Abstract
Early detection and easy continuous monitoring of emerging or re-emerging infectious, contagious or other diseases are of particular interest for controlling healthcare advances and developing effective medical treatments to reduce the high global cost burden of diseases in the backdrop of lack of awareness regarding advancing diseases. Under an ever-increasing demand for biosensor design reliability for early stage recognition of infectious agents or contagious diseases and potential proteins, nanoscale manufacturing designs had developed effective nanodynamic sensing assays and compact wearable devices. Dynamic developments of biosensor technology are also vital to detect and monitor advanced diseases, such as human immunodeficiency virus (HIV), hepatitis B virus (HBV), hepatitis C virus (HCV), diabetes, cancers, liver diseases, cardiovascular diseases (CVDs), tuberculosis, and central nervous system (CNS) disorders. In particular, nanoscale biosensor designs have indispensable contribution to improvement of health concerns by early detection of disease, monitoring ecological and therapeutic agents, and maintaining high safety level in food and cosmetics. This review reports an overview of biosensor designs and their feasibility for early investigation, detection, and quantitative determination of many advanced diseases. Biosensor strategies are highlighted to demonstrate the influence of nanocompact and lightweight designs on accurate analyses and inexpensive sensing assays. To date, the effective and foremost developments in various nanodynamic designs associated with simple analytical facilities and procedures remain challenging. Given the wide evolution of biosensor market requirements and the growing demand in the creation of early stage and real-time monitoring assays, precise output signals, and easy-to-wear and self-regulating analyses of diseases, innovations in biosensor designs based on novel fabrication of nanostructured platforms with active surface functionalities would produce remarkable biosensor devices. This review offers evidence for researchers and inventors to focus on biosensor challenge and improve fabrication of nanobiosensors to revolutionize consumer and healthcare markets.
Keywords: Dynamic biosensor designs, Nanomaterials, Nano chipset sensor device, Healthcare industry, Diseases screening, Virus detection
1. Introduction
Sensing and monitoring biological agents have gained increasing attention in biochemical and biomedical sectors particularly in early detection of specific diseases, identification of therapeutic agents, and recognition of vital biomolecular reactive species. The association of biosensors with daily life monitoring of advanced diseases, and biological agents have led to important concerns in the healthcare industry, environmental issues, and food processing [[1], [2], [3], [4]]. For instance, cardiovascular diseases (CVDs) are associated with cholesterol levels in blood. Metabolic disorders, such as diabetes, are commonly due to high levels of blood glucose or blood sugar, leading to problematic function, inability of usage, or insufficient production of the hormone insulin by pancreas inside the body. Furthermore, accumulation of blood and toxins overloading around the kidney; doses of chemicals and drugs, and alcoholic levels might lead to kidney damage (e.g. cancer, cysts, and kidney failure). Mental disorders (e.g. Parkinson's disease, schizophrenia, and Alzheimer's disease) are associated with disturbance in the level of neurotransmitters, particularly dopamine molecules [[3], [4], [5], [6], [7], [8], [9]]. Because of the significant increase in the activity of cancer and infectious diseases, which seriously threaten the human health worldwide, a particular concern is the ever-growing demand of rapid biosensor fabrication and fascinating evolution of non-invasive procedure. According to statistics published by the World Health Organization (WHO), CVD has become the leading cause of deaths globally (more than 17.5 million deaths in 2013 worldwide), causing great economic costs [9,10].
To date, diverse analyses associated with accessibility of biochemical and bioassay techniques are found to be overpriced analyses due to escalated work to obtain data. The most common analysis methodologies required repetitive screening steps in addition to reasonable robustness, cost adequacy, adaptability, and portability. For efficient detection and monitoring the stage of diseases, the development of biosensor technology design and material components remains a challenge. Therefore, low cost and simple biosensor designs are required for analysis with sufficient accuracy, ultratrace sensitivity, fast response, and high specificity. Wide, direct, and rapid detection ability for biomolecules, antibodies, enzymes, nucleic acids, and oligonucleotides is crucial for early detection of ailments. In this regard, fabrication of diverse and multidisciplinary biosensor designs is based primarily on controlling: (1) the appreciate vehicle material component platform scales; (2) efficient functionality of the surface-binding site; and (3) the transduction components that efficiently enabled recognizable signals [[11], [12], [13], [14], [15], [16], [17], [18]].
2. Nanoarchitect-led building blocks in biosensing design platforms for daily life monitoring of diseases
Biosensors have faced several challenges in regard to acceptability related to achievement of perfect sensitivity and selectivity of biological agents, and simple-to-utilize formats. The enormous potential advantage of biosensors motivated grasping all intents and objective of each possible analytical task running from medical diagnostics through drug discovery, food safety, military process control, and environmental monitoring for protection and security applications [5,[18], [19], [20], [21], [22]]. Accurate assessment and dose determination of certain biomolecules, antigens, proteins, and antibodies with high specificity and sensitivity play a pivotal part in monitoring and prescription of actively infectious cells and agents affecting human health [[23], [24], [25], [26], [27], [28], [29], [30]]. One of the vital effects of early recognition in light of biosensor use is that it essentially decreases the latent treatment costs for patients in advanced stages of numerous ailments.
Biosensor systems currently may have many limitations and drawbacks, such as slow response time, difficulty in obtaining meaningful assessment and read-out analyses, and burden to patients for several weeks before delivering the results [[28], [29], [30], [31], [32]]. In this context, developing vital, trustworthy, reliable, and economical biosensors with a robust sensing and recognition strategy for early detection of diseases and disorders such as hyperactivity disorders, excitatory blood pressure, cancers, and personal therapy is needed [[31], [32], [33], [34]]. For example, currently there are more than 347 million people worldwide suffering from diabetes – which figure is expected to leapfrog to 693 million by 2045, according to the International Diabetes Federation (IDF) – of which, 3.4 million died annually because of hypoglycemic disorders and severe diabetic snags [[35], [36], [37], [38]]. Therefore, controlling the glucose level in blood samples and appropriate level of insulin concentration is an urgent care for diabetics. Significant efforts have been devoted in developing nanobiosensors over the past decades to improve biosensors in terms of simplicity, accuracy, and cost effectiveness [[39], [40], [41], [42]].
An important consideration in the light of the advanced nanoscale architect-led building blocks in technology provides essential solutions for a significant number of day-to-day problems of the environment, energy, healthcare, and medicine. These nanomaterials showed evidence to be used as effective platforms for biomolecule immobilization with the desired orientation, high biological activity, and improved biosensor properties [[43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54]]. The developments in nano-/biotechnology, and bioengineering have been controlled and enabled the application of nanosensors/biosensors in numerous areas including industry, agriculture, environmental science, and pharmaceuticals [5,[55], [56], [57], [58], [59], [60], [61], [62], [63], [64]]. In this regard, nanostructured architect materials can provide a unique sensor design candidate that meets the high requirements of nanobiosensing assays due to their physical, chemical, mechanical, magnetic, optical, and electronic features (Scheme 1) [[65], [66], [67], [68], [69], [70]]. These nanocreation features led to a revolutionary new technology that can be used to fabricate nanodynamic biosensor design, leading to direct infection monitoring and high-throughput screening. Thus, the nanosized architect-led building-blocks-in biosensor device market is expected to offer unique, robust, and economic devices for real monitoring and detecting solutions (Scheme 1A) [4,43,[71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86]].
Scheme 1.
Illustration of a wide-range of molecules detection, potential targeting applications and diverse analyses of multiple types of biosensors. The nanostructured materials applied as a building platform of biosensors to enhance the monitoring of signals, selectivity and sensitive detection of ultra-trace targets (A). Biosensor building-blocks design orients toward daily life functionality and applicability in disease detection (B). The integration of nanobiosensors into smartphone or wearable handheld device for simultaneous screening, (C). The nanoscale architect-led materials fabricated with thermo-electric high-conductive sites, low-power consumption hybrid metal/non-metal /metalloid mesocrystal, atomic building arrangement, morphological structure disciplines, hierarchy, and orientation can act as key components in the creation of biosensor design of a high-performance output signals and rapid biosensing platforms (D).
In view of the literature, a variety of nanostructured metal oxides, metals or carbon-based materials, and the nanocomposite clusters were applied as a building platform of biosensors to enhance the monitoring signals of target analytes on their reactive binding-state surfaces [[87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98]]. The building-blocks-in sensor design may feature a solution to overcome shortcoming binding and selectivity and sensitive detection of ultra-trace targets still remains a challenge in this biosensing field (Scheme 1A) [4,43,[77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92]]. In early detection/biosensing assay, the technological major advance, understanding the role of hierarchical nanostructures and morphologies would offer super-efficient, high quality, and reliable building blocks of periodic biosensors for point-of-use monitoring diseases [[99], [100], [101], [102], [103]].
3. Biosensor building-blocks design merits toward daily life functionality and applicability in disease detection
In contemporary medical practice, intensive necessity of simple yet general biosensing designs of many dexterous and masterful analyses is frequently felt. Accurate analyses with visibly distinguishable and readable test results are unquestionable requirements to recognize actively infectious diseases, prevalence such as cancer, human immunodeficiency virus (HIV), tuberculosis (TB), hepatitis B virus (HBV), hepatitis C virus (HCV), diabetes, and symptoms of cardiovascular diseases (CVDs) (Scheme 1B). Crucially, the nanosized architect-led building blocks in nanobiosensor design is expected to offer high selectivity, accuracy, and fast and real-time responses, which are the challenges in the current stage of development in biosensor systems.
To boost online practices and visualize data analysis for the end users, the nanochipset sensor device is targeted to be integrated into a circuit diagram. The module integration would also improve the uncertainty management of biosensor analysis and other reliability drawbacks in terms of handling multisampling doses and collecting the data analyses of infected samples. Indeed, the logic gates in the integration of nanobiosensors into smartphone or wearable handheld device would enable low technical experience for operation, simultaneous screening, and reliable optimization analyses, and would reduce the prerequisite number of tested samples to ensure accurate calibration and then uncertainty quantification of analytics (Scheme 1C). The reliability-based nanosensor device is an easy-to-use module compared with the inherent modularity of the current analysis use of microscopic tools with much computational programs [[104], [105], [106]]. The nanoscale architect-led materials fabricated with thermo-electric high-conductive sites, low-power consumption hybrid metal/nonmetal/metalloid mesocrystal, atomic building arrangement, morphological structure disciplines, hierarchy, and orientation can act as key components in the creation of biosensor design of a high-performance output signals and rapid biosensing platforms (Scheme 1D).
This review will contribute to a better understanding of the potential applicability of biosensor structural design, signaling optimization, tolerance analysis methods, and cost-effective solutions of real-time monitoring of a range of disease types. This review provides the broad applicability and prevailing manufacture of sensor devices for early stage detecting and real-time analyses of actively infectious agents that affect human health. The diversity and specificity control over nano-biosensor structural designs based on nanoscale structural performance functions in terms of building arrangement, morphological structure, and hierarchical orientation will be highlighted. Among all commonly used techniques, the newly-developed nanodynamic robust designs for showing their reliable, cost effective, rapid, highly specific, and sensitive methodology are highlighted. Additionally, this review focused on reliability-based optimization of the developed nanobiosensors that will decrease the required expenses and professional trainers and technicians in developing countries.
4. Nanomaterials-based sensors for early detection of hepatitis B virus & hepatitis C virus
With an increase in a global demand to control the infectious liver diseases, it has highly important to develop hepatitis B virus (HBV) and hepatitis C virus (HCV) biosensing design disciplines to accomplish analytical optimization of individual health records in numerical monitoring simulations, accurate and early quantification, low-cost and sensitive analyses, extending component life reliability, and improving the rapid medication period and performance. HBV and HCV are hepatotropic infections that share comparative pathways of transmission and results that progress into hepatic fibrosis and hepatocellular carcinoma (HCC). HBV and HCV are the main infectious indications for chronically infected liver disease globally [107,108]. HBV is classified as a hepadnaviridae member, and its DNA genome is composed of a duplicate virus called pregenomic RNA. This infectious virus has chronically infected roughly 257 million individuals [107]. By contrast, HCV is categorized as a Hepacivirus of the Flaviviridae members that chronically infected around 80 million individuals, as statistically recorded in 2017 [108]. Therefore, regular analysis and monitoring progress are essential for HBV or HCV treatment care cascade. Early detection of antiviral and antibody of HBV and HCV infections in highly effective, sensitive and specific, well-tolerated and rapid response detection and monitoring is essential to reduce the high global burden imposed by these viruses and to control liver diseases.
The nucleic acid test was directly used for HBV and HCV DNA viral sensory in terms of quantitation and detection over a wide dynamic range of viral recognition. The analysis on hepatitis B and C antigens, DNA levels, and antibodies can also offer direct and/or indirect recognition of end-stage HBV and HCV infection and vaccine-effective immunity. Yao et al. [109,110] reported the modification of a quartz crystal microbalance electrode using rolling circle amplification-based enzymatic process can be utilized for anti-HBV detection. The results indicating the high sensitivity for the HBV real-time detection of 8.6 pg/L, without using polymerase chain reaction (PCR) [110].
Electrochemical electrode sensors built-on various electrode materials, including ferrocene, cobalt phenanthroline, and methylene blue, and CuO2 hollow microspheres were designed and employed for the detection and recognition of HBV and HCV [[111], [112], [113], [114]]. Selective electrochemical electrode sensors built-on nanoporous Au electrode were used for detecting and recognizing HBV genome by Ahangar et al. [115]. In this report, ferrocene was used to connect with DNA derived from the patient's blood to produce its electrochemical signal. The variability of structural building-block platforms using of nanoporous Au was improved the HBV biosensor sensitivity, specificity, and identification of healthy or HBV-infected individuals. The building-blocks of Au/rGO/hepB1S-probe biosensor were fabricated by consequence biofunctionalization steps of (i) modification of Au-electrode by reduced graphene oxide (rGO) or Au NPs, and then (ii) immobilization of an ssDNA probe (named as hepB1S) into Au electrode-modified rGO or AuNPs, respectively [116]. This proposed building assays based on AuNPs provided a 0.15 ng/μL detection limit. Electrochemical electrode biosensors based on Zeolite nanocrystals and MWCNT were constructed to detect the PCR of HBV [117]. The practical advantages of this structural nanobiosensor design showed 50 copies per mL as the detection limit of HBV genome, and high stability under storage for 4 weeks.
To explore a low-cost monitoring/detection assay for hepatitis B and C test based laboratory tools, a quick test strip for hepatitis B screening was designed to test three major serotypes of HBV infection using Au-NPs to generate the signal with detection limits of 0.5–0.1 μM (Fig. 1A) [118]. Additionally, the coupling of N-ethyl-N′-(3-(dimethylamino)propyl)carbo-diimide/N-hydroxy succinimide (EDC/NHS) and Anti-HBs antibodies was used to modify the electrode surface for electrochemical HBV examination in the range of 0.005–3 μM and LOD of 2.1 nM (Fig. 1A) [118].
Fig. 1.
(A) Schematic design of a rapid test strip and an electrochemical assay for hepatitis B surface antigen (HBsAg), hepatitis B surface antibody (Anti-HBs) and hepatitis B core antibody (Anti-HBc) [118]. (B) The design of the DNA detection assay using copper nanoclusters (a), mechanism of the colorimetric detection of DNA using peroxidase-like material (b), UV–vis spectra for selective detection of polynucleotide (c), and the uptake percentage of different targets, Reproduced with permission [119]. (C) A schematic diagram of a waveguide-mode biosensor and the reflectance spectra analysis for detection of antigen–antibody complexes [120].
Mao et al. [119] reported the possibility of using economic biosensor platforms such as Cu-nanoclusters, for the naked-eye colorimetric detection approach of HBV DNA with high sensitivity and detection limit of 12 × 109 molecules (Fig. 1B). Shimizu et al. [120] has reported the assessment of anti hepatitis virus antibody detection through a waveguide-mode biosensor (Fig. 1C). This proposed sensing method confirmed that significant reflectance signal was detected due to the interaction between antigen–antibody and anti hepatitis virus. In addition, the use of peroxidase such as aminoethyl carbazole-enhanced the colorimetric detection of anti-HBs around 300 times.
Self-assembly of the ssDNA probe on Au NPs/SWCNTs nanocomposite arrays electrochemically prepared was used as a biosensor platform for electrochemical detection of HBV virus and papillomavirus based on an impedance spectroscopy approach with LOD of 0.1 pM (Fig. 2A) [121]. The hepatitis B surface antigen (HBsAg) detection was used as an important marker to assess the detection of HBV infection. In this context, the modification of Au NPs by monoclonal hepatitis B surface antibody (HBsAb) was used to build a biosensor for HBV detection based on the HBsAg screening in physiological saline in the range of 0.01–1 IU/mL (Fig. 2B) [122]. Ehsani et al. [161] reported a luminol chemiluminescent immunoassay (CLIA) system based on CuO nanoparticles and CuO/MWCNT nanocomposite as a catalyst for the detection of HBV with detection range of 3.5 nM - 2.5 μM and 2.2 nM - 5.0 μM & LOD of 1.8 and 0.85 ng/mL, respectively (Fig. 2C), [123].
Fig. 2.
(A) Field emission scanning electron microscopy (FE-SEM) analysis of the biosensor platform based on gold nanoparticle–singlewall carbon nanotube composites (Au NPs/SWCNTs) arrays used in the detection of HBV and papilloma virus [121]. (B) Schematic diagram for HBV biosensor construction based on AuNPs modifications in different matrices (a), transverse electron microscopy (TEM) micrographs Au NPs used as a platform in the biosensor construction (b), and the experiment design validation in a 96-well microplate by enzyme-linked immunosorbent assay (ELISA) (c) [122]. (C) SEM analysis of MWCNTs/CuO-NPs (a), MWCNTs/CuO-NPs–COOH (b), and Ab-MWCNTs/CuO-NPs conjugates (c), and the developed mechanism of Ab-MWCNT/CuONPs in the chemiluminescence immunoassay detection of HBV [123].
The engineering design of magnetic quantum dots (MQD) microspheres has been reported as a host method for magnetic separation and immunoassay analysis for HBsAg and HBeAg (HB antigens) by Wang et al. [124]. The modification of MQD microspheres with hepatitis B e-antigen (HBeAb), and fluorescent probe phycoerythrin (PE) provides a good tool for quantifying HBsAg, HBeAg molecules based on fluorescence intensity changes (Fig. 3A a,b). The results confirmed high sensitivity and separation performance of the designed sensor platform, as evidenced by the change in HBsAg concentration from 500 nm to 3 nM (Fig. 3Ac). For selective detection of HCV, a biosensor was designed using a nanocomposite of AgNPs and thiol graphene quantum dots (GQD-SH) as a substrate for loading HCV antibodies to engage the antigen for electrochemical HCV detection in a the range of 0.05 pM–60 nM, and LOD of 3 fM (Fig. 3B) [125].
Fig. 3.
(A) Photographs depicting the separation process of the targeted molecules using magnetic quantum dots (MQDs) in aqueous suspension solution under excitation UV wavelength of 365 nm (a); SEM and laser confocal fluorescence microscopy images of MQD microspheres; and schematic design illustrating the magnetic capturing of hepatitis B surface antigen (HBsAg) and hepatitis B e-antigen (HBeAg) using MQD microspheres (c) [124]. (B) Selective electrochemical detection of HCV using AgNPs and GQD-SH nanocomposite as a substrate for HCV antibody loading [125]. (C) Schematic design for the HCV assay based on the ssDNA/MB@SiNPs/FTO electrode, Reproduced with permission [127].
The building-blocks-led design-nano-biosensors based on γ- and ε-MnO2 NPs modified by the electrochemical pencil graphite electrode were used for detecting the ledipasvir (LED), which is an HCV antiviral drug within detection range and limit of 0.025–3.60 μM, 4.50 nM, respectively, [126]. The sensor design built-in NPs-dressed dye molecules provided evidence of a great prospective in the biosensor applications. The objective function of biosensor design built-in the incorporation of silica/MB dye molecule-grafted fluorine-doped tin oxide (FTO) electrodes was led to offer HCV detection assay, as reported by Singhal et al. [127] and shown in Fig. 3C. The design structures of MB@SiNPs/FTO electrode engineering and utilization of ssDNA were offered an optimization assay for the HCV detection in serum sample. Considerable sensing performance in terms of a wide range of detection (101–106 copies mL−1) and detection limit of around 90 copies mL−1 was provided by such sensing assay.
