Abstract
Mycobacterium tuberculosis (Mtb), the etiologic agent of tuberculosis, must adapt to host‐associated environments during infection by modulating gene expression. Small regulatory RNAs (sRNAs) are key regulators of bacterial gene expression, but their roles in Mtb are not well understood. Here, we address the expression and function of the Mtb sRNA Mcr11, which is associated with slow bacterial growth and chronic infections in mice. We found that stable expression of Mcr11 requires multiple factors specific to TB‐complex bacteria, including the AbmR transcription factor. Bioinformatic analyses used to predict regulatory targets of Mcr11 identified 7–11 nucleotide regions with potential for direct base‐pairing with Mcr11 immediately upstream of Rv3282, fadA3, and lipB. mcr11‐dependent regulation of these genes was demonstrated using qRT‐PCR and found to be responsive to the presence of fatty acids. Mutation of the putative Mcr11 base‐pairing site upstream of lipB in a promoter reporter strain resulted in significant de‐repression of lipB expression, similar to that observed in mcr11‐deleted Mtb. These studies establish Mcr11’s roles in regulating growth and central metabolism in Mtb. Our finding that multiple TB‐complex‐specific factors are required for production of stable Mcr11 also emphasizes the need to better understand mechanisms of sRNA expression and stability in TB.
Keywords: gene regulation, lipoylation, RNA termination, RNA stability, sRNA targets
This study shows that the small non‐coding RNA Mcr11, which is highly expressed in slowly replicating Mtb and during mouse infection, directly regulates expression of target genes involved in central metabolism. Our unexpected finding that factors specific to TB complex mycobacteria are required for the stable production of Mcr11 further identifies sRNA stability as a new frontier for understanding gene expression in Mtb.
1. INTRODUCTION
Tuberculosis (TB) remains a major threat to global public health, with at least 10 million new, incident cases and 1.3 million deaths due to TB in 2017 (WHO, 2018). The basic biology of Mycobacterium tuberculosis (Mtb), the etiological agent of TB, is poorly understood despite its importance to the development of new therapeutic interventions. Mtb can adopt a specialized physiological state within host tissues, which renders the bacteria phenotypically drug resistant and viable despite extended periods of slow or non‐replicating persistence (NRP) (Gomez & McKinney, 2004). NRP and phenotypic drug resistance pose particular challenges for intervention, making it critical to understand the regulatory processes that enable Mtb to adapt to host conditions.
Bacterial and host factors that contribute to NRP and slow growth are still being defined (Bergkessel, Basta, & Newman, 2016). Host‐associated environmental cues that result in metabolic remodeling and a shift away from active growth toward a state of persistence include hypoxia, nitrosative stress, redox stress, nutrient starvation, as well as adaptation to cholesterol and fatty acid metabolism (Betts, Lukey, Robb, McAdam, & Duncan, 2002; Garton et al., 2008; Honaker, Dhiman, Narayanasamy, Crick, & Voskuil, 2010; Iona et al., 2016; Schubert et al., 2015; Shi et al., 2005). Lipoylated enzymes involved in the citric acid cycle, such as lipoamide dehydrogenase (Lpd) and dihydrolipoamide acyltransferase (DlaT), are necessary for Mtb survival in the host and viability during NRP (Bryk et al., 2008; Shi & Ehrt, 2006; Venugopal et al., 2011). However, factors that regulate these processes are not well understood.
Gene expression studies have provided critical insights into the regulation and function of proteins like transcription factors that modulate gene expression as Mtb adapts to the host environment during infection (Mvubu, Pillay, Gamieldien, Bishai, & Pillay, 2016; Schnappinger et al., 2003). The additional role of sRNAs in gene regulation is recognized in other bacteria (Waters & Storz, 2009), and several sRNAs whose expression is responsive to stress and/ or growth phase have been identified in mycobacteria (Arnvig et al., 2011; Arnvig & Young, 2009; DiChiara et al., 2010; Gerrick et al., 2018; Miotto et al., 2012; Moores, Riesco, Schwenk, & Arnvig, 2017; Namouchi et al., 2016; Pelly, Bishai, & Lamichhane, 2012; Solans et al., 2014; Tsai et al., 2013). It also has been observed that overexpression of some sRNAs leads to slow or delayed growth in mycobacteria (Arnvig & Young, 2009; Ignatov et al., 2015). Roles for the sRNAs Mcr7, MsrI and 6C/B11 in gene regulation in Mtb or Mycobacterium smegmatis (Msm) have been reported (Gerrick et al., 2018; Mai et al., 2019; Solans et al., 2014). However, the importance of these differentially expressed sRNAs to the adaptation and survival of Mtb during periods of stress has not been fully addressed.
The intergenic sRNA Mcr11 (ncRv11264Ac) is one of the best‐studied mycobacterial sRNAs, but its structure and function in TB‐complex mycobacteria is unknown (Arnvig et al., 2011; DiChiara et al., 2010; Girardin, Bai, He, Sui, & McDonough, 2018; Pelly et al., 2012). Expression of Mcr11 is regulated in response to advanced growth phase and levels of the universal second messenger 3′,5′‐cyclic adenosine monophosphate (cAMP) (Arnvig et al., 2011; DiChiara et al., 2010; Pelly et al., 2012). Additionally, Mcr11 is abundantly expressed by Mtb in the lungs of chronically infected mice (Pelly et al., 2012) as well as in hypoxic and non‐replicating Mtb (Ignatov et al., 2015). Transcriptional regulators of mcr11 expression include the product of the adjacent, divergently expressed gene abmR (Rv1265) (Girardin et al., 2018) and the cAMP‐binding transcription factor CRPMT (Arnvig et al., 2011).
Here, we report that cis‐acting, extended, native sequence 3′ to Mcr11 enhances transcriptional termination of Mcr11 in mycobacteria. Optimal Mcr11 termination efficiency needed the transcription factor AbmR and was regulated by growth phase in Mtb and Mycobacterium bovis BCG, but not in the fast‐growing saprophyte Mycobacterium smegmatis. We found that mcr11 regulates expression of fadA3, lipB and Rv3282, which contribute to central metabolic pathways associated with NRP and slow growth in Mtb. In addition, Mcr11 affected growth of both Mtb and BCG in hypoxic conditions without fatty acids. This study identifies TB complex‐specific cis and trans factors required for stable Mcr11 expression while providing a report of mcr11‐dependent regulation of gene targets in Mtb.
2. RESULTS
2.1. Modeling secondary structure of Mcr11
Secondary structural features of sRNAs greatly contribute to their function, and RNA folding is most strongly influenced by the nucleotide sequence of the RNA (Dubey, Baker, Romeo, & Babitzke, 2005; Hnilicova et al., 2014). Previous studies established two 5′ ends of Mcr11 at chromosomal positions 1413224 and 1413227 in Mycobacterium tuberculosis H37Rv (Arnvig et al., 2011; DiChiara et al., 2010), but the 3′ end of Mcr11 is poorly defined. Preliminary efforts to express Mcr11 based on size estimates from prior Northern blot experiments were not successful, despite the well‐mapped 5′ end of the sRNA. We reasoned that defining the precise boundaries of Mcr11 could help in identifying its function.
We mapped the 3′ end of Mcr11 to chromosomal positions 1413107 and 1413108 in Mycobacterium tuberculosis (Mtb) using 3′ rapid amplification of cNDA ends (RACE) and Sanger sequencing (Figure 1a). These 3′ ends are 120 and 119 nucleotides (nt) downstream from the most abundant previously mapped 5′ end at position 1413227 (DiChiara et al., 2010). Our mapped 3′ ends vary 3–4 nucleotides from the 3′ end at chromosomal position 1413111 inferred by deep sequencing (DeJesus et al., 2017), and are 13–14 nt shorter than the 3′ end estimated by cloning (DiChiara et al., 2010) (Figure 1a). These results indicate that Mcr11 is a transcript between 117 and 121 nt long, which is consistent with its observed size by Northern blot (Arnvig et al., 2011; DiChiara et al., 2010; Pelly et al., 2012).
Figure 1.
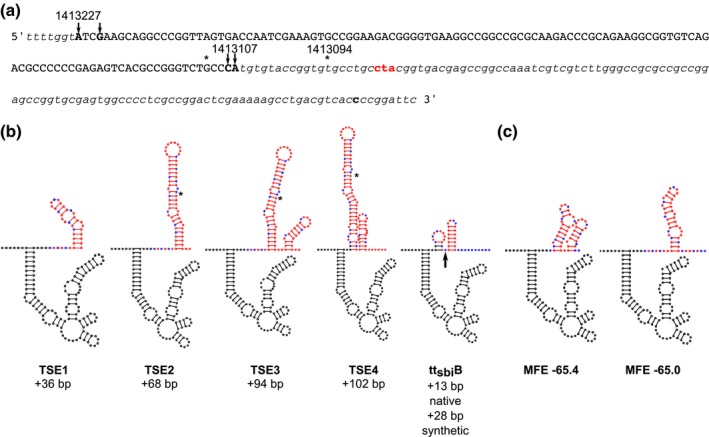
Secondary Structure modeling of 3′ sequences beyond the mapped 3′ end of Mcr11. (a) The DNA sequence of the mcr11 gene and extended 3′ sequence used for modeling experiments is shown. The mcr11 gene is shown in capital letters, and nucleotides where 5′ and 3′ boundaries have been mapped by RACE are indicated by arrows and shown in capitalized, bolded black text. Flanking sequences are in italics, and the Rv1264 stop codon in bolded red text. The last nucleotide on the 3′ side of mcr11 that was included in modeling experiments is in bold, lowercase black text. An asterisk indicates 3′ ends reported by DiChiara et al and DeJesus et al. Positions of mapped nucleotides on the Mtb chromosome are shown above the text. (b) Secondary structure diagrams of Mcr11 from 5′ position 1413227 and extended 3′ native sequences. The synthetic idealized intrinsic termination control (ttsbiB) is also modeled onto Mcr11 from the longest 3′ end reported at position 1413094; the last nucleotide of native sequence 3′ to Mcr11 is indicated by a black arrow. Black nucleotides indicate bases in the mapped boundaries of the mcr11 gene, red base pairs indicate base pairs beyond the mapped 3′ end of Mcr11 at position 1413107, except all uracils (Us) are shown in blue. An asterisk indicates that last nucleotide modeled in (c). (c). The secondary structure models with the lowest minimum free energies (MFE) of a shorter untested native Mcr11 TSE with the most U rich sequence at the 3′ end of the putative terminator. The MFE of each structure is shown below each diagram [Colour figure can be viewed at https://www.wileyonlinelibrary.com]
The 3′ ends of Mcr11 varied by 3–14 nt from previous estimates, so we considered the possibility that these variances are functionally significant. Several mycobacterial sRNAs have multiple reported 3′ ends, suggestive of Rho‐dependent termination and/or post‐transcriptional processing (Arnvig & Young, 2009; Miotto et al., 2012; Tsai et al., 2013). Rho‐dependent termination requires the association of the ATP‐hydrolyzing molecular motor Rho with the nascent RNA, which is typically cytosine‐enriched, guanosine‐depleted and unstructured at the 3′ end (Peters et al., 2009). In contrast, intrinsic, Rho‐independent termination is governed by the formation of a structured termination hairpin that is usually followed a poly‐U tail, and is highly dependent upon the sequence of the nascent RNA (Mooney & Landick, 2013; Ray‐Soni, Bellecourt, & Landick, 2016; Santangelo & Artsimovitch, 2011).
We modeled the secondary structure of Mcr11 with varying lengths of extended, native 3′ RNA sequence using RNA structure (Mathews, 2006) to reveal potential functional features. Mcr11 was predicted to be highly structured, with few single‐stranded (ss) regions available for potential base‐pairing with regulatory target RNA sequences (Figure 1b). Both mapped 5′ ends of mcr11 produced the same predicted secondary structure, as these 5′ nucleotides are expected to be unpaired (Figure 1b). Modeled structures of extended native sequence 3′ to our mapped ends of mcr11 revealed additional highly structured motifs with similarity to predicted Rho‐independent terminators (RITs) downstream of mcr11 in mycobacteria (Gardner, Barquist, Bateman, Nawrocki, & Weinberg, 2011; Mitra, Angamuthu, & Nagaraja, 2008). However, none included the long, characteristic poly‐U tail found in RITs of other bacteria (Ray‐Soni et al., 2016). A 10 nt region containing a sub‐optimal poly‐U tract was also identified by scanning sequence downstream of the mapped 3′ end of Mcr11 in the constructs that contained 3′ sequence elements (TSEs) 2–4 (Figure 1a). Modeling the secondary structure of this sequence revealed a potential stem loop structure with a 7nt trail containing four discontinuous Us (Figure 1c).
