Significance
Centromeres are the sites on the chromosome where kinetochores are assembled, a process that is required for faithful chromosome segregation. Nucleosomes in centromeric chromatin contain the histone H3 variant CENP-A. Here, we have identified the AAA+ ATPase Yta7/ATAD2 as a deposition factor for CENP-A at centromeres in yeast. Our findings indicate that Yta7 acts as a hexameric AAA+ ATPase that unfolds CENP-A/H4 and hands it over to Scm3/HJURP for incorporation into the centromeric nucleosome. Defects in this process result in kinetochore instability and chromosome segregation defects. The human homolog ATAD2 is frequently overexpressed in cancer cells, suggesting that it contributes to carcinogenesis by impairing chromosome segregation.
Keywords: centromere, Cse4, Ctf19, Chl4, Cbf1
Abstract
The AAA+ ATPase and bromodomain factor ATAD2/ANCCA is overexpressed in many types of cancer, but how it contributes to tumorigenesis is not understood. Here, we report that the Saccharomyces cerevisiae homolog Yta7ATAD2 is a deposition factor for the centromeric histone H3 variant Cse4CENP-A at the centromere in yeast. Yta7ATAD2 regulates the levels of centromeric Cse4CENP-A in that yta7∆ causes reduced Cse4CENP-A deposition, whereas YTA7 overexpression causes increased Cse4CENP-A deposition. Yta7ATAD2 coimmunoprecipitates with Cse4CENP-A and is associated with the centromere, arguing for a direct role of Yta7ATAD2 in Cse4CENP-A deposition. Furthermore, increasing centromeric Cse4CENP-A levels by YTA7 overexpression requires the activity of Scm3HJURP, the centromeric nucleosome assembly factor. Importantly, Yta7ATAD2 interacts in vivo with Scm3HJURP, indicating that Yta7ATAD2 is a cochaperone for Scm3HJURP. The absence of Yta7 causes defects in growth and chromosome segregation with mutations in components of the inner kinetochore (CTF19/CCAN, Mif2CENP-C, Cbf1). Since Yta7ATAD2 is an AAA+ ATPase and potential hexameric unfoldase, our results suggest that it may unfold the Cse4CENP-A histone and hand it over to Scm3HJURP for subsequent deposition in the centromeric nucleosome. Furthermore, our findings suggest that ATAD2 overexpression may enhance malignant transformation in humans by misregulating centromeric CENP-A levels, thus leading to defects in kinetochore assembly and chromosome segregation.
Centromeres are specialized chromatin structures that are the assembly site for kinetochores, the macromolecular machineries that connect centromeric chromatin to the microtubules. They are essential for the segregation of chromosomes to the daughter cell during mitosis (1). Proper kinetochore assembly is necessary to prevent errors in chromosome segregation, which can cause aneuploidy, a hallmark of cancer and infertility. The targeting of the kinetochore assembly to specific chromosomal regions is mediated by centromeric nucleosomes that contain the histone H3 variant CENP-A instead of canonical histone H3 (2, 3). The budding yeast Saccharomyces cerevisiae consists of a single nucleosome that contains the CENP-A homolog Cse4 (4, 5), which mediates the link to the inner kinetochore (1, 6).
Since centromeres are specified by the presence of CENP-A/Cse4 nucleosomes, the process of nucleosome assembly at the centromere is a key event that is essential for proper centromere function (7). Nucleosome assembly is performed by histone chaperones that associate with specific histones or histone variants and escort them to the site of nucleosome assembly (8). At the centromeres, Scm3 in yeast and its homolog HJURP in human cells deposit CENP-A/Cse4 in centromeric chromatin (9–13). They are chaperones that are dedicated to the centromeric histone H3 variant and ensure the correct spatiotemporal assembly of the centromeric nucleosome humans (14, 15).
Chromatin assembly of canonical H3/H4 during DNA replication is performed by chromatin assembly factor I (CAF-I) and the histone chaperone Asf1 (8). In this process, the histones are delivered to CAF-I by Asf1, and CAF-I deposits H3/H4 on nascent DNA (16). Asf1 in turn also has CAF-I-independent histone deposition functions both during and outside of replication. It helps to disassemble nucleosomes as the replicative helicase navigates through chromatin (17), and it performs transcription-coupled histone exchange (18). An open question is whether Scm3/HJURP has a partner chaperone that delivers histones to it, much like Asf1 hands over H3/H4 to CAF-I for deposition after DNA replication.
In this work, we describe a role for the histone chaperone Yta7/ATAD2 in depositing Cse4 at yeast centromeres. Yta7 (yeast Tat-binding analog) is an evolutionarily conserved protein that contains two AAA+ ATPase domains, which have similarity to unfoldases, and a bromodomain (19). It is a homolog of human ATAD2 (ATPase family AAA+ domain-containing protein 2) (20) that is frequently overexpressed in many cancer types and is associated with poor prognosis (21). Depending on the context, Yta7 has variously been described as an activator and a repressor of gene expression, suggesting that it may have both nucleosome assembly and disassembly activity (20). It localizes to the 5′ end of ∼600 genes in the yeast genome and helps evict histone H3 from genes upon their induction, thus indicating that it is an ATP-dependent nucleosome disassembly factor (22, 23). In the absence of Yta7, nucleosomes within the open reading frame (ORF) of genes are more densely packed, and the induction of gene expression is attenuated. In agreement with a role in transcription, cellular Yta7 is associated with Spt16 (24), a component of the histone chaperone complex FACT, which remodels H2A/H2B during transcription elongation (25).
Conversely, Yta7 has a more complex role in regulating the S-phase-specific induction of the histone gene locus HTA1-HTB1. It is localized to the promoter outside of S phase and restricts the association of the repressing histone chaperone Rtt106 to the promoter, thus acting as a “counterrepressor” (26, 27). During S phase, Yta7 is phosphorylated by Cdk1, which leads to its eviction from the histone gene promoter and to subsequent gene induction (26, 28). One interpretation of this observation is that Yta7, like other histone chaperones, also has nucleosome assembly activity and that this helps to maintain nucleosomes at the histone gene promoter and thus to repress transcription. A role for Yta7/ATAD2 proteins as histone deposition factors is reflected in the homolog in Schizosaccharomyces pombe, Abo1. The deletion of abo1+ results in a perturbed nucleosome organization on genes, and the gene expression signature of abo1∆ cells overlaps with that of histone deacetylases, indicating that Abo1 is a general repressor in S. pombe (29).
Yta7 furthermore has been described as a boundary factor that prevents the spreading of SIR heterochromatin across the tRNA boundary that lies next to the silent HMR locus (24, 30). Accordingly, Yta7 binds in vitro to reconstituted yeast heterochromatin, and it interacts in vivo with SIR heterochromatin (31). Yta7 purified from yeast is associated with all four core histones as well as with the histone variant H2A.Z (24, 28). Interestingly, the histone binding is not solely mediated by the bromodomain of Yta7 but also by its N-terminal region. Furthermore, unlike other bromodomains, the Yta7 bromodomain binds histones irrespective of their lysine acetylation status (32, 33).
