Abstract
Bacterial cyclic‐di‐GMP (c‐di‐GMP) production is associated with biofilm development and the switch from acute to chronic infections. In Pseudomonas aeruginosa, the diguanylate cyclase (DGC) SiaD and phosphatase SiaA, which are co‐transcribed as part of a siaABCD operon, are essential for cellular aggregation. However, the detailed functions of this operon and the relationships among its constituent genes are unknown. Here, we demonstrate that the siaABCD operon encodes for a signaling network that regulates SiaD enzymatic activity to control biofilm and aggregates formation. Through protein–protein interaction, SiaC promotes SiaD diguanylate cyclase activity. Biochemical and structural data revealed that SiaB is an unusual protein kinase that phosphorylates SiaC, whereas SiaA phosphatase can dephosphorylate SiaC. The phosphorylation state of SiaC is critical for its interaction with SiaD, which will switch on or off the DGC activity of SiaD and regulate c‐di‐GMP levels and subsequent virulence phenotypes. Collectively, our data provide insights into the molecular mechanisms underlying the modulation of DGC activity associated with chronic infections, which may facilitate the development of antimicrobial drugs.
Keywords: biofilm formation, crystal structure, phosphorylation, protein–protein interaction, Pseudomonas aeruginosa
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Post-translational Modifications, Proteolysis & Proteomics; Signal Transduction
Structural and functional analyses reveal how SiaB kinase and SiaC adaptor protein regulate SiaD diguanylate cyclase activity to modulate bacterial c‐di‐GMP production and biofilm formation.
Introduction
The intracellular messenger cyclic dimeric (3′–5′) GMP (cyclic di‐GMP or c‐di‐GMP) is a small molecule that mediates various aspects of the physiology of diverse environmental and pathogenic bacteria (Romling et al, 2013; Jenal et al, 2017). It was first described in 1986 as an allosteric factor that activated cellulose synthase in Acetobacter xylinum (Ross et al, 1987). To date, c‐di‐GMP has been shown to regulate the cell cycle, biofilm formation and dispersion, motility, virulence, and other processes (Romling et al, 2013; Jenal et al, 2017). C‐di‐GMP promotes biofilm growth by stimulating the production of exopolysaccharide (EPS) and adhesins (Simm et al, 2004; Jenal et al, 2017). C‐di‐GMP is synthesized by diguanylate cyclases (DGCs) harboring a GGDEF domain (Simm et al, 2004; Schmidt et al, 2005). Degradation of c‐di‐GMP is catalyzed by phosphodiesterases (PDE) with an EAL or HD‐GYP domain, which results in the production of 5‐phosphoguanylyl‐(3′,5′)‐guanosine (pGpG) or GMP (Simm et al, 2004). pGpG is further hydrolyzed by oligoribonucleases into two GMP molecules (Cohen et al, 2015; Orr et al, 2015).
The c‐di‐GMP regulatory system in Pseudomonas aeruginosa is highly sophisticated. The genome of P. aeruginosa PAO1 contains 17 genes with a predicted DGC domain, 5 with a PDE domain, and 16 with both domains (Kulasakara et al, 2006). SiaD contains a DGC domain that is essential for cell aggregation, a bacterial stress response that promotes the survival of P. aeruginosa in response to the toxic detergent sodium dodecylsulfate (SDS) (Klebensberger et al, 2009). This process requires the putative inner membrane‐located Ser/Thr phosphatase SiaA, which contains two transmembrane helices, a HAMP domain and a sigma factor PP2C‐like phosphatase domain. SiaA and SiaD also regulate the Psl polysaccharide, the CdrAB two‐partner secretion system, and the CupA fimbriae (Klebensberger et al, 2009). Recently, Chew et al (2018) have reported that SiaD is required for the competitiveness of P. aeruginosa during dual‐species biofilm development with Staphylococcus aureus. Additionally, SiaA and SiaD functionally interact with the RsmA post‐transcriptional regulatory system, which is known to control motility, biofilm formation, and cell aggregation (Brencic & Lory, 2009; Colley et al, 2016). SiaA and SiaD regulate the RsmA antagonist rsmZ during growth with SDS, whereas RsmA controls siaD transcription (Colley et al, 2016).
In all kingdoms of life, reversible protein phosphorylation controls the activity of enzymes, or the association of proteins with other proteins (Hunter, 1995). Phosphoryl groups, typically originating from ATP, are attached to amino acid side chains on protein by protein kinases, and are removed by protein phosphatases in response to specific cellular or environmental cues; therefore, reversible phosphorylation controls diverse biological processes (Taylor & Kornev, 2011). Phosphorylation and DGC activity crosstalk have been reported to control c‐di‐GMP production. The DGC activity of WspR is induced via phosphorylation by the Wsp chemosensory system in response to growth of P. aeruginosa on surface (Hickman et al, 2005). Active WspR then produces c‐di‐GMP to stimulate biofilm formation. In Bdellovibrio bacteriovorus, the DGC activity of DgcB is dependent on the sensory fork head‐associated (FHA) domain bound with the ligand from the N‐terminal region of DgcB itself (Meek et al, 2019). The MtG DGC activity in Moorella thermoacetica is modulated by the stressosome complex and the switch kinase MtT (Quin et al, 2012). Here, we report that SiaC and SiaB function together with SiaD and SiaA to regulate biofilm formation by modulating the intracellular c‐di‐GMP levels. We demonstrate that SiaCacts as a binding partner to regulate the DGC activity of SiaD. Structural and biochemical data show that SiaBis a kinase that phosphorylates SiaC and the phosphatase SiaA dephosphorylates it at the same site. The reversible phosphorylation of SiaC plays an essential role in the activity of SiaD and subsequent biofilm and aggregate formation. Taken together, this study uncovers SiaC as a key “switch” of the signaling pathway and elucidates a novel phospho‐signaling regulatory mechanism underlying the DGC activity by SiaC.
Results
SiaC and SiaB inversely regulates biofilm and aggregate formation through SiaD
SiaA and SiaD are essential for SDS‐induced formation of macroscopic aggregates in P. aeruginosa (Klebensberger et al, 2009). Furthermore, siaA (PA0172) and siaD (PA0169) are co‐transcribed with siaB (PA0171) and siaC (PA0170), which encode proteins with unknown functions (Klebensberger et al, 2009). This information led us to test whether SiaC and SiaB play a role in SDS‐induced aggregation. To this end, the deletion mutants of ΔsiaC and ΔsiaB were generated. Similar to the ΔsiaA and ΔsiaD mutants, phenotypic characterization of the ΔsiaC mutant during growth in M9 salt with SDS as the sole carbon source revealed the decreased formation of macroscopic aggregates compared to the PAO1 strain (Appendix Fig S1A; Klebensberger et al, 2009). However, the ΔsiaB mutant exhibited enhanced macroscopic aggregation in M9 salt with SDS, and expression of siaC or siaB in trans in the corresponding mutant restored formation of macroscopic aggregates (Appendix Fig S1A). SDS‐induced aggregation is linked to c‐di‐GMP, a ubiquitous bacterial second messenger that regulates the transition between motile and sessile biofilm lifestyles (Klebensberger et al, 2009). Therefore, we tested whether both SiaC and SiaB regulated the formation of biofilm. As shown in Fig 1A, the ability to form biofilm of ΔsiaC was drastically decreased compared to that of wild‐type parent. By contrast, deletion of siaB promoted biofilm formation, which was consistent with the SiaB‐mediated aggregation. Indeed, expression of siaC and siaB in trans in the corresponding mutant exhibited increased or reduced biofilm formation compared to that of wild‐type strain, respectively (Fig 1A). As expected, deletion of siaA or siaD impaired biofilm formation (Fig 1A). These data demonstrated that SiaA, SiaC, and SiaD positively, and SiaB negatively, regulate macroscopic aggregation and biofilm formation, suggesting the cooperative function of these proteins in regulating these processes.
Figure 1. SiaB and SiaC regulate biofilm formation by modulating intracellular c‐di‐GMP levels through SiaD.
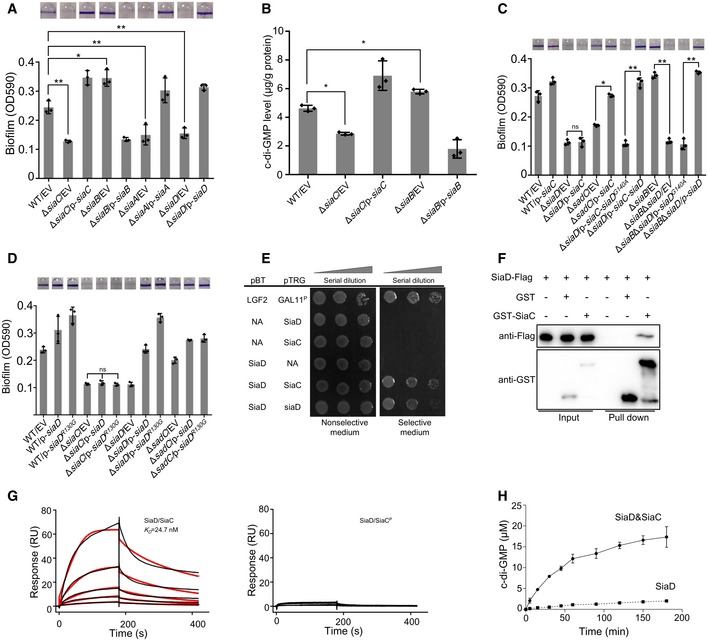
- Deletion of siaA, siaB, siaC, or siaD regulates biofilm formation. The biofilm formation of the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down).
- The intracellular levels of c‐di‐GMP in PAO1, ΔsiaC, ΔsiaB, and the corresponding complemented strains were detected by LC‐MS/MS.
- SiaB‐ and SiaC‐regulated biofilm formation is dependent on SiaD. Biofilm formation by the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down).
- SiaC is required for the function of SiaD. Biofilm formation by the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down).
- Bacterial two‐hybrid assay reveals an interaction between SiaC and SiaD. The recombinant strains harboring different vectors were separately streaked on nonselective and dual‐selective media (3‐amino‐1, 2, 4‐ triazole + streptomycin). NA represents the empty vector pBT or pTRG. The strain‐expressing LGF2 and Gal11P were used as positive controls.
- Pull‐down assays confirmed the interaction between SiaC and SiaD. Cell lysates of Pseudomonas aeruginosa containing pMM67EH‐siaD‐Flag were incubated with GST or GST‐SiaC individually, and protein complexes were captured by glutathione beads.
- SPR measurements of SiaC (left) or SiaCP (right) binding at varying concentrations to SiaD. SiaC specifically interacted with SiaD with a K D of 24.7 nM, whereas SiaCP failed to interact with SiaB. Shown are measured binding responses (black) and curve fits to a 1:1 interaction (red). Plots are representative of two experiments with similar results. RU, response units; K D, dissociation constant.
- Production of c‐di‐GMP by either SiaD or SiaD with SiaC at the indicated time‐point was determined by HPLC.
The c‐di‐GMP signal has been shown to be involved in P. aeruginosa macroscopic aggregation and biofilm formation (Klebensberger et al, 2009; Zhu et al, 2016a). Therefore, we sought to determine whether SiaB and SiaC influence the intracellular level of c‐di‐GMP. To this end, we measured the concentration of intracellular c‐di‐GMP via LC‐MS/MS, which showed that the ΔsiaC produced less but ΔsiaB had more c‐di‐GMP than the wild‐type strain (Fig 1B). The mRNA levels of cdrA, which is responsive to intracellular level of c‐di‐GMP (Baraquet et al, 2012), were also determined by quantitative reverse transcription‐polymerase chain reaction (qRT–PCR) and gain similar trends (Appendix Fig S1B). In addition, overexpression of a DGC SadC (for c‐di‐GMP biosynthesis) in ΔsiaC mutant restored its ability to form biofilm, and the elevated biofilm formation of ΔsiaB was abolished by overexpression of PDE PA2133 (phosphodiesterase, degradation of c‐di‐GMP) (Appendix Fig S1C). These results suggested that the regulation of biofilm formation by SiaC and SiaB is dependent on the intracellular concentrations of c‐di‐GMP.
