Synopsis
Today, it is widely accepted that proteins that lack highly defined, globular structures, 3D, termed “intrinsically disordered proteins (IDPs)”, play key roles in myriad biological processes. Our understanding of how intrinsic disorder mediates biological function is, however, incomplete. Here, we review disorder-mediated cell cycle regulation by two intrinsically disordered proteins, p21 and p27. A structural adaptation mechanism involving a stretchable, dynamic linker helix allows p21 to promiscuously recognize the various Cdk/cyclin complexes that regulate cell division. Disorder within p27 mediates transmission of an N-terminal tyrosine phosphorylation signal to a C-terminal threonine phosphorylation, constituting a signaling conduit. These mechanisms are mediated by folding upon binding p21/p27’s regulatory targets. However, residual disorder within the bound state contributes critically to these functional mechanisms. Our studies provide insights into how intrinsic protein disorder mediates regulatory processes and provide opportunities for designing drugs that target cancer-associated IDPs.
Keywords: Cell cycle regulator, intrinsically disordered proteins, p21, p27, tyrosine phosphorylation
INTRODUCTION
Over the past 15 years, there has been increasing awareness that globular 3D structure is not a prerequisite for protein function. While 3D protein structure is required for certain protein functions, another class of proteins lack stable secondary or ternary structure under physiological conditions and still perform important functions in cells. These proteins are termed intrinsically disordered proteins (IDPs). As the number of IDPs that play critical biological roles continues to grow, it has been proposed that the classic concept of structure-function relationships needs to be revised and extended to include IDPs 1,2. Despite broad recognition of the biological importance of IDPs, our understanding of disorder-mediated biological processes is incomplete. In this review, we provide a general introduction to IDPs and discuss how the intrinsic disorder of two prototypical IDPs, p21 and p27, mediate cell cycle regulation. These IDPs are of particular importance due to their critical roles in cell cycle regulation but also due to their potential as anticancer drug targets 3.
1. IDPs
1.1. Functional features of IDPs
IDPs are involved in various cellular functions, notably in processes involving signaling and regulation 4. The highly dynamic features of IDPs enable diverse functions in cells. Although detailed information about how intrinsic disorder of an individual protein is associated with its specific function is limited, several examples provide insight into “disorder-function relationship”.
Often, a single IDP performs multiple functions by associating with various targets to perform a specific function, which is mediated by its intrinsic disorder. For example, the cell cycle regulatory IDP, p21Cip1 (p21), binds several different cyclin-dependent kinase (Cdk)/cyclin complexes through structural adaptation to accommodate similar but topologically distinct binding sites 5.
Bioinformatics studies have shown that post-translational modifications (PTMs) predominantly occur within intrinsically disordered protein regions, which provides important regulatory mechanisms in cells 6,7. For example, the cell cycle regulatory IDP, p27Kip1 (p27), can switch the activity of Cdks from an inhibited state to an activated state through tyrosine phosphorylation 8.
Other biological processes mediated by intrinsic disorder are molecular movement and transport, as shown in, for example, motor proteins kinesin 9 and dynein 10, and FG-nups in the nuclear pore complex 11. Also, IDPs can serve as scaffolds for the assembly of multi-component, macromolecular complexes. As shown in several examples, axin 12, CBP 13 and BRCA1 14, multiple short interaction motifs in the disordered scaffold proteins 15 confer the ability to interact with and promote the co-assembly of many different partners.
1.2. Structural features of IDPs
IDPs have distinct primary structural features compared with those of globular proteins. Based on an analysis of the IDPs and intrinsically disordered regions deposited in the DisProt database 16, IDPs are primarily enriched in disorder-promoting residues (polar and/or charged amino acids: D, M, K, R, S, Q, P, and E) and are depleted in order-promoting residues (hydrophobic amino acids: C, W, Y, I, F, V, L, H, T, and N). Such distinct amino acid composition limits formation of highly defined protein structure by these proteins in isolation.
Although IDPs are highly disordered in solution, some retain partially populated secondary structure. For example, p21 and p27 in their free state do not exhibit completely random conformations but exhibit some degree of partially folded secondary structure, as evidenced by NMR spectroscopy, molecular dynamics (MD) simulations and circular dichroism (CD) spectroscopy 17–19.
Interestingly, in such many cases, nascent secondary structure in isolation is stabilized to form stable secondary structure upon binding to a target. This process is termed coupled folding and binding. In the case of p27, its partially populated secondary structure 18 is consistent with the secondary structure of its final bound state in p27/Cdk2/cyclin A crystal structure 20. This observation strongly suggests that the final bound state-like conformations, termed “intrinsically folded structural units” (IFSUs) when observed in isolation prior to binding 18, are populated and preferred for interaction with a binding partner. This is an example of conformational selection, one of two binding mechanism proposed to describe folding and binding by IDPs—conformational selection and induced folding—in which the target protein selects a conformation that is close to its bound state from the conformational ensemble of an IDP in its free state.
However, the partially populated secondary structure of an IDP does not always reflect the bound state. For the IDPs that adopt different conformations when bound to different partners, for example, the C-terminal region of p53 21, the secondary structure of the bound state and the intrinsic secondary structural propensity in the free state may differ. In this case, even if an IDP assumes a partially populated conformation consistent with binding to one of many targets, the generally flat energy landscape of IDPs enables other conformations to either be selected or induced to bind other targets. In the induced folding mechanism, an IDP binds to its target in a fully disordered state and is induced to fold upon binding to its target. For p27, only some regions exhibit IFSUs that suggests the conformational selection mechanism and other, highly disordered regions bind targets via the induced folding mechanism. Therefore, the p27 binding mechanism is likely a composite of the two extremes noted above and this complex behavior is likely exhibited in many other IDPs.
2. Functions of p21 and p27
2.1. p21 and p27 as complex regulators of cell cycle
Progression through the mammalian cell cycle is driven by the sequential activation of Cdk/cyclin complexes (Fig. 1A). Cdk activity is negatively regulated by the Cip/Kip protein family. The Cip/Kip family members, including p21, p27, and p57, associate with the full repertoire of Cdk/cyclin complexes and inhibit their kinase activities at the G1/S and G2/M checkpoints (Fig. 1A). The binding promiscuity of the Cip/Kip proteins is a functional advantage afforded by their disordered features.
