Abstract
Tartrate-resistant acid phosphatase 5 (TRAP/ACP5) has been shown to involve the development and prognosis of multiple tumors in previous studies; however, the mechanism in lung cancer is still unclear, and thus this study investigated the role of ACP5 in the progression of lung adenocarcinoma. After a series of in vitro and in vivo experiments, we observed that ACP5 expression was increased in lung adenocarcinomas (40/69, 57.97%); importantly, an increased ACP5 level was associated with patient age (p = 0.044) and lymph node metastasis (p = 0.0385). ACP5 overexpression significantly enhanced A549 and NCI-H1975 cell proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) and reduced cell apoptosis. Knocking down the expression of ACP5 could rescue the above cell phenotypes. Furthermore, enhancing ACP5 expression promoted lung adenocarcinoma cell hyperplasia and intrapulmonary metastasis in a mouse model. Additionally, mechanistic studies revealed that ACP5 might regulate p53 phosphorylation at Ser392, thereby enhancing the ubiquitination of p53, which then underwent degradation. Reducing the levels of p53 intensified the transcription of SMAD3, which promotes EMT in lung adenocarcinoma cells. In summary, the present study provides a theoretical basis and important scientific evidence on the key role of ACP5 in lung adenocarcinoma progression by inducing EMT via the regulation of p53/SMAD3 signaling.
Keywords: ACP5, lung adenocarcinoma, epithelial-mesenchymal transition, p53, SMAD3, metastasis
Introduction
Lung adenocarcinoma (LUAD) is a common and severe tumor that is a type of non-small-cell lung cancer (NSCLC). Although great advances have been made in the diagnosis and treatment of NSCLC, the average 5-year survival rate for LUAD is approximately 19%1, 2, 3 and has not improved in recent decades.4,5 Therefore, the molecular mechanisms underlying NSCLC progression and associated therapeutic targets have become areas of keen interest.6,7
It is well known that epithelial-mesenchymal transition (EMT) is a major driver of cancer progression. The process of EMT involves adhesion junctions and the loss of substrate polarity, the acquisition of mesenchymal characteristics, such as spindle-shaped cell morphology and reorganization of actin stress fibers, and an increase in movement, invasion, and resistance to apoptosis. Many molecules are involved in the processes of tumorigenesis, malignancy, and EMT progression, such as transforming growth factor β (TGF-β) and the tumor suppressor p53.8 TGF-β acts as a tumor promoter via the induction of EMT, which correlates with increased invasiveness, metastasis, and chemoresistance in tumor cells. When activated, TGF-β receptors phosphorylate SMAD2/3 and transfer the activated SMAD complex into nucleus to regulate target gene transcription.9,10 The tumor suppressor p53 plays an essential role in the cell cycle, DNA repair, and apoptosis and is the most commonly mutated gene in lung cancer; p53 is frequently inactivated by mutations or deletions during the process of EMT, although the underlying mechanism is not clear.11,12
Tartrate-resistant acid phosphatase 5 (TRAP/ACP5), a metalloprotein enzyme that belongs to the acid phosphatase family, is synthesized as a monomeric proenzyme (ACP5a, 35 kDa)13, 14, 15 and a disulfide-linked heterodimer (ACP5b) containing an N-terminal fragment of 20–23 kDa joined to a 16- to 17-kDa C-terminal part that originates from the posttranslational cleavage of the monomeric form and exhibits significantly increased phosphatase activity.15, 16, 17 In a previous study, lung cancer patients with high ACP5 expression had significantly worse overall survival than did patients with low ACP5 expression.18 In addition, ACP5 has been suggested to be downstream of forkhead box M1 (Foxm1), which is required for lung fibrosis and EMT in tumor metastasis.19, 20, 21 Additionally, ACP5 expression is significantly upregulated in cardiovascular disease, breast cancer, hepatocellular carcinoma, ovarian cancer, and melanoma, and it is a useful serum marker for extensive bone metastasis and is indicative of a poor prognosis in human melanoma patients.22, 23, 24, 25 These observations suggest that ACP5 may have an altered expression pattern in tumor progression and be related to EMT. However, the exact role of ACP5 in tumor pathoetiology, especially in LUAD pathoetiology, has not been fully addressed.
Herein, we report that altered expression of ACP5 regulated LUAD cell proliferation, apoptosis, and metastasis in vitro and in vivo. Mechanistic studies revealed that ACP5 regulated p53 phosphorylation, thereby enhancing the ubiquitination of p53, which then underwent degradation. Reducing the levels of p53 intensified the transcription of SMAD3 to promote LUAD cell EMT. Taken together, our data provide novel insights into the role of ACP5 in the pathogenesis of LUAD.
Results
ACP5 Expression Was Upregulated in LUAD and Increased ACP5 Expression Predicted Metastasis
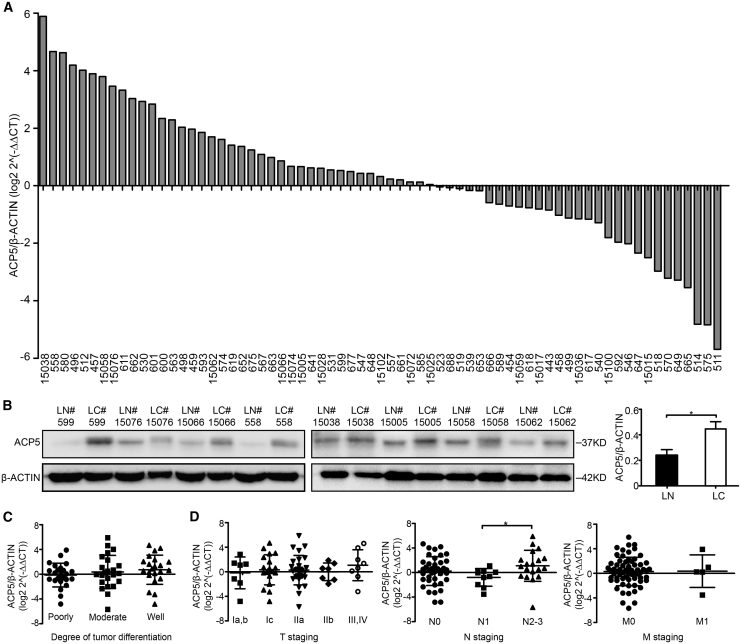
We first examined the expression of ACP5 in lung samples obtained from LUAD patients. ACP5 was expressed at low levels in the adjacent normal tissue samples, whereas higher levels of ACP5 were detected in the LUAD tissue samples (Figure 1A). Furthermore, the increased ACP5 protein expression in the LUAD tissue samples was also confirmed by western blot (Figure 1B).
Figure 1.
Expression of ACP5 in LUAD and Its Prognostic Significance in LUAD Patients
(A) The level of mRNA expression of ACP5 was compared between 69 pairs of LUAD tissue samples (LC) and adjacent non-tumor tissue samples (LN, >5 cm from the tumor edge). The ratio >0 signifies higher ACP5 mRNA expression in cancer tissue compared to non-tumor tissue and vice versa, and the numbers in the x axis are the patient numbers. (B) The protein expression of ACP5 was detected by western blot in eight paired LUAD samples and their adjacent non-tumor tissue samples. (C and D) The expression of ACP5 was correlated with the degree of tumor differentiation (C) and TNM stage (D). Each dot represents one patient sample. The results are expressed as the mean ± SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by independent Student’s t test.
To explore the function of ACP5 in LUAD, we evaluated the associations of ACP5 with the clinicopathological features of 69 LUAD patients. The results showed that ACP5 overexpression correlated with lymph node metastasis (N staging from N1 to N2, p = 0.0385, Figure 1D) and age (p = 0.044, Table 1), but not with tumor differentiation and the N staging from N0 to N1 (Figures 1C and 1D).
Table 1.
Correlation between ACP5 Expression and Clinicopathological Features in LUAD
| ACP5 |
||||
|---|---|---|---|---|
| All Cases | Low Expression | High Expression | p Valuea | |
| Sex | 0.334 | |||
| Male | 31 | 15 (21.73%) | 16 (23.19%) | |
| Female | 38 | 14 (20.29%) | 24 (34.78%) | |
| Ageb | 0.044 | |||
| ≤58 | 33 | 18 (26.09%) | 15 (21.74%) | |
| >58 | 36 | 11 (15.94%) | 25 (36.23%) | |
| Smoking | 0.863 | |||
| Never | 46 | 19 (27.54%) | 27 (39.13%) | |
| Current or past smoker | 23 | 10 (14.49%) | 13 (18.84%) | |
| Differentiation | 0.233 | |||
| Well | 22 | 6 (8.70%) | 16 (23.19%) | |
| Moderately | 23 | 11 (15.94%) | 12 (17.39%) | |
| Poorly | 24 | 12 (17.39%) | 12 (17.39%) | |
| Stage | ||||
| T staging | 0.886 | |||
| IA, IB | 7 | 4 (5.80%) | 3 (4.35%) | |
| IC | 19 | 7 (10.14%) | 12 (17.39%) | |
| IIA | 28 | 11 (15.94%) | 17 (24.63%) | |
| IIB | 7 | 3 (4.35%) | 4 (5.80%) | |
| III, IV | 8 | 4 (5.80%) | 4 (5.80%) | |
| N staging | 0.139 | |||
| 0 | 39 | 17 (24.84%) | 22 (31.88%) | |
| 1 | 11 | 7 (10.14%) | 4 (5.80%) | |
| 2–3 | 19 | 5 (7.25%) | 14 (20.29%) | |
| M staging | 0.389 | |||
| 0 | 64 | 28 (40.58%) | 36 (52.17%) | |
| 1 | 5 | 1 (1.45%) | 4 (5.80%) | |
LUAD, lung adenocarcinoma; T, tumor; N, node; M, metastasis.
The Pearson χ2 test or Fisher’s exact test was used for statistical analysis.
The mean age at diagnosis is 58.8 years in LUAD patients. Samples are divided into two groups based on the mean age.
