Abstract
Purpose:
5-hydroxymethylcytosine (5-hmC) is an epigenetic marker of open chromatin and active gene expression. We profiled 5-hmC with Nano-hmC-Seal technology using 10ng of plasma derived cell-free DNA (cfDNA) in blood samples from patients with neuroblastoma to determine its utility as a biomarker.
Experimental Design:
For the Discovery cohort, one hundred 5-hmC profiles were generated from 34 well children and 32 patients (27 high-risk, 2 intermediate-risk, and 3 low-risk) at various time points during the course of their disease. An independent Validation cohort encompassed 5-hmC cfDNA profiles (n = 29) generated from 21 patients (20 high-risk and 1 intermediate-risk). Metastatic burden was classified as high, moderate, low, or none per Curie Metaiodobenzylguanidine (MIBG) scores and percentage of tumor cells in bone marrow. Genes with differential 5-hmC levels between samples according to metastatic burden were identified using DESeq2.
Results:
Hierarchical clustering using 5-hmC levels of 347 genes identified from the Discovery cohort defined four clusters of samples that were confirmed in the Validation cohort and corresponded to high, high-moderate, moderate, and low/no metastatic burden. Samples from patients with increased metastatic burden had increased 5-hmC deposition on genes in neuronal stem cell maintenance and epigenetic regulatory pathways. Further, 5-hmC cfDNA profiles generated with 1,242 neuronal pathways genes were associated with subsequent relapse in the cluster of patients with predominantly low or no metastatic burden (sensitivity 65%, specificity 75.6%).
Conclusions:
cfDNA 5-hmC profiles in children with neuroblastoma correlate with metastatic burden and warrants development as a biomarker of treatment response and outcome.
Keywords: Neuroblastoma, epigenetics, 5-hydroxymethylcytosine, cell free DNA
Introduction
5-hydroxymethylcytosine (5-hmC), a stable intermediate of cytosine demethylation, is a marker of open chromatin and active gene expression. The development of a highly sensitive and robust sequencing technology (Nano-hmC-Seal) has allowed for the genome-wide profiling of 5-hmC using less than 10 ng of DNA (1). Converse to 5-methylcytosine which is a marker of decreased gene expression, the levels of 5-hmC accumulation in promoters, gene bodies and gene regulatory elements correlate with active gene expression (2). In neuroblastoma, we have shown that 5-hmC profiles categorize tumors by phenotype, are associated with survival, and identify underlying molecular pathways that drive tumor biology (3). While Nano-hmC-Seal was designed for use with DNA from frozen tumors, it has recently been adapted for profiling tumor-derived circulating cell-free DNA (cfDNA) (1,4,5).
Neuroblastoma, the most common extra-cranial solid tumor of childhood, is clinically and biologically heterogeneous (6). At diagnosis, patients are stratified as low-risk (LR), intermediate-risk (IR) or high-risk (HR) based on age, histology, stage, ploidy, and the presence or absence of MYCN-amplification (6). Although survival has improved with modern treatments (6), even with intensive, multi-modality approaches, disease progression or relapse occurs in approximately 40% of HR patients within 3 years from diagnosis (7). Patients undergo serial tumor evaluations with imaging studies and bone marrow tests following different courses of therapy and after completion of treatment to measure response and diagnose relapse. While response to treatment has been strongly associated with outcome (8), some drawbacks of standard tumor evaluation studies are the need for sedation and radiation exposure, limiting the feasibility of assessing response at frequent intervals. Furthermore, current evaluation methods lack the sensitivity to detect minimal residual disease that may ultimately lead to recurrence in some patients (9). Clinicians face challenges using standard imaging techniques as it remains difficult to distinguish malignant soft tumor masses from disease that has differentiated into benign neuroblastic masses (10).
cfDNA has increasingly been recognized as both a prognostic and predictive marker in cancer (11). In neuroblastoma, the quantity of cfDNA has been shown to correlate with tumor burden (12). Efforts to improve the precision of detecting neuroblastoma-derived cfDNA using somatic alterations have been described (13–15), but are limited by the low mutational burden in neuroblastoma (6) necessitating deep sequencing approaches or comparison to primary tumor. Nano-hmC-Seal has the potential to overcome these limitations by assessing 5-hmC genome-wide at low sequencing depth, independent of somatic alterations. Using genome-wide approaches are particularly important for evaluating pediatric tumors as it an imperative to maximize information from rare and precious samples (16). This technology has been shown to readily distinguish healthy adults from those with esophageal, gastric, hepatic, or colon cancer, even though cfDNA 5-hmC profiles significantly differ from 5-hmC profiles from prior tumor samples (1,4,5). Thus, we hypothesized that cfDNA-derived 5-hmC profiles would readily distinguish healthy children from those with neuroblastoma and also enable the evaluation of response to treatment over time. Furthermore, as 5-hmC is tightly associated with gene expression (3), we hypothesized that identification of genes with differential accumulation of this epigenetic mark would provide insight regarding the transcriptional and regulatory networks that drive aggressive tumor growth and treatment resistance.
Patients and Methods
Patients
The Discovery and Validation cohorts consisted of samples collected from children diagnosed with neuroblastoma between birth to 29 years old from the University of Chicago Comer Children’s Hospital and Lurie Children’s Hospital between August, 2016 through May, 2018, and March, 2019 through July, 2019, respectively. In addition, well child control samples were collected from children between birth to 10 years of age seen in the Comer Children’s General Pediatrics Clinic and were included in the Discovery cohort. Controls were excluded if they were acutely ill, had inflammatory or autoimmune conditions, or were taking inhaled or systemic steroids. Patient and tumor characteristics including date of diagnosis, stage, age, sex, ethnicity, MYCN-amplification status, treatment, date of relapse, and death were abstracted from electronic medical records and stored in a RedCap database. Clinical response to treatment was determined using the 2017 International Neuroblastoma Response Criteria (INRC) (17). Disease status (disease detected by standard imaging and/or bone marrow aspirate or biopsy versus no evidence of disease) at the time of blood sample collection was defined by the most recent preceding tumor evaluation. Metastatic disease burden at the time of collection was classified as high, moderate, low, or none according to the maximum amount of disease detected by Curie metaiodobenzylguanidine (MIBG) scores of metastatic bone or soft tissue sites and/or the percentage of neuroblastoma cells identified in bone marrow biopsies and aspirates. Curie scores of metastatic sites were classified as high (6–27), moderate (2–5), low (1), or none (0) (17,18). Bone marrow involvement was classified as high (> 10%), moderate (5–10%), low (1–4%), or none (0%) according to pathologist interpretation in the medical records (Table 1A) (17). Written informed consent was obtained from parents or guardians after the study was approved by the local Institutional Review Board according to the U.S. Common Rule ethical guidelines.