Figure 1. The kinase-inhibitory domains (KIDs) of p21 and p27 regulate cell cycle progression .
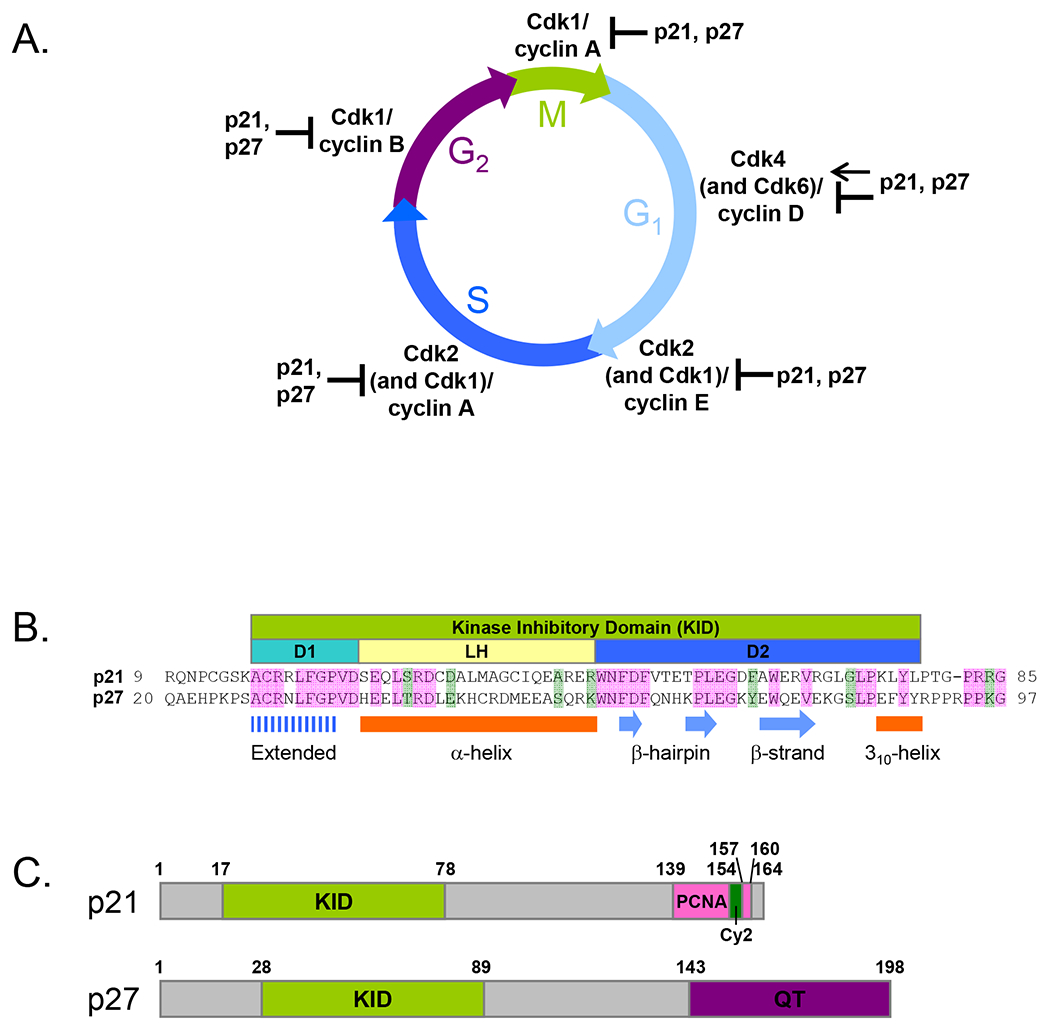
(A) p21 and p27 inhibit the activity of Cdk1(2)/cyclin E(A) and Cdk1/cyclin B(A) that are required for progression from G1 to S phase and for mitosis, respectively. During G1 phase, p21 and p27 mediate assembly and activation of Cdk4(6)/cyclin D in the cytoplasm, as well as inhibiting their activity in the nucleus. (B) Sequence alignment of the KIDs of p21 and p27. Residues identical in two sequences are shaded pink and similar residues are shaded green. The sub-domains are schematically represented as D1 (cyan), linker helix (LH) (light yellow), and D2 (blue). The secondary structure of p27-KID bound to Cdk2/cyclin A 20 is illustrated. (C) Domain organization in p21 and p27. The KID, PCNA-binding domain (PCNA), secondary cyclin–binding domain (Cy2), and QT domain (QT) are illustrated lime, pink, green, and purple, respectively.
Although p21 and p27 were first identified as negative regulators of the cell division cycle 22,23, later studies have demonstrated that they can positively regulate Cdk/cyclin complexes 8,24–28. Contrary to the nuclear roles of p21 and p27 in inhibiting Cdk/cyclin complexes, their cytoplasmic roles are to mediate the assembly and nuclear import of Cdk4(6)/cyclin D complexes 24–26,29. Furthermore, phosphorylation of p27 on Tyr 88 relieves the catalytic inhibition of Cdk2 8 and Cdk4 (6)27. As for p21, p21 phosphorylation on Tyr 77 also partially restores the kinase activity of Cdk2/cyclin A (M. Yoon, C. Park and R. Kriwacki, manuscript submitted), providing a mechanism to positively regulate Cdk2 activity. Moreover, it has been widely believed that the stoichiometry of p21 relative to Cdk/cyclin complexes determines whether a Cdk is active or not. At high levels of p21, Cdk2/cyclin A or Cdk4/cyclin D1 have been reported to be active even in the presence of p21 26,28.
Taken altogether, the mechanisms of regulation of Cdk activity by p21 and p27 are much more complicated than initially thought, depending on subcellular localization and tyrosine phosphorylation of p21 and p27, and the relative levels of p21 and Cdk/cyclin complexes.
