Abstract
Thermal springs are excellent locations for discovery of thermostable microorganisms and enzymes. In this study, we identify a novel thermotolerant bacterial strain related to Paenibacillus dendritiformis, denoted Paenibacillus sp. 3179, which was isolated from a thermal spring in East Greenland. A functional expression library of the strain was constructed, and the library screened for β‐d‐galactosidase and α‐l‐fucosidase activities on chromogenic substrates. This identified two genes encoding a β‐d‐galactosidase and an α‐l‐fucosidase, respectively. The enzymes were recombinantly expressed, purified, and characterized using oNPG (2‐nitrophenyl‐β‐d‐galactopyranoside) and pNP‐fucose (4‐nitrophenyl‐α‐l‐fucopyranoside), respectively. The enzymes were shown to have optimal activity at 50°C and pH 7–8, and they were able to hydrolyze as well as transglycosylate natural carbohydrates. The transglycosylation activities were investigated using TLC and HPLC, and the β‐d‐galactosidase was shown to produce the galactooligosaccharides (GOS) 6'‐O‐galactosyllactose and 3'‐O‐galactosyllactose using lactose as substrate, whereas the α‐l‐fucosidase was able to transfer the fucose moiety from pNP‐fuc to lactose, thereby forming 2'‐O‐fucosyllactose. Since enzymes that are able to transglycosylate carbohydrates at elevated temperature are desirable in many industrial processes, including food and dairy production, we foresee the potential use of enzymes from Paenibacillus sp. 3179 in the production of, for example, instant formula.
Keywords: galactooligosaccharides, Paenibacillus, transglycosylation, α‐l‐fucosidase, β‐d‐galactosidase
Characterization of novel thermotolerant, transglycosylating enzymes from Paenibacillus sp., which has been isloated from a thermal spring in Greenland. The enzymes are an alpha‐fucosidase and a beta‐galactosidase, respectively. Both are able to hydrolyse as well as transglycosylate natural substrates.
1. INTRODUCTION
Thermal springs are hot spots of microbial biodiversity that may be utilized within agriculture, food and feed industry, textile industry, and biotechnology (Gupta, Gupta, Capalash, & Sharma, 2017; Mahajan & Balachandran, 2017; Sahay et al., 2017). Thermostable enzymes from such environments have attracted increased interest from industry and academia due to the opportunities for high‐temperature processes and interesting molecular structures (DeCastro, Rodríguez‐Belmonte, & González‐Siso, 2016; Elleuche, Schäfers, Blank, Schröder, & Antranikian, 2015; Mehta, Singhal, Singh, Damle, & Sharma, 2016; Ribeiro, Sánchez, Hidalgo, & Berenguer, 2017; Urbieta et al., 2015). Especially within dairy production, processes at high or low temperatures are required due to low growth capabilities of food spoilers (Hervert, Martin, Boor, & Wiedmann, 2017; Ruusunen, Surakka, Korkeala, & Lindström, 2012). Furthermore, in some processes like oligosaccharide synthesis high temperatures are desired to obtain higher solubility of the reactants (Hassan et al., 2015; Yu & O'Sullivan, 2018). Galactooligosaccharides (GOS) in particular have been studied thoroughly (Singla & Chakkaravarthi, 2017). GOS are indigestible oligosaccharides, which have a documented positive effect on development and growth of beneficial gut bacteria such as bifidobacteria and lactobacilli in the human gastrointestinal tract (Gibson, Probert, Jan, Rastall, & Roberfroid, 2004; Knol, Scholtens, et al., 2005; Ladirat et al., 2014). Additionally, they have been shown to prevent adherence of pathogenic bacteria such as enteropathogenic Escherichia coli and Salmonella enterica to the cell walls of the colon (Knol, Boehm, et al., 2005; Searle et al., 2009; Shoaf, Mulvey, Armstrong, & Hutkins, 2006). Synthesis of oligosaccharides can be carried out by either chemical synthesis using acid hydrolysis, which is a relatively nonspecific, energy‐demanding, and time‐consuming process, or by enzymatic synthesis carried out by glycosyltransferases (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/4.html) or glycoside hydrolases (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/2/1.html). Glycoside hydrolases (GH) are the preferred catalysts due to high availability, natural hydrolytic function, ability to glycosylate a wide variety of hydroxyl group‐containing molecules, and the use of cheap and simple donor substrates (Crout & Vic, 1998; de Roode, Franssen, Padt, & Boom, 2003; Watt, Lowden, & Flitsch, 1997). One of the cheapest donor substrates for GOS synthesis is lactose (β‐d‐galactopyranosyl‐(1 → 4)‐d‐glucopyranose) which is a by‐product from cheese production. Great efforts have been spent on converting this low‐price disaccharide into carbohydrates of higher value, especially enzymatic conversion of lactose to lactobionic acid (Goderska, Szwengiel, & Czarnecki, 2014; Gutiérrez, Hamoudi, & Belkacemi, 2012), to the sweetener d‐tagatose (Jørgensen, Hansen, & Stougaard, 2004; Wanarska & Kur, 2012; Zhan et al., 2014) and to GOS (Arreola et al., 2014; Jørgensen, Hansen, & Stougaard, 2001; Splechtna et al., 2006). Synthesis of GOS through enzymatic transglycosylation results in linear oligomers of galactose (n = 2–6) joined by β1‐2, β1‐3, β1‐4, or β1‐6 linkages with a glucose at the reducing end (Ishikawa et al., 2015; Li et al., 2009; Torres, Gonçalves, Teixeira, & Rodrigues, 2010; Wu et al., 2013). Due to the risk of contamination by food‐spoiling bacteria during GOS production, focus has been on enzymes that are active at either high or low temperatures. Cold‐active enzymes that are able to catalyze the formation of GOS have been described from, for example, Pseudoalteromonas sp. 22b (Cieśliński et al., 2005) and Alkalilactibacillus ikkensis (Schmidt & Stougaard, 2010), whereas thermostable GOS‐producing enzymes have been described from, for example, Sulfolobus spp. (Park, Kim, Lee, & Oh, 2010; Park, Kim, Lee, Kim, & Oh, 2008), Halothermothrix orenii (Hassan et al., 2015), and hot environment metagenomes (Gupta, Govil, Capalash, & Sharma, 2012). In addition to production of GOS, where lactose is used as donor as well as acceptor, some β‐d‐galactosidases may use other acceptors than lactose (Díez‐Municio, Herrero, Olano, & Moreno, 2014). Mu, Chen, Wang, Zhang, and Jiang (2013) and Li et al. (2009) have described that a β‐d‐galactosidase from Bacillus circulans forms the trisaccharide lactosucrose (O‐β‐d‐galactopyranosyl‐(1 → 4)‐O‐α‐d‐glucopyranosyl‐(1 → 2)‐β‐d‐fructofuranoside) from lactose and sucrose, and Guerrero, Vera, Acevedo, and Illanes (2015), Wang, Yang, Hua, Zhao, and Zhang (2013), and Tang et al. (2011) have reported the enzymatic synthesis of the disaccharide lactulose (4‐O‐β‐d‐galactopyranosyl‐β‐d‐fructofuranose). GOS, lactosucrose, and lactulose all show prebiotic effects and are presumed to play an important role in stimulating growth of beneficial bifidobacteria in the human intestine (Gibson et al., 2004; Mu et al., 2013; Wang et al., 2013). Furthermore, previous studies have shown that several bifidobacterial species can grow on GOS as sole carbon source (Garrido et al., 2013; Gavlighi, Michalak, Meyer, & Mikkelsen, 2013; Rada et al., 2008).
Another group of prebiotics belong to the fucosylated oligosaccharides. l‐fucose (6‐deoxy‐l‐galactose) is a monosaccharide which is a component of many glycoproteins and glycolipids, and is often located at the nonreducing end of oligosaccharide chains via an α‐(1 → 2) linkage to a d‐galactosyl residue or via α‐(1 → 3), α‐(1 → 4), or α‐(1 → 6) linkage to N‐acetyl‐d‐glucosamine (reviewed by Becker & Lowe, 2003). In addition, fucosylated oligosaccharides are major components in human breast milk, while almost absent in bovine milk (Kunz, Kuntz, & Rudloff, 2014; Kunz & Rudloff, 1993; Kunz, Rudloff, Baier, Klein, & Strobel, 2000; Thurl, Henker, Taut, Tovar, & Sawatzki, 1993), which is the basis for infant formula. By supplementing bovine instant formula with fucosylated oligosaccharides as well as GOS, it has been shown that growth of bifidobacteria and lactobacilli in the fecal microflora of infants was improved (Boehm et al., 2002; Moro et al., 2002), and α‐linked fucosylated oligosaccharides have furthermore been shown to prevent infection with the pathogenic bacteria E. coli and Campylobacter jejuni (Morrow, Ruiz‐Palacios, Jiang, & Newburg, 2005). Fucosylated oligosaccharides such as 2'‐O‐fucosyllactose (2FL) and 3'‐O‐fucosyllactose (3FL) can be synthesized enzymatically through transglycosylation by the action of transfucosylating fucosidases (Lezyk et al., 2016; Zeuner et al., 2018) or by using fucosyltransferases, where an l‐fucose unit is transferred from guanosine 5'‐phosphate (GDP)‐l‐fucose to an acceptor substrate (Chin, Kim, Kim, Jung, & Seo, 2017; Chin, Kim, Lee, & Seo, 2015; Sprenger, Baumgärtner, & Albermann, 2017).
In this study, we screened hitherto unexplored hot springs in East Greenland for bacteria expressing thermostable glycoside hydrolases. One of the isolates, affiliated to Paenibacillus sp., was shown to produce a transglycosylating β‐d‐galactosidase and a transglycosylating α‐l‐fucosidase. The two enzymes were recombinantly expressed in E. coli, purified, and functionally characterized, and here, we report on the characterization of the β‐d‐galactosidase and the α‐l‐fucosidase from Paenibacillus sp. 3179.
2. MATERIALS AND METHODS
2.1. Bacterial strains and chemicals
All chemicals were purchased from Sigma‐Aldrich (St. Louis) unless otherwise stated. All enzymes were purchased from New England Biolabs (Ipswich) unless otherwise stated.
For construction of a genomic library and initial protein expression and characterization, the plasmid pUC18ΔlacZ (Schmidt & Stougaard, 2010) was used in E. coli TOP10 (F– mcrA Δ(mrr‐hsdRMS‐mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7,697 galU galK rpsL (StrR) endA1 nupG) (Thermo Fisher Scientific, Waltham, MA, USA). For enzyme expression, the cloning vector pET9a‐USER‐1 (Schultz‐Johansen et al., 2018) was used in the lactose‐deficient strain E. coli BL21(DE3)ΔlacZ, which was kindly provided by Professor Jin‐Ho Seo from Seoul National University, Seoul, the Republic of Korea.
2.2. Samples, isolation, and growth conditions
Samples were collected from a hot spring in Knighton Fjord in East Greenland (69°21'60.4”N; 24°39'67.1”W) in 2001, where more than 30 discharges were observed with temperatures around 54°C. Gravel samples from the bottom and the sides of the hot springs were plated directly on R2 agar, which contained per liter: 0.5 g yeast extract, 0.5 g Bacto peptone, 0.5 g casamino acids, 0.05 g MgSO4•7H2O, 0.1 g NaCl, 0.5 g soluble starch, 0.3 g sodium pyruvate, 0.3 g KH2PO4, and 20 g agar. The substrate was buffered to pH 7 with 0.01 M NaH2PO4/Na2HPO4 buffer. Plates were incubated at 50°C for 1–2 days, and bacterial colonies were isolated by re‐inoculating on R2 agar.