ACP5 Regulated LUAD Cell Proliferation, Migration, and Invasion
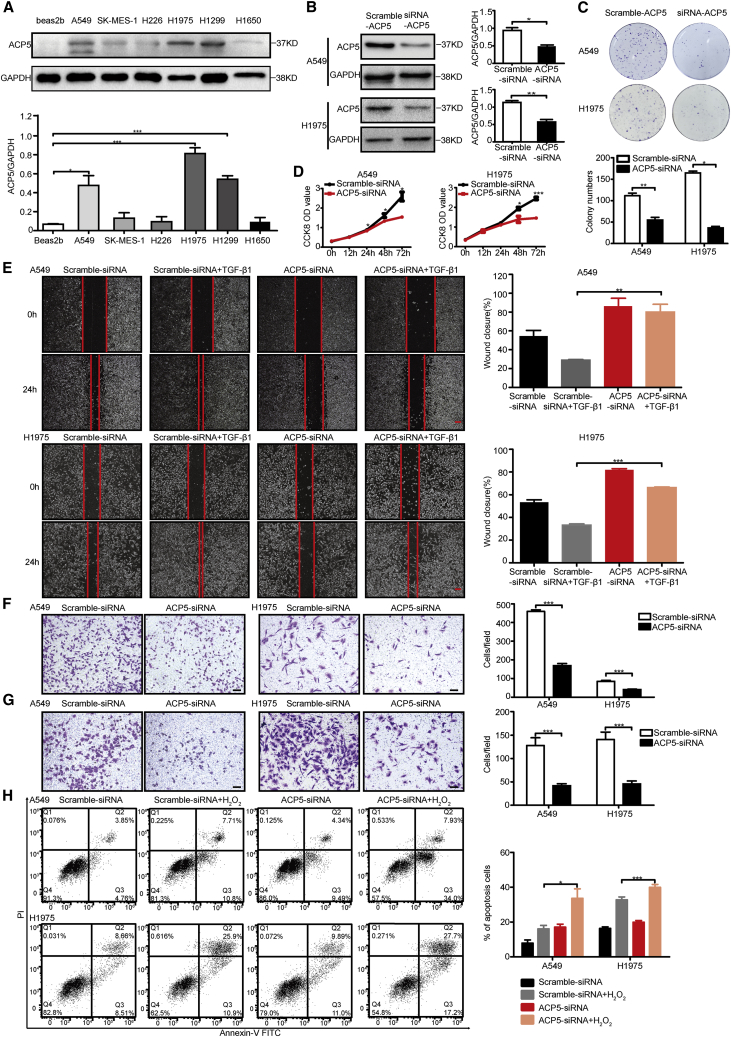
To evaluate the biological function of ACP5 in LUAD cells, ACP5 expression levels were detected in six LUAD cell lines and the Beas-2b cell line by western blot (Figure 2A). The results showed that the expression of ACP5 was higher in NCI-H1975, H1299, and A549 cells than in Beas-2b cells. As our previous data revealed an association between ACP5 and LUAD metastasis, we investigated the impact of ACP5 on LUAD cell motility, invasiveness, and apoptosis by knocking down ACP5 expression in A549 and NCI-H1975 cells by RNA interference. As expected, the small interfering RNA (siRNA) treatment efficiently knocked down ACP5 expression in A549 and NCI-H1975 cells (Figure 2B), which led to decreases in cell proliferation (Figures 2C and 2D) and wound-healing ability in the treated cells compared to control cells (Figure 2E). Migration and Matrigel invasion assays also demonstrated that the ablation of ACP5 markedly reduced the migration and invasion of both A549 and NCI-H1975 cells (Figures 2F and 2G). In addition, flow cytometry showed that inhibiting ACP5 could promote apoptosis induced by H2O2 in those cells (Figure 2H). Collectively, these results provided evidence that ACP5 knockdown could inhibit LUAD cell tumorigenicity in vitro.
Figure 2.
Silencing of ACP5 Inhibited Cell Proliferation, Migration, and Invasion and Promoted Apoptosis In Vitro
(A) ACP5 protein expression levels in different lung cancer cell lines were detected by western blot. (B) Efficient knockdown of ACP5 expression by siRNA in A549 and H1975 cells, as detected by western blot. (C) Silencing of ACP5 inhibits the colony formation of A549 and H1975 cells. (D) Cell Counting Kit-8 (CCK-8) assays were performed in A549 and H1975 cells silenced for ACP5. (E) Wound-healing assay investigated the migratory ability of ACP5 knockdown and control A549 cells (upper) or of H1975 cells (lower). Original magnification, ×40; scale bars, 500 μm. (F) Transwell assays to investigate the migratory ability of ACP5-silenced and control A549 cells (right) or of H1975 cells (left). Original magnification, ×100; scale bars, 100 μm. (G) Transwell assays to investigate the invasion ability of ACP5-silenced and control A549 cells (right) or of H1975 cells (left). Original magnification, ×100; scale bars, 100 μm. (H) Cell apoptosis of A549 and H1975 cells detected by a flow cytometry assay. The results are expressed as the mean ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by independent Student’s t test.
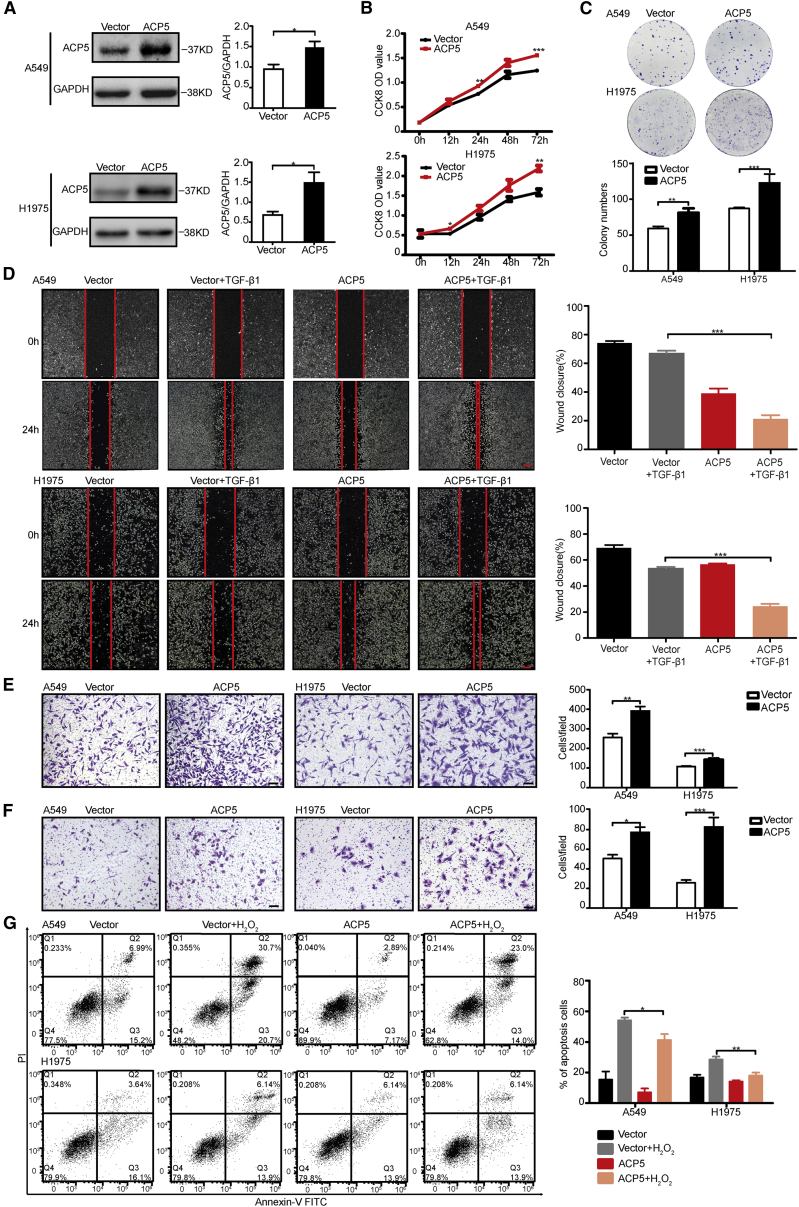
To further confirm whether ACP5 overexpression can promote cell growth, invasion, and apoptosis, a plasmid encoding ACP5 was transfected into A549 and NCI-H1975 cells, and then the expression of exogenous ACP5 was demonstrated by western blot (Figure 3A). Compared with control cells, these ACP5-transfected cells showed significantly increased cell proliferation, migration, and invasion (Figures 3B–3F). Flow cytometry also showed that the overexpression of ACP5 could inhibit apoptosis in A549 and NCI-H1975 cells (Figure 3G).
Figure 3.
ACP5 Promoted Cell Proliferation, Migration, and Invasion and Reduced Apoptosis In Vitro
(A) Western blot analysis showing the expression of exogenous ACP5 in ACP5-transfected A549 and H1975 cells. (B) Cell Counting Kit-8 (CCK-8) assays were performed in A549 and H1975 cells overexpressing ACP5. (C) Overexpression of ACP5 promotes the colony formation of A549 and H1975 cells. (D) Wound-healing assay investigated the migratory ability of ACP5 overexpression and control A549 (upper) or of H1975 cells (lower). Original magnification, ×40; scale bars, 20 μm. (E) Transwell assays to investigate the migratory ability of ACP5-overexpressed and control A549 cells (right) or of H1975 cells (left). Original magnification, ×100; scale bars, 100 μm. (F) Transwell assays to investigate the invasion ability of ACP5-overexpressed and control A549 cells (right) or of H1975 cells (left). Original magnification, ×100; scale bars, 100 μm. (G) Cell apoptosis of A549 and H1975 cells after overexpression of ACP5 assessed by a flow cytometry assay. The results are summarized as the mean ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by independent Student’s t test).
Taken together, these results indicate that ACP5 regulates LUAD cell proliferation, migration, invasion, and apoptosis.