There are striking parallels between Yta7 function in yeast and that of ATAD2 in higher eukaryotes, which is also termed ANCCA (AAA+ nuclear coregulator cancer-associated protein) (34). Originally described as a coactivator for various cancer-related transcription factors [e.g., estrogen and androgen receptor, MYC and E2F (35–37)], ATAD2 binds to the gene body of many genes throughout the genome, and reduction of ATAD2 levels results both in the up- and down-regulation of ATAD2-bound genes (38). The fact that ATAD2 has a bromodomain that binds acetylated lysines in histone tails (39, 40), combined with its characterization as an oncogenic factor, has sparked interest in ATAD2 as a pharmacological target for the treatment of cancer (41).
Here, we performed a genetic screen to identify regulators of centromere function in S. cerevisiae and identified Yta7 as a factor that cooperates with Scm3 to deposit Cse4 at centromeres in yeast. The deletion of YTA7 causes growth and chromosome segregation defects when combined with mutations in inner kinetochore components. Yta7 is enriched at centromeres, interacts with Cse4, and is required for the maintenance of normal Cse4 levels at centromeres. Yta7 furthermore interacts with the Cse4 chaperone Scm3, and Yta7-mediated Cse4 deposition requires functional Scm3. Altogether, our results suggest that Yta7 hands Cse4 to Scm3 to assemble CENP-A/Cse4-containing nucleosomes at the centromere.
Results
Identification of Yta7 as a Regulator of Centromere Function in Yeast.
To uncover regulators of centromere function, we made use of the observation that a mutation of arginine 37 of Cse4 (cse4-R37A), which is a methylation site, causes a synthetic growth defect when combined with a mutation in the gene encoding the kinetochore component Okp1CENP-Q [okp1-5 (42); the superscript indicates the mammalian ortholog] (6, 43) (Fig. 1A). We isolated mutations that suppressed the growth defect and identified the causative mutation by whole-genome sequencing. Of nine isolated independent candidates, eight carried an inactivating mutation in YTA7 [the ninth candidate carried a mutation in UBR2 (44)]. The subsequent analysis showed that the deletion of YTA7 (yta7∆) causes the phenotype. Also, yta7∆ partially suppressed the okp1-5 temperature sensitivity in a CSE4 background, indicating that it acts independently of Cse4-R37 methylation. The centromeric function of Yta7 required the AAA+ ATPase domain of Yta7, and the phenotype was specific for Yta7 and not shared by other histone chaperones (see SI Appendix, Fig. S1 for characterization of cse4-R37A okp1-5 yta7∆). Since the human Yta7 homolog ATAD2 is an oncogene and since cancer cells frequently arise from chromosome segregation defects (41), we surmised that investigation of a centromeric function of Yta7 in yeast will inform its role in carcinogenesis in humans.
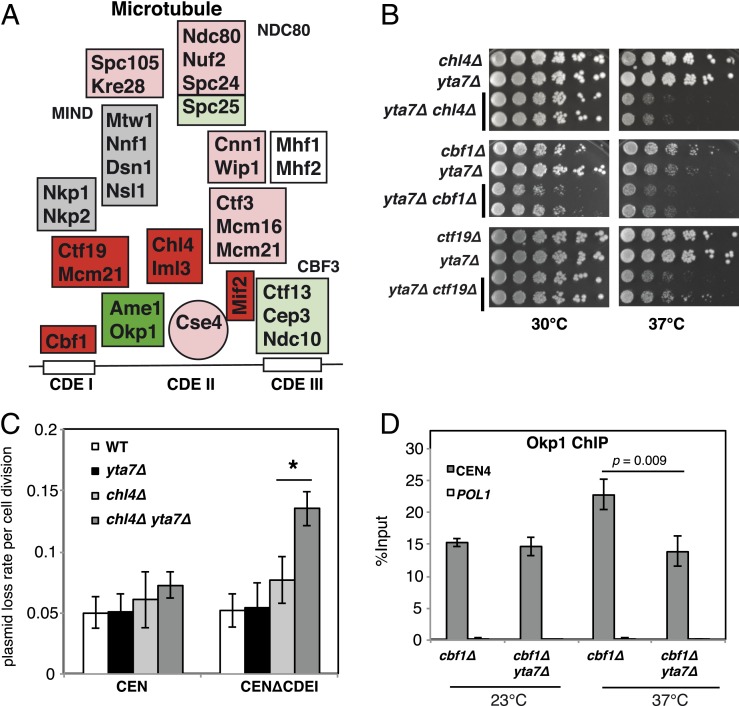
Fig. 1.
The deletion of YTA7 causes synthetic defects in centromere function and kinetochore assembly. (A) Schematic representation of the yeast kinetochore. Colors reflect the synthetic phenotype of yta7∆ with mutations in genes encoding kinetochore components (Table 1). For red or light red, yta7∆ causes a strong or moderate growth defect, respectively, when combined with mutation of the respective gene. For green or light green, yta7∆ causes strong or mild suppression, respectively. For gray, yta7∆ does not affect the growth phenotype. White indicates not tested. (B) Synthetic growth defects of yta7∆ with deletions of the genes encoding Cbf1 or the Ctf19 complex components Chl4 and Mcm21. Cells were spotted in serial dilutions on full medium and incubated for 3 d at 30 °C or 37 °C. (C) yta7∆ chl4Δ mutants showed a maintenance defect for a CEN6 plasmid lacking the CDEI element (CEN∆CDEI) at 30 °C. WT, wild-type. Error bars indicate means ± SD of three independent experiments. *P < 0.05. (D) Recruitment of CTF19 complex component Okp1 to the centromere was reduced by yta7∆ as measured by ChIP analysis. ChIP of 9xmyc-tagged Okp1 was performed in the indicated strains grown at the permissive temperature (23 °C) or shifted to the restrictive temperature (37 °C) for 4 h before ChIP. Enrichment of CEN4 and POL1 (as a control) relative to input is given. Values are the mean of at least three independent experiments ± SD (P value by Student’s t test).
To further explore the role of Yta7 in the regulation of centromeric function, we investigated genetic interactions of yta7Δ with mutations or deletions in genes encoding kinetochore components (Fig. 1A). Importantly, we observed a distinct pattern of genetic interactions of Yta7. yta7∆ showed strong genetic interactions with mutations or deletions in components of the inner kinetochore that act closer to the centromeric chromatin, for instance, yta7∆ combined with the absence of the CTF19 components Ctf19CENP-P, Mcm21CENP-O, or Chl4CENP-N (1, 45) (Fig. 1 A and B, SI Appendix, Fig. S2A, and Table 1). In contrast, it showed only slight or no defects in combination with mutations in components of the outer kinetochore [e.g., NDC80 (46) or MIND components (47)]. Furthermore, in the majority of combinations, yta7∆ caused an increased growth defect (Fig. 1A, red colors). The exceptions to this were okp1-5 (which we had used to identify yta7∆ as a suppressor) and ame1-4, whose temperature sensitivity was suppressed by yta7∆ (Fig. 1A, green colors). This finding was interesting in light of the fact that Ame1CENP-U and Okp1CENP-Q form a heterodimer within the CTF19 complex (45, 48) and directly interact with the Cse4 N terminus (6). Furthermore, yta7∆ weakly suppressed mutations in components of the DNA-binding CBF3 complex as well as in Spc25, an NDC80 component. While these genetic interactions are difficult to interpret, they may reflect physical interactions among proteins and protein complexes across the fully assembled kinetochore.