2.3. Screening for enzymatic activities
For screening of native bacterial isolates, the R2 agar was supplemented with 0.2% lactose and either 20 µg/ml 5‐bromo‐4‐chloro‐3‐indolyl β‐d‐galactopyranoside (X‐gal) for β‐d‐galactosidase activity or 20 µg/ml 5‐bromo‐4‐chloro‐3‐indolyl β‐d‐fucopyranoside (X‐fuc) for α‐l‐fucosidase activity. Cultures were incubated at 50°C overnight, and positive colonies were re‐inoculated until pure colonies with enzymatic activity were obtained.
Crude protein extract was made by bead beating of cells harvested from an overnight culture (50°C, 200 rpm) in a MP FastPrep Homogenizer (MP Biomedicals) at speed 5.5 for 3 × 25 s in 2‐ml Micro tubes PP (Sarstedt, Nümbrecht) supplemented with approx. 250 µl acid‐washed glass beads (212–300 µm, 425–600 µm, ratio 1:1). Extracts were tested for hydrolytic activity with 1 mM oNPG and 1 mM pNP‐fucose at 50°C until visible yellow color was developed (10–35 min). Reactions were stopped with 0.5 M Na2CO3, and OD405 was measured in an EL808 Ultra Microplate Reader (BioTek Instruments, Inc.,).
2.4. Characterization of bacterial growth
Bacterial growth experiments were carried out in triplicates in R2 broth supplemented with 0.2% lactose. Growth and enzyme activity was tested at 20, 28, 37, 50, and 60°C at 200 rpm. Optimal pH for growth was tested at pH 5.0, 5.6, 6.0 (50 mM citrate buffer), 7.0, 8.0 (50 mM Na‐phosphate buffer), 9.0, and 10.0 (50 mM glycine‐NaOH buffer) at 37°C, 200 rpm. Growth was monitored as OD600 using an UVmini 1,240 spectrophotometer (Shimadzu). Phenotypes were furthermore investigated on a nutrient‐poor substrate containing 0.8% (w/v) agar and 0.2% (w/v) peptone. After 48 hr incubation at 50°C, the agar plates were stained with 0.1% (w/v) Coomassie Brilliant Blue R‐250 dissolved in a solution of 20% EtOH (v/v) and 5% (v/v) acetic acid.
2.5. Construction and screening of genomic library
A genomic library was generated in E. coli TOP10 using the pUC18ΔlacZ plasmid. Genomic DNA from P. dendritiformis sp. 3179 was extracted using the Puregene Yeast/Bact. Kit B (Qiagen, Hilden) according to the manufacturer's protocol. The genomic DNA was partially digested with Sau3AI for 25 min at 37°C. The digest was applied to a 1% SeaPlaque® GTG agarose gel (Lonza, Visp), and fragments corresponding to 3–6 kb were cut out and purified using 1X GELase™ Reaction Buffer and GELase™ Enzyme (Epicentre) followed by precipitation with 70% EtOH.
The pUC18ΔlacZ vector was linearized by digestion with BamHI for 1 hr at 37°C, applied to a 1% SeaPlaque® GTG agarose gel, and purified using the GeneJET™ Gel Extraction Kit (Fermentas, Waltham, MA, USA) according to the manufacturer's protocol. The vector DNA was phosphorylated using the Shrimp Alkaline Phosphatase (New England Biolabs) for 45 min at 37°C followed by inactivation for 10 min at 65°C. Ligation of the partially digested genomic DNA and the digested vector was carried out using the T4 DNA Ligase at 16°C overnight. The ligation reaction was transformed into chemically competent E. coli TOP10. The transformation reaction was subsequently plated onto LB agar supplemented with 100 µg/ml ampicillin and either 20 µg/ml X‐gal + 0.3 mM isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) or 20 µg/ml X‐fuc. Plates were incubated for 37°C overnight, and blue colonies, indicating hydrolytic activity on the X‐linked substrates, were selected for further investigation.
2.6. Identification and cloning of enzyme‐coding genes
Plasmids from blue colonies were purified using the QIAprep® Spin Miniprep Kit (Qiagen), and inserts were sequenced at GATC Biotech (Constance). Sequences were analyzed for open reading frames (ORFs) using CLC Main Workbench v7.9.1 (Qiagen), translated into amino acid sequences, and blasted using the NCBI BLASTp Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). ORFs with the highest similarity to β‐galactosidases and α‐fucosidases were selected for further analysis. Two sequences of 1,749 bp and 1,281 bp corresponding to a β‐d‐galactosidase and α‐l‐fucosidase, respectively, were identified as best candidates and amplified with specific primers (PdenGal_USER_F: 5'‐GGCTTAAUATGACAACGCTTTCTTATAAC‐3', PdenGal_USER_R: 5'‐GGTTTAAUTTTCGATAAAGAAGGGGC‐3', PdenFuc_USER_F: 5'‐GGCTTAAUATGCGTCGGTCACG‐3', PdenFuc_USER_R: 5'‐GGTTTAAUTTCGCCATCCTCCA‐3'; uracil‐specific excision reagent (USER) overhangs are underlined). Reverse primers were designed without stop codons to utilize the 6xHis tag included in the cloning vector for protein purification.
The β‐d‐galactosidase gene was amplified using the following PCR protocol: 1 × 98°C for 30 s; 30 × (98°C for 10 s, 55°C for 20 s; and 72°C for 1 min); and 72°C for 10 min, while the gene encoding the α‐l‐fucosidase was amplified using the PCR protocol: 1x 98°C for 30 s; 30x (98°C for 10 s, 56°C for 20 s; and 72°C for 1 min 30 s); and 72°C for 10 min.
The cloning vector, pET9a‐USER‐1 vector, was prepared by digestion with PacI and nt.BbvCI at 37°C overnight followed by purification from a 1% SeaPlaque® GTG agarose gel using the GeneJET™ Gel Extraction Kit. The amplified ORFs were inserted into pET9a‐USER‐1 by including specific USER overhangs during the PCR (Table I) and ligating the PRC product into the vector by means of USER enzyme (New England Biolabs). The reaction was carried out at 37°C for 20 min followed by 25°C for 20 min and subsequently used for transformation into chemically competent E. coli BL21(DE3)ΔlacZ. Colonies were grown on LB agar supplemented with 50 µg/ml kanamycin (Km) at 37°C overnight. The plasmids from one randomly selected colony from each transformation were purified and verified by sequencing (as above).
2.7. Phylogenetic analyses
The 16S rRNA gene was analyzed against BLASTn (https://blast.ncbi.nlm.nih.goc/Blast.cgi), and the closest relatives were aligned using Clustal W (Thompson, Higgins, & Gibson, 1994). A phylogenetic tree was made in MEGA7 (Kumar, Stecher, & Tamura, 2016) using the maximum likelihood method based on the Tamura–Nei model (Tamura & Nei, 1993).
The two ORFs identified as a putative β‐d‐galactosidase and α‐l‐fucosidase, respectively, were analyzed using BLASTp against PDB to identify the phylogenetically closest relatives. When this only gave very few results, the search was expanded to the nonredundant (nr) protein database in combinations with the CAZy database. Sequences were aligned using Clustal W, and a phylogenetic tree was made using the maximum likelihood method based on the JTT matrix‐based model (Jones, Taylor, & Thornton, 1992).
2.8. Protein expression and purification
For initial transglycosylation experiments, the construct encoding the β‐d‐galactosidase gene was expressed and purified as described by Schmidt and Stougaard (2010) with minor modifications. In short, the gene containing the native SD sequence was amplified using specific primers β‐galFpUC (5'‐TCTCGGTACCAGAAGGAGAGAAGCCAATG‐3') and β‐galRpUC (5'‐CTCTAAGCTTATTTCGATAAAGAGGGGC‐3'), and cloned into the KpnI and HindIII site of pUC18∆LacZ. Overnight expression at 37°C in E. coli TOP10 cells was followed by protein extraction by bead beating (as described below), heating to 60°C for 15 min, and subsequent purification by anion exchange chromatography at pH 6.2 and eluted with a NaCl gradient, resulting in apparent homogeneity on a SDS gel.
For the remaining characterization and transglycosylation experiments, recombinant strains in E. coli BL21(DE3)ΔlacZ were used for protein expression. Cells were inoculated in 10 ml LB with 50 µg/ml Km and incubated at 37°C with 200 rpm overnight. The overnight culture was used to inoculate the expression culture in 100 ml LB with 50 µg/ml Km, which was incubated at 37°C with 200 rpm until OD600 ~ 0.3–0.4. The culture was then supplemented with 0.2 mM IPTG and 0.1% dl‐rhamnose to induce expression, and incubation was continued at room temperature with 200 rpm overnight to increase correct protein folding.
After overnight expression, cells were cooled on ice for 10 min and harvested by centrifugation for 10 min at 4,700 rpm, 4°C, and the supernatant was discarded. The cell pellet was resuspended in 10 ml binding buffer [20 mM sodium phosphate, 500 µM NaCl, and 20 mM imidazole, pH 7.4] and transferred to 50‐ml centrifuge tubes with screw caps and a gasket (VWR, Radnor) supplemented with approx. 2 ml acid‐washed glass beads (212–300 µm, 425–600 µm, ratio 1:1) for bead beating. Bead beating was carried out in an MP FastPrep Homogenizer at speed 4.0 for 3 × 25 s. The samples were incubated on ice for 1 min in between rounds. Samples were centrifuged for 5 min at 4,700 rpm to remove glass beads and cell debris, and the supernatant was transferred to a clean tube and kept on ice until purification.
Protein purification was carried out using the His GraviTrap Kit (GE Healthcare) following the manufacturer's protocol for purifications under native conditions. Elution was carried out using 500 mM imidazole. Expression was checked by SDS‐PAGE on RunBlue SDS 4%–12% Protein Gels (Expedeon).
2.9. Characterization of recombinant proteins
Concentration of purified enzyme was determined in a Bradford assay using BradfordUltra reagent (Expedeon Protein Solutions) with a dilution series of BSA (bovine serum albumin) as model protein. Absorbance was measured at 595 nm using an Epoch Microplate Spectrophotometer running Gen5 software ver. 3.03 (BioTek Instruments Inc.), and the slope was used to estimate the concentrations of purified enzymes.
To determine pH and temperature optimum, the purified β‐d‐galactosidase was characterized using 0.5 mM oNPG as a substrate, while the purified α‐l‐fucosidase was characterized using 0.5 mM pNP‐fucose. pH optimum was determined (with six replicates) at 50°C at pH 5.0, 5.6, and 6.0 (50 mM citrate buffer); pH 6.8, 7.4, and 8.0 (50 mM Na‐phosphate buffer); and pH 9.0, 10.0, and 10.7 (50 mM glycine‐NaOH buffer). Temperature optimum was determined (in triplicates) from 25 to 65°C for 5 min in 50 mM Na‐phosphate buffer at pH 7.4. OD420 was determined using an Epoch Microplate Spectrophotometer running Gen5 software ver. 3.03.
