Abstract
Sclerotinia stem rot is an economically important disease of canola (Brassica napus) and is caused by the fungal pathogen Sclerotinia sclerotiorum. This study evaluated the differential gene expression patterns of S. sclerotiorum during disease development on two canola lines differing in susceptibility to this pathogen. Sequencing of the mRNA libraries derived from inoculated petioles and mycelium grown on liquid medium generated approximately 164 million Illumina reads, including 95 million 75-bp-single reads, and 69 million 50-bp-paired end reads. Overall, 36% of the quality filter-passed reads were mapped to the S. sclerotiorum reference genome. On the susceptible line, 1301 and 1214 S. sclerotiorum genes were differentially expressed at early (8–16 hours post inoculation (hpi)) and late (24–48 hpi) infection stages, respectively, while on the resistant line, 1311 and 1335 genes were differentially expressed at these stages, respectively. Gene ontology (GO) categories associated with cell wall degradation, detoxification of host metabolites, peroxisome related activities like fatty acid ß-oxidation, glyoxylate cycle, oxidoreductase activity were significantly enriched in the up-regulated gene sets on both susceptible and resistant lines. Quantitative RT-PCR of six selected DEGs further validated the RNA-seq differential gene expression analysis. The regulation of effector genes involved in host defense suppression or evasion during the early infection stage, and the expression of effectors involved in host cell death in the late stage of infection provide supporting evidence for a two-phase infection model involving a brief biotrophic phase during early stages of infection. The findings from this study emphasize the role of peroxisome related pathways along with cell wall degradation and detoxification of host metabolites as the key mechanisms underlying pathogenesis of S. sclerotiorum on B. napus.
Introduction
Sclerotinia sclerotiorum (Lib.) de Bary, is a very efficient plant pathogen that affects a wide range of crops and is capable of infecting plant tissues above or below the soil surface. Diseases caused by this pathogen are favored by cool wet conditions [1]. In canola (Brassica napus L.), this pathogen is primarily responsible for causing Sclerotinia stem rot (SSR), a yield-reducing disease endemic to canola-producing areas worldwide. Each percent increase in SSR incidence can reduce potential canola yield by 0.5% [2]. Diseases caused by S. sclerotiorum are currently controlled by fungicides [3, 4], biological formulations [5, 6], and quantitative genetic disease resistance [7].
Understanding the molecular mechanisms employed by the pathogen during the infection process is essential for identifying novel targets for SSR management. As common with many broad-host range pathogens, the molecular aspects of S. sclerotiorum pathogenicity generally studied have concentrated on the roles of hydrolytic cell wall-degrading enzymes (CWDEs) [1, 8, 9]. S. sclerotiorum is known to produce several pectinases, including both endo- (Sspg1, Sspg3, Sspg5, and Sspg6) and exo- (Ssxpg1 and Ssxpg2) polygalacturonases [9]. Apart from polygalacturonases, gene disruption mutants of an arabinofuranosidase/β-xylosidase precursor (Ssaxp) and an endo-β-1, 4-xylanase (SsXyl1) showed either reduction or loss of virulence, indicating their importance as virulence factors [10, 11].
Oxalic acid (OA) has by far been the most studied S. sclerotiorum virulence factor to date [12, 13]. OA plays multiple roles in virulence of S. sclerotiorum, manipulating the host redox environment [13], inducing programmed cell death [14], detoxifying calcium, and mediating pH signaling [15, 16]. OA has long been considered an essential factor for pathogenicity of S. sclerotiorum. Recent studies using targeted mutants of the oxaloacetate acetylhydrolase gene (Ssoah1), responsible for biogenesis and accumulation of OA, showed that OA is required for virulence, but not essential for pathogenicity on all hosts [17, 18]. The creation of an acidic pH environment during host colonization rather than OA production per se has been suggested to be primary requirement for the colonization stage of pathogenesis [17].
Several studies have reported the role of secreted effector genes in pathogenicity or virulence of this pathogen. A small cysteine rich secreted cyanovirin-N-homology domain protein encoding gene SsCVNH [19], and a gene encoding small secreted hypothetical protein Ssv263 [20] were shown to be essential for full virulence of S. sclerotiorum. A compound appressorium formation related gene1 SsCaf1 [21] and an Rhs repeat-containing protein encoding gene Ss-Rhs1 [22] were shown to be required for host penetration and initial hyphal infection. Secreted effectors including an integrin-like protein SsITL [23], and a chorismate mutase SsCM1 [24] are known to suppress host resistance by interfering with jasmonic acid/ethylene signaling pathway and salicylic acid signaling pathways, respectively. Other secreted effectors, like the necrosis and ethylene-inducing peptides SsNep1 and SsNep2 [25], a small secreted virulence-related protein SsSSVP1 [26], and the cerato-platanin SsCP1 [27], are known to induce host cell death and necrosis.
Until recently, most of the reports involving S. sclerotiorum and canola, were limited to small scale EST studies [28, 29]; however, availability of the S. sclerotiorum genome sequence and the advances in sequencing technologies, particularly RNA-Seq have facilitated the study of global transcriptional changes occurring during pathogenesis [30, 31]. Transcriptome sequencing has been used to study the interaction of S. sclerotiorum with various crop hosts [19, 32–34] and recently with B. napus [35, 36] and B. oleracea [37]. In this study, for the first time we investigated the global transcriptional changes in S. sclerotiorum during infection of canola plants differing in their susceptibility to the pathogen.
In contrast to the widely accepted necrotrophic nature of the pathogen [1], a two-phase infection model involving a brief biotrophic or basic compatibility phase characterized by host resistance suppression and subverting of host defenses followed by a necrotrophic phase was proposed based on cytological and molecular and genomic evidences [38–40]. This brief initial phase appears to be partly facilitated by oxalic acid (OA) and by secreted effectors, which help S. sclerotiorum evade host recognition and suppress host defense signaling pathways [23, 24, 39]. The role of other effectors in avoiding host recognition and/or suppressing host defense responses is yet to be determined. We initiated this transcriptome sequencing study to gain better understanding of the molecular mechanisms underlying the interaction of S. sclerotiorum and canola and it provided insights into the temporal aspects of important mechanisms/pathways employed by S. sclerotiorum for successful infection of canola.
Materials and methods
Plant material and fungal strain
Two B. napus doubled haploid lines differing in their susceptibility to S. Sclerotiorum were used in this study. Both lines, NEP32 (susceptible) and NEP63 (resistant) were developed via microspore culture and were evaluated for their reaction to SSR [41]. The susceptible line NEP32 was derived from spring type canola variety Helga (PI649136), and the resistant line NEP63 was developed from a cross between Helga and winter type canola accession PI458940. When inoculated using the petiole inoculation technique [42], the susceptible line develops lesions on the stem that expand to >4 cm in length in length with 100% girdling within 8 days post inoculation, eventually leading to wilting and death. Within the same period, the average lesion size on the resistant line is limited to <1cm in length and <40% stem girdling, typically surrounded by purple margins suggesting accumulation of anthocyanins. The highly aggressive strain 1980 of S. sclerotiorum was used for inoculations and culture controls. Strain 1980 was chosen for this study as it is the genome sequenced strain [30, 31].
Plant growth and inoculation
The seeds of NEP32 and NEP63 were surface sterilized by soaking them in 3% NaOCl for one minute followed by immersion in 70% EtOH for one minute and were rinsed three times in sterilized deionized water for 1 minute. Seeds were planted in SunGro Sunshine mix #1 (Sun Gro Horticulture, MA) in 4 x 10 plastic plots with one seed per pot and plants were grown for 4 weeks in a growth chamber with a 16 h photoperiod, 21° C/16° C day/night temperature, and 60% relative humidity. Once germinated, seedlings were watered as necessary and fertilized once a week with 20-20-20 fertilizer. Four weeks after planting, plants were infected following an established petiole inoculation technique [42]. Briefly, the petiole of one leaf per plant was cut approximately 2.5 cm away from the stem and the stump was capped with PDA plugs containing hyphal tips of an actively growing 2 days-old S. sclerotiorum colony using 1 mL pipette tip. Inoculated petioles (≈ 2.5cm) were harvested at 8, 16, 24, and 48 hours post inoculation (hpi), flash frozen in liquid nitrogen and stored at -80° C until RNA extraction. Each treatment—time point consisted of three biological replicates; petioles collected from 5 individual plants were pooled to constitute one biological replicate.
RNA extraction, library preparation and sequencing
Total RNA was extracted from inoculated petioles using RNeasy mini kit, and mRNA was isolated using Oligotex mRNA mini kit (Qiagen Inc. Valencia, CA) following manufacturers’ instructions. cDNA libraries were constructed using NEBNext mRNA sample prep kit and NEBNext Multiplex Oligos kit (New England Biolabs, Ipswich, MA). Similar procedure was followed to construct cDNA libraries from S. sclerotiorum mycelia grown on PDB in 10cm petri plates for 48 h under the same conditions described above. For sequencing, RNA from 8 and 16 hpi were pooled and will be referred to as early infection stage (T1) henceforth. Similarly, RNA from 24 and 48 hpi were pooled and are referred to as late infection stage (T2). mRNA libraries were sequenced at the University of Minnesota Biomedical Genomics Center. Of the three biological replicates, two replicates per each treatment-time point were sequenced on Illumina GA-IIx platform (1X 76 bp Single Reads), while one replicate was sequenced on Illumina HiSeq 2500 platform (2X 50 bp Paired End).
Quality control and read mapping to the reference genome
Quality checks for the raw fastq files were conducted through a pipeline consisting of FastQC [43] and FastX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) to retain reads that were at least 50 bp long with a minimum quality score of 30. Reference genomes and the corresponding annotations for S. sclerotiorum and B. napus were downloaded from ensemble fungi (ftp://ftp.ensemblgenomes.org/pub/release-42/fungi/) and genoscope (http://www.genoscope.cns.fr/brassicanapus/data/), respectively. The most recent full genome sequence of S. sclerotiorum (Sclerotinia sclerotiorum_1980_uf_70_gca_001857865) [31] was used. Indexes for each reference genome were built and quality filter-passed reads were mapped to the reference genomes using HiSat2 [44]. Raw read counts mapped to each gene from the HiSat2 generated alignments were obtained using the featureCounts command [45] of the Subread package [46].
Differential gene expression analysis
Differential gene expression (DGE) analysis was conducted using edgeR package [47]. The raw count data were normalized with trimmed mean of means (TMM) normalization method implemented in edgeR [48]. Principal component analysis (PCA) was conducted to determine relatedness of the biological replicates. PCA plots were generated using scatterplot3d [49] package in R. Statistical analysis was performed using negative binomial distribution extended to generalized linear models [50]. Pairwise contrasts were performed following quasi-likelihood F tests [51]. A false discovery rate (FDR) cutoff of 0.05 was applied to account for multiple testing correction. A gene was considered as differentially expressed when the change in expression level was ≥ 2-fold (absolute value of log 2fold change (l2fc) ≥ 1), with an FDR-adjusted p-value < 0.05.
Functional classification and enrichment analyses of DEGs
Gene ontology (GO) enrichment analysis was performed in Blast2GO [52]. GO terms were assigned to S. sclerotiorum total genes list in Blast2GO, which were used as the background list for enrichment analysis. Fishers’ exact test implemented in Blast2GO was used to identify significantly enriched GO categories. A GO category was considered significantly enriched only when the p-value for that category was < 0.05 after applying FDR correction.
Differential gene expression data validation
The RNA-seq differential gene expression data was validated by performing qRT-PCR on six selected genes. Primers for qRT-PCR were designed using Primers-Blast [53]. QuantiTect reverse transcription kit (Qiagen Inc. Valencia, CA) was used to synthesize cDNA from total RNA. Real-time quantification was performed using two technical replications in a BioRad CFX96 Real-Time system using iTaq Universal SYBR Green Supermix with the following cycling conditions: 95° C for 30 s followed by 40 cycles of 95° C for 10 s and 60° C for 30 s. Expression levels of the DEGs were normalized against the S. sclerotiorum actin gene (sscle_14g099090) and the relative expression levels were calculated using the 2-ΔΔCt method.
Results
Disease development
There were no observable phenotypic differences between the susceptible and resistant canola lines at all sampling time points (Fig 1a). In both lines, necrotic lesions first appeared at point of inoculation (petiole tip) at 16 hpi and gradually spread along the length of the petiole by 48 hpi. Non-inoculated controls never showed necrotic lesion. In the susceptible line, by 72 hpi the lesions extended into stem. In the resistant line, the lesion growth was arrested at the nodes, surrounded by purple margin and did not extend into the stem. The differences in symptoms between the susceptible and resistant canola lines were clearly apparent four days post inoculation (Fig 1b).
Fig 1. Disease development on resistant and susceptible canola lines.
(a). Comparison of the appearance of representative inoculated petioles from both canola lines. The necrotic lesion spreads from the point of inoculation (i.e., petiole tip) along the length of the petiole. There are no apparent differences between NEP32 and NEP63 canola petioles. (b). Appearance of resistant (NEP 63) and susceptible (NEP32) canola four days post inoculation with S. sclerotiorum. Disease symptoms are more apparent in the susceptible line.
