Abstract
Objectives
To describe, for the first time (to the best of our knowledge), the genetic mechanisms of vancomycin resistance in clinical isolates of Clostridioides difficile ribotype 027.
Methods
Clinical isolates and laboratory mutants were analysed: genomically to identify resistance mutations; by transcriptional analysis of vanGCd, the vancomycin resistance operon encoding lipid II d-alanine-d-serine that is less bound by vancomycin than native lipid II d-alanine-d-alanine; by imaging of vancomycin binding to cell walls; and for changes in vancomycin bactericidal activity and autolysis.
Results
Vancomycin-resistant laboratory mutants and clinical isolates acquired mutations to the vanSR two-component system that regulates vanGCd. The substitutions impaired VanSR’s function, resulting in constitutive transcription of vanGCd. Resistance was reversed by silencing vanG, encoding d-alanine-d-serine ligase in the vanGCd operon. In resistant cells, vancomycin was less bound to the cell wall septum, the site where vancomycin interacts with lipid II. Vancomycin’s bactericidal activity was reduced against clinical isolates and laboratory mutants (64 and ≥1024 mg/L, respectively) compared with WT strains (4 mg/L). Truncation of the potassium transporter TrkA occurred in laboratory mutants, which were refractory to autolysis, accounting for their survival in high drug concentrations.
Conclusions
Ribotype 027 evolved first-step resistance to vancomycin by constitutively expressing vanGCd, which is otherwise silent. Experimental evolutions and bactericidal assays show that ribotype 027 can acquire mutations to drastically enhance its tolerance to vancomycin. Thus, further epidemiological studies are warranted to examine the extent to which vancomycin resistance impacts clinical outcomes and the potential for these strains to evolve higher-level resistance, which would be devastating.
Introduction
Clostridioides difficile infection (CDI) is the most common hospital-acquired infection in the USA and the leading cause of death due to gastroenteritis.1,2 Treatment response rates in patients with CDI, and in particular infections with the recent epidemic NAP1/027/BI, have decreased significantly, while the incidence of CDI has increased, making it an urgent public health threat.3,4 For >30 years, metronidazole and vancomycin have been the main treatments for CDI.5,6 Fidaxomicin was approved for CDI in 2010, but its clinical use has mainly been limited to recurrent infections.7 In the recent IDSA/SHEA 2017 guidelines for CDI management, metronidazole is no longer recommended as a first-line drug for adult CDI.8 Vancomycin usage is therefore likely to increase, which will inadvertently raise the selection pressure for the spread of vancomycin-resistant strains.
Vancomycin inhibits cell wall biosynthesis in dividing cells by binding to the C-terminal peptide of d-alanine-d-alanine (d-Ala-d-Ala) in the cell wall precursor lipid II and in the maturing cell wall.9,10C. difficile with reduced susceptibility to vancomycin has emerged11–14 and is defined by a breakpoint of >2 mg/L from EUCAST. For example, vancomycin-resistant C. difficile were reported from US surveillances by Tickler et al.11 (39.1% of 143 027/BI isolates from 2011 to 2013) and Snydman et al.12 (17.9% of 925 isolates from 2011 to 2012). Vancomycin concentration in stools is expected to be about 500–2000 mg/L [i.e. 500–2000× MIC for most strains (1 mg/L)] and is thought to mirror the drug’s concentration at the colonic site of infection.12,15,16 It is therefore anticipated that colonic concentrations of vancomycin will equally inhibit the growth of vancomycin-resistant strains even at higher MICs. However, this view is undermined by the fact that exact concentrations of vancomycin along the length of the colon are unknown. Furthermore, the genetic basis for resistance to vancomycin in clinical C. difficile is unknown, but could be the deciding factor on whether mutants with reduced susceptibility survive in colonic concentrations of vancomycin. Indeed, such is the case with VISA that survive in vancomycin concentrations ≥32-fold higher than the MICs.17,18
About 85% of C. difficile carry a functional vancomycin resistance vanG gene cluster (designated as vanGCd), which is closely related to the vanG operon in Enterococcus faecalis.19 Induction of this operon by vancomycin results in the production of an alternative lipid II bearing a d-alanine-d-serine (d-Ala-d-Ser) terminus that is ∼7-fold less bound by vancomycin compared with the d-Ala-d-Ala terminus.20,21 Expression of the operon in WT vancomycin-susceptible C. difficile leads to production of lipid II d-Ala-d-Ser. However, vancomycin-induced expression of vanGCd does not confer resistance in C. difficile, in contrast to E. faecalis, where induction of vanG causes resistance.19,22C. difficile vanGCd is therefore considered cryptic. Induction of vanG genes is thought to occur when the membrane-bound sensor kinase (VanSCd) detects vancomycin and, after undergoing ATP-dependent autophosphorylation, transfers its phosphoryl group to the cytoplasmic response regulator VanR.23 Phosphorylated VanR (VanR∼P) then binds upstream of van genes to increase transcription.23 In the absence of vancomycin, the VanS phosphatase activity attenuates gene expression by dephosphorylating VanR∼P.21 Several studies report that mutations to VanSR proteins cause constitutive expression or increased inducibility of van operons in enterococci.21,24 These mechanisms have not been reported in C. difficile. Herein, we describe the first, to the best of our knowledge, report of mutations to C. difficile VanSRCd that promote constitutive expression of vanGCd in vancomycin-resistant clinical isolates of ribotype 027. This phenotype was also recapitulated in laboratory mutants. We show that reduced susceptibility improves the in vitro survival of strains in vancomycin.
Materials and methods
Strains, growth and chemicals
The 027/BI strain C. difficile R20291 and 027/BI clinical isolates were cultivated in brain heart infusion (BHI) broth or agar at 37°C in a Whitley A35 anaerobic workstation (Don Whitley Scientific). Clinical strains in Table 1 were from hospitalized patients in the Texas Medical Center and a recent Israeli surveillance.13 Genotypes were assigned after genome sequencing, with ST1 indicative of ribotype 027. Vancomycin hydrochloride was from Sigma–Aldrich.
Table 1.
