Abstract
Epigallocatechin-3-Gallate (EGCG) has been extensively studied for its protective effect against cardiovascular disorders. This effect has been attributed to its action on multiple molecular pathways and transmembrane proteins, including the cardiac Nav1.5 channels, which are inhibited in a dose-dependent manner. However, the molecular mechanism underlying this effect remains to be unveiled. To this aim, we have characterized the EGCG effect on Nav1.5 using electrophysiology and molecular dynamics (MD) simulations. EGCG superfusion induced a dose-dependent inhibition of Nav1.5 expressed in tsA201 cells, negatively shifted the steady-state inactivation curve, slowed the inactivation kinetics, and delayed the recovery from fast inactivation. However, EGCG had no effect on the voltage-dependence of activation and showed little use-dependent block on Nav1.5. Finally, MD simulations suggested that EGCG does not preferentially stay in the center of the bilayer, but that it spontaneously relocates to the membrane headgroup region. Moreover, no sign of spontaneous crossing from one leaflet to the other was observed, indicating a relatively large free energy barrier associated with EGCG transport across the membrane. These results indicate that EGCG may exert its biophysical effect via access to its binding site through the cell membrane or via a bilayer-mediated mechanism.
Keywords: EGCG, Nav1.5, cellular electrophysiology, molecular dynamics, ion channels
1. Introduction
Ion channels are pore-forming transmembrane proteins involved with the transport of ions through cell membranes. Their activity is essential for the excitation-contraction coupling in cardiac cells. In this sense, the voltage-gated sodium channels generate the rapid action potential upstroke of cardiomyocytes and thereby play a crucial role in heart excitability and conduction.
Within the nine isoforms of voltage-gated sodium channels, Nav1.5 displays a preponderant expression in cardiac cells [1]. This isoform is composed of intracellular N and C terminal tails and four homologous domains (DI-DIV), each consists of six transmembrane segments (S1–S6). These domains fold together with their S5-S6 segments to build a highly selective Na+ pore [2,3].
Nav1.5 dysfunction is linked with an increasingly wide range of inherited cardiac arrhythmias [4,5]. Gain-of-function mutations of this channel are associated with many disorders such as the congenital long QT syndrome, multifocal ectopic Purkinje-related premature contractions (MEPPC), and exercise-induced polymorphic ventricular tachycardia [6,7,8,9,10,11]. Whereas the loss-of-function mutations are linked with Brugada syndrome, sick sinus syndrome, and cardiac conduction disease [4,5,12]. In the same line, mutations in genes encoding proteins that regulate, directly or indirectly, Nav1.5 function are associated with many cardiac arrhythmias [13]. This is true for many Nav1.5-interacting proteins such as Nav-β subunits, Plakophillin-2, and more recently for the nuclear proteins Lamin A/C [14,15,16,17]. As such, Nav1.5 channels become highly targeted by several drug classes, including anti-arrhythmic drugs [18], despite their associated side effects [19].
In this context, bioactive molecules extracted from medicinal plants are pledged to offer added values in preventing arrhythmic episodes. Epigallocatechin-3-Gallate (EGCG), the predominant polyphenol of green tea, was shown to be a promising natural alternative in the setting of cardiac arrhythmia [20,21].
Green tea extracts and EGCG have been extensively studied for their protective effects against cardiovascular disorders. Many authors have shown that green tea consumption is associated with a lower risk of heart disease and reduced cardiovascular mortality [22,23]. Moreover, several experimental studies revealed that EGCG reduces chronic ventricular remodeling after myocardial ischemia in rats [24], improves left ventricular dysfunction, suppresses myocardial inflammation in rat autoimmune myocarditis [25], modulates arrhythmogenic activity, and calcium homeostasis of rabbit left atrium [21], and inhibits angiotensin II-induced cardiomyocyte hypertrophy [26].
The cardiac activity of EGCG has been attributed to its effect on multiple intracellular molecular pathways and transmembrane proteins [20,21,22,26,27,28], including Nav1.5. This compound has shown a dose-dependent inhibitory effect on the cardiac sodium current INa [21,29,30]. However, the molecular and biophysical mechanism underlying this effect remains to be unveiled. To this aim, we have worked out to characterize the mechanisms underlying the EGCG effect on the cardiac sodium channel Nav1.5.
2. Results
2.1. Inhibitory Effect of EGCG on Human Nav1.5 Channels
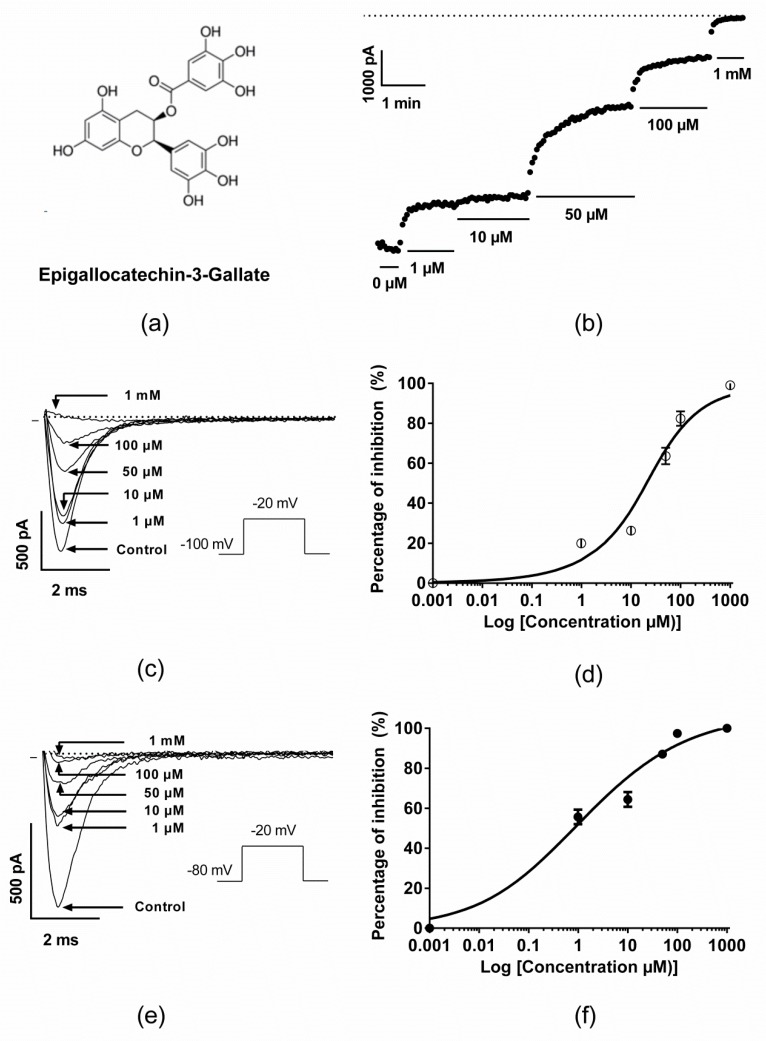
The pharmacological effects of EGCG (Figure 1a) on the cardiac sodium channel isoform were investigated using tsA201 cells transiently transfected with the human Nav1.5 channel in the presence of its regulatory subunit Nav-β1. The latter is known for its effect on increasing the functional expression of Nav1.5 channels in many heterologous systems [14]. INa currents were generated by 15 ms step depolarization to −20 mV from a holding potential of −100 or −80 mV at a frequency of 0.33 Hz. As previously reported by Kang et al. [30], EGCG superfusion induced a dose-dependent inhibition of the Nav1.5 channel. The IC50 for the EGCG inhibition was 31.2 ± 3.6 µM when measured from the −100 mV holding potential (Figure 1b–d) and 2.1 ± 1 µM when measured at −80 mV (Figure 1e,f). On the other hand, EGCG dose-dependent effect on the fast inactivation kinetics was investigated. This resulted in slowing the inactivation kinetics as long as the EGCG concentrations increased, mainly at 50 and 100 µM (Supplementary Figure S1).
