Abstract
Raw meats are sometimes contaminated with Campylobacter species from animal faeces, and meats have repeatedly been implicated in foodborne infections. This study evaluated the prevalence, virulence genes, antimicrobial susceptibility patterns, and resistance gene determinants in Campylobacter species isolated from retailed meat carcasses. A total of 248 raw meat samples were collected from butcheries, supermarkets, and open markets; processed for enrichment in Bolton broth; and incubated at 42 °C for 48 h in 10% CO2. Thereafter, the broths were streaked on modified charcoal cefoperazone deoxycholate agar (mCCDA) plates and incubated at the same conditions and for the same amount of time. After incubation, colonies were isolated and confirmed by Polymerase chain reaction using specific oligonucleotide sequences used for the identification of the genus Campylobacter, species, and their virulence markers. The patterns of antimicrobial resistance profiles of the identified isolates were studied by disk diffusion method against 12 antibiotics, and relevant resistance genes were assessed by PCR. From culture, 845 presumptive Campylobacter isolates were obtained, of which 240 (28.4%) were identified as genus Campylobacter. These were then characterised into four species, of which C. coli had the highest prevalence rate (22.08%), followed by C. jejuni (16.66%) and C. fetus (3.73%). The virulence genes detected included iam (43.14%), cadF (37.25%), cdtB (23.53%), flgR (18.63%), and flaA (1.96%), and some of the isolates co-harboured two to four virulence genes. Of the 12 antibiotics tested, the highest phenotypic resistance displayed by Campylobacter isolates was against clindamycin (100%), and the lowest level of resistance was observed against imipenem (23.33%). The frequency of resistance genes detected included catll (91.78%), tetA (68.82%), gyra (61.76%), ampC (55%), aac(3)-IIa (aacC2)a (40.98%), tetM (38.71%), ermB (18.29%), tetB (12.90%), and tetK (2.15%). There is a high incidence of Campylobacter species in meat carcasses, suggesting these to be a reservoir of campylobacteriosis agents in this community, and as such, consumption of undercooked meats in this community is a potential health risk to consumers.
Keywords: Campylobacter species, contamination, infection, meat, virulence
1. Introduction
In the last decade, there has been a global upsurge in the rate of Campylobacter infections [1,2], and Campylobacter has emerged as one of the most significant bacteria of public health importance [3]. Globally, Campylobacter infection is a significant zoonosis, considered to be the leading cause of bacterial foodborne infection [4]. This zoonotic infection is of great public health concern [5], with meats known as the major risk factor [6] due to consumption of undercooked poultry or red meats [7]. Worldwide, consumption of meats and their products is increasing, and this may be connected to meats’ high protein content and the recommendations of healthy nutrition [8]. Meat is one of the most nutritious food items and a regular component of the human diet. Moreover, countless products, such as burgers and sausages, are products from meat, and as a result of the central role that meats occupy in human nutrition, their hygienic value is very essential for public health, as consumption of poor-quality meats may cause infections [9]. Infections arising from consumption of contaminated undercooked meats or food remain a global threat to public health [10]. Foodborne disease is a major public health problem due to its increasing incidence worldwide [11,12,13] and its huge burden of morbidity and mortality caused by bacterial infections [14]. Campylobacter is among the regular foodborne bacteria pathogens that are responsible for most foodborne disease outbreaks [15,16,17].
Worldwide, Campylobacter is among the major pathogens that cause bacterial gastroenteritis [18]. Such bacteria are microaerophilic bacteria with respiratory-type metabolism [19], and several species are known to cause infections [20], with C. coli, C. lari, and C. jejuni being the most common species implicated in human infections [21]. Other species, such as C. upsaliensis, C. concisus, C. fetus, and C. ureolyticus, have also been reported to be implicated in human gastrointestinal infections and periodontitis [22]. The Campylobacter infectious dose is about 500 colony forming unit/g depending on the physical conditions of the individual or age [23], and the infection is caused by the virulence mechanisms that are involved in toxin production, flagellar motility, adhesion, and invasion of epithelial cells [24]. In general, the burden of Campylobacter foodborne disease remains significantly high across the world [25], and regular monitoring and examination of meats are necessary to maintain food safety standards [26].
Foodborne disease outbreak is defined as a food poisoning occurrence involving more than two persons epidemiologically connected to a common food source [27]. The shocking listeriosis outbreak that happened recently in South Africa, which took over 218 lives, highlights the importance of good food safety practices and food monitoring [28]. It is commonly accepted that the actual occurrence rate of foodborne disease is obviously higher than the documented data due to limited surveillance capacity and under-reporting, particularly in developing countries [29]. Campylobacter infection is an infection that is labelled “self-limiting”, which rarely requires antimicrobial treatment [30,31]. Nevertheless, antimicrobial treatment is needed in persisting or severe campylobacteriosis cases, immunocompromised patients, and cases of extragastrointestinal symptoms [32,33]. The antibiotics employed in the treatment of severe Campylobacter infections include azithromycin and erythromycin, ciprofloxacin, and tetracyclines [34]. Alternative drugs of choice for systemic campylobacteriosis treatment include gentamicin and ampicillin [35]. However, there are regular reports on the increasing rate of Campylobacter resistance to currently used antibiotics, including macrolides, quinolones, and fluoroquinolones, which represent a significant threat to public health that is of global concern [36]. In this paper, we report on the prevalence, virulence markers, and antimicrobial resistance of Campylobacter species in retailed fresh meat carcasses in two district municipalities in the Eastern Cape Province of South Africa as part of our larger study on reservoirs of antibiotic resistance in the environment.