Table 1A:
Classification of metastatic disease burden. Curie score excludes measurement of primary soft tissue disease.
| Curie Score | Bone marrow involvement (%) | Classification |
|---|---|---|
| 0 | 0 | None |
| 1 | 0–4 | Low |
| 0 | 1–4 | Low |
| 2–5 | 0–10 | Moderate |
| 0–1 | 5–10 | Moderate |
| > 5 | 0–100 | High |
| 0–27 | > 10 | High |
Blood collection and isolation of cfDNA
For all samples, 6–10 ml of blood was collected in EDTA tubes. Control samples from healthy children were obtained during the well child visit. High-risk (HR) patients were treated with intensive multi-modality strategies (6), and blood was collected at one or more of the following time points: diagnosis, after 2 cycles of induction therapy, prior to post-induction “bridge” therapy (19), prior to consolidation with myeloablative therapy and stem cell transplant, prior to immunotherapy, end of therapy, off therapy, at relapse, and during therapy for relapsed disease. For (low-risk) LR and (intermediate) IR patients, samples were collected at diagnosis, end of therapy, and/or off therapy (6). All blood samples were stored at 4°C and processed within two hours of collection. Plasma was obtained after centrifuging whole blood at 1350g for 12 minutes. Following centrifugation, the plasma layer was re-centrifuged at 1350g for 12 minutes and then at 13,500g for 5 minutes, and stored at −80°C. cfDNA was isolated from 1–2 ml of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Gaithersburg, MD) per manufacturer’s instructions with a final elution using 25 μl of IDTE pH 8.0 buffer.
Nano-hmC-Seal library preparation and sequencing
Nano-hmC-Seal libraries were constructed from 10 ng of cfDNA as described (2,20). Briefly, after ligation with sequencing adapters using the KAPA Hyper Prep Kit (KAPA Biosciences), cfDNA 5-hmC marks were subjected to T4 beta-glucosyltransferase enzymatic modification with UDP-N3-glucose, followed by subsequent chemical modification with biotin-PEG4-dibenzocyclooctyne (DBCO). Biotin-labeled cfDNA fragments were then pulled down using streptavidin M270 Dynabeads (Invitrogen), and PCR amplified to construct sequencing libraries which were purified with AMPure XP beads (Beckman Coulter) to construct libraries. Fifty base-pair, paired-end libraries were sequenced on an Illumina NextSeq 500. FASTQC v0.11.5 was used to assess sequence quality (21). Raw reads were processed with Trimmomatic (22) and aligned to hg19 with Bowtie2 v2.3.0 (23) using default settings. Aligned reads with Mapping Quality Score ≥10 were counted using featureCounts (24) of Subread using the gene flag and the gencode.v27lift37.annotation.gtf file from GENCODE (25).
Identification of genes with differential levels of 5-hmC modifications
Read counts of 5-hmC for the entire gene body were loaded into the DESeq2 v1.20.0 (26) package in R v3.6.0 with a differential 5-hmC model that adjusted for sex, batch and cohort. A False Discovery Rate (FDR) < 0.05 and ranking in the top or bottom 1% of fold-change differences was considered significant. To assess the robustness of differential 5-hmC levels in the complete Discovery cohort, we performed 10-fold cross validation by randomly selecting 80% of the samples from the Discovery cohort and repeating differential the 5-hmC analysis including data normalization steps performed by the DESeq function in DESeq2. For each loop of cross validation, we determined the percent of genes, using FDR < 0.05 and cutoffs of both 1 and 2% of top or bottom fold-change that were also found in the analysis of complete cohort. Principal component analysis of all samples was performed using FactoMineR v1.41 (27). ComplexHeatmap v1.20.0 was used to determine the distance matrix between samples for hierarchical clustering (28).
Generation of a model to confirm 5-hmC profiles correlate with metastatic burden
A model was trained on the Discovery cohort using elastic net regularization (5) limited to the top 347 genes with differential levels of 5-hmC identified to predict high/moderate or low/no metastatic burden. Feature selection using differential genes prior to model training was previously shown to increase the disease relevance of model trained of cfDNA 5-hmC profiles (1,5,29). Elastic net was implemented in the caret package v6.0–80 (30). Normalized batch- and gender-corrected read counts of the Discovery cohort for each of the 347 genes were treated as a feature in the model were tested on the independently processed Validation cohort. Models were trained with 10-fold cross validation across a range of alpha and lambda values from 0–1 optimized for area under the curve, where alpha controls for the relative proportion between the Ridge and Lasso penalty, and lambda controls for the overall strength of penalty.