Deletion of siaD reduces intracellular c‐di‐GMP levels (Colley et al, 2016), which regulates biofilm and SDS‐induced aggregation. To determine whether the functions of SiaB and SiaC were dependent on SiaD, the abilities to form biofilm of wild‐type parent and its derivatives were tested. Overexpression of siaC significantly promoted biofilm formation of wild‐type parent, whereas this effect was not observed in the ΔsiaD mutant, indicating that the functional SiaD was necessary for the function of SiaC (Fig 1C). Notably, deletion of another DGC SadC did not abolish the elevated biofilm formation caused by overexpression of SiaC (Fig 1C), suggesting that SiaC‐mediated regulation of biofilm formation is specifically dependent on SiaD. To further confirm that SiaC functions by modulating c‐di‐GMP through SiaD, plasmids overexpressing both siaC and siaD or siaD G140A [a siaD active site mutant (Colley et al, 2016)] were constructed and introduced into ΔsiaD mutant, respectively. Biofilm formation assay showed that the elevated biofilm formation by SiaC overexpression was lost in the presence of the siaD G140A (Fig 1C). We also revealed that the increased biofilm formation of ΔsiaB was abolished by further deletion of siaD gene, which was restored by overexpression of siaD but not siaD G140A gene (Fig 1C). Together, these results indicated that SiaC and SiaB regulate biofilm and aggregation formation by modulating c‐di‐GMP levels through SiaD.
SiaC is required for the DGC activity of SiaD via direct interaction
Since the biofilm formation of ΔsiaC is dependent on SiaD, the expression of siaD was determined in ΔsiaC mutant. qRT–PCR data showed that deletion of siaC reduced the transcript level of siaD (Appendix Fig S2A). To determine whether the defect in biofilm formation by ΔsiaC was due to the decreased expression level of siaD, plasmid overexpressing SiaD was introduced into ΔsiaC. However, our results showed that overexpression of SiaD restored the ability to form biofilm of ΔsiaD, but not that of ΔsiaC (Fig 1D). As expected, overexpression of SiaD in ΔsadC restored the biofilm phenotype (Fig 1D). To confirm that the siaD is sufficiently produced from the overexpression plasmid, the plasmid pUCP20‐siaD‐Flag was used to detect the transcription and translation of siaD. The function of SiaD was not affected by the C‐terminal Flag tag, as the biofilm formation of ΔsiaD was restored after introduction of siaD‐Flag (Appendix Fig S2B). qRT–PCR and Western blotting results showed that SiaD is sufficiently produced from the overexpression plasmid (Appendix Fig S2C). These results indicated that the function of SiaD is dependent on SiaC.
The failure of overexpressing SiaD to restore the biofilm formation by ΔsiaC prompted us to hypothesize that SiaC directly interacts with SiaD to regulate c‐di‐GMP production and biofilm formation. To test this, we performed a bacterial two‐hybrid (BTH) assay using SiaC as the bait. A specific two‐hybrid binding assay using the full‐length siaD gene construct showed robust growth of colonies on dual‐selective medium (Fig 1E), indicating that SiaC interacts with full‐length SiaD. The BTH assay suggested that SiaD could interact with itself (Fig 1E), consistent with the finding that GGDEF domains function as homodimers (Romling et al, 2013). To further validate the SiaC‐SiaD interaction in vitro, we performed a GST pull‐down assay using the purified GST‐SiaC (Appendix Fig S3A) and the lysates of PAO1 cells containing a C‐terminal Flag‐tagged SiaD plasmid. Western blot analysis showed that SiaD was retained with GST‐SiaC, but not GST (Fig 1F). Consistently, a GST pull‐down assay using GST‐SiaD (Appendix Fig S3A) and SiaC‐Flag demonstrated that GST‐SiaD, but not GST, interacts with SiaC‐Flag (Appendix Fig S3B). Further, the purified SiaC and SiaD proteins were incubated and the mixtures were subject to analysis by gel filtration chromatography using a Superdex 200 10/300 size exclusion column. Result showed that SiaC forms complexes with SiaD (Fig EV1A). We also performed label‐free surface plasmon resonance (SPR) binding assay and found that the purified SiaC binds to SiaD with a dissociation constant (K D) of 24.7 nM (Fig 1G). However, SiaC was unable to interact with itself (Appendix Fig S3C). Taken together, these results clearly revealed that SiaC directly interacts with SiaD.
Figure EV1. Interactions of SiaD with SiaC or SiaCP .
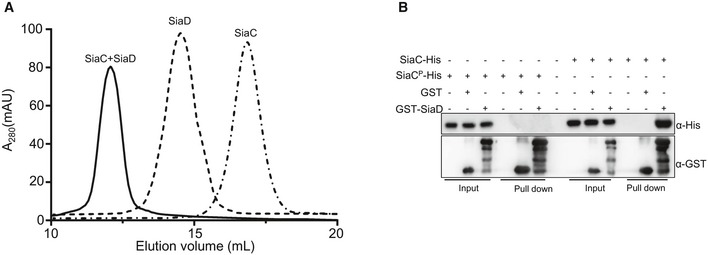
- Size exclusion chromatography elution profile of the purified SiaC and SiaD on a superdex 200 10/300 column.
- GST pull‐down assays showed that SiaD interacts with SiaC but not SiaCP. Purified SiaC or SiaCP was incubated with GST, GST‐PA0170 individually, and protein complexes were captured by glutathione beads.
Source data are available online for this figure.
The interaction between SiaD and SiaC prompted us to speculate that SiaC acts as a modulator to regulate DGC activity of SiaD. To evaluate the role of SiaC in modulation of the DGC activity of SiaD, enzymatic synthesis of c‐di‐GMP from GTP was performed in vitro using either SiaD or SiaD and SiaC. After incubation, the production of c‐di‐GMP was analyzed by high‐performance liquid chromatography (HPLC) analysis with retention times consistent with those of GTP and c‐di‐GMP standards. Surprisingly, GTP remained the primary compound in the reaction mix when GTP was incubated with SiaD alone even after 3 h, indicating that SiaD had weak DGC activity (Fig 1H). Interestingly, the production of c‐di‐GMP was significantly increased when GTP was incubated with both of SiaC and SiaD (Fig 1H), which accounts for the dependence of SiaD on SiaC in vivo (Fig 1D). Importantly, SiaC was unable to produce c‐di‐GMP (Appendix Fig S4A–C) or enhance the activity of another DGC WspR (Appendix Fig S4D–F; Hickman et al, 2005). Moreover, our data suggested that the SiaD stability was not influenced by deletion of SiaC (Appendix Fig S2D). Together, these results demonstrated that the direct interaction with SiaC is essential for activation of SiaD DGC activity.
SiaD harbors a conserved RXXD motif (I‐site), which is characteristic of c‐di‐GMP binding and allosteric regulation (Colley et al, 2016). Expression of the defective I‐site allele siaD R130G in ΔsiaD promotes bacterial aggregation compared to a strain complemented with the native siaD gene (Colley et al, 2016), indicating that I‐site mutation abolishes c‐di‐GMP binding and allosteric regulation. Therefore, we determined whether SiaC modulates the function of SiaD by repressing c‐di‐GMP‐mediated allosteric regulation. If so, biofilm formation by ΔsiaC should be restored by overexpression of siaD R130G. However, siaD R130G failed to restore the biofilm‐forming ability of ΔsiaC (Fig 1D and Appendix Fig S2B). In vitro HPLC analysis showed that SiaDR130G has some weak DGC activity, and the activity can be significantly enhanced by SiaC (Appendix Fig S2E). Therefore, SiaC seems unlikely to modulate the function of SiaD by repressing c‐di‐GMP‐mediated allosteric regulation.
SiaB interacts with SiaC to regulate aggregate and biofilm formation
As mentioned above, the hyperbiofilm phenotype of ΔsiaB mutant was due to the elevated intracellular c‐di‐GMP from SiaD (Fig 1C). In addition, SiaC is necessary for the DGC activity of SiaD (Fig 1H). These observations led us to hypothesize that SiaB might interact with SiaC to influence c‐di‐GMP and biofilm formation. To test this hypothesis, we performed a BTH assay using SiaB as the bait. Like the positive control, strain‐bearing pBT‐siaB and pTRG‐SiaC also grew robustly on dual‐selective medium (Fig 2A), indicating a direct interaction between SiaB and SiaC. However, the BTH assay indicated that SiaB does not interact with SiaD or SiaA (Fig 2A). To further confirm the interaction between SiaB and SiaC, in vitro pull‐down assays were performed. GST‐SiaB (Appendix Fig S3A), but not GST, was able to retain SiaC (Fig 2B). Consistently, interaction between the purified GST‐SiaC and SiaB‐Flag was observed (Fig EV2A). The gel filtration chromatography analysis also indicated the formation of SiaB‐SiaC complex (Fig EV2B). As determined by SPR experiment, SiaC can bind to SiaB with a K D of 40.1 nM (Fig 2C). Since both SiaB and SiaD interact with SiaC, we wondered whether the three proteins could form SiaB‐SiaC‐SiaD ternary complex. However, as confirmed by the pull‐down assays, the ternary complex was not formed (Appendix Fig S3D).
Figure 2. SiaA and SiaB regulate biofilm formation via interaction with SiaC.
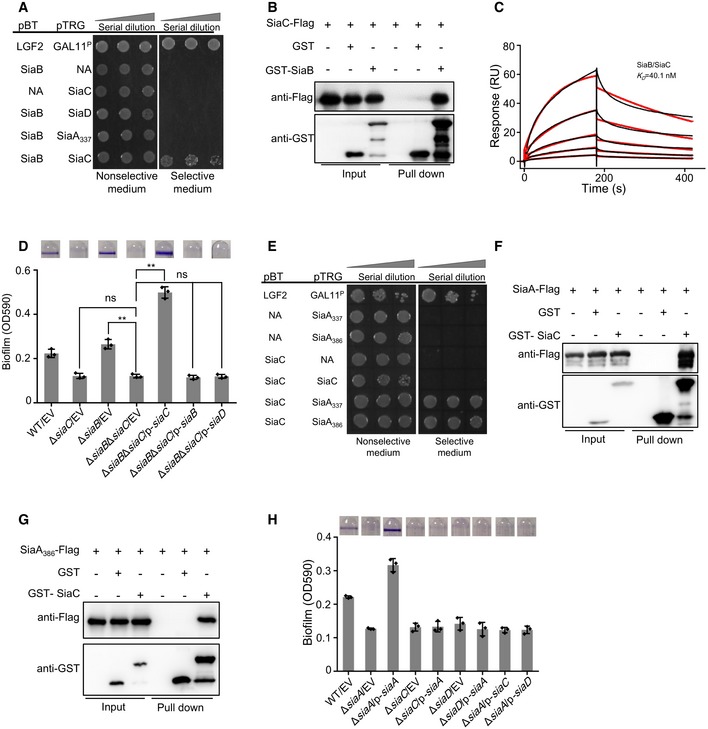
- Bacterial two‐hybrid assay reveals an interaction between SiaB and SiaC. The recombinant strains harboring different proteins were separately streaked on nonselective and dual‐selective media (3‐amino‐1, 2, 4‐ triazole + streptomycin). NA represents the empty vector pBT or pTRG. The strain‐expressing LGF2 and Gal11P were used as positive controls.
- Pull‐down assays confirmed the interaction between SiaB and SiaC. Cell lysates of Pseudomonas aeruginosa containing pMM67EH‐siaC‐Flag were incubated with GST or GST‐SiaB individually, and protein complexes were captured by glutathione beads.
- SPR measurements of SiaC binding at varying concentrations to SiaB. SiaC specifically interacted with SiaB with a K D of 40.1 nM. Shown are measured binding responses (black) and curve fits to a 1:1 interaction (red). Plots are representative of two experiments with similar results. RU, response units; K D, dissociation constant.
- SiaB‐mediated regulation of biofilm formation is dependent on SiaC. The biofilm formation of the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down).
- Bacterial two‐hybrid assay reveals an interaction between SiaC and SiaA PP2C‐like domain. NA represents the empty vector pBT or pTRG. The recombinant strains harboring different proteins were separately streaked on nonselective and dual‐selective media (3‐amino‐1, 2, 4‐ triazole + streptomycin). The strain‐expressing LGF2 and Gal11P were used as positive controls.
- Pull‐down assays confirmed the interaction between SiaC and SiaA. Cell lysates of Pseudomonas aeruginosa containing pMM67EH‐siaA‐Flag were incubated with GST or GST‐SiaC individually, and protein complexes were captured by glutathione beads.
