Introduction
Recent advances in high throughput omic technologies have greatly enhanced our knowledge of the molecular basis of complex lung diseases. In particular, with the advent of next generation sequencing, transcriptomic studies with RNA sequencing (RNA-seq) have yielded important insights into COPD and idiopathic pulmonary fibrosis (IPF) [1, 2]. However, most studies to date have been performed on whole lung tissue, such that many unique cell type-specific gene expression signatures, particularly those of alveolar epithelial cells (AECs), which play a key role in COPD and IPF, may have been diluted, if not missed altogether. Single-cell RNA-seq is rapidly gaining momentum as a strategy to profile individual cells [3, 4], but sequencing depth is typically much lower and cost much higher than that of bulk RNA-seq.
The challenges of procuring human lung tissue in a standardised way and isolating epithelial cells from cryopreserved samples, in addition to the requirement for RNA of adequate quantity and quality, have hampered the field’s ability to perform transcriptomic profiling in AECs. Using biobanked samples would significantly expand the pool of lung specimens for investigation and minimise the statistical biases related to batch processing of fresh specimens [5]. Protocols detailing lung digestion and isolation of specific cell populations have been described [3, 6, 7], but few studies have examined how cryopreservation alters lung epithelial cell viability, RNA quality and gene expression [8–10]. Here we demonstrate an example of successful RNA-seq of AECs isolated from biobanked lung cell suspensions. We discuss the technical factors that may influence the success of this methodology and how it may be adapted for a broad range of downstream applications.
Sample pipeline
Figure 1 depicts the overall methodology of our sample pipeline. Human explanted lungs were procured from donors with end-stage lung disease (COPD, IPF and non-IPF fibrosis) undergoing transplant or control lungs rejected for transplant. For IPF and COPD specimens, tissue was selected from visibly diseased parenchyma from any lobe. Lung tissue was digested and processed according to the protocol provided in the supplementary methods. Cell suspensions were sorted immediately or cryopreserved in liquid nitrogen until thawed later (figure 1a). Non-biobanked and biobanked cell suspensions were sorted according to a previously described protocol [7] to enrich for viable (DAPI−) non-haematopoietic (CD45−) type I and II AECs using two gates, EpCAM+/PDPNlow (P1 gate) and EpCAMhigh/PDPN− (P2 gate), respectively (figure 1b). A subset of sorted cells was fixed in paraformaldehyde, incubated with antibodies against surfactant protein C and aquaporin, and visualised using confocal microscopy (figure 1c). RNA was extracted from sorted