Moreover, the liver function disciplines have been widely adopted as blood receiver from other body organs, and as well as a pocket for drug metabolism and excretion. The main functional liver cells such as Kupffer astrocytes and hepatic stellate cells (HSCs) were usually considered for liver endocrine, metabolic and secretory functions, and for systematic responses to cirrhosis or cirrhosis [128]. Several relevant nanoscale biosensor formulations were recently developed for potential detection of liver diseases, and for their therapeutic decision making. In this regards, iron oxide NPs have been widely used for liver MRI (magnetic resonance imaging) with multicriteria optimization and sensitive analyses. The nanoscale structural design of concave octapod iron oxide NPs T2-negative associated with effective cores radii compared to real magnetization values was provided evidence of high sensitivity to MRI optimization analyses for early and accurate diagnosis of liver cancer, as reported by Zhao et al. [129]. In addition, the radioimmunoassay is considered as the best diagnostic indicator and sensitive test for cirrhosis in the path-dependent of early detection of liver injury. Fluorescence polarization immunoassay (FPIA) was developed for accurate detection of bile acids of cholyglycine, leading todiagnosis of liver disease [130]. Moreover, alanine aminotransferase enzyme has been used for development of sensitive electrochemical approach for early diagnosis and treatment of liver diseases [131]. Additionally, bound improvement of utilization of glutamate and alanine aminotransferase was found by their immunization into a design of highly sensitive nanocomposite-graphene biosensors [132]. The objective function of nanoscale architect-led building-blocks-in biosensor designs in early stage detection and then diagnostic imaging of liver diseases, and the evaluation of their merits in therapeutic decision making are summarized and shown in Table 1 [[133], [134], [135], [136], [137], [138], [139], [140], [141], [142], [143], [144], [145], [146], [147], [148], [149]].
Table 1.
Nanomaterials platform/carriers for diagnostic imaging and the liver carcinoma.
| Materials/Platform | Imaging Technique | Target | Ref | |
|---|---|---|---|---|
| Superparamagnetic iron oxide NPs | (30 nm) | Magnetic resonance imaging (MRI) | Liver and cancer of orthotopic HepG2 bearing mice | [133] |
| (20–250 nm) | Liver lesions of human | [134] | ||
| (10–100 nm) | Ischemia-reperfusion liver of Wistar rats | [135] | ||
| (50 nm) | Fibrosed liver of Wistar rats | [136] | ||
| (15 nm) | Liver cancer of VX2 rabbit bearing orthotopic N1S1 tumor | [137] | ||
| Gd2O3/SPION (14 nm) | Liver of BALB/c mice and cancer of HepG2 orthotopic tumor of nude mice | [138] | ||
| Gd-doped silica and gold (30 nm) | Cancer of BALB/c mice with colon cancer liver metastasis | [139] | ||
| Mn–SiO2 (25 nm) | Cancer of HepG2 orthotopic tumor mice | [140] | ||
| PEGylated Yb2O3:Er (170 nm) | Computed tomography (CT) | Liver of rats | [141] | |
| Oleic acid coated Bi2S3 nanodots (2–3 nm) | Liver of rats | [142] | ||
| ExiTron nano (110) | Liver and metastasis of mice with colon cancer liver metastasis | [143] | ||
| Lanthanide-doped Lu2O3 (115 nm) | UCLa/MRI/CT | Liver of C57BL/6 mice and rats | [144] | |
| Quantum dots linking to AFP antibody (4 nm) | Two-photon imaging technology | Cancer of nude mice | [145] | |
| Liposome (83.5–86.9 nm) | In-vivo fluorescence image system | Hepatic stellate cells of thioacetamide rats with liver fibrosis | [146] | |
| Nanogel Galactosylated (130 nm) | Doxorubicin (Drug) | Cancer of diethylnitrosamine-induced hepatocellular carcinoma rats | [147] | |
| Nanofiber (RGD peptide 10–20 nm) | Curcumin (Drug) | Cancer of mice bearing orthotopic transplant tumor | [148] | |
| Silica nanorattle (125 nm) | Docetaxel (drug) | Cancer of subcutaneous H22 bearing mice | [149] | |
Up-conversion luminescence (UCL); Gd2O3: gadolinium (III) oxide; HCC: hepatocellular carcinoma; Yb2O3: ytterbium (III) oxide; Bi2S3: bismuth (III) sulfide.
To date, given the inadequacy loophole and the complexity of hepatitis B and C tests particularly at the early stage infection, variable nanobiosensor designs should be proposed for effective, continuous monitoring HBV- and HCV-assays. Accurate HBV and HCV monitoring assays with rapid control and proficiency analysis are necessary to avoid false-negative monitoring results and to prevent transmission of the HBV and HCV infections for non-immune contacts and household partners. Therefore, early identification of individuals with chronic hepatitis B and C viral infection would enable on-time, effective point-of-care treatment to prevent or inhibit the onset of liver disease. Considering the low rate and poor quality of hepatitis test, and the high cost and complexity in diagnostics, the treatment of hepatitis B and C infection remains unattainable for most of the affected individuals in need. We expected that nanotailored hepatitis B- and hepatitis C-biosensor dynamics with quality control monitoring would help clarify to promote primary detection in a short delivery model, accurately discriminate active infection from inactive, and monitor the effectiveness response to HCV or HBV antiviral therapies.
5. Biosensor design for the early monitoring of cardiovascular diseases (CVDs)
Cardiovascular disease (CVD) is the leading cause of mortality rate among all diseases globally. Thus, designing biosensor-based devices is crucial for the successful monitoring of CVD and prognosis of stroke [[150], [151], [152]]. For an effective heart disease prediction, control over a cardiac-specific biosensing assay of cardiac troponin I (cTnI) is substantially necessary. However, troponin-based types of T, C, and I are proteins integrally generated in the skinny cardiac muscle fibers enabling cardiac muscle contraction. The concentration level of cardiac-specific troponin I or C in blood sample may identify the feasibly physiological symptom of individuals [[150], [151], [152]]. Generally, the quantities of troponin-based types in the blood are hardly detectable. The increase in the troponin quantities in blood is associated with the elevated level of cardiac muscle damage. In this context, developing vital, trustworthy, responsible, and economical biosensors with robust sensing and recognition strategy for the early detection of elevated level of cardiac-specific troponin I or C in blood sample is needed to avoid CVD.
Zhou et al. [153] designed an electrochemical nanobiomarker for the cTnI detection by using the sandwich-type Mn-doped CeO2 NPs, which were modified by Pt NPs (Pt@Mn–CeO2NPs), and for hemin/G-quadruplex (Fig. 4). The mechanistic approach was based on the production of H2O2 by l-cysteine, followed by the eletrocatalytic decomposition with hemin/G-quadruplex and Pt@Mn–CeO2 NPs (Fig. 4). Changes of the hemin electrochemical signal were used to determine the cTnI level that reflected the value of cardiovascular therapeutic. The Pt@Mn–CeO2 NPs electrode displayed a detection range of 0.5 pg/mL to 100 ng/mL and a low detection limit (0.15 pg/mL). AuNP-modified poly (dimethylsiloxane) (PDMS) producing a PDMS-AuNP nanocomposite film was used for cTnI colorimetric biosensor (Fig. 4B) [154]. Remote recognition monitoring based on PDMS-AuNP nanocomposite can be achieved through the connection between nanocomposite films with image digital transmission. In this context, cTnI optical immunoassay of cardiac troponin I based on fluoro-microbead guiding chip (FMGC) was investigated by Song et al. [155]. The cTnI-antibody was combined with the FMGC surface through the interaction with -3-dithiobis-propionic acid N-hydroxysuccinimide ester. Conventional fluorescence microscopy was employed to detect the concentrations of cTnI on the basis of the beads under immobilized counting on the FMGC. Moreover, cTnI has a wide range of detection in plasma samples (0.1–100 ng/mL).
Fig. 4.
Schematic design of the cardiovascular biomarker detection based on Pt@Mn–CeO2 3DN (A); and strategy for cardiovascular detection based on the silver enhancement (B), Reproduced with permission [153,154].
Nanoporous carbon nanofibers (CNFs) are designed through the pyrolysis process of the carbonized polyacrylonitrile (PAN)/AgNO3 blend with a diameter range of 130–190 nm, and around 30 nm of Ag NPs (Fig. 5 Aii-vii). The CVD biosensor was designed on the basis of a functional covalent of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide with the synthesized material that electronically modified ITO electrodes to detect triglycerides in the range of 25–500 mg/dL (Fig. 5 Ai) [156]. In addition, triacylglycerides can be detected with good sensitivity of 0.5 nm/mM and LOD of 17.71 mg/dl using long period fiber grating (LPG) with the biorecognition layer (Fig. 5B) [157]. 3D assembled ZnO@ZnS(ZB- wurtzite) and ZnO@ZnSWZ nanoheterostructure have nanorod and nanotubes shapes have been synthesized for cholesterol biosensing performance [196]. The biosensor based on ZnSZB nanotube and ZnO@ZnSZB nanoheterostructure show a wide range of detection with LOD of 0.44, and 0.08 mM, respectively (Fig. 5C), [158].
Fig. 5.
(A) Schematic design of the CVD biosensor construction using Nanoporous CNFs that prepared from the carbonization of PAN/AgNO3 (i), and the SEM and TEM images of the CNF materials (ii-vii) [156]. (B) LPG biosensor design for the triacylglycerides detection [157]. (C) Schematic design of cholesterol biosensor performance based on 3D assembled ZnO@ZnSZB and ZnO@ZnSWZ nanoheterostructure, and ZnSZB [158].
Larval zebrafish (Danio rerio), a 'vertebrate model organism,' can be used for the analysis of physiological factors including cardiovascular activity; this step is one of the striking strategies for detecting preclinical drug effects [159]. A fast and non-invasive pharmacologically induced assessment has been performed through quantification video stream analysis of the real-time imaging micro-echocardiography (iμEC) system for the temporal cardiac patterns (Fig. 6) [160]. Lab-on-a-chip iμEC system was used to determine the cardiovascular activity in the Zf larvae; a noteworthy benefit of this system is the video analysis of the heartbeat that can be possibly achieved simultaneously. Immobilization of micro-RNA as a probe on the Au electrode was used for CVD detection on the basis of electrical double layer (EDL)-gated aluminium gallium nitride (AlGaN)/gallium nitride (GaN) high electron mobility transistor [161]. The targeted biomolecule was hybridized with the micro-RNA 'miR −126, miR-21, and miR-208a' sensor, with a low detection limit of 1 fM. Significantly, for protective health activities, and remote, regular checkup and continuous monitoring of chronic heart disorders, wearable CVD biosensors are essential for the futuristic control of diseases and sudden heart attacks and strokes. The integration of innovative smart CVD touchless and wireless biosensor devices with reliable telemedicine sensing assay through Bluetooth or Wi-Fi would generate high-efficiency health record data, precise health monitoring, and medical and pharmaceutical database storages. The CVD sensor device enabled to create an in-depth biosensing assay with highly predictive analytics, early identification for rapidly initiative medication period [162].
Fig. 6.
Lab-on-a-Chip imaging micro-echocardiography (iμEC) system for assessment of cardiovascular activity in zebrafish larvae (A–C); D) Validation of the applicability of hydrodynamic entrapment of larval fish for cardiological analysis, Reproduced with permission [160].
6. Early-evaluation of blood sugar level for monitoring of diabetes
One of the critical medical issues associated with high blood glucose level is the metabolic disorder called diabetes, which is a silent killer. According to the International Diabetes Federation (IDF), more than 400 million people worldwide suffer from diabetes, leading to death [163]. Diabetes is usually associated with poorly controlled metabolisms, leading to heart attack, blindness, nerve damage for cognitive disability, and kidney failure. Early blood glucose level detection is considered the key diagnostic factor for investigation/follow-up and control to sustain the diabetes level in the human body [[164], [165], [166], [167]].
In general, glucose detection in the blood is considered one of the guaranteed tools in managing diabetes [168,169]. To avoid premature mortality and understand the trends in diabetes prevalence, a series of cost-effective analyses and detection methodologies for blood glucose level is particularly needed for real blood glucose test and primary health-care systems. Among a group of analytic sensory, these optical glucose biosensor techniques, such as absorptiometry or reflectometry, and fluorescence- and surface plasmon resonance (SPR)-based biosensors were perceived overall quality device for blood glucose level analysis. Addressing a key reaction mechanism leads to the generation of the optical signal output including the following [170,171]:
-
(i)
Change in the place of the intrinsic spectral positions (absorbance or fluorescence) as a result in interaction between the enzyme/or co-enzyme;
-
(ii)
Consumption or formation of enzyme metabolites possibly due to glucose oxidase (GOx). This process can be formed via: (1) the oxygen consumption; (2) the hydrogen peroxide (H2O2) produced; or (3) the acid produced in the reaction;
-
(iii)
Screening the H2O2 formation in the enzymatic reaction;
-
(iv)
Interaction of boronic acid with saccharides, such as glucose; and
-
(v)
Competitive binding of concanavalin A with glucose, which can label the carbohydrates, such as dextran and glycated protein.
Sanz et al. [171] reported that the glucose biosensor can be built on the combination of GOx–horseradish peroxidase (HRP). The mechanistic monitoring was based on the change in the ordinary spectrum of HRP (424) within glucose exposure as a result of the oxidation of the heme group in HRP by H2O2 (the byproduct of GOx and glucose reaction). The designed glucose biosensor produced by capturing HRP and GOx in a polyacrylamide gel surface exhibited long term stability and wide range of 0.1–300 mM.
Modification of the optical fiber biosensor by using poly (phenylboronic acid) (polyPBA) film onto the concentric area along the coverage fiber surface was enhanced the glucose detection in the range of 0–60 mM, displaying good reproducibility and stability in human serum [172]. Furthermore, modification of graphene QDs by phenylboronic acid receptor was employed as a non-enzymatic glucose optical sensor with a linear detection range of 4–40 mM and low detection limit of approximately 3.0 mM [173]. The glucose detection was associated with the determination of dissolved oxygen content reduction; however, the fluorescence intensity of the oxygen-sensitive membrane can be determined with changes in the glucose concentrations [174]. This glucose biosensing assay showed potential to determine effectively the glucose content in a beverage sample. Platinum nanoparticles (Pt NPs) were fabricated directly on the eggshell surface membrane, and then the GOx was simultaneously immobilized into Pt NPs/eggshell membrane to yield for glucose-sensing assay [175]. The glucose detection mechanism occurred on the base of reduction of the dissolved oxygen content. This glucose PtNPs/eggshell biosensor provided fast response time and detection limit and range of 5 and 10–225 μM, respectively.
Chemiluminescence (CL) flow glucose biosensor was fabricated by bi-enzyme networks. The real building-block engineering of CL biosensor was designated through three steps. First, silica sol–gel was dressed with glucose oxidase GOx and HRP and then incorporated into the surface of a clear quartz tube. Second, the luminol-hybrid lactate dehydrogenase (LDH) hybrid Mg–Al–CO3 LDHs were packed in the second half inside the clear quartz tube surface. Third, both bi-enzyme designs were made adjacent to a photomutiplier tube (Fig. 7) [176]. The mechanistic CL biosensor design was based on the enzymatic oxidation of glucose followed with luminol oxidation via the generated hydrogen peroxide (H2O2) to form a CL emission in the HRP presence [177]. An efficient CL glucose biosensor associated with the H2O2 enzymatically generated was fabricated by GOx immobilization on the Fe3O4–chitosan nanoparticle support by using glutaraldehyde as a cross-linking agent. Furthermore, the gold NPs enhanced the catalytic oxidation of luminol by H2O2 generated from the glucose-enzymatic reaction in the detection range and limit of 0.01–85 μM, and 43 μM, respectively (Fig. 8A) [178]. Importantly, the catalytic function of AuNPs on a luminol–H2O2 CL was significantly enhanced after AuNP crosslinking aggregation induced by (i) immunoreaction and (ii) the addition of appropriate antigen (Ag). A design performance via immunoreaction was played an important role in the effective aggregation of antibody (Ab)-modified AuNPs, leading to the enhancement of the catalytic CL reaction luminol–H2O2 and then the optimum sensing performance [179]. Building-blocks design of GOx/CNTs/AuNPs into Nafion film that supported the graphite surface offered an effective immobilization route for glucose CL biosensor [180]. Yu et al. [181] reported a triboelectrical nanogenerator (TENG) assembled along the ZnO nanowires (NWs). Compared with conventional biosensor design methodologies, this TENG design enhanced the self-powered glucose monitoring system (Fig. 8B).
Fig. 7.
Schematic design of chemiluminescence (CL) flow glucose biosensor fabricated by bienzyme network. The platform was formed using bienzyme and luminol-hybrid Mg–Al–CO3 LDHs. (A) The biosensing procedure of the glucose biosensor. (A-right & A-left down) the photo and schematic control of CL flow-through biosensing device, respectively. The mechanistic glucose biosensing process via an enzymatic oxidation of glucose using GOx, followed with luminol oxidation via H2O2 generated to yield CL emission in the presence of HRP (B), Reproduced with permission [176].
Fig. 8.
(A) Schematic design display the mechanism of glucose sensor using of the glucose oxidase (GOx) on the Fe3O4–chitosan nanoparticles for catalytic oxidation of glucose producing the H2O2 enzymatically generated. Furthermore, the Au NPs enhanced the catalytic oxidation of luminol via H2O2, Reproduced with permission [178]. (B) Schematic decoration of ZnO NW by GOx that surrounded by glucose under experimental conditions of i) no strain, and ii) compressive strain; iii) Scanning electron microscope (SEM) of low- and high-magnification of ZnO NWs; iv) I–V signaling of ZnO NWs-glucose biosensor, and the inset photos show digital and optical microscopy, respectively; v) I–t signaling of ZnO NWs-glucose biosensor as function of glucose concentrations, (v)-inset) The experimental set-up system [181].
Unlike some other sensor designs, there is considerable interest in high-throughput screening (HTS) and identification of diabetes [[182], [183], [184], [185]]. On the base of reliability design and optimization, the HTS manufacture campaigns were also considered in a therapeutic sector for diabetes. In this regard, the crypto-insulin reporter system was developed using a hybrid fluorescence polarization (FP)/internal Förster resonant energy transfer (FRET) biosensor. This robust structural biosensor design was built by using preproinsulin-mCherry protein (PPI-mCherry) and fluorescent protein (PPI-mCherry) for insulin dynamics checking in extreme events of pancreatic β-cells. Accordingly, the HTS optimization in scale-up industrial forming process would lead to generate 1782 FDA-approved design. The overall Z'factor of the optimized design fulfilled the requirement for HTS biosensor reliability [182]. On the basis of numerical HTS performance design, the HTS test variability would be considered as an effective tool for real identification and detection of other therapeutic agents in the way of the improvement of deterministic designs for the treatment of diabetes, obesity and cancer [185].
Wearable sensor devices are currently inevitable to regulate the analysis of the elevated level of blood glucose among people living with diabetes that required daily screening management. This biosensor design showed possibility of long-term management and superior blood sugar control and reduced the hypoglycemic episode frequency, which postponed the onset of diabetes complications [186]. Bandodkar et al. [187] reported the possibility of using flexible tattoo as an easy-to-wear platform for building an epidermal sensor device as a non-invasive glucose sensor (Fig. 9). The tattoo-based iontophoresis sensor showed effective diabetes monitoring with possibility to be applied for other physiologically non-invasive detection. This sensor design disciplines feasibly offered real possibility to be used as chemical markers for transcutaneous drug delivery. Continuous non-invasive glucose detection has been utilized without an external potentiostatic control by using carbon-paste-modified hollow microneedle-based self-powered biofuel cell (Fig. 10) [188,189]. The microneedle lattice was mechanically labeled, depicted, and developed to offer easy entrance to the human skin under normal pressure. This biodetector device generally can significantly determine the elevated levels of blood glucose and detect lactate and theophylline analytes. The device was utilized to gauge metabolites to help medications and biomarkers and to assume an imperative role in the chronic disease management [188,189].
Fig. 9.
(A, B) Systematic design of flexible tattoo as an easy-to-wear platform for building an epidermal diagnostic device as a non-invasive glucose sensor. (C) Mechanistic cycling and reaction of glucose during the real time monitoring of portable sensor device, Reproduced with permission [187].
Fig. 10.
A) Bioanode/cathode-based carbon electrode mates modified on hollow microneedle-based self-powered biofuel-cell. B) Configuration of carbon-fiber-metalized biofuel cell microneedles (a), and the power density curve as a function of glucose concentration (10–80 mM), Reproduced with permission [188]. C) The polymer (polycarbonate) microneedle arrays that sputtered with 50-nm platinum (a), microneedle micrographs recorded by scanning electrochemical microscope (b & c), optical coherence tomographic image (d), and a row of the microneedles entrenched in the tissue and recorded in 3D plugin projection (e), Reproduced with permission [189].
