Abstract
Antimicrobial resistance (AMR) poses a threat to public health. Clinical microbiology laboratories typically rely on culturing bacteria for antimicrobial-susceptibility testing (AST). As the implementation costs and technical barriers fall, whole-genome sequencing (WGS) has emerged as a ‘one-stop’ test for epidemiological and predictive AST results. Few published comparisons exist for the myriad analytical pipelines used for predicting AMR. To address this, we performed an inter-laboratory study providing sets of participating researchers with identical short-read WGS data from clinical isolates, allowing us to assess the reproducibility of the bioinformatic prediction of AMR between participants, and identify problem cases and factors that lead to discordant results. We produced ten WGS datasets of varying quality from cultured carbapenem-resistant organisms obtained from clinical samples sequenced on either an Illumina NextSeq or HiSeq instrument. Nine participating teams (‘participants’) were provided these sequence data without any other contextual information. Each participant used their choice of pipeline to determine the species, the presence of resistance-associated genes, and to predict susceptibility or resistance to amikacin, gentamicin, ciprofloxacin and cefotaxime. We found participants predicted different numbers of AMR-associated genes and different gene variants from the same clinical samples. The quality of the sequence data, choice of bioinformatic pipeline and interpretation of the results all contributed to discordance between participants. Although much of the inaccurate gene variant annotation did not affect genotypic resistance predictions, we observed low specificity when compared to phenotypic AST results, but this improved in samples with higher read depths. Had the results been used to predict AST and guide treatment, a different antibiotic would have been recommended for each isolate by at least one participant. These challenges, at the final analytical stage of using WGS to predict AMR, suggest the need for refinements when using this technology in clinical settings. Comprehensive public resistance sequence databases, full recommendations on sequence data quality and standardization in the comparisons between genotype and resistance phenotypes will all play a fundamental role in the successful implementation of AST prediction using WGS in clinical microbiology laboratories.
Keywords: antimicrobial resistance, antimicrobial-susceptibility testing, whole-genome sequencing, bioinformatics, carbapenem resistance
Data Summary
Sequence read files for all samples used in this study have been deposited in the European Nucleotide Archive under the project accession number PRJEB34513 and the following sample accession numbers: SAMEA5789893 (sample A-1), SAMEA5789894 (sample A-2), SAMEA5789895 (sample B-1), SAMEA5789896 (sample B-2), SAMEA5789897 (sample C-1), SAMEA5789898 (sample C-2), SAMEA5789899 (sample D), SAMEA5789900 (sample E), SAMEA5789901 (sample F), SAMEA5789902 (sample G).
Impact Statement.
Antimicrobial resistance (AMR) is now recognized as a worldwide public-health issue, and identifying those infections that are resistant to common antibiotics quickly and accurately is a leading priority. The improvement of molecular methods of analysing bacterial DNA, especially whole-genome sequencing (WGS), has raised the possibility of using it as a single assay that can identify the pathogen, antibiotic susceptibility and track transmission. In this study, we compared methods for predicting AMR from bacterial DNA sequences through an inter-laboratory study. This is, to the best of our knowledge, the first study of its kind to blind sets of participants to any contextual information on the samples they were analysing and they were free to choose any analytical pipeline they wanted. This led to variation among the methods used, but also variation in the results reported. Inter-laboratory studies such as these are useful as a precursor to the formal external quality-assurance schemes that come later when assays have been embedded into clinical service. We have shown that although there were discrepancies between results reported, these discrepancies could be traced back to problems such as sequence quality, database choice and user error, all of which can be addressed for WGS to fulfil its potential in clinical settings.
Introduction
Antimicrobial resistance (AMR) is a major, global, public-health threat, with projections of up to 10 million deaths per annum by 2050 [1]. The World Health Organization’s 2015 Global Action Plan on AMR identified diagnostics as a priority area for combating resistance [2]. Currently, most diagnostic AMR testing is phenotypic antimicrobial-susceptibility testing (AST) and is based on principles dating back to the early 20th century [3]. Molecular testing has facilitated the implementation of PCR assays that target key AMR mutations and genes [4, 5]. However, there remains an unmet need for truly rapid point-of-care AST [6, 7].
Whole-genome sequencing (WGS) is emerging as a routine clinical test that could be used to determine the bacterial species, undertake transmission tracking and identify multiple AMR-associated mutations and genes in a single assay [8–13]. Whilst the initial clinical roll-out of WGS has used cultured bacterial isolates, metagenomics and sequencing direct from clinical samples are future possibilities [14–16]. Resolving the challenges of AMR prediction using WGS for bacteria will provide key advances for the application of metagenomics as a clinical test.
There are currently a wide array of bioinformatics tools and pipelines to predict AMR from WGS data [17]. These have generally been developed by individual researchers and research groups, many with no clinical expertise, and mostly with the same basic principle of matching the input DNA sequence to entries in a reference database of known AMR-associated gene sequences. The testing of pipelines for AMR prediction is typically either performed in-house [18–20] or done ad hoc for specific research [21–24]. Often, these tools are not developed with clinical application or portability in mind. Currently, there are no higher-order reference materials (synthetic references that contain exact components of interest) that are available to validate these tools. Studies have reported good concordance between genotype and phenotype on datasets they have been applied to [9, 22, 25], but rarely address the factors underlying situations where different methods may produce discordant results and how this discordance should be resolved.
Gaining laboratory accreditation is an important, often essential, step for tests in clinical microbiology, but is less advanced for clinical bioinformatics due to its comparatively recent development. Bioinformatic reproducibility studies have been performed for clinically relevant bacterial sequence typing methods [26, 27]. However, while there have been intra-laboratory studies comparing methods of AMR prediction, there have been no comparisons of multiple methods at the inter-laboratory scale. As there is limited evidence of robust, reproducible analyses in bioinformatic prediction of AMR from clinical WGS data, adoption of these methods may be hampered in meeting the necessary accreditation.
This multi-centre study used genomic DNA sequences from clinical carbapenem-resistant organisms, specifically chosen to be of varying quality and complexity, to identify the range of methods used and contributors to discordant AMR predictions. Participants included a mixture of independent individuals and teams using non-commercial AMR prediction pipelines from research groups, hospital laboratories, public-health laboratories and clinical diagnostic companies. The observations made underpin our recommendations for future method developments.
Methods
Sample collection and WGS
For the purposes of this study, a panel of ten samples (A-1, A-2, B-1, B-2, C-1, C-2, D, E, F and G) were generated from seven clinical isolates (A, B, C, D, E, F and G). The bacteria were isolated between 2014 and 2017 from stool specimens from patients attending Great Ormond Street Hospital (GOSH), UK, or University Hospital Galway (UHG), Ireland. They represented six clinically relevant bacterial pathogens, including diverse Enterobacterales and also Acinetobacter baumannii , and contained six distinct families of carbapenemase genes (Table 1).