- Pull‐down assays confirmed the interaction between SiaC and SiaA PP2C‐like domain. Cell lysates of P. aeruginosa containing pMMB67EH‐siaA 386‐Flag were incubated with GST or GST‐SiaC individually, and protein complexes were captured by glutathione beads.
- The biofilm formation of the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down).
Figure EV2. SiaC interacts with SiaA or SiaB.
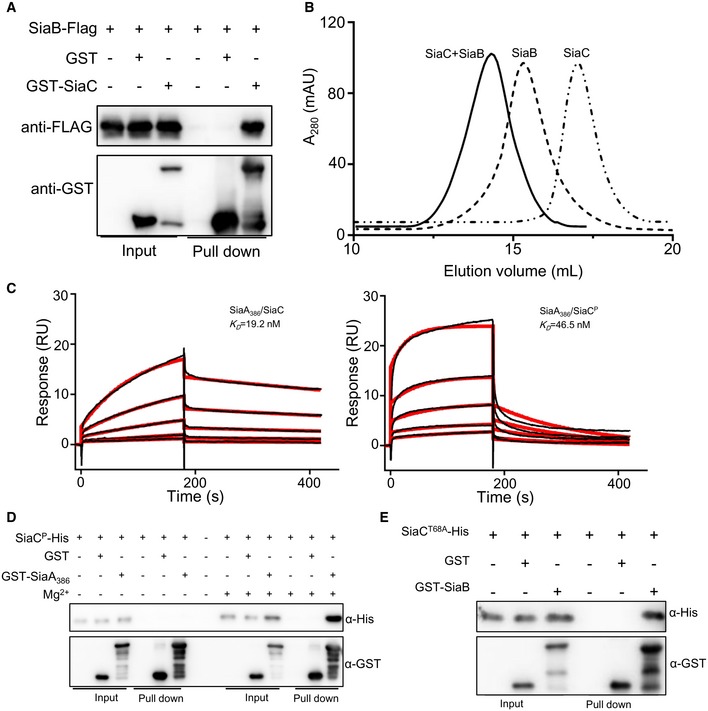
- GST pull‐down shows direct interaction between SiaC and SiaB. Cell lysates of Pseudomonas aeruginosa containing pMM67EH‐siaB‐Flag were incubated with GST and GST‐SiaC individually, and protein complexes were captured by glutathione beads.
- Size exclusion chromatography elution profile of the purified SiaB and SiaC on a superdex 200 10/300 column.
- SPR measurements of SiaC or SiaCP binding at varying concentrations to SiaA386. SiaC‐His or SiaCP‐His specifically interacted with SiaA386 with a K D of 19.2 or 46.5 nM. Shown are measured binding responses (black) and curve fits to a 1:1 interaction (red). Plots are representative of two experiments with similar results. RU, response units; K D, dissociation constant.
- GST pull‐down shows direct interaction between SiaCP and SiaA386. Purified SiaCP was incubated with GST and GST‐SiaA386 individually in buffers with or without 10 mM MgCl2, and protein complexes were captured by glutathione beads.
- GST pull‐down shows direct interaction between SiaCT68A and SiaB. Purified SiaCT68A‐His was incubated with GST, GST‐SiaB individually, and protein complexes were captured by glutathione beads.
Source data are available online for this figure.
To determine whether SiaC mediates the function of SiaB via direct interaction, we tested the biofilm phenotypes of P. aeruginosa and its derivates. Our results showed that deletion of siaC gene in the ΔsiaB mutant background abolished the hyperbiofilm formation (Fig 2D), suggesting that SiaB‐mediated biofilm regulation is dependent on its interaction with SiaC. As expected, the ΔsiaCΔsiaB double mutant complemented with SiaB formed biofilm similar to that of ΔsiaC (Fig 2D). Additionally, overexpression of SiaC in the ΔsiaCΔsiaB double mutant significantly promoted biofilm formation (Fig 2D). However, overexpression of siaD did not impact the biofilm phenotype of ΔsiaCΔsiaB strain (Fig 2D). We also determined the SDS‐induced aggregation by these strains and similar trends were observed (Appendix Fig S3E). Taken together, these results demonstrated that SiaC is essential for SiaB‐mediated repression of biofilm and aggregates formation.
SiaA regulates biofilm formation via direct interaction with SiaC
SiaA contains two transmembrane helices, PP2C‐like phosphatase and HAMP domains (Appendix Fig S5A), which are involved in signal transduction (Aravind & Ponting, 1999; Vijay et al, 2000). Deletion of siaA reduced biofilm formation, which prompted us to test whether SiaC physically interacts with SiaA. To determine the protein–protein interaction, and to test whether the HAMP and C‐terminal PP2C‐like phosphatase domains are sufficient to maintain the interaction with SiaC, we performed specific BTH binding assays using constructs encoding the free‐standing PP2C‐like phosphatase domain (SiaA protein lacking the N‐terminal 385 amino acids, SiaA386) or both the HAMP and PP2C‐like phosphatase domains (SiaA protein lacking the N‐terminal 336 amino acids, SiaA337). As evidenced by the robust growth of colonies on selective medium (Fig 2E), SiaC interacted with the SiaA PP2C‐like phosphatase domain. To further validate the interaction between SiaC and SiaA, GST pull‐down assays were performed using purified GST‐SiaC co‐incubated with lysate of PAO1 cells containing plasmids expressing Flag‐tagged SiaA and SiaA386, respectively. SiaA‐Flag was shown to be retained with GST‐SiaC but not GST (Fig 2F). Importantly, SiaA386‐Flag also interacted with GST‐SiaC (Fig 2G), indicating that SiaA interacts with SiaC through its PP2C‐like phosphatase domain. Notably, we observed no interaction between SiaD and SiaA (Appendix Fig S5B). Moreover, pull‐down assays showed that SiaA386‐SiaC‐SiaB formed a complex, whereas no SiaA386‐SiaC‐SiaD complex was observed (Appendix Fig S5C and D). Taken together, these data indicated that SiaA controls biofilm formation and SDS‐induced aggregation via direct interaction with SiaC.
To assess the functional interplay between SiaA and SiaC, phenotypic assays were performed. Biofilm assays showed that overexpression of siaA restored the ability to form biofilm of ΔsiaA, but not that of ΔsiaC or ΔsiaD (Fig 2H). Surprisingly, overexpression of siaC or siaD in ΔsiaA mutant did not promote the biofilm formation, which is in contrast to that observed in wild type (Fig 2H). These results indicated that SiaA functions upstream of SiaC‐SiaD and is necessary for their activity. These observations were confirmed by the SDS‐induced aggregations of these strains (Appendix Fig S5E). Collectively, these data suggested that SiaA is essential for the function of SiaC in vivo.
Reversible phosphorylation of SiaC by SiaB and SiaA regulates its function
A previous study showed that SiaC can be phosphorylated at Thr68 residue (Ravichandran et al, 2009). SiaA harbors a PP2C‐like phosphatase domain, and online homology modeling suggested that SiaB (Appendix Fig S6A) is similar to alpha‐ketoacid dehydrogenase kinase (PDB_ID: 3TZ5) with high confidence (92.1%) (Kelley et al, 2015). This led us to hypothesize that SiaB phosphorylates SiaC, whereas SiaA dephosphorylates it. To test this hypothesis, we assayed ATP hydrolysis activity; SiaB alone exhibited no ATP hydrolysis activity (Fig 3A). However, SiaB displayed ATP hydrolysis activity when purified SiaC was used as the substrate (Fig 3A). The activity of SiaB was independent of the presence of divalent cations, such as Mg2+ and Mn2+, as SiaB hydrolyzed ATP in the presence of ethylene diamine tetraacetic acid (EDTA; Appendix Fig S6B).
Figure 3. SiaB phosphorylates SiaC at threonine 68 residue and the phosphorylated SiaC is critical for biofilm formation.
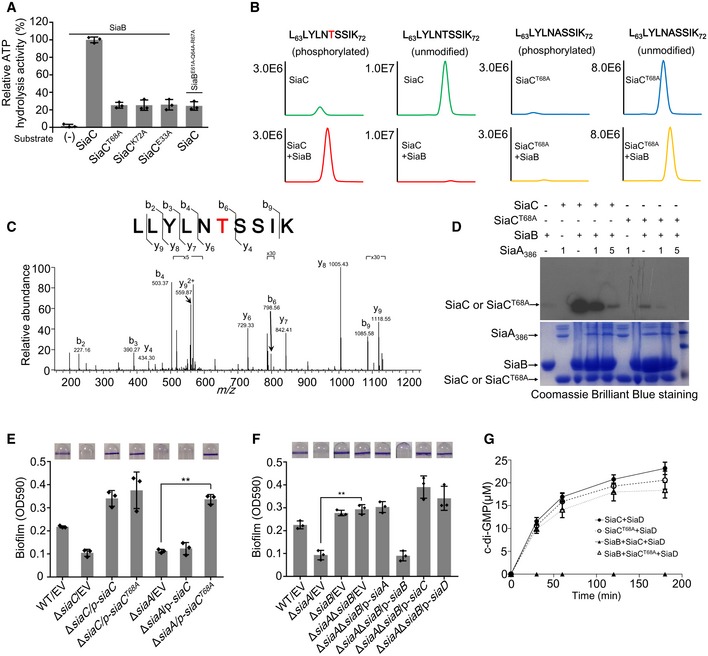
- The relative ATP hydrolysis activity of indicated sample was determined. After ATP hydrolysis reaction, the residual ATP concentration in the reaction buffer was determined by mixing the buffer with luciferase reagent. The emitted light was measured using a microplate luminometer. The amount of ATP hydrolyzed during reaction for each sample represents the ATP hydrolysis activity of each sample. The relative activity of each sample was normalized to that of the sample using SiaB and SiaC. (−) represents no substrate protein was added to reaction mixture.
- MS detection of SiaB peptide L63LYLNTSSIK72 that was covalently modified with one molecule of phosphate. Extracted ion chromatograms of the doubly protonated peptide are shown with peak intensities indicating the relative amount of either the modified or unmodified peptides.
- Determination of modification sites by collision‐induced dissociation (CID) analysis. The MS/MS spectrum of modified L63LYLNTSSIK72 is shown. The fragment ions b6 and y6 to y9 have a mass increase of 80 corresponding to phosphorylation, suggesting phosphorylation of Thr68.
- SiaB phosphorylates and SiaA dephosphorylates SiaC in vitro. SiaA or SiaAT68A was added to the reactions containing SiaB or SiaA386 and γ32P ATP. Phosphorylated products were separated by SDS–PAGE.
- Overexpression of SiaCT68A restored the ability of ΔsiaA to form biofilm. The biofilm formation of the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down).
- Deletion of ΔsiaB in the ΔsiaA background strain restored the ability of ΔsiaA to form biofilm. The biofilm formation of the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down).
- Production of c‐di‐GMP at the indicated time‐point by SiaD with SiaC or SiaCT68A was determined by HPLC. To evaluate the effect of SiaB‐mediated phosphorylation on SiaD DGC activity, SiaC or SiaCT68A was pretreated by SiaB in the presence of 1 mM ATP before reaction initiation.
Mass spectrometric analysis of SiaC in vitro in the presence of SiaB kinase confirmed the phosphorylation site at Thr68 (Fig 3B and C, and Table EV1), indicating that SiaB phosphorylates SiaC. Indeed, quantitative radioactive phosphorylation assays verified that SiaC was specifically phosphorylated by SiaB (Fig 3D). Next, we assessed the ability of SiaA to dephosphorylate SiaC. Because the expression and purification of full‐length SiaA is problematic, a truncated protein containing only the C‐terminal phosphatase domain of SiaA (SiaA386) was used. When present at sub‐stoichiometric ratios to SiaB, SiaA significantly decreased the level of phosphorylated SiaC (Fig 3D). SPR data showed that purified SiaC or SiaCP binds to SiaA386 with a K D of 19.2 nM or 46.5 nM (Fig EV2C), demonstrating slight difference for SiaA386‐SiaC and SiaA386‐SiaCP binding. The interaction between SiaCP and SiaA386 was also confirmed by pull‐down assays (Fig EV2D) in the presence of Mg2+. These data suggest that SiaA has protein phosphatase activity and dephosphorylates SiaC at the site phosphorylated by SiaB.