Sequencing and mapping
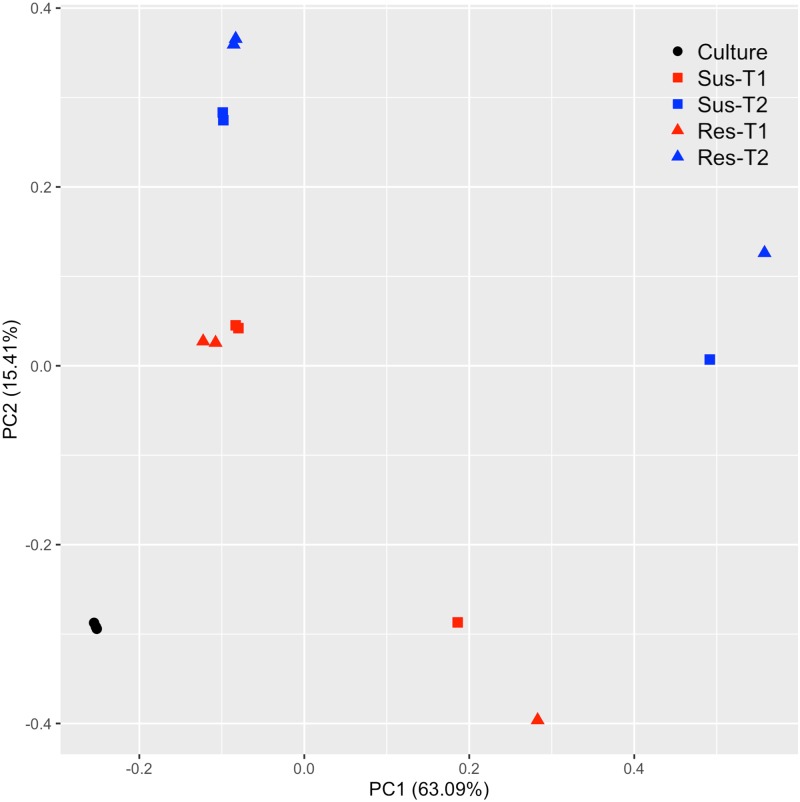
From the sequencing of in planta and in vitro cDNA libraries, approximately 164 million reads, comprising 95 million 76-bp-long reads, and 69 million 50-bp-long paired end reads were generated (Table 1). Approximately 87% of the reads from the in vitro libraries were mapped to the S. sclerotiorum genome. In contrast, 5.4–9.5% of the reads from the in planta libraries obtained from samples collected between 8 and 16 hpi, and approximately 38% of reads from the libraries obtained from samples collected between 24 and 48 hpi mapped to the reference S. sclerotiorum genome. On average, 0.6–1.5% of the reads mapped to S. sclerotiorum genome also mapped to B. napus genome, and these were excluded from the read counts for differential expression analysis. The percentage of reads from samples collected between 24 and 48 hpi that were mapped to the reference genome was 4 and 8 times larger than that of samples collected earlier for the susceptible and resistant lines, respectively. This difference was somewhat expected as it was apparent from the phenotypic observations that the fungal biomass at earlier stages was smaller compared to that at later stages (Fig 1a). Lower percentage of alignments at earlier phases of infection have also been observed in other pathogen-host interactions [34, 54, 55]. PCA results indicated that the differences in gene expression due to variability between replicates were small (9.15%, PC3, data not shown) compared to sample type (in vitro vs. in planta, PC1 63.1%) and time of sampling (T1 vs T2, PC2 15.4%) (Fig 2).
Table 1. Summary of the Illumina sequence reads obtained from Brassica napus plant inoculated with S. sclerotiorum and from mycelium of S. sclerotiorum isolate 1980 grown on potato dextrose agar.
| Time points | Total reads1 | Reads mapped to reference S. sclerotiorum genome (%) | |
|---|---|---|---|
| In Vitro | Culture | 41665593 | 36323396 (87.18) |
| NEP32 (Susceptible) | 8–16 hpi | 37771674 | 2021544 (5.35) |
| 24–48 hpi | 25360044 | 9642926 (38.02) | |
| NEP63 (Resistant) | 8–6 hpi | 28670528 | 2728735 (9.52) |
| 24–48 hpi | 31040632 | 11963775 (38.54) |
1 Number of reads represent total of three biological replications
Fig 2. Principal component analysis of transcriptome expression.
The PCA plot for RNA-Seq data shows the clustering of transcriptome by sample type (culture vs in planta) and time of sample collection (T1 vs. T2).
Differential gene expression analysis
DGE analysis was conducted to detect S. sclerotiorum transcriptome changes during pathogenesis of canola. A total of 1301 and 1214 S. sclerotiorum genes were found to be differentially expressed at early (8–16 hpi) and late (24–48 hpi) stages of infection, respectively, during infection of the susceptible line (S1 File).
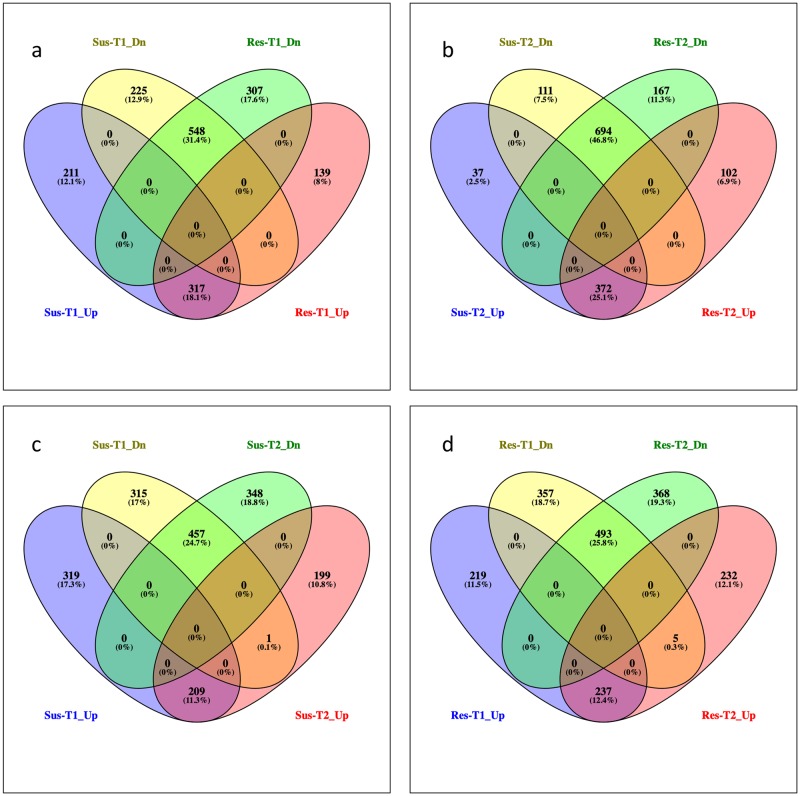
Of these, 528 and 773 genes were up- and down- regulated, respectively at T1, while 409 and 805 genes were up- and down- regulated, respectively at T2. When infecting the resistant line, 1311 and 1335 genes were differentially expressed at T1 and T2, respectively (Fig 3, S1 File). In this interaction, 456 and 474 genes were up-regulated, 855 and 861 genes were down-regulated at T1 and T2, respectively. At T1, 317 and 548 genes were common in up-regulated and down-regulated sets, respectively, between the susceptible and resistant interactions (Fig 3a). Similarly, there were 371 up-regulated and 694 down-regulated S. sclerotiorum genes that were common in both resistant and susceptible interactions at T2 (Fig 3b). When infecting the susceptible line, 209 up-regulated and 457 down-regulated genes were common between T1 and T2 (Fig 3c). The corresponding numbers when infecting the resistant line were 237 and 493 (Fig 3d). Regulation of an endo-glucanase gene, sscle_14g099920, shifted from significantly down-regulated at T1 to significant up-regulated at T2 in both susceptible and resistant interactions.
Fig 3. Venn diagrams showing differentially expressed Sclerotinia sclerotiorum genes during interaction with canola.
Venn diagram shows the number of common and unique genes at (a) T1 and (b) T2, between up and down regulated gene sets of NEP32 and NEP63. (c) and (d) shows the comparison of up and down regulated genes at T1 and T2 gene sets of NEP32 and NEP63, respectively.
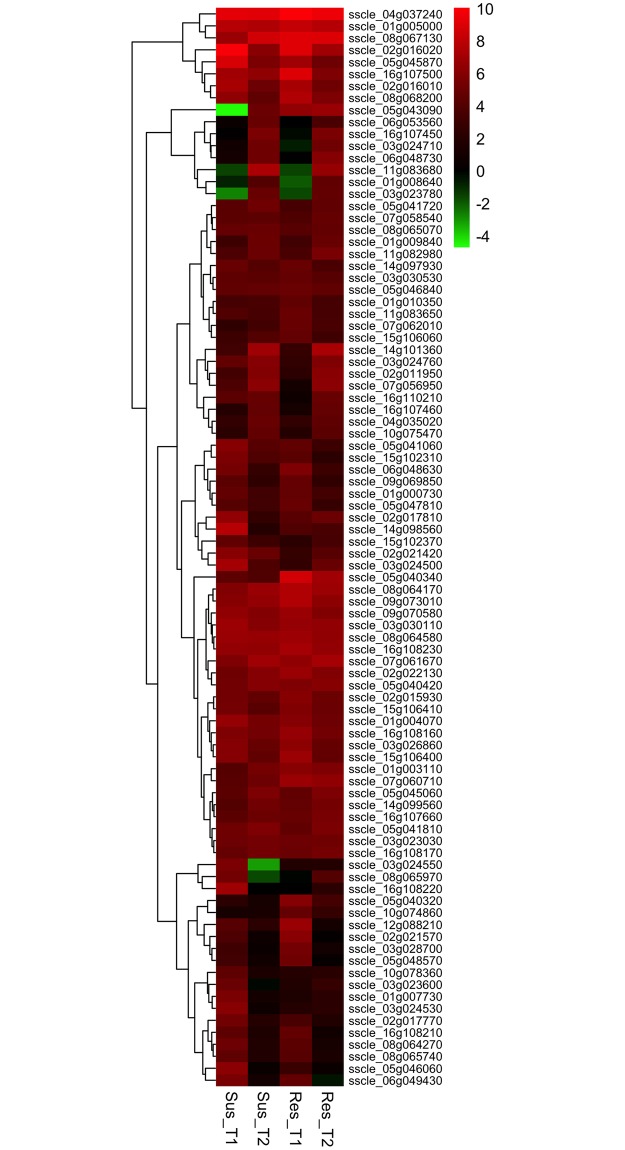
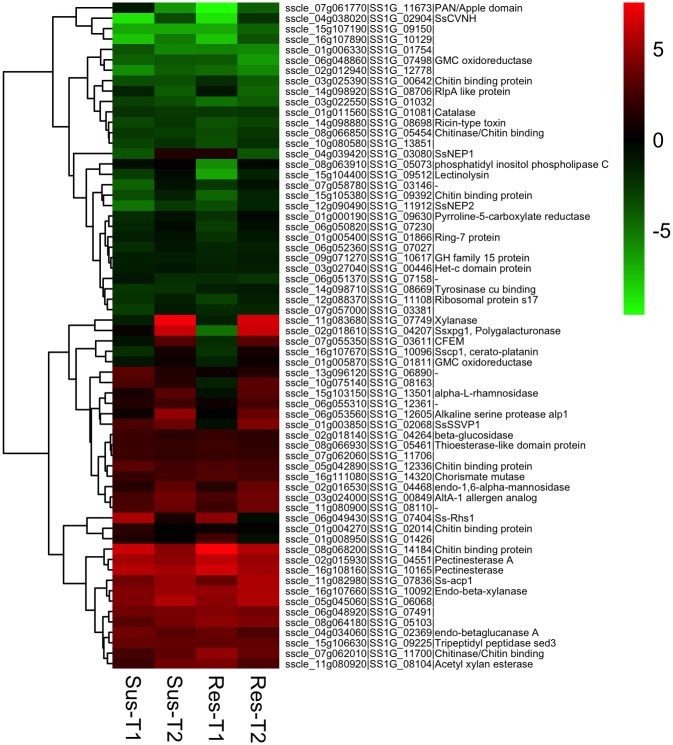
The fold-change in gene up-regulation ranged from 2 to 1053 (l2fc 1 to 10.04). In susceptible line, 12 and 8 genes were up-regulated over 100-fold at T1 and T2, respectively. In resistant line, 18 and 9 genes showed a similar 100-fold upregulation at T1 and T2, respectively. On average, approximately 50 genes were up-regulated by 25-fold (l2fc 4.64) or more in both susceptible and resistant lines at both time points. We made a comparison of the 50 most highly up-regulated genes from each of the interactions. Of these highly up-regulated genes, 22 were consistently up-regulated by 25-fold (l2fc 4.64) or more at both time points in both canola lines signifying their importance in pathogenesis (Fig 4 and S2 File). Five and 13 genes were common between susceptible and resistant lines at T1 and T2, respectively.
Fig 4. Heat maps showing expression patterns of the top 50 highly up-regulated Sclerotinia sclerotiorum genes from each treatment.
Genes are grouped according to hierarchical clustering based on their expression patternsT1 and T2 represents earlier (8 and 16 hpi) and later (24 and 48 hpi) time points of interaction. The color gradient represents the log2 fold change in gene expression (up-regulation (red), down-regulation (green), and no change (black)) compared to in vitro control.
GO categories and enrichment analysis
Functional characterization and gene functional enrichment analysis are powerful tools for analyzing DGE data to gain understanding of the important molecular pathways and functions underlying biological processes. Sclerotinia sclerotiorum DEGs were grouped according to their putative roles in biological processes (BP) and molecular functions (MF) as established by the Gene Ontology Consortium (http://www.geneontology.org/). S. sclerotiorum DEGs were assigned to the following GO classes: metabolic process, cellular process, cellular component organization, localization, biological regulation, response to stimulus, and signaling when interacting with susceptible and resistant lines at both time points.
GO enrichment analysis identified the key biological processes and molecular functions significantly enriched during pathogenesis. The up-regulated genes were significantly enriched with wide range of GO categories (S3 File). The significantly enriched categories included those involved in degradation of various cell wall components (including catabolism of cellulose (GO:0030245), xylan (GO:0045493), mannan (GO:0046355), pectin (GO:0045490)), peptidase activity (GO:0008233), oxidation-reduction processes (GO:0055114), response to xenobiotic stimulus (GO:0009410), fatty acid metabolic processes (GO:0006631), transmembrane transport (0055085), and binding (GO:0005488) activities.
