Abstract
Background
Antimicrobial resistance is an increasingly serious problem in public health globally. Monitoring resistance levels within healthcare and community settings is critical to combat its ongoing increase. This study aimed to describe the rates and molecular mechanisms of mupirocin resistance in clinical Staphylococcus aureus isolates from Tygerberg Hospital, and to describe its association with strain types.
Methods
We retrospectively selected 212 S. aureus isolates which were identified from blood samples and pus swabs during the years 2009–2011 and 2015–2017. The isolates were identified using conventional microbiological methods and genotyping was done using spa typing. Cefoxitin (30 μg) disc diffusion and the two disc strategy (5 μg and 200 μg) were used to determine susceptibility to methicillin and mupirocin, respectively. Isolates with high-level resistance were screened for the plasmid mediated genes mupA and mupB by PCR, and sequencing of the ileS gene was done for all isolates exhibiting low-level resistance to describe the mutations associated with this phenotype. Chi-square test was used to assess the associations between mupirocin resistance and S. aureus genotypes.
Results
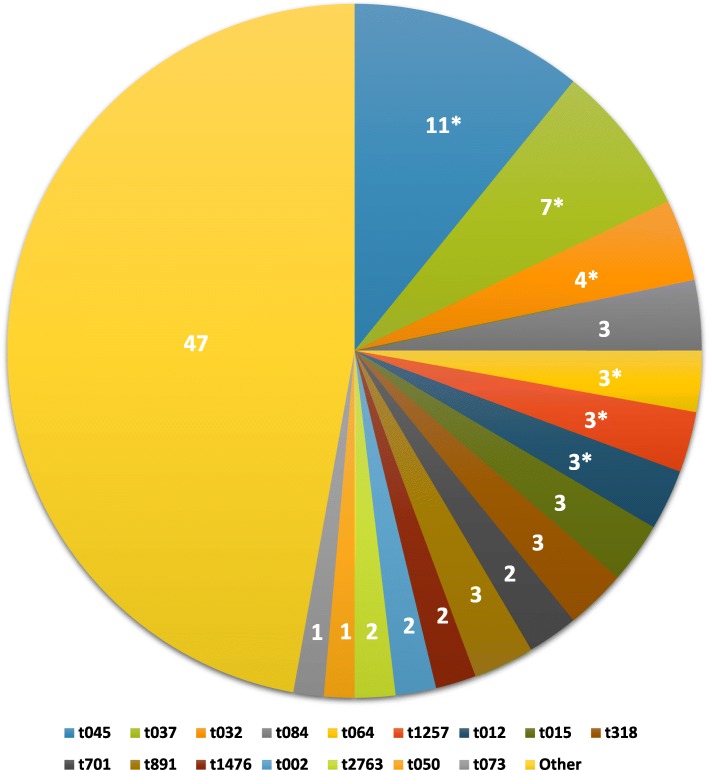
Of 212 S. aureus isolates, 12% (n = 25) were resistant to mupirocin, and 44% (n = 93) were methicillin resistant. Strain typing identified 73 spa types with spa t045 being the most predominant constituting 11% of the isolates. High-level mupirocin resistance was observed in 2% (n = 5), and low-level resistance in 9% (n = 20) of the isolates. The prevalence of high-level mupirocin resistance amongst MRSA and MSSA was 4 and 1% respectively, while the prevalence of low-level mupirocin resistance was significantly higher in MRSA (18%) compared to MSSA (3%), (p = 0.032). mupA was the only resistance determinant for high-level resistance, and the IleS mutation V588F was identified in 95% of the isolates which showed low-level resistance. A significant association was observed between spa type t032 and high-level mupirocin resistance, and types t037 and t012 and low-level resistance (p < 0.0001).
Conclusion
The study reported higher rates of low-level mupirocin resistance compared to high-level resistance, and in our setting, mupirocin resistance was driven by certain genotypes. Our study advocates for the continuous screening for mupirocin resistance in S. aureus in clinical settings to better guide treatment and prescribing practices.