2. Materials and Methods
2.1. Ethical Clearance
Ethical clearance was applied for on behalf of the study and granted by the University of Fort Harr Alice, South Africa research ethics committee with certificate reference no. OKO021IGW01.
2.2. Study Area
The samples were collected in Chris Hani and Amathole District Municipalities, Eastern Cape, South Africa, with geographical coordinates 31.8743° S, 26.7968° E and 32.5842° S, 27.3616° E, respectively, and Figure 1 is a map showing the study areas.
Figure 1.
Map of Chris Hani and Amathole District Municipalities in the Eastern Cape Province, South Africa.
2.3. Sample Collection
A total number of 258 meat samples (mutton, chicken, turkey, beef, and pork) were purchased from different retail markets, open markets, and butcheries in different locations and towns in Chris Hani and Amathole District Municipalities, Eastern Cape, South Africa. All the meat samples were aseptically packed into separate sterile plastic bags to prevent cross contamination and were transported to the laboratory for analysis in a cooler box with ice packs within six hours of collection.
2.4. Microbiological Analysis of the Meat Samples
The meat samples were analysed following ISO 10,272 guidelines for isolation and identification of Campylobacter species [37,38]. Briefly, 25 g portions of the meat samples were homogenised in 245 mL of buffered peptone water (M614-500G, (Vadhani, Mumbai, India)). Thereafter, 10 mL of the homogenate was added into 90 mL of Bolton selective enrichment broth (1.00068.0500 Merck), to which Bolton broth selective supplement (1.00079.0010 Merck) with 5% (v/v) defibrinated horse blood (JMS, Singapore) was added, and the resulting mixture was incubated at 42 °C for 48 h under microaerophilic conditions in 10% CO2 in an HF151UV CO2 incubator. After the 48 h incubation period, a loopful of the inoculum from the enriched broth was streaked on modified charcoal cefoperazone deoxycholate agar (mCCDA) plates containing antibiotic selective supplement (CCDA selective supplement 1.00071.0010) and incubated under the same conditions and for the same amount of time stated above. Thereafter, colonies suspected to be Campylobacter based on colony morphology were picked and re-streaked onto blood agar base plates supplemented with 7% (v/v) defibrinated horse blood, and the plates were incubated under the same conditions and for the same amount of time.
2.5. DNA Extraction
Template DNA for PCR assay was extracted following the process described by Sierra-Arguello et al. [39] with slight modification. Briefly, colonies isolated from the blood agar plates were grown in 5 mL of Tryptone Soya Broth (TSB) for 48 h at 42 °C under microaerobic conditions in a 10% CO2 incubator. After incubation, 1 mL of the broth was centrifuged at 12,800 rpm for 5 min, and the cells were suspended in 400 µL of sterile distilled water in sterile 1.5 mL Eppendorf tubes. The suspensions were boiled for 10 min at 100 °C in a heating block and allowed to cool, after which the suspensions were centrifuged at 12,800 rpm for 5 min and the supernatants were collected and stored at −20 °C until ready for use.
2.6. Molecular Identification of the Genus Campylobacter
A 439 bp part of the 16S rRNA gene was amplified using primer CAM220 F-GGTGTAGGATGAGACTATATA and CAM659 R-TTCCATCTGCCTCTCCC as reported by Moreno et al. [40]. A singleplex PCR assay was carried out in a 25 µL reaction volume (5 µL of the DNA, 12.5 µL master mix, 2 µL of primer, and 5.5 µL of nuclear free water), and the PCR cycling conditions were set at initial denaturation (95 °C for 5 min), followed by 33-cycle (94 °C for 1 min, 58 °C for 1 min, and 72 °C for 2 min), and the final extension was set at 72 °C for 2 min. Verification of the amplified PCR products was carried out by resolving them in 1.5% agarose gel stained with ethidium bromide at 135 volts for 30 min, which was detected under a short-wavelength UV light source; C. jejuni ATCC 33.560 was used as the positive control.
2.7. Molecular Classification of Campylobacter Species
PCR amplification was further carried out to delineate the isolates to the species level for the detection of C. lari, C. fetus, C. jejuni, and C. coli. The primer sets used for the detection of these species are as reported by Yamazaki-Matsune et al. [41].
2.8. Molecular Detection of Virulence Genes
The identified Campylobacter species were further screened for the presence of invasion genes (ciaB and iam), adherence genes (flaA and cadF), a toxin gene (cdtB) and flagella synthesis, and a modification gene (flgR). The primer sets for the detection of cdtB, flaA and cadF genes were used as reported by Modi et al. [24], iam gene [42], ciaB gene [43] and flgR) gene [44].