Table 1.
Inter-laboratory study sample characteristics
|
Study ID |
Isolate species |
Sequencing method |
Carbapenemase gene |
Median depth of coverage |
Comment |
|---|---|---|---|---|---|
|
A-1 |
NEBNext Ultra II+NextSeq 150 bp PE |
OXA-48-like |
190.2× |
Exact duplicate of A-2 |
|
|
A-2 |
NEBNext Ultra II+NextSeq 150 bp PE |
OXA-48-like |
190.2× |
Exact duplicate of A-1 |
|
|
B-1 |
Enterobacter cloacae complex |
NEBNext Ultra II+NextSeq 150 bp PE |
OXA-48-like |
1.4× |
Very low coverage duplicate of B-2 |
|
B-2 |
Enterobacter cloacae complex |
NEBNext Ultra II+NextSeq 150 bp PE |
OXA-48-like |
142.9× |
High coverage duplicate of B-1 |
|
C-1 |
Nextera DNA +HiSeq 100 bp PE |
OXA-48-like |
37.4× |
Same original isolate as C-2 |
|
|
C-2 |
NEBNext Ultra II+NextSeq 150 bp PE |
OXA-48-like |
156.4× |
Same original isolate as C-1 |
|
|
D |
NEBNext Ultra II+NextSeq 150 bp PE |
NDM |
83.5× |
|
|
|
E |
Nextera DNA +HiSeq 100 bp PE |
IMP |
20.6× |
|
|
|
F |
NEBNext Ultra II+NextSeq 150 bp PE |
VIM |
32.5× |
|
|
|
G |
NEBNext Ultra II+NextSeq 150 bp PE |
OXA-23-like and OXA-51-like |
22.2× |
|
PE, Paired end.
Phenotypic AST was performed at UHG and GOSH using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) disc diffusion method (http://www.eucast.org) and meropenem, ertapenem, cefotaxime, amikacin, gentamicin and ciprofloxacin. The isolates were confirmed as carbapenemase producers by PCR at a reference laboratory (Public Health England).
Total genomic DNA was extracted from isolate sweeps on an EZ1 Advanced XL instrument (Qiagen) using DNA Blood 350 µl kits with an additional bead beating step. For eight samples, the NEBNext Ultra II DNA library prep kit (New England Biolabs) and NextSeq (Illumina) 150 bp paired-end sequencing was used. For two samples, the Nextera DNA library prep kit (Illumina) and HiSeq 100 bp paired-end sequencing was used (Table 1). The fastq files were deposited in the European Nucleotide Archive (accession no. PRJEB34513).
Inter-laboratory study plan
Potential inter-laboratory participants were invited in an individual capacity, both in person and by email, at the meeting ‘Challenges and New Concepts in Antibiotics Research’, March 2018, at Institut Pasteur, France. Fifteen individuals were also emailed directly to participate in the study. From those invited, nine sets of participants agreed to take part in the study. We will refer to these sets as ‘participants’ throughout. These participants were labelled Lab_1 to Lab_9; ‘Lab’ is used as a catch-all term for an individual or team of participants, who came from a mixture of research groups, hospital laboratories, public-health laboratories and clinical diagnostic companies. All participants agreed to take part in a personal capacity using non-commercial pipelines under the condition of anonymity of the results. Each participant was not made aware who the other invited participants were at that stage.
Participants were sent ten paired fastq files (labelled AMRIL_1 to AMRIL_10) and were blinded to their contents. The samples included two exact duplicates A-1 and A-2 (renamed copies of the same fastq files). Two duplicates with different depths of coverage, B-1 and B-2 (sequenced from the same isolate, but with median read depths of 1.4× and 142.9×, respectively). Two samples sequenced from the same isolate, C-1 and C-2 (sequenced in two different laboratories using HiSeq and NextSeq, respectively). The remaining four samples, D, E, F and G, represented diverse bacterial species and carbapenemases.
Participants were asked to report a species identification for each pair of fastq files provided, as well as the presence of all AMR-associated genes present in that sample. They were asked, using the above data, to make a categorical prediction on whether that sample would be resistant to ciprofloxacin, gentamicin, amikacin and cefotaxime. Lastly, participants were asked to provide a description of the analysis pipeline they used.
Participant analyses
Participants returned results via an Excel spreadsheet (Tables S1–S10, available with the online version of this article). Results were collated for all species identifications and resistant or susceptible predictions from each participant. Collated AMR-associated genes had each name manually checked between each participant to identify minor differences in nomenclature used.
Individual methods are summarized in Table 2. Briefly, all participants used a unique combination of a number of tools to analyse the samples provided and report back results. For species identification, seven participants used a combination of command line tools Kraken [28], Kraken-HLL [29], mash [30], Centrifuge [31] and Kmerid (https://github.com/phe-bioinformatics/kmerid). Four participants also used the web-based tools wgsa (https://pathogen.watch/), blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and KmerFinder (https://cge.cbs.dtu.dk/services/KmerFinder/). All participants identified species from raw reads, apart from three participants that used assembled reads (Lab_2, Lab_5 and Lab_8). Lab_3 used both raw reads and assemblies to assign species ID using mash and wgsa, respectively. Six of the nine participating laboratories assembled the raw reads into a draft assembly before identifying AMR-associated genes. Only Lab_4, Lab_7 and Lab_9 used methods that required no assembly of the reads. Of those participants assembling their reads, SPAdes [32] was the most common assembler used, with five participants either using it directly or using one of two wrapper tools that contains it, Unicycler [33] or Shovill (https://github.com/tseemann/shovill). Lab_5 was the only participant to use the assembler A5-MiSeq [34]. Lab_6 was also unique as the only participant to use a commercial bioinformatics platform, Bionumerics (Applied Maths), to perform their analysis. For the identification of AMR-associated genes, ABRicate (https://github.com/tseemann/abricate) and rgi [35] were the most popular tools used, and both take assembled reads as input. The other assembly-based AMR gene identifiers used were c-SSTAR [36] and Resfinder (https://cge.cbs.dtu.dk/services/ResFinder/). Three tools were also used that took raw short reads as input and these were ariba [20], srst2 [37] and Genefinder (https://github.com/phe-bioinformatics/gene_finder). All participants used one or a combination of three AMR databases in their analysis, and these were card [35], Resfinder [18] and arg-annot [38]. The full methods, including command line parameters and software versions, can be found in Supplementary methods.