Enzyme kinetics was investigated at 50°C. Reactions of 200 µl [in 50 mM Na‐phosphate buffer pH 7.4] with a fixed amount of enzyme (18 µg for the α‐l‐fucosidase and 1,200 µg for the β‐d‐galactosidase) were incubated with increasing concentrations of the substrate (0.25–1 mM oNPG/pNP‐fucose). The product formation was monitored by measuring the absorbance at 420 nm every 30 s in real time by using a FLOUstar Omega microplate reader (BMG Labtech GmbH). Blanks contained buffer and substrate alone to represent the autohydrolysis of the substrate. All samples were performed in triplicates. Data were analyzed using the Omega Mars data analysis software ver. 3.20 (BMG Labtech). Molar extinction coefficients for oNP and pNP of 3.47 mM‐1 cm‐1 and 18.4 mM‐1 cm‐1, respectively (Bowers, McComb, Christensen, & Schaffer, 1980; Zeyer & Kocher, 1988), were used to calculate concentrations of the product. The steady‐state rates were determined from the slope of the initial (linear) part of the resulting progress curves. The experimental data points were fitted to the Michaelis–Menten equation to obtain the kinetic parameters Km and Vmax. Nonlinear regression and estimations of kinetic parameters were performed in the software OriginPro (OriginLab).
2.10. Transglycosylation
Transglycosylation by the β‐d‐galactosidase was tested using 10% (w/v) lactose as galactosyl donor and acceptor, and sucrose or fructose was used as additional galactosyl acceptors. The α‐l‐fucosidase was tested using 5–50 mM pNP‐fucose (dissolved in 10% dimethylformamide) as donor and 5% (w/v) lactose as acceptor. Reactions were incubated at 50°C for 0, 15, 30 min, 1, and 3 hr. Reactions were stopped at 99°C for 15 min. For transglycosylation reactions by the β‐d‐galactosidase using lactose, sucrose, and fructose as acceptors, a control reaction with inactivated enzyme (99°C for 15 min) was included.
Reactions were analyzed by thin‐layer chromatography (TLC) on TLC Silica gel 60 F254 (Merck, Darmstadt, Germany) developed in ethyl acetate:acetic acid:H2O (6:6:1, v/v/v). Saccharides were detected by 0.1M 2‐methylresorcinol dissolved in a 5% (v/v) solution of sulfuric acid in ethanol followed by heating of the silica gel.
Formation of oligosaccharides was analyzed by HPLC using an ICS‐5000 system (Dionex) with an AS autosampler and a pulsed amperometric detector (carbohydrate four‐potential waveform, sampling rate 2 Hz), with a gold electrode (Au) and an Ag/AgCl reference electrode. The column used was a Dionex CarboPac PA1 (250 × 4 mm, with 50 × 4 mm Guard, Thermo Scientific) maintained at 22°C. Eluents consisted of water and aqueous solutions of sodium hydroxide (NaOH) and sodium acetate (NaOAc), in the combinations: (a) water; (b) 1 M NaOH; (c) 500 mM NaOH + 750 mM NaOAc; and (d) 500 mM NaOAc. The flow: 1.0 ml/min, injection volume: 10 µl, and samples were maintained at 5°C. For identification of peaks, qualitative standards of 3'‐O‐galactosyllactose, 4'‐O‐galactosyllactose, and 6'‐O‐galactosyllactose were used.
3. RESULTS
3.1. Isolation and characterization of glycoside hydrolase‐producing isolate 3179
Water and sediment samples from hot springs in Knighton Fjord, East Greenland, were screened for isolates producing β‐d‐galactosidase and α‐l‐fucosidase enzymes on growth media supplemented with chromogenic substrates. One isolate, #3179, that showed good β‐d‐galactosidase and α‐l‐fucosidase activity was selected for further analysis.
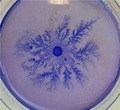
Phylogenetical analysis of the 16S rRNA gene sequence of isolate 3179 (GenBank accession number MK616148) showed a high degree of similarity to the genus Paenibacillus with 98.4% identity to Paenibacillus thiaminolyticus LMG 17412T, 97.8% identity to Paenibacillus dendritiformis DSM 18844T, and 94.7% identity to P. alvei DSM29T (Figure 1a). Culturing isolate 3179 on thin agar plates with peptone and subsequent staining with Coomassie Brilliant Blue showed a unique complex growth pattern (Figure 1b), which was similar to that reported for P. dendritiformis (Tcherpakov, Ben‐Jacob, & Gutnick, 1999). Thus, we assume that isolate 3179 was affiliated to the genus Paenibacillus, and in the following, the strain is denoted Paenibacillus sp. 3179.
Figure 1.
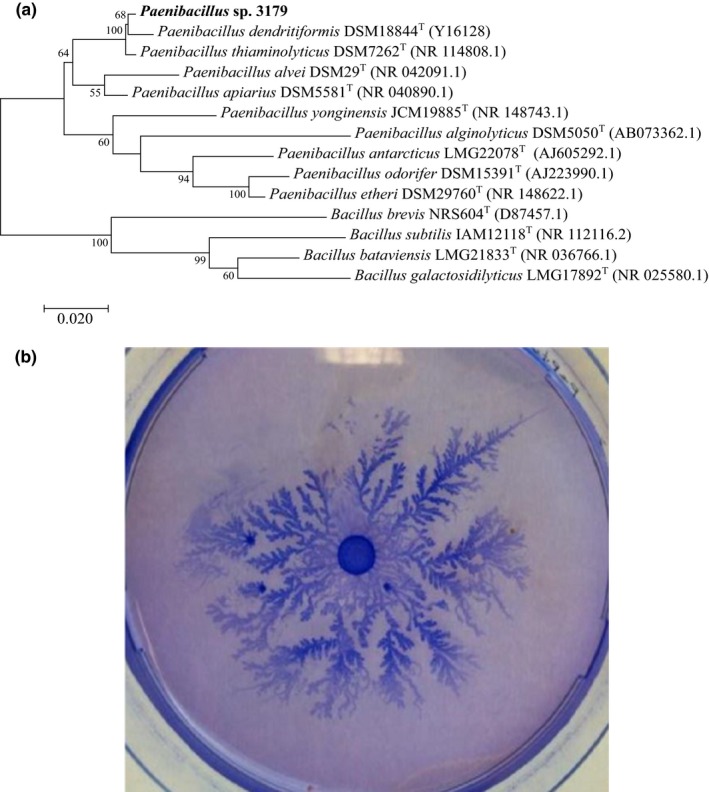
(a) Phylogeny of 16S rRNA genes generated using the maximum likelihood method based on the Tamura–Nei algorithm, showing the phylogenetic relationships between Paenibacillus sp. 3179 and related taxa. Numbers at nodes are bootstrap values shown as percentages of 100 replicates. (b). Paenibacillus sp. 3,170 cultured at 50°C for 48 hr on agar (0.8% w/v) with peptone (0.2% w/v) and subsequently stained with Coomassie Brilliant Blue and destained in ethanol (20% v/v) and acetic acid (5% v/v)
When Paenibacillus sp. 3179 was cultivated in lysogenic broth (LB) medium, growth optimum was determined to be at 50°C. No growth was observed at 28°C nor at 60°C. Optimal growth was observed at pH 8, and no growth was observed below pH 5.6; good growth was still maintained at pH 9–10.
3.2. Paenibacillus sp. 3179 crude extract contains α‐l‐fucosidase and β‐d‐galactosidase activity
A crude extract of the native Paenibacillus sp. 3179 was subjected to analysis of α‐l‐fucosidase and β‐d‐galactosidase hydrolytic and transglycosylation activity. No activity was observed in the membrane fraction, but the cytosolic fraction showed α‐l‐fucosidase and β‐d‐galactosidase hydrolytic activity as determined from hydrolysis of pNP‐fucose (4‐nitrophenyl‐α‐l‐fucopyranoside) and oNPG (2‐nitrophenyl‐β‐d‐galactopyranoside) substrates, respectively. In addition, the crude cytosolic extracts showed transgalactosylation activity using lactose as substrate as well as transfucosylation activity using pNP‐fucose as substrate (Figure 2).
Figure 2.
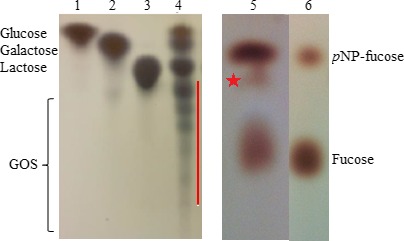
TLC analysis of hydrolysis and transglycosylation reactions of crude, intracellular extract from Paenibacillus sp. 3179. (a) Reaction with lactose; standard sugars in lanes 1–3; lane 4, extract from Paenibacillus sp. 3179 incubated with lactose for 16 hr at 37°C. (b) Reaction with pNP‐fucose; lane 5, extract from Paenibacillus sp. 3179 incubated with pNP‐fucose and fucose for 16 hr at 37°C; lane 6, pNP‐fucose and fucose. Transglycosylation products are shown with a red line (reaction with lactose) and a star (reaction with pNP‐fucose)
3.3. Identification of enzyme‐encoding genes
A functional expression library was constructed by inserting partially digested genomic DNA (gDNA) fragments from Paenibacillus sp. 3179 into plasmid pUC18ΔlacZ and transformed into E. coli TOP10. The library was screened for β‐galactosidase‐encoding genes by plating the expression library onto agar plates supplemented with the chromogenic substrate X‐gal. One inserted DNA fragment from a blue colony contained a 1,749 bp open reading frame (ORF) next to a seven bp putative Shine–Dalgarno sequence (AGGAGAG) located seven bp upstream of the ORF. BLASTp analysis of the translated ORF showed that the amino acid sequence displayed 92.5% identity to an uncharacterized putative β‐d‐galactosidase from P. dendritiformis (WP_133381740.1), 91.6% identity to a characterized β‐d‐galactosidase from P. thiaminolyticus (CAZ44333) (Benešová, Lipovová, Dvořáková, & Králová, 2010), and 79.7% identity to an uncharacterized putative β‐d‐galactosidase P. alvei (EPY10792.1) as its closest relatives (Figure 3a). A conserved domain search (CDD) (Marchler‐Bauer et al., 2015) showed the same domains in the Paenibacillus sp. 3179 β‐d‐galactosidase as in enzymes from P. thiaminolyticus and P. dendritiformis: a conserved GH35‐domain (Pfam01301) at position 10–322 and a β‐d‐galactosidase jelly roll domain (Pfam13364) at position 527–562. A BLAST search at UniProt (http://www.uniprot.org) confirmed the affiliation to the glycoside hydrolase family 35. A HMM search and in silico folding analysis over known structures showed that the Paenibacillus sp. 3179 β‐d‐galactosidase displayed high similarity to the characterized structure of the bgaC β‐d‐galactosidase from Bacillus circulans (4MAD) (5.6e‐95) (Henze et al., 2014): The Paenibacillus sp. 3179 amino acid sequence including the catalytic residues (nucleophile Glu233 and acid/base Glu157, according to the B. circulans numbering) could be superimposed onto the corresponding B. circulans structure, 4MAD (Figure A1).
Figure 3.
