Abstract
Background
Pseudomonas aeruginosa is an opportunistic bacterium that infects the airways of cystic fibrosis patients, surfaces of surgical and burn wounds, and indwelling medical devices. Patients are prone to secondary fungal infections, with Candida albicans being commonly co-isolated with P. aeruginosa. Both P. aeruginosa and C. albicans are able to form extensive biofilms on the surfaces of mucosa and medical devices.
Objectives
To determine whether the presence of C. albicans enhances antibiotic tolerance of P. aeruginosa in a dual-species biofilm.
Methods
Single- and dual-species biofilms were established in microtitre plates and the survival of each species was measured following treatment with clinically relevant antibiotics. Scanning electron microscopy and confocal microscopy were used to visualize biofilm structure.
Results
C. albicans enhances P. aeruginosa biofilm tolerance to meropenem at the clinically relevant concentration of 5 mg/L. This effect is specific to biofilm cultures and is dependent upon C. albicans extracellular matrix polysaccharides, mannan and glucan, with C. albicans cells deficient in glycosylation structures not enhancing P. aeruginosa tolerance to meropenem.
Conclusions
We propose that fungal mannan and glucan secreted into the extracellular matrix of P. aeruginosa/C. albicans dual-species biofilms play a central role in enhancing P. aeruginosa tolerance to meropenem, which has direct implications for the treatment of coinfected patients.
Introduction
The majority of infections in humans are polymicrobial in nature, with common diseases no longer considered to be caused by a single aetiological agent.1 The most prevalent polymicrobial infections include periodontitis, gastroenteritis, diabetic foot wounds, burn wounds and biofilm-associated infections.1,2
The genetic disease, cystic fibrosis (CF), is characterized by thickening of the mucus layer lining the endothelium of the respiratory tract, which provides an ideal environment for microbial colonization.3 Reduced mucociliary clearance enables these microorganisms to persist and form polymicrobial biofilms on the mucosa of the lower respiratory tract.1 The CF lung is a major site of interaction between Pseudomonas aeruginosa and Candida albicans.4,5 Around 70% of CF patients become chronically infected with P. aeruginosa by the age of 30,6 with C. albicans isolated from up to 75% of CF patients,7 although sputum samples are often contaminated with microbes from the upper respiratory tract and oral cavity.8 However, simultaneous colonization has been linked to severer clinical outcomes,9,10 due to accelerated decline in lung function and worsening of disease progression.10,11
Biofilms are structured communities of microbial cells ensnared within a matrix of extracellular polymeric substances.12,13 Biofilms are formed by bacterial and fungal species and an estimated 65%–80% of all microbial infections in humans are biofilm related.2,14 This has important clinical implications as the MICs of antimicrobials for biofilm cells can be 100–1000 times greater than for planktonic cells.15,16 Antimicrobial resistance in microorganisms poses an increasing challenge to public health worldwide,17,18 making biofilms a particularly relevant topic of research.
Previous work on interactions between P. aeruginosa and C. albicans has focused predominantly on physical and molecular interactions and their effects on growth, morphology and virulence.19–24 However, little is known of how their interactions affect antimicrobial drug efficacy. Studies on mono-species C. albicans and P. aeruginosa biofilms have linked biofilm extracellular matrix (ECM) material to antimicrobial drug inhibition. For example, the fungal polysaccharide β-1,3-glucan sequesters the antifungal fluconazole,25 whilst the P. aeruginosa exopolysaccharides Pel and Psl are implicated in the inhibition of various antibiotics, including tobramycin.26 Therefore, a greater understanding of the impact of this cross-kingdom interaction on antimicrobial tolerance is of great clinical importance.
Meropenem is a first-line antibiotic for treating Pseudomonas infections in the CF lung.27 Meropenem is a carbapenem β-lactam that targets PBPs within Gram-negative bacteria, causing inhibition of cell wall peptidoglycan synthesis, ultimately leading to osmotic lysis of bacterial cells.28,29 Meropenem is administered intravenously as a 15–30 min infusion of 1–2 g (adult dose), thrice daily for 2 weeks.30 When P. aeruginosa biofilms are treated with clinical doses of meropenem, only bacteria at the biofilm peripheries are killed, whilst cells closer to the base remain viable.31 In patients, the meropenem concentration found in epithelial lining fluid 1 h post-treatment is 5.3 mg/L,30 with the clinical breakpoint of Pseudomonas being >8 mg/L.32 Therefore, slight deviations in tolerance of P. aeruginosa to meropenem could impede clearance of the infection. Here, we observed that P. aeruginosa/C. albicans dual-species biofilms displayed enhanced tolerance to meropenem. This protection was provided through active secretion of fungal ECM components, specifically mannan and β-glucan. Therefore, co-colonization of P. aeruginosa and C. albicans within the CF lung may result in small reservoirs of protected P. aeruginosa, which could survive antimicrobial treatment and reseed the infection site.
Materials and methods
Strains and growth conditions
Strains of P. aeruginosa and Candida species used in this study are listed in Table 1. P. aeruginosa strains were maintained on, and cultured in, Miller-modified LB and C. albicans strains in yeast extract peptone dextrose (YPD) medium. Both were grown at 37°C, with aeration at 200 rpm. Antimicrobials (from Sigma–Aldrich, UK) were used at the following concentrations (mg/L): meropenem, 0, 1, 2.5, 5 and 10; ceftazidime, 0 and 5; ciprofloxacin, 0 and 0.05; tobramycin, 0 and 2; and fluconazole, 0, 250, 500, 750 and 1000.
Table 1.
