Abstract
Many early studies presented beneficial effects of polyunsaturated fatty acids (PUFA) on cardiovascular risk factors and disease. However, results from recent meta-analyses indicate that this effect would be very low or nil. One of the factors that may contribute to the inconsistency of the results is that, in most studies, genetic factors have not been taken into consideration. It is known that fatty acid desaturase (FADS) gene cluster in chromosome 11 is a very important determinant of plasma PUFA, and that the prevalence of the single nucleotide polymorphisms (SNPs) varies greatly between populations and may constitute a bias in meta-analyses. Previous genome-wide association studies (GWAS) have been carried out in other populations and none of them have investigated sex and Mediterranean dietary pattern interactions at the genome-wide level. Our aims were to undertake a GWAS to discover the genes most associated with serum PUFA concentrations (omega-3, omega-6, and some fatty acids) in a scarcely studied Mediterranean population with metabolic syndrome, and to explore sex and adherence to Mediterranean diet (MedDiet) interactions at the genome-wide level. Serum PUFA were determined by NMR spectroscopy. We found strong robust associations between various SNPs in the FADS cluster and omega-3 concentrations (top-ranked in the adjusted model: FADS1-rs174547, p = 3.34 × 10−14; FADS1-rs174550, p = 5.35 × 10−14; FADS2-rs1535, p = 5.85 × 10−14; FADS1-rs174546, p = 6.72 × 10−14; FADS2-rs174546, p = 9.75 × 10−14; FADS2-rs174576, p = 1.17 × 10−13; FADS2-rs174577, p = 1.12 × 10−12, among others). We also detected a genome-wide significant association with other genes in chromosome 11: MYRF (myelin regulatory factor)-rs174535, p = 1.49 × 10−12; TMEM258 (transmembrane protein 258)-rs102275, p = 2.43 × 10−12; FEN1 (flap structure-specific endonuclease 1)-rs174538, p = 1.96 × 10−11). Similar genome-wide statistically significant results were found for docosahexaenoic fatty acid (DHA). However, no such associations were detected for omega-6 PUFAs or linoleic acid (LA). For total PUFA, we observed a consistent gene*sex interaction with the DNTTIP2 (deoxynucleotidyl transferase terminal interacting protein 2)-rs3747965 p = 1.36 × 10−8. For adherence to MedDiet, we obtained a relevant interaction with the ME1 (malic enzyme 1) gene (a gene strongly regulated by fat) in determining serum omega-3. The top-ranked SNP for this interaction was ME1-rs3798890 (p = 2.15 × 10−7). In the regional-wide association study, specifically focused on the FADS1/FASD2/FADS3 and ELOVL (fatty acid elongase) 2/ELOVL 5 regions, we detected several statistically significant associations at p < 0.05. In conclusion, our results confirm a robust role of the FADS cluster on serum PUFA in this population, but the associations vary depending on the PUFA. Moreover, the detection of some sex and diet interactions underlines the need for these associations/interactions to be studied in all specific populations so as to better understand the complex metabolism of PUFA.
Keywords: polyunsaturated fatty acids, omega-3, omega-6, genetics, genome-wide association study, sex, Mediterranean population, heterogeneity, polymorphisms, metabolic syndrome
1. Introduction
Polyunsaturated fatty acids (PUFA) are fatty acids having more than one double bond on their carbon chain. Depending on the position of the first double bond close to the methyl end, they are classified as omega-6 or omega-3. These 2 families each encompass fatty acids with varying carbon chain length and degree of unsaturation. Supplemental Figure S1 shows PUFA biosynthesis, detailing the omega-6 and omega-3 pathways and the names of the main PUFA, as well as the elongation and the desaturation activities [1,2,3]. Briefly, the omega-6 PUFA linoleic acid (LA) and the omega-3 PUFA alpha-linolenic acid (ALA) from the diet are the key substrates that enter the pathways leading to the synthesis of the main long-chain PUFA. Long chain PUFA can be endogenously derived from (LA) or (ALA) by a consecutive series of desaturations (involving the Delta-6 desaturase encoded by the FADS2 (fatty acid desaturase 2) gene and the Delta-5 desaturase, encoded by the FADS1 (fatty acid desaturase 1) gene and elongation steps, as well as oxidation. Among the seven elongases, ELOVL2 (elongation of very long chain fatty acids 2) and ELOVL5 elongation of very long chain fatty acids 5) are PUFA-specific [4]. Docosahexaenoic acid (DHA) is a long-chain, highly unsaturated omega-3 fatty acid, synthesized from the ALA in this process. Both dietary and serum/plasma PUFA are a complex mix of different fatty acids. Circulating levels are determined by intake and by the biosynthesis pathways [5]. Omega-3 PUFA mainly exists in fish, marine oils, and other marine products, as well as in several plant sources [6,7,8,9]. Vegetable oils, nuts, seeds, meat, poultry, and cereal-based products are important sources of omega-6 PUFA [6,7,8,9].
Initial studies, both in humans and in animal models, showed several protective effects (hypotriglyceridemic, hypocholesterolemic, antithrombotic, anti-inflammatory, anti-arrhythmic, anti-hypertensive, improving vascular function and arterial compliance, etc.) of PUFA intake on cardiovascular risk [10,11,12,13,14,15,16]. This PUFA protective effect has been the focus of several dietary and pharmacological studies with PUFA on diverse cardiovascular risk factors as well as cardiovascular diseases for primary and secondary prevention. A detailed analysis of the findings of such studies is beyond the aim of the present work. There are many excellent reviews that cover these results [17,18,19,20,21,22,23,24,25,26,27,28,29]. However, the association between PUFA and cardiovascular risk factors and diseases has been surrounded by controversy [30,31,32,33,34,35] and the results of recent meta-analyses have been inconsistent [36,37,38,39,40,41].
Abdelhamid et al. [36] meta-analyzed 49 randomized, controlled trials (RCTs), examining the effects of total PUFA intake (including both diet or supplements) on lipids, adiposity, cardiovascular disease, and all-cause mortality. These authors concluded that increasing total PUFA intake probably reduces the risk of cardiovascular disease events slightly, but has little or no effect on all-cause or cardiovascular disease mortality [36]. Nevertheless, it is known that PUFA are a mix of different fatty acids. Early studies have already suggested that there could be differential effects of omega-6 and omega-3 PUFA on cardiovascular risk [42,43,44]. In general, it has been accepted that omega-3 PUFA helps decrease inflammation, whereas some omega-6 PUFA could promote inflammation [45,46,47,48,49]. The concept of a healthy omega-6 to omega-3 PUFA ratio in the diet, therefore, has been suggested [50,51]. Initially, the dietary content of omega-6 and omega-3 in the diet was well balanced, and a ratio close to 1:1 has been estimated for hunter–gatherer populations [52]. However, presently, Western diets are unbalanced worldwide due to an increase of dietary omega-6 and a decrease in omega-3. This results in a progressive shift in the omega-6/omega-3 ratio from 4:1 to >10:1 [53,54]. Although the optimal ratio has to be determined, an increased ratio may be harmful to human health and it has been related to metabolic dysfunction and higher cardiovascular risk in animal models and in human studies [48,49,50,51,52,53,54,55,56,57,58,59].
Nevertheless, recent studies suggest that omega-6 are not as harmful as initially thought [52,53]. Therefore, whether omega-6 and omega-3 PUFA oppositely contribute to a decrease in cardiovascular risk still remains debated. In a recent and comprehensive meta-analyses on the effects of omega-3 [37] and omega-6 PUFA [38] on cardiovascular risk, it has been pointed out that high quality evidence levels indicate that omega-3 PUFA has little or no effect on mortality or cardiovascular health. Likewise, the results indicated that increasing omega-6 PUFA only reduced myocardial infarction incidence but no other cardiovascular outcomes [39]. Similarly, Brown et al. [41] in an extensive meta-analysis including 83 RCTs analyzing the effect of PUFA for the prevention and treatment of type-2 diabetes, did not find any evidence that the omega-6/omega-3 ratio is important for diabetes or glucose metabolism.
Nonetheless, it has been claimed that the lack of conclusive evidence obtained in the meta-analyses may be due to several biases, including not only the poor quality of some of the meta-analyzed trials, but also other relevant factors such as the unmeasured role of genetic polymorphisms influencing the effect of dietary PUFA [1]. The potential role of FADS genetic polymorphism in determining the effects of dietary and pharmacological PUFA on several cardiovascular risk factors and outcomes, have been investigated by several studies [1,60,61,62,63,64,65]. Among them, a post-hoc analysis from the OMEGA-REMODEL RCT, investigating the effect of high-dose of omega-3 PUFA post-acute myocardial infarction [65] reported that the FADS2 genotype can predict the beneficial effects of the omega-3. Another issue in these meta-analyses is the lack of focus on potential sex-specific differences, as well as overall dietary pattern modulating PUFA interventions. Regarding sex, several human studies as well as research in animal models [66,67,68,69,70,71,72,73] have reported some differences between males and females on analyzing the PUFA effects on diverse cardiovascular risk phenotypes. However, more specific research remains to be done in order to better understand the potential differences. Likewise, the effect of the concomitant dietary pattern (Mediterranean diet, Nordic diet, Western diet, Chinese diet, etc.) on the PUFA effects (diet or supplements) on cardiovascular phenotypes in different populations has been subject to increased attention in recent years [74,75,76,77,78]. However, more research is needed to better understand the dietary pattern influence.
All these factors should be considered in any new RCTs to be conducted in the field, as well as in the corresponding meta-analyses. In terms of the genetic factors to be analyzed in novel studies related to PUFA, although several genome-wide association studies (GWASs) for the different fatty acid concentrations in numerous tissues have been carried out in several populations, mainly of European ancestry [79,80,81,82,83,84,85,86], the percentage of variability explained by the polymorphisms discovered is still low for the different PUFA. Thus, additional research remains to be done to discover more PUFA-related genes. Despite the strong associations at the GWAS level between the FADS1/2 locus in chromosome 11, and some fatty acids observed in all populations analyzed [79,80,81,82,83,84,85,86], some population-specific associations with other genetic variants and genes (i.e., TRIM58, MCM6, NTAN1/PDXDC1, PKD2L1, PCOLCE2, AGPAT4, HBS1L/MYB, LPCAT3) have also been described [79,81,82,84,86]. Specifically, in the GWAS carried out in Singaporean Chinese population [84] to test the homogeneity of previous findings in European ancestry populations, the authors found robust consistency for the main associations, but reported some heterogeneity for some loci (they were unable to detect the significant associations with ELOVL2 single nucleotide polymorphisms (SNPs), and the NTAN1/PDXDC1 for the corresponding PUFA). Moreover, in another GWAS undertaken in Chinese participants, followed by a trans-ethnic meta-analysis including European ancestry participants [86], confirmed previously reported loci in the different ethnicities and also reported some specific associations in Chinese participants. In this sense, several studies have warned of the importance of knowing the most relevant specific variants in each population in order to minimize bias, since the results SNPs identified in GWAS of specific populations, cannot be directly extrapolated to other populations [87,88,89]. In view of that, the vast majority of GWASs for PUFA levels have been carried out in populations participating in the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) Consortium [79,80,81,82,83], in Asian populations [84,86] and in Greenlandic Inuit subjects [85]. Although the CHARGE Consortium has analyzed one Italian population [79], no GWAS for serum PUFA has been carried out in Spain. Previous genetic analysis in this population has reported population-specific associations for some traits [90].
In general, GWASs for PUFA [79,80,81,82,83,84,85,86] have focused on serum/plasma or erythrocyte fatty acid levels. In these GWASs [79,80,81,82,83,84,85,86], a strong statistically significant signal has been consistently detected at the candidate region of the FADS1/FADS2 cluster, previously identified in initial studies [91,92,93,94]. FADS3 is the enigmatic third member of the FADS cluster [4]. It is important to bear in mind that the prevalence of the main SNPs in the FADS genes greatly varies among populations [95,96,97]. This reflects an evolutionary advantage of SNPs, enabling an active PUFA synthesis in response to drastic dietary differences [95,96,97,98]. Likewise, it has been suggested that there might be stronger selection pressure on the FADS1/FASD2 block in southern Europeans [95]. In Spain, the adherence to the Mediterranean diet pattern [99] is still relatively high in older subjects, but is decreasing in young people [100,101]. Although some studies specifically carried out in the Spanish Mediterranean population, have analyzed the association between several FADS and ELOVL candidate gene polymorphisms and PUFA concentrations [102,103,104,105,106], no study has investigated such associations at the GWAS level.
Therefore, the potential genetic differences in the Mediterranean population, the urgent need for greater diversity in genetic analysis, and the fact that in previous GWASs, sex-specific differences as well as dietary interactions have scarcely been analyzed, prompted us to carry out this study with the following aims: (1) To investigate, at the genome-wide level, the genetic variants most associated with serum PUFA concentrations (focusing on omega-3 and omega-6 PUFA, and also examining DHA and LA) in a Mediterranean population with metabolic syndrome; (2) To analyze, at the region-wide (RWAS) level, the associations between the SNPs in the selected candidate regions and serum PUFA concentrations in this population; and (3) To explore the modulating influence of sex and adherence to Mediterranean diet (as a whole dietary pattern) on these associations.
2. Materials and Methods
2.1. Study Design and Participants
We analyzed participants in the PREDIMED Plus-Valencia study [105]. The University of Valencia (located in the eastern Mediterranean coast), is one of the field centers of the multi-center PREDIMED Plus study, an ongoing primary prevention trial conducted in Spain [106]. A detailed description of the trial is available at http://predimedplus.com/. This trial was registered at https://doi.org/10.1186/ISRCTN89898870. Participants in the Valencia field center were recruited from several primary care health facilities. These participants were community-dwelling adults (men, 55–75 years; women, 60–75 years) with body-mass index (BMI) ranging from 27 to 40 kg/m2 and having metabolic syndrome [106]. In this recruitment center, the total number of randomized participants included in the PREDIMED Plus trial was 465. In the present work, we undertake the cross-sectional analyses of an ancillary project including all the participants in our center with complete data on serum PUFA concentrations at baseline and genome-wide genotyping, in addition to the other variables for adjustments (n = 426). The analyzed subjects did not differ significantly from our entire sample concerning the main variables. The Institutional Review Board of the Valencia University approved the study protocol (ethical approval code H1373255532771), and all participants provided written informed consent.
2.2. Baseline Clinical, Anthropometric, Biochemical, and Lifestyle Variables
In the baseline examination, we assessed socio-demographic variables, clinical and cardiovascular risk factors, and lifestyle variables by validated questionnaires as previously reported [106]. Anthropometric variables and blood pressure were determined by trained staff following the PREDIMED Plus operations protocol [107]. Weight and height were measured with calibrated scales and a wall-mounted stadiometer, respectively. BMI was calculated as the weight in kilograms divided by the height in meters squared. Waist circumference was measured midway between the lowest rib and the iliac crest, after normal exhalation, using an anthropometric tape. Blood pressure was measured with the use of a validated semiautomatic oscillometer (Omron HEM-705CP, Netherlands) while the participant was in a seated position for 5 min.
Blood samples were collected after a 12-h overnight fast. Fasting plasma glucose, total cholesterol, high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglyceride concentrations were measured as previously described [107]. Leisure-time physical activity was assessed using the validated REGICOR questionnaire [108], including questions to collect information on the type of activity, frequency (number of days), duration, and total leisure-time physical activity-related energy expenditure was estimated. The dietary pattern was assessed by a 17-item score capturing adherence to a traditional Mediterranean diet [109]. We have published the detailed version of this questionnaire as well as its association with quality of life and cardiovascular risk factors [109]. Briefly, traditional Mediterranean diet is characterized by a high intake of vegetables, fruits, legumes, fish, as well as fat from vegetable sources, including nuts, and a low intake of red meat and desserts. In the 17-item screener, a suitable consumption of typical traditional Mediterranean foods adds one point and a low consumption of foods not characteristic of the Mediterranean diet also adds one point. A higher score means higher adherence to the Mediterranean diet. Supplemental Table S1 shows the 17 detailed items and the corresponding scores.
2.3. Serum Fatty Acid Determinations
Serum was obtained from fasting blood samples as previously detailed [55]. Serum fatty acids were measured by proton nuclear magnetic resonance (NMR) spectroscopy as previously reported in detail [110,111]. This high—throughput NMR metabolomics platform (Nightingale Health Ltd., Helsinki, Finland), has been applied and validated in numerous studies [112,113]. The measurements of PUFA with this NMR platform were only undertaken on participants recruited at the PREDIMED Plus-Valencia field center. Currently, there are no participants from other field centers with these data determined. In this study, we used all the available measurements of serum fatty acids provided by the platform. These included total fatty acids, monounsaturated fatty acids (MUFA), saturated fatty acids (SFA), total PUFA, omega-3 PUFA, and omega-6 PUFA. We also included LA and DHA for being the two specific fatty acids that that platform reported separately. No additional PUFA were reported in this platform. The results for total PUFA, omega-3, omega-6, and LA and DHA are very robust and have been validated in many publications [110,111,112,113,114,115]. Although we focused our genetic analyses on PUFA, for descriptive reasons, SFA, MUFA, and total fatty acids were also reported. Serum fatty acids were measured as follows: After measuring the lipoprotein data from native serum samples, their lipids were extracted and another NMR spectrum measured (see reference [116] for details). In these lipid extracts, each fatty acid gives rise to a characteristic NMR resonance, the area of which is associated with their concentration. After further scaling to account for slight experimental variation in lipid acquisitions in the extraction procedure [116], the different PUFA concentrations were quantified in mmol/L, representing the true concentration in serum. Finally, serum fatty acids were expressed as a percentage of total fatty acids. Although both measures (absolute and relative) can be used [117], we have analyzed PUFA and percentage because in previous works, fatty acids have been commonly used as percentage of total, and we want to compare our results with those obtained in other populations.
2.4. Genome-Wide Genotyping
We isolated genomic DNA from the buffy coat samples with the MagNaPure LC DNA Isolation kit (ROCHE Diagnostics). We measured the concentration and the quality of the isolated DNA by the PicoGreen instrument (Invitrogen Corporation, Carlsbad, CA, USA). After DNA-quality control, high-density genotyping was performed in samples passing this control using the Infinium OmniExpress-24 v1.2 BeadChip genotyping array (Illumina Inc., San Diego, CA, USA) at the University of Valencia, Valencia. Genotyping was undertaken according to the manufacturer’s protocol with appropriate quality standards. The Infinium OmniExpress v1.2 BeadChip captures 713,599 markers. Allele detection and genotype calling were performed in the Genome Studio genotyping module (Illumina, Inc., San Diego, CA, USA). Data cleaning was performed using standard analysis pipelines implemented in the Python programing language using the Numpy library modules combined with PLINK [118,119]. From the initial full set of SNPs in the array, those not mapped on autosomal chromosomes were filtered out. In addition, SNPs with a minor allele frequency (MAF) <0.01 or those that deviated from expected Hardy–Weinberg equilibrium (p < 1.0 × 10−4) were removed. A total of 621,723 SNPs that passed the quality filter remained for further analysis.