Table 1.
Synthetic genetic interactions of yta7∆ with mutations in genes encoding kinetochore components
| Kinetochore component/complex | Human homolog* | Allele | Synthetic phenotype with yta7Ơ |
| CENP-A | CENP-A | cse4-103 | Slight enhancement of growth defect |
| CENP-C | CENP-C | mif2-3 | Enhanced growth defect |
| Cbf1 | — | cbf1∆ | Growth defect |
| COMA complex | CENP-Q | okp1-5 | Suppression of growth defect |
| CENP-U | ame1-4 | Suppression of growth defect | |
| CENP-P | ctf19∆ | Growth defect | |
| CENP-O | mcm21∆ | Growth defect | |
| Ctf19 complex | CENP-L | iml3∆ | Growth defect |
| CENP-N | chl4∆ | Growth defect | |
| CENP-I | ctf3∆ | Slight growth defect | |
| CENP-H | mcm16∆ | Slight growth defect | |
| CENP-K | mcm22∆ | Slight growth defect | |
| CENP-T | cnn1∆ | Slight growth defect | |
| CENP-W | wip1∆ | Slight growth defect | |
| — | nkp1∆ | — | |
| — | nkp2∆ | — | |
| Mtw1/MIND complex | Mis12 | mtw1-11 | — |
| PMF1 | nnf1-77 | — | |
| Dsn1 | dsn1-7, dsn1-8 | — | |
| NSL1 | nsl1-5, nsl1-6 | — | |
| Ndc80 complex | NDC80 | ndc80-1 | Slight enhancement of growth defect |
| NUF2 | nuf2-61 | Slight enhancement of growth defect | |
| SPC24 | spc24-1 | Slight enhancement of growth defect | |
| SPC25 | spc25-1 | Partial suppression of growth defect | |
| Knl1 complex | KNL1 | spc105-4 | Slight enhancement of growth defect |
| CBF3 complex | — | ctf13-3 | Partial suppression of growth defect |
| — | cep3-1, cep3-2 | Partial suppression of growth defect | |
| — | ndc10-1 | — | |
| — | ndc10-2 | Partial suppression of growth defect |
A dash (—) indicates no known homolog.
The combined defect of yta7∆ with the indicated mutations is given. A dash (—) indicates that yta7∆ does not affect the growth phenotype of the respective other mutation.
Altogether, these data showed that the function of Yta7 at the kinetochore was not restricted to Okp1 but that its absence caused defects in a broad range of kinetochore mutants, indicating an important function for Yta7 in maintaining centromere and kinetochore function.
The Absence of Yta7 Caused Defects in Kinetochore Recruitment and Centromere Function.
We next asked whether the synthetic growth defects of yta7∆ were caused by defects in cell-cycle progression and chromosome segregation. The CTF19 component Chl4CENP-N and Cbf1, which binds the CDEI element of yeast centromeres (49), were among the components that showed the strongest synthetic growth defects with yta7Δ when absent, suggesting an enhanced defect in kinetochore assembly and chromosome segregation (Fig. 1B and Table 1). To monitor chromosome segregation defects, we examined the stability of centromere-based plasmids (CEN) with or without the CDEI sequence of the centromere [CDEIΔ (50)] in chl4Δ, yta7Δ, and chl4Δ yta7Δ mutants. The double mutants displayed an increased rate of plasmid loss with the CDEI∆ plasmid (Fig. 1C), showing that the function of this centromere was severely compromised in chl4Δ yta7Δ. Moreover, chl4Δ yta7Δ accumulated with a 2N DNA content at the nonpermissive temperature, thus indicating a defect in the G2/M phase transition, which is consistent with a defect in kinetochore function (SI Appendix, Fig. S2B).
To test whether Yta7 is required for kinetochore assembly, we examined the recruitment of CTF19 component Okp1 to the centromere in cbf1Δ yta7Δ cells using chromatin immunoprecipitation (ChIP). At the restrictive temperatures, we observed a significant decrease in recruitment of Okp1CENP-Q to the centromere relative to cbf1Δ strain, which showed that Yta7 is required for proper localization of kinetochore proteins to the centromere in the absence of Cbf1 (Fig. 1D). Altogether, the chromosome segregation defects and impaired kinetochore assembly in yta7∆ cells suggest a role for Yta7 in the assembly of a functional kinetochore at the centromere.
yta7Δ causes an increased level of chromatin-incorporated histone H3 and increased nucleosomal density (22, 23). Several defects of yta7Δ are likely to be caused by excessive histone levels since they are suppressed by deleting one of two gene pairs encoding histone H3 and H4 (22, 51, 52). However, decreasing the dosage of histones H3 and H4 by deletion of one of the two H3–H4 histone gene pairs (hht1–hhf1Δ or hht2–hhf2Δ) did not restore the defects of chl4Δ yta7Δ (SI Appendix, Fig. S2C), suggesting that Yta7 has a specific role in centromere function that is distinct from its effect on histone H3 dosage.
Also, yta7∆ causes misregulation of gene expression (22). One possibility, therefore, was that yta7∆ affected chromosome segregation by changing the expression of a gene(s) encoding kinetochore components. However, no kinetochore-related genes were up- or down-regulated in yta7∆ (22), suggesting that it did not act indirectly by affecting the levels of a centromere component.
Yta7 Regulates Cse4 Levels at the Centromere.
We next sought to investigate by which mechanism Yta7 regulates centromere function. As an AAA+ domain protein, Yta7 has been shown to be involved in histone removal upon gene induction (22), suggesting that it has histone chaperone and chromatin remodeling activity. Our observation of a role for Yta7 at the kinetochore raised the question of whether it acted there by acting as a histone chaperone for Cse4 or histone H3.
To test this, we examined the amounts of Cse4 at the centromere in cbf1Δ or chl4Δ cells in the presence or absence of Yta7 by ChIP. cbf1∆ and chl4∆ were used because they cause synthetic growth defects with yta7∆ (Fig. 1B). Importantly, in the absence of Yta7, the level of Cse4 at the centromere was significantly reduced both at the permissive and the restrictive temperature in chl4∆ and cbf1∆ cells (Fig. 2A and SI Appendix, Fig. S3 A and B). Of note, this was not due to decreased levels of bulk Cse4 in yta7∆ (SI Appendix, Fig. S3C). This result indicated that Yta7 is required for the deposition of Cse4 at the centromere, arguing for a possible role of Yta7 as a Cse4 histone chaperone.