Figure 1.
Pharmacological effects of Epigallocatechin-3-Gallate (EGCG) on the cardiac sodium channel Nav1.5. (a) Chemical structure of EGCG. (b) Time course recording of the INa current in the presence of various concentrations of EGCG. The dashed line represents the zero current. (c) Representative traces of INa currents in the presence of various EGCG concentrations at holding potential −100 mV. (d) Dose-response effects of EGCG on the inhibition of INa peak currents (holding potential = −100 mV, IC50 = 31.2 ± 3.6 µM, n = 8). (e) Representative traces of INa currents in the presence of various EGCG concentrations at holding potential −80 mV. (f) Dose-response effects of EGCG on the inhibition of INa peak currents (holding potential = −80 mV, IC50 = 2,1 ± 1 µM, n = 5–7).
2.2. Effect of EGCG on the Voltage-Dependence Properties of Nav1.5 Channels
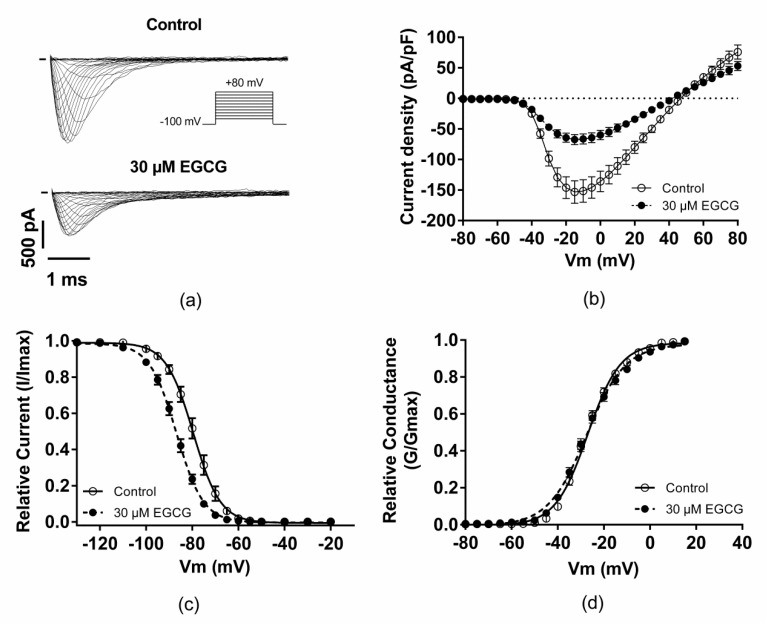
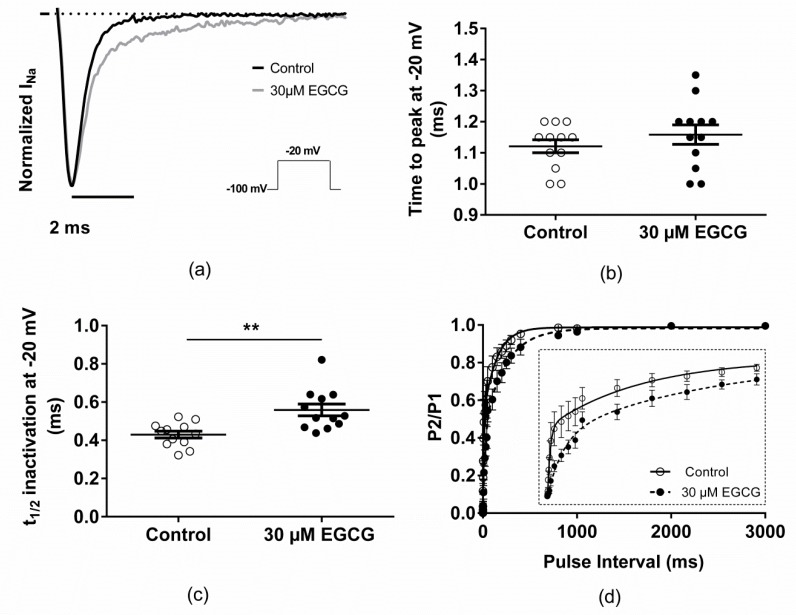
The second step of EGCG pharmacological characterization was to study the compound effect on Nav1.5 biophysics. For this aim, the voltage-dependence properties of Nav1.5 channels were investigated in the presence or absence of 30 µM EGCG. The superfusion of this compound inhibited the cardiac sodium channels (Figure 2a,b, Table 1), induced a shift of steady-state inactivation towards more negative potentials (Figure 2c, Table 1), slowed the inactivation kinetics (Figure 3a,c, Table 1), and delayed the recovery from fast inactivation (Figure 3d, Table 1). Moreover, EGCG did not modify the voltage-dependence of activation but significantly affected the activation curve slope (Figure 2d, Table 1).
Figure 2.
Effect of EGCG on the gating properties of Nav1.5 channels. (a) Representative traces of INa current in the presence or the absence of 30 µM of EGCG. (b) I/V relationship in the presence or the absence of EGCG. (c,d) Effect of EGCG on the voltage-dependence of inactivation and activation, respectively.
Table 1.