Vancomycin MICs and substitutions in VanSRCd for laboratory mutants and clinal isolates
| Strain or number of isolates with a mutation | MIC (mg/L) | Substitution in VanSR |
|
|---|---|---|---|
| VanSCd | VanRCd | ||
| Laboratory mutants | |||
| WT R20291 | 1 | – | – |
| WS2 | 8 | Arg314Leu | – |
| WS4 | 16 | Gly319Asp | – |
| Clinical isolates | |||
| 1a | 8 | Ser313Phe | – |
| 1b | 8 | Thr349Ile | – |
| 9c,d | 4–8e | – | Thr115Ala |
Eleven clinical isolates were analysed. They were from: the Texas Medical Center (aMT1470, bMT5006, cMT3678, cMT5055, cMT1349, cMT443, cMT201, cMT4887 and cMT3914); and Israel (d491858 and d490054).
Isolates from Israel had MICs of 8 mg/L, while those from the Texas Medical Center had MICs of 4 mg/L.
Susceptibility tests
MICs were determined on BHI broth or agar and Brucella agar with haemin (5 mg/L), vitamin K1 (10 mg/L) and 5% (v/v) sheep blood. Vancomycin ranged from 0.125 to 32 mg/L in doubling dilutions. Inocula of 105 cfu/spot or 105 cfu/mL were used. MICs were the lowest concentrations preventing visible growth after 24 h of incubation. Reduced susceptibility was interpreted according to the EUCAST breakpoint of vancomycin MIC >2 mg/L.
Selection of mutants via serial passage
Overnight R20291 colonies were resuspended in BHI broth (1 mL) to an OD600 of 0.8–1.0. At each passage, an aliquot of 0.01 mL (105–106 cfu/mL) was streaked onto BHI agars containing 0.25× to 8× MIC of vancomycin. After 48 h of incubation, all colonies from the highest concentration of vancomycin were resuspended into fresh broth to create a mixed population that was then passaged onto higher drug concentrations. There was no calculation of mutation frequencies at each passage step. The remaining suspension was stored as glycerol stocks.
Measurement of vancomycin’s bactericidal activity and autolysis
Bactericidal activity
Logarithmic cultures (OD600 of ∼0.2) were exposed to 0–1024 mg/L vancomycin and, after 24 h of incubation, viable counts were determined by serial dilution. Agars with ∼15% (w/v) charcoal were controls to test the antibiotic carry-over effect on viable counting.
Autolysis
This was performed25 on harvested mid-log cells (OD600 of ∼0.2) resuspended in 50 mM potassium buffer with 0.01% Triton X-100. Optical readings were recorded in a Biotek 2 plate reader under anaerobic conditions.
Imaging of changes in vancomycin cell wall binding
Vancomycin binding was visualized by staining with fluorescently labelled vancomycin (BODIPY® FL vancomycin, from Thermo Fisher). Cultures with an OD600 of ∼0.2 were mixed with 1 mg/L vancomycin and BODIPY® FL vancomycin for 20 min at 37°C. Next cells were centrifuged, washed three times with PBS and fixed with methanol at −20°C for 20 min, before washing with PBS. DNA was stained with DAPI (Sigma) at 2 mg/L for 2 min. Cells were aliquotted onto glass slides and air dried before fluorescence imaging with a DeltaVision Elite microscope.
Genome sequencing
Accession numbers are listed in Table S1 (available as Supplementary data at JAC Online).
General methodology
Multiplexed DNA libraries were sequenced by paired-end 2 × 150 bp on an Illumina NextSeq™500 platform at the Institute of Biosciences and Technology and University of Texas Dallas Genome Center. CLC Genomics Workbench version 12 was used for de novo assembly of paired-end reads of sequenced genomes. Assembled genomes were annotated with Rapid Annotation using Subsystem Technology (RAST). Sequence variations were identified using the ‘Quality-based variant detection’ tool in CLC Genomics Workbench with default parameters (≥10-fold coverage of the reference position and sequence variation ≥35% of mapped reads). Variations observed in ≥90% of mapped reads were manually analysed. Using the tool ‘Find low coverage’, regions with zero coverage were analysed, as they may represent possible deletions >1 nucleotide base pair.
Analysis of laboratory mutants
After de novo assembly of the R20291 parent genome in CLC Genomics Workbench, it was mapped to the reference genome for R20291 (FN545816.1) and annotated by RAST. Paired-end reads of R20291 mutants were de novo assembled, contigs were mapped to the parent genome and SNPs were detected. Genetic changes were confirmed by Sanger sequencing.
Analysis of clinical isolates
After assembly and annotation of paired-end reads, the genomes were subjected to MLST (https://pubmlst.org/cdifficile/), using the ‘MLST module’ in CLC Genomics Workbench, which assigned the clinical strains as ST1 clade-2, the ST of 027. Annotated genomes were mapped to R20291 and SNPs were confirmed as mentioned above.
Quantitative RT–PCR (qRT–PCR)
vanGCd transcription was assessed by qRT–PCR on cultures (OD600 of ∼0.2) before and after exposure to subinhibitory vancomycin (0.5 mg/L) for 15 min at 37°C. Bacterial RNAprotect™ reagent (QIAGEN) was added before RNA isolation with Qiagen’s RNeasy. qScript One-Step SYBR Green qRT–PCR Kit, ROX (Quantabio) and gene-specific primers were used to amplify genes in prepared cDNA in Applied Biosystems ViiA7. Transcripts were calculated using the comparative CT method (ΔΔCT method) and data were normalized to 16S rRNA. Primers are given in Table S2.
Genetic manipulations
Antisense RNA to vanGCd was expressed in vector pMSPT26 by anhydrotetracycline (0.25 mg/L). The 100 bp antisense spanned 50 bp upstream and downstream of the start codon and was synthesized and cloned into pMSPT by Genscript.
Homology modelling of the transcriptional regulator VanRCd: generation of the C. difficile VanRCd homology model
The homology model of R20291 VanRCd was generated from the response regulator PhoP (pdb ID 5ed4) of Mycobacterium tuberculosis.27 The model was built using Prime software in the 2018 Schrödinger modelling suite.28 Refinement was done by iterative energy calculations. Both monomer and homo-multimer DNA models were built using this algorithm with refinement of loop positions. Molecular dynamic simulations on a protein loop movement for WT and mutant VanRs are described in the Supplementary data available at JAC Online.
