We propose a statistical inference approach designed to detect the presence of collective behavior in cell motility experiments.
Abstract
Migration of cells can be characterized by two prototypical types of motion: individual and collective migration. We propose a statistical inference approach designed to detect the presence of cell-cell interactions that give rise to collective behaviors in cell motility experiments. This inference method has been first successfully tested on synthetic motional data and then applied to two experiments. In the first experiment, cells migrate in a wound-healing model: When applied to this experiment, the inference method predicts the existence of cell-cell interactions, correctly mirroring the strong intercellular contacts that are present in the experiment. In the second experiment, dendritic cells migrate in a chemokine gradient. Our inference analysis does not provide evidence for interactions, indicating that cells migrate by sensing independently the chemokine source. According to this prediction, we speculate that mature dendritic cells disregard intercellular signals that could otherwise delay their arrival to lymph vessels.
INTRODUCTION
Cell migration is a dynamic process, which may be characterized by two prototypical kinds of motion: a collective motion in which cells communicate with each other, e.g., by means of biochemical or mechanical signals, and an individual motion, where each cell migrates independently (1, 2). Among the notable examples of cell migration in pathological contexts are cancer cells during metastasis, where collective migration generally results from physical contacts between cells, which adhere to each other via specific adhesion molecules. Despite the fact that both individual and collective motions have been observed, for cancer cells, collective migration is believed to result in more efficient metastatic spreading (3, 4).
On the other hand, immune cells do not exhibit cell-cell adhesion during migration: For these cells, the existence of physical cell-to-cell communication is induced by specific signaling pathways (5) and is restricted to slow migratory phases. This direct cell-to-cell communication allows immune cells to share pathogenic information or to coordinate the arrival of other cells (6, 7), while it does not result in a collective motion. As a consequence, in the context of fast migration of immune cells, such cells have been suspected to migrate as single cells.
However, immune cells respond to a large variety of biochemical signals. They release cytokines, chemokines, and small molecules that control cell migration and may send signals to adjacent cells without the need of physical interactions (7, 8). This paracrine signaling might constitute an alternative mechanism to achieve cell-to-cell communication, which, as shown in other cellular systems (9), may ultimately lead to cellular coordination. As a result, the existence of these signaling mechanisms raises the question of whether immune cells may ultimately migrate “collectively,” regardless of physical interactions.
Among the difficulties in answering this question is the fact that immune cells are often guided by extracellular signals produced by tissues (10): As a result, they may be exchanging biochemical signals with each other but still appear to migrate individually because they all move toward the same signal source. In addition, the number of potential molecules that could be responsible for cell-to-cell signaling is so large that molecular perturbation approaches could not rule out the existence of cell-to-cell communication through some of these molecules. Moreover, molecular perturbation approaches rely on an a priori knowledge of the signaling involved and/or the development of new experimental tools, which are, in general, both time consuming and expensive.
In this study, we propose a statistical inference method to overcome the issues mentioned above. Unlike the classical molecular perturbation approaches, our procedure is statistically rather than biologically driven and does not rely on any a priori knowledge of the biochemical interactions among the migrating cells. Instead, our method leverages the statistical information stored in empirical observations on cell motility, e.g., statistics of cells’ speeds and directions of motion (11, 12). We designed this statistical inference method so as to take account of a variety of experimental sources of error that are specific to cell motility experiments, e.g., limited tracking resolution, missing trajectories, and tracking anomalies, and thus efficiently exploit the information contained in the cell-tracking data.
Given a set of data for a cell migration experiment, the resulting statistically inferred model allows one to single out cell-cell interactions. While the method cannot assess the nature of these interactions, e.g., the signaling protein that is responsible for it, it makes a clear prediction on the existence, or absence, of such interaction. In particular, the model allows us to tell apart a population of independent cells, which may appear to migrate collectively only because they all follow the same cue from a population that migrates in a truly collective manner.
We first tested the inference method on two benchmark datasets, i.e., synthetic cell trajectories generated from a mean-field model of interacting spins and from a non–mean-field model of self-propelled (SP) particles. In both cases, the method correctly reconstructs the microscopic features of the models, e.g., the spin-spin couplings, and the strength of the external signal to which the SP particles are subject.
Next, we applied the inference framework to two experiments: In the first, cells migrate toward a “wound” and physically interact with each other via cell-cell adhesion resulting from cellular crowding. The inference method predicts the existence of cell-cell interactions, and this prediction correctly reflects the presence of intercellular contacts in the experiment, thus yielding a proof of concept for our inference framework.
We then applied the inference method to a population of dendritic cells migrating in a chemokine gradient. Our inference analysis does not provide evidence for the existence of cell-cell interactions, indicating that dendritic cells migrate independently and sense only the chemokine source. This prediction is supported by an exhaustive analysis of the raw motional data, which indicates the absence of cell-cell correlations.
We then present the biological applications of our results and discuss how the absence of instantaneous cell-cell interactions detected by our inference method may constitute an indication of a strategy to efficiently trigger the immune response. Last, we discuss how our inference method may be applied to a wide variety of biological systems, where it could be used to reconstruct the maximum-entropy (ME) probability distribution while handling multiple sources of error.
RESULTS
ME methods
Here, we will describe the statistical inference method that we designed for the analysis of motional features of cell-tracking experiments. Given a dataset X = {x1, ⋯, xT} composed of multiple observations of a quantity x, ME models provide a fundamental principle to model and reconstruct the probability distribution P(x) from a limited number of empirical observations, which would be too small to reconstruct such distribution directly from the data. Specifically, given a set of features f1(x), f2(x), ⋯ related to the observable x, their experimental and model estimates are, respectively
| (1) |
| (2) |
The ME method then constructs P as the least-structured probability distribution that matches the experimental averages above. Given that the amount of “structure” in P is quantified by the entropy (13)
| (3) |
i.e., the higher S, the less structured P, the ME model is formulated in terms of the following constrained optimization problem
| (4) |
subject to
| (5) |
| (6) |
ME models have attracted growing interest in the past few years and have been used for a wide variety of domains and of biological systems. Notable examples are the inference of motional order in flocks of birds (14), collective behavior in networks of neurons (15, 16), interaction structures resulting from amino acid sequences in protein families (17, 18), and interaction structure of genetic networks (19).