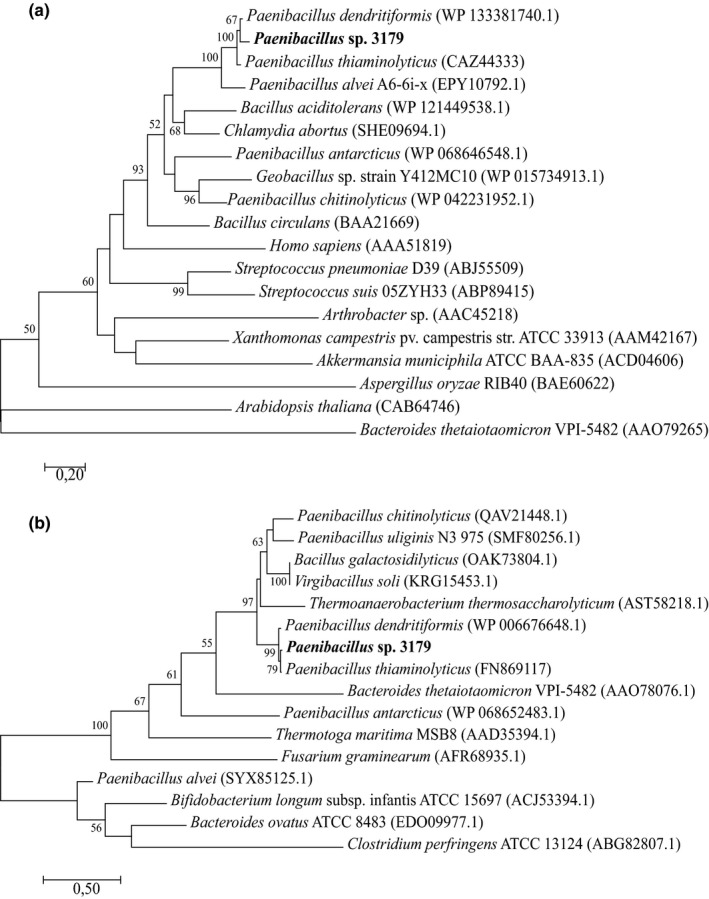
Phylogenetic analysis of (a) Paenibacillus sp. 3179 β‐d‐galactosidase and (b) Paenibacillus sp. 3179 α‐l‐fucosidase. Sequences that showed similarities to the Paenibacillus sp. 3179 enzymes were identified by BLASTp analysis against Protein Data Bank (PDB) and supplemented with closely related enzymes from the nonredundant (nr) protein sequence database. The latter were only selected if also identified among GH35 and GH29 family enzymes, respectively, in the CAZy database (http://www.cazy.org). Phylogenetic relationships were inferred the by maximum likelihood method based on the JTT matrix‐based model. The trees with the highest log likelihood are shown. Bootstrap values (n = 100) above 50 are shown. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7
The expression library was subsequently screened for α‐l‐fucosidase activity on agar plates containing the chromogenic substrate X‐fuc. DNA sequence analysis of an inserted DNA fragment from a blue colony showed a 1,281 bp ORF. BLASTp analysis showed that the amino acid sequence encoded by the ORF displayed 97.4% amino acid sequence identity to a characterized α‐l‐fucosidases from P. thiaminolyticus (aLfuk1, FN869117) (Benešová, Lipovová, Dvořáková, & Králová, 2013) and 97.2% sequence identity to a putative uncharacterized α‐l‐fucosidase from P. dendritiformis (WP_006676648.1) (Figure 3b). The conserved domain search revealed a conserved glycoside hydrolase superfamily D (GH‐D) domain (pfam01120), which comprises the families GH27, GH31, and GH36 containing enzymes with structurally and mechanistically similar activities, but none of them are known to show α‐l‐fucosidase activity. However, a BLAST analysis at UniProt showed that the most closely related α‐ l‐fucosidases all belong to GH29. Thus, we conclude that the α‐l‐fucosidase from Paenibacillus sp. 3179 belongs to GH29. An HMM search and modeling over the structure with highest similarity (2.2e‐86), which was an α‐l‐fucosidase from P. thiaminolyticus (6GN6) (Koval'ová, 2019), showed that the Paenibacillus sp. 3179 sequence including the catalytic residues (nucleophile Asp186 and acid/base Glu239, according to the P. thiaminolyticus numbering) could be superimposed onto the corresponding 6GN6 structure (Figure A1). The DNA sequences encoding the α‐l‐fucosidase and the β‐d‐galactosidase from Paenibacillus sp. 3179 have been uploaded to GenBank with the accession numbers MK625194 and MK625195, respectively.
3.4. Characterization of recombinant enzymes
The Paenibacillus sp. 3179 β‐d‐galactosidase and α‐l‐fucosidase were recombinantly produced in E. coli as 6xHis‐tagged enzymes. SDS‐PAGE showed that purified β‐d‐galactosidase and α‐L‐fucosidase migrated as 70 kDa and 50 kDa protein, respectively (calculated theoretical molecular weights 65.7 kDa and 49.5 kDa) (Figure A2).
Using oNPG as a substrate, the purified β‐d‐galactosidase showed optimal activity at 50°C and at pH 8.0 (Figure 4a). Stability was investigated at 37, 50, and 70°C, and the β‐d‐galactosidase was stable for up to 5 hr at 37°C or 50°C, while it was completely inactivated after 30 min at 70°C (data not shown). K m for the β‐d‐galactosidase with oNPG as substrate was estimated to be 0.57 ± 0.12 mM and Vmax = 0.11 ± 0.01 mmoles/min/mg enzyme (Table 1, Figure A3). The β‐d‐galactosidase was also tested with p‐nitrophenyl‐β‐d‐galactopyranoside (pNPG) as substrate, but no activity was detected (data not shown).
Figure 4.
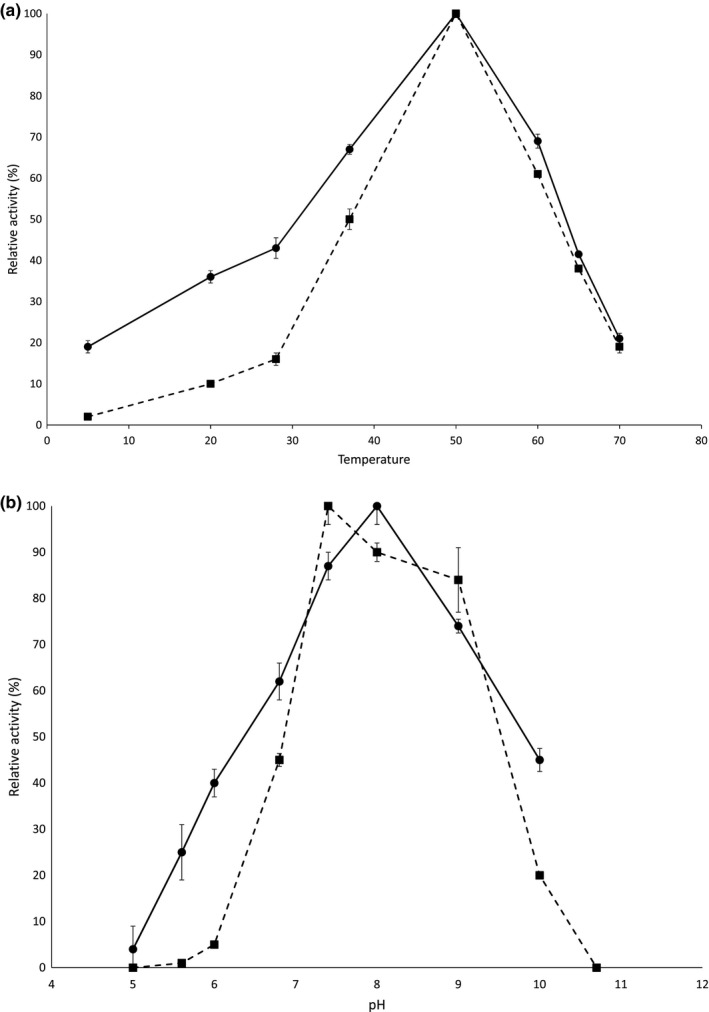
Temperature optimum determined at pH 9.0 (a) and pH optimum determined at 50°C (b) for Paenibacillus sp. 3179 β‐d‐galactosidase (solid line) and α‐l‐fucosidase (dashed line) based on reaction rates. Error bars indicate standard error (n = 3)
Table 1.
Characteristics of the β‐d‐galactosidase and α‐l‐fucosidase from Paenibacillus sp. 3179 compared to the closest related enzymes from P. thiaminolyticus (CCM 3,599)
| Enzyme | Km (mM) | Substrate | Temperature optimum (°C) | pH optimum | Ref. |
|---|---|---|---|---|---|
| Paenibacillus sp. 3179 β‐d‐galactosidase | 0.57 ± 0.12 | oNPG | 50 | 8.0 | This work |
| P. thiaminolyticus β‐d‐galactosidase | 250 | pNPG | 65 | 5.5 | a |
| Paenibacillus sp. 3179 α‐l‐fucosidase | 1.11 ± 0.75 | pNP‐l‐fucose | 50 | 7.4 | This work |
| P. thiaminolyticus α‐l‐fucosidase iso1 | 0.44 | pNP‐l‐fucose | 48 | 8.2 | b |
| P. thiaminolyticus α‐l‐fucosidase iso2 | 0.52 | pNP‐l‐fucose | 50 | 6.5 | c |
Benešová et al. (2010).
Benešová et al. (2013).
Benešová et al. (2015).
Activity of the purified α‐l‐fucosidase was similarly determined with pNP‐fucose as substrate. Optimal activity was observed at 50°C and at pH 7.4 (Figure 4b). Stability was investigated at 37°C, 50°C, and 70°C, and the α‐l‐fucosidase was stable for up to 5 hr at 37°C and 50°C, while it was completely inactivated after 30 min at 70°C (data not shown). K m with pNP‐fucose as substrate was estimated to be 1.11 ± 0.75 mM and Vmax = 3.88 ± 1.58 mmoles/min/mg enzyme (Table 1, Figure A3).
3.5. Transglycosylation of recombinant enzymes
Both of the investigated enzymes exhibited transglycosylation activity. Transglycosylation by the β‐d‐galactosidase was analyzed with lactose as galactosyl donor and lactose, sucrose, or fructose as accepting carbohydrates. When lactose was used as donor and acceptor, hydrolysis products (galactose and glucose) appeared in the TLC analysis together with unhydrolyzed lactose and galactooligosaccharides (GOS) (Figure 5a). Two of these GOS were identified as 6’‐O‐galactosyllactose and 3’‐O‐galactosyllactose by HPLC analysis (Figure 6a), and the enzyme was able to transform lactose into 6’‐galactosyllactose, 0.14 mol/mol, and 3’‐galactosyllactose, 0.027 mol/mol (Figure 6b). Hydrolysis and transgalactosylation were seen in the range from pH 5 to 8 (data not shown). When sucrose or fructose was used as acceptors, reddish spots appeared in the TLC analysis (white arrows in Figure 5b). In these transgalactosylation experiments, both dark blue/purple spots corresponding to GOS, lactose, galactose, and glucose and reddish spots were observed, indicating that transgalactosylation occurred with sucrose and fructose as galactosyl acceptors (Figure 5b, red and yellow arrows, respectively). Similarly, transglycosylation with the α‐l‐fucosidase was analyzed using pNP‐fucose as fucosyl donor and lactose as fucosyl acceptor. With TLC, an oligosaccharide corresponding in size to 2FL was detected (Figure 5c, black arrow). This finding was confirmed by HPLC, which showed that the formed oligosaccharide eluted as 2FL (Figure 6c). Further products were formed at about the same rate as 2FL, but these were not characterized further (Figure 6d).
Figure 5.