Keywords: Staphylococcus aureus, MRSA, Mupirocin, spa-typing, Antimicrobial resistance, South Africa
Introduction
Staphylococcus aureus is the second most frequent cause of nosocomial bloodstream infections worldwide [1]. S. aureus nasal carriage is a risk factor for subsequent infections, especially amongst surgical and dialysis patients [2, 3], and carriage of methicillin-resistant S. aureus (MRSA) is a particular problem in these high risk patients due to the limited therapeutic drugs available to treat post-operative infections [4]. Consequently, infection prevention strategies such as nasal decolonization are employed to minimize the occurrence of staphylococcal infection and reduce the risk of transmission within healthcare settings [5, 6]. In routine intensive care unit practice, universal decolonization has proven more effective in reducing nosocomial bloodstream infections caused by MRSA as well as any other pathogen, compared to a targeted approach [7]. In some settings, routine screening and decolonisation of MRSA carriers prior to hospital admission is also practiced [8]. The intranasal application of the antibiotic mupirocin (2%) 2–4 times daily for 4–7 days is an effective and affordable strategy for decolonization of MRSA, used alone or in combination with 4% chlorhexidine gluconate (CHG) based body wash [9, 10]. Mupirocin, also known as pseudomonic acid A, is naturally produced by Pseudomonas fluorescens, was first isolated in 1971 and first introduced into clinical practice in the United Kingdom in 1985 [5, 11]. It is also used as a topical agent to treat localised skin and soft tissue infections [10]. Mupirocin inhibits protein synthesis by binding to the bacterial isoleucyl-tRNA synthetase enzyme which is encoded by the ileS gene [11].
Resistance to mupirocin in S. aureus emerged largely due to long-term, unrestricted and unjustified use [12, 13]. Mupirocin resistance is phenotypically categorized into two levels based on the minimum inhibitory concentration (MIC); low-level resistance with MICs of 8–256 mg/ml, and high-level resistance with MICs > 512 mg/ml [4, 14]. The molecular mechanism of low-level mupirocin resistance involves point mutations in the ileS gene; V588F and V631F are two common mutations associated with this phenotype [15–17]. High level mupirocin resistance is mediated by the plasmid encoded genes mupA and mupB, which encode an alternative isoleucyl-tRNA synthetase (ileS2) that is not targeted by mupirocin [18, 19].
Mupirocin resistance in S. aureus, especially high-level resistance, is a serious clinical problem, since it is associated with failure of decolonization, especially among MRSA carriers [20]. This is of particular concern for infection prevention and control practices involved in the management of MRSA outbreaks and in the pre-admission management of surgical patients to minimize post-operative MRSA infections [10]. A recent systematic review describing mupirocin resistance in Africa highlighted the scarcity of data and advocated for the need for surveillance studies to monitor the levels of mupirocin resistance both in the community and healthcare settings [6]. Screening for mupirocin resistance has only been carried out in 12 of the 54 African countries, with an estimated pooled prevalence of 14% resistance amongst MRSA isolates. Only six studies have been conducted in South Africa reporting prevalences of 7–50%, 4–47%, and 0.9–23%, for overall mupirocin resistance, low-level and high-level mupirocin resistance, respectively, in MRSA [6]. There is a paucity of data describing associations between mupirocin resistance and the genetic background of S. aureus in Africa. However, studies from the United States as well as low-middle income countries such as Iran have reported associations between spa types t002, t008, and t064 and mupirocin resistance [21, 22]. Few studies have described mupirocin resistance in the public healthcare sector in the Western Cape of South Africa, however, they reported aggregated data from multiple provinces across South Africa, and mainly investigated resistance rates in MRSA isolates [23–25]. Therefore this study aimed to describe the rates of mupirocin resistance in S. aureus isolates collected at Tygerberg Hospital, to describe the associations between mupirocin resistance and the genotypes which are circulating in our setting, and to investigate the molecular mechanisms of low- and high-level mupirocin resistance.