Differential 5-hmC deposition gene set enrichment analysis
Differential 5-hmC deposition pre-ranked gene set enrichment analysis (GSEA) was performed with fgsea (31). For samples from patients with or without detectable disease, genes with significant differential 5-hmC enrichment regardless of fold change were ranked by p-value magnitude and 5-hmC effect sign, resulting in gene lists with 5-hmC upregulated genes at the top and downregulated genes at the bottom. An ‘ad hoc’ collection of gene sets was created from different sources, by prioritizing the inclusion of predicted and known human transcription factor (TF) targets (Supplementary Table 1). Gene sets were tested for significant extreme ranking in the sorted gene lists of genes with differential 5-hmC. Significance was assessed by n = 100,000 permutations of random gene sets, size matched to the actual gene set. In total, n = 34,823 non-redundant gene sets were partitioned into two main categories tested separately, named “miscellaneous” and “TF ChIP-Seq”. Partition criteria were based on their dissimilar mean gene set size. The “miscellaneous” gene set collection comprises n = 32,065 gene sets related to disease, drug, hormone and immunological processes categories, among others. Its mean gene set size is ~108 genes. The “TF ChIP-Seq” gene set collection comprises n = 2,758 gene sets corresponding to TF binding ChIP-Seq experiments across cell types and treatments (32). Its median gene set size is ~8,500 genes and there are between one and 184 experiments for 428 unique transcription factors. Significance was determined by FDR < 0.01. For each TF, the percent of TF ChIP-Seq gene sets reaching significance was determined for all TFs with at least five ChIP-Seq experiments in ENCODE. Visualization of enriched pathways present in MSigDB collections H, c1, c3, c5, c6, or c7 (33) was performed using EnrichmentMap v3.2 (34).
Results
Patient cohorts, blood sample collection, and cfDNA extraction
The Discovery cohort consisted of 35 healthy children and 32 children with neuroblastoma with blood samples collected between August, 2016 and May, 2018. The Validation cohort consisted of 21 children with neuroblastoma who had blood samples obtained between March, 2019 and July, 2019. As the primary goal of the study was to explore 5-hmC profiles at the sample level across batches collected and processed at different times, seven children were included in both the Discovery and Validation cohorts. 133 blood samples were collected in total, and cfDNA was successfully extracted from all but four samples from children in the Discovery cohort (one well child and three neuroblastoma patients who had serial sampling). The remaining 129 cfDNA samples were sequenced and passed quality control (library concentration and fragment analysis). The final Discovery cohort included 32 patients with LR (n = 3), IR (n = 2), and HR (n = 27) neuroblastoma at diagnosis and 34 well child controls (Table 1B; Supplementary Table 2). The final Validation cohort included 21 patients with IR (n = 1) and HR (n = 20) neuroblastoma. The 100 samples from patients in the Discovery cohort were comprised of one sample from each of the 34 controls and 66 samples from the 32 neuroblastoma patients. The Validation cohort was comprised of 29 samples from 21 neuroblastoma patients. Twenty-three patients had samples collected at more than one time point including diagnosis, during treatment, and follow up.
Table 1B:
Patient and Tumor Characteristics
| Feature | Discovery cohort (n = 32), % |
Validation cohort (n = 21a), % |
Healthy Controls (n = 34), % |
|---|---|---|---|
| Median age at diagnosis | 36.0 months | 48.0 months | 18.5 months |
| Age at diagnosis | |||
| ≤ 18 mo | 25 | 14.3 | 50 |
| 18 mo - 5 yr | 43.8 | 38.1 | 26.5 |
| > 5 yr | 31.2 | 47.6 | 23.5 |
| Sex | |||
| Male | 50 | 52.4 | 50 |
| Female | 50 | 47.6 | 50 |
| Race | |||
| White | 81.2 | 76.2 | 38.2 |
| Black | 6.3 | 23.8 | 44.1 |
| Asian | 3.1 | 0 | 5.9 |
| Unknown | 9.4 | 0 | 11.8 |
| Ethnicity | |||
| Non-Hispanic | 78.1 | 95.2 | 85.3 |
| Hispanic | 12.5 | 4.8 | 2.9 |
| Unknown | 9.4 | 0 | 11.8 |
| Sample collection time | |||
| Diagnosis | 28.1 | 4.8 | |
| During Therapy | 21.9 | 38.1 | |
| End or Off Therapy | 25 | 19 | |
| Relapse or progression | 25 | 38.1 | |
| INSS Stage | |||
| 4 | 71.9 | 61.9 | |
| 4S | 3.1 | 0 | |
| 3 | 12.5 | 33.3 | |
| 2 | 9.4 | 4.8 | |
| 1 | 3.1 | 0 | |
| Risk | |||
| High | 84.3 | 95.2 | |
| Intermediate | 6.3 | 4.8 | |
| Low | 9.4 | 0 | |
| MYCN | |||
| Non-Amplified | 71.9 | 66.7 | |
| Gain | 3.1 | 0 | |
| Amplified | 18.7 | 19 | |
| Unknown | 6.3 | 14.3 |
Abbreviations: INSS, International Neuroblastoma Staging System.
7 of these patients had samples in the Discovery cohort.
cfDNA 5-hmC profiles are associated with disease burden and disease status
5-hmC levels of each gene from the 100 cfDNA samples from the Discovery cohort were adjusted for patient sex, batch, and cohort and normalized with variance stabilizing transformation prior to principal component analysis (PCA). Principal component 1, which was significantly associated with metastatic disease burden (p = 7e-7 by one-way ANOVA; Table 1A), explained 84% of the variance among samples (Supplementary Figure 1). Samples from neuroblastoma patients with low or no detectable disease generally grouped closely with the healthy controls in principal component space.
The 66 samples from neuroblastoma patients in the Discovery cohort were reprocessed without the well controls, and re-normalized with variance stabilizing transformation. Nineteen cfDNA samples collected from neuroblastoma patients with disease detected clinically were compared to 47 samples from neuroblastoma patients who had no evidence of disease, identifying 347 genes with significantly different 5-hmC levels. Cross validation supported the robustness of this gene set (Supplementary Figures 2A, 2B). Using the Π1 statistic (3), we estimated that 92.6% of the 347 genes identified in the Discovery cohort also had significantly different 5-hmC levels between patients with or without evidence of disease in the 29 sample Validation cohort, which had been processed an analyzed independently. To further validate that 5-hmC levels on these genes inform metastatic burden in patients, we created a model based-classifier that applied elastic net regularization on differential gene lists as has been described (1,5,29). Using levels of 5-hmC deposition of these 347 genes from Discovery cohort samples, the model predicted metastatic burden in the Validation cohort (high/moderate versus low/no) with an AUC of 0.77, sensitivity of 70%, and specificity of 89.5%, again demonstrating the robustness of the identified genes. Of the 347 genes with differential 5-hmC levels in the Discovery cohort, those with increased 5-hmC deposition in samples with active disease were enriched for pathways of neuronal function (Supplementary Table 3). Genes with increased accumulation of 5-hmC from samples without active disease were enriched for pathways of peripheral blood monocyte function, presumably as the cfDNA was representative of non-tumor derived cfDNA.