Results
VanSRCd mutations associated with reduced susceptibility
Experimentally evolved mutants acquire mutations in the vancomycin sensor (VanSCd)
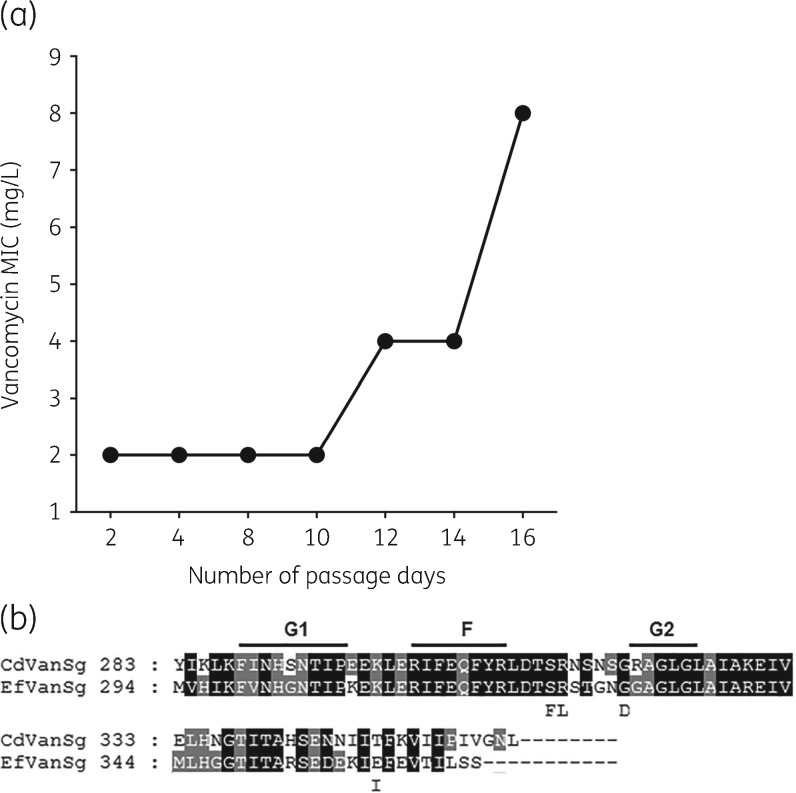
We first performed experimental evolutions using strain R20291 (MIC = 1 mg/L). By day 12 (i.e. fourth passage), mutants of R20291 grew beyond 2 mg/L, the EUCAST breakpoint (Figure 1a). By day 16, the population resisted up to 8 mg/L vancomycin. Isolated colonies (n = 12) had vancomycin MICs of 8–16 mg/L and, from this pool, two mutants (MICs = 8 and 16 mg/L) were genome sequenced. The first, designated as WS2 (MIC = 8 mg/L; Table 1), had an Arg314Leu substitution in VanSCd and a truncated potassium transporter (TrkA) due to a Gln26STOP mutation. The second, WS4 (MIC = 16 mg/L), carried a Gly319Asp substitution in VanSCd and the same Gln26STOP in TrkA. Comparison with enterococcal VanS homologues [i.e. VanS (C, D and B)] revealed that Arg314 and Gly319 occur in cytoplasmic F and G2 domains (Figure 1b). These domains accumulate mutations that increase expression of van operons by affecting the phosphatase activity of VanS.24,29 Disruptions to TrkA (e.g. Gln26STOP) were not seen in clinical strains, suggesting that it does not play a role in vanGCd-mediated resistance.
Figure 1.
(a) Serial evolution of R20291 showing time within which vancomycin-resistant mutants appeared. (b) Alignment of VanSCd with a VanS homologue from the E. faecalis vanG cluster; G1, F and G2 boxes (conserved motifs) are indicated above the alignment. Laboratory mutants had Arg314Leu and Gly319Asp substitutions, whereas Ser313Phe and Thr349Ile were found in clinical isolates. Except for position 349, all substitutions are located near the F and G2 boxes.
Clinical strains with substitutions in VanSCd
Strains MT1470 and MT5006 (Table 1), respectively, carried Ser313Phe and Thr349Ile substitutions in VanSCd (Table 1 and Figure 1b). Ser313Phe lies adjacent to Arg314 that was mutated in laboratory mutant WS2. Although Ile349 lies downstream of the G2 domain, it is probably associated with resistance, since WT Thr349 appears to be conserved in our BLAST analysis of C. difficile genomes in GenBank. Other polymorphisms in cell wall-associated genes are described and analysed in Table S3.
Clinical strains with an identical substitution in VanRCd
Independent clinical isolates (MICs = 4–8 mg/L) from the USA (n = 7) and Israel (n = 2) carried a Thr115Ala substitution in VanRCd. The mutation was confirmed by Sanger sequencing. BLASTing of ∼259 C. difficile strains in GenBank revealed that Thr115 is conserved in C. difficile VanRCd, suggesting that alanine is unlikely to be a drug-sensitive polymorphism. Additionally, WT Thr115 is present in E. faecalis VanR controlling the homologous vanG operon (e.g. accession no. Q6WRY8). Other polymorphisms in cell wall-associated genes are described and analysed in Table S3.