Table 2.
Summary of bioinformatic tools used for species identification and detecting AMR by each participant
|
Method step |
Lab_1a* |
Lab_1b* |
Lab_2 |
Lab_3 |
Lab_4 |
Lab_5 |
Lab_6 |
Lab_7 |
Lab_8 |
Lab_9 |
Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Species ID |
Kraken-HLL |
Kraken-HLL |
blast |
mash and wgsa |
Kraken |
KmerFinder (assembled contigs) |
KmerFinder (raw reads) |
Centrifuge |
Kraken |
Kmerid |
[28–31] |
|
Read assembly |
Shovill (SPAdes) |
Shovill (SPAdes) |
SPAdes |
Unicycler (SPAdes) |
No assembly |
A5-MiSeq |
Bionumerics |
No assembly |
Unicycler (SPAdes) |
No assembly |
[32–34] |
|
AMR identifier |
rgi |
c-SSTAR |
ABRicate |
rgi and Resfinder |
ariba |
rgi |
Bionumerics Escherichia coli genotyping plugin (blast) |
srst2 |
ABRicate |
Genefinder |
|
|
Reference database |
card |
Resfinder and arg-annot |
card |
card and Resfinder |
card and arg-annot |
card |
Resfinder |
arg-annot |
Resfinder |
card and Resfinder (manually curated) |
|
|
Sequence identity cut-off |
80% |
95% |
75% |
80 % (card) and 90 % (Resfinder) |
90% |
80% |
90% |
90% |
75% |
90% |
|
|
Breadth of coverage cut-off |
0% |
0% |
0% |
0 % (card) and 80 % (Resfinder) |
20% |
0% |
60% |
90% |
0% |
100% |
|
*Lab_1 provided two sets of results with two separate methods for AMR detection; these are referred to as Lab_1a and Lab_1b.
Results
Bacterial species identification
Four of the nine participants identified all species correctly from WGS data (Table 3). This included the low depth of coverage (1.4×) sample B-1, where we did not expect enough information for a correct call. Species misidentifications of D and B-2 at the genus level by Lab_5 is likely to be a human reporting error, as they correctly identified species in B-1 from a very low read depth. Lab_6 used the same web-based tool for species identification as Lab_5 (Kmerfinder; Center for Genomic Epidemiology), but one error was noted where raw sequence reads were input instead of assembled contiguous sequences (Table 3).
Table 3.
Species identification for each sample by each participant
|
Participant |
A-1 |
A-2 |
B-1 |
B-2 |
C-1 |
C-2 |
D |
E |
F |
G |
|---|---|---|---|---|---|---|---|---|---|---|
|
Reference |
KP |
KP |
ECl |
ECl |
KO |
KO |
KP |
EC |
CF |
AB |
|
Lab_1 |
KP |
KP |
ECl |
ECl |
KO |
KO |
KP |
EC |
CF |
AB |
|
Lab_2 |
KP |
KP |
– |
ECl |
KO |
KO |
KP |
EC |
CF |
AB |
|
Lab_3 |
KP |
KP |
Shigella phage SflV |
ECl |
KO |
KO |
KP |
EC |
Citrobacter sp. |
AB |
|
Lab_4 |
KP |
KP |
ECl |
ECl |
KO |
KO |
KP |
EC |
Citrobacter sp. |
AB |
|
Lab_5 |
KP |
KP |
ECl |
KP |
KO |
KO |
EC |
EC |
CF |
AB |
|
Lab_6 |
KP |
KP |
ECl |
ECl |
– |
KO |
Klebsiella sp. |
EC |
CF |
AB |
|
Lab_7 |
KP |
KP |
ECl |
ECl |
KO |
KO |
KP |
EC |
CF |
AB |
|
Lab_8 |
KP |
KP |
ECl |
ECl |
KO |
KO |
KP |
EC |
CF |
AB |
|
Lab_9 |
KP |
KP |
ECl |
ECl |
KO |
KO |
KP |
EC |
CF |
AB |
Missing data represent no results reported. Results highlighted in bold represent discrepancies.
AB, Acinetobacter baumannii ; CF, Citrobacter freundii ; EC, Escherichia coli ; ECl, Enterobacter cloacae ; KO, Klebsiella oxytoca ; KP, Klebsiella pneumoniae .
AMR gene identification
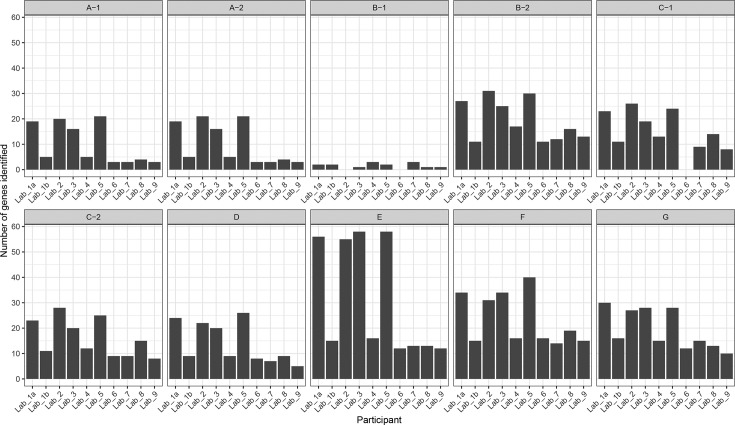
We compared the number of AMR-associated genes reported by each participant in each sample and found disparities in the total reported (Fig. 1). Lab_1 used two different methodologies for identifying AMR-associated genes; the results are referred to as Lab_1a and Lab_1b. The number of AMR-associated genes reported by each participant was affected by the choice of database used. Lab_1a, Lab_2, Lab_3 and Lab_5 all repeatedly reported the highest number of genes in each sample and all used the Comprehensive Antibiotic Resistance Database (card) as their reference database. This is due to card including many sequences from loosely AMR-associated efflux pump genes that are not found in the other databases. Lab_4 and Lab_9 also used card, but in combination with other databases and selectively reported genes. The number of AMR-associated genes reported by each participant was also found to be associated with sequence identity and breadth of coverage thresholds used to infer a ‘hit’. Both Lab_2 and Lab_8 used the lowest identity and breadth of coverage thresholds (75 % sequence identity and no breadth of coverage threshold), and Lab_2 consistently reported the highest number of AMR genes in each sample. While Lab_8 reported fewer AMR-associated genes than Lab_2, it did use ResFinder as its reference database rather than card, and reported the highest number of genes compared with other participants using the same database.
Fig. 1.
Number of AMR-associated genes identified in each sample by each team of participants.