2.5. Statistical Analysis
Student t-tests and ANOVA tests were applied to compare crude means of continuous variables. Triglyceride and fatty acid concentrations (in mmol/L) were log-transformed for the statistical analyses. Fatty acids, expressed as % (total fatty acid of interest to total fatty acids), reached a normal distribution, and were not log transformed. Chi-square tests were used to compare proportions. Finally, we analyzed the omega-6/omega-3 ratio and we log transformed this variable to achieve normality for statistical testing. We analyzed the associations using both crude models and adjusted multivariate regression models (general lineal models) including potential confounders.
For the GWAS, at the genome-wide level, we tested the association between all the SNPs in the array (excluding those indicated above according to the quality control issues) and serum concentrations (expressed as %) of the different PUFA. We fitted separate models for omega-3 PUFA, DHA, omega-6 PUFA, LA, and total PUFA. Statistical models were sequentially adjusted by sex, age, diabetes, and other covariates including BMI, smoking, medications, physical activity, and adherence to Mediterranean diet (17-item score). Additional adjustment did not change the statistical level of significance. Therefore, for the sake of simplicity, and to compare our results with the previous GWAS carried out in different populations [79,80,81,82,83,84,85,86], that were mainly adjusted for sex and age, unadjusted models (Model 1) and multivariate models sequentially adjusted for sex, age, and diabetes (Model 2), were presented in tables and figures. Additional adjustments were specifically presented when indicated. For the study of genome-wide interactions with selected factors, hierarchical statistical general linear models were fitted. These models include the main variables and the corresponding interaction terms. Gene*sex and gene*diet genome-wide interactions were analyzed. For the study of the interactions between the SNPs and adherence to Mediterranean diet, we categorized baseline adherence to the Mediterranean diet (17-item screening) into two groups based on the sample median (8 points), defining 2 groups as either “Low” adherence to Mediterranean diet (0–8 points), or “High” adherence to Mediterranean diet (9–17 points). This cut-off point has been previously used by our group in another genome-wide interaction study [105].
For GWAS, genetic association analyses were performed using PLINK v1.9 [118,119]. Additive genetic models (unadjusted and multivariate adjusted when indicated) were fitted using a genotype dosage (0, 1, or 2 copies of the variant allele). Regression coefficients for the minor allele were estimated. Likewise, determination coefficients (r2) were also estimated. To test gene*sex and gene*diet interactions at the genome-wide level, we used the PLINK GxE tool and showed the statistical significance on the interaction terms, as well as the regression coefficients for each stratum. Stratified analyses of interest were additionally carried out when indicated. We used the conventional threshold of p < 5 × 10−8 for genome-wide statistical significance. Likewise, according to the conventional GWAS rules, SNPs with p-values below 1 × 10−5 were also considered suggestive of genome-wide significance. As these thresholds are very conservative for a small sample size, in selected analyses, SNPs with p-values below 5 × 10−5 are also shown in the tables so that they may be compared with other studies that have analyzed similar associations and may be included in meta-analyses. SNPs were rank-ordered according to the minimum p-value in the genetic models.
We used R Qqman R library [120] and Haploview (version 4.2) [121] to create Manhattan plots and to calculate linkage disequilibrium among the SNPs of interest. Quantile–quantile plot comparing the expected and observed p-values [121] was performed in the R-statistical environment.
In addition to the GWAS approach, to explore potential associated loci (candidates based on previous knowledge) which might fail to be identified at the GWAS level due to the strict threshold for the whole genome approach (p < 5 × 10−8 for genome-wide statistical significance), we carried out additional RWASs. In these analyses, we previously selected a candidate region of the genome containing the most relevant genes previously associated with PUFA and analyzed the associations with all the SNPs present in the selected region. Two candidate regions were selected [4,94]: (A) the region in chromosome 11 containing the FASD1/FASD2/FASD3 cluster [94]; and (B) the region in chromosome 6 containing the ELOVL2 and ELOVL5 genes [4]. All the SNPs in the selected genes for each region, included in the Human OmniExpress array were analyzed for associations with the serum PUFA (omega-3, DHA, omega-6, LA, and total PUFA). The corresponding genotype data were extracted by a Python script, created by the authors. For selecting the available SNPs in these genes, we used another Python script, also created by the authors. For this procedure, we first used the batch query resource SNP Report with URL: https://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId, and provided the target gene-ID. The dbSNP-gene type file is used as input argument in the *.bat file for running the Python script. Finally, the Python script produces the corresponding PLINK-formatted couple *.ped and *.map files of genotypes for statistical testing. For the selected regions, we carried out unadjusted and multivariate adjusted analyses determining the serum PUFA, as previously detailed for the GWAS approach. Models adjusted for sex, age, and diabetes were presented. We also used PLINK v.1.9 [118,119] for these models. As the regional exploration is a replication analysis and our sample size is small, we considered a two-tailed p-value <0.05 as statistically significant for the RWAS associations.
3. Results
3.1. General Characteristics of the Population
Table 1 presents the demographic, lifestyle, and clinical characteristics of the study participants at baseline by sex. We analyzed 426 subjects, including 187 men and 239 women. These were all the subjects recruited and randomized in the Valencia field center having complete data of serum fatty acid determinations by NMR spectroscopy [110,111] and genome-wide genotyping available. These were individuals aged 65.1 ± 0.2 years, with metabolic syndrome. The average mean age of men and women was 64.0 ± 0.4 years and 66.0 ± 0.3 years (p < 0.001), respectively. BMI did not differ significantly in men and women. The women analyzed in this study were all post-menopausal women.
Table 1.
Demographic, clinical, lifestyle, and biochemical characteristics of the study participants at baseline according to sex.
| Variables | Total (n = 426) | Men (n = 187) | Women (n = 239) | p |
|---|---|---|---|---|
| Age (years) | 65.1 ± 0.2 | 64.0 ± 0.4 | 66.0 ± 0.3 | <0.001 |
| Weight (Kg) | 84.2 ± 0.7 | 92.5 ± 1.0 | 77.7 ± 0.6 | <0.001 |
| BMI (Kg/m2) | 32.3 ± 0.2 | 32.2 ± 0.3 | 32.3 ± 0.2 | 0.836 |
| Waist circumference (cm) | 105.8 ± 0.5 | 110.9 ± 0.6 | 101.8 ± 0.6 | <0.001 |
| SBP (mm Hg) | 141.9 ± 0.9 | 144.1 ± 1.4 | 140.1 ± 1.2 | 0.029 |
| DBP (mm Hg) | 81.0 ± 0.5 | 82.7 ± 0.7 | 79.6 ± 0.6 | 0.001 |
| Total cholesterol (mg/dL) | 196.4 ± 1.8 | 188.3 ± 2.9 | 202.7 ± 2.3 | <0.001 |
| LDL-C (mg/dL) | 124.9 ± 1.5 | 121.9 ± 2.4 | 127.2 ± 1.9 | 0.078 |
| HDL-C (mg/dL) | 51.6 ± 0.6 | 47.3 ± 0.8 | 55.0 ± 0.7 | <0.001 |
| Triglycerides (mg/dL) | 140.7 ± 2.9 | 137.6 ± 3.9 | 143.1 ± 4.1 | 0.334 |
| Fasting glucose (mg/dL) | 112.4 ± 1.3 | 113.5 ± 2.1 | 111.5 ± 1.6 | 0.441 |
| Total fatty acids (mmol/L) | 11.09 ± 0.10 | 10.63 ± 0.14 | 14.46 ± 0.14 | <0.001 |
| SFA (mmol/L) | 4.02 ± 0.04 | 3.85 ± 0.05 | 4.16 ± 0.05 | <0.001 |
| MUFA (mmol/L) | 3.09 ± 0.03 | 2.97 ± 0.04 | 3.20 ± 0.05 | <0.001 |
| PUFA (mmol/L) | 3.97 ± 0.04 | 3.81 ± 0.05 | 4.10 ± 0.05 | <0.001 |
| Omega-3 (mmol/L) | 0.46 ± 0.01 | 0.43 ± 0.01 | 0.48 ± 0.01 | <0.001 |
| Omega-6 (mmol/L) | 3.52 ± 0.03 | 3.38 ± 0.04 | 3.63 ± 0.04 | <0.001 |
| DHA (mmol/L) | 0.146 ± 0.002 | 0.140 ± 0.003 | 0.150 ± 0.003 | 0.013 |
| LA (mmol/L) | 2.88 ± 0.03 | 2.78 ± 0.04 | 2.97 ± 0.04 | 0.001 |
| SFA (%) 1 | 36.27 ± 0.06 | 36.25 ± 0.09 | 36.29 ± 0.08 | 0.722 |
| MUFA (%) 2 | 27.82 ± 0.09 | 27.85 ± 0.13 | 27.79 ± 0.13 | 0.754 |
| PUFA (%) 3 | 35.90 ± 0.11 | 35.89 ± 0.16 | 35.91 ± 0.15 | 0.950 |
| Omega-3 (%) 4 | 4.09 ± 0.03 | 4.04 ± 0.05 | 4.13 ± 0.05 | 0.182 |
| Omega-6 (%) 5 | 31.80 ± 0.10 | 31.85 ± 0.15 | 31.78 ± 0.14 | 0.723 |
| DHA (%) 6 | 1.30 ± 0.01 | 1.31 ± 0.02 | 1.30 ± 0.02 | 0.710 |
| LA (%) 7 | 26.04 ± 0.11 | 26.18 ± 0.18 | 25.90 ± 0.15 | 0.272 |
| Omega-6/Omega-3 ratio | 7.98 ± 0.07 | 8.07 ± 0.10 | 7.91 ± 0.09 | 0.254 |
| Type 2 diabetes: n, % | 160 (37.6) | 70 (37.4) | 90 (37.7) | 0.962 |
| Current smokers: n, % | 46 (10.8) | 28 (15.0) | 18 (7.5) | <0.001 |
| Antihypertensive drugs | 334 (78.4) | 148 (79.1) | 186 (77.8) | 0.742 |
| Hypolipidemic drugs | 274 (64.3) | 121 (64.7) | 153 (64.0) | 0.883 |
| Metformin treatment | 128 (30.0) | 56 (29.9) | 72 (30.1) | 0.968 |
| Insulin treatment | 19 (4.5) | 11 (5.9) | 8 (3.3) | 0.208 |
| Physical Activity (MET.min/wk) | 1724 ± 76 | 1950 ± 132 | 1547 ± 85 | 0.008 |
| Adherence to MedDiet (P17) 8 | 8.1 ± 0.1 | 7.8 ± 0.2 | 8.2 ± 0.2 | 0.145 |
Values are number and %. BMI indicates body mass index; MedDiet, Mediterranean diet; p: p-value for the comparisons (means or %) between men and women; PUFA: Polyunsaturated fatty acid; MUFA: Monounsaturated fatty acid; SFA: Saturated fatty acid; LA: Linoleic fatty acid; DHA: Docosahexaenoic fatty acid; MET: Metabolic equivalent. 1: Ratio of saturated fatty acid to total fatty acid. 2: Ratio of monounsaturated fatty acid to total fatty acid. 3: Ratio of polyunsaturated fatty acid to total fatty acid. 4: Ratio of omega-3 fatty acid to total fatty acid. 5: Ratio of omega-6 fatty acid to total fatty acid. 6: Ratio of docosahexaenoic fatty acid to total fatty acid. 7: Ratio of linoleic fatty acid to total fatty acid. 8: Quantitative 17-item questionnaire for adherence to Mediterranean diet.
Table 1 also shows total fatty acid concentrations in serum determined in the NMR platform as well as MUFA, SFA, PUFA, omega-3, and omega-6. In addition to these total profiles, the platform also reported specific measures for LA and DHA. Fatty acid concentrations were expressed in mmol/L as well as in % (ratio of the corresponding fatty acid to the total fatty acids). We used PUFA expressed as % for further statistical analyses.
Total PUFA was 35.9 ± % of the total fatty acids and did not differ between men and women. Similar percentages of omega-3 PUFA (4.09% total; p = 0.182 for sex differences) and omega-6 PUFA (31.8% total; p = 0.723 for sex differences were detected in men and women). Likewise, the percentage of DHA and LA did not differ per sex. We also calculated the omega-6 to omega-3 ratio (no units for the ratio). This ratio was 7.98 ± 0.07 for the whole population and no statistically significant differences were detected between men and women (p = 0.254).
In these subjects with metabolic syndrome, consumption of antihypertensive drugs was high (78.4%). Consumption of lipid-lowering drugs (64.3%), consisting of mainly statins, was also high. No significant differences by sex were detected regarding for antihypertensive, lipid-lowering, or antidiabetic medications. In addition to the main medications presented in Table 1, Supplemental Table S2 shows additional medications taken (self-reported) in this population. Consumption of vitamins and minerals was reported for 12% of the populations, with significant differences (p < 0.001) between men (5.9%) and women (16.7%). There were no participants taking fish oil supplements. These supplements mainly consisted of calcium, folic acid, vitamin D, and some multivitamins (currently we do not have detailed information about their composition).
Mean adherence to the Mediterranean diet was 8.1 ± 0.1 point in the 17-item score. No statistically significant differences were detected between men and women (p = 0.145). We defined 2 groups of adherence to the Mediterranean diet as either “Low” adherence (0–8 points), or “High” adherence (9–17 points). This cut-off point has been previously used by our group for genome-wide-diet interaction studies [105].
We next focused on the genes and SNPs associated with the serum PUFA concentrations for all the 6 PUFA variables analyzed (omega-3 PUFA, DHA, omega-6 PUFA, LA, total PUFA, and the omega-6 to omega-3 ratio), using both a GWASs approach and a RWASs approach as detailed in Methods.
3.2. GWAS for Serum Omega-3 PUFA (%) in this Population
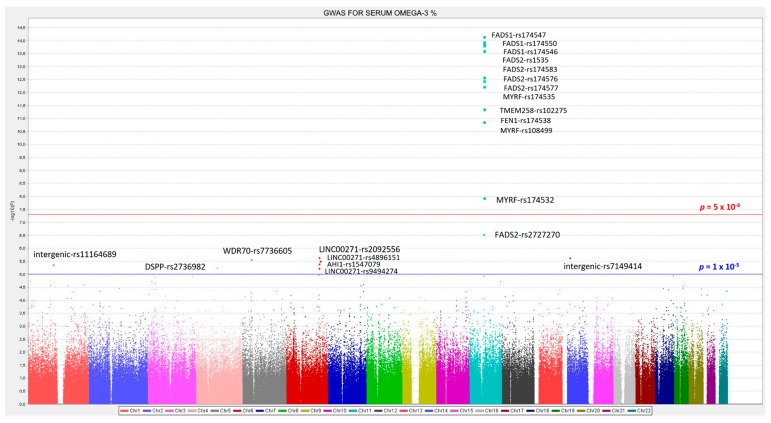
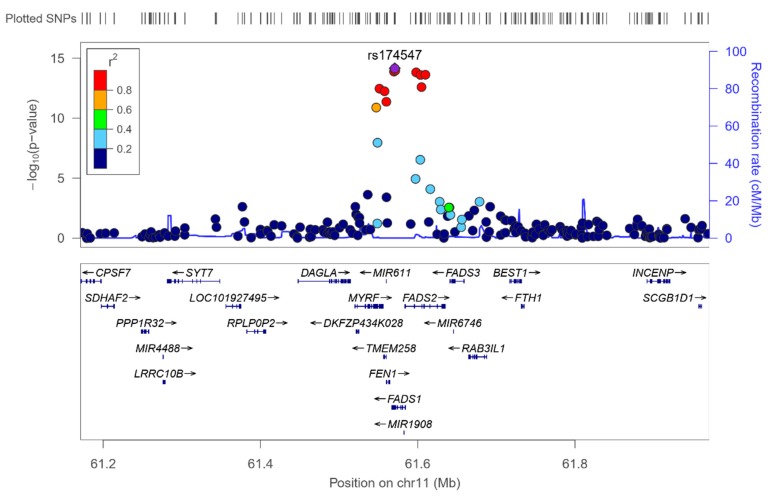
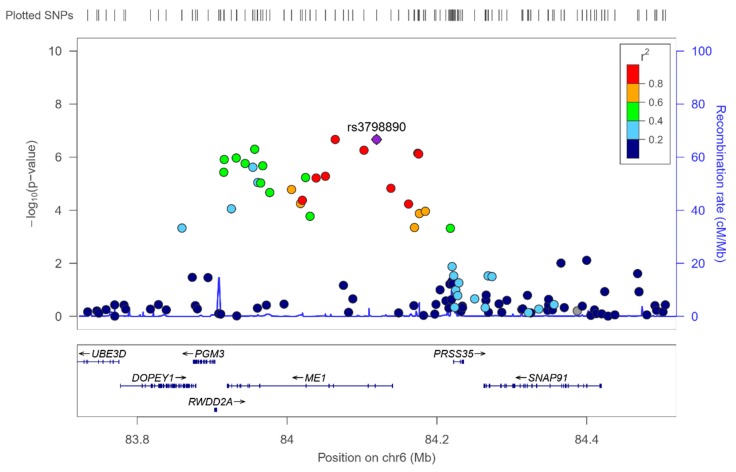
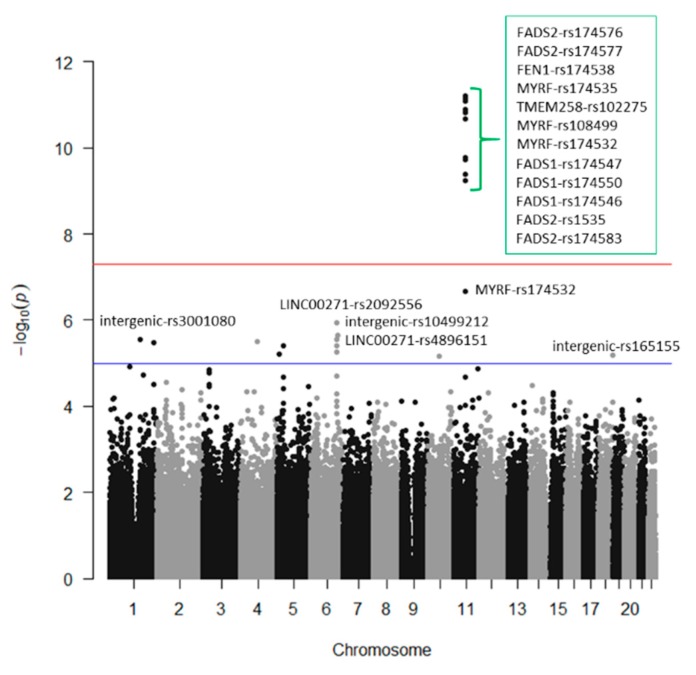
We analyzed the associations at the genome-wide level in the whole population among all the tested SNPs (after quality control filtering) in the Illumina Human OmniExpress array and the 6 PUFA determined. For omega-3 PUFA, we obtained several very strong associations at the genome-wide statistical level of significance (p < 5 × 10−8), replicating previous findings in other populations. Figure 1 presents the corresponding Manhattan plot showing the p-value (−log10 p) of each SNP analyzed as well as indicating both thresholds for significance (the GWAS p-value of significance and the p-value suggestive of significance; p < 5 × 10−5). Supplemental Figure S2 shows the corresponding Q–Q plot. The top-ranked SNP was the FADS1-rs174547 with p = 7.02 × 10−15 in the unadjusted model (Figure 2 shows the regional plot for this SNP).