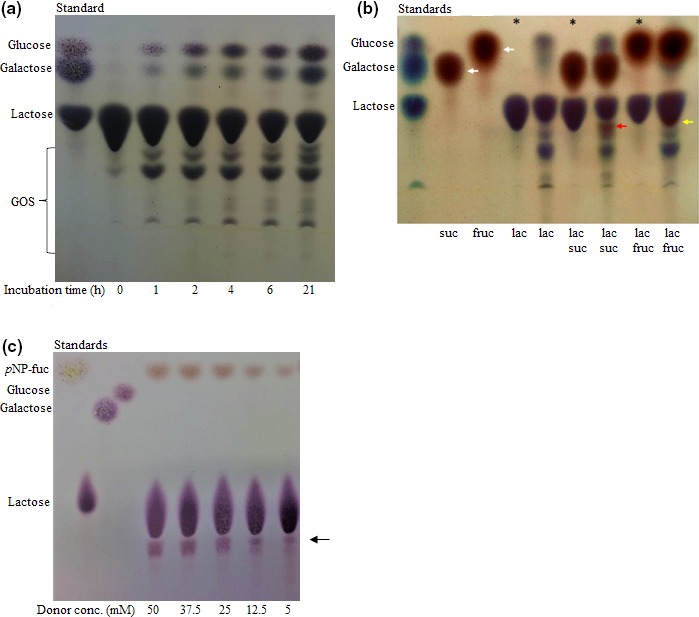
Transglycosylation activities by Paenibacillus sp. 3179 enzymes. (a) Transgalactosylation by Paenibacillus sp. 3179 β‐d‐galactosidase preliminarily purified over time (0–21 hr) with lactose as donor and acceptor. (b) Transgalactosylation by preliminarily purified Paenibacillus sp. 3179 β‐d‐galactosidase with lactose (lac) as donor, and sucrose (suc), fructose (fruc), and lactose (lac) as acceptors. White arrows show the acceptor molecules sucrose and fructose. Red and yellow arrows show the transglycosylation products using sucrose and fructose as galactosyl acceptors, respectively. * indicates where the enzyme was inactivated before addition of substrates. (c) Transfucosylation by Paenibacillus sp. 3179 α‐l‐fucosidase with increasing concentration of p‐nitrophenyl‐α‐l‐fucopyranoside as donor and lactose as acceptor. Black arrow shows the transglycosylation product corresponding in size to 2FL.
Figure 6.
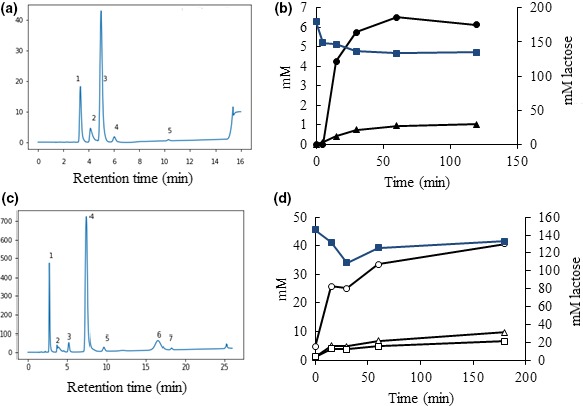
HPLC analyses of transglycosylation reactions. (a) Transgalactosylation profile after 4 hr with Paenibacillus sp. 3179 β‐d‐galactosidase using lactose as sole substrate. Peaks are identified as follows: 1: glucose and galactose monomers. 2: unidentified (not a part of the reaction, present in all samples). 3: lactose. 4:6’‐O‐galactosyl‐lactose (6‐gal‐lac). 5:3’‐O‐galactosyl‐lactose (3‐gal‐lac). After 120 min, 24% of the lactose has been converted and results in 0.14 mol/mol 6‐gal‐lac and 0.027 mol/mol 3‐gal‐lac. (b) Diagram showing the utilization of lactose (blue squares, secondary axis) and production of 6‐gal‐lac (black circles, primary axis) and 3‐gal‐lac (black triangles, primary axis) over time (120 min.). (c) Transfucosylation profile after 3 hr with Paenibacillus sp. 3179 α‐l‐fucosidase using pNP‐fucose as donor and lactose as acceptor. Peaks are identified as follows: 1: fucose. 2: unidentified (not a part of the reaction, present in all samples). 3: product of the reaction with increasing concentration over reaction time and a relative elution to 2FL and lactose corresponding retention of 3FL. 4: lactose. 5:2’‐O‐fucosyllactose (2FL). 6 and 7: unidentified. (d) Diagram showing utilization of lactose (blue squares, secondary axis) and production of monomeric fucose (open circles, primary axis), 2FL (open squares, primary axis), and the compound with retention corresponding to 3FL (peak 3 in panel C) (open triangles, primary axis) over time (180 min.)
4. DISCUSSION
A β‐d‐galactosidase and α‐l‐fucosidase producing strain, #3179, was isolated from a thermal spring in Greenland. 16S rRNA gene analysis showed that strain 3179 was affiliated to the genus Paenibacillus with P. dendritiformis and P. thiaminolyticus as closest relatives. Strain 3179 displayed dendritic growth similar to P. dendritiformis why we conclude that strain 3179 most likely could be classified as P. dendritiformis. However, as no other phylogenetical or chemotaxonomical analyses were carried out, we denote the strain Paenibacillus sp. 3179.
The two genes encoding a β‐d‐galactosidase and an α‐l‐fucosidase, respectively, were isolated and analyzed phylogenetically and functionally. Sequence analysis of the β‐d‐galactosidase showed close relationship to β‐galactosidases from P. dendritiformis (92.5% sequence identity) and from P. thiaminolyticus (91.6% identity) (Benešová et al., 2010). Further distantly related enzymes showed below 80% identity for hypothetical enzymes and below 55% identity for classified β‐galactosidase like Bacillus circulans BgaC (BAA21669) (Ito & Sasaki, 1997). All related β‐d‐galactosidase sequences were classified as GH35 according to the glycoside hydrolase classification system (Henrissat, 1991), and thus, the β‐d‐galactosidase from Paenibacillus sp. 3179 was hypothesized to belong to GH35, which primarily consists of β‐galactosidases (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/2/1/23.html), but also contain exo‐β‐glucosaminidases (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/2/1/165.html), exo‐β‐1,4‐galactanases (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/2/1/.html‐), and β‐1,3‐galactosidases (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/2/1/.html‐). GH35 enzymes can be found in more than 2,700 different organisms spanning archaea, bacteria, and eukaryotes, including plants and animals. GH35 β‐galactosidases have an (α/β)8 barrel as catalytic domain (http://www.cazypedia.org), and the mechanism is catalysis of the hydrolysis of terminal nonreducing β‐galactosyl residues in, for example, lactose, oligosaccharides, glycolipids, and glycoproteins. In silico analysis showed that the overall structure, including the catalytic residues, could be modeled over a known structure of a β‐d‐galactosidase from B. circulans (Figure A1), making it possible that the β‐d‐galactosidase from Paenibacillus sp. 3179 has the same functional activity as that from B. circulans.
The α‐l‐fucosidase sequence was analyzed in a similar manner. The closest related sequence was an α‐l‐fucosidase from P. thiaminolyticus (FN869117; 6GN6) (Benešová et al., 2013; Koval'ová et al., 2019). The phylogenetically closest related α‐l‐fucosidases were all shown to belong to GH29, which consists of exo‐acting α‐fucosidases (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/2/1/51.html) and α‐1 → 3/1 → 4‐l‐fucosidases (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/2/1/111.html). Enzymes in GH29 also consist of an (α/β)8‐like barrel domain plus a C‐terminal β‐sandwich domain (http://www.cazypedia.org) and function by classical retaining mechanisms involving double displacement via a covalent intermediate. In silico structural analysis showed that the α‐l‐fucosidase sequence from Paenibacillus sp. 3179 could be superimposed onto the corresponding 6GN6 structure, including catalytic residues, indicating a functional relationship (Figure A1).
Sequence analyses revealed that there are a lot of potentially unclassified β‐galactosidases and α‐fucosidases from strains within the Paenibacillus genus ranging in temperature profile from the psychrotolerant P. antarcticus which shows optimal growth at 10–15°C (Montes, Mercadé, Bozal, & Guinea, 2004) to the described thermotolerant Paenibacillus sp. 3179 with optimal growth at 50°C. However, since most of the proteins are only annotated as potential enzymes with no documented functions, the genus Paenibacillus appears to have a lot of potential for discovery of cold‐active as well as thermostable enzymes for a range of industrial applications.
Functional characterization of recombinant purified enzymes from Paenibacillus sp. 3179 produced by E. coli showed that both enzymes possessed hydrolytic as well as transglycosylating activities. Chromatography analyses showed that the β‐d‐galactosidase was able to form 6'‐O‐galactosyllactose and 3'‐O‐galactosyllactose using lactose as substrate, and the α‐l‐fucosidase was able to transfer the fucose moiety from pNP‐fuc to lactose, thereby forming 2FL and also indication of 3FL was observed. The chromatogram in Figure 6c identified the transglycosylation product 2FL and a peak (no 3), which relative to elution of 2FL and lactose corresponded to the retention of 3FL (Tan, Chai, & Zhang, 2015). This compound was observed in increasing concentration with reaction progress, indicating that 3FL was a product of the transglycosylation reaction. Enzyme catalyzing production of 2FL and 3FL is known, and analyses of these oligosaccharides are well established (Tan et al., 2015; Zeuner, Teze, Muschiol, & Meyer, 2019), but 6FL as an enzymatic transglycosylation product is not described and the present investigation cannot establish its presence. The compound corresponding to peak 6 was not identified, but increased concentration with reaction time indicated it was a product of the reaction.
In studies carried out on the β‐d‐galactosidase and two α‐l‐fucosidases from P. thiaminolyticus (Benešová et al., 2010, 2013, 2015), it is described that all three enzymes were able to transfucosylate a wide range of p‐nitrophenyl substrates and alcohols; however, little attention has been focused on transgalactosylation. In this study, we showed that the β‐d‐galactosidase from Paenibacillus sp. 3179 is able to transgalactosylate a range of natural substrates such as lactose, sucrose, and fructose, which can be of use for industrial purposes when, for example, upgrading disaccharides to oligosaccharides in food items. We furthermore showed that the α‐l‐fucosidase from Paenibacillus sp. 3179 is able to transfucosylate using lactose as acceptor, making it a good candidate for production of fucosylated oligosaccharides like 2FL for infant formula (Aldredge et al., 2013; Chen, 2015) due to its transfucosylating activity at elevated temperatures.
As the interest for thermostable transglycosylating enzymes has gained interest, similar enzymatic reactions have been reported. For example, Petrova and Kujumdzieva (2010) isolated several strains of thermotolerant yeasts from a range of milk products with transgalactosylation activities, and Fai, Silva, Andrade, Bution, and Pastore (2014) showed transgalactosylation activity by Pseudozyma tsukubaensis and Pichia kluyveri using lactose as a substrate. Lezyk et al. (2016) created a metagenome library from soil and discovered several putative α‐fucosidases, which were recombinantly expressed and proven to have hydrolytic as well as transfucosylating activities, potentially suitable for formation of fucosylated oligosaccharides for supplementing instant formula to mimic the natural composition of human breast milk.
5. CONCLUSIONS
In the current study, it was shown that the bacterium Paenibacillus sp. 3179 isolated from a thermal spring in East Greenland produced two thermostable enzymes, a β‐d‐galactosidase and an α‐l‐fucosidase, with hydrolytic as well as transglycosylating activities. It was confirmed that the β‐d‐galactosidase was able to produce 6'‐O‐galactosyllactose and 3'‐O‐galactosyllactose at its optimal temperature at 50°C and that the α‐l‐fucosidase was able to produce 2FL along with some uncharacterized oligosaccharides, speculated to include 3FL. Future studies will be aimed at characterizing the remaining oligosaccharides and to expand the range of investigated substrates for transglycosylation.
6. ETHICS STATEMENT
None required.
CONFLICTS OF INTEREST
None declared.