Because ΔsiaA was deficient in biofilm formation, whereas ΔsiaB exhibited enhanced biofilm formation, we speculated that the reversible phosphorylation of SiaC by SiaB and SiaA regulates the DGC activity of SiaD. To test this hypothesis, a strain with the point mutation T68A (Thr 68 to Ala) was tested for its ability to complement biofilm and aggregate formation by ΔsiaC and ΔsiaA. In contrast to wild‐type SiaC, SiaCT68A restored the ability of ΔsiaA to form biofilm and aggregates (Fig 3E and Appendix Fig S6C). In vitro GST pull‐down assay revealed that purified SiaCT68A was able to interact with GST‐SiaB (Fig EV2E). In addition to T68A, we also constructed a T68D mutant. Like T68A mutant, overexpression of SiaCT68D also restored the biofilm formation of ΔsiaA and ΔsiaC (Appendix Fig S6D). This result indicated that phosphorylation of SiaC by SiaB inhibits SiaC to promote biofilm formation in vivo. Next, to confirm the effect of SiaB on biofilm formation, we created a ΔsiaAΔsiaB double mutant. Although ΔsiaA was deficient in biofilm formation, further deletion of siaB (ΔsiaAΔsiaB) significantly promoted biofilm formation, similar to ΔsiaB (Fig 3F). Furthermore, HPLC was performed to evaluate the effect of SiaC phosphorylation on the activity of SiaD. Purified SiaC and SiaCT68A were pre‐incubated with SiaB before being added to the DGC reaction mixture. Pre‐incubation with SiaB rendered SiaC unable to promote the activity of SiaD (Fig 3G). However, the function of SiaCT68A was not influenced by SiaB (Fig 3G). Unlike native SiaC, both SPR and pull‐down assays revealed that SiaCP was unable to interact with SiaD (Figs 1G and EV1B). Taken together, these results provide strong evidence that SiaC is a physiological substrate for SiaB and SiaA. Furthermore, the antagonistic activity of SiaB and SiaA at Thr68 of SiaC modulates the function of that protein.
Overall structure of SiaB/SiaC complex
To understand the underlying basis for SiaC binding and phosphorylation by SiaB, we carried out crystallization studies. After extensively screening, we successfully obtained the crystals and solved the SiaB/SiaC complex structure at high resolution (2.1 Å, Appendix Table S1). The crystal belongs to P21 space group. The asymmetric unit contains four protein molecules, including two SiaB and two SiaC, assembled into a “Π” shape (Fig 4A). The two SiaB molecules exist as dimer (Appendix Fig S7), forming the top of the architecture, whereas the sides are formed by the two SiaC molecules. Both SiaB and SiaC are of α/β‐fold in nature (Fig EV3A and B). SiaB is composed of six α‐helices and five all anti‐parallel β‐sheets. The β‐strands form one flat β‐sheet, packed against α4 helix on one side. Conformations of the β‐sheet and α4 are stabilized by packing with helices α2, α3, α5, and α6. SiaC is composed of three α‐helices and six β‐strands. Except β2, the other five β‐strands are parallel to each other. Like SiaB, the β‐strands of SiaC also form one flat β‐sheet, which packed against helices α1–α3 on the same side.
Figure 4. The complex structure of SiaB/SiaC.
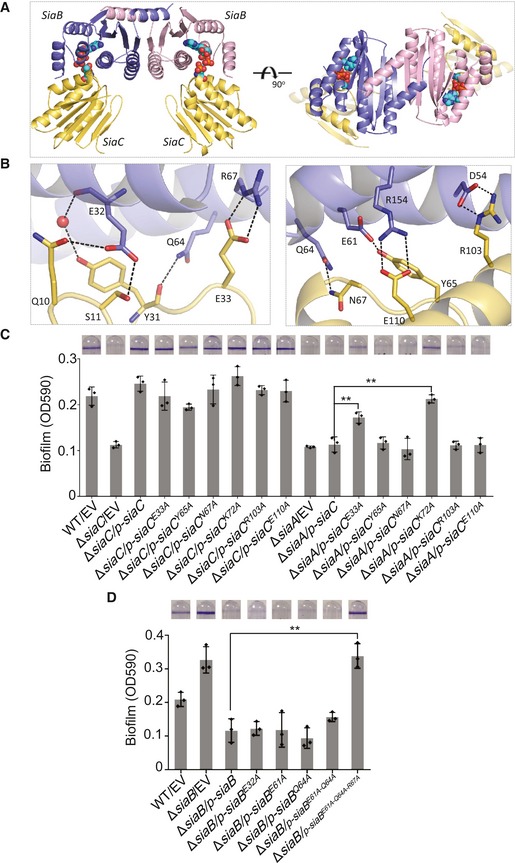
- The overall fold of SiaB/SiaC complex. The two SiaB molecules are shown as cartoons in light blue and pink, respectively. The two SiaC molecules are all shown as cartoons in yellow. ADP and TPO68 are shown as spheres in atomic colors (C, cyan; N, blue; O, red; P, orange).
- Detailed interactions between SiaB and SiaC. The interacting residues are showing as sticks. The C‐atoms of SiaB and SiaC residues are colored in light blue and yellow, respectively. H‐bonds are indicated by black dashed lines.
- SiaCE33A partially restored the biofilm formation of ΔsiaA. The biofilm formation by the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down).
- Triple mutation of E61A‐Q64A‐R67A is important for the function of SiaB. The biofilm formation by the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down).
Figure EV3. The structures of SiaB and SiaC.
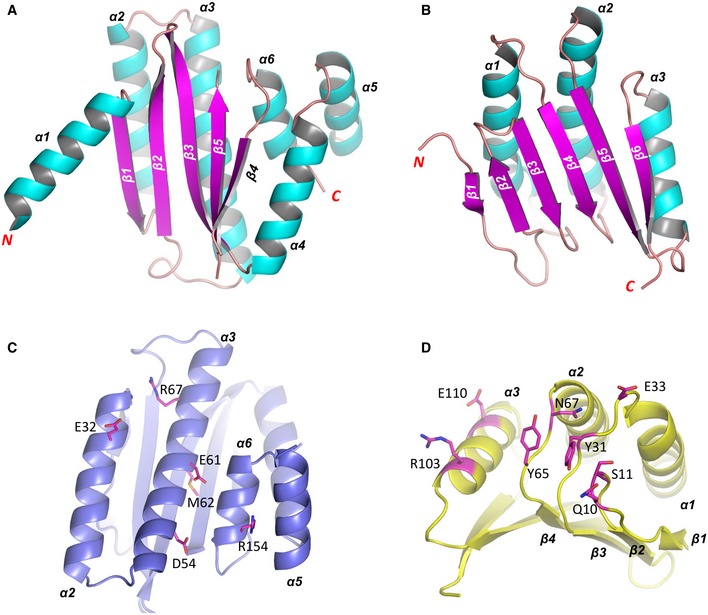
-
A, BThe overall fold of SiaB and SiaC monomers.
-
C, DThe relative orientations of residues involved in SiaB‐SiaC interactions.
The interactions between SiaB and SiaC are mainly mediated by the helices of SiaB (Fig EV3C) and the loops of SiaC (Fig EV3D). As depicted in Fig 4B, the side chain of Glu32 of SiaB forms two direct hydrogen bond (H‐bond) interactions with Gln10 and Ser11 of SiaC. Bridged one water molecule, the main chain of Glu32 of SiaB interacts with Tyr31 of SiaC, which further forms one H‐bond interaction with Gln64 of SiaB. Direct H‐bond interactions are also formed between the side chains of Glu61 and Gln64 of SiaB and Tyr65 and Asn67 of SiaC (Fig 4B). The SiaB/SiaC complex formation is further stabilized by three pairs of salt‐bridges, formed between Asp54, Arg67, and Arg154 of SiaB and Arg103, Glu33, and Glu110 of SiaC, respectively (Fig 4B).
To confirm their involvement in the formation of the SiaB‐SiaC complex, we performed a mutational analysis of a subset of these residues (E32A, E61A, Q64A, E61A‐Q64A, E61A‐Q64A‐R67A for SiaB and, E33A, R103A, E110A for SiaC) in a quantitative biofilm assay. We observed that the mutant SiaCE33A restored the ability of ΔsiaA to form biofilm, which suggested that Glu33 of SiaC is important for the phosphorylation of SiaC by SiaB (Fig 4C). Additionally, although none of the single mutations abolished the function of SiaB in vivo, the SiaBE61A‐Q64A‐R67A triple mutant failed to restore the ability of ΔsiaB to form biofilm to the level of the wild‐type strain (Fig 4D). Western blot analysis showed that the E61A‐Q64A‐R67A triple mutation did not influence SiaB expression (Appendix Fig S8). Consistently, the E61A‐Q64A‐R67A (SiaB) and E33A (SiaC) mutations significantly reduced the kinase activity of SiaC (Fig 3A). According to these data, the residues implicated in the formation of the SiaB‐SiaC complex are essential for the kinase activity of SiaB in vivo.
Basis for cofactor binding and phosphorylation
During the crystallization process of SiaB/SiaC complex, cofactor ATP was included in the protein sample. As supported by the clear electron density maps (Fig 5A), the phosphorylation reaction occurred, transforming ATP molecule into ADP. The nucleobase of ADP is bound in one deep pocket of SiaB (Fig 5B). The conformation of ADP nucleobase is stabilized by several different types of interactions (Fig 5C), including the direct H‐bond interaction between ADP N1 atom and the ND2 atom of Asn100, water‐mediated H‐bond interactions with the main chain of Met62 and side chain of Asn65, and hydrophobic stacking interactions with the side chains of Tyr69 and Leu110. The conformation of Leu110 is in turn stabilized by Phe174. The two phosphate groups of ADP sit next to the N‐terminus of α6 helix of SiaB, forming two H‐bonds [one between the O2A atom of ADP and the N atom of Leu149 and the another between the O2B atom of ADP and the N atom of Gly148 (Fig 5D)]. SiaB could bind to unphosphorylated SiaC (Fig 2C); however, SPR and pull‐down results showed that SiaB was unable to bind to SiaCP in the absence of ADP (Fig EV4A and B). We failed to purify protein SiaBL110A‐F174A or detect its kinase activity due to the insolubility of this protein when expressed in Escherichia coli. However, a biofilm formation assay using SiaB mutants showed that the L110A‐F174A double mutation abolished its function (Fig 5E). Compared to SiaB‐Flag, the expression level of SiaBL110‐F174A‐Flag was lower during biofilm formation (Appendix Fig S8). Altogether, these results indicated that the residues Leu110 and Phe174 are important for the function of SiaB.
Figure 5. ADP binding and phosphorylation.
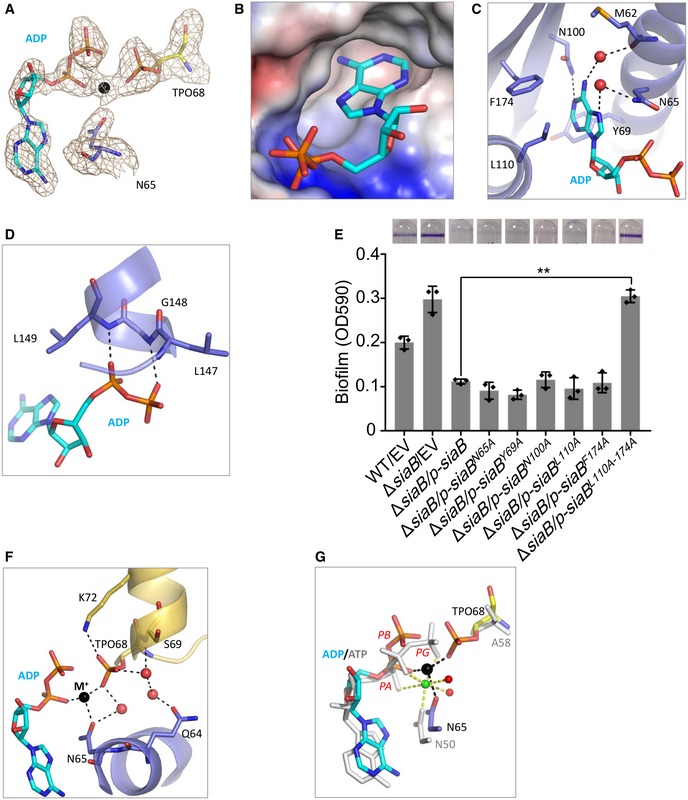
- F o−F c omit map of ADP and TPO68. The map was contoured at 2.5 σ level. ADP, TPO68 of SaiC, and Asn65 of SiaB are shown as sticks. Na+ ion is shown as black sphere.