2.9. Phenotypic Determination of Antimicrobial Resistance
Patterns of antimicrobial resistance of the identified Campylobacter species isolated from different meat types were studied using the Kirby–Bauer disk diffusion method according to Clinical and Laboratory Standard Institute (CLSI) 45] guidelines. The isolates were tested against 12 antibiotics regularly used in human and veterinary practices, comprising nine antimicrobial families, including tetracycline/doxycycline (30 μg), tetracycline (30 μg); penicillins/ampicillin (10 μg); macrolids/azithromycin (15 µg), erythromycin (15 μg); aminoglycoside/gentamicin (10 μg); lincomycin/clindamycin (2 μg); phenicols/chloramphenicol (30 μg); fluoroquinolones/ciprofloxacin (5 μg), levofloxacin (5 µg); cephalosporin/ceftriaxone (30 μg); and carbapenems/imipenem (10 µg). Briefly, 50 µL of the glycerol stock was suspended in 5 mL of Tryptone Soy Broth and incubated at 42 °C for 48 h in 10% CO2 in a CO2 incubator. After incubation, the broths were suspended in sterile normal saline solution, followed by adjustment of turbidity to 0.5 McFarland standard. The solutions were evenly spread using sterile cotton swabs on Müller Hinton agar plates supplemented with 5% defibrinated horse blood. After drying, antibiotic discs were dispensed using a disc-dispensing apparatus, and the plates were incubated at 42 °C for 24 h in a 10% CO2 incubator. C. jejuni (ATCC 33560) and C. fetus (ATCC 27374) were used as reference strains. The inhibition zones for tetracycline, doxycycline, ciprofloxacin, and erythromycin were interpreted following CLSI [45] breakpoints for Campylobacter. As there are no breakpoints available for ampicillin, azithromycin, gentamicin, clindamycin, chloramphenicol, levofloxacin, ceftriaxone, and imipenem for Campylobacter, the breakpoints established by CLSI [45] for Enterobacteriaceae were used for the interpretation of results.
2.10. Multiple Antibiotic Resistance (MAR) Index
For the determination of the multiple antibiotic resistance (MAR) index, the formula MAR = x/y, stated by Krumperman [46], was adopted where x = is the number of antibiotics to which the test isolate showed resistance and y = is the total number of antibiotics to which the test isolate has been evaluated for susceptibility.
2.11. Genotypic Assessment of Antibiotic Resistance Genes
Molecular screening of resistance genes was carried out on important resistance genes by both simplex and multiplex PCR assays on the isolates, which showed phenotypic resistance to the test antibiotics. The primer sets used for the detection of tetA, tetB, tetC and tetD genes were used as reported by Ng et al. [47], tetK and tetM genes [48], gyrA gene [49], ermB gene [50], catI and catII genes [51], (aac(3)-IIa (aacC2)a gene [52] and IMI, KPC, VIM and blaOXA-48-lik genes [53]. Verification of the amplified PCR products was carried out as stated above.
3. Results
3.1. Molecular confirmation of Campylobacter species
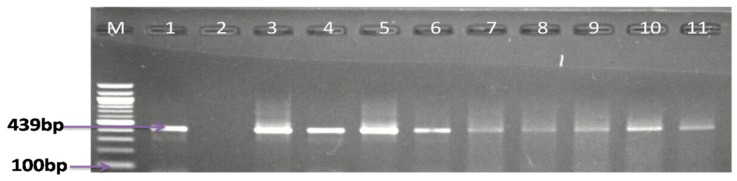
In the effort to isolate and detect Campylobacter species in meat samples, including pork, mutton, mutton offals, beef, beef offals, turkey, chicken, and chicken offals; the samples were subjected to both traditional culture and PCR techniques. From culture, a total of 845 presumptive isolates were obtained, of which 28.40% (208/845) were identified as belonging to the genus Campylobacter by PCR assay, of which 32.5% (208/640) were obtained from retail markets, 15.17% (22/145) from butcheries, and 16.67% (10/60) from open markets. The detailed results of the number of isolates from various meat types are shown in Table 1, while Figure 2 shows a representative gel image of the PCR confirmed genus Campylobacter.
Table 1.
Number of Campylobacter isolates identified in various meat types.
| Meat Types | No. of Samples | No. of Presumptive Campylobacter Isolates | No. of Isolates Identified as Genus Campylobacter |
|---|---|---|---|
| Turkey | 11 | 16 | 5 (31.25%) |
| Pork | 35 | 131 | 33 (25.19%) |
| Mutton | 22 | 9 | 4 (44.44%) |
| Mutton offals (heart) | 2 | 6 | 0 |
| Beef | 27 | 89 | 30 (33.71%) |
| Beef offals (intestine, kidney, and liver) | 31 | 126 | 39 (30.95%) |
| Chicken | 68 | 300 | 81 (27%) |
| Chicken offals (liver, gizzard, and heart) | 52 | 165 | 48 (29.09%) |
Figure 2.
