Undead neurons may underlie the evolution of new olfactory circuits.
Abstract
Programmed cell death (PCD) is widespread during neurodevelopment, eliminating the surpluses of neuronal production. Using the Drosophila olfactory system, we examined the potential of cells fated to die to contribute to circuit evolution. Inhibition of PCD is sufficient to generate new cells that express neural markers and exhibit odor-evoked activity. These “undead” neurons express a subset of olfactory receptors that is enriched for relatively recent receptor duplicates and includes some normally found in different chemosensory organs and life stages. Moreover, undead neuron axons integrate into the olfactory circuitry in the brain, forming novel receptor/glomerular couplings. Comparison of homologous olfactory lineages across drosophilids reveals natural examples of fate change from death to a functional neuron. Last, we provide evidence that PCD contributes to evolutionary differences in carbon dioxide–sensing circuit formation in Drosophila and mosquitoes. These results reveal the remarkable potential of alterations in PCD patterning to evolve new neural pathways.
INTRODUCTION
A fundamental way in which nervous systems evolve is through increases in the numbers of neurons (1, 2). Additional sensory neurons can enable higher sensitivity to environmental signals or lead to functional diversification to support acquisition of novel detection abilities (3). Increases in central neuron number might underlie diverse enhancements in cognitive abilities (4), such as parallel processing and memory storage.
The generation of more neurons could be achieved through greater production during development, by increasing the number and/or proliferation of neural precursor cells. This process appears to have contributed to neocortical expansion during primate evolution (5). Alternatively (or additionally), given the widespread occurrence of programmed cell death (PCD) during neural development (6, 7), prevention of this process can potentially yield a pool of new neurons. Consistent with this idea, genetic blockage of PCD in mice or Drosophila melanogaster results in the development of enlarged, albeit malformed, nervous systems (8, 9). In Caenorhabditis elegans lacking the CED-3 caspase (a key executioner of PCD), many of the surviving cells differentiate morphologically as neurons (10); moreover, one of these can partially compensate for the function of an experimentally ablated sister pharyngeal neuron (11).
Here, we examined the potential of PCD blockage to generate novel neural pathways in the D. melanogaster olfactory system. The principal olfactory organ in drosophilids, the third antennal segment (hereafter, antenna), is covered with ~400 porous sensory hairs (sensilla) of morphologically diverse classes (Fig. 1A). An individual sensillum derives from a single sensory organ precursor (SOP) cell that is specified in the larval antennal imaginal disc (12, 13). Each SOP gives rise to a short, fixed lineage of asymmetric cell divisions that produces eight terminal cells with distinct identities (13, 14) (Fig. 1B). Four adopt non-neural (“support cell”) fates and are involved in the construction of the hair among other roles. The other four cells can potentially differentiate as olfactory sensory neurons (OSNs), which each express a single (or, rarely, two) sensory receptor genes, develop ciliated dendrites that innervate the lumen of the sensillum hair, and project axons toward a specific glomerulus in the primary olfactory center (antennal lobe) in the brain. There are ~20 sensillum classes housing stereotyped combinations of OSNs (table S1) (15–17). Of these, only one class contains four neurons, with the others housing one, two, or three OSNs. The “missing” neurons are removed by spatially precise PCD ~22 to 32 hours after puparium formation (APF) (14, 18, 19), when OSN terminal fate is established.
Fig. 1. Inhibition of developmental PCD results in increased neuron numbers in the antenna.
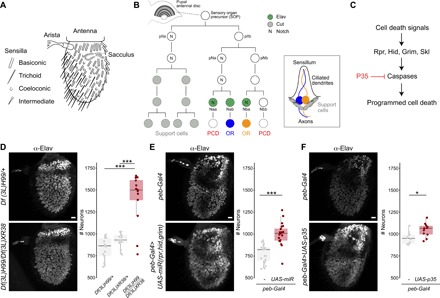
(A) Schematic of the D. melanogaster third antennal segment highlighting different sensory structures. (B) Schematic of the lineage of an antennal disc SOP cell that gives rise to a sensillum containing two neurons (illustrated on the right). The expression of a subset of molecular markers is shown: Elav is expressed in only three of four neural precursors; one of these (Naa) and the Elav-negative cell (Nbb) are eliminated by PCD. The lineage is based on data from (13, 14, 19). (C) Simplified summary of the PCD pathway in D. melanogaster, highlighting the elements relevant for this study. Several intermediate steps between the proapoptotic proteins (Rpr, Grim, Hid, and Skl) and the executioner caspases are not shown. (D) Representative images of anti-Elav immunofluorescence in whole-mount antennae from control [Df(3L)H99/+; the wild-type chromosome here and in other genotypes was derived from a w1118 parent] and PCD-deficient [Df(3L)H99/Df(3L)XR38] animals. Scale bar, 10 μm. Right: Quantifications of antennal neuron numbers of the indicated genotypes, including an additional control genotype [Df(3L)XR38/+] (n = 14, 14, and 13, respectively). Mixed sexes were analyzed; in all other experiments, female flies were used, except where noted otherwise. ***P = 0.0007216 for the comparison to Df(3L)H99/+ and P = 0.0013224 for the comparison to Df(3L)XR38/+ (Wilcoxon rank sum test, corrected for multiple comparisons using a Bonferroni correction). In this and subsequent panels, individual data points are shown, overlaid with boxes indicating the median and first and third quartiles of the data; whiskers show the limits of the distribution. (E) Representative images of anti-Elav immunofluorescence in whole-mount antennae from control (peb-Gal4/+) and PCD-blocked [peb-Gal4/+;UAS-miR(rpr,hid,grim)/+] animals. Scale bar, 10 μm. Right: Quantifications of neuron numbers of these genotypes. ***P = 2.4 × 10−7 (t test) (n = 19 and 21; control and PCD-blocked, respectively). (F) Representative images of anti-Elav immunofluorescence in whole-mount antennae from control (peb-Gal4/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+) animals. Scale bar, 10 μm. Right: Quantifications of neuron numbers of these genotypes. *P = 0.024 (t test) (n = 10 and 11; control and PCD-blocked, respectively).
Here, we demonstrate that prevention of cell death during the development of OSNs is sufficient to generate functional neurons that integrate within preexisting olfactory circuits. Some of these undead neurons represent novel cell types as reflected in their reproducible receptor expression pattern, soma location, and axonal projections. We also provide evidence for the evolutionary diversification of olfactory pathways through changes in the deployment of PCD, both within drosophilid species and between drosophilids and mosquitoes. We propose that cells normally fated to die represent an important evolutionary reserve for the generation of new neuron types and neural circuits.
RESULTS
Inhibition of PCD results in increased neuron number in the antenna
To block PCD during OSN development, we first used animals bearing deletions in the tandem cluster of proapoptotic genes [reaper (rpr), head involution defective (hid), grim, and sickle (skl)], which are critical for promoting developmentally regulated PCD (Fig. 1C) (20, 21). Homozygous chromosomal deficiencies that span the entire cluster cause embryonic lethality. However, a transheterozygous combination of two deletions [Df(3L)H99/Df(3L)XR38], which removes both copies of rpr and one copy each of hid, grim, and skl, allowed recovery of a few viable adults. Immunofluorescence on whole-mount antennae with an antibody against a neural nuclear marker, Elav, revealed a clear increase in the number of labeled cells in mutant animals compared with controls (Fig. 1D), indicating that new neurons develop when cell death is prevented.
PCD might be impaired in these mutant animals at any stage of olfactory system development, including during SOP specification in the antennal disc. To selectively block PCD in terminal OSN precursors (Fig. 1B), we down-regulated expression of rpr, hid, and grim simultaneously by transgenic RNAi from ~18-hour APF using the pebbled-Gal4 (peb-Gal4) driver, which is broadly expressed in postmitotic cells in these lineages. Blockage of OSN-specific PCD also led to a significant increase in Elav-positive cells (Fig. 1E). The number of extra Elav-positive cells observed in this experiment [~200 to 300, recognizing the limits of automated neuron counting in nuclei-dense antennal tissue (fig. S1)] is in line with estimates of the total number of potential “undead” neurons (~300 to 400) (table S1). We further confirmed the role of the canonical PCD pathway in the antenna through expression of the baculoviral caspase inhibitor P35 (Fig. 1C) with the same driver. The peb-Gal4>UAS-p35 (hereafter, “PCD-blocked”) animals displayed higher numbers of Elav-positive cells compared with a peb-Gal4 (control) genotype (Fig. 1F), consistent with a caspase-dependent PCD pathway in this sensory organ.