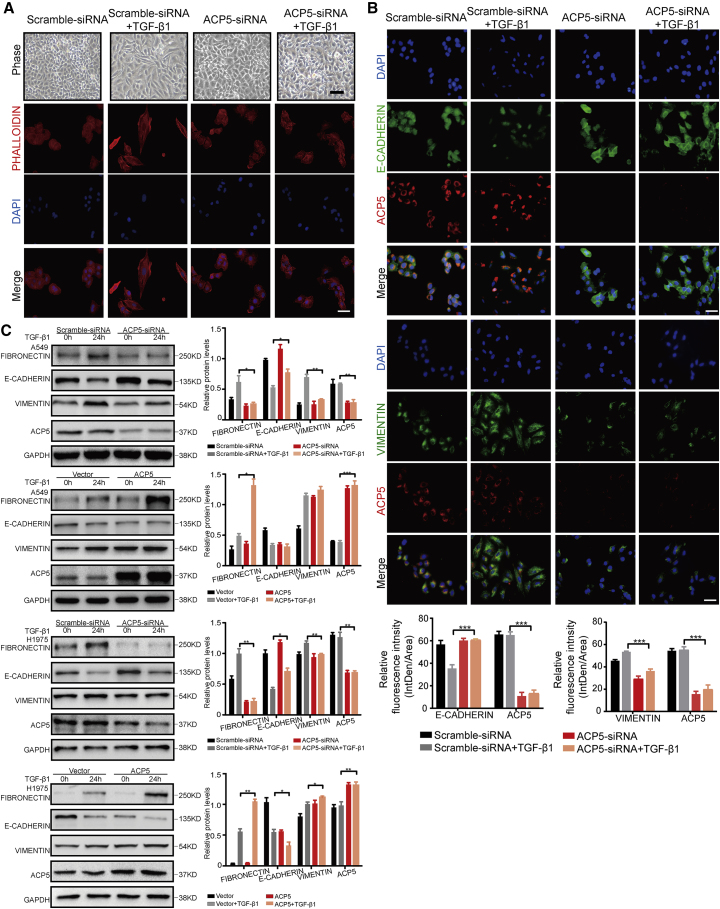
ACP5 Induced EMT in LUAD Cells
EMT is an important mechanism driving tumor invasion and metastasis. Filamentous actin was assembled into thick parallel bundles throughout TGF-β1-treated A549 cells. In contrast, in untreated A549 cells, filamentous actin filaments were tightly associated with cell-cell contact, showing a pericellular plasma membrane distribution that was visualized by rhodamine-phalloidin staining. The majority of the TGF-β1-treated A549 cells underwent EMT-like elongation to become spindle-shaped cells, and knocking down the expression of ACP5 may attenuate the EMT induced by TGF-β1, as manifested by less spindle-shaped cells (Figure 4A). To further investigate whether the ablation of ACP5 impacted the progression of EMT, we next conducted co-immunostaining of A549 with ACP5 and EMT markers (E-cadherin, vimentin) or transcription factors (ZEB2 and SNAIL1). As expected, the expression of E-cadherin was decreased following TGF-β1 induction in scramble siRNA groups (Figure 4B). Interestingly, knocking down the expression of ACP5 could recover the expression of E-cadherin to normal control levels. Furthermore, the immunostaining of vimentin, ZEB2, and SNAIL1 all revealed that ACP5 knockdown likely attenuated the expression of EMT markers and transcription factors (Figure 4B; Figure S1A), indicating that reducing the expression of ACP5 may blunt the process of EMT induced by TGF-β1. Moreover, we next conducted western blot in the cell homogenates for EMT markers (fibronectin, vimentin, and E-cadherin) and transcription factors (ZEB2 and SNAIL1). Consistent with above data, silencing ACP5 expression significantly reducing the expression of fibronectin, vimentin, ZEB2 and SNAIL1, and increasing the expression of E-cadherin compared with scramble siRNA after TGF-β1 treatment in A549 and H1975 cells (Figure 4C; Figure S1B). In addition, overexpressing ACP5 by plasmid abolished the protection conferred by ACP5 silencing as evidenced by higher expression of fibronectin, vimentin, ZEB2, and SNAIL1 and lower E-cadherin levels in H1975 cells and partially in A549 cells (Figure 4C; Figure S2B). Taken together, these results suggest that ACP5 is associated with EMT, which likely contributes to tumor metastasis in LUAD.
Figure 4.
ACP5 Regulated EMT in LUAD Cells
(A) The morphology of A549 cells treated with TGF-β1 (10 ng/mL) for different time intervals was visualized by phase-contrast microscopy. Original magnification, ×200; scale bars, 100 μm. An immunofluorescence staining assay showed the results of rhodamine-phalloidin staining. Actin microfilaments were stained by rhodamine-conjugated phalloidin. (B) Immunofluorescence staining assays were used to evaluate the expressions of E-cadherin and vimentin upon ACP5 knockdown in A549 cells. Cell nuclei were visualized by DAPI. Original magnification, ×400; scale bars, 50 μm. (C) ACP5 in A549 and H1975 cells were knocked down or overexpressed treated with or without 10 ng/mL TGF-β1 for 24 h. The levels of fibronectin, E-cadherin, and vimentin were determined by western blot. The results are summarized as the mean ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by independent Student’s t test.
ACP5 Regulated the Expression of p53
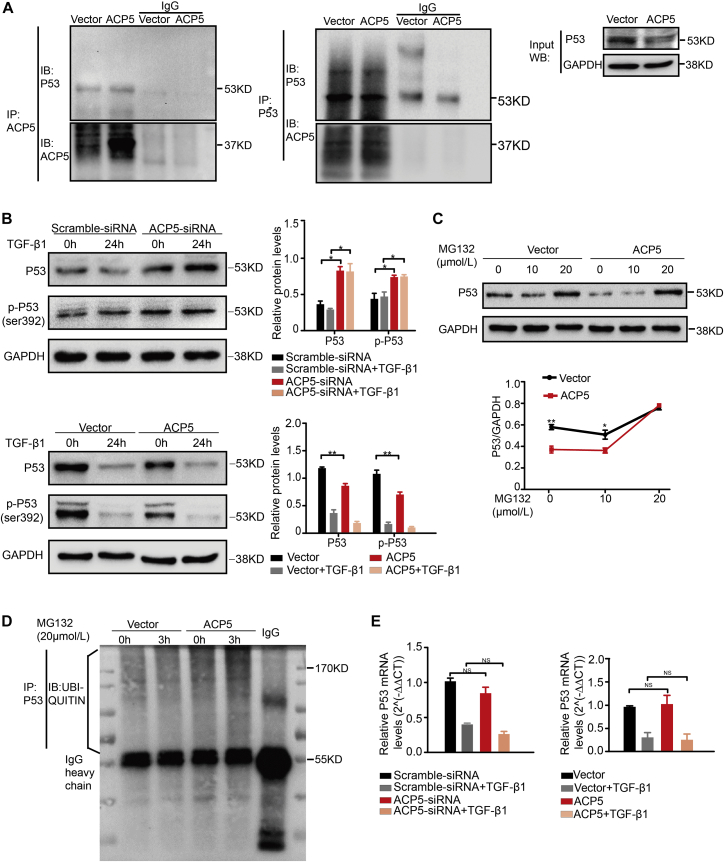
Given the notion that p53 played an important role in the progression of human cancers, especially in LUAD pathogenesis, we examined p53 expression in A549. Then, we observed that the expression of p53 was significantly attenuated with the stimulation of TGF-β1 for 24 h. Interestingly, the levels of p53 were markedly increased in ACP5-silenced A549 cells following TGF-β1 treatment, whereas in ACP5-transfected A549 cells, the expression of p53 was blunted (Figure 5B). Additionally, the confocal assay showed that ACP5 and p53 were both located in the cytoplasm, especially when overexpressing ACP5 (Figure S2), indicating that an association between ACP5 and p53 may have existed during the process of EMT. Thus, we next conducted co-immunoprecipitation (coIP) analyses to confirm this hypothesis. Indeed, ACP5 and p53 were validated to be reciprocally immunoprecipitated in A549 (Figure 5A).
Figure 5.
ACP5 Regulated the Protein Level of p53
(A) coIP (co-immunoprecipitation) and IB (immunoblot) showed that ACP5 interacts with p53. (B) Western blot showed the expression of p53 and p-p53 (Ser392) in A549 cells transfected with ACP5 or the empty vector. (C) Western blot showed the expression of p53 after overexpressing ACP5 with or without MG132 (20 μmol/L). (D) p53 ubiquitination was observed in ACP5-transfected A549 cells by immunoprecipitation assay. (E) The mRNA level of p53 in A549 cells was evaluated by quantitative real-time PCR. The results are summarized as the mean ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by independent Student’s t test. NS, no significant difference between two groups, as analyzed by Student’s t test.
As a kind of a phosphatase, ACP5 can dephosphorylate many kinds of sites on proteins. There is feeble evidence that ACP5 could dephosphorylate the Ser392 of p53,26 and phosphorylation of this site may be related to the stability of p53.27,28 The above results prompted us to examine the impact of ACP5 on the phosphorylation of Ser392 on p53 following TGF-β1 stimulation in A549 cells. Remarkably, the phosphorylation of p53 at the site of Ser392 was enhanced after silencing ACP5 expression. Meanwhile, overexpression of ACP5 could blunt the phosphorylation of p53 at the site of Ser392 (Figure 5B). Given that phosphorylation of Ser392 on p53 may impact the stability of p53, we next adopted western blot to observe the levels of p53 in A549 cells. In line with the co-immunostaining data (Figure S2), the levels of p53 were increased in ACP5-silenced cells. As expected, the opposite expression pattern for p53 was found in ACP5-transfected cells (Figure 5B).
To investigate whether decreased levels of p53 observed in ACP5-transfected cells were dependent on the stability of p53, we adopted MG132, a kind of proteasome inhibitor, to observe the degradation of p53 induced by ACP5. Indeed, a western blot assay showed that MG132 could reverse the downregulation of p53 expression in ACP5-transfected cells (Figure 5C). To further dissect the mechanisms by which ACP5 regulated the degradation of p53, we detected the levels of ubiquitination for p53 by an immunoprecipitation assay. Interestingly, the ubiquitination of p53 levels was increased after MG132 stimulation for 3 h. Importantly, the levels of ubiquitination for p53 were significantly enhanced in ACP5-transfected A549 cells (Figure 5D). Furthermore, quantitative real-time PCR was carried out to further confirm the effect of p53 induced by ACP5 due to the stability rather than transcription. Indeed, we failed to detect perceptible differences for the expression of p53 mRNA in ACP5-silenced cells or ACP5-overexpressed cells (Figure 5E).