Agarose gel electrophoresis image of PCR-confirmed genus Campylobacter. Lane M: molecular marker (100 bp); lane 1: positive control (C. jejuni ATCC 33560); lane 2: negative control; and lanes 3−11: positive Campylobacter isolates (439 bp).
3.2. Molecular Characterisation of Campylobacter Species
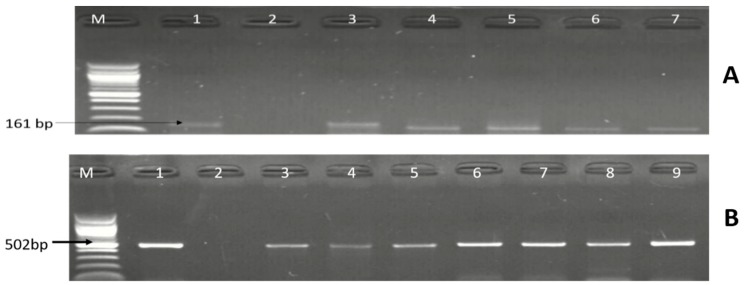
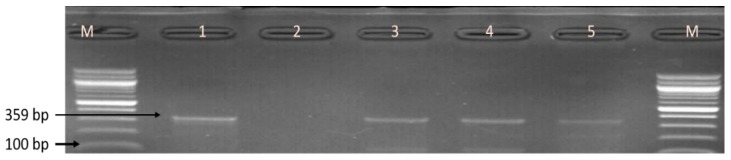
The 240 isolates identified as belonging to the genus Campylobacter were further delineated into four Campylobacter species by PCR technique: 53 (22.08%) isolates were identified as C. coli, 40 (16.66%) as C. jejuni, and 9 (3.75%) as C. fetus, whereas C. lari was not detected. A summary of the numbers of Campylobacter species identified in the meat types is shown in Table 2, while Figure 3 and Figure 4 are representative gel electrophoresis images of PCR-confirmed C. jejuni, C. coli, and C. fetus, respectively.
Table 2.
Summary of Campylobacter species identified in the meat types.
| Meat Typologies | C. jejuni | C. coli | C. lari | C. fetus | No. of Campylobacter Species Detected in the Meat Types |
|---|---|---|---|---|---|
| Pork | 4 | 9 | 0 | 3 | 16 |
| Beef | 3 | 0 | 0 | 2 | 5 |
| Beef offals | 10 | 15 | 0 | 1 | 26 |
| Chicken | 9 | 16 | 0 | 2 | 27 |
| Chicken offals | 14 | 5 | 0 | 1 | 20 |
| Mutton | 0 | 0 | 0 | 0 | 0 |
| Mutton offals | 0 | 3 | 0 | 0 | 3 |
| Turkey | 0 | 5 | 0 | 0 | 5 |
| Total | 40 | 53 | 0 | 9 | 102 |
Figure 3.
Gel (A) is a representative gel electrophoresis image of PCR-confirmed C. jejuni. Lane M: DNA ladder (100 bp); lane 1: positive control (C. jejuni ATCC 33560); lane 2: negative control; lanes 3–7: positive C. jejuni isolates (161 bp); and (B) is a representative gel image of PCR-confirmed C. coli. Lane M: DNA ladder (100 bp); lane 1: positive control (C. coli ATCC 33559); lane 2: negative control; and lane 3–9: positive C. coli isolates (502 bp).
Figure 4.
Gel picture of some PCR-identified C. fetus. Lane M: molecular weight marker (100 bp); lane 1: positive control (C. fetus ATCC 27374); lane 2: negative control; lanes 3–5: positive C. fetus isolates (359 bp).
3.3. Molecular Detection of Virulence Genes
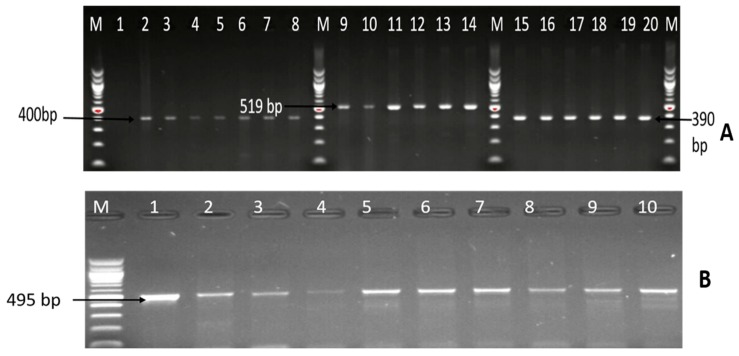
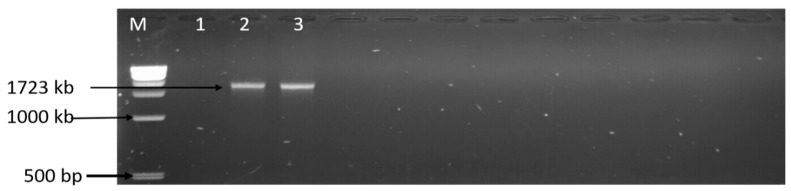
Among the 102 isolates characterised as C. coli, C. fetus, and C. jejuni; the presence of six virulence genes (iam, cdtB, ciaB, cadF, flgR, and flaA) were assessed by PCR. The virulence genes detected included iam (43.14%), cadF (37.25%), cdtB (23.53%), flgR (18.63%), and flaA (1.96%), and the detailed results are shown in Table 3. Moreover, 26 (25.49%) isolates co-harboured two virulence genes, 9 (8.82%) isolates co-harboured three virulence genes, and three (3.94%) isolates co-harboured four different virulence genes, and the patterns of distribution of the virulence makers co-harboured in the isolates are shown in Table 4, while Figure 5 and Figure 6 are gel electrophoresis images of PCR-confirmed cadF, iam, flgR, cdtB, and flaA genes.