Undead OSNs are functional
To determine whether these additional Elav-positive cells are functional neurons, we performed single-sensillum electrophysiological recordings. We focused on one class of sensillum, antennal trichoid 1 (at1), that houses a single OSN in wild-type animals, due to PCD of the other three potential neurons in the lineage (19). This OSN expresses the odorant receptor OR67d, which detects the pheromone 11-cis vaccenyl acetate (cVA) (22). The at1 sensilla are easily recognized by their sparse basal (spontaneous) pattern of spikes of single amplitude and the robust train of spikes elicited by cVA but not by other odors (Fig. 2, A and D). In PCD-blocked animals, these sensilla often contain additional spikes of smaller amplitude (Fig. 2, A to C), suggesting the presence of one or more extra active OSNs [pike amplitude is a characteristic property of antennal sensory neurons (23)]. Moreover, exposure to a blend of food-derived odors [which activate many different ORs (24)] led to responses of the undead neurons in about one-third of the tested sensilla (Fig. 2, D and E). The nonresponding, but spiking, undead neurons might express a receptor activated by other stimuli. While this variability in responsiveness could reflect a stochastic fate of these cells, our analysis of the location and wiring of undead neurons presented below argues against this interpretation. We therefore suspect that it is attributable to the PCD-blocking method not being fully efficient, resulting in random rescue of distinct undead OSN types in different at1 sensilla. Regardless, these observations indicate that blocking PCD can lead to the development of functional OSNs.
Fig. 2. Undead OSNs are functional.
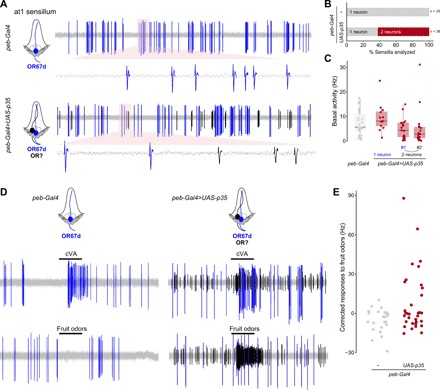
(A) Representative extracellular electrophysiological traces of basal activity from neurons in an at1 sensillum of control (peb-Gal4/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+) animals. Automatically detected spikes (see Materials and Methods) from the neuron expressing OR67d are shown in blue, and those of the additional, undead neuron(s) in black, as schematized in the cartoons on the left (cells fated to die are shown with dashed outlines). (B) Quantifications of the proportion of sensilla containing one neuron (gray) or two (or more) neurons (red) in control (peb-Gal4/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+) animals. (C) Quantifications of the basal activity of the indicated neurons for the control and PCD-blocked genotypes. (D) Representative electrophysiological traces from at1 sensillum recordings in control (peb-Gal4/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+) animals upon stimulation with a 0.5-s pulse (black horizontal bar) of the pheromone cVA [10−2 dilution (v/v) in paraffin oil] or a mix of fruit odors [butyl acetate, ethyl butyrate, 2-heptanone, hexanol, isoamyl acetate, pentyl acetate; each odor at 10−2 dilution (v/v) in paraffin oil]. Automatically detected spikes from the neuron expressing OR67d are shown in blue, and those of the undead neuron(s) in black. (E) Quantifications of odor-evoked responses to fruit odors (see Materials and Methods) in control (peb-Gal4/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+) animals (n = 25 and 34, respectively).
Undead neurons express a subset of olfactory receptor genes
To identify the receptor genes expressed by undead OSNs, we performed comparative transcriptomics of whole antennae of control and PCD-blocked animals by RNA sequencing (RNA-seq). As a positive control, we first examined the changes in transcript levels of rpr, hid, grim, and skl, reasoning that inhibition of PCD downstream in the pathway should lead to the presence of undead cells expressing mRNAs for these proapoptotic genes (Fig. 1C). Three of these genes showed significantly higher expression levels in PCD-blocked antennae (Fig. 3A and table S2).
Fig. 3. Undead neurons express a subset of olfactory receptor genes.
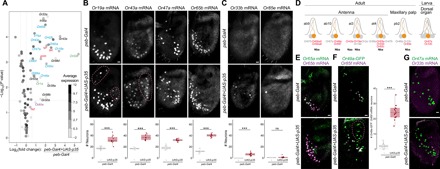
(A) Gene expression differences measured by RNA-seq (see Materials and Methods) between control and PCD-blocked antennae. The volcano plot shows the differential expression (on the x axis) of D. melanogaster Or, Ir, and Gr gene transcripts (each gene represented by a dot), as well as the four proapoptotic genes (grim, rpr, hid, and skl; red labels), plotted against the statistical significance (on the y axis). The mean expression level of individual genes across all samples is shown by the shading of the dot, as indicated by the gray scale on the right [units: log2(counts per million)]. Only chemosensory genes showing a >1.5-fold increase in PCD-blocked antennae are labeled: Blue labels indicate genes whose expression in the antenna has previously been demonstrated by RNA in situ hybridization; magenta and green labels indicate receptors normally only expressed in the adult maxillary palps and larval dorsal organ, respectively; and black labels indicate receptors that are expressed in gustatory organs. The horizontal dashed line indicates a false discovery rate threshold of 5%. Data for all Or, Ir, Gr, and proapoptotic genes are provided in table S2. (B) Representative images of RNA FISH for the indicated Or genes in whole-mount antennae of control (peb-Gal4/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+) animals. Scale bar, 10 μm. Quantifications of neuron numbers are shown at the bottom. *** indicates Or19a P = 6.526 × 10−5 (t test) [n = 17 and 10 (control and PCD-blocked, respectively)], Or43a P = 5.888 × 10−7 (t test) (n = 10 and 10), Or47a P = 3.088 × 10−10 (t test) (n = 13 and 13), Or65b P = 2.2 × 10−16 (t test) (n = 17 and 11) (see fig. S2A for additional examples). The pink dashed lines encircle cells in PCD-blocked antennae that express the visualized Ors outside their usual spatial domain (see also figs. S2B and S3A). Comparison of the variation in OSN number between control and PCD-blocked antennae indicated only one neuron type—of those in this panel and in figs. S2A and S4—displayed greater variance in PCD-blocked antennae (Or19a P = 0.01; F test for equality of variance, with Bonferroni correction for multiple comparisons). (C) Representative images of RNA FISH for the indicated Or genes in whole-mount antennae of control (peb-Gal4/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+) animals. Scale bar, 10 μm. Quantifications of neuron numbers are shown at the bottom. *** indicates Or33a P = 1.812 × 10−7 (t test) (n = 23 and 23), ns indicates Or85e P = 0.053 (Wilcoxon rank sum test) (n = 12 and 22). We never detected any Or85e mRNA-positive neurons in control antennae but frequently detected one (or more) labeled cells in PCD-blocked antennae. (D) Schematic summarizing the normal olfactory organ/sensillum expression pattern of the subset of Or genes with higher expression in PCD-blocked antennae that display coexpression in wild-type neurons (highlighted in red; receptor genes showing no changes in transcript levels are in gray). The neuronal precursor identity of these OSNs is shown below. We represent Or69aA and Or69aB as distinct receptors here because, although they share 3′ exons, they are transcribed from different promoters and encode receptors with different odor specificities (15, 56). (These isoforms were not, however, distinguished in the RNA-seq and RNA FISH analyses). (E) Representative images of combined RNA FISH for Or65a (green) and Or65b (magenta) in whole-mount antennae of control (peb-Gal4/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+) animals (n = 4 and 5, respectively), showing coexpression of these receptors in both endogenous and undead neurons. Scale bar, 10 μm. (F) Representative images of RNA FISH for Or85f and anti-GFP in whole-mount antennae of control (peb-Gal4/+;Or49a-GFP/+) and PCD-blocked (peb-Gal4/+;Or49a-GFP/UAS-p35) animals. Scale bar, 10 μm. Quantifications of coexpression are shown to the right. *** indicates Or49a-GFP+/Or85f mRNA− population P = 5.48 × 10−12 (t test) (n = 16 and 15; control and PCD-blocked, respectively). The pink dashed line encircles cells in the PCD-blocked antenna that express Or49a-GFP outside the usual spatial domain (see also fig. S2B). We used an Or49a-GFP reporter due to our inability to reliably detect Or49a transcripts in situ; the higher number of Or49a-GFP+/Or85f mRNA− negative cells is not an artifact of the detection method, as an Or85f-GFP reporter revealed a similarly limited increase in neuron number (fig. S3B). (G) Representative images of combined RNA FISH for Or47a (green) and Or33b (magenta) in whole-mount antennae of control (peb-Gal4/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+) animals. In control animals, Or33b is coexpressed with Or47a in the larval dorsal organ and is never detected in the antenna. In PCD-blocked antennae, Or33b- and Or47a-expressing undead neurons are almost completely nonoverlapping: 4% of Or33b-positive undead OSNs weakly coexpress Or47a (n = 79 cells from 10 antennae). Scale bar, 10 μm.