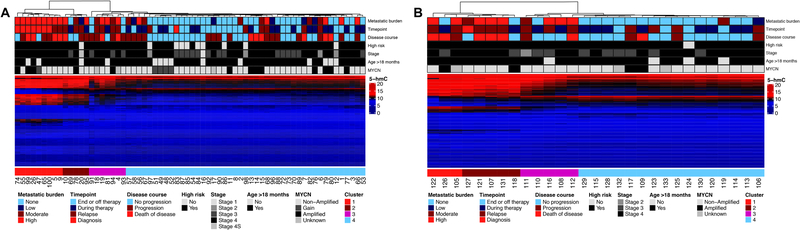
Using the 347 identified genes with differential 5-hmC levels, hierarchical clustering of samples from neuroblastoma patients in the Discovery cohort revealed four primary clusters that grouped samples according to metastatic disease burden and timepoint of sample collection (i.e., at diagnosis, during treatment, off therapy, and relapse) (Figure 1A). The same four clusters were also identified in the independently processed and analyzed Validation cohort using the same gene set (Figure 1B) and when all 95 samples from neuroblastoma patients were processed by cohort, but clustered together using the same parameters (Supplementary Figure 3).
Figure 1: cfDNA 5-hmC profiles are associated with disease burden and disease status in both the Discovery and Validation cohorts.
A) Hierarchical clustering of the Discovery cohort using 347 genes identified in the Discovery cohort with differential 5-hmC between samples from those with or without active disease. B) Hierarchical clustering of the Validation cohort using the same gene set and clustering parameters. Cluster 1 is enriched for patients with high metastatic disease burden. Clusters 2 and 3 are enriched for patients with moderate to high metastatic disease burden. Cluster 4 is enriched for patients with low or no metastatic disease burden.
Cluster 1 was comprised of cfDNA collected from 11 samples from 11 patients with high metastatic disease burden (7 newly diagnosed and 4 at the time of relapse, Table 1A, Supplementary Table 4) and one sample from a patient who was in a complete remission but developed progressive disease within a month of sample collection. Clusters 2 and 3 were comprised of cfDNA samples from patients with high (n = 11), moderate (n = 5), or low (n = 1), or no (n = 5) disease burden. Samples from patients classified as high or moderate metastatic disease burden in Clusters 2 and 3 primarily comprised of recently diagnosed patients during induction therapy, newly identified recurrent disease with few metastases or, widely metastatic recurrence on therapy. Cluster 4 samples were predominantly from patients with low (n = 6) or no (n = 43) metastatic disease burden, though 12 samples were from patients with moderate or high metastatic disease burden (Supplementary Table 5).
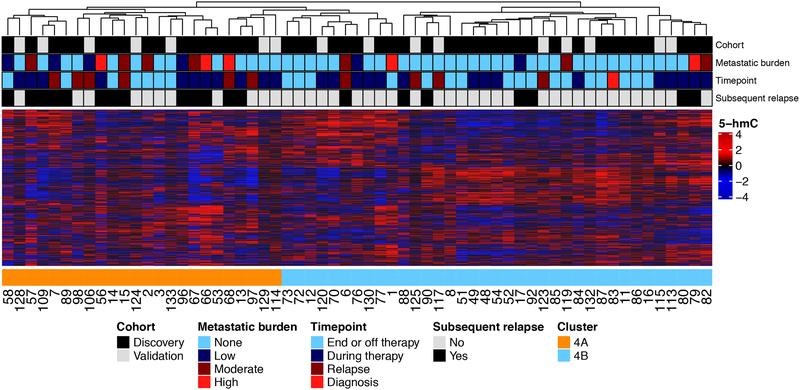
Because Cluster 4 included samples from patients of low/no metastatic disease burden as well as some from patients with moderate/high metastatic burden, we next sought to determine if pathway analysis of the cfDNA 5-hmC profiles could be used to further stratify Cluster 4 samples according to outcome. Based on the finding that samples from patients with high metastatic burden had increased 5-hmC deposition on genes of neuronal pathways, we performed hierarchical clustering of the 61 samples in Cluster 4 using genes from several GO pathways of neural development (Supplementary Table 6). Samples from the Discovery and Validation cohorts were processed together to remove batch effect as that signal was stronger than that from any small amount of neuroblastoma derived cfDNA. The 1,242 gene neural tube development pathway had the highest combined sensitivity and specificity for delineating these samples into two distinct subsets, Clusters 4A and 4B, according to subsequent event status (Figure 2). Of the 20 Cluster 4A samples, 13 (65%) were from patients who had a subsequent event. In contrast, 31 (75.6%) of the 41 Cluster 4B samples were collected from patients who did not have a subsequent event (sensitivity 65.0%, 95% CI: 40.8–84.6%, specificity 75.6%, 95% CI: 59.7–87.3%).
Figure 2: Cluster 4 samples are subdivided into those with minimal disease burden and those without.
1,242 genes from the GO pathway of neuronal systems processes distinguishes Cluster 4 patients with some metastatic disease burden (Cluster 4A) from those without (Cluster 4B). The sensitivity and specificity for identifying patients who subsequently relapsed was 65% (95% CI: 40.8–84.6%) and 75.6% (95% CI: 59.7–87.3%), respectively.