In the specific problem under consideration in this study, the positions of N moving cells are imaged and tracked in time with a camera on a pixel grid (see Fig. 1). Given that the precision with which the cell position is determined cannot exceed the pixel size, at any instant of time t, every cell, labeled by index i, is assigned a nominal position ri(t), which coincides with the center of the pixel (see section S5 for details). The nominal cell positions ri(t) and ri(t + Δt) at time t and at a subsequent observation t + Δt, respectively, yield the velocity vi(t) = [ri(t + Δt) − ri(t)]/Δt, where we use boldface for vector quantities. We compute the direction of motion, i.e., the normalized velocity, of each cell at time t
| (7) |
which we rewrite in terms of its polar angle as
| (8) |
Fig. 1. Motivation for ME models with bound constraints in cell-tracking experiments.
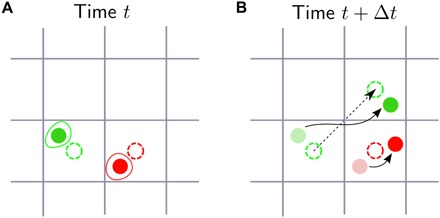
(A) Two cells (whose boundaries are the red and green closed curves) and their centers of mass (red and green disks) are tracked through a grid of pixels (gray). Each center is assigned a nominal position, i.e., the center of the pixel where it is located (dashed red and green circles, respectively). (B) At a subsequent time, the green cell has moved to a neighboring pixel (curved black arrow). The nominal position of the cell has changed, and its displacement is the vector difference between the nominal position in (B) and in (A) (dashed black arrow), resulting in a well-defined direction of motion. Because the red cell has moved within the pixel, its nominal position is the same as in (A), its nominal displacement is null, and its direction of motion is not defined, thus leading to an uncertainty in all physical observables that involve directions of motion.
We then obtain the full set of motional directions of the population, St = {s1(t), ⋯, sN(t)}, which we regard as the empirical observations for the ME problem, i.e., xt = St (14). We select as features the average pairwise correlation and polarization
| (9) |
| (10) |
respectively, where Np ≡ N(N − 1)/2 is the number of cell pairs, and in the classical ME approach, the ME distribution is obtained by solving the optimization problem (Eqs. 4 to 6) (see section S1.1 for details).
We recall the fundamental difference between correlation, C, and cell-cell interaction, which will be denoted by J (14, 20). A nonzero correlation does not necessarily imply a nonzero interaction—a population of cells attracted by an external source may display a nonzero correlation C, even if cells do not interact with each other. In this regard, the ME method allows one to extract cell-cell interactions, which are, in general, encoded in a nontrivial way in the motional data, e.g., correlation and polarization. If the resulting ME distribution factors out as the product of distribution of independent directions of motion, we obtain that cells behave independently; if not, we conclude that an interaction between cells exists. The feature choice (Eq. 9) sets the type of interaction that the ME method will probe. For instance, the experimental average 〈〉ex of Eq. 9 involves directions of motion si(t) · sj(t) of cells i and j evaluated at the same instant of time t: It follows that the resulting cell-cell interaction inferred by the ME model will necessarily be an instantaneous one, i.e., its propagation time is much shorter than all other time scales (15). Alternative types of interactions could be probed by choosing other features, e.g., by introducing a lag between times at which si and sj are evaluated.
Despite its wide use in a variety of systems, the ME method above may suffer from a fundamental limitation when applied to data affected by strong uncertainties (14, 15, 18). If the empirical data contain significant errors or a limited amount of information, complete satisfaction of the equality constraint is known to be too strict a criterion and may lead to data overfitting (21, 22). A prototypical example of this issue comes from ME models for language modeling (22), where the observations x = (w, w′) are pairs of consecutive words, w and w′, in a corpus of text, which constitutes the dataset X. Given two words, e.g., “saint” and “George,” we consider as features the frequency with which George occurs in the text , and the frequency of the bigram “saint George,” , where the indicator function is one if both the conditions in its argument are satisfied, and zero otherwise. Given a limited amount of empirical information, e.g., a short corpus of text where the word George occurs only after saint, if we impose these constraints in their equality form (Eq. 5), it is straightforward to show that P(w, George) = 0 if w ≠ ‘ saint′. These zero-frequency events in the ME model may not only cause numerical instability in ME estimation (21) but also result in poor performance of the ME model in a variety of applications, e.g., text recognition, where any word pair (w, George) in which w ≠ ‘ saint′ would not be recognized as a bigram.
The effect of data uncertainties may be even more dramatic in cell-tracking experiments. As shown in Fig. 1, if the cell motion is slow compared to the rate at which the observations are collected, the nominal position r(t + Δt) at time t + Δt may coincide with r(t), and the direction of motion (Eq. 7) is not defined. As a result, any empirical average that involves the directions of motion, e.g., the polarization (Eq. 10), will be affected by an error and will not be uniquely determined from the data, thus making the classical ME formulation pointless.
A potential workaround for this issue would be to measure the positions at intervals larger than Δt in such a way that two subsequent measurements of the cell position r(t) lie in different bins for all cells and times: The resulting empirical averages could then be analyzed with the classical ME formulation (see section S1.1). However, this strategy would throw away a large number of the original measurements r(t), r(t + Δt), ⋯ and thus use only a fraction of the experimental data available. In what follows, we discuss a ME formulation with bound constraints (MEb) (21, 22) that efficiently exploits all the information contained in the experimental data (see section S1.2 for details). In addition to the uncertainty above on the directions of motion, this MEb method may be used to handle a variety of other experimental sources of error, such as missing tracks, tracking anomalies, and others.
Let us suppose that, because of the experimental uncertainty described in Fig. 1, the tracking data do not provide a precise value for the empirical averages but a confidence interval in which these averages lie: Namely, if we let each unknown direction of motion si vary between 0 and 2π, then 〈C〉ex and 〈M〉ex will fluctuate between a lower bound and an upper bound, which define a confidence interval. As a result, in the MEb approach, we introduce explicitly such confidence interval by smoothening (22) the equality constraints in the ME model (Eqs. 4 to 6): The equality constraint (Eq. 5) is replaced by
| (11) |
where are the lower and upper bounds for the empirical average of feature fi, respectively (21).
Statistical inference analysis
In what follows, we will describe the main features of the MEb method and refer to section S1.2 for details. The joint distribution of velocities resulting from the MEb construction has the shape of a Boltzmann distribution
| (12) |
with Hamiltonian
| (13) |
where J reflects the “interaction” between cell velocities, the “external field” ℋ represents the overall tendency of the cells to flow in one particular spatial direction, and the partition function Z ensures that P is normalized.