Constitutive expression of vanGCd is associated with reduced vancomycin susceptibility
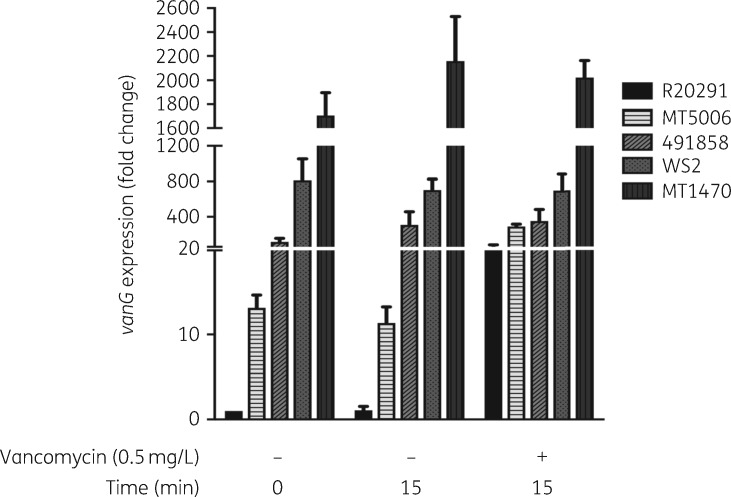
Compared with parental R20291, WS2 (Arg314Leu) constitutively transcribed vanGCd (Figure 2 and Figure S1). Without vancomycin, WS2 transcript levels for vanG (d-Ala-d-Ser ligase) and vanT (serine racemase) were on average 1032- and 813-fold higher than those of R20291; vanG and vanT are the first and last genes in the vanGCd operon.19 No notable increase in vanGCd expression occurred upon exposing WS2 to vancomycin (0.5 mg/L) for 15 min. In contrast, vancomycin induced substantial expression in R20291 (Figure 2 and Figure S1). Constitutive expression of vanGCd was seen in vancomycin-resistant clinical isolates carrying either VanRCd or VanSCd substitutions (Figure 2). These findings are consistent with reports that substitutions in VanSRCd alter regulation of cognate van genes.30
Figure 2.
Transcriptional analysis of vanGCd of various strains in the presence and absence of vancomycin (0.5 mg/L). Transcription in WT R20291 is induced only by vancomycin exposure for 15 min, whereas transcription was independent of vancomycin exposure in the isogenic laboratory mutant (WS2) and clinical isolates (MT5006, 491858 and MT1470).
Genetic validation of constitutive expression
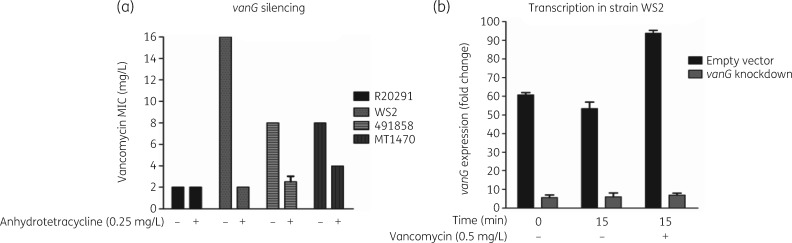
Association between constitutive expression and resistance was further tested by determining whether gene silencing of vanGCd reinstated susceptibility. The antisense (100 bp) was designed to target the ribosome binding site of d-Ala-d-Ser ligase encoded by vanG. Antisense induction by anhydrotetracycline (0.25 mg/L) reduced vancomycin MICs from 8–16 mg/L to 2–4 mg/L for the laboratory mutant WS2 and clinical strains MT1470 and 491858 (Figure 3a). qRT–PCR analysis of WS2 showed decreases of vanG’s mRNA transcripts by 8.93- and 13.40-fold, with or without vancomycin (Figure 3b). As control, antisensing had no impact on the vancomycin susceptibility of WT R20291. These findings substantiate the role of constitutive vanGCd expression in reducing susceptibility to vancomycin.
Figure 3.
Reversion of vancomycin resistance by antisense to vanG (d-Ala-d-Ser ligase). (a) Antisense induction increased the susceptibility of resistant strains to vancomycin (laboratory mutant WS2 and clinical isolates 491858 and MT1470); there was no effect on WT R20291. (b) Cellular confirmation that antisense decreased transcripts of vanG in the presence or absence of vancomycin, suggesting attenuation of constitutive resistance.
Expression of vanGCd alters binding of vancomycin to the cell wall
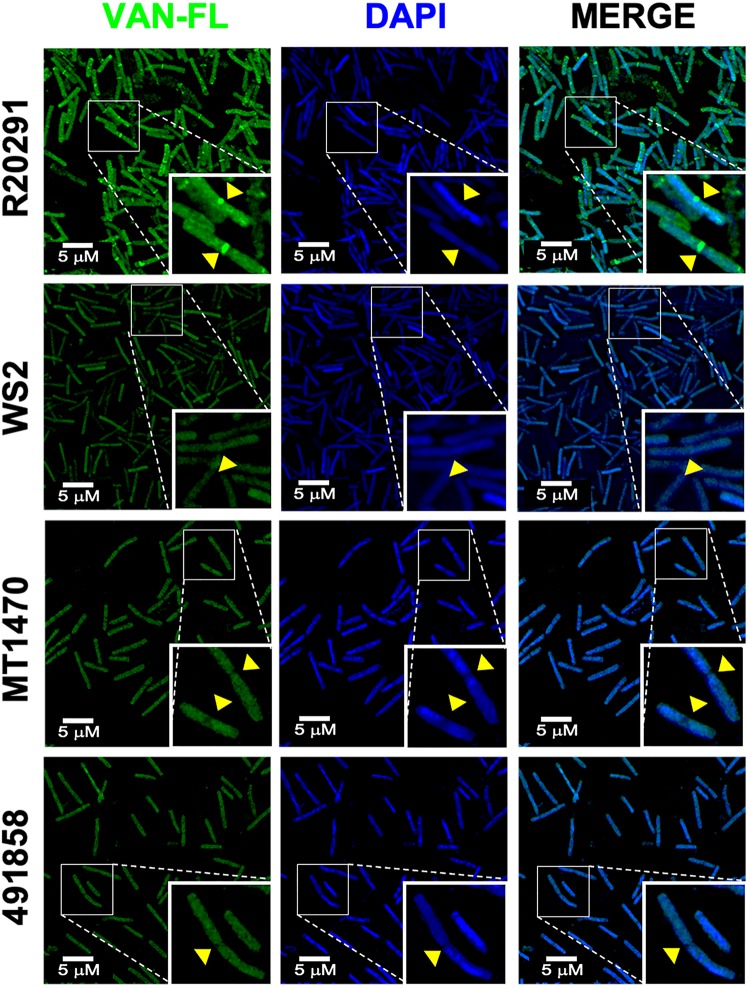
As expected, the septum and maturing cell walls of logarithmic R20291 were intensely stained by fluorescently labelled vancomycin (Figure 4). Staining was marked at the division septum, the point where lipid II accumulates and vancomycin is most active. In contrast, against vancomycin-resistant strains (WS2, MT1470 and 491858), notable reductions in vancomycin binding occurred, especially evident at the division septum.