Table 3.
Percentage distribution pattern of detected virulence genes in the identified Campylobacter species.
| Virulence Genes Screened |
Campylobacter Species | ||
|---|---|---|---|
| C. jejuni (%) | C. coli (%) | C. fetus (%) | |
| iam | 7 (6.86) | 35 (34.31) | 2 (1.96) |
| cadF | 4 (3.92) | 34 (33.33) | 0 |
| flgR | 11 (10.78) | 8 (7.84) | 0 |
| cdtB | 6 (5.88) | 17 (16.67) | 1(0.98) |
| flaA | 0 | 2 (1.96) | 0 |
| ciaB | 0 | 0 | 0 |
Table 4.
Patterns of occurrence of multiple virulence genes in the identified species.
| Pattern of Multiple Virulence Genes | Number of Campylobacter Species | Total Number | ||||
|---|---|---|---|---|---|---|
| C. coli | C. fetus | C. jejuni | - | |||
| 1 | iam, cadF | 16 | - | 1 | - | 17 |
| 2 | iam, flgR | 2 | - | - | - | 2 |
| 3 | iam, cdtB | 1 | - | - | - | 1 |
| 4 | cadF, flgR | - | - | 1 | - | 1 |
| 5 | cadF, cdtB | 4 | - | - | - | 4 |
| 6 | flgR, cdtB | 1 | - | - | - | 1 |
| 7 | iam, cadF, flaA | 1 | - | - | - | 1 |
| 8 | iam, cadF, cdtB | 7 | - | - | - | 7 |
| 9 | cadF, cdtB, flgR | 1 | - | - | - | 1 |
| 10 | iam, cadF, flgR, cdtB | 2 | - | 1 | - | 3 |
Figure 5.
Image (A) is a representative gel picture of the PCR-confirmed cadF, iam, and flgR genes. Lane M: molecular marker (100 bp); lane 1: negative control; lanes 2–8: positive Campylobacter isolates that harboured the cadF gene (400 bp); lanes 9–14: positive Campylobacter isolates that harboured the iam gene (519 bp); lanes 15–20: positive Campylobacter isolates that harboured the flgR gene (390 bp); and (B) is a gel image of the PCR-confirmed cdtB gene. Lane M: DNA ladder (100bp); lanes 1–10: positive Campylobacter isolates that harboured the cdtB gene (495 bp).
Figure 6.
Gel image of the PCR-confirmed flaA gene. Lane M: DNA ladder (1 kb); lane 1: negative control; lanes 2–3: positive Campylobacter isolates that harboured the flaA gene (1723 kb).
3.4. Antibiotic Phenotypic Characteristics of the Identified Campylobacter Species
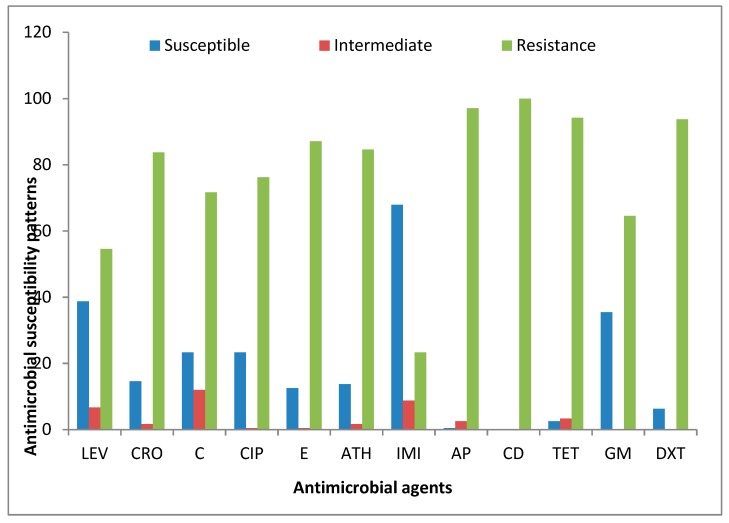
A total of 240 PCR-confirmed genus Campylobacter isolates were profiled for their possible phenotypic resistance against 12 antibiotics belonging to nine antimicrobial families. Astonishingly, all the isolates displayed the highest phenotypic resistance against clindamycin (100%). The Campylobacter isolates recovered from meat carcasses also displayed high phenotypic resistance against ampicillin (97.08%), tetracycline (94.17%), doxycycline (93.75%), erythromycin (87.03%), azithromycin (84.58%), ceftriaxone (83.75%), ciprofloxacin (76.25%), chloramphenicol (71.67%), gentamicin (64.58%), and levofloxacin (54.58%), and the lowest level of resistance was observed against imipenem (23.33%). Table 5 shows the detailed phenotypic resistance patterns observed in the isolates tested against the antibiotics, while Figure 7 is the interpreted antimicrobial susceptibility result according to CLSI [45] guidelines.