2.2. Non-cell cycle functions of p21 and p27
In addition to their roles in cell cycle regulation, p21 and p27 are involved in other cellular functions, many of which are independent of Cdk/cyclin complexes. For example, p21 and p27 regulate transcription by directly binding to transcription factors, in addition to their ability to indirectly suppress transcription of cell cycle-related genes through inhibition of Cdk/cyclin complexes. p27 directly interacts with Neurogenin-2 (Ngn-2) via its N-terminal region and promotes transcription of its target genes, whereas p21 directly interacts with E2F 30, c-Myc31, and signal transducer and activator of transcription 3 (STAT3) 32 and inhibits their transcriptional activities.
Both p21 and p27 enhance cell migration by inhibiting the Rho/ROCK/LIMK/Cofilin signaling pathway 33 in the cytoplasm. These functions require phosphorylation on specific residues to enforce cytoplasmic localization (e.g., Thr157 34 or Thr198 35 on p27; Thr 145 on p21 36). Cytoplasmic p27 binds to RhoA and inhibits its activation by guanine-nucleotide exchange factors (GEFs), resulting in decreased actin stress fiber and focal-adhesion formation and subsequent increased cell migration 35. In contrast, cytoplasmic p21 binds to ROCK, a downstream target of RhoA, and inhibits its kinase activity, resulting in decreased actin stress fiber formation 37.
The unique C-terminal region of p21 directly associates with DNA polymerase δ processivity factor (PCNA) and inhibits its activity during DNA replication by blocking other DNA replication factors from binding PCNA 38. The C-terminal PCNA-binding region of p21 is overlapped with other interacting proteins, for example, the E7 oncoprotein of human papilloma virus 16 (HPV-16) 39 and c-Myc 31.
Interestingly, p21 has conflicting roles in apoptosis, having been demonstrated to both promote and inhibit programmed cell death. The pro-apoptotic activity of p21 has been attributed to both p53-dependent and p53-independent regulation of the apoptotic effector protein, Bax 40. On the contrary, cytoplasmic p21 protects cells against apoptosis both by directly binding to pro-apoptotic proteins, including procaspase 3, caspase 8, caspase 10, stress-activated protein kinases (SAPKs) and apoptosis signal-regulating kinase 1 (ASK1), and inhibiting their pro-apoptotic activities, and by suppressing the induction of pro-apoptotic genes by Myc and E2F1, thus inhibiting their transcription activity 41.
Other known p21 functions include regulation of DNA repair, senescence, cell differentiation, stem renewal and commitment 41–43. Furthermore, it is likely that other roles remain to be elucidated. It is remarkable that relatively small proteins such as p21 and p27 are entangled in such complex interaction networks. Their disordered features enable promiscuous protein-protein interactions within complex functional networks.
2.3. p21 and p27 as dual regulators of oncogenesis
p21 was first identified as a mediator of p53 tumor suppressor function by serving as a downstream effector of p53-dependent inhibition of cell cycle progression through inhibition of Cdk/cyclin complexes at the G1/S and G2/M phase transitions and PCNA during S phase. However, such anti-proliferative activity of p21 was challenged by the later finding that it can also serve as an oncogene both by promoting cell growth and by inhibiting apoptosis, as discussed above.
Now it appears that the oncogenic function of p21 is associated with its cytoplasmic localization. As discussed above, when p21 is phosphorylated on Thr 145 by Akt, which lies downstream of the anti-apoptotic signaling protein PI3K, p21 is localized to the cytoplasm 36. Subsequently, cytoplasmic p21 inhibits the activity of pro-apoptotic proteins, mediates the assembly and activation of Cdk4(6)/cyclin D, and increases cell migration, invasion and metastasis by inhibiting the kinase activity of ROCK.
Interestingly, cytoplasmic mislocalization is also associated with the oncogenic activity of p27. Phosphorylation of p27 on Thr 157, Ser 10, or Thr 198 leads to cytoplasmic localization of p27, resulting in proliferation of cancer cells 34, promoting migration of hepatocellular carcinoma cells 44, or RhoA-dependent promotion of cell migration 35, respectively.
Furthermore, Grimmler, et al. 8, discovered that Tyr 88 of p27 was phosphorylated by the BCR-ABL fusion oncoprotein in chronic myelogenous leukemia (CML) cells and that this modification was associated with p27 ubiquitination and 26S proteasome-dependent degradation. As will be discussed in more detail below, phosphorylation of p21 on Tyr 77 may also lead to oncogenesis through a similar mechanism.
Taken altogether, although p21 and p27 exhibit tumor-suppressing activity by inhibiting Cdk/cyclin complexes in the nucleus, they also show oncogenic activity by altered subcellular localization and degradation.
3. Disorder-mediated cell cycle regulation by p21 and p27
3.1. Primary structure of p21 and p27
The kinase-inhibitory function of p21 and p27 is mediated by a highly homologous N-terminal KID (Fig. 1B). The KID consists of 3 sub-domains, D1, linker helix (LH), and D2. Sub-domain D1, spanning 10 residues at the N-terminus of the KID, interacts with cyclin A through the RxL cyclin binding motif. The ~30 residues at the C-terminus of the KID, termed the D2 sub-domain, bind Cdk2 and inhibit its catalytic activity. The D1 and D2 domains are connected by sub-domain LH, which adopts a dynamic, helical conformation.
In contrast to conservation within the KIDs, the C-terminal domains of these proteins are more diverse, which is thought to mediate the unique aspects of their functions. The sequences of these proteins are rich in signaling and interaction motifs that encode diverse functionality within relatively short amino acid sequences. Different motifs are utilized differently in the three proteins, giving rise to their distinct biological roles.
3.2. Induced structure of p21 and p27 in the Cdk/cyclin-bound state
Highly disordered, isolated p27 19,45, is induced to fold upon binding to its target Cdk2/cyclin A 20. In 1996, Russo, et al., reported the crystal structure of the p27-KID/Cdk2/cyclin A ternary complex (Fig. 2A), providing structural insight into the mechanism by which p27 inhibits Cdk2 activity. Sub-domain D1 binds in an extended conformation on the surface of cyclin A, whereas sub-domain D2 forms a β-hairpin and an intermolecular β-sheet on the surface of the N-terminal lobe of Cdk2. Sub-domain LH forms a 22 residue-long α-helix, spanning the ~40 Å gap between Cdk2 and cyclin A. These structural features reveal that p27 inhibits Cdk2/cyclin A in three different ways. First, the RxL motif within sub-domain D1 blocks the substrate binding site on cyclin A. Second, sub-domain D2 displaces the first β-strand of Cdk2, remodeling the catalytic cleft. Finally, the inhibitory 310-helix occupies the ATP binding pocket of Cdk2.