Figure 1.
Manhattan plot for the genome-wide association study (GWAS) of serum omega-3 fatty acid (%) in the whole population. Associations were obtained from the genetic additive model and p-values were expressed as −log10 (p-value). The red line represents the threshold for GWAS statistical significance (−log10 (5 × 10−8)). The blue line represents the threshold for suggestive GWAS significance (−log10 (1 × 10−5)).
Figure 2.
Regional plot for the top-ranked SNP rs174547 for serum omega-3 fatty acid, located in the FADS1 gene, on chromosome 11. Results in the whole population.
Table 2 presents more information of the top-ranked SNPs both in the crude (Model 1) and in the adjusted model for sex, age, and diabetes (Model 2). The variance explained by the hit SNP was high (13.4%) and this SNP remained as the top-ranked in the model adjusted for sex, age, and diabetes. Its MAF was also relatively high (0.298) and the effect (regression coefficient beta was −0.366 in the adjusted model). As we fitted an additive model, it means that each minor allele (C-allele) was associated with a decrease of the % of serum omega-3 PUFA. Other SNPs in the FADS1 and FADS2 genes also reached the GWAS level of significance even after adjustment for sex, age, and diabetes (FADS1-rs174550, p = 5.35 × 10−14; FADS2-rs1535, p = 5.85 × 10−14; FADS1-rs174546, p = 6.72 × 10−14; FADS2-rs174546, p = 9.75 × 10−14; FADS2- rs174576, p = 1.17 × 10−13; FADS2-rs174577, p = 1.12 × 10−12). Also, at the genome-wide level of significance, we detected other genes, close to the FADS1/FADS2 cluster. These genes and SNPs being: MYRF (myelin regulatory factor)-rs174535, p = 1.49 × 10−12; TMEM258 (transmembrane protein 258)-rs102275, p = 2.43 × 10−12; FEN1 (flap structure-specific endonuclease 1)-rs174538, p = 1.96 × 10−11; MYRF-rs108499, p = 4.21 × 10−11; MYRF-rs174532, p = 1.20 × 10−08. Previous studies also have detected these SNPs as top-ranked in different populations (31, 37, 38). The SNPs near the FADS1/FADS2 locus, were also highly associated with the FADS1/FASD2 SNPs.
Table 2.
Top-ranked single nucleotide polymorphisms (SNPs) in the genome-wide association study (GWAS) for serum omega-3 fatty acid concentrations (%) in the whole population.
| Unadjusted 1 | Adjusted 2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Gene | Position 3 | Allele | MAF | Beta 1 | SE 1 | R2 (1) | p 1 | Chr | SNP | Gene | MAF | Beta 2 | p 2 |
| 11 | rs174547 | FADS1 | 61570783 | C | 0.298 | −0.372 | 0.046 | 0.134 | 7.02 × 10−15 | 11 | rs174547 | FADS1 | 0.298 | −0.366 | 3.34 × 10−14 |
| 11 | rs174550 | FADS1 | 61571478 | C | 0.298 | −0.369 | 0.046 | 0.132 | 1.10 × 10−14 | 11 | rs174550 | FADS1 | 0.298 | −0.364 | 5.35 × 10−14 |
| 11 | rs174546 | FADS1 | 61569830 | T | 0.298 | −0.368 | 0.046 | 0.131 | 1.34 × 10−14 | 11 | rs1535 | FADS2 | 0.322 | −0.356 | 5.85 × 10−14 |
| 11 | rs1535 | FADS2 | 61597972 | G | 0.322 | −0.361 | 0.045 | 0.131 | 1.51 × 10−14 | 11 | rs174546 | FADS1 | 0.298 | −0.362 | 6.72 × 10−14 |
| 11 | rs174583 | FADS2 | 61609750 | T | 0.369 | −0.352 | 0.044 | 0.129 | 2.38 × 10−14 | 11 | rs174583 | FADS2 | 0.369 | −0.346 | 9.75 × 10−14 |
| 11 | rs174576 | FADS2 | 61603510 | A | 0.363 | −0.352 | 0.045 | 0.129 | 2.49 × 10−14 | 11 | rs174576 | FADS2 | 0.363 | −0.346 | 1.17 × 10−13 |
| 11 | rs174577 | FADS2 | 61604814 | A | 0.392 | −0.339 | 0.045 | 0.119 | 2.56 × 10−13 | 11 | rs174577 | FADS2 | 0.392 | −0.332 | 1.12 × 10−12 |
| 11 | rs174535 | MYRF | 61551356 | C | 0.340 | −0.344 | 0.046 | 0.117 | 3.56 × 10−13 | 11 | rs174535 | MYRF | 0.340 | −0.339 | 1.49 × 10−12 |
| 11 | rs102275 | TMEM258 | 61557803 | C | 0.493 | −0.338 | 0.045 | 0.116 | 5.79 × 10−13 | 11 | rs102275 | TMEM258 | 0.493 | −0.332 | 2.43 × 10−12 |
| 11 | rs174538 | FEN1 | 61560081 | A | 0.282 | −0.346 | 0.048 | 0.107 | 4.25 × 10−12 | 11 | rs174538 | FEN1 | 0.282 | −0.339 | 1.96 × 10−11 |
| 11 | rs108499 | MYRF | 61547237 | T | 0.288 | −0.332 | 0.048 | 0.103 | 1.31 × 10−11 | 11 | rs108499 | MYRF | 0.288 | −0.327 | 4.21 × 10−11 |
| 11 | rs174532 | MYRF | 61548874 | A | 0.100 | 0.267 | 0.046 | 0.074 | 1.12 × 10−08 | 11 | rs174532 | MYRF | 0.100 | 0.267 | 1.20 × 10−08 |
| 11 | rs2727270 | FADS2 | 61603237 | T | 0.163 | −0.402 | 0.077 | 0.060 | 2.88 × 10−07 | 11 | rs2727270 | FADS2 | 0.163 | −0.391 | 8.01 × 10−07 |
| 6 | rs2092556 | LINC00271 | 135899344 | C | 0.188 | 0.409 | 0.085 | 0.052 | 2.23 × 10−06 | 6 | rs2092556 | LINC00271 | 0.188 | 0.413 | 1.85 × 10−06 |
| 14 | rs7149414 | intergenic | 31237170 | A | 0.133 | 0.381 | 0.080 | 0.052 | 2.31 × 10−06 | 4 | rs2736982 | DSPP | 0.326 | 0.220 | 2.09 × 10−06 |
| 5 | rs7736605 | WDR70 | 37642044 | A | 0.059 | 0.775 | 0.163 | 0.051 | 2.72 × 10−06 | 5 | rs7736605 | WDR70 | 0.059 | 0.782 | 2.26 × 10−06 |
| 6 | rs10499212 | intergenic | 140027475 | T | 0.100 | 0.642 | 0.136 | 0.051 | 3.11 × 10−06 | 14 | rs7149414 | intergenic | 0.133 | 0.382 | 2.31 × 10−06 |
| 6 | rs4896151 | LINC00271 | 135829796 | T | 0.141 | 0.398 | 0.085 | 0.049 | 3.93 × 10−06 | 1 | rs11164689 | intergenic | 0.065 | 0.500 | 3.09 × 10−06 |
| 1 | rs11164689 | intergenic | 103603452 | T | 0.065 | 0.491 | 0.106 | 0.049 | 4.32 × 10−06 | 6 | rs4896151 | LINC00271 | 0.141 | 0.402 | 3.42 × 10−06 |
| 6 | rs1547079 | AHI1 | 135749202 | C | 0.177 | 0.389 | 0.085 | 0.047 | 5.91 × 10−06 | 6 | rs1547079 | AHI1 | 0.177 | 0.634 | 4.69 × 10−06 |
| 6 | rs9494274 | LINC00271 | 0.234 | 0.394 | 7.95 × 10−06 | ||||||||||
Chr: Chromosome. SNP: Single nucleotide polymorphism. 1: Model 1, unadjusted general linear model (GLM). SNPs were tested in an additive model (0, 1, or 2 minor alleles). 2: Model 2, GLM adjusted for sex, age, and diabetes. Beta: Indicates the regression coefficients per one minor allele. SE: Standard error of Beta. Beta 1: indicates the regression coefficients for the unadjusted general linear model. Beta 2: indicates the regression coefficients for the adjusted general linear model for sex, age, and diabetes. R2: Determination coefficient. 3: Base position in the chromosome (Homo Sapiens GRCh37.p13 genome build used in Illumina HumanOmniExpress-24 BeadChip). SNPs with p-values < 1 × 10−5 are listed (n = 426 subjects analyzed).
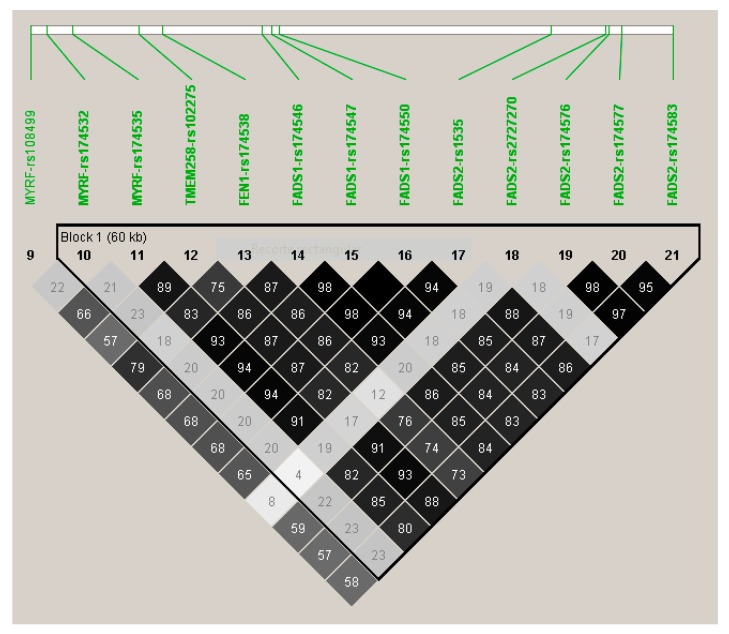
Figure 3 shows the LD (using r2) among the top-ranked SNPs. Apart from the MYRF-rs174532, and the FADS2-rs2727270, the other SNPs were highly correlated among them and with the lead SNP (FADS1-rs174547).
Figure 3.
Linkage disequilibrium (r2) among the top-ranked SNPs (p < 5 × 10−8, in the model adjusted for sex, age, and diabetes) for the genome-wide association study on serum omega-3 fatty acid.
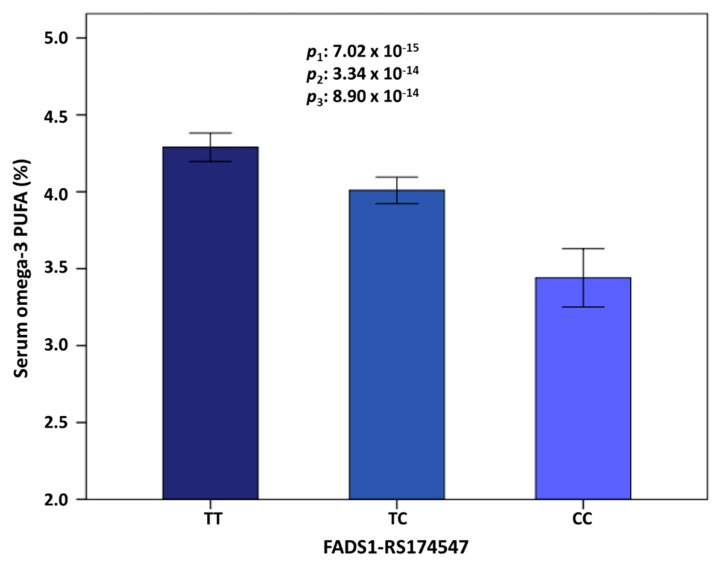
Figure 4 shows the means of omega-3 PUFA (%) depending on the top-ranked FADS1-rs174547 polymorphism. In this Mediterranean population, the T allele was the major allele (this prevalence highly varies depending on the population studied). Minor allele carriers presented significantly lower serum omega-3 PUFA concentrations (TT: 4.29% ± 0.04; TC: 4.09% ± 0.05 and CC: 3.44% ± 0.1). This strong genetic association did not vary significantly even after multivariate association (p = 7.02 × 10−15 in the unadjusted model; p = 3.35 × 10−14 in the model adjusted for sex, age, and diabetes and p = 8.90 × 10−14, in the model adjusted for sex, age, diabetes, BMI, medications (including antihypertensive drugs, lipid lowering drugs, and insulin and oral antidiabetic medication), smoking, physical activity, and adherence to the Mediterranean diet. A previous GWAS in the Framingham study also showed a slight effect of the dietary adjustments in the SNP-PUFA associations [122]. Additional adjustment of the model containing all of these variables for more medications (including drug for pain and fever, anti-platelet drugs, tranquilizers-sedative-hypnotics, and vitamins/minerals), did not substantially change the statistical significance of the association between the FADS1-rs174547 polymorphism and serum omega-3 concentrations (p = 2.90 × 10−13).
Figure 4.
Serum omega-3 PUFA concentrations (%) depending on the FADS1-rs174547 genotype in the whole population. Genotype prevalence was: TT 49.3% (n = 212), TC 41.0% (n = 172), and CC 9.7% (n = 42). The minor allele (C) was associated with lower concentrations in all the adjusted models. 1: Unadjusted model; 2: Model adjusted for sex, age, and diabetes; 3: Model adjusted for sex, age, diabetes, body mass index, medications (antihypertensive, hypolipidemic, and antidiabetic drugs), smoking, physical activity, and adherence to Mediterranean diet. Values are means by genotype. Error bars: 2 × Standard Error.
In addition to the SNPs found at the genome-wide level of significance, we also detected several SNPs at the suggestive p-value of GWAS significance (p < 5 × 10−5). Among these genes, we would like to highlight the signal at the AHI1 (Jouberin) gene, located on chromosome 6, close to the MYB (proto-oncogene, transcription factor) gene and the HBS1L (like translational GTPase) gene, both of which have previously been reported in the Framingham study as new genes associated with PUFA [69]. However, no association signals at the GWAS level, or at the suggestive p-value of GWAS significance, were found for the ELOVL candidate genes on chromosome 6 for omega-3 PUFA in our Mediterranean population.
We next explored gene*sex interaction in determining serum omega-3 PUFA concentrations at the genome-wide level and only found a statistically significant interaction at the genome-wide level of statistical significance between the rs17097464-intergenic at chromosome 12 and sex (p = 1.13 × 10−8). The MAF for this SNP was 0.193. In addition to this hit, we found 7 other SNPs (rs11081343, rs1402508, rs8052428, rs4732314, rs897476, rs6499260, and rs11754354) reaching p-values < 1 × 10−5 for the interaction terms with sex. Future studies should investigate these loci in more detail.
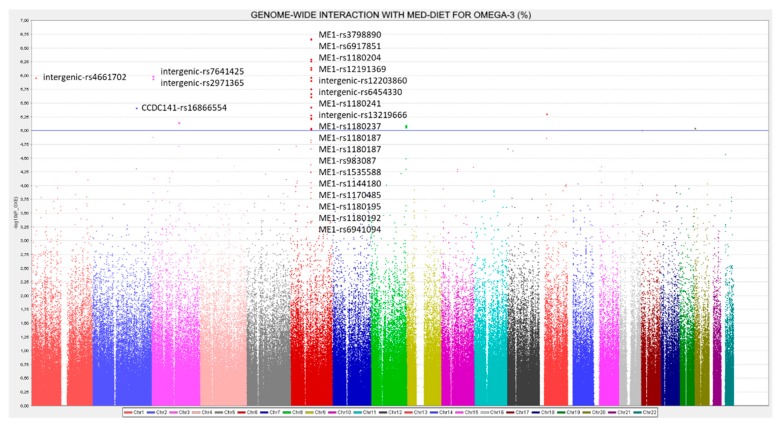
In another exploratory analysis, we investigated gene* Mediterranean diet interactions in determining serum omega-3 PUFA concentrations. Two Mediterranean diet strata were considered (low and high adherence to Mediterranean diet). Figure 5 shows the Manhattan plot for these interactions.
Figure 5.
Manhattan plot for the genome-wide interaction with adherence to Mediterranean diet in determining serum omega-3 fatty acid (%) in the whole population. p-values for the gene*diet interaction terms were obtained in the additive model for the SNPs and diet expressed as dichotomous (low and high adherence to the Mediterranean diet) and expressed as −log10 (p-value). The blue line represents the threshold for suggestive GWAS significance (−log10 (1 × 10−5)).
13 SNPs in the ME1 (malic enzyme 1) gene significantly interacted (at p < 1 × 10−5) with adherence to Mediterranean diet with p-values ranging from 2.15 × 10−7 to 9.4 × 10−6 (see Supplemental Table S3 for the detailed SNPs, betas, MAF, genes, and p-values for the corresponding interaction terms). Supplemental Figure S3 shows the Q–Q plot for these interactions, indicating a good performance. The ME1 gene is located on chromosome 6 and it is a plausible candidate for gene-diet interactions, taking into account that this gene is involved in fatty acids synthesis in the liver [123,124] and its modulation by the dietary fat content has been reported [124,125]. Figure 6 shows the regional plot for the top-ranked SNPs, ME1-rs3798890 (p = 2.15 × 10−7) in the genome-wide interaction analysis with Mediterranean diet. For this SNP as well as for the other SNPs top-ranked in the region, Supplemental Table S3 shows the interaction effect and the corresponding beta in the two strata of Mediterranean diet. Thus, when adherence to Mediterranean diet is low, the minor allele is associated with an increase in serum omega-3 PUFA concentrations, whereas when adherence to Mediterranean diet is high, the minor allele is associated with a decrease in serum omega-3 PUFA concentrations. This interaction effect might be similar to the previously reported positive selection for the FADS cluster [95], where depending on the diet, the prevalence of the allele resulting in the generation of higher PUFA concentrations, changed in the different populations.