The Hamiltonian (Eq. 13) is the one of the mean-field XY model—a statistical-mechanical model originally introduced to describe ferromagnetic systems (23). In the XY model, the larger J, the higher the energetic cost for si and sj to be misaligned; similarly, the larger H, the higher the energy cost for si to misalign with respect to the direction of the external field. The mean-field structure of the Hamiltonian (Eq. 13) follows from the choice of the feature (Eq. 9), which involves an average over all cell pairs. This mean-field structure makes the model analytically tractable: Its partition function (eq. S32) can be expressed exactly in terms of a one-dimensional integral and Bessel functions even for a finite number of cells N (see section S1.2).
The solution of the MEb problem is determined by a set of equality and inequality conditions, also denoted by bound constraints, known as the Karush-Kuhn-Tucker (KKT) conditions (24, 25). Given that each of the three parameters that appear in P, i.e., J and the two components of H, can be either positive, negative, or zero, we obtain a set of candidate MEb solutions, where each solution corresponds to a sign configuration of the parameters above. The MEb solution is then given by the solution with the largest entropy, and that satisfies all equality and inequality constraints (see the “Statistical inference analysis: Wound-healing experiment” and “Statistical inference analysis: Dendritic cell experiment” sections for details).
Tests of the ME method with bounds on synthetic data
To test the predictive capabilities of the MEb method, we generate synthetic data for a system of N moving units that evolve according to a given dynamics. We will consider two different models for the synthetic dynamics: The XY model and a model of SP particles. We denote by P the set of parameters that determine the dynamics, generate samples of the configurations of the N elements for different choices of P, and analyze them with the MEb.
XY model
As shown in the “Statistical inference analysis” section, the Hamiltonian (Eq. 13) of the MEb model coincides with the one of the mean-field XY model. As a result, the simplest fundamental test for the MEb method consists of generating samples of the spin configurations with the XY model for a given choice of the model parameters P = {J, H}, analyzing these configurations with the MEb model, and inferring back the same values of the model parameters.
To achieve this, for a given set of parameters P = {J, H}, we initialize the XY model in a configuration S and let it evolve with a single spin-flip Glauber dynamics, reaching equilibration after a large enough number of spin flips. Then, we collect T = 100 configuration samples S1, ⋯, ST, with which we compute the configuration averages 〈C(S)〉ex and 〈M(S)〉ex (see Eq. 1). Given that the averages above are estimated from a finite number of samples, they are subjected to a statistical error. We thus estimate the confidence interval for these averages in terms of this statistical error, which we denote by σ, setting (see Eq. 11 and section S1.2 for details). Last, we use these bounds in the MEb method and obtain the inferred values of the interaction and external field, Jinf and Hinf, respectively.
The procedure above is repeated Q = 100 times for each parameter configuration, resulting in a mean value and error bar for Jinf, Hinf (see fig. S1): Overall, the agreement between the inferred and original parameter values is very good, even for small N.
SP model
In the SP model, we consider N particles moving on a plane with no boundaries. The dynamics of the particles’ velocities and positions is set by the following rules
| (14) |
| (15) |
where Θ is a normalization operator Θ(y) = y/∣ y∣ which keeps the velocity modulus fixed at ∣v∣ = v0, and the sum in Eq. 14 runs over the nc nearest neighbors of particle i, according to a topological definition of distance. The first term in the square bracket in the right-hand side of Eq. 14 makes particles that are close to each other move in the same direction; the second term involves an interaction force fij, which accounts for excluded volume (vide infra); and the third term represents a uniform gradient in the direction of the unit vector h. The fourth term represents noise and includes a random unit vector ηi(t), which is independent for each particle and instant of time. This noise term physically represents the uncertainty with which particle i feels the force fij exerted by particle j, which appears in the second term: Given that the second term involves a sum over the nc nearest neighbors of particle i, the noise term is proportional to nc (26).
The parameters α, β, γ set the magnitude of the terms which they multiply. The distance-dependent force fij acts along the direction between i and j, eij = rij/∣ rij∣, with rij = ri − rj, and is defined as
with ra, rb, re, r0 suitable length scales. More precisely, rb represents the particle size, and r0 sets the interaction range in such a way that particles are insensitive to each other when their distance exceeds r0. The radii ra and re set the magnitude and direction of the force fij, which is attractive and repulsive when rij > re and rij < re, respectively.
We set the parameters α, β, γ, nc, ra, rb, re, r0 and the direction of the field h, and then we initialize the system by placing all the particles in a two-dimensional area and by associating to each particle a (normalized) velocity given by the angle θi, i = 1, …, N randomly and uniformly drawn in [0,2π] (see Eq. 8). Next, we let the particles move according to Eqs. 14 and 15. In an effort to emulate the positional uncertainty due to the camera pixels in the experiments above, for each simulation, we assumed that the positions are affected by an error σ = 0.02 and rounded off the cell positions according to this error (see section S5 for details). The numerical value of σ has been chosen so as to obtain a fraction of nondefined directions of motion roughly comparable with the experimental ones. After a large enough number of iterations, we collect the resulting configurations of velocities vi(t), t = 1, ⋯, T, the related normalized velocities, and the lower and upper bounds for 〈C〉 and 〈M〉, which we then used in the MEb method.
We would like to stress that, unlike the XY model where the Hamiltonian has the same form and parameter structure as the MEb model (Eq. 13), for the SP model, a direct comparison between P = {α, β, γ} and the output Jinf, Hinf of the MEb is no longer possible. In particular, the former model is built out of mean-field interactions, while the latter involves a non–mean-field interaction structure. Despite the fact that there is no one-to-one mapping between the parameters of the two models, the interaction parameter J in the MEb Hamiltonian (Eq. 13) can be related to the parameter α in the SP dynamics, which sets the strength of the alignment between particle velocities. In fact, the inferred value of J appears to be positively correlated with α (see fig. S2), indicating that the MEb model correctly captures the tendency of SP particles to align their directions of motion. More precisely, in fig. S2, we set γ = 0 and vary α, and we correctly infer a vanishing field H, while the inferred value of J is non-null. However, we observe that Jinf seems to depend also on N, and its dependence on α appears to be nonlinear. On the other hand, by setting α = 1.2 and varying γ by fixing h directed in the direction −π/4, we correctly infer a linear relation between Hinf and γ, while Jinf still exhibits a slight dependence on N and γ.
In addition, we performed the inference analysis of fig. S2 with a mean-field SP model with nc = N − 1, where each particle interacts with all the other ones, and found that the resulting level of accuracy of the inference analysis is similar to the one for an SP model with short-range interactions, i.e., fig. S2. This indicates that the accuracy of the inference analysis for the SP model is not directly related to the mean-field structure of the MEb model (Eq. 13), but it may depend on a variety of features of the SP model, e.g., its out-of-equilibrium dynamics, topological interaction structure, and the form of interparticle forces.
