Abstract
In our previous studies, enhancer of zeste homolog 2 (EZH2) has been proven to be a key oncogenic driver in oral squamous cell carcinoma (OSCC). However, the regulatory mechanisms on EZH2 remain poorly understood in OSCC. Here, through multi-transcriptomics, bioinformatics analysis, and quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), the co-expression network of long noncoding RNA RC3H2 (RC3H2), microRNA-101-3p (miR-101-3p), and EZH2 were screened and validated as a competing endogenous RNA (ceRNA) mechanism in OSCC. Silencing of RC3H2 inhibited OSCC cell proliferation, colony formation, migration, and invasion in vitro and reduced the expression of EZH2 and H3K27Me3, whereas RC3H2 overexpression significantly promoted OSCC cell growth, colony formation, migration, invasion, and xenograft tumor growth in vivo and increased the expression of EZH2 and H3K27Me3. A fluorescence in situ hybridization (FISH) assay verified that RC3H2 was predominately localized to the cytoplasm. RNA pull-down and luciferase activity assays showed that miR-101-3p was physically bound to RC3H2 as well as EZH2, and its inhibitor reversed the inhibitory effect of RC3H2 knockdown on progression of OSCC. Taken together, our findings demonstrate that RC3H2 as completive endogenous RNA sponging miR-101-3p targets EZH2 and facilitates OSCC cells’ malignant behavior.
Keywords: EZH2, lncRNA RC3H2, microRNA-101-3p, proliferation, migration, invasion, metastasis, competitive endogenous RNA, oral squamous cell carcinoma
Introduction
Oral squamous cell carcinoma (OSCC) is the one of most common cancers, accounting for over 90% of malignant lesions in the oral cavity,1 with approximately 300,000 new cases reported every year around the world.2 Though great progresses in treatments including surgery, chemotherapy, and radiotherapy was achieved in the past several years, 5-year overall survival of patients with OSCC was still about 50%. Hence, the underlying molecular mechanism of OSCC initiation and progression was fully illuminated to decrease the morbidity and mortality of patients with OSCC.
Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of polycomb repressive complex 2 (PRC2), a highly conserved histone methyltransferase that methylates lysine-27 of histone H3 (H3-K27).3 It has been demonstrated that EZH2 is involved in methylation and silencing of a subset of genes implicated in cell differentiation, suggesting that it may play a key role in cell differentiation and maintenance of adult stem cell populations.4 In our prior studies, EZH2 was identified as a key oncogenic driver that promotes the malignant transformation of oral leukoplakia and the progression of head and neck squamous cell carcinoma (HNSCC).5,6 However, the potential mechanisms of regulating EZH2 in OSCC remain to be fully elucidated.
Long noncoding RNAs (lncRNAs) are one class of transcripts with more than 200 nucleotides, including antisense, intergenic, and intronic transcripts and pseudogenes.7,8 Accumulating evidence has revealed that lncRNAs play an important role in regulating genes at the transcriptional, posttranscriptional, and chromosomal levels8 and participate in biological processes of tumor cell proliferation, differentiation, invasion, and metastasis. For example, antisense lncRNAs can hybridize to their specific pre-mRNA, which leads to alternatively spliced mRNAs or the production of endogenous small interfering RNAs (siRNAs) through Dicer.9 lncRNAs also acts as a sponge that can silence miRNA expression.10 Moreover, lncRNAs interact with proteins, which results in dysregulated protein activity and cellular localization.11 Additionally, lncRNAs can be transferred to small ncRNAs and act as endo-siRNAs or microRNAs (miRNAs).12 In our previous studies, decreased expression of lncRNA PTENP1 was found to promote malignant behaviors, and its reduced expression of lncRNA PTENP1 associated with the poor survival of patients with HNSCC.13 Furthermore, a three-lncRNA signature was identified as a novel biomarker panel for the accurate prognosis prediction of patients with HNSCC,8 suggesting that lncRNAs play a vital role as predictive biomarkers or therapeutic targets in OSCC initiation and progression.
In this study, multi-transcriptomics, bioinformatics analysis, and qRT-PCR were performed to screen and validate the co-expression network of RC3H2, miR-101-3p, and EZH2, which was regarded as the candidate competing endogenous RNA (ceRNA) mechanism in the development of OSCC. To further verify the hypothesis, the expression level of RC3H2 and the biological function of RC3H2 were evaluated by qRT-PCR, loss of function, and gain of functions in vitro and in vivo. The ceRNA mechanism was further tested by luciferase activity, RNA pull-down, FISH, and rescue assays in vitro.
Results
The Co-expression Network among EZH2, RC3H2, and miR-101-3p Expression Was Screened and Validated by Multi-transcriptome Analysis, Bioinformatics Analysis, and qRT-PCR in OSCCs
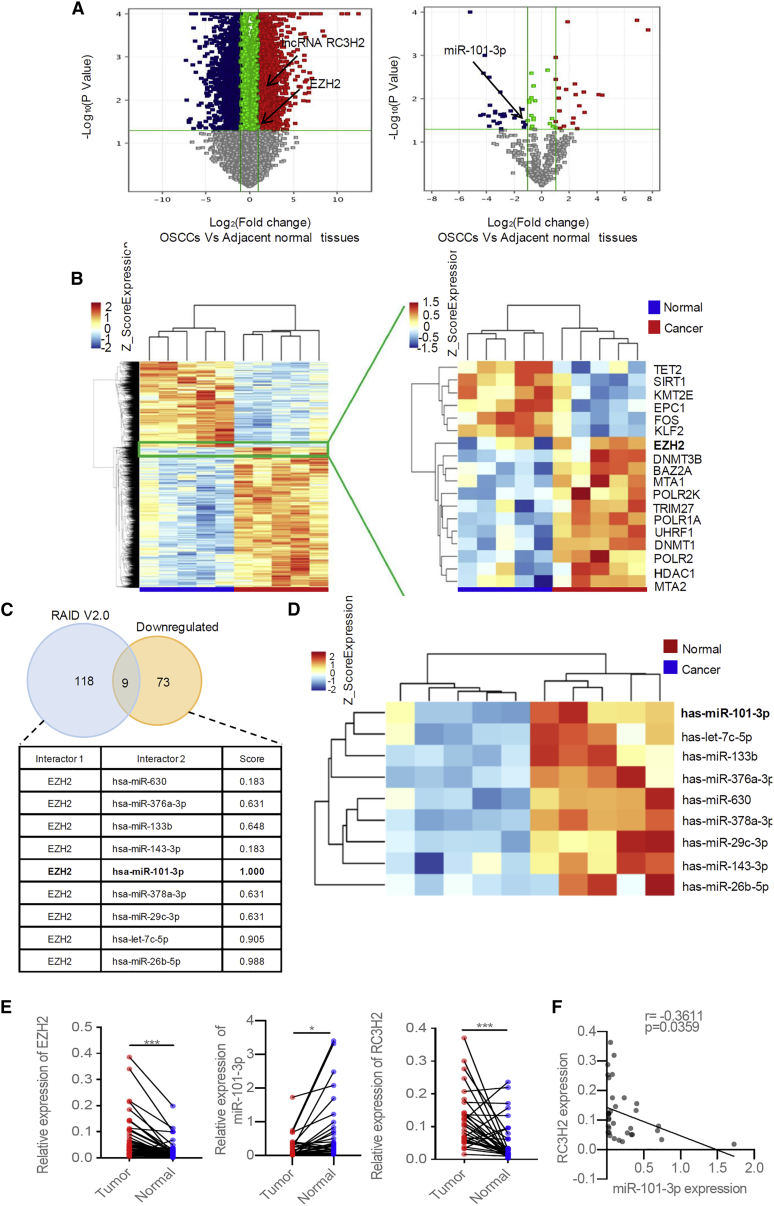
To identify the co-expression network among EZH2, lncRNAs, and miRNAs of OSCCs, we performed multi-transcriptome analysis (Affymetrix human OE lncRNA microarray and Agilent human miRNA v21.0 8 × 60 K) and bioinformatics analysis using five paired OSCCs and the corresponding adjacent normal tissues (Figures 1A and 1B). The clinical characteristic of the five OSCC patients used for microarray assay and bioinformatics analysis is shown in Table S1. 70 upregulated lncRNAs were verified as the co-expression network of EZH2, including RC3H2 (Figure S1). Additionally, nine remarkably downregulated more than 2-fold miRNAs targeting EZH2 were identified by Venn analysis. Of them, miR-101-3p was predicted to be a binding site with RC3H2 (http://www.rna-society.org/raid2/index.html) (Figure 1C), suggesting it was the candidate ceRNA mechanism in OSCCs. The relationship among RC3H2, miR-101-3p, and EZH2 was fully predicted as shown in Figure S2. Furthermore, the expression of RC3H2, EZH2 and miR-101-3p were assessed by qRT-PCR in 34 paired OSCCs and adjacent normal tissues. The EZH2 and RC3H2 expression levels were significantly elevated in OSCCs as compared to adjacent normal tissues, whereas miR-101-3p expression was dramatically reduced in OSCCs (Figure 1E). RC3H2 expression was negatively correlated with miR-101-3p expression in OSCCs (Figure 1F). No relationship among RC3H2, EZH2, and miR-101-3p levels and the clinical parameters, including patient age (≥60 years and <60 years), gender (male and female), smoking history (yes and no), T stage (T1–T2 and T3–T4), lymph node status (pN positive and pN negative), tumor-lymph node-metastasis (TNM) stage (I–II and III–IV), perineural invasion (yes and no) or extracellular spread (ECS) (yes and no) was found in our study (Table S2).