Figure 6.
Regional plot for the top-ranked SNP rs3798890 (hit in the genome-wide interaction study with Mediterranean diet in determining serum omega-3 fatty acid), located in the ME1 gene, on chromosome 6. Results for the whole population.
3.3. GWAS for Serum DHA Concentrations (%) in this Population
Figure 7 depicts the Manhattan plot for the GWAS on serum DHA concentrations (%) in this population and Table 3 shows the SNPs, MAFs, betas, determination coefficients, and p-values for the top-ranked SNPs in the models.
Figure 7.
GWAS for serum docosahexaenoic fatty acid (%) in the whole population.
Table 3.
Top-ranked SNPs in the GWAS for docosahexaenoic fatty acid concentrations (%) in the whole population.
| Unadjusted 1 | Adjusted 2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Gene | Position 3 | Allele | MAF | Beta 1 | SE 1 | R2 (1) | p 1 | Chr | SNP | Gene | MAF | Beta 2 | p 2 |
| 11 | rs174547 | FADS1 | 61570783 | C | 0.298 | −0.122 | 0.017 | 0.106 | 6.12 × 10−12 | 11 | rs174547 | FADS1 | 0.298 | −0.121 | 1.28 × 10−11 |
| 11 | rs174550 | FADS1 | 61571478 | C | 0.298 | −0.121 | 0.017 | 0.105 | 7.35 × 10−12 | 11 | rs174550 | FADS1 | 0.298 | −0.121 | 1.50 × 10−11 |
| 11 | rs174546 | FADS1 | 61569830 | T | 0.298 | −0.121 | 0.017 | 0.104 | 8.24 × 10−12 | 11 | rs174546 | FADS1 | 0.298 | −0.120 | 1.66 × 10−11 |
| 11 | rs1535 | FADS2 | 61597972 | G | 0.322 | −0.118 | 0.017 | 0.103 | 1.29 × 10−11 | 11 | rs1535 | FADS2 | 0.322 | −0.117 | 2.87 × 10−11 |
| 11 | rs174583 | FADS2 | 61609750 | T | 0.369 | −0.115 | 0.017 | 0.102 | 1.54 × 10−11 | 11 | rs174583 | FADS2 | 0.369 | −0.114 | 3.34 × 10−11 |
| 11 | rs174576 | FADS2 | 61603510 | A | 0.363 | −0.115 | 0.017 | 0.101 | 2.11 × 10−11 | 11 | rs174576 | FADS2 | 0.363 | −0.114 | 4.42 × 10−11 |
| 11 | rs174577 | FADS2 | 61604814 | A | 0.392 | −0.109 | 0.017 | 0.092 | 1.66 × 10−10 | 11 | rs174538 | FEN1 | 0.282 | −0.117 | 3.40 × 10−10 |
| 11 | rs174538 | FEN1 | 61560081 | A | 0.282 | −0.117 | 0.018 | 0.092 | 1.77 × 10−10 | 11 | rs174577 | FADS2 | 0.392 | −0.109 | 3.47 × 10−10 |
| 11 | rs174535 | MYRF | 61551356 | C | 0.340 | −0.111 | 0.017 | 0.091 | 1.91 × 10−10 | 11 | rs174535 | MYRF | 0.340 | −0.111 | 3.89 × 10−10 |
| 11 | rs102275 | TMEM258 | 61557803 | C | 0.493 | −0.108 | 0.017 | 0.088 | 4.14 × 10−10 | 11 | rs102275 | TMEM258 | 0.493 | −0.108 | 8.79 × 10−10 |
| 11 | rs108499 | MYRF | 61547237 | T | 0.288 | −0.112 | 0.018 | 0.087 | 5.76 × 10−10 | 11 | rs108499 | MYRF | 0.288 | −0.111 | 1.22 × 10−09 |
| 11 | rs174532 | MYRF | 61548874 | A | 0.100 | 0.089 | 0.017 | 0.061 | 2.19 × 10−07 | 11 | rs174532 | MYRF | 0.100 | 0.088 | 3.43 × 10−07 |
| 6 | rs2092556 | LINC00271 | 135899344 | C | 0.188 | 0.154 | 0.031 | 0.055 | 1.13 × 10−06 | 6 | rs2092556 | LINC00271 | 0.188 | 0.154 | 1.24 × 10−06 |
| 6 | rs10499212 | intergenic | 140027475 | T | 0.100 | 0.239 | 0.050 | 0.052 | 2.27 × 10−06 | 4 | rs2736982 | DSPP | 0.326 | 0.081 | 2.25 × 10−06 |
| 1 | rs3001080 | intergenic | 163434750 | G | 0.426 | −0.082 | 0.017 | 0.050 | 2.90 × 10−06 | 6 | rs10499212 | intergenic | 0.100 | 0.238 | 2.64 × 10−06 |
| 6 | rs4896151 | LINC00271 | 135829796 | T | 0.141 | 0.148 | 0.031 | 0.050 | 2.90 × 10−06 | 6 | rs4896151 | LINC00271 | 0.141 | 0.149 | 3.04 × 10−06 |
| 4 | rs2736982 | DSPP | 88534235 | A | 0.326 | 0.079 | 0.017 | 0.050 | 3.13 × 10−06 | 1 | rs3001080 | intergenic | 0.426 | −0.081 | 3.57 × 10−06 |
| 1 | rs16856236 | DISC1 | 232160058 | G | 0.062 | 0.188 | 0.040 | 0.050 | 3.38 × 10−06 | 6 | rs1547079 | AHI1 | 0.177 | 0.146 | 3.91 × 10−06 |
| 6 | rs1547079 | AHI1 | 135749202 | C | 0.177 | 0.146 | 0.031 | 0.049 | 3.85 × 10−06 | 1 | rs16856236 | DISC1 | 0.062 | 0.186 | 4.01 × 10−06 |
| 5 | rs7736605 | WDR70 | 37642044 | A | 0.059 | 0.280 | 0.060 | 0.049 | 3.86 × 10−06 | 5 | rs7736605 | WDR70 | 0.059 | 0.281 | 4.05 × 10−06 |
| 6 | rs9494274 | LINC00271 | 135880480 | G | 0.234 | 0.135 | 0.029 | 0.048 | 5.55 × 10−06 | 5 | rs9312754 | CTNND2 | 0.261 | 0.123 | 5.24 × 10−06 |
| 5 | rs9312754 | CTNND2 | 11088028 | C | 0.261 | 0.122 | 0.027 | 0.047 | 6.18 × 10−06 | 6 | rs9494274 | LINC00271 | 0.234 | 0.135 | 6.35 × 10−06 |
| 18 | rs165155 | intergenic | 75341905 | C | 0.163 | −0.110 | 0.024 | 0.047 | 6.53 × 10−06 | 18 | rs165155 | intergenic | 0.163 | −0.109 | 8.42 × 10−06 |
| 10 | rs16911516 | intergenic | 59751257 | C | 0.086 | −0.337 | 0.074 | 0.047 | 7.04 × 10−06 | 10 | rs16911516 | intergenic | 0.086 | −0.336 | 8.92 × 10−06 |
| 1 | rs11164689 | intergenic | 0.065 | 0.175 | 9.18 × 10−06 | ||||||||||
Chr: Chromosome. SNP: Single nucleotide polymorphism. 1: Model 1, unadjusted general linear model (GLM). SNPs were tested in an additive model (0, 1, or 2 minor alleles). 2: Model 2, GLM adjusted for sex, age and diabetes. Beta: Indicates the regression coefficients per one minor allele. SE: Standard error of beta. Beta 1: indicates the regression coefficients for the unadjusted general linear model. Beta 2: indicates the regression coefficients for the adjusted general linear model for sex, age, and diabetes. R2: Determination coefficient. 3: Base position in the chromosome (Homo Sapiens GRCh37.p13 genome build used in Illumina HumanOmniExpress-24 BeadChip). SNPs with p < 5 × 10−5 are listed.
.
Supplemental Figure S4 presents the corresponding Q–Q plot. In general, a similar pattern of genes and polymorphisms were detected for DHA and total omega-3 and we also identified several associations at the genome-wide level of significance for DHA. Three SNPs in the FADS1 gene (rs174547, rs174550, and rs174546) were the hits, having p-values of 1.28 × 10−11, 1.50 × 10−11, and 1.66 × 10−11). These SNPs were in very high LD. Likewise, 4 SNPs in the FADS2 with p-values ranging from 2.87 × 10−11 to 3.47 × 10−10; 1SNP in FEN1-rs174538 (p = 3.47 × 10−10), 1 SNP in TMEM258 (p = 8.79 × 10−10), and 2 SNPs in MYRF, with p-values of 3.89 × 10−10 and 1.22 × 10−9, were statistically significant at the genome-wide level. Despite their significance, the genetic effect for DHA was lower than that observed for total omega-3. Here, the top-ranked SNP (the FADS1), only explained the 10.6% of the variability, versus a 13.4%, explained by the same SNP for total omega-3. This indicates that here are other omega-3 fatty acids (not determined in this project) for which the genetic association effect should be higher.
In the exploratory analysis of the genome-wide interaction by sex in determining serum DHA concentrations, we found as top-ranked the same SNP that reached statistical significance at the GWAS level for the omega-3 PUFA. In this case, the interaction between the intergenic-rs17097464 on chromosome 12 and sex was 1.31 × 10−7. Likewise, consistent gene-sex interactions were found at p < 1 × 10−5 with several genes also detected at this level for the analysis on omega-3 PUFA. Among these genes, similar interaction results at (p < 1 × 10−5) have been found with LOC105372018-rs8091414; NHEJ1 (non-homologous end joining factor 1)-rs1402508 and the intergenic-rs11081343.
Regarding the exploratory analysis of genome-wide interactions with adherence to Mediterranean diet, we also detected several interactions with the ME1 gene in determining serum DHA concentrations. The top-ranked SNP for gene*Mediterranean diet interactions in the ME1 was the rs1180204-ME1 at p = 5.34 × 10−7.
3.4. GWAS for Serum Omega-6 Concentrations (%) in this Population
Supplemental Figure S5 displays the Manhattan plot for the GWAS associations in the whole population in determining serum omega-6 concentrations. Unlike that observed in GWAS for omega-3 PUFA, we did not detect any statistically significant association at the GWAS level for omega-6 PUFA. We only detected some associations at p < 5 × 10−5. Supplemental Figure S6 shows the corresponding Q–Q plot and Supplemental Table S4 [2], presents the top ranked SNPs for the omega-6 GWAS, both in the unadjusted and in the multivariate adjusted model. We can see that the genetic effect is lower for omega-6 PUFA compared to omega-3 PUFA in this population. The top-ranked SNP for omega-6 PUFA only accounted for 4.9% of variability in serum omega-6 concentrations, compared to 13.4% for omega-3 PUFA. Furthermore, top-ranked SNPs were not polymorphisms in the FADS genes, but in other genes. After adjustment for age, sex, and diabetes, the top-ranked SNP (rs4491485; p = 1.97 × 10−6) is located in the ISG20 (interferon stimulated exonuclease gene 20) gene, followed by an intergenic SNP (rs2551402) in chromosome 18 and another SNP (rs949037; p = 4.84 × 10−6) located in the BCL2 (BCL2 apoptosis regulator) gene, also in chromosome 18.
When we analyzed gene–sex interactions at the genome-wide level on omega-6 PUFA concentrations, we found no interaction at the GWAS significance level. However, the most significant interaction was obtained with the DNTTIP2 (deoxynucleotidyltransferase terminal interacting protein 2) gene located at chromosome 1. The top ranked SNP was the DNTTIP2-rs3747965 at p = 1.07 × 10−7. Other SNPs in the DNTTIP2 gene also achieved statistical significance at p < 5 × 10−5. Regarding, the exploratory analysis of gene*Mediterranean diet interactions on serum omega-6 concentrations, the top-ranked SNP was the LOC105376941-rs977905 at p = 1.17 × 10−6.
3.5. GWAS for Serum LA Concentrations (%) in this Population
Supplemental Figure S7 depicts the Manhattan plot for the GWAS on serum LA concentrations, and Supplemental Figure S8 shows the corresponding Q–Q plot. Two SNPs, the rs5994479 at the C22orf42 and the DAB1 (DAB adaptor protein 1) presented association at the GWAS level. This gene, involved in brain signaling in the endothelium, is an integrative signal essential for neuro-glia-vascular communication [126]. Supplemental Table S5 shows the descriptors and coefficients of these SNPs. In the model adjusted for sex, age, and diabetes, the significance achieved by each of them was 5.25 × 10−10 for rs5994479 and 5.69 × 10−10 for DAB1-rs11207162. Although the variability explained by the top-ranked SNP is greater for LA (9.5%) than for total omega-6, it is still lower than for DHA. It is also striking that none of the candidate genes in the FADS or ELOVL genes are among the top-ranked.
In the genome-wide interaction study to analyze the interactions with sex, we obtained several significant associations at the p < 1 × 10−5 with SNPs in the DNTTIP2 gene. This is highly consistent with that observed for total omega-6. Likewise, the top ranked gene–sex interaction for serum LA concentrations was observed with the gene DNTTIP2-rs3747965 at 1.04 × 10−7 level. For the genome-wide interactions with adherence to Mediterranean diet on serum LA, the top-ranked SNP was LOC105374410-rs1906292 at p = 8.88 × 10−7. This SNP was also the hit for omega-6 PUFA.
3.6. GWAS for Serum Total PUFA Concentrations (%) in this Population
In the GWAS for total PUFA, we only detected 4 statistically significant SNPs at p < 5 × 10−5 in the sex-, age-, and diabetes-adjusted model. None of them reached the level of GWAS significance and the p-values were higher than 3.12 × 10−6. This may reflect the greater heterogeneity of this variable. Among the most significant SNPs are those already detected as top-ranked for omega-6 or LA, as omega-6 are the most abundant PUFA. These SNPs were: Intergenic-rs7047109 (3.12 × 10−6), DNAH11 (dynein heavy chain 11, axonemal)-rs10256021 (p = 4.8 × 10−6); ISG20-rs4491485 (p = 7.18 × 10−6) and intergenic-rs252181 (p = 9.69 × 10−6).
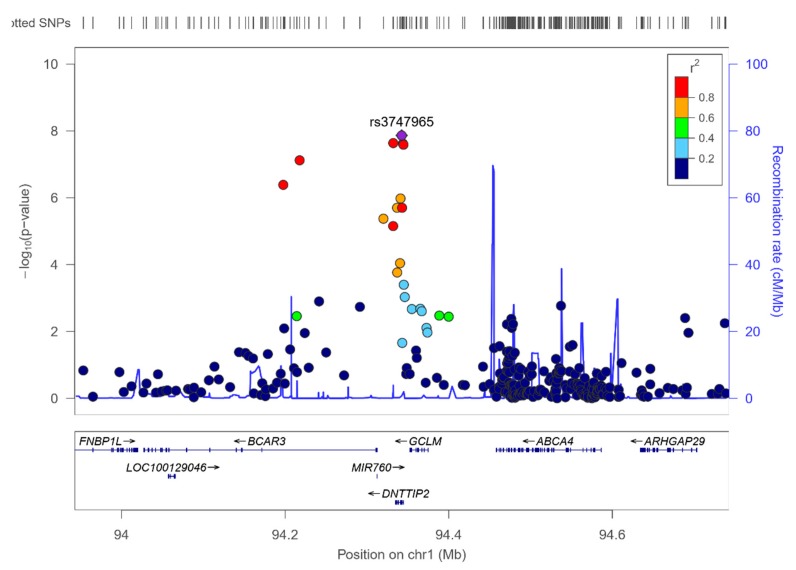
Interestingly, in the study of genome-wide interaction with sex, the top-ranked SNP determining the concentrations of total PUFA was the DNTTIP2-rs-3747965, already obtained in the previous analyses for omega-6 and for LA, but this time, reaching the level of statistical significance of GWAS (p = 1.36 × 10−8). Other SNPs in the DNTTIP2 gene and nearby, also reached the genome-level of significance. Supplemental Figure S9 shows the Manhattan plot for the gene*sex interaction terms in determining serum total PUFA, and Figure 8, shows the regional plot for the top-ranked SNP DNTTIP2-rs-3747965.
Figure 8.
Regional plot for the top-ranked SNP rs3747965 (hit in the genome-wide interaction study with sex in determining serum polyunsaturated fatty acids), located in the DNTTIP2 gene, on chromosome 1. Results for the whole population.
According to this interaction, the minor allele of this SNP (MAF: 0.345) was associated with reduced PUFA in females (beta: −0.634) and with increased PUFA in males (beta: 1.158).
3.7. GWAS for the Omega-6 to Omega 3 Ratio
We also explored at the GWAS level the SNPs associated with the omega-6 to omega-3 ratio in this population. Supplemental Table S6 shows the top-ranked SNPs associated with this ratio (log transformed). Both in the unadjusted and in the multivariate adjusted models (for sex, age, and diabetes), several SNPs in the chromosome 11 (FADS1/2 cluster) showed statistically significant associations with the omega-6/omega-3 ratio at the GWAS level. The top-ranked SNP was the FADS1-rs174547 T > C in both models. Specifically, in the multivariate adjusted model, the minor allele of this SNP was associated with a higher omega-6/omega-3 ratio (B = 0.041; p = 2.19 × 10−14).
Supplemental Figure S10 shows the means of the serum omega-6/omega-3 ratio (log transformed) depending on the top-ranked FADS1-rs174547 polymorphism. Homozygous subjects for the minor allele (CC) presented significantly higher ratio. This strong genetic association changed little even after multivariate adjustment (p = 3.90 × 10−15 in the unadjusted additive model; p = 2.19 × 10−14 in the model adjusted for sex, age, and diabetes and p = 9.81 × 10−14, in the model adjusted for sex, age, diabetes, BMI, medications (including antihypertensive drugs, lipid lowering drugs, and insulin and oral antidiabetic medication), smoking, physical activity, and adherence to the Mediterranean diet. Additional adjustment of the model containing all of these variables for more medications (including drug for pain and fever, anti-platelet drugs, tranquilizers-sedative-hypnotics, and vitamins/minerals), did not substantially change the statistical significance of the association (p = 1.71 × 10−13). Furthermore, we analyzed the interaction between the adherence to the Mediterranean diet and the SNPs at the genome-wide level in determining the serum omega-6/omega-3 ratio (Supplemental Figure S11). We obtained similar results for this interaction than those described for the serum omega-3 PUFA concentrations. Thus, the top-ranked gene interacting with adherence to the Mediterranean diet was the ME1 gene. More studies are needed to replicate and to characterize this interaction.