Last, we present an additional test of the MEb method. For a given parameter configuration α = 1.2, β = 4, γ = 2.2, and nc = 9, we performed Q = 100 simulations of the SP with N = 60 particles and collected T = 360 snapshots of their positions. For each simulation, we rounded off the cell positions as discussed above and, by using this positional uncertainty, we obtained the lower and upper bounds for the feature averages 〈C〉 and 〈M〉, which we used to infer J and H with the MEb. Furthermore, we considered the connected correlation function between motional correlation and polarization 〈CM〉 − 〈C〉〈M〉. We compute this quantity both from the inferred MEb model above and from the data and compare them in fig. S3. The MEb model is based on the average correlations and polarization 〈C〉 and 〈M〉 only, not on their cross correlation 〈CM〉 − 〈C〉〈M〉: As a result, the MEb prediction for the connected correlation involves no free parameters. First, fig. S3 shows a good agreement between model and empirical connected correlation. Second, the MEb method reproduces quantitatively the negative correlation between the x and y components of 〈CM〉 − 〈C〉〈M〉 across multiple simulations—the larger the x component, the smaller the y component. Overall, the analysis above indicates that the MEb method, which is based on averages of empirical features only, is able to describe and capture also more complex statistical properties, e.g., the cross correlation between two of such experimental features (14). Given the satisfactory results of the MEb on these benchmark datasets, in what follows, we will present the experimental data of our study, discuss their statistical features, and analyze them with the MEb.
Experiments
In the wound-healing experiment, a population of human cancerous epithelial cells migrates in a planar device in which we realized a wound (see Fig. 2 and the “Wound-healing experiment” section for details). In the dendritic cell experiment, cells move in a spatially varying concentration of chemokines built along the horizontal axis of the device (see Fig. 3 and the “Dendritic cell experiment” section for details).
Fig. 2. Wound-healing experiment.
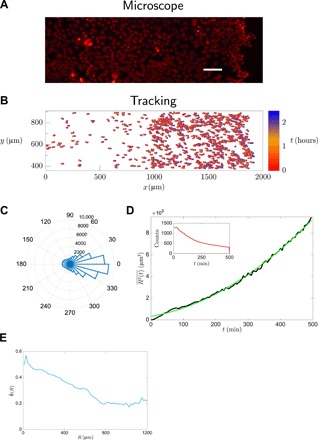
(A) Fluorescence image showing the nuclei (red, H2B-mCherry) of HeLa cells migrating toward a wound located at the right edge of the image (scale bar, 100 μm). (B) Tracked cell trajectories: The instantaneous position of each cell is marked with a circle, and the respective time is specified by the color code. Only a few representative cells are shown for clarity. (C) Polar histograms for the angle θ and (D) mean-squared displacement versus time show that the motion is affected by a strong bias, which yields a mean-squared displacement growing quadratically in time: Experimental data are shown in dark color, and the best fit y = p1 + p2t + p3t2 with p1 = 33, p2 = 0.5, p3 = 0.01 is shown in bright color. The inset in (D) shows the number of tracks recorded at each instant of time to figure out the statistically significant time window. (E) Angle pairwise correlation for the wound-healing experiment.
Fig. 3. Dendritic cell experiment.
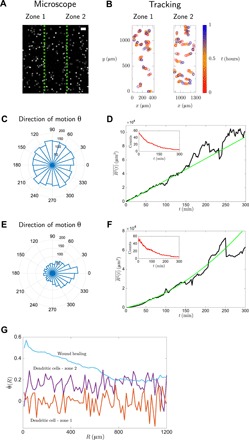
(A) Microscope image of cells showing chemokine-poor and chemokine-rich regions, i.e., zones 1 and 2, respectively, separated by green dashed lines (scale bar, 100 μm). (B) Tracked cell trajectories in zones 1 and 2: The instantaneous position of each cell is marked with a circle, and the respective time is specified by the color code. Only a few representative cells are shown for clarity. Cells in zone 1 (C and D) move isotropically and diffusively (best fit y = p1 + p2t with p1 = − 90, p2 = 31), while cells in zone 2 (E and F) feel a drift and move ballistically (y = p1 + p2t + p3t2 with p1 = − 306, p2 = 99, p3 = 0.6). The insets in (D) and (F) show the number of tracks recorded at each time. (G) Angle pairwise correlation for the three experimental instances above.
Analysis of cellular trajectories
For both experiments, the data consists of the nominal coordinates ri(t), t = 1, …, Ti, where the length Ti of track i is cell dependent because the tracks may lie in the observation window for different periods, and the time lapse between two measurements is Δt = 5 min and Δt = 2 min for the wound-healing and dendritic cell experiment, respectively (see the “Wound-healing experiment” and “Dendritic cell experiment” sections for details). Therefore, tracks are regarded as discrete-time random walks, where the ith walker at time t takes a step Δri(t) = ri(t + Δt) − ri(t), with length Δri(t) = ∣ Δri(t)∣ and velocity vi(t). In addition to the Cartesian coordinate system above, we describe the motion in a polar system where, at the tth time step, the ith track performs a step of length Δri(t) in the direction described by the angle θi(t) with respect to the horizontal axis.
In the remainder of this section, we introduce the fundamental motional features of the cells tracked in the two experiments, in view of the statistical analysis of the “Statistical inference analysis: Wound-healing experiment” and “Statistical inference analysis: Dendritic cell experiment” sections. First, we introduce the displacement of cell i, i.e., Ri(t) = ri(t) − ri(0) and its square average over all available tracks
| (16) |
where the normalization Nt is the number of cells whose tracks have length larger or equal than t, i.e., . Also, we introduce the pairwise correlation of the angle θ versus the distance R between cells, i.e.,
| (17) |
where the normalization NR is the total number of pairs at distance R, namely, .
