Abstract
Cost-effectiveness analyses often rely on cohort state-transition models (cSTMs). The cohort trace is the primary outcome of cSTMs, which captures the proportion of the cohort in each health state over time (state occupancy). However, the cohort trace is an aggregated measure that does not capture information about the specific transitions among health states (transition dynamics). In practice, these transition dynamics are crucial in many applications, such as incorporating transition rewards or computing various epidemiological outcomes that could be used for model calibration and validation (e.g. disease incidence and lifetime risk). In this manuscript, we propose an alternative approach to compute and store cSTMs outcomes that capture both state occupancy and transition dynamics. This approach produces a multidimensional array from which both the state occupancy and the transition dynamics can be recovered. We highlight the advantages of the multidimensional array over the traditional cohort trace and provide potential applications of the proposed approach with an example coded in R to facilitate the implementation of our method.
Keywords: cost-effectiveness analysis, decision modeling, health economics, matrices, R project, state-transition models, transition dynamics, transition rewards, multidimensional arrays, tensors
State-transition models (STM) are decision models commonly used in cost-effectiveness analysis (CEA) to estimate economic and health outcomes of different strategies over time in discrete time cycles.[1, 2] In a cohort STM (cSTM), the disease dynamics are captured by distributing a closed cohort among a mutually exclusive and collectively exhaustive set of health states.[2, 3, 4] The cohort trace is the primary outcome of cSTMs, which comprises the proportion of the cohort in each health state over time (i.e., it summarizes state occupancy).[5, 1] A limitation of the cohort trace is that it does not keep track of the transitions among health states over time (i.e., the transition dynamics of the cohort). As a consequence, it can only be used to capture outcomes that result from residing in a state for a full cycle by applying the so-called state rewards and does not contain a mechanism to assign transition rewards, which are applied only when specific transitions occur. It also limits the type of epidemiological outcomes that can be obtained from cSTMs. For example, obtaining incidence of a disease requires knowledge of the proportion of the population transitioning from a subset of states without disease to the state(s) representing the disease of interest.
To overcome the limitations of the cohort trace, we propose a multidimensional array-based approach that serves as a full summary of cSTM dynamics that complements the already useful cohort trace. The proposed approach, called the dynamics-array approach, allows modelers to efficiently calculate all measures of interest that rely on transition dynamics and at the same time to aggregate all model dynamics into a standard cohort trace.
We start by providing a formal definition of cSTM components and the cohort trace. We complement this standard notation with a description of the detailed transition dynamics. Then, we introduce the multidimensional-array structure and show how it can be easily generated. In addition, we illustrate its use to compute a measure of interest that depends on transitions among health states. Finally, we demonstrate the dynamics-array approach with an illustrative example of a cSTM programmed in R [9, 10] and compare this implementation with the traditional cohort trace approach in a simulation study. The R code is provided in the supplementary material and in GitHub https://github.com/DARTH-git/state-transition-model-dynamics).
Traditional cohort trace approach
We denote the distribution of the cohort across ns health states in a cSTM at the beginning of cycle t for all t in 0,…, nt as the state vector mt of dimensions 1 × ns. That is, each element in mt represents the proportion of the cohort in health state i = 1,…, ns at time t. Thus, mt is written as
| (1) |
where the initial state vector m0 contains the distribution of the cohort across all ns health states at the start of the simulation. The simulation begins at cycle 0 and all transitions are assumed to happen at the end of each cycle. The probability of transitioning from health state i to health state j at the end of cycle t is denoted as p[i,j,t]. This means that p[i,j,t] determines how the population will be distributed in cycle t + 1. The collection of transition probabilities across the model states over the time horizon forms the time-dependent state transition probability matrix, Pt of dimensions ns × ns,
| (2) |
For any t, all rows of Pt must sum to one. Note that if Pt is equal for all t times, equation (2) becomes a time-homogeneous transition probability matrix, where Pt = P.
The state vector at cycle t + 1, mt+1, is then obtained by the inner product between the state vector at cycle t, mt, and the corresponding transition probability matrix Pt, such that
| (3) |
Stacking the state vectors by rows for all t = 0,…, nt results in the full cohort trace matrix, M, of dimensions (nt + 1) × ns, where each row is a state vector (−mt−), resulting in
| (4) |
Together, the state vectors mt, the transition probability matrices Pt and the cohort trace M in equations (1), (2) and (4), respectively, represent the three main components of a cSTM.