We next queried the transcript levels for all chemosensory receptors, comprising Or, Ionotropic receptor (Ir), and Gustatory receptor (Gr) gene families (tables S1 and S2). Of the receptors previously detected in antennal neurons in situ (15, 16, 25), we found that 10/36 Ors, 1/17 Irs, and 0/3 Grs displayed a >1.5-fold increase in expression, suggesting that only subsets of these receptors are expressed in undead neurons (Fig. 3A and tables S2 and S3).
To validate these transcriptomic data, we visualized the neuronal expression of several of the Ors in situ. Transcripts for all of those tested by RNA fluorescence in situ hybridization (FISH) or validated Or promoter-CD8:green fluorescent protein (GFP) (hereafter, Or-GFP) reporters (15) were detected in more neurons in PCD-blocked antennae compared with controls (Fig. 3B and figs. S2A and S3). In some cases, these neurons were found only within the same region of the antenna as the endogenous OSNs (e.g., Or42b, Or47a, Or65a, Or65b, and Or85f), while in others (e.g., Or19a, Or43a, and Or49a), undead neurons were observed in novel, but reproducible, locations (Fig. 3B and figs. S2 and S3A). The variance in the number of neurons expressing a particular receptor was not significantly different between control and PCD-blocked antennae for almost all populations (Fig. 3B legend). Together, these observations suggest that undead neurons display consistent, rather than stochastic, receptor expression patterns.
Notably, many of the other receptors displaying increases in transcript levels normally act in other chemosensory organs, including one Or (Or85e) of the maxillary palp (a distinct olfactory appendage of insects), two larval Ors (Or1a and Or33b), and seven Grs, which function in various gustatory organs (Fig. 3A and tables S2 and S3). In situ analysis confirmed the presence of transcripts for the palp-specific Or85e and the larval-specific Or33b in populations of undead neurons in PCD-blocked antennae (Fig. 3C). These observations suggest that undead neurons can provide a cellular substrate to allow switching of receptor expression between sensory organs and/or life stages during evolution.
For the vast majority of receptor genes, RNA levels were either unchanged or slightly down-regulated in PCD-blocked antennae (Fig. 3A and table S2). Consistently, in situ analysis by FISH or immunofluorescence of several antennal receptors revealed only a very small increase (Ir75c), no change (e.g., Or13a, Or67d, Gr63a, Ir40a, and Ir75b), or a decrease (Or22a) in the size of the corresponding neuron populations (fig. S4). The latter, unexpected phenotype raises the possibility that undead neurons affect (directly or indirectly) the specification and/or survival of certain populations of neurons.
Coexpressed receptor genes are overrepresented in undead neurons
We examined the properties that characterize the small subset of receptors that are predominant in undead neurons. These genes are normally expressed in neurons housed in diverse sensillum types: basiconic (e.g., Or42b), trichoid (e.g., Or65a), intermediate (e.g., Or19a), and coeloconic (e.g., Ir75d). By contrast, most of these receptors (including 9 of 10 Ors) are expressed in OSNs derived from the Nba precursor cell; the remaining Or (Or43a) and the sole Ir (Ir75d) are expressed in Naa-derived OSNs (Figs. 1B and 3D and table S3). This pattern suggests that undead neurons—which are largely Naa derived (Fig. 1B)—preserve gene regulatory networks that are more similar to Nba cells than Nab cells, possibly reflecting the shared terminal Notch activity in Nba and Naa precursors (Fig. 1B) (13).
Of the 13 Ors detected in undead neurons (including those from other olfactory organs), 10 are normally coexpressed with other Or genes (Fig. 3D and table S3), most of which are relatively recent receptor duplicates. This enrichment (P = 1.101 × 10−4, chi-squared test; table S4) is notable given the rarity of receptor coexpression within this repertoire (15, 25). While some of these receptors retain their coexpression properties in undead OSNs [e.g., Or65a and Or65b (Fig. 3E)], this is not always the case. For example, Or19a, but not the coexpressed Or19b, displays up-regulation by RNA-seq (table S2). Similarly, while Or49a-GFP and Or85f are always coexpressed in control antennal OSNs, we detected in PCD-blocked antennae a population of undead neurons that expresses Or49a-GFP but not Or85f (Fig. 3F). Last, the larval receptor gene Or33b is coexpressed with Or47a at this life stage (26), but in PCD-blocked antennae, the novel Or33b undead neurons display only very rare expression of Or47a (Fig. 3G).
Why normally coexpressed receptors are overrepresented in undead neurons remains unknown. Nevertheless, these observations reveal that the regulatory regions of coexpressed receptor genes are, in some cases, different enough to support independent expression under the control of the gene regulatory networks of undead neurons. This phenomenon provides an intriguing link between the generation of novel OSNs and the segregation of coexpressed receptors into distinct OSN populations.
Undead OSNs form novel receptor/glomerular couplings in the brain
We next investigated whether undead OSNs project their axons to the antennal lobe. We first broadly labeled these neurons using an enhanced green fluorescent protein (EGFP) gene trap allele of grim [grimMI03811(EGFP)], in which the fluorophore should report on the expression pattern of this proapoptotic gene. In control animals, grimMI03811(EGFP) expression was detected only at background levels across the antenna (Fig. 4A); this is expected, as cells that induce Grim (and so EGFP) expression are fated to die. By contrast, in PCD-blocked antennae, EGFP was present in many soma (Fig. 4A), which presumably represent the undead neurons previously observed with Elav antibodies (Fig. 1F). In the brains of these animals, we observed that EGFP-labeled processes innervate multiple glomeruli of the antennal lobe (Fig. 4B), indicating that undead neurons can extend axons to the primary olfactory center. Antennal deafferentation experiments confirmed that the glomerular signals in PCD-blocked animals were entirely due to the contribution of OSNs (Fig. 4B).
Fig. 4. Undead OSNs form novel receptor/glomerular couplings in the brain.