Table 5.
Antibiotic resistance patterns of Campylobacter isolates isolated from meat carcasses.
| No | Multiple Antimicrobial Resistance Profile | No of Isolates | Total | MAR Index | ||
|---|---|---|---|---|---|---|
| C. coli | C. jejuni | C. fetus | ||||
| 1 | C-CD-AP | 1 | - | - | 1 | 0.25 |
| 2 | C-E-ATH-CD-AP | 1 | - | - | 1 | 0.42 |
| 3 | CRO-C-E-CD-AP | - | - | 1 | 1 | 0.42 |
| 4 | E-ATH-CD-T-DXT-AP | - | 1 | - | 1 | 0.5 |
| 5 | LEV-CRO-CIP-ATH-CD-AP | - | - | 1 | 1 | 0.5 |
| 6 | CRO-C-CIP-E-ATH-CD-AP | - | 1 | - | 1 | 0.58 |
| 7 | CRO-E-ATH-CD-T-DXT-AP | - | 1 | - | 1 | 0.58 |
| 8 | CRO-C-CIP-E-ATH-CD-AP | - | 1 | - | 1 | 0.58 |
| 9 | C-E-ATH-CD-T-DXT-AP | 1 | - | - | 1 | 0.58 |
| 10 | LEV-CRO-C-CIP-E-ATH-CD-AP | 1 | - | - | 1 | 0.67 |
| 11 | CRO-C-E-ATH-T-GM-DXT-AP | 1 | - | - | 1 | 0.67 |
| 12 | CRO-C-E-ATH-CD-T-DXT-AP | 1 | 1 | - | 2 | 0.67 |
| 13 | CRO-CIP-E-ATH-CD-T-DXT-AP | 1 | 1 | - | 2 | 0.67 |
| 14 | CRO-E-ATH-CD-T-GM-DXT-AP | - | 1 | - | 1 | 0.75 |
| 15 | CRO-C-CIP-E-ATH-CD-T-DXT-AP | - | - | 1 | 1 | 0.75 |
| 16 | CRO-CIP-E-ATH-IMI-CD-T-DXT-AP | - | - | 1 | 1 | 0.75 |
| 17 | LEV-CRO-CIP-E-ATH-CD-T-DXT-AP | - | 1 | - | 1 | 0.75 |
| 18 | CRO-C-CIP-ATH-CD-T-GM-DXT-AP | 1 | - | - | 1 | 0.75 |
| 19 | LEV-CRO-C-CIP-E-ATH-CD-T-DXT | 1 | - | - | 1 | 0.75 |
| 20 | CRO-C-E-ATH-CD-T-GM-DXT-AP | 2 | 1 | 3 | 0.75 | |
| 21 | LEV-CRO-C-CIP-E-ATH-CD-T-AP | - | - | 1 | 1 | 0.75 |
| 41 | LEV-CRO-C-CIP-E-ATH-CD-GM-DXT-AP | - | 1 | - | 1 | 0.83 |
| 22 | LEV-CRO-C-CIP-E-ATH-CD-T-DXT-AP | 6 | 8 | - | 14 | 0.83 |
| 23 | LEV-CRO-CIP-E-ATH-CD-T-GM-DXT-AP | 1 | - | - | 1 | 0.83 |
| 24 | CRO-C-CIP-E-ATH-CD-T-GM-DXT-AP | - | 1 | - | 1 | 0.83 |
| 25 | LEV-CRO-CIP-E-ATH-CD-T-GM-DXT-AP | - | 1 | - | 1 | 0.83 |
| 26 | LEV-CRO-C-CIP-E-ATH-CD-T-GM-DXT-AP | 8 | 8 | - | 16 | 0.92 |
| 27 | CRO-C-CIP-E-ATH-IMI-CD-T-GM-DXT-AP | - | 1 | - | 1 | 0.92 |
| 28 | LEV-CRO-C-CIP-E-ATH-IMI-CD-GM-DXT-AP | - | 1 | - | 1 | 0.92 |
| 29 | LEV-CRO-C-CIP-E-ATH-IMI-CD-T-GM-DXT-AP | 14 | 3 | - | 17 | 1 |
Figure 7.
Antimicrobial susceptibility patterns of Campylobacter isolates recovered from meat carcasses sold in Eastern Cape, South Africa. Levofloxacin (LEV), ciprofloxacin (CIP), azithromycin (ATH), imipenem (IMI), ampicillin (AP), clindamycin (CD), tetracycline (TET), ceftriaxone (CRO), chloramphenicol (C), erythromycin (E), gentamicin (GM), and doxycycline (DXT).