2.2. Measurement of TSE function in Msm
We tested the impact of different mcr11 TSEs on the transcriptional termination of Mcr11 using promoter:mcr11:TSE:GFPv reporter constructs (Figure 2a,b). We reasoned that transcriptional termination levels could be inferred using GFPv fluorescence as a relative measure of transcriptional read‐through beyond the mcr11 gene (Figure 2a). The relative percentage of termination for each TSE was calculated by dividing the observed fluorescence of each Pmcr11:TSE:GFPv strain by the observed fluorescence of an mcr11 promoter‐only:GFPv reporter construct (Pmcr11:GFPv), subtracting the product from 1, and multiplying the result by 100%. Promoterless GFPv served as a negative assay control, and an mcr11‐independent promoter:GFPv fusion construct served as a positive assay control. A positive intrinsic termination control was generated by fusing mcr11 and a small amount of native 3′ trailing sequence to a synthetic RIT (ttsbiB) that has been shown to function at ~ 98% efficiency with Mtb RNAP in vitro (Figure 1b) (Czyz, Mooney, Iaconi, & Landick, 2014).
Figure 2.
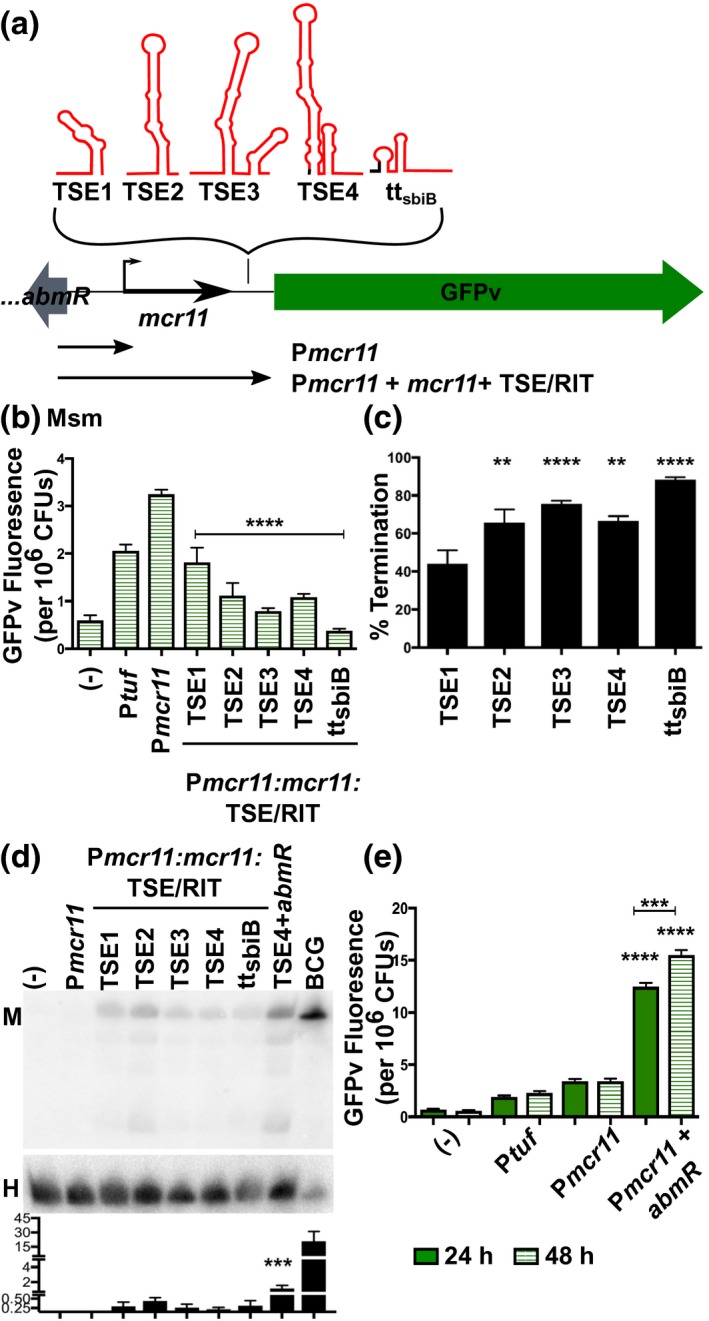
TSEs decrease transcriptional read‐through of Mcr11‐GFPv reporters, but Mcr11 is not robustly expressed in Msm, even in the presence of abmR from Mtb. (a). A schematic representation of GFPv fluorescence‐based reporter assay to determine the functionality of TSEs, and a synthetic idealized intrinsic termination control (ttsbiB). (b). GFPv fluorescence assay used to measure promoter activity and read‐through of TSEs of mcr11 in late stationary phase Msm, which lacks a native mcr11 locus. The promoter Ptuf served as a positive control. The various TSE constructs tested are indicated underneath the corresponding bar. Statistical comparisons are relative to Pmcr11. (c). The % termination of constructs tested in (b). Statistical comparisons are relative to TSE1. (d). Northern blot analysis of Mcr11 expression (M) in Msm with various mcr11 + TSE constructs. About 10 µg of total Msm RNA was loaded, whereas 3 µg of the BCG positive control was loaded. The HisT tRNA (H) was used as a loading control. The bar chart displays the normalized levels of Mcr11 as determined by densitometry. Statistical comparison is between TSE4 + abmR and TSEs 1–4 and ttsbiB. (e). GFPv fluorescence assay used to measure promoter activity in Msm. Statistical comparisons are relative to Pmcr11 or between 24 h and 28 h as indicated. Fluorescence is normalized to the OD620 of each sample. Results are the means of three biological replicates. Statistical analysis conducted with an unpaired, two‐tailed Student’s t‐test (e) or by one‐way ANOVA with Bonferroni correction for multiple comparisons (b, c, d). Asterisks indicate significance as follows: *p < .05, **p < .01, ***p < .001, ****p < .0001 [Colour figure can be viewed at https://www.wileyonlinelibrary.com]
Our initial termination experiments measuring TSE efficiency were performed in Msm, because it lacks an mcr11 orthologue and thus has no basal Mcr11 expression. The inclusion of any TSE sequence at the 3′ end of mcr11 resulted in decreased transcriptional read‐through of the mcr11:GFPv fusion reporter, as compared to the relative levels of GFPv fluorescence for Pmcr11 alone (Figure 2b). This result suggests that all TSEs supported transcriptional termination to varying degrees. TSEs 2–4 had greater termination efficiencies than TSE1, but the positive ttsbiB intrinsic termination control exhibited the strongest mean termination efficiency at 88% (Figure 2c). Paraformaldehyde‐fixed duplicates of each sample were subjected to flow cytometry analysis to determine if the mean GFPv fluorescence observed in our plate‐based assay was reflective of the fluorescence observed in individual cells, or if there were populations of cells with varying levels of fluorescence. A single, homogenous population of fluorescent cells was observed for each reporter, demonstrating that mean fluorescence measured in the plate reader assay was representative of the fluorescence in individual cells (Supplemental Figure 2). From these data, we concluded that mean fluorescence reflects relative termination efficiency for each TSE reporter.
Northern blot analysis was performed to further evaluate the expression of Mcr11 in Msm. We noted that the size of Mcr11 did not vary in Msm despite the presence of TSEs with various lengths, suggesting that TSEs are rapidly processed of the mature sRNA if they are transcribed (Figure 2d). Larger forms of unprocessed Mcr11 were not observed, but we also noted poor transfer of high molecular weight RNAs (such as rRNA), due to the high percentage of acrylamide in the gel mixture. Mcr11 was also present at very low levels in Msm when compared to a BCG control for which only 1/3 the amount of total RNA present in the Msm samples was loaded in the gel (Figure 2d). These results indicate reduced Mcr11 expression and/or stability in Msm compared to BCG.
2.3. TB complex‐specific factors are required for stable Mcr11 expression
Mcr11 expression varies across growth phases in TB complex mycobacteria (DiChiara et al., 2010; Girardin et al., 2018), so we examined the effect of growth phase on mcr11 expression in Msm. Fluorescence from the GFPv transcriptional reporters was measured at mid‐log and late stationary phases. Surprisingly, we observed that Pmcr11 activity was not increased by advancing growth phase in Msm (Figure 2e), suggesting lack of a positive regulatory factor.
Previously, we showed that the divergently expressed DNA‐binding protein AbmR is a growth phase‐responsive activator of mcr11 expression (Girardin et al., 2018). Although the Msm orthologue (MSMEG_5010) of abmR displays both high amino acid sequence identity (68.75%) and similarity (83.51%) to AbmR (Girardin et al., 2018), we reasoned that abmR could have functionality that the Msm orthologue lacks. Thus, we added a copy of the Mtb abmR locus upstream of the Pmcr11:GFPv fusion sequences and re‐tested the activity of Pmcr11 in response to growth phase in Msm. The addition of abmR significantly increased Pmcr11 activity in Msm, and rendered Pmcr11 activity responsive to growth phase (Figure 2e), as previously observed in BCG and Mtb (Girardin et al., 2018). Inclusion of abmR also significantly increased the amount of stable Mcr11 detected by Northern blot in Msm (Figure 2d), indicating that Mcr11 is unstable even when abundantly transcribed in Msm. Together, these results show that Mtb abmR regulates mcr11 expression at the transcriptional level, but is not sufficient for production of high levels of stable Mcr11 expression in a non‐native environment.
Further studies in TB‐complex mycobacteria showed that the termination efficiencies of mcr11 TSEs in mid‐log phase BCG were similar to those observed in Msm (Figure 3), but only BCG showed enhanced mcr11 TSE termination efficiencies in late stationary phase (Supplemental Table 3A and B). This growth phase‐dependent enhancement of termination efficiency in BCG was strongest for TSE1, which increased from approximately 40%–70% (Figure 3c). Overall termination efficiencies of mcr11 TSEs were also greater in virulent Mtb (H37Rv) than in BCG or Msm (Figure 3d‐f). Deletion of abmR caused a small but significant decrease in mcr11 termination efficiencies from all constructs, with the TSE1 short hairpin being the most strongly affected (Supplemental Figure 3).
Figure 3.
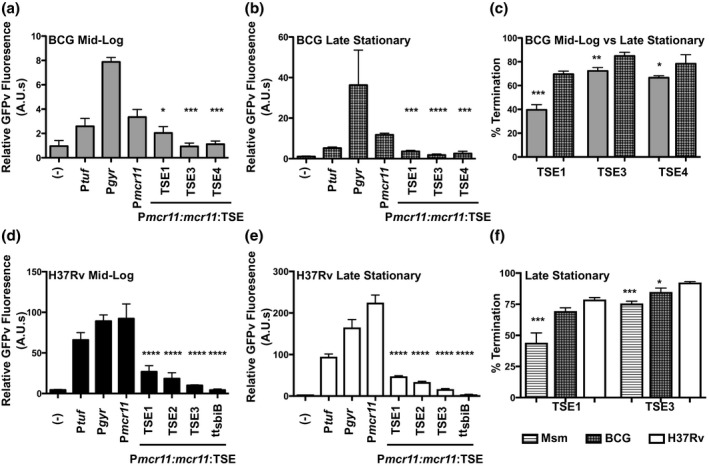
Efficiency of TSEs is positively regulated in response to growth phase in BCG and function significantly better in Mtb. (a) GFPv fluorescence assay used to measure promoter activity and read‐through of the TSEs of mcr11 in mid‐log phase BCG in hypoxic (1.3% O2, 5% CO2), shaking conditions. A promoterless (−) construct was used as a negative control and the promoters Ptuf and Pgyr served as positive controls. The various TSE constructs tested are indicated underneath the corresponding bar. Statistical comparisons were made to Pmcr11. (b) As in (a), but assayed in late stationary phase. (c) A comparison of % termination observed in mid‐log phase (solid bars) and late stationary phase (hatched bars) BCG. Statistical comparisons were made between mid‐log and stationary phase. (d) GFPv fluorescence assay used to measure promoter activity and read‐through of the TSEs of mcr11 in mid‐log phase Mtb in hypoxic (1.3% O2, 5% CO2), shaking conditions. A promoterless (−) construct was used as a negative control and the promoters Ptuf and Pgyr serve as positive controls. The various TSE constructs tested are indicated underneath the corresponding bar. Statistical comparisons are made to Pmcr11. Fluorescence is normalized to the OD620 of each sample. (e) As in (d), but assayed in late stationary phase. Statistical comparisons are made to Pmcr11. (f) A comparison of % termination observed in late stationary phase Msm, BCG and Mtb. Statistical comparisons are made to Mtb. Results are the means of three biological replicates. Statistical analysis conducted with a one‐way ANOVA with Bonferroni correction for multiple comparisons. Asterisks indicate significance as follows: * p < .05, ** p < .01, *** p < .001, **** p < .0001
We used Northern blot analyses to evaluate the size of Mcr11 produced from various mcr11 TSE constructs in Δmcr11 strains of BCG and Mtb during late stationary phase (Figure 4a,b). Robust levels of Mcr11 were produced from constructs containing native mcr11 TSEs in both BCG and Mtb. As was observed in experiments with Msm, Mcr11 size remained constant despite its expression with TSEs of different lengths in BCG or Mtb (Figure 4a,b). This size restriction is consistent with precise termination or an RNA processing event that produces discrete 3′ ends. The levels of Mcr11 produced were independent of their TSE in late stationary phase BCG and Mtb, but the Mcr11 constructs produced lower levels in BCG (Figure 4a). Conversely, the levels of Mcr11 generated from Mcr11 constructs in Mtb were more comparable to those observed in the Wt strain (Figure 4b). These data further support the conclusion that species‐specific factors have a significant impact on the expression and stability of mature Mcr11.