3.8. RWASs for Serum PUFA Concentrations
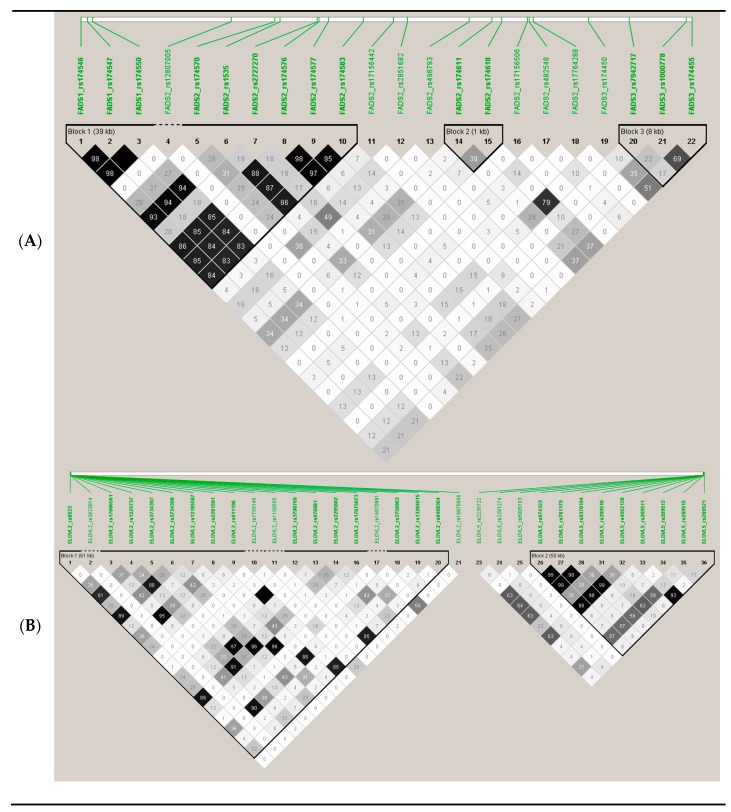
To explore potential associated loci which might fail to be identified due to the strict threshold for GWAS associations, we carried out a RWAS investigating 2 regions previously reported to be genetic determinants of the SNPs analyzed. Figure 9 shows the SNPs analyzed in region A (FADS cluster) and in region B (ELOVL2/ELOVL5).
Figure 9.
Genes, SNPs, and linkage disequilibrium (r2) among the SNPs selected for the regional-wide association studies (RWASs), based on previous knowledge of the relevance of this region for the associations between its SNPs and PUFA concentrations: (A) region containing all the SNPs in/near the cluster FADS1/FADS2/FADS3 (chromosome 11) in the Illumina HumanOmniexpress array; (B) region containing all the SNPs in/near the cluster ELOV2-ELOV5 (chromosome 6) in the Illumina HumanOmniexpress array.
Supplemental Table S7 presents the SNPs and the associations obtained in the RWAS analysis for serum omega-3 at the FADS1/FADS2/FADS3 region. We obtained statistically significant associations at p < 0.05 with 15 SNPs (models adjusted for sex, age, and diabetes), including SNPs detected at the GWAS level and other SNPs detected at the nominal level of significance. In addition to the FADS1 and FADS2 SNPs, we detected a significant association (p = 0.012) with the FADS3-rs174450 SNP and with the FADS3-rs174455 (p = 0.027). Supplemental Table S8 shows the corresponding RWAS associations with DHA. Similar results were obtained, and 12 SNPs were detected at p < 0.05. However, no statistically significant association even at this level with FADS3 SNPs was observed.
For omega-6 PUFA, the FADS1/FADS2/FADS3 region was poorly associated with their serum levels in this population even after considering the nominal p-value. Supplemental Table S9 shows these associations. Only 2 SNPs FADS2-rs174618 and FADS2-rs174611, reached the significance at p = 0.037 and p = 0.047, respectively. However, for LA concentrations, 11 SNPs in the analyzed FADS1/FADS2/FADS3 region, reached statistical significance at p < 0.05 (Table 4). The most significant SNP was the FADS2-rs174611 (p = 0.0005).
Table 4.
Top-ranked SNPs in the RWAS for the FADS1/FADS2/FADS3 region in determining serum linoleic fatty acid concentrations (%) in the whole population.
| Adjusted 1 | |||||||
|---|---|---|---|---|---|---|---|
| Chr | SNP | Gene | Position2 | Alleles | MAF | Beta | p |
| 11 | rs174611 | FADS2 | 61627881 | C | 0.116 | 0.679 | 4.86 × 10−04 |
| 11 | rs174618 | FADS2 | 61629322 | C | 0.349 | 0.550 | 1.21 × 10−03 |
| 11 | rs174547 | FADS1 | 61570783 | C | 0.298 | 0.527 | 2.78 × 10−03 |
| 11 | rs174583 | FADS2 | 61609750 | T | 0.369 | 0.507 | 2.88 × 10−03 |
| 11 | rs174546 | FADS1 | 61569830 | T | 0.298 | 0.521 | 3.19 × 10−03 |
| 11 | rs174550 | FADS1 | 61571478 | C | 0.298 | 0.512 | 3.82 × 10−03 |
| 11 | rs174576 | FADS2 | 61603510 | A | 0.363 | 0.493 | 3.83 × 10−03 |
| 11 | rs174577 | FADS2 | 61604814 | A | 0.392 | 0.491 | 3.89 × 10−03 |
| 11 | rs174450 | FADS3 | 61641542 | T | 0.435 | 0.481 | 5.15 × 10−03 |
| 11 | rs1535 | FADS2 | 61597972 | G | 0.322 | 0.482 | 5.49 × 10−03 |
| 11 | rs17156442 | FADS2 | 61614023 | T | 0.098 | 0.976 | 8.04 × 10−03 |
| 11 | rs174455 | FADS3 | 61656117 | A | 0.407 | 0.401 | 1.93 × 10−02 |
| 11 | rs1000778 | FADS3 | 61655305 | A | 0.477 | 0.329 | 7.15 × 10−02 |
| 11 | rs174570 | FADS2 | 61597212 | T | 0.228 | 0.219 | 3.84 × 10−01 |
| 11 | rs2727270 | FADS2 | 61603237 | T | 0.163 | 0.236 | 4.08 × 10−01 |
| 11 | rs482548 | FADS2 | 61633182 | T | 0.030 | 0.216 | 4.19 × 10−01 |
| 11 | rs17764288 | FADS2 | 61633736 | A | 0.003 | 0.520 | 5.00 × 10−01 |
| 11 | rs12807005 | FADS2 | 61591059 | A | 0.006 | 0.357 | 6.30 × 10−01 |
| 11 | rs2851682 | FADS2 | 61616012 | G | 0.149 | 0.133 | 6.75 × 10−01 |
| 11 | rs498793 | FADS2 | 61624705 | T | 0.313 | −0.048 | 7.77 × 10−01 |
| 11 | rs7942717 | FADS3 | 61647288 | G | 0.097 | 0.040 | 8.83 × 10−01 |
Chr: Chromosome. SNP: Single nucleotide polymorphism. 1: Model 2, general linear model (GLM) adjusted for sex, age, and diabetes: SNPs were tested in an additive model (0, 1, or 2 minor alleles). Beta: Indicates the regression coefficients per one minor allele for the adjusted general linear model for sex and age. MAF: Minor allele frequency. 2: Base position in the chromosome (Homo Sapiens GRCh37.p13 genome build used in Illumina HumanOmniExpress-24 BeadChip). SNPs with p < 5 × 10-5 are listed.
For total PUFA, the RWAS at the FADS1/FADS2/FADS3 region revealed 6 SNPs (rs2727270, rs174547, rs174550, rs174546, rs1535, and rs174570) statistically associated with serum levels at p < 0.05.
For region B, consisting of SNPs in ELOVL2 and ELOVL5, we did not detect any statistically significant association with serum omega-3 PUFA (Supplemental Table S10). Similar results were obtained for DHA (no statistically significant associations). However, for omega-6 PUFA (Supplemental Table S11) and LA (Supplemental Table S12), some statistically significant associations involving ELOVL2 SNPs were found. For total PUFA concentrations, no statistically significant associations with region B SNPs were found.
4. Discussion
In this first GWAS carried out in a Mediterranean population composed of subjects with metabolic syndrome in Spain, we confirmed the large contribution of the FASD1 gene cluster (chromosome 11) on total serum omega-3 PUFA [94]. Despite the limited sample size, our findings show robust statistically significant associations with p < 5 × 10−8 for several SNPs in/near the FASD1 and FASD2 genes. The top-ranked SNP was the FADS1-rs174547 T > C. This SNP was strongly associated with serum omega-3 concentrations even after additional adjustment for covariates (p = 3.34 × 10−14 in the model adjusted for sex, age, and diabetes). Further adjustments for medications, diet, and other lifestyle variables did not substantially change the statistical significance of the association. Likewise, some previous GWAS [81,83,96] only adjusted the main statistical models for age and sex (and recruitment site in multicenter studies) and further evaluated the effect of additional adjustment for other covariates (dietary intake, physical activity, and BMI). After these additional adjustments, the PUFA-SNP associations changed little, in agreement with our results testing sequential adjustments. In our population, the minor allele of the FADS1-rs174547 T > C top-ranked SNP (C allele) was associated with decreased serum omega-3 PUFA concentrations. This is consistent with functional studies showing a functional association between the FADS1-rs174547 and delta-5 desaturase activity [127]. Moreover, a recent study examining the effects of aging and FADS polymorphisms on long chain PUFA biosynthetic capacity via direct quantification [128] has reported for the first time that aging especially decreases the long chain PUFA biosynthetic capacity regulated by the FADS1-rs174547 SNP in FADS1 C allele carriers [128]. The fact of studying an elderly population (subjects aged 55 to 75 years) is another factor that may have contributed to the differences detected between our study and others previously carried out [79,80,81,82,83,84,85,86,93,98].
In our GWAS, in addition to the association with serum omega-3 PUFA, we have studied the association with serum concentrations of DHA, an omega-3 fatty acid derived primarily from fish. For DHA we have also obtained statistically significant associations at the GWAS level with several SNPs in the genes FADS1 and FADS2 in agreement with the previous GWAS analyzing DHA concentrations, with FADS1-rs174547 SNP also being the top-ranked.
However, we did not detect statistically significant associations between the FADS1/FADS2 cluster SNPs at the GWAS level with total serum omega-6 PUFA or LA (omega-6). This result differs from other GWAS studies [79,80,81,82,83,84,85,86]. The lack of association at the GWAS level of the FADS1/FADS2 SNPs with serum omega-6 may be due to several factors. One of them may be related to the age of our population. We have studied older subjects (mean age 65 years old), and it has been reported that aging decreases biosynthetic activity regulated by the FADS1 polymorphisms [128], so potentially decreasing the magnitude of the genetic associations at this locus. Along these lines, another factor influencing the activity of the FADS1/FADS2/FADS3 desaturases is the ethnic background [95]. Our Spanish Mediterranean population may differ from the majority of the other populations included in the previously published GWAS [80,81,82,83,84,85,86]. Moreover, several environmental factors may contribute to the differences. Among them, the dietary pattern may play a relevant role. The existing GWAS have been mainly carried out in American populations [81,83,96] as well as in Asian populations [84,86] having different diets. The Spanish Mediterranean population has been characterized over many years as one that follows the so-called traditional Mediterranean diet which is rich in vegetables, fruit, fish, olive oil and nuts, and low in red meats, butter, pastries and processed products [99]. Nowadays, the younger population is turning away from the Mediterranean diet, but a fairly high adherence, on average, to the traditional Mediterranean diet can still be detected in the elderly, although variability in adherence also exists within this group [99,101]. Among the populations included in the previous GWAS for circulating PUFA, the one that most resembles our population is the INCHIANTI study [79], focused on an Italian Mediterranean population. In the INCHIANTI study, a GWAS was carried out on plasma levels of six omega-3 and omega-6 PUFA in 1075 participants. In this study, the strongest evidence for global associations was found at the FADS1/FADS2/FADS3 cluster. This is similar to our results. However, in the INCHIANTI study, the associations were mainly observed for some omega-6 PUFA [79]. Thus, the SNP with the most significant association was rs174537 near FADS1, in determining plasma arachidonic acid (AA) concentrations (p = 5.95 × 10−46). In our study, we did not analyze the association with AA, because the NMR platform used for measuring serum fatty acids [110,111,112,113,114,115,116] did not provide the separate concentration of AA. Therefore, we cannot compare the results and this is a limitation of our study in comparison with other studies providing a more comprehensive profile of fatty acids determined in other platforms (80,81,83,84,86). However, in INCHIANTI, total omega-3 or total omega-6 PUFA were not analyzed. In the INCHIANTI paper [79], the authors compared their results with those obtained in the GOLDN (Genetics of Lipid Lowering Drugs and Diet Network) study including American subjects (n = 1076) recruited in Minneapolis, MN and Salt Lake City, UT. For some associations, replications were found, but other associations were population specific. This observation was in agreement with our initial hypothesis regarding the interest in analyzing specific populations to better understand the associations. Again, diet could be an important driver, taking into account the differences in the diet consumed when comparing INCHIANTY and the GOLDN populations. Additionally, the authors, pointed out that the tissue selected for PUFA determination may be another cofounder when results are compared. In GOLDN, PUFA were measured in erythrocyte membranes and in INCHIANTI, PUFA were measured in plasma [79]. It has been reported that erythrocytes and plasma may reflect two slightly different pools of fatty acids [129]. In general, it is accepted that plasma fatty acids reflect a short term fatty acid intake whereas erythrocyte levels reflect a more long term intake. This limitation is also present in the studies meta-analyzing populations with different fatty acid determinations and may have an influence when comparing results [130]. Moreover, an additional limitation, even in studies analyzing plasma fatty acids, is the type of measurement. Thus, Wu et al. [83] in the meta-analysis that they carried out in the CHARGE consortium outlined this potential limitation, specifically indicating that the INCHIANTI cohort measured total plasma fatty acids while the other 4 cohorts (ARIC, CARDIA, CHS, and MESA), measured plasma phospholipid fatty acids, and whether the scale of these different measurements is completely the same is unclear.
Another factor that may be related to some differences among studies when comparing GWASs is the medication use in the different populations. In previous GWAS [79,80,81,82,83,84,85,86], no adjustment for medications has been reported. In our study, we fitted statistical models sequentially adjusted for sex, age, and diabetes in order to compare our results with the previously published studies having similar adjustment. Additionally, we adjusted our main results for medications and this adjustment changed little the associations. However, an influence of lipid-lowering drugs (mainly statins) on PUFA levels has been reported in other studies [131,132]. Therefore, additional analysis with a large sample size are needed to better understand the role (and potential gene*statins interactions) of the medications in determining PUFA concentrations.
In addition to these limitations, sample size is another limitation for our study. Considering the strict threshold for statistically significant results at the GWAs level, some potential associations with relevant loci, might fail to be identified. Therefore, we conducted a RWAS, focused on previously identified regions considered as relevant for the genetic associations with PUFA [4,94]. The selected regions were the FADS1/FADS2/FADS3 [94] cluster and the ELOVL2/ELOVL5 [4] region. After testing the SNPs in the candidate genes in these regions, we detected statistically significant associations with LA concentrations that were not detected as significant signals at the GWAS level, supporting the associations between some FADS1/FADS2/FADS3 SNPs and LA concentrations also in this population, but of a lower magnitude than the associations found for DHA or total omega-3 PUFA. Regarding the ELOVL region [4], although other studies have reported associations with several SNPs in these genes [79,81,82,83,84], in this Mediterranean population, the specific associations have been smaller, and even at the RWAS level, no significant associations have been detected for omega-3 PUFA. Only a few ELOVL2 SNPs presented significant associations at p < 0.05 with total serum omega-6 and LA.
These results emphasize the need to carry out specific studies to know which genetic variants are most relevant in each population, because the results of the most significant associations in a population or in a meta-analysis are not always those that are actually observed in a different population [89]. In the case of the genetics of serum PUFA, it is known that due to the relevance that these fatty acids have for the functioning of the organism [94], the prevalence of the different genetic variants in the FADS1/FASD2 genes has been adapting to changes in the diet, with the aim of achieving the highest concentrations of long chain PUFA [96]. Several studies have analyzed the details of this so-called positive selection [95,96,97,98], and currently, the prevalence of genetic variants and the overall effect of enzymes may be different depending on the population analyzed [95,96,97,98].
Some GWAS have considered dietary intake in adjustments carried out in the statistical analysis, indicating a modest influence [122]. However, to better understand dietary modulation, a gene*diet interaction analysis should be carried out. Although some GWAS and various candidate gene studies have examined the interaction between several dietary components (food or nutrients) and the genetic variants in determining PUFA concentrations [1,60,83,129,130,131,132,133,134,135], the interaction with a whole dietary pattern such as the Mediterranean diet pattern [99], has not been comprehensively investigated. In an exploratory study, taking into account our limited sample size, we explored gene* Mediterranean diet interactions at the genome-wide level in determining serum PUFA. Two categories (low and high adherence to Mediterranean diet) were analyzed using the same approach that we previously carried out in another genome-wide-interaction study with Mediterranean diet in the same population in determining plasma bilirubin concentrations [105]. The most relevant gene*Mediterranean diet interaction was found for the ME1 gene in determining serum omega-3 concentrations. Several SNPs in this gene, having different MAF, reached statistical significance for the interaction term at p < 5 × 10−5. According to this interaction, the association between the ME1 genotypes and serum omega-3 depends on the adherence to the Mediterranean diet pattern. Moreover, when we further analyzed the Mediterranean diet interaction at the GWAS level in determining the serum omega-6/omega-3 ratio, we also obtained the ME1 as the top-ranked gene, replicating the results for omega-3 PUFA. This is the first time that this interaction is reported in humans and subsequent studies are needed to replicate and characterize the modulation. However, existing literature initially supports a potential mechanism behind this interaction. The ME1 gene encodes a cytosolic, NADP-dependent enzyme that produces NADPH for fatty acid biosynthesis [124]. Likewise, the ME1 gene has been implicated in PUFA biosynthesis [136,137,138]. Furthermore, in animal models, the ME1 gene expression is strongly regulated by the dietary fat content [125,139]. Fat content of the low or high adherence group (high adherence characterized by a high fish intake, nuts, olive oil, and a low intake of red meat and other products rich in SFA) may modulate the effects of the ME1 polymorphisms on serum omega-3 PUFA (as well as on the omega-6/omega-3 ratio), contributing to explain the statistical signal. Our findings are exploratory and additional replication studies are needed to confirm this interaction.