In what follows, we will discuss shortly the results of the analysis of motional data and refer to section S4 for details. In the wound-healing experiment, the trajectories show a strong bias along the horizontal direction, evidenced by a sharply peaked polar histogram for the angle θ (Fig. 2C) and a ballistic-like mean-squared displacement (Fig. 2D). In addition, the angle pairwise correlation among cells is non-null, and it decays with respect to cell-cell distance (Fig. 2G). In the dendritic cell experiment, we found different behaviors depending on the proximity to the chemokine-rich region (27). In chemokine-free region (zone 1), cells exhibit an isotropic random walk with a uniform polar histogram for the angle of migration θ (Fig. 3C) and a diffusive-like mean-squared displacement (Fig. 3D). On the other hand, in the region where there is a chemokine gradient (zone 2), cells perform a biased walk with a peaked polar histogram for the angle θ (Fig. 3E) and a ballistic-like mean-squared displacement (Fig. 3F). Moreover, Fig. 3G shows that angle pairwise correlation is absent in zone 1 and non-null in zone 2 and that, in both cases, no statistically significant dependence on R is found. Figure 3G shows that the correlation decay with distance is peculiar to the wound-healing experiment, and it suggests that diverse migratory mechanisms may be at work in the two experiments, possibly indicating the existence of a collective behavior in the wound-healing experiment. In this regard, in fig. S4, we show a visual comparison between the cell tracks of the wound-healing and dendritic cell experiment, demonstrating that a naked-eye inspection of the tracks does not allow to confirm or to rule out the existence of a collective behavior in either of the two experiments. In fact, in both experiments, cells appear to be moving toward a source, and the existence of genuine collective effects is encoded in their trajectories in a nontrivial way. To unveil the presence of these mechanisms, in what follows, we will leverage the MEb method and find out whether cell motion is simply gradient driven or whether a collective migration is also at play.
Statistical inference analysis: Wound-healing experiment
To analyze the cell trajectories with the MEb, we estimated the uncertainty on cell positions resulting from the finite pixel size (see Fig. 1 and section S5 for details), incorporated this uncertainty in the lower and upper bounds of empirical averages as described in the “ME methods” section, and used the MEb with these bounds.
First, we observe that the spread of the correlation average due to the experimental error, i.e., , can be as large as ∼50% (see table S1). Such a large spread indicates that it would be pointless to rely on the empirical averages only by using the classical ME method (14), thus demonstrating the need for a MEb formulation. The MEb solution is depicted in Fig. 4: A visual inspection of the full set of candidate solutions shows that there is a unique solution that has the largest entropy per cell s = S/N and which satisfies all constraints within numerical precision. As shown in Fig. 4, the results of the MEb analysis are J = 1.1, H = (0.018,0). Because J and H are multiplied by O(N) terms in the Hamiltonian (Eq. 13), these parameters should be considered to be significantly different from zero as long as they are of order unity: It follows that the MEb solution yields an interaction parameter J significantly different from zero, thus indicating the presence of a collective behavior in the cell migration of Fig. 2. In addition, the nonzero value of the x component of the external field may be ascribed to the attractive effect of the wound located on the right edge of the observation window (see Fig. 2A). Last, the null y component of the external field reflects the spatial homogeneity of the experiment with respect to the vertical direction.
Fig. 4. Analysis of the wound-healing experiment via ME method with bound constraints.
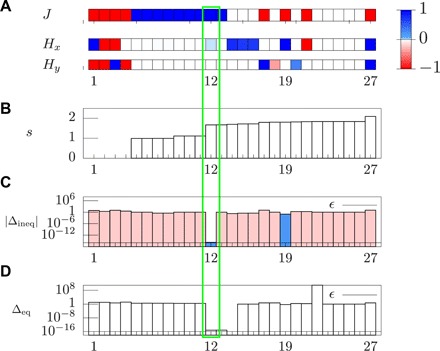
(A) Full set of solutions of the KKT conditions in order of increasing entropy from left to right. Each solution is labeled with an integer shown on the abscissa, and the corresponding value of J (top), Hx (middle), and Hy (bottom) is represented with the color in the box. (B) Entropy per cell for each solution. (C) Modulus of the relative residual Δineq of the inequality KKT conditions shown for each solution, where residuals that are positive and negative are marked in blue and red, respectively. (D) Modulus of relative residual Δeq of the equality KKT conditions, shown for each solution. The numerical precision used in the calculation, ϵ, is marked in (C) and (D). Only the solutions with Δineq ≥ 0 [blue columns in (C)] are admissible, i.e., solutions 12 and 19. Among these two solutions, solution 12 is the only one with Δeq ∼ ϵ, see (D), and it thus constitutes the only admissible, ME solution (green rectangle). In (B), unphysical solutions with negative probability are ruled out and not shown.
It is important to point out that the result J ≠ 0 is consistent with the analysis of motional data of the “Analysis of cellular trajectories” section and section S4.1. The analysis shows that the empirical distribution of the cell velocities is exponential in the y direction, but it markedly differs from an exponential distribution in x direction, thus indicating that cells do not migrate as independent units. The existence of a cell-cell cooperation is also indicated by the analysis of angle correlations between cell pairs, , which decays with the intercellular distance R (see Fig. 2E) and by the pairwise correlation coefficient between the directions of motion, , whose mean is clearly different from zero (see fig. S5F).
Statistical inference analysis: Dendritic cell experiment
Proceeding along the lines of the wound-healing experiment, we estimated the uncertainty on cell positions resulting from the finite pixel size as described in section S5. This uncertainty results in broad confidence intervals for the feature averages: Indeed, table S2 shows that the relative spread for the empirical average of the correlation can be as large as 100% and similarly for the polarization averages, thus confirming the need for the MEb approach.
Figures 5 and 6 show the MEb solution for the dendritic cell experiment in the high- and low-density case and chemokine-poor and chemokine-rich regions, i.e., zones 1 and 2, respectively, and the resulting values of J and H are shown in table S2. First, we observe that the y component of the external field H is very small and that the x component vanishes in zone 1, while it is different from zero in zone 2 for all densities: This dependence of the inferred external field on the zone reflects the chemokine gradient built in the device in the horizontal direction (see Fig. 3).
Fig. 5. Statistical inference analysis of the dendritic cell experiment with high cell density with the ME method with bound constraints.
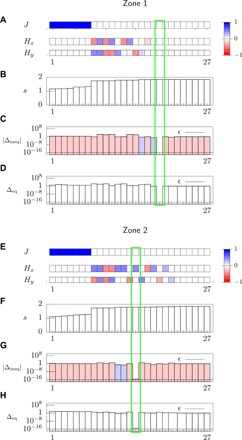
(A to D) Analysis for data in zone 1, where we use the same notation and the same procedure to select the ME solution as in Fig. 4. (E to H) Analysis for zone 2. The numerical values of J and H for the MEb solutions are shown in table S2.
Fig. 6. Statistical inference analysis of the dendritic cell experiment with low cell density with the ME method with bound constraints.