GO enrichment analysis also provided insights into temporal aspects of pathogenesis. Genes involved in transcriptional reprogramming, e.g. gene expression (GO:0006396), ribosome biogenesis (GO:0042254), translation (GO:0006412), and cellular amino acid metabolic processes (GO:0006520) were enriched at the early infection stage, T1, in the up-regulated gene set indicating rapid transcriptional changes to adopt to pathogenic phase from in vitro phase. At the later infection stage, T2, many GO categories were overrepresented in the up-regulated set compared to the earlier stage. The significantly enriched up-regulated GO categories at later stage can be broadly grouped into enzymes involved in degradation of cell wall components (cell wall-degrading enzymes CWDE) and proteins, catabolism/detoxification of xenobiotic compounds and peroxisome associated pathways including peroxisome biogenesis, fatty acid catabolism and glyoxylate cycle.
Differential gene expression data validation
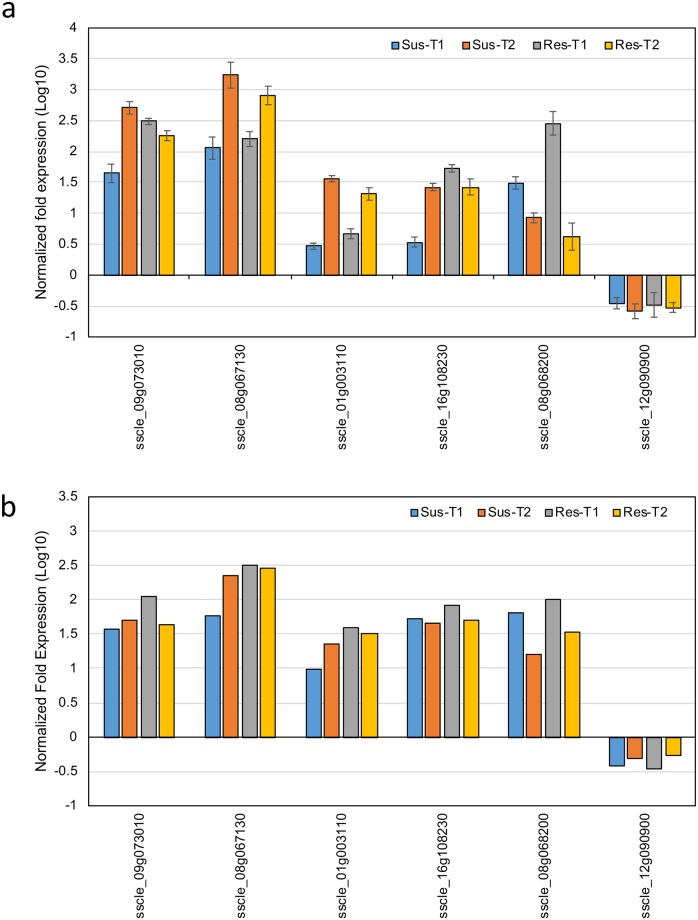
DGE data from RNA-seq analysis was validated by performing quantitative RT-PCR on five up-regulated DEGs and one down-regulated DEG. A list of the genes used for validation, their putative functions, and the primer sequences are presented in S4 File. The expression patterns of the six genes agreed with DGE data, thus validating the results of DGE analysis (Fig 5).
Fig 5. qRT-PCR validation of the relative expression levels of selected S. sclerotiorum differentially expressed genes.
Expression profiles of six S. sclerotiorum genes as determined by a. qRT-PCR and b. RNA-Seq.
Discussion
Both functional class enrichment analysis and the expression patterns of the highly up-regulated genes indicated that successful pathogenicity of S. sclerotiorum depends on cell wall degradation, detoxification and host defense evasion.
CWDE and proteolytic enzymes
The importance of cell wall degrading activity was emphasized by the fact that the genes involved in this activity were both among the highly up-regulated and constituted many significantly enriched up-regulated GO categories. Approximately one third (29/90) of the highly up-regulated (top 50 genes up-regulated by 25-fold (l2fc 4.64) or more) S. sclerotiorum genes are involved in cell wall degrading enzymatic activities. Similarly, the significantly enriched CWDE GO categories included: hydrolases (GO:0016787) acting on: O-glucosyl bonds (GO:0004553), including cellulase (GO:0008810), glucosidase (GO:0015926), galactosidase (GO:0015925), polygalacturonase (GO:0004650), alpha-L-arabinofuranosidase (GO:0046556), beta-mannosidase (GO:0004567); ester bonds (GO:0016788), including aspartyl esterase (GO:0045330), lipase (GO:0016298), cutinase (GO:0050525), pectinesterase (GO: 0030599); peptidase (GO: 0008233), including serine-type endopeptidase (GO: 0004252), tripeptidyl-peptidase (GO:0008240) and serine-type carboxypeptidase (GO: 0004185) activities. The genome of S. sclerotiorum has a significantly large repository of both plant cell wall and fungal cell wall active enzymes [19]. Many of these enzymes were up-regulated in the current study (Table 2). Consistent with our findings, previous comparative transcriptome analyses [19, 28–30, 34–36] have reported that a large number of cell wall-degrading enzymes were up-regulated during pathogenesis, while at least one functional study demonstrated that disruption of the cell wall-degrading enzyme arabinofuranosidase/xylosidase (SS1G_02462) resulted in decreased virulence on canola [10].
Table 2. Changes in expression levels of S. sclerotiorum genes involved in degradation of plant cell wall components during interaction with susceptible (Sus) and resistant (Res) canola lines at early (T1, 8–16 hpi) and late (T2, 24–48 hpi).
| gene_id | version1id | Sus-T1 | Sus-T2 | Res-T1 | Res-T2 | Activity | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| l2fc | fdr | l2fc | fdr | l2fc | fdr | l2fc | fdr | |||
| sscle_10g077670 | SS1G_08493 | 0.89 | 0.788 | -1.05 | 0.196 | -0.87 | 0.366 | -0.96 | 0.223 | Cellulose |
| sscle_03g023660 | SS1G_00891 | 1.66 | 0.383 | 1.67 | 0.402 | 1.73 | 0.373 | 1.23 | 0.556 | |
| sscle_01g008530 | SS1G_01485 | -0.47 | 0.653 | -1.00 | 0.230 | -2.23 | 0.057 | -1.50 | 0.065 | |
| sscle_15g105560 | SS1G_09365 | 0.59 | 0.670 | -2.74 | 0.036 | 0.40 | 0.821 | -2.42 | 0.040 | |
| sscle_07g056950 | SS1G_03387 | 3.84 | 0.145 | 6.23 | 0.032 | 1.12 | 0.816 | 6.22 | 0.030 | |
| sscle_14g099920 | SS1G_08837 | -3.12 | 0.038 | 2.29 | 0.048 | -3.59 | 0.025 | 2.48 | 0.032 | |
| sscle_08g063000 | SS1G_04945 | 3.37 | 0.058 | 1.95 | 0.260 | 2.97 | 0.084 | 1.80 | 0.293 | |
| sscle_01g005720 | SS1G_01828 | 0.52 | 0.571 | 2.16 | 0.008 | 0.92 | 0.269 | 2.50 | 0.003 | |
| sscle_01g001690 | SS1G_09821 | -5.48 | 0.420 | 1.64 | 0.526 | -5.48 | 0.370 | -1.12 | 0.728 | |
| sscle_04g039100 | SS1G_03041 | 1.76 | 0.201 | 1.58 | 0.236 | 1.86 | 0.188 | 1.40 | 0.267 | |
| sscle_03g028060 | SS1G_00321 | 0.37 | 0.882 | 0.41 | 0.812 | -0.80 | 0.715 | 0.84 | 0.571 | |
| sscle_03g026840 | SS1G_00471 | -1.56 | 0.010 | -1.19 | 0.028 | -1.34 | 0.028 | -1.36 | 0.013 | |
| sscle_05g045320 | SS1G_06037 | -2.75 | 0.227 | -1.04 | 0.546 | -4.02 | 0.126 | -0.38 | 0.855 | |
| sscle_04g033820 | SS1G_02334 | 2.78 | 0.007 | 4.24 | 0.000 | 1.88 | 0.049 | 4.30 | 0.000 | |
| sscle_14g101360 | SS1G_09020 | 3.33 | 0.143 | 6.97 | 0.013 | 2.82 | 0.270 | 7.29 | 0.010 | |
| sscle_03g023650 | SS1G_00892 | 2.80 | 0.007 | 4.36 | 0.000 | 2.62 | 0.011 | 4.37 | 0.000 | |
| sscle_14g102070 | SS1G_09118 | 2.08 | 0.386 | -1.19 | 0.573 | -4.65 | 0.251 | -0.77 | 0.711 | |
| sscle_04g033090 | SS1G_02245 | -3.23 | 0.007 | -3.34 | 0.002 | -4.05 | 0.006 | -3.88 | 0.001 | |
| sscle_10g080790 | SS1G_13872 | -11.04 | 0.061 | -5.78 | 0.039 | -7.68 | 0.085 | -5.41 | 0.036 | |
| sscle_04g035060 | SS1G_02501 | -2.53 | 0.017 | -2.15 | 0.023 | -3.92 | 0.004 | -1.90 | 0.035 | |
| sscle_14g100430 | SS1G_08907 | -1.66 | 0.003 | -2.54 | 0.000 | -1.98 | 0.001 | -2.60 | 0.000 | |
| sscle_11g082770 | SS1G_07863 | -2.22 | 0.328 | -0.85 | 0.668 | -3.56 | 0.189 | -1.34 | 0.447 | |
| sscle_08g064580 | SS1G_05151 | 6.75 | 0.002 | 6.66 | 0.001 | 6.89 | 0.002 | 6.41 | 0.002 | |
| sscle_08g064310 | SS1G_05118 | 0.57 | 0.478 | -0.64 | 0.438 | 0.03 | 0.978 | -1.50 | 0.051 | |
| sscle_01g007050 | SS1G_01662 | 2.89 | 0.001 | 3.47 | 0.000 | 3.19 | 0.000 | 3.23 | 0.000 | |
| sscle_15g105550 | SS1G_09366 | 2.72 | 0.000 | 3.96 | 0.000 | 2.10 | 0.001 | 3.76 | 0.000 | |
| sscle_05g043280 | SS1G_06304 | -4.06 | 0.037 | -0.26 | 0.890 | -3.96 | 0.056 | 0.13 | 0.949 | |
| sscle_11g082920 | SS1G_07847 | -2.34 | 0.259 | -0.44 | 0.804 | 0.43 | 0.875 | 0.49 | 0.771 | |
| sscle_06g051500 | SS1G_07146 | 1.56 | 0.653 | 3.24 | 0.222 | -3.14 | 0.487 | 3.07 | 0.236 | |
| sscle_03g030980 | SS1G_13255 | -3.78 | 0.018 | -1.59 | 0.137 | -2.69 | 0.063 | -2.00 | 0.057 | |
| sscle_08g066260 | SS1G_05368 | 2.70 | 0.184 | 2.68 | 0.154 | 0.81 | 0.799 | 2.44 | 0.185 | |
| sscle_06g053450 | SS1G_12622 | -1.19 | 0.239 | 1.34 | 0.163 | -1.41 | 0.208 | 1.62 | 0.085 | |
| sscle_06g051350 | SS1G_07162 | 2.01 | 0.018 | 1.97 | 0.019 | 1.53 | 0.058 | 1.79 | 0.027 | |
| sscle_14g102160 | SS1G_09129 | 2.69 | 0.009 | 3.78 | 0.001 | 1.98 | 0.039 | 3.57 | 0.001 | |
| sscle_02g018140 | SS1G_04264 | 2.30 | 0.007 | 1.69 | 0.029 | 2.15 | 0.009 | 1.72 | 0.025 | |
| sscle_03g022640 | SS1G_01021 | 2.25 | 0.088 | 3.50 | 0.012 | 1.50 | 0.330 | 3.16 | 0.018 | |
| sscle_16g108170 | SS1G_10167 | 4.78 | 0.018 | 5.31 | 0.011 | 4.99 | 0.015 | 5.40 | 0.010 | Pectin |
| sscle_09g070580 | SS1G_10698 | 6.42 | 0.002 | 5.98 | 0.002 | 6.74 | 0.001 | 5.83 | 0.002 | |
| sscle_05g046840 | SS1G_05832 | 4.62 | 0.006 | 4.76 | 0.005 | 4.98 | 0.004 | 4.60 | 0.005 | |
| sscle_04g035440 | SS1G_02553 | -2.04 | 0.363 | 2.58 | 0.064 | -3.43 | 0.187 | 2.81 | 0.040 | |
| sscle_02g018610 | SS1G_04207 | 0.27 | 0.978 | 6.25 | 0.063 | -4.95 | 0.702 | 6.39 | 0.054 | |
| sscle_05g040500 | SS1G_12057 | 0.41 | 0.762 | 0.31 | 0.953 | -0.74 | 0.612 | 0.36 | 0.783 | |
| sscle_07g055890 | SS1G_03540 | 0.18 | 0.939 | 2.30 | 0.084 | -2.82 | 0.190 | 1.82 | 0.162 | |
| sscle_16g107500 | SS1G_10071 | 7.06 | 0.002 | 6.39 | 0.003 | 9.02 | 0.001 | 5.80 | 0.005 | |
| sscle_11g085640 | SS1G_14449 | 1.42 | 0.064 | 2.36 | 0.005 | 1.72 | 0.030 | 2.56 | 0.003 | |
| sscle_02g015930 | SS1G_04551 | 5.34 | 0.006 | 4.68 | 0.010 | 6.08 | 0.003 | 5.07 | 0.005 | |
| sscle_03g027970 | SS1G_00332 | 3.67 | 0.002 | 3.75 | 0.001 | 3.46 | 0.003 | 3.56 | 0.002 | |
| sscle_07g057800 | SS1G_03286 | 4.10 | 0.076 | 4.10 | 0.082 | 4.12 | 0.074 | 3.93 | 0.087 | |
| sscle_03g026860 | SS1G_00468 | 6.05 | 0.000 | 4.90 | 0.000 | 6.32 | 0.000 | 4.83 | 0.000 | |
| sscle_02g019490 | SS1G_04095 | -1.26 | 0.187 | 0.18 | 0.892 | -1.06 | 0.280 | 0.67 | 0.507 | |
| sscle_05g040420 | SS1G_12048 | 5.26 | 0.001 | 6.03 | 0.000 | 5.82 | 0.001 | 6.08 | 0.000 | |
| sscle_12g089930 | SS1G_11992 | 3.16 | 0.099 | 4.27 | 0.012 | 4.23 | 0.015 | 4.27 | 0.010 | |
| sscle_01g002020 | SS1G_09857 | -0.86 | 0.836 | 3.77 | 0.046 | -1.30 | 0.725 | 3.98 | 0.033 | |
| sscle_15g103150 | SS1G_13501 | 0.99 | 0.287 | 3.20 | 0.002 | -0.81 | 0.501 | 3.16 | 0.002 | |