AUTHOR CONTRIBUTION
Mariane Thøgersen: data curation—lead, formal analysis—lead, investigation—equal, methodology—equal, writing—original draft—equal, and writing—review and editing—equal; Stefan Christensen: data curation—supporting, formal analysis—supporting, and writing—review and editing—supporting; Morten Jepsen: data curation—supporting; Lars Pedersen: data curation—supporting, formal analysis—supporting, and writing—review and editing—supporting; and Peter Stougaard: conceptualization—lead, funding acquisition—lead, methodology—lead, project administration—lead, writing—original draft—equal, and writing—review and editing—equal.
ACKNOWLEDGMENTS
We thank Professor Jin‐Ho Seo from Seoul National University, Seoul, The Republic of Korea, for kindly providing us with the lactose‐deficient E. coli BL21(DE3)ΔlacZ and Susanne Iversen for her excellent technical skills. This work was supported by The Danish Council for Strategic Research under grant no. 1308‐00014B / 0652‐20140008.
APPENDIX 1.
Figure A1.
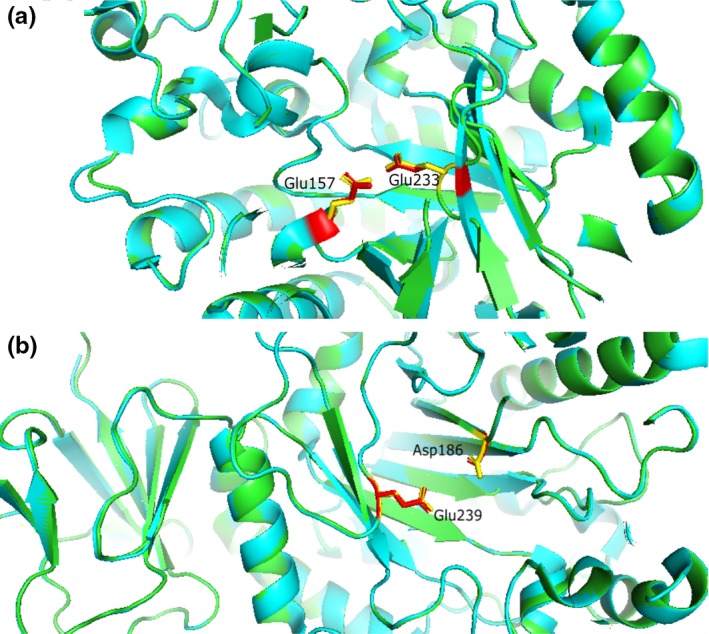
Three dimension structure of the catalytic residues of (a) the Paenibacillus sp. 3179 β‐d‐galactosidase (amino acid backbone in green, nucleophile Glu and acid/base Glu in yellow) superimposed onto the corresponding B. circulans structure, 4MAD, (amino acid backbone in blue, nucleophile Glu233 and acid/base Glu157 (B. circulans numbering) in red), and (b) the Paenibacillus sp. 3179 α‐l‐fucosidase sequence (amino acid backbone in green) including the catalytic residues (nucleophile Asp and acid/base Glu in yellow) superimposed onto the corresponding P. thiaminolyticus structure 6GN6 (amino acid backbone in blue, nucleophile Asp186 and acid/base Glu239, P. thiaminolyticus numbering, in red)
Figure A2.
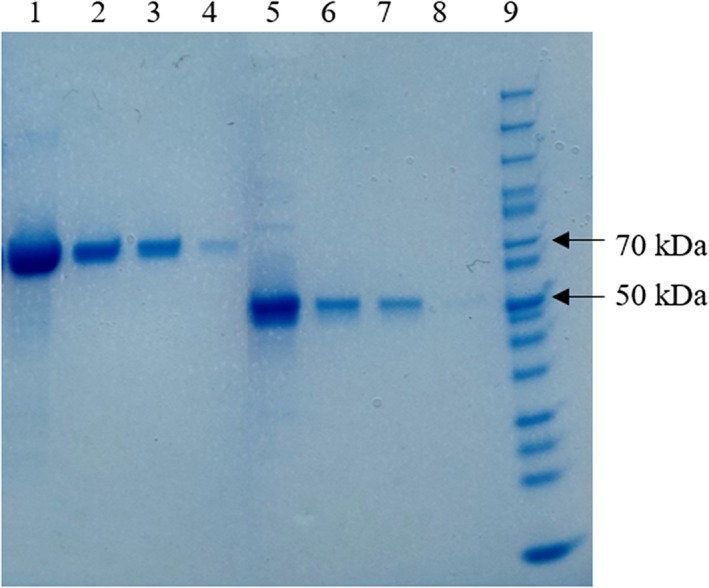
SDS‐PAGE of Ni‐NTA‐purified enzymes. Lanes 1–4: Paenibacillus sp. 3179 β‐d‐galactosidase at approx. 70 kDa [1:1,200 µg; 2:240 µg; 3:120 µg; 4:24 µg]. Lanes 5–8: Paenibacillus sp. 3179 α‐l‐fucosidase at approx. 50 kDa [5:450 µg; 6:90 µg; 7:45 µg; 8:9 µg]. Lane 9:5 µl PageRuler Unstained Protein Ladder (Thermo Fisher Scientific #26614). Enzyme concentrations were determined by Bradford assay with BSA as standard protein
Figure A3.
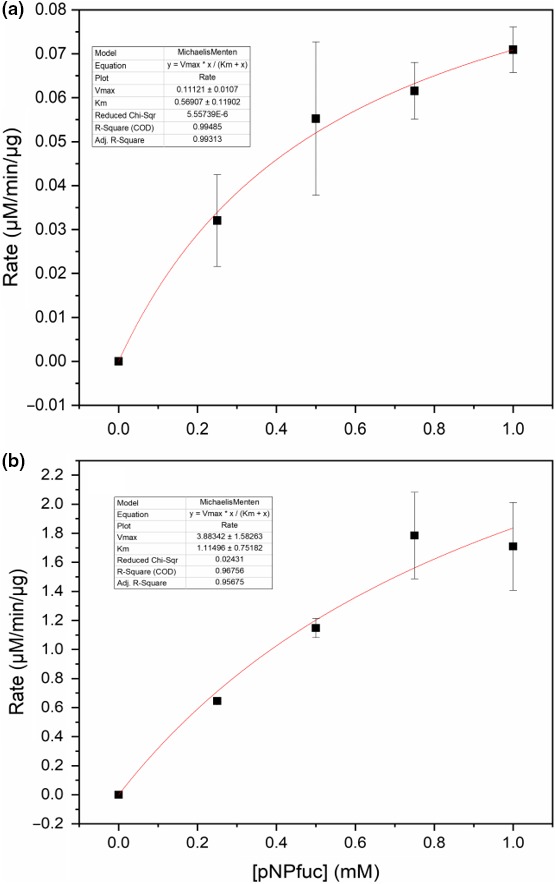
Nonlinear regression and estimation of kinetic parameters for (a) the Paenibacillus sp. 3179 β‐d‐galactosidase and (b) the Paenibacillus sp. 3179 α‐l‐fucosidase. The experimental data points (black squares) were fitted to the Michaelis–Menten equation (red lines) to obtain the kinetic parameters Km and Vmax. The two curves did not reach full saturation, which is reflected in the relatively large errors on the parameters. Therefore, the values given here are rough estimates of the kinetic parameters
Thøgersen MS, Christensen SJ, Jepsen M, Pedersen LH, Stougaard P. Transglycosylating β‐d‐galactosidase and α‐l‐fucosidase from Paenibacillus sp. 3179 from a hot spring in East Greenland. MicrobiologyOpen. 2020;9:e980 10.1002/mbo3.980
DATA AVAILABILITY STATEMENT
All data generated in this study are available from the corresponding author upon reasonable request.
The 16S rRNA gene sequence is available at https://www.ncbi.nlm.nih.gov/genbank/ under accession number MK616148, and the sequences encoding the β‐d‐galactosidase and an α‐l‐fucosidase under accession numbers MK625195 and MK625194, respectively.
REFERENCES
- Aldredge, D. L. , Geronimo, M. R. , Hua, S. , Nwosu, C. C. , Lebrilla, C. B. , & Barile, D. (2013). Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology, 23, 664–676. 10.1093/glycob/cwt007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arreola, S. L. , Intanon, M. , Suljic, J. , Kittl, R. , Pham, N. H. , Kosma, P. , … Nguyen, T. H. (2014). Two β‐galactosidases from the human isolate Bifidobacterium breve DSM 20213: Molecular cloning and expression, biochemical characterization and synthesis of galacto‐oligosaccharides. PLoS ONE, 9, 1–13. 10.1371/journal.pone.0104056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D. J. , & Lowe, J. B. (2003). Fucose: Biosynthesis and biological function in mammals. Glycobiology, 13, 41R–53R. 10.1093/glycob/cwg054 [DOI] [PubMed] [Google Scholar]
- Benešová, E. , Lipovová, P. , Dvořáková, H. , & Králová, B. (2010). β‐D‐galactosidase from Paenibacillus thiaminolyticus catalyzing transfucosylation reactions. Glycobiology, 20, 442–451. 10.1093/glycob/cwp196 [DOI] [PubMed] [Google Scholar]
- Benešová, E. , Lipovová, P. , Dvořáková, H. , & Králová, B. (2013). α‐L‐fucosidase from Paenibacillus thiaminolyticus: Its hydrolytic and transglycosylation abilities. Glycobiology, 23, 1052–1065. 10.1093/glycob/cwt041 [DOI] [PubMed] [Google Scholar]
- Benešová, E. , Lipovová, P. , Krejzová, J. , Kovaľová, T. , Buchtová, P. , Spiwok, V. , & Králová, B. (2015). Alpha‐L‐fucosidase isoenzyme iso2 from Paenibacillus thiaminolyticus . BMC Biotechnology, 15, 1–7. 10.1186/s12896-015-0160-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm, G. , Lidestri, M. , Casetta, P. , Jelinek, J. , Negretti, F. , Stahl, B. , & Marini, A. (2002). Supplementation of a bovine milk formula with an oligosaccharide mixture increases counts of faecal bifidobacteria in preterm infants. Archives of Disease in Childhood. Fetal and Neonatal Edition, 86, F178–F181. 10.1136/fn.86.3.F178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, G. N. , McComb, R. B. , Christensen, R. G. , & Schaffer, R. (1980). High‐purity 4‐nitrophenol: Purification, characterization, and specifications for use as a spectrophotometric reference material. Clinical Chemistry, 26, 724–729. [PubMed] [Google Scholar]
- Chen, X. (2015). Human milk oligosaccharides (hmos): Structure, function, and enzyme‐catalyzed synthesis. Advances in Carbohydrate Chemistry and Biochemistry, 72, 113–190. 10.1016/bs.accb.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, Y. W. , Kim, J. Y. , Kim, J. H. , Jung, S. M. , & Seo, J. H. (2017). Improved production of 2′‐fucosyllactose in engineered Escherichia coli by expressing putative α‐1,2‐fucosyltransferase, WcfB from Bacteroides fragilis . Journal of Biotechnology, 257, 192–198. 10.1016/j.jbiotec.2016.11.033 [DOI] [PubMed] [Google Scholar]
- Chin, Y. W. , Kim, J. Y. , Lee, W. H. , & Seo, J. H. (2015). Enhanced production of 2’‐fucosyllactose in engineered Escherichia coli BL21star(DE3) by modulation of lactose metabolism and fucosyltransferase. Journal of Biotechnology, 210, 107–115. 10.1016/j.jbiotec.2015.06.431 [DOI] [PubMed] [Google Scholar]
- Cieśliński, H. , Kur, J. , Białkowska, A. , Baran, I. , Makowski, K. , & Turkiewicz, M. (2005). Cloning, expression, and purification of a recombinant cold‐adapted β‐D‐galactosidase from antarctic bacterium Pseudoalteromonas sp. 22b. Protein Expression and Purification, 39, 27–34. 10.1016/j.pep.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Crout, D. H. , & Vic, G. (1998). Glycosidases and glycosyl transferases in glycoside and oligosaccharide synthesis. Current Opinion in Chemical Biology, 2, 98–111. 10.1016/S1367-5931(98)80041-0 [DOI] [PubMed] [Google Scholar]
- de Roode, B. M. , Franssen, M. C. R. , van der Padt, A. , & Boom, R. M. (2003). Perspectives for the industrial enzymatic production of glycosides. Biotechnology Progress, 19, 1391–1402. 10.1021/bp030038q [DOI] [PubMed] [Google Scholar]
- DeCastro, M. E. , Rodríguez‐Belmonte, E. , & González‐Siso, M. I. (2016). Metagenomics of thermophiles with a focus on discovery of novel thermozymes. Frontiers in Microbiology, 7, 1–21. 10.3389/fmicb.2016.01521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez‐Municio, M. , Herrero, M. , Olano, A. , & Moreno, F. J. (2014). Synthesis of novel bioactive lactose‐derived oligosaccharides by microbial glycoside hydrolases. Microbial Biotechnology, 7, 315–331. 10.1111/1751-7915.12124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleuche, S. , Schäfers, C. , Blank, S. , Schröder, C. , & Antranikian, G. (2015). Exploration of extremophiles for high temperature biotechnological processes. Current Opinion in Microbiology, 25, 113–119. 10.1016/j.mib.2015.05.011 [DOI] [PubMed] [Google Scholar]
- Fai, A. E. C. , da Silva, J. B. , de Andrade, C. J. , Bution, M. L. , & Pastore, G. M. (2014). Production of prebiotic galactooligosaccharides from lactose by Pseudozyma tsukubaensis and Pichia kluyveri . Biocatalysis and Agricultural Biotechnology, 3, 343–350. 10.1016/j.bcab.2014.04.005 [DOI] [Google Scholar]
- Garrido, D. , Ruiz‐Moyano, S. , Jimenez‐Espinoza, R. , Eom, H.‐J. , Block, D. E. , & Mills, D. A. (2013). Utilization of galactooligosaccharides by Bifidobacterium longum subsp. infantis isolates. Food Microbiology, 33, 262–270. 10.1016/j.fm.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavlighi, H. A. , Michalak, M. , Meyer, A. S. , & Mikkelsen, J. D. (2013). Enzymatic depolymerization of gum tragacanth: Bifidogenic potential of low molecular weight oligosaccharides. Journal of Agriculture and Food Chemistry, 61, 1272–1278. 10.1021/jf304795f [DOI] [PubMed] [Google Scholar]