Figure 4.
Analysis of changes in vancomycin binding. Cell walls were labelled with fluorescently labelled vancomycin (VAN-FL) and DNA with DAPI. Qualitatively, there is a reduction in VAN-FL staining intensity in the resistant cells compared with WT R20291. There was a lack of staining of the cell wall septum in vancomycin-resistant strains (WS2, MT1470 and 491858) compared with the WT. This is indicative of less binding by vancomycin to lipid II d-Ala-d-Ser at the division septum compared with lipid II d-Ala-d-Ala in R20291. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Structural explanation of VanRCd-mediated resistance
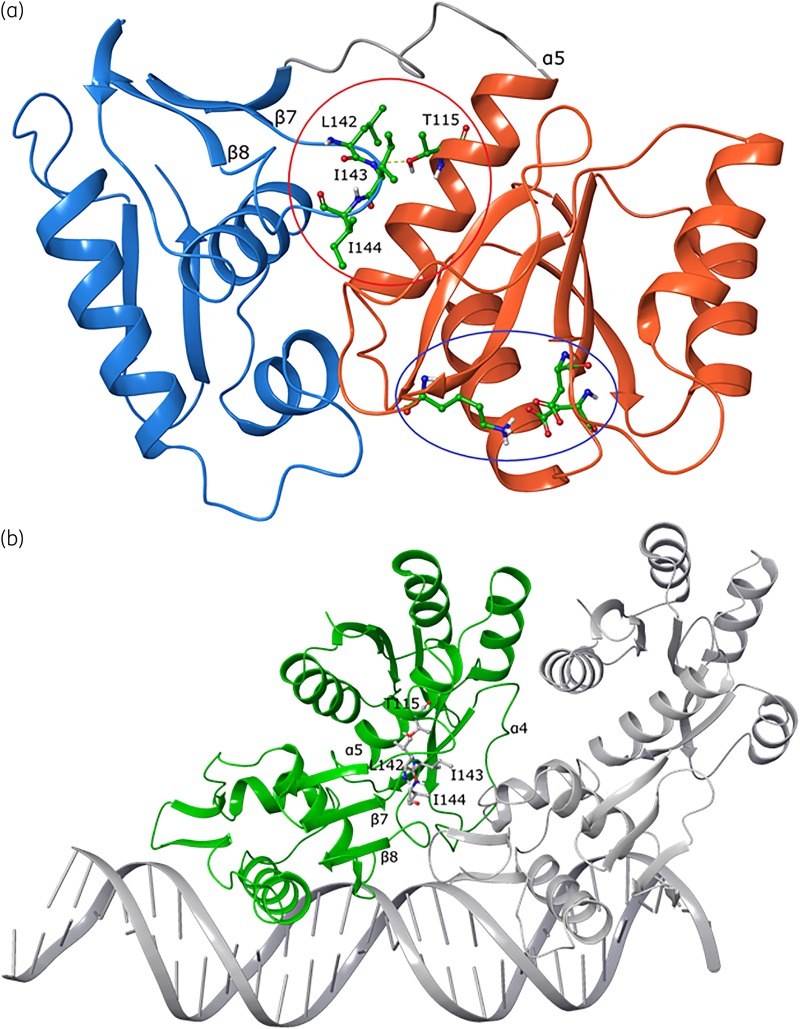
To predict the effect of Ala115, we developed a homology model of VanRCd from M. tuberculosis PhoP bound to DNA.27 Both VanR and PhoP are OmpR family regulators that probably share similar mechanisms of DNA interaction.31,32 VanR consists of a C-terminal effector domain that interacts with DNA and an N-terminal receiver domain containing the phosphorylation site acted on by VanS kinase/phosphatase. Upon phosphorylation, the two domains are brought together by a conformational change from an extended to a closed conformation, resulting in dimerization of VanR, producing its DNA binding conformation for transcription.23 In the monomeric model of C. difficile VanRCd, position 115 is located on the large α-helix (α5) of the receiver domain, ∼20 Å from the phosphorylation site (Figure 5a). In the DNA-bound dimer structure (Figure 5b), the substitution site is seen to interact with a conserved flexible loop located between two β-strands (β7 and β8) of the effector domain (Figure S2). This flexible loop is composed of several hydrophobic residues (e.g. Leu142, Ile143 and Ile144), which directly engage position 115 (Figure S3). Thus, we hypothesize that the flexible loop and position 115 are involved in stabilizing the dimeric conformation of VanRCd for DNA engagement. Furthermore, the change from polar threonine to non-polar alanine may facilitate energetically favourable hydrophobic contact with the lipophilic loop residues. In testing this hypothesis, our molecular dynamic simulations revealed that large loop movements (amino acids 141–149) in the Thr115 model were significantly decreased with substitution of Ala115 (Figure S4), suggesting better stabilization for DNA interaction.
Figure 5.
Structural analysis of mutant VanRCd in monomeric and DNA binding conformations built from the M. tuberculosis PhoP structure. (a) Monomeric VanRCd shows the DNA binding effector domain in blue ribbons, with the key helix that engages DNA at the far left. The receiver domain is shown in orange ribbons, with the conserved phosphorylation site (lysine and aspartic acid residues) shown as balls and sticks with green carbons (indicated by the blue oval). The resistance-conferring Thr115 substitution site is on the receiver domain and its interaction with a key loop containing lipophilic residues (Leu142, Ile143 and Ile144) on the effector domain is shown; the loop is indicated by the red circle and the residues are shown as balls and sticks with green carbons. (b) Interaction of VanRCd (monomer A, green) and second monomer B (PhoP, grey) with a segment of DNA. Shown as ball and stick representations are the Thr115 amino acid site on the receiver domain and interacting residues from the adjacent loop on the effector domain.