Bacterial and fungal strains used in this study
| Strain | Common name | Genotype | Reference/source |
|---|---|---|---|
| P. aeruginosa strains | |||
| ATCC 15692 | PAO1 | WT | ATCC |
| Midlands 1 | Midlands 1 | clinical isolate | 48 |
| C. albicans strains | |||
| SC5314 | SC5314 | type strain | 56 |
| NGY152 | NGY152 | ura3Δ::imm34/ura3Δ::imm434; RPS1/rps1Δ::URA3 | 57 |
| NGY355 | pmr1Δ | ura3Δ::imm434/ura3Δ::imm434; pmr1Δ::hisG/pmr1Δ::hisG; RPS10/rps10Δ::URA3 | 58 |
| NGY356 | pmr1Δ + PMR1 | ura3Δ::imm434/ura3Δ::imm434; pmr1Δ::hisG/pmr1Δ::hisG; RPS1/rps1Δ::CIp10-PMR1 | 58 |
| CDH15 | mnn4Δ | ura3Δ::imm434/ura3Δ::imm434; mnn4Δ::hisG/mnn4Δ::hisG; RPS10::URA3 | 57 |
| CDH13 | mnn4Δ + MNN4 | ura3Δ::imm434/ura3Δ::imm434; mnn4Δ::hisG/mnn4Δ::hisG; RPS10:: [CIp10-MNN4-URA3]n | 57 |
| NGY582 | mnn2Δ | ura3Δ::imm434/ura3Δ::imm434; mnn2Δ::dpi200/mnn2Δ::dpi200; RPS1/rps1Δ::CIp10 | 46 |
| NGY583 | mnn2Δ + MNN2 | ura3Δ::imm434/ura3Δ::imm434; mnn2Δ::dpl200/mnn2Δ::dpl200; RPS1/rps1Δ::CIp10-MNN2 | 46 |
| NGY600 | Δmnn2–26 | ura3Δ::imm434/ura3Δ::imm434; mnn2Δ::dpl200/mnn2Δ::dpl200; mnn22Δ::dpl200/mnn2Δ::dpl200; mnn23Δ::dpl200/mnn23Δ::dpl200; mnn24Δ::dpl200/mnn24Δ::dpl200; mnn26Δ::dpl200/mnn26Δ::dpl200; mnn21Δ::dpl200/mnn21Δ::dpl200; RPS1/rps1Δ::CIp10 | 46 |
| NGY337 | mnt1Δ/mnt2Δ | ura3Δ::imm434/ura3Δ::imm434; mnt1Δ::hisG/mnt1Δ::hisG; mnt2Δ:: hisG/mnt2Δ::hisG; RPS10/rps10Δ::CIp10 | 55 |
| NGY335 | mnt1Δ/mnt2Δ + MNT1 | ura3Δ::imm434/ura3Δ::imm434; mnt1Δ::hisG/mnt1Δ::hisG; mnt2Δ:: hisG/mnt2Δ::hisG; RPS10/rps10Δ::CIp10-MNT1 | 55 |
| Non-albicans Candida strains | |||
| WU284 | C. dubliniensis | WT | 59 |
| CAY676 | C. tropicalis | type strain | ATCC |
| CLIB214 | C. parapsilosis | type strain | 60 |
| AM16/0701 | C. krusei | clinical isolate | D. MacCallum, University of Aberdeen, Scotland, UK |
| ATCC 2001 | C. glabrata | type strain | ATCC |
Formation of dual-species biofilms
Biofilms were grown in 96-well plates as previously described.33 Briefly, cultures were washed twice in PBS and P. aeruginosa cultures diluted to OD600 of 0.2 and Candida strains diluted to 1 × 106 cells/mL in Mueller–Hinton broth (MHB) or DMEM supplemented with 1% l-glutamine. P. aeruginosa (2.4 × 106) and Candida (1 × 105) cells were incubated statically in flat-bottom 96-well plates for 2 h at 37°C to enable attachment, then non-adhered cells were removed and replaced with fresh medium. After 24h, the medium was replaced with fresh medium containing the appropriate amount of antimicrobial, or vehicle control, for an additional 18 h. To disrupt biofilms, medium was replaced with 100 μL of PBS containing 50 mg/L DNase and incubated at 37°C for 1 h. Biofilms were detached using a water bath sonicator, serially diluted 1 in 10 in PBS and plated onto cetrimide agar (to determine viable P. aeruginosa cfu) and YPD agar supplemented with 100 mg/L tetracycline (to determine viable Candida cfu). Experiments were performed with three technical and at least three biological replicates.
Formation of P. aeruginosa biofilms in the presence of dead C. albicans or ECM components
To inactivate C. albicans, stationary-phase cultures were washed with PBS and cells either heat-killed at 100°C in PBS for 1 h or fixed in 1 mL of 4% paraformaldehyde (PFA) at room temperature for 1 h. Cells were then washed with PBS and diluted in MHB to 1 × 106 cells/mL. Subsequently, biofilms were established and quantified as above.
Mono-species P. aeruginosa biofilms were established and grown for 24 h in MHB supplemented, at either 0 or 24 h, with 0.25 mg/mL glucan (from Saccharomyces cerevisiae), laminarin (from Laminaria digitata), mannan (from S. cerevisiae) or chitosan. All polysaccharides were obtained from Sigma–Aldrich, UK. After 24 h, medium was replaced with MHB containing the appropriate amount of antibiotic and biofilms were subsequently incubated and quantified as above.
Scanning electron microscopy of biofilms
Biofilms were prepared for scanning electron microscopy using a previously published protocol34 with modifications. Single- and dual-species biofilms were grown on cell culture-treated plastic coverslips (Thermo Fisher Scientific) in 24-well plates for 24 h, after which the MHB medium was replaced with MHB with or without 5 mg/L meropenem. At 48 h, coverslips were washed twice with PBS and samples fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer for 1 h, at 4°C. Samples were dehydrated using increasing ethanol concentrations (50%, 70%, 90% and 100%) twice for 15 min each. Ethanol was replaced with liquid CO2 and heated up to the critical point to dry the samples. Each coverslip was mounted on a stub and sputter-coated with platinum. Scanning electron microscopy images were captured using a Philips XL30 ESEM-FEG environmental scanning electron microscope.
Confocal microscopy of biofilms
Single- and dual-species biofilms with or without 5 mg/L meropenem were grown as above, scaled up to a final volume of 6 mL, in 6-well plates. Medium was replaced with PBS containing 5 mg/L propidium iodide (stains dead cells), 1 μM Syto 9 (dyes DNA) and 3 mg/L calcofluor white (stains fungal cell wall chitin) and incubated at 4°C in the dark for 1 h. Biofilms were then fixed by adding 4% PFA, incubated at 4°C in the dark for 1 h and then washed twice with PBS. Confocal microscopy was performed using a Leica SP8 system equipped with a Leica DM6 upright microscope, a ×40/0.80 objective and 402, 488 and 561 nm lasers. Biofilms were imaged directly in wells with a water-dipping lens. Z-stack scans were taken at two or three different areas within each well and processed with Fiji and LASx software.
Planktonic assay
P. aeruginosa (2.4 × 107) and C. albicans (1 × 106) cells were added to 14 mL vent-capped culture tubes in a final volume of 2 mL MHB. Cultures were incubated for 3 h at 37°C with aeration at 200 rpm; the appropriate amount of antibiotic was added and cultures incubated for an additional 18 h. Cultures were sonicated in a water bath sonicator and serially diluted and plated for viable counts. Experiments were performed with three technical and four biological replicates.
Determination of P. aeruginosa susceptibility to meropenem
MICs of meropenem were determined for P. aeruginosa strains according to the standardized broth microdilution method using MHB.35 Concentrations of meropenem used were 0, 1, 2, 4, 8, 16 and 32 mg/L. MICs were determined for P. aeruginosa strains using cells grown on LB agar and cells recovered from biofilms. MICs were the lowest concentrations of meropenem that caused no visible growth.
Statistical analysis
Statistical analyses were done using GraphPad Prism 8.0.0 software. Data were analysed using two-way ANOVA and Holm–Sidak’s multiple comparisons test.