Effect of EGCG on the gating properties of Nav1.5 channels.
| Control | 30 µM EGCG | |
|---|---|---|
| Peak current densities at −20 mV (pA/pF) | −146.2 ± 17.8; n = 12 | −64.3 ± 7.3 ***; n = 12 |
| V1/2 activation (mV) | −27.4 ± 0.8; n = 12 | −27.4 ± 1; n = 12 |
| Activation slope (mV) | 6.7 ± 0.2; n = 12 | 8 ± 2 ***; n = 12 |
| Time to peak at −20 mV (ms) | 1.12 ± 0.02; n = 12 | 1.16 ± 0.03; n = 12 |
| V1/2 inactivation (mV) | −79.4 ± 1.3; n = 13 | −87 ± 0.9 ***; n = 13 |
| Inactivation slope (mV) | 5 ± 0.2; n = 13 | 5.5 ± 0.2 *; n = 13 |
| t1/2 inactivation at −20 mV (ms) | 0.43 ± 0.02; n = 12 | 0.56 ± 0.03 **; n = 12 |
| Recovery from fast inactivation (ms) | ||
| τfast,1 | 5.1 ± 0.9; n = 5 | 24.3 ± 7.1 *; n = 5 |
| τfast,2 | 110 ± 28.6; n = 5 | 233.7 ± 37.6 *; n = 5 |
* p < 0.05, ** p < 0.01, *** p < 0.001.
Figure 3.
Effect of EGCG on INa kinetics. (a) Representative traces of normalized INa current in the presence or the absence of 30 µM of EGCG. INa currents were normalized to the maximal peak current measured, in each condition, at −20 mV. EGCG effect on INa time to peak (b), inactivation kinetics (c), and recovery from fast inactivation (d; inset, zoom on the interval between 0 and 300 ms).
2.3. EGCG did not Produce any Use-Dependent Blockade of Nav1.5 Channels
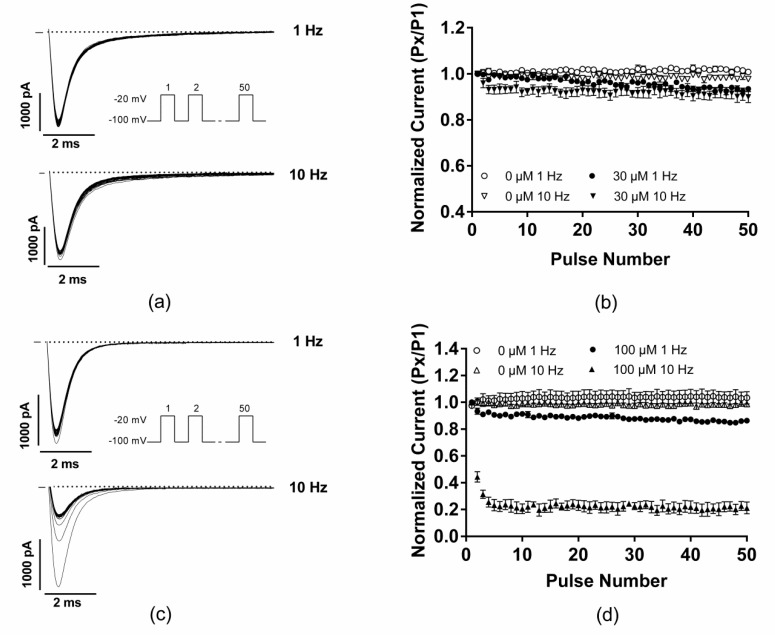
To characterize the potential use-dependent block (UDB) of EGCG on INa current, the effect of rapid pulsing on Nav1.5 was investigated via the application of a series of 50 15 ms depolarizing pulses from −100 to −20 mV at various stimulation rates (1 and 10 Hz). After the establishment of the whole-cell configuration, cells were allowed to stabilize before the first run of the UDB protocol in the absence of EGCG. Then, EGCG was perfused for 3 min while the membrane potential was held at −100 mV, followed by the UDB protocol under 30 µM EGCG.
After the application of EGCG, INa currents were slightly but significantly reduced by 7% and 8% compared with the control when pulsing at 1 and 10 Hz, respectively (Figure 4a,b, Table 2). These results suggest that there is little or no use-dependent blockade of EGCG on Nav1.5 related currents.
Figure 4.
Use-dependent block protocol in the presence or absence of 30 µM EGCG. (a) Representative trace of INa currents at 1 and 10 Hz in the presence of 30 µM of EGCG. (b) Normalized INa current versus pulse number in the presence or absence of 30 µM of EGCG at 1 and 10 Hz. (c) Representative trace of INa currents at 1 and 10 Hz in the presence of 100 µM of Mexiletine. (d) Normalized INa current versus pulse number in the presence or absence of 100 µM of mexiletine at 1 and 10 Hz.
Table 2.
Characterization of the use-dependent block of EGCG.
| UDB-1 Hz | UDB-10 Hz | |
|---|---|---|
| EGCG (30 µM) | 0.93 ± 0.01 **, n = 5 | 0.90 ± 0.02 *, n = 5 |
| Mexiletine (100 µM) | 0.86 ± 0.01 ***, n = 4 | 0.21 ± 0.04 ***, n = 4 |
* p < 0.05, ** p < 0.01, *** p < 0.001 versus control.
To validate our experimental conditions, especially the used UDB protocol, mexiletine has been used as a positive control. As shown in Figure 4c,d, the application of 100 µM mexiletine significantly reduces the INa current by 17% and 77% compared with the control when pulsing at 1 and 10 Hz, respectively. These findings demonstrated the ability of the applied UDB protocol to characterize the use-dependent block of a given drug.
2.4. MD Simulations Suggest that EGCG does not Spontaneously Permeate Cell Membranes
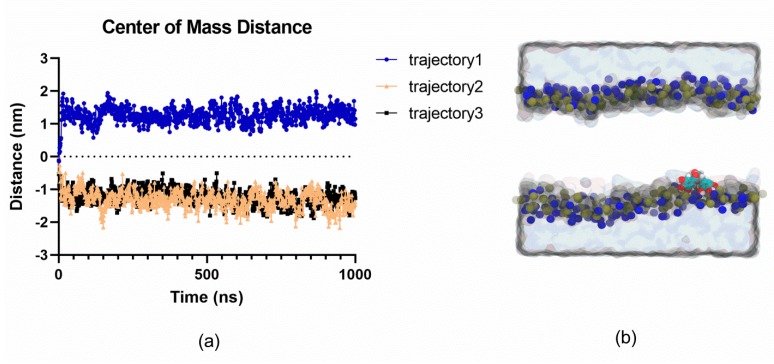
To obtain a first insight into the mechanism through which EGCG may exert its effect on Nav1.5, the interaction between EGCG and a model lipid membrane that mimics the environment in which Nav1.5 is found, was characterized. To this effect, molecular dynamics (MD) simulations of a system containing EGCG initially positioned at the center of a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer, were carried out. Molecular dynamics simulations enable us to investigate the time-dependent dynamical and structural properties of systems at the atomistic level. Three 1 µs simulations suggested that EGCG did not preferentially stay in the center of the bilayer, but that it spontaneously and consistently relocated to the membrane headgroup region (Figure 5). Over the course of the simulations, no signs of spontaneous crossing from one leaflet to the other were observed, indicative of a relatively large free energy barrier associated with EGCG transport across the membrane, that is not easily crossed on the microsecond timescale. This finding is consistent with the molecule’s amphiphilic nature. These results indicate that EGCG may not spontaneously permeate the membrane and become able to block the Nav1.5 channel via the entrance to the internal cavity via the inner pore mouth.
