ppGpp0 strains in LB showing deficiencies in segregation and filamentation. Download FIG S5, TIF file, 2.6 MB (2.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The number of chromosomes per cell is growth rate dependent and independent of (p)ppGpp. Shown are flow cytometry profiles for WT and (p)ppGpp-deficient strains grown in rich (LB plus 0.2% glucose) or poor (M9 medium with the Σ set plus 0.2% glucose) medium. The inset shows standards used to associate the intensity peaks with the number of chromosomes. The light gray vertical bars identify the chromosomal number per cell. The numerical distribution of these data is shown in Table 1. Download FIG S6, TIF file, 1.5 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The effect of (p)ppGpp on DNA replication initiation is not due to effects on central metabolism or dnaA expression. (A) ori/ter ratios are related to various growth rates obtained with strains deficient in tktB, pgi, or pta and grown in different media. The dashed line represents the correlation between ori/ter ratios and growth rates observed for the WT, as shown in Fig. 1. A chi-square goodness-of-fit test was used to determine that the samples follow the WT pattern shown in Fig. 1, with 88%, 90.2%, and 91.6% probabilities for tktB, pgi, and pta samples, respectively. Error bars represent the SD from 2 biological replicates and 3 technical replicates. (B) Strains MG1655 and CF10237 were grown in the same media as the ones for Fig. 4. The expression level of the dnaA gene was determined by RT-PCR. Error bars represent the SD from 2 biological replicates and 3 technical replicates. The values plotted are normalized to the value for the fastest-growing WT strain. The P values were calculated by comparing the values to the highest growth rate (LB) for each strain. All samples are significantly different, with a P value of <0.01. Download FIG S7, TIF file, 0.6 MB (654.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Bacterial strains and plasmids used in this study. Download Table S2, PDF file, 0.1 MB (126.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Oligonucleotides used in this study. Download Table S3, PDF file, 0.01 MB (91.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.