All isolates included in this study were carbapenem resistant. The reporting of carbapenemase genes from WGS from all participants matched the reference PCR result in 91 % of cases (91/100) (Table 4). Eight of the ten misidentifications occurred in the very low depth of coverage sample B-1, as would be expected. Differences between reported gene variants of bla IMP were seen in sample E. Five participants reported bla IMP-1, whereas the other five reported bla IMP-34. This discrepancy exactly matched the reference database used with those who reported bla IMP-1 having used card and those who reported bla IMP-34 having used either ResFinder or arg-annot. While the sequences for bla IMP-34 included in each database are identical, the choice of bla IMP-1 reference sequence included in both databases only share 85 % sequence identity. This is due to card’s bla IMP-1 reference sequence being isolated from a Pseudomonas aeruginosa integron [National Center for Biotechnology Information (NCBI) accession no.: AJ223604] and arg-annot’s reference sequence from an A. baumannii integron (NCBI accession no.: HM036079). While there is variation at the nucleotide level, both encode the same IMP-1 enzyme.
Table 4.
Carbapenemase genes identified for each sample by each participant and the reference laboratory PCR (Ref PCR)
|
Participant |
A-1 |
A-2 |
B-1* |
B-2 |
C-1 |
C-2 |
D |
E |
F |
G |
|---|---|---|---|---|---|---|---|---|---|---|
|
Ref PCR† |
OXA-48-like |
OXA-48-like |
OXA-48-like |
OXA-48-like |
OXA-48-like |
OXA-48-like |
NDM |
IMP |
VIM |
OXA-23-like+OXA-51-like |
|
Lab_1a‡ |
OXA-48 |
OXA-48 |
– |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-1 |
VIM-4 |
OXA-23+OXA-66 |
|
Lab_1b‡ |
OXA-48 |
OXA-48 |
OXA-48 |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-34 |
VIM-4 |
OXA-23+OXA-66 |
|
Lab_2 |
OXA-48 |
OXA-48 |
– |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-1 |
VIM-4 |
OXA-23+OXA-66 |
|
Lab_3 |
OXA-48 |
OXA-48 |
– |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-1 |
VIM-4 |
OXA-23+OXA-66 |
|
Lab_4 |
OXA-48 |
OXA-48 |
– |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-34+IMP-9 |
VIM-4 |
OXA-23+OXA-66 |
|
Lab_5 |
OXA-48 |
OXA-48 |
– |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-1 |
VIM-4 |
OXA-23 |
|
Lab_6 |
OXA-48 |
OXA-48 |
– |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-34 |
VIM-4 |
OXA-23+OXA-66 |
|
Lab_7 |
OXA-48 |
OXA-48 |
OXA-48 |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-34 |
VIM-4 |
OXA-23+OXA-66 |
|
Lab_8 |
OXA-48 |
OXA-48 |
– |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-34 |
VIM-4 |
OXA-23+OXA-66 |
|
Lab_9 |
OXA-48 |
OXA-48 |
OXA-405 |
OXA-48 |
OXA-181 |
OXA-181 |
NDM-1 |
IMP-1 |
VIM-4 |
OXA-23+OXA-66 |
*Missing data represent no results reported. Results highlighted in bold represent discrepancies.
†Specific carbapenemase PCR results for each sample.
‡Lab_1 provided different results using two separate methods; these are referred to as Lab_1a and Lab_1b.
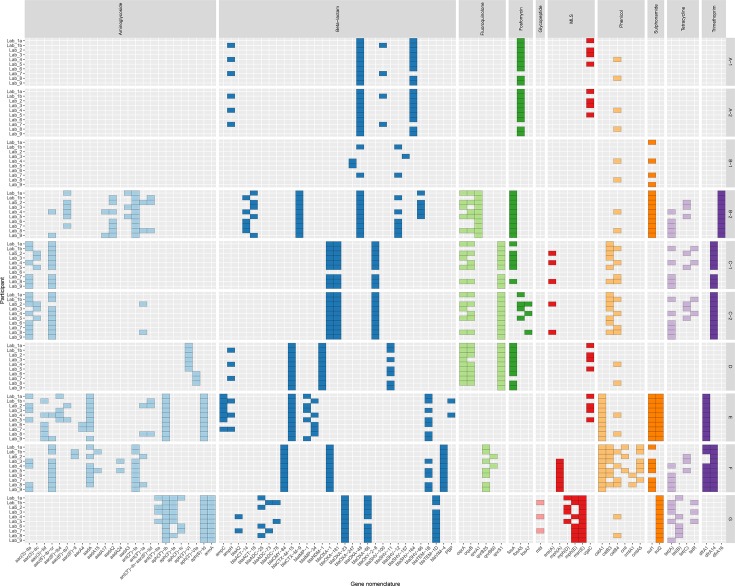
We compared all AMR-associated genes identified by each participant in each sample. As previously noted, the largest discrepancies were the 55 efflux pump gene sequences that were present only in card (Fig. S1). To understand the other factors influencing discordant reporting, we removed these genes that were only present in one database from our comparisons (Fig. 2). A pairwise comparison between all participants found that two sets of participants only reported the exact same genes within a sample in 2 % (18/900) of cases. Fourteen of these cases occurred when analysing the two identical samples (A-1 and A-2; Fig. 2). Although there was little agreement between participants for genes identified in A-1 and A-2, there was complete within-participant concordance across both samples, exhibiting reproducibility within each analysis pipeline. No two participants reported the exact same combination of gene variants in samples B-2, C-1, D, F and G. There were many clear examples where participants assigned different gene variants to the same sequence data where the reference sequences only differed by a few single nucleotides. This can be seen in Fig. 2 amongst samples that contained tetracycline-resistance genes [tet(A), tet(B) and tet(C)], some aminoglycoside modifying enzyme gene variants [aac(3)-IIa and aac(3)-IIc] and β-lactamases (bla ACT-14 and bla ACT-18). We also observed differences between the same participants analysing samples from the same original isolate. Due to the very low read depth, the genes reported in B-1 bore little resemblance to B-2 across all participant results. However, even in the samples from the same isolates with sufficient sequencing depth (C-1 and C-2), we observed differences in the genes identified in four out of nine participants. This suggests that resequencing, and even small increases in read length and quality, can produce variation in results. It is worth noting that all but one of these differences were additional genes identified in C-2, which had a higher read depth than B-2 (156 vs 37× median read depth). The additional genes in C-2 included ant(3′′)−Ia (Lab_2 and Lab_8), fosA7 (Lab_2 and Lab_8) and tet(C) (Lab_3), but the reported reference breadth of coverage of ant(3′′)−Ia and fosA7 was low (17 and 75 %, respectively) and the sequence similarity between the purported tet(C) sequence and the reference was also low (75%). We also found no systematic differences in genes present or absent between those participants that used tools that required assembly of short reads first and those that took unassembled short reads as input (Lab_4, Lab_7 and Lab_9, ariba, srst2 and Genefinder, respectively).