Figure 5.
Molecular dynamics simulations show the preferential localization of EGCG in the headgroup region of a POPC membrane. (a) Localization of EGCG molecules along 1-microsecond simulation runs with respect to the bilayer center (0 nm). (b) Representative snapshot highlighting the preferential localization of the EGCG in the headgroup region of a POPC bilayer (phosphate and nitrogen atoms are shown as blue and brown spheres, respectively, the solution is shown as a grey surface and lipid tails are omitted for clarity. The EGCG molecule is shown as cyan, red, and white spheres).
3. Discussion
The present study aimed to characterize the biophysical mechanism underlying the EGCG effect on the cardiac sodium channel Nav1.5.
Our results showed that EGCG inhibited the INa current in a concentration-dependent manner either at a holding potential of −100 or at −80 mV. The IC50 of EGCG was much lower at a holding potential of −80 mV compared to −100 mV, (IC50 = 32.4 ± 3.6 µM and 2.1 ± 1 µM at −100 mV and −80 mV, respectively). This demonstrates that the membrane holding potential affects the affinity of EGCG to Nav1.5, suggesting that EGCG interacts with a higher affinity to the inactivated state of the cardiac sodium channel. Similar results have been reported by Kang et al., showing a dose and state-dependent inhibition of INa current in the presence of EGCG [30]. However, the obtained IC50 were different from those published by these authors (100 µM and 24.6 µM at holding potentials of −90 mV and −70 mV, respectively [30]). These discrepancies are likely related to different experimental conditions used in the two studies. Kang et al., have stably transfected Nav1.5 in HEK293 cells in the absence of its regulatory subunit Nav-β1, investigated the pharmacological effect of ECGC at 35 °C, and used −90 or −70 mV as holding potentials to generate INa currents. In contrast to these conditions, we have transiently transfected tsA-201 cells with Nav1.5 in the presence of Nav-β1, recorded INa currents at room temperature (23–25 °C), and used −100 or −80 mV as holding potentials to generate INa currents. These protocol changes may induce noticeable differences. Indeed, since Nav-βs are strong modulators of Nav1.5 electrophysiological properties [14], the sensitivity of the cardiac sodium channel towards EGCG may be affected by the presence or absence of Nav-β1. Furthermore, conducting experiments at elevated temperatures may induce the degradation of EGCG and its epimerization to gallocatechin-3-gallate (GCG) [31], which could trigger different biophysical modifications of INa current as it was shown in rat dorsal root ganglion neurons [32]. On the other hand, under the same conditions, the activity of EGCG on Nav1.5 could be altered as it was described for other INa inhibitors such as flecainide, ajmaline, and ranolazine [33,34].
The biophysical mechanism of Nav1.5 inhibition by EGCG was further investigated by characterizing the biophysical gating properties of these channels in the presence of 30 µM EGCG. Under these conditions, EGCG shifts the voltage-dependence of inactivation curve toward more negative potentials by 7.6 mV, slows the inactivation kinetics, and delays the recovery from fast inactivation, indicating that EGCG binds to the inactivated state of Nav1.5. However, EGCG had no effect on the voltage-dependence of activation and significantly affected the activation curve slope. The modification of the activation slope may be caused by a more efficient clamp of the membrane potential in the presence of 30 µM EGCG. Indeed, even with optimal compensation for series resistance (Rs), the remaining uncompensated Rs combined with large and rapid INa currents let a voltage error that could produce a loss of the voltage clamp. Thus, a reduced sodium current magnitude, as it has been noticed in the presence of EGCG, could decrease the resulting series resistance error and improve the voltage clamp quality.
On the other hand, the observed shift of the steady-state of inactivation is consistent with what has been previously described for Nav1.5 [30]. One should note that the EGCG effects on the other biophysical properties were mainly investigated on the neuronal Nav channels [32,35,36]. These published results are in concordance with the EGCG effect on the fast inactivation kinetics, and recovery from inactivation of the cardiac sodium channels.
With regards to the activation process, Kim et al. observed that 1µM EGCG did not affect the voltage-dependence of activation of tetrodotoxin-sensitive and resistant INa currents in rat dorsal root ganglion neurons [36], which was consistent with the results of the present study studying the cardiac sodium current. In contrast, Deng et al. showed a negative shift of the activation curve in the presence of EGCG in primary cultures of rat hippocampal neurons [35]. From our perspective, these discrepancies could be related to the quality of the membrane potential clamp. Indeed, based on the sodium current traces presented in Deng’s study, a sub-optimal voltage-clamp quality of these currents is observed, which makes any interpretation difficult [35].
Furthermore, our investigations suggested that there is little or no use-dependent block of EGCG on Nav1.5. The absence of the UDB could be explained by the slight modification of the recovery from fast inactivation. Indeed, in the presence of EGCG, the almost “normal” recovery from fast inactivation prevents an additional progressive accumulation of inactivated Nav1.5, which represents the preferential binding state of EGCG. These findings are consistent with those obtained in other studies testing the effect of gallocatechin-3-gallate on the tetrodotoxin-resistant voltage-gated sodium channels in rat dorsal root ganglion neurons [32].
In a first attempt to characterize the molecular mechanism through which EGCG may exert its effect on Nav1.5, we investigated how EGCG interacts with the lipidic environment surrounding Nav1.5. Indeed, relatively hydrophobic compounds such as EGCG have been hypothesized to act on membrane proteins either via direct interaction, via the effect on the membrane, or both. Voltage-gated sodium channels, in particular, have fenestrations lining their pore domain, which enable modulators to access the pore via the membrane. We thus sought to gain a first insight into how Nav channels may be modulated by EGCG by characterizing where this compound localized preferentially in a membrane. This was done by carrying out MD simulations, which provided a detailed atomistic view of the behavior of EGCG when it was placed in a model lipidic membrane: Calculations were initiated with EGCG located at the center of the POPC bilayer. The obtained results suggest that EGCG is spontaneously relocated to the membrane headgroup region.