Figure 4.
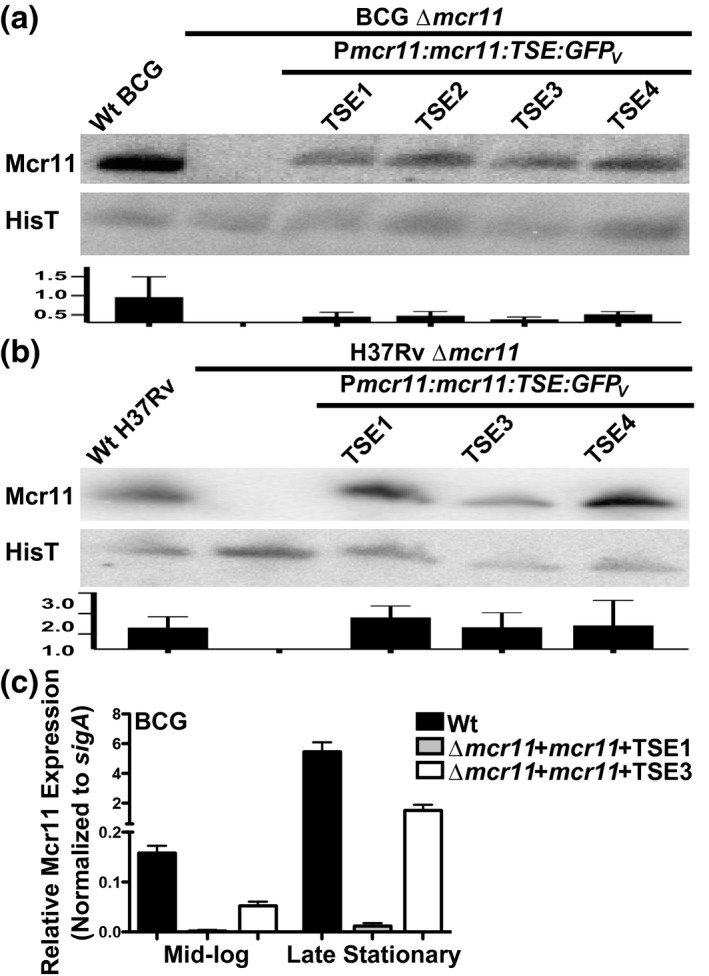
Different Mcr11 TSEs do not alter the size of stable Mcr11. Northern blot analysis of Mcr11 expression in (a) BCG or (b) H37Rv in hypoxic (1.3% O2, 5% CO2) late stationary phase. The tRNA HisT is used as a loading control. Results of densitometry analysis are presented below the corresponding blot. (c) qPCR analysis of Mcr11 expression in mid‐log and late stationary phase BCG, normalized to sigA expression. Results representative of 2–3 independent repeats
As advancing growth phase significantly improved the termination efficiencies of TSEs in BCG (Figure 3c), we tested the effects of growth phase on the expression of mcr11. We expected that increased termination efficiencies would correlate with higher levels of mature Mcr11 in TB complex mycobacteria. Mcr11 levels were measured by qRT‐PCR (Figure 4c) in BCGΔmcr11 strains carrying ectopic copies of either mcr11:TSE1 or mcr11:TSE3. Both constructs showed growth phase‐dependent increases in expression of stable Mcr11 (Figure 4c). However, the striking increase in Mcr11 produced from TSE3 compared to TSE1 observed by qPCR was not observed by Northern blot analyses of late stationary phase bacteria (Figure 4a). While qPCR has a lower limit of detection than Northern blot analyses, the results suggest that the TSE contributes to the stability of larger, unprocessed transcript products not detected by the Northern blot probe and/or that the secondary structures of the transcripts significantly impact their reverse transcription to cDNA. Together, these data indicate that multiple trans‐acting factors specific to the genetic background of TB‐complex mycobacteria facilitate the transcriptional termination and production of stable Mcr11.
2.4. Mcr11 size is unaffected by the presence of a transgene
We considered the possibility that expression of mcr11 in the context of an mRNA with a translated, stable transgene such as GFPv could contribute to the stability or processing of Mcr11. Production of Mcr11 from mcr11:TSE1 constructs was compared to Mcr11 levels from a similar construct in which GPFv is not immediately downstream of TSE1 (Supplemental Figure 4). The size of the Mcr11 transcript and Mcr11 levels were unaffected by the GFPv location (Supplemental Figure 4), indicating that GFPv did not contribute to the production or processing of stable Mcr11 in these experiments. This conclusion is also consistent with the low levels of Mcr11 observed in Msm despite the presence of the GFPv transgene (Figure 2d).
Stress conditions can modulate the efficiency of RITs (Morita, Ueda, Kubo, & Aiba, 2015) and induce the appearance of multiple, different size products of a single sRNA in Mtb (Arnvig & Young, 2009; Namouchi et al., 2016). However, neither BCG nor Mtb displayed significant stress‐induced changes in the termination efficiencies of mcr11 TSEs (Supplemental Table 3) when subjected to nitrosative stress (DETA‐NO), ATP depletion (BDQ), DNA damage (OFX), or transcriptional stress (RIF). These data demonstrate that mcr11 TSEs are not affected by stress.
2.5. Identification of Mcr11 regulatory targets
Having identified sequence features required for robust ectopic production of Mcr11 in complemented Δmcr11 strains, we considered mcr11 function. A bioinformatic search of potential regulatory targets of Mcr11 using TargetRNA (Tjaden, 2008) and TargetRNA2 (Kery, Feldman, Livny, & Tjaden, 2014) identified abmR as the top‐scoring hit (Figure 5a) (Supplemental Table 4). We measured the relative abundance of AbmR protein by Western blot analysis, and found that it was decreased relative to Wt in BGCΔmcr11 and MtbΔmcr11 (Figure 5b,c). However, recent reports indicate that the abmR mRNA is a leaderless transcript that lacks a 5′ UTR (Cortes et al., 2013), so the region of direct base‐pair complementarity with the Mcr11 sRNA and the 5′ end of abmR predicted by TargetRNA is not likely to be present in the abmR mRNA (Cortes et al., 2013). We also previously showed that the mcr11 gene overlaps a substantial portion of the DNA sequences identified as the upstream enhancer and promoter regions for abmR transcription (Figure 5a) (Gazdik, Bai, Wu, & McDonough, 2009; Girardin et al., 2018).
Figure 5.
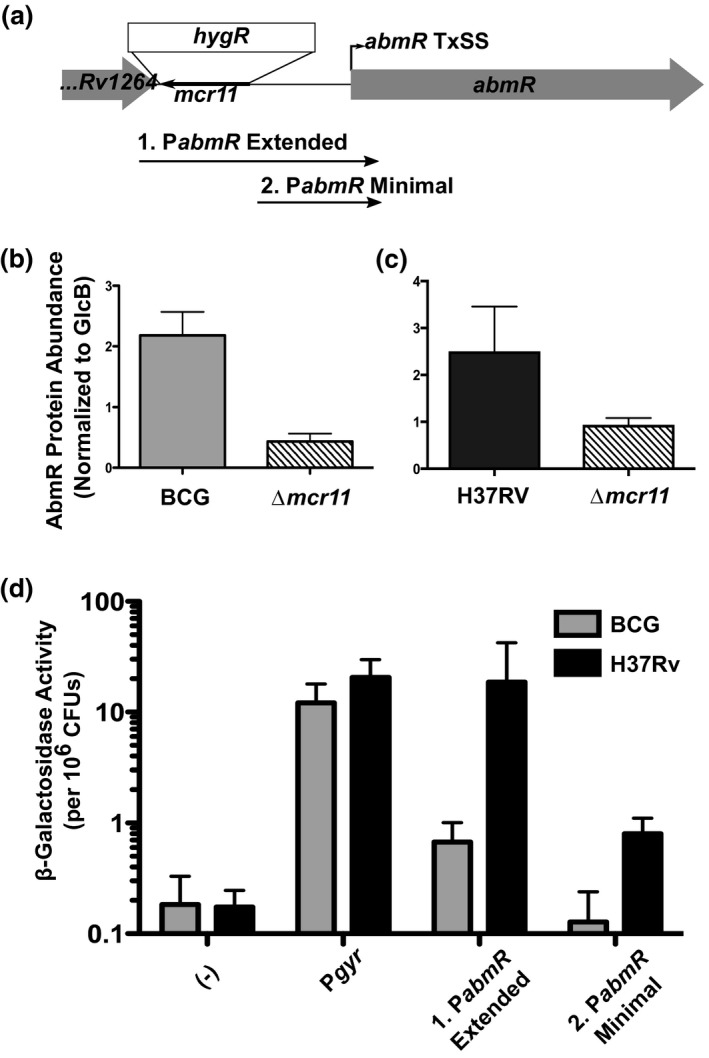
Δmcr11 has a disrupted abmR promoter, resulting in reduced expression of AbmR protein in BCG and Mtb. (a) The mcr11 locus with the position of the hygromycin knockout cassette is shown. The regions of DNA used to create promoter:lacZ fusions are shown. Fragment (1) includes the Rv1264‐abmR intergenic region, which includes the mcr11 locus. Fragment (2) includes the mcr11‐Rv1265 sequence that is available in the Δmcr11 strain. (b) Quantification of Western blot analysis from BCG grown to late‐log phase in ambient, shaking conditions. (c) Quantification of Western blot analysis from Mtb grown to late‐log phase in hypoxic (1.3% O2, 5% CO2), shaking conditions. AbmR was detected with poly‐clonal anti‐sera, and levels were normalized to GlcB levels, as detected by a monoclonal antibody. (d) β‐galactosidase activity assays using the promoter:lacZ fusions shown in (a) in Wt BCG (gray bars) and MTB (black bars) grown to late‐log phase in ambient, shaking conditions. Fluorescence is normalized to the OD620 of each sample. An asterisk *indicates p‐value < .05 using an unpaired Student’s t‐test. Results representative of 2–3 biological repeats
We addressed the possibility that mcr11 deletion has cis rather than trans effects on abmR expression due to disruption of abmR regulatory sequences. Promoter:lacZ reporter studies were used to compare PabmR activity in the presence and absence of flanking mcr11 sequences in Wt BCG and Mtb backgrounds that express Wt levels of Mcr11. Transcriptional activity from the minimal abmR promoter lacking adjacent sequence in the mcr11 gene was much lower than that from the extended abmR promoter that includes contiguous mcr11 gene sequences (Figure 5d). These assays were conducted in the Wt backgrounds of BCG and Mtb with comparable levels of native mcr11 expression in all matched strains, indicating that sequence overlapping the mcr11 gene was required in cis for full abmR promoter activity. We conclude that the reduced expression of abmR in Δmcr11 strains is likely due to the loss of cis sequence‐based abmR regulatory elements within the mcr11 gene rather than a trans‐acting regulatory effect of Mcr11 on abmR expression. Additionally, abmR and mcr11 promoters were both substantially more active in Mtb than in BCG, suggesting that pathogen‐specific factors impact the expressivity of both genes.