Likewise, genetic modulation by sex may be another relevant factor explaining some differences in the effect of the genetics of PUFA. Some PUFA are specifically required for reproduction [104]. It has been reported that PUFA concentrations are higher in females than in males during pregnancy and there are some studies showing sex-specific effects for some PUFA in determining several outcomes [68,69,70,71,72,73,74,140,141]. Although this is still an emerging field, the promotion of research investigating sex-specific differences [142] could provide more information regarding sex-specific effects on gene*sex interactions in determining PUFA levels for a more personalized nutrition. Our genome-wide gene*sex interaction study, has revealed a statistically significant gene*sex interaction involving the DNTTIP2 gene (rs3747965) in determining serum total PUFA (mainly omega-6 PUFA), at the genome-wide level of significance (p < 5 × 10−8). Again, this is the first time that this statistical interaction has been reported, and more studies are needed to replicate and characterize this interaction. The DNTTIP2 gene has the alternative name of “estrogen receptor-binding protein gene”, as well as the short name: “TdIF2” (terminal deoxynucleotidyltransferase interacting factor 2). The TdIF2/estrogen receptor α-binding protein (ERBP) is a multifunctional nucleolar protein. It promotes ribosomal RNA transcription by still partially unknown mechanisms [143]. Thus, there is a potential link between the SNP in this gene and estrogens [143], explaining the potential sex-specific interaction on PUFA. The literature examining the DNTTIP2 is scarce, but in a recent GWAS, the DNTTIP2 gene has been linked to ischemic stroke [144], an outcome potentially linked to the PUFA modulations. As stated before, this finding is only exploratory, and more research has to be done for replication and characterization. Overall, our novel findings provide a roadmap for further studies analyzing gene*sex and gene*diet interactions on PUFA concentrations in different populations.
5. Conclusions
In conclusion, our results allowed us to define the genetic variants most associated with serum PUFA concentrations in a Mediterranean population with metabolic syndrome and to confirm the strong association between the FADS1/FADS2 locus and omega-3 PUFA (specifically DHA) in this population. The associations of this locus with omega-6 were much weaker, but also present at the RWAS level. Likewise, we have observed a strong genetic influence of the FADS1/FADS2 cluster in determining the omega-6/omega-3 ratio, at the GWAS level, in this population. We have also detected some new interactions with sex (on omega-6 PUFA) and adherence to the Mediterranean diet (on omega-3 PUFA), which will have to be replicated and functionally characterized in other studies. This underlines the need for these variables to be studied in all specific populations, so as to better understand the complex metabolism of PUFA and their repercussions on health.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6643/12/2/310/s1: Supplemental Figure S1: Conversion flow of omega-3 and omega-6 fatty acids, respectively modulated by FADS1/FASD2 and ELOVL2/ELOVL5 enzymes, from linoleic acid to β-docosapentaenoic acid and from α-linolenic acid to docosahexaenoic acid; Supplemental Table S1: Quantitative 17-item questionnaire for Adherence to Mediterranean diet; Supplemental Table S2: Other medications of the study population by sex; Supplemental Figure S2: Q–Q plot for the GWAS on serum omega-3 fatty acid concentrations (%) in the whole population; Supplemental Table S3: Genome-wide interaction study with adherence to Mediterranean diet in determining serum omega-3 fatty acid concentrations (%) in the whole population. Top-ranked SNP*diet interaction terms; Supplemental Figure S3: Q–Q plot for the genome-wide interaction study with adherence to Mediterranean diet in determining serum omega-3 fatty acid concentrations (%) in the whole population; Supplemental Figure S4: Q–Q plot for the GWAS on total docosahexaenoic fatty acid concentrations (%) in the whole population; Supplemental Figure S5. Manhattan plot for the GWAs results for the serum omega-6 fatty acids (%) in the whole population; Supplemental Figure S6. Q–Q plot for the GWAS on serum omega-6 fatty acid concentrations (%) in the whole population; Supplemental Table S4: Top-ranked SNPs in the GWAS for serum omega-6 fatty acids concentrations (%) in the whole population; Supplemental Figure S7: Manhattan plot for the GWAS results for the linoleic fatty acids (%) in the whole population; Supplemental Figure S8: Q–Q plot for the GWAS on linoleic fatty acid concentrations (%) in the whole population; Supplemental Table S5: Top-ranked SNPs in the GWAS for linoleic fatty acids concentrations (%) in the whole population; Supplemental Figure S9. Manhattan plot for the genome-wide interaction with adherence to Mediterranean diet in determining polyunsaturated fatty acids (%) in the whole population; Supplemental Table S6: Top-ranked SNPs in the GWAS for the serum omega-6/omega-3 PUFA ratio in the whole population; Supplemental Table S7: SNPs in the RWAS for the FADS1/FADS2/FADS3 region in determining serum omega-3 fatty acid concentrations (%) in the whole population; Supplemental Table S8: SNPs in the RWAS for the FADS1/FADS2/FADS3 region in determining serum docosahexaenoic fatty acid concentrations (%) in the whole population; Supplemental Table S9: SNPs in the RWAS for the FADS1/FADS2/FADS3 region in determining serum omega-6 fatty acid concentrations (%) in the whole population; Supplemental Table S10: SNPs in the RWAS for the ELOVL2/ELOVL5 region in determining serum omega-3 fatty acid concentrations (%) in the whole population; Supplemental Table S11: SNPs in the RWAS for the ELOVL2/ELOVL5 region in determining serum omega-6 fatty acid concentrations (%) in the whole population; Supplemental Table S12: SNPs in the RWAS for the ELOVL2/ELOVL5 region in determining linoleic fatty acid concentrations (%) in the whole population Supplemental Figure S10. Means of serum omega-6/omega-3 PUFA ratio depending on the FADS1-rs174547 genotype in the whole population; Supplemental Figure S11. Manhattan plot for the GWAs interaction results between the top-ranked SNPs and the adherence to the Mediterranean diet in determining serum omega-6/omega-3 PUFA ratio in the whole population.
Author Contributions
The authors’ responsibilities were as follows: Conceptualization, O.C., J.V.S., R.E., J.M.O. and D.C.; Data curation, E.M.A., R.B., J.I.G., I.M.G.-A., J.B.R.-S., E.C.P. and C.O.-A.; Formal analysis, O.C., J.V.S. and D.C.; Funding acquisition, O.C., R.E. and D.C.; Investigation, J.V.S., E.M.A., R.B., J.I.G., I.M.G.-A., V.Z.-M., J.B.R.-S., E.P. and C.O.-A.; Methodology, J.M.O. and D.C.; Project administration, O.C., E.M.A. and D.C.; Software, O.C.; Resources, E.M.A. and C.O.-A.; Supervision, J.M.O., J.V.S. and D.C.; Writing—original draft, O.C., and D.C.; Writing—review & editing, O.C., D.C., J.V.S., V.Z.-M., R.E. and J.M.O. All authors read and approved the final manuscript.
Funding
This study was partially funded, by the Spanish Ministry of Health (Instituto de Salud Carlos III) and the Ministerio de Economía y Competitividad-Fondo Europeo de Desarrollo Regional (FEDER) (grants CIBER 06/03, PI06/1326, PI13/00728, PI16/00366, SAF2016-80532-R and FPU 18/01703); the University Jaume I (grants P1-1B2013-54 and COGRUP/2016/06); the Rei Jaume I Award for Medical Research 2018; the Fundació La Marató de TV3 (grant 538/U/2016); and the Generalitat Valenciana (grants PROMETEO2017/017, AEST/2018/044 and APOSTD/2019/136) and the US Department of Agriculture, Agriculture Research Service (grant 8050-51000-098-00D).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chilton F., Dutta R., Reynolds L., Sergeant S., Mathias R., Seeds M. Precision Nutrition and Omega-3 Polyunsaturated Fatty Acids: A Case for Personalized Supplementation Approaches for the Prevention and Management of Human Diseases. Nutrients. 2017;9:1165. doi: 10.3390/nu9111165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park H.G., Park W.J., Kothapalli K.S.D., Brenna J.T. The fatty acid desaturase 2 (FADS2) gene product catalyzes Δ4 desaturation to yield n-3 docosahexaenoic acid and n-6 docosapentaenoic acid in human cells. FASEB J. 2015;29:3911–3919. doi: 10.1096/fj.15-271783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim O., Lee S., An W. Impact of Blood or Erythrocyte Membrane Fatty Acids for Disease Risk Prediction: Focusing on Cardiovascular Disease and Chronic Kidney Disease. Nutrients. 2018;10:1454. doi: 10.3390/nu10101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J.Y., Kothapalli K.S.D., Brenna J.T. Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr. Opin. Clin. Nutr. Metab. Care. 2016;19:103–110. doi: 10.1097/MCO.0000000000000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astorg P., Bertrais S., Laporte F., Arnault N., Estaquio C., Galan P., Favier A., Hercberg S. Plasma n-6 and n-3 polyunsaturated fatty acids as biomarkers of their dietary intakes: a cross-sectional study within a cohort of middle-aged French men and women. Eur. J. Clin. Nutr. 2008;62:1155–1161. doi: 10.1038/sj.ejcn.1602836. [DOI] [PubMed] [Google Scholar]
- 6.Li D., Wahlqvist M.L., Sinclair A.J. Advances in n-3 polyunsaturated fatty acid nutrition. Asia Pac. J. Clin. Nutr. 2019;28:1–5. doi: 10.6133/apjcn.201903_28(1).0001. [DOI] [PubMed] [Google Scholar]
- 7.Meyer B.J., Mann N.J., Lewis J.L., Milligan G.C., Sinclair A.J., Howe P.R.C. Dietary intakes and food sources of omega-6 and omega-3 polyunsaturated fatty acids. Lipids. 2003;38:391–398. doi: 10.1007/s11745-003-1074-0. [DOI] [PubMed] [Google Scholar]
- 8.Ortega Anta R.M., González Rodríguez L.G., Villalobos Cruz T.K., Perea Sánchez J.M., Aparicio Vizuete A., López Sobaler A.M. Food sources and adequacy of intake of omega 3 and omega-6 fatty acids in a representative sample of Spanish adults. Nutr. Hosp. 2013;28:2236–2245. [PubMed] [Google Scholar]
- 9.González-Rodríguez L.G., Aparicio A., López-Sobaler A.M., Ortega R.M. Omega 3 and omega 6 fatty acids intake and dietary sources in a representative sample of Spanish adults. Int. J. Vitam. Nutr. Res. 2013;83:36–47. doi: 10.1024/0300-9831/a000143. [DOI] [PubMed] [Google Scholar]
- 10.Dyerberg J., Bang H.O., Stoffersen E., Moncada S., Vane J.R. Eicosapentaenoic acid and prevention of thrombosis and atherosclerosis? Lancet. 1978;2:117–119. doi: 10.1016/S0140-6736(78)91505-2. [DOI] [PubMed] [Google Scholar]
- 11.Demaison L., Moreau D. Dietary n-3 polyunsaturated fatty acids and coronary heart disease-related mortality: A possible mechanism of action. Cell. Mol. Life Sci. 2002;59:463–477. doi: 10.1007/s00018-002-8439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbige L.S. Fatty acids, the immune response, and autoimmunity: A question of n-6 essentiality and the balance between n-6 and n-3. Lipids. 2003;38:323–341. doi: 10.1007/s11745-003-1067-z. [DOI] [PubMed] [Google Scholar]
- 13.Nageswari K., Banerjee R., Menon V.P. Effect of saturated, omega-3 and omega-6 polyunsaturated fatty acids on myocardial infarction. J. Nutr. Biochem. 1999;10:338–344. doi: 10.1016/S0955-2863(99)00007-8. [DOI] [PubMed] [Google Scholar]
- 14.Bang H.O., Dyerberg J., Sinclair H.M. The composition of the Eskimo food in north western Greenland. Am. J. Clin. Nutr. 1980;33:2657–2661. doi: 10.1093/ajcn/33.12.2657. [DOI] [PubMed] [Google Scholar]
- 15.Salonen J.T., Salonen R., Penttilä I., Herranen J., Jauhiainen M., Kantola M., Lappeteläinen R., Mäenpää P.H., Alfthan G., Puska P. Serum fatty acids, apolipoproteins, selenium and vitamin antioxidants and the risk of death from coronary artery disease. Am. J. Cardiol. 1985;56:226–231. doi: 10.1016/0002-9149(85)90839-2. [DOI] [PubMed] [Google Scholar]
- 16.Kang J.X., Leaf A. The cardiac antiarrhythmic effects of polyunsaturated fatty acid. Lipids. 1996;31:S41–S44. doi: 10.1007/BF02637049. [DOI] [PubMed] [Google Scholar]
- 17.Nordøy A., Marchioli R., Arnesen H., Videbaek J. N-3 polyunsaturated fatty acids and cardiovascular diseases. Lipids. 2001;36:S127–S129. doi: 10.1007/s11745-001-0695-7. [DOI] [PubMed] [Google Scholar]
- 18.Mozaffarian D., Ascherio A., Hu F.B., Stampfer M.J., Willett W.C., Siscovick D.S., Rimm E.B. Interplay between different polyunsaturated fatty acids and risk of coronary heart disease in men. Circulation. 2005;111:157–164. doi: 10.1161/01.CIR.0000152099.87287.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavie C.J., Milani R.V., Mehra M.R., Ventura H.O. Omega-3 polyunsaturated fatty acids and cardiovascular diseases. J. Am. Coll. Cardiol. 2009;54:585–594. doi: 10.1016/j.jacc.2009.02.084. [DOI] [PubMed] [Google Scholar]
- 20.Mori T.A. Omega-3 fatty acids and hypertension in humans. Clin. Exp. Pharmacol. Physiol. 2006;33:842–846. doi: 10.1111/j.1440-1681.2006.04451.x. [DOI] [PubMed] [Google Scholar]
- 21.Rupp H. Omacor (prescription omega-3-acid ethyl esters 90): From severe rhythm disorders to hypertriglyceridemia. Adv. Ther. 2009;26:675–690. doi: 10.1007/s12325-009-0045-2. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z.-H., Emma-Okon B., Remaley A.T. Dietary marine-derived long-chain monounsaturated fatty acids and cardiovascular disease risk: A mini review. Lipids Health Dis. 2016;15:201. doi: 10.1186/s12944-016-0366-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schunck W.-H., Konkel A., Fischer R., Weylandt K.-H. Therapeutic potential of omega-3 fatty acid-derived epoxyeicosanoids in cardiovascular and inflammatory diseases. Pharmacol. Ther. 2018;183:177–204. doi: 10.1016/j.pharmthera.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Yates C.M., Calder P.C., Ed Rainger G. Pharmacology and therapeutics of omega-3 polyunsaturated fatty acids in chronic inflammatory disease. Pharmacol. Ther. 2014;141:272–282. doi: 10.1016/j.pharmthera.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Saini R.K., Keum Y.-S. Omega-3 and omega-6 polyunsaturated fatty acids: Dietary sources, metabolism, and significance—A review. Life Sci. 2018;203:255–267. doi: 10.1016/j.lfs.2018.04.049. [DOI] [PubMed] [Google Scholar]
- 26.Darwesh A.M., Sosnowski D.K., Lee T.Y., Keshavarz-Bahaghighat H., Seubert J.M. Insights into the cardioprotective properties of n-3 PUFAs against ischemic heart disease via modulation of the innate immune system. Chem. Biol. Interact. 2019;308:20–44. doi: 10.1016/j.cbi.2019.04.037. [DOI] [PubMed] [Google Scholar]
- 27.Schwab U., Lauritzen L., Tholstrup T., Haldorssoni T., Riserus U., Uusitupa M., Becker W. Effect of the amount and type of dietary fat on cardiometabolic risk factors and risk of developing type 2 diabetes, cardiovascular diseases, and cancer: A systematic review. Food Nutr. Res. 2014;58:25145. doi: 10.3402/fnr.v58.25145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marangoni F., Agostoni C., Borghi C., Catapano A.L., Cena H., Ghiselli A., La Vecchia C., Lercker G., Manzato E., Pirillo A., et al. Dietary linoleic acid and human health: Focus on cardiovascular and cardiometabolic effects. Atherosclerosis. 2020;292:90–98. doi: 10.1016/j.atherosclerosis.2019.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Simonetto M., Infante M., Sacco R.L., Rundek T., Della-Morte D. A Novel Anti-Inflammatory Role of Omega-3 PUFAs in Prevention and Treatment of Atherosclerosis and Vascular Cognitive Impairment and Dementia. Nutrients. 2019;11:2279. doi: 10.3390/nu11102279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maki K., Dicklin M. Omega-3 Fatty Acid Supplementation and Cardiovascular Disease Risk: Glass Half Full or Time to Nail the Coffin Shut? Nutrients. 2018;10:864. doi: 10.3390/nu10070864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tortosa-Caparrós E., Navas-Carrillo D., Marín F., Orenes-Piñero E. Anti-inflammatory effects of omega 3 and omega 6 polyunsaturated fatty acids in cardiovascular disease and metabolic syndrome. Crit. Rev. Food Sci. Nutr. 2017;57:3421–3429. doi: 10.1080/10408398.2015.1126549. [DOI] [PubMed] [Google Scholar]
- 32.Harris W.S. Are n-3 fatty acids still cardioprotective? Curr. Opin. Clin. Nutr. Metab. Care. 2013;16:141–149. doi: 10.1097/MCO.0b013e32835bf380. [DOI] [PubMed] [Google Scholar]
- 33.Mozaffarian D., Wu J.H.Y. Omega-3 fatty acids and cardiovascular disease: Effects on risk factors, molecular pathways, and clinical events. J. Am. Coll. Cardiol. 2011;58:2047–2067. doi: 10.1016/j.jacc.2011.06.063. [DOI] [PubMed] [Google Scholar]
