Abstract
Background:
Sudden cardiac death occurs in ~220,000 U.S. adults annually, the majority of whom have no prior symptoms or cardiovascular diagnosis. Rare pathogenic DNA variants in any of 49 genes can predispose to four important causes of sudden cardiac death – cardiomyopathy, coronary artery disease, inherited arrhythmia syndrome, and aortopathy or aortic dissection.
Objectives:
This study assessed the prevalence of rare pathogenic variants in sudden cardiac death cases versus controls, and the prevalence and clinical importance of such mutations in an asymptomatic adult population.
Methods:
We performed whole exome sequencing in a case-control cohort of 600 adult-onset sudden cardiac death cases and 600 matched controls from 106,098 participants of 6 prospective cohort studies. Observed DNA sequence variants in any of 49 genes with known association to cardiovascular disease were classified as pathogenic or likely pathogenic by a clinical geneticist blinded to case status. In an independent population of 4,525 asymptomatic adult participants of a prospective cohort study, we performed whole genome sequencing and determined the prevalence of pathogenic or likely pathogenic variants and prospective association with cardiovascular death.
Results:
Among the 1,200 sudden cardiac death cases and controls, we identified 5,178 genetic variants and classified 14 as pathogenic or likely pathogenic. These 14 variants were present in 15 individuals, all of whom had suffered sudden cardiac death – corresponding to a pathogenic variant prevalence of 2.5% in cases and 0% in controls (p<0.0001). Among the 4,525 participants of the prospective cohort study, 41 (0.9%) carried a pathogenic or likely pathogenic variant and these individuals had 3.24-fold higher risk of cardiovascular death over a median follow-up of 14.3 years (p=0.02).
Conclusions:
Gene sequencing identifies a pathogenic or likely pathogenic variant in a small but potentially important subset of adults suffering sudden cardiac death; these variants are present in ~1% of asymptomatic adults.
Keywords: sudden cardiac death, gene sequencing, genomic medicine
CONDENSED ABSTRACT
Rare pathogenic DNA variants in any of 49 genes can predispose to important causes of adult-onset sudden cardiac death. We performed gene sequencing and blinded, clinical-grade variant classification of 5,725 individuals and observe: 1) significant enrichment of pathogenic variants in adult-onset sudden cardiac death cases as compared to matched controls, and 2) about 1% of asymptomatic adults carry a pathogenic DNA variant, and these individuals are at substantially increased risk of cardiovascular death in subsequent years. Taken together, these results lay the scientific foundation for the inclusion of gene sequencing into sudden cardiac death assessment and prevention.
Introduction
A primary goal of genomic medicine is to identify asymptomatic individuals at high inborn risk for a serious health consequence to enable targeted screening or prevention (1). This approach is particularly appealing for sudden cardiac death, a heritable and irreversible outcome that afflicts approximately 220,000 U.S. adults annually and often has a devastating impact on the surviving family and the community (2).
Recent advances in human genetics have identified 49 genes that harbor pathogenic variants predisposing to any of four important causes of sudden cardiac death – cardiomyopathy, coronary artery disease, inherited arrhythmia syndrome, and aortopathy or aortic dissection (3-5). If identified, risk stratification of pre-symptomatic individuals harboring such variants can be performed using tests already available in clinical practice and, in cases where high-risk features are recognized, evidence-based therapies to attenuate risk for each of the four conditions have been demonstrated (6-10).
A prior study conducted in the Dutch population assessed the prevalence of 6 prespecified genetic mutations using a genotyping approach, noting a prevalence of 1.1% in sudden cardiac death cases versus 0.4% in controls (11). This result suggests potential utility for gene sequencing studies, which enable ascertainment of all variants affecting the coding sequence of a given gene.
Previous gene sequencing studies have focused primarily on children and young adults who suffered sudden cardiac death unexplained by autopsy; these studies have suggested a rare predisposing genetic variant in 13 – 28% of afflicted individuals (12-15). However, the vast majority of sudden cardiac death occurs in middle-aged or elderly adults, most of whom do not undergo autopsy (2,16). Moreover, most prior studies have relied on retrospective case referral that can lead to ascertainment bias (12,14,15), implemented variable gene sequencing approaches (13,15), and did not assess the potential pathogenicity of observed variants using current clinical standards (12-14,17,18). Most importantly, all have lacked a control group subjected to identical genetic sequencing and variant classification (12-15).
We set out to overcome these previous limitations by using a matched control group, systematic gene sequencing, and blinded clinical-grade classification of variant pathogenicity. We first determined the extent to which pathogenic variants are enriched in adult-onset sudden cardiac death cases as compared to matched controls, and second, quantified the prevalence and prognostic importance of such variants in a multiethnic population of asymptomatic middle-aged adults.
Methods
Study populations
We assembled a nested case-control cohort for sudden cardiac death based on 6 previously described prospective studies. These studies included four randomized clinical trials – Physician’s Health Study I and II (19,20), Women’s Health Study (21), and Women’s Antioxidant Cardiovascular Study (22) – and two prospective cohort studies, the Health Professionals Follow-up Study (23) and Nurses’ Health Study (24). Together, these cohorts included a total of 38,975 men and 67,093 women with stored blood samples. Additional details on enrollment criteria and study design are available in Online Table 1. For each case, a control was selected that matched as closely as possible based on study cohort, age at blood sample (± 1 year), race/ethnicity, smoking status (current, never, or past), and presence or absence of atherosclerotic cardiovascular disease (myocardial infarction, coronary revascularization, angina, or stroke) at time of blood sample collection.
A multiethnic prospective cohort study of adults was derived from the Multi-Ethnic Study of Atherosclerosis (MESA) study, which enrolled individuals free of known cardiovascular disease or symptoms in the United States between 2000 and 2002 (25).
Sudden cardiac death ascertainment (Nested case-control study)
All 6 prospective studies employed similar methods to document the timing and mechanism of cardiovascular deaths as definite or probable sudden cardiac death as previously described (26) – including collection and review of medical records (inclusive of emergency medical services, emergency room, hospital, and autopsy reports as available) and standardized interviews with next-of-kin or witnesses to obtain detailed descriptions of the circumstances surrounding the death. Additional details are provided in the Online Appendix.
Incident cardiovascular death and intermediate phenotype ascertainment in Multi-Ethnic Study of Atherosclerosis (MESA) study
Within the MESA study, incident cardiovascular deaths were adjudicated based on review of death certificates, hospitalization records, and next-of-kin interviews. Death adjudication was performed by an independent endpoints committee without access to genetic data. Intermediate cardiovascular phenotypes, including lipid, electrocardiographic, and imaging phenotypes were ascertained by MESA core lab investigators (additional details available in the Online Appendix). Clinical hypercholesterolemia was defined as measured LDL-cholesterol ≥ 130 mg/dl or use of statin medications (27). Prolonged corrected QT interval was defined ≥ 450 milliseconds in men or ≥ 460 milliseconds in women (28). Left ventricular ejection fraction values were considered reduced if <52% in men or < 54% in women and hyperdynamic if >72% in men or > 74% in women (29).