Dynamics-array approach
The trace matrix M aggregates transitions from all the states to a specific state, thus loses details of the transition dynamics. We propose to use a multidimensional array, A, of dimensions ns × ns × (nt + 1) to store the proportion of the cohort that transitions between any two health states in each cycle over the time horizon. This array can be thought of as a set of two-dimensional matrices stacked along a third dimension that represents time. Below, we illustrate how to compute A from the three main components, the state vector mt, the transition probability matrices Pt and the cohort trace M, of cSTMs.
A0 represents the first "slice" of A. We compute A0 as a matrix containing the initial state vector m0 in its diagonal and 0s in the off-diagonal, such that
| (5) |
Each subsequent (t + 1)th "slice" of A is obtained by multiplying a diagonal matrix of mt, denoted as diag(mt), by Pt, such that
| (6) |
The resulting elements of the tth slice of A, At for t > 0, are
| (7) |
where a[i,j,t] is the proportion of the cohort that transitions from state i to state j between cycles t – 1 and t, generated via
| (8) |
where m[t–1,i] is the proportion of the cohort in state i at cycle t – 1 and p[i,j,t–1] the corresponding transition probability of transitioning from state i to state j at the end of cycle t – 1. In other words, A stores the transition dynamics of a simulated cohort in a cSTM.
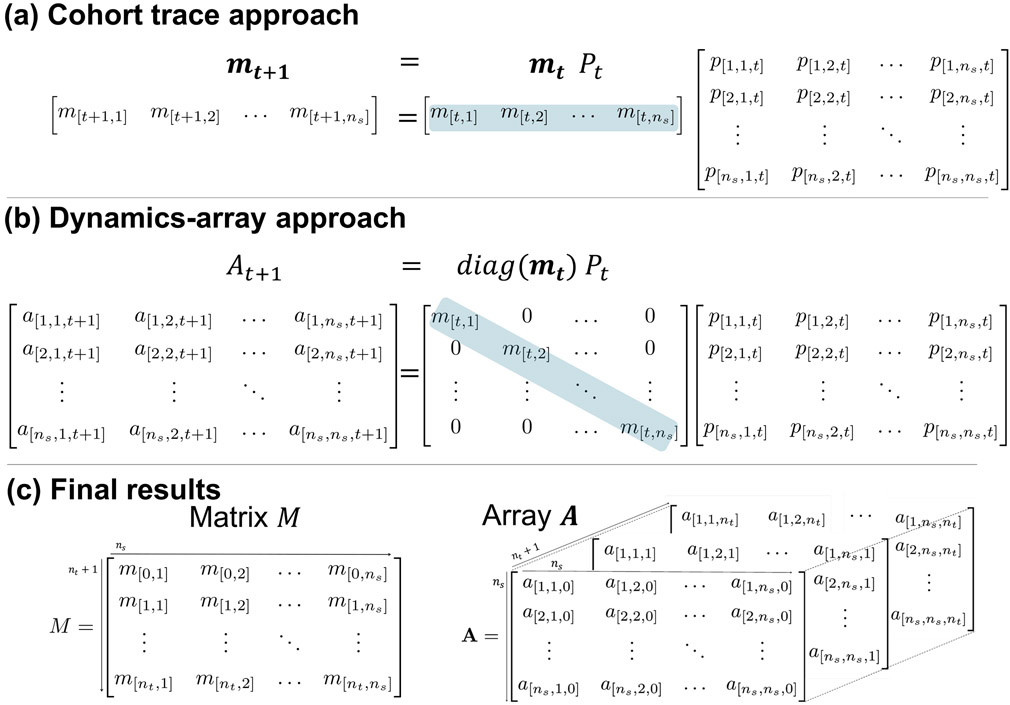
Figure 1 presents graphically the computation involved in both the traditional cohort trace approach (a) and the dynamics-array approach (b) and shows the structures of the resulting cohort trace M and dynamics-array A (c).
Figure 1:
(a) The cohort trace approach computes each row vector mt+1 of the cohort trace M, where mt+1 describes the distribution of the simulated cohort among the different health states at time t + 1. mt+1 results from multiplying the state vector mt (gray) by the transition probability matrix Pt. (b) The dynamics-array approach computes matrix At+1 containing information regarding the transition dynamics of the simulated cohort at time t + 1. The state vector mt is highlighted (gray) to emphasize that the information in both approaches is identical. (c) Shows the resulting matrix M and array A of the approaches (a) and (b), respectively.
In R, it takes only a few lines of code to generate A complementary to M (Box 1).