The predicted re-localization of EGCG to the membrane head group is consistent with the results of Salazar et al., which have demonstrated that EGCG modifies the activity of a membrane-bound protein upon binding to the phospholipid head groups of the red blood cell membranes [37]. Similarly, Ingólfsson et al. have shown that the biological effects of green tea catechins could be attributed to multiple molecular interactions because the catechins accumulate at the bilayer/solution interface and modify bilayer properties that can influence gramicidin channel function [38]. However, MD simulations results cannot exclude the possibility that EGCG exerts its biophysical effect via access to its binding site, on Nav1.5, through the cell membrane. Indeed, as shown for local anesthetics and antiarrhythmic drugs [39], the EGCG blocking effect could also be controlled by the channel’s fenestrations.
The current findings support the hypothesis that EGCG could be used as an anti-arrhythmic drug in the setting of Nav1.5 gain-of-function mutations; as has been suggested by Boukhabza et al. These authors have predicted that EGCG may limit the cardiac cells hyperexcitability related to the presence of Nav1.5-R222Q and Nav1.5-I141V mutants [20]. On the other hand, the present study demonstrated that the membrane holding potential affects the affinity of EGCG to Nav1.5, suggesting that EGCG interacts with a higher affinity to the inactivated state of the cardiac sodium channel. Thus, we could expect an atrial-selective block of Nav1.5 channels due to a more depolarized resting membrane potential in the atria compared to the ventricles. Then, a higher beneficial effect of EGCG could be observed in the setting of atrial hyperexcitability phenotypes.
Nonetheless, it remains somehow challenging to interpret the clinical relevance of our results even in the context of cardiac hyperexcitability phenotypes. Many in vitro studies have shown that the effective concentrations of EGCG to target relevant pathophysiological pathways are typically ranged from 1 to 1000 µmol/L. However, the pharmacological disposition of EGCG in humans indicates that its serum concentration is in the nanomolar scale [40,41,42,43,44,45,46,47,48]. In addition, EGCG is well known for its chemical instability under physiological conditions due to its easy degradation and metabolism [31,49,50]. Therefore, any potential impact of EGCG intake may reflect the combined effects of EGCG and its metabolites.
In conclusion, the present study suggests that EGCG may modulate the biophysical properties of Nav1.5 via access to its binding site through the cell membrane or via a bilayer-mediated mechanism. These results constitute a first step towards the characterization of the biophysical mechanism underlying the inhibitory action of EGCG on Nav1.5. However, the exact molecular interactions underlying these effects are still unknown and remain to be studied.
4. Materials and Methods
4.1. Cell Culture and Transfection
TsA-201 cells were cultured at 37 °C in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS), 4 mM glutamine, and a cocktail of streptomycin-penicillin antibiotics in a humidified atmosphere of 5% CO2 and 95% air. All cell medium components, except glutamine (Sigma-Aldrich), were purchased from Gibco (Fisher Scientific, Zurich, Switzerland).
For electrophysiological experiments, tsA-201 cells were transiently transfected with DNA complexed to JetPEI (Polyplus-transfection) according to the manufacturer’s instructions. DNA concentrations were 1 µg of pCDN3.1-Nav1.5 and 1 µg of pIRES-hβ1-CD8. The resulting Nav1.5 protein was a splice variant lacking glutamine at position 1077 (Q1077del). Eight hours after transfection, the cells were isolated and seeded in plastic Petri dishes at low density.
4.2. Cellular Electrophysiology
Twenty-four hours after transfection, the resulting sodium current (INa) was recorded at room temperature (23–25 °C), under whole-cell voltage-clamp conditions with Axon PClamp 10 software through an A/D converter (Digidata 1440A) using an Axopatch 200B (all Axon Instruments Molecular Devices Corp., CA, USA). Series resistances were compensated (up to 90% compensation), and the residual capacitive currents were canceled using a P/4 protocol. The cells were bathed with an extracellular solution containing (in mmol/L): NaCl 50, NMDG-Cl 80, CsCl 5, MgCl2 1.2, CaCl2 2, HEPES 10, glucose 5. The pH was adjusted to 7.4 with CsOH. Glass pipettes (tip resistance: 1.5 to 3 MΩ) were filled with an intracellular medium containing (in mmol/L): Na2ATP 5, CsAsp 70, CsCl 60, EGTA 11, CaCl2 1, MgCl2 1, HEPES 10. pH was adjusted to 7.2 with CsOH.
Activation properties of Nav1.5 channels were determined from I/V relationships by normalizing the sodium current peak to the driving force. Steady-state inactivation was measured with a standard double-pulse protocol. Cells were held at −100 mV before the application of 500 ms conditioning pulses, ranging from −130 to −20 mV. Then, a 20 ms test pulse to −20 mV was applied. The measured current was then normalized to the maximum peak current recorded after conditioning pulses (I/Imax) and plotted as a function of conditioning step voltage. Activation and inactivation curves were fit to a Boltzmann equation.
For the pharmacological investigation, the peak current amplitudes at different EGCG concentrations were subtracted from the current obtained with the control solution and were normalized to the control value:
| Inhibition Percentage (%) = [(Icontrol − IEGCG)/Icontrol] × 100 |
IC50 was calculated according to Quest GraphTM IC50 Calculator (Quest GraphTM IC50 Calculator). AAT Bioquest, Inc., 24 Apr. 2019, https://www.aatbio.com/tools/ic50-calculator).
4.3. Chemicals
Epigallocatechin-3-Gallate (EGCG, Sigma-Aldrich, CAS Number 989-51-5) was dissolved directly into the external solutions. Fresh EGCG solutions were prepared before each experiment. During the same day, EGCG solutions were renewed every 5 h to avoid compound degradation (e.g., brown discoloration of solutions).