Results
C. albicans increases the tolerance of P. aeruginosa to meropenem in dual-species biofilms
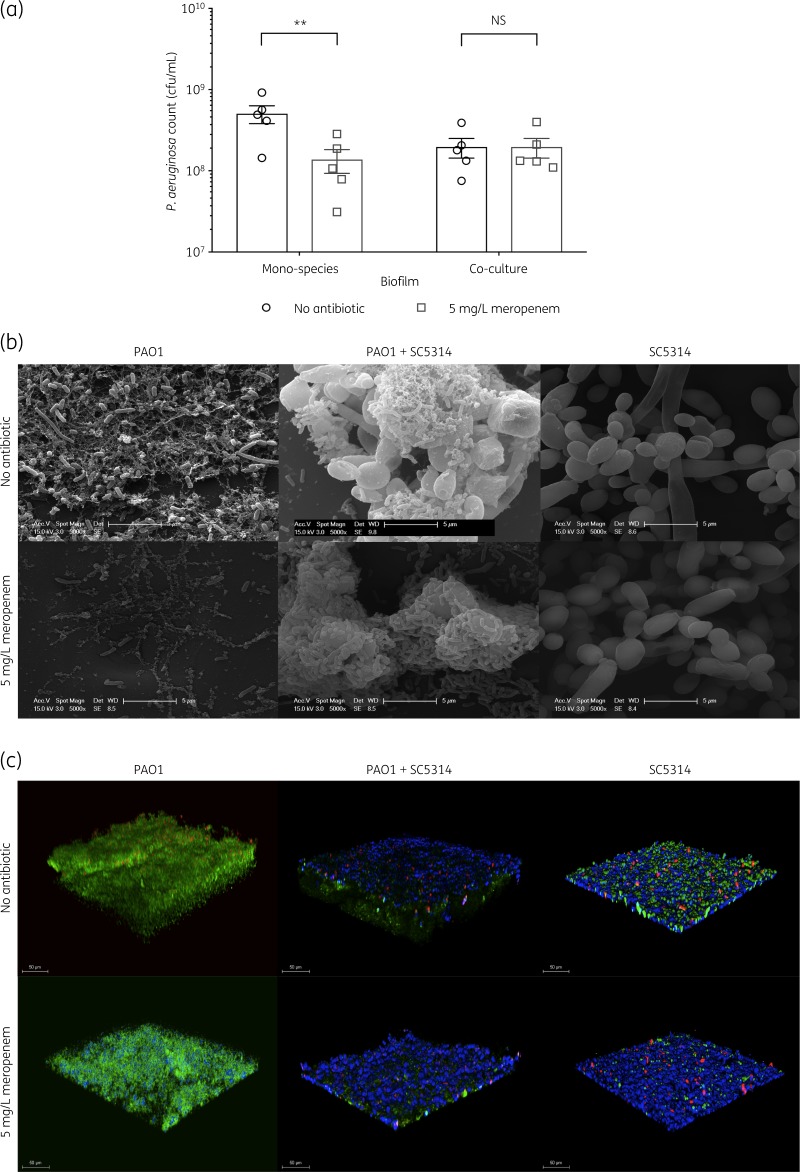
To determine whether the presence of C. albicans within a P. aeruginosa biofilm can enhance tolerance of P. aeruginosa to meropenem, preformed mono-species (P. aeruginosa) and dual-species (P. aeruginosa/C. albicans) biofilms were treated with meropenem and P. aeruginosa survival quantified by viable counts. Given that the concentration of meropenem in the lung immediately after administration is between 5 and 6 mg/L,30 this drug concentration was the focus of the study. The viability of P. aeruginosa mono-species biofilms was reduced to 25.35% when treated with 5 mg/L meropenem, indicating P. aeruginosa biofilm cells are susceptible to meropenem. Fewer P. aeruginosa cells were recovered from dual-species biofilms, which is likely due to nutrient competition. However, in the presence of C. albicans, meropenem was non-effective against P. aeruginosa in both MHB (Figure 1a) and DMEM (Figure S1, available as Supplementary data at JAC Online), indicating that this inter–kingdom interaction negatively affects meropenem efficacy.
Figure 1.
C. albicans increases the tolerance of P. aeruginosa to meropenem in a dual-species biofilm. (a) Preformed 24 h biofilms were incubated for 18 h in MHB containing no antibiotic or 5 mg/L meropenem. Data are the mean ± SEM from five biological replicates. Data were analysed using two-way ANOVA and Holm–Sidak’s multiple comparisons test (**P < 0.01). NS, not significant. (b) Scanning electron microscopy analysis of biofilms. Meropenem treatment of mono-species P. aeruginosa biofilms results in death of bacterial cells, whilst the presence of C. albicans in the dual-species biofilm enhances meropenem tolerance; the tight association of P. aeruginosa cells to fungal surfaces is visible. C. albicans alone is unaffected by meropenem. (c) 3D reconstructions of biofilms from confocal z-stacks. Red indicates propidium iodide stain (dead cells), green indicates Syto 9 dye (DNA) and blue indicates calcofluor white stain (chitin).
To visualize the structure of the biofilms, samples were analysed by scanning electron microscopy and confocal microscopy. Mono-species P. aeruginosa biofilms were significantly reduced in the presence of meropenem, while in the meropenem-treated dual-species biofilms, significant levels of P. aeruginosa colonized the fungal hyphae (Figure 1b), confirming the cfu data. In agreement with this, the biofilm thickness was lower in meropenem-treated dual-species biofilms (Figure 1c and Figure S2), indicating dense packing of bacterial cells against fungal hyphae, creating a more compact biofilm structure. Therefore, the presence of C. albicans enhances the tolerance of P. aeruginosa to meropenem.
To identify whether this dual-species interaction had any impact on antifungal resistance, biofilms were treated with fluconazole. C. albicans cells in dual-species biofilms showed similar susceptibility levels to fluconazole as untreated controls at all tested concentrations. Therefore, the presence of P. aeruginosa does not affect the antifungal activity of fluconazole under the tested conditions (Figure S3).
Meropenem tolerance is not maintained following subculture of P. aeruginosa biofilm cells
The selective pressure from antibiotic use increases the likelihood of cells developing resistance. The sessile nature and close proximity of biofilm cells promotes cell–cell interactions,36 increasing horizontal gene transfer and mutation frequencies relative to planktonic cells.37 Furthermore, the presence of C. albicans increases P. aeruginosa mutation rates.20 To determine whether the observed increase in meropenem tolerance of P. aeruginosa was due to selection for resistance mutations, the meropenem MICs for cells recovered from both P. aeruginosa mono- and dual-species biofilms that had been treated with 5 mg/L meropenem, or untreated, were determined by standard broth microdilution MIC assay and compared with the MIC for the starter culture. The MIC for P. aeruginosa under all tested conditions was 4 mg/L, suggesting that P. aeruginosa cells recovered from treated biofilms were not resistant to meropenem and, therefore, the observed increased tolerance was unlikely due to selection for resistance mutations.