Survival advantage associated with reduced susceptibility to vancomycin
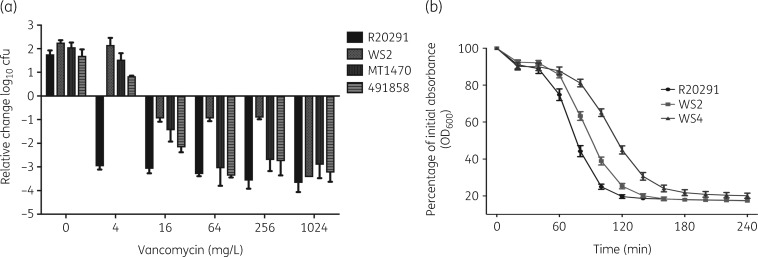
We exposed WT and resistant strains to the same incremental concentrations of drug between 1 and 1024 mg/L. As shown in Figure 6(a), vancomycin at ≥4 mg/L killed ∼3 log of R20291. In contrast, against its isogenic mutant WS2, only 0.82–1.11 log were killed by vancomycin concentrations of 16–256 mg/L. The difference in killing between R20291 and WS2 was statistically significant by unpaired t-tests (P < 0.01). However, both strains were equally killed by vancomycin at 1024 mg/L. Interestingly, against the resistant strain WS4, vancomycin at 256 and 1024 mg/L only killed 1.53 ± 0.34 and 1.49 ± 0.35 log, respectively. Against the clinical strains, vancomycin’s bactericidal activity (i.e. killing of ≥3 log) started at 64 mg/L. Responses to autolysis were further assessed for isogenic strains, showing a correlation with survival in vancomycin (Figure 6b); mutants WS2 and especially WS4 were refractory to autolysis compared with R20291.
Figure 6.
Alterations in bactericidal activity of vancomycin against laboratory mutants and clinical strains. (a) Against laboratory mutant WS2, vancomycin was only bactericidal (3 log reduction) at 1024 mg/L, but was bactericidal against clinical strains (MT1470 and 491858) at 64 mg/L. (b) Laboratory mutants WS2 and WS4 were less autolytic than their isogenic parent R20291. The reduced autolysis of WS2 and WS4 could contribute to tolerance to vancomycin. The MIC for WS2 is 8 mg/L and the MBC for WS2 is ∼1024 mg/L, while the lesser autolytic strain WS4 has an MIC of 16 mg/L and an MBC of >1024 mg/L.
Discussion
We report for the first time (to the best of our knowledge) mechanisms of vancomycin resistance in clinical C. difficile, focusing on ribotype 027 since reduced susceptibility to vancomycin is common in this lineage.11,12 Our results show that ribotype 027 has resolved how to acquire vancomycin resistance by constitutively expressing vanGCd, following mutations to VanSRCd. Without these mutations, vanGCd is phenotypically silent and does not exhibit vancomycin resistance.22 To explain why vanGCd is cryptic, Ammam et al.19 proposed that high cellular concentrations of UDP-MurNAc-pentapeptide[d-Ala] may be masking lesser abundant UDP-MurNAc-pentapeptide[d-Ser]. This is evident from the vanB mechanism, where increased production of d-Ala-d-lactate worsens vancomycin MICs for Enterococcus faecium.19,33 By extension, constitutive vanGCd may enrich the d-Ala-d-Ser peptidoglycan precursor pool, to reduce masking by d-Ala-d-Ala precursors. This may explain why we observed reduced binding of vancomycin to septa of resistant strains. However, measurement of peptidoglycan precursors will be required to validate these observations.
Even from the limited number of strains studied, ribotype 027 shows it can accumulate different resistance mutations in vanSRCd. Indeed, two clinical isolates (MT1470 and MT5006) from Houston (TX, USA) had different VanSCd substitutions, as was the case for two laboratory mutants. Nonetheless, clonal dissemination cannot be ruled out, as two isolates from Israel and seven from Houston had the same substitution in VanRCd. While these observations are new to C. difficile, constitutive van resistance mechanisms are established in enterococci. For example, Panesso et al.29 reported constitutive VanC resistance in Enterococcus gallinarum, associated with substitutions in VanSC as follows: Asp312Asn, Asp312Ala and Gly320Ser, corresponding to amino acid positions Asp311 and Gly319 in C. difficile (Figure 1b). In enterococci, the molecular basis for VanS-mediated constitutivity is due to mutations affecting the enzyme’s ATP-binding domain. This is thought to diminish its phosphatase activity, required to deplete VanR∼P that may arise from phosphorylation by cellular acetyl phosphates or alternative histidine kinases.24,34
Less studied is how VanR mutations cause constitutive expression.30 Interestingly, in E. faecium, a Gly140Glu substitution in VanR, from vanD genes, was thought to lock the effector domain into a DNA binding conformation.30 Gly140 is homologous to Gly141 in C. difficile and lies next to residues of the conserved flexible loop seen in C. difficile VanRCd and other OmpR regulators.30–32 Through molecular dynamics, we predict that the Thr115Ala change in VanRCd rigidified the effector domain for better DNA binding, resulting from improved interaction between position 115 and hydrophobic residues in the flexible loop. Experimental validation will, however, be required, as alternative or complementary theories exist, such as Ala115 playing a role in extending the half-life of VanR∼P or reducing dephosphorylation rates. In addition to vanGCd resistance, we anticipate that clinical C. difficile isolates are capable of acquiring other mechanisms of first-step vancomycin resistance. Indeed, Sanger sequencing of 10 colonies (MICs = 4 mg/L), from the mixed culture obtained at earlier timepoints of the passage (Figure 1a), did not show mutations in TrkA or VanS. This suggests that other mechanisms also arose during our in vitro evolution. This view is supported by Leeds et al.,35 who analysed vancomycin-resistant C. difficile, from in vitro evolutions, showing two mutants carried either a single mutation to RNA polymerase subunit C or three mutations to peptidoglycan glycosyltransferase (MurG), l-serine dehydrogenase (SdaB) and putative signal transduction phosphoesterase (CD3659). Furthermore, a recent report of a phenotypically silent vanB homologue carried by non-toxigenic C. difficile36 might be the first warning for potential emergence of transferable high-level vancomycin resistance, which would be disastrous for vancomycin’s use to treat CDI.