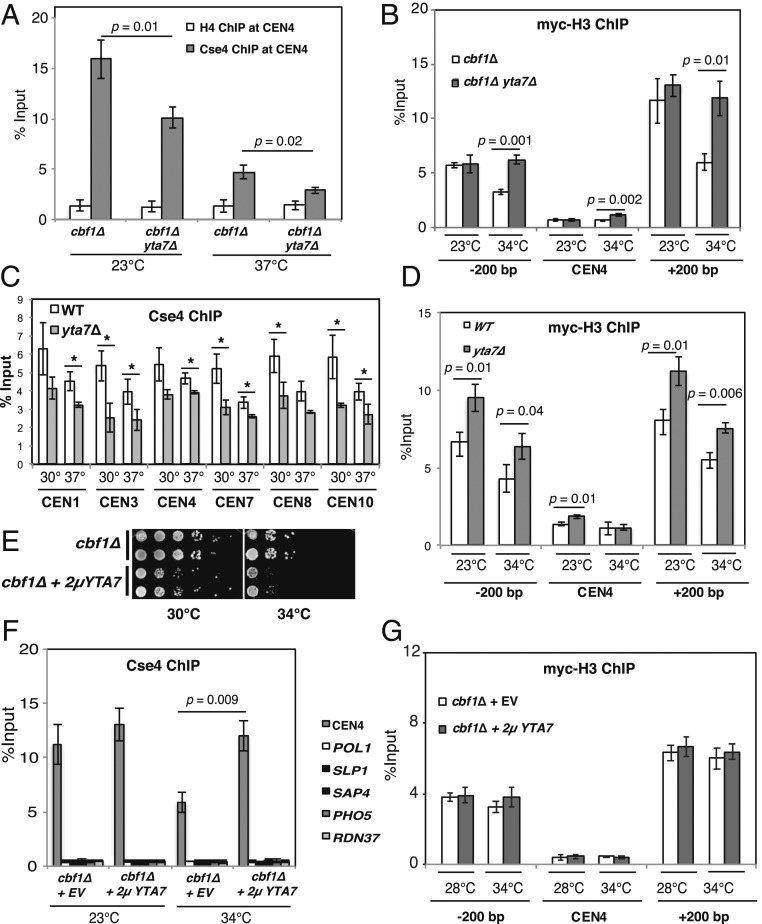
Fig. 2.
Yta7 regulates Cse4 levels at the centromere. (A) yta7∆ caused a reduction of centromere-bound Cse4 at CEN4 in cbf1Δ cells at both the permissive (23 °C) and restrictive (37 °C) temperatures as measured by ChIP analysis. ChIP was performed with α-HA (for Cse4) and, as a control, with an α-H4 antibody. (B) The levels of histone H3 at the centromere proper were only slightly increased in yta7∆. ChIP of myc-tagged H3 (myc-HHT2) using an α-myc antibody was used to analyze the association of H3 at CEN4 and regions located 200 bp to the left or right of CEN4. Cells were grown at 23 °C or shifted to 34 °C for 4 h prior to ChIP. Representation as in Fig. 1D. (C) yta7∆ caused reduced Cse4 levels in otherwise wild-type cells at several centromeres at 30 °C and 37 °C. Representation as in A. *P < 0.05. (D) yta7∆ caused a slight increase of H3 levels at CEN4, whereas H3 levels were strongly increased in regions surrounding the centromere. ChIP of H3 (myc-HHT2) was conducted as in B. (E) Overexpression of YTA7 caused a growth defect in the cbf1Δ strain. Serial dilution of cbf1∆ with an empty vector or a 2µYTA7 plasmid was spotted on selective medium and grown at indicated temperatures for 3 d. (F) Overexpression of YTA7 increased the levels of centromere-associated Cse4 in cbf1Δ. Representation as in A. (G) Histone H3 levels at the centromere were unaffected by YTA7 overexpression. ChIP of H3 (myc-HHT2) to the centromeric region on chromosome IV in cbf1Δ was conducted as in B. Values for ChIP in A, D, F, and G are the mean of at least three independent experiments, with error bars indicating the SD (P values by Student’s t test).
Since chromatin-associated levels of histone H3 are known to increase in yta7∆ (22, 23), our above observation of a decrease in centromeric Cse4 levels raised the question of whether this was a direct effect of Yta7 in depositing Cse4 or whether it was the indirect consequence of higher H3 levels at the centromere in yta7∆. To test this, we measured the levels of a myc-tagged version of histone H3 (myc-HHT2) in cells carrying myc-H3 as the only H3 source (in order to circumvent potential unspecific ChIP with α-H3 antibodies). In cbf1∆ cells, we found the levels of H3 at the nucleosomes flanking CEN4 to be high compared to CEN4 itself and to be reduced when cells were grown at 34 °C compared to 23 °C (Fig. 2B). The absence of Yta7 reversed this reduction of H3 levels, which was in agreement with the previously described role of Yta7 in histone H3 removal (22). Furthermore, as previously reported (22), yta7∆ led to increased levels of H3 at a region unrelated to the centromere, the POL1 gene (SI Appendix, Fig. S3D). At CEN4 itself, H3 levels, as expected (53), were very low compared to the flanking regions but were slightly increased in cbf1∆ yta7∆ cells at the restrictive temperature. However, this was to a degree that seems insufficient to explain the strong loss of Cse4 that we observed at the centromere in cbf1∆ yta7∆ (Fig. 2A). Indeed, H3 levels were not increased at five other centromeres examined (SI Appendix, Fig. S3E), indicating that yta7∆ does not increase H3 levels at many centromeres. Taken together, these results argue that Yta7 is directly required for Cse4 deposition at the centromere, rather than indirectly increasing Cse4 levels by removing H3.
We also investigated whether yta7∆ reduces centromeric Cse4 levels at kinetochores that were not weakened by the deletion of CBF1 and CHL4. Importantly, although yta7∆ cells did not show a growth defect, Cse4 levels were reduced both at 30 °C and 37 °C at several centromeres (Fig. 2C). Furthermore, this reduction in Cse4 was only accompanied by a mild increase of centromeric H3 at CEN4 but not at other centromeres (Fig. 2D and SI Appendix, Fig. S3F). Altogether, this showed that Yta7 deposits Cse4 at wild-type centromeres but that Yta7-dependent changes in centromeric Cse4 levels only have phenotypic consequences when the centromere is compromised by mutations in genes encoding other kinetochore components.