Importantly, in advanced diabetic assay, the biosensor design was used to detect the level of acetone in human respiration as a results of the relationship between the acetone level and abnormal concentrations of blood glucose level, as reported by Guo et al. [190]. In addition, the analysis of tear glucose was used as an alternative and a non-invasive blood–glucose detector. Further investigation on the detection of the tear glucose concentrations in anesthetized rabbits showed noteworthy correlation between the glucose level in the tear and blood, indicating the precision of electrochemical sensor-based tear glucose determinations [191]. A microflow injection analysis connected with passive pumping may show evidence as a low-cost technique in conducting tear glucose analyses in a wide linear detection range and limit of 0.075–7.5 mmol/L and 22.2 μmol/L, respectively, due to the cost effect of the tear glucose analyzers engineering [192].
For the daily life monitoring of diabetes, the internet of things (IoT) connected from platform devices offer a practical solution for diabetic retinopathy, promote the mobility and privacy of users, and deliver self-regulating or real-time-controllable detection analysis [[193], [194], [195], [196]]. Smartphone-based colorimetric analysis systems were designated for point-of-care blood glucose monitoring applications and for project image improvement via direct focusing the light from the light-emitting diode of a smartphone and in perfectly diffused light conditions (Fig. 11) as reported by Chen et al. [194]. Gold-ZnO modified thin-film electrode was fabricated on a flexible substrate to detect alcohol and real-time monitoring of glucose in sweat samples. The biosensor design offered detection ranges of alcohol and glucose of 0.01–100 and 0.01–50 mg/dL, respectively. A wearable glucose sensor with biocompatible features based on modified flexible stainless steel has been developed by Yoon et al. [197].
Fig. 11.
Visual analysis systems that controlled with smartphones for point-of-care blood glucose monitoring applications; a) smartphone captured images and (b) corresponding gray intensity plot profile; c) smartphone captured glucose testing on paper strips, Reproduced with permission [194].
7. Nanostructured-based platform for human immunodeficiency virus (HIV) detection
HIV could effectively spread through weakening body fluid functions and attacking the immune system, particularly CD4 lymphocyte or T helper cells, causing acquired immune deficiency syndrome (AIDS). Typically, HIV has two main categories: HIV-1, the globally known virus; and HIV-2, a less dominant and pathogenic category [[198], [199], [200], [201]]. For instance, 39 million deaths were mounted because of HIV/AIDS virus since its discovery. In addition, 1–2 million individuals with HIV-2 infection were reported [200]. To date, no effective medication is available for this illness; early detection and specific monitoring assay of HIV infection are an essential aspect to treat AIDS [202].
For selective and sensitive monitoring of HIV-1 gene recognition in the range of 3.0 fM to 0.3 nM with a detection limit of 0.3 fM, europium sulfide nanocrystals (EuS NCs) as signal-producing compounds were applied on the biosensor building-blocks designs based on molecularly imprinted polymer electrochemiluminescence (MIP-ECL) [203]. The EuS NC luminophore was highly sensitive and specific, and the signaling assay was stable, leading to the development of MIP-ECL sensors for wide-range recognition of DNA biomarkers. In addition, MIP electrochemical sensor was designed through the surface building block of multiwalled carbon nanotubes (MWCNTs) and polyacrylic acid (PAA) onto a glassy carbon electrode for fast, simple, and sensitive HIV-p24 detection Fig. 12C [204]. The designed electrode showed evidence of wide detection range and limit of 1.0 × 10−4 ng/cm3 to 2 ng/ cm3 and of 0.083 pg/ cm3, respectively.
Fig. 12.
(A) Schematic design of the Graphene-based materials nanoscale sensor platform for HIV detection and CVD diseases [205]. (B) HIV detection based on PC biosensor microplate wells nanostructured subwavelength coated with TiO2, with surface modification by different biomolecules including functionalization process, including 3-mercaptopropyltrimethoxysilane (3-MPS), N-gamma-Maleimidobutyryl-oxysuccinimide ester (GMBS), NeutrAvidin, anti-gp120 and bovine serum albumin (BSA) [206]. (C) Construction of an electrochemical biosensor using modified GCE with MWCNTs, biological molecules, and polyacrylamide for the of HIV-p24 determination in human serum [204].
Graphene-based materials were used to develop a nanoscale sensor platform for HIV detection and associated diseases [205]. Effective amino graphene modification with anti-p24 and antitcardiac troponin 1 (anti-cTn1) were used for electrochemical detection of HIV, and cardiovascular diseases, respectively, in the range of 1 fg/mL to 1 μg/mL, and LOD of 100 fg for HIV, and 10 fg/mL for CVD Fig. 12A [205]. Shafiee ei al [206]. reported the possibility of quantifying HIV from biological samples including serum and PBS samples in the range of 104–108 copies/mL using nanostructured photonic crystals (PC) biosensor microplate wells coated with TiO2 and modified by immobilized biomolecular layers Fig. 12B [206]. A label-free, liquid-ion gate for building field-effect transistor (FET) immunosensor approach using p-type NiO-modified single-stranded DNA (ssDNA) thin film was used for the biorecognition of HIV-1 Fig. 13A [207]. The proposed biosensor exhibited high sensitive HIV/DNA recognition at a wide-range linearity of 1.0 nM–10.0 nM and a detection limit up to 0.3 nM. Moreover, HIV in saliva and leukocytosis in physiological samples has been successfully detected by using biosensor platform of magneto-nanosensor array smartphone Fig. 13B [208].
Fig. 13.
(A) Schematic design of the A label-free, liquid-ion gate for building field-effect transistor (FET) immunosensor approach (i), which fabricated via modification of p-type nickel oxide (NiO) with single strand DNA (ssDNA) thin film for biorecognition of HIV-1/DNA (ii) [207]. (B) Magneto-nanosensor array smartphone for HIV detection in physiological samples [208]. (C) Schematic diagrams detailing the fabrication steps and performance of amperometric immunosensor that can be fabricated by anti-p24 Ab immobilization into the Au-electrode-modified AuNPs, MWCNTs, and an acetone-extracted propolis (AEP) film membrane for sensitive detection of HIV-1 p24 Ag [215].
For the object to provide a means for easily detecting of HIV-1 DNA, Long et al. [209] reported a colorimetric detection assay of HIV DNA biomarker with high sensitivity using multi-amplification nanofibrous sensing membrane Fig. 14A. The surface modification of nanofibrous membrane platform using the protein and DNA identification probes enhance the performance of the optical and regenerative sensing properties. Pan et al. [210] reported the fabrication of sensitive and selective fluorescent sensor for the ultra-trace detection (1.2 nmol/L) of HIV-1 DNA. This biosensor design was successfully built by using zinc-methylimidazolate framework-8 as a platform and ssDNA as a surface probe modifier (Fig. 14 B-i). The optimal design was led to the use of different metal-organic frameworks (MOFs) in the early detection and real-time monitoring of HIV-1 DNA target. In the same context, novel Zn(II)-MOFs were modified by FAM and then labeled probe ssDNA as a fluorescence quenching, providing potential sensing system for HIV double-stranded DNA (ds-DNA) detection, (Fig. 14B-ii) [211]. Sun et al. [212] reported the HIV-1 fluorescence recovery that was fabricated by modifying water-stable polymers such as zwitterionic poly-zinc-carboxylate with 6-carboxyfluorescein (6-FAM)-labeled ss-DNA probe (signified as P-DNA). The biosensor design confirmed that the fluorometric sensing approach of HIV-1 ds-DNA, and the fluorescence recovery process were a concentration-dependent assay with a linear range of 0–60 nM and a detection limit of 7.4 nM.
Fig. 14.
(A) Optical HIV biosensor based on the multi-amplification nanofibrous sensing membrane platform [209]. (B) Schematic design of the fluorescence mechanism of the HIV target based on ZIF-8 for the fluorescence recovery efficiency of P-DNA@ZIF-8 (A). Representation of the fluorescence mechanism of the HIV target based on Zn-MOFs fluorescence design (B-a), and the fluorescence performance in terms of simple recognition, selectivity and sensitivity B (b-c), Reproduced with permission [210,211].
Immobilization of an antibody fragment into Au NPs and then on the (indium tin oxide) ITO electrode surface by a self-assembly process-assisted gold–thiol interaction was applied for the electrochemical detection of HIV-1 [213]. The designed electrodes were tested against diverse HIV-1 concentrations. The biosensor design provided clear evidence about the successful detection of HIV-1 in the range of 600 fg/mL to 375 pg/mL. A chitosan/Fe3O4 nanocomposite was used as an electrochemical platform for HIV-1 detection [214]. Kheiri et al. [215] reported that the amperometric immunosensor was fabricated by p24 antibody (anti-p24 Ab) immobilization into the Au-electrode-modified Au NPs, MWCNTs, and an acetone-extracted propolis film membrane, leading to the sensitive detection of p24 antigen from HIV-1, as shown in Fig. 13C. The designed immunosensor showed a wide range of sensitivity responses of 0.01–60.00 ng/mL and a detection limit of 0.0064 ng/mL.
Toward the ultrasensitive recognition of HIV, a controlled design of multiple building-blocks sensor has been implemented experimentally. For instance, an impedimetric HIV-1 gene biosensor was fabricated following this sequence of (i) the impregnation of ssDNA into a glassy carbon-surface electrode, and then modification of electrode by graphene–Nafion film composites. The proposed sensing design optimization enabled the HIV detection in the range of 1.0 × 10−13 M to 1.0 × 10−10 M and detection limit up to 2.3 × 10−14 M [216]. Furthermore, non-faradic electrochemical sensor was fabricated onto an electrode surface by using biotin in 5′ position-binding streptavidin that decorated magnetic NPs. This deterministic structural design offered a sensitive detection assay of 50 and 160 pmol hepatitis B virus (HBV) DNA and HIV DNA, respectively [217]. The design integration of toehold-mediated strand displacement reaction/cruciform DNA crystal enabled fabrication of ultrasensitive biochemical sensor. The processing optimization was used to quantify HIV-related DNA in the range of 1 pM–100 nM and in a detection limit of 0.21 pM [218]. Silica NP-enhanced dynamic-spin microcantilever was used for the picomolar detection level of HBV biosensing assay that targeted DNA of 243-mer nucleotides [219]. Zhu et al. [220] established a fluorescent-sensing approach in which nicking enzyme signal amplification associated with triplex formation can be used for a particular HIV dsDNA detection within the range of 100 pM–200 nM and up to 65 pM level. In the context of improving sensing performance for HBV gene detection, quantum dot-DNA (QD-DNA) has been fabricated by modifying CdSe/ZnS QDs with 3-MPA, DNA targets, and Cy5-modified surfaces [221].
8. Wearable biosensor devices for touchscreen monitoring of HIV-transmitted infections
Design improvements for daily routine monitoring of HIV level are rather considered if the HIV sensing system could be modulated into wearable and compact devices. The optimization and integration of HIV optical biosensor devices into mobile phones is an area of interest to target for HIV patients. A selective visualization assay has been achieved through impregnating an antibody (for p24 antigen) on the interior surface of plastic microchips associated with micro-pit arrays (μPACs). The chipset arrays were designed by using transparent copolymer sheets of cyclic olefin (COC), as shown in (Fig. 15) [222]. Toward the accurate analysis for HIV-positive individuals, the deterministic data information on the screen of mobile phone was transferred to a server for read-out and quantitative analysis. To fulfill the requirement of both improving finite analysis and minimizing its uncertainty and variability under daily observance, constraint conditions for building effective designs that can offer updated, diverse HIV immunosensor technology for point-of-care, real-time, and rapid HIV detection laboratory should be solved by methodological optimization.
Fig. 15.
(A) Schematic design of HIV optical biosensor into mobile phone through the impregnation of antibody (for p24 antigen) on the interior surface of plastic micro-chips associated with micro-pit array (μPACs). (B) For specific determination of proteins, pixel intensities were recorded along the μPAC, Reproduced with permission [222].
For future optimization and miniaturization of industrial sensor design for touchscreen monitoring of HIV-transmitted infections, deformation path-independent process throughout all stages from fashioning sensor device to its modulation into smartphone should be considered. Along with all dynamic biosensor designs for HIV detection, development of fast and reliable point-of-care biosensors with suitable detection tools that could significantly affect HIV-transmitted infections is challenging. Additionally, developing micro- and nanoscale devices to obtain daily point-of-care sensing and managements by HIV-infected individuals in an hourly response order remains a major concern. A personal, touchscreen monitoring sensor-built-in nanodevice, one of the latest advances in detection tools, can potentially fulfill the premise of HIV detection in real-time and more accurate readings. The wearable point-of-care nanodevices may provide authentic means of HIV infection data collection and monitoring of survivals in an easily deliverable, highly sensible and specific, and graphical inspection to professionals for simultaneously engaged investigation and rapid decision-based therapy.
9. Early-stage detection of central nervous system disorders (CNS)
To date, building-blocks engineering of early stage detection and monitoring assays for neurochemical signals associated with turbulent neuronal disorders has gained an ever increasing significance. The CNS disorders usually associated with a number of diseases such as, Alzheimer's disease (AD), Parkinson's disease (PD), stroke, and epilepsy. Statistically significant difference indicates that the damage rate in CNS has dramatically increased in world's top aging society, creating a new value in the fields of monitoring, diagnosis, and treatment along with continuous optimization and progress of new structural sensor design technologies. More crucial efforts for nanoscale architect-led building-blocks-in sensor optimization problems of early stage detection/monitoring of neurochemical signals related to dementia or AD at different stages of the service life-age cycles are needed to minimize the uncontrollable variations and constraints related to CNS disorders [[223], [224], [225]].
For an engineering sensor perspective, well-designed development of monitoring methodologies for screen diagnosed CNS disorders, diseases at early stages and before the onset of symptoms that are usually seen in patients with neurological illness is a noteworthy challenge. Due to the turbulent balance associated with the development of brain disorders, neurochemical signals-generated in the brain are of primary concern to deterministic life-stages of CNS disorders [[226], [227], [228]]. Neuron silicon electrodes were applied to detect neurochemical signals in live animal cell lines of CNS [[226], [227], [228], [229], [230]].
An optimal design of AD biomarker detection assay is critical for the early diagnosis of AD diseases. A wide variety of nanoscale architect-led building-blocks-in engineering designs for biomarker monitoring of accumulated Aβ (amyloid beta) peptide molecules are key broadening the effective and early stage detection of AD pathophysiology [231,232]. The surface modification of NPs with high-affinity compounds has great potential to sensitive and accurate detection of various peptide molecules such as Aβ-binding peptides, leading to fast diagnosis of AD diseases. Demeritte et al. [231] reported that the generation of the plasmon coupling along with the core–shell plasmonic NPs and graphene oxide (GO) surface interfaces was enhanced the SERS platform sensitivity of AD biomarker design during the detection of Aβ molecule with a limit of detection (LOD) of 100 fg/mL. A significant progress in the optimal amyloid β 42 detection designs was reported by Kim et al. [232]. The buildup sensor design was based on integration of superparamagnetic iron oxide NPs in planar microfluidic channels and used to identify as a low concentration level of amyloid β 42 to 23.8 pg/mL.
For Parkinson's disease 'PD' sensory-physical activity formulation, the design optimization of the inertial measurement unit (IMU) was tested by Hsu et al. [233]. These well-developed sensory approaches were based on the fabrication of multiple wearable sensors, leading to daily monitoring analyses and reliable indentation of gait abnormalities, and then classification stages of patient's symptoms with neurological disorders. A possible device approach for triaxial accelerometer-based body-worn monitors (BWM) has been reported by Din et al. [234]. A set of reliability experiments of BWM design offered evidence of deterministic PD optimization with the following:
-
(i)
Reproduction of 14 clinically relevant essential gait characteristics,
-
(ii)
Highly sensitive detection of slight changes in gait pattern for both aging and pathology, and
-
(iii)
Possibility distribution of an equipped corridor with systematic values for general movement control and evaluation stages of PD diseases.
With an increasing demand for more control monitoring and rational approaches of analytical assays and designs, still it becomes highly important to acquire an optimal sensor design for sensitive signaling of neural activity in the brain, and for early stage diagnosis of CNS disorders.
10. Nanobiosensor response towards early stage detection of common diseases
A wide range of detection models with cancerous diseases have been estimated and specified for clinical use in the past few years worldwide. With increases in disease-specific mortality and morbidity rate and the limitation of cancer monitoring, early detection, monitoring, and regulatory decision-making treatment of cancer are crucial in the subsequent control of effective, real-time therapy. In medical use system, evidence-based, cost-effective, and affordable biomarkers were applied to express the changes in the portions correlated to risk or progression of cancerous diseases in tissues or in the blood. Therefore, a fabrication design of an effective cancer biomarker or biosensor that can be clinically sensitive and specific to cells, enzymes, genes, and hormones is of particular interest for early detection [[235], [236], [237], [238], [239]]. Tumor markers, such as carcinoembryonic antigen (CEA), and hepatocellular carcinoma α-fetoprotein (AFP), were detected a wide range of cancer types affecting the colon, lung, ovarian, and breast cells (Table 2) [[235], [236], [237], [238], [239], [240], [241], [242], [243], [244], [245], [246], [247], [248], [249], [250], [251], [252], [253], [254], [255], [256], [257], [258], [259], [260], [261], [262], [263], [264], [265], [266], [267], [268], [269], [270], [271]]. Nanodevice-based graphene- or CNT-modified paper platforms enabled meticulous and reliable monitoring of tumor-associated antigens (CEA) quantification during the cancer progression and assigned early detection and screening of unmarked cancer metastases [[235], [236], [237]].
Table 2.
Nanostructured materials-based for cancer screening.
| Materials | Target | detection limit | Ref |
|---|---|---|---|
| AuNPs/SsDNA/HRP | Optical immunoassay of CEA | 12 ng/L | [237] |
| QDs | 20 ng/L | [238] | |
| CdTe/CdS QDs/antibodies (goat antimouse IgG) | 250 fM | [239] | |
| 3D microfluidic paper-based immunodevice | 0.02 ng/mL | [240] | |
| PdNPs/Fe3O4@C | 1.7 pg/mL | [241] | |
| High luminescent streptavidin modified QDs (SA-QDs) | Optical immunoassay of AFP | 3.9 ng/mL | [242] |
| HRP-Ab2-AuNPs | 0.2 fg/chip | [243] | |
| integrated microfluidic bead-based ELISA | APOA1 | 9.16 ng/mL | [244] |
| PDMS microfluidic device | Bladder cancer TNF | 3.1 pg/mL | [245] |
| o-aminophenol (OAP)/CEA antibody (CEAAb) -GSH monolayer-AuNP (CEAAb–AuNP) | Eelectrochemical immunoassay of CEA | 100 ng/L | [246] |
| HRP-conjugated anti-CEA -NGGNs/PB | 10 ng/L | [247] | |
| AuNPs/nickel hexacyanoferrates nanoparticles (NiHCFNPs) | 100 ng/L | [248] | |
| flower-like AuNPs sensor & Au–Ag BMNPs | 0.3 pg/mL | [249] | |
| μPADs | 0.5 ng/mL | [250] | |
| Au/CS-MWCNT-Thi/anti-CEA | 0.5 pg/mL | [251] | |
| Au/l-Cys/anti-CEA//MWCNT/PDDA/HRP/ConA/HRP-anti-CEA | 18 pg/mL | [252] | |
| HRP/Nano-Au hollow microsphere | 1.5 ng/L | [254] | |
| HRP-O-phenylenediamine-H2O2 | Eelectrochemical immunoassay of AFP | 0.01 ng/mL | [253] |
| A chitosan–AuNP | 0.01 ng/mL | [235] | |
| interdigitated electrode (IDE)/PDMS | 0.01 ng/L | [236] | |
| graphene sheets (GS)/chitosan (CS)/PANI layer/(AuNPs) | BCR/ABL in CML | 2.11 pM | [255] |
| AuNPs/5′-[γ-thio] triphosphate (ATP-S) | CML | 10 ng/L | [256] |
| SWNT/5 nm GSH-AuNP | Interleukin-6 (IL-6) in serum | 10 ng/L | [257] |
| Au NPs | Human acute leukemia CCL-119 |
30 cells per 3 μL | [60] |
| Double-strand DNA (dsDNA)/AuNPs/PB | CA15-3 (human serum) | 600 ng/L | [259] |
| Nano-Au hollow sphere | Lung Cancer | 1–10 ng/L | [260] |
| perylene bisimide (PBI)/cholesterol (Chol) | 15 ppb | [261] | |
| μPADs | Pancreatic cancer CA-199 |
0.17 U/mL | [250] |
| PoP-ECL device | 0.0055 U/mL | [262] | |
| 3D microfluidic paper-based immunodevice | 0.06 U/mL | [240] | |
| HRP-O-phenylenediamine-H2O2 | Breast cancer CA153 |
0.05 ng/mL | [253] |
| AuNR@CA15-3 antibody | 0.1 ng/L | [263] | |
| hydrazine –AuNP–aptamer bioconjugate/anti-HER2/DPB | 26 cells per milliliter | [264] | |
| ZnCQDs | [265] | ||
| N, B/N, and S-doped GQDs | HeLa and MCF-7 cell | [266] | |
| G/Fe3O4/AuNPs | HeLa cells | 8 cells per milliliter | [267] |
| G-C3N4 nanosheets & Ag-PAMAM-luminol NCs | HL-60 cancer cells | 150 cells | [268] |
| FA-GAM-OA | Cancer cell | 5 cells per milliliter | [269] |
| DNA nanotetrahedron (NTH)/carboxyfluorescein (FAM) -TLS11a aptamer –HRP/PtNPs | cytosensing of liver cancer cells | 3 cells per milliliter | [270] |
Abbreviations: Bladder cancer apolipoprotein A1 (APOA1);Cadmium telluride (CdTe); Cadmium sulfide (CdS); Chronic myelogenous leukemia (CML); Microfluidic paper-based analytical device (μPADs); Enzyme-linked immunosorbent assay (ELISA); Glutathione (GSH) monolayer; The enwrapped graphene nanocomposites (NGGNs); Prussian Blue (PB); Poly(methyl methacrylate) (PMMA); Poly(ethyleneimine) (PEI); A pen-on-paper electrochemiluminescence (PoP-ECL); 2,5-bis(2-thienyl)-1H-pyrrole-1-(p-benzoic acid) (DPB); Zinc-co-adsorbed carbon quantum dots (ZnCQDs); Graphene quantum dots (GQDs); Ag-PAMAM-luminol nanocomposites (Ag-PAMAM-luminol NCs); Folic acid (FA); Octadecylamine (OA); Graphene aerogel microspheres (GAMs); Poly (dimethylsiloxane) (PDMS); Carboxyfluorescein (FAM) functionalized TLS11a aptamer and horseradish peroxidase (HRP); Poly(3,4-ethylenedioxythiophene):poly(styrenesulfonate) (PEDOT:PSS); Concanavalin A (conA); Poly(diallyldimethylammonium chloride) protected Prussian blue NPs (PDDA-PB); ssDNA/Au/reduced graphene oxide nanoribbons (RGONR); N-(aminobutyl)-N-(ethylisoluminol) (ABEI); Polypyrrole (PPy); Graphene quantum dot (GQD); Reduced graphene oxide doped with metal-organic framework (MOF-rGO); Polyaniline PANI; Paper-based analytical devices (PAD); Oligo-methoxy-phenyl-acetonitrile” (Fc-ac-OMPA); Multiwalled carbon nanotubes (MWCNTs).