Methods
This study took place at Tygerberg Hospital, a 1384 bed tertiary academic hospital that serves a population of approximately 1.9 million in the Western Cape, South Africa. S. aureus isolates were identified at the National Health Laboratory Service (NHLS) microbiology laboratory as part of the diagnostic investigation of patients presenting to the hospital. Our Biobank was retrospectively searched for any S. aureus isolates which had been collected from clinical specimens between the years 2009 and 2017. A subset of these isolates was randomly selected to be included in this study. Information on the date of collection and the type of clinical specimen were retrieved from laboratory records. The study was approved by the Health Research Ethics Committee of Stellenbosch University (Reference number N14/06/065).
Bacterial identification and strain typing
The selected S. aureus isolates were identified by standard microbiological methods such as Gram morphology, catalase, mannitol fermentation and DNase activity, and methicillin susceptibility was determined using cefoxitin disc diffusion, according to the CLSI guidelines [26]. Confirmed S. aureus isolates were stored at − 80 °C. All S. aureus isolates were genotyped using spa typing as described previously [27].
Mupirocin susceptibility testing
The stored isolates were streaked on Tryptone Blood Agar (Diagnostic Media Products, South Africa). Overnight cultures were used to perform mupirocin susceptibility testing on Muller-Hinton Sensitivity medium using two different mupirocin containing discs: a 5 μg disc to detect low-level mupirocin resistance and a 200 μg disc to detect high-level mupirocin resistance (Mast diagnostic group, United Kingdom). Zones of inhibition were interpreted according to the CLSI guidelines [26].
Molecular mechanisms of mupirocin resistance
S. aureus isolates which exhibited high-level mupirocin resistance were screened for the presence of the plasmid mediated mupA and mupB genes using the primers and conditions published previously [28]. For low-level resistance, a 450 bp region in the ileS gene was amplified using the modified primers mupLL-F1 5’CCGGAATTAAGTTTCCCAGC-3′ and mupLL-R 5′ CAAAGTTTTCATAGTTGTTAATCGT3’ [29]. Sanger sequencing was done to describe the presence of point mutations within the ileS gene.
Statistical analysis
Statistical analysis was done using STATA version 12 (StataCorp LLC, USA). The difference in the rates of mupirocin resistance between MRSA and MSSA was determined using z-test. Chi-square test was used to assess the associations between strain type and mupirocin resistance. Statistical significance was defined as p-values of < 0.05.
Results
Rates of mupirocin resistance
We included 212 S. aureus isolates based on the availability in the Biobank. Of these, 93 were collected between 2009 and 2011, and 119 between 2015 and 2017. Ninety-two percent (n = 194) of the isolates were from blood cultures and 8% (n = 18) were isolated from pus swabs. Forty four percent (n = 93) of the isolates were MRSA. Strain typing was successful for 180 isolates and 73 spa types were identified.
Mupirocin resistance was observed in 12% (n = 25) of the isolates; five (2%) and 20 (9%) exhibited high-level and low-level resistance, respectively. The prevalence of mupirocin resistance was significantly different between MRSA (23%; n = 21), and MSSA (3%; n = 4) isolates (p = 0.04) (Fig. 1).
Fig. 1.
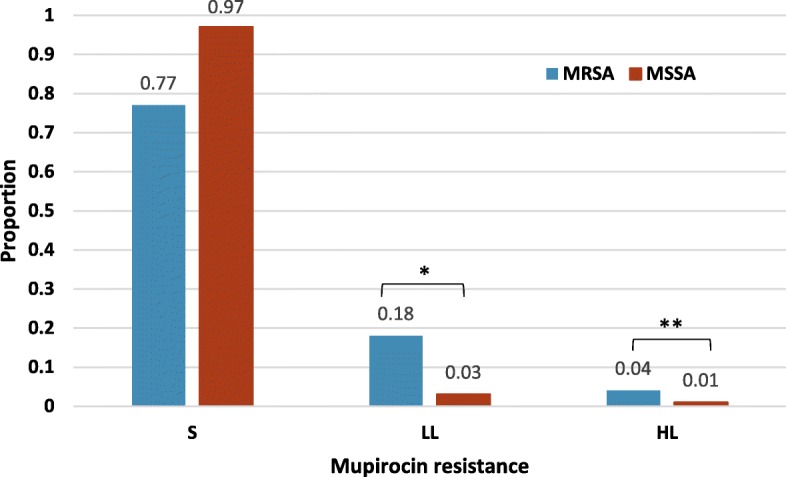
Distribution of mupirocin resistance among S. aureus isolates categorized by methicillin-susceptibility. S: mupirocin susceptible; LL: Low-level mupirocin resistance; HL: high-level mupirocin resistance. The asterisks show the significant differences in the resistance rates between MSSA and MRSA. * p-value =0.032, ** p-value < 0.005
This was consistent for low- and high-level mupirocin resistance; with low level resistance rates of 18% (n = 17) among MRSA and 3% (n = 3) among MSSA (p = 0.032), and high-level mupirocin resistance rates of 4% (n = 4) and 1% (n = 1) among MRSA and MSSA, respectively (p < 0.005).