Increased tolerance of P. aeruginosa to meropenem is specific to dual-species biofilms
Enhanced survival of P. aeruginosa as a result of interactions with C. albicans has previously been observed in planktonic cultures, through inter–kingdom communication via secreted metabolites.21 To determine whether the observed increase in meropenem tolerance of P. aeruginosa was specific to biofilms, P. aeruginosa susceptibility to meropenem in the presence of C. albicans was tested during planktonic growth. At all meropenem concentrations tested, there was no significant difference in P. aeruginosa survival whether in the presence or absence of C. albicans (Table 2), suggesting that C. albicans-mediated protection is biofilm specific.
Table 2.
Increased P. aeruginosa meropenem tolerance in the presence of C. albicans is biofilm specific
| Meropenem (mg/L) | Planktonic culture | P. aeruginosa count (cfu/mL), mean ± SEM | P |
|---|---|---|---|
| 0 | mono-species | 4.00 × 109 ± 4.14 × 108 | 0.9040 |
| co-culture | 3.81 × 109 ± 2.78 × 108 | ||
| 1 | mono-species | 2.19 × 109 ± 1.63 × 108 | 0.3218 |
| co-culture | 1.64 × 109 ± 2.97 × 108 | ||
| 2.5 | mono-species | 7.36 × 108 ± 8.39 × 107 | 0.9650 |
| co-culture | 8.12 × 108 ± 1.57 × 108 | ||
| 5 | mono-species | 8.75 × 107 ± 2.82 × 107 | 0.9650 |
| co-culture | 1.56 × 108 ± 4.92 × 107 |
Planktonic cultures were grown for 3 h and subsequently incubated for 18 h in MHB containing 0, 1, 2.5 or 5 mg/L meropenem. Data are the mean ± SEM from four biological replicates. Data were analysed using two-way ANOVA and Holm–Sidak’s multiple comparisons test.
Increased P. aeruginosa tolerance to meropenem is dependent on fungal viability
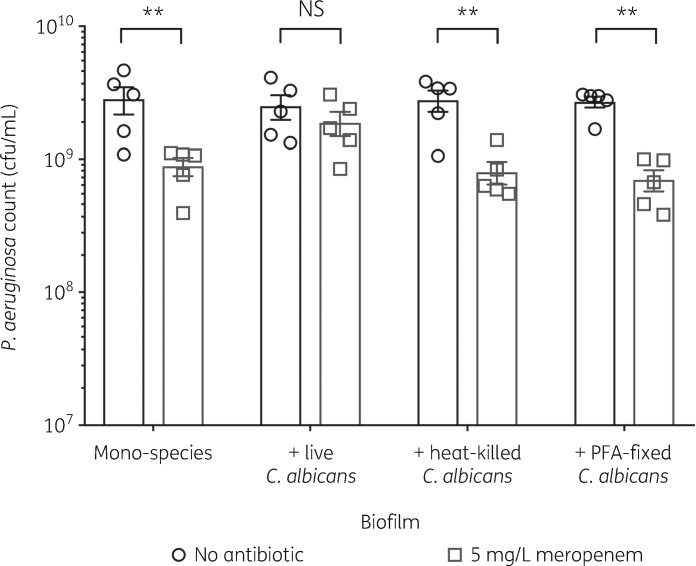
Production of biofilm ECM is an active process, involving secretion of glycoproteins, polysaccharides, lipids and nucleic acids.2 To determine whether the protective effect of C. albicans is mediated by an active or passive mechanism, P. aeruginosa biofilms were grown in the presence of either heat-killed C. albicans (disrupts the fungal cell wall, denatures proteins and causes cell lysis) or by fixing the C. albicans cells in 4% PFA (maintains cell structure). However, in the presence of heat-killed or PFA-fixed C. albicans, P. aeruginosa remained susceptible to meropenem (Figure 2), suggesting that C. albicans actively protects P. aeruginosa from meropenem.
Figure 2.
Increased tolerance to meropenem is dependent on fungal viability. Preformed 24 h biofilms were incubated for 18 h in MHB containing no antibiotic or 5 mg/L meropenem. Data are the mean ± SEM from five biological replicates. Data were analysed using two-way ANOVA and Holm–Sidak’s multiple comparisons test (**P <0.01). NS, not significant.
Candida dubliniensis also enhances P. aeruginosa meropenem tolerance in dual-species biofilms
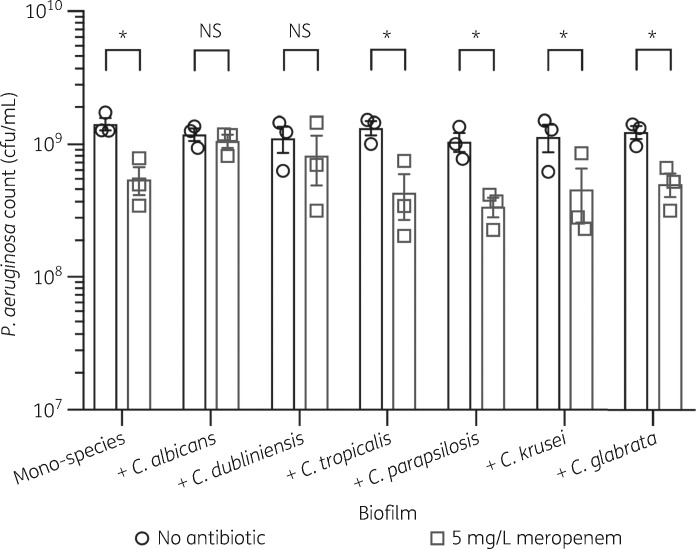
Only a few species of the Candida genus are associated with disease in humans, including C. albicans, C. dubliniensis, Candida tropicalis, Candida parapsilosis, Candida krusei and Candida glabrata.38 To determine whether other Candida species can also protect P. aeruginosa from meropenem, biofilms were grown in the presence of these clinically relevant non-albicans Candida (NAC) species. Of the NAC species tested, only C. dubliniensis increased the tolerance of P. aeruginosa to meropenem (Figure 3). C. dubliniensis is phylogenetically most closely related to C. albicans,39 indicating that C. albicans and C. dubliniensis may enhance P. aeruginosa meropenem tolerance in a similar manner.
Figure 3.
C. dubliniensis also enhances tolerance of P. aeruginosa to meropenem in dual-species biofilms. Preformed 24 h biofilms were incubated for 18 h in MHB containing no antibiotic or 5 mg/L meropenem. Data are the mean ± SEM from three biological replicates. Data were analysed using two-way ANOVA and Holm–Sidak’s multiple comparisons test (*P < 0.05). NS, not significant.