Moreover, (MTB) mycobacterium tuberculosis can lead to paramount dangers and an infectious TB disease affecting roughly 10.4 million individuals globally. The WHO pronounced in 2015 that treatment for TB, 'a worldwide issue,' is necessary [[272], [273], [274], [275], [276]]. Numerous SPR-based biosensors were applied to monitor protein–protein/DNA binding in the MTB-infected area [272,273]. Various biosensing designs were reported to enhance the function of the proposed assay for sensitive and selective monitoring of MTB or anti-TB drug resistance, as shown in (Table 3) [[276], [277], [278], [279], [280], [281], [282], [283], [284], [285], [286], [287], [288], [289], [290], [291], [292]]. Within the present review stage, an ever increasing demand for deployment of robust nanobiosensor designs, real-time monitoring analyses, nanodynamic tools and techniques, and wearable read-out devices is crucial for early stage recognition of infectious agents or contagious diseases and potential proteins.
Table 3.
An overview of the various nanostructured materials used to detect TB.
| Materials | LOD | Ref |
|---|---|---|
| carboxylic acid-functionalized magnetic nanoparticles (CAMNPs) | 8 CFU/μL | 276 |
| ssDNA/AuNPs/RGONR | 0.1 fM | 277 |
| Fc-ac-OMPA/MWCNTs | 0.08 fmol/L | 278 |
| Dextrin coated AuNPs | 0.01 ng/μL | 279 |
| ZrO2/Au | 0.065 ng/μL | 280 |
| MWCNTs/PPy/PAMAN/ferrocene | 0.3 fM | 281 |
| spaNQ/PAMAM/PPy/Fe3O4 | 1 fM | 282 |
| Fe3O4@Ag/GQD | 0.33 ng/mL | 283 |
| ABEI functionalized AuNPs | 3.3 × 10−16M | 284 |
| 3D graphene/Au NRs | 10 fM | 285 |
| MOF-rGO | 3.3 × 10−5 ng/mL | 286 |
| rGO–AuNPs –PANI | – | 287 |
| luminol functionalized AGNPs | 0.03 fmol/L | 288 |
| Zr(IV) based metal-organic framework (MOF) | 10 fg/mL | 289 |
| Organic light-emitting diode (OLED) | 63 pg/mL | 290 |
| PAD- pyrrolidinyl PNA (acpcPNA) | 1.24 nM | 291 |
| AuNP-labeled staphylococcal protein A immunosensor (Au-SPA) | 5.3 ng/mL | 292 |
In this regard, the HTS system introduced two performance sets of major optimization designs of MTB acetyltransferase Eis inhibitors that cause resistance to anti-TB drug kanamycin (KAN) [293]. To screen the anti-MTB drugs, mycobacterium tuberculosis glutamine synthetase was studied by using HTS to ascertain the binding position targeting inhibitors of ATP and to produce several active classes of mycobacterium glutamine compounds including 2-tert-butyl-4,5-diarylimidazoles. The proposed method would lead to identify the mycobacterium tuberculosis glutamine synthetase inhibitors in sub-μM concentrations, indicating the formation of promising anti-MTB MIC (i.e. minimum inhibitory concentration values) [294]. The HTS strategy enabled a detection of MTB drugs using a complete cell line assay. Two proteins have been defined as targets for many chemically unrelated small molecules such as decaprenylphosphoryl-β-d-ribose 2-epimerase (DprE1) enzyme for the biosynthesis of the cell wall, and mycobacterial membrane protein large 3 (MmpL3) exporters of newly synthesized cell wall mycolic acids. This strategy provided a powerful tool for screening and target identification such as MTB and antibiotic detection [295]. The HTS system based on thermal shift test was used to screen the binding interaction of protein–ligand detection. The optimal analyses of the HTS strategy would also offered a high-precision strategy for examination and identification of enzyme such as isocitrate lyase (ICL), as a vital agent for MTB survival and inhibition within macrophages through latent TB. The HTS virtual system has significantly led to the discovery of many ICL inhibitors with catalytic activity [296].
11. Conclusion and future viewpoint
The current review presents the key feature of the prospective vision of nanobiosensor development toward the early and easily continuous diagnosis and monitoring of advanced diseases to identify the appropriate care and treatment and to reduce further transmission. Hence, we report the incredible endeavors providing effective nanodynamic tools and biosensor devices for the intrinsic early stage recognition of infectious agents and potential biomolecules, and then powerful counseling for the inhibition or prevention of diseases, including HIV, HBV, HCV, liver diseases, diabetes, CNS, TB, CVD and cancer. To date, performance evidence of easy continuous monitoring, and the detection accuracy test of chronically infectious diseases still needs more advanced nanoscale architect-led building-blocks-in biosensor design optimizations to permit early stage recognition and sensitive reliability analyses. Reliability-based biosensor design is the gateway for accurate access to prevention, inhibition, and treatment amenities. Therefore, it is a crucial to control a distinctive sensitivity analysis and actual-time of response gradient to avoid a widespread occurrence of an infectious disease in a community at an intense time. The biosensor accuracy in terms of reliable sensitivity information with multiple samples of individuals should be considered. Special emphasis in the structural sensor design optimization would offer reliable and adequate safeguard evidence to ensure confidentiality and validation of biosensing assays.
In a commercial market's view, nanoscale biosensor tailored into personal, wearable, and touchable devices for individuals and increasingly delivered at home remains challenging. The path-dependence of the sensitive devices would lead to sustainable development of low-cost IoT-based platforms. Simple biosensor models would offer a cost-effective and powerful solution to detect the clinical care point of diseases. Smartphone-based visual analysis and managing human wellness in customized environments will be the potential future investigation in nanobiosensing, see (Table 4).
Table 4.
Initiative commercial market's biosensor devices.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Kirsch J., Siltanen C., Zhou Q., Revzin A., Simonian A. Chem. Soc. Rev. 2013;42:8733–8768. doi: 10.1039/c3cs60141b. [DOI] [PubMed] [Google Scholar]
- 2.Gaikwad P.S., Banerjee R. Analyst. 2018;143:1326–1348. doi: 10.1039/c7an01771e. [DOI] [PubMed] [Google Scholar]
- 3.Wu X.Y., Chai Y.Q., Yuan R., Su H.L., Han J. Analyst. 2013;138:1060–1066. doi: 10.1039/c2an36506e. [DOI] [PubMed] [Google Scholar]
- 4.Emran M.Y., Shenashen M.A., Morita H., El-Safty S.A. Adv. Healthc Mater. 2018;7(16) doi: 10.1002/adhm.201701459. [DOI] [PubMed] [Google Scholar]
- 5.Riether C., Schürch C., Ochsenbein A. Cell Death Differ. 2015;22:187–198. doi: 10.1038/cdd.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng H., Wang X., Wei H. Anal. Chem. 2015;87:8889–8895. doi: 10.1021/acs.analchem.5b02014. [DOI] [PubMed] [Google Scholar]
- 7.Knowles T.P., Dobson M. Vendruscolo C.M. Nat. Rev. Mol. Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 8.Wei G., Su Z., Reynolds N.P., Arosio P., Hamley I.W., Gazit E., Mezzenga R. Chem. Soc. Rev. 2017;46:4661–4708. doi: 10.1039/c6cs00542j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu S., Yu T., Wang Y., Liang L., Chen Y., Xu F., Wang S. Analyst. 2017;142:3309–3321. doi: 10.1039/c7an00847c. [DOI] [PubMed] [Google Scholar]
- 10.Cho N.H., Shaw J.E., Karuranga S., Huang Y., da Rocha Fernandes J.D., Ohlrogge A.W., Malanda B. Diabetes Res. Clin. Pract. 2018;138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Chen G., Jiang H., Li Z., Wang Xuemei. Analyst. 2013;138:4427–4435. doi: 10.1039/c3an00438d. [DOI] [PubMed] [Google Scholar]
- 12.Zhang N., Ruan Y.F., Zhang L.B., Zhao W.W., Xu J.J., Chen H.Y. Anal. Chem. 2018;90(3):2341–2347. doi: 10.1021/acs.analchem.7b04862. [DOI] [PubMed] [Google Scholar]
- 13.Meng Z., Stolz R.M., Mendecki L., Mirica K.A. Chem. Rev. 2019;119(1):478–598. doi: 10.1021/acs.chemrev.8b00311. [DOI] [PubMed] [Google Scholar]
- 14.Farandos N.M., Yetisen A.K., Monteiro M.J., Lowe C.R., Yun S.H. Adv. Healthcare Mater. 2015;4:792–810. doi: 10.1002/adhm.201400504. [DOI] [PubMed] [Google Scholar]
- 15.Moon H.G., Jung Y., Jun D., Park J.H., Chang Y.W., Park H.H., Kang C.Y., Kim C., Kaner R.B. ACS Sens. 2018;3(3):661–669. doi: 10.1021/acssensors.7b00955. [DOI] [PubMed] [Google Scholar]
- 16.Giljohann D.A., Mirkin C.A. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronkainen N.J., Halsall H.B., Heineman W.R. Chem. Soc. Rev. 2010;39:1747–1763. doi: 10.1039/b714449k. [DOI] [PubMed] [Google Scholar]
- 18.Li N., Su X., Lu Y. Analyst. 2015;140:2916–2943. doi: 10.1039/c4an02376e. [DOI] [PubMed] [Google Scholar]
- 19.Turner A.P.F. Chem. Soc. Rev. 2013;42:3184–3196. doi: 10.1039/c3cs35528d. [DOI] [PubMed] [Google Scholar]
- 20.Kimmel D.W., LeBlanc G., Meschievitz M.E., Cliffel D.E. Anal. Chem. 2012;84(2):685–707. doi: 10.1021/ac202878q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabowska I., Sharma N., Vasilescu A., Iancu M., Badea G., Boukherroub R., Ogale S., Szunerits S. ACS Omega. 2018;3(9):12010–12018. doi: 10.1021/acsomega.8b01558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govindhan M., Adhikari B.-R., Chen A. RSC Adv. 2013;4:63741–63760. [Google Scholar]
- 23.Li L., Wang Y., Pan L., Shi Y., Cheng W., Shi Y., Yu G. Nano Lett. 2015;15(2):1146–1151. doi: 10.1021/nl504217p. [DOI] [PubMed] [Google Scholar]
- 24.Dhand C., Das M., Datta M., Malhotra B.D. Biosens. Bioelectron. 2011;26:2811–2821. doi: 10.1016/j.bios.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L., Dong W.-F., Sun H.-B. Nanoscale. 2013;5:7664–7684. doi: 10.1039/c3nr01616a. [DOI] [PubMed] [Google Scholar]
- 26.Perumal V., Hashim U. J. Appl. Biomed. 2014;12:1–15. [Google Scholar]
- 27.Kim J., Campbell A.S. J. Wang. Talanta. 2018;177:163–170. doi: 10.1016/j.talanta.2017.08.077. [DOI] [PubMed] [Google Scholar]
- 28.Mosciatti T., Bonacchi S., Gobbi M., Ferlauto L., Liscio F., Giorgini L., Orgiu E., Samorì P. ACS Appl. Mater. Interfaces. 2016;8(10):6563–6569. doi: 10.1021/acsami.5b12430. [DOI] [PubMed] [Google Scholar]
- 29.Silvestri D., Sonato A., Ruffato G., Meneghello A., Antognoli A., Cretaio E., Dettin M., Zamuner A., Casarin E., Zacco G., Romanato F., Morpurgo M. Anal. Methods. 2015;7:4173–4180. [Google Scholar]
- 30.Mogha N.K., Sahu V., Sharma R.K., Masram D.T. J. Mater. Chem. B. 2018;6:5181–5187. doi: 10.1039/c8tb01604f. [DOI] [PubMed] [Google Scholar]
- 31.Fu L.-H., Qi C., Lin J., Huang P. Chem. Soc. Rev. 2018;47:6454–6472. doi: 10.1039/c7cs00891k. [DOI] [PubMed] [Google Scholar]
- 32.Emran M.Y., Shenashen M.A., Abdelwahab A.A., Abdelmottaleb M., El-Safty S.A. New J. Chem. 2018;42:5037–5044. [Google Scholar]