| sscle_02g016000 | SS1G_04541 | 0.13 | 0.943 | 0.14 | 0.940 | 0.33 | 0.850 | 0.80 | 0.538 | |
| sscle_10g075640 | SS1G_08229 | -6.87 | 0.013 | -3.25 | 0.082 | -4.81 | 0.036 | -3.00 | 0.099 | |
| sscle_06g052290 | SS1G_07039 | 1.32 | 0.349 | -0.04 | 0.982 | 0.64 | 0.727 | 0.95 | 0.455 | |
| sscle_02g015920 | SS1G_04552 | 3.23 | 0.039 | 1.38 | 0.379 | 3.70 | 0.024 | 1.24 | 0.437 | |
| sscle_05g041650 | SS1G_12191 | 1.27 | 0.370 | 3.85 | 0.016 | 1.05 | 0.483 | 3.97 | 0.012 | Xylan |
| sscle_16g107660 | SS1G_10092 | 4.37 | 0.000 | 5.07 | 0.000 | 4.89 | 0.000 | 5.54 | 0.000 | |
| sscle_09g074790 | SS1G_03618 | -3.94 | 0.675 | 3.29 | 0.256 | -3.94 | 0.619 | 2.28 | 0.460 | |
| sscle_11g083680 | SS1G_07749 | -1.52 | 0.906 | 7.39 | 0.039 | -1.52 | 0.879 | 6.63 | 0.052 | |
| sscle_08g064500 | SS1G_05140 | -1.88 | 0.118 | -1.16 | 0.287 | -2.66 | 0.057 | -1.31 | 0.218 | |
| sscle_11g080920 | SS1G_08104 | 2.29 | 0.076 | 3.93 | 0.004 | 4.20 | 0.004 | 2.84 | 0.019 | |
| sscle_08g066710 | SS1G_05434 | 0.32 | 0.882 | 2.92 | 0.028 | 0.10 | 0.968 | 2.87 | 0.027 | |
| sscle_03g024710 | SS1G_00746 | 0.88 | 0.758 | 5.18 | 0.016 | -0.67 | 0.864 | 5.23 | 0.013 | |
| sscle_05g045850 | SS1G_05977 | 1.15 | 0.451 | 3.90 | 0.008 | 0.49 | 0.832 | 3.82 | 0.007 | |
| sscle_10g075470 | SS1G_08208 | 2.23 | 0.043 | 4.69 | 0.000 | 1.93 | 0.106 | 4.35 | 0.000 | |
| sscle_10g074850 | SS1G_08118 | -1.88 | 0.072 | 0.04 | 0.972 | -3.48 | 0.024 | -0.14 | 0.918 | |
| sscle_15g105540 | SS1G_09367 | -1.11 | 0.248 | 0.75 | 0.458 | -1.80 | 0.101 | 0.57 | 0.588 | |
| sscle_07g060690 | SS1G_11535 | -1.33 | 0.255 | 0.86 | 0.359 | -1.52 | 0.200 | 0.77 | 0.412 | |
| sscle_02g015110 | SS1G_04662 | -3.85 | 0.001 | -2.62 | 0.002 | -6.69 | 0.000 | -2.53 | 0.002 | |
| sscle_07g056960 | SS1G_03386 | 0.22 | 0.935 | 3.28 | 0.019 | -1.86 | 0.428 | 2.95 | 0.028 | |
| sscle_11g082440 | SS1G_07904 | 0.70 | 0.485 | 3.55 | 0.000 | -2.29 | 0.084 | 3.52 | 0.000 | |
| sscle_04g034810 | SS1G_02462 | 2.71 | 0.022 | 4.10 | 0.002 | 1.12 | 0.387 | 4.29 | 0.002 | |
| sscle_07g055410 | SS1G_03602 | 2.13 | 0.085 | 3.65 | 0.006 | 1.76 | 0.161 | 3.50 | 0.007 | |
| sscle_01g010480 | SS1G_01216 | -5.59 | 0.033 | -3.31 | 0.067 | -11.82 | 0.019 | -3.27 | 0.059 | Arabinogalactans |
| sscle_07g061080 | SS1G_11585 | 0.64 | 0.830 | 3.64 | 0.034 | -0.65 | 0.820 | 4.02 | 0.020 | |
| sscle_04g035910 | SS1G_02618 | -2.48 | 0.008 | 0.05 | 0.962 | -4.89 | 0.001 | -0.01 | 0.993 | |
| sscle_09g069470 | SS1G_10842 | 3.20 | 0.001 | 3.54 | 0.000 | 1.65 | 0.045 | 3.31 | 0.001 | |
| sscle_12g091670 | SS1G_11763 | -1.51 | 0.137 | -0.18 | 0.974 | -2.33 | 0.053 | -0.35 | 0.713 | |
| sscle_01g007810 | SS1G_01572 | -0.35 | 0.859 | 1.44 | 0.257 | -3.06 | 0.147 | 1.07 | 0.412 | |
| sscle_09g074570 | SS1G_03647 | 1.51 | 0.126 | 2.96 | 0.008 | 0.61 | 0.612 | 2.69 | 0.012 | |
| sscle_04g037140 | SS1G_02781 | 1.57 | 0.078 | 1.42 | 0.111 | 1.23 | 0.174 | 1.35 | 0.122 | |
| sscle_01g002110 | SS1G_09866 | 0.29 | 0.925 | 2.49 | 0.188 | -2.66 | 0.397 | 2.28 | 0.219 | |
| sscle_12g090430 | SS1G_11922 | -5.34 | 0.569 | 3.63 | 0.186 | -5.34 | 0.514 | 3.64 | 0.171 | |
| sscle_01g010330 | SS1G_01238 | -0.43 | 0.834 | -2.29 | 0.112 | -0.51 | 0.790 | -2.08 | 0.147 | |
| sscle_04g035930 | SS1G_02620 | -1.36 | 0.358 | 0.95 | 0.452 | -4.00 | 0.058 | 0.84 | 0.512 | |
| sscle_09g069260 | SS1G_10867 | 0.15 | 0.932 | -1.92 | 0.117 | 1.30 | 0.344 | -1.65 | 0.152 | Mannan |
| sscle_02g016530 | SS1G_04468 | 1.53 | 0.261 | 3.46 | 0.013 | 1.54 | 0.344 | 3.65 | 0.009 | |
| sscle_02g011730 | SS1G_12937 | 0.98 | 0.550 | 0.14 | 0.944 | 0.58 | 0.816 | -0.08 | 0.968 | |
| sscle_08g064250 | SS1G_05110 | -3.21 | 0.004 | -2.91 | 0.001 | -3.15 | 0.007 | -2.94 | 0.001 | |
| sscle_07g061030 | SS1G_11579 | -0.57 | 0.733 | -0.67 | 0.654 | -0.11 | 0.960 | -1.99 | 0.132 | |
| sscle_15g106590 | SS1G_09229 | -2.94 | 0.054 | -0.38 | 0.815 | -5.67 | 0.013 | -0.40 | 0.809 | |
| sscle_03g026560 | SS1G_00505 | -3.52 | 0.009 | -3.19 | 0.003 | -2.14 | 0.081 | -2.13 | 0.016 | |
| sscle_02g019080 | SS1G_04148 | -1.66 | 0.011 | -0.54 | 0.365 | -2.63 | 0.002 | -0.73 | 0.202 | |
| sscle_01g004220 | SS1G_02022 | -3.61 | 0.002 | -1.18 | 0.147 | -4.42 | 0.002 | -1.30 | 0.105 | |
| sscle_02g018660 | SS1G_04200 | -0.04 | 0.989 | -2.10 | 0.233 | -4.71 | 0.177 | -0.84 | 0.656 | |
| sscle_01g009600 | SS1G_01334 | -2.64 | 0.030 | -1.83 | 0.045 | -2.90 | 0.037 | -1.24 | 0.151 | |
Detoxification of xenobiotic compounds
Genes with known roles in detoxification and host defense evasion constituted the next group of highly up-regulated genes after the CWDEs (Table 3). These include: two laccases (sscle_03g023030, sscle_02g021570), nitrilase (sscle_16g108230), brassinin glycosyltransferase (sscle_01g003110, SsBGT1), two glutathione-S-transferases (GST), two cytochrome P450 monooxygenases, and three transporter genes, including two major facilitator superfamily transporters, three S-adenosyl methionine (SAM) dependent methyltransferases including a thiol methyltransferase.
Table 3. Changes in expression levels of S. sclerotiorum genes involved in detoxification of xenobiotic compounds and peroxisome associated pathways during interaction with susceptible (Sus) and resistant (Res) canola lines at early (T1, 8–16 hpi) and late (T2, 24–48 hpi).
| gene_id | version 1 id | Sus-T1 | Sus-T2 | Res-T1 | Res-T2 | Activity/pathway | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| l2fc | fdr | l2fc | fdr | l2fc | fdr | l2fc | fdr | |||
| sscle_01g007350 | SS1G_01627 | 1.00 | 0.572 | -1.74 | 0.267 | -0.75 | 0.714 | -1.71 | 0.269 | Laccase |
| sscle_03g023030 | SS1G_00974 | 5.13 | 0.007 | 5.34 | 0.005 | 5.15 | 0.007 | 5.17 | 0.006 | |
| sscle_02g018680 | SS1G_04196 | -1.96 | 0.121 | -4.09 | 0.003 | -2.52 | 0.085 | -4.53 | 0.001 | |
| sscle_08g064260 | SS1G_05112 | -1.26 | 0.435 | -3.28 | 0.029 | -5.94 | 0.038 | -3.48 | 0.017 | |
| sscle_12g090390 | SS1G_11927 | -3.80 | 0.067 | -4.05 | 0.018 | -2.53 | 0.171 | -3.18 | 0.030 | |
| sscle_02g021570 | SS1G_13036 | 3.80 | 0.034 | 0.67 | 0.744 | 6.15 | 0.003 | 0.19 | 0.939 | |
| sscle_13g092370 | SS1G_06365 | 0.75 | 0.711 | -1.91 | 0.341 | -1.74 | 0.461 | -0.21 | 0.936 | |
| sscle_01g003110 | SS1G_09997 | 4.10 | 0.001 | 5.42 | 0.000 | 6.13 | 0.000 | 5.82 | 0.000 | glucosyltransferase |
| sscle_02g016980 | SS1G_04416 | 0.30 | 0.709 | 1.22 | 0.057 | 1.34 | 0.060 | 1.28 | 0.043 | |
| sscle_03g023640 | SS1G_00894 | 0.90 | 0.292 | -0.62 | 0.484 | 0.06 | 0.968 | -0.34 | 0.727 | |
| sscle_05g046370 | SS1G_05901 | -1.47 | 0.243 | -1.26 | 0.249 | -3.60 | 0.039 | -1.30 | 0.215 | |
| sscle_08g062730 | SS1G_04910 | -0.14 | 0.951 | 0.41 | 0.802 | -0.19 | 0.937 | 0.52 | 0.733 | |
| sscle_11g081870 | SS1G_07979 | -0.80 | 0.134 | -0.31 | 0.584 | -1.09 | 0.066 | -0.42 | 0.423 | |
| sscle_12g088170 | SS1G_11129 | 0.80 | 0.210 | 0.69 | 0.279 | 0.34 | 0.662 | 0.64 | 0.316 | |
| sscle_15g103340 | SS1G_13524 | -3.05 | 0.064 | -4.36 | 0.012 | -4.70 | 0.033 | -4.34 | 0.010 | |
| sscle_15g106380 | SS1G_09252 | -2.18 | 0.070 | -2.86 | 0.008 | -2.71 | 0.039 | -2.54 | 0.010 | |
| sscle_10g079920 | SS1G_13754 | -4.52 | 0.002 | -0.28 | 0.838 | -4.38 | 0.002 | 0.40 | 0.761 | cyanide hydratase |
| sscle_16g108230 | SS1G_10174 | 6.55 | 0.000 | 6.41 | 0.000 | 7.17 | 0.000 | 6.51 | 0.000 | |
| sscle_01g007130 | SS1G_01652 | -3.22 | 0.021 | -0.84 | 0.402 | -3.23 | 0.053 | -1.03 | 0.286 | |
| sscle_07g060330 | SS1G_11485 | -0.17 | 0.859 | 1.02 | 0.130 | -0.68 | 0.391 | 0.98 | 0.142 | |
| sscle_01g008520 | SS1G_01487 | -2.53 | 0.020 | -2.66 | 0.006 | -5.99 | 0.002 | -3.52 | 0.001 | |
| sscle_15g104110 | SS1G_09546 | -1.08 | 0.132 | -0.49 | 0.455 | -1.70 | 0.050 | -0.69 | 0.256 | |
| sscle_16g108980 | SS1G_10281 | -0.42 | 0.519 | -0.54 | 0.339 | -1.24 | 0.070 | -0.17 | 0.818 | |
| sscle_03g030680 | SS1G_13215 | -0.73 | 0.628 | 0.25 | 0.863 | -0.49 | 0.772 | 0.06 | 0.973 | |
| sscle_13g096730 | SS1G_14415 | 0.43 | 0.823 | 0.49 | 0.743 | -1.01 | 0.580 | 0.33 | 0.837 | |
| sscle_01g005000 | SS1G_01918 | 7.74 | 0.000 | 7.51 | 0.000 | 8.20 | 0.000 | 7.70 | 0.000 | glutathione-S-transferase |
| sscle_16g107740 | SS1G_10108 | -3.67 | 0.016 | -4.00 | 0.004 | -3.39 | 0.029 | -4.99 | 0.001 | |
| sscle_10g075490 | SS1G_08210 | -1.61 | 0.069 | -0.59 | 0.471 | -2.60 | 0.021 | -0.72 | 0.358 | |
| sscle_08g062750 | SS1G_04914 | -0.91 | 0.419 | -0.99 | 0.311 | -1.24 | 0.305 | -1.39 | 0.125 | |
| sscle_06g051110 | SS1G_07195 | -0.49 | 0.398 | 0.66 | 0.237 | -0.81 | 0.167 | 0.79 | 0.142 | |
| sscle_13g096550 | SS1G_14440 | 0.25 | 0.892 | 0.72 | 0.588 | 1.29 | 0.297 | 1.17 | 0.317 | |
| sscle_15g104750 | SS1G_09479 | -0.33 | 0.830 | 0.38 | 0.749 | -0.60 | 0.661 | 0.73 | 0.480 | |
| sscle_10g075830 | SS1G_08258 | -0.87 | 0.457 | 0.09 | 0.954 | 1.71 | 0.092 | 0.20 | 0.892 | |
| sscle_06g053300 | SS1G_12640 | 1.97 | 0.006 | 2.36 | 0.001 | 1.83 | 0.009 | 2.24 | 0.001 | |
| sscle_08g067590 | SS1G_05554 | -1.46 | 0.102 | -0.59 | 0.498 | -1.69 | 0.082 | -0.34 | 0.729 | |
| sscle_01g008020 | SS1G_01545 | -2.18 | 0.038 | -1.68 | 0.048 | -1.99 | 0.068 | -1.70 | 0.036 | |
| sscle_13g094100 | SS1G_06623 | -0.19 | 0.876 | -0.48 | 0.589 | -0.66 | 0.485 | -0.29 | 0.771 | |
| sscle_01g004960 | SS1G_01922 | -0.06 | 0.962 | -0.31 | 0.757 | 0.24 | 0.835 | -0.01 | 0.990 | |