4.4. Molecular Dynamics Simulations
The 3D structure of the EGCG molecule was obtained from PubChem Database ((−)-Epigallocatechin gallate, CID = 65064). The system containing an EGCG molecule placed at the center of a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine membrane and plunged in a 0.1 M NaCl solution was prepared using the CHARMM-GUI server [51,52,53]. The lipid and ions interactions were described using the CHARMM36 (Chemistry at Harvard Macromolecular Mechanics) force field [54], whereas water molecules were modeled using the TIP3P model [55]. Simulations were performed using the NPT ensemble, where the temperature and the pressure were maintained at 310 K and 1 atm, respectively, using Parinello–Rahaman pressure coupling [56] and Nose-Hoover temperature coupling [57]. The cut-off value used to compute non-bonded interactions was set to 12 Å and long-range electrostatic interactions were computed using the particle mesh Ewald method [58]. Three replicas 1 µs simulations were run using the GROMACS package version 2018.4 on our local high-performance computing cluster [59]. Distances between the COM (Center of Mass) of EGCG and the membrane were calculated by using the gmx distance analysis tool of GROMACS. EGCG was parameterized using the SWISS-PARAM software [60].
4.5. Data Analysis and Statistical Methods
Currents were analyzed with Clampfit software (Axon Instruments, San Jose, CA, USA). Data were analyzed using a combination of pClamp10, Excel (Microsoft), Igor Pro (WaveMetrics), and Prism (graphpad).
Comparisons between groups were performed with a two-tailed paired Student’s t-test. Data were expressed as mean ± SEM. A p-value < 0.05 was considered significant.
5. Conclusions
In the current study, we have characterized the effect of epigallocatechin-3-gallate on the cardiac sodium channel. This compound shows a higher affinity for the inactivated state of Nav1.5 without any effect on its activation process. In addition, molecular dynamics simulations have predicted the re-localization of EGCG to the membrane headgroup region, suggesting that EGCG may not spontaneously permeate the cell membrane and may exert it effects on Nav1.5 by a bilayer mediated mechanism.
Acknowledgments
The authors gratefully acknowledge Jean-Sébastien Rougier and Maria Essers, Institute of Biochemistry and Molecular Medicine, University of Bern; and Marina Kasimova, Science for Life Laboratory, Department of Applied Physics, KTH Royal Institute of Technology, Solna, Sweden, for their support. L.D. acknowledges support from SciLifeLab. The MD simulations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at PDC Centre for High-Performance Computing (PDC-HPC).
Supplementary Materials
The following are available online, Figure S1: EGCG dose-dependent effect on the fast inactivation kinetics. (a) Representative traces of normalized INa current in the presence of various EGCG concentrations. (b) EGCG effect on INa inactivation kinetics.
Author Contributions
Conceptualization, M.-Y.A. and H.A.; methodology, M.-Y.A., L.D., and H.A; software, H.K., M.-Y.A., and L.D.; validation, M.-Y.A., L.D., and H.A.; formal analysis, M.-Y.A. and L.D.; investigation, H.K., M.-Y.A., and L.D.; resources, H.A.; data curation, M.-Y.A. and L.D.; writing—original draft preparation, H.K., M.-Y.A., L.D., and H.A.; writing—review and editing, H.K., M.-Y.A., L.D., and H.A.; visualization, M.-Y.A.; supervision, M.-Y.A. and H.A.; project administration, M.-Y.A. and H.A.; funding acquisition, M.-Y.A. and H.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the leading House from the Middle East and North Africa, Switzerland, grant number Seed Money 2018 -09.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of Epigallocatechin-3-Gallate are available from the authors.
References
- 1.Chevalier M., Vermij S.H., Wyler K., Gillet L., Keller I., Abriel H. Transcriptomic analyses of murine ventricular cardiomyocytes. Sci. Data. 2018;5:180170. doi: 10.1038/sdata.2018.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naylor C.E., Bagneris C., DeCaen P.G., Sula A., Scaglione A., Clapham D.E., Wallace B.A. Molecular basis of ion permeability in a voltage-gated sodium channel. Embo J. 2016;35:820–830. doi: 10.15252/embj.201593285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payandeh J., Scheuer T., Zheng N., Catterall W.A. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savio-Galimberti E., Argenziano M., Antzelevitch C. Cardiac Arrhythmias Related to Sodium Channel Dysfunction. Handb. Exp. Pharmacol. 2018;246:331–354. doi: 10.1007/164_2017_43. [DOI] [PubMed] [Google Scholar]
- 5.Wilde A.A.M., Amin A.S. Clinical Spectrum of SCN5A Mutations: Long QT Syndrome, Brugada Syndrome, and Cardiomyopathy. Jacc. Clin. Electrophysiol. 2018;4:569–579. doi: 10.1016/j.jacep.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Laurent G., Saal S., Amarouch M.Y., Beziau D.M., Marsman R.F., Faivre L., Barc J., Dina C., Bertaux G., Barthez O., et al. Multifocal ectopic Purkinje-related premature contractions: A new SCN5A-related cardiac channelopathy. J. Am. Coll. Cardiol. 2012;60:144–156. doi: 10.1016/j.jacc.2012.02.052. [DOI] [PubMed] [Google Scholar]
- 7.Swan H., Amarouch M.Y., Leinonen J., Marjamaa A., Kucera J.P., Laitinen-Forsblom P.J., Lahtinen A.M., Palotie A., Kontula K., Toivonen L., et al. Gain-of-function mutation of the SCN5A gene causes exercise-induced polymorphic ventricular arrhythmias. Circ. Cardiovasc. Genet. 2014;7:771–781. doi: 10.1161/CIRCGENETICS.114.000703. [DOI] [PubMed] [Google Scholar]
- 8.Yu S., Li G., Huang C.L., Lei M., Wu L. Late sodium current associated cardiac electrophysiological and mechanical dysfunction. Pflug. Arch. Eur. J. Physiol. 2018;470:461–469. doi: 10.1007/s00424-017-2079-7. [DOI] [PubMed] [Google Scholar]