TargetRNA also predicted multiple putative targets of Mcr11 regulation, including three genes within operons involved in central metabolic processes and cell division: lipB (encodes lipoate‐protein ligase B, needed for lipoate biosynthesis) (Figure 6a), Rv3282 (encodes a conserved hypothetical protein with homology to Maf septum‐site inhibition protein) (Figure 6b) and fadA3 (encodes a beta‐ketoacyl coenzyme A thiolase) (Figure 6c). The intergenic spacing between putative target gene fadA3 and the preceding gene (Rv1075c) is identical to that of the intergenic spacing between Rv2216 and lipB (Figure 6a,c). However, fadA3 is likely to be transcribed independently of Rv1075c (Cortes et al., 2013). fadA3, lipB and Rv3282 were selected for follow‐up because they are associated with growth and central metabolism of Mtb and contain predicted Mcr11 base‐pairing sequences that are within known mRNA transcript boundaries. Each of these target transcripts has the potential to interact with Mcr11 through a 7–11 nt continuous base‐pairing region adjacent to a shorter stretch of gapped or imperfect base‐pairing (Figure 6a,c). A region from nt 39–55 of Mcr11 that is located within an unpaired loop of multiple modeled secondary structures of Mcr11 (Supplemental Figure 5) was predicted to interact with these mRNA targets (Figure 6d, Supplemental Table 4).
Figure 6.
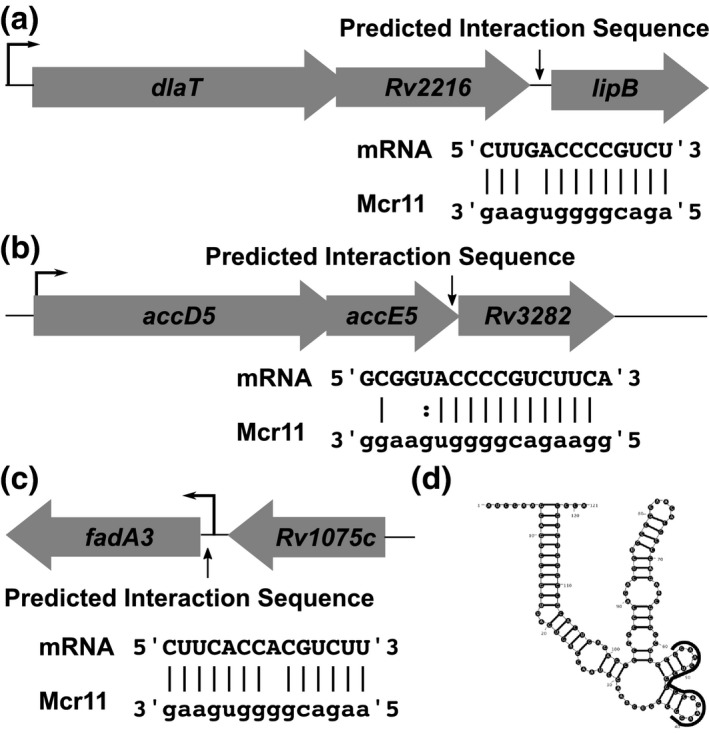
Bioinformatic modeling of Mcr11 targets reveals potential regulatory targets that are involved in central metabolism and cell division. (a) The organization of the dlaT‐lipB locus, with the position and potential base‐pairing interactions between the mRNA and Mcr11 is shown below. The transcriptional start site of the operon is shown with a thin black arrow. (b) As in (a), but for the accD5‐Rv3282 locus. Dashes indicate Watson–Crick base pairs, dots indicate non‐Watson–Crick base pairs, and a blank space indicates no interaction between bases. (c) As in (a) and (b), but for the Rv1074c/fadA3 locus. (d) The MFE secondary structure of Mcr11 as predicted by RNAStructure, with the portion of the sRNA predicted to interact with targets is shown in (a‐c) outlined in black
2.6. Fatty acids affect Mcr11‐dependent regulation of target genes in Mtb
The lipB operon includes dlaT, which encodes DlaT, the E2 component of pyruvate dehydrogenase (PDH) and the peroxynitrite reductase/peroxidase (PNR/P) complex in Mtb. The function of DlaT is modulated by lipoylation, which is dependent upon lipoate biosynthesis by LipB (Spalding & Prigge, 2010; Venugopal et al., 2011). BkdC (also called PdhC or Rv2495c) is a component of the branched chain keto‐acid dehydrogenase (BCKADH) complex in Mtb that also requires lipoylation for activity. Disruption of components in any of these complexes can cause growth defects in Mtb, some of which are dependent upon the nutrient mixture present in the growth media (Shi & Ehrt, 2006; Venugopal et al., 2011). The genes upstream of Rv3282 are accD5 and accE5, which are needed for a long chain aceyl‐CoA carboxylase enzymatic complex that generates substrates for fatty acid and mycolic acid biosynthesis (Bazet Lyonnet et al., 2014, 2017). fadA3 is expected to have a role in lipid degradation through beta‐oxidation, although its role in the metabolic processes of Mtb has not been well defined.
Thus, we reasoned that media components, such as fatty acids, may impact the regulation of fadA3 and the dlaT‐Rv2216‐lipB and/or accD5‐accE5‐Rv3282 operons. We used qRT‐PCR to measure expression of predicted Mcr11‐regulatory targets fadA3, lipB and Rv3282 in Wt versus Δmcr11 strains. Gene expression was queried in stationary phase BCG and Mtb grown in media with (+OA) or without (−OA) added fatty acids to assess the impact of Mcr11 regulatory function in these conditions. Tween‐80® is a hydrolysable detergent that releases a substantial amount of fatty acids (primarily oleic acid), and so was replaced with the non‐hydrolysable detergent Tyloxapol in media lacking fatty acids. The mcr11 complements included TSE4 with or without an intact copy of abmR. Levels of phoP, a response regulator expected to be independent of mcr11 and abmR, and pknA, a serine‐threonine protein kinase required for growth, were also measured.
Expression of Rv3282, Rv2216, lipB and fadA3 was significantly de‐repressed in MtbΔmcr11 compared to Wt Mtb in the absence, but not the presence, of fatty acids (Figure 7a,b). In contrast, levels of lipB and pknA expression decreased in the mcr11 Mtb mutant relative to Wt when fatty acids were present. Complementation of mcr11 fully (Rv3282) or partially (Rv2216, lipB, fadA3) restored expression to Wt levels (Figure 7a). The expression of accD5 (Rv3280) was significantly de‐repressed and accE5 (Rv3281) trended toward de‐repression in MtbΔmcr11, and complementation partially restored Wt expression levels of both genes (Supplemental Figure 6a). Expression of dlaT and phoP was not altered in Mtb (Figure 7a). From these data, we concluded that Mcr11‐mediated regulation of Rv3282, Rv2216, lipB and fadA3 expression in Mtb is affected by the levels of fatty acids in the media. In contrast, no Mcr11‐dependent regulation of lipB was observed in BCG, although trend of higher Rv3282 expression in BCGΔmcr11 relative to Wt BCG in the absence of fatty acids was observed (Figure 7c,d). These data demonstrate that mcr11‐dependent regulation of specific target genes is responsive to the fatty acid content of the culture media, and suggest that Mcr11 has a role in regulating the central metabolism of Mtb.
Figure 7.
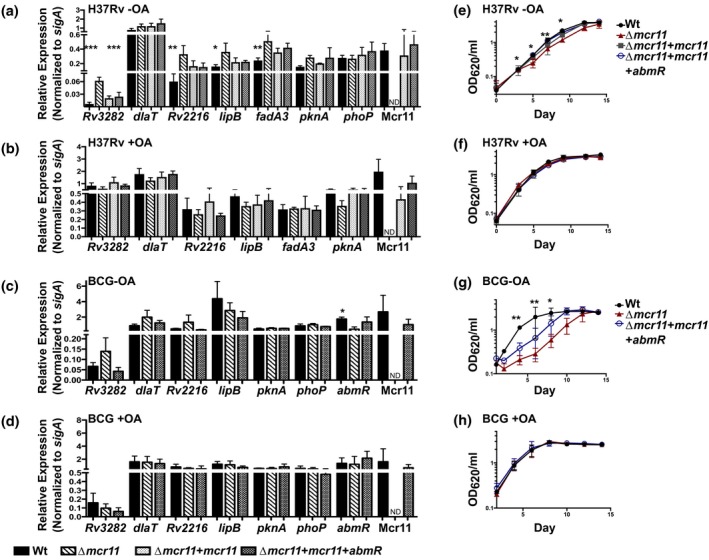
Δmcr11 strains of BCG and Mtb are defective for growth in fatty‐acid depleted media and predicted regulatory targets of Mcr11 are dysregulated at the mRNA level. (a) Mtb was grown for 12 days in under hypoxic (1.3% O2, 5% CO2), shaking conditions in −OA media (7H9 + 0.2% glycerol, 10% ADC and 0.05% Tween‐80). Gene expression was measured by qRT‐PCR and normalized to the reference gene sigA. Comparison made of each strain versus Δmcr11. Complementation strains included a single‐copy of mcr11 with TSE3 fused to GFPv or a single‐copy of the abmR locus and mcr11 with TSE4. (b) Mtb was grown for 7 days in under hypoxic, shaking conditions in + OA media (7H9 + 0.2% glycerol, 10% OADC and 0.05% Tween‐80). Gene expression was measured by qRT‐PCR and normalized to the reference gene sigA. (c) BCG was grown for 12 days in under hypoxic, shaking conditions in ‐OA media. Gene expression was measured by qRT‐PCR and normalized to the reference gene sigA. (d) BCG was grown for 12 days in under hypoxic, shaking conditions in + OA media. Gene expression was measured by qRT‐PCR and normalized to the reference gene sigA. (e) Growth curve of Mtb grown in hypoxic, shaking conditions in −OA media. Growth was surveyed by measuring the optical density at 620 nm (OD620). (f) Growth curve of Mtb grown in hypoxic, shaking conditions in + OA media. Growth was surveyed by measuring OD620. (g) As in (E), but with BCG. H. As in (F), but with BCG. Results are the means of three biological replicates. Statistical comparisons made of each strain versus Δmcr11 using a one‐way ANOVA with Bonferroni correction for multiple comparisons. Asterisks indicate significance as follows: *p < .05, **p < .01, ***p < .001, ****p < .0001 [Colour figure can be viewed at https://www.wileyonlinelibrary.com]
2.7. Mcr11 is required for optimal growth of BCG and Mtb without fatty acids
Optimal growth of Mtb on carbohydrate‐based carbon sources requires appropriate dlaT expression (Shi & Ehrt, 2006; Venugopal et al., 2011), while lipB, accD5 and accE5 are essential for Mtb growth (Sassetti, Boyd, & Rubin, 2003; Sassetti & Rubin, 2003). Mutations in Rv3282 delay Mtb growth, even in nutrient rich media (Sassetti et al., 2003). Based on our observed regulation of these genes by Mcr11, we hypothesized that Δmcr11 deleted strains would exhibit a growth defect when forced to utilize carbohydrate carbon sources for growth in media lacking a source of fatty acids.
BCGΔmcr11 was severely growth‐lagged and MtbΔmcr11 was moderately growth delayed compared to Wt bacteria when grown in media lacking added fatty acids (Figure 7e‐h). In contrast, no growth differences were observed between Wt and Δmcr11 mutant Mtb or BCG in media containing fatty acids. Complementation of mutant strains with mcr11 partially complemented growth in BCG and fully restored growth to Wt levels in Mtb (Figure 7g,e). From these data, we conclude that mcr11 has a role in the central metabolism and growth of BCG and Mtb.
2.8. The bioinformatically predicted Mcr11 base‐pairing sequence is required for the repression of lipB expression in Mtb
The necessity of the putative base‐pairing interaction between Mcr11 and the lipB mRNA was tested by constructing a promoter:GFPv fusion reporter construct with or without disrupted potential base‐pairing sequences in the lipB mRNA (Figure 8a) (Vasudeva‐Rao & McDonough, 2008). The bases in the lipB promoter that were mutagenized were selected using TargetRNA to determine the minimal change required to disrupt the putative base‐pairing interaction between the mRNA and Mcr11. Mtb was grown for 14 days in hypoxic, shaking conditions in media lacking fatty acids and expression from the Wt and mutagenized lipB internal promoters was measured using a GFPv fluorescence assay across growth phase. The Wt and mutant lipB promoters showed similar expression in early log phase, but the mutant promoter was significantly de‐repressed at later time points (Figure 8b). The trend of de‐repression observed in the mutagenized lipB promoter was similar to the de‐repression of lipB expression that occurred in MtbΔmcr11 grown in the same condition (Figure 7a). These data demonstrate that the bioinformatically predicted Mcr11 base‐pairing sequences are required for the repression of lipB expression in Mtb by Mcr11, and they provide evidence for an mRNA‐Mcr11 base‐pairing mechanism of Mcr11‐mediated repression of lipB expression in Mtb.
Figure 8.