- Surface presentation of the ADP binding pocket.
- Detailed interactions between ADP and SiaB. ADP and the interacting residues are shown as sticks.
- Interactions between ADP and residues Leu149 and Gly148. ADP and the interacting residues are shown as sticks.
- Leu110 and Phe174 residues are important for SiaB function. The biofilm formation by the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down). Data represent the means and SDs of three biological replicates. **P < 0.01 based on one‐way ANOVA test.
- Conformation of TPO68 and coordination of Na+ ion. Na+ ion and water molecules are shown as spheres in black and red, respectively.
- Comparison of the cofactors bound at the active sites of SiaB/SiaC and BsSpoIIAB/SpoIIAA complexes. For SiaB/SiaC complex, ADP, TPO68 of SaiC, and Asn65 of SiaB are all shown as sticks in atomic colors (O, red; N, blue), whereas their C‐atoms are colored in cyan, yellow, and light blue, respectively. For BsSpoIIAB/SpoIIAA complex, ATP, Ala58 (mutation of Ser58) of SpoIIAA, and Asn50 of SpoIIAB are all colored in white. Mg2+ ion and its coordinating water molecules are shown as spheres in green and red, respectively.
Figure EV4. SiaB interacts with SiaCP in the presence of ADP .
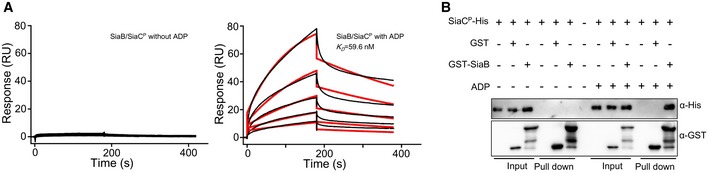
- SPR measurements of SiaCP binding at varying concentrations to SiaB with or without 2 mM ADP. SiaCP‐His specifically interacted with SiaB with a K D of 59.6 nM in the presence of ADP. Shown are measured binding responses (black) and curve fits to a 1:1 interaction (red). Plots are representative of two experiments with similar results. RU, response units; K D, dissociation constant.
- GST pull‐down shows direct interaction between SiaCP and SiaB in the presence of 2 mM ADP. Purified SiaCP was incubated with GST and GST‐SiaB individually in buffers supplemented with or without 2 mM ADP, and protein complexes were captured by glutathione beads.
Source data are available online for this figure.
Thr68 of SiaC is the targeting residue of SiaB. In the complex structure (Fig 5F), Thr68 has been phosphorylated and was termed as TPO68 hereafter. The phosphate group of TPO68 forms one H‐bond interaction with the side chain NZ atom of Lys72 of SiaC. Via one water molecules, the TPO68 phosphate group interacts with the main chain N atom of Ser69 of SiaC. Water‐mediated interactions are also formed between TPO68 phosphate group and the side chains of Gln64 and Asn65 of SiaB. The structural data suggested that Lys72 of SiaC may play an important role in SiaC phosphorylation. To further confirm this observation, we performed biofilm assay and found that SiaCK72A restores the ability of ΔsiaA to form biofilm, indicating that the mutated protein is present in vivo predominantly in the unphosphorylated state (Fig 4C). Additionally, the ATP hydrolysis activity of SiaB was significantly impaired when SiaCK72A was used as the substrate (Fig 3A). In addition to ADP, one well‐defined metal ion was observed in the complex and coordinated with the O1A atom of ADP, the O2P atom of TPO68, and the OD1 atom of Asn65 of SaiB. Through sample and crystallization condition analysis, we speculated that this was likely to be a Na+ ion captured in the structure. Notably, our ATP hydrolysis assay showed that SiaB is still active in the ammonium phosphate buffer without Na+ (Appendix Fig S6E), suggesting that Na+ ion is not strictly required for the ATP hydrolysis activity of SiaB. However, cations (Na+, NH4+, or others) could still play certain role in the function of SiaB, such as stabilizing the SiaB‐SiaC complex.
Dali search program showed that SiaB shares similar fold with some kinase proteins. The two closest matches, SpoIIAB from Bacillus stearothermophilus (Masuda et al, 2004) and SpoIIAB from Bacillus subtilis (Campbell et al, 2002), all gave a Z‐score of 11.5. The root mean square deviation (rmsd) values between SiaB and the two SpoIIAB proteins are around 2.8 Å. However, the overall sequence similarities between SiaB and SpoIIAB proteins are very low (< 10%). As depicted in Fig EV5A, SiaB can superimpose with BsSpoIIAB at the β‐strand, α2, α3, and α6 regions, whereas it is very different from BsSpoIIAB at α1, α4, and α5 regions. α1 is involved in dimerization of SiaB (Appendix Fig S7), but it is absent in SpoIIAB proteins. Instead of α‐helices, the region corresponding to α4 and α5 of SiaB folds into one short α‐turn followed by one extended loop in BsSpoIIAB (Fig EV5B). The loop was termed as ATP‐lid previously; it contains one conserved arginine residue, which interacts with the γ‐phosphate and stabilizes the conformation of ATP in the BsSpoIIAB/SpoIIAA complex structure. SpoIIAB can catalyze the phosphorylation of a Ser residue of SpoIIAA. BsSpoIIAB/SpoIIAA and our SiaB/SiaC structures represent the reaction states prior to and after phosphorylation, respectively. The orientations of the nucleobases of ADP in SiaB/SiaC complex and ATP in the BsSpoIIAB/SpoIIAA complex are very similar (Fig 5G). In the BsSpoIIAB/SpoIIAA complex, one Mg2+ was captured. In addition to ATP, the Mg2+ ion also coordinates with the side chain of Asn50. Asn50 of BsSpoIIAB corresponds to Asn65 of SiaB, and they adopt similar conformations in the two structures. Owing to the different reaction states, the conformations of the phosphate groups are slightly different. However, the relative orientations between ATP (or ADP) and the targeting residues (Thr or Ser) are similar, suggesting that SiaB and BsSpoIIAB may follow a similar mechanism in catalysis.
Figure EV5. Structural analysis of SiaB and SiaC.
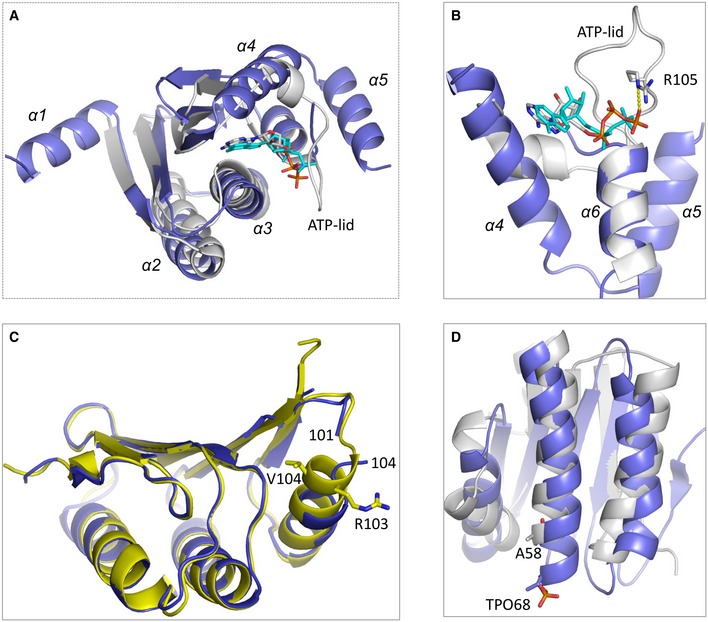
-
A, BComparison of SiaB and BsSpoIIAB. SiaB and BsSpoIIAB are shown as cartoon in blue and white, respectively. ADP in the SiaB/SiaC complex is shown as stick in cyan. ATP in the BsSpoIIAB/BsSpoIIAA complex is shown as sticks in atomic color (C, white; N, blue; O, red; P, orange).
-
CSuperposition of SiaC in the complex and apo‐structures, which are colored in yellow and blue, respectively.
-
DSuperposition of SiaC and BsSpoIIAA, which are colored in blue and white, respectively.
Besides SiaB/SiaC complex, one apo‐SiaC structure was also solved in this study (Appendix Table S1). Indicated by the very low rmsd value (0.3 Å), the overall conformations of SiaC are virtually identical in the complex and apo‐structures. However, structural superposition could reveal some conformational changes of SiaC, especially at residue 102–104 region (Fig EV5C). Arg103 of SiaC is involved in the binding with SiaB (Fig 4B). Both Glu102 and Arg103 are disordered in the apo‐structure, whereas they fold into the N‐terminus of α3 in the complex structure. Interestingly, superposition also revealed some structural similarity between SiaC and BsSpoIIAA, especially at their β‐sheet and α2 region (Fig EV5D). The rmsd value between SiaC and BsSpoIIAA is 2.9 Å and the Dali search Z‐score between the two proteins is 7.9. As described above, the phosphate group of TPO68 interacts with Lys72 of SiaC in SiaB/SiaC complex structure. However, such lysine residue is not present in BsSpoIIAA. Taken together, these results demonstrate that SiaB is a unique kinase that plays an important role for the activity of SiaC.
Discussion
The putative Ser/Thr phosphatase SiaA and the regulation of the c‐di‐GMP level by SiaD have been proposed to modulate sodium dodecyl sulfate‐induced macroscopic aggregation (Klebensberger et al, 2009). We report here that the direct interactions among SiaD, SiaC, SiaB, and SiaA constitute a signaling network that regulates biofilm formation by modulating the intracellular c‐di‐GMP level. Within this network, the direct interaction of SiaC with SiaD regulates the DGC activity of the latter, which increases c‐di‐GMP production and biofilm formation. Furthermore, the biochemical and structural data revealed that SiaB is a kinase with several unique characteristics. Both SiaB and the phosphatase SiaA reverse‐phosphorylate SiaC, which is important for regulating the DGC activity of SiaD. Based on our data, we propose a model in which SiaC plays a central role via its interactions with the other three proteins (Fig 6). Under normal conditions (such as in rich media), the kinase SiaB maintains SiaC phosphorylation, preventing SiaC‐SiaD interaction, hampering activation of the DGC activity of SiaD. Under stress conditions (such as sodium dodecyl sulfate, SDS), SiaA phosphatase is activated by an unknown signal and most SiaC remains unphosphorylated, leading to SiaD activation and c‐di‐GMP production, thus promoting the formation of aggregates and biofilm. In addition, the extensive distribution of homologues of these proteins among bacterial taxa suggests the importance of this system (Appendix Fig S9).
Figure 6. Proposed model of the SiaABCD signaling network involved in regulation of biofilm and aggregate formation.
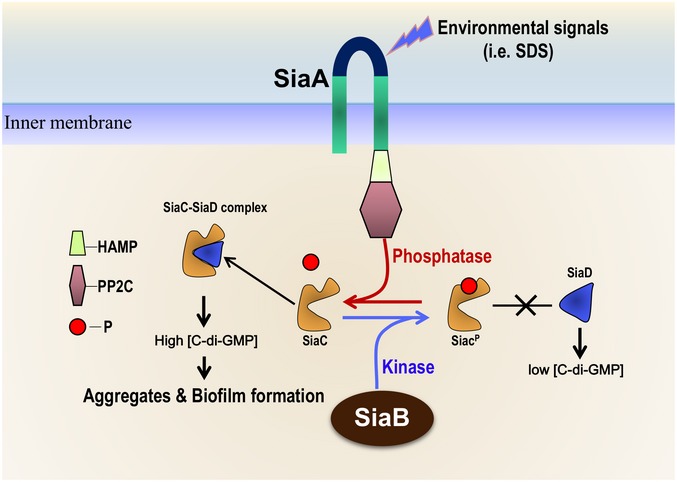
Under normal conditions (such as in rich media), the protein kinase SiaB dominates and maintains most SiaC phosphorylated, which failed to bind to SiaD or induce the DGC activity of SiaD; therefore, c‐di‐GMP is insufficient to activate the biofilm and aggregate formation. Once under stress conditions (such as SDS stimulation), SiaA phosphatase is activated and most SiaC stays unphosphorylated; in this situation, the unphosphorylated SiaC promotes the DGC activity of SiaD and produces sufficient c‐di‐GMP, which leads to aggregate and biofilm formation.