Correlation between mupirocin resistance and strain types
Figure 2 shows the spa type distribution in the isolate collection, with spa types identified in ≤2 isolates (n = 100) grouped as “others”. Type t045 was the most common, representing 11% of isolates, followed by t037 which constituted 7% of the isolate collection. Of the 16 most common spa types (n = 112), six contained only MRSA isolates (Fig. 2). Mupirocin resistance was represented in nine different spa types (Table 1). A significant association was noted between spa types t012 and t037 and low-level mupirocin resistance, while the spa type t032 was associated with high-level mupirocin resistance (p < 0.0001). Although all the isolates with spa type t045 were MRSA, they were all mupirocin susceptible. Seventy six percent of susceptible isolates belonged to spa types in which no resistant isolates were detected (Table 1).
Fig. 2.
Distribution of the spa types identified in all the clinical S. aureus isolates. The data shown are the percentages of the different spa types. The asterisks indicate the spa types that contained only MRSA isolates. “Others” represent the spa types with ≤2 isolates
Table 1.
Correlation between S. aureus genotypes and mupirocin resistance
| Spa type | High-level N (%) |
Low-level N (%) |
Susceptible N (%) |
Molecular mechanism | Methicillin resistance | P-value |
|---|---|---|---|---|---|---|
| t037 | 0 (0) | 11 (55) | 5 (3) | V588F | MRSA | < 0.0001 |
| t012 | 0 (0) | 5 (25) | 1 (0.5) | V588F | MRSA | < 0.0001 |
| t032 | 3 (60) | 0 (0) | 5 (3) | mupA | MRSA | < 0.0001 |
| t127 | 1 (20) | 0 (0) | 0 (0) | mupA | MSSA | – |
| t1467 | 1 (20) | 0 (0) | 0 (0) | mupA | MRSA | – |
| t891 | 0 (0) | 1 (5) | 5 (3) | V588F | MSSA | – |
| t1517 | 0 (0) | 1 (5) | 1 (0.5) | V588F | MSSA | – |
| t2360 | 0 (0) | 1 (5) | 1 (0.5) | V588F | MSSA | – |
| t073 | 0 (0) | 1 (5) | 2 (1) | S570A | MSSA & MRSA | – |
| Mupirocin susceptiblespatypes | 0 (0) | 0 (0) | 167 (88.5) | – | MSSA & MRSA | – |
| Total | 5 (100) | 20 (100) | 187 (100) | – | – | – |
Only significant p-values (Chi-square test) are shown in the table.
Molecular mechanisms of mupirocin resistance
The mupA gene was detected in all five high-level mupirocin resistant isolates and the amplification product confirmed by Sanger sequencing. mupB was not detected, however the absence could not be confirmed as a positive control was not available. The absence of mupA and mupB was also noted in all isolates with low-level mupirocin resistance. The IleS V588F mutation was detected in 95% (n = 19) of the isolates with low-level resistance. One isolate (spa type t073) carried a possible novel ileS mutation, S570A.
Discussion
Monitoring the levels of antimicrobial resistance within healthcare settings is critical to combat the ongoing increase in resistance. In the current study we sought to describe the rates and molecular mechanisms of mupirocin resistance at Tygerberg Hospital, and to determine possible associations between mupirocin resistance and specific S. aureus lineages.