Fig. 2.
Presence of AMR-associated genes in each sample by each team of participants. Genes are organized and coloured by the class of antibiotics their resistance is associated with. Genes are only shown here if reported by more than one participant and if they were present in more than one reference database used. MLS, Macrolide lincosamide and streptogramin.
Phenotypic and genotypic resistance concordance
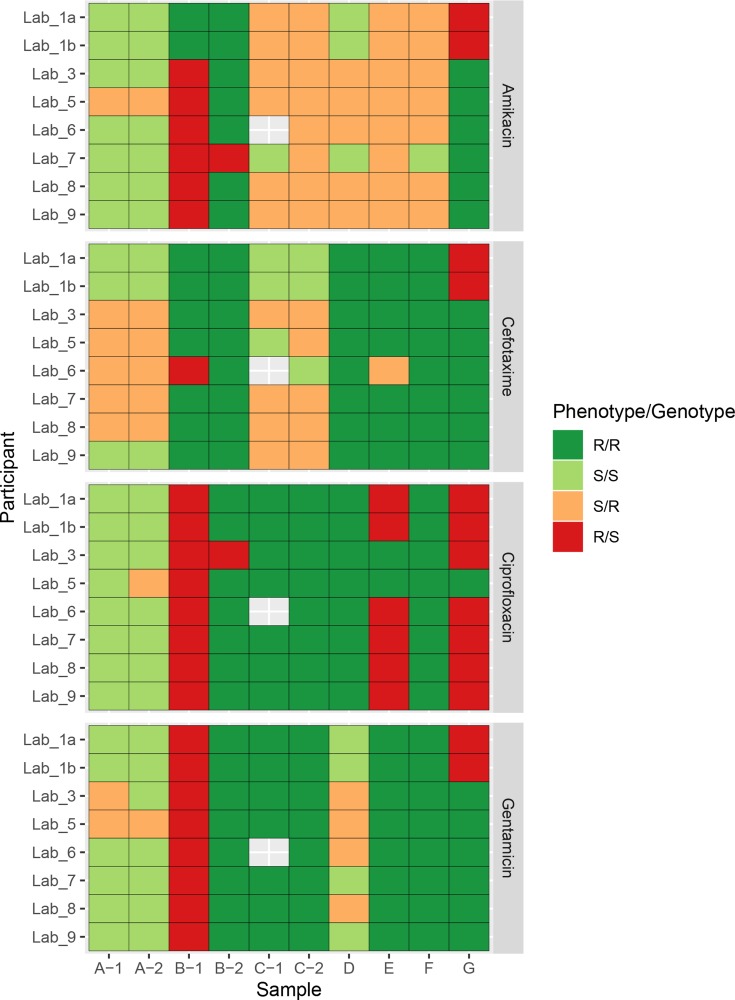
Given the differences in the AMR-associated genes identified in the samples by each participant, we also compared predictions of antibiotic resistance to phenotypic AST results and each other. Two participants (Lab_2 and Lab_4) did not submit any results for phenotypic resistance prediction, so were not included in the subsequent analysis. A pairwise comparison between genotypic prediction results reported by all participants, on all antibiotics and samples, showed an overall consensus of 79 % (864/1092, Fig. 3). This varied depending on the antibiotic tested with the highest pairwise reporting consensus of 88 % (240/273) between participants for ciprofloxacin and the lowest pairwise reporting consensus of 72 % (197/273) for cefotaxime, which could be understandable given the different complexities of the resistance mechanisms involved. When we compared results from each participant with the phenotypic AST results, we found an overall sensitivity of 76 % and specificity of 50 %. The overall number of false positives was 64/316 (20 %) and the overall number of false negatives was 44/316 (14 %). Lab_5 had the highest number of false positives (14/40) and lowest number of false negatives (3/40), whereas Lab_1 had the lowest number of false positives (4/40) but the highest number of false negatives (7/40). Broken down by antibiotic, the highest consensus between phenotype and genotype was gentamicin (78%, 62/79) and the lowest amikacin (43 % 34/79). As expected, there was little agreement between predictions within the very low read depth sample (B-1) and most participants predicted a susceptible isolate due to missing data when in fact it was resistant by phenotypic AST. However, when analysing the same isolate at an appropriate higher read depth (B-2), there was near perfect concordance between participant reported genotypes and the resistance phenotype, with only two discrepant results reported by Lab_3 (ciprofloxacin) and Lab_7 (amikacin). Lab_3 also reported different results between the two identical samples (A-1 and A-2), where A-1 was reported as resistant and A-2 was reported as sensitive. As there were no differences in the gene content reported in either sample by this participant (Fig. 2), this is likely to be due to a human reporting error. We also identified a single discrepancy between amikacin resistance predicted by Lab_7 between samples C-1 and C-2, which both were sequenced from the same isolate. C-1 was reported as sensitive but C-2 was reported as resistant, and the phenotypic AST result was sensitive; however, there was no difference in the reported gene content in both samples by Lab_7, so it is also another likely human reporting error. Excluding the extremely low depth sample, B-1, there were only 2/30 cases where no laboratory correctly predicted the phenotypic AST result. Both of these results were an incorrect resistance prediction for amikacin in C-2 and E, but as noted earlier the prediction from Lab_7 for C-2 was likely human error.
Fig. 3.
Concordance between phenotypic AST result and the genotypic prediction from WGS data. Results are presented separately for each participant, sample and antibiotic. Each tile is coloured based on whether both the resistant phenotype and genotype agreed (R/R); both phenotype and genotype predicted sensitive (S/S); major errors where the phenotype was sensitive, but the genotype was resistant (S/R); and very major errors where the phenotype was resistant, but the genotype was sensitive (R/S). Missing cells represent a result not reported.