Figure 1.
The Expression and Relationship of RC3H2, EZH2, and miR-101-3p in OSCCs
(A) Volcano plots used to visualize the differential expression between two different groups; x axis, log2 ratio of the level of lncRNA, mRNA, and miRNA expression between normal and tumor tissues; y axis, the false discovery rate value (−log10 transformed) of the lncRNAs, mRNAs, and miRNAs. (B) EZH2 was screened to be upregulated in OSCCs using an Affymetrix OE lncRNA microarray and hierarchical clustering analysis (fold change, FC > 2.0, p < 0.05). Red indicates high expression; blue indicates low expression. (C) The nine microRNAs including miR-101-3p were selected by a Venn analysis between RAID v2.0 predicted targeted EZH2 miRNAs (blue circle) and downregulated miRNAs (light red circle) derived from Agilent human miRNA microarray analysis. The interaction score is also shown. (D) The heatmap of the nine miRNAs was shown in OSCCs by hierarchical clustering analysis (FC > 2.0, p < 0.05). (E) The expression level of EZH2, miR-101-3p, and RC3H2 in 34 paired OSCCs and adjacent normal tissues were verified by qRT-PCR, respectively (***p < 0.001 versus *p < 0.05). (F) The remarkably negative association between RC3H2 expression and miR-101-3p expression was found by expression correlation analysis.
RC3H2 Physically Interacted with miR-101-3p and Its Subcellular Predominate Localization to the Cytoplasm of OSCC Cells
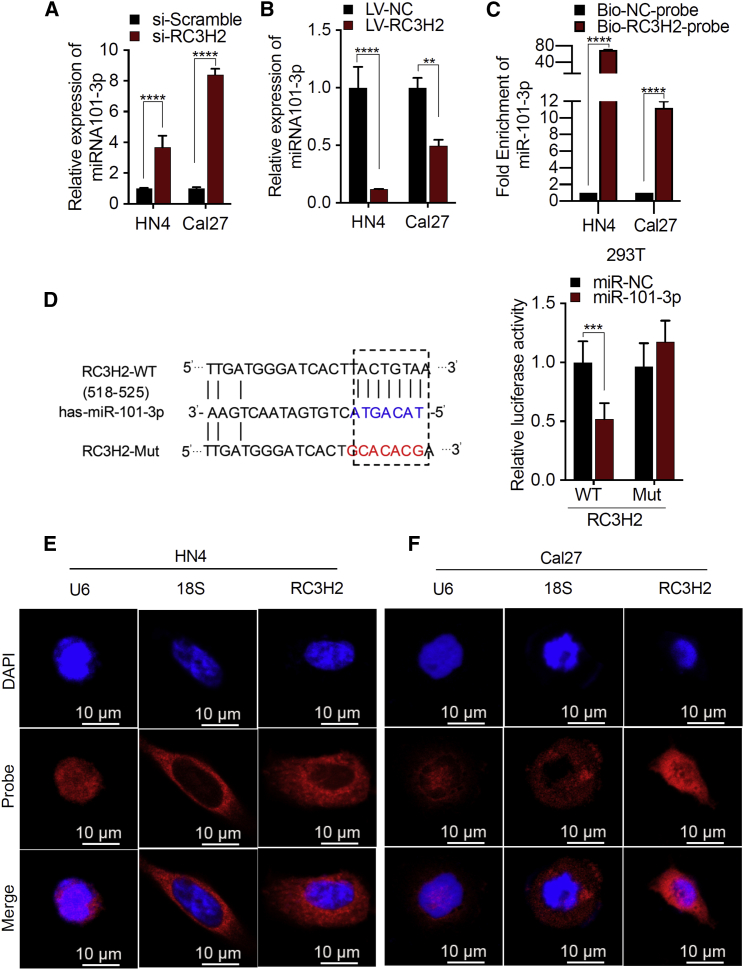
To further validate the relationship between miR-101-3p and RC3H2, the expression level of miR-101-3p was evaluated by qRT-PCR with RC3H2 knockdown or RC3H2 overexpression in OSCC cells, respectively. Upon silencing of RC3H2 in Cal27 and HN4 cells, the expression level of miR-101-3p was increased, whereas miR-101-3p expression level was decreased in OSCC cells when RC3H2 was overexpressed (Figures 2A and 2B). Furthermore, an RNA pull-down assay was applied to determine the direct interaction of miR-101-3p and RC3H2 by using 5′ biotin-linked RNAs. Our results showed that miR-101-3p was enriched to biotinylated RC3H2 probes (Figure 2C). Finally, a bioinformatics analysis was used to predict miR-101-3p as the downstream target of RC3H2. To further identify whether miR-101-3p was a possible target of RC3H2, a wild-type or mutated sequence containing the miR-101-3p binding sequence was used to construct a luciferase reporter vector (Figure 2D). The luciferase reporter vector was transfected into 293T cells combined with or without the miR-101-3p mimic. The luciferase reporter analysis showed that miR-101-3p inhibited the luciferase activity in 293T cells transfected with RC3H2-wild-type (WT) reporter vectors, but not in cells transfected with the corresponding RC3H2-mutant (Mut) reporter vectors (Figure 2D), suggesting that miR-101-3p was the target of RC3H2. To further identify the association between miR-101-3p and RC3H2, the subcellular localization of miR-101-3p was also detected in OSCC cells by fluorescence in situ hybridization (FISH) assay. Our results indicated that the miR-101-3p expression was decreased in OSCC transfected with lentivirus (LV)-RC3H2, whereas miR-101-3p expression was increased in OSCC transfected with si-RC3H2 (Figure S3). In order to characterize the precise role of RC3H2 in the progression of OSCC, the subcellular localization of RC3H2 was detected in OSCC cells by FISH assay. The findings showed that RC3H2 was predominantly localized to the cytoplasm of HN4 and Cal27 cells (Figures 2E and 2F).
Figure 2.
RC3H2 Directly Bound to miR-101-3p and Its Mainly Subcellular Localized to Cytoplasm
(A) qRT-PCR was used to determine the expression of miR-101-3p in HN4 and Cal27 cells transfected with si-RC3H2. (B) qRT-PCR was used to determine the expression of miR-101-3p in HN4 and Cal27 cells infected with LV-RC3H2. (C) qRT-PCR analysis of miR-101-3p following RNA pull-down assays with RC3H2 probes in HN4 and Cal27 cells. (D) StarBase version v2.0 results showing the sequence of RC3H2 with highly conserved putative miR-101-3p binding sites and construction of the RC3H2-Mut vector, which changed TACTGTAA into GCACACGA from nucleotides 518 to 525. miR-101-3p mimic considerably reduced the luciferase activity of the RC3H2-WT luciferase reporter vector compared with negative control, while miR-101-3p mimic did not have any impact on the luciferase activity of RC3H2-Mut-transfected cells. (E) FISH analysis of RC3H2 in HN4 cells. (The nuclei were stained with DAPI, and 18S rRNA was used as a cytoplasmic marker. Scale bars, 10 μm.) (F) FISH analysis of RC3H2 in Cal27 cells. (The nuclei were stained with DAPI, and 18S rRNA was used as a cytoplasmic marker. Scale bars, 10 μm.)