If Yta7 is a Cse4 deposition factor, one prediction is that higher levels of Yta7 cause higher levels of Cse4 at the centromere, which is expected to cause a growth defect in kinetochore mutants. In agreement with this, we found that the overexpression of YTA7 caused a pronounced growth defect in several kinetochore mutants (e.g., cbf1∆, ctf19∆, Fig. 2E and SI Appendix, Fig. S4). Importantly, YTA7 overexpression in cbf1Δ cells resulted in higher Cse4 levels at the centromere at the nonpermissive temperature but not a control region, the POL1 gene (Fig. 2F). It also did not increase Cse4 levels at other regions that show Cse4 misincorporation upon overexpression (54, 55). As above, the increase in centromeric Cse4 was not reflected in a reciprocal decrease of H3 levels at the centromere or at the flanking nucleosomes (Fig. 2G), and YTA7 overexpression did not increase H3 levels at the unrelated POL1 gene (SI Appendix, Fig. S3G). Altogether, these data showed that Yta7 is responsible for the deposition of Cse4 at the centromere and that it does so independently of its effect on H3 removal.
yta7∆ Does Not Affect Transcription of cenRNA.
Centromere activity, which ensures accurate chromosome segregation, requires an optimal level of centromeric long noncoding RNAs (cenRNAs) (56, 57). Since yta7∆ has been implicated in transcription of the histone genes (27, 28, 32), we asked whether the centromeric defect arising in the absence of Yta7 could be the result of altered cell-cycle-specific transcription through the centromere, which may interfere with Cse4 loading. Importantly, cbf1Δ increases cenRNA level, consistent with the disruption of centromere activity in these cells (57). However, the level of centromeric transcription was not further elevated in cbf1Δ yta7Δ, showing that the modulation of centromeric activity by Yta7 was not mediated through misregulation of cenRNA (SI Appendix, Fig. S3H).
Yta7 Interacts In Vivo with Cse4 and Localizes to the Centromere.
All data so far pointed toward a direct role for Yta7 in depositing Cse4 at the centromere. If so, Yta7 can be hypothesized to interact with Cse4 within the cell. We therefore tested for possible protein–protein interactions between Yta7 and Cse4 using coimmunoprecipitation (co-IP). In agreement with the above notion, we found that Yta7 associates with Cse4 in vivo in that Cse4 coimmunoprecipitated with Yta7 (Fig. 3A) and vice versa (Fig. 3B).
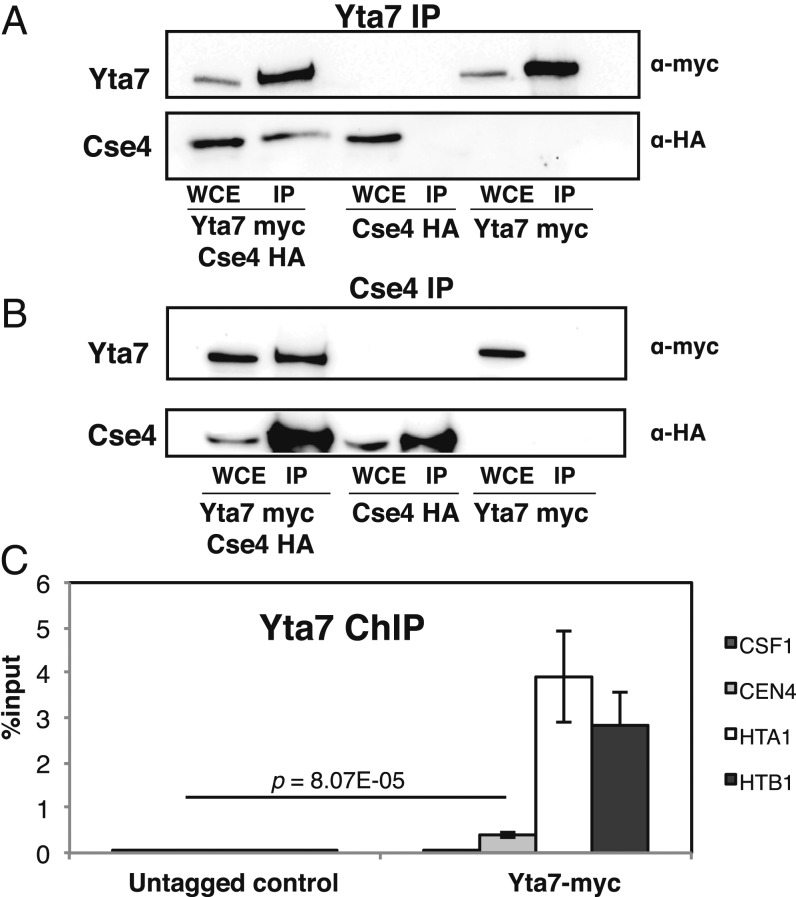
Fig. 3.
Yta7 interacts in vivo with Cse4 and is associated with centromeric chromatin. (A and B) In vivo interaction of Yta7 with Cse4 was determined by coimmunoprecipitation. Yta7-myc (A) or Cse4-HA (B) was precipitated from whole-cell extracts (WCEs) from cells with both proteins tagged (Left) or only Cse4 (Middle) or Yta7 (Right) tagged using α-myc (A) or α-HA agarose (B), and WCE and precipitates (IP) were subjected to Western blotting with α-myc antibody and α-HA antibody. (C) Yta7 bound to centromeres as measured by ChIP. ChIP analysis was performed in wild-type (untagged) and Yta7-9xmyc cells. Enrichment at CEN4 relative to input is given (means ± SD, n = 3, P values by Student’s t test).
We furthermore tested whether Yta7 is associated with centromeric sequences in the cell. Indeed, we found a mild enrichment of Yta7 at centromeric sequences (Fig. 3C; compare to no-tag control and an unrelated region, CSF1), although the levels were lower than those of Yta7 detected at the known binding sites of Yta7 at the histone genes HTA1 and HTB1 (23, 28). Altogether, we conclude that Yta7 associates with Cse4 and is present at the point centromere of budding yeast centromeres, which is consistent with a direct role for Yta7 in Cse4 deposition at the centromere.
Yta7 Cooperates with Scm3 to Deposit Cse4 at the Centromere.
Cse4 has previously been shown to be deposited at the centromere by the chaperone Scm3 (9, 58). Our observation that YTA7 overexpression increases centromeric Cse4 levels raised the question of whether or not this depended on Scm3 activity. We tested this by measuring the association of Cse4 at the centromere by ChIP in cells in which Scm3 was inactivated. This was achieved by placing SCM3 under control of the glucose-repressible GALS promoter (GALSpr-SCM3). Importantly, while Cse4 levels at the centromere were increased by YTA7 overexpression under SCM3-expressing conditions (galactose), this increase was not seen when SCM3 was inactivated by growing the cells in glucose (Fig. 4A and SI Appendix, Fig. S5A). This result showed that the deposition of Cse4 at the centromeres by Yta7 depended on functional Scm3.