Serial 5-hmC profiles associate with clinical response to treatment in neuroblastoma patients
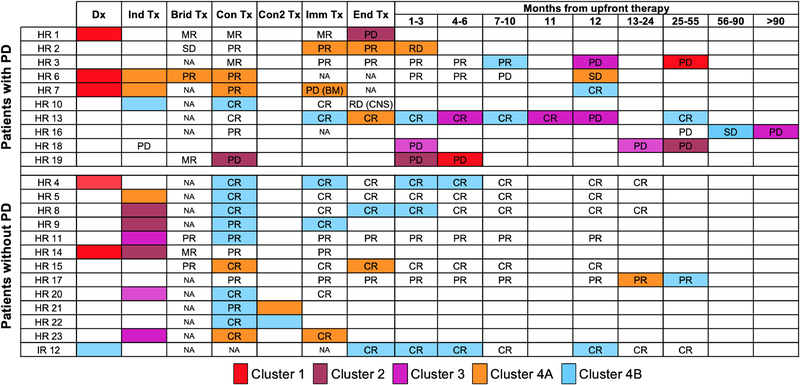
We next investigated changes in 5-hmC profiles over the course of treatment. More than one cfDNA sample was collected from twenty-three neuroblastoma patients, twenty-two of whom were classified as HR at diagnosis. For each patient, the serial 5-hmC profile cluster assignments changed over time and paralleled changes in metastatic disease burden and response to therapy (Figure 3). Samples collected at the time of diagnosis from six HR patients all grouped in Cluster 1, whereas the one diagnostic sample from an IR patient with localized disease was in Cluster 4B. After two cycles of induction therapy, all seven HR patients had profiles assigned to Cluster 2, 3, 4A, or 4B, suggesting patients had less metastatic disease burden and were responding to therapy.
Figure 3: 5-hmC profiling matches clinical response by INRC.
Each patient with two or more samples in the Discovery and Validation cohorts are shown. 5-hmC Cluster assignments are highlighted in the box and the INRC response classification at that time point is written. All cfDNA was collected at the beginning of the timepoint. Clinical response was determined using the 2017 INRC comparing each timepoint to diagnosis or prior assessment for relapsed patients. Abbreviations: Tx; Therapy, Dx; Diagnosis, Ind; Induction, Brid; Bridge therapy per ANBL1221, Con; first consolidative transplant, Con2; second consolidative transplant Imm; Immunotherapy, Fu; follow up, SD; Stable Disease, MR; Minor Response, PR; Partial Response, CR: Complete Response, PD; Progressive Disease, HR; high-risk, IR; intermediate-risk, NA; Not applicable.
5-hmC profiles from patients at the end of induction therapy appear to have prognostic potential. Of the 14 patients who had cfDNA for analysis at the end of induction, 4 were in Cluster 4A, 9 were in 4B, and one had progressive disease and was in Cluster 2. Two of the 4 patients stratified to Cluster 4A subsequently relapsed, suggesting that the 4A epigenomic cfDNA profile reflects aggressive residual disease. In contrast, the 5-hmC profile that defines Cluster 4B likely reflects little to no residual neuroblastoma, as eight of the nine patients grouped in Cluster 4B continue to survive without an event.
Patient 13’s 5-hmC profile pattern is one illustrative example of the potential for 5-hmC to detect relapse early in patients who are being monitored off of therapy. Samples obtained at six months and one month prior to relapse, which occurred a year off therapy, assigned Patient 13 into Cluster 2, even though the child was in a clinical complete response (CR) per the INRC (17).
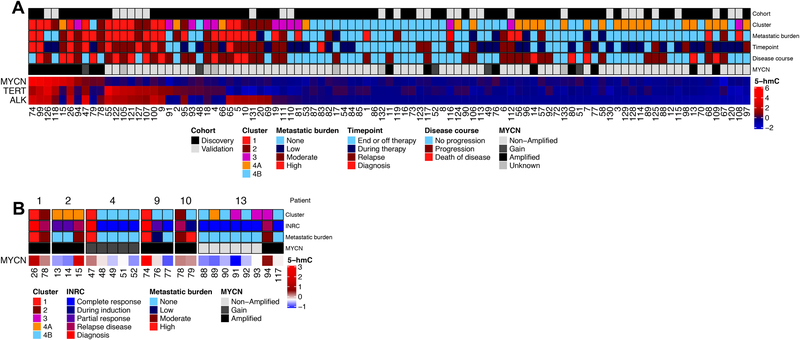
Increased 5-hmC levels observed at known oncogenic drivers in cfDNA
To further refine the ability of Nano-hmC-Seal to identify minimal amounts of residual disease, we next focused on the MYCN, TERT, and ALK genes, established oncogenic drivers in neuroblastoma (35,36). High levels of 5-hmC deposition on MYCN were detected in Cluster 1, 2, and 3 samples from patients with tumors known to have MYCN amplification or gain (Figure 4A). Of note, there were two samples from Cluster 4B which had high 5-hmC levels on MYCN, both of which were from patients who subsequently relapsed. Patient 13 presented with non-MYCN amplified disease but had increased 5-hmC deposition on MYCN detected at relapse (Figure 4B). While rare, discordant MYCN status in relapsed and diagnostic tumors has been described (37) and clinical sequencing of relapsed tumor tissue from this patient confirmed somatic acquisition of MYCN-amplification. All but one Cluster 1, 2, or 3 samples from patients with non-MYCN amplified disease had higher 5-hmC levels on TERT and/or ALK, providing further evidence that incorporating the 5-hmC status of known oncogenes may improve the sensitivity and specificity of Nano-hmC-Seal to identify patients with residual disease who are at high risk of relapse.
Figure 4: Candidate gene analysis of 5-hmC on MYCN, TERT, and ALK.
A) Ranking of 95 samples according to normalized 5-hmC levels of these three genes. B) Normalized 5-hmC for MYCN over time for 5 patients with tumors having MYCN-amplification or gain and more than one sample collected.