- 33.Ma F., Li C-c., Zhang C-y. J. Mater. Chem. B. 2018;6:6173–6190. doi: 10.1039/c8tb01869c. [DOI] [PubMed] [Google Scholar]
- 34.Emran M.Y., Shenashen M.A., Mekawy M., Azzam A.M., Akhtar N., Gomaa H., Selim M.M., Faheem A., El-Safty S.A. Sensor. Actuator. B Chem. 2018;259:114–124. [Google Scholar]
- 35.WHO . 2013. World Health Organization.http://www.who.int/mediacentre/factsheets/fs312/en/ [Google Scholar]
- 36.Kottmann J., Rey J.M., Luginbuehl J., Reichmann E., Sigrist M.W. Biomed. Optic Express. 2012;3:667–680. doi: 10.1364/BOE.3.000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nalini S., Nandini S., Reddy M.B.M., Suresh G.S., Melo J.S., Neelagund S.E., Kumar H.N.N., Shanmugam S. RSC Adv. 2016;6:60693–60703. [Google Scholar]
- 38.Jijie R., Barras A., Boukherroub R., Szunerits S. J. Mater. Chem. B. 2017;5:8653–8675. doi: 10.1039/c7tb02529g. [DOI] [PubMed] [Google Scholar]
- 39.Wang N., Burugapalli K., Song W., Halls J., Moussy F., Ray A., Zheng Y. Biomaterials. 2013;34:889–901. doi: 10.1016/j.biomaterials.2012.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vashist S.K. Anal. Chim. Acta. 2012;750:16–27. doi: 10.1016/j.aca.2012.03.043. [DOI] [PubMed] [Google Scholar]
- 41.Boss C., Meurville E., Sallese J.-M., Ryser P. Biosens. Bioelectron. 2011;30:223–228. doi: 10.1016/j.bios.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 42.El-Safty S.A., Shenashen M.A., Ismael M., Khairy M. Adv. Funct. Mater. 2012;22:3013–3021. [Google Scholar]
- 43.Solanki P.R., Kaushik A., Agrawal V.V., Malhotra B.D. NPG Asia Mater. 2011;3(1):17–24. [Google Scholar]
- 44.Emran M.Y., Shenashen M.A., Abdelwahab A.A., Abdelmottaleb M., Khairy M., El-Safty S.A. Electrocatalysis. 2018;9(4):514–525. [Google Scholar]
- 45.Azzam A.M., Shenashen M.A., Mostafa B.B., Kandeel W.A., El-Safty S.A. Environ. Prog. Sustain. Energy. 2019;38:S260–S266. [Google Scholar]
- 46.Emran M., El-Safty S.A., Shenashen M.A., Minowa T. Sensor. Actuator. B Chem. 2019;284:456–467. [Google Scholar]
- 47.Doong R., Shih H. Biosens. Bioelectron. 2010;25:1439–1446. doi: 10.1016/j.bios.2009.10.044. [DOI] [PubMed] [Google Scholar]
- 48.Lucisano J.Y., Routh T.L., Lin J.T., Gough D.A. Trans. Biomed. Eng. 2017;64:1982–1993. doi: 10.1109/TBME.2016.2619333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El-Safty S.A., Shenashen M.A., Ismael M., Khairy M. Chem. Commun. 2012;48:6708–6710. doi: 10.1039/c2cc30725a. [DOI] [PubMed] [Google Scholar]
- 50.Farandos N.M., Yetisen A.K., Monteiro M.J., Lowe C.R., Yun S.H. Adv. Healthcare Mater. 2015;4:792–810. doi: 10.1002/adhm.201400504. [DOI] [PubMed] [Google Scholar]
- 51.Guo X., Kuhlmann J., Heineman W.R. Environmental Analysis by Electrochemical Sensors and Biosensors. Springer; 2014. p. 283. [Google Scholar]
- 52.Emran M.Y., Shenashen M.A., Morita H., El-Safty S.A. Biosens. Bioelectron. 2018;109(2018):237–245. doi: 10.1016/j.bios.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 53.Selim M.S., Yang H., El-Safty S.A., Fatthallah N.A., Shenashen M.A., Wang F.Q., Huang Y. Colloids Surfaces A. 2019;570:518–530. [Google Scholar]
- 54.Gomaa H., El-Safty S.A., Shenashen M.A., Yamaguchi H., Alamoudi A.S., Abdelmottaleb M., Cheira M.F. ACS Sustain. Chem. Eng. 2018;6(11):13813–13825. [Google Scholar]
- 55.Ji C., Walton J., Su Y., Tella M. Anal. Chim. Acta. 2010;670:84–91. doi: 10.1016/j.aca.2010.04.051. [DOI] [PubMed] [Google Scholar]
- 56.El-Sewify I.M., Shenashen M.A., Shahat A., Selim M.M., Khalil M.M.H., El-Safty S.A. Microchem. J. 2018;139:24–33. [Google Scholar]
- 57.Hahn Y.-B., Ahmad R., Tripathy N. Chem. Commun. 2012;48:10369–10385. doi: 10.1039/c2cc34706g. [DOI] [PubMed] [Google Scholar]
- 58.Shenashen M.A., Kawada S., Selim M.M., Morsy W.M., Yamaguchi H., Alhamid A.A., Ohashi N., Ichinose I., El-Safty S.A. Nanoscale. 2017;9:7947–7959. doi: 10.1039/c7nr01092c. [DOI] [PubMed] [Google Scholar]
- 59.Gomaa H., Shenashen M.A., Yamaguchi H., Alamoudi A.S., El-Safty S.A. Green Chem. 2018;20:1841–1857. [Google Scholar]
- 60.Mohammed M.-I., Desmulliez M.P.Y. Lab Chip. 2011;11:569–595. doi: 10.1039/c0lc00204f. [DOI] [PubMed] [Google Scholar]
- 61.Shenashen M.A., Akhtar N., Selim M.M., Morsy W.M., Yamaguchi H., Kawada S., Alhamid A.A., Ohashi N., Ichinose I., Alamoudi A.S., El-Safty S.A. Chem. Asian J. 2017;12(15):1952–1964. doi: 10.1002/asia.201700666. [DOI] [PubMed] [Google Scholar]
- 62.Derbalah A., El-Safty S.A., Shenashen M.A., Abdel Ghany N. ChemPlusChem. 2015;80(7):1119–1126. doi: 10.1002/cplu.201500098. [DOI] [PubMed] [Google Scholar]
- 63.Shenashen M.A., Derbalah A., Hamza A., Mohamed A., El Safty S.A. Pest Manag. Sci. 2017;73(6):1121–1126. doi: 10.1002/ps.4420. [DOI] [PubMed] [Google Scholar]
- 64.El-Safty S.A., Shenashen M.A., Ismail A.A. Chem. Commun. 2012;48:9652–9654. doi: 10.1039/c2cc34788a. [DOI] [PubMed] [Google Scholar]
- 65.Castillo J., Gáspár S., Leth S., Niculescu M., Mortari A., Bontidean I., Soukharev V., Dorneanu S.A., Ryabov A.D., Csöregi E. Sensor. Actuator. B Chem. 2004;102(2):179–194. [Google Scholar]
- 66.Shenashen M.A., El-Safty S.A., Shahat A. J. Hazard Mater. 2013;244–245:726–735. doi: 10.1016/j.jhazmat.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 67.Justino C.I.L., Rocha-Santos T.A., Duarte A.C. Trac. Trends Anal. Chem. 2010;29(10):1172–1183. [Google Scholar]
- 68.El-Safty S.A., Shenashen M.A. Sensor. Actuator. B Chem. 2013;183:58–70. [Google Scholar]
- 69.El-Safty S.A., Shenashen M.A. A. Shahat. small. 2013;9(13):2288–2296. doi: 10.1002/smll.201202407. [DOI] [PubMed] [Google Scholar]
- 70.Khairy M., El-Safty S.A., Shenashen M.A., Elshehy E.A. Nanoscale. 2013;5(17):7920–7927. doi: 10.1039/c3nr02403b. [DOI] [PubMed] [Google Scholar]
- 71.Mao S., Chang J., Pu H., Lu G., He Q., Zhang H., Chen J. Chem. Soc. Rev. 2017;46:6872–6904. doi: 10.1039/c6cs00827e. [DOI] [PubMed] [Google Scholar]
- 72.El-Safty S.A., Ismail A.A., Matsunaga H., Nanjo H., Mizukami F. J. Phys. Chem. C. 2008;112:4825–4835. [Google Scholar]
- 73.Wang G., Morrin A., Li M., Liu N., Luo X. J. Mater. Chem. B. 2018;6:4173–4190. doi: 10.1039/c8tb00817e. [DOI] [PubMed] [Google Scholar]
- 74.Gomaa H., Khalifa H., Selim M., Shenashen M., Kawada S., Alamoudi A.S., Azzam A., Alhamid A., El-Safty S.A. ACS Sustain. Chem. Eng. 2017;5:10826–10839. [Google Scholar]
- 75.Khalifa H, El-Safty S.A., Reda A., Shenashen M.A., Selim M.M., Elmarakbi A., Metawa H.A. Nano-Micro Lett. 2019;11:84. doi: 10.1007/s40820-019-0315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li X., Shenashen M.A., Wang X., Ito A., Taniguchi A., EI-Safty S.A. Sci. Rep. 2017;7(1):16749. doi: 10.1038/s41598-017-12446-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Israr-Qadir M., Jamil-Rana S., Nur O., Willander M. Sensors. 2017;17:1645. doi: 10.3390/s17071645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shenashen M.A., El-Safty S.A., Elshehy E.A. Analyst. 2014;139(24):6393–6405. doi: 10.1039/c4an00980k. [DOI] [PubMed] [Google Scholar]
- 79.Willander M., Khun K., Ibupoto Z.H. Sensors. 2014;14:8605–8632. doi: 10.3390/s140508605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hassen D., El-Safty S.A., Tsuchiya K., Chatterjee A., Elmarakbi A., Shenashen M.A., Sakai M. Sci. Rep. 2016;6:24330. doi: 10.1038/srep24330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y., Wu C., Zhou X., Wu X., Yang Y., Wu H., Guo S., Zhang J. Nanoscale. 2013;5:1816–1819. doi: 10.1039/c3nr33954h. [DOI] [PubMed] [Google Scholar]
- 82.Selim M.S., El-Safty S.A., El-Sockary M.A., Hashem A.I., Elenien O.M.A., EL-Saeed A.M., Fatthallah N.A. Mater. Des. 2016;101:218–225. [Google Scholar]
- 83.El-Safty S.A., Khairy M., Shenashen M.A., Elshehy E.A., Warkocki W., Sakai M. Nano Research. 2015;8(10):3150–3163. [Google Scholar]
- 84.Akhtar N., Emran M.Y., Shenashen M.A., Khalifa H., Osaka T., Faheem A., Homma T., Kawarada H., El-Safty S.A. J. Mater. Chem. B. 2017;5:7985–8799. doi: 10.1039/c7tb01803g. [DOI] [PubMed] [Google Scholar]
- 85.Chang H., Wang X., Shiu K.-K., Zhu Y., Wang J., Li Q., Chen B., Jiang H. Biosens. Bioelectron. 2013;41:789–794. doi: 10.1016/j.bios.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 86.Yu Q., Eptein H., Engstrom R., Walker D. Anal. Methods. 2017;9:3895–3907. doi: 10.1111/gcb.13632. [DOI] [PubMed] [Google Scholar]
- 87.Sun Y., Luo M., Meng X., Xiang J., Wang L., Ren Q., Guo S. Anal. Chem. 2017;89:3761–3767. doi: 10.1021/acs.analchem.7b00248. [DOI] [PubMed] [Google Scholar]
- 88.Emran M.Y., Mekawy M., Akhtar N., Shenashen M.A., El-Sewify I.M., Faheem A., El-Safty S.A. Biosens. Bioelectron. 2018;100:122–131. doi: 10.1016/j.bios.2017.08.050. [DOI] [PubMed] [Google Scholar]
- 89.Kannan P.K., Late D.J., Morgan H., Rout C.S. Nanoscale. 2015;7:13293–13312. doi: 10.1039/c5nr03633j. [DOI] [PubMed] [Google Scholar]
- 90.Emran M.Y., Shenashen M.A., Abdelwahab A.A., Khalifa H., Mekawy M., Abdelmottaleb N. Akhtar M., El-Safty S.A. J. Appl. Electrochem. 2018;48(5):529–542. [Google Scholar]
- 91.Zhu J., Chen M., He Q., Shao L., Wei S., Guo Z. RSC Adv. 2013;3:22790–22824. [Google Scholar]
- 92.Akhtar N., El-Safty S.A., Abdelsalam M.E., Shenashen M.A., Kawarada H. Biosens. Bioelectron. 2016;77:656–665. doi: 10.1016/j.bios.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 93.Xu H., Xia C., Wang S., Han F., Akbari M.K., Hai Z., Zhuiykov S. Sensor. Actuator. B Chem. 2018;267:93–103. [Google Scholar]
- 94.Chen A., Xu L., Zhang X., Yang Z., Yang S. ACS Appl. Mater. Interfaces. 2016;8:33765–33774. doi: 10.1021/acsami.6b11088. [DOI] [PubMed] [Google Scholar]
- 95.Yu Z., Li H., Lu J., Zhang X., Liu N., Zhang X. Electrochim. Acta. 2015;158:264–270. [Google Scholar]
- 96.Ahmad R., Tripathy N., Ahn M.-S., Bhat K.S., Mahmoudi T., Wang Y., Yoo J.-Y., Kwon D.-W., Yang H.-Y. Hahn. Sci. Rep. 2017;7:5715. doi: 10.1038/s41598-017-06064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barkaoui S., Haddaoui M., Dhaouadi H., Raouafi N., Touati F. J. Solid State Chem. 2015;228:226–231. [Google Scholar]
- 98.Cui Z., Yin H., Nie Q., Qin D., Wu W., He X. J. Electroanal. Chem. 2015;757:51–57. [Google Scholar]
- 99.McKeating K.S., Aubé A., Masson J.-F. Analyst. 2016;141:429–449. doi: 10.1039/c5an01861g. [DOI] [PubMed] [Google Scholar]
- 100.Chang L., Hu J., Chen F., Chen Z., Shi J., Yang Z., Li Y., Lee L.J. Nanoscale. 2016;8:3181–3206. doi: 10.1039/c5nr06694h. [DOI] [PubMed] [Google Scholar]
- 101.Xu J.-J., Zhao W.-W., Song S., Fan C., Chen H.-Y. Chem. Soc. Rev. 2014;43:1601–1611. doi: 10.1039/c3cs60277j. [DOI] [PubMed] [Google Scholar]
- 102.Zhang W., Guo S., Carvalho W.S.P., Jiang Y., Serpe M.J. Anal. Methods. 2016;8:7847–7867. [Google Scholar]