cell suspensions and purified. RNA quality and quantity were measured with a bioanalyser. RNA-seq was performed on 11 biobanked control samples. The Illumina TruSeq RNA Access Library Prep kit was used for library preparation. Purified libraries were validated on TapeStation and quantitated with Qubit. FastQC was used to visualise aggregate Phred scores from demultiplexed sample FastQ files. Sequence quality was assessed using Phred scores, mapping rate, and RNA composition. R package Rsubreads [11] and DESeq2 [12] were used to quantify and normalise gene expression counts. Statistical inference testing was performed using linear regression models in DESeq2, controlling for age, gender and cell type percentage.
FIGURE 1.
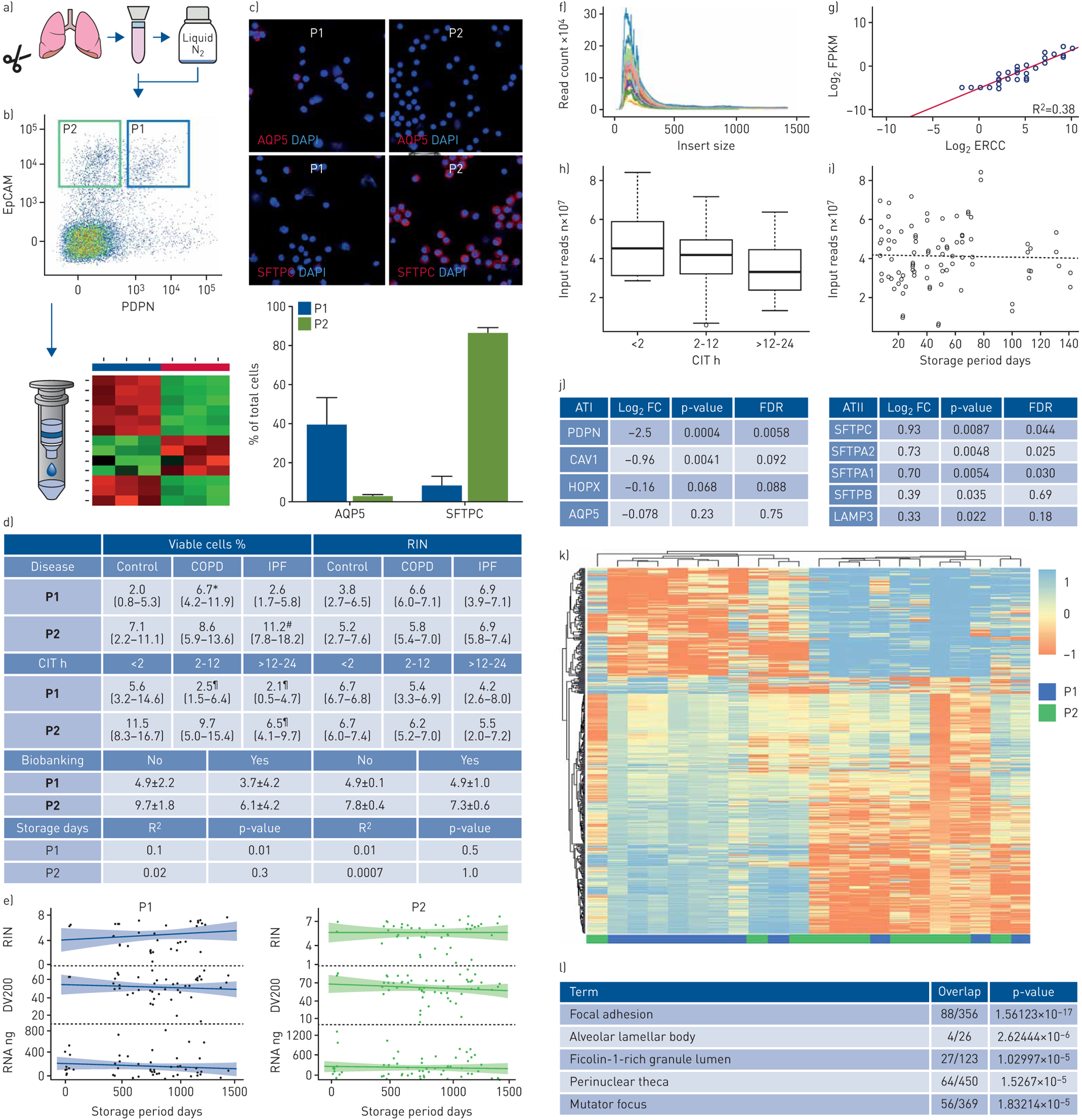
Isolation and RNA-seq of human alveolar epithelial cells (AECs). a) 52 human lung explants (20 control, 10 COPD and 22 idiopathic pulmonary fibrosis (IPF)) were enzymatically digested into cell suspensions that were biobanked in liquid nitrogen or flow-sorted immediately. b) Non-biobanked (n=6) and biobanked (n=52) cell suspensions from control and diseased lungs were flow-sorted using two gates, P1 and P2, to enrich for type I and II AECs (ATI and ATII), respectively. c) AQP5 and SFTPC expression in paired P1- and P2-gated biobanked control cells was quantified by immunofluorescence (n=3). d and e) Effects of cold ischaemic time (CIT), biobanking and storage duration (median 726 days, P25-P75 441–1021) on yield of viable P1- and P2-gated cells (as a percentage of total pre-sorted cells) and quality of RNA extracted from control and diseased lungs (n=52). RNA-seq was performed on paired P1- and P2-gated biobanked control samples (n=11). Quality control metrics demonstrated an adequate median insert size (f) and a high level of agreement between observed and expected concentrations of the target sequences (g), as well as a significant correlation of input size with CIT (h) but not storage duration (i) (p=0.03 and 0.79, respectively). j) Gene expression of canonical markers for ATI (left) and ATII (right) was significantly elevated in P1- and P2-gated cell populations, respectively, although not all comparisons reached statistical significance (e.g. for AQP5). Log2 FC reflects fold change of gene expression in P2- versus P1-gated samples. k) Hierarchical clustering of the top 650 genes revealed good separation of P1- and P2-gated samples from biobanked specimens. l) Pathway enrichment analysis with gene ontology terms was performed on differentially expressed P2-gated cells. *: p<0.05 versus control and IPF; #: p<0.05 versus control; ¶: p<0.05 versus <2 h. FC: fold change; FPKM: fragments per kilobase of transcript per million mapped reads; ERCC: external RNA control consortium spike-in; FDR: false discovery rate; RIN: RNA integrity number.