Box 1: R code to iteratively generate the cohort trace M and the dynamics array A.
1 for (t in 1:n.t) { # loop through the number of cycles 2 m_M [t + 1, ] <- m_M [t, ] %*% m_P # estimate the state vector for the next cycle (t + 1) 3 a_A [, , t + 1] <- diag (m_M [t, ]) %*% m_P # estimate the transition dynamics at t + 1 4 }
The cohort trace M can be computed from A by obtaining the t-th row of M, mt, summing each of the columns of At as follows:
| (9) |
where 1 is a vector of ones of dimension ns × 1. Although M can be obtained from A, we prefer to compute both M and A simultaneously. Once generated, both M and A can be exported as data objects that contain all the information about the cSTM dynamics, which can then be used in future calculation of outcomes of interest.
Applying state and transition rewards
One of the main advantages of A over M is the ability to incorporate transition rewards. Here, we demonstrate how to apply both state and transition rewards (e.g. utilities or cost) to the cSTM by using the dynamics array, A. Let Rt be a reward matrix of dimensions ns × ns that contains both state and transition rewards:
| (10) |
where r[i,j,t] is the reward associated with transitioning from state i to state j at the end of cycle t. When j = i, r[i,i,t] is the reward associated with staying in the ith health state at cycle t. That is, the off-diagonal entries of Rt store the transition rewards and the diagonal of Rt stores the state rewards for cycle t. Note that if Rt is equal for all t times (i.e., neither state nor transition rewards vary over time), equation (10) becomes a time-homogeneous rewards matrix, where Rt = R.
The state and transition rewards can be applied to the model dynamics by element-wise multiplication between At and Rt, indicated by the ⊙ sign, which produces the matrix of outputs at cycle t, Yt. Formally,
| (11) |
With this approach, the state rewards are accounted for at the beginning of the cycle and the transition rewards -assumed to happen at the end of the cycle- are accounted for at the next cycle.
In R, applying these rewards required one additional line of code compared to Box 1, as shown in line 5 of Box 2.
Box 2: R code to apply time-invariant state and transition rewards to the model dynamics stored in array A.
1 for (t in 1:n.t) { # loop through the number of cycles 2 m_M [t + 1, ] <- m_M [t, ] %*% m_P # estimate the state vector for the next cycle (t + 1) 3 a_A [, , t +1] <- diag (m_M [t, ]) %*% m_P # estimate the transition dynamics at t + 1 4 # element-wise-multiplication of array A with the rewards matrices to apply both state and transition rewards 5 a_Y [, , t + 1] <- a_A [, , t + 1] * m_R 6 }
The total rewards for each health state at cycle t, rt, is obtained by summing the rewards across all j = 1,…, ns health states.
| (12) |
Implementation in R using an illustrative example
To facilitate the implementation of the dynamics-array approach, we demonstrate its use with a stylistic healthy-sick-dead 3-state time-homogeneous cSTM example coded in R.[10] The model is used to simulate a cohort of 70-year-old individuals to compute their expected costs and quality-adjusted life years (QALYs) accrued over their remaining lifetime accounting for several transition rewards. Accounting for transition rewards with the traditional cohort trace approach is possible, however this requires creating additional temporary health states that keep track of the transitions. For our simple 3-state model this already requires two additional temporary states. In more complex models, accounting for transition rewards will result in state explosion and consequently, it is more likely to make errors while coding these models. The explanation of the state expansion and the R code for the traditional cohort trace approaches for our stylistic model can be found in the supplementary material and on GitHub (https://github.com/DARTH-git/state-transition-model-dynamics). GitHub also provides some code that shows that both approaches give identical results.