Fungal cell wall polysaccharides enhance P. aeruginosa tolerance to meropenem
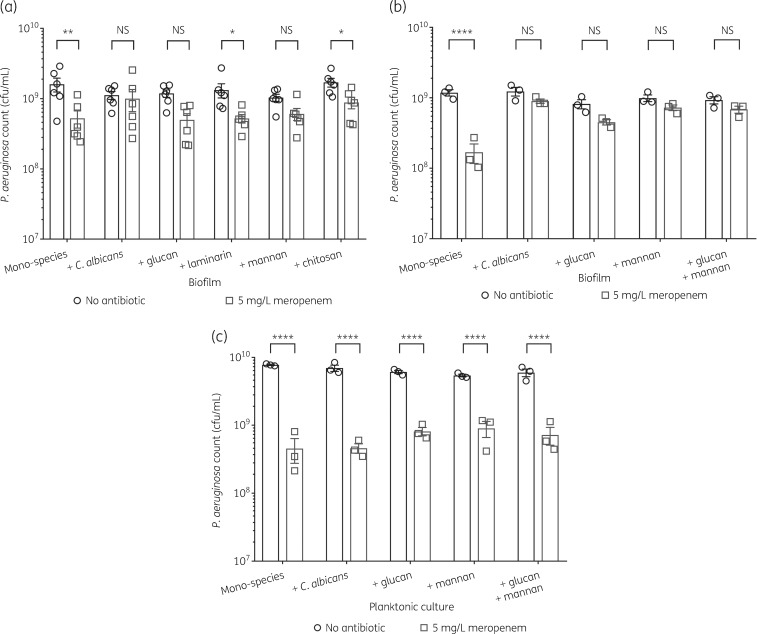
The secretion of ECM polymers, specifically polysaccharides, by C. albicans biofilm cells is linked to increased antifungal resistance of fungal biofilms.40,41 However, there is increasing evidence that fungal ECM polysaccharides also contribute to antibiotic resistance in dual-species fungal/bacterial biofilms.34,42,43 To determine whether secreted fungal cell wall polymers play a role in protecting P. aeruginosa, mono-species P. aeruginosa biofilms were grown in the presence of purified fungal cell wall polysaccharides, including glucan (a mix of β-1,3-glucan and β-1,6-glucan), laminarin (an isoform of β-1,3-glucan), mannan and chitosan (deacetylated chitin). Both mannan and glucan enhanced tolerance of P. aeruginosa to meropenem in biofilms (Figure 4a), but not in planktonic cultures (Figure 4c). To determine whether mannan and glucan have independent effects, P. aeruginosa biofilms were supplemented with glucan and mannan in combination. However, no additive effect was observed (Figure 4b). Mannan and glucan protected P. aeruginosa when added to mature biofilms at the same time as meropenem (Figure S4), suggesting that the polysaccharides may sequester or inhibit the activity of the drug. Therefore, C. albicans actively secretes mannan and/or glucan into the biofilm ECM, which protects P. aeruginosa from meropenem.
Figure 4.
Mannan and glucan enhance P. aeruginosa biofilm tolerance to meropenem. Fungal polysaccharides were used at a final concentration of 0.25 mg/mL. (a) Mannan and glucan enhance P. aeruginosa biofilm tolerance to 5 mg/L meropenem. Preformed 24 h biofilms were incubated for 18 h in MHB containing no antibiotic or 5 mg/L meropenem. Data are the mean ± SEM from six biological replicates. (b) The effects of mannan and glucan are not additive. (c) Enhanced meropenem tolerance from mannan and glucan is biofilm specific. Planktonic cultures supplemented with exogenous mannan and/or glucan were grown for 3 h and subsequently incubated for 18 h in MHB containing 5 mg/L meropenem. Data are the mean ± SEM from three biological replicates. Data were analysed using two-way ANOVA and Holm–Sidak’s multiple comparisons test (*P < 0.05, **P < 0.01 and ****P < 0.0001). NS, not significant.
C. albicans cell wall glycosylation is important for protection against meropenem
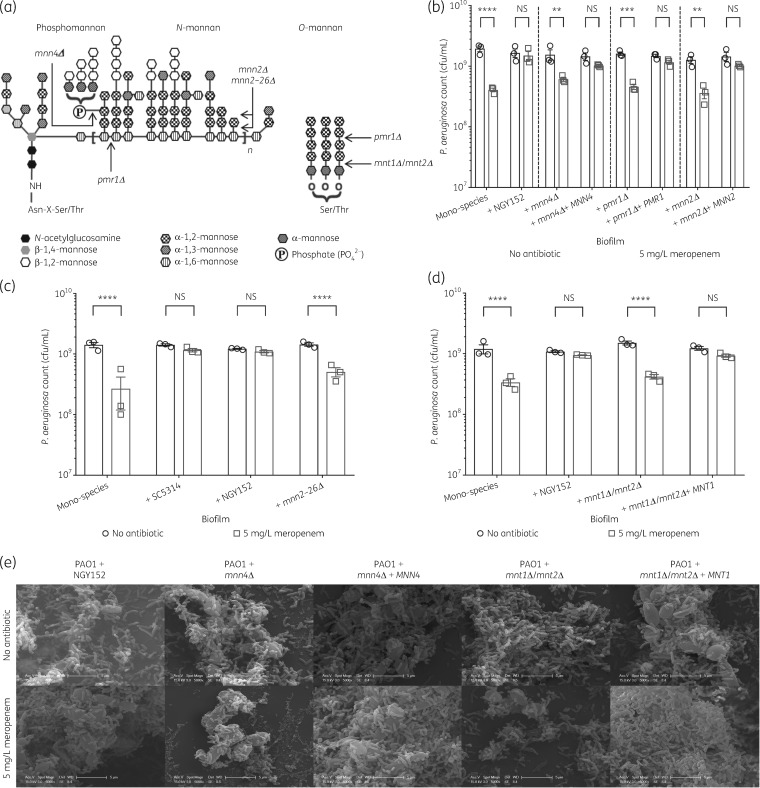
To further investigate the role of fungal mannan in meropenem tolerance, the ability of C. albicans cell wall glycosylation mutants to protect P. aeruginosa was quantified. C. albicans has two major forms of mannan, the extensively branched N-linked mannan and the simple, linear O-linked mannan. Deletion of genes involved in key glycosylation steps results in the incorporation of altered mannan epitopes in the cell wall (Figure 5a) and within the ECM.44–46 To elucidate the role of these glycosylation structures in protecting P. aeruginosa from meropenem, mutants defective in general protein glycosylation (pmr1Δ, ATPase required for transporting Ca2+ and Mn2+ ions into the Golgi), N-mannan phosphomannan incorporation (mnn4Δ), N-mannan side chain elaboration (mnn2Δ, mnn2–26Δ) and O-mannan biosynthesis (mnt1Δ/mnt2Δ) were tested for their ability to protect P. aeruginosa from meropenem. Deletion of genes required for N-mannan biosynthesis (mnn4, mnn2–26 and pmr1) reduced the ability of C. albicans to protect P. aeruginosa from meropenem (Figure 5b and c). Scanning electron microscopy analysis showed very few bacterial cells surviving meropenem treatment in dual-species biofilms with C. albicans glycosylation mutants (Figure 5e) but bacteria that did survive were closely adhered to the fungal cell surface, suggesting cell–cell adherence may play a role in meropenem tolerance. In agreement with this, deletion of genes involved in O-mannan biosynthesis (mnt1/mnt2), which have previously been shown to be involved in bacterial attachment to C. albicans,47 had the greatest impact on P. aeruginosa protection (Figure 5d and e). This indicates that protection of P. aeruginosa requires full elaboration of fungal mannan.