Taken together, these data provide evidence that ACP5 dephosphorylates p53 at the site of Ser392, which accelerates the ubiquitination and degradation of p53 in LUAD cells.
ACP5 Regulated the Expression of SMAD3 by p53
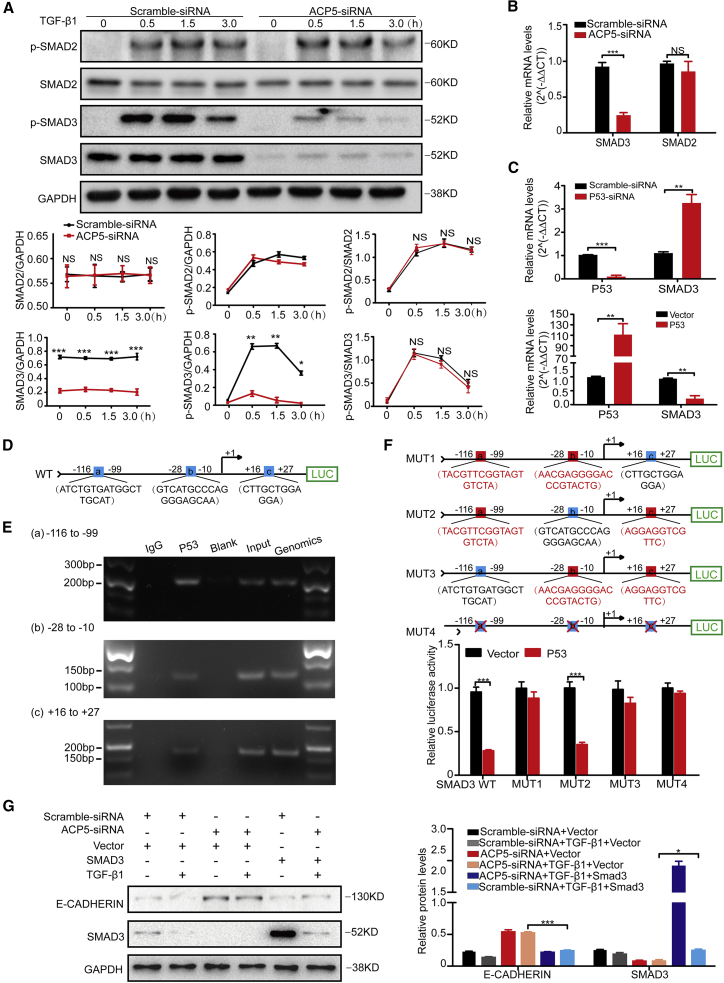
The TGF-β/SMAD pathway is critical for the progression of lung cancer and the associated cellular changes, especially in the process of EMT. Based on this information, we suspected that ACP5 may affect EMT by regulating this signaling pathway. Interestingly, we observed that knocking down ACP5 expression obviously decreased the expression of phosphorylated (p-)SMAD3 but did not affect the expression of p-SMAD2 or other signaling pathway molecules related to TGF-β1, such as ERK, p38, AKT, and β-catenin (Figure S3). Surprisingly, we found that the expression of total SMAD3 was decreased by silencing ACP5 expression (Figure 6A). Quantitative real-time PCR was further carried out and revealed a significantly lower expression of SMAD3 in ACP5-silenced A549 cells (Figure 6B).
Figure 6.
ACP5 Regulated the Expression of SMAD3 by p53
(A) Knockdown of the expression of ACP5 attenuated the levels of SMAD3 and p-SMAD3. Upper panel: Representative western blot results for SMAD3, p-SMAD3, SMAD2, and p-SMAD2 at different time points of TGF-β1 stimulation. Lower panel: Graphs showing the data with three replications. (B) Quantitative real-time PCR results for SMAD3 and SMAD2 in ACP5-silenced A549 cells after TGF-β1 stimulation. (C) p53 inhibited expression of SMAD3. Upper panel: mRNA levels of SMAD3 in A549 cells after knockdown of the expression of p53. Lower panel: mRNA levels of SMAD3 in A549 cells after overexpression of p53. (D) Schematic illustrations of the three binding sites (the blue boxes) of p53 (site a, −116 to −99 bp; site b, −28 to −10 bp; site c, +16 to +27 bp, with the SMAD3 transcription start site as +1) to the SMAD3 promoter as predicted by the Lasagna databases (https://biogrid-lasagna.engr.uconn.edu/lasagna_search/). (E) ChIP-PCR assays using antibody specific for p53 to prove that p53 binds to SMAD3 promoter. (F) Results for SMAD3 promoter luciferase reporter assays. Upper panel: Schematic results showing the mutant plasmids (MUT1–MUT3) of these three binding sites. Each of the mutant plasmids (MUT1–MUT3) of these three binding sites maintained one normal binding site (blue box) and two mutant binding sites (red boxes), and all binding sites were deleted in MUT4 (blue boxes with red cross). Lower panel: Result for promoter reporter assays. (G) After silencing ACP5 for 24 h, we transfected SMAD3 plasmid into the A549 cells for 24 h and then stimulated cells with TGF-β (10 ng/mL) for 24 h and collected the cells for western blot, and overexpression of SMAD3 reversed the process of EMT after silencing ACP5 in A549 cells. The results are summarized as the mean ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by independent Student’s t test.
ACP5 was a phosphatase mainly distributed in the cytoplasm,13,14 and there was less possibility for ACP5 transferring into the nucleus and directly regulating the transcriptional expression of SMAD3. Based on the above data indicating that p53 could act as a transcription factor and that ACP5 blunted p53 protein levels, we hypothesized that p53 may transcribe the expression of SMAD3. Indeed, it was noted that p53 strongly regulated the expression of SMAD3 by quantitative real-time PCR analysis (Figure 6C). Meanwhile, in silico analysis of the SMAD3 promoter identified three potential p53 binding sites in the SMAD3 gene (a, b, and c; Figure 6D). Chromatin immunoprecipitation (ChIP) was next conducted, and the resulting products were used to amplify the SMAD3 promoter regions flanking the above-indicated three potential p53 binding sites. Indeed, p53 manifested binding activity to the SMAD3 promoter in three regions (−99 to −116 bp, −10 to −28 bp, and +16 to +27 bp, with transcriptional start site as +1, Figure 6E). SMAD3 promoter reporter assays were next employed to confirm these findings. Four mutated SMAD3 promoter reporters (MUT1, MUT2, MUT3, and MUT4) and wild-type (WT) SMAD3 promoter reporter were constructed (Figure 6F). As expected, the luciferase activity of SMAD3-WT was distinctly decreased after co-transfection with p53 plasmid. Furthermore, luciferase activity was restored by all of the mutants except for MUT2 (Figure 6F), indicating that p53 binds to the region (−10 to −28 bp) of the SMAD3 promoter and represses SMAD3 expression. Consistently, overexpression of SMAD3 could effectively reverse the process of EMT after silencing ACP5 in A549 cells (Figure 6G). Taken together, these data provide evidence that ACP5/p53/SMAD3 signaling is responsible for ACP5-induced metastasis in LUAD cells.
ACP5 Promoted Xenograft Tumor Growth and Metastatic Potential in Mouse Models
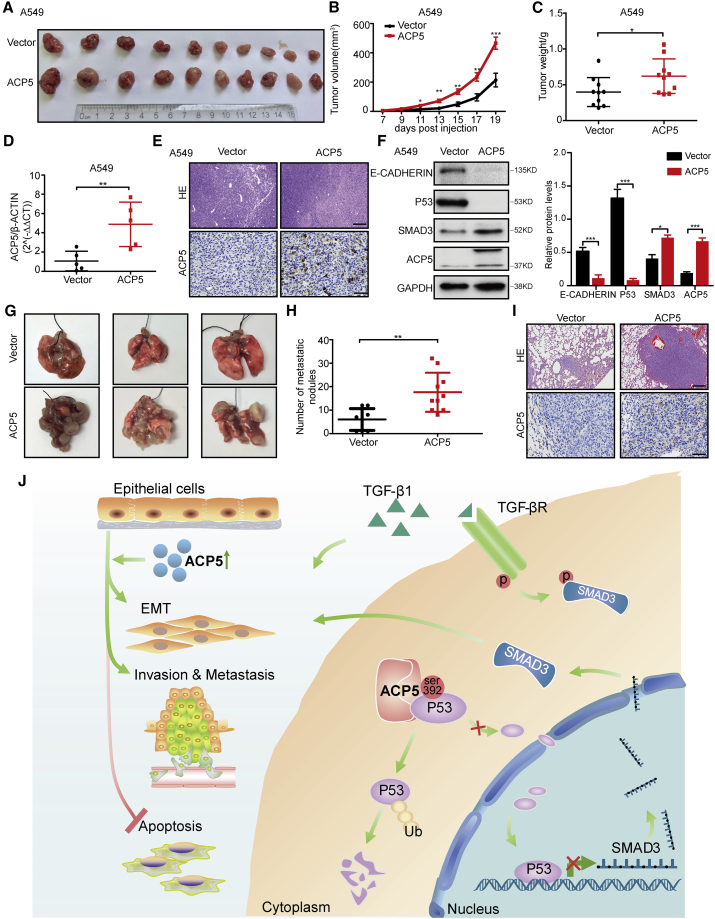
To assess whether the overexpression of ACP5 could enhance tumorigenesis in vivo, a xenograft tumor mouse model was established by subcutaneously injecting A549-Vec or A549-ACP5 cells into the right dorsal flank of nude mice, and this model was also established with NCI-H1975-Vec and NCI-H1975-ACP5 cells. At the end of the experiments, the xenograft tumors were isolated and weighed, which showed increased tumor size and weight in the mice with ACP5 overexpression (Figures 7A–7C; Figures S4A–S4C). As expected, significantly higher expression of ACP5 was detected in the group with ACP5 overexpression than that of the control group in xenograft tumor tissues (Figure 7D; Figure S4D). The results for hematoxylin and eosin (H&E) staining confirmed the tumor formation results (Figure 7E; Figure S4E). Furthermore, the expression of ACP5 in the xenograft tumors developed from ACP5-transfected cells was confirmed by immunohistochemical staining and western blot in tumor tissue samples that originated from xenograft mice (Figure 7F; Figure S4F). In line with in vitro data, the expression of E-cadherin and p53 in the A549-ACP5 group and H1975-ACP5 group was decreased, but that of SMAD3 was increased. To evaluate whether ACP5 could promote tumor metastasis in vivo, we injected A549-Vec or A549-ACP5 cells intravenously into nude mice via the tail vein. Five weeks after injection, the mice were sacrificed and the metastatic nodules formed on the lung surfaces were examined. As shown in Figures 7G and 7H, after calculating the numbers of the metastatic nodules with naked eyes, we found that the mice injected with A549-Vec cells formed fewer nodules on the lung surfaces than did the mice injected with A549-ACP5 cells. The pathology of metastatic nodules on the surfaces of the mouse lungs was confirmed by H&E staining, while the expression level of ACP5 in the nodules was also verified by immunohistochemical staining (Figure 7I). Therefore, our data demonstrated that high ACP5 expression enhanced tumor growth and metastasis in vivo, which was consistent with our in vitro and clinical findings.