Figure 2. p27-KID binds to Cdk2/Cyclin A through a two-step mechanism.
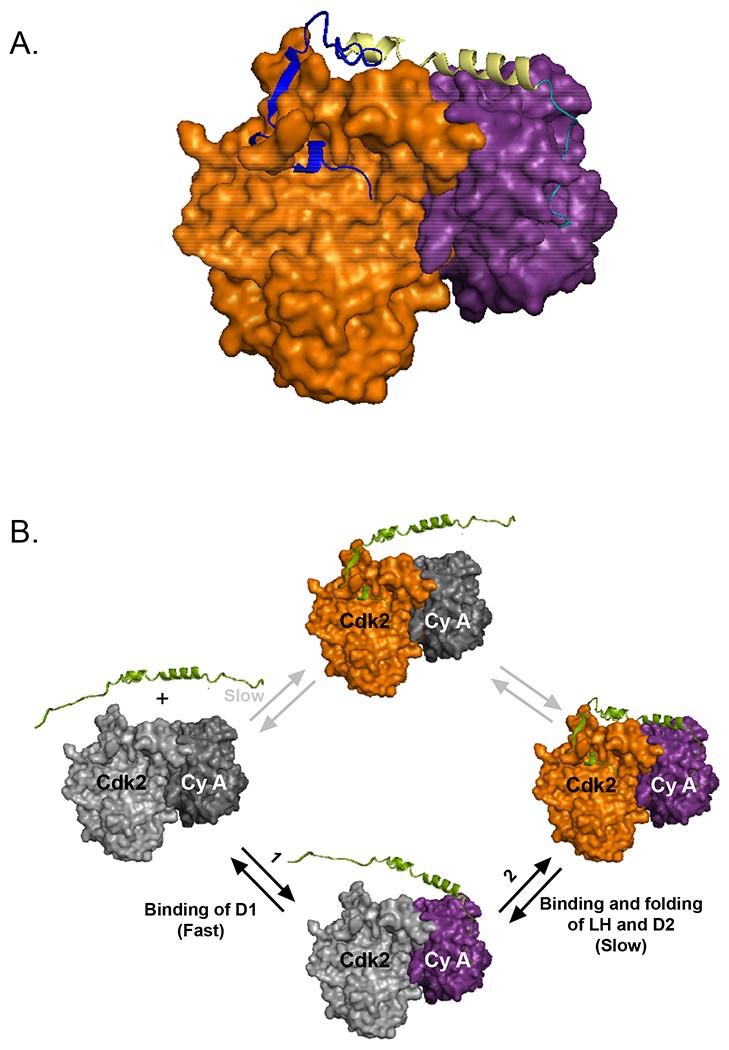
(A) Crystal structure of p27-KID in complex with Cdk2 (orange) and cyclin A (magenta); PDB accession number 1JSU. Sub-domain D1 (cyan) binds and inhibits substrate binding to cyclin A, while sub-domain D2 (blue) binds Cdk2, inserting a 310 helix into the ATP binding pocket of Cdk2. Sub-domain LH (light yellow) connects the other two sub-domains. (B) Sequential mechanism of p27-KID association with Cdk2/Cyclin A; first, the highly disordered and dynamic RxL motif within sub-domain D1 rapidly associates with cyclin A; next, sub-domain LH fully folds, positioning sub-domain D2 near Cdk2; finally, sub-domain extensively folds upon binding and remodeling Cdk2 (adapted and modified from Lacy, et al. 45). Cdk2 and Cyclin A are colored as in panel A.
The NMR secondary 13Cα chemical shift values of p21-KID bound to Cdk2/cyclin A indicate that the secondary structure of bound p21-KID is generally consistent with that of p27-KID complexed with Cdk2/cyclin A in the crystal structure 5. This suggests that the sequence similarity between p21- and p27-KID leads to structural similarity in the Cdk/cyclin-bound state, and indicates that the two disordered proteins have similar intrinsic abilities to experience induced folding.
3.3. Nascent secondary structure of p21 and p27 in their free states
Despite being highly disordered, p21 and p27 do not completely lack secondary structure and exhibit partially folded secondary structure in their free states. CD spectra of p27 19 and p21 17 indicate a partially formed α-helix, within the LH sub-domain, as evidenced by the 13Cα secondary chemical shift values 45 (For p21, M. Yoon, C. Park and R. Kriwacki, unpublished results). In contrast to the nascent helical conformation of the LH sub-domain, only sub-domain D1 exhibits negative {1H}-15N heteronuclear NOE (HetNOE) values consistent with a high degree of flexibility 45. Interestingly, this nascent secondary structure of p27 45 is similar to that of p27-KID bound to Cdk2/cyclin A, suggesting that, at least for the motionally restricted regions of p27, the conformational selection mechanism provides a thermodynamic advantage by reducing the entropic penalty associated with folding upon binding. Furthermore, the 13Cα and 13C’ secondary chemical shift analyses of free p21-KID (M. Yoon, C. Park, and R. Kriwacki, unpublished results) demonstrate that the nascent secondary structure of p21-KID is generally consistent with that of its Cdk2/cyclin A-bound structure 5, again suggesting a significant role for the conformational selection mechanism in this coupled folding and binding process. Taken together, p21 and p27 may derive thermodynamic advantage by assuming highly populated conformations, even before associating with their targets, that are similar to their conformations when bound to Cdk2/cyclin A.
3.4. Sequential binding mechanism of p27
Lacy, et al., provided insights into the mechanism associated with the binding of p27-KID to the Cdk2/cyclin A complex in terms of thermodynamics and kinetics, using isothermal titration calorimetry (ITC) 45 and surface plasmon resonance (SPR) 46. In the sequential binding mechanism identified in these studies (Fig. 2B), the highly dynamic D1 sub-domain rapidly binds to cyclin A though its RxL cyclin binding motif, and then LH sub-domain folds into an α-helix, finally followed by folding of three IFSUs, a β-hairpin, an intermolecular β-strand and a 310-helix within the D2 sub-domain. Rapid association of D1 sub-domain with cyclin A may be due to the large capture radius of disordered p27 through the “fly casting” mechanism 47. This fast initial step is followed by the slow association of less flexible, D2 sub-domain with the surface of Cdk2. Furthermore, this sequential binding mechanism, which combines induced fit and conformational selection mechanism, is also observed for binding to the Cdk4/cyclin D1 complex (L. Ou, B. Waddell, R. Kriwacki, unpublished results).