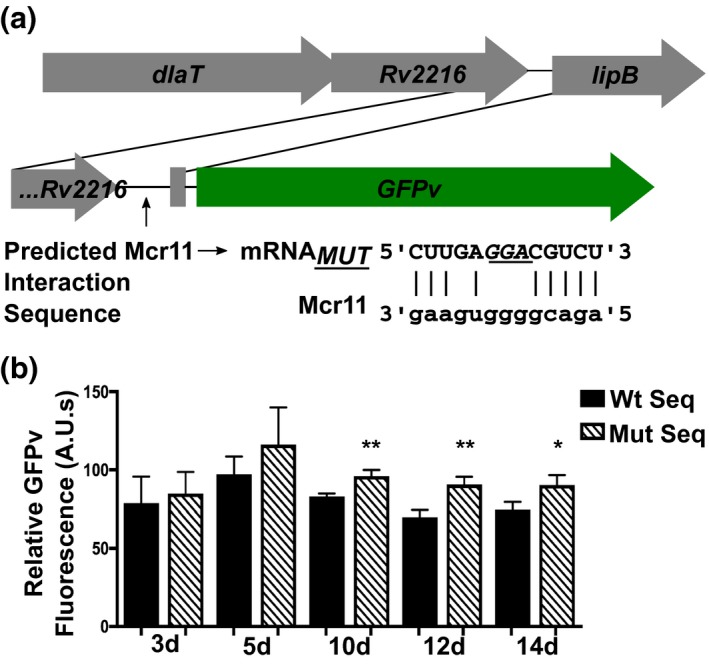
The bioinformatically predicted Mcr11 base‐pairing sequence is required for the repression of lipB expression in Mtb. (a) The organization of the dlaT‐lipB locus, with the internal promoter:GFPv fusion reporter construct and the disrupted potential base‐pairing interactions between the lipB mRNA and Mcr11 is shown below. The bases in the lipB promoter that were mutagenized are indicated in underlined italics. (b) Mtb was grown for 14 days in under hypoxic (1.3% O2, 5% CO2), shaking conditions in −OA media (7H9 + 0.2% glycerol, 10% ADC and 0.05% Tyloxapol). Expression from the lipB internal promoter was measured using a GFPv fluorescence assay used to measure promoter activity at the indicated time points. Fluorescence is normalized to the OD620 of each sample. Expression from the lipB promoter with wild‐type sequence is shown in black bars and expression from the mutagenized promoter is shown in slashed bars. Results are the means of three biological replicates. Statistical analysis conducted with a two‐tailed Student’s T‐test to compare the expression of the lipB promoter with wild‐type sequence to the mutagenized promoter at each time point. Asterisks indicate significance as follows: *p < .05, **p < .01, ***p < .001, ****p < .0001 [Colour figure can be viewed at https://www.wileyonlinelibrary.com]
3. DISCUSSION
This work identifies genes associated with central metabolism in Mtb as regulatory targets of the sRNA Mcr11. We also demonstrated that transcriptional termination and stable production of Mcr11 are enhanced by extended native sequence 3′ to mcr11 along with the product of the divergently transcribed adjacent gene, abmR (Figure 9). The regulation of central metabolism in Mtb is consistent with mcr11 expression profiles in response to growth phase and in vivo infection (Girardin et al., 2018; Pelly et al., 2012). However, the additional requirements for TB complex specific factors for stable Mcr11 expression were unexpected, and may have broader implications for understanding sRNA expression in Mtb.
Figure 9.
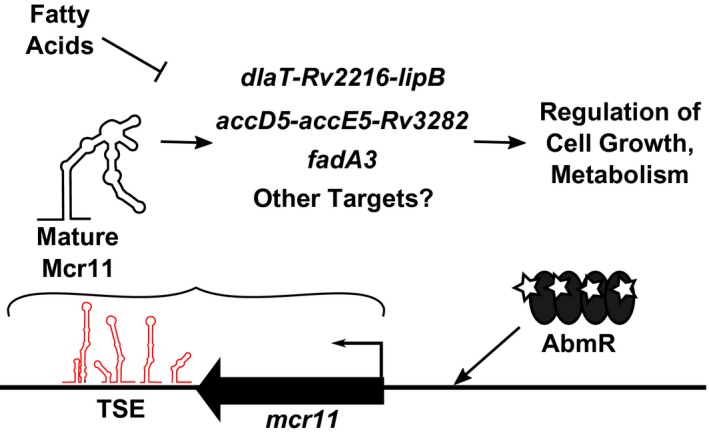
Model of Mcr11 function in Mtb. Expression of mcr11 is activated by advancing growth phase and the ATP‐binding transcription factor AbmR. Native 3′ sequence elements (TSEs, predicted secondary structures shown in red) promote the transcriptional termination of Mcr11 (shown in black). Mcr11 regulates the expression of genes involved in the central metabolism and growth of Mtb through base‐pairing interaction between Mcr11 and target mRNAs. This regulation and the importance of Mcr11 for optimal growth of Mtb is affected by the presence of fatty acids [Colour figure can be viewed at https://www.wileyonlinelibrary.com]
3.1. Mechanism of mcr11 transcriptional termination
Characterization of the factors required for efficient termination and stable expression of Mcr11 was a prerequisite for the complementation studies that confirmed Mcr11 regulatory targets. Protein coding mRNAs often can tolerate variable amounts of 5′ and 3′ flanking sequence because the signals for protein expression are provided immediately upstream and within the open reading frame (ORF). In contrast, expression of functional sRNAs may be more dependent on RNA chaperones and/or processing factors, as well as cis‐acting sequence elements at their transcriptional boundaries that are difficult to define at (Chae et al., 2011; Moores et al., 2017; Morita et al., 2015; Kriner, Sevostyanova, & Groisman, 2016; Shiflett et al., 2003). Although RNA chaperones have yet to be identified in mycobacteria, the roles of extended native 3′ sequences for expression of mycobacterial sRNAs warrant further investigation to determine whether mcr11 is exceptional or representative of a larger group of sRNAs with regards to expression and stability requirements (Ishikawa, Otaka, Maki, Morita, & Aiba, 2012; Johnson et al., 2014; Moores et al., 2017; Regnier & Hajnsdorf, 2013; Sauer & Weichenrieder, 2011; Shiflett et al., 2003; Olejniczak & Storz, 2017).
A recent study of Rho function in Mtb (Botella, Vaubourgeix, Livny, & Schnappinger, 2017) found that depletion of Rho did not impact transcriptional boundaries at predicted RITs (Gardner et al., 2011; Mitra et al., 2008), demonstrating a clear separation in the populations of transcripts terminated by RITs and Rho‐dependent mechanisms. While we found that TSEs promote expression of Mcr11, their underlying mechanism remains unclear and our data do not precisely fit either the current RIT or Rho‐dependent termination models (Table 1). We propose a model in which mcr11 processing occurs immediately following, or concurrent with, Rho‐dependent termination.
Table 1.
Comparison of transcription termination types
| Features | Rho‐Dependent | Intrinsic | Mcr11 |
|---|---|---|---|
| Predicted hairpin in 3' sequence | ✔ | ✔ | |
| Unstructured, C‐rich 3' sequence | ✔ | ||
| Poly U‐tract in 3' sequence | ✔ | ||
| Extrinsic factor(s) required | ✔ | ✔ | |
| Transcription factor may impact functionality | ✔ | ✔ | ✔ |
Rho‐independent termination is considered ‘intrinsic′ because extrinsic accessory factors are not required for function (Ray‐Soni et al., 2016). Our results clearly indicate that production of stable Mcr11 requires factors other than the TSEs (Figures 2d and 4). The species‐specific effects we observed for mcr11 termination measured by the GFPv reporter assays suggest that bacterial‐specific factors affect the efficiency of mcr11 termination itself, although the effects on Mcr11 stability are more striking (Figures 2, 3, 4).
Rho utilization (rut) sites are degenerate and difficult to predict using bioinformatics approaches, but transcripts terminated by Rho tend to have C‐enriched, G‐depleted 3′ ends that are thought to be unstructured (Peters et al., 2009). It is not clear if there is a rut site within mcr11, as the sequences downstream of the 3′ end of Mcr11 are predicted to be highly structured and are rich in both G and C (Figure 1). A run of 6Cs (nucleotides 93–98) occurs within Mcr11, but this region may be highly structured (Figure 1). In contrast, Mcr11 TSEs 2–4, which contributed strongly to the termination of Mcr11, all possess C‐rich loops in their predicted secondary structures that could be rut sites (Figure 1). Rho‐significant regions (RSRs) reported in the Botella et al study (Botella et al., 2017) include an RSR that begins 6 nt downstream of the mapped 3′ end of Mcr11, that is characterized as a region of general antisense transcription. The presence of this RSR is consistent with the possibility that mcr11 expression is Rho‐terminated. Mcr10 (ncRv1157a) is also proximal to an RSR (Botella et al., 2017). Future work is needed to address the importance of Rho‐dependent termination for mycobacterial sRNAs.
In Escherichia coli, multiple trans‐acting factors are known to modulate termination and anti‐termination of Rho‐terminated sRNAs (Rabhi et al., 2011). AbmR, an ATP‐responsive DNA‐binding transcription factor that activates mcr11 expression (Girardin et al., 2018), was found here to also positively regulate termination efficiency of mcr11 (Supplemental Figure 3). Despite the high similarity between abmR and its Msm orthologue (Girardin et al., 2018), MSM_5010 failed to strongly activate mcr11 expression and Mcr11 transcripts were unstable in Msm, even though provision of abmR in trans enhanced Mcr11 levels. It is possible that AbmR interacts directly with RNAP or Rho to affect termination, or recruits an as yet undefined trans‐acting factor that modulates Mcr11 termination in Mtb. For example, Rv1222 is a mycobacterial DNA‐binding protein that was found to slow the rate of RNA synthesis through direct interaction with RNAP, and AbmR may function in a similar manner to promote termination at the mcr11 locus (Rudra, Prajapati, Banerjee, Sengupta, & Mukhopadhyay, 2015).
The discrete 3′ end of mature Mcr11 observed by Northern blot is consistent with a processed or precisely terminated RNA, as transcription through TSEs would otherwise result in significantly longer RNA products. Processing of the 3′ ends of mycobacterial sRNAs has been proposed (Tsai et al., 2013), and a recent report identified a hypoxia‐regulated mycobacterial sRNA that is extensively processed at its 3′ end (Moores et al., 2017). The observed size variants of specific sRNA species in response to host‐associated stress conditions (Arnvig & Young, 2009) provide further evidence for processing of sRNAs in mycobacteria. Recent work has demonstrated that Rho terminated transcripts have processed 3′ ends immediately downstream of a stable stem‐loop (Dar & Sorek, 2018). While few RNA processing enzymes of mycobacteria have been well characterized, the Mtb genome encodes many known RNases with varying sequence specificities (Abendroth et al., 2014; Taverniti, Forti, Ghisotti, & Putzer, 2011; Uson, Ordonez, & Shuman, 2015; Zeller et al., 2007; Zhu et al., 2008, 2006). We speculate that unidentified TB‐complex RNA chaperones and/or modifying enzymes contribute to Mcr11 stability, and that their absence in Msm results in Mcr11 degradation.
3.2. Regulation of predicted targets of Mcr11
Oleic acid is the main fatty acid present in rich mycobacterial media formulations, provided either directly as oleic acid or in the hydrolyzable, non‐ionic detergent Tween‐80®. The presence of Tween‐80® can alter acid resistance (Vandal, Pierini, Schnappinger, Nathan, & Ehrt, 2008), enhances the growth of mycobacteria when combined with glucose or glycerol (Lofthouse et al., 2013; Schaefer & Lewis, 1965) and increases the uptake of glucose by BCG (Lofthouse et al., 2013; Schaefer & Lewis, 1965). While we observed no increase in sensitivity to acid in BCGΔmcr11 or BCGΔabmR (data not shown), mcr11 was required for growth on media lacking added fatty acids. Nutrient rich media is often used in batch culture experimentation with Mtb, and the potential confounding effect of multiple nutrient sources on characterizing the essentiality and function of gene products has recently gained appreciation (DeJesus et al., 2017; Griffin et al., 2011). Future studies characterizing nutrient uptake in Mtb and BCG strains of Δmcr11 or ΔabmR will further our understanding of the nutrient‐related growth defects of these strains.
Dihydrolipoamide acyltransferase (DlaT) and BkdC, a component of the Lpd‐dependent branched chain keto‐acid dehydrogenase (BCKADH), are critical for Mtb pathogenesis and known to be lipoylated in Mtb (Venugopal et al., 2011). Lipoylation in Mtb is presumed to depend solely on lipoate synthesis by the enzymes LipA and the Mcr11 target LipB, as scavenging and import pathways are apparently lacking (Spalding & Prigge, 2010). Rv3282 has homology to Maf, a septum inhibition protein conserved across all domains of life (Hamoen, 2011; Tchigvintsev et al., 2013). Despite mcr11‐dependent regulation of Rv3282 in the absence of fatty acids, MtbΔmcr11 did not have a filamentous cell morphology (data not shown) and the function of Rv3282 in Mtb has not been defined. The regulation of Mcr11’s targets was responsive to the fatty acid content of the growth media, and it will be important to determine if the observed growth defects of MtbΔmcr11 are due to the dysregulation of fadA3, lipB and Rv3282, or if there are additional targets of Mcr11 regulation that account for this phenotype (Figure 9). Additionally, the role of Mcr11 in supporting the growth and persistence of Mtb in response to nutrient availability and growth arrest should be explored.