- 9.Mann S.A., Castro M.L., Ohanian M., Guo G., Zodgekar P., Sheu A., Stockhammer K., Thompson T., Playford D., Subbiah R., et al. R222Q SCN5A mutation is associated with reversible ventricular ectopy and dilated cardiomyopathy. J. Am. Coll. Cardiol. 2012;60:1566–1573. doi: 10.1016/j.jacc.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 10.Amarouch M.Y., Swan H., Leinonen J., Marjamaa A., Lahtinen A.M., Kontula K., Toivonen L., Widen E., Abriel H. Antiarrhythmic Action of Flecainide in Polymorphic Ventricular Arrhythmias Caused by a Gain-of-Function Mutation in the Nav 1.5 Sodium Channel. Ann. Noninvasive Electrocardiol. 2016;21:343–351. doi: 10.1111/anec.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amarouch M.Y., Abriel H. Cellular hyper-excitability caused by mutations that alter the activation process of voltage-gated sodium channels. Front. Physiol. 2015;6:45. doi: 10.3389/fphys.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saber S., Amarouch M.Y., Fazelifar A.F., Haghjoo M., Emkanjoo Z., Alizadeh A., Houshmand M., Gavrilenko A.V., Abriel H., Zaklyazminskaya E.V. Complex genetic background in a large family with Brugada syndrome. Physiol. Rep. 2015:3. doi: 10.14814/phy2.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyle J.W., Makielski J.C. Diseases caused by mutations in Nav1.5 interacting proteins. Card. Electrophysiol. Clin. 2014;6:797–809. doi: 10.1016/j.ccep.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edokobi N., Isom L.L. Voltage-Gated Sodium Channel beta1/beta1B Subunits Regulate Cardiac Physiology and Pathophysiology. Front. Physiol. 2018;9:351. doi: 10.3389/fphys.2018.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salvarani N., Crasto S., Miragoli M., Bertero A., Paulis M., Kunderfranco P., Serio S., Forni A., Lucarelli C., Dal Ferro M., et al. The K219T-Lamin mutation induces conduction defects through epigenetic inhibition of SCN5A in human cardiac laminopathy. Nat. Commun. 2019;10:2267. doi: 10.1038/s41467-019-09929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerrone M., Lin X., Zhang M., Agullo-Pascual E., Pfenniger A., Chkourko Gusky H., Novelli V., Kim C., Tirasawadichai T., Judge D.P., et al. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 2014;129:1092–1103. doi: 10.1161/CIRCULATIONAHA.113.003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neubauer J., Rougier J.S., Abriel H., Haas C. Functional implications of a rare variant in the sodium channel beta1B subunit (SCN1B) in a 5-month-old male sudden infant death syndrome case. Heartrhythm Case Rep. 2018;4:187–190. doi: 10.1016/j.hrcr.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Hoeijen D.A., Blom M.T., Tan H.L. Cardiac sodium channels and inherited electrophysiological disorders: An update on the pharmacotherapy. Expert Opin. Pharmacother. 2014;15:1875–1887. doi: 10.1517/14656566.2014.936380. [DOI] [PubMed] [Google Scholar]
- 19.Mankad P., Kalahasty G. Antiarrhythmic Drugs: Risks and Benefits. Med Clin. North. Am. 2019;103:821–834. doi: 10.1016/j.mcna.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Boukhabza M., El Hilaly J., Attiya N., El-Haidani A., Filali-Zegzouti Y., Mazouzi D., Amarouch M.Y. In Silico Evaluation of the Potential Antiarrhythmic Effect of Epigallocatechin-3-Gallate on Cardiac Channelopathies. Comput. Math. Methods Med. 2016;2016:7861653. doi: 10.1155/2016/7861653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang J.H., Chang S.L., Hong P.D., Chen P.N., Hsu C.H., Lu Y.Y., Chen Y.C. Epigallocatechin-3-gallate modulates arrhythmogenic activity and calcium homeostasis of left atrium. Int. J. Cardiol. 2017;236:174–180. doi: 10.1016/j.ijcard.2017.01.090. [DOI] [PubMed] [Google Scholar]
- 22.Eng Q.Y., Thanikachalam P.V., Ramamurthy S. Molecular understanding of Epigallocatechin gallate (EGCG) in cardiovascular and metabolic diseases. J. Ethnopharmacol. 2018;210:296–310. doi: 10.1016/j.jep.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 23.Kuriyama S., Shimazu T., Ohmori K., Kikuchi N., Nakaya N., Nishino Y., Tsubono Y., Tsuji I. Green tea consumption and mortality due to cardiovascular disease, cancer, and all causes in Japan: The Ohsaki study. Jama. 2006;296:1255–1265. doi: 10.1001/jama.296.10.1255. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki J., Ogawa M., Maejima Y., Isobe K., Tanaka H., Sagesaka Y.M., Isobe M. Tea catechins attenuate chronic ventricular remodeling after myocardial ischemia in rats. J. Mol. Cell. Cardiol. 2007;42:432–440. doi: 10.1016/j.yjmcc.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki J., Ogawa M., Futamatsu H., Kosuge H., Sagesaka Y.M., Isobe M. Tea catechins improve left ventricular dysfunction, suppress myocardial inflammation and fibrosis, and alter cytokine expression in rat autoimmune myocarditis. Eur. J. Heart Fail. 2007;9:152–159. doi: 10.1016/j.ejheart.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y., Hu Y., Wu J., Wen J., Li S., Zhang L., Zhang J., Li Y., Li J. Epigallocatechin-3-gallate inhibits angiotensin II-induced cardiomyocyte hypertrophy via regulating Hippo signaling pathway in H9c2 rat cardiomyocytes. Acta Biochim. Et Biophys. Sin. 2019;51:422–430. doi: 10.1093/abbs/gmz018. [DOI] [PubMed] [Google Scholar]
- 27.Al Hroob A.M., Abukhalil M.H., Hussein O.E., Mahmoud A.M. Pathophysiological mechanisms of diabetic cardiomyopathy and the therapeutic potential of epigallocatechin-3-gallate. Biomed. Pharmacother. 2019;109:2155–2172. doi: 10.1016/j.biopha.2018.11.086. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q., Hu L., Chen L., Li H., Wu J., Liu W., Zhang M., Yan G. (−)-Epigallocatechin-3-gallate, the major green tea catechin, regulates the desensitization of beta1 adrenoceptor via GRK2 in experimental heart failure. Inflammopharmacology. 2018;26:1081–1091. doi: 10.1007/s10787-017-0429-x. [DOI] [PubMed] [Google Scholar]
- 29.Wei H., Meng Z. Epigallocatechin-3-gallate protects Na+ channels in rat ventricular myocytes against sulfite. Cardiovasc. Toxicol. 2010;10:166–173. doi: 10.1007/s12012-010-9075-x. [DOI] [PubMed] [Google Scholar]
- 30.Kang J., Cheng H., Ji J., Incardona J., Rampe D. In vitro electrocardiographic and cardiac ion channel effects of (-)-epigallocatechin-3-gallate, the main catechin of green tea. J. Pharmacol. Exp. Ther. 2010;334:619–626. doi: 10.1124/jpet.110.169391. [DOI] [PubMed] [Google Scholar]