Biologic profiling of genes with 5-hmC enrichment in cfDNA
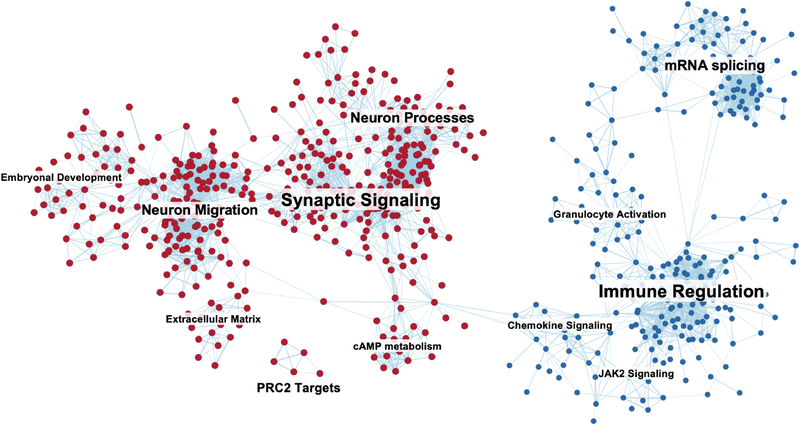
To discover 5-hmC-profile derived biological pathways in neuroblastoma, we next examined all genes (n = 11,446) with significantly different 5-hmC levels between samples from patients with or without any clinically detectable disease in the Discovery cohort. Of the 34,823 pathways evaluated, ranked fGSEA analysis showed significant enrichment of 1,974 pathways at FDR < 0.01 (Figure 5, Supplementary Table 7), whereas no pathways were enriched when the ranking of each gene was randomized. In the analysis, we incorporated target genes from publicly available transcription factor (TF) ChIP data (32). This revealed that in samples from patients with clinically detectable disease, 5-hmC was enriched on target genes of three informative groups of transcription factors; 1) stem cell maintenance, 2) CTCF and the cohesin complex, 3) PRC2 complex (Supplementary Table 8).
Figure 5: Pathway clusters with significant enrichment of genes with differential 5-hmC in blood from patients with active disease compared to those without.
Each node represents an MSigDB pathway with FDR < 0.01. Pathways enriched in patients with clinically detectable disease are shown in red and those from those without detectable disease in blue. Edge similarity cutoff of 0.375.
Discussion
Utilizing highly sensitive Nano-hmC-Seal technology to map whole genome 5-hmC in circulating cfDNA collected from healthy children and neuroblastoma patients at the time of initial diagnosis, during treatment, off-therapy, and/or after relapse, we identified distinct and consistent profiles in both Discovery and Validation cohorts that correlated with metastatic disease burden. PCA demonstrated patterns of 5-hmC deposition on cfDNA collected from patients with high metastatic disease was distinct from cfDNA profiles from healthy children, whereas the profiles identified in samples from patients with low or no metastatic disease clustered more closely with those from healthy children. Although robust diagnostic cancer-associated 5-hmC signatures have previously been identified in cfDNA from patients with colorectal, gastric, pancreatic, liver or thyroid cancer, to our knowledge, this is the first study in which Nano-hmC-Seal has been used to assess metastatic disease burden and determine changes in 5-hmC profiles over time.
Response to therapy is clearly one of the most important prognostic indicators in all malignancies, and the presence of minimal residual disease after initial treatment has been incorporated into risk stratification and therapeutic decision making for multiple cancers (38–40). Several groups have evaluated techniques to improve sensitivity for detecting minimal disease including immune profiling, expression panels from blood and bone marrow, and cell free nucleotides (41,42). These studies do not report sensitivity and specificity for detecting minimal disease status making direct comparisons to the present findings difficult. Additionally, limitations of sample stability and generalizability have hindered the application of these methods in the clinic. Although more patients need to be evaluated, our studies suggest that 5-hmC profiles of cfDNA collected at the end of induction are prognostic of outcome.
Real time patient monitoring of disease response during the course of treatment may be possible by serially collecting blood samples for cfDNA 5-hmC profiles. We identified two patients with no evidence of disease using standard clinical tests who had detectable circulating tumor DNA prior and subsequently relapsed, suggesting Nano-hmC-Seal could also be developed as a non-invasive test to monitor for recurrent neuroblastoma. 5-hmC profiles may also be able to distinguish malignant soft tumor masses from disease that has differentiated into benign neuroblastic masses (10). While validation with a prospective patient cohort is needed to confirm these findings, profiling of 5-hmC in cfDNA could be an informative addition to clinical disease evaluation and has the potential to decrease the frequency of imaging evaluations and the need for invasive procedures during follow-up.
Chromosomal copy number alterations are commonly detected in neuroblastoma and patterns of chromosomal aberrations are prognostic of survival (6). Others have identified MYCN-amplification in cfDNA collected from patients with neuroblastoma, measured at diagnosis or relapse (13,14). In our study, all Cluster 1, 2, and 3 samples from patients with MYCN-amplified tumors had high levels of 5-hmC readily identifiable on MYCN. Further, evaluation of 5-hmC accumulation on MYCN in cfDNA may provide additional prognostic information. As 5-hmC levels correlate with gene expression (3,20) the elevated 5-hmC on TERT and ALK from Clusters 1, 2, and 3, but not Cluster 4 demonstrate the ability to identify aggressive tumor biology (35,36) using cfDNA.
An additional strength of this approach is the broad applicability of 5-hmC profiles to identify tumor biology and the ability to use cfDNA to detect metastatic disease without the need for prior knowledge of a patient’s tumor genomics. We were able to incorporate functional gene pathways to further delineate patients with residual tumor burden with subsequent risk for relapse from those with no detectable disease with a sensitivity of 65% and specificity of 70%. We expect significant improvement of the sensitivity and specificity of this approach by analyzing additional patient samples with advanced feature selection tools such as elastic net (1,4). Furthermore, these results demonstrate an increase in 5-hmC on genes in neuronal networks which is consistent with expression profiling of independent neuroblastoma tumors examined with RNAseq and Nano-hmC-Seal (3,43–45). Larger numbers of samples will also be needed to explore differences in 5-hmC profiles between different biologic subsets of neuroblastoma and between newly diagnosed and relapsed patients.