| sscle_11g083650 | SS1G_07752 | 3.97 | 0.001 | 3.54 | 0.001 | 4.93 | 0.000 | 3.56 | 0.001 | |
| sscle_01g004270 | SS1G_02014 | 1.66 | 0.043 | 0.03 | 0.979 | 0.51 | 0.600 | 0.06 | 0.961 | Chitin Binding or LysM effector |
| sscle_02g014090 | SS1G_04786 | -0.48 | 0.735 | -0.70 | 0.534 | -0.70 | 0.604 | -0.30 | 0.822 | |
| sscle_03g024480 | SS1G_00772 | 2.35 | 0.012 | 1.53 | 0.067 | 2.35 | 0.015 | 1.11 | 0.174 | |
| sscle_03g025390 | SS1G_00642 | -3.63 | 0.037 | -3.63 | 0.011 | -2.16 | 0.223 | -4.09 | 0.005 | |
| sscle_05g042890 | SS1G_12336 | 3.25 | 0.001 | 2.49 | 0.004 | 2.89 | 0.003 | 2.57 | 0.003 | |
| sscle_06g054140 | SS1G_12513 | -8.68 | 0.010 | -5.19 | 0.003 | -4.41 | 0.030 | -4.31 | 0.005 | |
| sscle_06g054180 | SS1G_12509 | -10.20 | 0.032 | -10.20 | 0.009 | -10.20 | 0.033 | -10.20 | 0.007 | |
| sscle_08g068200 | SS1G_14184 | 6.39 | 0.018 | 4.59 | 0.049 | 7.56 | 0.006 | 5.74 | 0.015 | |
| sscle_15g105380 | SS1G_09392 | -3.52 | 0.028 | -1.60 | 0.192 | -4.45 | 0.028 | -1.67 | 0.158 | |
| sscle_15g107050 | SS1G_09169 | -1.39 | 0.605 | -4.37 | 0.071 | -0.52 | 0.902 | -3.87 | 0.072 | |
| sscle_07g062010 | SS1G_11700 | 2.54 | 0.020 | 3.40 | 0.002 | 4.87 | 0.000 | 3.59 | 0.001 | |
| sscle_08g066850 | SS1G_05454 | -2.69 | 0.077 | -2.89 | 0.031 | -3.45 | 0.056 | -2.59 | 0.033 | |
| sscle_08g066840 | SS1G_05453 | -2.36 | 0.282 | -1.11 | 0.533 | -7.05 | 0.067 | -2.04 | 0.215 | |
| sscle_05g041720 | SS1G_12200 | 4.27 | 0.005 | 5.19 | 0.001 | 3.51 | 0.018 | 4.62 | 0.002 | |
| sscle_03g027760 | SS1G_00355 | 2.33 | 0.013 | 2.99 | 0.003 | 1.83 | 0.041 | 2.89 | 0.003 | nitronate monooxygenase |
| sscle_12g087340 | SS1G_11235 | -1.62 | 0.374 | 1.97 | 0.222 | -3.25 | 0.157 | 2.04 | 0.198 | |
| sscle_11g085500 | SS1G_14466 | 1.85 | 0.064 | 2.00 | 0.049 | 1.66 | 0.095 | 2.00 | 0.045 | |
| sscle_09g069190 | SS1G_10881 | 0.57 | 0.702 | 0.17 | 0.914 | -1.42 | 0.384 | 0.20 | 0.897 | |
| sscle_04g038190 | SS1G_02923 | 0.03 | 0.976 | 2.06 | 0.007 | -1.13 | 0.189 | 2.19 | 0.005 | fatty acid beta-oxidation |
| sscle_16g108660 | SS1G_10238 | 4.00 | 0.004 | 3.30 | 0.008 | 3.58 | 0.006 | 3.21 | 0.009 | |
| sscle_03g028710 | SS1G_00237 | -0.31 | 0.727 | 1.42 | 0.045 | 0.05 | 0.970 | 1.65 | 0.020 | |
| sscle_07g059730 | SS1G_11414 | 0.81 | 0.238 | 2.21 | 0.004 | 1.24 | 0.092 | 2.26 | 0.003 | |
| sscle_12g090420 | SS1G_11923 | -0.37 | 0.773 | 1.59 | 0.101 | 0.13 | 0.943 | 1.56 | 0.101 | |
| sscle_09g069100 | SS1G_10890 | -0.38 | 0.793 | 1.35 | 0.231 | -1.93 | 0.198 | 1.36 | 0.217 | |
| sscle_10g076540 | SS1G_08354 | 0.62 | 0.573 | 1.61 | 0.098 | -0.38 | 0.785 | 1.97 | 0.042 | |
| sscle_14g099760 | SS1G_08821 | 0.64 | 0.479 | 0.83 | 0.363 | 0.23 | 0.835 | 1.02 | 0.246 | |
| sscle_02g018210 | SS1G_04249 | 1.89 | 0.276 | 1.27 | 0.406 | 1.29 | 0.459 | 2.42 | 0.069 | |
| sscle_05g041330 | SS1G_12152 | -0.16 | 0.925 | -0.53 | 0.690 | 0.29 | 0.840 | -0.54 | 0.690 | |
| sscle_11g086520 | SS1G_14001 | 0.69 | 0.554 | 0.48 | 0.707 | 0.32 | 0.820 | 0.41 | 0.764 | |
| sscle_09g069850 | SS1G_10796 | 4.43 | 0.042 | 2.48 | 0.220 | 4.78 | 0.035 | 2.54 | 0.198 | oxalic acid |
| sscle_14g099710 | SS1G_08814 | 0.20 | 0.862 | 0.91 | 0.231 | 1.01 | 0.239 | 1.13 | 0.121 | |
| sscle_10g075560 | SS1G_08218 | 2.18 | 0.010 | 1.87 | 0.020 | 2.04 | 0.013 | 2.12 | 0.010 | |
| sscle_08g062640 | SS1G_04900 | 0.29 | 0.601 | 1.20 | 0.017 | 0.40 | 0.476 | 1.35 | 0.008 | glyoxylate cycle |
| sscle_08g063200 | SS1G_04975 | 3.50 | 0.001 | 4.20 | 0.000 | 3.72 | 0.001 | 4.39 | 0.000 | |
| sscle_08g067810 | SS1G_05583 | 1.29 | 0.047 | 2.78 | 0.001 | 2.06 | 0.005 | 2.92 | 0.000 | |
| sscle_04g038190 | SS1G_02923 | 0.03 | 0.976 | 2.06 | 0.007 | -1.13 | 0.189 | 2.19 | 0.005 | Acetyl-CoA acetyl transferase |
| sscle_09g069100 | SS1G_10890 | -0.38 | 0.793 | 1.35 | 0.231 | -1.93 | 0.198 | 1.36 | 0.217 | |
| sscle_10g075460 | SS1G_08207 | 3.01 | 0.001 | 3.07 | 0.001 | 2.47 | 0.003 | 3.13 | 0.000 | |
| sscle_05g041330 | SS1G_12152 | -0.16 | 0.925 | -0.53 | 0.690 | 0.29 | 0.840 | -0.54 | 0.690 | |
Laccases are multicopper oxidase enzymes that are known to detoxify phenolic compounds by oxidizing them [56]. Two of the seven predicted laccase genes in the S. sclerotiorum genome were up-regulated. Sscle_02g021570 (sslacc6) was up-regulated only at T1 (15–70 fold-change, l2fc 3.80–6.15). In contrast, Sslacc2 (sscle_03g023030) was consistently up-regulated (35–40 fold-change, l2fc 5.13–5.34) at both time points (Table 3). A similar expression pattern was observed in soybean–S. sclerotiorum interaction by Westrick et al. [34]. In B. cinerea, laccase gene BcLCC2, along with BcAtrB, an ABC transporter, was required for detoxification of the antifungal phenolic antibiotic 2,4-diacetylphloroglucinol [57]. Significantly enriched GO category xenobiotic metabolic processes (GO:0006805) also represented a catechol 1,2-dioxygenase gene, sscle_04g037100, involved in detoxification of host phenolic compounds. Catechol dioxygenases are induced in response to the phenolics produced by host plants [58]. The catechol dioxygenase gene CCHD1 was induced by maize phenolics [59], and was shown to be a virulence factor in the spruce pathogen Endoconidiophora polonica [60].
In addition to phenolic compounds, Brassica spp. are known to produce a wide range of phytoalexins, plant secondary metabolites that are elicited by biotic/ abiotic stress [61]; and in response to fungal pathogen attacks [62–64]. Production of phytoalexins has been considered a resistance determinant in some host-pathogen interactions [65]. The metabolism of these strongly antifungal compounds by pathogenic fungi, both in vitro and in planta, to less toxic compounds has been well researched [61]. Detoxification of cruciferous phytoalexins by Sclerotinia sclerotiorum involves glucosylation [66], a mechanism unusual for plant pathogens. Through genome mining and transcriptional profiling, Sexton et al. [67] identified several candidate glucosyltransferases including a brassinin glucosyl transferase (SsBGT1, sscle_01g003110). Consistent with Sexton et al. [67], the SsBGT1 gene was 17–70 fold up-regulated (l2fc 4.10–6.13) in our study (Table 3). The increased activity of this gene has also been reported by other researchers during infection of canola and soybean [34, 35]. Brassicas also are known to produce phytoanticipins like cyanogenic glucosides and glucosinolates [61, 68] which are produced as part of normal plant metabolism and could have strong antimicrobial properties. Non-toxic glucosinolates, upon cellular injury are hydrolyzed to produce highly toxic isothiocyanates (ITC) and nitriles [68]. In phytopathogenic fungi, nitrilases or cyanide hydratases play a role in detoxifying HCN to the less fungitoxic formamide [69, 70], which some fungi can utilize as a nitrogen source [71, 72]. In our study, the cyanide hydratase gene sscle_16g108230 was up-regulated by 85–145 folds (l2fc 6.41–7.17) at both time points. Cyanide hydratases/nitrilases, detected in the secretomes of B. cinerea [73], were up-regulated during infection of brassica hosts by A. brassicicola, L. maculans and S. sclerotiorum [35, 72, 74]. In L. maculans, increase in cyanide hydratase gene expression was induced in the presence of potassium cyanide or derivatives of brassica glucosinolates [72]. In our study, another cyanate hydratase gene, sscle_07g060330, was up-regulated at late stage only (> 2-fold in susceptible (l2fc 1.02), ≈ 2-fold (l2fc 0.98) in resistant line). Late activation of this gene was also observed by Seifbarghi et al. [35].
Glutathione-S-transferases (GSTs), are well known for their detoxification activity of xenobiotics and endogenous toxic compounds in fungi by their conjugation to glutathione [75, 76]. A GST gene, sscle_01g005000 was highly up-regulated (182–294 fold, l2fc 7.51–8.20) in our study (Table 3). Similarly high levels of up-regulation of this gene were reported during infection of canola [35] and soybean [34]. Sscle_11g083650, another GST, a membrane-associated protein in eicosanoid and glutathione metabolism (MAPEG), was also up-regulated by 12–30-fold (l2fc 3.54–4.93) in our study. Another GST, sscle_06g053300 was also consistently upregulated (4–5-fold, l2fc 1.83–2.36) (Table 3). In A. brassicicola, an ITC-inducible MAPEG class GST, AbMAPEG1, was required for full virulence on B. oleracea [77]. Deletion of two other A. brassicicola ITC-inducible GSTs, AbGSOT1 and AbUre2pB1, resulted in both hyper-susceptibility to ITC as well as impairment in pathogenicity [77]. These observations emphasize the importance of cyanogenic compound detoxification during pathogenesis.
SAM-dependent methyltransferases catalyze the transfer of methyl groups from SAM to diverse range of substrates [78]. In our study, three SAM-dependent methyltransferases, sscle_09g073010 (68–192-fold, l2fc 6.08–7.59), sscle_15g106060 (8.5–27-fold, l2fc 3.09–4.74) and sscle_08g065070 (19–30-fold, l2fc 4.24–4.92), were highly up-regulated (S2 File). In wood degrading fungus Phanerochaete chrysosporium, SAM transferases were involved in detoxifying phenolics [79]. In Brassica and Arabidopsis, thiol methyltransferases are known to detoxify glucosinolates [80].