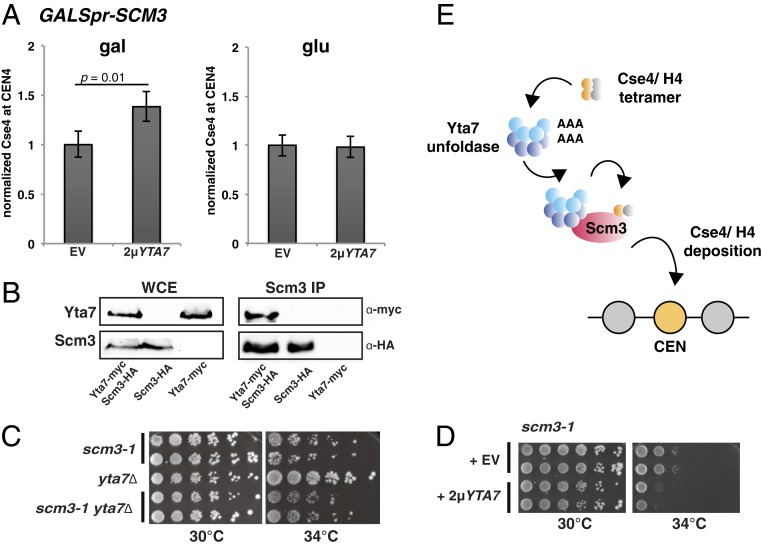
Fig. 4.
Genetic and physical interaction of Yta7 and Scm3. (A) Scm3 is required for Cse4 deposition at the centromere upon YTA7 overexpression. ChIP of HA-Cse4 was conducted in cells with GALSpr-SCM3 and YTA7 overexpression (2µYTA7) or vector control (EV). Cells were grown either in galactose (SCM3 on, Left) or glucose (SCM3 off, Right), and HA-Cse4 was precipitated with α-HA agarose. Values are normalized to the vector control. Means ± SD, n = 3 (P values by Student’s t test) are shown. (B) Yta7 interacts with Scm3 in vivo. Whole-cell extracts (WCEs) from indicated strains were used to carry out immunoprecipitation of Scm3-HA with α-HA agarose. α-HA and α-myc antibodies were used to detect Scm3 and Yta7 on the Western blot, respectively. We did not observe the reciprocal IP or Scm3 with Yta7 because the Scm3 levels were substantially lower than those of Yta7. (C) yta7∆ did not affect the temperature sensitivity of scm3-1. The strains were spotted on full medium and incubated for 3 d at the indicated temperatures. (D) YTA7 overexpression enhanced the temperature sensitivity of scm3-1. scm3-1 containing either an empty vector (EV) or 2µYTA7 was spotted on selective medium and incubated for 3 d at the indicated temperatures. (E) Model of the function of Yta7 at the centromere. Since Yta7 has two AAA+ domains (AAA), we postulate that it constitutes a double hexameric ring. In this model, Yta7 unfolds Cse4/H4 tetramers and hands them to Scm3HJURP for incorporation into the centromeric nucleosome. Cse4/H4 binds to Scm3 as a dimer.
To further validate this conclusion, Scm3 was inactivated by using the temperature-sensitive allele scm3-1 (SCM3 is an essential gene) (59). At the permissive temperature of scm3-1, YTA7 overexpression resulted in increased centromeric Cse4 levels. However, this increase was not seen when Scm3 was inactivated at the restrictive temperature (SI Appendix, Fig. S5B), confirming the notion that Yta7-mediated Cse4 deposition requires Scm3.
If Yta7 and Scm3 cooperate in depositing Cse4 at centromeres, this indicates that they may interact with each other. We tested this by investigating coimmunoprecipitation of Yta7 with Scm3. Indeed, we found that Yta7 could be coprecipitated with Scm3 (Fig. 4B), which thus showed that the two chaperones interact with each other within the cell.
The requirement of Scm3 for Yta7-mediated Cse4 deposition reveals an epistatic relationship between YTA7 and SCM3. We therefore investigated genetic interactions between YTA7 and SCM3. yta7∆ did not enhance the growth defect of scm3-1, which is in agreement with epistasis of YTA7 and SCM3 (Fig. 4C). However, YTA7 overexpression enhanced the growth defect of scm3-1 (Fig. 4D). Since centromeric Cse4 levels are unaffected under this condition (SI Appendix, Fig. S5B), this suggests that YTA7 overexpression affects scm3-1 temperature sensitivity for reasons other than affecting centromeric Cse4 levels.
Discussion
The faithful incorporation of the H3 variant CENP-A/Cse4 into the centromeric nucleosome is crucial for establishing centromere identity and recruiting the inner kinetochore complexes to centromeric DNA in order to build a functional kinetochore. Here, we report that the AAA+ ATPase protein Yta7 deposits the centromeric H3 variant Cse4 at the centromeres in the yeast Saccharomyces cerevisiae. Importantly, Yta7-mediated Cse4 deposition requires a functional Scm3 chaperone. We therefore propose that Yta7 functions as a Cse4 cochaperone that hands Cse4 over to Scm3 for incorporation into centromeric chromatin (Fig. 4E). Interestingly, Yta7 has two AAA+ ATPase domains, suggesting that it might be a complex with two hexameric rings that unfolds histones in its central pore in an ATP-dependent fashion, as is the case for AAA+ ATPases of the proteasome (60). Of note, the Yta7 homolog Abo1 from S. pombe was recently shown to consist of a stack of two hexameric AAA+ rings and to deposit H3/H4 onto DNA (61). We hypothesize that Yta7 unfolds the Cse4/H4 tetramer in order for it to bind to Scm3 as a heterodimer and that Scm3 then incorporates Cse4/H4 into the centromeric nucleosome. Yta7 thus functions analogously to Asf1, which hands newly synthesized H3/H4 to CAF-I for chromatin assembly after DNA replication and thus ensures sufficient H3/H4 supply in this process (8). Perhaps Yta7 likewise serves to orchestrate the supply of Cse4/H4 during the assembly of the centromeric nucleosome.
Several lines of evidence indicate that Yta7 directly acts to deposit Cse4 at the centromere, rather than by removing histone H3 and thus indirectly affecting Cse4 levels. First, we observed an interaction between Yta7 and Cse4 by co-IP, suggesting direct involvement of Yta7 in delivering Cse4 to chromatin. Second, Yta7 is associated with centromeric DNA sequences, albeit at lower levels than at the HTA1-HTB1 promoter, a known Yta7 binding region. Yta7/ATAD2 most likely is not a stoichiometric subunit of the kinetochore in yeast or higher eukaryotes (47, 48, 62–64). Interestingly, however, a recent study using BioID (proximity-dependent biotin identification) to map CENP-A-associated factors identified ATAD2 as 1 out of more than 300 proteins (65). One possibility therefore is that the association of Yta7/ATAD2 with the centromere is transient or that it is temporally restricted. Third, our investigation of H3 levels at the centromere and at noncentromeric sites shows that the changes in Cse4 levels are not accompanied by a compensative increase or decrease in H3 levels, thus demonstrating that changes in Cse4 levels are not an indirect consequence of Yta7-mediated changes in H3.