- 103.Heikenfeld J., Jajack A., Rogers J., Gutruf P., Tian L., Pan T., Li R., Khine M., Kim J., Wang J., Kim J. Nanoscale, Lab Chip. 2018;18:217–248. doi: 10.1039/c7lc00914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ma Z., Li S., Wang H., Cheng W., Li Y., Pan Yi Shi L. J. Mater. Chem. B. 2019;7:173–197. doi: 10.1039/c8tb02862a. [DOI] [PubMed] [Google Scholar]
- 105.Chen K., Gao W., Emaminejad S., Kiriya D., Ota H., Nyein H.Y., Takei K., Javey A. Adv. Mater. 2016;28(22):4397–4414. doi: 10.1002/adma.201504958. [DOI] [PubMed] [Google Scholar]
- 106.Campbell A.S., Jayoung K., Joseph W. Curr. Opin. Electrochem. 2018;10:126–135. doi: 10.1016/j.coelec.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Khan M., Imam H., Siddiqui A. Liver Res. 2018;2(3):146–156. doi: 10.1016/j.livres.2018.09.002. ; M. Levrero, T. Pollicino, J. Petersen, L. Belloni, G. Raimondo, M. Dandri. J. Hepatol., 51 (2009) 581-592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ramirez S. J. Bukh. Antiviral Research. 2018;158:264–287. doi: 10.1016/j.antiviral.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 109.Yao C., Xiang Y., Deng K., Xia H., Fu W. Sensor. Actuator. B Chem. 2013;181:382–387. [Google Scholar]
- 110.Yao C., Zhu T., Tang J., Wu R., Chen Q., Chen M., Zhang B., Huang J., Fua W. Biosens. Bioelectron. 2008;23(6):879–885. doi: 10.1016/j.bios.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 111.Chen C.C., Lai Z.L., Wang G.J., Wu C.Y. Biosens. Bioelectron. 2016;77:603–608. doi: 10.1016/j.bios.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 112.Erdem A., Kerman K., Meric B., Akarca U.S., Ozsoz M. Electroanalysis. 1999;11:586–587. (199906)11:8<586::AID-ELAN586>3.0.CO;2-J. [Google Scholar]
- 113.Meric B., Kerman K., Ozkan D., Kara P., Erensoy S., Akarca U.S., Mascini M., Ozsoz M. Talanta. 2002;56(2013):837–846. doi: 10.1016/s0039-9140(01)00650-6. [DOI] [PubMed] [Google Scholar]
- 114.Zhu H., Wang J., Xu G. Cryst. Growth Des. 2008;9:633–638. [Google Scholar]
- 115.Ahangar L.E., Mehrgardi M.A. Bioelectrochemistry. 2017;117:83–88. doi: 10.1016/j.bioelechem.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 116.Oliveira D.A., Silva J.V., Flauzino J.M.R., Castro A.C.H., Moço A.C.R., Soares M.M.C.N., Madurro J.M., Brito-Madurroa A.G. Anal. Biochem. 2018;549:157–163. doi: 10.1016/j.ab.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 117.Narang J., Singhal C., Malhotra N., Narang S., PN A.K., Gupta R., Kansal R., Pundir C.S. Biosens. Bioelectron. 2016;86:566–574. doi: 10.1016/j.bios.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 118.Akkapinyo C., Khownarumit P., Waraho-Zhmayev D., Poo-arporn R.P. Anal. Chim. Acta. 2020;1095:162–171. doi: 10.1016/j.aca.2019.10.016. [DOI] [PubMed] [Google Scholar]
- 119.Mao X., Liu S., Yang C., Liu F., Wang K., Chen G. Anal. Chim. Acta. 2016;909:101–108. doi: 10.1016/j.aca.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 120.Shimizu T., Tanaka T., Uno S., Ashiba H., Fujimaki M., Tanaka M., Awazu K., Makishima M. J. Biosci. Bioeng. 2017;123(6):760–764. doi: 10.1016/j.jbiosc.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 121.Wang S., Li L., Jin H., Yang T., Bao W., Huang S., Wang J. Biosens. Bioelectron. 2013;41:205–210. doi: 10.1016/j.bios.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 122.Wang X., Li Y., Wang H., Fu Q., Peng J., Wang Y., Du J., Zhou Y., Zhana L. Biosens. Bioelectron. 2013;26(2):404–410. doi: 10.1016/j.bios.2010.07.121. [DOI] [PubMed] [Google Scholar]
- 123.Ehsani M., Chaichi M.J., Hosseini S.N. Sensor. Actuator. B Chem. 2017;247:319–328. [Google Scholar]
- 124.Wang G., Leng Y.i, Guo H., Song S., Jiang Z., Yuan X., Wang X., Sun K., Sun K., Dou H. J. Mater. Chem. B. 2014;2:8310–8313. doi: 10.1039/c4tb01672f. [DOI] [PubMed] [Google Scholar]
- 125.Valipour A., Roushani M. Biosens. Bioelectron. 2017;89(2):946–951. doi: 10.1016/j.bios.2016.09.086. [DOI] [PubMed] [Google Scholar]
- 126.Abdel-aal F.A.M., Rageh A.H., Said M.I., Saleh G.A. Anal. Chim. Acta. 2018;1038:29–40. doi: 10.1016/j.aca.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 127.Singhal C., Ingle A., Chakraborty D., PN A.K., Pundir C.S. J. Narang. Int. J. Biological Macromole. 2017;98:84–93. doi: 10.1016/j.ijbiomac.2017.01.093. [DOI] [PubMed] [Google Scholar]
- 128.Wang H., Thorling C.A., Liang X., Bridle K.R., Grice J.E., Zhu Y., Crawford D.H.G., Xu Z.P., Liu X., Roberts M.,S. J. Mater. Chem. B. 2015;3:939–958. doi: 10.1039/c4tb01611d. ; F. Hamel , M. Grondin , F. Denizeau , D. A. Averill-Bates and F. Sarhan , Biotechnol. Bioeng., 95 (2006) 661 -670. [DOI] [PubMed] [Google Scholar]
- 129.Zhao Z., Zhou Z., Bao J., Wang Z., Hu J., Chi X., Ni K., Wang R., Chen X., Chen Z., Gao J. Nat. Commun. 2013;4:2266. doi: 10.1038/ncomms3266. [DOI] [PubMed] [Google Scholar]
- 130.Shen D., Zheng J., Cui X., Chena Y., He Q., Lva R., Li Z., Zhao S. Sensor. Actuator. B Chem. 2018;256:846–852. [Google Scholar]
- 131.Cour J.B.l., Generelli S., Barbe L., Guenat O.T. Sens. Actuators, B. 2016;228:360–365. [Google Scholar]
- 132.Jiang D., Li C., Liu T., Li L., Chu Z., Jin W., Ren X. Sensor. Actuator. B Chem. 2017;241:860–867. [Google Scholar]
- 133.Zhao Z., Zhou Z., Bao J., Wang Z., Hu J., Chi X., Ni K., Wang R., Chen X., Chen Z., Gao J. Nat. Commun. 2013;4:2266. doi: 10.1038/ncomms3266. [DOI] [PubMed] [Google Scholar]
- 134.Tanimoto A., Kuribayashi S. Eur. J. Radiol. 2006;58:200–216. doi: 10.1016/j.ejrad.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 135.Murakami T., Kim T., Takamura M., Shimizu J., Hori M., Dono K., Takachi K., Kato N., Miyazawa T., Sakon M., Monden M., Nakamura H. Dig. Dis. Sci. 2001;46:148–155. doi: 10.1023/a:1005585101620. [DOI] [PubMed] [Google Scholar]
- 136.Saraswathy A., Nazeer S.S., Nimi N., Arumugam S., Shenoy S.J., Jayasree R.S. Carbohydr. Polym. 2014;101:760–768. doi: 10.1016/j.carbpol.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 137.Mouli S.K., Tyler P., McDevitt J.L., Eifler A.C., Guo Y., Nicolai J., Lewandowski R.J., Li W., Procissi D., Ryu R.K., Wang Y.A., Salem R., Larson A.C., Omary R.A. ACS Nano. 2013;7:7724–7733. doi: 10.1021/nn4023119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou Z., Huang D., Bao J., Chen Q., Liu G., Chen Z., Chen X., Gao J. Adv. Mater. 2012;24:6223–6228. doi: 10.1002/adma.201203169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tian X., Shao Y., He H., Liu H., Shen Y., Huang W., Li L. Nanoscale. 2013;5:3322–3329. doi: 10.1039/c3nr00170a. [DOI] [PubMed] [Google Scholar]
- 140.Kim S.M., Im G.H., Lee D.G., Lee J.H., Lee W.J., Lee I.S. Biomaterials. 2013;34:8941–8948. doi: 10.1016/j.biomaterials.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 141.Liu Z., Li Z., Liu J., Gu S., Yuan Q., Ren J., Qu X. Biomaterials. 2012;33:6748–6757. doi: 10.1016/j.biomaterials.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 142.Ai K., Liu Y., Liu J., Yuan Q., He Y., Lu L. Adv. Mater. 2011;23:4886–4891. doi: 10.1002/adma.201103289. [DOI] [PubMed] [Google Scholar]
- 143.Boll H., Figueiredo G., Fiebig T., Nittka S., Doyon F., Kerl H.U., Nolte I., Forster A., Kramer M., Brockmann M.A. Acad. Radiol. 2013;20:1137–1143. doi: 10.1016/j.acra.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 144.Liu Z., Dong K., Liu J., Han X., Ren J., Qu X. Small. 2014;10:2429–2438. doi: 10.1002/smll.201303909. [DOI] [PubMed] [Google Scholar]
- 145.Yu X., Chen L., Deng Y., Li K., Wang Q., Li Y., Xiao S., Zhou L., Luo X., Liu J., Pang D. J. Fluoresc. 2007;17:243–247. doi: 10.1007/s10895-007-0163-7. [DOI] [PubMed] [Google Scholar]
- 146.Li F., Li Q.H., Wang J.Y., Zhan C.Y., Xie C., Lu W.Y. J. Contr. Release. 2012;159:261–270. doi: 10.1016/j.jconrel.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 147.Wu J., Sun T.-M., Yang X.-Z., Zhu J., Du X.-J., Yao Y.-D., Xiong M.-H., Wang H.-X., Wang Y.-C., Wang J. Biomater Sci. 2013;1:1143–1150. doi: 10.1039/c3bm60099h. [DOI] [PubMed] [Google Scholar]
- 148.Liu J., Liu J., Xu H., Zhang Y., Chu L., Liu Q., Song N., Yang C. Int. J. Nanomed. 2014;9:197–207. doi: 10.2147/IJN.S55875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li L., Tang F., Liu H., Liu T., Hao N., Chen D., Teng X., He J. ACS Nano. 2010;4:6874–6882. doi: 10.1021/nn100918a. [DOI] [PubMed] [Google Scholar]
- 150.Lu S., Yu T., Wang Y., Liang L., Chen Y., Xu F., Wang S.Q. Analyst. 2017;142:3309–3321. doi: 10.1039/c7an00847c. [DOI] [PubMed] [Google Scholar]
- 151.Castellano J.M., Narula J., Castillo J., Fuster V. Rev. Esp. Cardiol. 2014;67:724–730. doi: 10.1016/j.rec.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 152.Singal S., Srivastava A.K., Dhakate S., Biradar A.M. Rajesh. RSC Adv. 2015;5:74994–75003. [Google Scholar]
- 153.Zhou Y.-C., Zhao M., Yu Y.-Q., Lei Y.-M., Chai Y.-Q., Yuan R., Zhuo Y. Sensor. Actuator. B Chem. 2017;246:1–8. [Google Scholar]
- 154.Wu W.-Y., Bian Z.-P., Wang W., Wang W., Zhu J.-J. Sensor. Actuator. B Chem. 2010;147:298–303. [Google Scholar]
- 155.Song S.Y., Han Y.D., Kim K., Yang S.S., Yoon H.C. Biosens. Bioelectron. 2011;26:3818–3824. doi: 10.1016/j.bios.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 156.Mondal K., Ali Md A., Singh C., Sumana G., Malhotra B.D., Sharma A. Sensor. Actuator. B Chem. 2017;246:202–214. [Google Scholar]
- 157.Baliyan A., Sital S., Tiwari U., Gupta R., Sharma E.K. Biosens. Bioelectron. 2016;79:693–700. doi: 10.1016/j.bios.2015.12.089. [DOI] [PubMed] [Google Scholar]
- 158.Giri A.K., Charan C., Ghosh S.C., Shahi V.K., Panda A.B. Sensor. Actuator. B Chem. 2016;229:14–24. [Google Scholar]
- 159.Parker T., Libourel P.-A., Hetheridge M.J., Cumming R.I., Sutcliffe T.P., Goonesinghe A.C., Ball J.S., Owen S.F., Chomis Y., Winter M.J., Pharma J. Toxicol. Mech. Methods. 2014;69(1):30–38. doi: 10.1016/j.vascn.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 160.Fuad N.M., Kaslin J., Wlodkowic D. Sensor. Actuator. B Chem. 2018;256:1131–1141. [Google Scholar]
- 161.Chen Y.-W., Kuo W.-C., Tai T.-Y., Hsu C.-P., Sarangadharan I., Pulikkathodi A.K., Wang S.-L., Sukesan R., Lin H.-Y., Kao K.-W., Hsu C.-L., Chen C.C., Wang Y.-L. Sensor. Actuator. B Chem. 2018;262:365–370. [Google Scholar]
- 162.Bandodkar A.J., Jeerapan I., Wang J. ACS Sens. 2016;1(5):464–482. [Google Scholar]
- 163.Zhu H., Li L., Zhou W., Shao Z., Chen X. J. Mater. Chem. B. 2016;4:7333–7349. doi: 10.1039/c6tb02037b. [DOI] [PubMed] [Google Scholar]
- 164.Kim J., Kim M., Lee M.S., Kim K., Ji S., Kim Y.T., Park J., Na K., Bae K.H., Kyun Kim H., Bien F., Young Lee C., Park J.U. Nat. Commun. 2017;8:14997. doi: 10.1038/ncomms14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pappa A.M., Curto V.F., Braendlein M., Strakosas X., Donahue M.J., Fiocchi M., Malliaras G.G., Owens R.M. Adv. Healthcare Mater. 2016;5:2295–2302. doi: 10.1002/adhm.201600494. [DOI] [PubMed] [Google Scholar]