- 34.Rimm E.B., Appel L.J., Chiuve S.E., Djoussé L., Engler M.B., Kris-Etherton P.M., Mozaffarian D., Siscovick D.S., Lichtenstein A.H. On behalf of the American Heart Association Nutrition Committee of the Council on Lifestyle and Cardiometabolic Health; Council on Epidemiology and Prevention; Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; and Council on Clinical Cardiology. Seafood Long-Chain n-3 Polyunsaturated Fatty Acids and Cardiovascular Disease: A Science Advisory From the American Heart Association. Circulation. 2018;138:e35–e47. doi: 10.1161/CIR.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sacks F.M., Lichtenstein A.H., Wu J.H.Y., Appel L.J., Creager M.A., Kris-Etherton P.M., Miller M., Rimm E.B., Rudel L.L., Robinson J.G., et al. Dietary Fats and Cardiovascular Disease: A Presidential Advisory from the American Heart Association. Circulation. 2017;136:e1–e23. doi: 10.1161/CIR.0000000000000510. [DOI] [PubMed] [Google Scholar]
- 36.Abdelhamid A.S., Martin N., Bridges C., Brainard J.S., Wang X., Brown T.J., Hanson S., Jimoh O.F., Ajabnoor S.M., Deane K.H., et al. Polyunsaturated fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2018;11:CD012345. doi: 10.1002/14651858.CD012345.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdelhamid A.S., Brown T.J., Brainard J.S., Biswas P., Thorpe G.C., Moore H.J., Deane K.H., AlAbdulghafoor F.K., Summerbell C.D., Worthington H.V., et al. Omega-3 fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2018;11:CD003177. doi: 10.1002/14651858.CD003177.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hooper L., Al-Khudairy L., Abdelhamid A.S., Rees K., Brainard J.S., Brown T.J., Ajabnoor S.M., O’Brien A.T., Winstanley L.E., Donaldson D.H., et al. Omega-6 fats for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2018;11:CD011094. doi: 10.1002/14651858.CD011094.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imamura F., Micha R., Wu J.H.Y., de Oliveira Otto M.C., Otite F.O., Abioye A.I., Mozaffarian D. Effects of Saturated Fat, Polyunsaturated Fat, Monounsaturated Fat, and Carbohydrate on Glucose-Insulin Homeostasis: A Systematic Review and Meta-analysis of Randomised Controlled Feeding Trials. PLoS Med. 2016;13:e1002087. doi: 10.1371/journal.pmed.1002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balk E.M., Adam G.P., Langberg V., Halladay C., Chung M., Lin L., Robertson S., Yip A., Steele D., Smith B.T., et al. Omega-3 Fatty Acids and Cardiovascular Disease: An Updated Systematic Review. Agency for Healthcare Research and Quality; Rockville, MD, USA: 2016. [DOI] [PubMed] [Google Scholar]
- 41.Brown T.J., Brainard J., Song F., Wang X., Abdelhamid A., Hooper L. Omega-3, omega-6, and total dietary polyunsaturated fat for prevention and treatment of type 2 diabetes mellitus: Systematic review and meta-analysis of randomised controlled trials. BMJ. 2019:l4697. doi: 10.1136/bmj.l4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeffery N.M., Newsholme E.A., Calder P.C. Level of polyunsaturated fatty acids and the n-6 to n-3 polyunsaturated fatty acid ratio in the rat diet alter serum lipid levels and lymphocyte functions. Prostaglandins Leukot. Essent. Fatty Acids. 1997;57:149–160. doi: 10.1016/S0952-3278(97)90005-3. [DOI] [PubMed] [Google Scholar]
- 43.Wijendran V., Hayes K.C. Dietary n-6 and n-3 fatty acid balance and cardiovascular health. Annu. Rev. Nutr. 2004;24:597–615. doi: 10.1146/annurev.nutr.24.012003.132106. [DOI] [PubMed] [Google Scholar]
- 44.Andrioli G., Carletto A., Guarini P., Galvani S., Biasi D., Bellavite P., Corrocher R. Differential effects of dietary supplementation with fish oil or soy lecithin on human platelet adhesion. Thromb. Haemost. 1999;82:1522–1527. [PubMed] [Google Scholar]
- 45.Calder P.C. Omega-3 fatty acids and inflammatory processes. Nutrients. 2010;2:355–374. doi: 10.3390/nu2030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calder P.C. Omega-3 fatty acids and inflammatory processes: from molecules to man. Biochem. Soc. Trans. 2017;45:1105–1115. doi: 10.1042/BST20160474. [DOI] [PubMed] [Google Scholar]
- 47.Patterson E., Wall R., Fitzgerald G.F., Ross R.P., Stanton C. Health implications of high dietary omega-6 polyunsaturated Fatty acids. J. Nutr. Metab. 2012;2012:539426. doi: 10.1155/2012/539426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Innes J.K., Calder P.C. Omega-6 fatty acids and inflammation. Prostaglandins Leukot. Essent. Fatty Acids. 2018;132:41–48. doi: 10.1016/j.plefa.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 49.DiNicolantonio J.J., O’Keefe J.H. Importance of maintaining a low omega–6/omega–3 ratio for reducing inflammation. Open Heart. 2018;5:e000946. doi: 10.1136/openhrt-2018-000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simopoulos A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008;233:674–688. doi: 10.3181/0711-MR-311. [DOI] [PubMed] [Google Scholar]
- 51.Russo G.L. Dietary n-6 and n-3 polyunsaturated fatty acids: From biochemistry to clinical implications in cardiovascular prevention. Biochem. Pharmacol. 2009;77:937–946. doi: 10.1016/j.bcp.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 52.Eaton S.B., Eaton S.B., Konner M.J., Shostak M. An evolutionary perspective enhances understanding of human nutritional requirements. J. Nutr. 1996;126:1732–1740. doi: 10.1093/jn/126.6.1732. [DOI] [PubMed] [Google Scholar]
- 53.Griffin M.D., Sanders T.A.B., Davies I.G., Morgan L.M., Millward D.J., Lewis F., Slaughter S., Cooper J.A., Miller G.J., Griffin B.A. Effects of altering the ratio of dietary n-6 to n-3 fatty acids on insulin sensitivity, lipoprotein size, and postprandial lipemia in men and postmenopausal women aged 45-70 y: the OPTILIP Study. Am. J. Clin. Nutr. 2006;84:1290–1298. doi: 10.1093/ajcn/84.6.1290. [DOI] [PubMed] [Google Scholar]
- 54.Griffin B.A. How relevant is the ratio of dietary n-6 to n-3 polyunsaturated fatty acids to cardiovascular disease risk? Evidence from the OPTILIP study. Curr. Opin. Lipidol. 2008;19:57–62. doi: 10.1097/MOL.0b013e3282f2e2a8. [DOI] [PubMed] [Google Scholar]
- 55.Dunbar B.S., Bosire R.V., Deckelbaum R.J. Omega 3 and omega 6 fatty acids in human and animal health: An African perspective. Mol. Cell. Endocrinol. 2014;398:69–77. doi: 10.1016/j.mce.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Shrestha N., Sleep S.L., Cuffe J.S., Holland O.J., Perkins A.V., Yu Yau S., McAinch A.J., Hryciw D.H. Role Of Omega-6 and Omega-3 fatty acids in fetal programming. Clin. Exp. Pharmacol. Physiol. 2019 doi: 10.1111/1440-1681.13244. [DOI] [PubMed] [Google Scholar]
- 57.Kaliannan K., Li X.-Y., Wang B., Pan Q., Chen C.-Y., Hao L., Xie S., Kang J.X. Multi-omic analysis in transgenic mice implicates omega-6/omega-3 fatty acid imbalance as a risk factor for chronic disease. Commun. Biol. 2019;2:276. doi: 10.1038/s42003-019-0521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steffen B.T., Guan W., Stein J.H., Tattersall M.C., Kaufman J.D., Sandfort V., Szklo M., Tsai M.Y. Plasma n-3 and n-6 Fatty Acids Are Differentially Related to Carotid Plaque and Its Progression: The Multi-Ethnic Study of Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2018;38:653–659. doi: 10.1161/ATVBAHA.117.310366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Virtanen J.K. Randomized trials of replacing saturated fatty acids with n-6 polyunsaturated fatty acids in coronary heart disease prevention: Not the gold standard? Prostaglandins Leukot. Essent. Fatty Acids. 2018;133:8–15. doi: 10.1016/j.plefa.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 60.Simopoulos A.P. Genetic variants in the metabolism of omega-6 and omega-3 fatty acids: Their role in the determination of nutritional requirements and chronic disease risk. Exp. Biol. Med. 2010;235:785–795. doi: 10.1258/ebm.2010.009298. [DOI] [PubMed] [Google Scholar]
- 61.Dumont J., Huybrechts I., Spinneker A., Gottrand F., Grammatikaki E., Bevilacqua N., Vyncke K., Widhalm K., Kafatos A., Molnar D., et al. FADS1 genetic variability interacts with dietary α-linolenic acid intake to affect serum non-HDL-cholesterol concentrations in European adolescents. J. Nutr. 2011;141:1247–1253. doi: 10.3945/jn.111.140392. [DOI] [PubMed] [Google Scholar]
- 62.Cormier H., Rudkowska I., Paradis A.-M., Thifault E., Garneau V., Lemieux S., Couture P., Vohl M.-C. Association between polymorphisms in the fatty acid desaturase gene cluster and the plasma triacylglycerol response to an n-3 PUFA supplementation. Nutrients. 2012;4:1026–1041. doi: 10.3390/nu4081026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hellstrand S., Sonestedt E., Ericson U., Gullberg B., Wirfält E., Hedblad B., Orho-Melander M. Intake levels of dietary long-chain PUFAs modify the association between genetic variation in FADS and LDL-C. J. Lipid Res. 2012;53:1183–1189. doi: 10.1194/jlr.P023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brayner B., Kaur G., Keske M.A., Livingstone K.M. FADS Polymorphism, Omega-3 Fatty Acids and Diabetes Risk: A Systematic Review. Nutrients. 2018;10:758. doi: 10.3390/nu10060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwong R.Y., Heydari B., Ge Y., Abdullah S., Fujikura K., Kaneko K., Harris W.S., Jerosch-Herold M., Antman E.M., Seidman J.G., et al. Genetic profiling of fatty acid desaturase polymorphisms identifies patients who may benefit from high-dose omega-3 fatty acids in cardiac remodeling after acute myocardial infarction-Post-hoc analysis from the OMEGA-REMODEL randomized controlled trial. PLoS ONE. 2019;14:e0222061. doi: 10.1371/journal.pone.0222061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Goede J., Verschuren W.M.M., Boer J.M.A., Kromhout D., Geleijnse J.M. Gender-specific associations of marine n-3 fatty acids and fish consumption with 10-year incidence of stroke. PLoS ONE. 2012;7:e33866. doi: 10.1371/journal.pone.0033866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferdouse A., Leng S., Winter T., Aukema H.M. Dietary n-6 and n-3 PUFA alter the free oxylipin profile differently in male and female rat hearts. Br. J. Nutr. 2019;122:252–261. doi: 10.1017/S0007114519001211. [DOI] [PubMed] [Google Scholar]
- 68.Lauritzen L., Sørensen L.B., Harsløf L.B., Ritz C., Stark K.D., Astrup A., Dyssegaard C.B., Egelund N., Michaelsen K.F., Damsgaard C.T. Mendelian randomization shows sex-specific associations between long-chain PUFA-related genotypes and cognitive performance in Danish schoolchildren. Am. J. Clin. Nutr. 2017;106:88–95. doi: 10.3945/ajcn.117.152595. [DOI] [PubMed] [Google Scholar]
- 69.Da Boit M., Sibson R., Sivasubramaniam S., Meakin J.R., Greig C.A., Aspden R.M., Thies F., Jeromson S., Hamilton D.L., Speakman J.R., et al. Sex differences in the effect of fish-oil supplementation on the adaptive response to resistance exercise training in older people: A randomized controlled trial. Am. J. Clin. Nutr. 2017;105:151–158. doi: 10.3945/ajcn.116.140780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ip W.T.K., McAlindon A., Miller S.E., Bell J.R., Curl C.L., Huggins C.E., Mellor K.M., Raaijmakers A.J.A., Bienvenu L.A., McLennan P.L., et al. Dietary omega-6 fatty acid replacement selectively impairs cardiac functional recovery after ischemia in female (but not male) rats. Am. J. Physiol. Heart Circ. Physiol. 2016;311:H768–H780. doi: 10.1152/ajpheart.00690.2015. [DOI] [PubMed] [Google Scholar]
- 71.Balogun K.A., Cheema S.K. Dietary Omega-3 Fatty Acids Prevented Adipocyte Hypertrophy by Downregulating DGAT-2 and FABP-4 in a Sex-Dependent Fashion. Lipids. 2016;51:25–38. doi: 10.1007/s11745-015-4105-x. [DOI] [PubMed] [Google Scholar]
- 72.Ghasemifard S., Hermon K., Turchini G.M., Sinclair A.J. Metabolic fate (absorption, β-oxidation and deposition) of long-chain n-3 fatty acids is affected by sex and by the oil source (krill oil or fish oil) in the rat. Br. J. Nutr. 2015;114:684–692. doi: 10.1017/S0007114515002457. [DOI] [PubMed] [Google Scholar]
- 73.Childs C.E., Kew S., Finnegan Y.E., Minihane A.M., Leigh-Firbank E.C., Williams C.M., Calder P.C. Increased dietary α-linolenic acid has sex-specific effects upon eicosapentaenoic acid status in humans: Re-examination of data from a randomised, placebo-controlled, parallel study. Nutr. J. 2014;13:113. doi: 10.1186/1475-2891-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin H., Nicodemus-Johnson J. Gender and Age Stratified Analyses of Nutrient and Dietary Pattern Associations with Circulating Lipid Levels Identify Novel Gender and Age-Specific Correlations. Nutrients. 2018;10:1760. doi: 10.3390/nu10111760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zou X.-G., Huang Y.-H., Li H.-Y., Xu T.-C., Fan Y.-W., Li J., Deng Z.-Y. Effects of Chinese Dietary Pattern of Fat Content, n-6/n-3 Polyunsaturated Fatty Acid Ratio, and Cholesterol Content on Lipid Profile in Rats. Biomed. Res. Int. 2018;2018:4398086. doi: 10.1155/2018/4398086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Petersson S., Philippou E., Rodomar C., Nikiphorou E. The Mediterranean diet, fish oil supplements and Rheumatoid arthritis outcomes: Evidence from clinical trials. Autoimmun. Rev. 2018;17:1105–1114. doi: 10.1016/j.autrev.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 77.Adamsson V., Cederholm T., Vessby B., Risérus U. Influence of a healthy Nordic diet on serum fatty acid composition and associations with blood lipoproteins—Results from the NORDIET study. Food Nutr. Res. 2014;58:24114. doi: 10.3402/fnr.v58.24114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muzsik A., Bajerska J., Jeleń H.H., Walkowiak J., Krzyżanowska-Jankowska P., Chmurzynska A. FADS1 and FADS2 polymorphism are associated with changes in fatty acid concentrations after calorie-restricted Central European and Mediterranean diets. Menopause. 2019;26:1415–1424. doi: 10.1097/GME.0000000000001409. [DOI] [PubMed] [Google Scholar]
- 79.Tanaka T., Shen J., Abecasis G.R., Kisialiou A., Ordovas J.M., Guralnik J.M., Singleton A., Bandinelli S., Cherubini A., Arnett D., et al. Genome-Wide Association Study of Plasma Polyunsaturated Fatty Acids in the InCHIANTI Study. PLoS Genet. 2009;5:e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lemaitre R.N., Tanaka T., Tang W., Manichaikul A., Foy M., Kabagambe E.K., Nettleton J.A., King I.B., Weng L.-C., Bhattacharya S., et al. Genetic Loci Associated with Plasma Phospholipid n-3 Fatty Acids: A Meta-Analysis of Genome-Wide Association Studies from the CHARGE Consortium. PLoS Genet. 2011;7:e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guan W., Steffen B.T., Lemaitre R.N., Wu J.H.Y., Tanaka T., Manichaikul A., Foy M., Rich S.S., Wang L., Nettleton J.A., et al. Genome-Wide Association Study of Plasma N6 Polyunsaturated Fatty Acids Within the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. Circ. Cardiovasc. Genet. 2014;7:321–331. doi: 10.1161/CIRCGENETICS.113.000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tintle N.L., Pottala J.V., Lacey S., Ramachandran V., Westra J., Rogers A., Clark J., Olthoff B., Larson M., Harris W., et al. A genome-wide association study of saturated, mono- and polyunsaturated red blood cell fatty acids in the Framingham Heart Offspring Study. Prostaglandins Leukot. Essent. Fatty Acids. 2015;94:65–72. doi: 10.1016/j.plefa.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu J.H.Y., Lemaitre R.N., Manichaikul A., Guan W., Tanaka T., Foy M., Kabagambe E.K., Djousse L., Siscovick D., Fretts A.M., et al. Genome-Wide Association Study Identifies Novel Loci Associated with Concentrations of Four Plasma Phospholipid Fatty Acids in the De Novo Lipogenesis Pathway: Results From the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium. Circ. Cardiovasc. Genet. 2013;6:171–183. doi: 10.1161/CIRCGENETICS.112.964619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dorajoo R., Sun Y., Han Y., Ke T., Burger A., Chang X., Low H.Q., Guan W., Lemaitre R.N., Khor C.-C., et al. A genome-wide association study of n-3 and n-6 plasma fatty acids in a Singaporean Chinese population. Genes. Nutr. 2015;10:53. doi: 10.1007/s12263-015-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fumagalli M., Moltke I., Grarup N., Racimo F., Bjerregaard P., Jorgensen M.E., Korneliussen T.S., Gerbault P., Skotte L., Linneberg A., et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349:1343–1347. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 86.Hu Y., Li H., Lu L., Manichaikul A., Zhu J., Chen Y.-D.I., Sun L., Liang S., Siscovick D.S., Steffen L.M., et al. Genome-wide meta-analyses identify novel loci associated with n-3 and n-6 polyunsaturated fatty acid levels in Chinese and European-ancestry populations. Hum. Mol. Genet. 2016;25:1215–1224. doi: 10.1093/hmg/ddw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.PAGE Consortium. Yoneyama S., Yao J., Guo X., Fernandez-Rhodes L., Lim U., Boston J., Buzková P., Carlson C.S., Cheng I., et al. Generalization and fine mapping of European ancestry-based central adiposity variants in African ancestry populations. Int. J. Obes. 2017;41:324–331. doi: 10.1038/ijo.2016.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duncan L., Shen H., Gelaye B., Meijsen J., Ressler K., Feldman M., Peterson R., Domingue B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019;10:3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ortega-Azorín C., Coltell O., Asensio E.M., Sorlí J.V., González J.I., Portolés O., Saiz C., Estruch R., Ramírez-Sabio J.B., Pérez-Fidalgo A., et al. Candidate Gene and Genome-Wide Association Studies for Circulating Leptin Levels Reveal Population and Sex-Specific Associations in High Cardiovascular Risk Mediterranean Subjects. Nutrients. 2019;11:2751. doi: 10.3390/nu11112751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schaeffer L., Gohlke H., Müller M., Heid I.M., Palmer L.J., Kompauer I., Demmelmair H., Illig T., Koletzko B., Heinrich J. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum. Mol. Genet. 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 92.Malerba G., Schaeffer L., Xumerle L., Klopp N., Trabetti E., Biscuola M., Cavallari U., Galavotti R., Martinelli N., Guarini P., et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43:289–299. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- 93.Rzehak P., Heinrich J., Klopp N., Schaeffer L., Hoff S., Wolfram G., Illig T., Linseisen J. Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. Br. J. Nutr. 2008;101:20–26. doi: 10.1017/S0007114508992564. [DOI] [PubMed] [Google Scholar]