3.5. Binding promiscuity mechanism of p21
Recently, Wang, et al., reported the structural mechanism by which p21 promiscuously binds to various Cdk/cyclin complexes 5 (Fig. 3). Unlike the D1 and D2 sub-domains, the LH sub-domain of p21 exhibits sequence variation among Cip/Kip family members and highly dynamic features, as evidenced by analysis of HetNOE values. Inspection of B-factors for this region of p27 in the complex crystal structure revealed increased disorder, in contrast to a high degree of order observed for sub-domains D1 and D2 20. Furthermore, in this crystal structure, the LH sub-domain of p27 exhibited a conformation corresponding to a stretched α-helix, elongated over its 22 residue length by about 4 Å. Finally, Wang, et al. 5, deduced that helix stretching was responsible for the dynamic features of p21 when bound to Cdk2/cyclin A, and that this “structural adaptation” by both p21 and p27 was associated with their ability to promiscuously bind to the entire Cdk/cyclin repertoire.
Figure 3. The stretchable linker helix of p21 mediates promiscuous binding to the various Cdk/cyclin complexes.
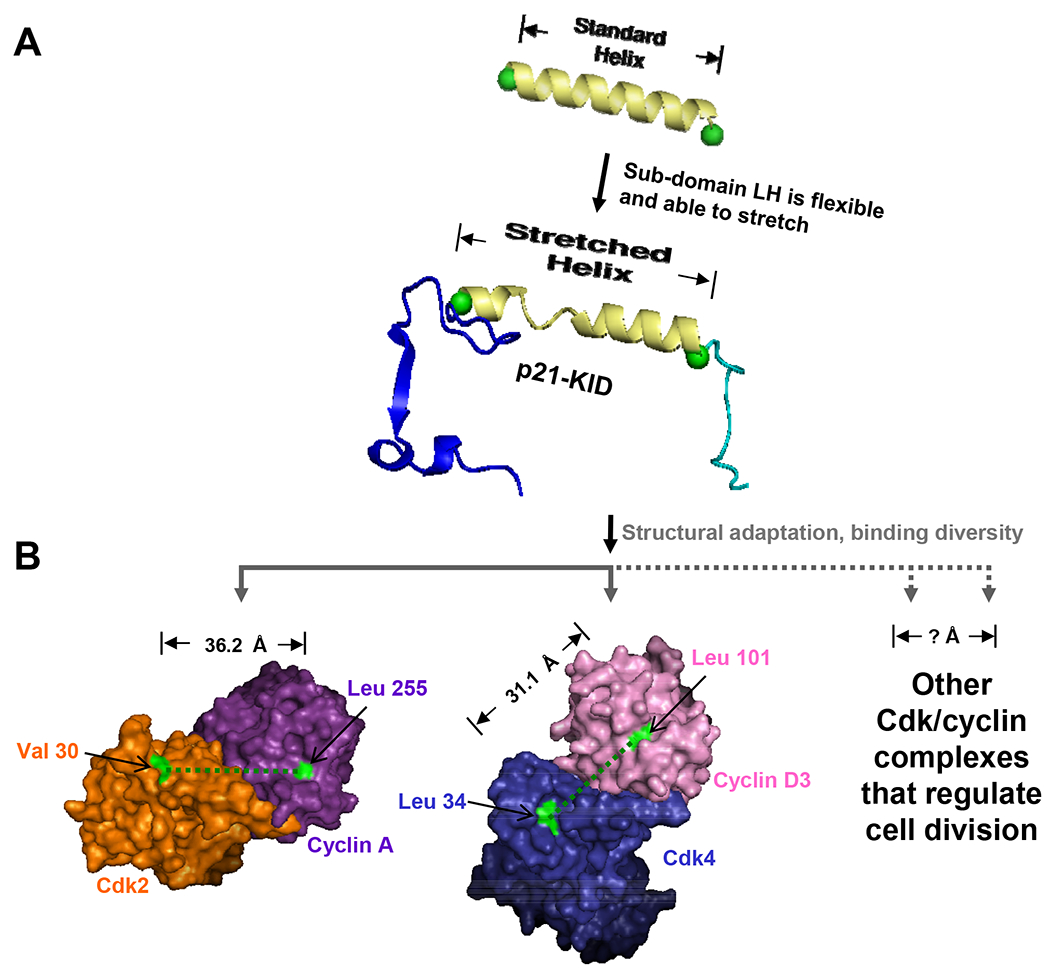
(A) Comparison of a standard α-helix, 22 residues in length, with the LH sub-domain of p21 that structurally adapts through helix stretching to accommodate binding to Cdk2/cyclin A. (B) The binding of p21 to different Cdk/cyclin complexes requires structural adaptation by the LH sub-domain through helix stretching/contraction and pivoting; the distance between conserved features of the p21 binding sites within the Cdk and cyclin subunits are illustrated for Cdk2/cyclin A (orange/magenta) and Cdk4/cyclin D3 (blue/pink). The locations of and distance between Val 30 and Leu 255, and Leu 34 and Leu 101, respectively, within these two complexes are illustrated. (adapted and modified from Wang, et al. 5).
Experiments with p21 variants with different length LH sub-domains further confirmed this hypothesis. Also, the crystal structures of Cdk4/cyclin D1 48 and Cdk4/cyclin D3 49 revealed that the LH sub-domain would have to contract and pivot in order for the D1 and D2 sub-domains to bind conserved surfaces on the D-type cyclins and Cdk4, respectively (Figure 3). The lack of tertiary contacts between the different sub-domains of the p21 and p27 KIDs creates a flat energy landscape that enables structural adaptation while folding upon binding to Cdk/cyclin complexes.