Key observations
To investigate the impact of technical variables on cell viability and RNA quality, we examined the effect of disease, cold ischaemic time (CIT, defined as the period from lung explantation to the first step of tissue digestion) and use/duration of cryopreservation on the yield of viable P1- and P2-gated epithelial cells and the RNA Integrity Number (RIN) and DV200, standard metrics of RNA quality (figure 1d and e). Data on warm ischaemic times (period from cross-clamping of aorta to lung explantation) were not routinely available. There was a relative abundance of P2-gated cells in IPF lungs compared to COPD and control lungs, possibly reflecting aberrant alveolar re-epithelialisation, a key pathological feature of IPF [13]. P2-gated samples also yielded a higher quantity of RNA (427±395 ng versus 241±197 ng per mL of total cell suspension). Fresh cell suspensions had more viable total cells, but there was no significant difference in the percentage of P1 or P2-gated cells compared to biobanked cells. Longer CIT and storage duration (median 726 days, interquartile range 441–1021) generally affected the yield and RNA integrity of P1-gated cells more than P2-gated cells. This was not surprising, as type I AECs are more vulnerable and less abundant than type II AECs [14], which serve as facultative progenitor cells for alveolar regeneration in response to lung injury [15, 16]. While RIN varied widely among samples, no significant differences between non-biobanked and biobanked sorted cells were observed (figure 1d). Moreover, there were no strong correlations (r2 >0.9) of RIN with disease, CIT or storage times, although there was a trend towards lower RNA quality as CIT and storage time increased.
To determine whether RNA obtained from biobanked samples using this enrichment strategy was adequate for transcriptomic profiling, RNA-seq of P1- and P2-gated cells from control lungs was performed using exome capture sequencing. Overall quality of sequence data was high, with a high read mapping frequency (>90% for all samples), adequate insert size (median 248 bp) (figure 1f), high level of agreement between observed and expected concentrations of target sequences (figure 1g) and no duplicate transcripts (data not shown). There was a significant correlation of input reads with CIT but not storage duration (p=0.03 and 0.79, respectively) (figure 1h and i). To assess whether our cell sorting approach reliably partitioned type I and type II AECs, we compared differential gene expression of paired P1- and P2-gated biobanked samples, focusing on signature markers associated with these cell types. Genes related to type I and type II AECs were upregulated in P1- and P2-gated samples, respectively, although differential gene expression did not reach statistical significance for AQP5 (figure 1j). This was consistent with the relative expression of AQP5 in the two cell populations as determined by immunofluorescence; among P1-gated cells, only 39.4±13.7% stained positive for AQP5, while among P2-gated cells, 86.1±2.8% stained positive for SFTPC (figure 1c). The comparatively low differential expression of AQP5 in P1-gated cells likely reflects admixture by other cell types, as well as potential transdifferentiation of ATII to ATI cells. However, unbiased hierarchical clustering demonstrated overall good segregation of P1- and P2-gated samples (figure 1k). Gene ontology enrichment analysis revealed increased regulation of genes related to focal adhesion and lamellar bodies in P2-gated cells (figure 1l).
Implications for current and future applications
Our work provides a sample pipeline for isolating a targeted cell population from cryopreserved human lung explants for large-scale molecular analysis. After enriching for AECs from lung cell suspensions using an established flow-sorting protocol, we performed RNA-seq on biobanked control samples using exome capture sequencing. Our approach was limited by lower sample numbers, the use of imperfect cell markers which likely contributed to admixture in our AT1-gated cells, and a restricted set of comparisons in our analysis. Despite these challenges, as well as the significant variability in source collection and cryopreservation times, input RNA was still of adequate quality to generate transcriptome libraries that identify biologically relevant signatures, paving the way for the next step of profiling our diseased samples as well as collaborative studies with other groups.
High throughput molecular studies have yielded valuable insights into lung pathobiology, yet many studies are limited by low sample number and lack validation cohorts [17]. Individuals diagnosed with the same disease may have widely disparate clinical courses and responses to therapies, underscoring the need to generate biorepositories with comprehensive cellular and molecular phenotyping that can facilitate personalised approaches to disease classification and management. Organ explants obtained during transplantation or diagnostic surgical lung biopsies are a valuable source of human tissue. However, obtaining good quality lung specimens remains challenging due to the unpredictable timing of transplants and vulnerability to ischaemic injury, particularly in epithelial cells. Prospectively biobanking lung tissue and systematically enriching for cell populations of interest such as AECs, which are at the core of the pathogenesis of diseases such as IPF and COPD, can facilitate biologically and statistically rigorous analyses of samples by minimising batch variability while providing an entrance into investigating lung architecture and function at multiple levels of regulation even beyond the transcriptome, be it the epigenome, proteome or metabolome. Our methodology validates the use of prospectively biobanking digested lung suspensions, but alternative approaches, such as biobanking whole tissue and then digesting and sorting cells, could also be considered and systematically studied.