Comparison of methods using a simulation study
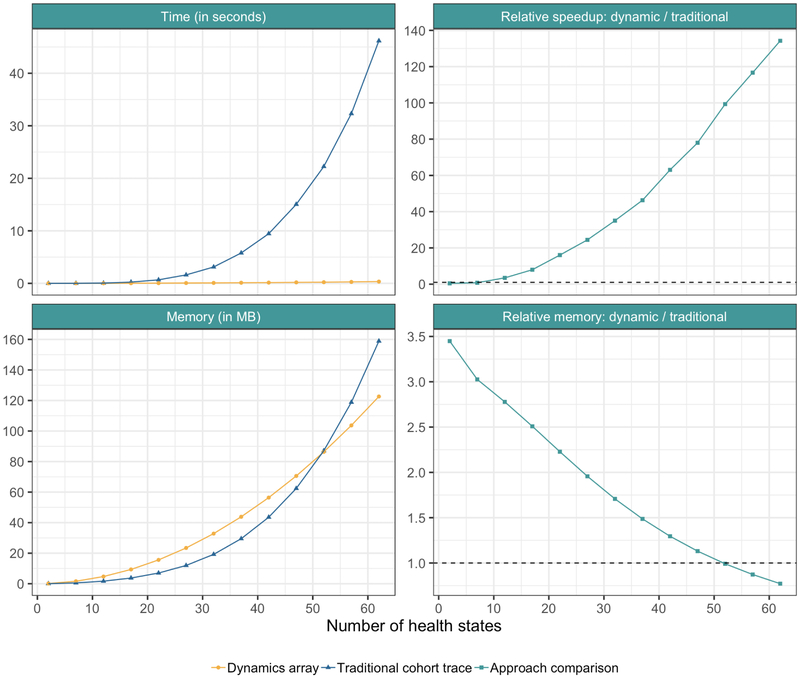
We conducted a simulation study to compare the computation time and memory requirements of the dynamics-array and traditional cohort trace approaches. We created a full factorial (13 × 110) design of experiment with the number of health states (from 2 to 62 with increments of 5 states) and the number of cycles (from 12 to 1320 with increments of 12 cycles) in a cSTM as the factors of the simulation study. We ran this full factorial experiment 10 times and took the average of the required time and memory to smooth out the variations in the computation time of R. Correcting for variation in R computation time is important, because the total required time is small (<50 seconds) and even a small variation (e.g. 3-5 seconds) could affect our results. Figure 2 shows that the dynamics-array approach is both faster (140-times with 62 health states, top part Figure 2) and requires less memory than the traditional cohort trace approach as the number of states increases (bottom part Figure 2). A more detailed description of the output of the simulation study is included in the supplementary material and the code is available on GitHub.
Figure 2:
Computation time and memory storage of the two approaches as a function of the number of states when running the model for 1,320 cycles. The top left panel shows the absolute computation time in seconds of both approaches. The top right panel shows the relative speedup of the dynamics-array approach compared to the traditional cohort trace approach. The horizontal line at y-axis equals 0 indicates when the two approached are equally fast. The bottom left panel shows the absolute memory storage in megabytes (MB) of the two approached, while the bottom right panel shows the relative required memory of the dynamics-array approach compared to the traditional cohort trace approach. The horizontal line at y-axis equals 1 indicate when both approached required the same memory storage. Above the line the traditional cohort trace requires less memory, while below the line the dynamics-array approach requires less memory. All results are based on the average of 10 simulations. This was done to smooth out the variations caused by the computation time of R.
Estimation of epidemiological measures
By obtaining A, it is possible to compute epidemiological outcomes that otherwise would not have been easily derived from M. For example, obtaining incidence and lifetime risk from M would require creating additional steps, variables or health states. Epidemiological outcomes could be used as outputs of simulation models for calibration or validation purposes. A full exposition of computing epidemiological measures from A is case-specific and is beyond the scope of this brief report. However, we illustrate the potential application of our approach with an example below.
Consider a cSTM with ns > 3 health states. We are interested in calculating a ratio et of those that transition from health state 2 to health state ns at cycle t to those that make the transition to health state ns from health states 1, 2 and 3. Using the dynamics-array approach, the ratio et can be computed as
| (13) |
With the traditional cohort trace approach, calculating this ratio would require adding three temporary health states to distinguish those that transition to ns from the health states 1, 2 and 3.
Discussion
We propose a multidimensional-array approach to overcome a limitation of the cohort trace produced by cSTM in not being able to store transition dynamics. The practical application of our approach involves adding a simple step to the traditional cohort trace approach that stores all transitions among health states over time in multidimensional array A. We described the multidimensional-array approach for a general cSTM where transitions are allowed from any state to others within a cycle, but our approach can also be applied to models that only allow one-state transition in a cycle (i.e., where the pi,j,t are zero for those transitions that are not allowed).
Traditionally, researchers have dealt with this limitation of the cSTM cohort trace by creating temporary health states that collect the state-to-state transition information. However, as we showed in our method comparison section, this solution can quickly complicate a model and result in an explosion of the number of health states. Using an individual-based microsimulation STM is another alternative [1], with considerable implications on computational time.[11]
Another method that explicitly keeps track of state-to-state transitions is through a discretely integrated condition-event (DICE) simulation.[8, 7] DICE is a modeling technique that can free up some of the Markov restrictions that makes it possible to explicitly include many events occurring at various times. Although DICE simulation is a well-structured method, and the authors of the DICE papers provided very useful supplementary files to apply the method, we see the dynamics-array approach as a relatively simpler method to compute than DICE to overcome the limitation of the cohort trace on applying transition rewards and generating all the epidemiological outcomes of interest.