Vancomycin concentrations in patient stools are presumed to mirror those in the colon of CDI patients,16 but it is not known if the drug is homogeneously or heterogeneously distributed along the colon’s various segments. In a study of vancomycin faecal pharmacokinetics, Gonzales et al.16 reported that vancomycin stool concentrations decrease with high stool frequencies (≥4/day) in 15 patients given 125 mg of vancomycin q6h; conversely, the association between stool frequencies and vancomycin concentration was not confirmed in the study by Thabit and Nicolau.37 Both studies reflect the need for better ways to analyse factors that influence the deposition of vancomycin in the colon, especially in severe CDI. If there are intestinal niches where low drug concentrations might occur, this could favour the survival of mutants with low levels of resistance. Our analysis of vancomycin’s bactericidal activity leads us to anticipate two possible clinical scenarios. Firstly, low-level resistant mutants are unlikely to cause clinical failure during therapy, because they become growth inhibited at low concentrations. Conversely, these mutants may have a survival advantage under suboptimal concentrations that can occur in some patients.38 Secondly, based on laboratory mutants, it may be possible for some mutants to better survive high optimal concentrations. Such mutants might be a source for spores that contribute to endogenous recurrence. In the case of laboratory mutants, increased survival in vancomycin also correlated with reductions in autolysis. We speculate that loss of TrkA allowed laboratory mutants to switch from potassium to other osmoprotectants (e.g. amino acids and carbohydrates) that bacteria prefer to use for maintaining turgor pressure to prevent cell lysis.39–41 These results suggest that C. difficile has the potential to acquire genetic changes that mediate survival in high concentrations of vancomycin. Interestingly, lower rates of autolysis in some VISA are also associated with survival in vancomycin concentrations ≥32-fold higher than the MICs.17,18 We propose that measurement of MBCs rather than MICs may be a better way to evaluate if reduced susceptibility to vancomycin in C. difficile increases the risk for recurrent CDI, following vancomycin therapy. However, animal efficacy and pharmacokinetic studies in CDI rodent models will be required, to support our in vitro observations, to directly evaluate survival of resistant strains in colonic concentrations of vancomycin. The results of such animal experiments along with clinical epidemiological surveys will improve current limited understanding of the extent to which vancomycin-resistant C. difficile could influence treatment outcomes.
In conclusion, this study explored the genetic basis and survivability of vancomycin-resistant C. difficile, from an in vitro perspective, using a small sample set of laboratory mutants and clinical strains from two geographical locations. It suggests that further studies are needed to explore cell wall-associated metabolism in ribotype 027, as it appears that this organism may be evolving genetic changes in cell wall-associated genes (Table S3), for which the impact on its physiology and cell wall-active antibiotics is unknown. We also suggest that C. difficile with reduced susceptibility to vancomycin should not be ignored, as their increasing prevalence coupled with growing use of vancomycin may well provide a gateway for accumulation of mutations that produce higher-level resistance.
Supplementary Material
Acknowledgements
We are grateful to Dr Wenwen Huo, presently at Tufts University, for genome coverage quality analysis on initial sequences.
Funding
This work was in part funded by grants R56AI126881 and R01AI139261 to J.G.H. and R01AI116610 to K.L.P. from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health, with additional funds from the Texas A&M Health Science Center. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Transparency declarations
None to declare.
References
- 1. Hall AJ, Curns AT, McDonald LC. et al. The roles of Clostridium difficile and norovirus among gastroenteritis-associated deaths in the United States, 1999–2007. Clin Infect Dis 2012; 55: 216–23. [DOI] [PubMed] [Google Scholar]
- 2. Lessa FC, Mu Y, Bamberg WM. et al. Burden of Clostridium difficile infection in the United States. N Engl J Med 2015; 372: 825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weiner LM, Fridkin SK, Aponte-Torres Z. et al. Vital signs: preventing antibiotic-resistant infections in hospitals – United States, 2014. MMWR Morb Mortal Wkly Rep 2016; 65: 235–41. [DOI] [PubMed] [Google Scholar]
- 4. Shah DN, Chan FS, Kachru N. et al. A multi-center study of fidaxomicin use for Clostridium difficile infection. Springerplus 2016; 5: 1224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen SH, Gerding DN, Johnson S. et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 2010; 31: 431–55. [DOI] [PubMed] [Google Scholar]
- 6. Johnson S, Louie TJ, Gerding DN. et al. Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: results from two multinational, randomized, controlled trials. Clin Infect Dis 2014; 59: 345–54. [DOI] [PubMed] [Google Scholar]
- 7. Vivancos-Gallego MJ, Jimenez-Lopez MA, Gioia F. et al. A scoring system for prescribing fidaxomicin in Clostridium difficile infection. Enferm Infecc Microbiol Clin 2018; 36: 34–7. [DOI] [PubMed] [Google Scholar]
- 8. McDonald LC, Gerding DN, Johnson S. et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018; 66: 987–94. [DOI] [PubMed] [Google Scholar]
- 9. Hu Q, Peng H, Rao X.. Molecular events for promotion of vancomycin resistance in vancomycin intermediate Staphylococcus aureus. Front Microbiol 2016; 7: 1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hubbard BK, Walsh CT.. Vancomycin assembly: nature's way. Angew Chem Int Ed Engl 2003; 42: 730–65. [DOI] [PubMed] [Google Scholar]
- 11. Tickler IA, Goering RV, Whitmore JD. et al. Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States, 2011 to 2013. Antimicrob Agents Chemother 2014; 58: 4214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Snydman DR, McDermott LA, Jacobus NV. et al. U.S.-based national sentinel surveillance study for the epidemiology of Clostridium difficile-associated diarrheal isolates and their susceptibility to fidaxomicin. Antimicrob Agents Chemother 2015; 59: 6437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adler A, Miller-Roll T, Bradenstein R. et al. A national survey of the molecular epidemiology of Clostridium difficile in Israel: the dissemination of the ribotype 027 strain with reduced susceptibility to vancomycin and metronidazole. Diagn Microbiol Infect Dis 2015; 83: 21–4. [DOI] [PubMed] [Google Scholar]