Figure 5.
C. albicans cell wall glycosylation is important for protection against meropenem. (a) Schematic diagram representing the structure of N-mannan (including phosphomannan) and O-mannan of C. albicans.44,46 The points of truncation of the mutants used in this study are indicated by arrows. The pmr1Δ mutant causes loss of a Golgi Ca2+/Mn2+-ATPase, affecting numerous mannosyltransferases, so the extent of truncation of the α-1,6-mannose backbone is variable.44,45 (b) N-mannan glycosylation is important for protection against meropenem. Preformed 24 h biofilms were incubated for 18 h in MHB containing no antibiotic or 5 mg/L meropenem. The N-mannan glycosylation mutants (mnn4Δ, pmr1Δ or mnn2Δ) inhibit the ability of C. albicans to protect P. aeruginosa. Tolerance to meropenem is restored in reconstituted control strains. (c) The mnn2–26Δ sextuple mutant, in which only the unsubstituted α-1,6-mannose backbone of N-mannan remains, inhibits the ability of C. albicans to protect P. aeruginosa. (d) O-mannan glycosylation is important for protection against meropenem. The mnt1Δ/mnt2Δ double mutant inhibits the ability of C. albicans to protect P. aeruginosa. Meropenem tolerance is restored when MNT1 is reconstituted. Data are the mean ± SEM from three biological replicates. Data were analysed using two-way ANOVA and Holm–Sidak’s multiple comparisons test (**P < 0.01, ***P < 0.001 and ****P < 0.0001 in panels b, c and d). NS, not significant. (e) Scanning electron microscopy analysis of biofilms. Deletion of genes required for fungal N-mannan biosynthesis (mnn4) or O-mannan biosynthesis (mnt1/mnt2) reduced the ability of C. albicans to protect P. aeruginosa from meropenem, as indicated by the reduction in the number of bacterial cells following meropenem treatment; the majority of surviving bacteria are in close contact with fungal cells. When the genes (MNN4 or MNT1) are reconstituted, the protective effect is restored, as evidenced by the abundance of P. aeruginosa cells coating the fungi in the meropenem-treated samples.
C. albicans does not enhance P. aeruginosa tolerance to other antibiotics
To determine whether the presence of C. albicans also affects tolerance of P. aeruginosa biofilm cells to other clinically relevant antibiotics, mono- and dual-species biofilms were treated with 5 mg/L ceftazidime, 0.05 mg/L ciprofloxacin, 2 mg/L tobramycin or a combination of 5 mg/L meropenem and 2 mg/L tobramycin. However, the presence of C. albicans did not provide protection against these antimicrobial treatments (Figure S5a and b), suggesting that the mechanism by which C. albicans confers enhanced tolerance is likely due to the chemical structure of meropenem.
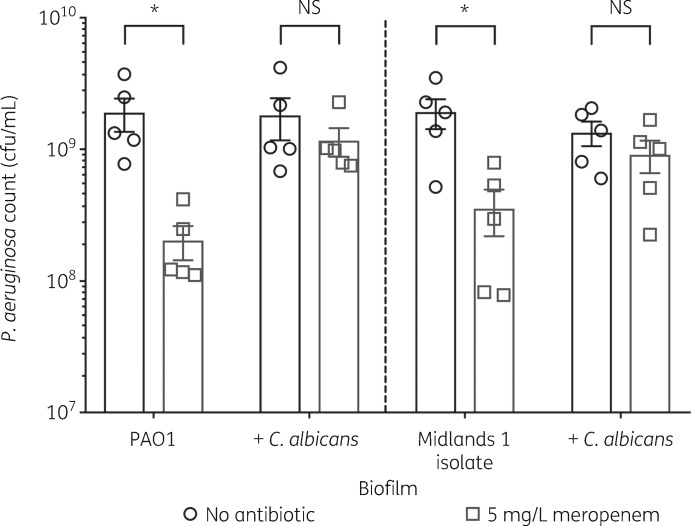
C. albicans protects P. aeruginosa CF isolates from meropenem
To explore the clinical relevance of the above findings, the ability of C. albicans to increase meropenem tolerance of clinical CF isolates was tested. Mono-species biofilms of the Midlands 1 CF isolate48 were susceptible to meropenem. However, P. aeruginosa survival was unaffected during growth in a dual-species biofilm with C. albicans (Figure 6), suggesting that in CF patients, co-colonization with C. albicans may increase P. aeruginosa tolerance to meropenem.
Figure 6.
C. albicans enhances meropenem tolerance of a P. aeruginosa CF isolate. Preformed 24 h biofilms were incubated for 18 h in MHB containing no antibiotic or 5 mg/L meropenem. Data are the mean ± SEM from five biological replicates. Data were analysed using two-way ANOVA and Holm–Sidak’s multiple comparisons test (*P < 0.05). NS, not significant.
Discussion
P. aeruginosa and C. albicans are commonly co-isolated from the sputum of CF patients,20 where the thickened mucus layer lining the endothelium of the lower respiratory tract provides an ideal environment for biofilm formation.3,7 Chronic P. aeruginosa colonization in the CF lung is correlated with increased likelihood of C. albicans colonization, indicating a synergistic interaction that leads to a greater decline in lung function.10,11 Here, we show that C. albicans significantly enhanced P. aeruginosa biofilm tolerance to 5 mg/L meropenem in both a laboratory strain and a CF isolate. Although the protective effect was relatively small (<1 log10 change in P. aeruginosa cfu/mL), this may still have clinical relevance. For example, the clinical breakpoint of P. aeruginosa for meropenem is >8 mg/L;32 this increased tolerance could push the required meropenem concentration over the clinical breakpoint, categorizing the infection as meropenem resistant. This has direct implications for treatment of CF patients who present with coinfection. Standard doses of meropenem may become insufficient for treating P. aeruginosa infection, with combination antibiotic/antifungal therapy being a potentially more effective therapeutic option.