3.6. p21 and p27 as flexible signaling conduits
It has been well known that the Ser/Thr phosphorylation of p21 and p27 regulate their stability, activity, and subcellular localization. For example, phosphorylation of p27 Thr 187 50 or p21 Ser 130 51,52 by the Cdk2/cyclin E (A) complexes creates phosphodegrons, which are required for recognition by the SCFSkp2 E3 ubiquitin ligase complex for ubiquitination and subsequent proteasomal degradation of p21 and p27. Interestingly, Cdk2 phosphorylates p21 or p27 when bound to these so-called inhibitors. In other words, p21 or p27 serves as both an inhibitor and a substrate of Cdk2. This paradox was resolved by a previous study that demonstrated a molecular mechanism by which the activity and stability of p27 is regulated 8. In the two step phosphorylation mechanism (Fig. 4), p27 can be phosphorylated on Tyr 88 within the inhibitory 310-helix by Abl and Lyn when bound to Cdk2/cyclin A, leading to the restoration of Cdk2 activity by ejecting the 310-inhibitory helix from the active site of Cdk2. Subsequently, this reactivated Cdk2 phosphorylates p27 on Thr187 in the highly flexible C-terminal tail of p27 53 within the same ternary complex via a pseudo-unimolecular mechanism, resulting in the ubiquitination by SCFSkp2 E3 ligase and subsequent degradation.
Figure 4. p27 serves as a signaling conduit that controls cell cycle progression.
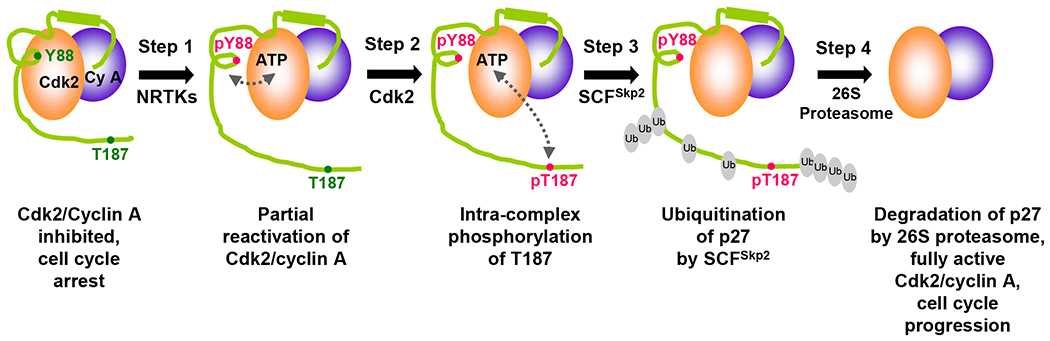
Cell cycle is arrested by p27 inhibition of Cdk/cyclin complexes. The inhibitory effect is released through phosphorylation of p27 at Tyr 88 by NRTKs (Step 1). The now liberated kinase active site is able to phosphorylate p27 at Thr 187 (Step 2), which in turn recruits the SCFSkp2 ubiquitin ligase complex (Step 3). Poly-ubiquitinated p27 is finally degraded by the 26S proteasome (Step 4). Note that the ubiquitination pattern shown in the figure is solely for illustrative purposes. Figure adapted and modified from Galea, et al.57.
Furthermore, we reasoned that, due to the functional and sequence similarities between p21 and p27, the same two-step phosphorylation mechanism may be applied also for p21. Indeed, there are critical Tyr and Ser/Thr residues that can participate in this signaling cascade in p21, Tyr 77 and Ser 130, equivalent to Tyr 88 and Thr 187 in p27, respectively. Also, the intrinsic disorder within the CTD of p21 54 may allow p21 to conduct the Tyr 77 signal within the KID to Ser 130 signal within the CTD, as observed for p27. Furthermore, as discussed above, phosphorylation of Ser 130 has been reported to be a signal for p21 ubiquitination and 26S proteosomal degradation 51,52. As expected, our biochemical and NMR studies show that p21 can be phosphorylated on Tyr77 and that the Tyr77-phosphorylated p21 couples the signal for Ser 130 phosphorylation by partially restoring the Cdk2 activity (M. Yoon, C. Park and R. Kriwacki, manuscript submitted).
This molecular mechanism illustrates how the intrinsic disorder of p21 and p27 allows them to function as flexible signaling conduits, enabling transmission of an intramolecular N-terminal Tyr phosphorylation signal to a C-terminal Ser/Thr phsophorylation site, followed by inter-molecular ubiquitination signaling.
CONCLUDING REMARKS
We have reviewed how the intrinsic disorder of p21 and p27 mediates cell cycle regulation and have highlighted functional advantages associated with disorder. p21 promiscuously binds to various Cdk/cyclin complexes through a helix stretching, structural adaptation mechanism. Also, p27 serves as a flexible signaling conduit by transmitting tyrosine phosphorylation signals to a threonine degradation signal through the protein, with this mechanism facilitated by flexibility and disorder. Although these studies provide molecular insights into disorder-function relationship for p21 and p27 in cell cycle regulation, it remains to be elucidated how disorder mediates other cellular functions, for example, transcriptional regulation and control of apoptosis. Also, it would be interesting to address how p21 or p27 utilizes the same (as for cell cycle regulation) or different disordered regions to mediate their various functions and to compare these new functional mechanisms with those known to mediate cell cycle regulation.
More complex than initially found, p21 and p27 serve not only as negative regulators of cell cycle but also as assembly factors for and activators of several Cdk/cyclin complexes. This pro-proliferation effect for p21, due to the positive regulation of Cdk/cyclin complexes together with anti-apoptotic activity, makes p21 as an attractive therapeutic target in many cancers 55. We believe that the aforementioned studies involving disorder-mediated cell cycle regulation provide a structural basis for the design of anti-cancer drugs that target p21 or p27. Additionally, it may be possible to utilize the concepts associated with disorder-function relationships that have emerged from studies of p21 and p27 to develop molecules that target Cdk/cyclin complexes; such molecules hold promise against cancer as the Cdks are valid anti-cancer drug targets 56. The highly specific, potent peptide-based drug inhibiting Cdk activity may be designed using the intrinsically disordered features of p21 and p27. We believe that this “disorder-based drug design” will open a new door for designing and developing anti-cancer drugs, as more intrinsic disorder is found in cancer-associated proteins 4.