Mcr11‐dependent regulation of lipB, fadA3 and Rv3282 expression was expected to occur through base‐paring with the mRNA, and we demonstrated that the predicted base‐pairing sequences were required for repression of lipB expression (Figure 8). The sRNA RyhB is known to differentially regulate expression of individual genes within the iscRSUA operon, resulting in down‐regulation of the icsSUA genes while maintaining expression of iscR (Desnoyers, Morissette, Prevost, & Masse, 2009). This mechanism requires an intra‐cistronic base‐pairing dependent translational blockade of iscS, followed by recruitment of RNaseE and selective degradation of the 3′ end of the mRNA. The position of two putative Mcr11 base‐pairing sites within target gene operons raises multiple possibilities for mcr11‐mediated regulation of co‐transcribed genes that will be an intriguing topic for future study. We noted that the mcr11 target interaction site within the intergenic region of Rv2216 and lipB had identical spacing to that of Rv1075c and fadA3, although the significance of this architecture is unknown.
This study shows that the transcriptional termination and stability of the sRNA Mcr11 are enhanced by extended, native 3′ sequence elements (TSEs) in TB complex mycobacteria. The role of AbmR in mcr11 expression was extended from that of a transcriptional activator to include enhancement of mcr11 transcriptional termination. Our observation that bacterial species‐specific factors govern sRNA stability in mycobacteria may also extend to other sRNAs. Combined use of bioinformatic and molecular tools established fatty acid responsive, mcr11‐dependent gene regulation in Mtb and provides a versatile strategy for the continued search of sRNA function in pathogenic mycobacteria. Future work defining the precise roles of the sRNA Mcr11 in regulating the growth and metabolism of Mtb will greatly advance our understanding of this important pathogen.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains and growth conditions
Mycobacterium tuberculosis H37Rv (Mtb) (ATCC 25618) and Mycobacterium bovis BCG (BCG) (Pasteur strain, Trudeau Institute) were grown on 7H10 agar (Difco) supplemented with 10% oleic acid‐albumin‐dextrose‐catalase (OADC) (Becton Dickson and Company) and 0.01% cycloheximide or in Middlebrook 7H9 liquid medium (Difco) supplemented with 10% (vol/vol) OADC, 0.2% (vol/vol) glycerol and 0.05% (vol/vol) Tween‐80 (Sigma‐Aldrich) in physiologically relevant shaking, hypoxic conditions (1.3% O2, 5% CO2) (Florczyk, McCue, Stack, Hauer, & McDonough, 2001) for promoter::reporter fusion assays and Northern blot analysis. Mycobacterium smegmatis mc2155 (Msm) (ATCC 700,048) was grown in Middlebrook 7H9 liquid medium (Difco) supplemented with 10% (vol/vol) OADC, 0.2% (vol/vol) glycerol and 0.05% (vol/vol) Tween‐80 (Sigma‐Aldrich), shaking in ambient air for promoter::reporter fusion assays and Northern blot analysis. All experiments were started from low‐passage frozen stocks. For cloning experiments, Escherichia coli strains were grown on Luria‐Bertani (Difco) agar plates or in liquid broth. Growth media and plates were supplemented with 50 μg/mL of hygromycin, 25 μg/mL of kanamycin (Sigma‐Aldrich) for selection of clones. All bacterial cultures were incubated at 37°C.
4.2. Mutant strain construction
Knockout strains of mcr11 in Mtb and BCG genetic backgrounds were generated using homologous recombination (Bardarov et al., 2002) to replace nucleotides 1–100 with a hygromycin resistance cassette. Disruption of mcr11 was confirmed with polymerase chain reaction (PCR) (Supplemental Figure 1b,c) and Northern blot analysis (Supplemental Figure 1d,e.). Several complementation constructs were created to restore Mcr11 expression. Sequence from the corrected abmR (Rv1265) start site (Gazdik et al., 2009) to the end of the Rv1264 ORF was amplified by PCR and cloned downstream of the efTu (Rv0685) promoter in the multi‐copy plasmid pMBC280, creating pMBC1211. PCR was used to amplify varying lengths of sequence from the end of the Rv1264 ORF through the end of the abmR ORF, and they were cloned it into pMBC409, which contains a kanamycin resistance marker, to create pMBC2040, pMBC2041 and pMBC2042 (Supplemental Figure 1a). Complementation plasmids were sequenced to verify that the desired sequences had been correctly incorporated and then used to transform Mtb and BCG by electroporation. A complete list of plasmids used in this study is shown in Supplemental Table 1 and primers are shown in Supplemental Table 2.
4.3. Mapping the 3′ end of Mcr11
RNA was harvested from late‐log phase Mtb and the 3′ end of Mcr11 was mapped using the miScript Reverse Transcription Kit (Qiagen) (Beauregard et al., 2013) and a gene‐specific primer. The reaction product was cloned into pCRΙΙ‐TOPO vector (Life Technologies) and 10 positive clones were subjected to Sanger sequencing. The sequences were mapped back to the chromosome of Mtb.
4.4. Bioinformatic modeling of Mcr11
Secondary structure modeling of Mcr11 and putative native intrinsic terminators was performed using RNAstructure using default parameters (Mathews, 2006). A synthetic terminator (ttsbiB) (Huff, Czyz, Landick, & Niederweis, 2010) was modeled onto Mcr11 as well. Secondary structure modeling of Mcr11 was performed using RNAstructure (Mathews, 2006). Using the previously published 5′ and 3′ boundaries of Mcr11, minimum free energy (MFE) and centroid structures for Mcr11 derived from CentroidFold (using the CONTRAfold inference engine), Mfold, NUPACK, RNAfold and RNAStructure algorithms were created (Ding, Chan, & Lawrence, 2004; Mathews, 2006; Sato, Hamada, Asai, & Mituyama, 2009; Zadeh et al., 2011; Zuker, 2003). Base‐pair drawings of predicted RNA secondary structures were either directly exported from the software used, or created with Visualization Applet for RNA (VARNA) (Darty, Denise, & Ponty, 2009) and simplified line drawings were made in Inkscape (inkscape.org).
Regulatory targets of Mcr11 in Mtb were predicted using TargetRNA (Tjaden, 2008) and TargetRNA2 algorithms (Kery et al., 2014), set to default parameters (Table3). Where possible, the position of predicted regions of base‐paring between Mcr11 and a putative regulatory target was evaluated against known 5′ ends of mRNAs in Mtb (Cortes et al., 2013) (Table 3).
4.5. Promoter:reporter fusion assays
Promoter:lacZ fusions were created to compare the relative transcriptional activities from DNA sequences spanning the intergenic space between Rv1264 and abmR, and mcr11 and abmR (Supplemental Figure 2). The promoters of gyrB (Rv0005) and tuf (Rv0685) were used as positive controls, and a promoterless construct was used as a negative control. The relevant constructs were transformed into wild‐type (Wt) strains of BCG and Mtb and assayed for β‐galactosidase activity in late‐log phase cultures grown in ambient, shaking conditions by adding 5‐acetylaminofluoresceindi‐β‐D‐galactopyranoside (C2FDG; Molecular Probes) and measuring fluorescence in a Cytofluor 4000 fluorometer (PerSeptive Biosystems) as described previously (Vasudeva‐Rao & McDonough, 2008).
Constructs including the mcr11 promoter, Mcr11 sequence and various potential intrinsic terminator sequences were fused to the reporter green fluorescent protein Venus (GFPv) on an integrating plasmid as previously described (Girardin et al., 2018). A GFPv fluorescence assay was used to measure promoter activity and read‐through of the putative intrinsic terminators of Mcr11 in Msm, BCG and Mtb.
At mid‐log phase and late stationary phase, aliquots of recombinant strains were collected and gently sonicated (setting 4, 4 pulses of 5′′ on time interspersed by 5′ off time) using a Virsonic 475 Ultrasonic Cell Disrupter with a cup horn attachment (VirTis Company) before duplicate samples were diluted 1:1 in fresh media. The level of fluorescence in arbitrary units from the GFPv reporter strain was detected using the CytoFluor Multi‐Well Plate Reader Series 4,000 (PerSeptive Biosystems) at 485 nm excitation and 530 nm emission. The optical density (OD) at 620 nm was read using a Tecan Sunrise® microplate reader and fluorescence levels were normalized to 106 bacteria. A promotorless (‐) construct was used as a negative control and the promoters Ptuf and Pgyr served as positive controls. The % termination was calculated by dividing the observed GFPv fluorescence in arbitrary units of each indicated terminator construct by the amount of GFPv fluorescence in arbitrary units measured from the mcr11 promoter.
Late stationary phase reporter strains of BCG and Mtb were diluted with fresh media to an OD620 nm of 1.25 in a 12‐well plate, and then subjected to a variety of cellular stressors for a 24 hr period in shaking, hypoxic conditions (1.3% O2, 5% CO2). Cells were treated with 250 μM diethyltriamine‐NO (DETA‐NO) (Sigma‐Aldrich), 1 μg per mL of ofloxacin (OFX), 0.28 μg per mL of rifampacin (RIF), 53 μg per ml of bedaquiline (BDQ), or 0.1% dimethyl sulfoxide (DMSO) vehicle control. GFPv fluorescence in arbitrary units per 106 bacteria (by measured OD at 620 nm) was monitored as described above, and percent termination was calculated relative to Pmcr11 controls.
4.6. Fluorescence‐activated cell sorting analysis
Fluorescence‐activated cell sorting analysis was performed on Msm samples that had been fixed with 4% paraformaldehyde (PFA) in phosphate‐buffered saline (PBS) at pH 7.0 for 30 min at room temperature. Fixation was quenched pH‐adjusted glycine, and cells were washed three times with PBS. Cells were diluted approximately 1:500 in DPBS and subjected to FACS analysis with a FACS Calibur (Becton Dickson) as previously described (Purkayastha, McCue, & McDonough, 2002). Data were collected for 20,000 events per sample and analyzed in CellQuest software (Becton Dickenson).
4.7. RNA isolation
Total RNA was harvested from Msm, BCG and Mtb after cultures were treated 0.5 M final GTC solution (5.0 M guanadinium isothiocyanate, 25 mM sodium citrate, 0.5% sarkosyl, and 0.1 M 2‐β‐mercaptoethanol) and pelleted at 4°C. Pelleted cells were resuspended in TRIzol Reagent (Invitrogen) and cells were disrupted with 0.1 mm zirconia‐silica beads (BioSpec Products) and three 70‐s high‐speed pulse treatments in a bead‐beater (BioSpec Products). RNA was recovered from lysates with Direct‐zol Mini Prep columns (Zymo) per the manufacturer’s protocol. RNA was eluted from the column with nuclease free‐water, treated with DNaseI (Qiagen) for 20 min at room temperature, and isopropanol precipitated. One μg of total RNA was screened for DNA contamination by PCR using primers KM1309 and KM1310 internal to the sigA ORF. The quality of each RNA sample was assessed by running 1.0 μg of DNA‐free RNA on a 0.1% (weight/vol) SDS‐agarose gel and visualizing intact 23S, 16S, and 5S ribosomal RNA bands after staining with ethidium bromide (Sigma‐Aldrich).
4.8. Northern blots
High quality DNA‐free total RNA isolated from Msm, BCG and Mtb was used for Northern blot analysis of Mcr11 expression. About 3–10 μg RNA was separated on a 10% 8 M urea PAGE run at a constant current of 20 mA for 1–1.5 hr. The gel was electroblotted onto a Hybond N (Millipore) nylon membrane using a wet transfer system (Bio‐Rad Laboratories) as previously described (Girardin, in prep). Blots were UV cross‐linked and baked at 80°C for 2 hr prior to pre‐hybridization at 42°C for 1 hr in Rabid‐Hyb Buffer (GE Healthcare Life Sciences). Hybridization with α‐32P‐ATP end‐labeled DNA oligo probes was performed at 42°C for 2–16 hr. Blots were washed per the manufacturer’s protocol and exposed to phosphor‐screens for visualization. Densitometry was completed using ImageQuant software.