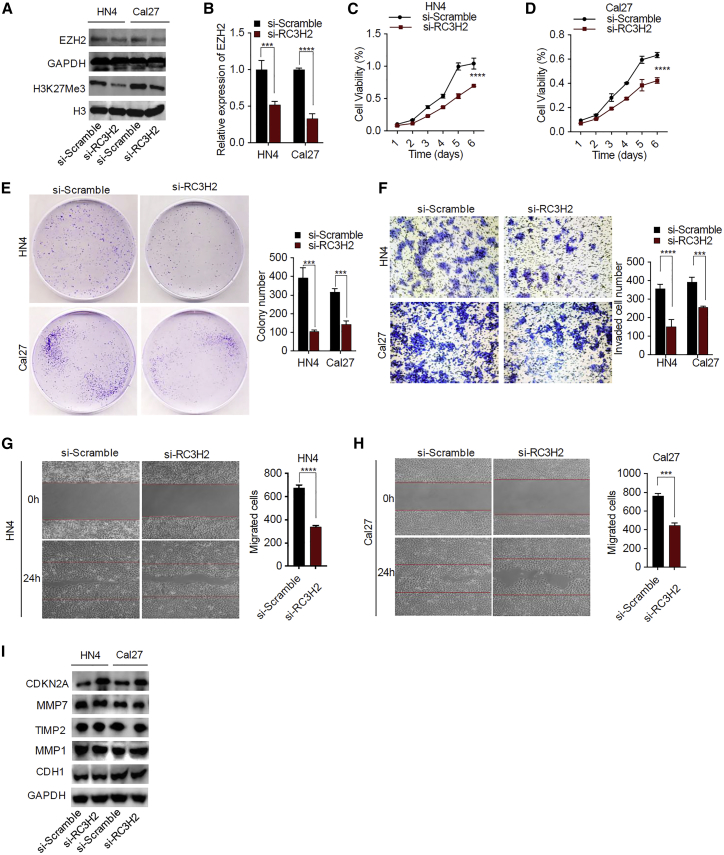
Knockdown of RC3H2 Suppressed the Proliferation, Colony Formation, Migration, and Invasion In Vitro and Reduced the Expression of H3K27Me3 as well as Increased Expression of CDKN2A
To evaluate the biological function of RC3H2 knockdown in OSCC cells, siRNAs against RC3H2 were synthesized (5′-CTCTGTAACCTGAAATGAA-3′), and the silencing effect of siRNAs against RC3H2 were tested by qRT-PCR in HN4 and Cal27 cells. Upon RC3H2 knockdown, the transcriptional level of EZH2 as well as the post-translational level of EZH2 was dramatically decreased in OSCC cells by qRT-PCR analysis and western blotting assays (Figures 3A and 3B). Additionally, the expression level of H3K27Me3 were correspondingly reduced in OSCC cells with RC3H2 knockdown (Figure 3A). Knockdown of RC3H2 significantly inhibited cell proliferation at 72 h (p < 0.01) and 96 h (p < 0.001) as well as colony-formation abilities (p < 0.01) compared to the control group in the HN4 and Cal27 cells (Figure 3C). Meanwhile, the migration and invasion abilities were decreased in HN4 and Cal27 cells with the RC3H2 knockdown group as compared to the control group by wound-healing and Transwell assays (p < 0.01) (Figures 3F–3H). Additionally, the expression level of CDKN2A protein was increased in OSCC cells upon RC3H2 knockdown. However, there were no significant alterations in the expression levels of MMP1, MMP7, TIMP2, and CDH1 proteins after RC3H2 silencing (Figure 3I).
Figure 3.
RC3H2 Knockdown Inhibited OSCC Cell Growth, Migration, and Invasion In Vitro
(A) Western blot analysis was used to confirm the protein expression of EZH2 and H3K27Me3 in HN4 and Cal27 cells with si-RC3H2 or si-Scramble. (B) qRT-PCR was used to determine the mRNA expression of EZH2 in HN4 and Cal27 cells transfected with si-RC3H2 or si-Scramble. (C and D) The cell proliferation of HN4 (C) and Cal27 (D) cells transfected with si-RC3H2 or si-Scramble were evaluated by CCK-8 assays. (E) The colony-formation abilities of HN4 and Cal27 cells transfected with si-RC3H2 or si-Scramble were determined by colony-formation assays. (F) The invasion ability of HN4 and Cal27 cells transfected with si-RC3H2 or si-Scramble was performed by Transwell assays. (G and H) The migration behavior of HN4 (G) and Cal27 (H) cells transfected with si-RC3H2 or si-Scramble was assessed by wound-healing assays. Data are presented as mean ± SD from three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). (I) Western blot analysis was used to confirm the protein expression of DKN2A, MMP1, MMP7, TIMP2, and CDH1 in HN4 and Cal27 cells with si-RC3H2 or si-Scramble.
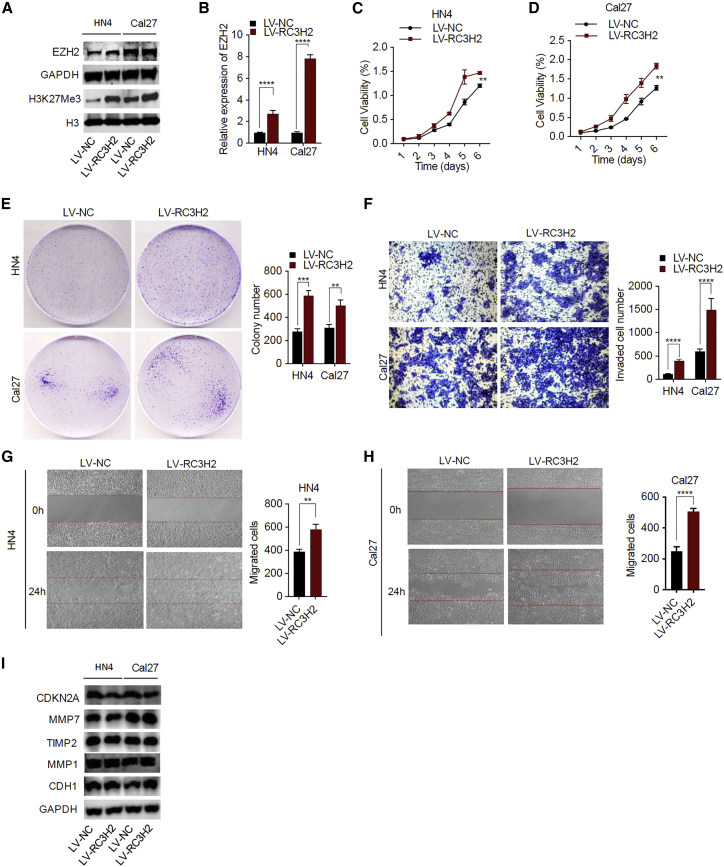
RC3H2 Overexpression Promotes Cell Proliferation, Migration, and Invasion in OSCC Cells In Vitro and Increases the Expression of H3K27Me3 as well as Decreases the Expression of CDKN2A
In HN4 and Cal27 cells, RC3H2 overexpression significantly increased the expression level of EZH2 as compared with the control group, as shown in western blotting (Figure 4A) and qRT-PCR assays (Figure 4B). In addition, H3K27Me3 levels were also found to be notably increased in OSCC cells with overexpression of RC3H2 (Figure 4A). Cell proliferation was remarkably increased in the RC3H2 overexpression group at 72 h and 96 h (p < 0.001) compared to the control group (Figures 4C and 4D). The number of colony formations were dramatically increased with RC3H2 overexpression compared to the control (p < 0.01) (Figure 4F). The counts of invasive cells were also promoted in RC3H2 overexpression group compared to the control group both in HN4 cells (p < 0.001) and Cal27 cells (p < 0.001) (Figure 4F). Meanwhile, cell migration rate was increased in the RC3H2 overexpression group compared with the control group in HN4 cells (p < 0.001) and Cal27 cells (p < 0.01) (Figures 4G and 4H). In addition, the expression level of CDKN2A protein was decreased in OSCC cells with RC3H2 overexpression. However, there were no significant alterations in the expression levels of MMP1, MMP7, TIMP2, and CDH1 proteins after RC3H2 overexpression (Figure 4I).
Figure 4.