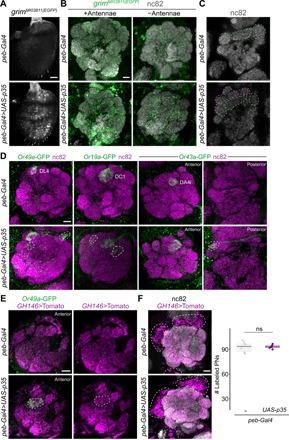
(A) Representative images of anti-GFP immunofluorescence in whole-mount antennae of control [peb-Gal4/+;;grimMI03811(EGFP)/+] and PCD-blocked [peb-Gal4/+; UAS-p35/+;grimMI03811(EGFP)/+] animals. Blind scoring by two independent observers of antennae as control (n = 10) or PCD-blocked (n = 9) was 100% accurate. Scale bar, 10 μm. (B) Representative images of combined anti-GFP and nc82 immunofluorescence in whole-mount brains of control (peb-Gal4/+;;grimMI03811(EGFP)/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+;grimMI03811(EGFP)/+) animals with antennae intact (left) or excised (right). Blind categorization of brains as control (n = 9) or PCD-blocked (n = 12) was 85% accurate (two independent observers). Scale bar, 10 μm. (C) Representative images of nc82 immunofluorescence in whole-mount brains of control (peb-Gal4/+;;grimMI03811(EGFP)/+) and PCD-blocked (peb-Gal4/+;UAS-p35/+;grimMI03811(EGFP)/+) animals. Equivalent glomeruli found in antennal lobes of control and PCD-blocked animals are labeled for reference with dashed blue lines; potential novel glomerular structures (or displaced original glomeruli) in PCD-blocked animals are shown with dashed pink lines. Further planes of the same brains are shown in fig. S5A. Blind categorization of brains as control (n = 11) or PCD-blocked (n = 13)—visualizing only the nc82 channel of confocal stacks of animals containing a diversity of fluorescent reporters in the genetic background; see genotypes below—was 85% accurate (two independent observers). Scale bar, 10 μm. (D) Representative images of combined anti-GFP and nc82 immunofluorescence in whole-mount brains of control (peb-Gal4/+;Or49a-GFP/Or49a-GFP, peb-Gal4/+;;Or19a-GFP/+, peb-Gal4/+;Or43a-GFP/+) and PCD-blocked (peb-Gal4/+;Or49a-GFP/Or49a-GFP,UAS-p35, peb-Gal4/+;UAS-p35/+;Or19a-GFP/+, peb-Gal4/+;Or43a-GFP/UAS-p35) animals. Blind categorization of brains as control or PCD-blocked—visualizing only the GFP channel of confocal stacks and based exclusively on additional glomerular innervation patterns—was, for each reporter, respectively, Or49a-GFP: 85% (n = 10 and 10; control and PCD-blocked, respectively); Or19a-GFP: 89.5% (n = 20 and 18); and Or43a-GFP: 93.9% (n = 19 and 15). Scale bar, 10 μm. Additional representative images are provided in fig. S5B. (E) Representative images of combined anti-GFP, anti–red fluorescent protein (Tomato), and nc82 immunofluorescence in whole-mount brains of control (peb-Gal4/+;Or49a-GFP/Or49a-GFP;GH146-QF,QUAS-Tomato/+) and PCD-blocked (peb-Gal4/+;Or49a-GFP/Or49a-GFP,UAS-p35;GH146-QF,QUAS-Tomato/+) animals. The dashed line encircles the novel Or49a-GFP–labeled glomerulus (i.e., not the normal DL4 glomerulus, which is not visible in this anterior plane of the antennal lobe). Scale bar, 10 μm. (F) Representative images of PN soma (bounded by the dashed lines) labeled by GH146>Tomato in whole-mount brains of control (peb-Gal4/+;Or49a-GFP/Or49a-GFP;GH146-QF,QUAS-Tomato/+) and PCD-blocked (peb-Gal4/+;Or49a-GFP/Or49a-GFP,UAS-p35;GH146-QF,QUAS-Tomato/+) animals. Scale bar, 10 μm. Quantifications of labeled PN numbers are shown to the right. ns indicates P = 0.819 (t test) (n = 6 and 5, control and PCD-blocked, respectively).
Analysis of the overall architecture of the antennal lobe in control and PCD-blocked animals, as visualized with the synaptic marker nc82 (Bruchpilot), revealed substantial morphological differences (Fig. 4C and fig. S5A), including less distinct boundaries between certain glomeruli and putatively novel regions of neuropil. To test whether these differences reflect the innervation patterns of populations of undead OSNs, we examined the projections of neurons expressing reporters for several of the Or populations that increase in size in PCD-blocked antennae: Or49a-GFP, Or19a-GFP, and Or43a-GFP (Fig. 4D and fig. S5B). In control animals Or49a-GFP neurons project to a single glomerulus, DL4, as previously described (15). In PCD-blocked animals, labeled axons projected to DL4, as well as to a second, more anterior, glomerulus-like structure, and occasionally to a more medial location (Fig. 4D and fig. S5B). These presumably correspond to the wild-type Or49a neuron population and the undead neurons that express this reporter [but not Or85f (Fig. 3F)], respectively. Similarly, endogenous neurons expressing Or19a-GFP project to a single glomerulus, DC1, whereas undead Or19a-GFP–expressing neurons target two additional regions of neuropil (Fig. 4D and fig. S5B). Last, endogenous Or43a-GFP OSNs project to DA4l, while the undead neurons labeled by this reporter innervate a distinct posterolateral glomerulus (Fig. 4D and fig. S5B).
The reproducibility of projection patterns of undead neurons of a given type—together with the characteristic location of undead neuron soma in the antenna (Fig. 3B and figs. S2 and S3A)—is consistent with undead neurons adopting a relatively constrained, rather than completely random, developmental fate. The global changes in glomerular boundaries make it difficult to distinguish whether the axons of undead neurons in these three cases target novel glomeruli or partially/completely overlap with preexisting glomeruli (i.e., those formed by other populations of endogenous neurons). Nevertheless, the differences in projection patterns of undead and endogenous neurons that express the same receptor highlight that undead neurons may allow novel coupling between receptor expression and glomerular target, an essential step during the evolution of new olfactory pathways.
We investigated whether undead OSN axons can potentially synapse with second-order projection neurons (PNs), which carry olfactory signals to higher brain regions. We combined the Or-GFP transgenic reporters with a genetic reporter for most uniglomerular PNs (GH146-QF>QUAS-Tomato) in control and antennal PCD-blocked flies. In the novel Or49a-GFP–labeled glomerulus, GH146-labeled processes were detected (Fig. 4E). The undead neuron glomeruli innervated by Or19a-GFP and Or43a-GFP did not overlap with GH146-positive PNs (fig. S5C), but we suspect that this absence is due to incomplete coverage of PNs by the GH146-QF driver (we note that it does not label the endogenous Or19a/DC1 glomerulus) (fig. S5C). The nc82 immunoreactivity is present in the undead neuron glomeruli of all three OSN classes (Fig. 4D), implying the formation of synapses between these sensory neurons and central circuit elements. Novel connectivity of undead OSNs does not result from the production of additional PNs (Fig. 4F). This result suggests that there is no mechanism to match OSN and PN numbers and that the observed innervation derives from local recruitment of PN dendrites in the antennal lobe during synapse formation. How new PN classes dedicated to novel OSN populations evolve remains unknown.
Natural examples of evolved differences in OSN numbers
Our demonstration that inhibition of PCD is sufficient to allow the development of new functional OSN populations that integrate into the olfactory circuitry is consistent with the hypothesis that modulation of cell death patterns during evolution can be a mechanism to create olfactory channels. While the variation in OSN number per sensillum within D. melanogaster (table S1) implies that different SOP lineages have distinct regulation of PCD, we wondered whether we could identify examples of divergent deployment of PCD across shorter evolutionary time scales by comparing numbers of neurons in homologous olfactory sensilla in different drosophilids. Previous cross-species analyses that surveyed a subset of basiconic and coeloconic sensilla in a limited range of drosophilids found no differences in neuron numbers (although receptor tuning properties can vary substantially) [e.g., (27, 28)]. However, all the sensilla characterized contain two or more neurons, likely making it challenging to identify an additional spike amplitude.
We therefore performed a broader electrophysiological screening in 26 drosophilid species, focusing on the at1 sensillum class for ease of neuronal spike amplitude sorting (fig. S6). While at1 sensilla of most species house a single cVA-responsive neuron (Fig. 5, A and B), similar to D. melanogaster, we identified nine species in which this sensillum class houses two neurons of distinct spike amplitudes (Fig. 5C), only one of which is cVA responsive. The lack of genomic data and genetic tools for these species currently precludes further molecular analysis, but we assume that the cVA-responsive at1 neuron expresses an OR67d ortholog and the partner neuron a distinct receptor of still-unknown sensory specificity. We have unfortunately not been able to determine the receptor(s) expressed in undead neurons in D. melanogaster at1 sensilla (Fig. 2), but, given the phylogenetic distance between these drosophilids, it is possible—if not likely—that these natural additional at1 neurons express receptors not even present in D. melanogaster.
Fig. 5. Examples of naturally occurring extra neurons in at1 sensilla.