Abbreviations:
- CD
circular dichroism
- Cdk
cyclin-dependent kinase
- CTD
C-terminal domain
- GEFs
guanine-nucleotide exchange factors
- HetNOE
heteronuclear Overhauser effect
- HPV-16
human papilloma virus 16
- IDPs
intrinsically disordered proteins
- IFSUs
intrinsically folded structural units
- ITC
isothermal titration calorimetry
- KID
kinase-inhibitory domain
- LH
linker helix
- NLS
nuclear localization signal
- NMR
nuclear magnetic resonance
- NRTK
non-receptor tyrosine kinase
- PCNA
DNA polymerase δ processivity factor
Contributor Information
MI-KYUNG YOON, St. Jude Children’s Research Hospital, Department of Structural Biology, 262 Danny Thomas Place, Memphis TN, 38105, USA.
DIANA M. MITREA, St. Jude Children’s Research Hospital, Department of Structural Biology, 262 Danny Thomas Place, Memphis TN, 38105, USA
LI OU, St. Jude Children’s Research Hospital, Department of Structural Biology, 262 Danny Thomas Place, Memphis TN, 38105, USA.
RICHARD W. KRIWACKI, St. Jude Children’s Research Hospital, Department of Structural Biology, 262 Danny Thomas Place, Memphis TN, 38105, USA.
REFERENCES
- 1.Wright PE & Dyson HJ Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 293, 321–331, (1999). [DOI] [PubMed] [Google Scholar]
- 2.Chouard T Structural biology: Breaking the protein rules. Nature 471, 151–153, (2011). [DOI] [PubMed] [Google Scholar]
- 3.Funk JO & Galloway DA Inhibiting CDK inhibitors: new lessons from DNA tumor viruses. Trends Biochem Sci 23, 337–341, (1998). [DOI] [PubMed] [Google Scholar]
- 4.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z & Dunker AK Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol 323, 573–584, (2002). [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Fisher JC, Mathew R, Ou L, Otieno S, Sublet J, Xiao L, Chen J, Roussel MF & Kriwacki RW Intrinsic disorder mediates the diverse regulatory functions of the Cdk inhibitor p21. Nat Chem Biol 7, 214–221, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z & Dunker AK The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32, 1037–1049, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gnad F, Gunawardena J & Mann M PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 39, D253–260, (2011). 3013726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, Jakel H, Kullmann M, Kriwacki RW & Hengst L Cdk-Inhibitory Activity and Stability of p27(Kip1) Are Directly Regulated by Oncogenic Tyrosine Kinases. Cell 128, 269–280, (2007). [DOI] [PubMed] [Google Scholar]
- 9.Hyeon C & Onuchic JN Mechanical control of the directional stepping dynamics of the kinesin motor. Proc Natl Acad Sci U S A 104, 17382–17387, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan JL, Song Y & Barbar E Structural dynamics and multi-region interactions in dynein-dynactin recognition. J Biol Chem 2011, 39349–39359, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, Gopinathan A, Lau EY, Colvin ME, Uversky VN & Rexach MF A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics 9, 2205–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noutsou M, Duarte AM, Anvarian Z, Didenko T, Minde DP, Kuper I, de Ridder I, Oikonomou C, Friedler A, Boelens R, Rudiger SG & Maurice MM Critical scaffolding regions of the tumor suppressor Axin1 are natively unfolded. J Mol Biol 405, 773–786, (2011). [DOI] [PubMed] [Google Scholar]
- 13.Dyson HJ & Wright PE Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6, 197–208., (2005). [DOI] [PubMed] [Google Scholar]
- 14.Mark WY, Liao JC, Lu Y, Ayed A, Laister R, Szymczyna B, Chakrabartty A & Arrowsmith CH Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein-protein and protein-DNA interactions? J Mol Biol 345, 275–287, (2005). [DOI] [PubMed] [Google Scholar]
- 15.Davey NE, Trave G & Gibson TJ How viruses hijack cell regulation. Trends Biochem Sci 36, 159–169, (2011). [DOI] [PubMed] [Google Scholar]
- 16.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z & Dunker AK DisProt: the Database of Disordered Proteins. Nucleic Acids Res 35, D786–793, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kriwacki RW, Hengst L, Tennant L, Reed SI & Wright PE Structural studies of p21(waf1/cip1/sdi1) in the free and Cdk2-bound state: Conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. USA 93, 11504–11509, (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sivakolundu SG, Bashford D & Kriwacki RW Disordered p27(Kip1) Exhibits Intrinsic Structure Resembling the Cdk2/Cyclin A-bound Conformation. J. Mol. Biol 353, 1118–1128, (2005). [DOI] [PubMed] [Google Scholar]
- 19.Bienkiewicz EA, Adkins JN & Lumb KJ Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1). Biochem. 41, 752–759, (2002). [DOI] [PubMed] [Google Scholar]
- 20.Russo AA, Jeffrey PD, Patten AK, Massague J & Pavletich NP Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382, 325–331, (1996). [DOI] [PubMed] [Google Scholar]
- 21.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN & Dunker AK Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 9 Suppl 1, S1, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper JW, Adami GR, Wei N, Keyomarsi K & Elledge SJ The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816, (1993). [DOI] [PubMed] [Google Scholar]
- 23.Toyoshima H & Hunter T p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78, 67–74, (1994). [DOI] [PubMed] [Google Scholar]
- 24.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM & Sherr CJ The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J 18, 1571–1583., (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alt JR, Gladden AB & Diehl JA p21(Cip1) Promotes cyclin D1 nuclear accumulation via direct inhibition of nuclear export. J Biol Chem 277, 8517–8523, (2002). [DOI] [PubMed] [Google Scholar]
- 26.LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A & Harlow E New functional activities for the p21 family of Cdk inhibitors. Genes Dev. 11, 847–862, (1997). [DOI] [PubMed] [Google Scholar]