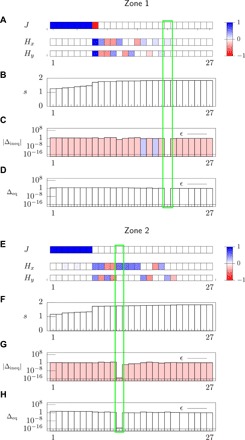
(A to D) Analysis for data in zone 1, where we use the same notation and the same procedure to select the ME solution as in Fig. 4. (E to H) Analysis for zone 2. The numerical values of J and H for the MEb solutions are shown in table S2.
Second, for all densities and zones, the MEb indicates that the interaction parameter, J, is null. This result is supported by the motional data analysis of the “Analysis of cellular trajectories” section. In fact, Fig. 3G demonstrates that the angle correlations between cell pairs, , do not depend on the intercellular distance, and figs. S7F and S9F indicate that the pairwise correlation coefficient between the directions of motion, , has zero mean. Overall, these results are markedly different from the ones obtained for the wound-healing experiment in the “Statistical inference analysis: Wound-healing experiment” section, and they indicate that these dendritic cells migrate individually in the chemokine gradient.
DISCUSSION
Motivated by the recent ubiquitous applications of inference methods to biological systems (14–19) and by the importance of cell migration phenomena in both physiological and pathological contexts (3, 4), we proposed a statistical inference method to detect and single out cell-cell interactions in a population of migrating cells. In fact, while there is a variety of cases of collectively moving biological entities that do not have direct physical interactions, it is often unclear whether migrating cells, such as immune cells, could interact remotely, e.g., by means of diffusible factors or other intercellular signals, and thus display collective behaviors. This issue is particularly important when cells migrate in the same direction toward an external attractor, e.g., a biochemical signal: It is difficult to tell whether cells all go in the same direction independently of each other simply because they are all attracted by the same cue or whether they also interact with each other through some signals.
To address this question, we proposed a statistical inference method specifically designed for cell-tracking experiments, which is capable of handling the empirical uncertainties specific to tracking processes, such as the errors resulting from finite camera resolution, missing tracks, and others. In addition, the mean-field structure of our inferred statistical model allows an explicit solution even for a finite number of cells—a feature that may prove to be particularly useful for, say, experiments on lab-on-a-chip technology, where the number of tracked cells can be small.
To check the soundness of our inference approach, we first tested it on synthetic datasets, i.e., trajectories generated from an XY model of spins, and on tracks generated from a non–mean-field SP model of particles. We find, overall, a good agreement between the original and inferred values of the model parameters, e.g., the spin-spin couplings and the strength of the external field to which the SP particles are subject.
Building on the results above, we applied our method to two prototypical cell migration experiments: Mesenchymal migration toward a wound and amoeboid migration of immune cells, i.e., dendritic cells, following a chemokine gradient. The inference analysis gives strong evidence of intercellular interactions for the wound-healing experiment, which served as a stereotypical case of collective migration, thus providing a positive control for detection of cell-cell interactions by our method. As far as the dendritic cell experiment is concerned, immune cells release molecules toward the extracellular milieu, which could steer the migration of adjacent cells (7). The release of vesicles or small molecules to extracellular milieu as a mechanism of paracrine cell communication allows coordinated migration in a contact-independent manner in other cellular systems (9, 28). Similarly, dendritic cells release adenosine triphosphate (ATP), which acts in an autocrine manner (8), but it is unknown whether it can affect the migration of adjacent cells. Nonetheless, our inference method did not provide evidence of cell-cell interactions during dendritic cell chemotaxis, revealing that these cells move independently toward the gradient.
From the statistical inference standpoint, the results above support the validity of a mean-field ME model (Eq. 13). First, the ME with bound constraints yields qualitatively correct results for the SP model, which contains non–mean-field interactions, and the nonzero interaction inferred in the wound-healing experiment correctly mirrors the existence of actual cell-cell contacts. Second, the prediction of a null interaction in the dendritic cell experiment is in agreement with the statistical analysis in the “Analysis of cellular trajectories” section (see the absence of a correlation decay with distance in Fig. 3G). In sum, these results indicate that the mean-field ME model yields, at the least, qualitatively correct results on the existence or absence of cell-cell interactions.
In addition, it is important to point out that the statistical inference method that we proposed detects cell-cell interactions that are instantaneous, i.e., whose propagation time is much shorter than all other time scales: The observables that have been chosen in the ME method (Eq. 9) involve directions of motion si(t) · sj(t) of cells i and j evaluated at the same instant of time t (see the “ME methods” section). Despite the fact that such instantaneous interactions have been used in a variety of statistical inference studies for biological systems (14, 15), other types of interactions may be present in cell migration experiments. For example, cell i may locally release a chemical compound along its migratory path, and cell j may cross the former path of i at a later time and thus feel a delayed interaction with i mediated by this compound. The implementation of a time-lagged ME model goes beyond the scope of this work, but it may be studied by introducing, in the present model, features in which the directions of motion si and sj appear at different instants of time (29). The existence of a time-lagged interaction would denote a type of collective migration that is markedly different from the one addressed in this study. While instantaneous interactions characterize a genuinely collective behavior, time-lagged interactions would be rather an indirect effect due to the fact that cell i alters the environment in which cell j moves. As a result, the absence of instantaneous cell-cell interactions resulting from our analysis indicates the lack of a genuinely collective behavior in the dendritic cell experiment.
An additional interesting extension of our analysis would consist of introducing further observables in the ME model. For instance, while the correlation and polarization (Eqs. 9 and 10) involve averages over the entire cell population, one could restrict these averages to cells lying in a specific spatial region, e.g., zone 1 or 2 in the dendritic cell experiment. While this complex feature structure goes beyond the minimal ME model that we propose in our analysis, it may provide additional insights on cell-cell interactions in relation to their proximity to the signaling source.
From a biological standpoint, our statistical method may provide unprecedented insights on the strategy by which dendritic cells perform their function. Indeed, to trigger the immune response, dendritic cells need to uptake an antigen in the peripheral tissues and then quickly reach the lymph nodes via the lymphatic vessels (30): In this regard, our statistical inference analysis suggests that dendritic cells individually follow signals from the lymphatic vessels (i.e., CCL21 chemokine), disregarding any other signal. It is conceivable that such absence of cell-cell interactions during dendritic cell migration may correspond to a mechanism to undergo their main function. In fact, dendritic cell migration markedly differs from the one of other immune cells, e.g., neutrophils, which, when receiving an attractive signal from a pathogen, emit secondary signals to attract other neutrophils on site (31). For dendritic cells, only the cells activated directly by the pathogen should leave the tissue to reach the lymph nodes: As a result, the fact that these cells should not emit such signal to attract other cells allows a natural interpretation of our statistical inference results. Last, the individual migratory mechanism that we inferred may allow the cells to disregard not only intercellular but also external signals that might delay their arrival toward the lymphatic vessels and ultimately to the lymph nodes, thus constituting a strategy to reach the vessels as efficiently as possible. Further studies should be done in this direction to corroborate these hypotheses.