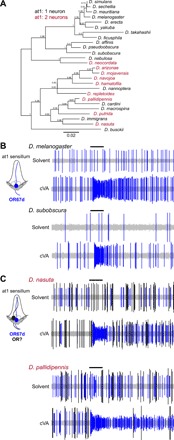
(A) Phylogeny of 26 drosophilid species, representing most of the Drosophila genus subgroups, based on the protein sequences of housekeeping loci (see Materials and Methods). Species names are colored to reflect the presence of one or two neurons in at1 sensilla. Numbers next to the tree nodes indicate the support values. The scale bar for branch length represents the number of substitutions per site. (B) Representative electrophysiological traces from recordings of at1 sensilla of the indicated drosophilid species (n = 5 per species) upon stimulation with a 0.5-s pulse (black horizontal bar) of solvent (dichloromethane) or cVA [10−2 dilution (v/v)]. A single cVA-responsive neuron (known or assumed to express OR67d orthologs) is detected (shown in blue), as schematized in the cartoon on the left. (C) Representative electrophysiological traces from recordings of at1 sensilla of the indicated drosophilid species (n = 5 per species) upon stimulation with a 0.5-s pulse (black horizontal bar) of solvent (dichloromethane) or cVA [10−2 dilution (v/v)]. Two classes of spike are detected: a cVA-responsive neuron (assumed to express OR67d orthologs) (shown in blue) and a second neuron with a larger spike amplitude, which does not respond to cVA (shown in black). The inferred sensilla organization is shown in the cartoon on the left.
Although we cannot exclude that the extra at1 neuron in these nine species is due to an extra cell division, it would be an unprecedented property of an SOP lineage to have only one of the four terminal cells undergo an additional division. The most plausible explanation for the at1 phenotype in these species is that it reflects a change in fate from PCD to a functional OSN to permit formation of a novel olfactory channel. Mapping the species whose at1 sensilla house more than one OSN onto a phylogenetic tree revealed that the acquisition of an additional neuron (i.e., a potential change in PCD patterning) has occurred independently multiple times during the evolution of the drosophilid clade (Fig. 5A). This observation suggests that the diversification in sensilla development has a relatively simple—and potentially common—genetic basis.
PCD can explain an evolutionary difference in carbon dioxide–sensing neuron formation in drosophilids and mosquitoes
Differences in neuron numbers within homologous sensilla have been described in the maxillary palp of more divergent dipteran species. In D. melanogaster (and other drosophilids), palp sensilla each house two Or OSNs (29). By contrast, in mosquitoes, these sensilla house three neurons, comprising two Or-expressing OSNs and the carbon dioxide (CO2)–sensing neuron (expressing CO2 receptor subunits encoded by Gr genes) (Fig. 6A) (30, 31). In drosophilids, CO2-sensing neurons (expressing orthologous Gr genes) are confined to the antenna (Fig. 6A) (32, 33).
Fig. 6. PCD can explain an evolutionary difference in CO2-sensing neuron formation in D. melanogaster and mosquitoes.
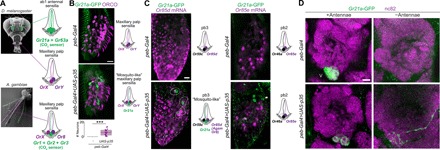
(A) Left: Scanning electron micrographs of D. melanogaster and Anopheles gambiae heads indicating the two olfactory organs: antennae (blue) and maxillary palps (magenta) [adapted from (3), with permission]. Right: Schematic illustrating that, in D. melanogaster, CO2-sensing neurons (green) are located in antennal ab1 sensilla and maxillary palp sensilla house only two Or neurons. By contrast, in A. gambiae (and other mosquitoes), CO2-sensing neurons are located in the maxillary palp, housed together with two Or neurons. (B) Representative images of combined anti-ORCO (the OR coreceptor, which labels all OR neurons) and anti-GFP immunofluorescence in whole-mount maxillary palps of control (peb-Gal4/+;Gr21a-GFP/+) and PCD-blocked (peb-Gal4/+;Gr21a-GFP/UAS-p35) animals. Scale bar, 10 μm. Quantifications of Gr21a-GFP–positive neuron numbers are shown at the bottom; *** indicates P = 6.7 × 10−9 (Wilcoxon rank sum test) [n = 22 and n = 21 (control and PCD-blocked, respectively)]. Schematic of inferred maxillary palp sensilla organization in each genotype is illustrated on the right. (C) Representative images of combined RNA FISH for the indicated Or genes (magenta) and anti-GFP immunofluorescence (green) in whole-mount maxillary palps of control (peb-Gal4/+;Gr21a-GFP/+) and PCD-blocked (peb-Gal4/+;Gr21a-GFP/UAS-p35) animals. Schematic of the inferred maxillary palp sensilla organization in each genotype is illustrated on the right. GFP-positive neurons pair with Or85d OSNs in pb3 sensilla (encircled by dashed lines) but not Or85e OSNs in pb2 sensilla (arrowheads). (D) Representative images of combined anti-GFP and nc82 immunofluorescence in whole-mount brains of control (peb-Gal4/+;Gr21a-GFP/+) and PCD-blocked (peb-Gal4/+;Gr21a-GFP/UAS-p35) animals with antennae intact (left) or excised (right). Blind categorization of brains from animals lacking antennae as control (n = 19) or PCD-blocked (n = 17) (visualizing only the GFP channel of confocal stacks and based exclusively on the presence of GFP-labeled axons) was 94.4% accurate. Scale bar, 10 μm. Additional representative images are provided in fig. S7.
To investigate whether PCD can account for differences in palp sensilla neuron organization, we examined a reporter of the CO2 receptor subunit GR21a in control and PCD-blocked D. melanogaster. Gr21a-GFP is expressed in the antenna but not the palps of control animals (Fig. 6B), consistent with previous observations (32, 33). Remarkably, in PCD-blocked palps, this reporter labels several neurons (Fig. 6B). Costainings with RNA probes for maxillary palp Ors revealed that these Gr21a-GFP–positive neurons are paired specifically with OSNs expressing Or85d in the pb3 sensillum class (Fig. 6C). This is notable because Or85d is the D. melanogaster receptor that is most closely related to mosquito Or8 (34), which is one of the two Ors expressed in mosquito palp sensilla (Fig. 6A). Thus, prevention of PCD creates sensilla in D. melanogaster maxillary palps that molecularly resemble sensillar organization in the homologous mosquito sensory organ.
Last, we examined where these palp Gr21a-GFP–expressing undead neurons project. In control animals, antennal Gr21a-GFP neurons project unilaterally to the V glomerulus (Fig. 6D), as described (32, 33). In PCD-blocked animals, Gr21a-GFP neurons (comprising both antennal and undead palp neuron populations) also converge on the V glomerulus. However, we could detect commissural projections and/or ectopic sites of innervation adjacent to this glomerulus (Fig. 6D). Antennal deafferentation allowed us to selectively visualize Gr21a-GFP undead palp neurons: in controls, as expected, no innervations are labeled, while in PCD-blocked animals, GFP-positive palp axons project bilaterally and run among and occasionally branch within the medial glomeruli of the antennal lobe before terminating within (or very near) the V glomerulus (Fig. 6D and fig. S7). The projection pattern of these undead neurons is partly reminiscent of mosquito CO2-sensing palp neurons, which project bilaterally to medial glomeruli (35, 36).
These results indicate that changes in PCD patterning can explain, in part, an evolutionary difference between drosophilids and mosquitoes for this important olfactory pathway. Additional genetic modifications must have occurred, for example, to promote high-level expression of CO2-receptor genes (which we have not yet been able to detect by RNA FISH) and to fully distinguish the axonal projection patterns of these neurons. Such developmental differences may be controlled—directly or indirectly—by the transcription factor Prospero and/or the microRNA, mir-279, whose loss in D. melanogaster leads to formation of ectopic palp CO2 neurons (37, 38).
DISCUSSION
We have shown that prevention of PCD is sufficient to allow development of functional OSNs that integrate into the extant olfactory circuitry of D. melanogaster. We have also provided evidence that species-specific deployment of PCD contributes to differences in olfactory pathway organization across drosophilids and mosquitoes.