Gene sequencing
Whole exome sequencing was performed on 1,200 samples of the sudden cardiac death nested case-control study and whole genome sequencing was performed on the 4,525 MESA study participants at the Broad Institute of MIT and Harvard (Cambridge, MA, USA). Additional details of gene sequencing methods are available in the Online Appendix.
Filtering and classification of variants
We searched for pathogenic or likely pathogenic variants in 49 prespecified genes with known relevance to sudden cardiac death-related conditions – cardiomyopathy (14 genes), coronary artery disease (3 genes), arrhythmia syndromes (24 genes), or aortopathy/aortic dissection (8 genes) – listed in Online Table 2. We derived this list of 49 genes by extracting those related to cardiovascular disease in which pathogenic variants were deemed clinically actionable by the American College of Medical Genetics and Genomics (ACMG) or the Geisinger Health System, and additionally include the gene encoding titin (TTN), in which inactivating mutations are a leading cause of inherited cardiomyopathy (3-5).
Analysis was restricted to the protein-coding regions and canonical splice sites for each of the genes. We used bioinformatic filtering to first narrow the list of observed variants to candidate variants: predicted loss-of-function variants – those that lead to a premature stop codon (nonsense), disrupt canonical splice consensus sequence, exon splicing, or cause an inactivating frameshift (30); variants previously annotated as pathogenic or likely pathogenic in the ClinVar genetics database (31); and rare missense variants – those present with an allele frequency < 0.0001 in each racial subpopulation of the gnomAD Genome Aggregation Database, a publicly available population allele frequency database of 123,136 exomes (30).
Candidate variants were then further narrowed to those meeting American College of Medical Genetics and Genomics (ACMG)/Association of Molecular Pathology (AMP) pathogenic or likely pathogenic criteria by an American Board of Genetics and Genomics (AMBGG)-certified clinical laboratory geneticist within the Partners HealthCare Laboratory for Molecular Medicine who was blinded to case versus control status or any other phenotype information (17,18).
Statistical Analysis
Comparison of the number of observed genetic variants per individual in sudden cardiac death cases versus controls was performed with an unpaired t-test. 95% confidence interval for pathogenic variant prevalence was assessed using a binomial test. Association of pathogenic mutation status with sudden cardiac death case status was assessed using a Fisher’s exact test. The relationship of pathogenic variant status to incident cardiovascular death was determined using a Cox proportional-hazard model adjusted for age, sex, and race. Statistical analyses were performed with the use of R software, version 3.4 (R Project for Statistical Computing).
Results
We performed whole exome sequencing of 600 adult-onset sudden cardiac death cases and 600 matched controls. Mean age at time of blood draw was 64 years and 66% were male. Among cases, mean age at sudden cardiac death was 72 years (range 48 – 98; Table 1). Genetic background, as quantified by principal components of ancestry, was similar between cases and controls (Online Figure 1).
Table 1.
Characteristics of sudden cardiac death cases and controls
| Characteristics | Sudden cardiac death cases (N = 600) |
Matched controls (N = 600) |
|---|---|---|
| Age at time of blood sample – years | 64 (9) | 64 (9) |
| Age at sudden death – years | 72 (9) | N/A |
| Male sex | 395 (66%) | 395 (66%) |
| White race | 581 (97%) | 585 (98%) |
| ASCVD at time of blood sample | 145 (24%) | 145 (24%) |
| ASCVD at time of sudden death | 232 (39%) | N/A |
| Smoking status | ||
| Never | 225 (38%) | 230 (38%) |
| Former | 275 (46%) | 283 (47%) |
| Current | 94 (16%) | 85 (14%) |
| Study cohort | ||
| Physicians Health Study I | 156 | 156 |
| Physicians Health Study II | 112 | 112 |
| Health Professionals Follow-up Study | 127 | 127 |
| Nurses’ Health Study | 126 | 126 |
| Women’s Antioxidant Cardiovascular Study | 41 | 41 |
| Women’s Health Study | 38 | 38 |
Data are mean SD, or n (%).
ASCVD – atherosclerotic cardiovascular disease; N/A – Not applicable;
Among the 600 sudden cardiac death cases, 145 (24%) had been diagnosed with atherosclerotic cardiovascular disease at time of study enrollment and an additional 87 (15%) were diagnosed prior to sudden cardiac death. Chart review suggested a diagnosis of congestive heart failure or cardiomyopathy prior to death in an additional 36 individuals (6%). However, the majority (N = 332; 55%) had not been diagnosed with atherosclerotic cardiovascular disease or congestive heart failure at the time they suffered sudden cardiac death.
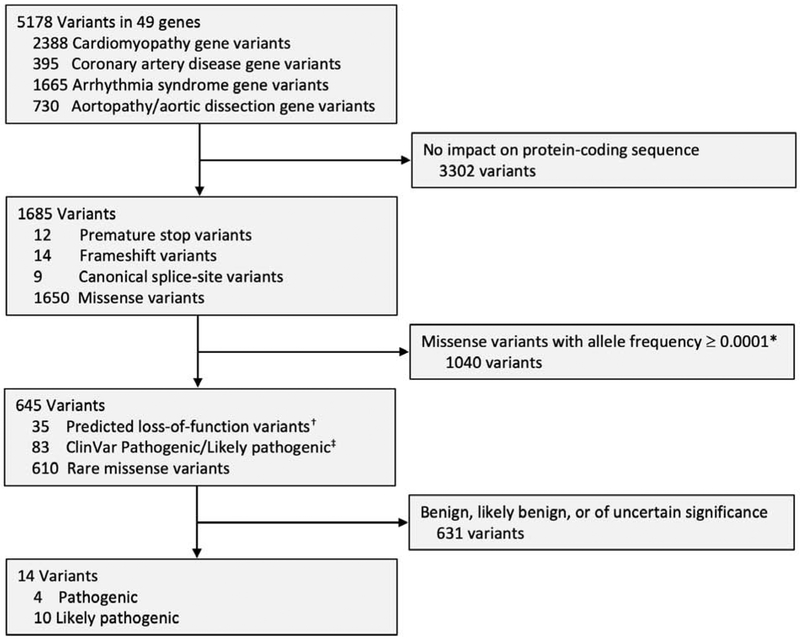
Analysis of the 49 sudden cardiac death-related genes from whole exome sequencing data identified a total of 5,178 genetic variants present in at least one of these 1,200 individuals. Synonymous DNA sequence variants – single base pair changes that have no impact on the amino acid sequence of the protein – are often used as a negative control in genetic case-control studies. 1,096 of the identified 5,178 genetic variants were annotated as synonymous variants (Online Table 3). Consistent with expectations, the average number of synonymous variants in these 49 genes did not differ between cases and controls, mean 38.4 versus 38.3 variants per person respectively (p = 0.99).