Lysine motif, LysM, secreted effectors are proteins [81, 82] that contribute to mask the presence of plant pathogenic fungi in plant tissues by binding to chitin on the fungal cell walls [83–85] and interfering in this way with the plant’s ability to detect it [86–88]. In our study, two chitin-binding domain protein genes, sscle_08g068200 (24–189-fold, l2fc 4.60–7.56) and sscle_05g041720 (11–36-fold, l2fc 3.51–5.19) were found to be highly up-regulated. In addition to these two highly up-regulated genes, two other genes with chitin binding domain (sscle_01g004270 and sscle_07g062010) and a LysM effector gene (sscle_03g024480) were also up-regulated (Table 3). Regulation of four of these six genes followed a similar pattern, with highest up-regulation at T1 and gradually decreasing at T2. Up-regulation of LysM and chitin binding domain genes also were reported in S. sclerotiorum interactions with B. napus [35] and G. max [34], respectively.
Oxidative burst, characterized by a rapid and transient accumulation of reactive oxygen species (ROS) is one of the first plant defense responses to pathogen invasion [89], creating oxidative stress conditions hostile for the pathogens. Thus, coping with ROS is essential for pathogen survival and successful infection of the host. Fungal pathogens have evolved various mechanisms for tolerating or scavenging ROS, including peroxidases, catalases, superoxide dismutases (sod), and NADPH oxidases (nox). In our study, three peroxidases, sscle_01g000730 (10–26-fold, l2fc 3.35–4.73), sscle_08g065740 (3–24-fold, l2fc 1.43–4.59), and sscle_04g035020 (6–29-fold, l2fc 2.58–4.86) were highly up-regulated (Table 3). Nitronate monooxygenases (NMOs), FMN enzymes are known to play important role in oxidative detoxification of nitroalkanes [90]. Recently, Marroquin-Guzman et al. [91] showed that NMOs are involved in reactive nitrogen species (RNS) stress tolerance and suppressing host immune responses by maintaining cell redox status. In our study, two NMO genes, sscle_03g027760 and sscle_11g085500, were consistently up-regulated throughout the course of infection in both susceptible and resistant lines. A third NMO gene, sscle_12g087340 was down-regulated at T1 and up-regulated at only T2. There is evidence suggesting a brief biotrophic phase of S. sclerotiorum during the infection process and thus we speculate that apart from nitro-oxidative stress protection, S. sclerotiorum’s nitronate monooxygenases might have a similar role in suppressing host defenses during its interaction with canola plants.
S. sclerotiorum is known to employ OA for suppressing host defenses by manipulating host-redox environment [13]. OA, by far has been the most studied S. sclerotiorum virulence factor to date. In addition to manipulating the host redox environment, OA also plays a key role in virulence by acting in multiple ways: induction of programmed cell death [14], calcium chelating, and mediating pH signaling [15, 16]. We examined the expression of genes involved in OA metabolism. The gene sscle_10g075560, an oxaloacetate acetylhydrolase (OAH, Ssoah1) that is a key enzyme responsible for OA biogenesis and accumulation [92], was consistently up-regulated during infection (Table 3). The oxalate decarboxylase (OxDC) gene sscle_09g069850 (Ss-odc2), was also up-regulated in concert with Ssoah1. OxDC genes play an important role by preventing OA from being accumulated in fungal cell and thus protect the pathogen from its detrimental effects [93]. A similar expression pattern for Ssoah1 and Ss-odc2 was observed during infection of B. napus and P. vulgaris, respectively [33, 35], but not on G. max [34]. However, another OxDc gene, Ss-odc1 (sscle_14g099710) was not differentially expressed (1–2 -fold change, l2fc 0.20–1.13). GO terms involved in production of OA precursors and oxidation- reduction process (GO:0055114) were also found to be significantly enriched in the DE gene sets.
Peroxisome associated pathways
In addition to the above two broad classes, genes associated with peroxisomal pathways were consistently found to be significantly enriched in the up-regulated genes. The important GO categories significantly enriched/overrepresented in this broad group are: peroxisome organization (GO:0007031) including protein targeting (GO:0006625) and protein import (GO:0016558) into peroxisome matrix, fatty acid β-oxidation (GO:0006635) including and glyoxylate cycle (GO:0006097) (S3 File). Peroxisome-related metabolic functions are shown to be essential for pathogenic development of several plant pathogenic fungi [94]. Peroxisome biogenesis proteins, known as peroxins or PEX genes are involved in peroxisome biogenesis. PEX genes have been shown to be essential for pathogenicity/ virulence in the fungal pathogens M. oryzae [95–97], C. orbiculare [98–100] and A. alternata [101]. Up-regulation of large number of PEX genes in our study suggests a possible important role of these genes for S. sclerotiorum pathogenicity/ virulence.
Fatty acid β-oxidation is a lipid metabolic pathway for degrading long chain fatty acids for nutrient and energy generation [102, 103]. This is an enzyme mediated, four-step process pathway which results in acetyl-CoA, which can be fed into glyoxylate cycle or transported to mitochondria for energy generation through citric acid cycle. The carnitine acetyl transferase gene pth2, an appressorium-associated gene which catalyzes the transportation of acetyl-CoA is required for rice infection by M. oryzae [104]. Ss-pth2, a S. sclerotiorum ortholog of the pth2 gene was found to be essential for host colonization [105], suggesting an essential role for peroxisomal pathways for host colonization and disease development. This gene (sscle_03g031670) was up-regulated during early and late stages of infection in both susceptible and resistant interactions in this study.
The glyoxylate cycle is important for gluconeogenesis, generation of glucose under nutrient scarce condition by assimilating the Acetyl-CoA generated via fatty acid β-oxidation. Isocitrate lyase (ICL1) and Maleate synthetase (MSL1) are two important enzymes of glyoxylate cycle [102]. ICL1 was found to be essential for the pathogenicity of another canola pathogen L. maculans [106]; and was shown to be important for full virulence of M. oryzae [107] and C. orbiculare [108]. Sclerotinia sclerotiorum malate synthase gene (mls1) was shown be conditionally essential for fatty acid metabolism and pathogenicity on tomato [109].
Secreted effectors
Secreted effector proteins play a key role in pathogenesis. Putative effector candidate genes were identified in the S. sclerotiorum genome based on bio-informatic analysis [31, 110]. We compared the expression of these S. sclerotiorum putative effector genes to determine specific temporal changes in their regulation. In total, 64 putative effector candidates were differentially regulated, of which 37 genes were up-regulated in at least one time point (Fig 6). The majority of these genes were consistently up-regulated in both susceptible and resistant lines at both time points. The up-regulated effector genes were mostly involved in CWDE activity, a non-aspartyl acid protease (acp1, sscle_11g082980), Rhs repeat containing protein (Ss-rhs1, sscle_06g049430), chorismate mutase (SsCm1, sscle_16g111080), and two chitin binding domain proteins (sscle_08g068200, sscle_11g082980). The gene acp1 is induced in presence of cell walls and its expression is regulated by carbon, nitrogen starvation and pH [111]. Up-regulation of this gene increased from 12.5 -fold (l2fc 3.64) at T1 to 48 -fold (l2fc 5.58) at T2. A similar expression pattern was observed in S. sclerotiorum interactions with P. vulgaris, B. napus and G. max [33–35]. Ss-Rhs1 was shown to be important for sclerotial development and initial infection on B. napus and Arabidopsis [22]. This gene was highly expressed during initial stages of sclerotial development and hyphal infection [22]. Ss-Rhs1 was found to be highly up-regulated during the early infection stage (24 hpi) compared to the late infection (48–96 hpi) stage on G. max [34], implying a possibly important role during early infection process. Similarly, we also observed 26–48 -fold up-regulation (l2fc 4.72–5.56) of Ss-Rhs1 only at T1 in both susceptible and resistant lines, respectively. In contrast, the gene SsCm1 was up-regulated by 4–8.5 -fold (l2fc 2.02–3.08) in our study, which is similar to previous reports of high levels of expression in interactions with B. napus [24], and G. max [34] during early stages of infection. The biotrophic pathogen Ustilago maydis chorismate mutase gene cmu1, was shown to decrease SA levels in infected host tissue during infection of Zea mays, contributing to the virulence [112]. We speculate that SsCm1 is likely involved in host manipulation like the cmu1 in U. maydis–Z. mays interaction by interfering with SA signaling in B. napus. The role of chitin-binding proteins and LysM effectors in avoiding host recognition was discussed earlier. However, a few effector candidates (11) exhibited a shift in their regulation from down-regulation at T1 to up-regulation at T2. Interestingly four of these genes coding for a cerato-platanin (SsCP1, sscle_16g107670), CFEM domain containing (sscle_07g055350), a small secreted virulence-related protein SsSSVP1 (sscle_01g003850, in resistant), and a necrosis and ethylene-inducing peptides SsNep1 (sscle_04g039420, in susceptible) are known to function as necrotrophic effectors. Cerato-platanins are fungal specific, small secreted cysteine rich proteins known to function both as elicitors of plant defenses and as well as effectors contributing to virulence [113] by inducing localized necrosis of host tissue. In S. sclerotiorum, SsCP1 was shown to contribute to its virulence by directly interacting with pathogenesis-related protein PR1 [27]. In our study Sscp1 was down-regulated at T1 by 4.9 -fold (l2fc -2.39) and up-regulated 1.8 -fold (l2fc 0.85) by T2. The CFEM, conserved fungal extracellular membrane proteins domain is a fungal specific domain containing eight conserved cysteine residues [114, 115] and are proposed to have role in fungal pathogenesis. The CFEM protein, pth11 in M. oryzae is essential for appressoria development and pathogenesis [116, 117]. Kou et al. [117] showed that deletion of pth11 results in disruption of redox homeostasis and thus affects appressorium formation during pathogenesis. In B. cinerea, a closely related broad-host-range necrotroph, BcCFEM1, a CFEM containing gene, plays a key role in stress resistance and virulence [118]. BcCFEM1 also has a potential elicitor role. Gene Sscle_07g055350 was 4.4 -fold down-regulated (l2fc -2.13) at T1, followed by 9 -fold upregulation (l2fc 3.18) at T2. A similar expression pattern (up-regulation only at late infection) was observed by Seifbarghi et al [35] and Guyon et al. [110] during infection of canola and soybean [34] by S. sclerotiorum and by Thatcher et al. [119] during Fusarium oxysporum and Medicago truncatula interaction. SsSSVP1, another characterized S. sclerotiorum virulence factor, is a small cysteine rich secreted protein essential for full virulence. SsSSVP1 interferes with the host mitochondrial respiratory pathway by interacting with the QCR8 subunit of cytochrome b-c1 complex, resulting in plant cell death [26]. Consistent with our study, expression of SsSSVP1 was not detected until late infection stage (96 hpi) in B. napus [35]. SsNep1 and SsNep2 are shown to induce necrosis in host plants [25]. In this study, SsNep1 was not up-regulated until T2 (2.5 -fold up-regulation, l2fc 1.35), with gradual increase from early infection (15 -fold down-regulation, l2fc -3.89). A similar expression pattern was also observed for SsNep2 (-33 –-3 -fold change (l2fc -5.07 –-1.76) from T1 –T2). These two genes were induced mid—late stages of infection in B. napus and G. max [34, 35].
Fig 6. Heat maps showing expression patterns of Sclerotinia sclerotiorum effector genes.
Genes are grouped according to hierarchical clustering based on their expression patterns. T1 and T2 represents earlier (8 and 16 hpi) and later (24 and 48 hpi) time points of interaction. The color gradient represents the log2 fold change in gene expression (up-regulation (red), down-regulation (green), and no change (black)) compared to in vitro control.
In contrast to the widely accepted necrotrophic nature of S. sclerotiorum, recent molecular and cytological evidences suggest a two-phase model involving a brief biotrophic or basic compatibility phase characterized by host suppression and subverting of host defenses following by necrotrophic phase [38]. The temporal differential expression patterns of the effector candidates in our study supports the two-phase infection model [38–40] proposed for S. sclerotiorum. Biotrophic effectors like chorismate mutase (SsCm1), chitin binding proteins, LysM, and genes involved in ROS and RNS scavenging are up-regulated at early infection phase. Effectors known to induce necrosis like SsCp1, SsSSVp1, SsNep1, SsNep2 were either not up-regulated until late infection and/or down-regulated at early stages of infection. A similar trend in expression of necrotrophic effectors was observed by Westrick et al in S. sclerotiorum—G. max interaction [34]. Two characterized effector genes SsCVNH [19] and SsITL [23] were significantly down-regulated at both time points, whereas another characterized effector gene, Ssggt2 (sscle_09g068730), encoding γ-glutamyl transpeptidase was consistently up-regulated (2–3 -fold, l2fc 1.12–1.65) at both time points.