4.9. Generation of mycobacterial cell lysates and Western blotting
Strains of BCG and Mtb were grown in the desired condition, and pelleted and washed twice in ice‐cold Dulbecco’s phosphate‐buffered saline, calcium and magnesium free (DPBS‐CMF), with 0.2% protease inhibitor (Sigma‐Aldrich). Cell pellets were resuspended in 1/25 volume lysis buffer (0.3% SDS, 200 mM DTT, 28 mM Tris‐Hcl, 22 mM Tris‐Base and 1% protease inhibitor cocktail) and disrupted by two rounds of high‐powered sonication at 4°C with Virsonic 475 Ultrasonic Cell Disrupter with a cup horn attachment (VirTis Company) interspersed with 10 freeze‐thaw cycles as previously described (Girardin et al., 2018). Lysate was cleared by centrifugation and the cleared lysate was heat‐killed at 95°C before quantification with a NanoDrop 2000 (Thermo Scientific).
A total of 30 μg protein of each sample was separated by 12%–15% Tris‐glycine SDS‐PAGE, and gels were immunoblotted on Immobilon‐P membranes (Millipore) for 1h at 1 mA/cm2 using a wet transfer system (Bio‐Rad Laboratories). Blots were blocked and proved with 1° antibody in 5% milk (vol/vol) in 50 mM Tris‐buffered saline with 0.05% (vol/vol) Tween‐20 (Fisher Scientific). Monoclonal antibodies against Mtb GlcB (Gene Rv1837c), Clone α‐GlcB (produced in vitro NR‐13799) was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH. Polyclonal mouse AbmR anti‐serum was previously generated in‐house (Girardin et al., 2018). Primary antibodies were detected with peroxidase conjugated goat secondary antibody and enhanced chemiluminescence (ECL) Western blotting detection reagent (Thermo Scientific).
4.10. Growth curve analysis
The growth of BCG and Mtb in hypoxic (1.3% O2, 5% CO2), shaking conditions was monitored by measuring the optical density at 620 nm (OD620) of gently sonicated aliquots of cells with a Tecan Sunrise® microplate reader. Cultures were grown in vented T25 tissue culture flasks (Corning), and time points were in single or multi‐day intervals. Growth comparisons between Middlebrook 7H9 + 0.2% glycerol, 10% OADC and 0.05% Tween‐80 (+OA) or in Middlebrook 7H9 + 0.2% glycerol, 10% ADC and 0.05% Tyloxapol (−OA) were made.
DATA SHARING
The data that support the findings of this study are available in the supplementary material of this article.
CONFLICTS OF INTEREST
None declared.
Supporting information
Supinfo
ACKNOWLEDGMENTS
We are very grateful to Dr. Guangchun Bai and Damen Schaak for generating the mcr11 knockout strains of BCG and Mtb used in this study. We also thank Dr. Joseph Wade for helpful discussions, and gratefully acknowledge the Wadsworth Center Applied Genomics Technologies Core for DNA sequencing and the Wadsworth Center Media and Tissue Culture Core for media preparation. This work was supported in part by National Institutes of Health grants R01AI063499 (KAM) and T32AI055429 (RCG), with additional funding from Potts Memorial Foundation (RCG).
Girardin RC, McDonough KA. Small RNA Mcr11 requires the transcription factor AbmR for stable expression and regulates genes involved in the central metabolism of Mycobacterium tuberculosis . Mol Microbiol. 2020;113:504–520. 10.1111/mmi.14436
REFERENCES
- Abendroth, J. , Ollodart, A. , Andrews, E. S. , Myler, P. J. , Staker, B. L. , Edwards, T. E. , … Grundner, C. (2014). Mycobacterium tuberculosis Rv2179c protein establishes a new exoribonuclease family with broad phylogenetic distribution. Journal of Biological Chemistry, 289, 2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnvig, K. B. , Comas, I. , Thomson, N. R. , Houghton, J. , Boshoff, H. I. , Croucher, N. J. , … Young, D. B. (2011). Sequence‐based analysis uncovers an abundance of non‐coding RNA in the total transcriptome of Mycobacterium tuberculosis . PLoS Path, 7, e1002342 10.1371/journal.ppat.1002342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnvig, K. B. , & Young, D. B. (2009). Identification of small RNAs in Mycobacterium tuberculosis . Molecular Microbiology, 73, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardarov, S. , Bardarov, S., Jr. , Pavelka, M. S., Jr., Sambandamurthy, V. , Larsen, M. , Tufariello, J. , … Jacobs, W. R., Jr. (2002). Specialized transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology, 148, 3007–3017. 10.1099/00221287-148-10-3007 [DOI] [PubMed] [Google Scholar]
- Bazet Lyonnet, B. , Diacovich, L. , Cabruja, M. , Bardou, F. , Quemard, A. , Gago, G. , & Gramajo, H. (2014). Pleiotropic effect of AccD5 and AccE5 depletion in acyl‐coenzyme A carboxylase activity and in lipid biosynthesis in mycobacteria. PLoS ONE, 9, e99853 10.1371/journal.pone.0099853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazet Lyonnet, B. , Diacovich, L. , Gago, G. , Spina, L. , Bardou, F. , Lemassu, A. , … Gramajo, H. (2017). Functional reconstitution of the Mycobacterium tuberculosis long‐chain acyl‐CoA carboxylase from multiple acyl‐CoA subunits. FEBS Journal, 284, 1110–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauregard, A. , Smith, E. A. , Petrone, B. L. , Singh, N. , Karch, C. , McDonough, K. A. , & Wade, J. T. (2013). Identification and characterization of small RNAs in Yersinia pestis . RNA Biology, 10, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergkessel, M. , Basta, D. W. , & Newman, D. K. (2016). The physiology of growth arrest: Uniting molecular and environmental microbiology. Nature Reviews Microbiology, 14, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts, J. C. , Lukey, P. T. , Robb, L. C. , McAdam, R. A. , & Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Molecular Microbiology, 43, 717–731. [DOI] [PubMed] [Google Scholar]
- Botella, L. , Vaubourgeix, J. , Livny, J. , & Schnappinger, D. (2017). Depleting Mycobacterium tuberculosis of the transcription termination factor Rho causes pervasive transcription and rapid death. Nature Communications, 8, 14731 10.1038/ncomms14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk, R. , Gold, B. , Venugopal, A. , Singh, J. , Samy, R. , Pupek, K. , … Nathan, C. (2008). Selective killing of nonreplicating mycobacteria. Cell Host & Microbe, 3, 137–145. 10.1016/j.chom.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, H. , Han, K. , Kim, K. S. , Park, H. , Lee, J. , & Lee, Y. (2011). Rho‐dependent termination of ssrS (6S RNA) transcription in Escherichia coli: Implication for 3′ processing of 6S RNA and expression of downstream ygfA (putative 5‐formyl‐tetrahydrofolate cyclo‐ligase). Journal of Biological Chemistry, 286, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes, T. , Schubert, O. T. , Rose, G. , Arnvig, K. B. , Comas, I. , Aebersold, R. , & Young, D. B. (2013). Genome‐wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis . Cell Reports, 5, 1121–1131. 10.1016/j.celrep.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyz, A. , Mooney, R. A. , Iaconi, A. , & Landick, R. (2014). Mycobacterial RNA polymerase requires a U‐tract at intrinsic terminators and is aided by NusG at suboptimal terminators. Mbio, 5, e00931 10.1128/mBio.00931-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar, D. , & Sorek, R. (2018). High‐resolution RNA 3′ends mapping of bacterial Rho‐dependent transcripts. Nucleic Acids Research, 46, 6797–6805. 10.1093/nar/gky274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty, K. , Denise, A. , & Ponty, Y. (2009). VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics, 25, 1974–1975. 10.1093/bioinformatics/btp250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus, M. A. , Gerrick, E. R. , Xu, W. , Park, S. W. , Long, J. E. , Boutte, C. C. , … Ioerger, T. R. (2017). Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon m utagenesis, MBio, 8(1), e02133–16. 10.1128/mBio.02133-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers, G. , Morissette, A. , Prevost, K. , & Masse, E. (2009). Small RNA‐induced differential degradation of the polycistronic mRNA iscRSUA. EMBO Journal, 28, 1551–1561. 10.1038/emboj.2009.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiChiara, J. M. , Contreras‐Martinez, L. M. , Livny, J. , Smith, D. , McDonough, K. A. , & Belfort, M. (2010). Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis . Nucleic Acids Research, 38, 4067–4078. 10.1093/nar/gkq101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. , Chan, C. Y. , & Lawrence, C. E. (2004). Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Research, 32, W135–141. 10.1093/nar/gkh449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey, A. K. , Baker, C. S. , Romeo, T. , & Babitzke, P. (2005). RNA sequence and secondary structure participate in high‐affinity CsrA‐RNA interaction. RNA, 11, 1579–1587. 10.1261/rna.2990205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florczyk, M. A. , McCue, L. A. , Stack, R. F. , Hauer, C. R. , & McDonough, K. A. (2001). Identification and characterization of mycobacterial proteins differentially expressed under standing and shaking culture conditions, including Rv2623 from a novel class of putative ATP‐binding proteins. Infection and Immunity, 69, 5777–5785. 10.1128/IAI.69.9.5777-5785.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, P. P. , Barquist, L. , Bateman, A. , Nawrocki, E. P. , & Weinberg, Z. (2011). RNIE: Genome‐wide prediction of bacterial intrinsic terminators. Nucleic Acids Research, 39, 5845–5852. 10.1093/nar/gkr168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton, N. J. , Waddell, S. J. , Sherratt, A. L. , Lee, S. M. , Smith, R. J. , Senner, C. , … Barer, M. R. (2008). Cytological and transcript analyses reveal fat and lazy persister‐like bacilli in tuberculous sputum. PLoS Medicine, 5, e75 10.1371/journal.pmed.0050075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdik, M. A. , Bai, G. , Wu, Y. , & McDonough, K. A. (2009). Rv1675c (cmr) regulates intramacrophage and cyclic AMP‐induced gene expression in Mycobacterium tuberculosis‐complex mycobacteria. Molecular Microbiology, 71, 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrick, E. R. , Barbier, T. , Chase, M. R. , Xu, R. , Francois, J. , Lin, V. H. , … Fortune, S. M. (2018). Small RNA profiling in Mycobacterium tuberculosis identifies MrsI as necessary for an anticipatory iron sparing response. Proceedings of the National Academy of Sciences of the United States of America, 115, 6464–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin, R. C. , Bai, G. , He, J. , Sui, H. , & McDonough, K. A. (2018). AbmR (Rv1265) is a Novel transcription factor of Mycobacterium tuberculosis that regulates host cell association and expression of the non‐coding small RNA Mcr11. Molecular Microbiology, 110(5), 811–830. 10.1111/mmi.14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, J. E. , & McKinney, J. D. (2004). M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb), 84, 29–44. 10.1016/j.tube.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Griffin, J. E. , Gawronski, J. D. , Dejesus, M. A. , Ioerger, T. R. , Akerley, B. J. , & Sassetti, C. M. (2011). High‐resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Path, 7, e1002251 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen, L. W. (2011). Cell division blockage: But this time by a surprisingly conserved protein. Molecular Microbiology, 81, 1–3. 10.1111/j.1365-2958.2011.07693.x [DOI] [PubMed] [Google Scholar]
- Hnilicova, J. , Jirat Matejckova, J. , Sikova, M. , Pospisil, J. , Halada, P. , Panek, J. , & Krasny, L. (2014). Ms1, a novel sRNA interacting with the RNA polymerase core in mycobacteria. Nucleic Acids Research, 42, 11763–11776. 10.1093/nar/gku793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honaker, R. W. , Dhiman, R. K. , Narayanasamy, P. , Crick, D. C. , & Voskuil, M. I. (2010). DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. Journal of Bacteriology, 192, 6447–6455. 10.1128/JB.00978-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff, J. , Czyz, A. , Landick, R. , & Niederweis, M. (2010). Taking phage integration to the next level as a genetic tool for mycobacteria. Gene, 468, 8–19. 10.1016/j.gene.2010.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatov, D. V. , Salina, E. G. , Fursov, M. V. , Skvortsov, T. A. , Azhikina, T. L. , & Kaprelyants, A. S. (2015). Dormant non‐culturable Mycobacterium tuberculosis retains stable low‐abundant mRNA. BMC Genomics, 16, 954 10.1186/s12864-015-2197-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iona, E. , Pardini, M. , Mustazzolu, A. , Piccaro, G. , Nisini, R. , Fattorini, L. , & Giannoni, F. (2016). Mycobacterium tuberculosis gene expression at different stages of hypoxia‐induced dormancy and upon resuscitation. Journal of Microbiology (Seoul, Korea), 54, 565–572. 10.1007/s12275-016-6150-4 [DOI] [PubMed] [Google Scholar]