Twelve percent of the isolates selected for this study exhibited resistance to mupirocin, and the prevalence of mupirocin resistance was significantly higher in MRSA (23%) than MSSA (3%). This trend is consistent with other studies across South Africa; 17% mupirocin resistance was described in MRSA and 2% in MSSA isolates collected from 13 public academic healthcare centres in Gauteng, KZN, Free-State and Western Cape in the years 2010–2012 [24]. The association between MRSA and resistance to a wide range of other antibiotics is well-established [30], and is considered a serious problem due to the limited available therapeutic drugs. In keeping with this, a study from the private sector in the Western Cape of South Africa, which investigated S. aureus isolates from all clinical specimens, reported 28% mupirocin resistance and a strong association between methicillin resistance and mupirocin resistance, and worryingly, co-resistance to mupirocin and fusidic acid (also a topical antibiotic used for skin infections) [30]. This highlights the need for stewardship of topical antimicrobial agents. The authors also recommended that in private healthcare sectors, antibiotic susceptibility testing should be performed before prescribing mupirocin to patients. We could not correlate the level of resistance with the use of mupirocin in our setting due to the limited access to this information. In our setting, mupirocin is used primarily for decolonisation as part of the management of MRSA outbreaks, although it is also sporadically used as a therapeutic agent for minor skin infections. This practice is under review as part of the hospital’s antimicrobial stewardship programme.
We reported only 2% high-level resistance as opposed to 23% in private healthcare sectors in the Western Cape, which suggests higher use of mupirocin in the private healthcare sector. Conversely, low-level resistance was 9% in our study compared to 4% in private healthcare sectors [30]. It is worth noting that more than 90% of the isolates in our study were from blood cultures compared to only 0.2% in the study from the private sector, where 77% of the isolates were from skin swabs. Evidence suggest that although invasive and nosocomial isolates (such as blood cultures) may be associated with a denser antimicrobial history, their mupirocin resistance rates are lower [23, 30, 31]. Of note, in previous studies, mupirocin resistance was highly associated within certain clinical practices such as plastic surgery, dermatology, and general medicine [30, 31]. Unfortunately, due to the retrospective nature of our study, we were unable to correlate our isolates with any clinical practices. This, as well as the issue of the overrepresentation of blood culture isolates should be addressed in future studies.
Clinically, high-level mupirocin resistance is associated with decolonization failure, however, recent studies have shown that low-level resistance is also associated with reduced effectiveness of mupirocin in eradicating MRSA carriage [17, 32, 33]. We detected higher rates of low-level mupirocin resistance than high-level resistance. The mupA gene was the only resistance determinant observed in the five high-level resistant isolates, which is consistent with what is commonly described world-wide [17, 22, 34]. The mupA gene is usually carried on plasmids, however a previous study identified a chromosomally encoded mupA gene in isolates with low-level resistance [35]. In our study neither mupA nor mupB were detected in the low-level resistant isolates. The ileS mutation V588F was the most common mechanism of low-level resistance, detected in 95% of low level resistant isolates, and is strongly linked to this phenotype [17]. In-vitro studies have shown that the presence of a single mutation (including the common ileS V588F) may have a small effect on bacterial growth rate. These mutations could occur after a single cycle of exposure to mupirocin, which provides selective pressure for low-level resistant MRSA strains within settings where mupirocin is commonly used [16, 17]. This could explain the high rate of low-level mupirocin resistance in our setting. A possibly novel ileS mutation, S570A, was identified in one of the low-level resistant isolates. However, further investigation is warranted since not all mutations within the ileS gene translate to phenotypic resistance [17], and only a small region of the ileS gene was sequenced in this study.
Certain strains were associated with mupirocin resistance; spa types t037 (linked to multilocus sequence type clonal complex (CC) 8) and t012 (linked to CC5) had strong associations with low-level resistance (mainly harbouring the mutation V588F), and t032 (linked to CC22) was associated with high-level (mupA) resistance. Studies within South Africa and even within Africa either did not correlate strain typing data with mupirocin resistance, or used typing methods which are not comparable across different laboratories [25, 36, 37]. In Africa, a single study from Ghana reported a mupirocin resistant isolate belonging to the spa type t4805 from a healthcare worker in Korle Bu Teaching Hospital [38]. In the United States, low-level resistance was predominantly reported amongst spa types linked to CC5 and CC8 [21, 39], consistent with the findings of our study. Furthermore, these genetic backgrounds (CC5 and CC8) are more prone to developing mutations within the ileS even following a single short treatment course with mupirocin [17]; which is consistent with the findings of this study.