A potential limitation of the use of A is the additional computation needed when building the model. However, for many applications this may be a minor limitation given the matrix-based computational efficiency of current computers. Another potential limitation is the additional storage memory required to store A, which could become a limitation in systems with limited memory. This could be an issue for computationally complex models with multiple states. However, the benefit of using A for large models is that all the complexity in the model dynamics is summarized into a compact structure which makes it relatively simple to extract information or to apply new rewards without re-running the model.
In conclusion, structuring the output of cSTMs using the dynamics-array is an efficient, simple and convenient approach to summarize the model dynamics. This simple structure allows applying state and transition rewards and obtaining epidemiological measures, while still being able to obtain and display the conventional cohort trace.
Supplementary Material
Contributor Information
Eline M. Krijkamp, Department of Epidemiology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Fernando Alarid-Escudero, Drug Policy Program, Center for Research and Teaching in Economics, (CIDE)-CONACyT, Aguascalientes, Ags., Mexico, 20313.
Eva A. Enns, Division of Health Policy and Management, University of Minnesota School of Public Health, Minneapolis, MN, USA 55455
Petros Pechlivanoglou, Child Health Evaluative Sciences, The Hospital for Sick Children, Toronto, ON, Canada, Institute of Health Policy Management and Evaluation, University of Toronto, ON, Canada.
M.G. Myriam Hunink, Departments of Epidemiology and Radiology, Erasmus University Medical Center, Rotterdam, The Netherlands, Center of Health Decision Sciences, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
Alan Yang, Child Health Evaluative Sciences, The Hospital for Sick Children, Toronto, ON, Canada.
Hawre J. Jalal, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA
References
- [1].Siebert U, Alagoz O, Bayoumi AM, Jahn B, Owens DK, Cohen DJ and Kuntz KM. State-Transition Modeling: A Report of the ISPOR-SMDM Modeling Good Research Practices Task Force-3. Value Health, Sep-Oct;15(6): 812–20, 2012. [DOI] [PubMed] [Google Scholar]
- [2].Kuntz KM, Russell BL, Owens DK, Sanders GD, Trikalions TA and Salomon JA. Decision Models in Cost-Effectiveness Analysis In Cost-Effectiveness in Health and Medicine, Neumann PJ, Ganiats TG, Russell LB, Sanders GD, Siegel JE, Chapter 5, pages 105–36. Oxford University Press, New York, second edition, 2017. [Google Scholar]
- [3].Beck JR and Pauker SG. The markov process in medical prognosis. Med Decis Making, Dec;3(4):419–58, 1983. [DOI] [PubMed] [Google Scholar]
- [4].Iskandar R A theoretical foundation for state-transition cohort models in health decision analysis. PLoS One, Dec;13(12): 1–11, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sonnenberg FA and Beck JR. Markov models in medical decision making: A practical guide. Med Decis Making, Oct-Dec;13(4): 322–38, 1993. [DOI] [PubMed] [Google Scholar]
- [6].Bouter LM, Zielhuis GA and Zeegers MPA. Textbook of Epidemiology Bohn Stafleu van Loghum, 2018. [Google Scholar]
- [7].Caro JJ and Möller J. Adding Events to a Markov Model Using DICE Simulation Med Decis Making, February;38(2):235–45, 2018. [DOI] [PubMed] [Google Scholar]
- [8].Caro JJ Discretely Integrated Condition Event (DICE) Simulation for Pharmacoeconomics PharmacoEconomics, July;34(7): 665–72, 2016. [DOI] [PubMed] [Google Scholar]
- [9].Jalal HJ, Pechlivanoglou P, Krijkamp EM, Alarid-Escudero F, Enns EA and Hunink MGM. An Overview of R in Health Decision Sciences. Med Decis Making, October;37(7): 735–46, 2017. [DOI] [PubMed] [Google Scholar]
- [10].R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2013. [Google Scholar]
- [11].Krijkamp EM, Alarid-Escudero F, Enns EA, Jalal HJ, Hunink MGM and Pechlivanoglou P. Microsimulation Modeling for Health Decision Sciences Using R: A Tutorial. Med Decis Making, April;38(3): 400–22, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.