Overall, the statistical inference method presented in this analysis may be applied not only to cell-tracking experiments but also to a wide variety of biological systems, some of which have attracted growing interest in the past few years. Notable examples are the inference of motional order in flocks of birds (14), collective behavior in networks of neurons (15, 16), interaction patterns resulting from amino acid sequences in protein families (17, 18), and interaction structure of genetic networks (19). In all these instances, both the experimental and data analysis procedure may introduce a variety of errors in the estimates of experimental features, which can be naturally handled with the ME method with bound constraints presented in this analysis.
MATERIALS AND METHODS
Wound-healing experiment
For the wound-healing experiment, human cancerous epithelial cells (HeLa) from adenocarcinoma were used. HeLa cells stably transfected with H2B-mCherry that allows live imaging of the nucleus were cultured in DMEM (Dulbecco’s modified Eagle’s medium containing glutamax), 10% fetal bovine serum, and 1% penicillin-streptomycin. Once confluence was reached, HeLa H2B cells were detached with trypsin and 0.5 × 106 cells were plated in a FluoroDish 24 hours before the experiment. Then, a monolayer was formed, and four scratches were done (two in the vertical axis and two in the horizontal one) using a 200-μl pipette tip. Medium was changed twice after the scratching to remove detached and dead cells. The dishes were left in the incubator at 37∘C for 4 hours before starting the experiment. Phase-contrast images were acquired every 5 min overnight with a DMi8 inverted microscope (Leica), at 37∘C with 5% CO2 atmosphere using a 10× dry objective [numerical aperture (NA) 0.40 phase], binning 2. In addition to phase contrast, mCherry fluorescence was also imaged.
Dendritic cell experiment
Bone marrow–derived dendritic cells (BMDCs) were prepared as previously described (8). Briefly, mouse bone marrow precursors were obtained from wild-type C57/B6 mice and were differentiated in vitro for 10 days with granulocyte-macrophage colony-stimulating factor–containing culture medium, obtained from transfected J558 cells. At day 10, cells were stimulated with a pulse bacterial lipopolysaccharide (LPS; 100 ng/ml), as previously described (32). The migration experiments in collagen gels were performed as previously described (27). LPS-stimulated BMDCs were mixed with bovine collagen type I at 3 mg/ml and loaded in a custom-made chamber of polydimethylsiloxane. After 30 min of incubation at 37∘C, gel polymerization was reached and samples were bathed with a medium containing CCL21 (200 ng/ml). Last, imaging was done as for the wound-healing experiment, but the period of acquisition was 2 min.
Supplementary Material
Acknowledgments
We thank L. Caccianini, G. Colazzo, L. Del Mercato, J.-F. Joanny, A. S. Kumar, I. Lavi, G. Maruccio, P. Šulc, and A. Zilman for valuable conversations. Funding: A.B. acknowledges Università del Salento and INFN for financial support. M.C. and M.P. acknowledge support from CNRS and Institut Curie. This research was supported by Sapienza University of Rome, Progetto Ateneo RG11715C7CC31E3D and RM118164368D6841; Association Nationale pour la Recherche (MOTILE project, ANR-16-CE13-0009) attributed to P.V.; and “Institut Pierre-Gilles de Gennes” (“Laboratoire d’excellence,” “Investissements d’avenir” program ANR-10-IDEX-0001-02 PSL and ANR-10-LABX-31). Author contributions: E.A., A.B., M.C., M.P., and P.V. conceived the study. E.A. and M.C. developed the theory. P.J.S. designed and performed the experiments. E.A., A.B., and M.C. wrote most of the paper. M.P. and P.J.S. contributed to the revisions. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaay2103/DC1
Section S1. ME models
Section S2. Tests of ME method with bound constraints
Section S3. Visual comparison between experiments
Section S4. Analysis of motional data
Section S5. Estimate of positional uncertainty
Fig. S1. Test of ME method with bound constraints on synthetic data for the XY model.
Fig. S2. Test of ME method with bound constraints on synthetic data for the SP model.
Fig. S3. Consistency test of ME method with bound constraints.
Fig. S4. Visual comparison between tracks of the wound-healing and dendritic cell experiment.
Fig. S5. Statistics of motional data for the wound-healing experiment.
Fig. S6. Empirical distributions for cell velocities in the wound-healing experiment.
Fig. S7. Statistics of motional data for zone 1 in the dendritic cell experiment.
Fig. S8. Empirical distributions for cell velocities for zone 1 in the dendritic cell experiment.
Fig. S9. Statistics of motional data for zone 2 in the dendritic cell experiment.
Fig. S10. Empirical distributions for cell velocities for zone 2 in the dendritic cell experiment.
Fig. S11. Estimate of the positional error.
Table S1. Confidence intervals for the empirical averages in the wound-healing experiment.
Table S2. Confidence intervals for the empirical averages in the dendritic cell experiment.