We next used bioinformatics filters to identify those variants that were predicted to cause loss-of-function, were extremely rare missense variants, or were previously annotated as likely to be pathogenic in the ClinVar genetics database. This filtering narrowed the list to 645 variants.
A clinical laboratory geneticist who was blinded to case/control status evaluated these 645 variants according to ACMG/AMP criteria (Figure 1) – 631 were classified as benign, likely benign, or variants of uncertain significance. Cases and controls carried a similar number of these variants, mean 0.79 versus 0.75 variants per person respectively (p = 0.40). The remaining 14 variants were classified as pathogenic or likely pathogenic, including 11 predicted loss-of-function variants and 3 missense variants (Figure 1). Importantly, while loss-of function variants can often be interpreted sole on the basis of predicted effect on DNA sequence, missense variants require prior evidence in the literature for disease association. The three missense variants with adequate evidence based on published pedigrees and segregation to support classification as pathogenic or likely pathogenic were the p.Arg141Gln variant in TNNI3 predisposing to hypertrophic cardiomyopathy and the p.Trp87Gly and p.Trp483Arg variants in LDLR predisposing to familial hypercholesterolemia, present in participants P6, P7, P8, and P10 respectively.(32-36) Additional details on evidence used in support of these annotations for each of the 14 pathogenic or likely pathogenic variants is provided in Online Table 4.
Figure 1.
Filtration and classification of variants identified by whole exome sequencing in the sudden cardiac death case-control cohort. * Missense variants filtered out if present with allele frequency ≥0.0001 in any racial subpopulation of the Genome Aggregation Database, a publicly available allele frequency database derived from 123,136 exome sequences (30). † Predicted loss-of-function variants do not necessarily all meet current standards to be classified as pathogenic or likely pathogenic. Representative examples include variants in the gene encoding titin (TTN) that occur in exons not found in cardiac-specific isoforms(44), variants in the gene encoding apolipoprotein B (APOB) that are associated with lower risk of hypercholesterolemia and coronary artery disease(45), and variants in the gene encoding tyrosine-protein phosphatase (DSP2), where only those variants that have previously been proven to segregate with disease are classified as pathogenic or likely pathogenic (3). ‡ The ClinVar database provides open access to variant classifications shared by many individuals and clinical laboratories. While an important means of communicating and coordinating efforts, many variant pathogenicity assertions in ClinVar do not warrant this designation based on current ACMG/AMP criteria (46).
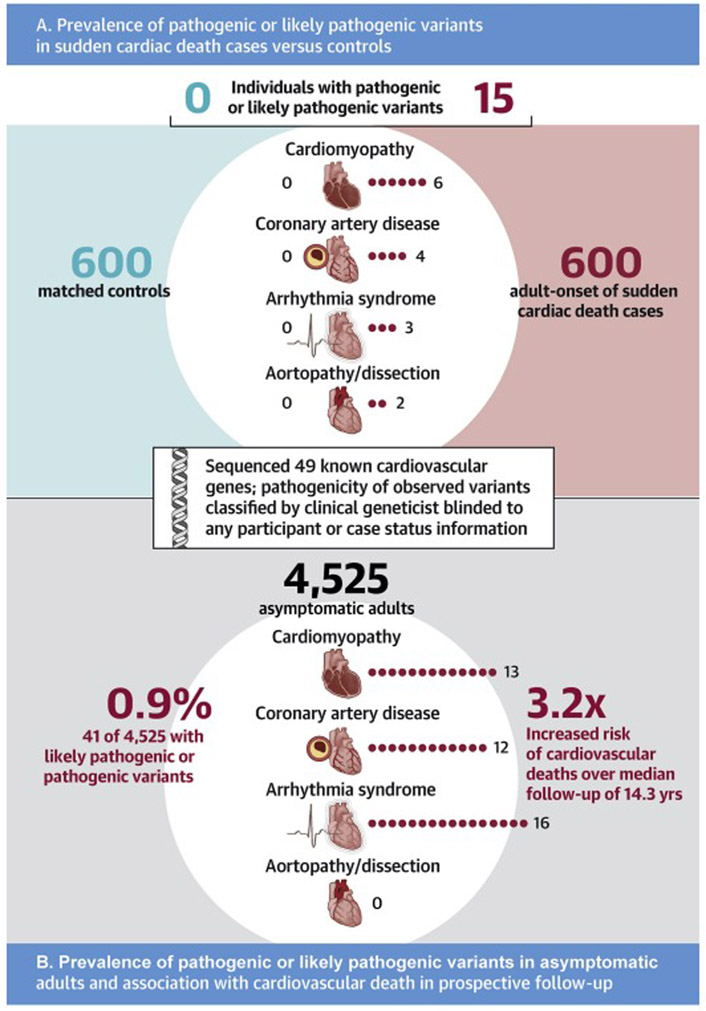
A total of 15 individuals harbored one of these 14 variants, with 13 variants observed in only one individual and the p.Trp87Gly LDLR missense variant observed in two individuals, participants P7 and P8. Subsequent unblinding of sudden cardiac death status revealed that all 15 (100%) of the individuals harboring a pathogenic or likely pathogenic variant were sudden cardiac death cases and none were controls. This represented a pathogenic or likely pathogenic variant prevalence of 2.5% (95%CI 1.4 to 4.1%) in cases and 0% (95%CI 0 to 0.6%) in controls (p <0.0001). Baseline characteristics among sudden cardiac death cases in whom a pathogenic or likely pathogenic variant was identified were similar to the remainder of the cases with respect to age, sex, and history of cardiovascular disease (Online Table 5). Of the 15 individuals harboring a pathogenic or likely pathogenic variant who suffered sudden cardiac death, 8 (53%) had no prior diagnosis of atherosclerotic cardiovascular disease or congestive heart failure. These 15 cases included 6 with a variant related to cardiomyopathy, 4 related to coronary artery disease, 3 related to an inherited arrhythmia syndrome, and 2 related to aortopathy or aortic dissection (Central Illustration). Variants in the TTN gene predisposing to dilated cardiomyopathy and the LDLR gene related to familial hypercholesterolemia were the most common, present in 5 and 4 sudden cardiac death cases respectively (Table 2).