- Gibson, G. R. , Probert, H. M. , Jan, V. L. , Rastall, R. A. , & Roberfroid, M. B. (2004). Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. Nutrition Research Reviews, 17, 259–275. 10.1093/jn/125.6.1401 [DOI] [PubMed] [Google Scholar]
- Goderska, K. , Szwengiel, A. , & Czarnecki, Z. (2014). The utilization of Pseudomonas taetrolens to produce lactobionic acid. Applied Biochemistry and Biotechnology, 173, 2189–2197. 10.1007/s12010-014-1024-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero, C. , Vera, C. , Acevedo, F. , & Illanes, A. (2015). Simultaneous synthesis of mixtures of lactulose and galacto‐oligosaccharides and their selective fermentation. Journal of Biotechnology, 209, 31–40. 10.1016/j.jbiotec.2015.06.394 [DOI] [PubMed] [Google Scholar]
- Gupta, R. , Govil, T. , Capalash, N. , & Sharma, P. (2012). Characterization of a glycoside hydrolase family 1 β‐D‐galactosidase from hot spring metagenome with transglycosylation activity. Appl Biochemistry Biotechnol, 168, 1681–1693. 10.1007/s12010-012-9889-z [DOI] [PubMed] [Google Scholar]
- Gupta, V. , Gupta, N. , Capalash, N. , & Sharma, P. (2017). Bio‐prospecting bacterial diversity of hot springs in Northern Himalayan region of India for laccases. Indian J Microbiol, 57, 285–291. 10.1007/s12088-017-0656-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, L. F. , Hamoudi, S. , & Belkacemi, K. (2012). Lactobionic acid: A high value‐added lactose derivative for food and pharmaceutical applications. International Dairy Journal, 26, 103–111. 10.1016/j.idairyj.2012.05.003 [DOI] [Google Scholar]
- Hassan, N. , Nguyen, T. H. , Intanon, M. , Kori, L. D. , Patel, B. K. C. , Haltrich, D. , … Tan, T. C. (2015). Biochemical and structural characterization of a thermostable β‐glucosidase from Halothermothrix orenii for galacto‐oligosaccharide synthesis. Applied Microbiology and Biotechnology, 99, 1731–1744. 10.1007/s00253-014-6015-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. The Biochemical Journal, 280, 309–316. 10.1042/bj2800309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze, M. , You, D. J. , Kamerke, C. , Hoffmann, N. , Angkawidjaja, C. , Ernst, S. , … Elling, L. (2014). Rational design of a glycosynthase by the crystal structure of β‐D‐galactosidase from Bacillus circulans (BgaC) and its use for the synthesis of N‐acetyllactosamine type 1 glycan structures. Journal of Biotechnology, 191, 78–85. 10.1016/j.jbiotec.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Hervert, C. J. , Martin, N. H. , Boor, K. J. , & Wiedmann, M. (2017). Survival and detection of coliforms, Enterobacteriaceae, and gram‐negative bacteria in Greek yogurt. Journal of Dairy Science, 100, 950–960. 10.3168/jds.2016-11553 [DOI] [PubMed] [Google Scholar]
- Ishikawa, K. , Kataoka, M. , Yanamoto, T. , Nakabayashi, M. , Watanabe, M. , Ishihara, S. , & Yamaguchi, S. (2015). Crystal structure of β‐D‐galactosidase from Bacillus circulans ATCC 31382 (BgaD) and the construction of the thermophilic mutants. FEBS Journal, 282, 2540–2552. 10.1111/febs.13298 [DOI] [PubMed] [Google Scholar]
- Ito, Y. , & Sasaki, T. (1997). Cloning and characterization of the gene encoding a novel β ‐galactosidase from Bacillus circulans . Bioscience, Biotechnology, and Biochemistry, 61, 1270–1276. 10.1016/0378-1119(95)00758-X [DOI] [PubMed] [Google Scholar]
- Jones, D. T. , Taylor, W. R. , & Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences, 8, 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Jørgensen, F. , Hansen, O. C. , & Stougaard, P. (2001). High‐efficiency synthesis of oligosaccharides with a truncated β‐D‐galactosidase from Bifidobacterium bifidum . Applied Microbiology and Biotechnology, 57, 647–652. https://doi.org/10.1007%252Fs00253-001-0845-z [DOI] [PubMed] [Google Scholar]
- Jørgensen, F. , Hansen, O. C. , & Stougaard, P. (2004). Enzymatic conversion of D‐galactose to D‐tagatose: Heterologous expression and characterisation of a thermostable L‐arabinose isomerase from Thermoanaerobacter mathranii . Applied Microbiology and Biotechnology, 64, 816–822. 10.1007/s00253-004-1578-6 [DOI] [PubMed] [Google Scholar]
- Knol, J. , Boehm, G. , Lidestri, M. , Negretti, F. , Jelinek, J. , Agosti, M. , … Mosca, F. (2005). Increase of faecal bifidobacteria due to dietary oligosaccharides induces a reduction of clinically relevant pathogen germs in the faeces of formula‐fed preterm infants. Acta Paediatrica Supplement, 94, 31–33. 10.1111/j.1651-2227.2005.tb02152.x [DOI] [PubMed] [Google Scholar]
- Knol, J. , Scholtens, P. , Kafka, C. , Steenbakkers, J. , Gro, S. , Helm, K. , … Wells, J. (2005). Colon microflora in infants fed formula with galacto‐ and fructo‐oligosaccharides: More like breast‐fed infants. Journal of Pediatric Gastroenterology and Nutrition, 40, 36–42. 10.1097/00005176-200501000-00007 [DOI] [PubMed] [Google Scholar]
- Koval’ová, T. , Kovaľ, T. , Benešová, E. , Vodičková, P. , Spiwok, V. , Lipovová, P. , & Dohnálek, J. (2019). Active site complementation and hexameric arrangement in the GH family 29; a structure–function study of α‐L‐fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus . Glycobiology, 29, 59–73. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, C. , Kuntz, S. , & Rudloff, S. (2014). Bioactivity of human milk oligosaccharides In Moreno F. J., & Sanz M. L. (Eds.), Food Oligosaccharides: Production, Analysis and Bioactivity (pp. 5–20). Hoboken, NJ: John Wiley & Sons Ltd. [Google Scholar]
- Kunz, C. , & Rudloff, S. (1993). Biological functions of oligosaccharides in human milk. Acta Paediatrica, 82, 903–912. 10.1111/j.1651-2227.1993.tb12597.x [DOI] [PubMed] [Google Scholar]
- Kunz, C. , Rudloff, S. , Baier, W. , Klein, N. , & Strobel, S. (2000). Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annual Review of Nutrition, 20, 699–722. 10.1146/annurev.nutr.20.1.699 [DOI] [PubMed] [Google Scholar]
- Ladirat, S. E. , Schuren, F. H. J. , Schoterman, M. H. C. , Nauta, A. , Gruppen, H. , & Schols, H. A. (2014). Impact of galacto‐oligosaccharides on the gut microbiota composition and metabolic activity upon antibiotic treatment during in vitro fermentation. FEMS Microbiology Ecology, 87, 41–51. [DOI] [PubMed] [Google Scholar]
- Lezyk, M. , Jers, C. , Kjaerulff, L. , Gotfredsen, C. H. , Mikkelsen, M. D. , & Mikkelsen, J. D. (2016). Novel α‐L‐fucosidases from a soil metagenome for production of fucosylated human milk oligosaccharides. PLoS ONE, 11 10.1371/journal.pone.0147438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Wang, H. , Lu, L. , Li, Z. , Xu, X. , & Xiao, M. (2009). Purification and characterization of a novel β‐D‐galactosidase with transglycosylation activity from Bacillus megaterium 2–37‐4‐1. Applied Biochemistry and Biotechnology, 158, 192–199. 10.1007/s12010-008-8310-4 [DOI] [PubMed] [Google Scholar]
- Mahajan, G. B. , & Balachandran, L. (2017). Sources of antibiotics: Hot springs. Biochemical Pharmacology, 134, 35–41. 10.1016/j.bcp.2016.11.021 [DOI] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Derbyshire, M. K. , Gonzales, N. R. , Lu, S. , Chitsaz, F. , Geer, L. Y. , … Bryant, S. H. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Research, 43, D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, R. , Singhal, P. , Singh, H. , Damle, D. , & Sharma, A. K. (2016). Insight into thermophiles and their wide‐spectrum applications. 3 Biotech, 6, 1–9. 10.1007/s13205-016-0368-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes, M. J. , Mercadé, E. , Bozal, N. , & Guinea, J. (2004). Paenibacillus antarcticus sp. nov., a novel psychrotolerant organism from the Antarctic environment. International Journal of Systematic and Evolutionary Microbiology, 54, 1521–1526. 10.1099/ijs.0.63078-0 [DOI] [PubMed] [Google Scholar]
- Moro, G. , Minoli, I. , Mosca, M. , Fanaro, S. , Jelinek, J. , Stahl, B. , & Boehm, G. (2002). Dosage‐related bifidogenic effects of galacto‐ and fructooligosaccharides in formula‐fed term infants. Journal of Pediatric Gastroenterology and Nutrition, 34, 291–295. 10.1097/00005176-200203000-00014 [DOI] [PubMed] [Google Scholar]
- Morrow, A. L. , Ruiz‐Palacios, G. M. , Jiang, X. , & Newburg, D. S. (2005). Human‐milk glycans that inhibit pathogen binding protect breast‐feeding infants against infectious diarrhea. Journal of Nutrition, 135, 1304–1307. 10.1093/jn/135.5.1304 [DOI] [PubMed] [Google Scholar]
- Mu, W. , Chen, Q. , Wang, X. , Zhang, T. , & Jiang, B. (2013). Current studies on physiological functions and biological production of lactosucrose. Applied Microbiology and Biotechnology, 97, 7073–7080. 10.1007/s00253-013-5079-3 [DOI] [PubMed] [Google Scholar]
- Park, A. R. , Kim, H. J. , Lee, J. K. , & Oh, D. K. (2010). Hydrolysis and transglycosylation activity of a thermostable recombinant β‐glycosidase from Sulfolobus acidocaldarius . Applied Biochemistry and Biotechnology, 160, 2236–2247. 10.1007/s12010-009-8705-x [DOI] [PubMed] [Google Scholar]