RC3H2 Overexpression Facilitated OSCC Cell Growth, Migration, and Invasion In Vitro
(A) Western blot analysis was used to confirm the protein expression of EZH2 and H3K27Me3 in HN4 and Cal27 cells stably transduced with LV-RC3H2 or LV-NC. (B) qRT-PCR was used to determine the mRNA expression of EZH2 in HN4 and Cal27 cells stably transduced with LV-RC3H2 or LV-NC. (C and D) The cell proliferation of HN4 (C) and Cal27 (D) cells stably transduced with LV-RC3H2 or LV-NC were determined by CCK-8 assays. (E) The colony-forming abilities of HN4 and Cal27 cells stably transduced with LV-RC3H2 or LV-NC were determined by colony-formation assays. (F) The invasion abilities of HN4 and Cal27 cells stably transduced with LV-RC3H2 or LV-NC were performed by Transwell assays. (G and H) The migration behavior of HN4 (G) and Cal27 (H) cells stably transduced with LV-RC3H2 or LV-NC was assessed by wound-healing assays. Data are presented as mean ± SD from three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). (I) Western blot analysis was used to confirm the protein expression of DKN2A, MMP1, MMP7, TIMP2, and CDH1 in HN4 and Cal27 cells with si-RC3H2 in HN4 and Cal27 cells stably transduced with LV-RC3H2 or LV-NC.
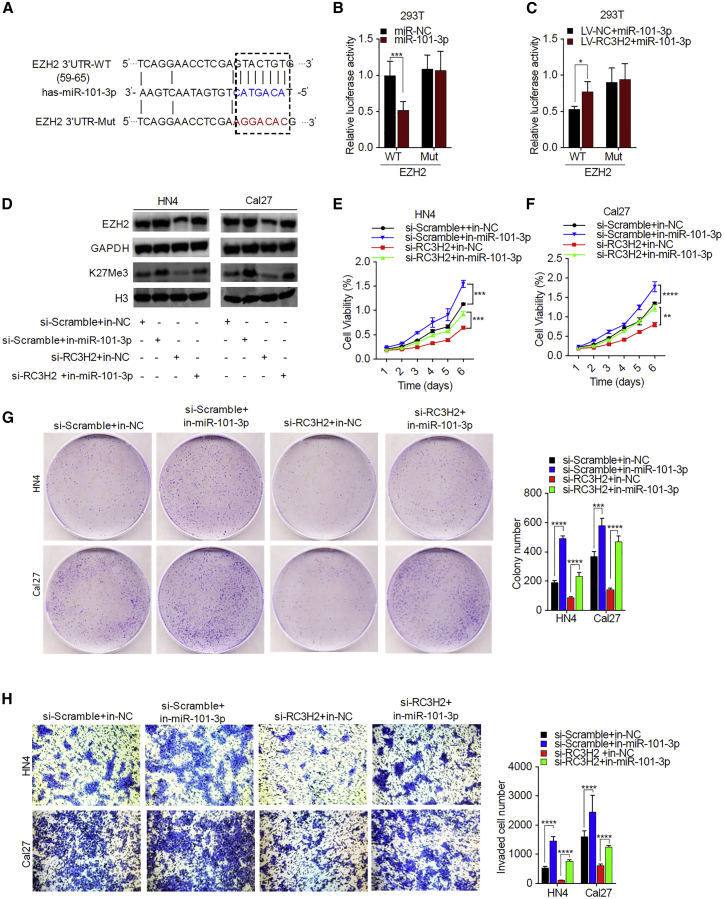
Suppression of miR-101-3p Attenuated the RC3H2 Knockdown-Induced Inhibitory Effects on OSCC Cells by Targeting EZH2
To verify that EZH2 is a direct target of miR-101-3p (Figure 5A), a luciferase assay with the 3′ UTR-WT or 3′ UTR-Mut of EZH2 was first performed in 293T cells. miR-101-3p mimics remarkably reduced the luciferase activity of 3′ UTR-WT of EZH2 but not that of 3′ UTR-Mut of EZH2 in 293T cells, suggesting EZH2 is one of the target genes for miR-101-3p (Figure 5B). Next, 293T cells stably expressed with RC3H2 or negative control (NC) were cotransfected with the 3′ UTR-WT or 3′ UTR-Mut EZH2 vectors and miR-101-3p mimics, respectively. Our findings showed that the miR-101-3p mimics partially reversed the inhibitory effects on the luciferase activity in stably expressed RC3H2 293T cells transfected with the 3′ UTR-WT of the EZH2 vector due to the sponging effect of RC3H2 on miR-101-3p (Figure 5C). Western blot analysis showed that silencing of RC3H2 significantly decreased the posttranscriptional levels of EZH2 and H3K27Me3, whereas miR-101-3p inhibitor partially reversed the inhibitory effect of RC3H2 knockdown and induced the increased expression levels of EZH2 and H3K27Me3 in HN4 and Cal27 cells (Figure 5D). RC3H2 knockdown remarkably inhibited cell proliferation, colony-formation ability, migration, and invasion, while the miR-101-3p inhibitor partially recovered RC3H2 knockdown-induced cell proliferation (Figures 5E and 5F), colony-formation ability (Figure 5G), and invasion in HN4 and Cal27 cells (Figure 5H).
Figure 5.
miR-101-3p Inhibitor Reversed the Inhibitory Effect of RC3H2 Knockdown on Malignant Behaviors of OSCC Cells
(A) miRWalk v2.0 predicts the sequence of EZH2 with highly conserved putative miR-101-3p binding sites and construction of the EZH2-Mut vector, which changed GTACTGTG into AGGACAC from nucleotides 59 to 65. (B) Downregulation of the reporter gene with the wild-type region from EZH2-WT was apparent, whereas no effect on the EZH2-Mut was detected. (C) RC3H2 overexpression partially reversed miR-101-3p mimic-induced reduction of the luciferase activity of the EZH2-WT luciferase reporter vector compared with negative control. (D) The downregulation of EZH2 and H3K27Me3 protein levels by RC3H2 silencing was partially reversed through miR-101-3p inhibitor in HN4 and Cal27 cells. (E and F) The cell proliferation of HN4 (E) and Cal27 (F) cells co-transfected with si-Scramble plus in-NC, si-RC3H2 plus in-NC, si-Scramble plus in-miR-101-3p, or si-RC3H2 plus in-miR-101-3p were evaluated by CCK-8 assays. (G) The colony-forming abilities of HN4 and Cal27 cells co-transfected with si-Scramble plus in-NC, si-RC3H2 plus in-NC, si-Scramble plus in-miR-101-3p, or si-RC3H2 plus in-miR-101-3p were determined by colony-formation assays. (H) The invasion abilities of HN4 and Cal27 cells co-transfected with si-Scramble plus in-NC, si-RC3H2 plus in-NC, si-Scramble plus in-miR-101-3p, or si-RC3H2 plus in-miR-101-3p were performed by Transwell assays. Data are presented as mean ± SD from three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
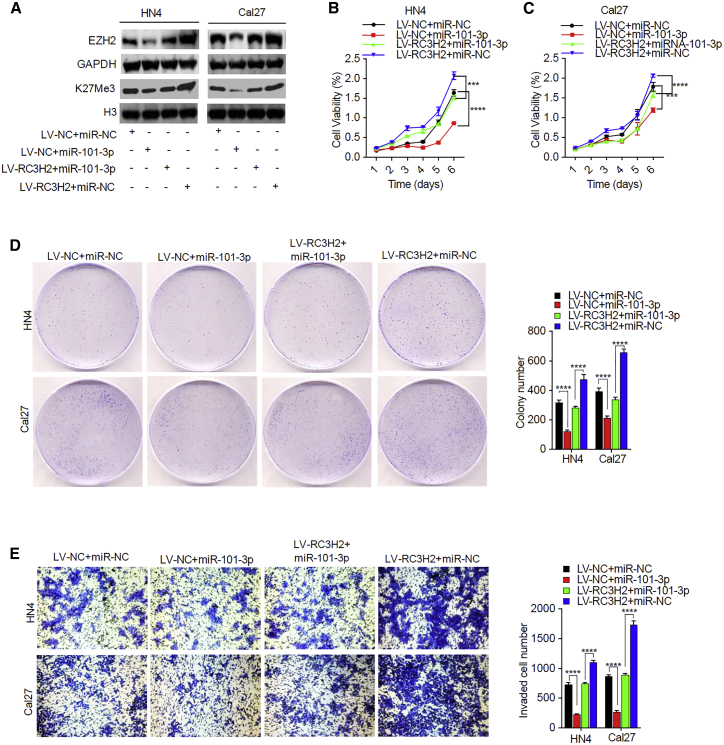
Overexpression of miR-101-3p Attenuates the RC3H2-Induced Aggressive Effects in OSCC Cells by Targeting EZH2
Overexpression of RC3H2 significantly increased the expression levels of EZH2 and H3K27Me3, whereas miR-101-3p mimic partially reversed RC3H2 overexpression-induced EZH2 and K27Me3 expression in OSCC cells, as shown by western blot analysis (Figure 6A). RC3H2 overexpression significantly promoted cell proliferation, colony-formation ability, and invasion in HN4 and Cal27 cells, while miR-101-3p mimic partially inhibited the EZH2 overexpression-mediated cell proliferation (Figures 6B and 6C), colony-formation ability (Figure 6D), and invasion (Figure 6E), suggesting that RC3H2 affects cell proliferation, invasion, and colony-formation ability by sponging miR-101-3p-modulating EZH2 in OSCC cells.