- 166.Kim J., Valdés-Ramírez G., Bandodkar A.J., Jia W., Martinez A.G., Ramírez J., Mercier P., Wang J. Analyst. 2014;139:1632–1636. doi: 10.1039/c3an02359a. [DOI] [PubMed] [Google Scholar]
- 167.Cao X., Halder A., Tang Y., Hou C., Wang H., Duus J.Ø., Chi Q. Mater. Chem. Front. 2018;2:1944–1986. [Google Scholar]
- 168.Zhu Z., Garcia-Gancedo L., Flewitt A.J., Xie H., Moussy F., Milne W.I. Sensors. 2012;12:5996–6022. doi: 10.3390/s120505996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Inzucchi S.E., Bergenstal R.M., Buse J.B., Diamant M., Ferrannini E., Nauck M., Peters A.L., Tsapas A., Wender R., Matthews D.R. Diabetes Care. 2015;38:140–149. doi: 10.2337/dc14-2441. [DOI] [PubMed] [Google Scholar]
- 170.Steiner M.-S., Duerkop A., Wolfbeis O.S. Chem. Soc. Rev. 2011;40:4805–4839. doi: 10.1039/c1cs15063d. [DOI] [PubMed] [Google Scholar]
- 171.Sanz V., de Marcos S. J. Galban. Biosens. Bioelectron. 2007;22:956–964. doi: 10.1016/j.bios.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 172.Sun X., Li N., Zhou B., Zhao W., Liu L., Huang C., Ma L., Kost A.R. Optic Commun. 2018;416:32–35. [Google Scholar]
- 173.Shehab M., Ebrahim S., Soliman M. J. Lumin. 2017;184:110–116. [Google Scholar]
- 174.Choi M.M.F., Pang W.S.H., Xiao D., Wu X. Analyst. 2001;126:1558–1563. [Google Scholar]
- 175.Liu W., Wu H., Li B., Dong C., Choi M.M.F., Shuang S. Anal. Methods. 2013;5:5154–5160. [Google Scholar]
- 176.Wang Z., Liu F., Lu C. Biosens. Bioelectron. 2012;38:284–288. doi: 10.1016/j.bios.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 177.Li B., Lan D., Zhang Z. Anal. Biochem. 2008;374:64–70. doi: 10.1016/j.ab.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 178.Chaichi M.J., Ehsani M. Sensor. Actuator. B Chem. 2016;223:713–722. [Google Scholar]
- 179.Qi Y., Xiu F.-R., Li B. Anal. Biochem. 2014;449:1–8. doi: 10.1016/j.ab.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 180.Zargoosh K., Chaichi M.j., Shamsipur M., Hossienkhani S., Asghari S. M. Qandalee. Talanta. 2012;93:37–43. doi: 10.1016/j.talanta.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 181.Yu R., Pan C., Chen J., Zhu G., Wang Z.L. Adv. Funct. Mater. 2013;23:5868–5874. [Google Scholar]
- 182.Yi N.Y., He Q., Caligan T.B., Smith G.R., Forsberg L.J., Brenman J.E., Sexton J.Z. Assay Drug Dev. Technol. 2015;13(9):558–569. doi: 10.1089/adt.2015.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Luo Y., Zhang X., Li Y., Deng Jiu, Li X., Qu Y., Lu Y., Liu T., Gao Z., Lin B. RSC Adv. 2018;8:25409–25416. doi: 10.1039/c8ra04040k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jung D.W., Ha H.H., Zheng X., Chang Y.T., Williams D.R. Mol. Biosyst. 2011;7:346–358. doi: 10.1039/c0mb00089b. [DOI] [PubMed] [Google Scholar]
- 185.Jo A., Park J., Park S.B. Chem. Commun. 2013;49:5138–5140. doi: 10.1039/c3cc41529e. [DOI] [PubMed] [Google Scholar]
- 186.Tierney M.J., Tamada J.A., Potts R.O., Jovanovic L., Garg S. Biosens. Bioelectron. 2001;16(9–12):621–629. doi: 10.1016/s0956-5663(01)00189-0. [DOI] [PubMed] [Google Scholar]
- 187.Bandodkar A.J., Jia W., Yardımcı C., Wang X., Ramirez J., Wang J. Anal. Chem. 2015;87(1):394–398. doi: 10.1021/ac504300n. [DOI] [PubMed] [Google Scholar]
- 188.Valdés-Ramírez G., Li Y.-C., Kim J., Jia W., Bandodkar A.J., Nuñez-Flores R., Millercd P.R., Wu S.-Y., Narayan R., Windmiller J.R., Polsky R., Wang J. Electrochem. Commun. 2014;47:58–62. [Google Scholar]
- 189.Sharma S., Saeed A., Johnson C., Gadegaard N., Cass A.E.G. Sens. Bio-Sens. Res. 2017;13:104–108. doi: 10.1016/j.sbsr.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Guo D., Zhang D., Zhang L., Lu G. Sensor. Actuator. B Chem. 2012;173:106–113. [Google Scholar]
- 191.Peng B., Lu J., Balijepalli A.S., Major T.C., Cohan B.E., Meyerhoff M.E. Biosens. Bioelectron. 2013;49:204–209. doi: 10.1016/j.bios.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 192.Agustini D., Bergamini M.F., Marcolino-Junior L.H. Biosens. Bioelectron. 2017;98:161–167. doi: 10.1016/j.bios.2017.06.035. [DOI] [PubMed] [Google Scholar]
- 193.Jebaseeli T.J., Durai C.A.D., Peter J.D. Inform. Syst.; 2019. Sustainable Computing. In Press. [Google Scholar]
- 194.Chen G., Fang C., Chai H.H., Zhou Y., Li W.Y., Yu L. Sensor. Actuator. B Chem. 2019;281:253–261. [Google Scholar]
- 195.Guo J., Huang X., Ma X. Sensor. Actuator. B Chem. 2018;275:446–450. [Google Scholar]
- 196.Bhide A., Muthukumar S., Prasad S. Biosens. Bioelectron. 2018;117:537–545. doi: 10.1016/j.bios.2018.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yoon H., Xuan X., Jeong S. J. Y. Park. Biosens. Bioelectron. 2018;117:267–275. doi: 10.1016/j.bios.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 198.Cohen J. Science. 2012;337:168–217. doi: 10.1126/science.337.6091.168. [DOI] [PubMed] [Google Scholar]
- 199.Kim E.Y., Stanton J., Korber B.T., Krebs K., Bogdan D., Kunstman K., Wu S., Phair J.P., Mirkin C.A., Wolinsky S.M. Nanomedicine. 2008;3:293–303. doi: 10.2217/17435889.3.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Visseaux B., Damond F., Matheron S., Descamps D., Charpentier C. Infect. Genet. Evol. 2016;46:233–240. doi: 10.1016/j.meegid.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 201.Saez-Cirion A., Manel N. Annu. Rev. Immunol. 2018;36:193–220. doi: 10.1146/annurev-immunol-051116-052155. [DOI] [PubMed] [Google Scholar]
- 202.Respess R.A., Cachafeiro A., Withum D., Fiscus S.A., Newman D., Branson B., Varnier O.E., Lewis K., Dondero T.J. J. Clin. Microbiol. 2005;43:506–508. doi: 10.1128/JCM.43.1.506-508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Babamiri B., Salimi A., Hallaj R. Biosens. Bioelectron. 2018;117:332–339. doi: 10.1016/j.bios.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 204.Ma Y., Shen X.-L., Zeng Q., Wang H.-S., Wang L.-S. Talanta. 2017;164:121–127. doi: 10.1016/j.talanta.2016.11.043. [DOI] [PubMed] [Google Scholar]
- 205.Islam S., Shukla S., Bajpai V.K., Han Y.K., Huh Y.S., Kumar A., Ghosh A., Gandhi S. Biosens. Bioelectron. 2019;126:792–799. doi: 10.1016/j.bios.2018.11.041. [DOI] [PubMed] [Google Scholar]
- 206.Shafiee H., Lidstone E.A., Jahangir M., Inci F., Hanhauser E., Henrich T.J., Kuritzkes D.R., Cunningham B.T., Demirci U. Sci. Rep. 2014;4:4116. doi: 10.1038/srep04116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Majd S.M., Salimi A., Astinchap B. Sensor. Actuator. B Chem. 2018;266:178–186. [Google Scholar]
- 208.Ng E., Yao C., Shultz T.O., Ross-Howe S., Wang S.X. Biol. Med. 2019;16:10–19. doi: 10.1016/j.nano.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 209.Long Y., Zhou C., Wang C., Cai H., Yin C., Yang Q., Xiao D. Sci. Rep. 2016;6:23949. doi: 10.1038/srep23949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pan Y., Zhan S., Xia F. Anal. Biochem. 2018;546:5–9. doi: 10.1016/j.ab.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 211.Zhao H.-Q., Qiu G.-H., Liang Z., Li M.-M., Sun B., Qin L., Yang S.-P., Chen W.-H., Chen J.-X. Anal. Chim. Acta. 2016;922:55–63. doi: 10.1016/j.aca.2016.03.054. [DOI] [PubMed] [Google Scholar]
- 212.Sun B., Zhao H.-Q., Xie B.-P., Bai Li-P., Jiang Z.-H., Chen J.-X. J. Inorg. Biochem. 2017;176(2017):17–23. doi: 10.1016/j.jinorgbio.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 213.Lee J.-H., Oh B.-K. J-W. Choi. Biosens. Bioelectron. 2013;49:531–535. doi: 10.1016/j.bios.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 214.Tran L.D., Nguyen B.H., Hieu N.V., Tran H.V., Nguyen H.L., Nguyen P.X. Mater. Sci. Eng. C. 2011;31:477–485. [Google Scholar]
- 215.Kheiri F., Sabzi R.E., Jannatdoust E., Shojaeefar E., Sedghi H. Biosens. Bioelectron. 2011;26(11):4457–4463. doi: 10.1016/j.bios.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 216.Gong Q., Wang Y., Yang H. Biosens. Bioelectron. 2017;89:565–569. doi: 10.1016/j.bios.2016.02.045. [DOI] [PubMed] [Google Scholar]
- 217.Hassen W.M., Chaix C., Abdelghani A., Bessueille F., Leonard D., Jaffrezic-Renault N. Sensor. Actuator. B Chem. 2008;134:755–760. [Google Scholar]
- 218.Hu Y., Li H., Li J. J. Electroanal. Chem. 2018;822:66–72. [Google Scholar]
- 219.Chaad B.H., Lee S.-M., Park J.C., Hwang K.S., Kim S.K., Lee Y.-S., Ju B.-K., Kim T.S. Biosens. Bioelectron. 2009;25(1):130–135. doi: 10.1016/j.bios.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 220.Zhu H., Zhang M., Zou L., Li R., L Ling. Talanta. 2017;173:9–13. doi: 10.1016/j.talanta.2017.05.043. [DOI] [PubMed] [Google Scholar]
- 221.Wang X., Lou X., Wang Y., Guo Q., Fang Z., Zhong X., Mao H., Jin Q., Wu L., Zhao H. J. Zhao. Biosens. Bioelectron. 2010;25(8):1934–1940. doi: 10.1016/j.bios.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 222.Li F., Li H., Wang Z., Wu J., Wang W., Zhou L., Xiao Q., Pu Q. Sensor. Actuator. B Chem. 2018;271:189–194. [Google Scholar]
- 223.Ajetunmobi A., Prina-Mello A., Volkovab Y., Corvin A., Tropea D. Prog. Neurobiol. 2014;123:18–36. doi: 10.1016/j.pneurobio.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 224.Li X., Ma M., Xin X., Tang Y., Zhao G., Xiao X. RSC Adv. 2019;9:16701–16712. doi: 10.1039/c9ra01605h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhang W., Wang W., Yu D.X., Xiao Z., He Z. Nanomedicine. 2018;13(18) doi: 10.2217/nnm-2018-0163. [DOI] [PubMed] [Google Scholar]
- 226.Fekete Z. Sensor. Actuator. B Chem. 2015;215:300–315. [Google Scholar]
- 227.Frey O., van der Wal P.D., Spieth S., Brett O., Seidl K., Paul O., Ruther P., Zengerle R., de Rooij N.F. J. Neural. Eng. 2011;8 doi: 10.1088/1741-2560/8/6/066001. [DOI] [PubMed] [Google Scholar]
- 228.Pongrácz A., Fekete Z., Márton G., Bérces Z., Ulbert I., Fürjes P. Sens. Actuators, B. 2013;189:97. [Google Scholar]
- 229.Jack C.R., Jr., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Nordberg A., Rinne J.O., Kadir A., Langstrom B. Nat. Rev. Neurol. 2010;6:78–87. doi: 10.1038/nrneurol.2009.217. [DOI] [PubMed] [Google Scholar]
- 231.Demeritte T., Nellore B.P.V., Kanchanapally R., Sinha S.S., Pramanik A., Chavva S.R., Ray P.C. ACS Appl. Mater. Interfaces. 2015;7:13693–13700. doi: 10.1021/acsami.5b03619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kim C.-B., Lim E.-G., Shin S.W., Krause H.J., Hong H. Biosens. Bioelectron. 2016;83:293–299. doi: 10.1016/j.bios.2016.04.076. [DOI] [PubMed] [Google Scholar]
- 233.Hsu W.-C., Sugiarto T., Lin Y.-J., Yang F.-C., Lin Z.-Y., Sun C.-T., Hsu C.-L., Chou K.-N. Sensors. 2018;18(10):3397. doi: 10.3390/s18103397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Din S.D., Godfrey A., Rochester L. IEEE J. Biomed. Health Inform. 2016;20:838–847. doi: 10.1109/JBHI.2015.2419317. [DOI] [PubMed] [Google Scholar]