A future challenge will be to understand the natural mechanisms controlling PCD in OSN lineages to address how this process is selectively suppressed (or activated) during evolution to create (or eliminate) OSNs. Our RNA-seq dataset provides an entry point to answer this question by identifying candidate genes expressed highly in cells normally fated to die, similar to the proapoptotic factors. Given the timing and precise stereotypy of PCD within the olfactory lineages (13, 14, 19), it is also possible that PCD is induced through mechanisms similar to those that define the fate of OSNs that survive and express specific receptor genes.
One key observation of our study is that undead neuron populations do not necessarily exhibit functional or anatomical properties that match those of existing OSNs, as exemplified by their expression of receptor genes not normally transcribed in antennal neurons and by the existence of new receptor/glomerular couplings. These traits presumably reflect properties of undead OSNs’ inherent (although normally “hidden”) gene regulatory networks and reveal the evolutionary potential of cells fated to die to differentiate as neurons with novel functions and wiring patterns. Future work should reveal how alterations in PCD patterning combine with other genetic changes (e.g., to refine receptor expression and neuronal projections) to form new, precisely segregated olfactory pathways. Given the widespread occurrence of PCD throughout the nervous systems of insects and other animals (6, 7), the contribution of undead neurons to neural circuit evolution is likely to extend to many different brain regions in diverse species (39).
MATERIALS AND METHODS
Drosophila culture
Flies were maintained at 25°C in 12-hour light:12-hour dark conditions, except where noted. D. melanogaster strains were cultured on a standard cornmeal diet; other drosophilid species were grown on food sources as indicated in table S5 (for recipes, see http://blogs.cornell.edu/drosophila/recipes). Care of experimental animals was in accordance with institutional guidelines. Published mutant and transgenic D. melanogaster are described in table S5. Df(3L)H99/Df(3L)XR38 (and their controls) were cultured at 22°C to increase the recovery of adult offspring of the desired genotype. For most histological experiments, only female flies were analyzed to avoid confounding variation due to known sexual dimorphisms (17). Mixed sexes were used for Df(3L)H99/Df(3L)XR38 flies in Fig. 1D due to the limitation in the recovery of this genotype, as well as for anti-IR75b and anti-IR75c immunofluorescence in fig. S4 (there is no sexual dimorphism in the numbers of these OSNs). For histological experiments, flies were 1 to 12 days old. Animals subjected to antennal deafferentation (and control intact flies) were left for 10 days after surgery to permit degeneration of OSN axons. The experiments in Fig. 5 and fig. S6 were carried out with 8- to 15-day-old female flies.
Histology and image analysis
Whole-mount antennal immunofluorescence and RNA FISH were performed essentially as described (40), except samples were incubated with RNA probes for at least 40 hours at 60°C, and anti–digoxigenin (DIG)–peroxidase (POD) (or anti–fluorescein-POD) antibodies for at least 48 hours at 4°C. For maxillary palp histology, whole mouthparts (with palps attached) were dissected from the head in 4% paraformaldehyde and fixed for 3 hours at 4°C, followed by the same staining protocols as for antennae; before mounting of palps, excess mouthpart tissue was removed. For combined RNA FISH/anti-GFP immunofluorescence, tissues were treated as for RNA FISH but including anti-GFP antibody together with the anti–DIG-POD; after RNA visualization, a second incubation with anti-GFP (for 24 hour at room temperature) was performed. Whole-mount brain immunofluorescence was performed essentially as described (41). Primary and secondary antibodies are listed in table S6. Sources and/or construction details of templates for RNA probes are provided in table S7. Imaging was performed on a Zeiss confocal microscope LSM710 or LSM880 using a 40× oil immersion objective.
For automated counting of Elav-positive cell bodies, confocal stacks were imported into Fiji (42) and passed through a median three-dimensional (3D) filter of radius 1 in all dimensions. Images were subsequently thresholded using the 3D iterative thresholding plugin (43), and cells were automatically counted using the 3D object counter (44). For counting of OSN numbers expressing specific olfactory receptor genes, confocal stacks were imported into Fiji (42), and cell counting was performed manually using the Cell Counter plugin of ImageJ. Analyses of OSN numbers expressing specific olfactory receptor genes and of morphological differences of the antennal lobes were performed by experimenters who were blind to the genotype using RandomNames.bat (https://github.com/DavidOVM/File-Name-Randomizer/blob/master/RandomNames.bat) to encode image names.
Electrophysiology
Single-sensillum recordings were performed and analyzed essentially as described (45, 46). The at1 sensilla were identified on the basis of their morphology and characteristic distal distribution on the antenna; they could also be clearly distinguished from the only other trichoid sensillum class, at4, which houses three OSNs (fig. S6). Chemical stimuli and solvents are described in table S8. For the experiments in Fig. 2, neuron activity was recorded for 10 s, starting 3 s before a stimulation period of 0.5 s. For the experiments in Fig. 5 and fig. S6, neuron activity was recorded for 6 s, starting 2 s before a stimulation period of 0.5 s.
Traces were analyzed by automated sorting of spike amplitudes in AutoSpike (Syntech); representative traces presented in the figures were further processed in Adobe Illustrator CS (Adobe systems, San Jose, CA). Basal neuron activity was quantified by counting spontaneous spikes in a 10-s recording window without stimulus. Odor-evoked activity was quantified by counting spikes in a 0.5-s window during odor stimulation, subtracting from this the number of spikes in a 0.5-s recording window before stimulation, and multiplying the result by two. For the solvent-corrected quantifications in Fig. 2E, the responses to solvent (paraffin oil) were subtracted from the responses to the odor. In Fig. 2B, sensilla were classified as having two neurons if two different spike amplitudes were automatically detected and/or corrected responses to the fruit odor mix were >20 Hz.
RNA-seq and analysis
Antennal RNA was extracted from three biological replicates of control (peb-Gal4/+;Or49a-GFP/+) and PCD-blocked (peb-Gal4/+;Or49a-GFP/UAS-p35) animals. (The increased numbers of neurons labeled by Or49a-GFP was noted in preliminary studies, and we therefore incorporated this transgene into the genotypes used in these experiments as an internal control; see below.) For each pair of biological replicates, ~200 animals were grown under identical conditions, and RNA was extracted in parallel using 2- to 5-day-old flies, as described (19). RNA quality was assessed on a fragment analyzer (Advanced Analytical Technologies, Inc.); all RNAs had an RNA quality number of 9.8 to 10. From 100 ng of total RNA, mRNA was isolated with the NEBNext Poly(A) mRNA Magnetic Isolation Module. RNA-seq libraries were prepared from the mRNA using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs). Cluster generation was performed with the resulting libraries using the Illumina TruSeq PE Cluster Kit v4 reagents and sequenced on the Illumina HiSeq 2500 using TruSeq SBS Kit v4 reagents (Illumina). Sequencing data were demultiplexed using the bcl2fastq Conversion Software (v2.20, Illumina).
Purity-filtered reads were adapter and quality trimmed with Cutadapt (v1.8 (47)). Reads matching to ribosomal RNA sequences were removed with fastq_screen (v0.11.1). Remaining reads were further filtered for low complexity with Reaper (v15-065) (48). Reads were aligned to the D. melanogaster BDGP6.92 genome using STAR (v2.5.3a) (49). The number of read counts per gene locus was summarized with htseq-count (v0.9.1) (50) using D. melanogaster BDGP6.92 gene annotation. The quality of the RNA-seq data alignment was assessed using RSeQC (v2.3.7) (51).
Statistical analysis was performed for genes in R (v3.5.3). Genes with low counts were filtered out according to the rule of one count per million in at least one sample. Library sizes were scaled using TMM normalization (edgeR package v3.24.3) (52) and log transformed with limma cpm function (Limma package v3.38.3) (53).
Differential expression was computed with limma for paired samples by fitting the six samples into a linear model and performing the comparison PCD-blocked antennae versus controls. Comparison of read number for GFP (encoded by the Or49a-GFP transgene) was performed by mapping reads to the GFP sequence with Bowtie2 (54): control antennal RNA—139 ± 9.5 reads per sample (mean ± SD); PCD-blocked antennal RNA—227 ± 8.7 reads per sample.