Discussion
In this study, we have shown that participants using different choices of bioinformatics pipelines reported different AMR-associated gene variants when given identical mixed quality bacterial isolate WGS datasets. This led to differences in the reporting of predicted resistance phenotypes. We observed good concordance for genotypic-resistance predictions between participants, but poor concordance with phenotypic AST results. A similar trend has previously been seen in a study of Staphylococcus aureus genomes [39]. Concordance in phenotype prediction differed for different antibiotic classes. Good concordance was seen comparing WGS with AST results for gentamicin, but for amikacin concordance was poor. This may be due to the fact that amikacin is not affected by the action of most aminoglycoside-modifying enzymes [40]. Previous studies predicting antimicrobial susceptibility from WGS data have reported sensitivities of 96 and 99 % against phenotypic AST as a benchmark [21, 22], compared with an overall sensitivity of 76 % in this inter-laboratory study. It should be noted, however, that some of the data used in this study were purposefully very low quality, with some of the clinical isolates deliberately chosen to be difficult to characterize. Similar mixed quality data tested using current clinical AST phenotyping may also result in equivalent discrepancies. However, our aim here was to document the range of bioinformatics approaches being used and identify plausible contributors to discordant results reported between participants working on the same data, in order to provide useful recommendations and direct future work.
We identified three stages of analysis that contributed to discrepancies in predictions: the quality of the sequence data used, the bioinformatic methods (choice of database or software used) and the interpretation of those results. Where single gene calling is required (e.g. presence of a carbapenemase), results are mainly affected by sequence quality. However, once multiple genes are involved, all three analytical issues become important. We found the largest contributors to discrepant results between the gene variants reported in each sample and the phenotypic resistance predictions were the sample sequence quality, read depth and the choice of reference resistance-gene database. Samples must be sequenced to a sufficient depth as well as sufficient breadth of coverage for the expected size of the genome, usually inferred by mapping to a suitable reference genome, of at least above 90 %. Based on our own experience and these results, we recommend 30× depth as a lower limit. This also tends to be a default setting for many read assembly tools, but generally most samples should have a higher depth of coverage than this for meaningful prediction. Some participants did flag that they would not normally analyse the low depth of coverage samples (<30×, samples B-1, E and G) and if those samples are excluded from this analysis sensitivity in comparison to phenotypic AST rises from 76 to 98 %. This is highly encouraging as it suggests that as long as the sequence data produced is of sufficient depth and quality (e.g. current Illumina error rates) genotypic prediction of resistance phenotype can be comparable to AST. However, we also note that many sets of participants provided little information on their employment of quality control and filtering steps. Our results, therefore, suggest an increased emphasis on data quality control is highly relevant to improving sensitivity. Conversely, we have observed the choice of sequencer and DNA library preparation method has a small effect on closely related gene variants, but little discernible effect on the inference of resistance phenotype.
Some participants ran the same set of read data against different reference databases and merged the results, which led to different gene variants being reported at the same loci. In practice, different variants of the same gene may not always result in a different clinically relevant phenotype. However, we also found reference sequences in different databases for same gene variant can differ by 15 % nucleotide identity (bla IMP-1 in card and arg-annot). If precise identification of gene variants is required, we would strongly recommend avoiding this, as it effectively leads to ‘double-dipping’ using the same reads. Multiple reference databases could be used, but after screening for reads that have already been assigned a hit against one of the databases. This would avoid multiple different genes reported at the same genomic loci. However, it would be better to merge the different reference databases and remove the redundant sequences before comparisons are made against the test data. Sequence identity, and to lesser extent breadth of coverage cut-offs, should be kept high when comparing test data to a reference database. Based on this study, we would recommend using a sequence identity cut-off of at least 90%, in combination with an up to date curated reference resistance-gene database. Although lowering of these thresholds does identify more candidate genes within a sample, many were false negatives; thus, not improving concordance with phenotypic AST results in this study.
There is an overwhelming need for a standardized, centralized database that integrates the current knowledge base for linking genotype with resistance phenotype and is not linked to a single research group, as previously suggested [10]. There is also a growing need regarding computational reproducibility [41, 42]. This would deal with many of the issues we have raised, such as which sequences to include and what gene nomenclature to use. With strict version control, such a resource would allow greater integration of results and be an invaluable tool for larger epidemiological studies. Currently, databases are being built for organisms such as for Mycobacterium tuberculosis , though this is a less challenging organism for genotype–phenotype predictions due to it being highly clonal and lacking an accessory genome [43, 44]. A recent publication of a new protein-based database also obtained high concordance (98.4%) between genotype and phenotype for four food-borne pathogens [45]. However, for other clinically relevant organisms there are limited resources.
Participants in this study included a mixture of individuals and teams involved in AMR prediction in a variety of settings. A potential criticism is that we did not restrict these settings to those routinely predicting AMR phenotype for clinical use, meaning that some participants were attempting analyses they did not usually perform. However, the fact that AMR phenotype prediction from WGS is not yet routine in most clinical laboratories was the very reason for undertaking this study. Clinical laboratories at the moment do not have the tools or knowledge to make good phenotypic resistance calls from genotypic data. This is evident from the fact that two participants in this study did not report any phenotypic resistance predictions as they felt they could find no valid method for doing so. At this point in time, many research laboratories use these methods to track specific resistance genes or one specific resistance mechanism, rather than building tools for the broad detection of AMR in bacteria for clinical purposes. We found in this study that there was particularly low concordance between participants reporting sensitive isolates compared with phenotypic AST. The problem with the inference of phenotype from genotype is that the information either is not known at all or is expert knowledge restricted to single laboratories working on specific bacteria. In addition to this, although the identification of the presence of genes is performed in a systematic way, the prediction of resistance is still performed in an ad hoc manner by scientists and, therefore, subject to user error given the same set of genes. Once again, M. tuberculosis is providing the first example of the need for a defined decision tree when working from the presence of genes or gene variants to the prediction of phenotypic drug resistance [46]. Interpretation and reporting of this genotypic data will need to be subjected to the same level of scrutiny as current tests if it is to form part of an accredited laboratory service within the healthcare service.
A limitation of this study is that we focused on the use of short-read sequence data, which produces sequences far shorter than the length of genes being identified. However, we feel this is more reflective of the WGS data that is more routinely generated in clinical laboratories at this point in time. If these short reads need to be assembled into longer contiguous sequences, we found it essential to use an actively developed short-read assembler such as SPAdes (http://cab.spbu.ru/software/spades/). Web-hosted tools that provide a ‘black box’ solution to assembly and identifying resistance from uploaded WGS data should be avoided if possible, because of the lack of interpretability. Tools are needed that are open source, designed for clinical purpose and can be subjected to thorough troubleshooting when erroneous results arise [47]. To this end, permanently employed bioinformaticians are required, who can provide expert interpretation of the results and update approaches as necessary. In this study, tools that either require assembled contigs (ABRicate) and those that take unassembled short reads (srst2 and ariba) were capable of producing very similar results with no notable effects alone on the predication of phenotypic resistance. This holds promise for rapid phenotypic predictions, as genome assembly is one of the largest bottlenecks in computational analysis time.