Figure 7.
ACP5 Promoted Xenograft Tumor Growth and Enhanced Metastatic Potential in Mouse Models
(A–F) Nude mice were subcutaneously injected with A549 cells stably overexpressed for ACP5 (A549-ACP5) or control (A549-Vec) cells. (A) Images of the tumor lumps from the indicated groups at the endpoint of the experiment. n = 10 mice per group. (B) Tumor formation in nude mice treated with A549-ACP5 or A549-Vec cells was monitored at the indicated time points. (C) Tumor weights were measured at the last time point. (D) Quantitative real-time PCR for ACP5 expression in xenograft tumor tissues of A549-Vec (n = 5) and A549-ACP5 groups (n = 5). (E) Representative H&E staining images (top panel) (original magnification, ×100; scale bars, 200 μm) and IHC images of ACP5 staining (bottom panel) (original magnification, ×400; scale bars, 50 μm) in sections of xenograft tumor of A549 cells. (F) The expression of E-cadherin, p53, SMAD3, and ACP5 was detected by western blot in xenograft tumors from the A549-Vec (n = 5) and A549-ACP5 (n = 5) groups. (G–I) A549-ACP5 or A549-Vec cells were intravenously injected into nude mice and lung tissues were isolated after 5 weeks (A549-ACP5, n = 8; A549-vector, n = 10). (G) Representative lung images showing metastases in the lung. (H) Number of the metastatic nodules visible to the naked eye in the lung of mice. (I) H&E staining (original magnification, ×100; scale bars, 200 μm) and IHC staining for ACP5 (original magnification, 400; scale bars, 50 μm) performed with sections of metastatic lung nodules. (J) Schematic illustration of the role of ACP5 in LUAD. The results are expressed as the mean ± SD.∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by independent Student’s t test.
Discussion
In the present study, we demonstrated the expression of ACP5 in LUAD patients and the correlations between ACP5 and clinical information of the patients. Specifically, enhancing the expression ACP5 in A549 or NCI-H1975 promoted tumor growth and metastasis in vivo. In addition, in vitro, ACP5 was shown to be involved in the regulation of EMT, migration, invasion, and apoptosis in LUAD cells. Mechanistic studies revealed that ACP5 interacted with p53 in the cytoplasm and dephosphorylated the phosphorylation site (Ser392) of p53, which then promoted the ubiquitination and degradation of p53, leading to reduced amounts of p53 in the nucleus, and blunting the transcriptional repressive effect of p53 on SMAD3, by which it blunts the TGF-β signal pathway (Figure 7J), as SMAD3 has been reported to play a key role in the processes of EMT and tumor metastasis and to act as an important transcription factor in tumors. These results provide not only novel insights into the understanding of the pathogenesis underlying LUAD, but also evidence that targeting ACP5 regulates the p53/SMAD3 pathway, which may have great potential for the treatment of LUAD in clinical settings.
The human ACP5 gene maps to chromosome 19p13.2 and spans approximately 3 kb, and the ACP5 protein is mainly found in the cytoplasm. High ACP5 expression correlates with reduced tumor- and metastasis-free survival in malignant melanoma. Although there is no direct evidence of ACP5 involvement in tumor metastasis, all ACP5-interacting proteins have been reported to have direct associations with tumor invasiveness and metastasis.29, 30, 31 These findings underscore a potentially important role for ACP5 in an underlying biological mechanism involved in the development and progression of LUAD. In addition, the molecular mechanisms underlying ACP5 functions are also poorly understood, and knowledge concerning the cellular targets of ACP5 is very limited. Notably, these findings identified the potential of ACP5 and provided further links between ACP5 and cancer metastasis.
ACP5 expression was found to be increased in human LUAD tissue samples compared to adjacent noncancerous tissue samples. Consistent with a previous study, which showed that ACP5 was significantly overexpressed in human LUAD tissue samples compared to adjacent noncancerous tissue samples and significantly associated with differentiation, lymph node metastasis status, and TNM (tumor, node, metastasis) stage, found that patients with elevated ACP5 expression had shortened survival.18 We also demonstrated that ACP5 was overexpressed in LUAD samples and that the overexpression of ACP5 was associated with lymph node metastasis and age in patients with LUAD.
In our study, first, series of in vitro and in vivo assays were conducted to clarify the biological functions of ACP5 in the regulation of LUAD cell motility and invasiveness. The results showed that the ectopic overexpression of ACP5 could promote LUAD, and our study differed from other studies by focusing on the relationship between ACP5 and EMT as well as the mechanism of LUAD. These results indicate that ACP5 plays a role in promoting the development of cancer-promoting biological behaviors in cancer cells.
Metastasis represents a multistep cell biology process termed the invasion-metastasis cascade, which includes EMT.32 EMT, which is initially involved in normal embryonic morphogenesis, plays pivotal roles in tumor invasion and metastasis during tumor progression by suppressing the expression of epithelial markers or inducing the expression of mesenchymal markers.33, 34, 35 A close relationship between ACP5 and Foxm1, which is considered to be a master regulator of tumor metastasis that functions by inducing EMT, has been uncovered, and interestingly, in our study, we found that the overexpression of ACP5 had a significant impact on EMT in vitro and in vivo. Thus, LUAD cells overexpressing ACP5 probably undergo EMT to increase motility and invasiveness.
Previous studies have found that the activation of the TGF-β/SMAD3 pathway and p53 suppression are closely associated with lung tumorigenesis; in addition, TGF-β can negatively regulate the expression of p53.9,36, 37, 38, 39, 40, 41 SMAD proteins have been implicated in TGF-β-mediated signal transduction. Accumulating evidence showed that altering SMAD3 expression could affect the EMT.42 In our study, ACP5 knockdown resulted in downregulated SMAD3 expression, by which it blunted the TGF-β signal to attenuate the EMT. In previous studies, ACP5 was shown to interact with TGF-β receptor interacting protein-1 (TRIP-1) intracellularly, thereby activating TGF-β receptor type II (TβR2) and osteoblast differentiation through the SMAD2/3 pathway at sites of prior bone resorption. ACP5a interaction with TRIP-1 has also been demonstrated in mouse preadipocytes.26 Also, in our study, knocking down ACP5 seems to weaken the response to TGF-β, and overexpression of ACP5 aggravates the effects of TGF-β, altogether suggesting that ACP5 also targets the TGF-β pathway, which further confirms the previous work.26 However, to date, the molecular mechanisms by which ACP5 regulates EMT to promote cancer cell migration and invasiveness have not been completely elucidated. In this study, for the first time, we observed that ACP5 might be upstream of p53 and further explained the underlying mechanism involving p53 and SMAD3. Our dual-luciferase reporter and ChIP-PCR assays explained this mechanism at the molecular level. Regarding the relationship between p53 and TGF-β, we observed that the decrease in the p53 level caused by TGF-β1 stimulation with knocking down ACP5 expression in cells was not as obvious as that observed with ACP5 transfected into cells. We hypothesize that this finding results from the regulation of p53 requiring the participation of ACP5 and TGF-β, but the relationship between ACP5 and TGF-β remains to be studied. In regard to SMAD3, we overexpressed SMAD3 by transfecting a plasmid into ACP5-silenced A549 cells, which could reverse the expression pattern of E-cadherin. These results suggested that SMAD3 was indeed regulated by ACP5 in the promotion of EMT.
More importantly, our results showed that the opposite change in the expression of SMAD3 and p53 occurred after overexpressing ACP5; in particular, these changes could be observed in the tumor tissue of mice subjected to subcutaneous injection by western blot analysis. In vivo, in subcutaneous injection and intravenous injection mouse models, the overexpression of ACP5 resulted in significant increases in tumor growth and the number of metastatic lesions in the lungs. All of these findings strongly supported the conclusion that the overexpression of ACP5 plays important roles in LUAD invasion and metastasis. Therefore, the role of ACP5 in promoting EMT might be carried out through the p53/SMAD3 signaling pathway, ultimately driving LUAD cell motility and invasiveness.