A moderated t test was used for the comparison of a subset of 83 detectably expressed D. melanogaster genes including Or, Ir, and Gr genes as well as the four proapoptotic genes (rpr, hid, grim, and skl). For multiple testing correction, the P values were adjusted by the Benjamini-Hochberg method, which controls the false discovery rate. The volcano plot (Fig. 3A) was generated in R by plotting the log2(fold change PCD-blocked versus control) against the −log(P value). Data points were shaded according to mean expression value across all samples.
Phylogenetics
Phylogenetic analysis of drosophilid species was conducted using six housekeeping proteins, encompassing two encoded by nuclear genes (Adh and Xdh), and four by mitochondrial genes (COI, COII, COIII, and ND2). Amino acid sequences of each species were obtained from UniProt (https://www.uniprot.org; accession numbers are listed in table S9) and concatenated in Geneious (v11.0.5). A multiple sequence alignment of 2939 positions was generated using MAFFT (v7.309) with E-INS-i parameters and scoring matrix 200 PAM/K = 2 (55). The final tree was reconstructed using a maximum likelihood approach with the GTR + G + I model of nucleotide substitution and 1000 rate categories of sites in FastTree (v2.1.5). The tree was visualized and processed in Geneious (v11.0.5).
Statistics
Statistical analyses and plotting were made in RStudio (v1.1.463 R Foundation for Statistical Computing, Vienna, Austria, 2005; R-project.org), except for the RNA-seq analyses (described above). Normality was first assessed on datasets using a Shapiro test. If both datasets (i.e., control and PCD-blocked) for a given experiment were normally distributed, a two-sided t test was performed; otherwise, a Wilcoxon rank sum test was performed. For the analysis of the enrichment of the subset of coexpressed Ors (within the entire Or repertoire) in undead neurons, we performed a chi-squared test using a contingency table where the expected frequencies were calculated on the basis of the normal expression pattern of all Ors across all olfactory organs in D. melanogaster (table S4).
Supplementary Material
Acknowledgments
We are grateful to I. Alali and M. Erdogmus for technical assistance; D. Williams and S. Pop for sharing flies and discussions; R. Álvarez-Ocaña for sharing information on neuron numbers; and M. Alenius, L. Luo, L. Vosshall, the Bloomington Drosophila Stock Center (NIH P40OD018537), and the Developmental Studies Hybridoma Bank (NICHD of the NIH, University of Iowa) for reagents. We thank R. Arguello and members of the Benton laboratory for comments on the manuscript. Funding: L.L.P.-G. was supported by a FEBS Long-Term Fellowship and an ERC Starting Independent Researcher Grant (802531). M.A.K., B.S.H., and M.K. are supported by the Max Planck Society. Research in R.B.’s laboratory is supported by the University of Lausanne, an ERC Consolidator Grant (615094), and the Swiss National Science Foundation. Author contributions: L.L.P.-G. and R.B. conceived the project. All authors contributed to the experimental design, analysis, and interpretation of results. Experimental contributions were as follows: L.L.P.-G. (Figs. 2 and 4A and figs. S1 and S4), A.F.S. [Figs. 1 (D to F) and 4 (B, C, E, and F) and figs. S1, S2, S3B, S4, and S5A], M.A.K. (Fig. 5 and fig. S6), S.C. (Figs. 3 and 6, B and C, and figs. S2, S3A, and S4), and R.B. (Figs. 4D and 6D and figs. S5, B and C, and S7). K.B. and S.P. performed RNA-seq data analysis. R.B., L.L.P.-G., and A.F.S. wrote the paper with input from all the other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. RNA-seq data are available in GEO (accession GSE128725).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaaz7238/DC1
Fig. S1. Automated quantification of Elav-positive antennal neurons.
Fig. S2. Undead neurons express a subset of olfactory receptor genes and can be found in novel, reproducible locations.
Fig. S3. Or-GFP reporters faithfully recapitulate receptor expression in undead neurons.
Fig. S4. Many sensory neuron populations show little or no increase, or a decrease, in size in PCD-blocked antennae.
Fig. S5. Undead OSNs form novel receptor/glomerular couplings in the brain.
Fig. S6. Electrophysiological distinction of at1 and at4 sensilla.
Fig. S7. Projections of Gr21a-GFP–expressing undead neurons from the maxillary palps.
Table S1. Estimated potential pool of undead neurons in the antenna.
Table S2. Comparison of chemosensory receptor and proapoptotic factor transcript abundance in control and PCD-blocked antennae.
Table S3. In situ expression properties of chemosensory genes up-regulated in PCD-blocked antennae.
Table S4. Statistical analysis of the enrichment of coexpressed Ors in undead neurons.
Table S5. Drosophila strains.
Table S6. Antibodies.
Table S7. RNA FISH probes.
Table S8. Odors.
Table S9. Accession numbers of the housekeeping protein sequences used to reconstruct the drosophilid phylogenetic tree.
REFERENCES AND NOTES
- 1.Herculano-Houzel S., Catania K., Manger P. R., Kaas J. H., Mammalian brains are made of these: A dataset of the numbers and densities of neuronal and nonneuronal cells in the brain of glires, primates, scandentia, eulipotyphlans, afrotherians and artiodactyls, and their relationship with body mass. Brain Behav. Evol. 86, 145–163 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Strausfeld N. J., Sinakevitch I., Brown S. M., Farris S. M., Ground plan of the insect mushroom body: Functional and evolutionary implications. J. Comp. Neurol. 513, 265–291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramdya P., Benton R., Evolving olfactory systems on the fly. Trends Genet. 26, 307–316 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Fang W. Q., Yuste R., Overproduction of neurons is correlated with enhanced cortical ensembles and increased perceptual discrimination. Cell Rep. 21, 381–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilsch-Bräuninger M., Florio M., Huttner W. B., Neocortex expansion in development and evolution—From cell biology to single genes. Curr. Opin. Neurobiol. 39, 122–132 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Dekkers M. P., Nikoletopoulou V., Barde Y. A., Cell biology in neuroscience: Death of developing neurons: New insights and implications for connectivity. J. Cell Biol. 203, 385–393 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi Y., Miura M., Programmed cell death in neurodevelopment. Dev. Cell 32, 478–490 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Kuida K., Haydar T. F., Kuan C. Y., Gu Y., Taya C., Karasuyama H., Su M. S. S., Rakic P., Flavell R. A., Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell 94, 325–337 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Rogulja-Ortmann A., Luer K., Seibert J., Rickert C., Technau G. M., Programmed cell death in the embryonic central nervous system of Drosophila melanogaster. Development 134, 105–116 (2007). [DOI] [PubMed] [Google Scholar]
- 10.White J. G., Southgate E., Thompson J. N., On the nature of undead cells in the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 331, 263–271 (1991). [Google Scholar]
- 11.Avery L., Horvitz H. R., A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell 51, 1071–1078 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues V., Hummel T., Development of the Drosophila olfactory system. Adv. Exp. Med. Biol. 628, 82–101 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Endo K., Karim M. R., Taniguchi H., Krejci A., Kinameri E., Siebert M., Ito K., Bray S. J., Moore A. W., Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nat. Neurosci. 15, 224–233 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Endo K., Aoki T., Yoda Y., Kimura K.-i., Hama C., Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nat. Neurosci. 10, 153–160 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Couto A., Alenius M., Dickson B. J., Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15, 1535–1547 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Benton R., Vannice K. S., Gomez-Diaz C., Vosshall L. B., Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabe V., Baschwitz A., Dweck H. K. M., Lavista-Llanos S., Hansson B. S., Sachse S., Elucidating the neuronal architecture of olfactory glomeruli in the Drosophila antennal lobe. Cell Rep. 16, 3401–3413 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Sen A., Kuruvilla D., Pinto L., Sarin A., Rodrigues V., Programmed cell death and context dependent activation of the EGF pathway regulate gliogenesis in the Drosophila olfactory system. Mech. Dev. 121, 65–78 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Chai P. C., Cruchet S., Wigger L., Benton R., Sensory neuron lineage mapping and manipulation in the Drosophila olfactory system. Nat. Commun. 