Conclusion
Although the method of selecting the S. aureus isolates from the biobank limited our ability to calculate the overall prevalence of mupirocin resistance, our data provided a baseline overview of the rates of resistance to mupirocin at Tygerberg Hospital. We reported high rates of low-level mupirocin resistance, driven by spa types t012 and t037, and low rates of high-level resistance, associated with spa type t032. Our study advocates for the continuous screening for mupirocin resistance in S. aureus from a wide range of clinical specimens in order to monitor resistance rates and to inform prescribing practices.
Acknowledgements
The authors would like to thank the staff of the NHLS microbiology laboratory at Tygerberg Hospital for assisting with isolate identification and storage. We also thank Mr. Mohammed Fadul for assistance with statistical analyses. SA was supported by the Claude-Leon Post-doctoral Research fellowship.
Authors’ contributions
SA, MNF and AW conceptualised the study design, TL and SMA performed the experiments. SA, TL, AW and MNF contributed to data analysis and interpretation. SA drafted the initial manuscript and all authors contributed to and approved the final manuscript.
Funding
This research was supported by a grant from the NHLS Research Trust.
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
Ethics approval and consent to participate
The works has been approved by the Health Research Ethics Committee of Stellenbosch University (Reference number N14/06/065).
Consent for publication
The study does not contain any individual person’s data, therefore, consent for publication is not required.
Competing interests
The authors have none to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shima M. Abdulgader, Email: sabdulgader@sun.ac.za
Tshepiso Lentswe, Email: 19442793@sun.ac.za.
Andrew Whitelaw, Email: awhitelaw@sun.ac.za.
Mae Newton-Foot, Email: maen@sun.ac.za.
References
- 1.Wertheim H, Melles DC, Vos MC, Van Leeuwen W, Van Belkum A, Verbrugh HA, et al. The role of nasal carriage in Staphylococcus aureusStaphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 2.Ziakas PD, Anagnostou T, Mylonakis E. The prevalence and significance of methicillin-resistant Staphylococcus aureus colonization at admission in the general ICU setting: a meta-analysis of published studies. Crit Care Med. 2014;42:433–444. doi: 10.1097/CCM.0b013e3182a66bb8. [DOI] [PubMed] [Google Scholar]
- 3.Zacharioudakis IM, Zervou FN, Ziakas PD, Mylonakis E. Meta-analysis of methicillin-resistant Staphylococcus aureus colonization and risk of infection in Dialysis patients. J Am Soc Nephrol. 2014;25:1–11. doi: 10.1681/ASN.2013091028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel JB, Gorwitz RJ, Jernigan JA. Antimicrobial Resistance: Mupirocin Resistance. Clin Infect Dis. 2009;49:935–941. doi: 10.1086/605495. [DOI] [PubMed] [Google Scholar]
- 5.Cookson B, Lacey R, Noble W, Reeves D, Wise R, Redhead R. Mupirocin-resistant Staphylococcus aureus. Lancet. 1990;335:1095. doi: 10.1016/0140-6736(90)92667-7. [DOI] [PubMed] [Google Scholar]
- 6.Shittu AO, Kaba M, Abdulgader SM, Ajao YO, Abiola MO, Olatimehin AO. Mupirocin-resistant Staphylococcus aureus in Africa: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2018;7:1–16. doi: 10.1186/s13756-018-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, et al. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med. 2013;368:2255–2265. doi: 10.1056/NEJMoa1207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harbarth S, Fankhauser C, Schrenzel J, Christenson J, Gervaz P, Bandiera-Clerc C, et al. Universal screening for methicillin-resistant Staphylococcus aureus at hospital admission and nosocomial infection in surgical patients. JAMA. 2008;299:1149–1157. doi: 10.1001/jama.299.10.1149. [DOI] [PubMed] [Google Scholar]
- 9.Ammerlaan HSM, JW KJA, HFL W, Nouwen JL, MJM B. Eradication of methicillin-resistant Staphylococcus aureus carriage: a systematic review. Clin Infect Dis. 2009;48:922–930. doi: 10.1086/597291. [DOI] [PubMed] [Google Scholar]