- 14. Liao CH, Ko WC, Lu JJ. et al. Characterizations of clinical isolates of Clostridium difficile by toxin genotypes and by susceptibility to 12 antimicrobial agents, including fidaxomicin (OPT-80) and rifaximin: a multicenter study in Taiwan. Antimicrob Agents Chemother 2012; 56: 3943–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Freeman J, Vernon J, Morris K. et al. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect 2015; 21: 248.e9–16. [DOI] [PubMed] [Google Scholar]
- 16. Gonzales M, Pepin J, Frost EH. et al. Faecal pharmacokinetics of orally administered vancomycin in patients with suspected Clostridium difficile infection. BMC Infect Dis 2010; 10: 363.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cazares-Dominguez V, Cruz-Cordova A, Ochoa SA. et al. Vancomycin tolerant, methicillin-resistant Staphylococcus aureus reveals the effects of vancomycin on cell wall thickening. PLoS One 2015; 10: e0118791.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones RN. Microbiological features of vancomycin in the 21st century: minimum inhibitory concentration creep, bactericidal/static activity, and applied breakpoints to predict clinical outcomes or detect resistant strains. Clin Infect Dis 2006; 42 Suppl 1: S13–24. [DOI] [PubMed] [Google Scholar]
- 19. Ammam F, Meziane-Cherif D, Mengin-Lecreulx D. et al. The functional vanGCd cluster of Clostridium difficile does not confer vancomycin resistance. Mol Microbiol 2013; 89: 612–25. [DOI] [PubMed] [Google Scholar]
- 20. Arthur M, Reynolds P, Courvalin P.. Glycopeptide resistance in enterococci. Trends Microbiol 1996; 4: 401–7. [DOI] [PubMed] [Google Scholar]
- 21. Depardieu F, Bonora MG, Reynolds PE. et al. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol Microbiol 2003; 50: 931–48. [DOI] [PubMed] [Google Scholar]
- 22. Ammam F, Marvaud JC, Lambert T.. Distribution of the vanG-like gene cluster in Clostridium difficile clinical isolates. Can J Microbiol 2012; 58: 547–51. [DOI] [PubMed] [Google Scholar]
- 23. Depardieu F, Courvalin P, Kolb A.. Binding sites of VanRB and σ70 RNA polymerase in the vanB vancomycin resistance operon of Enterococcus faecium BM4524. Mol Microbiol 2005; 57: 550–64.15978084 [Google Scholar]
- 24. Depardieu F, Courvalin P, Msadek T.. A six amino acid deletion, partially overlapping the VanSB G2 ATP-binding motif, leads to constitutive glycopeptide resistance in VanB-type Enterococcus faecium. Mol Microbiol 2003; 50: 1069–83. [DOI] [PubMed] [Google Scholar]
- 25. Wydau-Dematteis S, El Meouche I, Courtin P. et al. Cwp19 is a novel lytic transglycosylase involved in stationary-phase autolysis resulting in toxin release in Clostridium difficile. MBio 2018; 9: e00648-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marreddy RKR, Wu X, Sapkota M. et al. The fatty acid synthesis protein enoyl-ACP reductase II (FabK) is a target for narrow-spectrum antibacterials for Clostridium difficile infection. ACS Infect Dis 2019; 5: 208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He X, Wang L, Wang S.. Structural basis of DNA sequence recognition by the response regulator PhoP in Mycobacterium tuberculosis. Sci Rep 2016; 6: 24442.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrödinger Release 2018-3: Prime. Schrödinger, LLC, 2018.
- 29. Panesso D, Abadia-Patino L, Vanegas N. et al. Transcriptional analysis of the vanC cluster from Enterococcus gallinarum strains with constitutive and inducible vancomycin resistance. Antimicrob Agents Chemother 2005; 49: 1060–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Depardieu F, Kolbert M, Pruul H. et al. VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother 2004; 48: 3892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson VL, Wu T, Stock AM.. Structural analysis of the domain interface in DrrB, a response regulator of the OmpR/PhoB subfamily. J Bacteriol 2003; 185: 4186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buckler DR, Zhou Y, Stock AM.. Evidence of intradomain and interdomain flexibility in an OmpR/PhoB homolog from Thermotoga maritima. Structure 2002; 10: 153–64. [DOI] [PubMed] [Google Scholar]
- 33. Arthur M, Depardieu F, Reynolds P. et al. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol 1996; 21: 33–44. [DOI] [PubMed] [Google Scholar]
- 34. Wright GD, Holman TR, Walsh CT.. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 1993; 32: 5057–63. [DOI] [PubMed] [Google Scholar]
- 35. Leeds JA, Sachdeva M, Mullin S. et al. In vitro selection, via serial passage, of Clostridium difficile mutants with reduced susceptibility to fidaxomicin or vancomycin. J Antimicrob Chemother 2014; 69: 41–4. [DOI] [PubMed] [Google Scholar]
- 36. Knight DR, Androga GO, Ballard SA. et al. A phenotypically silent vanB2 operon carried on a Tn1549-like element in Clostridium difficile. mSphere 2016; 1: e00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thabit AK, Nicolau DP.. Impact of vancomycin faecal concentrations on clinical and microbiological outcomes in Clostridium difficile infection. Int J Antimicrob Agents 2015; 46: 205–8. [DOI] [PubMed] [Google Scholar]
- 38. Johnson S, Homann SR, Bettin KM. et al. Treatment of asymptomatic Clostridium difficile carriers (fecal excretors) with vancomycin or metronidazole. A randomized, placebo-controlled trial. Ann Intern Med 1992; 117: 297–302. [DOI] [PubMed] [Google Scholar]
- 39. Sleator RD, Hill C.. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 2002; 26: 49–71. [DOI] [PubMed] [Google Scholar]
- 40. Dinnbier U, Limpinsel E, Schmid R. et al. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch Microbiol 1988; 150: 348–57. [DOI] [PubMed] [Google Scholar]
- 41. Czarny TL, Perri AL, French S. et al. Discovery of novel cell wall-active compounds using PywaC, a sensitive reporter of cell wall stress, in the model gram-positive bacterium Bacillus subtilis. Antimicrob Agents Chemother 2014; 58: 3261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.