Central Illustration. Gene sequencing to identify pathogenic variants in sudden cardiac death cases versus controls, and in asymptomatic adults.
A. In an adult-onset sudden cardiac death case-control cohort, whole exome sequencing identified pathogenic or likely pathogenic variants in 15 (2.5%) of cases versus 0 (0%) of controls (p<0.0001). The number of carriers of a pathogenic or likely pathogenic variant observed in each of four important conditions underlying sudden cardiac death is displayed according to case versus control status. Test of statistical significance was performed for the combined variant counts using a Fisher’s exact test. B. Among 4,525 asymptomatic adult participants of the Multi-Ethnic Study of Atherosclerosis free of known cardiovascular disease, whole genome sequencing identified 41 (0.9%) with a pathogenic or likely pathogenic variant – the number of carriers of a pathogenic or likely pathogenic variant observed in each of four important conditions underlying sudden cardiac death is displayed. Over a median follow-up of 14.3 years, these 41 carriers had a 3.24-fold (p=0.02) increased risk of incident cardiovascular death, as assessed in a Cox proportional hazards model adjusted for age, sex, and race.
Table 2.
Sudden Cardiac Death Case/Control Status and Clinical Characteristics of Carriers of a Pathogenic or Likely Pathogenic Mutation
| Case ID |
Age*/ Sex |
Case / Control |
Known ASCVD† |
Known CHF‡ |
Variant § | Gene (Variant Type) |
Amino acid or cDNA change |
Associated disease | Classification using ACMG criteria |
|---|---|---|---|---|---|---|---|---|---|
| Cardiomyopathy variants | |||||||||
| P1 | 64M | Case | No | No | 2:179417587:C>A |
TTN Premature stop |
p.Gly27446Ter | Dilated cardiomyopathy | Likely pathogenic |
| P2 | 81M | Case | No | Yes | 2:179433438:G>T |
TTN Premature stop |
p.Tyr23239Ter | Dilated cardiomyopathy | Likely pathogenic |
| P3 | 64F | Case | Yes | No | 2:179444577:T>G |
TTN Splice site |
c.59645-2A>C | Dilated cardiomyopathy | Likely pathogenic |
| P4 | 75M | Case | No | No | 2:179486244:G>A |
TTN Premature stop |
p.Arg12535Ter | Dilated cardiomyopathy | Likely pathogenic |
| P5 | 61M | Case | No | Yes | 2:179604601:TTC>T |
TTN Frameshift |
p.Glu4215ArgfsTer7 | Dilated cardiomyopathy | Likely pathogenic |
| P6 | 78F | Case | No | No | 19:55665525:C>T |
TNNI3 Missense |
p.Arg141Gln | Hypertrophic cardiomyopathy | Likely pathogenic |
| Coronary artery disease variants | |||||||||
| P7 | 78M | Case | Yes | No | 19:11213408:T>G |
LDLR Missense |
p.Trp87Gly | Familial hypercholesterolemia | Pathogenic |
| P8 | 70M | Case | No | No | 19:11213408:T>G |
LDLR Missense |
p.Trp87Gly | Familial hypercholesterolemia | Pathogenic |
| P9 | 69F | Case | Yes | Yes | 19:11221435:C>T |
LDLR Premature stop |
p.Arg350Ter | Familial hypercholesterolemia | Pathogenic |
| P10 | 73F | Case | No | No− | 19:11224299:T>C |
LDLR Missense |
p.Trp483Arg | Familial hypercholesterolemia | Likely pathogenic |
| Arrhythmia syndrome variants | |||||||||
| P11 | 53F | Case | No | No | 6:7580603:C>T |
DSP Premature stop |
p.Gln1394Ter | Arrhythmogenic right ventricular cardiomyopathy | Likely pathogenic |
| P12 | 60M | Case | No | No | 7:150647424:G>A |
KCNH2 Premature stop |
p.Arg744Ter | Long QT syndrome | Pathogenic |
| P13 | 67M | Case | Yes | Yes | 7:150655169:CG>C |
KCNH2 Frameshift |
p.Pro298ArgfsTer62 | Long QT syndrome | Likely pathogenic |
| Aortopathy/ aortic dissection variants | |||||||||
| P14 | 75F | Case | No | No | 2:189855742:C>T |
COL3A1 Premature stop |
p.Arg271Ter | Vascular Ehlers-Danlos syndrome | Pathogenic |
| P15 | 80F | Case | Yes | No | 15:48892335:C>T |
FBN1 Splice site |
c.442+1G>A | Marfan syndrome | Likely pathogenic |
Age refers to age at sudden cardiac death event.
Known ASCVD (atherosclerotic cardiovascular disease) prior to sudden cardiac death based upon review of medical records
Known CHF (congestive heart failure) or cardiomyopathy prior to sudden cardiac death based upon review of medical records
Variant is described based on’ chromosome:position reference allele>alternate allele’ formatting, with chromosome positions based on the GRCh37 genome assembly
Although definitive confirmation that a given variant was the ‘cause’ of a sudden cardiac death is typically not possible, we next reviewed available health records of the 15 cases who harbored a pathogenic or likely pathogenic variant to assess concordance of the variant and mechanism of death. Of these, 6 of the 15 (40%) were deemed probably consistent, 2 (13%) possibly consistent, and 7 (47%) uncertain (Online Table 6). Representative examples of probably consistent annotations include a history of congestive heart failure and ventricular tachycardia in a participant harboring a TTN cardiomyopathy variant (participant P2) and a history of lipid-lowering medication use and exertional angina in a participant harboring a LDLR familial hypercholesterolemia variant (participant P10). Other scenarios – such as a report of neck discomfort leading up to sudden cardiac death in a participant with a FBN1 aortopathy/aortic dissection variant were deemed possibly consistent (participant P15). Cases were deemed uncertain owing to lack of autopsy or relevant tests performed in clinical care prior to sudden cardiac death event.
Through analysis of the same 49 genes from whole genome sequencing data and using an identical variant classification procedure, we next determined the prevalence of pathogenic or likely pathogenic variants in 4,525 adults free of known cardiovascular disease derived from the MESA prospective cohort study. Average participant age was 61 years (range 44—84) and 2,200 (49%) were male. 1,837 (41%) of the participants were white, 1,098 (24%) black, 1,004 (22%) Hispanic, and 586 (13%) Asian.