Figure 6.
miR-101-3p Mimic Attenuated the RC3H2 Overexpression-Induced Aggressive Behaviors of OSCC Cells
(A) The upregulation of EZH2 and H3K27Me3 protein levels by RC3H2 overexpression was partially reversed through miR-101-3p mimic in HN4 and Cal27 cells. (B and C) The cell proliferation of HN4 (B) and Cal27 (C) cells co-transduced with LV-NC plus miR-NC, LV-RC3H2 plus miR-NC, LV-NC plus miR-101-3p mimic, or LV-RC3H2 plus miR-101-3p mimic were evaluated by CCK-8 assays. (D) The colony-forming abilities of HN4 and Cal27 cells co-transduced with LV-NC plus miR-NC, LV-RC3H2 plus miR-NC, LV-NC plus miR-101-3p mimic, or LV-RC3H2 plus miR-101-3p mimic were determined by colony-formation assays. (E) The invasion abilities of HN4 and Cal27 cells co-transduced with LV-NC plus miR-NC, LV-RC3H2 plus miR-NC, LV-NC plus miR-101-3p, mimic or LV-RC3H2 plus miR-101-3p mimic were performed by Transwell assays. Data are presented as mean ± SD from three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Ectopic Expression of RC3H2 Promoted Xenograft Tumor Growth and Metastasis in Nude Mice
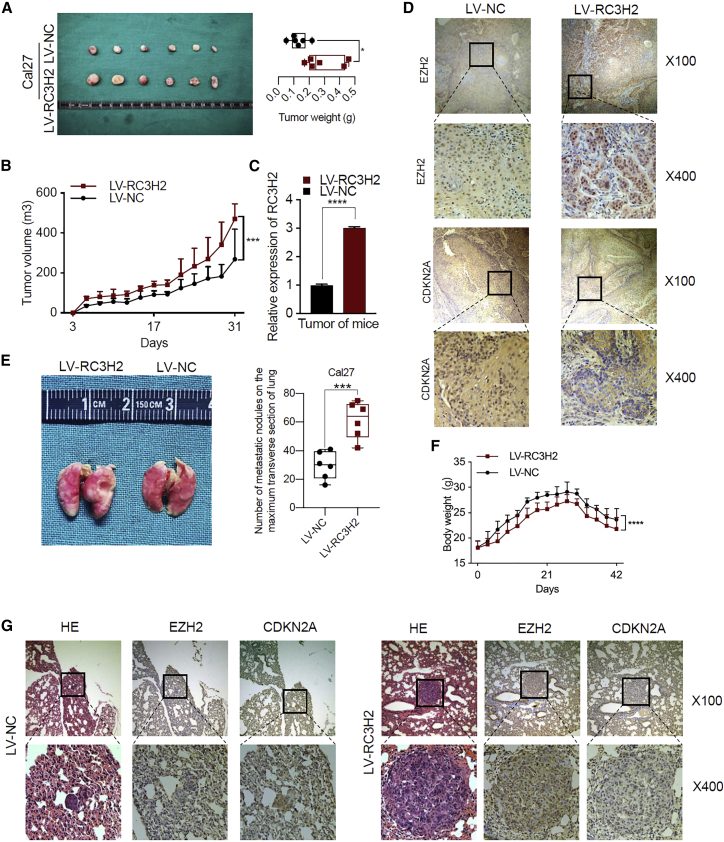
Since RC3H2 expression facilitates the malignant properties of OSCC cells in vitro, we also evaluated the effect of RC3H2 on the tumorigenicity in a xenograft model. For this purpose, Cal27 cells stably transfected with LV-RC3H2 or LV-NC were injected subcutaneously into the left and right dorsal flanks of nude mice, respectively.
Tumor sizes were monitored by a caliper in every 3 days. After 4 weeks, the animals were executed, and the tumor weights were measured. The results showed that the tumor growth and tumor weight were significantly increased in RC3H2-overexpressing cells as compared to control cells (p < 0.001 versus p < 0.05, respectively) (Figures 7A and 7B). The transcriptional expression of RC3H2 was significantly increased in LV-RC3H2-injected subcutaneous xenografts compared to that of LV-NC-injected xenografts (p < 0.001) (Figure 7C). Furthermore, the xenograft tumors with RC3H2 overexpression possessed a significantly increased nuclear EZH2 and decreased CDKN2A labeling index in the Cal27 cell line xenograft (Figure 7D).
Figure 7.
RC3H2 Promoted OSCC Cell Growth and Metastasis In Vivo
(A) The volumes and weights of tumors from Cal27 cells stably transduced with LV-NC or LV-RC3H2 tumor-bearing nude mice are shown; n = 6/group, *p < 0.05. (B) The tumor growth curves of tumors from Cal27 cells stably transduced with LV-NC or LV-RC3H2 tumor-bearing nude mice are shown. **p < 0.01. (C) The levels of RC3H2 expression in tumor tissues formed from Cal27 cells stably transduced with LV-NC or LV-RC3H2 as determined by qRT-PCR. ****p < 0.0001. (D) Representative immunohistochemical staining for EZH2 and CDKN2A were conducted in Cal27 cells stably transduced with LV-NC or LV-RC3H2 xenografts each group (upper row indicates original magnification, ×100, and lower row indicates original magnification, ×400). (E) Representative images of the lungs of mice inoculated with Cal27 cells stably transduced with RC3H2 or NC by tail-vein injection for 8 weeks and the number of metastatic nodules on the maximum transverse section of lung was calculated;***p < 0.001. (F) The body weights of Cal27 cells stably transduced with LV-NC or LV-RC3H2 tumor-bearing nude mice are shown; n = 6/group, ****p < 0.0001. (G) H&E staining and IHC of EZH2 and CDKN2A expression in lung tissues with tumor colonization (upper row indicates original magnification, ×100, and lower row indicates original magnification, ×400).
For the animal metastasis assay, Cal27 cells stably overexpressing LV-RC3H2 or LV-NC were injected into the tail vein of mice. After 7 weeks, the mice were sacrificed, and the metastatic nodules of lungs were detected by picric acid. The results showed that the metastatic nodules were significantly increased in RC3H2-overexpressing cells as compared to control cells (p < 0.001) (Figure 7E). The body weight was significantly decreased in RC3H2-overexpressing cells as compared to control cells (p < 0.0001) (Figure 7F). Furthermore, the xenograft lungs with RC3H2 overexpression possessed a significantly increased nuclear EZH2 and decreased CDKN2A labeling index in the Cal27 cell line xenograft (Figure 7G).
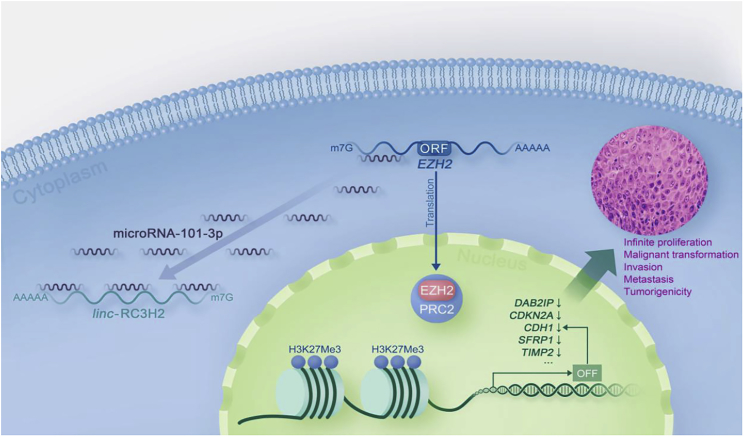
Conclusively, our findings reveal the regulatory mechanism of EZH2 by the sponging interaction between RC3H2 and miR-101-3p in OSCCs (Figure 8).
Figure 8.