- 27.Blain SW Switching cyclin D-Cdk4 kinase activity on and off. Cell Cycle 7, 892–898, (2008). [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Hannon GJ & Beach D p21-copntaining cyclin kinases exist in both active and inactive states. Genes & Development 8, 1750–1758, (1994). [DOI] [PubMed] [Google Scholar]
- 29.Ou L, Waddell MB & Kriwacki RW Mechanism of Cell Cycle Entry Mediated by the Intrinsically Disordered Protein p27(Kip1). ACS Chem Biol, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delavaine L & La Thangue NB Control of E2F activity by p21Waf1/Cip1. Oncogene 18, 5381–5392, (1999). [DOI] [PubMed] [Google Scholar]
- 31.Kitaura H, Shinshi M, Uchikoshi Y, Ono T, Iguchi-Ariga SM & Ariga H Reciprocal regulation via protein-protein interaction between c-Myc and p21(cip1/waf1/sdi1) in DNA replication and transcription. J Biol Chem 275, 10477–10483, (2000). [DOI] [PubMed] [Google Scholar]
- 32.Coqueret O & Gascan H Functional interaction of STAT3 transcription factor with the cell cycle inhibitor p21WAF1/CIP1/SDI1. J Biol Chem 275, 18794–18800, (2000). [DOI] [PubMed] [Google Scholar]
- 33.Besson A, Gurian-West M, Schmidt A, Hall A & Roberts JM p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev 18, 862–876. Epub 2004 Apr 2012, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang J, Zubovitz J, Petrocelli T, Kotchetkov R, Connor MK, Han K, Lee JH, Ciarallo S, Catzavelos C, Beniston R, Franssen E & Slingerland JM PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat. Med 8, 1153–1160., (2002). [DOI] [PubMed] [Google Scholar]
- 35.Larrea MD, Hong F, Wander SA, da Silva TG, Helfman D, Lannigan D, Smith JA & Slingerland JM RSK1 drives p27Kip1 phosphorylation at T198 to promote RhoA inhibition and increase cell motility. Proc Natl Acad Sci U S A 106, 9268–9273, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou BP, Liao Y, Xia W, Spohn B, Lee M-H & Hung M-C Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in Her-2/neu-overexpressing cells. Nat. Cell Biol 3, 245–252, (2001). [DOI] [PubMed] [Google Scholar]
- 37.Tanaka H, Yamashita T, Asada M, Mizutani S, Yoshikawa H & Tohyama M Cytoplasmic p21(Cip1/WAF1) regulates neurite remodeling by inhibiting Rho-kinase activity. J Cell Biol 158, 321–329., (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waga S, Hannon GJ, Beach D & Stillman B The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 369, 574–578, (1994). [DOI] [PubMed] [Google Scholar]
- 39.Funk JO, Waga S, Harry JB, Espling E, Stillman B & Galloway DA Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev 11, 2090–2100, (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghanem L & Steinman R A proapoptotic function of p21 in differentiating granulocytes. Leukemia research 29, 1315–1323, (2005). [DOI] [PubMed] [Google Scholar]
- 41.Dotto GP p21(WAF1/Cip1): more than a break to the cell cycle? Biochim Biophys Acta 1471, M43–56, (2000). [DOI] [PubMed] [Google Scholar]
- 42.Abbas T & Dutta A p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9, 400–414, (2009). Pmc2722839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Besson A, Dowdy SF & Roberts JM CDK inhibitors: cell cycle regulators and beyond. Dev Cell 14, 159–169, (2008). [DOI] [PubMed] [Google Scholar]
- 44.McAllister SS, Becker-Hapak M, Pintucci G, Pagano M & Dowdy SF Novel p27(kip1) C-terminal scatter domain mediates Rac-dependent cell migration independent of cell cycle arrest functions. Mol Cell Biol 23, 216–228, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L & Kriwacki RW p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat. Struct. Mol. Biol 11, 358–364, (2004). [DOI] [PubMed] [Google Scholar]
- 46.Lacy ER, Wang Y, Post J, Nourse A, Webb W, Mapelli M, Musacchio A, Siuzdak G & Kriwacki RW Molecular Basis for the Specificity of p27 Toward Cyclin-dependent Kinases that Regulate Cell Division. J. Mol. Biol 349, 764–773, (2005). [DOI] [PubMed] [Google Scholar]
- 47.Shoemaker BA, Portman JJ & Wolynes PG Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci U S A 97, 8868–8873., (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Day PJ, Cleasby A, Tickle IJ, O’Reilly M, Coyle JE, Holding FP, McMenamin RL, Yon J, Chopra R, Lengauer C & Jhoti H Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci U S A 106, 4166–4170, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takaki T, Echalier A, Brown NR, Hunt T, Endicott JA & Noble ME The structure of CDK4/cyclin D3 has implications for models of CDK activation. Proc Natl Acad Sci U S A 106, 4171–4176, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vlach J, Hennecke S & Amati B Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J 16, 5334–5344, (1997). 1170165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu H, Nie L & Maki CG Cdk2-dependent Inhibition of p21 stability via a C-terminal cyclin-binding motif. J Biol Chem 280, 29282–29288, (2005). [DOI] [PubMed] [Google Scholar]
- 52.Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M & Hershko A Role of the SCFSkp2 Ubiquitin Ligase in the Degradation of p21Cip1 in S Phase. J. Biol. Chem 278, 25752–25757, (2003). [DOI] [PubMed] [Google Scholar]
- 53.Galea CA, Nourse A, Wang Y, Sivakolundu SG, Heller WT & Kriwacki RW Role of intrinsic flexibility in signal transduction mediated by the cell cycle regulator, p27 Kip1. J Mol Biol 376, 827–838, (2008). 2350195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon MK, Venkatachalam V, Huang A, Choi BS, Stultz CM & Chou JJ Residual structure within the disordered C-terminal segment of p21(Waf1/Cip1/Sdi1) and its implications for molecular recognition. Protein Sci 18, 337–347, (2009). 2708053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss RH p21Waf1/Cip1 as a therapeutic target in breast and other cancers. Cancer Cell 4, 425–429, (2003). [DOI] [PubMed] [Google Scholar]
- 56.Collins I & Garrett MD Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr Opin Pharmacol 5, 366–373, (2005). [DOI] [PubMed] [Google Scholar]
- 57.Galea CA, Wang Y, Sivakolundu SG & Kriwacki RW Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry 47, 7598–7609, (2008). 2580775. [DOI] [PMC free article] [PubMed] [Google Scholar]