Our phenotypic analysis of yta7∆ shows multiple genetic interactions with mutations or deletions in genes encoding components of the CTF19 complex (47) and with Mif2/CENP-C (66). It is interesting to note that we observe, among the CTF19 components, the most pronounced defects of yta7∆ with chl4∆ and iml3∆. Both respective proteins are located at the inner surface of the Y-shaped CTF19 complex, which is the yeast equivalent of the constitutive centromere-associated network (CCAN) in higher eukaryotes and clamps onto the Cse4-containing nucleosome (45, 67). We hypothesize that the absence of Chl4CENP-N or Iml3CENP-L reduces the affinity of the CTF19 complex for the nucleosome, such that the reduced incorporation of Cse4 in yta7∆ cells leads to a further destabilization, which compromises kinetochore function and leads to aberrant chromosome segregation. Notably, YTA7 overexpression and the accompanying increase in centromeric Cse4 levels also cause a growth defect, indicating that the “right level” of centromeric Cse4 is required for optimal kinetochore assembly and that both too high and too low levels are detrimental.
Interestingly, YTA7 also shows pronounced defects when combined with cbf1∆. Cbf1 is a DNA-binding factor that binds as a dimer and induces DNA bending (68). We hypothesize that its absence changes the path of entry and exit of the CEN DNA into the Cse4 nucleosome, thus altering the geometry the nucleosome and possibly its interaction with CTF19/CCAN. This defect is then compounded when centromeric Cse4 levels are altered by yta7∆ or YTA7 overexpression.
Our discovery of the involvement of Yta7 in centromere function in S. cerevisiae raises the question of whether its homolog ATAD2/ANCCA in humans has similar functions. ATAD2 is frequently amplified in cancer cells and has been classified as a candidate oncogene and a therapeutic target for several types of malignancies (20, 41). Its characterization as a general facilitator of chromatin function in transcription (38) and replication (69) has revealed striking similarities to Yta7 function in yeast, but it has been unclear whether its coregulator function is the predominant mechanism of malignant transformation. We speculate that its centromeric function is also conserved and that ATAD2 drives carcinogenesis not only via its function as a transcriptional regulator but also by misincorporation of CENP-A into chromatin and subsequent chromosome missegregation, thus contributing to the genetic instability of cancer cells. The involvement of ATAD2 in centromere function in higher eukaryotes seems likely, given that ATAD2 has been identified in a proteomics screen for CENP-A-associated proteins (65). Our work, therefore, points toward possible pathways of ATAD2 function in tumor cell proliferation and survival that merit further investigation. It furthermore informs efforts in the development of ATAD2 inhibitors, which so far have focused on the inhibition of its bromodomain and their use for tumor therapy (41).
Materials and Methods
Yeast Strains, Plasmids, and Methods.
The Saccharomyces cerevisiae strains and plasmids used in this study are listed in SI Appendix, Tables S1 and S2, respectively. Yeast cells were grown and manipulated using standard genetic techniques (70). Gene deletions and epitope-tagged alleles were constructed at the endogenous loci using standard PCR-based integration and confirmed by PCR and sequence analysis (71). All epitope tagging was confirmed by immunoblotting. GALSpr-SCM3 was generated by PCR-based integration (72). Strains with the integration showed a strong growth defect on the glucose medium.
The plasmid loss rate was measured in strains carrying a CEN6 plasmid containing either a wild-type centromere or a centromere lacking the CDEI element (CEN∆CDEI, pAE1771) as previously described (73).
Ultraviolet Mutagenesis and Isolation of Suppressor Mutations.
To screen for suppressor mutants of cbf1Δ okp1-5 (AEY6115), cells were plated on full medium (yeast peptone dextrose [YPD]), exposed to 4,500 µJ ultraviolet light cm−2 and incubated in the dark at 34 °C until colonies became visible. The isolated suppressor mutants that survived the restrictive temperature (34 °C) were backcrossed to an unmutagenized mating partner (cse4-R37A) to eliminate any bystander mutations. Genomic DNA was isolated from pooled temperature-resistant or temperature-sensitive cbf1Δ okp1-5 segregants obtained from the backcross (five from each set). Identification of the causative mutations was carried out using whole-genome sequencing from these pools. Samples were sequenced with an Illumina NextSEq 500 instrument as 150 base pairs (bp) paired read runs. Reads were mapped to the S. cerevisiae reference genome, and single-nucleotide polymorphisms that were only presented in the temperature-resistant isolates were analyzed.
Chromatin Immunoprecipitation.
ChIP was carried out essentially as described (6). In brief, cells were grown to exponential phase, cross-linked with formaldehyde. Chromatin was extracted from spheroplasts and digested to mononucleosomes with micrococcal nuclease, and proteins were precipitated with α-myc antibody or α-HA agarose beads. After extensive washing and subsequent reverse cross-linking, DNA was recovered using Chelex 100 resin. Samples were subsequently analyzed by quantitative real-time PCR. Primer sequences are available upon request. More details on ChIP are provided in SI Appendix.
More methods are available in SI Appendix, Supplementary Materials and Methods.
Data Availability Statement.
All data discussed in the paper can be found within the main text and SI Appendix.
Supplementary Material
Acknowledgments
We thank Phil Hieter, Amine Nourani, Jasper Rine, and Elmar Schiebel for reagents and Silke Steinborn and Josta Hamann for technical assistance. This work was supported by Deutsche Forschungsgemeinschaft (Grants EH237/12-1 and 14-1) and Humboldt-Universität zu Berlin.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917814117/-/DCSupplemental.