10, 643 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee G., Sehgal R., Wang Z., Nair S., Kikuno K., Chen C.-H., Hay B., Park J. H., Essential role of grim-led programmed cell death for the establishment of corazonin-producing peptidergic nervous system during embryogenesis and metamorphosis in Drosophila melanogaster. Biol. Open 2, 283–294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto-Teixeira F., Konstantinides N., Desplan C., Programmed cell death acts at different stages of Drosophila neurodevelopment to shape the central nervous system. FEBS Lett. 590, 2435–2453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha T. S., Smith D. P., A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci. 26, 8727–8733 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallem E. A., Ho M. G., Carlson J. R., The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Hallem E. A., Carlson J. R., Coding of odors by a receptor repertoire. Cell 125, 143–160 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Fishilevich E., Vosshall L. B., Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol. 15, 1548–1553 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Fishilevich E., Domingos A. I., Asahina K., Naef F., Vosshall L. B., Louis M., Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr. Biol. 15, 2086–2096 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Stensmyr M. C., Dekker T., Hansson B. S., Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc. Biol. Sci. 270, 2333–2340 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prieto-Godino L. L., Rytz R., Cruchet S., Bargeton B., Abuin L., Silbering A. F., Ruta V., Dal Peraro M., Benton R., Evolution of acid-sensing olfactory circuits in drosophilids. Neuron 93, 661–676.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Dweck H. K., Ebrahim S. A. M., Khallaf M. A., Koenig C., Farhan A., Stieber R., Weißflog J., Svatoš A., Grosse-Wilde E., Knaden M., Hansson B. S., Olfactory channels associated with the Drosophila maxillary palp mediate short- and long-range attraction. eLife 5, e14925 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMeniman C. J., Corfas R. A., Matthews B. J., Ritchie S. A., Vosshall L. B., Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell 156, 1060–1071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu T., Qiu Y. T., Wang G., Kwon J. Y., Rutzler M., Kwon H. W., Pitts R. J., van Loon J. J. A., Takken W., Carlson J. R., Zwiebel L. J., Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 17, 1533–1544 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones W. D., Cayirlioglu P., Grunwald Kadow I., Vosshall L. B., Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Kwon J. Y., Dahanukar A., Weiss L. A., Carlson J. R., The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 104, 3574–3578 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews B. J., Dudchenko O., Kingan S. B., Koren S., Antoshechkin I., Crawford J. E., Glassford W. J., Herre M., Redmond S. N., Rose N. H., Weedall G. D., Wu Y., Batra S. S., Brito-Sierra C. A., Buckingham S. D., Campbell C. L., Chan S., Cox E., Evans B. R., Fansiri T., Filipović I., Fontaine A., Gloria-Soria A., Hall R., Joardar V. S., Jones A. K., Kay R. G. G., Kodali V. K., Lee J., Lycett G. J., Mitchell S. N., Muehling J., Murphy M. R., Omer A. D., Partridge F. A., Peluso P., Aiden A. P., Ramasamy V., Rašić G., Roy S., Saavedra-Rodriguez K., Sharan S., Sharma A., Smith M. L., Turner J., Weakley A. M., Zhao Z., Akbari O. S., Black W. C. IV, Cao H., Darby A. C., Hill C. A., Johnston J. S., Murphy T. D., Raikhel A. S., Sattelle D. B., Sharakhov I. V., White B. J., Zhao L., Aiden E. L., Mann R. S., Lambrechts L., Powell J. R., Sharakhova M. V., Tu Z., Robertson H. M., McBride C. S., Hastie A. R., Korlach J., Neafsey D. E., Phillippy A. M., Vosshall L. B., Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563, 501–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riabinina O., Task D., Marr E., Lin C. C., Alford R., O’Brochta D. A., Potter C. J., Organization of olfactory centres in the malaria mosquito Anopheles gambiae. Nat. Commun. 7, 13010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghaninia M., Hansson B. S., Ignell R., The antennal lobe of the African malaria mosquito, Anopheles gambiae – innervation and three-dimensional reconstruction. Arthropod Struct. Dev. 36, 23–39 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Hartl M., Loschek L. F., Stephan D., Siju K. P., Knappmeyer C., Kadow I. C. G., A new Prospero and microRNA-279 pathway restricts CO2 receptor neuron formation. J. Neurosci. 31, 15660–15673 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cayirlioglu P., Kadow I. G., Zhan X., Okamura K., Suh G. S. B., Gunning D., Lai E. C., Zipursky S. L., Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319, 1256–1260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pop S., Chen C.-L., Sproston C. J., Kondo S., Ramdya P., Williams D. W., Patterned cell death sculpts functional and adaptive neural networks in flies. bioRxiv, 626465 (2019). [Google Scholar]
- 40.Saina M., Benton R., Visualizing olfactory receptor expression and localization in Drosophila. Methods Mol. Biol. 1003, 211–228 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Sánchez-Alcañiz J. A., Zappia G., Marion-Poll F., Benton R., A mechanosensory receptor required for food texture detection in Drosophila. Nat. Commun. 8, 14192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ollion J., Cochennec J., Loll F., Escude C., Boudier T., TANGO: A generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolte S., Cordelières F. P., A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Benton R., Dahanukar A., Electrophysiological recording from Drosophila olfactory sensilla. Cold Spring Harb. Protoc. 2011, 824–838 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Olsson S. B., Hansson B. S., Electroantennogram and single sensillum recording in insect antennae. Methods Mol. Biol. 1068, 157–177 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10.14806/ej.17.1.200 (2011). [Google Scholar]
- 48.Davis M. P. A., van Dongen S., Abreu-Goodger C., Bartonicek N., Enright A. J., Kraken: A set of tools for quality control and analysis of high-throughput sequence data. Methods 63, 41–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L., Wang S., Li W., RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lebreton S., Borrero-Echeverry F., Gonzalez F., Solum M., Wallin E. A., Hedenström E., Hansson B. S., Gustavsson A. L., Bengtsson M., Birgersson G., Walker W. B. III, Dweck H. K. M., Becher P. G., Witzgall P., A Drosophila female pheromone elicits species-specific long-range attraction via an olfactory channel with dual specificity for sex and food. BMC Biol. 15, 88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dweck H. K. M., Ebrahim S. A. M., Thoma M., Mohamed A. A. M., Keesey I. W., Trona F., Lavista-Llanos S., Svatoš A., Sachse S., Knaden M., Hansson B. S., Pheromones mediating copulation and attraction in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 112, E2829–E2835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silbering A. F., Bell R., Münch D., Cruchet S., Gomez-Diaz C., Laudes T., Galizia C. G., Benton R., Ir40a neurons are not DEET detectors. Nature 534, E5–E7 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Larsson M. C., Domingos A. I., Jones W. D., Chiappe M. E., Amrein H., Vosshall L. B., Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Benton R., Vannice K. S., Vosshall L. B., An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–293 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaaz7238/DC1
Fig. S1. Automated quantification of Elav-positive antennal neurons.
Fig. S2. Undead neurons express a subset of olfactory receptor genes and can be found in novel, reproducible locations.
Fig. S3. Or-GFP reporters faithfully recapitulate receptor expression in undead neurons.
Fig. S4. Many sensory neuron populations show little or no increase, or a decrease, in size in PCD-blocked antennae.
Fig. S5. Undead OSNs form novel receptor/glomerular couplings in the brain.
Fig. S6. Electrophysiological distinction of at1 and at4 sensilla.
Fig. S7. Projections of Gr21a-GFP–expressing undead neurons from the maxillary palps.
Table S1. Estimated potential pool of undead neurons in the antenna.
Table S2. Comparison of chemosensory receptor and proapoptotic factor transcript abundance in control and PCD-blocked antennae.
Table S3. In situ expression properties of chemosensory genes up-regulated in PCD-blocked antennae.
Table S4. Statistical analysis of the enrichment of coexpressed Ors in undead neurons.
Table S5. Drosophila strains.
Table S6. Antibodies.
Table S7. RNA FISH probes.
Table S8. Odors.
Table S9. Accession numbers of the housekeeping protein sequences used to reconstruct the drosophilid phylogenetic tree.