A Proposed Model Illustrating the Regulatory Role of RC3H2 in Promoting Cell Proliferation and Migration by Sponging miR-101-3p to Affect EZH2 Expression
Discussion
Accumulative evidence has shown that lncRNAs play an important role in human disease progression14, 15, 16 and various kinds of cancer initiation and development.17, 18, 19 Dysregulated lncRNAs have been found to participate in some biological processes of cancer cell proliferation, differentiation, invasion, and metastasis in OSCCs.8 For instance, the expression of lncRNA CASC2 was found to be significantly decreased in OSCC tissues as compared to adjacent normal tissues, and ectopic expression of CASC2 inhibited OSCC cell proliferation, invasion, and migration by downregulating CDK1, suggesting CASC2 was a tumor suppressor.20 lncRNA FAL1 was detected to be overexpressed in OSCCs as compared with adjacent normal tissues and promoted the progression of OSCC by targeting CRKL as a sponge of miRNA-761.21 In the present study, the upregulation of RC3H2 was identified by Affymetrix human OE lncRNA microarray in OSCCs and was further verified by qRT-PCR in 34 OSCC samples and paired adjacent normal tissues. However, the association among the expression levels of RC3H2, miR-101-3p, EZH2, and clinicopathological factors has not been found, probably due to the small size cohort of OSCC patients in this study. Silencing or overexpression of RC3H2 dramatically inhibited or increased OSCC cell proliferation, colony-formation ability, migration, and invasion behaviors. Thus, RC3H2 was regarded as an oncogenic role in malignant progression of OSCC in the present study.
A widespread interaction network of ceRNA has been found to be involved in OSCC carcinogenesis, in which lncRNAs could regulate mRNAs by sponging targeting miRNAs.21,22 Previous studies have reported that lncRNA MALAT1 could modulate STAT3 expression by sponging miR-125b in OSCC.23 lncRNA NEAT1 facilitates proliferation and invasion of OSCC cells by regulating the miR-365/RGS20 axis.24 The other findings also demonstrated that lncRNA H1 promotes cell proliferation and invasion by targeting EZH2 by sponging miR-138 in OSCC.25 In OSCC, miR-101-3p has been proven to be a tumor suppressor and inhibits the growth, migration, and invasion of cancer cells by targeting oncogenic genes, such as CX chemokine receptor 7,26 ZEB1,27 EZH2,28 etc. Our results revealed that RC3H2 physically binds with miR-101-3p by using bioinformatics analysis, a luciferase reporter assay, and an RNA pull-down assay. miR-101-3p was further detected to be significantly downregulated and negatively correlated with the expression level of RC3H2 in OSCCs by qRT-PCR.
Yet increasing evidence indicates that EZH2 has been regarded as a driver oncogene in different human cancer types, including OSCC.29 In our prior studies, EZH2 has been found to promote malignant behaviors via cell cycle dysregulation in non-small cell lung cancer.30 EZH2 has also been identified as a key oncogenic driver promoting the malignant transformation of oral leukoplakia and the progression of HNSCC.5,6 Furthermore, X-linked FHL1 has been verified as a novel target downstream gene for EZH2 and a promising therapeutic target in HNSCC.31 Definitively, a covalently bound inhibitor for EZH2 was screened and identified through chromatin immunoprecipitation (ChIP)-mediated EZH2 degradation.29 In the present study, our findings demonstrated that the suppression of miR-101-3p attenuated the RC3H2 knockdown-induced inhibitory effects on OSCC cells by targeting EZH2. Similarly, overexpression of miR-101-3p attenuated the RC3H2 overexpression-induced malignant behaviors in OSCC cells by targeting EZH2, suggesting that the ceRNA existed among RC3H2, miR-101-3p, and EZH2 in OSCCs.
In summary, our results first demonstrated that RC3H2 functions as a ceRNA to regulate EZH2 expression by sponging miR-101-3p in OSCC. Our findings further revealed a novel RC3H2/miR-101-3p/EZH2 axis in OSCC, and RC3H2 may represent a potential therapeutic target for OSCC.
Materials and Methods
Patients and Specimens
OSCC tissues and their paired adjacent normal tissues were obtained from the Shanghai Sharing Platform for the Tissue and Bioinformatics Database of Oral Maxillofacial Tumors (http://mdl.shsmu.edu.cn/OMNDB/page/home/home_en.jsp), which was established by the Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, and the Shanghai Institute of Stomatology (Shanghai, China). All tissue samples used for the Sharing Platform were collected from the Department of Oral and Maxillofacial-Head and Neck Oncology, Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine. The study was approved by the Ethics Committee of Shanghai Ninth People’s Hospital affiliated with Shanghai Jiao Tong University, School of Medicine.
Affymetrix OE lncRNA Microarray Analysis
The Affymetrix OE lncRNA microarray was performed on an Affymetrix gene chip command console (version 4.0, Affymetrix) software by OEbiotech (Shanghai, P.R. China). In brief, total RNA was quantified by the NanoDrop ND-2000 (Thermo Scientific), and the RNA’s integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies). The sample labeling, microarray hybridization, and washing were performed according to the manufacturer’s instructions. In brief, total RNAs were transcribed to double-strand cDNAs and then synthesized cRNAs. Then, second-cycle cDNAs were synthesized from cRNAs. Followed fragmentation and biotin labeling, the second-cycle cDNAs were hybridized onto the microarray. After washing and staining, the arrays were scanned by the Affymetrix Scanner 3000 (Affymetrix). Differentially expressed genes and lncRNAs were then identified through fold change. The threshold set for up- and downregulated genes was a fold change ≥ 2.0.
Agilent Human miRNA, Release 21.0 Analysis (8*60 K, Design ID: 070156)
Total RNA was quantified by the NanoDrop ND-2000 (Thermo Scientific), and the RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies). The sample labeling, microarray hybridization, and washing were performed based on the manufacturer’s standard protocols. In brief, total RNA was dephosphorylated, denaturated, and then labeled with cyanine-3-CTP. After purification, the labeled RNAs were hybridized onto the microarray. After washing, the arrays were scanned with the Agilent scanner G2505C (Agilent Technologies). Feature extraction software (v10.7.1.1, Agilent Technologies) was used to analyze array images to get raw data. Next, Genespring software (v13.1, Agilent Technologies) was employed to finish the basic analysis with the raw data. To begin with, the raw data were normalized with the quantile algorithm. The probes that at least 100% of samples in any one condition out of two conditions have flags in “Detected” were chosen for further data analysis. Differentially expressed miRNAs were then identified through fold change as well as p value calculated using t test. The threshold set for up- and downregulated genes was a fold change ≥ 2.0 and a p value ≤ 0.05. Target genes of differentially expressed miRNAs were the intersection predicted with databases (Targetscan, microRNA.org, PITA).
Cell Lines and Cell Culture
OSCC cell line HN4 (Research Resource Identifiers: CVCL_5515) cells were kindly provided by Prof. Li Mao from the University of Maryland Dental School (MD, USA), Cal27 (ATCC: CRL-2095) and 293T (ATCC: CRL3216) cells were purchased from the American Type Culture Collection (USA). Cal27, HN4, and 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO-BRL, USA) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL), penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37°C in a humidified 5% CO2 atmosphere.
RNA Extraction and qRT-PCR
Total RNA from tissues and cells was extracted using TRIzol reagent (Takara, Japan) and reverse-transcribed into cDNA by a PrimeScript RT reagent kit (Takara, Japan). Then, miRNA cDNA was synthesized by a miRcute plus miRNA first-strand cDNA synthesis kit (TIANGEN, China). Real-time PCR was performed using SYBR premix Ex Taq reagent kit (Takara, Japan) or a miRcute miRNA qPCR kit (TIANGEN, China) through the 7500 real-time PCR system (Applied Biosystems, USA) according to manufacturer’s instructions. The mRNA levels were normalized against GAPDH levels, and miRNA levels were normalized against U6 levels. The primer sequences used in the present study are listed in Table S3.
Cell Transfection
The siRNAs against RC3H2 (si-RC3H2), si-Scramble, or miR-101-3p mimics (miR-101-3p), miR-101-3p inhibitors (in-miR-101-3p), NC mimics (miR-NC) and NC inhibitor (in-NC) (Ribobio, China) were transfected into cells using Lipofectamine RNAiMAX (Invitrogen, USA) at a final concentration of 100 nM. At 24 h after incubating, the cells were harvested and we performed the corresponding experiments. The siRNAs used in our study were designed and synthesized by Guangzhou RiboBio (Guangzhou, China).