A total of 15,348 variants were present in any of the 49 genes in at least one of the 4,525 MESA participants. This list was narrowed by bioinformatics filtering to 5,419 variants. A clinical laboratory geneticist who was blinded to phenotype status evaluated these 5,419 variants, and classified 39 as pathogenic or likely pathogenic according to ACMG/AMP criteria, including 15 predicted loss-of-function variants and 24 missense variants (Online Figure 2). Only one of these 39 variants, a splice site mutation in the TTN gene predisposing to cardiomyopathy, overlapped with the variants observed in the sudden cardiac death nested case-control dataset. A summary of the evidence used in support of these annotations for each of these 39 variants is provided in Online Table 6.
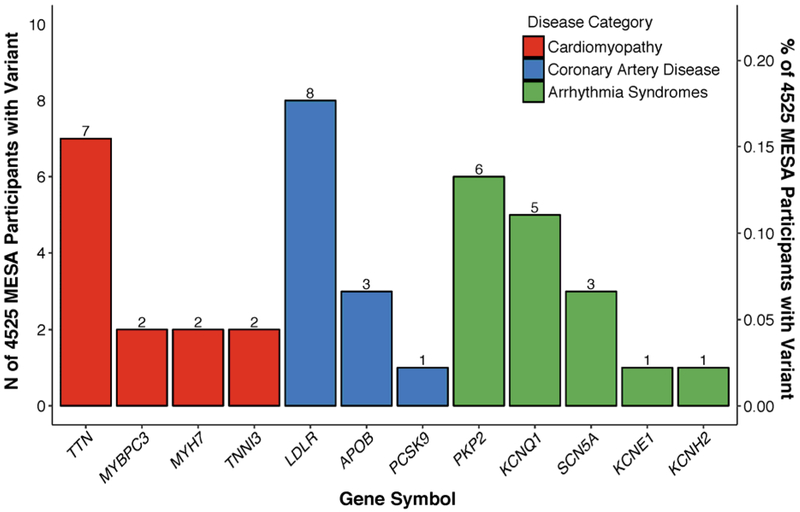
The 39 variants occurred in 12 distinct genes and, in aggregate, were present in 41 (0.91%) of the MESA participants (Figure 2). Individuals harboring these variants included 13 participants with a mutation related to cardiomyopathy, 12 related to coronary artery disease, 16 related to an inherited arrhythmia syndrome, and 0 related to aortopathy or aortic dissection (Central Illustration).
Figure 2. Pathogenic or likely pathogenic variants in a prospective cohort study of adults free of known cardiovascular disease according to gene.
Shown are the number of pathogenic or likely pathogenic variants observed in 4,525 participants of the Multi-Ethnic Study of Atherosclerosis free of known cardiovascular disease according to gene and disease category.
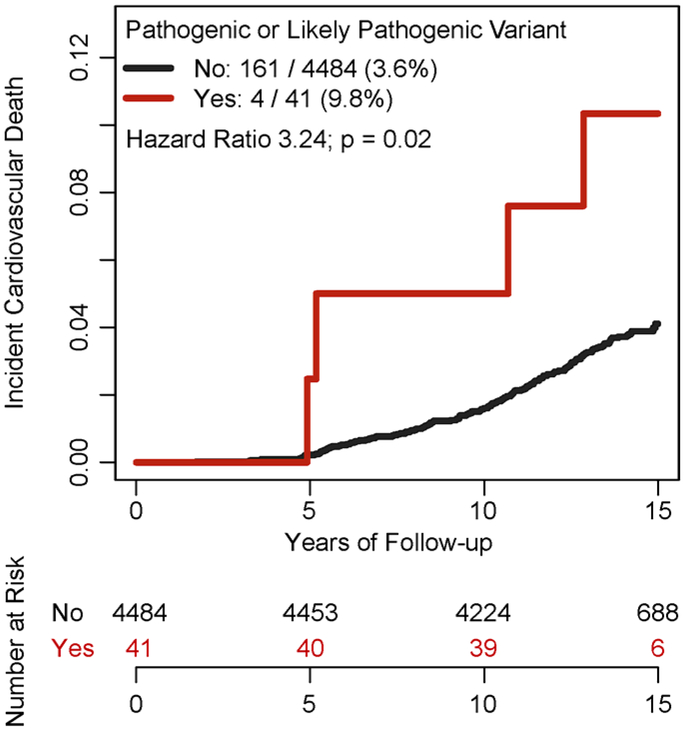
Cardiovascular death occurred in 165 of 4525 (3.6%) participants over a median follow-up of 14.3 years (Q1-Q3 13.7 to 14.8 years), including 4 of 41 (9.8%) with a pathogenic or likely pathogenic variant and 161 of 4483 (3.6%) without such a variant (Figure 3). In a survival analysis adjusted for age, sex, and race, participants with a pathogenic or likely pathogenic variant were at significantly increased risk of cardiovascular death compared to noncarriers – hazard ratio 3.24 (95%CI 1.20 – 8.79; p = 0.02).
Figure 3. Association of pathogenic or likely pathogenic variant carrier status with incident cardiovascular death in a prospective cohort study of adults free of known cardiovascular disease.
Among 4,525 adult participants of the Multi-Ethnic Study of Atherosclerosis, whole genome sequencing identified 41 (0.9%) with a pathogenic or likely pathogenic variant. Over a median follow-up of 14.3 years, these 41 carriers had a 3.24-fold (95% confidence interval 1.20 – 8.79; p=0.02) increased risk of incident cardiovascular death, as assessed in a Cox proportional hazards model adjusted for age, sex, and race.
We next assessed the extent to which pathogenic variants associated with expected intermediate phenotypes. Among 12 carriers of a familial hypercholesterolemia mutation predisposing to coronary artery disease, average LDL cholesterol was 165 mg/dl versus 124 mg/dl in the remainder of the population (p = 0.01) and 9 of the 12 (75%) met clinical criteria for hypercholesterolemia as compared with 45% of noncarriers (p = 0.04). Among 8 carriers of a long QT syndrome variant predisposing to arrhythmia, mean QT interval corrected for heart rate was 457 milliseconds versus 417 milliseconds in the remainder of the population (p = 0.01) and 4 of the 8 (50%) met clinical criteria for prolonged QT interval versus 4.2% in the remainder of the population (p = 0.0002). Left ventricular ejection fraction was available in 4 patients with dilated cardiomyopathy mutations – mean value was 64% in mutation carriers versus 69% in the remainder of the population (p = 0.31), with no carriers meeting clinical criteria for reduced ejection fraction. Among 3 carriers of a hypertrophic cardiomyopathy mutation, mean ejection fraction was 79% versus 69% in the remainder of the population (p = 0.04), with all three carriers (100%) meeting clinical criteria for hyperdynamic contractile function versus 29.4% of the remainder of the population (p = 0.03). Taken together, these results suggest that clinical assessment of asymptomatic carriers of pathogenic variants is likely to reveal high-risk intermediate features in a meaningful portion of individuals.