3.5. Assessment of Resistance Determinants
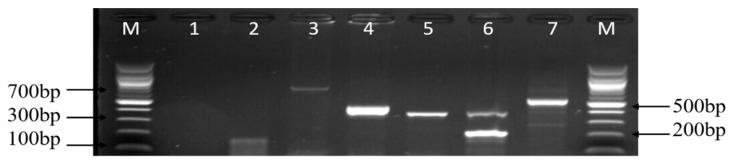
The resistance genes screened among the phenotypic resistant-Campylobacter isolates are listed in Table S3, and the choice for the selection of these genes was centred on their high phenotypic resistance rates. Thus, 12 antibiotic resistance genes were screened for probable detection of resistance genes among the identified Campylobacter species and also to determine the pattern of occurrence of multiple resistance genes in the isolates. From the PCR results obtained, the order of the frequency level of the resistance genes detected was as follows: catll (91.78%), tetA (68.82%), gyra (61.76%), ampC (55%), aac(3)-IIa (aacC2)a (40.98%), tetM (38.71%), ermB (18.29%), tetB (12.90%), and tetK (2.15%). In contrast, the IMI, KPC, VIM, blaOXA-48-like, catl, tetC, tetD, and tetK genes were not detected. Figure 8 and Figure 9 are representative gel images of the amplified PCR products of the assessed antibiotic resistance genes. Similarly, the patterns of the level of detected multiple resistance genes are shown in Table 6.
Figure 8.
A representative gel image of various amplified antimicrobial resistance genes of Campylobacter isolates. Lanes M: DNA ladder (100 bp); lane 1: negative control; lane 2: tetM gene (158 bp); lane 3: aac(3)-IIa (aacC2)a gene (740 bp); lane 4: gyrA gene (441 bp); lane 5: ermB gene (320 bp); lane 6: tetA gene (201 bp) and tetB gene (359 bp); and lane 7: ampC gene (530 bp).
Figure 9.
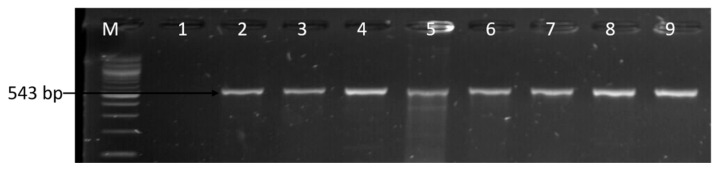
Gel image of the PCR-confirmed catll gene. Lane M: DNA ladder (100 bp); lane 1: negative control; and lanes 2–9, Campylobacter isolates that harboured the catll gene (543 bp).
Table 6.
Distribution and pattern of multiple antibiotic resistance of Campylobacter species isolated from meat carcasses.
| No | Distribution Pattern of Antibiotic Resistance Determinants | No of Isolates | Total | ||
|---|---|---|---|---|---|
| C. coli | C. jejuni | C. fetus | |||
| 1 | tetA, catII | 1 | - | 3 | 4 |
| 2 | tetM, catII | 2 | - | - | 2 |
| 3 | ampC, catII | 1 | - | - | 1 |
| 4 | catII, ermB | 1 | - | - | 1 |
| 5 | tetA, ampC | - | - | 1 | 1 |
| 6 | tetA, catII, gyra | - | - | 1 | 1 |
| 7 | tetA, tetB, ampC | - | 1 | - | 1 |
| 8 | tetA, ampC, catII | 2 | 2 | - | 4 |
| 9 | tetA, catII, ermB | 1 | - | - | 1 |
| 10 | tetM, ampC, catII | 1 | - | - | 1 |
| 11 | tetA, tetK, aac(3)-IIa (aacC2)a | - | 1 | - | 1 |
| 12 | ampC, catII, aac(3)-IIa (aacC2)a | - | 1 | - | 1 |
| 13 | tetM, catII, aac(3)-IIa (aacC2)a | 2 | - | - | 2 |
| 14 | tetA, ampC, catII, gyrA | 2 | 1 | 1 | 4 |
| 15 | tetA, ampC, catII, ermB | - | 2 | - | 2 |
| 16 | tetA, tetM, catII, ermB | 2 | - | - | 2 |
| 17 | tetA, tetB, ampC, gyrA | - | 1 | - | 1 |
| 18 | tetA, tetB, ampC, catII | 1 | 1 | - | 2 |
| 19 | tetA, tetM, tetK, catII | - | 1 | - | 1 |
| 20 | tetA, catII, ermB, gyrA | - | 1 | - | 1 |
| 21 | tetA, tetB, catII, ermB | 1 | - | - | 1 |
| 22 | tetA, tetM, ampC, catII | 1 | - | - | 1 |
| 23 | tetA, tetM, ampC, gyrA | 1 | - | - | 1 |
| 24 | tetM, ampC, catII, gyrA | 1 | - | - | 1 |
| 25 | tetM, catII, gyrA, aac(3)-IIa (aacC2)a | 1 | - | - | 1 |
| 26 | tetM, ampC, gyrA, aac(3)-IIa (aacC2)a | 1 | - | - | 1 |
| 27 | tetA, ampC, catII, aac(3)-IIa (aacC2)a | 1 | 3 | - | 4 |
| 28 | tetA, tetM, ampC, catII, gyrA | 3 | 1 | - | 4 |
| 29 | tetA, tetB, tetM, catII, gyrA | 1 | - | - | 1 |
| 30 | tetA, tetB, ampC, catII, gyrA | - | 3 | - | 3 |
| 31 | tetA, ampC, catII, ermB, gyrA | - | 2 | - | 2 |
| 32 | tetA, tetM, ampC, catII, aac(3)-IIa (aacC2)a | 1 | - | - | 1 |
| 33 | tetA, tetM, catII, gyrA, aac(3)-IIa (aacC2)a | 1 | - | - | 1 |
| 34 | tetM, ampC, catII, gyrA, aac(3)-IIa (aacC2)a | 1 | - | - | 1 |
| 35 | tetA, ampC, catII, gyrA, aac(3)-IIa (aacC2)a | 1 | 2 | - | 3 |