Lentiviral Transduction and Screening of Stable Strains
RC3H2 lentiviral expression vectors (LV-RC3H2) and NC (LV-NC) were constructed by HanYin Biotechnology. The RC3H2 lentiviral expression vector (LV-RC3H2) conferred puromycin resistance. Lentiviral transduction was performed following the manufacturer’s instructions. After 72 h of transfection, the culture medium was mixed with puromycin at a final concentration of 3–10 μg/mL. After being cultured with puromycin and passaged 2–3 times, the stably stained cells were screened.
Western Blot Analysis
Cells were collected at the indicated times in SEMS lysis buffer (Beyotime, China). The protein concentration was determined by a BCA protein assay kit (Beyotime, China). Equal amounts of proteins were separated using 10% polyacrylamide gels and transferred to 0.22-μm polyvinylidene fluoride (PVDF) membranes (Merck Millipore, USA). The membrane was soaked in 10% skim milk in PBS for 1 h at room temperature and incubated with primary antibody overnight at 4°C. Then, the membranes were probed with infrared (IR) dye-labeled secondary antibodies, and signals were detected using an Odyssey Infrared Imaging System (Biosciences, USA). GAPDH was used as a control. Antibodies (1:1,000) against EZH2, H3K27Me3, H3, and GAPDH were purchased from Cell Signaling Technology (Boston, MA, USA). The quantitative analysis of western blot was performed by ImageJ (NIH, Bethesda, MD, USA).
Cell Counting Kit-8 (CCK-8) Analysis
Cells transfected for 24 h with siRNA or stably lentivirus-transduced cells were seeded into 96-well plates at a density of 1,000 cells per well in triplicate. The cells were harvested, and 10 μL of CCK-8 reagent (Dojindo, Kumamoto, Japan) was added to 100 μL of culture medium. The cells were subsequently incubated for 2 h at 37°C, and the optical density was measured at 450 nm using a microplate reader (SpectraMax i3, Molecular Devices, USA).
Colony-Forming Assay
Cells transfected for 24 h with siRNAs or lentivirus-transduced stable cells were seeded into 6-well plates at a density of 1,000 cells per well and incubated for 10–14 days to form cell colonies. The colonies were fixed and stained, and those with more than 50 cells were counted under a dissecting microscope.
Wound-Healing Assay
Cells transfected for 24 h with siRNAs or lentivirus-transduced stable cells were plated in 6-well plates and cultured to confluency. Then, tumor cells were serum-starved and scraped with a P200 tip (0 h), washed with PBS, and cultured with serum-free DMEM. Five nonoverlapping field images were obtained at 24 h.
Transwell Invasion Assay
Cell invasion assays were performed using 24-well Transwell chambers with 8-μm porosity polycarbonate filters and Transwell insert chambers (Corning, USA) coated with Matrigel (BD Biosciences, USA). A total of 150 μL of cell suspension in serum-free medium was added into each upper chamber, while 600 μL of DMEM supplemented with 10% FBS was added to the lower chambers as a chemoattractant. After incubating for 24–36 h, the migrated or invaded cells were fixed with 4% paraformaldehyde (Sangon Biotech) for 15 min and stained with 1% crystal violet (Beyotime) for 30 min. After the cells on the upper surface of the filter were removed, at least five randomly selected microscopic fields of fixed cells per filter were imaged using an inverted phase-contrast microscope. The cells were counted, and the average was calculated.
FISH Assay
Fluorescence-labeled probes (RiboBio) for RC3H2, 18S rRNA, and U6 RNA were designed and synthesized. The FISH experiments were performed using the Ribo fluorescent in situ hybridization kit (RiboBio). Images were obtained with a TCS SP2 laser-scanning confocal microscope (Leica Microsystems, Germany).
Cy3-labeled miR-101-3p probes (QIAGEN, Germany) was also synthesized. The cells were then prepared for immunofluorescence as described above. The FISH experiments were performed using miRCURY LNA miRNA ISH buffer set (QIAGEN, Germany). Images were obtained with a TCS SP2 laser-scanning confocal microscope (Leica Microsystems, Germany).
Luciferase Reporter Assay
pmirGLO-RC3H2-WT or pmirGLO-RC3H2-Mut were cotransfected with hsa-miR-101-3p mimics or miR-NC into 293T cells using Lipofectamine 2000 reagent (Invitrogen, USA) according to the manufacturer’s instructions. Moreover, 293T cells infected with LV-NC and LV-RC3H2 were cotransfected with pmirGLO-EZH2-WT or pmirGLO-EZH2-Mut and hsa-miR-101-3p mimics in a 24-well dish using Lipofectamine 2000 reagent (Invitrogen, USA). Then, cells were cultured for 24 h. Finally, luciferase activity was assessed using a dual-luciferase reporter kit (Promega, USA). The relative firefly luciferase activity was normalized to Renilla luciferase activity.
RNA Pull-Down Assay
A biotinylated RNA pull-down assay was performed using a Target RNA purification kit (ZEHENG Biotech, Shanghai) according to the manufacturer’s instructions. In brief, Cal27 and HN4 cells stably overexpressing RC3H2 were crosslinked with 1% formaldehyde for 10 min, equilibrated in glycine buffer for 5 min, washed in cold PBS 3 times, scraped in 1 mL lysis buffer, and then incubated for 10 min. The cell samples were sonicated and then centrifuged. The supernatant was transferred to a 2-mL tube, and 50 μL was used for input analysis. The supernatant was incubated with RC3H2 biotin probes (RiboBio) and a negative probe at 4°C overnight with rotation, and then 100 μL streptavidin magnetic beads was used for 1 h with rotation. The bead-sample mixture was washed two times, and 10% of the mixture was collected to verify the efficiency of target RNA purification; the remaining 90% was used to purify RNA and test the miRNA enrichment. After subsequent washes and RNA purification, the pull-down complexes were assayed by qRT-PCR.
Immunohistochemical Analysis
In brief, paraffin-embedded 3-μm-thick sections were deparaffinized, rehydrated, and submerged into EDTA buffer for heat-induced antigen retrieval. The sections immersed into 0.3% hydrogen peroxide for blocking endogenous peroxidase activity, blocked with 5% goat serum albumin, incubated with primary antibodies at 4°C overnight, and performed using the DAKO ChemMate envision kit/HRP (Dako-Cytomation, USA) according the manufacturer’s instructions. The sections were then counterstained with hematoxylin, dehydrated, cleared, and mounted. The tissue sections exhibiting brown in cytoplasm, nucleus, or membrane were considered positive.
Xenograft Mouse Experiments and In Vivo Metastasis Assay
The animal experiments, implemented in 4-week-old BALB/c nude mice (Shanghai Laboratory Animal Center), were performed in accordance by the ethical standards and national guidelines. Cal27 cells (1 × 106) stably overexpressing LV-RC3H2 or LV-NC were subcutaneously injected into the left and right dorsal flanks of six mice. Tumor sizes were monitored by a caliper in every 3 days. The tumor volume was measured using the following formula: tumor volume = 0.5 × length × width2. After 4 weeks, the mice were killed for collecting tumor samples and measuring tumor weights.
The pulmonary metastasis model was described in our previous study.32 In brief, Cal27 cells (1 × 106) stably overexpressing LV-RC3H2 or LV-NC were injected into tail vein of six mice. After 7 weeks, the mice were killed for harvesting lung samples. The metastatic nodules formed on the lung surfaces were detected by picric acid and neutral aldehyde staining.
Statistical Analysis
Paired t tests were used to assess the expression differences between OSCC and matched normal tissue. The Kruskal-Wallis tests and Mann-Whitney U tests were used to analyze the association among EZH2, miR-101-3p, RC3H2 levels, and clinical parameters. The significance of the differences between groups was analyzed using one-way ANOVA or Student’s t test. All statistical analyses were performed using SPSS 17.0 (SPSS, Chicago, IL, USA). All values were two sided, and p < 0.05 was considered statistically significant.
Author Contributions
W.C., W.T.C, and Z.H.R conceived and designed the study. K.W., Y.Y.J, B.L.Z, Y.L and F.X. conducted the experiments. J.J.Z, X.W., M.Y., Q.X., W.K.Z analyzed the data. W.C. and K.W. wrote the manuscript. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Natural Science Foundation of China (81972589, 81672745, and 81874126), the Shanghai Municipal Science and Technology Commission Funded Project Grants (no. 18DZ2291500), and the Shanghai Anticancer Association EYAS PROJECT (SACA-CY1B06). We would like to thank Dr. Qidong Zu (OE Biotech, Inc., Shanghai, China; https://www.oebiotech.com/) for assistance with the bioinformatics analysis of Affymetrix OE lncRNA microarray and Agilent Human miRNA microarray.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.02.006.