- 10.Poovelikunnel T., Gethin G., Humphreys H. Mupirocin resistance: clinical implications and potential alternatives for the eradication of MRSA. Journal of Antimicrobial Chemotherapy. 2015;70(10):2681–2692. doi: 10.1093/jac/dkv169. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz FJ, Jones ME. Antibiotics for treatment of infections caused by MRSA and elimination of MRSA carriage. What are the choices? Int J Antimicrob Agents. 1997;9:1–19. doi: 10.1016/S0924-8579(97)00027-7. [DOI] [PubMed] [Google Scholar]
- 12.Conly JM, Johnston BL. Mupirocin - Are we in danger of losing it? Can. J. Infect. Dis. Pulsus Group Inc.; 2002. p. 157–9. [DOI] [PMC free article] [PubMed]
- 13.Hetem D.J., Bonten M.J.M. Clinical relevance of mupirocin resistance in Staphylococcus aureus. Journal of Hospital Infection. 2013;85(4):249–256. doi: 10.1016/j.jhin.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Hodgson J E, Curnock S P, Dyke K G, Morris R, Sylvester D R, Gross M S. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrobial Agents and Chemotherapy. 1994;38(5):1205–1208. doi: 10.1128/AAC.38.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonio M, McFerran N, Pallen MJ. Mutations affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:438–442. doi: 10.1128/AAC.46.2.438-442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurdle JG, O’Neill AJ, Ingham E, Fishwick C, Chopra I. Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques. Antimicrob Agents Chemother. 2004;48:4366–4376. doi: 10.1128/AAC.48.11.4366-4376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee AS, Gizard Y, Empel J, Bonetti EJ, Harbarth S, François P. Mupirocin-induced mutations in ileS in various genetic backgrounds of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2014;52:3749–3754. doi: 10.1128/JCM.01010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbart J, Perry CR, Slocombe B. High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob Agents Chemother. 1993;37:32–38. doi: 10.1128/AAC.37.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seah C, Alexander DC, Louie L, Simor A, Low DE, Longtin J, et al. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:1916–1920. doi: 10.1128/AAC.05325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel JB, Gorwitz RJ, Jernigan JA. Mupirocin Resistance. Clin Infect Dis. 2009;49:935–941. doi: 10.1086/605495. [DOI] [PubMed] [Google Scholar]
- 21.Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persinga DH. Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States hospitals. Antimicrob Agents Chemother. 2012;56:1324–1330. doi: 10.1128/AAC.05804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goudarzi M, Kobayashi N, Hashemi A, Fazeli M, Navidinia M. Genetic variability of methicillin resistant Staphylococcus aureus strains isolated from burns patients. Osong Public Health Res Perspect. 2019;10:170–176. doi: 10.24171/j.phrp.2019.10.3.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marais E, Aithma N, Perovic O, Oosthuysen WF, Musenge E, Dusé AG. Antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolates from South Africa. South African Med J. 2009;99:170–173. [PubMed] [Google Scholar]
- 24.Perovic O, Iyaloo S, Kularatne R, Lowman W, Bosman N, Wadula J, et al. Prevalence and trends of Staphylococcus aureus bacteraemia in hospitalized patients in South Africa, 2010 to 2012: laboratory-based surveillance mapping of antimicrobial resistance and molecular epidemiology. PLoS One. 2015;10:1–14. doi: 10.1371/journal.pone.0145429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh-Moodley A, Perovic O. Characterisation of Staphylococcus aureus bloodstream isolates from Gauteng and Western cape provinces, South Africa, 2016 and 2017. Public Heal Surveill Bull. 2018;16:99–106. [Google Scholar]
- 26.CLSI. M02-A12 Performance Standards for Antimicrobial Disk. Clsi. 2015.