SiaC seems to be a key player in the signaling pathway. SiaD alone exhibited weak DCG activity (Fig 1H), consistent with a previous report of the lack of c‐di‐GMP in an extract of a siaD‐overexpressing strain (Kulasakara et al, 2006). Additionally, direct interaction of SiaC with SiaD regulates the DGC activity of the latter, consistent with the observation in vivo that siaC is required for the SiaD‐mediated promotion of biofilm formation (Fig 1D). In P. aeruginosa, the DGC WspR is activated by phosphorylation and the activity of SadC is controlled by its transmembrane domain and the presence of oxygen (Hickman et al, 2005; Schmidt et al, 2016; Zhu et al, 2016a). Post‐translational modulation of the activity of SiaD by its direct interaction with SiaC is in agreement with these reports. However, it is unlikely that direct interaction with SiaC stabilizes SiaD or represses the allosteric inhibition by c‐di‐GMP (Appendix Fig S2D and Fig 1D). The mechanism by which the interaction between SiaC and SiaD regulates the DGC activity of the latter is unclear. The SiaD‐SiaC interaction may alter the conformation of SiaD to promote its binding to GTP, thus producing c‐di‐GMP. Further studies of the underlying molecular mechanisms are needed.
Protein phosphorylation is a ubiquitous regulatory mechanism that affects multiple cellular processes (Hunter, 1995; Taylor & Kornev, 2011). During surface‐bound growth of P. aeruginosa, the hybrid response regulator/DGC, WspR, is activated via phosphorylation by the Wsp chemosensory system (Hickman et al, 2005; Guvener & Harwood, 2007). Activation of DGC HsbD in P. aeruginosa (Valentini et al, 2016) and BgrR in Sinorhizobium meliloti (Baena et al, 2019) is accomplished via phosphorylation by the individual binding partner. In this study, we revealed a similar signaling network for modulation of SiaD activity. Given the wide distribution of homologues of SiaC (Appendix Fig S9), the activation model for DGC revealed here should be universal among bacteria.
SiaC is modified post‐translationally by the SiaB kinase and SiaA phosphatase. No interaction between SiaB and SiaD was observed (Fig 2A); therefore, SiaB cannot directly modify SiaD. Additionally, because the GST pull‐down data showed that SiaB can interact with SiaCT68A (Fig EV2E), it seems unlikely that SiaB prevents the activation of SiaD by sequestering SiaC. Several lines of evidence indicate that reversible phosphorylation of SiaC by SiaB and SiaA is essential for the modulation of SiaD DGC activity. First, the ΔsiaA mutant was deficient in biofilm formation, including in the presence of a plasmid overexpressing siaC or siaD, whereas further deletion of SiaB (ΔsiaAΔsiaB) significantly promoted biofilm formation (Fig 3F). Second, overexpression of SiaC restored the ability of ΔsiaA to form biofilm when the target residue of phosphorylation, Thr68, was substituted for alanine (Fig 3E). Third, phosphorylated SiaC was unable to interact with SiaD (Figs 1G and EV1B). Finally, HPLC showed that pre‐incubation with the kinase SiaB inhibits the promotion of the DGC activity of SiaD by SiaC, whereas the function of SiaCT68A was not affected by SiaB (Fig 3G). Moreover, we solved the crystal structure of SiaB/SiaC complex, demonstrating SiaB is a kinase and can phosphorylate the Thr68 residue of SiaC. Consistently, the mass spectrometric quantitative radioactive phosphorylation assays showed that the kinase SiaB specifically phosphorylate SiaC. Though SiaB shares some similarities in folding and catalytic mechanism with other protein kinases, it possesses some very unique structural features, especially at the ATP binding region. Many protein kinases contain a ATP‐lid loop, which is flexible and can undergo large conformational changes upon the binding (or dissociation) of cofactor ATP (or ADP), whereas SiaB has two rigid helices (α4 and α5) surrounding the ATP binding region. Compared to other kinases, SiaB lacks the ATP‐interacting Arg residue within the loop region. However, our structure and in vitro data suggested that Lys72 of SiaC plays a critical role in the SiaB‐catalyzed phosphorylation reaction. Taken together, we conclude that SiaB is a unique kinase and it may follow a substrate‐mediated mechanism in catalysis.
SiaA harbors a PP2C‐like phosphatase domain, a typical output domain in bacterial transmembrane receptors that respond to energy stress, such as RsbP of B. subtilis (Vijay et al, 2000). SiaA and SiaD might respond to the extracellular polysaccharide Psl in mixed‐species biofilms (Irie et al, 2012; Chew et al, 2018). However, overexpression of SiaC or SiaA in the ΔpslBCD mutant displays the similar biofilm phenotype as the wild‐type parent, indicating activation of SiaD by SiaC in vivo is independent on the polysaccharide Psl, at least in the present experiment conditions (Appendix Fig S10). The direct interaction between SiaC and the PP2C‐like domain of SiaA and the results of radioactive phosphorylation assays suggest that SiaC is the downstream target of SiaA. Because SiaA is essential for SiaC‐mediated promotion of the formation of biofilm and aggregates, dephosphorylation by SiaA should be important for the activation of SiaD by SiaC.
In conclusion, we identified a novel signaling network that regulates biofilm formation by modulating the activity of SiaD and the intracellular level of c‐di‐GMP, wherein SiaC switches the DGC activity of SiaD “on” and “off” after receiving an upstream signal from SiaB or SiaA. P. aeruginosa may use this signaling network to rapidly respond and adapt to environmental changes.
Materials and Methods
Bacterial strains and culture conditions
Bacterial strains, plasmids, and primers used are listed in Appendix Tables S2 and S3. Strains were incubated in LB medium or M9 salt (6.78 g/l Na2HPO4, 3 g/l KH2PO4, 0.5 g/l NaCl, 1.0 g/l NH4Cl) with succinate (10 mM) or SDS (0.1%) as the sole carbon source. When needed, appropriate antibiotics were supplemented: tetracycline (100 μg/ml) and carbenicillin (150 μg/ml) for P. aeruginosa, and tetracycline (20 μg/ml) and carbenicillin (100 μg/ml) for E. coli (Sangon life sciences).
Plasmids construction
To construct mini‐CTX‐lacZ‐siaD‐Flag plasmid, the DNA fragments containing siaA promoter, siaD coding region, and Flag‐tag encoding sequence were generated by overlap PCR using mini‐siaD‐F1/R1 and mini‐siaD‐F2/R2 primer pairs. The DNA fragments were then cloned into mini‐CTX‐lacZ vector (Becher & Schweizer, 2000). The purified PCR products amplified by pUC‐siaD‐F/pUC‐siaD‐R primer pair were digested with BamH1 and HindIII enzymes and cloned into pUCP20 plasmid (West et al, 1994), yielding pUC‐siaD plasmid. Plasmids pUCP‐siaA, pUCP‐siaC, and pUCP‐siaB were constructed similarly. The pMMB67EH‐Flag vector was constructed based on pMMB67EH to overexpress Flag‐tagged genes. Vector pMMB67EH was digested with HindIII before blunted by T4 DNA polymerase (Transgene, Beijing, China). The blunted fragment was then digested with PstI enzyme. DNA fragments containing Flag tag coding sequence were amplified by primers flag‐up and flag‐dn, digested with PstI and EcoRV and cloned into the PstI‐digested pMMB67EH. For construction of pMMB67EH‐siaD‐Flag plasmid, the purified PCR products amplified by pMM‐siaD‐F/pMM‐siaD‐R were digested with BamH1 and SalI enzymes and cloned into the BamH1‐XhoI digested pMMB67EH‐Flag plasmid fragment. Accordingly, the coding sequences of siaC, siaB, siaA, siaA 337, and siaA 386 were amplified with the corresponding primer pairs and cloned into the pMMB67EH‐Flag vector after enzymatic digestion.
Construction of Pseudomonas aeruginosa deletion mutants
In‐frame deletion mutants were generated as described previously (Hoang et al, 1998). To construct the siaC deletion mutant, PCRs were performed to amplify sequences upstream (1,724) and downstream (1,499) of the intended deletion with primer pairs siaC‐H1F/H1R and siaC‐H2F/H2R. The homologous arms were digested and cloned into EcoRI/HindIII‐digested vector pEX18Tc, yielding pEX‐siaC. The resulting plasmid was transformed into PAO1 with selection for tetracycline‐resistant first homologous recombinants. Colonies were then streaked on LB agar plates with 10% sucrose to select second homologous recombinants. And the ΔsiaC mutant was identified by PCR using primers siaC‐H1F and siaC‐H2R. For generating ΔsiaA, ΔsiaD, ΔsiaB, ΔsiaCΔsiaB deletion mutants, similar strategy was applied. All resultant mutants were verified by PCR and DNA sequencing.
Biofilm formation assay
Biofilm formation assay was performed as described previously with modification (Zhu et al, 2016b). Briefly, overnight cultures grown in LB medium were diluted (1:1,000) into 1‐ml fresh LB medium in glass tube and incubated statically at 25°C for 22 h. Biofilms attached to the sides of the glass tubes were washed twice gently with sterile water and stained with crystal violet (CV). Quantification of biofilm biomass was performed by dissolution of the CV‐stained biofilm with ethanol, and the CV solution was measured at an absorbance of OD590 nm. The assay was performed at least three times with a minimum of three replicates for each strain.
SDS‐induced macroscopic aggregation
Macroscopic aggregation was performed as previously described with slight modification (Klebensberger et al, 2009). Briefly, overnight cultures of bacterial strains were diluted (1:100) into 2 ml fresh M9 salt with 10 mM succinate or 0.1% SDS as sole carbon source. Macroscopic aggregation was tested after 12‐h growth in glass tubes on a rotary shaker at 180 rpm at 37°C. The aggregations were gently transferred into 24‐well plate for photographing.
RNA extraction and real‐time quantitative PCR
Overnight bacterial cultures were sub‐cultured in fresh LB medium until stationary phase (OD600 = 2.0). The bacterial strains were collected by centrifugation at 12,000 g for 2 min. Total RNA was isolated with RNAprep pure cell/bacteria kit (Tiangen Biotech, Beijing, China). cDNA was synthesized from each RNA sample using a PrimerScript Reverse Transcriptase (TaKaRa, Dalian, China) with random primer, then subjected to qRT–PCR using SYBR Premix Ex Taq II (TaKaRa). The 30s ribosomal protein gene rpsL was used as an internal control.
Bacterial two‐hybrid experiments
The bacterial two‐hybrid assays were performed as previously described (Zhao et al, 2016). Briefly, the bait and prey plasmids were co‐transformed into E. coli reporter strain before plated onto selective and nonselective media. After grown on M9 + His‐deficient medium containing 5 mM 3‐amino‐1,2,4‐triazole (3‐AT) for 24–48 h at 30°C, colonies appeared on these plates were selected. Positive colonies were streaked on M9+ His‐deficient medium containing 5 mM 3‐AT and 12.5 μg/ml streptomycin and subject to gradient dilution and plate inoculation.
Protein purification
To purify GST‐SiaA386, GST‐SiaB, GST‐SiaC, and GST‐SiaD, the pGEX6p‐siaA 386, pGEX6p‐1‐siaB, pGEX6p‐1‐siaC, or pGEX6p‐1‐siaD was transformed into E. coli BL21 (DE3), respectively. Bacteria were cultured at 37°C in LB medium to an OD600 of ~ 0.6 before shifted to 16°C and induced with 0.5 mM IPTG and then cultivated for an additional 20 h at 16°C. Harvested cells were resuspended in GST buffer A (50 mM Tris–HCl, pH 8.0 150 mM NaCl) and sonicated, and proteins were purified with the 5‐ml GSTrap HP column (GE Healthcare) using GST buffer B (50 mM Tris–HCl, pH 8.0 150 mM NaCl, 10 mM glutathione). For purification of Sumo‐SiaD, Sumo‐SiaDR130G, Sumo‐SiaC, and Sumo‐SiaB proteins, the corresponding plasmid was constructed and proteins were purified with 5‐ml HisTrap HP column (GE Healthcare, USA) using His buffer A (10 mM Tris–HCl, pH 8.0, 500 mM NaCl, 1 mM DTT) and His buffer B (10 mM Tris–HCl, pH 8.0, 500 mM NaCl, 1 mM DTT, 500 mM imidazole) according to manufacture's instructions. The purified proteins were then digested by UlpI protease to remove the Sumo. For size exclusion chromatography, the purified Sumo‐SiaD and Sumo‐SiaC or Sumo‐SiaB and Sumo‐SiaC were mixed for 1 h before UlpI protease digestion. The samples were loaded onto a Ni‐NTA resin column again, and the flow‐through containing the target proteins was collected. The samples were concentrated and passed through a 10/300 Superdex 200 size exclusion column (GE Healthcare, USA) equilibrated with SEC buffer (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM DTT).