- 31.Wang R., Zhou W., Jiang X. Reaction kinetics of degradation and epimerization of epigallocatechin gallate (EGCG) in aqueous system over a wide temperature range. J. Agric. Food Chem. 2008;56:2694–2701. doi: 10.1021/jf0730338. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., Jia Y.Y., Guo J.L., Liu P.Q., Jiang J.M. Effects of (-)-gallocatechin-3-gallate on tetrodotoxin-resistant voltage-gated sodium channels in rat dorsal root ganglion neurons. Int. J. Mol. Sci. 2013;14:9779–9789. doi: 10.3390/ijms14059779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdelsayed M., Ruprai M., Ruben P.C. The efficacy of Ranolazine on E1784K is altered by temperature and calcium. Sci. Rep. 2018;8:3643. doi: 10.1038/s41598-018-22033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Battrawy I., Lang S., Zhao Z., Akin I., Yucel G., Meister S., Patocskai B., Behnes M., Rudic B., Tulumen E., et al. Hyperthermia Influences the Effects of Sodium Channel Blocking Drugs in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. PLoS ONE. 2016;11:e0166143. doi: 10.1371/journal.pone.0166143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng H.M., Yin S.T., Yan D., Tang M.L., Li C.C., Chen J.T., Wang M., Ruan D.Y. Effects of EGCG on voltage-gated sodium channels in primary cultures of rat hippocampal CA1 neurons. Toxicology. 2008;252:1–8. doi: 10.1016/j.tox.2008.07.053. [DOI] [PubMed] [Google Scholar]
- 36.Kim T.H., Lim J.M., Kim S.S., Kim J., Park M., Song J.H. Effects of (−) epigallocatechin-3-gallate on Na(+) currents in rat dorsal root ganglion neurons. Eur. J. Pharmacol. 2009;604:20–26. doi: 10.1016/j.ejphar.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 37.Salazar P.B., de Athayde Moncorvo Collado A., Canal-Martinez V., Minahk C.J. Differential inhibition of human erythrocyte acetylcholinesterase by polyphenols epigallocatechin-3-gallate and resveratrol. Relevance of the membrane-bound form. BioFactors. 2017;43:73–81. doi: 10.1002/biof.1322. [DOI] [PubMed] [Google Scholar]
- 38.Ingolfsson H.I., Koeppe R.E., 2nd, Andersen O.S. Effects of green tea catechins on gramicidin channel function and inferred changes in bilayer properties. Febs Lett. 2011;585:3101–3105. doi: 10.1016/j.febslet.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gamal El-Din T.M., Lenaeus M.J., Zheng N., Catterall W.A. Fenestrations control resting-state block of a voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 2018;115:13111–13116. doi: 10.1073/pnas.1814928115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henning S.M., Niu Y., Lee N.H., Thames G.D., Minutti R.R., Wang H., Go V.L., Heber D. Bioavailability and antioxidant activity of tea flavanols after consumption of green tea, black tea, or a green tea extract supplement. Am. J. Clin. Nutr. 2004;80:1558–1564. doi: 10.1093/ajcn/80.6.1558. [DOI] [PubMed] [Google Scholar]
- 41.Chow H.H., Cai Y., Hakim I.A., Crowell J.A., Shahi F., Brooks C.A., Dorr R.T., Hara Y., Alberts D.S. Pharmacokinetics and safety of green tea polyphenols after multiple-dose administration of epigallocatechin gallate and polyphenon E in healthy individuals. Clin. Cancer Res. 2003;9:3312–3319. [PubMed] [Google Scholar]
- 42.Ullmann U., Haller J., Decourt J.P., Girault N., Girault J., Richard-Caudron A.S., Pineau B., Weber P. A single ascending dose study of epigallocatechin gallate in healthy volunteers. J. Int. Med Res. 2003;31:88–101. doi: 10.1177/147323000303100205. [DOI] [PubMed] [Google Scholar]
- 43.Lee M.J., Maliakal P., Chen L., Meng X., Bondoc F.Y., Prabhu S., Lambert G., Mohr S., Yang C.S. Pharmacokinetics of tea catechins after ingestion of green tea and (-)-epigallocatechin-3-gallate by humans: Formation of different metabolites and individual variability. Cancer Epidemiol. Biomark. Prev. 2002;11:1025–1032. [PubMed] [Google Scholar]
- 44.Narumi K., Sonoda J., Shiotani K., Shigeru M., Shibata M., Kawachi A., Tomishige E., Sato K., Motoya T. Simultaneous detection of green tea catechins and gallic acid in human serum after ingestion of green tea tablets using ion-pair high-performance liquid chromatography with electrochemical detection. J. Chromatography. Banalytical Technol. Biomed. Life Sci. 2014;945–946:147–153. doi: 10.1016/j.jchromb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Lambert J.D., Lee M.J., Lu H., Meng X., Hong J.J., Seril D.N., Sturgill M.G., Yang C.S. Epigallocatechin-3-gallate is absorbed but extensively glucuronidated following oral administration to mice. J. Nutr. 2003;133:4172–4177. doi: 10.1093/jn/133.12.4172. [DOI] [PubMed] [Google Scholar]
- 46.Yang C.S., Wang X., Lu G., Picinich S.C. Cancer prevention by tea: Animal studies, molecular mechanisms and human relevance. Nat. Reviews. Cancer. 2009;9:429–439. doi: 10.1038/nrc2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambert J.D., Lee M.J., Diamond L., Ju J., Hong J., Bose M., Newmark H.L., Yang C.S. Dose-dependent levels of epigallocatechin-3-gallate in human colon cancer cells and mouse plasma and tissues. Drug Metab. Dispos. Biol. Fate Chem. 2006;34:8–11. doi: 10.1124/dmd.104.003434. [DOI] [PubMed] [Google Scholar]
- 48.Nagle D.G., Ferreira D., Zhou Y.D. Epigallocatechin-3-gallate (EGCG): Chemical and biomedical perspectives. Phytochemistry. 2006;67:1849–1855. doi: 10.1016/j.phytochem.2006.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krupkova O., Ferguson S.J., Wuertz-Kozak K. Stability of (-)-epigallocatechin gallate and its activity in liquid formulations and delivery systems. J. Nutr. Biochem. 2016;37:1–12. doi: 10.1016/j.jnutbio.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 50.Li N., Taylor L.S., Ferruzzi M.G., Mauer L.J. Kinetic study of catechin stability: Effects of pH, concentration, and temperature. J. Agric. Food Chem. 2012;60:12531–12539. doi: 10.1021/jf304116s. [DOI] [PubMed] [Google Scholar]
- 51.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 52.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu E.L., Cheng X., Jo S., Rui H., Song K.C., Davila-Contreras E.M., Qi Y., Lee J., Monje-Galvan V., Venable R.M., et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klauda J.B., Venable R.M., Freites J.A., O’Connor J.W., Tobias D.J., Mondragon-Ramirez C., Vorobyov I., MacKerell A.D., Jr., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chemistry. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 56.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 57.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. doi: 10.1063/1.447334. [DOI] [Google Scholar]
- 58.Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 59.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 60.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.