- 27.Harmsen D, Claus HH, Witte W, Rothgänger J, Turnwald D, Vogel U, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahmoudi S, Mamishi S, Mohammadi M, Banar M, Ashtiani MTH, Mahzari M, et al. Phenotypic and genotypic determinants of mupirocin resistance among Staphylococcus aureus isolates recovered from clinical samples of children: an Iranian hospital-based study. Infect Drug Resist. 2019;12:137–143. doi: 10.2147/IDR.S185610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagant C, Deplano A, Nonhoff C, De Mendonça R, Roisin S, Dodémont M, et al. Low prevalence of mupirocin resistance in Belgian Staphylococcus aureus isolates collected during a 10 year nationwide surveillance. J Antimicrob Chemother. 2016;71:266–267. doi: 10.1093/jac/dkv286. [DOI] [PubMed] [Google Scholar]
- 30.Wasserman E., Orth H., Senekal M., Harvey K. High prevalence of mupirocin resistance associated with resistance to other antimicrobial agents inStaphylococcus aureusisolated from patients in private health care, Western Cape. Southern African Journal of Infectious Diseases. 2014;29(4):126–132. doi: 10.1080/23120053.2014.11441586. [DOI] [Google Scholar]
- 31.Zinn CS, Westh H, Rosdahl VT. An international multicenter study of antimicrobial resistance and typing of hospital Staphylococcus aureus isolates from 21 laboratories in 19 countries or states. Microb Drug Resist. 2004;10:160–168. doi: 10.1089/1076629041310055. [DOI] [PubMed] [Google Scholar]
- 32.Walker ES, Vasquez JE, Dula R, Bullock H, Sarubbi FA. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus : does Mupirocin remain effective? Infect Control Hosp Epidemiol. 2003;24:342–346. doi: 10.1086/502218. [DOI] [PubMed] [Google Scholar]
- 33.Simor AE, Phillips E, McGeer A, Konvalinka A, Loeb M, Devlin HR, et al. Randomized controlled trial of Chlorhexidine Gluconate for washing, intranasal Mupirocin, and rifampin and doxycycline versus no treatment for the eradication of methicillin-resistant Staphylococcus aureus colonization. Clin Infect Dis. 2007;44:178–185. doi: 10.1086/510392. [DOI] [PubMed] [Google Scholar]
- 34.Khoshnood S, Heidary M, Asadi A, Soleimani S, Motahar M, Savari M, et al. A review on mechanism of action, resistance, synergism, and clinical implications of mupirocin against Staphylococcus aureus. Biomed Pharmacother. 2019;109:1809–1818. doi: 10.1016/j.biopha.2018.10.131. [DOI] [PubMed] [Google Scholar]
- 35.Ramsey MA, Bradley SF, Kauffman CA, Morton TM. Identification of chromosomal location of mupA gene, encoding low-level mupirocin resistance in staphylococcal isolates. Antimicrob Agents Chemother. 1996;40:2820–2823. doi: 10.1128/AAC.40.12.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shittu AO, Lin J. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province. South Africa BMC Infect Dis. 2006;6:125. doi: 10.1186/1471-2334-6-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swe Swe Han K, Naidoo N, Jaglal P. Molecular epidemiology of a suspected methicillin-resistant Staphylococcus aureus outbreak in a renal unit of a central academic hospital in KwaZulu-Natal, South Africa. South African J Infect Dis. 2015;30:6–10. [Google Scholar]
- 38.Egyir B, Guardabassi L, Nielsen SS, Larsen J, Addo KK, Newman MJ, et al. Prevalence of nasal carriage and diversity of Staphylococcus aureus among inpatients and hospital staff at Korle Bu teaching hospital, Ghana. J Glob Antimicrob Resist. 2013;1:189–193. doi: 10.1016/j.jgar.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Tickler IA, Goering RV, Mediavilla JR, Kreiswirth BN, Tenover FC. Continued expansion of USA300-like methicillin-resistant Staphylococcus aureus (MRSA) among hospitalized patients in the United States. Diagn Microbiol Infect Dis. 2017;88:342–347. doi: 10.1016/j.diagmicrobio.2017.04.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.