Other limitations of this study include our focus on acquired genes rather than point mutations or many of the other resistance mechanisms found in bacteria (e.g. target site modifications and efflux pumps). We also only required reporting on categorical resistance predictions. Furthermore, because our focus was on WGS, and although we validated AST at two independent laboratories, we did not investigate potential variability and discordance in phenotypic prediction. More work needs to be done on the prediction of minimum inhibitory concentrations (MICs) from WGS data before it can be implemented in laboratories. This will be aided by more systematic reporting of accompanying MIC data when making WGS data available.
We have outlined recommendations for improving the current state of prediction of AMR from WGS data. Some of these recommendations, such as a standardized database and better dissemination of phenotype/genotype relationships, cannot be implemented immediately. However, current pipelines can be improved right now by robust quality control of starting sequence reads to make sure that the genome breadth of coverage is high (>90 %) and that there is sufficient depth of coverage (>30×). We also recommended that running the same sequence read data set against multiple databases should be avoided due to the erroneous results, and that sequence identity between the predicted and reference AMR genes should be higher than 90 % to avoid non-specific hits. We found little difference between the results of participants depending on what reference database they chose to use, between which Illumina short-read sequencer was used and whether they used assembly or assembly-free methods.
In conclusion, we have identified some of the current contributors to discrepancies in predicting AMR-associated genes and phenotypes from bacterial isolate WGS data. We have provided recommendations for improving the current reporting of results. Despite its clear potential, even after accounting for poor sequence data, we found that the current public methods, in particular databases, are not adequate ‘off-the-shelf’ tools for the prediction of AMR from bacterial WGS data as a universal clinical test at this point in time.
Data bibliography
1. Doyle, RM. Sequence read files for all samples used in this study have been deposited in the European Nucleotide Archive under the project accession number PRJEB34513 (2019).
Supplementary Data
Funding information
This work was supported by the UK National Measurement System and the European Metrology Programme for Innovation and Research (EMPIR) joint research project (HLT07) ‘AntiMicroResist’, which has received funding from the EMPIR programme co-financed by the participating states and the European Union’s Horizon 2020 research and innovation programme. A.C.P. received funding from the European Union’s Horizon 2020 research and innovation programme ‘New Diagnostics for Infectious Diseases’ (ND4ID) under the Marie Skłodowska-Curie grant agreement no. 675412. These funding bodies had no influence on the design of the study, collection, analysis and interpretation of data, nor the writing of the manuscript.
Acknowledgements
The authors thank the biomedical scientist teams for sample collection and processing. We also thank the Pathogen Informatics Group at the Wellcome Sanger Institute, UK, for their contributions to the study.
Author contributions
R.M.D., D.M.O., K.A.H. and J.F.H. conceived and designed the study. S.D.A., S.B., T.C., A.C.P., M.C., E.D.B., M.J.E., E.M., Y.M., T.P.T.N., J.P., L.P.S., R.A.S., A.V., L.V. and N.W. all performed the initial participant analyses, and are listed in alphabetical order. Only those on the author list from participating institutions contributed to this analysis. R.M.D. performed all secondary analyses and drafted the manuscript with assistance from D.M.O., J.M.-G., J.F.H. and K.A.H. All authors read and approved the final manuscript.
Conflicts of interest
A.C.P. and A.V.B. are employees of bioMérieux, a company developing, marketing and selling tests in the infectious disease domain. The company had no influence on the design and execution of the clinical study, neither did the company influence the choice of the diagnostic tools used during the clinical study. The opinions expressed in the manuscript are the authors', which do not necessarily reflect company policies. M.J.E. and N.W. are members of Public Health England’s AMRHAI Reference Unit, which has received financial support for conference attendance, lectures, research projects or contracted evaluations from numerous sources, including: Accelerate Diagnostics, Achaogen Inc., Allecra Therapeutics, Amplex, AstraZeneca UK Ltd, AusDiagnostics, Basilea Pharmaceutica, Becton Dickinson Diagnostics, bioMérieux, Bio-Rad Laboratories, British Society for Antimicrobial Chemotherapy, Cepheid, Check-Points B.V., Cubist Pharmaceuticals, the Department of Health, Enigma Diagnostics, European Centre for Disease Prevention and Control, Food Standards Agency, GlaxoSmithKline Services Ltd, Helperby Therapeutics, Henry Stewart Talks, IHMA Ltd, Innovate UK, Kalidex Pharmaceuticals, Melinta Therapeutics, Merck Sharpe and Dohme Corp., Meiji Seika Pharma Co. Ltd, Mobidiag, Momentum Biosciences Ltd, Neem Biotech, National Institute for Health Research, Nordic Pharma Ltd, Norgine Pharmaceuticals, Rempex Pharmaceuticals Ltd, Roche, Rokitan Ltd, Smith and Nephew UK Ltd, Shionogi and Co. Ltd, Trius Therapeutics, VenatoRx Pharmaceuticals, Wockhardt Ltd and the World Health Organization. All other authors declare that they have no competing interests and have performed the work in an individual capacity.
Ethical statement
All investigations were performed in accordance with the hospitals’ research governance policies and procedures. No specific ethical approval was required, as no patient samples nor identifiable data were used. The project was registered as a research study. All participants gave consent to take part in this study.
Footnotes
Abbreviations: AMR, antimicrobial resistance; AST, antimicrobial-susceptibility testing; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
The datasets generated in this study are available in the European Nucleotide Archive under accession number PRJEB34513 (https://www.ebi.ac.uk/ena/browser/view/PRJEB34513) and the following sample accession numbers: SAMEA5789893 (sample A-1), SAMEA5789894 (sample A-2), SAMEA5789895 (sample B-1), SAMEA5789896 (sample B-2), SAMEA5789897 (sample C-1), SAMEA5789898 (sample C-2), SAMEA5789899 (sample D), SAMEA5789900 (sample E), SAMEA5789901 (sample F), SAMEA5789902 (sample G).