Our findings raise a number of intriguing questions. First, the detailed regulatory mechanisms remain unclear, and we do not know the specific sites of p53 recognized by ACP5, but it is likely that p53 is directly downstream of ACP5. There may be other molecules participating in these processes, and this area of research will be indispensable in our future work. Second, we could not get commercial antibodies for ACP5b, and herein we detected the total expression of ACP5. In our further studies, we will identify the roles of ACP5a and ACP5b, respectively, in the pathogenesis of LUAD. Third, to date, we could not find an efficient specific inhibitor of ACP5. In a previous study, the authors found that the small chemical enzyme inhibitor 5-phenylnicotinic acid/CD13 could inhibit cell migration and invasion in ACP5-overexpressing MDA-MB-231 breast cancer cells; however, they concluded that CD13 was efficient as an ACP5 inhibitor for the ACP5b isoform but was not able to inhibit the ACP5a isoform.43 In the future, we expect that better inhibitors of ACP5 will be developed, and then drugs could be used to solve clinical problems.
In conclusion, this report demonstrated that ACP5 played an important role in the development and progression of LUAD. The overexpression of ACP5 in LUAD was significantly associated with tumor metastasis in LUAD patients. The underlying mechanism was revealed by functional and mechanistic studies, which showed that ACP5 regulated EMT to promote cancer cell motility and tumor metastasis through p53/SMAD3 signaling. Furthermore, our findings provide insight into ACP5, which may serve as a clinically useful diagnostic and prognostic biomarker and a potential therapeutic target in LUAD.
Materials and Methods
Tissue Samples
In total, 138 fresh tissues samples including LUAD (n = 69) and paired adjacent normal lung (n = 69, >5 cm from the tumor edge) samples were obtained from Tongji Hospital (Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China). No patient had received chemotherapy or radiotherapy before surgery. The study protocol was approved by the Institutional Review Board of Tongji Hospital, and written informed consent was obtained from each patient. All procedures were conducted in accordance with the Declaration of Helsinki and International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS).
Cell Lines and Cell Culture
The human cell lines Beas-2b, A549, NCI-H1650, NCI-H1299, NCI-H226, and SK-MES-1 were obtained from Zhong Qiao Xin Zhou Biotechnology (Shanghai, China). The cell line NCI-H1975 was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cell lines A549, H1299, NCI-H1975, and NCI-H1650 were identified by STR (Jianda, Suzhou, China). Cells were routinely cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and maintained in a humidified incubator with 5% CO2 at 37°C and were tested for mycoplasma regularly.
Reagents and Chemicals
Recombinant human TGF-β1 was purchased from PeproTech (Rocky Hill, NJ, USA), reconstituted in RPMI 1640 medium containing 10% FBS, and utilized at a final concentration of 10 ng/mL. MG132 was purchased from Cayman Chemical (Ann Arbor, MI, USA), dissolved in DMSO.
RNA Interference and ACP5 Overexpression
siRNAs specific for ACP5 and a corresponding scrambled siRNA were purchased from RiboBio (Guangzhou, China) and then transiently transfected into A549 and NCI-H1975 cells using Lipofectamine 3000 (Invitrogen, Shanghai, China) according to the manufacturer’s protocol. The siRNA for ACP5 was performed by targeting the following sequence in ACP5 mRNA: 5′-GACACTATGTGGCAACTCA-3′. When the cells reached 30%–50% confluence, siRNA transfection was performed using Lipofectamine 3000 (Invitrogen, Shanghai, China). Plasmid-ACP5 and a vector were purchased from GeneChem (Shanghai, China), and 2 μg of purified DNA was complexed with a transfection reagent and applied to the cells. Forty-eight hours after transfection, the cells were analyzed by quantitative real-time PCR and western blot.
A549 and NCI-H1975 cells were transfected with the plasmid containing ACP5 and vector purchased from Shanghai GeneChem (Shanghai, China) by Lipofectamine 3000 (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. For the establishment of an A549-ACP5 and H1975-ACP5 cell lines stably expressing ACP5, a lentivirus containing ACP5 and the associated vector were kindly gifted by Dr. Jiang Dingsheng. After 24 h of transfection, the cells were maintained in RPMI 1640 medium containing 2 μg/mL puromycin (InvivoGen, San Diego, CA, USA). After 3 weeks, the resistant colonies stably transfected with ACP5 were pooled.
Western Blot
Western blot analysis was performed according to a standard protocol.44 The primary antibodies included anti-ACP5 (Gentex, CA, USA, 1:1,000), anti-fibronectin, anti-E-cadherin, anti-vimentin (Proteintech, Wuhan, China, 1:1,000), anti-p38, anti-ERK, anti-p-ERK, anti-AKT, anti-β-catenin, anti-SMAD2/3, anti-p-SMAD2, anti-p-SMAD3, anti-p53, anti-p53 (Ser392), anti-ubiquitin (Cell Signaling Technology, Danvers, MA, USA, 1:1,000), anti-GAPDH, and anti-β-actin (Abcam, Cambridge, MA, USA, 1:3,000) antibodies. Detection was performed using a chemiluminescent substrate system (Bio-Rad, Hercules, CA, USA). The gray values were analyzed with ImageJ software.
Quantitative Real-Time PCR
Total RNA was extracted from tissue samples or cells by TRIzol (Takara, Dalian, China). Single-stranded cDNA was synthesized from total RNA using a PrimeScript RT reagent kit (Takara, Dalian, China). Quantitative real-time PCR was performed using SYBR Green mix (Takara, Dalian, China) under the following conditions: 30 s at 95°C for initial denaturation, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s on an Applied Biosystems 7500 real-time PCR system (Thermo Fisher Scientific, Waltham, MA). The RNA levels of β-actin were used to normalize the data. The primer sequences were as follows: ACP5 forward, 5′-GCT GTC CTG GCT CAA GAA AC-3′, reverse, 5′-CCC ACG CCA TTC TCA TCT TG-3′; SMAD2 forward, 5′-CGT CCA TCT TGC CAT TCA CG-3′, reverse, 5′-CTC AAG CTC ATC TAA TCG TCC TG-3′; SMAD3 forward, 5′-GCG TGC GGC TCT ACT ACA TC-3′, reverse, 5′-GCA CAT TCG GGT CAA CTG GTA-3′; p53 forward, 5′-TTC CTT TGG GTG CCC TGA A-3′, reverse, 5′-GGC AGA ATC TGT CCA TCT TTC G-3′; and ACTIN forward, 5′-AGC GAG CAT CCC CCA AAG TT-3′, reverse, 5′-GGG CAC GAA GGC TCA TCAT T-3′.
Cell Proliferation Assay
A proliferation assay was carried out using Cell Counting Kit-8 (CCK-8, Promotor, Wuhan, China) according to the manufacturer’s protocol.45 The absorbance was recorded at 450 nm with a universal microplate reader (ELx800, BioTek Instruments, Winooski, VT, USA).
As for colony formation assay, we seeded 500 or 1,000 transfected cells in each well of a six-well plate and maintained for 9 or 14 days in different cell lines. The colonies were then fixed with methanol, stained with 0.5% crystal violet, and counted under a microscope.
Flow Cytometry
Apoptosis in transfected cells was analyzed by flow cytometry using a Becton Dickinson LSR flow cytometer with CellQuest software (BD Biosciences, San Diego, CA, USA). We used 300 nM H2O2 for 24 h to induce apoptosis after transfection for 48 h, and then cells were collected and resuspended in 500 μL of binding buffer of every tube. Next, 5 μL of annexin V and 5 μL of propidium iodide (Keygen Biotech, Nanjing, China) were added to every tube for 15 min in the dark simultaneously or separately (as the control to debug the machine) and then analyzed to identify the late apoptotic cells, early apoptotic cells, and viable cells, and the proportion of apoptotic cells was calculated.
Wound-Healing Assay
For a wound-healing assay, cells were grown in a six-well plate until 95% confluent, and the monolayers were scratched with a pipette tip. Cell migration was recorded at 0 and 24 h after scratching. Three independent experiments were performed.
Cell Invasion and Migration Assays
Invasion and migration assays were performed using chambers from Corning Life Sciences (Corning, MA, USA) with or without Matrigel according to the manufacturer’s protocol.46 Briefly, cell suspensions prepared in medium containing 2% FBS were added into the upper chambers, while medium containing 20% FBS was placed in the lower chambers to function as a chemoattractant. The cells were incubated for 24 h at 37°C and allowed to migrate into or invade through the membrane filter. Afterward, the nonmigratory and noninvasive cells on the upper surface were gently removed with a cotton swab. Then, the cells that had invaded or migrated to the lower surface of the membrane were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA). The cells in four randomly selected microscopic fields were counted and imaged. Both experiments were repeated independently in triplicate.
Immunohistochemistry (IHC) Analysis
Immunohistochemical staining was performed according to the protocol in a previous report (Boster Biological Technology, Wuhan, China).18 Rabbit primary antibodies against human ACP5 (Abcam, Cambridge, MA, USA, 1:100), p53 (Cell Signaling Technology, Danvers, MA, USA, 1:50), and SMAD3 (Cell Signaling Technology, Danvers, MA, USA, 1:100) were added to tissue sections and incubated overnight at 4°C in a moist chamber. Then, the slides were incubated with horseradish peroxidase-conjugated secondary antibodies (goat anti-rabbit or mouse) at 37°C for 30 min. After washing in phosphate-buffered saline (PBS), diaminobenzidine (DAB)-H2O2 was used for a color reaction. Finally, all of the slides were counterstained with hematoxylin, dehydrated, and mounted.
Immunofluorescence Analysis
Cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 5 min. The samples were washed with PBS and blocked with 5% BSA in PBS. Subsequently, the cells were incubated with primary antibodies against mouse-originated vimentin antibody (R&D Systems, Minneapolis, MN, USA, 1:100), mouse-originated E-cadherin antibody (R&D Systems, Minneapolis, MN, USA, 1:100), mouse-originated p53 antibody (Cell Signaling Technology, Danvers, MA, USA, 1:50), and rabbit-originated ACP5 antibody (Proteintech, Wuhan, China, 1:100) overnight at 4°C and then incubated with an Alexa Fluor 488-labeled anti-mouse antibody and an Alexa Fluor 594-conjugated anti-rabbit antibody (Abbkine, Redlands, CA, USA, 1:400) for 1 h, as instructed. For SNAIL1 and ZEB2, we used rabbit-originated antibody (Proteintech, Wuhan, China, 1:100), and the mouse-originated ACP5 antibody (Abnova, Taipei, China, 1:100) was used. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min.