To purify SiaC‐His and SiaCT68A‐His, the pET28a‐siaC and pET28a‐siaC T68A were constructed and proteins were purified with 5‐ml HisTrap HP column (GE Healthcare) using His buffer A and His buffer B according to manufacture's instructions. To purify phosphorylated SiaC‐His (SiaCP‐His), 10 mg purified SiaC‐His protein was treated with 2 mg purified SiaB for 4 h at room temperature in the presence of 1 mM ATP. After incubation, SiaCP‐His was separated by a 10/300 Superdex 200 size exclusion column equilibrated with SEC buffer.
GST pull‐down assay
To verify the interaction between SiaC and SiaD, 100 μg purified GST‐SiaC was mixed with 20‐μl prewashed glutathione magnetic beads (Promega, Madison, USA) on a rotator for 2 h at 4°C before washed three times by reaction buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.1% Triton X‐100). To avoid non‐specific interaction, the beads were then blocked with 5% skim milk (m/v) 1 h at 4°C before washed three times by reaction buffer. SiaD‐Flag expressed in PAO1 harboring pMMB67EH‐siaD‐Flag was cultured in LB medium with 0.5 mM IPTG and 30‐ml stationary phase bacterial cultures were collected and lysed by sonication (Scientz Biotechnology, Ningbo, China) in reaction buffer. After addition of cell lysate to the pre‐blocked beads, binding was allowed to proceed on a rotator for 4 h at 4°C. The beads were then washed five times with reaction buffer. Retained proteins were detected by Western blotting after SDS–PAGE. To determine SiaA‐SiaC, SiaB‐SiaC interactions, a similar strategy was applied.
To test the interaction between SiaB and SiaC or SiaCT68A, 50 μg purified GST‐SiaB was mixed with 15‐μl prewashed glutathione magnetic beads on a rotator for 2 h at 4°C before washed three times by reaction buffer. To avoid non‐specific interaction, the beads were then blocked with 5% skim milk (m/v) 1 h at 4°C before addition of purified SiaC‐His, SiaCP‐His, or SiaCT68A‐His. After 6 times washing by reaction buffer, samples were subject to Western blotting assays. To test the interaction between SiaD and SiaC or SiaCP, a similar strategy was applied using purified GST‐SiaD, SiaC‐His, and SiaCP‐His. For SiaB‐SiaCP interaction, experiments were performed in reaction buffer with or without 2 mM ADP. For SiaA‐SiaCP interaction, experiments were performed in reaction buffer with or without 10 mM MgCl2.
To test the formation of SiaA‐SiaC‐SiaB, 50 μg purified GST‐SiaA386 was mixed with 15‐μl prewashed glutathione magnetic beads on a rotator for 2 h at 4°C before washed three times by reaction buffer. To avoid non‐specific interaction, the beads were then blocked with 5% skim milk (m/v) 1 h at 4°C before addition of purified SiaC‐His. After 2‐h incubation, cell lysates of DH5α containing SiaB‐Flag were added and incubated for another 2 h. After six times washing by reaction buffer, samples were subjected to Western blotting assays. To test formation of SiaA‐SiaC‐SiaD (or SiaB‐SiaC‐SiaD), a similar strategy was applied using GST‐SiaA386 (or GST‐SiaB), SiaC‐His, and cell lysates of DH5α‐containing SiaD‐Flag.
Surface plasmon resonance (SPR) experiments
Surface plasmon resonance (SPR) experiments were performed to measure the binding kinetic parameters between proteins. After the activation of the surface using NHS and EDC (1:1, v/v), the protein was immobilized on a CM5 sensor chip (GE Healthcare) by using standard amine‐coupling at 25°C with buffer PBS‐P (20 mM phosphate buffer, 2.7 mM NaCl, 137 mM KCl, 0.05% surfactant P‐20, pH 7.4). The protein concentration was fixed at 50 ng/μl, and the immobilization level of protein was about 5,000 RU (response unit, 1 RU response value is roughly equivalent to 1 pg/mm2 change of the concentration of the bound substance on the chip surface). After coupling, unreacted NHS ester groups were blocked with ethanolamine. After equilibration by running buffer (10 mM Tris–HCl, 150 mM NaCl, 0.05% surfactant P‐20, pH 7.5), different concentrations of analytes were serially injected into the channel to evaluate the binding kinetic parameters. A reference channel was only activated and blocked to eliminate compound unspecific binding to the surface of the chip. An extra wash with 2 mM NaCl was added to remove the last remaining sample in the pipeline. The on‐rates and the off‐rates of the compounds were obtained by fitting the data sets to 1:1 Langmuir binding model using the Biacore T200 Evaluation Software (GE Healthcare). For interaction of SiaA with SiaC or SiaCP, SiaA386 was immobilized and experiments were performed in buffer with 10 mM MgCl2. For SiaB‐SiaCP interactions, SiaB was immobilized and experiments were performed in buffers with or without 2 mM ADP.
SiaD stabilization assays
Overnight culture of wild‐type PAO1, siaC mutant harboring pUCP20‐siaD‐Flag plasmid was sub‐cultured into fresh LB broth to an OD600 of 1.5, followed by treatment with 50 μg/ml streptomycin. Bacterial samples were collected at 0, 30, 60, 90, 120, 180 min, and subjected to SDS–PAGE and Western blotting.
Western blot analysis
Samples were subjected to electrophoresis on 12% or 15% SDS–PAGE and transferred onto polyvinylidene difluoride membranes. After blocking with 5% (w/v) skim milk in TBST buffer (50 mM Tris, 150 mM NaCl, 0.05% Tween‐20, pH 7.4), membranes were incubated with the appropriate primary antibody: anti‐Flag (Sigma, Germany), anti‐GST (Zhongshan Golden Bridge Biotechnology, Beijing, China), anti‐RNA pol α (Biolegend, California, USA). After three times wash in TBST buffer, the membranes were incubated with horseradish peroxidase‐conjugated Rabbit (TransGen, Beijing, China, for GST) or Mouse (TransGen, for Flag and RNAP) antibodies. Signals were detected using the ECL kit following the manufacture's specified protocol.
Quantification of c‐di‐GMP production
Synthesis of c‐di‐GMP was carried out in 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, and 5 mM MgCl2. Subsequently, 3.5 μM SiaD alone or 3.5 μM SiaD with 7 μM SiaC was added to the reaction mixture for 1 h at room temperature. Reactions using 0.35 μM WspR alone or 0.35 μM SiaD with 7 μM siaC for 10 min were used as a control. To test the effect of SiaB on the SiaD activation, 7 μM SiaC or SiaCT68A was pre‐incubated with 2 μM SiaB in the presence of ATP (1 mM) for 2 h at room temperature. Reactions were initiated by the addition of 50 μM GTP to the mixture and allowed to incubate 3 h at 37°C. Reactions were terminated by heating the samples at 95°C for 5 min. Precipitated proteins were removed by centrifugation after which the supernatant was filtered through a 0.22‐mm membrane. 15 μl of sample was loaded for high‐performance liquid chromatography (Shimadzu, Japan), with 254 nm as detection wavelength. Symmetry C18 Column (4.6 mm × 25 cm) (Waters) was used with solvent A (10 mM ammonium acetate in water) and solvent B (10 mM ammonium acetate in methanol) at a flow rate of 0.2 ml/min. Eluent gradient is as follows: 0–9 min with 1% B; 9–14 min with 15% B; 14–19 min with 25% B; 19–35 min with 1% B. For SiaDR130G, a similar strategy was applied.
Quantification of intracellular c‐di‐GMP level
Extraction of c‐di‐GMP was conducted as previously described with some modifications (Chen et al, 2015). Bacterial culture was prepared in ABTGC growth medium in 37°C shaker, with three replicates for each strain. Bacterial cells were harvested at early stationary stage for c‐di‐GMP extraction. Briefly, cell pellets were washed by pre‐chilled 1 mM ammonium acetate for twice and were soaked in extraction buffer (acetonitrile:methanol:H2O 2:2:1) overnight. Thereafter, three cycles of snap freeze–thaw and ice water sonication were carried out to extract raw metabolites. High‐speed centrifugation (1,080 g at 4°C for 20 min) was applied to separate metabolites from the raw extracts. The supernatant of each sample was concentrated by speed‐vacuum centrifugation for LC‐MS/MS analysis, and the pellets were used for protein quantification and sample normalization.
The c‐di‐GMP in samples and c‐di‐GMP standard were analyzed in negative full scan mode with liquid chromatography coupled to hybrid quadrupole‐Orbitrap mass spectrometer (Thermo Scientific Q Exactive). Waters BEH C18 column (2.1 × 50 mm, 1.7 μm) was used to separate 5 μl samples with phase A (10 mM ammonium formate and 0.1% formic acid in H2O) and phase B (0.1% formic acid in MeOH) as mobile phases, and the flow rate was 0.3 ml/min. The gradient was as follows: Phase B was increased to 60% in 4.5 min from 3% at initial, then held for 0.5 min, returned back to 3% and held for 1.2 min. The capillary temperature of HESI source was 320°C, and the spray voltages were both 3 KV in negative mode. The sheath gas and aux gas were 30 and 10 psi separately. The standard c‐di‐GMP at 1 μg/ml was injected after every six injections as quality control. Data were analyzed using Xcalibur 2.2 software (Thermo Scientific).
Determination of ATP hydrolysis activity
10 μM SiaB or its mutants, 30 μM SiaC or its mutants, and 30 μM ATP were added to reaction buffer (10 mM Tris–HCl, pH 8.0, 10 mM NaCl) at room temperature. After 40 min, the concentration of residual ATP was determined using ATP assay kit (Beyotime Biotechnology, Beijing, China; Su et al, 2014). Briefly, the residual ATP concentration in the reaction buffer was determined by mixing the buffer with luciferase reagent. The emitted light was measured using a microplate luminometer. The amount of ATP hydrolyzed during reaction for each sample represents the activity for each sample. The relative activity of each sample was normalized to that of the sample using SiaB and SiaC. 1 mM EDTA was added to reaction buffer when indicated. To test whether Na+ is essential for the ATP hydrolysis of SiaB, purified SiaB and SiaC was desalted by ammonium phosphate buffer (50 mM ammonium phosphate, pH 7.5) before reaction initiation.
Mass spectrum
Upon SDS–PAGE fractionation, the band of interest was excised and subjected to in‐gel trypsin digestion as previously described1. Nanoflow reversed‐phase LC separation was carried out on an EASY‐nLC 1200 system (Thermo Scientific). The capillary column (75 μm × 150 mm) with a laser‐pulled electrospray tip (Model P‐2000, Sutter Instruments) was home‐packed with 5 μm, 100 Å Magic C18AQ silica‐based particles (Michrom BioResources Inc., Auburn, CA). The mobile phase was composed of Solvent A (97% H2O, 3% ACN, and 0.1% FA) and Solvent B (20% H2O, 80% ACN and 0.1% FA). The LC separation was carried out at room temperature with the following gradient: Solvent B was started at 7% for 3 min and then raised to 40% over 40 min; subsequently, Solvent B was rapidly increased to 90% in 2 min and maintained for 10 min before 100% Solvent A was used for column equilibration. Eluted peptides from the capillary column were electrosprayed directly into a linear ion trap mass spectrometer (LTQ Velos Pro, Thermo Scientific) for MS and MS/MS analyses in a data‐dependent acquisition mode. One full MS scan (m/z 400‐1200) was acquired, and then, MS/MS analyses were performed on the 10 most intense ions. The selected ions were fragmented by collision‐induced dissociation (CID) in the ion trap with the following parameters: ≥ +2 precursor ion charge, 2 Da precursor ion isolation window, and 35% normalized collision energy. Dynamic exclusion was set with repeat duration of 24 s and exclusion duration of 12 s. Peptides and proteins were assigned by Mascot (2.3.02).
In vitro radioactive phosphorylation assays
For autokinase activity assays, full‐length SiaB was incubated with 10 μCi [γ‐32P] ATP (PerkinElmer) in 20 μl of reaction buffer (10‐mM Tris–HCl, pH 7.4, 50‐mM KCl, 5‐mM MgCl2, 10% glycerol) for 1–5 min at room temperature.