Conclusions
This is the first study that examine global transcriptional changes in S. sclerotiorum during infection of canola plants differing in their susceptibility to the pathogen. The findings from this study emphasize the role of peroxisome related pathways, in addition to the cell wall degradation and detoxification of host metabolites as the key mechanisms underlying pathogenesis of S. sclerotiorum on canola. Further, temporal changes in expression pattern of several functional classes of genes, like expression of genes involved in avoiding host recognition or suppressing host defenses at early infection stage (Chitin binding domains, LysM effectors, ROS scavenging) and late onset of expression of necrosis inducing effectors (cerato-platanin, SsSSVP1, CFEM domain, SsNep1 and SsNep2 genes etc.) provided support for the proposed two-phase infection strategy involving a brief biotrophic phase during early infection. Functional analysis of these genes would provide further insight on the events that lead to disease development and colonization of plant tissues.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All sequence files are available from the NCBI SRA database under BioProject accession number PRJNA601001. All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
National Sclerotinia Initiative USDA-ARS (NACA) award numbers 58-5442-0-243 and 58-3060-5-036 awarded to LdR. https://www.ars.usda.gov/plains-area/docs/white-mold-research/research/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bolton MD, Thomma BPHJ, Nelson BD. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Molecular Plant Pathology. 2006;7(1):1–16. 10.1111/j.1364-3703.2005.00316.x [DOI] [PubMed] [Google Scholar]
- 2.del Río LE, Bradley CA, Henson RA, Endres GJ, Hanson BK, McKay K, et al. Impact of Sclerotinia stem rot on yield of canola. Plant Disease. 2007;91(2):191–4. 10.1094/PDIS-91-2-0191 [DOI] [PubMed] [Google Scholar]
- 3.Bardin SD, Huang HC. Research on biology and control of Sclerotinia diseases in Canada. Canadian Journal of Plant Pathology. 2001;23(1):88–98. [Google Scholar]
- 4.Morton JG, Hall R. Factors determining the efficacy of chemical control of white mold in white bean. Canadian Journal of Plant Pathology. 1989;11(3):297–302. [Google Scholar]
- 5.Jones EE, Whipps JM. Effect of inoculum rates and sources of Coniothyrium minitans on control of Sclerotinia sclerotiorum disease in glasshouse lettuce. European Journal of Plant Pathology. 2002;108(6):527–38. [Google Scholar]
- 6.Reeleder RD. The use of yeasts for biological control of the plant pathogen Sclerotinia sclerotiorum. BioControl. 2004;49(5):583–94. [Google Scholar]
- 7.Lu G. Engineering Sclerotinia sclerotiorum resistance in oilseed crops. African Journal of Biotechnology. 2003;2(12):509–16. [Google Scholar]
- 8.Dallal Bashi Z, Rimmer SR, Khachatourians GG, Hegedus DD. Factors governing the regulation of Sclerotinia sclerotiorum cutinase A and polygalacturonase 1 during different stages of infection. Canadian Journal of Microbiology. 2012;58(5):605–16. 10.1139/w2012-031 [DOI] [PubMed] [Google Scholar]
- 9.Li R, Rimmer R, Buchwaldt L, Sharpe AG, Séguin-Swartz G, Hegedus DD. Interaction of Sclerotinia sclerotiorum with Brassica napus: cloning and characterization of endo- and exo-polygalacturonases expressed during saprophytic and parasitic modes. Fungal Genetics and Biology. 2004;41(8):754–65. 10.1016/j.fgb.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 10.Yajima W, Liang Y, Kav NNV. Gene Disruption of an Arabinofuranosidase/β-Xylosidase precursor decreases Sclerotinia sclerotiorum virulence on canola tissue. Molecular Plant-Microbe Interactions. 2009;22(7):783–9. 10.1094/MPMI-22-7-0783 [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Jifen X, Jiao D, Yuheng Y, Chaowei B, Ling Q. Disruption of the gene encoding endo-β-1, 4-xylanase affects the growth and virulence of Sclerotinia sclerotiorum. Frontiers in Microbiology. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu L, Li G, Jiang D, Chen W. Sclerotinia sclerotiorum: An evaluation of virulence theories. Annu Rev Phytopathol. 2018;56:311–38. 10.1146/annurev-phyto-080417-050052 [DOI] [PubMed] [Google Scholar]
- 13.Williams B, Kabbage M, Kim H-J, Britt R, Dickman MB. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLOS Pathogens. 2011;7(6):e1002107 10.1371/journal.ppat.1002107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim KS, Min J-Y, Dickman MB. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Molecular Plant-Microbe Interactions. 2008;21(5):605–12. 10.1094/MPMI-21-5-0605 [DOI] [PubMed] [Google Scholar]
- 15.Rollins JA. The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Molecular Plant-Microbe Interactions. 2003;16(9):785–95. 10.1094/MPMI.2003.16.9.785 [DOI] [PubMed] [Google Scholar]
- 16.Liang X, Liberti D, Li M, Kim Y-T, Hutchens A, Wilson R, et al. Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Molecular Plant Pathology. 2015;16(6):559–71. 10.1111/mpp.12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Xiang M, White D, Chen W. pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ Microbiol. 2015;17(8):2896–909. 10.1111/1462-2920.12818 [DOI] [PubMed] [Google Scholar]
- 18.Li J, Zhang Y, Yu PL, Pan H, Rollins JA. Introduction of large sequence inserts by CRISPR-Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen Sclerotinia sclerotiorum. MBio. 2018;9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyu X, Shen C, Fu Y, Xie J, Jiang D, Li G, et al. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Scientific Reports. 2015;5:15565 10.1038/srep15565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Y, Yajima W, Davis MR, Kav NNV, Strelkov SE. Disruption of a gene encoding a hypothetical secreted protein from Sclerotinia sclerotiorum reduces its virulence on canola (Brassica napus). Canadian Journal of Plant Pathology. 2013;35(1):46–55. [Google Scholar]
- 21.Xiao X, Xie J, Cheng J, Li G, Yi X, Jiang D, et al. Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol Plant Microbe Interact. 2014;27(1):40–55. 10.1094/MPMI-05-13-0145-R [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, Xiao J, Zhu W, Yang Y, Mei J, Bi C, et al. Ss-Rhs1, a secretory Rhs repeat-containing protein, is required for the virulence of Sclerotinia sclerotiorum. Mol Plant Pathol. 2017;18(8):1052–61. 10.1111/mpp.12459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu W, Wei W, Fu Y, Cheng J, Xie J, Li G, et al. A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLOS ONE. 2013;8(1):e53901 10.1371/journal.pone.0053901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nováková M, Sašek V, Dobrev PI, Valentová O, Burketová L. Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum—reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol Biochem. 2014;80:308–17. 10.1016/j.plaphy.2014.04.019 [DOI] [PubMed] [Google Scholar]
- 25.Dallal Bashi Z, Hegedus DD, Buchwaldt L, Rimmer SR, Borhan MH. Expression and regulation of Sclerotinia sclerotiorum necrosis and ethylene-inducing peptides (NEPs). Molecular Plant Pathology. 2010;11(1):43–53. 10.1111/j.1364-3703.2009.00571.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyu X, Shen C, Fu Y, Xie J, Jiang D, Li G, et al. A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 2016;12(2):e1005435 10.1371/journal.ppat.1005435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang G, Tang L, Gong Y, Xie J, Fu Y, Jiang D, et al. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytologist. 2018;217(2):739–55. 10.1111/nph.14842 [DOI] [PubMed] [Google Scholar]
- 28.Li R, Rimmer R, Buchwaldt L, Sharpe AG, Séguin-Swartz G, Coutu C, et al. Interaction of Sclerotinia sclerotiorum with a resistant Brassica napus cultivar: expressed sequence tag analysis identifies genes associated with fungal pathogenesis. Fungal Genetics and Biology. 2004;41(8):735–53. 10.1016/j.fgb.2004.03.001 [DOI] [PubMed] [Google Scholar]
- 29.Sexton AC, Cozijnsen AJ, Keniry A, Jewell E, Love CG, Batley J, et al. Comparison of transcription of multiple genes at three developmental stages of the plant pathogen Sclerotinia sclerotiorum. FEMS Microbiology Letters. 2006;258(1):150–60. 10.1111/j.1574-6968.2006.00212.x [DOI] [PubMed] [Google Scholar]
- 30.Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, et al. Genomic Analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLOS Genetics. 2011;7(8):e1002230 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derbyshire M, Denton-Giles M, Hegedus D, Seifbarghy S, Rollins J, van Kan J, et al. The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biology and Evolution. 2017;9(3):593–618. 10.1093/gbe/evx030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuang X, McPhee KE, Coram TE, Peever TL, Chilvers MI. Rapid transcriptome characterization and parsing of sequences in a non-model host-pathogen interaction; pea-Sclerotinia sclerotiorum. BMC Genomics. 2012;13(1):668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira MB, de Andrade RV, Grossi-de-Sá MF, Petrofeza S. Analysis of genes that are differentially expressed during the Sclerotinia sclerotiorum–Phaseolus vulgaris interaction. Frontiers in Microbiology. 2015;6:1162 10.3389/fmicb.2015.01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westrick NM, Ranjan A, Jain S, Grau CR, Smith DL, Kabbage M. Gene regulation of Sclerotinia sclerotiorum during infection of Glycine max: on the road to pathogenesis. BMC Genomics. 2019;20(1):157 10.1186/s12864-019-5517-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seifbarghi S, Borhan MH, Wei Y, Coutu C, Robinson SJ, Hegedus DD. Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genomics. 2017;18(1):266 10.1186/s12864-017-3642-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng Q, Xie Q, Chen F, Zhou X, Zhang W, Zhang J, et al. Transcriptome analysis of Sclerotinia sclerotiorum at different infection stages on Brassica napus. Current Microbiology. 2017;74(10):1237–45. 10.1007/s00284-017-1309-8 [DOI] [PubMed] [Google Scholar]
- 37.Ding Y, Mei J, Chai Y, Yu Y, Shao C, Wu Q, et al. Simultaneous transcriptome analysis of host and pathogen highlights the interaction between Brassica oleracea and Sclerotinia sclerotiorum. Phytopathology. 2019;109(4):542 10.1094/PHYTO-06-18-0204-R [DOI] [PubMed] [Google Scholar]
- 38.Liang X, Rollins JA. Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology. 2018;108(10):1128–40. 10.1094/PHYTO-06-18-0197-RVW [DOI] [PubMed] [Google Scholar]
- 39.Kabbage M, Williams B, Dickman MB. Cell Death Control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLOS Pathogens. 2013;9(4):e1003287 10.1371/journal.ppat.1003287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabbage M, Yarden O, Dickman MB. Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle. Plant Science. 2015;233(Supplement C):53–60. [DOI] [PubMed] [Google Scholar]
- 41.Su Y. Double haploid production of canola (Brassica napus L.) with improved resistance to Sclerotinia sclerotiorum via microspore culture: Thesis (M.S.)—North Dakota State University, 2009.; 2009.
- 42.Zhao J, Peltier AJ, Meng J, Osborn TC, Grau CR. Evaluation of sclerotinia stem rot resistance in oilseed Brassica napus using a petiole inoculation technique under greenhouse conditions. Plant Disease. 2004;88(9):1033–9. 10.1094/PDIS.2004.88.9.1033 [DOI] [PubMed] [Google Scholar]
- 43.Andrews S. FastQC A quality control tool for high throughput sequence data. http://www.bioinformaticsbabrahamacuk/projects/fastqc/.