CoIP Assay
Immunoprecipitation was performed according to a standard protocol.47 Anti-ACP5 and anti-p53 antibodies were used to form immune complexes with the ACP5 and p53 proteins in lysates and were immunoprecipitated with magnetic beads (Cell Signaling Technology, Danvers, MA, USA). Finally, equivalent protein samples were subjected to western blot analysis for ACP5 and p53.
Luciferase Assay
Plasmids encoding p53 in the GV141 vector were purchased from GeneChem (Shanghai, China). Both WT and mutant SMAD3 genes were synthesized by chemical methods and cloned downstream of the luciferase gene in GV238 luciferase vectors that were obtained from GeneChem (Shanghai, China). These plasmids and a Renilla luciferase plasmid (GeneChem, Shanghai, China), which was used as a normalization control, were cotransfected into cells using Lipofectamine 3000 (Invitrogen, Shanghai, China). 293T cells (8 × 103 per well) were seeded in 96-well plates the day before transfection, and the cells were cotransfected with 1.25 μg of GV238-SMAD3 and 0.75 μg of p53 DNA when they reached 70%–90% confluence. In total, 50 ng of the expression plasmid (Renilla) was cotransfected as a transfection efficiency control. After 24 h, the cells were harvested, washed three times with PBS, and lysed in 20 μL of passive lysis buffer (PLB) according to the manufacturer’s instructions (Promega, Madison, WI, USA). Cell debris was removed by centrifugation, and the supernatant was used for a luciferase assay read with SpectraMax single-mode microplate readers (Molecular Devices, Sunnyvale, CA, USA).
ChIP and RNA Analyses of Solid Tissue and Cultured Cells
Chromatin immunoprecipitation (ChIP) assays were conducted using a ChIP assay kit (Biyuntian, Shanghai, China) as previously described.48 Quantitative real-time PCR was then performed using the primers as follows: site a forward, 5′-GCCTCAGGACCACTCTTGTA-3′, reverse, 5′- CTCAGAGGAGTCAGTTGTCTTAAA-3′; site b forward, 5′-ATCCAGAGTGCGTGGTGTTT-3′, reverse, 5′-CCAGCAAGCACAAGCTCATA-3′; site c forward, 5′-GGAGCAAACGGCCTGAAATA-3′, reverse, 5′-GCGAGCGGCGGCAATAAAT-3′.
Xenograft Formation and an In Vivo Metastasis Assay
Male BALB/c nude mice (4–5 weeks old) were purchased from GemPharmatech (Nanjing, China). All animals were housed in a specific pathogen-free animal facility at the Tongji Medical College under a 12-h light/12-h dark photocycle and provided with feed and water ad libitum. All experimental procedures were approved by the Animal Care and Use Committee of Tongji Hospital. A xenograft tumor growth model was established by subcutaneous injection of A549/H1975-ACP5 cells or control cells (2 × 106, suspended in PBS) into the right dorsal flank. Tumor volume (TV) was measured by calipers and calculated with the following formula: TV (mm3) = (L × W2)/2 (L, long diameter; W, wide diameter). After sacrifice, the tumors were excised for subsequent assays. To establish an intravenous mouse model, 5 × 106 A549-ACP5 cells or control cells were injected intravenously via the tail vein. After 5 weeks, the mice were sacrificed, and the tumor nodules formed on the lung surfaces were counted. The lungs were collected and embedded in paraffin for further H&E staining and IHC analysis.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism (San Diego, CA, USA) or SPSS 21.0 (IBM, Armonk, NY, USA). The correlations between ACP5 expression and clinicopathological features of LUAD patients were analyzed by the χ2 test or Fisher’s exact test. Other data are expressed as the mean ± SEM, and an independent Student’s t test was applied to analyze the statistical significance of differences between two groups. p <0.05 was considered statistically significant. For comparisons between multiple groups, one-way ANOVA followed by a Tukey’s test was performed. p <0.05 was considered significant. All data were tested for normalization before analysis.
Author Contributions
W.X. and Y.W. designed, edited, and oversaw the experiments of this study. Y.H., J.Y., Q.W., L.Z., and X.C. conducted the experiments, data analysis, and critical discussions of the results. Y.C., D.J., J.Z., and Y.X. provided material support and study supervision. All authors contributed to the writing and editing of the manuscript and approved the final draft of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the Wuhan Young and Middle-Aged Medical Key Talents Training Project, the Clinical Research Physician Program of Tongji Medical College, and by the Integrated Innovative Team for Major Human Diseases Program of Tongji Medical College (HUST).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.01.010.
Contributor Information
Yi Wang, Email: xinyiwy2012@163.com.
Weining Xiong, Email: xiongdoctor@qq.com.
Supplemental Information
References
- 1.Imielinski M., Berger A.H., Hammerman P.S., Hernandez B., Pugh T.J., Hodis E., Cho J., Suh J., Capelletti M., Sivachenko A. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller K.D., Siegel R.L., Lin C.C., Mariotto A.B., Kramer J.L., Rowland J.H., Stein K.D., Alteri R., Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 4.Kasinski A.L., Slack F.J. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72:5576–5587. doi: 10.1158/0008-5472.CAN-12-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenzweig K.E., Gomez J.E. Concurrent chemotherapy and radiation therapy for inoperable locally advanced non-small-cell lung cancer. J. Clin. Oncol. 2017;35:6–10. doi: 10.1200/JCO.2016.69.9678. [DOI] [PubMed] [Google Scholar]
- 6.Wilkins A., McDonald F., Harrington K., Melcher A. Radiotherapy enhances responses of lung cancer to CTLA-4 blockade. J. Immunother. Cancer. 2019;7:64. doi: 10.1186/s40425-019-0542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emoto K., Eguchi T., Tan K.S., Takahashi Y., Aly R.G., Rekhtman N., Travis W.D., Adusumilli P.S. Expansion of the concept of micropapillary adenocarcinoma to include a newly recognized filigree pattern as well as the classical pattern based on 1468 stage I lung adenocarcinomas. J. Thorac. Oncol. 2019;14:1948–1961. doi: 10.1016/j.jtho.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C.J., Chao C.H., Xia W., Yang J.Y., Xiong Y., Li C.W., Yu W.H., Rehman S.K., Hsu J.L., Lee H.H. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakai S., Ohhata T., Kitagawa K., Uchida C., Aoshima T., Niida H., Suzuki T., Inoue Y., Miyazawa K., Kitagawa M. Long noncoding RNA ELIT-1 acts as a Smad3 cofactor to facilitate TGFβ/Smad signaling and promote epithelial-mesenchymal transition. Cancer Res. 2019;79:2821–2838. doi: 10.1158/0008-5472.CAN-18-3210. [DOI] [PubMed] [Google Scholar]
- 10.Chen H.T., Liu H., Mao M.J., Tan Y., Mo X.Q., Meng X.J., Cao M.T., Zhong C.Y., Liu Y., Shan H., Jiang G.M. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Cancer. 2019;18:101. doi: 10.1186/s12943-019-1030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greathouse K.L., White J.R., Vargas A.J., Bliskovsky V.V., Beck J.A., von Muhlinen N., Polley E.C., Bowman E.D., Khan M.A., Robles A.I. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawarada Y., Inoue Y., Kawasaki F., Fukuura K., Sato K., Tanaka T., Itoh Y., Hayashi H. TGF-β induces p53/Smads complex formation in the PAI-1 promoter to activate transcription. Sci. Rep. 2016;6:35483. doi: 10.1038/srep35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun P., Sleat D.E., Lecocq M., Hayman A.R., Jadot M., Lobel P. Acid phosphatase 5 is responsible for removing the mannose 6-phosphate recognition marker from lysosomal proteins. Proc. Natl. Acad. Sci. USA. 2008;105:16590–16595. doi: 10.1073/pnas.0807472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayman A.R., Macary P., Lehner P.J., Cox T.M. Tartrate-resistant acid phosphatase (Acp 5): identification in diverse human tissues and dendritic cells. J. Histochem. Cytochem. 2001;49:675–684. doi: 10.1177/002215540104900601. [DOI] [PubMed] [Google Scholar]
- 15.Ljusberg J., Wang Y., Lång P., Norgård M., Dodds R., Hultenby K., Ek-Rylander B., Andersson G. Proteolytic excision of a repressive loop domain in tartrate-resistant acid phosphatase by cathepsin K in osteoclasts. J. Biol. Chem. 2005;280:28370–28381. doi: 10.1074/jbc.M502469200. [DOI] [PubMed] [Google Scholar]
- 16.Zenger S., Ek-Rylander B., Andersson G. Biogenesis of tartrate-resistant acid phosphatase isoforms 5a and 5b in stably transfected MDA-MB-231 breast cancer epithelial cells. Biochim. Biophys. Acta. 2010;1803:598–607. doi: 10.1016/j.bbamcr.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 17.Sträter N., Jasper B., Scholte M., Krebs B., Duff A.P., Langley D.B., Han R., Averill B.A., Freeman H.C., Guss J.M. Crystal structures of recombinant human purple acid phosphatase with and without an inhibitory conformation of the repression loop. J. Mol. Biol. 2005;351:233–246. doi: 10.1016/j.jmb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 18.Gao Y.L., Liu M.R., Yang S.X., Dong Y.J., Tan X.F. Prognostic significance of ACP5 expression in patients with lung adenocarcinoma. Clin. Respir. J. 2018;12:1100–1105. doi: 10.1111/crj.12637. [DOI] [PubMed] [Google Scholar]
- 19.Xia L., Huang W., Tian D., Chen Z., Zhang L., Li Y., Hu H., Liu J., Chen Z., Tang G. ACP5, a direct transcriptional target of FoxM1, promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Oncogene. 2014;33:1395–1406. doi: 10.1038/onc.2013.90. [DOI] [PubMed] [Google Scholar]