The protective effect of C. albicans was biofilm specific and dependent upon fungal ECM components. This is consistent with other reports where C. albicans ECM components have been shown to provide protection against ofloxacin and vancomycin in dual-species biofilms with Escherichia coli and Staphylococcus aureus, respectively.42,43 However, in contrast to these studies, where C. albicans ECM components provide protection against a range of antimicrobials, C. albicans ECM components only increased P. aeruginosa tolerance to meropenem. This suggests that the mechanism by which C. albicans ECM components enhance meropenem tolerance may be different to those proposed for other antibiotics. Considering the effect was not seen for other β-lactams (i.e. ceftazidime) this suggests the protective mechanism may depend on chemical structure or ability to bind mannan or β-glucan, rather than the mode of action.
Previously, mannan and β-glucan have been shown to bind and sequester antimicrobials, limiting their diffusion through biofilms.43 Therefore, it is possible that the actual concentration of meropenem within dual-species biofilms is significantly lower. Similar interactions have been observed in dual-species biofilms where Streptococcus mutans exopolysaccharides bind and sequester fluconazole, reducing its efficacy against C. albicans.49 Alternatively, mannans or glucans may coat bacterial cells, providing a physical barrier that impedes drug permeation, supporting the proposed mechanism by which S. aureus is protected from vancomycin.42
Although C. albicans remains clinically the most commonly isolated Candida species, the prevalence of NAC species is increasing.50C. dubliniensis was the only NAC species that protected P. aeruginosa from meropenem. This finding is of clinical relevance as, although less common than C. albicans, the prevalence of C. dubliniensis within CF patients ranges from 2.6% to 39.0% and there are cases of C. dubliniensis being co-isolated with P. aeruginosa from the lower respiratory tracts of CF patients.51C. dubliniensis is the most closely related NAC species to C. albicans and, as a result, their biofilms are structurally similar,38,39 with networks of yeast and hyphal cells embedded in a comparable ECM.52 Although the other NAC species produce biofilms, the composition of their ECM is considerably different53,54 and their biofilms lack hyphae, which are important for bacterial attachment. Scanning electron microscopy confirmed that most bacteria in the treated dual-species biofilms were attached to fungal hyphae, suggesting that this interaction is important for protection. This hypothesis is supported by the fact that removal of O-mannan, which is required for bacterial binding,47,55 reduced the ability of C. albicans to protect P. aeruginosa. However, given that purified carbohydrates were able to provide similar protection to C. albicans, it would suggest that ECM composition is the major contributing factor providing antimicrobial protection.
In conclusion, secreted C. albicans ECM polysaccharides protect P. aeruginosa by reducing the efficacy of meropenem. Clinically, this could result in persistent bacterial infection due to pockets of protected cells, which may then acquire true resistance as a result of continued exposure to subMIC concentrations of antibiotics. This highlights the importance of early diagnosis of dual-species biofilm infections, so that more efficacious therapeutic options, such as combination antibiotic/antifungal therapy, can be considered.
Supplementary Material
Acknowledgements
We would like to thank Paul Stanley and Theresa Morris, at the University of Birmingham’s Centre for Electron Microscopy, for assistance with scanning electron microscopy sample preparation and imaging. We thank Neil Gow, for providing the C. albicans mannan glycosylation mutant strains, and Donna MacCallum, for providing the C. krusei strain.
Funding
F.A. is supported by the Wellcome Trust Antimicrobials and Antimicrobial Resistance (AAMR) doctoral training programme (108876/Z/15/Z). Work in the laboratory of J.M.A.B. is supported by a David Phillips Fellowship to J.M.A.B. (BB/M02623X/1). Work in the laboratory of R.A.H. is supported by an MRC Career Development Award (MR/L00903X/1) and the BBSRC (BB/R00966X/1).
Transparency declarations
None to declare.
References
- 1. Peters BM, Jabra-Rizk MA, O’May GA. et al. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev 2012; 25: 193–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lohse MB, Gulati M, Johnson AD. et al. Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol 2018; 16: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Muthig M, Hebestreit A, Ziegler U. et al. Persistence of Candida species in the respiratory tract of cystic fibrosis patients. Med Mycol 2010; 48: 56–63. [DOI] [PubMed] [Google Scholar]
- 4. Dixon EF, Hall RA.. Noisy neighbourhoods: quorum sensing in fungal-polymicrobial infections. Cell Microbiol 2015; 17: 1431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Brien S, Fothergill JL.. The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol Lett 2017; 364: fnx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sherrard LJ, Tunney MM, Elborn JS.. Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 2014; 384: 703–13. [DOI] [PubMed] [Google Scholar]
- 7. Williams C, Ranjendran R, Ramage G.. Pathogenesis of fungal infections in cystic fibrosis. Curr Fungal Infect Rep 2016; 10: 163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rana A, Sharma A, Pandey G.. Diagnostic value of sputum Gram’s stain and sputum culture in lower respiratory tract infections in a tertiary care hospital. Int J Curr Microbiol App Sci 2017; 6: 4310–4. [Google Scholar]
- 9. Hamet M, Pavon A, Dalle F. et al. Candida spp. airway colonization could promote antibiotic-resistant bacteria selection in patients with suspected ventilator-associated pneumonia. Intensive Care Med 2012; 38: 1272–9. [DOI] [PubMed] [Google Scholar]
- 10. Dhamgaye S, Qu Y, Peleg AY.. Polymicrobial infections involving clinically relevant Gram-negative bacteria and fungi. Cell Microbiol 2016; 18: 1716–22. [DOI] [PubMed] [Google Scholar]
- 11. Gileles-Hillel A, Shoseyov D, Polacheck I. et al. Association of chronic Candida albicans respiratory infection with a more severe lung disease in patients with cystic fibrosis. Pediatr Pulmonol 2015; 50: 1082–9. [DOI] [PubMed] [Google Scholar]
- 12. Lindsay AK, Hogan DA.. Candida albicans: molecular interactions with Pseudomonas aeruginosa and Staphylococcus aureus. Fungal Biol Rev 2014; 28: 85–96. [Google Scholar]
- 13. Flemming HC, Wingender J.. The biofilm matrix. Nat Rev Microbiol 2010; 8: 623–33. [DOI] [PubMed] [Google Scholar]
- 14. Van Acker H, Van Dijck P, Coenye T.. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol 2014; 22: 326–33. [DOI] [PubMed] [Google Scholar]
- 15. Mah TF. Biofilm-specific antibiotic resistance. Future Microbiol 2012; 7: 1061–72. [DOI] [PubMed] [Google Scholar]
- 16. Hill D, Rose B, Pajkos A. et al. Antibiotic susceptibilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J Clin Microbiol 2005; 43: 5085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/.