Discussion
In this study, we show that pathogenic or likely pathogenic variants in any of 49 cardiovascular disease genes are present in a small but potentially important subset of adults suffering sudden cardiac death. Second, we determine that such variants are present in about 1% of an adult population free of known cardiovascular disease and conferred a more than three-fold increased risk of cardiovascular death in prospective follow-up.
This analysis builds upon previous efforts to study the genetic basis of sudden cardiac death in several key ways (11-15). First, we emphasize the careful adjudication of sudden cardiac death in 600 adults from prospective cohort studies and the largest gene sequencing analysis of this phenotype to date. Second, cases were ascertained solely based upon suffering sudden cardiac death, eliminating the potential referral bias caused by the requirement of an unrevealing autopsy or referral from a medical examiner. Third, a certified clinical laboratory geneticist performed clinical-grade assessment of variant pathogenicity in both cases and matched controls blinded to any phenotype information. Fourth, we extend our variant annotation approach used in our sudden cardiac death nested case control analysis to a population-based sample of 4,525 asymptomatic adults.
Gene sequencing to identify individuals with a pathogenic variant at scale in the population is increasingly practical as the costs of sequencing have declined rapidly. At least one U.S. health care system has committed to returning pathogenic variants related to cardiovascular disease to patients and their health care providers,(4) and a similar commitment has been embraced as a core tenet of the All of Us Research Program, a U.S. National Institutes of Health sponsored study that plans to enroll over one million participants in coming years (37).
Explicit demonstration that gene sequencing reduces the risk of sudden cardiac death is unlikely to be attainable given low absolute event rates. However, identification of pathogenic mutation carriers prior to disease onset may already provide a clinically actionable opportunity for targeted screening or prevention. Each of the four sudden cardiac death etiologies considered have a strong evidence base for treatment considerations, ranging from aggressive LDL cholesterol lowering for those with familial hypercholesterolemia to beta-blocker therapy for those with long QT syndrome (6-10). Below, we highlight several key challenges to widespread implementation into clinical practice.
First, the current variant classification procedure may underestimate pathogenic variant prevalence, deeming some variants as of uncertain significance because they currently have insufficient evidence for disease association. This is because assessing the pathogenicity of a given missense variant relies heavily on previous observations of the variant in family-based studies and co-segregation with disease and detailed manual curation of this evidence (17,18). The labor-intensive nature of clinical-grade variant classification remains an important barrier to rapid scaling of genome interpretation for large segments of the population. However, efforts are ongoing to enhance variant annotation by incorporating larger and more diverse population allele frequency databases,(38) promote data sharing and best practices (18,39), and develop new technology or machine learning algorithms to enable high-throughput functional assessment of variants in a given gene (40,41).
Second, current clinical care is largely designed to treat patients with disease, not healthy individuals with inherited susceptibility. Few health systems house the necessary physician and genetic counseling expertise to effectively manage healthy individuals who harbor pathogenic risk variants. A larger workforce trained in both genome interpretation as well as the psychosocial or healthcare cost implications of genetic risk disclosure is needed to realize the full potential of genomic medicine (1).
Third, current guidelines to report variants as ‘pathogenic’ may lead to misinterpretation by patients and their health care providers as a predetermined destiny to manifest disease, but whether genetic risk ultimately manifests itself as disease is related to a large number of genetic and non-genetic factors (17). For example, although familial hypercholesterolemia variants confer a 4-fold increased risk of coronary artery disease, about 25% of individuals who harbor such a variant do not meet clinical criteria for hypercholesterolemia (42). Similarly, despite conferring a more than 10-fold increased risk of dilated cardiomyopathy, many individuals with a loss-of-function TTN mutation have a normal left ventricular ejection fraction (43). We thus suggest improving genetic risk disclosure and describing the risk conferred by variants in a ‘probabilistic’ manner rather than a dichotomy of ‘pathogenic’ versus not.
Study Limitations
Our analysis, as with previous gene sequencing studies of sudden cardiac death, was limited in the ability to definitively link an individual’s demise to an identified pathogenic or likely pathogenic variant (11-15). However, the enrichment of such variants in sudden cardiac death cases as compared to a matched control group suggests significant attributable risk. Second, our sudden cardiac death case-control cohort was comprised primarily of white individuals. Third, individual pathogenic or likely pathogenic variants are exceedingly rare and were thus considered in aggregate to confirm enrichment in sudden cardiac death cases. Fourth, our analysis of asymptomatic adults free of known cardiovascular disease was performed in the middle-aged MESA cohort, raising the potential for an underestimation of pathogenic variant prevalence due to survival bias. Fifth, the association of pathogenic variants with intermediate risk features in MESA participants was limited to phenotypes available in the cohort.
Conclusions
Gene sequencing confirms significant enrichment of pathogenic variants in sudden cardiac death cases as compared to controls, and such variants are present in approximately 1% of asymptomatic adults free of known cardiovascular disease. We believe these results lay the scientific foundation for the integration of gene sequencing to identify asymptomatic individuals with pathogenic variants into routine clinical practice, with the ultimate goal of preventing a sudden cardiac death tragedy that might have been anticipated.
Supplementary Material
CLINICAL PERSPECTIVES.
Competency in Medical Knowledge: Gene sequencing identifies pathogenic variants in a small subset of adults suffering sudden cardiac death that are also present in ~1% of asymptomatic individuals.
Translational Outlook: Additional research is needed to assess the utility of integrating genetic testing into risk assessment algorithms for prevention of sudden cardiac death in adults.
Acknowledgments:
We are indebted to the studies and participants who provided biological samples and data for this analysis and members of the Broad Institute’s Pattern data visualization team – Bang Wong and Mariya Khan – for graphical and visual design assistance with the central illustration.