- Park, H. Y. , Kim, H. J. , Lee, J. K. , Kim, D. , & Oh, D. K. (2008). Galactooligosaccharide production by a thermostable β‐D‐galactosidase from Sulfolobus solfataricus . World Journal of Microbiology & Biotechnology, 24, 1553–1558. 10.1007/s11274-007-9642-x [DOI] [Google Scholar]
- Petrova, V. Y. , & Kujumdzieva, A. V. (2010). Thermotolerant yeast strains producers of galacto‐oligosaccharides. Biotechnology and Biotechnological Equipment, 24, 1612–1619. 10.2478/V10133-010-0014-6 [DOI] [Google Scholar]
- Rada, V. , Nevoral, J. , Trojanová, I. , Tomá Nková, E. , Mehilová, M. Š. , & Killer, J. (2008) Growth of infant faecal bifidobacteria and clostridia on prebiotic oligosaccharides in in vitro conditions. Anaerobe, 14(4), 205–208. 10.1016/j.anaerobe.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Ribeiro, A. L. , Sánchez, M. , Hidalgo, A. , & Berenguer, J. (2017). Stabilization of enzymes by using thermophiles In Barredo J.‐L., & Herráiz I. (Eds.), Microbial Steroids: Methods and Protocols ‐ Methods in Molecular Biology (pp. 297–312). New York, NY: Springer New York; 10.1007/978-1-4939-7183-1_21 [DOI] [PubMed] [Google Scholar]
- Ruusunen, M. , Surakka, A. , Korkeala, H. , & Lindström, M. (2012). Clostridium tyrobutyricum strains show wide variation in growth at different NaCl, pH, and temperature conditions. Journal of Food Protection, 75, 1791–1795. 10.4315/0362-028X.JFP-12-109 [DOI] [PubMed] [Google Scholar]
- Sahay, H. , Yadav, A. N. , Singh, A. K. , Singh, S. , Kaushik, R. , & Saxena, A. K. (2017). Hot springs of Indian Himalayas: Potential sources of microbial diversity and thermostable hydrolytic enzymes. 3 Biotech, 7, 1–11. 10.1007/s13205-017-0762-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M. , & Stougaard, P. (2010). Identification, cloning and expression of a cold‐active β‐D‐galactosidase from a novel Arctic bacterium, Alkalilactibacillus ikkense . Environmental Technology, 31, 1107–1114. 10.1080/09593331003677872 [DOI] [PubMed] [Google Scholar]
- Schultz‐Johansen, M. , Bech, P. K. , Hennessy, R. C. , Glaring, M. A. , Barbeyron, T. , Czjzek, M. , & Stougaard, P. (2018). A novel enzyme portfolio for red algal polysaccharide degradation in the marine bacterium Paraglaciecola hydrolytica S66T encoded in a sizeable polysaccharide utilization locus. Frontiers in Microbiology, 9, 1–15. 10.3389/fmicb.2018.00839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, L. E. J. , Best, A. , Nunez, A. , Salguero, F. J. , Johnson, L. , Weyer, U. , … La Ragione, R. M. (2009). A mixture containing galactooligosaccharide, produced by the enzymic activity of Bifidobacterium bifidum, reduces Salmonella enterica serovar Typhimurium infection in mice. Journal of Medical Microbiology, 58, 37–48. 10.1099/jmm.0.004390-0 [DOI] [PubMed] [Google Scholar]
- Shoaf, K. , Mulvey, G. L. , Armstrong, G. D. , & Hutkins, R. W. (2006). Prebiotic galactooligosaccharides reduce adherence of enteropathogenic Escherichia coli to tissue culture cells. Infection and Immunity, 74, 6920–6928. 10.1128/IAI.01030-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla, V. , & Chakkaravarthi, S. (2017). Applications of prebiotics in food industry: A review. Food Science and Technology International, 23, 649–667. 10.1177/1082013217721769 [DOI] [PubMed] [Google Scholar]
- Splechtna, B. , Nguyen, T. H. , Steinböck, M. , Kulbe, K. D. , Lorenz, W. , & Haltrich, D. (2006). Production of prebiotic galacto‐oligosaccharides from lactose using β‐galactosidases from Lactobacillus reuteri . Journal of Agriculture and Food Chemistry, 54, 4999–5006. 10.1021/jf053127m [DOI] [PubMed] [Google Scholar]
- Sprenger, G. A. , Baumgärtner, F. , & Albermann, C. (2017). Production of human milk oligosaccharides by enzymatic and whole‐cell microbial biotransformations. Journal of Biotechnology, 258, 79–91. 10.1016/j.jbiotec.2017.07.030 [DOI] [PubMed] [Google Scholar]
- Tamura, K. , & Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10, 512–526. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- Tan, J. , Chai, F. , & Zhang, C. (2015). Determination of human milk oligosaccharides in human breast milk by HPAE‐PAD with on‐line sample cleanup. Costumer Application Note 119, Abbott Nutrition / Thermo Fisher Scientific, 1–6. [Google Scholar]
- Tang, L. , Li, Z. A. , Dong, X. X. , Yang, R. J. , Zhang, J. H. , & Mao, Z. G. (2011). Lactulose biosynthesis by β‐D‐galactosidase from a newly isolated Arthrobacter sp. Journal of Industrial Microbiology and Biotechnology, 38, 471–476. 10.1007/s10295-010-0897-0 [DOI] [PubMed] [Google Scholar]
- Tcherpakov, M. , Ben‐Jacob, E. , & Gutnick, D. L. (1999). Paenibacillus dendritiformis sp. nov., proposal for a new pattern‐forming species and its localization within a phylogenetic cluster. International Journal of Systematic Bacteriology, 49, 239–246. 10.1099/00207713-49-1-239 [DOI] [PubMed] [Google Scholar]
- Thompson, J. D. , Higgins, D. G. , & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acid Research, 22, 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurl, S. , Henker, J. , Taut, H. , Tovar, K. , & Sawatzki, G. (1993). Variations of neutral oligosaccharides and lactose in human milk during the feeding. Zeitschrift Fur Ernahrungswissenschaft, 32, 262–269. 10.1007/BF01611164 [DOI] [PubMed] [Google Scholar]
- Torres, D. P. M. , Gonçalves, M. D. P. F. , Teixeira, J. A. , & Rodrigues, L. R. (2010). Galacto‐oligosaccharides: Production, properties, applications, and significance as prebiotics. Compr Rev Food Sci Food Saf, 9, 438–454. 10.1111/j.1541-4337.2010.00119.x [DOI] [PubMed] [Google Scholar]
- Urbieta, M. S. , Donati, E. R. , Chan, K. G. , Shahar, S. , Sin, L. L. , & Goh, K. M. (2015). Thermophiles in the genomic era: Biodiversity, science, and applications. Biotechnology Advances, 33, 633–647. 10.1016/j.biotechadv.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Wanarska, M. , & Kur, J. (2012). A method for the production of D‐tagatose using a recombinant Pichia pastoris strain secreting β‐D‐galactosidase from Arthrobacter chlorophenolicus and a recombinant L‐arabinose isomerase from Arthrobacter sp. 22c. Microbial Cell Factories, 11, 1–15. 10.1186/1475-2859-11-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Yang, R. , Hua, X. , Zhao, W. , & Zhang, W. (2013). Enzymatic production of lactulose and 1‐lactulose: Current state and perspectives. Applied Microbiology and Biotechnology, 97, 6167–6180. 10.1007/s00253-013-4998-3 [DOI] [PubMed] [Google Scholar]
- Watt, G. M. , Lowden, P. A. , & Flitsch, S. L. (1997). Enzyme‐catalyzed formation of glycosidic linkages. Current Opinion in Structural Biology, 7, 652–660. 10.1016/S0959-440X(97)80074-7 [DOI] [PubMed] [Google Scholar]
- Wu, Y. , Yuan, S. , Chen, S. , Wu, D. , Chen, J. , & Wu, J. (2013). Enhancing the production of galacto‐oligosaccharides by mutagenesis of Sulfolobus solfataricus β‐galactosidase. Food Chemistry, 138, 1588–1595. 10.1016/j.foodchem.2012.11.052 [DOI] [PubMed] [Google Scholar]
- Yu, L. , & O’Sullivan, D. J. (2018). Immobilization of whole cells of Lactococcus lactis containing high levels of a hyperthermostable β‐D‐galactosidase enzyme in chitosan beads for efficient galacto‐oligosaccharide production. Journal of Dairy Science, 101, 2974–2983. 10.3168/jds.2017-13770 [DOI] [PubMed] [Google Scholar]
- Zeuner, B. , Muschiol, J. , Holck, J. , Lezyk, M. , Gedde, M. R. , Jers, C. , … Meyer, A. S. (2018). Substrate specificity and transfucosylation activity of GH29 α‐L‐fucosidases for enzymatic production of human milk oligosaccharides. N Biotechnol, 41, 34–45. 10.1016/j.nbt.2017.12.002 [DOI] [PubMed] [Google Scholar]
- Zeuner, B. , Teze, D. , Muschiol, J. , & Meyer, A. S. (2019). Synthesis of human milk oligosaccharides: Protein engineering strategies for improved enzymatic transglycosylation. Molecules, 24(2033), 1–22. 10.3390/molecules24112033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyer, J. , & Kocher, H. P. (1988). Purification and characterization of a bacterial nitrophenol oxygenase which converts ortho‐nitrophenol to catechol and nitrite. Journal of Bacteriology, 170, 1789–1794. 10.1128/jb.170.4.1789-1794.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, Y. , Xu, Z. , Li, S. , Liu, X. , Xu, L. , Feng, X. , & Xu, H. (2014). Coexpression of β‐D‐galactosidase and L‐arabinose isomerase in the production of D‐tagatose: A functional sweetener. Journal of Agriculture and Food Chemistry, 62, 2412–2417. 10.1021/jf4042485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated in this study are available from the corresponding author upon reasonable request.
The 16S rRNA gene sequence is available at https://www.ncbi.nlm.nih.gov/genbank/ under accession number MK616148, and the sequences encoding the β‐d‐galactosidase and an α‐l‐fucosidase under accession numbers MK625195 and MK625194, respectively.