References
- 1.Musacchio A., Desai A., A molecular view of kinetochore assembly and function. Biology (Basel) 6, E5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Earnshaw W. C., Migeon B. R., Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 92, 290–296 (1985). [DOI] [PubMed] [Google Scholar]
- 3.Moreno-Moreno O., Torras-Llort M., Azorín F., Variations on a nucleosome theme: The structural basis of centromere function. BioEssays 39, 1600241 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Camahort R., et al. , Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 35, 794–805 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wisniewski J., et al. , Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. eLife 3, e02203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anedchenko E. A., et al. , The kinetochore module Okp1CENP-Q/Ame1CENP-U is a reader for N-terminal modifications on the centromeric histone Cse4CENP-A. EMBO J. 38, e98991 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zasadzińska E., Foltz D. R., Orchestrating the specific assembly of centromeric nucleosomes. Prog. Mol. Subcell. Biol. 56, 165–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond C. M., Strømme C. B., Huang H., Patel D. J., Groth A., Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camahort R., et al. , Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26, 853–865 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Stoler S., et al. , Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. U.S.A. 104, 10571–10576 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pidoux A. L., et al. , Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33, 299–311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dechassa M. L., Wyns K., Luger K., Scm3 deposits a (Cse4-H4)2 tetramer onto DNA through a Cse4-H4 dimer intermediate. Nucleic Acids Res. 42, 5532–5542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shivaraju M., Camahort R., Mattingly M., Gerton J. L., Scm3 is a centromeric nucleosome assembly factor. J. Biol. Chem. 286, 12016–12023 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunleavy E. M., et al. , HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Foltz D. R., et al. , Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyler J. K., et al. , The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555–560 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Groth A., et al. , Regulation of replication fork progression through histone supply and demand. Science 318, 1928–1931 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Rufiange A., Jacques P. E., Bhat W., Robert F., Nourani A., Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27, 393–405 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Schnall R., et al. , Identification of a set of yeast genes coding for a novel family of putative ATPases with high similarity to constituents of the 26S protease complex. Yeast 10, 1141–1155 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Cattaneo M., et al. , Lessons from yeast on emerging roles of the ATAD2 protein family in gene regulation and genome organization. Mol. Cells 37, 851–856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boussouar F., Jamshidikia M., Morozumi Y., Rousseaux S., Khochbin S., Malignant genome reprogramming by ATAD2. Biochim. Biophys. Acta 1829, 1010–1014 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Lombardi L. M., Ellahi A., Rine J., Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc. Natl. Acad. Sci. U.S.A. 108, E1302–E1311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lombardi L. M., Davis M. D., Rine J., Maintenance of nucleosomal balance in cis by conserved AAA-ATPase Yta7. Genetics 199, 105–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tackett A. J., et al. , Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 169, 35–47 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurova K., Chang H. W., Valieva M. E., Sandlesh P., Studitsky V. M., Structure and function of the histone chaperone FACT–Resolving FACTual issues. Biochim. Biophys. Acta. Gene Regul. Mech. 1861, 892–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fillingham J., et al. , Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35, 340–351 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Zunder R. M., Rine J., Direct interplay among histones, histone chaperones, and a chromatin boundary protein in the control of histone gene expression. Mol. Cell. Biol. 32, 4337–4349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurat C. F., et al. , Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein. Genes Dev. 25, 2489–2501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gal C., et al. , Abo1, a conserved bromodomain AAA-ATPase, maintains global nucleosome occupancy and organisation. EMBO Rep. 17, 79–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jambunathan N., et al. , Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 171, 913–922 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zukowski A., et al. , Proteomic profiling of yeast heterochromatin connects direct physical and genetic interactions. Curr. Genet. 65, 495–505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gradolatto A., et al. , Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics 179, 291–304 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gradolatto A., et al. , A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol. Cell. Biol. 29, 4604–4611 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou J. X., Revenko A. S., Li L. B., Gemo A. T., Chen H. W., ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc. Natl. Acad. Sci. U.S.A. 104, 18067–18072 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou J. X., et al. , Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 69, 3339–3346 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Revenko A. S., Kalashnikova E. V., Gemo A. T., Zou J. X., Chen H. W., Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol. Cell. Biol. 30, 5260–5272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciró M., et al. , ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 69, 8491–8498 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Morozumi Y., et al. , Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells. J. Mol. Cell Biol. 8, 349–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harner M. J., Chauder B. A., Phan J., Fesik S. W., Fragment-based screening of the bromodomain of ATAD2. J. Med. Chem. 57, 9687–9692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poncet-Montange G., et al. , Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2. Biochem. J. 466, 337–346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hussain M., et al. , ATAD2 in cancer: A pharmacologically challenging but tractable target. Expert Opin. Ther. Targets 22, 85–96 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Ortiz J., Stemmann O., Rank S., Lechner J., A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13, 1140–1155 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samel A., Cuomo A., Bonaldi T., Ehrenhofer-Murray A. E., Methylation of CenH3 arginine 37 regulates kinetochore integrity and chromosome segregation. Proc. Natl. Acad. Sci. U.S.A. 109, 9029–9034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samel A., Nguyen T. K. L., Ehrenhofer-Murray A. E., Defects in methylation of arginine 37 on CENP-A/Cse4 are compensated by the ubiquitin ligase complex Ubr2/Mub1. FEMS Yeast Res., 17, fox009 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Yan K., et al. , Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature 574, 278–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wigge P. A., Kilmartin J. V., The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152, 349–360 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Wulf P., McAinsh A. D., Sorger P. K., Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17, 2902–2921 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hornung P., et al. , A cooperative mechanism drives budding yeast kinetochore assembly downstream of CENP-A. J. Cell Biol. 206, 509–524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai M., Davis R. W., Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61, 437–446 (1990). [DOI] [PubMed] [Google Scholar]
- 50.Hegemann J. H., Shero J. H., Cottarel G., Philippsen P., Hieter P., Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 2523–2535 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins S. R., et al. , Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Costanzo M., et al. , The genetic landscape of a cell. Science 327, 425–431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lochmann B., Ivanov D., Histone H3 localizes to the centromeric DNA in budding yeast. PLoS Genet. 8, e1002739 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hildebrand E. M., Biggins S., Regulation of budding yeast CENP-A levels prevents misincorporation at promoter nucleosomes and transcriptional defects. PLoS Genet. 12, e1005930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hewawasam G., et al. , Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40, 444–454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohkuni K., Kitagawa K., Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr. Biol. 21, 1695–1703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ling Y. H., Yuen K. W. Y., Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 116, 6270–6279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shivaraju M., et al. , Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell 150, 304–316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luconi L., Araki Y., Erlemann S., Schiebel E., The CENP-A chaperone Scm3 becomes enriched at kinetochores in anaphase independently of CENP-A incorporation. Cell Cycle 10, 3369–3378 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Bard J. A. M., et al. , Structure and function of the 26S proteasome. Annu. Rev. Biochem. 87, 697–724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho C., et al. , Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone. Nat. Commun. 10, 5764 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ranjitkar P., et al. , An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40, 455–464 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foltz D. R., et al. , The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Hori T., et al. , CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135, 1039–1052 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Remnant L., et al. , In vitro BioID: Mapping the CENP-A microenvironment with high temporal and spatial resolution. Mol. Biol. Cell 30, 1314–1325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meluh P. B., Koshland D., Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6, 793–807 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hinshaw S. M., Harrison S. C., The structure of the Ctf19c/CCAN from budding yeast. eLife 8, e44239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niedenthal R. K., Sen-Gupta M., Wilmen A., Hegemann J. H., Cpf1 protein induced bending of yeast centromere DNA element I. Nucleic Acids Res. 21, 4726–4733 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koo S. J., et al. , ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget 7, 70323–70335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sherman F., Getting started with yeast. Methods Enzymol. 194, 3–21 (1991). [DOI] [PubMed] [Google Scholar]
- 71.Longtine M. S., et al. , Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998). [DOI] [PubMed] [Google Scholar]
- 72.Janke C., et al. , A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004). [DOI] [PubMed] [Google Scholar]
- 73.McNally F. J., Rine J., A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 5648–5659 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper can be found within the main text and SI Appendix.