Contributor Information
Zhenhu Ren, Email: ren.zhenhu@outlook.com.
Wantao Chen, Email: chenwantao196323@sjtu.edu.cn.
Wei Cao, Email: caowei561521@shsmu.edu.cn.
Supplemental Information
References
- 1.Bhatia N., Lalla Y., Vu A.N., Farah C.S. Advances in optical adjunctive AIDS for visualisation and detection of oral malignant and potentially malignant lesions. Int. J. Dent. 2013;2013:194029. doi: 10.1155/2013/194029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salazar C., Nagadia R., Pandit P., Cooper-White J., Banerjee N., Dimitrova N., Coman W.B., Punyadeera C. A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cell Oncol. (Dordr.) 2014;37:331–338. doi: 10.1007/s13402-014-0188-2. [DOI] [PubMed] [Google Scholar]
- 3.Simon J.A. Transcription. Sweet silencing. Science. 2009;325:45–46. doi: 10.1126/science.1177264. [DOI] [PubMed] [Google Scholar]
- 4.Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 5.Cao W., Younis R.H., Li J., Chen H., Xia R., Mao L., Chen W., Ren H. EZH2 promotes malignant phenotypes and is a predictor of oral cancer development in patients with oral leukoplakia. Cancer Prev. Res. (Phila.) 2011;4:1816–1824. doi: 10.1158/1940-6207.CAPR-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao W., Feng Z., Cui Z., Zhang C., Sun Z., Mao L., Chen W. Up-regulation of enhancer of zeste homolog 2 is associated positively with cyclin D1 overexpression and poor clinical outcome in head and neck squamous cell carcinoma. Cancer. 2012;118:2858–2871. doi: 10.1002/cncr.26575. [DOI] [PubMed] [Google Scholar]
- 7.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao W., Liu J.N., Liu Z., Wang X., Han Z.G., Ji T., Chen W.T., Zou X. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. doi: 10.1016/j.oraloncology.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 9.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebert M.S., Sharp P.A. Emerging roles for natural microRNA sponges. Curr. Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Röther S., Meister G. Small RNAs derived from longer non-coding RNAs. Biochimie. 2011;93:1905–1915. doi: 10.1016/j.biochi.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 13.Liu J., Xing Y., Xu L., Chen W., Cao W., Zhang C. Decreased expression of pseudogene PTENP1 promotes malignant behaviours and is associated with the poor survival of patients with HNSCC. Sci. Rep. 2017;7:41179. doi: 10.1038/srep41179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F., Chen Q., Chen F., Wang J., Gong R., He B. The lncRNA ENST00000608794 acts as a competing endogenous RNA to regulate PDK4 expression by sponging miR-15b-5p in dexamethasone induced steatosis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864:1449–1457. doi: 10.1016/j.bbalip.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Chinnappan M., Gunewardena S., Chalise P., Dhillon N.K. Analysis of lncRNA-miRNA-mRNA Interactions in Hyper-proliferative Human Pulmonary Arterial Smooth Muscle Cells. Sci. Rep. 2019;9:10533. doi: 10.1038/s41598-019-46981-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo H., Liu J., Ben Q., Qu Y., Li M., Wang Y., Chen W., Zhang J. The aspirin-induced long non-coding RNA OLA1P2 blocks phosphorylated STAT3 homodimer formation. Genome Biol. 2016;17:24. doi: 10.1186/s13059-016-0892-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao S., Lin L., Xia X., Wu H. lncRNA SPRY4-IT1 Regulates Cell Proliferation and Migration by Sponging miR-101-3p and Regulating AMPK Expression in Gastric Cancer. Mol. Ther. Nucleic Acids. 2019;17:455–464. doi: 10.1016/j.omtn.2019.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Huang D., Wei Y., Zhu J., Wang F. Long non-coding RNA SNHG1 functions as a competitive endogenous RNA to regulate PDCD4 expression by sponging miR-195-5p in hepatocellular carcinoma. Gene. 2019;714:143994. doi: 10.1016/j.gene.2019.143994. [DOI] [PubMed] [Google Scholar]
- 19.Fang Z., Zhao J., Xie W., Sun Q., Wang H., Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by sunppressing miR-184 expression. Cancer Med. 2017;6:2897–2908. doi: 10.1002/cam4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing H.B., Qiu H.M., Li Y., Dong P.F., Zhu X.M. Long noncoding RNA CASC2 alleviates the growth, migration and invasion of oral squamous cell carcinoma via downregulating CDK1. Eur. Rev. Med. Pharmacol. Sci. 2019;23:4777–4783. doi: 10.26355/eurrev_201906_18060. [DOI] [PubMed] [Google Scholar]
- 21.Ye J., Jiao Y. LncRNA FAL1 promotes the development of oral squamous cell carcinoma through regulating the microRNA-761/CRKL pathway. Eur. Rev. Med. Pharmacol. Sci. 2019;23:5779–5786. doi: 10.26355/eurrev_201907_18316. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L.M., Ju H.Y., Wu Y.T., Guo W., Mao L., Ma H.L., Xia W.Y., Hu J.Z., Ren G.X. Long non-coding RNA ANRIL promotes tumorgenesis through regulation of FGFR1 expression by sponging miR-125a-3p in head and neck squamous cell carcinoma. Am. J. Cancer Res. 2018;8:2296–2310. [PMC free article] [PubMed] [Google Scholar]
- 23.Chang S.M., Hu W.W. Long non-coding RNA MALAT1 promotes oral squamous cell carcinoma development via microRNA-125b/STAT3 axis. J. Cell. Physiol. 2018;233:3384–3396. doi: 10.1002/jcp.26185. [DOI] [PubMed] [Google Scholar]
- 24.Huang G., He X., Wei X.L. lncRNA NEAT1 promotes cell proliferation and invasion by regulating miR-365/RGS20 in oral squamous cell carcinoma. Oncol. Rep. 2018;39:1948–1956. doi: 10.3892/or.2018.6283. [DOI] [PubMed] [Google Scholar]
- 25.Hong Y., He H., Sui W., Zhang J., Zhang S., Yang D. Long non-coding RNA H1 promotes cell proliferation and invasion by acting as a ceRNA of miR-138 and releasing EZH2 in oral squamous cell carcinoma. Int. J. Oncol. 2018;52:901–912. doi: 10.3892/ijo.2018.4247. [DOI] [PubMed] [Google Scholar]
- 26.Hui Y., Li Y., Jing Y., Feng J.Q., Ding Y. miRNA-101 acts as a tumor suppressor in oral squamous cell carcinoma by targeting CX chemokine receptor 7. Am. J. Transl. Res. 2016;8:4902–4911. [PMC free article] [PubMed] [Google Scholar]
- 27.Wu B., Lei D., Wang L., Yang X., Jia S., Yang Z., Shan C., Yang X., Zhang C., Lu B. MiRNA-101 inhibits oral squamous-cell carcinoma growth and metastasis by targeting zinc finger E-box binding homeobox 1. Am. J. Cancer Res. 2016;6:1396–1407. [PMC free article] [PubMed] [Google Scholar]
- 28.Li B., Xie D., Zhang H. MicroRNA-101-3p advances cisplatin sensitivity in bladder urothelial carcinoma through targeted silencing EZH2. J. Cancer. 2019;10:2628–2634. doi: 10.7150/jca.33117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Cao W., Zhang J., Yan M., Xu Q., Wu X., Wan L., Zhang Z., Zhang C., Qin X. A covalently bound inhibitor triggers EZH2 degradation through CHIP-mediated ubiquitination. EMBO J. 2017;36:1243–1260. doi: 10.15252/embj.201694058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao W., Ribeiro Rde.O., Liu D., Saintigny P., Xia R., Xue Y., Lin R., Mao L., Ren H. EZH2 promotes malignant behaviors via cell cycle dysregulation and its mRNA level associates with prognosis of patient with non-small cell lung cancer. PLoS ONE. 2012;7:e52984. doi: 10.1371/journal.pone.0052984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao W., Liu J., Xia R., Lin L., Wang X., Xiao M., Zhang C., Li J., Ji T., Chen W. X-linked FHL1 as a novel therapeutic target for head and neck squamous cell carcinoma. Oncotarget. 2016;7:14537–14550. doi: 10.18632/oncotarget.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q., Wu K., Qin X., Yu Y., Wang X., Wei K. LASP1 promotes proliferation, metastasis, invasion in head and neck squamous cell carcinoma and through direct interaction with HSPA1A. J. Cell. Mol. Med. 2020;24:1626–1639. doi: 10.1111/jcmm.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.