- 18. Holmes AH, Moore LSP, Sundsfjord A. et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016; 387: 176–87. [DOI] [PubMed] [Google Scholar]
- 19. Lopez-Medina E, Fan D, Coughlin LA. et al. Candida albicans inhibits Pseudomonas aeruginosa virulence through suppression of pyochelin and pyoverdine biosynthesis. PLoS Pathog 2015; 11: e1005129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trejo-Hernández A, Andrade-Domínguez A, Hernandez M. et al. Interspecies competition triggers virulence and mutability in Candida albicans-Pseudomonas aeruginosa mixed biofilms. ISME J 2014; 8: 1974–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen AI, Dolben EF, Okegbe C. et al. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog 2014; 10: e1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bandara HM, Cheung BPK, Watt RM. et al. Pseudomonas aeruginosa lipopolysaccharide inhibits Candida albicans hyphae formation and alters gene expression during biofilm development. Mol Oral Microbiol 2013; 28: 54–69. [DOI] [PubMed] [Google Scholar]
- 23. Cugini C, Morales DK, Hogan DA.. Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 2010; 156: 3096–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hogan DA, Vik A, Kolter R.. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 2004; 54: 1212–23. [DOI] [PubMed] [Google Scholar]
- 25. Taff HT, Nett JE, Zarnowski R. et al. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog 2012; 8: e1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hall CW, Mah TF.. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 2017; 41: 276–301. [DOI] [PubMed] [Google Scholar]
- 27.Cystic Fibrosis Trust. Antibiotic Treatment for Cystic Fibrosis: Report of the UK Cystic Fibrosis Trust Antibiotic Working Group, 3rd Edition. 2009. https://www.cysticfibrosis.org.uk/~/media/documents/the-work-we-do/care/consensus-docs-with-new-address/anitbiotic-treatment.ashx?la=en.
- 28. Fernandez R, Amador P, Prudêncio C.. β-Lactams: chemical structure, mode of action and mechanisms of resistance. Rev Med Microbiol 2013; 24: 7–17. [Google Scholar]
- 29. Papp-Wallace KM, Endimiani A, Taracila MA. et al. Carbapenems: past, present, and future. Antimicrob Agents Chemother 2011; 55: 4943–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicolau DP. Pharmacokinetic and pharmacodynamic properties of meropenem. Clin Infect Dis 2008; 47 Suppl 1: S32–40. [DOI] [PubMed] [Google Scholar]
- 31. Haagensen JA, Verotta D, Huang L. et al. New in vitro model to study the effect of human simulated antibiotic concentrations on bacterial biofilms. Antimicrob Agents Chemother 2015; 59: 4074–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 8.1. 2018. http://www.eucast.org/.
- 33. Merritt JH, Kadouri DE, O’Toole GA.. Growing and analyzing static biofilms. Curr Protoc Microbiol 2005; Chapter 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Harriott MM, Noverr MC.. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother 2009; 53: 3914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 2001; 48: 5–16. [DOI] [PubMed] [Google Scholar]
- 36. Savage VJ, Chopra I, O’Neill AJ.. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob Agents Chemother 2013; 57: 1968–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olsen I. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis 2015; 34: 877–86. [DOI] [PubMed] [Google Scholar]
- 38. Whibley N, Gaffen SL.. Beyond Candida albicans: mechanisms of immunity to non-albicans Candida species. Cytokine 2015; 76: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Papon N, Courdavault V, Clastre M. et al. Emerging and emerged pathogenic Candida species: beyond the Candida albicans paradigm. PLoS Pathog 2013; 9: e1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taff HT, Mitchell KF, Edward JA. et al. Mechanisms of Candida biofilm drug resistance. Future Microbiol 2013; 8: 1325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nett J, Lincoln L, Marchillo K. et al. Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother 2007; 51: 510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kong EF, Tsui C, Kucharikova S. et al. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. MBio 2016; 7: e01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. De Brucker K, Tan Y, Vints K. et al. Fungal β-1,3-glucan increases ofloxacin tolerance of Escherichia coli in a polymicrobial E. coli/Candida albicans biofilm. Antimicrob Agents Chemother 2015; 59: 3052–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sheth CC, Hall R, Lewis L. et al. Glycosylation status of the C. albicans cell wall affects the efficiency of neutrophil phagocytosis and killing but not cytokine signaling. Med Mycol 2011; 49: 513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murciano C, Moyes DL, Runglall M. et al. Candida albicans cell wall glycosylation may be indirectly required for activation of epithelial cell proinflammatory responses. Infect Immun 2011; 79: 4902–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hall RA, Bates S, Lenardon MD. et al. The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans. PLoS Pathog 2013; 9: e1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brand A, Barnes JD, Mackenzie KS. et al. Cell wall glycans and soluble factors determine the interactions between the hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol Lett 2008; 287: 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scott FW, Pitt TL.. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J Med Microbiol 2004; 53: 609–15. [DOI] [PubMed] [Google Scholar]
- 49. Kim D, Liu Y, Benhamou RI. et al. Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J 2018; 12: 1427–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sardi JC, Scorzoni L, Bernardi T. et al. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol 2013; 62: 10–24. [DOI] [PubMed] [Google Scholar]
- 51. Wahab AA, Taj-Aldeen SJ, Kolecka A. et al. High prevalence of Candida dubliniensis in lower respiratory tract secretions from cystic fibrosis patients may be related to increased adherence properties. Int J Infect Dis 2014; 24: 14–9. [DOI] [PubMed] [Google Scholar]
- 52. Ramage G, Vande Walle K, Wickes BL. et al. Biofilm formation by Candida dubliniensis. J Clin Microbiol 2001; 39: 3234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cavalheiro M, Teixeira MC.. Candida biofilms: threats, challenges, and promising strategies. Front Med (Lausanne) 2018; 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Silva S, Negri M, Henriques M. et al. Adherence and biofilm formation of non-Candida albicans Candida species. Trends Microbiol 2011; 19: 241–7. [DOI] [PubMed] [Google Scholar]
- 55. Munro CA, Bates S, Buurman ET. et al. Mnt1p and Mnt2p of Candida albicans are partially redundant α-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J Biol Chem 2005; 280: 1051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sobel JD. Vaginitis. N Engl J Med 1997; 337: 1896–903. [DOI] [PubMed] [Google Scholar]
- 57. Hobson RP, Munro CA, Bates S. et al. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J Biol Chem 2004; 279: 39628–35. [DOI] [PubMed] [Google Scholar]
- 58. Bates S, MacCallum DM, Bertram G. et al. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J Biol Chem 2005; 280: 23408–15. [DOI] [PubMed] [Google Scholar]
- 59. Morschhäuser J, Ruhnke M, Michel S. et al. Identification of CARE-2-negative Candida albicans isolates as Candida dubliniensis. Mycoses 1999; 42: 29–32. [DOI] [PubMed] [Google Scholar]
- 60. Zwolinska-Wcislo M, Budak A, Trojanowska D. et al. Fungal colonization of the stomach and its clinical relevance. Mycoses 1998; 41: 327–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.