REFERENCES AND NOTES
- 1.van Helvert S., Storm C., Friedl P., Mechanoreciprocity in cell migration. Nat. Cell Biol. 20, 8–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark A. G., Vignjevic D. M., Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 36, 13–22 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Carmona-Fontaine C., Matthews H. K., Kuriyama S., Moreno M., Dunn G. A., Parsons M., Stern C. D., Mayor R., Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 456, 957–961 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayor R., Carmona-Fontaine C., Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 20, 319–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sáez P. J., Shoji K. F., Aguirre A., Sáez J. C., Regulation of hemichannels and gap junction channels by cytokines in antigen-presenting cells. Mediators Inflamm. 2014, 742734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreisel D., Nava R. G., Li W., Zinselmeyer B. H., Wang B., Lai J., Pless R., Gelman A. E., Krupnick A. S., Miller M. J., In vivo two-photon imaging reveals monocyte-dependent neutrophil extravasation during pulmonary inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 18073–18078 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majumdar R., Steen K., Coulombe P. A., Parent C. A., Non-canonical processes that shape the cell migration landscape. Curr. Opin. Cell Biol. 57, 123–134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sáez P. J., Vargas P., Shoji K. F., Harcha P. A., Lennon-Dumenil A. M., Sáez J. C., Atp promotes the fast migration of dendritic cells through the activity of pannexin 1 channels and p2x7 receptors. Sci. Signal. 10, eaah7107 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Kriebel P. W., Majumdar R., Jenkins L. M., Senoo H., Wang W., Ammu S., Chen S., Narayan K., Iijima M., Parent C. A., Extracellular vesicles direct migration by synthesizing and releasing chemotactic signals. J. Cell Biol. 217, 2891–2910 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiberio L., Del Prete A., Schioppa T., Sozio F., Bosisio D., Sozzani S., Chemokine and chemotactic signals in dendritic cell migration. Cell. Mol. Immunol. 15, 346–352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agliari E., Biselli E., De Ninno A., Schiavoni G., Gabriele L., Gerardino A., Mattei F., Barra A., Businaro L., Cancer driven dynamics of immune cells in a microfluidic enviroment. Sci. Rep. 4, 6639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polini A., Del Mercato L., Barra A., Zhang Y. S., Calabi F., Gigli G., Towards the development of human immune-system-on-a-chip platforms. Drug Discov. Today 24, 517–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shannon C. E., A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 (1948). [Google Scholar]
- 14.Bialek W., Cavagna A., Giardina I., Mora T., Silvestri E., Viale M., Walczak A. M., Statistical mechanics for natural flocks of birds. P. Natl. Acad. Sci. U.S.A. 109, 4786–4791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneidman E., Berry II M. J., Segev R., Bialek W., Weak pairwise correlations imply strongly correlated network states in a neural population. Nature 440, 1007–1012 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shlens J., Field G. D., Gaulthier J. L., Grivich M. I., Petrusca D., Sher A., Litke A. M., Chichilnisky E. J., The structure of multi-neuron firing patterns in primate retina. J. Neurosci. 26, 8254–8266 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seno F., Trovato A., Banavar J. R., Maritan A., Maximum entropy approach for deducing amino acid interactions in proteins. Phys. Rev. Lett. 100, 078102 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Weigt M., White R. A., Szurmant H., Hoch J. A., Hwa T., Identification of direct residue contacts in protein-protein interaction by message passing. Proc. Natl. Acad. Sci. U.S.A. 106, 67–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lezon T. R., Banavar J. R., Cieplak M., Maritan A., Fedoroff N. V., Using the principle of entropy maximization to infer genetic interaction networks from gene expression patterns. Proc. Natl. Acad. Sci. U.S.A. 103, 19033–19038 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cocco S., Monasson R., Weigt M., From principal component to direct coupling analysis of coevolution in proteins: Low-eigenvalue modes are needed for structure prediction. PLOS Comput. Biol. 9, e1003176 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazama J., Tsujii J., Maximum entropy models with inequality constraints: A case study on text categorization. Mach. Learn. 60, 159–194 (2005). [Google Scholar]
- 22.Chen S. F., Rosenfeld R., A survey of smoothing techniques for ME models. IEEE Trans. Acoust. Speech Audio Process. 8, 37–50 (2000). [Google Scholar]
- 23.K. Huang, Statistical Mechanics (John Wiley & Sons, ed. 2, 1987). [Google Scholar]
- 24.H. W. Kuhn, A. W. Tucker, Nonlinear programming, in Second Berkeley Symposium on Mathematical Statistics and Probability (University of California Press, 1951); https://projecteuclid.org/euclid.bsmsp/1200500213.
- 25.W. Karush, “Minima of functions of several variables with inequalities as side constraints,” thesis, University of Chicago (1939). [Google Scholar]
- 26.Grégoire G., Chaté H., Onset of collective and cohesive motion. Phys. Rev. Lett. 92, 025702 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Sáez P. J., Barbier L. M., Attia R., Thiam H.-R., Piel M., Vargas P., Leukocyte migration and deformation in collagen gels and microfabricated constrictions. Methods Mol. Biol. 1749, 361–373 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Ma L., Li Y., Peng J., Wu D., Zhao X., Cui Y., Chen L., Yan X., Du Y., Yu L., Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 25, 24–38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang A., Jackson D., Hobbs J., Chen W., Smith J. L., Patel H., Prieto A., Petrusca D., Grivich M. I., Sher A., Hottowy P., Dabrowski W., Litke A. M., Beggs J. M., A maximum entropy model applied to spatial and temporal correlations from cortical networks in vitro. J. Neurosci. 28, 505–518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sáez P. J., Sáez J. C., Lennon-Duménil A. M., Vargas P., Role of calcium permeable channels in dendritic cell migration. Curr. Opin. Immunol. 52, 74–80 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Futosi K., Fodor S., Mócsai A., Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int. Immunopharmacol. 17, 638–650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bretou M., Sáez P. J., Sanséau D., Maurin M., Lankar D., Chabaud M., Spampanato C., Malbec O., Barbier L., Muallem S., Maiuri P., Ballabio A., Helft J., Piel M., Vargas P., Lennon-Duménil A.-M., Lysosome signaling controls the migration of dendritic cells. Sci. Immunol. 2, eaak9573 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Jaynes E. T., Information theory and statistical mechanics. Phys. Rev. 106, 620 (1957). [Google Scholar]
- 34.M. Abramowitz, I. A. Stegun, Handbook of Mathematical Functions: With Formulas, Graphs, and Mathematical Tables (National Bureau of Standards, 1972). [Google Scholar]
- 35.J. Zinn-Justin, Quantum Field Theory and Critical Phenomena (Clarendon Press, 1996). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaay2103/DC1
Section S1. ME models
Section S2. Tests of ME method with bound constraints
Section S3. Visual comparison between experiments
Section S4. Analysis of motional data
Section S5. Estimate of positional uncertainty
Fig. S1. Test of ME method with bound constraints on synthetic data for the XY model.
Fig. S2. Test of ME method with bound constraints on synthetic data for the SP model.
Fig. S3. Consistency test of ME method with bound constraints.
Fig. S4. Visual comparison between tracks of the wound-healing and dendritic cell experiment.
Fig. S5. Statistics of motional data for the wound-healing experiment.
Fig. S6. Empirical distributions for cell velocities in the wound-healing experiment.
Fig. S7. Statistics of motional data for zone 1 in the dendritic cell experiment.
Fig. S8. Empirical distributions for cell velocities for zone 1 in the dendritic cell experiment.
Fig. S9. Statistics of motional data for zone 2 in the dendritic cell experiment.
Fig. S10. Empirical distributions for cell velocities for zone 2 in the dendritic cell experiment.
Fig. S11. Estimate of the positional error.
Table S1. Confidence intervals for the empirical averages in the wound-healing experiment.
Table S2. Confidence intervals for the empirical averages in the dendritic cell experiment.