Funding: Funding support was provided by an institutional grant from the Broad Institute of MIT and Harvard (BroadIgnite, to A.V.K.), grant 1K08HG010155 from the National Human Genome Research Institute (to A.V.K.) a Hassenfeld Scholar Award from Massachusetts General Hospital (to A.V.K.), an institutional grant from the Broad Institute of MIT and Harvard (BroadNext10, to A.P. and S.K.), and a sponsored research agreement from IBM Research (to A.V.K., A.P., and S.K.). The sudden cardiac death case-control cohort was supported by grants HL-03783, HL-26490, HL-34595, HL-34594, HL-35464, HL-043851, HL-46959, HL-099355 and HL-080467 from the National Heart, Lung, and Blood Institute and CA-167552, CA-186107, CA-49449CA-34944, CA-40360, CA-47988, CA-55075, CA-87969, and CA-97193 from the National Cancer Institute. The sudden death confirmation was supported by grants from the NIH (HL-068070) and Novo Nordisk Foundation, Copenhagen, Denmark. The MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. Whole genome sequencing of the MESA cohort was funded through the Trans-Omics for Precision Medicine (TOPMed) Program of the National Heart, Lung, and Blood Institute. General study coordination was provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1). Funding agencies had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Abbreviations
- DNA
deoxyribonucleic acid
- MESA
Multi-Ethnic Study of Atherosclerosis
- ACMG
American College of Medical Genetics and Genomics
- AMP
Association of Molecular Pathology
- AMBGG
American Board of Genetics and Genomics
- SD
standard deviation
- ASCVD
atherosclerotic cardiovascular disease
- CHF
congestive heart failure
Footnotes
Disclosures: A.V.K. has served as a consultant for Color Genomics and Navitor Pharmaceuticals and reports a patent related to a genetic risk predictor (20190017119). E.S.L serves on the Board of Directors for Codiak BioSciences and Neon Therapeutics, and serves on the Scientific Advisory Board of F-Prime Capital Partners and Third Rock Ventures; he is also affiliated with several non-profit organizations including serving on the Board of Directors of the Innocence Project, Count Me In, and Biden Cancer Initiative, and the Board of Trustees for the Parker Institute for Cancer Immunotherapy. He has served and continues to serve on various federal advisory committees. K.N. is an employee of IBM Research. A.P. is a Venture Partner at GV, a subsidiary of Alphabet Corporation. C.M.A. has served as a consultant to Myocardia and as an advisory board member for Roche Diagnostics, and has received grant support from St. Jude Medical, Roche Diagnostics, and Abbott. S.K. is an employee of Verve Therapeutics, and holds equity in Verve Therapeutics, Maze Therapeutics, Catabasis, and San Therapeutics. He is a member of the scientific advisory boards for Regeneron Genetics Center and Corvidia Therapeutics; he has served as a consultant for Acceleron, Eli Lilly, Novartis, Merck, Novo Nordisk, Novo Ventures, Ionis, Alnylam, Aegerion, Haug Partners, Noble Insights, Leerink Partners, Bayer Healthcare, Illumina, Color Genomics, MedGenome, Quest, and Medscape; he reports patents related to a method of identifying and treating a person having a predisposition to or afflicted with cardiometabolic disease (20180010185) and a genetics risk predictor (20190017119). The remaining authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Amit V. Khera, Center for Genomic Medicine, Cardiology Division, Massachusetts General Hospital, Harvard Medical School, Boston, MA; Cardiovascular Disease Initiative, Broad Institute of MIT and Harvard, Cambridge, MA.
Heather Mason-Suares, Laboratory for Molecular Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Deanna Brockman, Center for Genomic Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA.
Minxian Wang, Cardiovascular Disease Initiative, Broad Institute of MIT and Harvard, Cambridge, Massachusetts.
Martin J. VanDenburgh, Divisions of Preventive Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Ozlem Senol-Cosar, Laboratory for Molecular Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Candace Patterson, Center for Genomic Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA; Cardiovascular Disease Initiative, Broad Institute of MIT and Harvard, Cambridge, MA
Christopher Newton-Cheh, Center for Genomic Medicine, Cardiology Division, Massachusetts General Hospital, Harvard Medical School, Boston, MA.
Seyedeh M. Zekavat, Cardiovascular Disease Initiative, Broad Institute of MIT and Harvard, Cambridge, MA.
Julie Pester, Divisions of Preventive Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Daniel I. Chasman, Divisions of Preventive Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Christopher Kabrhel, Department of Emergency Medicine, Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Majken K. Jensen, Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
JoAnn E. Manson, Divisions of Preventive Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
J. Michael Gaziano, Cardiovascular Medicine, Division of Aging, Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Kent D. Taylor, The Institute for Translational Genomics and Population Sciences, Departments of Pediatrics and Medicine, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA.
Nona Sotoodehnia, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, Washington.
Wendy S. Post, Division of Cardiology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Stephen S. Rich, Center for Public Health Genomics, University of Virginia, Charlottesville, Virginia.
Jerome I. Rotter, The Institute for Translational Genomics and Population Sciences, Departments of Pediatrics and Medicine, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA.
Eric S. Lander, Cardiovascular Disease Initiative, Broad Institute of MIT and Harvard, Cambridge, MA.
Heidi L. Rehm, Center for Genomic Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA; Cardiovascular Disease Initiative, Broad Institute of MIT and Harvard, Cambridge, MA.
Kenney Ng, Center for Computational Health, IBM Research, Cambridge, Massachusetts.
Anthony Philippakis, Cardiovascular Disease Initiative, Broad Institute of MIT and Harvard, Cambridge, MA.
Matthew Lebo, Laboratory for Molecular Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Christine M. Albert, Divisions of Preventive Medicine, Cardiovascular Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Sekar Kathiresan, Verve Therapeutics, Cambridge, Massachusetts.