- 44.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nature methods. 2015;12(4):357–60. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y, Smyth G, Shi W. featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. arXivorg. 2013;30(7). [DOI] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic acids research. 2013;41(10):e108–e. 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome biology. 2010;11(3):R25–R. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin M, Uwe L. Scatterplot3d—an R package for Visualizing Multivariate Data. Journal of Statistical Software. 2003;8(11). [Google Scholar]
- 50.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic acids research. 2012;40(10):4288–97. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lun AT, Chen Y, Smyth GK. It's DE-licious: A recipe for differential expression analyses of rna-seq experiments using quasi-likelihood methods in edgeR. Methods Mol Biol. 2016;1418:391–416. 10.1007/978-1-4939-3578-9_19 [DOI] [PubMed] [Google Scholar]
- 52.Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. International Journal of Plant Genomics. 2008;2008:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawahara Y, Oono Y, Kanamori H, Matsumoto T, Itoh T, Minami E. Simultaneous RNA-Seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLOS ONE. 2012;7(11):e49423 10.1371/journal.pone.0049423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong W, Chen N, Liu T, Zhu J, Wang J, He X, et al. Large-scale transcriptome analysis of cucumber and Botrytis cinerea during Infection. PLOS ONE. 2015;10(11):e0142221 10.1371/journal.pone.0142221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shraddha R, Shekher S, Sehgal M, Kamthania A, Kumar A. Laccase: microbial sources, production, purification, and potential biotechnological applications. Enzyme research. 2011;2011(1):217861-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schouten A, Maksimova O, Cuesta-Arenas Y, van den Berg G, Raaijmakers JM. Involvement of the ABC transporter BcAtrB and the laccase BcLCC2 in defence of Botrytis cinerea against the broad-spectrum antibiotic 2,4-diacetylphloroglucinol. Environ Microbiol. 2008;10(5):1145–57. 10.1111/j.1462-2920.2007.01531.x [DOI] [PubMed] [Google Scholar]
- 58.Shalaby S, Horwitz BA. Plant phenolic compounds and oxidative stress: integrated signals in fungal–plant interactions. Current Genetics. 2015;61(3):347–57. 10.1007/s00294-014-0458-6 [DOI] [PubMed] [Google Scholar]
- 59.Shanmugam V, Ronen M, Shalaby S, Larkov O, Rachamim Y, Hadar R, et al. The fungal pathogen Cochliobolus heterostrophus responds to maize phenolics: novel small molecule signals in a plant-fungal interaction. Cellular Microbiology. 2010;12(10):1421–34. 10.1111/j.1462-5822.2010.01479.x [DOI] [PubMed] [Google Scholar]
- 60.Wadke N, Kandasamy D, Vogel H, Lah L, Wingfield BD, Paetz C, et al. The bark-beetle-associated fungus, Endoconidiophora polonica, utilizes the phenolic defense compounds of its host as a carbon source. Plant Physiology. 2016;171(2):914 10.1104/pp.15.01916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pedras MSC, Yaya EE, Glawischnig E. The phytoalexins from cultivated and wild crucifers: Chemistry and biology. Natural Product Reports. 2011;28(8):1381–405. 10.1039/c1np00020a [DOI] [PubMed] [Google Scholar]
- 62.Pedras MSC, Zheng Q-A, Gadagi RS, Rimmer SR. Phytoalexins and polar metabolites from the oilseeds canola and rapeseed: Differential metabolic responses to the biotroph Albugo candida and to abiotic stress. Phytochemistry. 2008;69(4):894–910. 10.1016/j.phytochem.2007.10.019 [DOI] [PubMed] [Google Scholar]
- 63.Dahiya JS, Rimmer SR. Phytoalexin accumulation in tissues of Brassica napus inoculated with Leptosphaeria maculans. Phytochemistry. 1988;27(10):3105–7. [Google Scholar]
- 64.Storck M, Sacristán Maria D. The role of phytoalexins in the seedling resistance to Leptosphaeria maculans in some crucifers. Zeitschrift für Naturforschung C 1995. p. 15. [Google Scholar]
- 65.Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. The Plant Journal. 2003;35(2):193–205. 10.1046/j.1365-313x.2003.01794.x [DOI] [PubMed] [Google Scholar]
- 66.Pedras MSC, Hossain M. Metabolism of crucifer phytoalexins in Sclerotinia sclerotiorum: detoxification of strongly antifungal compounds involves glucosylation. Organic & Biomolecular Chemistry. 2006;4(13):2581–90. [DOI] [PubMed] [Google Scholar]
- 67.Sexton AC, Minic Z, Cozijnsen AJ, Pedras MSC, Howlett BJ. Cloning, purification and characterisation of brassinin glucosyltransferase, a phytoalexin-detoxifying enzyme from the plant pathogen Sclerotinia sclerotiorum. Fungal Genetics and Biology. 2009;46(2):201–9. 10.1016/j.fgb.2008.10.014 [DOI] [PubMed] [Google Scholar]
- 68.Osbourn AE. Preformed antimicrobial compounds and plant defense against fungal attack. The Plant Cell. 1996;8(10):1821 10.1105/tpc.8.10.1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fry WE, Evans PH. Association of formamide hydro-lyase with fungal pathogenicity to cyanogenic plants. Phytopathology. 1977;67(8):1001–6. [Google Scholar]
- 70.Wang P, VanEtten HD. Cloning and properties of a cyanide hydratase gene from the phytopathogenic fungus Gloeocercospora sorghi. Biochemical and Biophysical Research Communications. 1992;187(2):1048–54. 10.1016/0006-291x(92)91303-8 [DOI] [PubMed] [Google Scholar]
- 71.Barclay M, Tett VA, Knowles CJ. Metabolism and enzymology of cyanide/metallocyanide biodegradation by Fusarium solani under neutral and acidic conditions. Enzyme and Microbial Technology. 1998;23(5):321–30. [Google Scholar]
- 72.Sexton AC, Howlett BJ. Characterisation of a cyanide hydratase gene in the phytopathogenic fungus Leptosphaeria maculans. Molecular and General Genetics. 2000;263(3):463–70. 10.1007/s004380051190 [DOI] [PubMed] [Google Scholar]
- 73.González-Fernández R, Aloria K, Valero-Galván J, Redondo I, Arizmendi JM, Jorrín-Novo JV. Proteomic analysis of mycelium and secretome of different Botrytis cinerea wild-type strains. Journal of Proteomics. 2014;97(Supplement C):195–221. [DOI] [PubMed] [Google Scholar]
- 74.Cramer RA, Lawrence CB. Identification of Alternaria brassicicola genes expressed in planta during pathogenesis of Arabidopsis thaliana. Fungal Genetics and Biology. 2004;41(2):115–28. 10.1016/j.fgb.2003.10.009 [DOI] [PubMed] [Google Scholar]
- 75.Morel M, Ngadin AA, Droux M, Jacquot JP, Gelhaye E. The fungal glutathione S-transferase system. Evidence of new classes in the wood-degrading basidiomycete Phanerochaete chrysosporium. Cell Mol Life Sci. 2009;66(23):3711–25. 10.1007/s00018-009-0104-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gullner G, Komives T, Király L, Schröder P. Glutathione S-Transferase enzymes in plant-pathogen interactions. Front Plant Sci. 2018;9:1836 10.3389/fpls.2018.01836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Calmes B, Morel-Rouhier M, Bataillé-Simoneau N, Gelhaye E, Guillemette T, Simoneau P. Characterization of glutathione transferases involved in the pathogenicity of Alternaria brassicicola. BMC Microbiol. 2015;15:123 10.1186/s12866-015-0462-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Struck AW, Thompson ML, Wong LS, Micklefield J. S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem. 2012;13(18):2642–55. 10.1002/cbic.201200556 [DOI] [PubMed] [Google Scholar]
- 79.Jeffers M, McRoberts W, Harper D. Identification of a phenolic 3-O-methyltransferase in the lignin-degrading fungus Phanerochaete chrysosporium. Microbiology. 1997;143(6):1975–81. [DOI] [PubMed] [Google Scholar]
- 80.Attieh J, Kleppinger-Sparace K, Nunes C, Sparace S, Saini H. Evidence implicating a novel thiol methyltransferase in the detoxification of glucosinolate hydrolysis products in Brassica oleracea L. Plant, Cell and Environment. 2000;23(2):165. [Google Scholar]
- 81.de Jonge R, Thomma BPHJ. Fungal LysM effectors: extinguishers of host immunity? Trends in Microbiology. 2009;17(4):151–7. 10.1016/j.tim.2009.01.002 [DOI] [PubMed] [Google Scholar]
- 82.Kombrink A, Thomma BPHJ. LysM effectors: secreted proteins supporting fungal life. PLOS Pathogens. 2013;9(12):e1003769 10.1371/journal.ppat.1003769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takahara H, Hacquard S, Kombrink A, Hughes HB, Halder V, Robin GP, et al. Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytologist. 2016;211(4):1323–37. 10.1111/nph.13994 [DOI] [PubMed] [Google Scholar]
- 84.de Jonge R, Peter van Esse H, Kombrink A, Shinya T, Desaki Y, Bours R, et al. Conserved Fungal LysM Effector Ecp6 Prevents Chitin-Triggered Immunity in Plants. Science. 2010;329(5994):953 10.1126/science.1190859 [DOI] [PubMed] [Google Scholar]
- 85.Mentlak TA, Kombrink A, Shinya T, Ryder LS, Otomo I, Saitoh H, et al. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. The Plant Cell. 2012;24(1):322 10.1105/tpc.111.092957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shibuya N, Minami E. Oligosaccharide signalling for defence responses in plant. Physiological and Molecular Plant Pathology. 2001;59(5):223–33. [Google Scholar]
- 87.Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci U S A. 2006;103(29):11086–91. 10.1073/pnas.0508882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fesel PH, Zuccaro A. β-glucan: Crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genetics and Biology. 2016;90:53–60. 10.1016/j.fgb.2015.12.004 [DOI] [PubMed] [Google Scholar]
- 89.Wojtaszek P. Oxidative burst: An early plant response to pathogen infection. Biochem J. 1997;322 (Pt 3):681–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Francis K, Nishino SF, Spain JC, Gadda G. A novel activity for fungal nitronate monooxygenase: Detoxification of the metabolic inhibitor propionate-3-nitronate. Archives of Biochemistry and Biophysics. 2012;521(1):84–9. [DOI] [PubMed] [Google Scholar]
- 91.Marroquin-Guzman M, Hartline D, Wright JD, Elowsky C, Bourret TJ, Wilson RA. The Magnaporthe oryzae nitrooxidative stress response suppresses rice innate immunity during blast disease. Nature Microbiology. 2017;2:17054 10.1038/nmicrobiol.2017.54 [DOI] [PubMed] [Google Scholar]
- 92.Liang X, Moomaw EW, Rollins JA. Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Molecular Plant Pathology. 2015;16(8):825–36. 10.1111/mpp.12239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heard S, Brown NA, Hammond-Kosack K. An interspecies comparative analysis of the predicted secretomes of the necrotrophic plant pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLOS ONE. 2015;10(6):e0130534 10.1371/journal.pone.0130534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kubo Y. Function of Peroxisomes in Plant-Pathogen Interactions In: del Río LA, editor. Peroxisomes and their Key Role in Cellular Signaling and Metabolism. Dordrecht: Springer Netherlands; 2013. p. 329–45. [DOI] [PubMed] [Google Scholar]
- 95.Wang J, Li L, Zhang Z, Qiu H, Li D, Fang Y, et al. One of three pex11 family members is required for peroxisomal proliferation and full virulence of the rice blast fungus Magnaporthe oryzae. PLOS ONE. 2015;10(7):e0134249 10.1371/journal.pone.0134249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Z-Y, Soanes DM, Kershaw MJ, Talbot NJ. Functional analysis of lipid metabolism in magnaporthe grisea reveals a requirement for peroxisomal fatty acid β-oxidation during appressorium-mediated plant infection. Molecular Plant-Microbe Interactions. 2007;20(5):475–91. 10.1094/MPMI-20-5-0475 [DOI] [PubMed] [Google Scholar]
- 97.Goh J, Jeon J, Kim KS, Park J, Park S-Y, Lee Y-H. The PEX7-mediated peroxisomal import system is required for fungal development and pathogenicity in Magnaporthe oryzae. PLOS ONE. 2011;6(12):e28220 10.1371/journal.pone.0028220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kimura A, Takano Y, Furusawa I, Okuno T. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. The Plant Cell. 2001;13(8):1945 10.1105/TPC.010084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fujihara N, Sakaguchi A, Tanaka S, Fujii S, Tsuji G, Shiraishi T, et al. Peroxisome biogenesis factor pex13 is required for appressorium-mediated plant infection by the anthracnose fungus Colletotrichum orbiculare. Molecular Plant-Microbe Interactions. 2010;23(4):436–45. 10.1094/MPMI-23-4-0436 [DOI] [PubMed] [Google Scholar]
- 100.Kubo Y. Appressorium function in Colletotrichum orbiculare and prospect for genome based analysis In: Pérez-Martín J, Di Pietro A, editors. Morphogenesis and Pathogenicity in Fungi. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. p. 115–31. [Google Scholar]
- 101.Imazaki A, Tanaka A, Harimoto Y, Yamamoto M, Akimitsu K, Park P, et al. Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen Alternaria alternata. Eukaryotic Cell. 2010;9(5):682–94. 10.1128/EC.00369-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen X-L, Wang Z, Liu C. Roles of peroxisomes in the rice blast fungus. BioMed Research International. 2016;2016:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK. Peroxisomal β-oxidation—A metabolic pathway with multiple functions. Biochimica et Biophysica Acta (BBA)—Molecular Cell Research. 2006;1763(12):1413–26. [DOI] [PubMed] [Google Scholar]
- 104.Ramos-Pamplona M, Naqvi NI. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Molecular Microbiology. 2006;61(1):61–75. 10.1111/j.1365-2958.2006.05194.x [DOI] [PubMed] [Google Scholar]
- 105.Liberti D, Rollins JA, Dobinson KF. Peroxysomal carnitine acetyl transferase influences host colonization capacity in Sclerotinia sclerotiorum. Molecular Plant-Microbe Interactions. 2013;26(7):768–80. 10.1094/MPMI-03-13-0075-R [DOI] [PubMed] [Google Scholar]
- 106.Idnurm A, Howlett BJ. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryotic Cell. 2002;1(5):719–24. 10.1128/EC.1.5.719-724.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Z-Y, Thornton CR, Kershaw MJ, Debao L, Talbot NJ. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Molecular Microbiology. 2003;47(6):1601–12. 10.1046/j.1365-2958.2003.03412.x [DOI] [PubMed] [Google Scholar]
- 108.Asakura M, Okuno T, Takano Y. Multiple contributions of peroxisomal metabolic function to fungal pathogenicity in Colletotrichum lagenarium. Applied and Environmental Microbiology. 2006;72(9):6345–54. 10.1128/AEM.00988-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liberti D, Grant SJ, Benny U, Rollins JA, Dobinson KF. Development of an agrobacterium tumefaciens mediated gene disruption method for Sclerotinia sclerotiorum. Canadian Journal of Plant Pathology. 2007;29(4):394–400. [Google Scholar]
- 110.Guyon K, Balagué C, Roby D, Raffaele S. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics. 2014;15(1):336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Poussereau N, Creton S, Billon-Grand G, Rascle C, Fevre M. Regulation of acp1, encoding a non-aspartyl acid protease expressed during pathogenesis of Sclerotinia sclerotiorum. Microbiology. 2001;147(Pt 3):717–26. 10.1099/00221287-147-3-717 [DOI] [PubMed] [Google Scholar]
- 112.Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, et al. Metabolic priming by a secreted fungal effector. Nature. 2011;478(7369):395–8. 10.1038/nature10454 [DOI] [PubMed] [Google Scholar]
- 113.Pazzagli L, Seidl-Seiboth V, Barsottini M, Vargas WA, Scala A, Mukherjee PK. Cerato-platanins: elicitors and effectors. Plant Sci. 2014;228:79–87. 10.1016/j.plantsci.2014.02.009 [DOI] [PubMed] [Google Scholar]
- 114.Kulkarni RD, Kelkar HS, Dean RA. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends in Biochemical Sciences. 2003;28(3):118–21. 10.1016/S0968-0004(03)00025-2 [DOI] [PubMed] [Google Scholar]
- 115.Zhang Z-N, Wu Q-Y, Zhang G-Z, Zhu Y-Y, Murphy RW, Liu Z, et al. Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi. Scientific Reports. 2015;5:13032 10.1038/srep13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.DeZwaan TM, Carroll AM, Valent B, Sweigard JA. Magnaporthe grisea Pth11 is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. The Plant Cell. 1999;11(10):2013–30. 10.1105/tpc.11.10.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kou Y, Tan YH, Ramanujam R, Naqvi NI. Structure–function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytologist. 2017;214(1):330–42. 10.1111/nph.14347 [DOI] [PubMed] [Google Scholar]
- 118.Zhu W, Wei W, Wu Y, Zhou Y, Peng F, Zhang S, et al. BcCFEM1, a CFEM Domain-Containing Protein with putative GPI-anchored site, is involved in pathogenicity, conidial production, and stress tolerance in Botrytis cinerea. Frontiers in Microbiology. 2017;8:1807 10.3389/fmicb.2017.01807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thatcher LF, Williams AH, Garg G, Buck S-AG, Singh KB. Transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. medicaginis during colonisation of resistant and susceptible Medicago truncatula hosts identifies differential pathogenicity profiles and novel candidate effectors. BMC Genomics. 2016;17(1):860 10.1186/s12864-016-3192-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All sequence files are available from the NCBI SRA database under BioProject accession number PRJNA601001. All relevant data are within the manuscript and its Supporting Information files.