These findings also highlight the role of epigenetic regulators in neuroblastoma biology, as cfDNA from patients with active disease had 5-hmC accumulation on genes regulated by PRC2 and CTCF/cohesin complexes, which has not been previously demonstrated in other malignancies using the same technology (1,2,5,46). CTCF/cohesin has been shown to regulate topologically associated domains (TADs) for organizing larger scale chromatin structures (47) and have a direct interaction with both trimethylation of histone 3 at lysine 27 (H3K27me3), 5-mC, and 5-hmC (48,49). BORIS, a germ-cell-specific paralogue of CTCF was recently shown to be a regulator of resistance to ALK inhibition in neuroblastoma (50). Additional studies are needed to elucidate if these findings are unique to neuroblastoma or are more generalizable to 5-hmC profiling from multiple tumor types.
Although this study is limited by the number of samples evaluated and its retrospective design, we demonstrate that Nano-hmC-Seal can be used to profile 5-hmC in cfDNA collected from children with neuroblastoma. The technology is highly sensitive, using a cost-effective sequencing approach that requires < 10 ng of DNA input and no specialized biospecimen handling. We identified and validated that 5-hmC cfDNA profiles correlate with disease burden and sequential samples can inform response to treatment and detect early relapse. Further, whole genome analysis of 5-hmC provides insight regarding tumor biology and transcriptomic networks. If these results are confirmed in prospective studies, this methodology has the potential for being rapidly integrated into the clinic as cfDNA 5-hmC is a stable marker allowing for patients to be profiled at any time point during treatment without prior knowledge of the primary tumor’s biology or genomics. Efforts are underway to test the ability of cfDNA 5-hmC profiles to monitor treatment response in a prospective clinical trial.
Supplementary Material
Statement of Translational Relevance.
We investigated the prognostic value of the DNA modification 5-hydroxymethylcytosine (5-hmC) in neuroblastoma using Nano-hmC-Seal, a revolutionary, low-cost, genome-wide technology that requires minimal input DNA, to serially profile 5-hmC in cell-free DNA (cfDNA) collected from Discovery (n = 32) and Validation (n = 21) cohorts of neuroblastoma patients and 34 well child controls. In this study, cfDNA 5-hmC profiles correlated with metastatic disease burden independent of underlying tumor biology. We were also able to use these profiles to distinguish patients with low or no appreciable metastatic disease using standard tumor imaging and bone marrow studies, who subsequently relapsed from those who remained in remission. Analysis of transcriptional networks regulated by these epigenomic modifications may lead to a deeper understanding of the pathways that drive resistance to treatment. Efforts are underway to test the ability of cfDNA 5-hmC profiles to monitor treatment response in a prospective clinical trial.
Acknowledgments
This work was supported in part by the Alex’s Lemonade Stand Foundation (S.L. Cohn); a kind gift from Barry & Kimberly Fields (S.L. Cohn and C. He); Neuroblastoma Children’s Cancer Society (S.L. Cohn); the Children’s Neuroblastoma Cancer Foundation (S.L. Cohn); the Matthew Bittker Foundation (S.L. Cohn); the Super Jake Foundation (S.L. Cohn); and the Elise Anderson Neuroblastoma Research Fund (S.L. Cohn). Also, supported by the National Institutes of Health K08CA226237 (M.A. Applebaum), and the Ludwig Center at the University of Chicago (C. He). C. He is a Howard Hughes Medical Institute Investigator. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. We thank the Center for Research Informatics of the University of Chicago for use of the Gardner High-Performance Computing cluster and the Cancer Center Support Grant (P30 CA014599) for support of the Genomics Core Facility.
Footnotes
Conflict of Interests Statement
CH and WZ are shareholders of Shanghai Epican Genetech Co. Ltd. that licensed 5-hmC-Seal from the University of Chicago. CH is a scientific founder and scientific advisory board member of Accent Therapeutics, Inc.
References
- 1.Li W, Zhang X, Lu X, You L, Song Y, Luo Z, et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res 2017;27(10):1243–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han D, Lu X, Shih AH, Nie J, You Q, Xu MM, et al. A Highly Sensitive and Robust Method for Genome-wide 5hmC Profiling of Rare Cell Populations. Molecular cell 2016;63(4):711–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Applebaum MA, Barr EK, Karpus J, Nie J, Zhang Z, Armstrong AE, et al. 5-Hydroxymethylcytosine Profiles Are Prognostic of Outcome in Neuroblastoma and Reveal Transcriptional Networks That Correlate With Tumor Phenotype. JCO Precision Oncology 2019(3):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian X, Sun B, Chen C, Gao C, Zhang J, Lu X, et al. Circulating tumor DNA 5-hydroxymethylcytosine as a novel diagnostic biomarker for esophageal cancer. Cell Res 2018;28(5):597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai J, Chen L, Zhang Z, Zhang X, Lu X, Liu W, et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, et al. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J Clin Oncol 2015;33(27):3008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JR, Kreissman SG, London WB, Naranjo A, Cohn SL, Hogarty MD, et al. Effect of Tandem Autologous Stem Cell Transplant vs Single Transplant on Event-Free Survival in Patients With High-Risk Neuroblastoma: A Randomized Clinical Trial. JAMA 2019;322(8):746–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinto N, Naranjo A, Hibbitts E, Kreissman SG, Granger MM, Irwin MS, et al. Predictors of differential response to induction therapy in high-risk neuroblastoma: A report from the Children’s Oncology Group (COG). Eur J Cancer 2019;112:66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung IY, Feng Y, Gerald W, Cheung NK. Exploiting gene expression profiling to identify novel minimal residual disease markers of neuroblastoma. Clin Cancer Res 2008;14(21):7020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villablanca JG, Ji L, Shapira-Lewinson A, Marachelian A, Shimada H, Hawkins RA, et al. Predictors of response, progression-free survival, and overall survival using NANT Response Criteria (v1.0) in relapsed and refractory high-risk neuroblastoma. Pediatr Blood Cancer 2018;65(5):e26940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med 2018;379(18):1754–65. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Wang L, Su Y, Yue Z, Xing T, Zhao W, et al. Plasma cell-free DNA quantification is highly correlated to tumor burden in children with neuroblastoma. Cancer Medicine 2018;7(7):3022–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chicard M, Colmet-Daage L, Clement N, Danzon A, Bohec M, Bernard V, et al. Whole-Exome Sequencing of Cell-Free DNA Reveals Temporo-spatial Heterogeneity and Identifies Treatment-Resistant Clones in Neuroblastoma. Clin Cancer Res 2018;24(4):939–49. [DOI] [PubMed] [Google Scholar]