- 235.Xu T., Ma J., Ma Z. New J. Chem. 2015;39:1006–1012. [Google Scholar]
- 236.Jin Y., Mao H., Jin Q., Zhao J. Anal. Methods. 2016;8:4861–4866. [Google Scholar]
- 237.Liu M., Jia C., Huang Y., Lou X., Yao S., Jin Q., Zhao J., Xiang J. Analyst. 2010;135:327. doi: 10.1039/b916629g. [DOI] [PubMed] [Google Scholar]
- 238.Jokerst J.V., Raamanathan A., Christodoulides N., Floriano P.N., Pollard A.A., Simmons G.W., Wong J., Gage C., Furmaga W.B., Redding S.W., McDevitt J.T. Biosens. Bioelectron. 2009;24:3622–3659. doi: 10.1016/j.bios.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hu M., Yan J., He Y., Lu H., Weng L., Song S., Fan C., Wang L. ACS Nano. 2009;4:488–494. doi: 10.1021/nn901404h. [DOI] [PubMed] [Google Scholar]
- 240.Ge L., Wang S., Song X., Ge S., Yu J. Lab Chip. 2012;12:3150–3158. doi: 10.1039/c2lc40325k. [DOI] [PubMed] [Google Scholar]
- 241.Liang L., Ge S., Li L., Liu F., Yu J. Anal. Chim. Acta. 2015;862:70–76. doi: 10.1016/j.aca.2014.12.050. [DOI] [PubMed] [Google Scholar]
- 242.Yu X., Xia H.-S., Sun Z.-D., Lin Y., Wang K., Yu J., Tang H., Pang D.-W., Zhang Z.-L. Biosens. Bioelectron. 2013;41:129–136. doi: 10.1016/j.bios.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 243.Zhang H., Liu L., Fu X., Zhu Z. Biosens. Bioelectron. 2013;42:23–30. doi: 10.1016/j.bios.2012.10.076. [DOI] [PubMed] [Google Scholar]
- 244.Lin Y.-H., Wang C.-C., Lei K.F. Biomed. Microdevices. 2014;16:199–207. doi: 10.1007/s10544-013-9822-4. [DOI] [PubMed] [Google Scholar]
- 245.Diercks A.H., Ozinsky A., Hansen C.L., Spotts J.M., Rodriguez D.J., Aderem A. Anal. Biochem. 2009;386:30–35. doi: 10.1016/j.ab.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tang H., Chen J., Nie L., Kuang Y., Yao S. Biosens. Bioelectron. 2007;22:1061. doi: 10.1016/j.bios.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 247.Zhong Z., Wu W., Wang D., Wang D., Shan J., Qing Y., Zhang Z. Biosens. Bioelectron. 2010;25:2379–2383. doi: 10.1016/j.bios.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 248.Yuan Y.R., Yuan R., Chai Y.Q., Zhuo Y., Miao X.M. J. Electroanal. Chem. 2009;626:6. [Google Scholar]
- 249.Sun G., Ding Y.-n., Ma C., Zhang Y., Ge S., Yu J., Song X. Electrochim. Acta. 2014;147:650–656. [Google Scholar]
- 250.Ge L., Yan J., Song X., Yan M., Ge S., Yu J. Biomaterials. 2012;33:1024–1031. doi: 10.1016/j.biomaterials.2011.10.065. [DOI] [PubMed] [Google Scholar]
- 251.Xu H., Wang Y., Wang L., Song Y., Luo J., Cai X. Nanomaterials. 2016;6:132. doi: 10.3390/nano6070132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Yang P., Li X., Wang L., Wu Q., Chen Z., Lin X. J. Electroanal. Chem. 2014;732:38–45. [Google Scholar]
- 253.Wu Y., Xue P., Hui K.M., Kang Y. Biosens. Bioelectron. 2014;52:180–187. doi: 10.1016/j.bios.2013.08.039. [DOI] [PubMed] [Google Scholar]
- 254.Tang D., Ren J. Anal. Chem. 2008;80:8064–8070. doi: 10.1021/ac801091j. [DOI] [PubMed] [Google Scholar]
- 255.Wang L., Hua E., Liang M., Ma C., Liu Z., Sheng S., Liu M., Xie G., Feng w. Biosens. Bioelectron. 2014;51:201–207. doi: 10.1016/j.bios.2013.07.049. [DOI] [PubMed] [Google Scholar]
- 256.Kerman K., Kraatz H.B. Biosens. Bioelectron. 2009;24:1484. doi: 10.1016/j.bios.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 257.Munge B.S., Krause C.E., Malhotra R., Patel V., Silvio Gutkind J., Rusling J.F. Electrochem. Commun. 2009;11:1009. doi: 10.1016/j.elecom.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Liu W., Wei H., Lin Z., Mao S., Lin J.-M. Biosens. Bioelectron. 2011;28:438–442. doi: 10.1016/j.bios.2011.07.067. [DOI] [PubMed] [Google Scholar]
- 259.Yang Y., Zhong Z., Liu H., Zhu T., Wu J., Li M., Wang D. Electroanalysis. 2008;20:2621. [Google Scholar]
- 260.Chon H., Lee S., Son S.W., Oh C.H., Choo J. Anal. Chem. 2009;81:3029–3034. doi: 10.1021/ac802722c. [DOI] [PubMed] [Google Scholar]
- 261.Shang C., Wang G., He M., Chang X., Fan J., Liu K., Peng H., Fang Y. Sensor. Actuator. B Chem. 2017;241:1316–1323. [Google Scholar]
- 262.Yang H., Kong Q., Wang S., Xu J., Bian Z., Zheng X., Ma C., Ge S., Yu J. Biosens. Bioelectron. 2014;61:21–27. doi: 10.1016/j.bios.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 263.Chen S., Zhao Q., Zhang L., Wang L., Zeng Y., Huang H. Sensor. Actuator. B Chem. 2015;221:1391–1397. [Google Scholar]
- 264.Zhu Y., Chandra P., Shim Y.-B. Anal. Chem. 2013;85(2):1058–1064. doi: 10.1021/ac302923k. [DOI] [PubMed] [Google Scholar]
- 265.Qiu Y., Zhou B., Yang X., Long D., Hao Y., Yang P. ACS Appl. Mater. Interfaces. 2017;9(20):16848–16856. doi: 10.1021/acsami.7b02793. [DOI] [PubMed] [Google Scholar]
- 266.Campbell E., Hasan Md T., Rodriguez R.G., Akkaraju G.R., Naumov A.V. Biomater. Sci. Eng. 2019;59:4671–4682. doi: 10.1021/acsbiomaterials.9b00603. [DOI] [PubMed] [Google Scholar]
- 267.Gu W.L., Deng X., Gu X.X., Jia X.F., Lou B.H., Zhang X.W. Anal. Chem. 2015;87(3):1876–1881. doi: 10.1021/ac503966u. [DOI] [PubMed] [Google Scholar]
- 268.Wang Y.-Z., Hao N., Feng Q.M., Shi H.W., Xu J.J., Chen H.Y. Biosens. Bioelectron. 2016;77:76–82. doi: 10.1016/j.bios.2015.08.057. [DOI] [PubMed] [Google Scholar]
- 269.Ruiyi L., Fangchao C., Haiyan Z., Xiulan S., Zaijun L. Biosens. Bioelectron. 2018;119:156–162. doi: 10.1016/j.bios.2018.07.060. [DOI] [PubMed] [Google Scholar]
- 270.Sun D., Lu J., Luo Z., Zhang L., Liu P., Chen Z. Biosens. Bioelectron. 2018;120:8–14. doi: 10.1016/j.bios.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 271.Spencer A., Spruell C., Nandi S., Wong M., Creixell M., Baker A.B. Lab Chip. 2016;16:142–152. doi: 10.1039/c5lc00994d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Gui W.-J., Lin S.-Q., Chen Y.-Y., Zhang X.-E., Bi L.-J., Jiang T. Biochem. Biophys. Res. Commun. 2011;405(2):272–277. doi: 10.1016/j.bbrc.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 273.Barik S., Sureka K., Mukherjee P., Basu J., Kundu M. Mol. Microbiol. 2010;75(3):592–606. doi: 10.1111/j.1365-2958.2009.07008.x. [DOI] [PubMed] [Google Scholar]
- 274.Chutichetpong P., Cheeveewattanagul N., Srilohasin P., Rijiravanich P., Chaiprasertd A., Surareungchai W. Anal. Chim. Acta. 2018;1025:108–117. doi: 10.1016/j.aca.2018.04.064. [DOI] [PubMed] [Google Scholar]
- 275.Miodek A., Mejri N., Gomgnimbou M., Sola C., Korri-Youssoufi H. Anal. Chem. 2015;87(18):9257–9264. doi: 10.1021/acs.analchem.5b01761. [DOI] [PubMed] [Google Scholar]
- 276.Liang G., Chen H., Zhang S., Wu W., Kong J. Analyst. 2012;137:675–679. doi: 10.1039/c1an15897j. [DOI] [PubMed] [Google Scholar]
- 277.Mogha N.K., Sahu V., Kishore Sharma R., Masram D.T. J. Mater. Chem. B. 2018;6:5181–5187. doi: 10.1039/c8tb01604f. [DOI] [PubMed] [Google Scholar]
- 278.Bizid S., Blili S., Mlika R., Said A.H., Korri-Youssoufi H. Talanta. 2018;184:475–483. doi: 10.1016/j.talanta.2018.03.025. [DOI] [PubMed] [Google Scholar]
- 279.Torres-Chavolla E., Alocilja E.C. Biosens. Bioelectron. 2011;26:4614–4618. doi: 10.1016/j.bios.2011.04.055. [DOI] [PubMed] [Google Scholar]
- 280.Das M., Sumana G., Nagarajan R., Malhotra B.D. Appl. Phys. Lett. 2010;96:133703. [Google Scholar]
- 281.Miodek A., Mejri N., Gomgnimbou M., Sola C., Korri-Youssoufi H. Anal. Chem. 2015;87:9257–9264. doi: 10.1021/acs.analchem.5b01761. [DOI] [PubMed] [Google Scholar]
- 282.Haddaoui M., Sola C., Raouafi N., Korri-Youssoufi H. Biosens. Bioelectron. 2019;128:76–82. doi: 10.1016/j.bios.2018.11.045. [DOI] [PubMed] [Google Scholar]
- 283.Tufa L.T., Oh S., Tran V.T., Kim J., Jeong K.-J., Park T.J., Kim H.J. J. Lee. Electrochimica Acta. 2018;290:369–377. [Google Scholar]
- 284.Yu X., Chai Y., Jiang J., Cui H. J. Photochem. Photobiol. Chem. 2012;241:45–51. [Google Scholar]
- 285.Perumalag V., Saheed M.S.M., Mohamed N.M., Saheed M.S.M., Murthe S.S., Gopinath S.C.B., Chiu J.M. Biosens. Bioelectron. 2018;116:116–122. doi: 10.1016/j.bios.2018.05.042. [DOI] [PubMed] [Google Scholar]
- 286.Li L., Yuan Y., Chen Y., Zhang P., Bai Y., Bai L. Mikrochim. Acta. 2018;185(8):379. doi: 10.1007/s00604-018-2884-5. [DOI] [PubMed] [Google Scholar]
- 287.Liu C., Jiang D., Xiang G., Liu L., Liu F., Pu X. Analyst. 2014;139:5460–5465. doi: 10.1039/c4an00976b. [DOI] [PubMed] [Google Scholar]
- 288.He Y., Liu D., He X., Cui H. Chem. Commun. 2011;47:10692–10694. doi: 10.1039/c1cc14389a. [DOI] [PubMed] [Google Scholar]
- 289.Li N.g, Huang X., Sun D., Yu W., Tan W., Luo Z., Chen Z. Microchimica Acta. 2018;(12):185. doi: 10.1007/s00604-018-3081-2. [DOI] [PubMed] [Google Scholar]
- 290.Prabowo B.A., Chang Y.-F., Lai H.-C., Alom A., Pal P., Lee Y.-Y., Chiu N.-F., Hatanaka K., Su L.-C., Liu K.-C. Sensor. Actuator. B Chem. 2018;254:742–748. [Google Scholar]
- 291.Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Anal. Chim. Acta. 2018;1044:102–109. doi: 10.1016/j.aca.2018.07.045. [DOI] [PubMed] [Google Scholar]
- 292.Wang L., Leng C., Tang S., Lei J., Ju H. Biosens. Bioelectron. 2012;38:421–424. doi: 10.1016/j.bios.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 293.Garzan A., Willby M.J., Green K.D., Tsodikov O.V., Posey J.E., Garneau-Tsodikova S. ACS Med. Chem. Lett. 2016;712:1219–1221. doi: 10.1021/acsmedchemlett.6b00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Gising J., Nilsson M.T., Odell L.R., Yahiaoui S., Lindh M., Iyer H., Sinha A.M., Srinivasa B.R., Larhed M., Mowbray S.L., Karlén A. J. Med. Chem. 2012;556:2894–2898. doi: 10.1021/jm201212h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Stanley S.A., Grant S.S., Kawate T., Iwase N., Shimizu M., Wivagg C., Silvis M., Kazyanskaya E., Aquadro J., Golas A., Fitzgerald M., Dai H., Zhang L., Hung D.T. ACS Chem. Biol. 2012;78:1377–1384. doi: 10.1021/cb300151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Bhusal R.P., Patel K., Kwai B.X.C., Swartjes A., Bashiri G., Reynisson J., Sperry J., Leung I.K.H. Med. Chem. Commun. 2017;8:2155–2163. doi: 10.1039/c7md00456g. [DOI] [PMC free article] [PubMed] [Google Scholar]



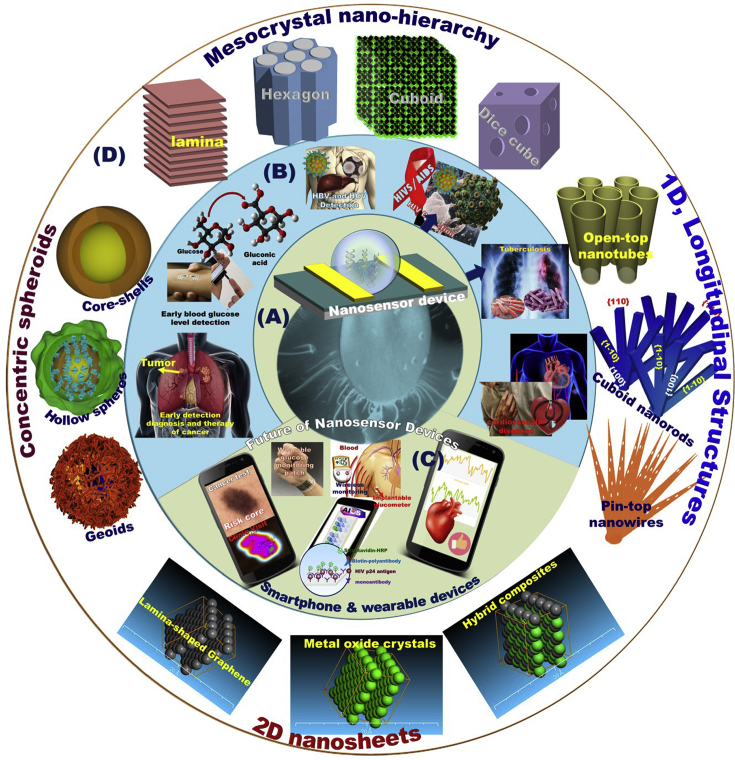
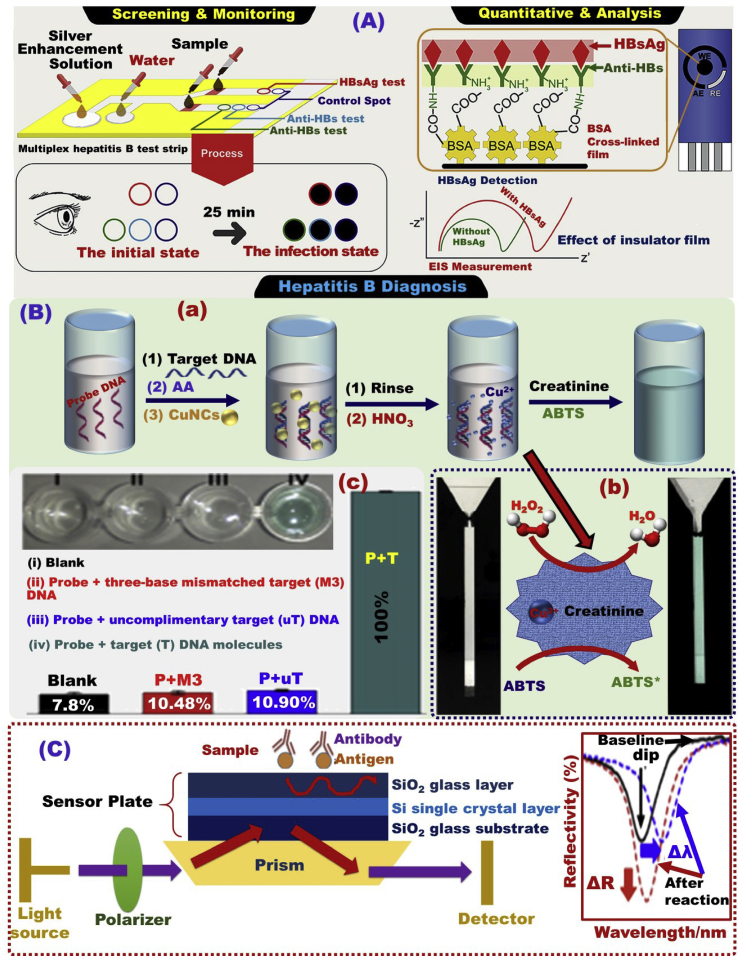
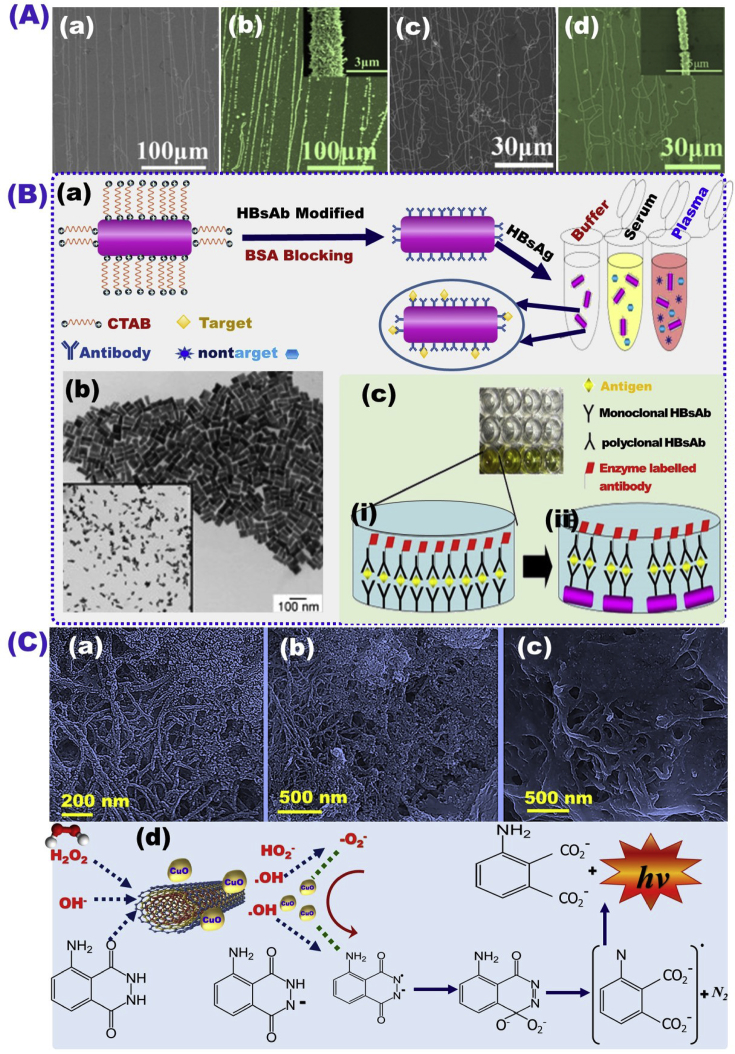
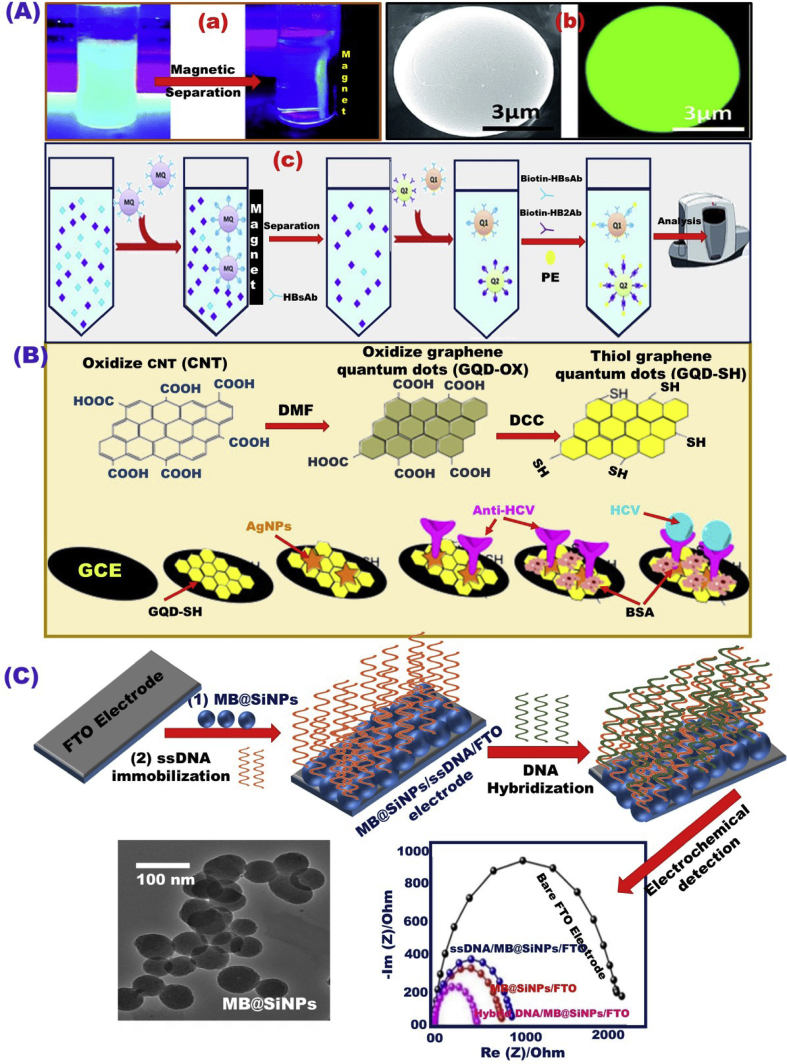
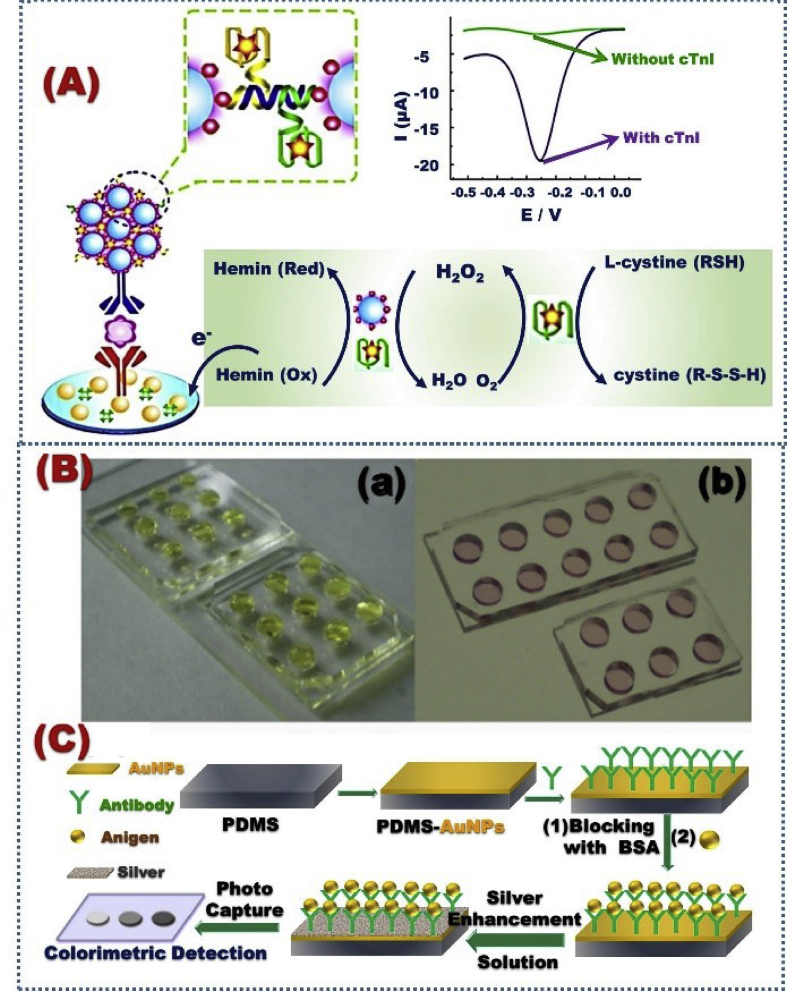
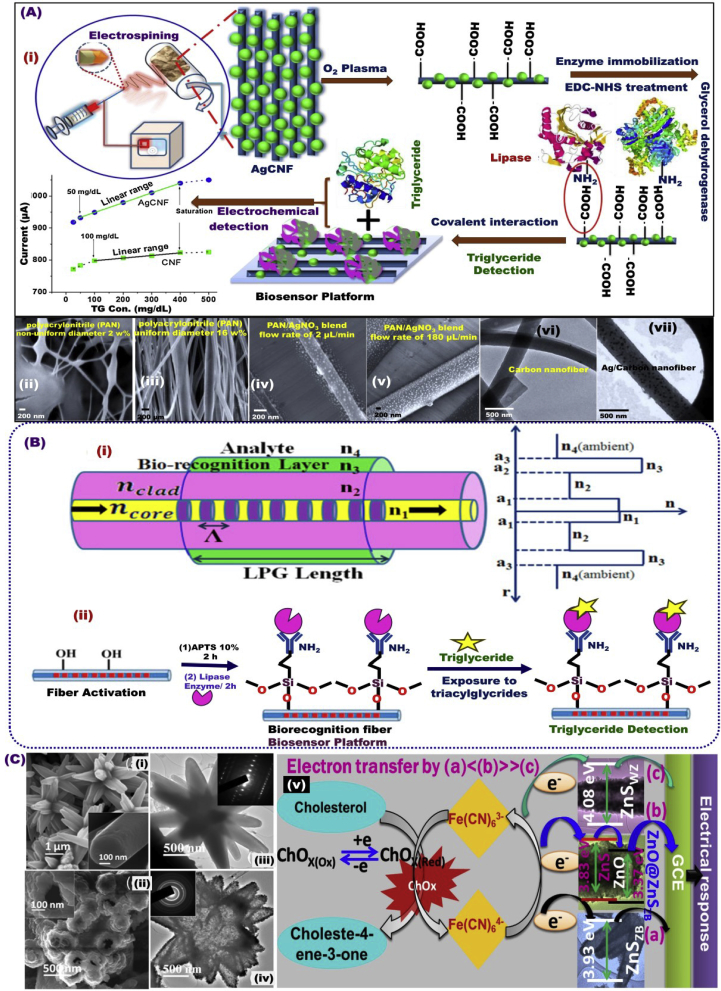

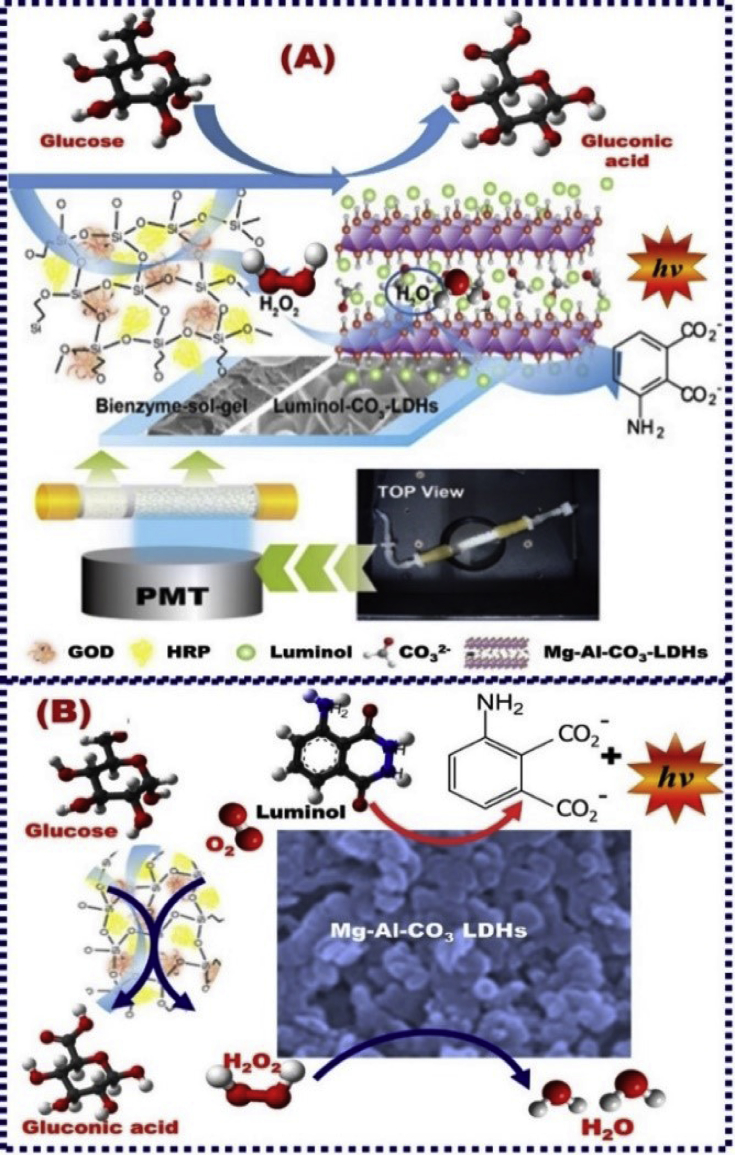
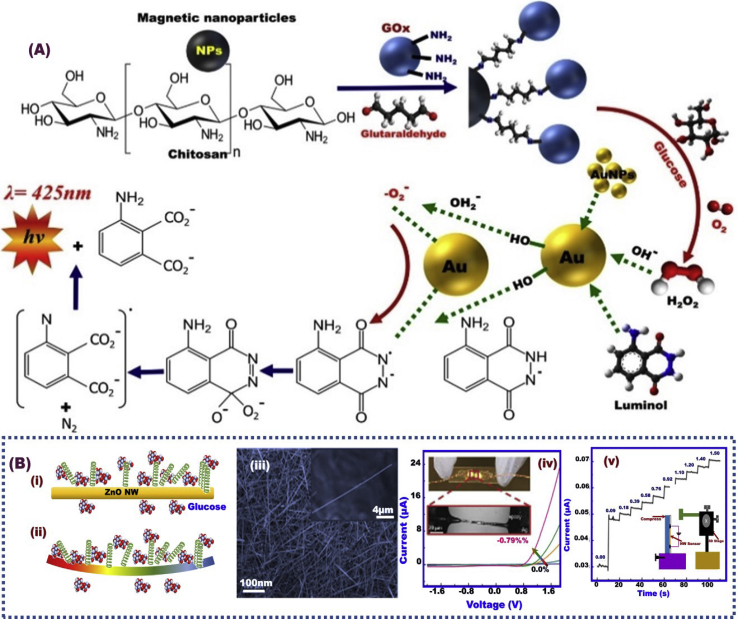
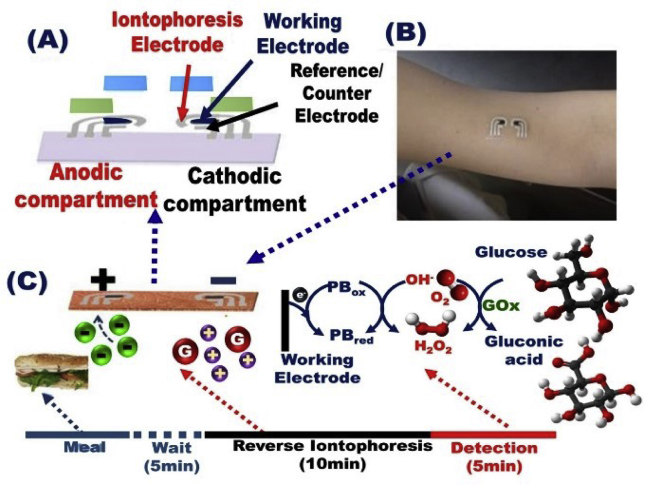
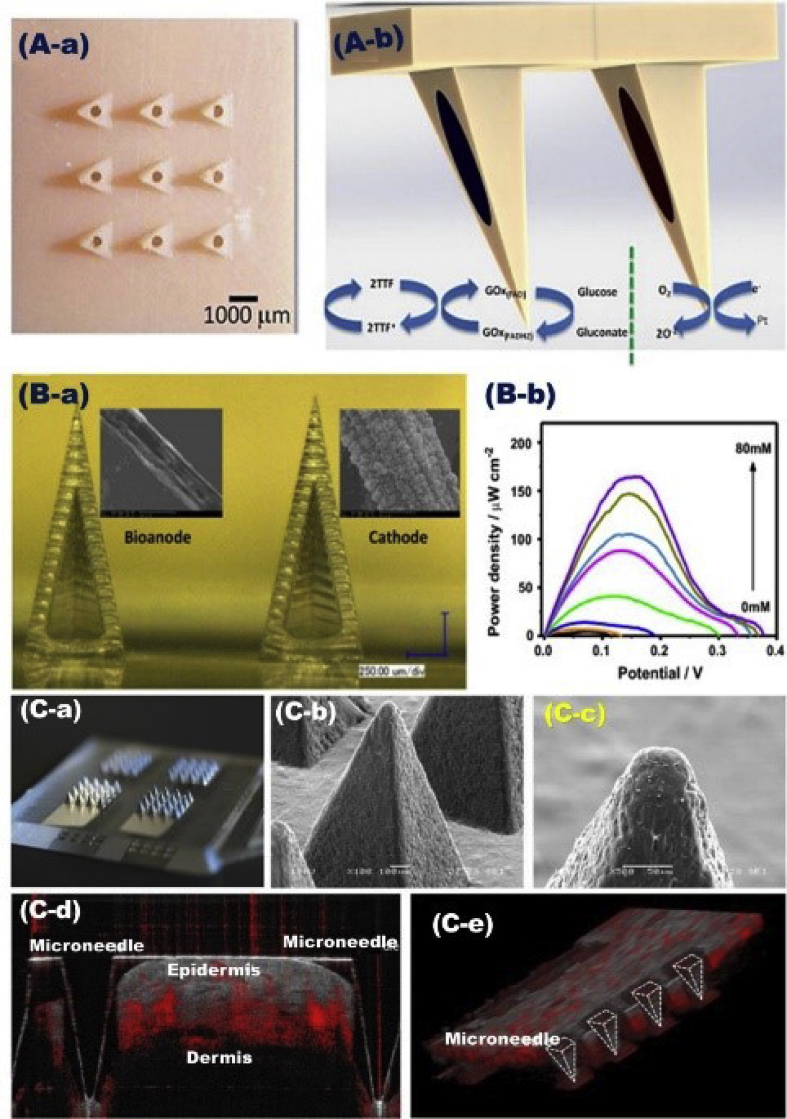
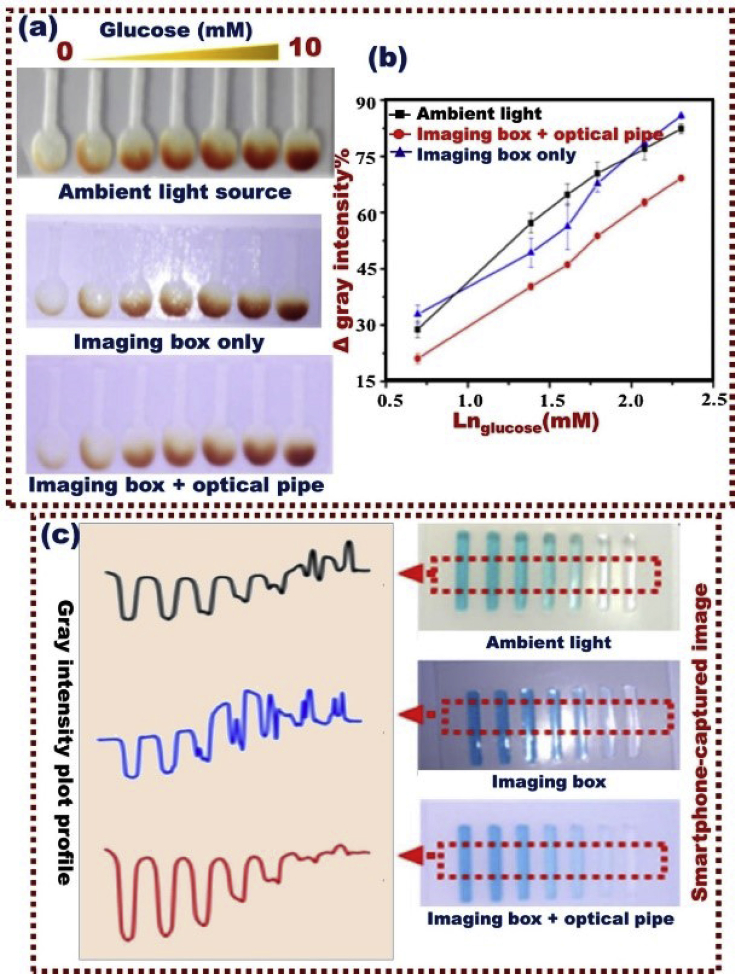
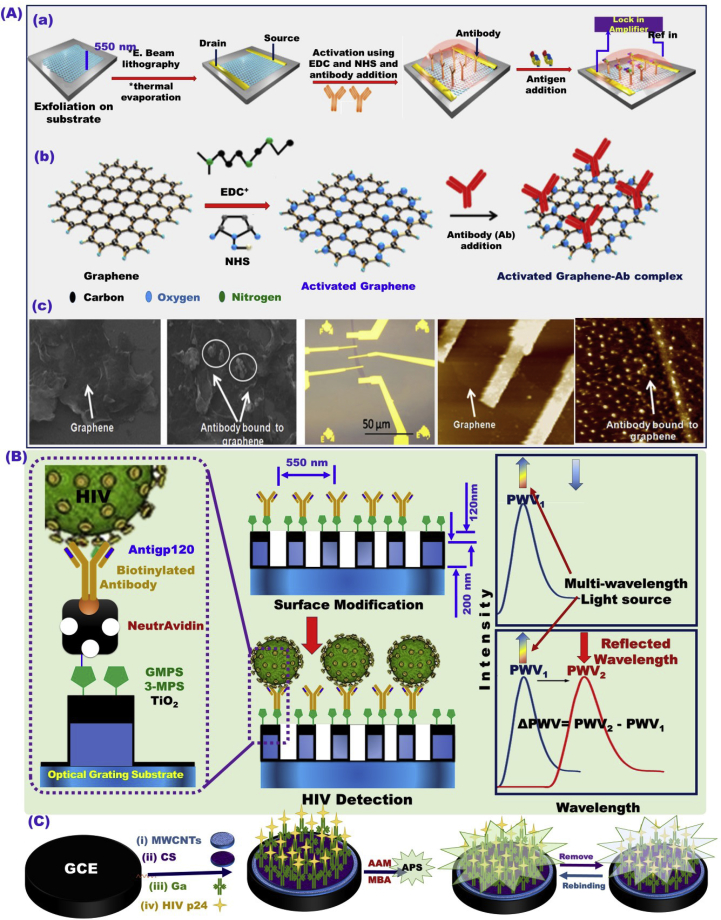
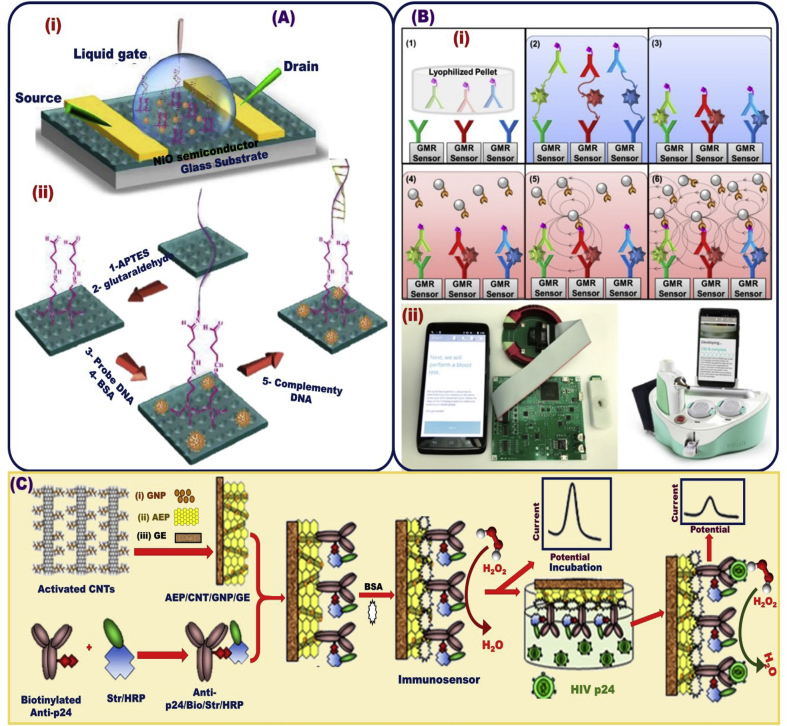
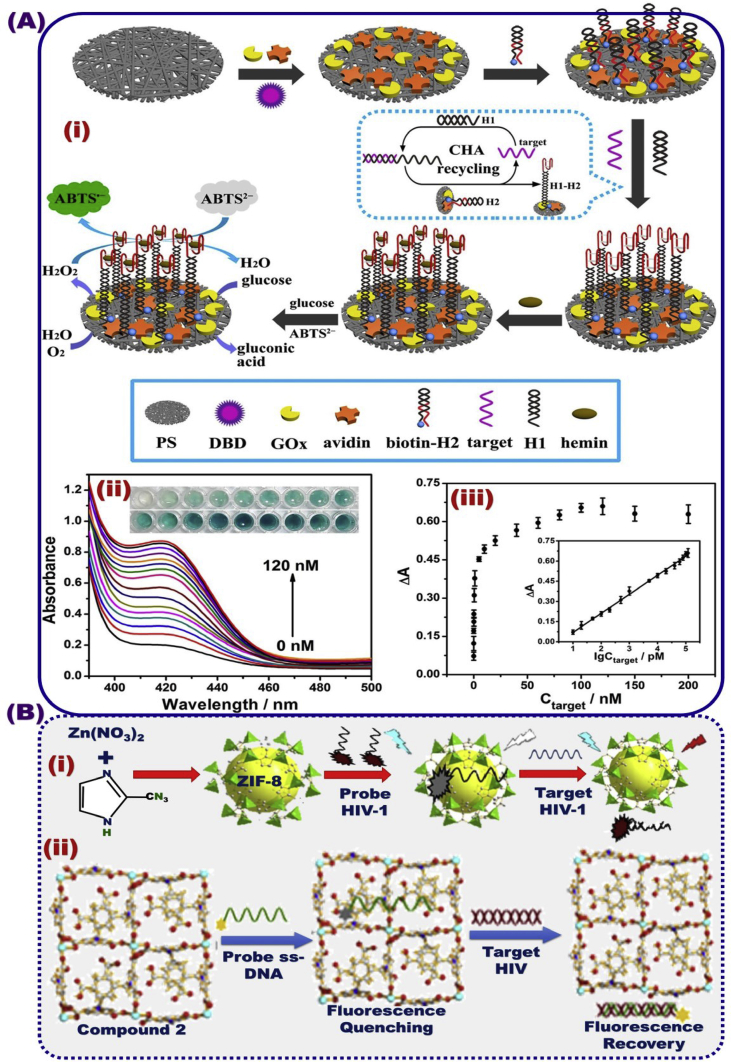
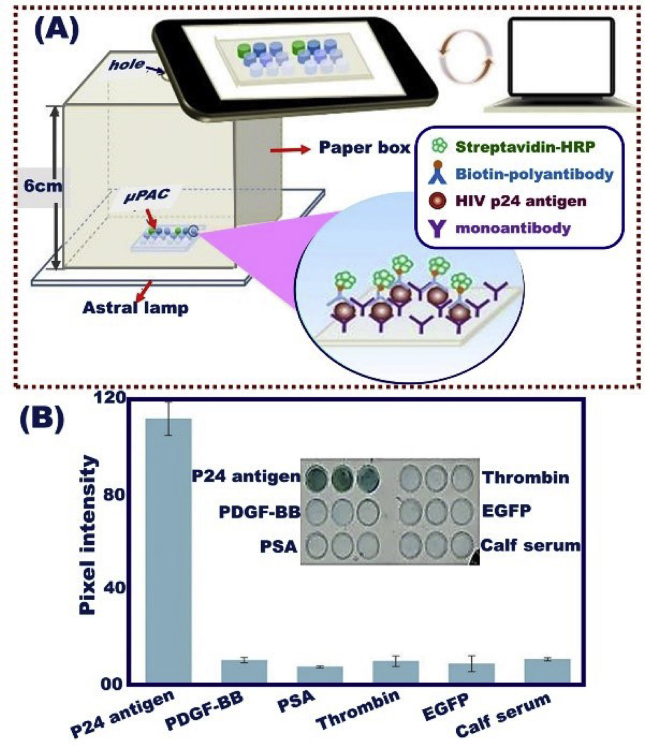









 0
0 1
1