To detect the phosphotransferase activity of SiaB toward SiaC/SiaCT68A, and the phosphatase activity of SiaA, SiaB was autophosphorylated as above, and then, 10 μM of purified SiaC/SiaCT68A and SiaA was added into the reaction mixture, which was incubated at room temperature for the appropriate time. The reaction was stopped with 5 × SDS–PAGE loading buffer. The phosphorylated proteins were separated by 12% SDS–PAGE. After electrophoresis, SDS–PAGE gels were dried by gel dryer and exposed to a X‐OMAT BT Film (Kodak) for 3 h. After autoradiography, gels were stained by Coomassie Brilliant Blue to check the protein amounts.
Crystallization and data collection
Both SiaC and Se‐substituted SiaB/SiaC samples in gel filtration buffer were concentrated to 10 mg/ml. The crystallization samples of SiaB/SiaC were prepared by mixing SiaB/SiaC and 2 mM ATP. The crystallization conditions were identified at 18°C using the Gryphon crystallization robot system from Art Robbins Instrument company and crystallization kits from Hampton Research Company. The apo‐SiaC crystals were grown in buffer composed of 0.4 M NaH2PO4/1.6 M K2HPO4, 0.1 M imidazole pH 8.0, 0.2 M NaCl. The crystals of SiaB/SiaC complex were grown in 0.1 M Bis‐Tris pH 6.5, 25% PEG 3350. All crystals were cryoprotected using their mother liquor supplemented with 25% glycerol and snap‐frozen in liquid nitrogen. X‐ray diffraction data (Appendix Table S1) were collected on beamline BL17U and BL19U at the Shanghai Synchrotron Radiation Facility (SSRF). Data processing was carried out using the HKL2000 or XDS programs (Minor et al, 2006).
Structure determination and refinement
The phase of Se‐substituted SiaB/SiaC structure was determined by single‐wavelength anomalous diffraction (SAD) method (Giacovazzo & Siliqi, 2004) using the anomalous signal of Se‐ atoms with AutoSol program (Terwilliger et al, 2009) embedded in the Phenix suite (Adams et al, 2002). The initial model was refined using Refmac5 program (Murshudov et al, 2011) of CPP4i suite (Potterton et al, 2003). The apo‐SiaC structure was solved by molecular replacement method; the SiaC molecule of the complex was used as search model. The model was manually built using COOT (Emsley & Cowtan, 2004) and refined using either Refmac5 or phenix.refine programs (Afonine et al, 2012). During refinement, 5% of randomly selected data was set aside for free R‐factor cross‐validation calculations. The 2F o –F c and F o –F c electron density maps were regularly calculated and used as guides for building the missing amino acids and solvent molecules with COOT. The structural refinement statistics are summarized in Appendix Table S1.
Author contributions
GC and HL designed research. GC, YZ, JP, ML, WH, and YX performed the experiments. GC, CY, XD, and TW analyzed data; GC, JG, and HL wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Acknowledgements
We are grateful to BL17U and BL19U beamline staff at the Shanghai Synchrotron Radiation Facility for their expert assistance during X‐ray diffraction data collection. This work was supported the National Natural Science Foundation of China (31622003, 31670080, and 31870060 to HL and 31700064 to GC), Natural Science Basic Research Program of Shaanxi (2019JQ‐134), the Program for Changjiang Scholars and Innovative Research Team in University (IRT1174 and ITR_15R55). We thank other members of the Liang laboratory for providing reagents, general support and critical reading of the manuscript. We thank Jing Wang and Qian Wang of Beijing University for help in SPR experiments.
The EMBO Journal (2020) 39: e103412
Data availability
The atomic coordinates and structure factors for the reported crystal structures of SiaB‐SiaC‐ADP complex and SiaC have been deposited in the Protein Data Bank (PDB) under accession numbers 6KKO (http://www.rcsb.org/pdb/explore/explore.do?structureId=6KKO) and 6KKP (http://www.rcsb.org/pdb/explore/explore.do?structureId=6KKP).
References
- Adams PD, Grosse‐Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Afonine PV, Grosse‐Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Ponting CP (1999) The cytoplasmic helical linker domain of receptor histidine kinase and methyl‐accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett 176: 111–116 [DOI] [PubMed] [Google Scholar]
- Baena I, Perez‐Mendoza D, Sauviac L, Francesch K, Martin M, Rivilla R, Bonilla I, Bruand C, Sanjuan J, Lloret J (2019) A partner‐switching system controls activation of mixed‐linkage beta‐glucan synthesis by c‐di‐GMP in Sinorhizobium meliloti . Environ Microbiol 21: 3379–3391 [DOI] [PubMed] [Google Scholar]
- Baraquet C, Murakami K, Parsek MR, Harwood CS (2012) The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c‐di‐GMP. Nucleic Acids Res 40: 7207–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher A, Schweizer HP (2000) Integration‐proficient Pseudomonas aeruginosa vectors for isolation of single‐copy chromosomal lacZ and lux gene fusions. Biotechniques 29: 948–950, 952 [DOI] [PubMed] [Google Scholar]
- Brencic A, Lory S (2009) Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72: 612–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Masuda S, Sun JL, Muzzin O, Olson CA, Wang S, Darst SA (2002) Crystal structure of the Bacillus stearothermophilus anti‐sigma factor SpoIIAB with the sporulation sigma factor sigmaF. Cell 108: 795–807 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yuan M, Mohanty A, Yam JK, Liu Y, Chua SL, Nielsen TE, Tolker‐Nielsen T, Givskov M, Cao B et al (2015) Multiple diguanylate cyclase‐coordinated regulation of pyoverdine synthesis in Pseudomonas aeruginosa . Environ Microbiol Rep 7: 498–507 [DOI] [PubMed] [Google Scholar]
- Chew SC, Yam JKH, Matysik A, Seng ZJ, Klebensberger J, Givskov M, Doyle P, Rice SA, Yang L, Kjelleberg S (2018) Matrix polysaccharides and siad diguanylate cyclase alter community structure and competitiveness of Pseudomonas aeruginosa during dual‐species biofilm development with Staphylococcus aureus . MBio 6: e00585‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Mechold U, Nevenzal H, Yarmiyhu Y, Randall TE, Bay DC, Rich JD, Parsek MR, Kaever V, Harrison JJ et al (2015) Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa . Proc Natl Acad Sci USA 112: 11359–11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley B, Dederer V, Carnell M, Kjelleberg S, Rice SA, Klebensberger J (2016) SiaA/D interconnects c‐di‐GMP and RsmA signaling to coordinate cellular aggregation of Pseudomonas aeruginosa in response to environmental conditions. Front Microbiol 7: 179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Giacovazzo C, Siliqi D (2004) Phasing via SAD/MAD data: the method of the joint probability distribution functions. Acta Crystallogr D Biol Crystallogr 60: 73–82 [DOI] [PubMed] [Google Scholar]
- Guvener ZT, Harwood CS (2007) Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic‐di‐GMP in response to growth on surfaces. Mol Microbiol 66: 1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS (2005) A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA 102: 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff‐Schweizer RR, Kutchma AJ, Schweizer HP (1998) A broad‐host‐range Flp‐FRT recombination system for site‐specific excision of chromosomally‐located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212: 77–86 [DOI] [PubMed] [Google Scholar]
- Hunter T (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80: 225–236 [DOI] [PubMed] [Google Scholar]
- Irie Y, Borlee BR, O'Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR (2012) Self‐produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa . Proc Natl Acad Sci USA 109: 20632–20636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Reinders A, Lori C (2017) Cyclic di‐GMP: second messenger extraordinaire. Nat Rev Microbiol 15: 271–284 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebensberger J, Birkenmaier A, Geffers R, Kjelleberg S, Philipp B (2009) SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa . Environ Microbiol 11: 3073–3086 [DOI] [PubMed] [Google Scholar]
- Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y et al (2006) Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis‐(3′‐5′)‐cyclic‐GMP in virulence. Proc Natl Acad Sci USA 103: 2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Murakami KS, Wang S, Anders Olson C, Donigian J, Leon F, Darst SA, Campbell EA (2004) Crystal structures of the ADP and ATP bound forms of the Bacillus anti‐sigma factor SpoIIAB in complex with the anti‐anti‐sigma SpoIIAA. J Mol Biol 340: 941–956 [DOI] [PubMed] [Google Scholar]
- Meek RW, Cadby IT, Moynihan PJ, Lovering AL (2019) Structural basis for activation of a diguanylate cyclase required for bacterial predation in Bdellovibrio. Nat Commun 10: 4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006) HKL‐3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62: 859–866 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, Lee VT (2015) Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic‐di‐GMP turnover. Proc Natl Acad Sci USA 112: E5048–E5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr 59: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Quin MB, Berrisford JM, Newman JA, Basle A, Lewis RJ, Marles‐Wright J (2012) The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling. Structure 20: 350–363 [DOI] [PubMed] [Google Scholar]
- Ravichandran A, Sugiyama N, Tomita M, Swarup S, Ishihama Y (2009) Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non‐pathogenic Pseudomonas species . Proteomics 9: 2764–2775 [DOI] [PubMed] [Google Scholar]
- Romling U, Galperin MY, Gomelsky M (2013) Cyclic di‐GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77: 1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger‐Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH et al (1987) Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325: 279–281 [DOI] [PubMed] [Google Scholar]
- Schmidt AJ, Ryjenkov DA, Gomelsky M (2005) The ubiquitous protein domain EAL is a cyclic diguanylate‐specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187: 4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Hammerbacher AS, Bastian M, Nieken KJ, Klockgether J, Merighi M, Lapouge K, Poschgan C, Kolle J, Acharya KR et al (2016) Oxygen‐dependent regulation of c‐di‐GMP synthesis by SadC controls alginate production in Pseudomonas aeruginosa . Environ Microbiol 18: 3390–3402 [DOI] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U (2004) GGDEF and EAL domains inversely regulate cyclic di‐GMP levels and transition from sessility to motility. Mol Microbiol 53: 1123–1134 [DOI] [PubMed] [Google Scholar]
- Su B, Ji YS, Sun XL, Liu XH, Chen ZY (2014) Brain‐derived neurotrophic factor (BDNF)‐induced mitochondrial motility arrest and presynaptic docking contribute to BDNF‐enhanced synaptic transmission. J Biol Chem 289: 1213–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Kornev AP (2011) Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci 36: 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse‐Kunstleve RW, Afonine PV, Zwart PH, Hung LW (2009) Decision‐making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr 65: 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini M, Laventie BJ, Moscoso J, Jenal U, Filloux A (2016) The diguanylate cyclase HsbD intersects with the HptB regulatory cascade to control Pseudomonas aeruginosa biofilm and motility. PLoS Genet 12: e1006354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay K, Brody MS, Fredlund E, Price CW (2000) A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis . Mol Microbiol 35: 180–188 [DOI] [PubMed] [Google Scholar]
- West SE, Schweizer HP, Dall C, Sample AK, Runyen‐Janecky LJ (1994) Construction of improved Escherichia‐Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa . Gene 148: 81–86 [DOI] [PubMed] [Google Scholar]
- Zhao J, Yu X, Zhu M, Kang H, Ma J, Wu M, Gan J, Deng X, Liang H (2016) Structural and molecular mechanism of CdpR involved in quorum‐sensing and bacterial virulence in Pseudomonas aeruginosa . PLoS Biol 14: e1002449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Liu C, Liu S, Cong H, Chen Y, Gu L, Ma LZ (2016a) Membrane association of SadC enhances its diguanylate cyclase activity to control exopolysaccharides synthesis and biofilm formation in Pseudomonas aeruginosa . Environ Microbiol 18: 3440–3452 [DOI] [PubMed] [Google Scholar]
- Zhu M, Zhao J, Kang H, Kong W, Liang H (2016b) Modulation of type III secretion system in Pseudomonas aeruginosa: involvement of the PA4857 gene product. Front Microbiol 7: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Data Availability Statement
The atomic coordinates and structure factors for the reported crystal structures of SiaB‐SiaC‐ADP complex and SiaC have been deposited in the Protein Data Bank (PDB) under accession numbers 6KKO (http://www.rcsb.org/pdb/explore/explore.do?structureId=6KKO) and 6KKP (http://www.rcsb.org/pdb/explore/explore.do?structureId=6KKP).