In our study, we demonstrated the impact of ischaemic time and cryopreservation on AEC viability and RNA quality. RIN scores higher than seven are typically recommended for RNA-seq, which is commonly performed using poly(A) tail selection to enrich mRNA. Poly(A) libraries may greatly underrepresent the target transcriptome in degraded RNA [18]. As our samples had lower mean RIN scores, we adopted exome capture technology, a validated alternate approach of enriching cDNA transcripts after the main enzymatic steps of library construction rather than at the RNA stage. Using excess complementary capture probes at multiple positions enables transcript recovery even without poly(A) tails [18, 19]. RNA-seq on biobanked samples achieved standard quality control metrics, suggesting that exome capture sequencing is a good alternative in settings where RNA quality is reduced, such as clinical or formaldehyde-fixed paraffin-embedded samples. The limited availability of fresh tissue often leads to sporadic processing of individual samples, introducing biases from technical/operator variability that are mitigated when processing cryopreserved specimens in bulk. Importantly, our study showed similar RINs between non-biobanked and biobanked specimens. While we did not determine the effect of biobanking on transcriptomic signatures, other studies have demonstrated that cryopreservation does not significantly alter gene expression in cell lines or lung cancer tissue, including those involved in inflammatory or immune responses, with over 2 h from resection to freezing [8–10].
Having validated our methodology with RNA-seq of control AECs, we can thus proceed with bulk RNA-seq and single-cell gene expression studies to identify key transcriptomic signatures in chronic parenchymal lung disease. While RNA-seq’s most widely used application has been gene expression profiling, its capacity for single base pair resolution enables characterisation of other facets of the human genome, including non-coding RNA species, regulatory elements, allele-specific expression, alternative splicing and gene fusions [20]. All of these may have unprecedented implications on our understanding of complex lung diseases and the development of precision therapies.
Harnessing such applications requires sufficient sequencing depth to ensure accuracy, but this frequently limits the number of transcripts that can be read in each sample. One strategy to maximise read depth in heterogeneous organs such as the lung is to isolate targeted populations of cells, which we achieved using a straightforward method of enriching AECs from whole lung tissue. However, admixture by other cell types in our sorted populations does highlight the challenges of distinguishing AEC subtypes solely using positive/negative selection with established markers, particularly as there may be loss of these markers associated with disease pathogenesis. Other markers of type II cells have been identified that could be used for more targeted enrichment [21]. Lung cells may transdifferentiate [22] and type II cells can transform into type I cells under various conditions [21, 23, 24]. Integrating agnostic sequencing approaches such as single-cell RNA-seq or single-nucleus RNA-seq (Nuc-seq), which obviates the need for live cells [25], might effectively deal with cell admixture and uncover novel cell types and molecular signatures, including more specific cell surface markers. While our study has several technical limitations as discussed, it presents methodological concepts that can be refined and widely adapted for diverse applications, facilitating large-scale, deep molecular phenotyping of complex organ diseases.
Supplementary Material
Acknowledgements:
A subset of control lungs was provided by the International Institute for the Advancement of Medicine.
Support statement: This work was supported by NHLBI grant P01HL114501 (G.R. Washko, A.M.K. Choi, B.A. Raby and I.O. Rosas). Funding information for this article has been deposited with the Crossref Funder Registry.
Footnotes
Conflict of interest: S.G. Chu has nothing to disclose. S. Poli De Frias has nothing to disclose. Y. Sakairi reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. R.S. Kelly reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. R. Chase reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. K. Konishi reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. A. Blau reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. E. Tsai reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. K. Tsoyi has nothing to disclose. R.F. Padera reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. L.M. Sholl reports grants from National Institutes of Health (P01HL114501), during the conduct of the study; personal fees for consultancyfrom Foghorn Therapeutics, outside the submitted work. H.J. Goldberg reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. H.R. Mallidi reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. P.C. Camp reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. S.Y. El-Chemaly has nothing to disclose. M.A. Perrella reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. A.M.K. Choi reports grants from National Institutes of Health (P01HL114501, R01HL132198, P01HL108801, R01HL055330, R01HL133801), during the conduct of the study. G.R. Washko reports grants from NIH and BTG Interventional Medicine, grants from and consultancy/advisory board work for Boehringer Ingelheim, consultancy for Genentech, Regeneron and GlaxoSmithKline, consultancy/data monitoring committee work for PulmonX, advisory board work for ModoSpira and Toshiba, grants from and consultancy for Janssen Pharmaceuticals, outside the submitted work; is founder and co-owner of Quantitative Imaging Solutions; and G.R Washko’s spouse works for Biogen, which is focused on developing therapies for fibrotic lung disease. B.A. Raby reports grants from National Institutes of Health (P01HL114501), during the conduct of the study. I.O. Rosas reports grants from National Institutes of Health (P01HL114501), during the conduct of the study.