- 94.Koletzko B., Reischl E., Tanjung C., Gonzalez-Casanova I., Ramakrishnan U., Meldrum S., Simmer K., Heinrich J., Demmelmair H. FADS1 and FADS2 Polymorphisms Modulate Fatty Acid Metabolism and Dietary Impact on Health. Annu. Rev. Nutr. 2019;39:21–44. doi: 10.1146/annurev-nutr-082018-124250. [DOI] [PubMed] [Google Scholar]
- 95.Ye K., Gao F., Wang D., Bar-Yosef O., Keinan A. Dietary adaptation of FADS genes in Europe varied across time and geography. Nat. Ecol. Evol. 2017;1:0167. doi: 10.1038/s41559-017-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harris D.N., Ruczinski I., Yanek L.R., Becker L.C., Becker D.M., Guio H., Cui T., Chilton F.H., Mathias R.A., O’Connor T.D. Evolution of Hominin Polyunsaturated Fatty Acid Metabolism: From Africa to the New World. Genome Biol. Evol. 2019;11:1417–1430. doi: 10.1093/gbe/evz071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buckley M.T., Racimo F., Allentoft M.E., Jensen M.K., Jonsson A., Huang H., Hormozdiari F., Sikora M., Marnetto D., Eskin E., et al. Selection in Europeans on Fatty Acid Desaturases Associated with Dietary Changes. Mol. Biol. Evol. 2017;34:1307–1318. doi: 10.1093/molbev/msx103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ameur A., Enroth S., Johansson Å., Zaboli G., Igl W., Johansson A.C.V., Rivas M.A., Daly M.J., Schmitz G., Hicks A.A., et al. Genetic Adaptation of Fatty-Acid Metabolism: A Human-Specific Haplotype Increasing the Biosynthesis of Long-Chain Omega-3 and Omega-6 Fatty Acids. Am. J. Hum. Genet. 2012;90:809–820. doi: 10.1016/j.ajhg.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Corella D., Coltell O., Macian F., Ordovás J.M. Advances in Understanding the Molecular Basis of the Mediterranean Diet Effect. Annu. Rev. Food Sci. Technol. 2018;9:227–249. doi: 10.1146/annurev-food-032217-020802. [DOI] [PubMed] [Google Scholar]
- 100.Vilarnau C., Stracker D.M., Funtikov A., da Silva R., Estruch R., Bach-Faig A. Worldwide adherence to Mediterranean Diet between 1960 and 2011. Eur. J. Clin. Nutr. 2019;72:83–91. doi: 10.1038/s41430-018-0313-9. [DOI] [PubMed] [Google Scholar]
- 101.Serra-Majem L., Román-Viñas B., Sanchez-Villegas A., Guasch-Ferré M., Corella D., La Vecchia C. Benefits of the Mediterranean diet: Epidemiological and molecular aspects. Mol. Aspects Med. 2019;67:1–55. doi: 10.1016/j.mam.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 102.De la Garza Puentes A., Montes Goyanes R., Chisaguano Tonato A.M., Torres-Espínola F.J., Arias García M., de Almeida L., Bonilla Aguirre M., Guerendiain M., Castellote Bargalló A.I., Segura Moreno M., et al. Association of maternal weight with FADS and ELOVL genetic variants and fatty acid levels- The PREOBE follow-up. PLoS ONE. 2017;12:e0179135. doi: 10.1371/journal.pone.0179135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Salas Lorenzo I., Chisaguano Tonato A.M., de la Garza Puentes A., Nieto A., Herrmann F., Dieguez E., Castellote A.I., López-Sabater M.C., Rodríguez-Palmero M., Campoy C. The Effect of an Infant Formula Supplemented with AA and DHA on Fatty Acid Levels of Infants with Different FADS Genotypes: The COGNIS Study. Nutrients. 2019;11:602. doi: 10.3390/nu11030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chisaguano A.M., Montes R., Pérez-Berezo T., Castellote A.I., Guerendiain M., Bustamante M., Morales E., García-Esteban R., Sunyer J., Franch À., et al. Gene Expression of Desaturase (FADS1 and FADS2) and Elongase (ELOVL5) Enzymes in Peripheral Blood: Association with Polyunsaturated Fatty Acid Levels and Atopic Eczema in 4-Year-Old Children. PLoS ONE. 2013;8:e78245. doi: 10.1371/journal.pone.0078245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Coltell O., Asensio E.M., Sorlí J.V., Barragán R., Fernández-Carrión R., Portolés O., Ortega-Azorín C., Martínez-LaCruz R., González J.I., Zanón-Moreno V., et al. Genome-Wide Association Study (GWAS) on Bilirubin Concentrations in Subjects with Metabolic Syndrome: Sex-Specific GWAS Analysis and Gene-Diet Interactions in a Mediterranean Population. Nutrients. 2019;11:90. doi: 10.3390/nu11010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Martínez-González M.A., Buil-Cosiales P., Corella D., Bulló M., Fitó M., Vioque J., Romaguera D., Martínez J.A., Wärnberg J., López-Miranda J., et al. Cohort Profile: Design and methods of the PREDIMED-Plus randomized trial. Int. J. Epidemiol. 2019;48:387–388. doi: 10.1093/ije/dyy225. [DOI] [PubMed] [Google Scholar]
- 107.Salas-Salvadó J., Díaz-López A., Ruiz-Canela M., Basora J., Fitó M., Corella D., Serra-Majem L., Wärnberg J., Romaguera D., Estruch R., et al. Effect of a Lifestyle Intervention Program with Energy-Restricted Mediterranean Diet and Exercise on Weight Loss and Cardiovascular Risk Factors: One-Year Results of the PREDIMED-Plus Trial. Diabetes Care. 2018;42:777–788. doi: 10.2337/dc18-0836. [DOI] [PubMed] [Google Scholar]
- 108.Molina L., Sarmiento M., Peñafiel J., Donaire D., Garcia-Aymerich J., Gomez M., Ble M., Ruiz S., Frances A., Schröder H., et al. Validation of the Regicor Short Physical Activity Questionnaire for the Adult Population. PLoS ONE. 2017;12:e0168148. doi: 10.1371/journal.pone.0168148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Galilea-Zabalza I., Buil-Cosiales P., Salas-Salvadó J., Toledo E., Ortega-Azorín C., Díez-Espino J., Vázquez-Ruiz Z., Zomeño M.D., Vioque J., Martínez J.A., et al. Mediterranean diet and quality of life: Baseline cross-sectional analysis of the PREDIMED-PLUS trial. PLoS ONE. 2018;13:e0198974. doi: 10.1371/journal.pone.0198974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Soininen P., Kangas A.J., Würtz P., Tukiainen T., Tynkkynen T., Laatikainen R., Järvelin M.-R., Kähönen M., Lehtimäki T., Viikari J., et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134:1781–1785. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- 111.Tukiainen T., Kettunen J., Kangas A.J., Lyytikäinen L.-P., Soininen P., Sarin A.-P., Tikkanen E., O’Reilly P.F., Savolainen M.J., Kaski K., et al. Detailed metabolic and genetic characterization reveals new associations for 30 known lipid loci. Hum. Mol. Genet. 2012;21:1444–1455. doi: 10.1093/hmg/ddr581. [DOI] [PubMed] [Google Scholar]
- 112.Würtz P., Kangas A.J., Soininen P., Lawlor D.A., Davey Smith G., Ala-Korpela M. Quantitative Serum Nuclear Magnetic Resonance Metabolomics in Large-Scale Epidemiology: A Primer on -Omic Technologies. Am. J. Epidemiol. 2017;186:1084–1096. doi: 10.1093/aje/kwx016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jelenkovic A., Bogl L.H., Rose R.J., Kangas A.J., Soininen P., Ala-Korpela M., Kaprio J., Silventoinen K. Association between serum fatty acids and lipoprotein subclass profile in healthy young adults: Exploring common genetic and environmental factors. Atherosclerosis. 2014;233:394–402. doi: 10.1016/j.atherosclerosis.2013.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deelen J., Kettunen J., Fischer K., van der Spek A., Trompet S., Kastenmüller G., Boyd A., Zierer J., van den Akker E.B., Ala-Korpela M., et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat. Commun. 2019;10:3346. doi: 10.1038/s41467-019-11311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jääskeläinen O., Solje E., Hall A., Katisko K., Korhonen V., Tiainen M., Kangas A.J., Helisalmi S., Pikkarainen M., Koivisto A., et al. Low Serum High-Density Lipoprotein Cholesterol Levels Associate with the C9orf72 Repeat Expansion in Frontotemporal Lobar Degeneration Patients. J. Alzheimers Dis. 2019;72:127–137. doi: 10.3233/JAD-190132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Inouye M., Kettunen J., Soininen P., Silander K., Ripatti S., Kumpula L.S., Hämäläinen E., Jousilahti P., Kangas A.J., Männistö S., et al. Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol. Syst. Biol. 2010;6:441. doi: 10.1038/msb.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sergeant S., Ruczinski I., Ivester P., Lee T.C., Morgan T.M., Nicklas B.J., Mathias R.A., Chilton F.H. Impact of methods used to express levels of circulating fatty acids on the degree and direction of associations with blood lipids in humans. Br. J. Nutr. 2016;115:251–261. doi: 10.1017/S0007114515004341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 121.Turner S.D. Qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014 doi: 10.1101/005165. [DOI] [Google Scholar]
- 122.Kalsbeek A., Veenstra J., Westra J., Disselkoen C., Koch K., McKenzie K.A., O’Bott J., Vander Woude J., Fischer K., Shearer G.C., et al. A genome-wide association study of red-blood cell fatty acids and ratios incorporating dietary covariates: Framingham Heart Study Offspring Cohort. PLoS ONE. 2018;13:e0194882. doi: 10.1371/journal.pone.0194882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Koronowicz A.A., Banks P., Szymczyk B., Leszczyńska T., Master A., Piasna E., Szczepański W., Domagała D., Kopeć A., Piątkowska E., et al. Dietary conjugated linoleic acid affects blood parameters, liver morphology and expression of selected hepatic genes in laying hens. Br. Poult. Sci. 2016;57:663–673. doi: 10.1080/00071668.2016.1192280. [DOI] [PubMed] [Google Scholar]
- 124.Al-Dwairi A., Brown A.R., Pabona J.M.P., Van T.H., Hamdan H., Mercado C.P., Quick C.M., Wight P.A., Simmen R.C.M., Simmen F.A. Enhanced Gastrointestinal Expression of Cytosolic Malic Enzyme (ME1) Induces Intestinal and Liver Lipogenic Gene Expression and Intestinal Cell Proliferation in Mice. PLoS ONE. 2014;9:e113058. doi: 10.1371/journal.pone.0113058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang Y., Ye T., Liu C., Fang F., Chen Y., Dong Y. Maternal high-fat diet during pregnancy and lactation affects hepatic lipid metabolism in early life of offspring rat. J. Biosci. 2017;42:311–319. doi: 10.1007/s12038-017-9675-8. [DOI] [PubMed] [Google Scholar]
- 126.Segarra M., Aburto M.R., Cop F., Llaó-Cid C., Härtl R., Damm M., Bethani I., Parrilla M., Husainie D., Schänzer A., et al. Endothelial Dab1 signaling orchestrates neuro-glia-vessel communication in the central nervous system. Science. 2018;361:eaao2861. doi: 10.1126/science.aao2861. [DOI] [PubMed] [Google Scholar]
- 127.Suhre K., Shin S.-Y., Petersen A.-K., Mohney R.P., Meredith D., Wägele B., Altmaier E., CARDIoGRAM. Deloukas P., Erdmann J., et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sasaki H., Sueyasu T., Tokuda H., Ito M., Kaneda Y., Rogi T., Kawashima H., Horiguchi S., Kawabata T., Shibata H. Aging and FADS1 polymorphisms decrease the biosynthetic capacity of long-chain PUFAs: A human trial using [U-13C] linoleic acid. Prostaglandins Leukot. Essent. Fatty Acids. 2019;148:1–8. doi: 10.1016/j.plefa.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 129.Liu F., Li Z., Lv X., Ma J. Dietary n-3 Polyunsaturated Fatty Acid Intakes Modify the Effect of Genetic Variation in Fatty Acid Desaturase 1 on Coronary Artery Disease. PLoS ONE. 2015;10:e0121255. doi: 10.1371/journal.pone.0121255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marklund M., Wu J.H.Y., Imamura F., Del Gobbo L.C., Fretts A., de Goede J., Shi P., Tintle N., Wennberg M., Aslibekyan S., et al. Biomarkers of Dietary Omega-6 Fatty Acids and Incident Cardiovascular Disease and Mortality. Circulation. 2019;139:2422–2436. doi: 10.1161/CIRCULATIONAHA.118.038908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nyalala J.O., Wang J., Dang A., Faas F.H., Smith W.G. Hypertriglyceridemia and hypercholesterolemia: Effects of drug treatment on fatty acid composition of plasma lipids and membranes. Prostaglandins Leukot. Essent. Fatty Acids. 2008;78:271–280. doi: 10.1016/j.plefa.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 132.Risé P., Marangoni F., Galli C. Regulation of PUFA metabolism: Pharmacological and toxicological aspects. Prostaglandins Leukot. Essent. Fatty Acids. 2002;67:85–89. doi: 10.1054/plef.2002.0403. [DOI] [PubMed] [Google Scholar]
- 133.Juan J., Huang H., Jiang X., Ardisson Korat A.V., Song M., Sun Q., Willett W.C., Jensen M.K., Kraft P. Joint effects of fatty acid desaturase 1 polymorphisms and dietary polyunsaturated fatty acid intake on circulating fatty acid proportions. Am. J. Clin. Nutr. 2018;107:826–833. doi: 10.1093/ajcn/nqy025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gillingham L.G., Harding S.V., Rideout T.C., Yurkova N., Cunnane S.C., Eck P.K., Jones P.J. Dietary oils and FADS1-FADS2 genetic variants modulate [13C] α-linolenic acid metabolism and plasma fatty acid composition. Am. J. Clin. Nutr. 2013;97:195–207. doi: 10.3945/ajcn.112.043117. [DOI] [PubMed] [Google Scholar]
- 135.Hallmann J., Kolossa S., Gedrich K., Celis-Morales C., Forster H., O’Donovan C.B., Woolhead C., Macready A.L., Fallaize R., Marsaux C.F.M., et al. Predicting fatty acid profiles in blood based on food intake and the FADS1 rs174546 SNP. Mol. Nutr. Food. Res. 2015;59:2565–2573. doi: 10.1002/mnfr.201500414. [DOI] [PubMed] [Google Scholar]
- 136.Zhu B.-H., Zhang R.-H., Lv N.-N., Yang G.-P., Wang Y.-S., Pan K.-H. The Role of Malic Enzyme on Promoting Total Lipid and Fatty Acid Production in Phaeodactylum tricornutum. Front. Plant Sci. 2018;9:826. doi: 10.3389/fpls.2018.00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mühlroth A., Li K., Røkke G., Winge P., Olsen Y., Hohmann-Marriott M., Vadstein O., Bones A. Pathways of Lipid Metabolism in Marine Algae, Co-Expression Network, Bottlenecks and Candidate Genes for Enhanced Production of EPA and DHA in Species of Chromista. Marine Drugs. 2013;11:4662–4697. doi: 10.3390/md11114662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rovadoscki G.A., Pertile S.F.N., Alvarenga A.B., Cesar A.S.M., Pértille F., Petrini J., Franzo V., Soares W.V.B., Morota G., Spangler M.L., et al. Estimates of genomic heritability and genome-wide association study for fatty acids profile in Santa Inês sheep. BMC Genom. 2018;19:375. doi: 10.1186/s12864-018-4777-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Benítez R., Fernández A., Isabel B., Núñez Y., De Mercado E., Gómez-Izquierdo E., García-Casco J., López-Bote C., Óvilo C. Modulatory Effects of Breed, Feeding Status, and Diet on Adipogenic, Lipogenic, and Lipolytic Gene Expression in Growing Iberian and Duroc Pigs. Int. J. Med. Sci. 2017;19:22. doi: 10.3390/ijms19010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wathes D.C., Abayasekara D.R.E., Aitken R.J. Polyunsaturated Fatty Acids in Male and Female Reproduction. Biol. Reprod. 2007;77:190–201. doi: 10.1095/biolreprod.107.060558. [DOI] [PubMed] [Google Scholar]
- 141.Feltham B.A., Balogun K.A., Cheema S.K. Perinatal and postweaning diets high in omega-3 fatty acids have age- and sex-specific effects on the fatty acid composition of the cerebellum and brainstem of C57BL/6 mice. Prostaglandins Leukot. Essent. Fatty Acids. 2019;148:16–24. doi: 10.1016/j.plefa.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 142.Corella D., Coltell O., Portolés O., Sotos-Prieto M., Fernández-Carrión R., Ramirez-Sabio J., Zanón-Moreno V., Mattei J., Sorlí J., Ordovas J. A Guide to Applying the Sex-Gender Perspective to Nutritional Genomics. Nutrients. 2018;11:4. doi: 10.3390/nu11010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fukada T., Shibata S., Ueda T., Sasaki K., Shimoida Y., Senda-Murata K., Sugimoto K. Characterization of nucleolar localization and exclusion signals in terminal deoxynucleotidyltransferase interacting factor 2/estrogen receptor α-binding protein. Biosci. Biotechnol. Biochem. 2019;83:1255–1262. doi: 10.1080/09168451.2019.1591265. [DOI] [PubMed] [Google Scholar]
- 144.Yamada Y., Kato K., Oguri M., Horibe H., Fujimaki T., Yasukochi Y., Takeuchi I., Sakuma J. Identification of nine genes as novel susceptibility loci for early-onset ischemic stroke, intracerebral hemorrhage, or subarachnoid hemorrhage. Biomed. Rep. 2018;9:8–20. doi: 10.3892/br.2018.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.