- 20.Ren X., Shah T.A., Ustiyan V., Zhang Y., Shinn J., Chen G., Whitsett J.A., Kalin T.V., Kalinichenko V.V. FOXM1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Mol. Cell. Biol. 2013;33:371–386. doi: 10.1128/MCB.00934-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balli D., Ustiyan V., Zhang Y., Wang I.C., Masino A.J., Ren X., Whitsett J.A., Kalinichenko V.V., Kalin T.V. Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. EMBO J. 2013;32:231–244. doi: 10.1038/emboj.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lång P., Andersson G. Differential expression of monomeric and proteolytically processed forms of tartrate-resistant acid phosphatase in rat tissues. Cell. Mol. Life Sci. 2005;62:905–918. doi: 10.1007/s00018-005-4550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janckila A.J., Lin H.F., Wu Y.Y., Ku C.H., Yang S.P., Lin W.S., Lee S.H., Yam L.T., Chao T.Y. Serum tartrate-resistant acid phosphatase isoform 5a (TRACP5a) as a potential risk marker in cardiovascular disease. Clin. Chim. Acta. 2011;412:963–969. doi: 10.1016/j.cca.2011.01.035. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y.Y., Chao T.Y., Liu H.Y., Huang T.C., Chen J.H., Dai M.S., Janckila A., Lai S.W., Chang P.Y. The correlation between a chronic inflammatory marker Tartrate-resistant acid phosphatase 5a with cancer cachexia. J. BUON. 2015;20:325–331. [PubMed] [Google Scholar]
- 25.Shih K.C., Janckila A.J., Lee W.J., Chou Y.C., Huang C.J., Kwok C.F., Ho L.T., Chao T.Y. Effects of bariatric weight loss surgery on glucose metabolism, inflammatory cytokines, and serum tartrate-resistant acid phosphatase 5a in obese Chinese adults. Clin. Chim. Acta. 2016;453:197–202. doi: 10.1016/j.cca.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Reithmeier A., Panizza E., Krumpel M., Orre L.M., Branca R.M.M., Lehtiö J., Ek-Rylander B., Andersson G. Tartrate-resistant acid phosphatase (TRAP/ACP5) promotes metastasis-related properties via TGFβ2/TβR and CD44 in MDA-MB-231 breast cancer cells. BMC Cancer. 2017;17:650. doi: 10.1186/s12885-017-3616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox M.L., Meek D.W. Phosphorylation of serine 392 in p53 is a common and integral event during p53 induction by diverse stimuli. Cell. Signal. 2010;22:564–571. doi: 10.1016/j.cellsig.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 28.Huang C., Ma W.Y., Maxiner A., Sun Y., Dong Z. p38 kinase mediates UV-induced phosphorylation of p53 protein at serine 389. J. Biol. Chem. 1999;274:12229–12235. doi: 10.1074/jbc.274.18.12229. [DOI] [PubMed] [Google Scholar]
- 29.Suwinski R., Giglok M., Galwas-Kliber K., Idasiak A., Jochymek B., Deja R., Maslyk B., Mrochem-Kwarciak J., Butkiewicz D. Blood serum proteins as biomarkers for prediction of survival, locoregional control and distant metastasis rate in radiotherapy and radio-chemotherapy for non-small cell lung cancer. BMC Cancer. 2019;19:427. doi: 10.1186/s12885-019-5617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klement J.D., Paschall A.V., Redd P.S., Ibrahim M.L., Lu C., Yang D., Celis E., Abrams S.I., Ozato K., Liu K. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J. Clin. Invest. 2018;128:5549–5560. doi: 10.1172/JCI123360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo J., Yang W.L., Pak D., Celestino J., Lu K.H., Ning J., Lokshin A.E., Cheng Z., Lu Z., Bast R.J. Osteopontin, macrophage migration inhibitory factor and anti-interleukin-8 autoantibodies complement CA125 for detection of early stage ovarian cancer. Cancers (Basel) 2019;11:596. doi: 10.3390/cancers11050596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang B., Wu N., Guan R., Pang L., Li X., Li S., Tang L., Guo Y., Chen J., Sun D. Overexpression of RCC2 enhances cell motility and promotes tumor metastasis in lung adenocarcinoma by inducing epithelial-mesenchymal transition. Clin. Cancer Res. 2017;23:5598–5610. doi: 10.1158/1078-0432.CCR-16-2909. [DOI] [PubMed] [Google Scholar]
- 33.Risolino M., Mandia N., Iavarone F., Dardaei L., Longobardi E., Fernandez S., Talotta F., Bianchi F., Pisati F., Spaggiari L. Transcription factor PREP1 induces EMT and metastasis by controlling the TGF-β-SMAD3 pathway in non-small cell lung adenocarcinoma. Proc. Natl. Acad. Sci. USA. 2014;111:E3775–E3784. doi: 10.1073/pnas.1407074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y., Ahn Y.H., Chen Y., Tan X., Guo L., Gibbons D.L., Ungewiss C., Peng D.H., Liu X., Lin S.H. ZEB1 sensitizes lung adenocarcinoma to metastasis suppression by PI3K antagonism. J. Clin. Invest. 2014;124:2696–2708. doi: 10.1172/JCI72171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan X., Banerjee P., Liu X., Yu J., Gibbons D.L., Wu P., Scott K.L., Diao L., Zheng X., Wang J. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J. Clin. Invest. 2018;128:1267–1282. doi: 10.1172/JCI97225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kudinov A.E., Deneka A., Nikonova A.S., Beck T.N., Ahn Y.H., Liu X., Martinez C.F., Schultz F.A., Reynolds S., Yang D.H. Musashi-2 (MSI2) supports TGF-β signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc. Natl. Acad. Sci. USA. 2016;113:6955–6960. doi: 10.1073/pnas.1513616113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Till J.E., Yoon C., Kim B.J., Roby K., Addai P., Jonokuchi E., Tang L.H., Yoon S.S., Ryeom S. Oncogenic KRAS and p53 loss drive gastric tumorigenesis in mice that can be attenuated by e-cadherin expression. Cancer Res. 2017;77:5349–5359. doi: 10.1158/0008-5472.CAN-17-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepherd F.A., Lacas B., Le Teuff G., Hainaut P., Jänne P.A., Pignon J.P., Le Chevalier T., Seymour L., Douillard J.Y., Graziano S., LACE-Bio Collaborative Group Pooled analysis of the prognostic and predictive effects of TP53 comutation status combined with KRAS or EGFR mutation in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J. Clin. Oncol. 2017;35:2018–2027. doi: 10.1200/JCO.2016.71.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang D., Cheng D., Tu Q., Yang H., Sun B., Yan L., Dai H., Luo J., Mao B., Cao Y. HUWE1 controls the development of non-small cell lung cancer through down-regulation of p53. Theranostics. 2018;8:3517–3529. doi: 10.7150/thno.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J., Feng M., Liu D., Liu H., Shi M., Zhang J., Qu J. Antagonism between HTRA3 and TGFβ1 contributes to metastasis in non-small cell lung cancer. Cancer Res. 2019;79:2853–2864. doi: 10.1158/0008-5472.CAN-18-2507. [DOI] [PubMed] [Google Scholar]
- 41.Filipczak P.T., Leng S., Tellez C.S., Do K.C., Grimes M.J., Thomas C.L., Walton-Filipczak S.R., Picchi M.A., Belinsky S.A. p53-Suppressed oncogene TET1 prevents cellular aging in lung cancer. Cancer Res. 2019;79:1758–1768. doi: 10.1158/0008-5472.CAN-18-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki K., Masugi Y., Effendi K., Tsujikawa H., Hiraoka N., Kitago M., Shinoda M., Itano O., Tanabe M., Kitagawa Y., Sakamoto M. Upregulated SMAD3 promotes epithelial-mesenchymal transition and predicts poor prognosis in pancreatic ductal adenocarcinoma. Lab. Invest. 2014;94:683–691. doi: 10.1038/labinvest.2014.53. [DOI] [PubMed] [Google Scholar]
- 43.Krumpel M., Reithmeier A., Senge T., Baeumler T.A., Frank M., Nyholm P.G., Ek-Rylander B., Andersson G. The small chemical enzyme inhibitor 5-phenylnicotinic acid/CD13 inhibits cell migration and invasion of tartrate-resistant acid phosphatase/ACP5-overexpressing MDA-MB-231 breast cancer cells. Exp. Cell Res. 2015;339:154–162. doi: 10.1016/j.yexcr.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Zhu J., Zhang L., Zhang Z., He L., Mou Y., Deng Y., Cao Y., Yang P., Su Y. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor α positive feedback loop in M2 macrophages. J. Allergy Clin. Immunol. 2017;140:1550–1561.e8. doi: 10.1016/j.jaci.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y.H., Wang Y., Yusufali A.H., Ashby F., Zhang D., Yin Z.F., Aslanidi G.V., Srivastava A., Ling C.Q., Ling C. Cytotoxic genes from traditional Chinese medicine inhibit tumor growth both in vitro and in vivo. J. Integr. Med. 2014;12:483–494. doi: 10.1016/s2095-4964(14)60057-1. [DOI] [PubMed] [Google Scholar]
- 46.Wang X., Xu W., Zhan P., Xu T., Jin J., Miu Y., Zhou Z., Zhu Q., Wan B., Xi G. Overexpression of geranylgeranyl diphosphate synthase contributes to tumour metastasis and correlates with poor prognosis of lung adenocarcinoma. J. Cell. Mol. Med. 2018;22:2177–2189. doi: 10.1111/jcmm.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X., Lai Q., Chen C., Li N., Sun F., Huang W., Zhang S., Yu Q., Yang P., Xiong F. Both conditional ablation and overexpression of E2 SUMO-conjugating enzyme (UBC9) in mouse pancreatic beta cells result in impaired beta cell function. Diabetologia. 2018;61:881–895. doi: 10.1007/s00125-017-4523-9. [DOI] [PubMed] [Google Scholar]
- 48.Yao Y., Wang Y., Zhang Z., He L., Zhu J., Zhang M., He X., Cheng Z., Ao Q., Cao Y. Chop deficiency protects mice against bleomycin-induced pulmonary fibrosis by attenuating M2 macrophage production. Mol. Ther. 2016;24:915–925. doi: 10.1038/mt.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.