References
- 1.O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. London: Review on Antimicrobial Resistance; 2016. [Google Scholar]
- 2.World Health Organization Global Action Plan on Antimicrobial Resistance ( http://www.who.int/iris/bitstream/10665/193736/1/9789241509763_eng.pdf) Geneva: World Health Organization; 2015. [DOI] [PubMed] [Google Scholar]
- 3.Fleming A. Classics in infectious diseases: on the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae by Alexander Fleming, reprinted from the British Journal of Experimental Pathology 10:226-236, 1929. Rev Infect Dis. 1980;2:129–139. [PubMed] [Google Scholar]
- 4.Archer GL, Pennell E. Detection of methicillin resistance in staphylococci by using a DNA probe. Antimicrob Agents Chemother. 1990;34:1720–1724. doi: 10.1128/AAC.34.9.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marlowe EM, Novak-Weekley SM, Cumpio J, Sharp SE, Momeny MA, et al. Evaluation of the Cepheid Xpert MTB/RIF assay for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J Clin Microbiol. 2011;49:1621–1623. doi: 10.1128/JCM.02214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hays JP, Mitsakakis K, Luz S, van Belkum A, Becker K, et al. The successful uptake and sustainability of rapid infectious disease and antimicrobial resistance point-of-care testing requires a complex 'mix-and-match' implementation package. Eur J Clin Microbiol Infect Dis. 2019;38:1015–1022. doi: 10.1007/s10096-019-03492-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Belkum A, Bachmann TT, Lüdke G, Lisby JG, Kahlmeter G, et al. Developmental roadmap for antimicrobial susceptibility testing systems. Nat Rev Microbiol. 2019;17:51–62. doi: 10.1038/s41579-018-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Török ME, Peacock SJ. Rapid whole-genome sequencing of bacterial pathogens in the clinical microbiology laboratory – pipe dream or reality? J Antimicrob Chemother. 2012;67:2307–2308. doi: 10.1093/jac/dks247. [DOI] [PubMed] [Google Scholar]
- 9.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, et al. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother. 2013;68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 10.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect. 2017;23:2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 11.Tagini F, Greub G. Bacterial genome sequencing in clinical microbiology: a pathogen-oriented review. Eur J Clin Microbiol Infect Dis. 2017;36:2007–2020. doi: 10.1007/s10096-017-3024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossen JWA, Friedrich AW, Moran-Gilad J, ESCMID Study Group for Genomic and Molecular Diagnostics (ESGMD) Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect. 2018;24:355–360. doi: 10.1016/j.cmi.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Moran-Gilad J. How do advanced diagnostics support public health policy development? Euro Surveill. 2019;24:1900068. doi: 10.2807/1560-7917.ES.2019.24.4.1900068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Votintseva AA, Bradley P, Pankhurst L, Del Ojo Elias C, Loose M, et al. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol. 2017;55:1285–1298. doi: 10.1128/JCM.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle RM, Burgess C, Williams R, Gorton R, Booth H, et al. Direct whole-genome sequencing of sputum accurately identifies drug-resistant Mycobacterium tuberculosis faster than MGIT culture sequencing. J Clin Microbiol. 2018;56:e00666-18. doi: 10.1128/JCM.00666-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charalampous T, Kay GL, Richardson H, Aydin A, Baldan R, et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37:783. doi: 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- 17.Hendriksen RS, Bortolaia V, Tate H, Tyson GH, Aarestrup FM, et al. Using genomics to track global antimicrobial resistance. Front Public Health. 2019;7:242. doi: 10.3389/fpubh.2019.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clausen PTLC, Zankari E, Aarestrup FM, Lund O. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J Antimicrob Chemother. 2016;71:2484–2488. doi: 10.1093/jac/dkw184. [DOI] [PubMed] [Google Scholar]
- 20.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:mgen.0.000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, et al. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother. 2013;68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, et al. WGS accurately predicts antimicrobial resistance in Escherichia coli . J Antimicrob Chemother. 2015;70:2763–2769. doi: 10.1093/jac/dkv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemon JK, Khil PP, Frank KM, Dekker JP. Rapid Nanopore sequencing of plasmids and resistance gene detection in clinical isolates. J Clin Microbiol. 2017;55:3530–3543. doi: 10.1128/JCM.01069-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greig DR, Dallman TJ, Hopkins KL, Jenkins C. MinION nanopore sequencing identifies the position and structure of bacterial antibiotic resistance determinants in a multidrug-resistant strain of enteroaggregative Escherichia coli . Microb Genom. 2018;4:mgen.0.000213. doi: 10.1099/mgen.0.000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allix-Béguec C, Arandjelovic I, Bi L, Beckert P, Bonnet M, et al. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med. 2018;379:1403–1415. doi: 10.1056/NEJMoa1800474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aires-de-Sousa M, Boye K, de Lencastre H, Deplano A, Enright MC, et al. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J Clin Microbiol. 2006;44:619–621. doi: 10.1128/JCM.44.2.619-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellmann A, Andersen PS, Bletz S, Friedrich AW, Kohl TA, et al. High interlaboratory reproducibility and accuracy of next-generation-sequencing-based bacterial genotyping in a ring trial. J Clin Microbiol. 2017;55:908–913. doi: 10.1128/JCM.02242-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitwieser FP, Baker DN, Salzberg SL. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018;19:198. doi: 10.1186/s13059-018-1568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tritt A, Eisen JA, Facciotti MT, Darling AE. An integrated pipeline for de novo assembly of microbial genomes. PLoS One. 2012;7:e42304. doi: 10.1371/journal.pone.0042304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, et al. Card 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Man TJB, Limbago BM. SSTAR, a stand-alone easy-to-use antimicrobial resistance gene predictor. mSphere. 2016;1:e00050-15. doi: 10.1128/mSphere.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology Labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mason A, Foster D, Bradley P, Golubchik T, Doumith M, et al. Accuracy of different bioinformatics methods in detecting antibiotic resistance and virulence factors from Staphylococcus aureus whole-genome sequences. J Clin Microbiol. 2018;56:e01815-17. doi: 10.1128/JCM.01815-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez M, Tolmasky M. Amikacin: uses, resistance, and prospects for inhibition. Molecules. 2017;22:2267. doi: 10.3390/molecules22122267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garijo D, Kinnings S, Xie L, Xie L, Zhang Y, et al. Quantifying reproducibility in computational biology: the case of the tuberculosis drugome. PLoS One. 2013;8:e80278. doi: 10.1371/journal.pone.0080278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loman N, Watson M. So you want to be a computational biologist? Nat Biotechnol. 2013;31:996–998. doi: 10.1038/nbt.2740. [DOI] [PubMed] [Google Scholar]
- 43.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, et al. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flandrois J-P, Lina G, Dumitrescu O. MUBII-TB-DB: a database of mutations associated with antibiotic resistance in Mycobacterium tuberculosis . BMC Bioinformatics. 2014;15:107. doi: 10.1186/1471-2105-15-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis . Eur Respir J. 2017;50:1701354. doi: 10.1183/13993003.01354-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balloux F, Brønstad Brynildsrud O, van Dorp L, Shaw LP, Chen H, et al. From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol. 2018;26:1035–1048. doi: 10.1016/j.tim.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.