REFERENCES
- 1.Green ED, Guyer MS, National Human Genome Research I. Charting a course for genomic medicine from base pairs to bedside. Nature 2011;470:204–13. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin EJ, Virani SS, Callaway CW et al. Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation 2018;137:e67–e492. [DOI] [PubMed] [Google Scholar]
- 3.Kalia SS, Adelman K, Bale SJ et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017;19:249–255. [DOI] [PubMed] [Google Scholar]
- 4.Dewey FE, Murray MF, Overton JD et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 2016;354. [DOI] [PubMed] [Google Scholar]
- 5.Herman DS, Lam L, Taylor MR et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med 2012;366:619–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hershberger RE, Givertz MM, Ho CY et al. Genetic Evaluation of Cardiomyopathy-A Heart Failure Society of America Practice Guideline. J Card Fail 2018;24:281–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Priori SG, Wilde AA, Horie M et al. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm 2013;10:1932–63. [DOI] [PubMed] [Google Scholar]
- 8.Gersh BJ, Maron BJ, Bonow RO et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol 2011;58:2703–38. [DOI] [PubMed] [Google Scholar]
- 9.Nordestgaard BG, Chapman MJ, Humphries SE et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. European heart journal 2013;34:3478–90a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiratzka LF, Bakris GL, Beckman JA et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM Guidelines for the diagnosis and management of patients with thoracic aortic disease. A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology,American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons,and Society for Vascular Medicine. J Am Coll Cardiol 2010;55:e27–e129. [DOI] [PubMed] [Google Scholar]
- 11.Milano A, Blom MT, Lodder EM et al. Sudden Cardiac Arrest and Rare Genetic Variants in the Community. Circ Cardiovasc Genet 2016;9:147–53. [DOI] [PubMed] [Google Scholar]
- 12.Tester DJ, Medeiros-Domingo A, Will ML, Haglund CM, Ackerman MJ. Cardiac channel molecular autopsy: insights from 173 consecutive cases of autopsy-negative sudden unexplained death referred for postmortem genetic testing. Mayo Clin Proc 2012;87:524–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagnall RD, Weintraub RG, Ingles J et al. A Prospective Study of Sudden Cardiac Death among Children and Young Adults. N Engl J Med 2016;374:2441–52. [DOI] [PubMed] [Google Scholar]
- 14.Torkamani A, Muse ED, Spencer EG et al. Molecular Autopsy for Sudden Unexpected Death. JAMA 2016;316:1492–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahrouchi N, Raju H, Lodder EM et al. Utility of Post-Mortem Genetic Testing in Cases of Sudden Arrhythmic Death Syndrome. Journal of the American College of Cardiology 2017;69:2134–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton JL, Underwood J. Clinical, educational, and epidemiological value of autopsy. Lancet 2007;369:1471–1480. [DOI] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly MA, Caleshu C, Morales A et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen's Inherited Cardiomyopathy Expert Panel. Genet Med 2018;20:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steering Committee of the Physicians' Health Study Research Group. Final report on the aspirin component of the ongoing Physicians' Health Study. N Engl J Med 1989;321:129–35. [DOI] [PubMed] [Google Scholar]
- 20.Sesso HD, Buring JE, Christen WG et al. Vitamins E and C in the prevention of cardiovascular disease in men: the Physicians' Health Study II randomized controlled trial. JAMA 2008;300:2123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ridker PM, Cook NR, Lee IM et al. A randomized trial of low-dose aspirin in the primary prevention of cardiovascular disease in women. N Engl J Med 2005;352:1293–304. [DOI] [PubMed] [Google Scholar]
- 22.Bassuk SS, Albert CM, Cook NR et al. The Women's Antioxidant Cardiovascular Study: design and baseline characteristics of participants. J Womens Health (Larchmt) 2004;13:99–117. [DOI] [PubMed] [Google Scholar]
- 23.Grobbee DE, Rimm EB, Giovannucci E, Colditz G, Stampfer M, Willett W. Coffee, caffeine, and cardiovascular disease in men. N Engl J Med 1990;323:1026–32. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg L, Hennekens CH, Rosner B, Belanger C, Rothman KJ, Speizer FE. Oral contraceptive use in relation to nonfatal myocardial infarction. Am J Epidemiol 1980;111:59–66. [DOI] [PubMed] [Google Scholar]
- 25.Bild DE, Bluemke DA, Burke GL et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. American journal of epidemiology 2002;156:871–81. [DOI] [PubMed] [Google Scholar]
- 26.Newton-Cheh C, Cook NR, VanDenburgh M, Rimm EB, Ridker PM, Albert CM. A common variant at 9p21 is associated with sudden and arrhythmic cardiac death. Circulation 2009;120:2062–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundy SM, Stone NJ, Bailey AL et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: Executive Summary. J Am Coll Cardiol. 2018. DOI: 10.1016/j.jacc.2018.11.002 [DOI] [Google Scholar]
- 28.Rautaharju PM, Surawicz B, Gettes LS et al. AHA/ACCF/HRS recommendations for the standardization and interpretation of the electrocardiogram: part IV: the ST segment, T and U waves, and the QT interval: a scientific statement from the American Heart Association Electrocardiography and Arrhythmias Committee, Council on Clinical Cardiology; the American College of Cardiology Foundation; and the Heart Rhythm Society. Endorsed by the International Society for Computerized Electrocardiology. J Am Coll Cardiol 2009;53:982–91. [DOI] [PubMed] [Google Scholar]
- 29.Lang RM, Badano LP, Mor-Avi V et al. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur Heart J Cardiovasc Imaging 2015;16:233–70. [DOI] [PubMed] [Google Scholar]
- 30.Lek M, Karczewski KJ, Minikel EV et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landrum MJ, Lee JM, Benson M et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curila K, Benesova L, Penicka M et al. Low prevalence and variable clinical presentation of troponin I and troponin T gene mutations in hypertrophic cardiomyopathy. Genet Test Mol Biomarkers 2009;13:647–50. [DOI] [PubMed] [Google Scholar]
- 33.Kapplinger JD, Landstrom AP, Bos JM, Salisbury BA, Callis TE, Ackerman MJ. Distinguishing hypertrophic cardiomyopathy-associated mutations from background genetic noise. J Cardiovasc Transl Res 2014;7:347–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin R, Latten M, Hart P et al. Genetic diagnosis of familial hypercholesterolaemia using a rapid biochip array assay for 40 common LDLR, APOB and PCSK9 mutations. Atherosclerosis 2016;254:8–13. [DOI] [PubMed] [Google Scholar]
- 35.Ward AJ, O'Kane M, Young I, Nicholls DP, Nevin NC, Graham CA. Three novel mutations in the EGF precursor homology domain of the low-density lipoprotein receptor gene in Northern Irish patients with familial hypercholesterolemia. Hum Mutat 1995;6:254–6. [DOI] [PubMed] [Google Scholar]
- 36.Leitersdorf E, Tobin EJ, Davignon J, Hobbs HH. Common low-density lipoprotein receptor mutations in the French Canadian population. J Clin Invest 1990;85:1014–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.All of Us Research Program Investigators, Denny JC, Rutter JL et al. The "All of Us" Research Program. N Engl J Med 2019;381:668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whiffin N, Minikel E, Walsh R et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med 2017;19:1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehm HL, Berg JS, Brooks LD et al. ClinGen--the Clinical Genome Resource. N Engl J Med 2015;372:2235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majithia AR, Tsuda B, Agostini M et al. Prospective functional classification of all possible missense variants in PPARG. Nat Genet 2016;48:1570–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Findlay GM, Daza RM, Martin B et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature 2018;562:217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khera AV, Won HH, Peloso GM et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J Am Coll Cardiol 2016;67:2578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schafer S, de Marvao A, Adami E et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat Genet 2017;49:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts AM, Ware JS, Herman DS et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med 2015;7:270ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peloso GM, Nomura A, Khera AV et al. Rare Protein-truncating Variants in APOB, Lower LDL-C, and Protection Against Coronary Heart Disease. Circ Genom Precis Med 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang S, Lincoln SE, Kobayashi Y, Nykamp K, Nussbaum RL, Topper S. Sources of discordance among germ-line variant classifications in ClinVar. Genet Med 2017;19:1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.