- 14.Van Roy N, Van Der Linden M, Menten B, Dheedene A, Vandeputte C, Van Dorpe J, et al. Shallow Whole Genome Sequencing on Circulating Cell-Free DNA Allows Reliable Noninvasive Copy-Number Profiling in Neuroblastoma Patients. Clin Cancer Res 2017;23(20):6305–14. [DOI] [PubMed] [Google Scholar]
- 15.Combaret V, Iacono I, Bellini A, Brejon S, Bernard V, Marabelle A, et al. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med 2015;4(4):540–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Applebaum MA, Desai AV, Glade Bender JL, Cohn SL. Emerging and investigational therapies for neuroblastoma. Expert Opin Orphan Drugs 2017;5(4):355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JR, Bagatell R, Cohn SL, Pearson AD, Villablanca JG, Berthold F, et al. Revisions to the International Neuroblastoma Response Criteria: A Consensus Statement From the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol 2017;35(22):2580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanik GA, Parisi MT, Naranjo A, Nadel H, Gelfand MJ, Park JR, et al. Validation of Postinduction Curie Scores in High-Risk Neuroblastoma: A Children’s Oncology Group and SIOPEN Group Report on SIOPEN/HR-NBL1. J Nucl Med 2018;59(3):502–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mody R, Naranjo A, Van Ryn C, Yu AL, London WB, Shulkin BL, et al. Irinotecan-temozolomide with temsirolimus or dinutuximab in children with refractory or relapsed neuroblastoma (COG ANBL1221): an open-label, randomised, phase 2 trial. Lancet Oncol 2017;18(7):946–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature biotechnology 2011;29(1):68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews S 2013. FastQC: A quality control application for high throughput sequence data. <http://www.bioinformatics.babraham.ac.uk/projects/fastqc/>.
- 22.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 25.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47(D1):D766–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lê S, Josse J, Husson F. FactoMineR: An R Package for Multivariate Analysis. 2008 2008;25(1):18. [Google Scholar]
- 28.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32(18):2847–9. [DOI] [PubMed] [Google Scholar]
- 29.Coffman LG, Pearson AT, Frisbie LG, Freeman Z, Christie E, Bowtell DD, et al. Ovarian Carcinoma-Associated Mesenchymal Stem Cells Arise from Tissue-Specific Normal Stroma. Stem Cells 2019;37(2):257–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn M Building Predictive Models in R Using the caret Package. 2008 2008;28(5):26. [Google Scholar]
- 31.Sergushichev AA. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv 2016:060012. [Google Scholar]
- 32.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27(12):1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS one 2010;5(11):e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet 2015;47(12):1411–4. [DOI] [PubMed] [Google Scholar]
- 36.George RE, Sanda T, Hanna M, Frohling S, Luther W 2nd, Zhang J, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 2008;455(7215):975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theissen J, Boensch M, Spitz R, Betts D, Stegmaier S, Christiansen H, et al. Heterogeneity of the MYCN oncogene in neuroblastoma. Clin Cancer Res 2009;15(6):2085–90. [DOI] [PubMed] [Google Scholar]
- 38.Eckert C, Biondi A, Seeger K, Cazzaniga G, Hartmann R, Beyermann B, et al. Prognostic value of minimal residual disease in relapsed childhood acute lymphoblastic leukaemia. Lancet 2001;358(9289):1239–41. [DOI] [PubMed] [Google Scholar]
- 39.van Dongen JJ, Seriu T, Panzer-Grumayer ER, Biondi A, Pongers-Willemse MJ, Corral L, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 1998;352(9142):1731–8. [DOI] [PubMed] [Google Scholar]
- 40.Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 2007;25(28):4414–22. [DOI] [PubMed] [Google Scholar]
- 41.Marachelian A, Villablanca JG, Liu CW, Liu B, Goodarzian F, Lai HA, et al. Expression of Five Neuroblastoma Genes in Bone Marrow or Blood of Patients with Relapsed/Refractory Neuroblastoma Provides a New Biomarker for Disease and Prognosis. Clin Cancer Res 2017;23(18):5374–83. [DOI] [PubMed] [Google Scholar]
- 42.Zeka F, Decock A, Van Goethem A, Vanderheyden K, Demuynck F, Lammens T, et al. Circulating microRNA biomarkers for metastatic disease in neuroblastoma patients. JCI Insight 2018;3(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei JS, Kuznetsov IB, Zhang S, Song YK, Asgharzadeh S, Sindiri S, et al. Clinically Relevant Cytotoxic Immune Cell Signatures and Clonal Expansion of T-Cell Receptors in High-Risk MYCN-Not-Amplified Human Neuroblastoma. Clin Cancer Res 2018;24(22):5673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Applebaum MA, Jha AR, Kao C, Hernandez KM, DeWane G, Salwen HR, et al. Integrative genomics reveals hypoxia inducible genes that are associated with a poor prognosis in neuroblastoma patients. Oncotarget 2016;7(47):76816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valentijn LJ, Koster J, Haneveld F, Aissa RA, van Sluis P, Broekmans ME, et al. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci U S A 2012;109(47):19190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiu BC, Zhang Z, You Q, Zeng C, Stepniak E, Bracci PM, et al. Prognostic implications of 5-hydroxymethylcytosines from circulating cell-free DNA in diffuse large B-cell lymphoma. Blood Adv 2019;3(19):2790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017;169(5):930–44 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu M, Zhao GN, Lv X, Liu G, Wang LY, Hao DL, et al. CTCF controls HOXA cluster silencing and mediates PRC2-repressive higher-order chromatin structure in NT2/D1 cells. Mol Cell Biol 2014;34(20):3867–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 2011;43(7):630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debruyne DN, Dries R, Sengupta S, Seruggia D, Gao Y, Sharma B, et al. BORIS promotes chromatin regulatory interactions in treatment-resistant cancer cells. Nature 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.