This article has supplementary material available from erj.ersjournals.com
References
- 1.Schafer MJ, White TA, Iijima K, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun 2017; 8: 14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusko RL, Brothers JF 2nd, Tedrow J, et al. Integrated genomics reveals convergent transcriptomic networks underlying chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2016; 194: 948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y, Mizuno T, Sridharan A, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI insight 2016; 1: e90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reyfman PA, Walter JM, Joshi N, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med 2019; 199: 1517–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conesa A, Madrigal P, Tarazona S, et al. A survey of best practices for RNA-seq data analysis. Genome Biol 2016; 17: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujino N, Kubo H, Suzuki T, et al. Isolation of alveolar epithelial type II progenitor cells from adult human lungs. Lab Invest 2011; 91: 363–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujino N, Kubo H, Ota C, et al. A novel method for isolating individual cellular components from the adult human distal lung. Am J Respir Cell Mol Biol 2012; 46: 422–430. [DOI] [PubMed] [Google Scholar]
- 8.Baatz JE, Newton DA, Riemer EC, et al. Cryopreservation of viable human lung tissue for versatile post-thaw analyses and culture. In vivo 2014; 28: 411–423. [PMC free article] [PubMed] [Google Scholar]
- 9.Guillaumet-Adkins A, Rodriguez-Esteban G, Mereu E, et al. Single-cell transcriptome conservation in cryopreserved cells and tissues. Genome Biol 2017; 18: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caboux E, Paciencia M, Durand G, et al. Impact of delay to cryopreservation on RNA integrity and genome-wide expression profiles in resected tumor samples. PLoS One 2013; 8: e79826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013; 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecker L, Thannickal VJ. Nonresolving fibrotic disorders: idiopathic pulmonary fibrosis as a paradigm of impaired tissue regeneration. Am J Med Sci 2011; 341: 431–434. [DOI] [PubMed] [Google Scholar]
- 14.Ward HE, Nicholas TE. Alveolar type I and type II cells. Aust N Z J Med 1984; 14: Suppl. 3, 731–734. [PubMed] [Google Scholar]
- 15.Uhal BD. Cell cycle kinetics in the alveolar epithelium. Am J Physiol 1997; 272: L1031–L1045. [DOI] [PubMed] [Google Scholar]
- 16.Selman M, Pardo A. Role of epithelial cells in idiopathic pulmonary fibrosis: from innocent targets to serial killers. Proc Am Thorac Soc 2006; 3: 364–372. [DOI] [PubMed] [Google Scholar]
- 17.Vukmirovic M, Kaminski N. Impact of transcriptomics on our understanding of pulmonary fibrosis. Front Med 2018; 5: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cieslik M, Chugh R, Wu YM, et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res 2015; 25: 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sigurgeirsson B, Emanuelsson O, Lundeberg J. Sequencing degraded RNA addressed by 3’ tag counting. PLoS One 2014; 9: e91851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y, Gao S, Muegge K, et al. Advanced applications of RNA sequencing and challenges. Bioinform Biol Insights 2015; 9: Suppl. 1, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barkauskas CE, Cronce MJ, Rackley CR, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 2013; 123: 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng D, Soh BS, Yin L, et al. Differentiation of club cells to alveolar epithelial cells in vitro. Sci Rep 2017; 7: 41661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danto SI, Shannon JM, Borok Z, et al. Reversible transdifferentiation of alveolar epithelial cells. Am J Respir Cell Mol Biol 1995; 12: 497–502. [DOI] [PubMed] [Google Scholar]
- 24.Beers MF, Moodley Y. When is an alveolar type 2 cell an alveolar type 2 cell? A conundrum for lung stem cell biology and regenerative medicine. Am J Respir Cell Mol Biol 2017; 57: 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Waters J, Leung ML, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 2014; 512: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