- Ishikawa, H. , Otaka, H. , Maki, K. , Morita, T. , & Aiba, H. (2012). The functional Hfq‐binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U‐rich sequence and followed by a 3′ poly(U) tail. RNA, 18, 1062–1074. 10.1261/rna.031575.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C. M. , Chen, Y. , Lee, H. , Ke, A. , Weaver, K. E. , & Dunny, G. M. (2014). Identification of a conserved branched RNA structure that functions as a factor‐independent terminator. Proceedings of the National Academy of Sciences of the United States of America, 111, 3573–3578. 10.1073/pnas.1315374111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kery, M. B. , Feldman, M. , Livny, J. , & Tjaden, B. (2014). TargetRNA2: Identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Research, 42, W124–129. 10.1093/nar/gku317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriner, M. A. , Sevostyanova, A. , & Groisman, E. A. (2016). Learning from the leaders: Gene regulation by the transcription termination factor Rho. Trends in Biochemical Sciences, 41, 690–699. 10.1016/j.tibs.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofthouse, E. K. , Wheeler, P. R. , Beste, D. J. , Khatri, B. L. , Wu, H. , Mendum, T. A. , … McFadden, J. (2013). Systems‐based approaches to probing metabolic variation within the Mycobacterium tuberculosis complex. PLoS ONE, 8, e75913 10.1371/journal.pone.0075913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai, J. , Rao, C. , Watt, J. , Sun, X. , Lin, C. , Zhang, L. , & Liu, J. (2019). Mycobacterium tuberculosis 6C sRNA binds multiple mRNA targets via C‐rich loops independent of RNA chaperones. Nucleic Acids Research, 47, 4292–4307. 10.1093/nar/gkz149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, D. H. (2006) RNA secondary structure analysis using RNAstructure. Current protocols in bioinformatics, 13, 12.6.1–12.6.14. 10.1002/0471250953.bi1206s13 [DOI] [PubMed] [Google Scholar]
- Miotto, P. , Forti, F. , Ambrosi, A. , Pellin, D. , Veiga, D. F. , Balazsi, G. , … Cirillo, D. M. (2012). Genome‐wide discovery of small RNAs in Mycobacterium tuberculosis . PLoS ONE, 7, e51950 10.1371/journal.pone.0051950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, A. , Angamuthu, K. , & Nagaraja, V. (2008). Genome‐wide analysis of the intrinsic terminators of transcription across the genus Mycobacterium. Tuberculosis (Edinb), 88, 566–575. 10.1016/j.tube.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Mooney, R. A. , & Landick, R. (2013). Building a better stop sign: Understanding the signals that terminate transcription. Nature Methods, 10, 618–619. 10.1038/nmeth.2527 [DOI] [PubMed] [Google Scholar]
- Moores, A. , Riesco, A. B. , Schwenk, S. , & Arnvig, K. B. (2017). Expression, maturation and turnover of DrrS, an unusually stable, DosR regulated small RNA in Mycobacterium tuberculosis . PLoS ONE, 12, e0174079 10.1371/journal.pone.0174079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, T. , Ueda, M. , Kubo, K. , & Aiba, H. (2015). Insights into transcription termination of Hfq‐binding sRNAs of Escherichia coli and characterization of readthrough products. RNA, 21, 1490–1501. 10.1261/rna.051870.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mvubu, N. E. , Pillay, B. , Gamieldien, J. , Bishai, W. , & Pillay, M. (2016). Canonical pathways, networks and transcriptional factor regulation by clinical strains of Mycobacterium tuberculosis in pulmonary alveolar epithelial cells. Tuberculosis (Edinb), 97, 73–85. 10.1016/j.tube.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Namouchi, A. , Gomez‐Munoz, M. , Frye, S. A. , Moen, L. V. , Rognes, T. , Tonjum, T. , & Balasingham, S. V. (2016). The Mycobacterium tuberculosis transcriptional landscape under genotoxic stress. BMC Genomics, 17, 791 10.1186/s12864-016-3132-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak, M. , & Storz, G. (2017). ProQ/FinO‐domain proteins: Another ubiquitous family of RNA matchmakers? Molecular Microbiology, 104, 905–915. 10.1111/mmi.13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelly, S. , Bishai, W. R. , & Lamichhane, G. (2012). A screen for non‐coding RNA in Mycobacterium tuberculosis reveals a cAMP‐responsive RNA that is expressed during infection. Gene, 500, 85–92. 10.1016/j.gene.2012.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, J. M. , Mooney, R. A. , Kuan, P. F. , Rowland, J. L. , Keles, S. , & Landick, R. (2009). Rho directs widespread termination of intragenic and stable RNA transcription. Proceedings of the National Academy of Sciences of the United States of America, 106, 15406–15411. 10.1073/pnas.0903846106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha, A. , McCue, L. A. , & McDonough, K. A. (2002). Identification of a Mycobacterium tuberculosis putative classical nitroreductase gene whose expression Is coregulated with that of the acr gene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infection and Immunity, 70, 1518–1529. 10.1128/IAI.70.3.1518-1529.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi, M. , Espeli, O. , Schwartz, A. , Cayrol, B. , Rahmouni, A. R. , Arluison, V. , & Boudvillain, M. (2011). The Sm‐like RNA chaperone Hfq mediates transcription antitermination at Rho‐dependent terminators. EMBO Journal, 30, 2805–2816. 10.1038/emboj.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray‐Soni, A. , Bellecourt, M. J. , & Landick, R. (2016). Mechanisms of bacterial transcription termination: All good things must end. Annual Review of Biochemistry, 85, 319–347. 10.1146/annurev-biochem-060815-014844 [DOI] [PubMed] [Google Scholar]
- Regnier, P. , & Hajnsdorf, E. (2013). The interplay of Hfq, poly(A) polymerase I and exoribonucleases at the 3' ends of RNAs resulting from Rho‐independent termination: A tentative model. RNA Biology, 10, 602–609. 10.4161/rna.23664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra, P. , Prajapati, R. K. , Banerjee, R. , Sengupta, S. , & Mukhopadhyay, J. (2015). Novel mechanism of gene regulation: The protein Rv1222 of Mycobacterium tuberculosis inhibits transcription by anchoring the RNA polymerase onto DNA. Nucleic Acids Research, 43, 5855–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo, T. J. , & Artsimovitch, I. (2011). Termination and antitermination: RNA polymerase runs a stop sign. Nature Reviews Microbiology, 9, 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti, C. M. , Boyd, D. H. , & Rubin, E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Molecular Microbiology, 48, 77–84. 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- Sassetti, C. M. , & Rubin, E. J. (2003). Genetic requirements for mycobacterial survival during infection. Proceedings of the National Academy of Sciences of the United States of America, 100, 12989–12994. 10.1073/pnas.2134250100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K. , Hamada, M. , Asai, K. , & Mituyama, T. (2009). CENTROIDFOLD: A web server for RNA secondary structure prediction. Nucleic Acids Research, 37, W277–280. 10.1093/nar/gkp367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, E. , & Weichenrieder, O. (2011). Structural basis for RNA 3'‐end recognition by Hfq. Proceedings of the National Academy of Sciences of the United States of America, 108, 13065–13070. 10.1073/pnas.1103420108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, W. B. , & Lewis, C. W. Jr (1965). Effect of oleic acid on growth and cell structure of mycobacteria. Journal of Bacteriology, 90, 1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger, D. , Ehrt, S. , Voskuil, M. I. , Liu, Y. , Mangan, J. A. , Monahan, I. M. , … Schoolnik, G. K. (2003). Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. Journal of Experimental Medicine, 198, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, O. T. , Ludwig, C. , Kogadeeva, M. , Zimmermann, M. , Rosenberger, G. , Gengenbacher, M. , … Aebersold, R. (2015). Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis . Cell Host & Microbe, 18, 96–108. 10.1016/j.chom.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Shi, L. , Sohaskey, C. D. , Kana, B. D. , Dawes, S. , North, R. J. , Mizrahi, V. , & Gennaro, M. L. (2005). Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proceedings of the National Academy of Sciences of the United States of America, 102, 15629–15634. 10.1073/pnas.0507850102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, S. , & Ehrt, S. (2006). Dihydrolipoamide acyltransferase is critical for Mycobacterium tuberculosis pathogenesis. Infection and Immunity, 74, 56–63. 10.1128/IAI.74.1.56-63.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett, P. R. , Taylor‐McCabe, K. J. , Michalczyk, R. , “Pete” Silks, L. A. , & Gupta, G. (2003). Structural studies on the hairpins at the 3′ untranslated region of an anthrax toxin gene. Biochemistry, 42, 6078–6089. [DOI] [PubMed] [Google Scholar]
- Solans, L. , Gonzalo‐Asensio, J. , Sala, C. , Benjak, A. , Uplekar, S. , Rougemont, J. , … Cole, S. T. (2014). The PhoP‐dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis . PLoS Path, 10, e1004183 10.1371/journal.ppat.1004183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding, M. D. , & Prigge, S. T. (2010). Lipoic acid metabolism in microbial pathogens. Microbiology and Molecular Biology Reviews, 74, 200–228. 10.1128/MMBR.00008-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverniti, V. , Forti, F. , Ghisotti, D. , & Putzer, H. (2011). Mycobacterium smegmatis RNase J is a 5′‐3′ exo‐/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Molecular Microbiology, 82, 1260–1276. 10.1111/j.1365-2958.2011.07888.x [DOI] [PubMed] [Google Scholar]
- Tchigvintsev, A. , Tchigvintsev, D. , Flick, R. , Popovic, A. , Dong, A. , Xu, X. , … Yakunin, A. F. (2013). Biochemical and structural studies of conserved Maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides. Chemistry & Biology, 20, 1386–1398. 10.1016/j.chembiol.2013.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden, B. (2008). TargetRNA: A tool for predicting targets of small RNA action in bacteria. Nucleic Acids Research, 36, W109–W113. 10.1093/nar/gkn264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C. H. , Baranowski, C. , Livny, J. , McDonough, K. A. , Wade, J. T. , & Contreras, L. M. (2013). Identification of novel sRNAs in mycobacterial species. PLoS ONE, 8, e79411 10.1371/journal.pone.0079411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uson, M. L. , Ordonez, H. , & Shuman, S. (2015). Mycobacterium smegmatis HelY Is an RNA‐activated ATPase/dATPase and 3'‐to‐5' helicase that unwinds 3'‐tailed RNA duplexes and RNA:DNA hybrids. Journal of Bacteriology, 197, 3057–3065. 10.1128/JB.00418-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandal, O. H. , Pierini, L. M. , Schnappinger, D. , Nathan, C. F. , & Ehrt, S. (2008). A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis . Nature Medicine, 14, 849–854. 10.1038/nm.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudeva‐Rao, H. M. , & McDonough, K. A. (2008). Expression of the Mycobacterium tuberculosis acr‐coregulated genes from the DevR (DosR) regulon is controlled by multiple levels of regulation. Infection and Immunity, 76, 2478–2489. 10.1128/IAI.01443-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal, A. , Bryk, R. , Shi, S. , Rhee, K. , Rath, P. , Schnappinger, D. , … Nathan, C. (2011). Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host & Microbe, 9, 21–31. 10.1016/j.chom.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, L. S. , & Storz, G. (2009). Regulatory RNAs in bacteria. Cell, 136, 615–628. 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2018). Global tuberculosis report 2018. Geneva: World Health Organization 2018. [Google Scholar]
- Zadeh, J. N. , Steenberg, C. D. , Bois, J. S. , Wolfe, B. R. , Pierce, M. B. , Khan, A. R. , … Pierce, N. A. (2011). NUPACK: Analysis and design of nucleic acid systems. Journal of Computational Chemistry, 32, 170–173. 10.1002/jcc.21596 [DOI] [PubMed] [Google Scholar]
- Zeller, M. E. , Csanadi, A. , Miczak, A. , Rose, T. , Bizebard, T. , & Kaberdin, V. R. (2007). Quaternary structure and biochemical properties of mycobacterial RNase E/G. The Biochemical Journal, 403, 207–215. 10.1042/BJ20061530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. , Phadtare, S. , Nariya, H. , Ouyang, M. , Husson, R. N. , & Inouye, M. (2008). The mRNA interferases, MazF‐mt3 and MazF‐mt7 from Mycobacterium tuberculosis target unique pentad sequences in single‐stranded RNA. Molecular Microbiology, 69, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. , Zhang, Y. , Teh, J. S. , Zhang, J. , Connell, N. , Rubin, H. , & Inouye, M. (2006). Characterization of mRNA interferases from Mycobacterium tuberculosis . Journal of Biological Chemistry, 281, 18638–18643. [DOI] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research, 31, 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supinfo