| 36 | tetA, tetM, ampC, catII, ermB, gyrA | 2 | 1 | - | 3 |
| 37 | tetA, tetB, tetM, ampC, catII, ermB | 1 | - | - | 1 |
| 38 | tetA, tetM, ampC, catII, gyrA, aac(3)-IIa (aacC2)a | - | 1 | - | 1 |
| 39 | tetA, tetB, tetK, ampC, ermB, aac(3)-IIa (aacC2)a | - | 1 | - | 1 |
| 40 | tetA, tetB, ampC, catII, gyrA, aac(3)-IIa (aacC2)a | - | 1 | - | 1 |
| 41 | tetA, tetM, ampC, catII, gyrA, aac(3)-IIa (aacC2)a | 2 | - | - | 2 |
| 42 | tetA, tetM, ampC, ermB, gyrA, aac(3)-IIa (aacC2)a | - | 1 | - | 1 |
| 42 | tetA, tetM, ampC, catII, gyrA, aac(3)-IIa (aacC2)a | 1 | 1 | - | 2 |
| 43 | tetA, tetM, ampC, catII, ermB, gyrA, aac(3)-IIa (aacC2)a | - | 1 | - | 1 |
| 44 | tetA, tetB, tetM, ampC, catII, gyrA, aac(3)-IIa (aacC2)a | 1 | - | - | 1 |
4. Discussion
Reports on the prevalence, virulence marker, and antimicrobial resistance genes in Campylobacter isolates recovered from retailed meat samples are well documented in some parts of the world, but limited information is available in some provinces in South Africa, particularly in the Eastern Cape Province. Hence, this study aimed to address the prevalence and characterise the identified Campylobacter species, virulence genes, and resistance genes in Campylobacter isolates recovered from meat carcasses. Campylobacter species are major bacteria foodborne enteropathogens that are regularly spread to humans through the consumption of contaminated food including meats [54]. In this study, a high rate of Campylobacter was detected in 240 (28.40%) isolates from meat carcasses, and this gives valuable insight into the possible risks of foodborne infection to humans. Studies carried out in Italy by Stella et al. [55], in Malaysia [56], in China [57], in France [58], and in South Korea [59] also detected Campylobacter in meat samples with detection rates of 34.10%, 50.9%, 48.9%, 76%, and 31.67%, respectively, and our findings are in line with their reports. Other studies carried out in Spain by García-Sánchez et al. [60], in Pakistan by Nisar et al. [3], in Yangzhou, China by Zou et al. [61], and in Northern Poland by Andrzejewska et al. [62] also detected Campylobacter in meat samples, and these results are also akin with their reports. According to Seliwiorstow et al. [63], different sampling sources make an impact on the occurrence rates of Campylobacter, which indicate the risk factors associated with handling. The occurrence rates of Campylobacter detected in the different market sources are in the order of 32.5% (retail markets), 16.67% (open markets), and 15.17% (butcheries). The highest occurrence rates of the genus Campylobacter were also detected in isolates from mutton (44.4%), followed by beef (34%), turkey (31.3%), beef offals (31%), chicken offals (29%), chicken (27%), and pork (25.2%). The occurrence rate of Campylobacter in mutton samples was much higher than that in other meat types, and this result is in agreement with the report of Maktabi et al. [64].
In the present study, the high detection rate of Campylobacter was observed in isolates from beef carcasses, and this result also corroborates the reports of Kashoma et al. [65] and Premarathne et al. [66]. The high rates of Campylobacter detection in turkey samples observed in this study also support those reported by Noormohamed and Fakhr [67] in Oklahoma, USA (17%), and Szosland-Fałtyn et al. in Poland (47.37%) [68]. The 240 isolates identified belonging to the genus Campylobacter were then characterised into four species, of which 53 (22.08%) were identified as C. coli, 40 (16.66%) as C. jejuni, and 9 (3.73%) as C. fetus, whereas C. lari was not detected. Higher rates of C. coli and C. jejuni were detected than other Campylobacter species, and our findings corroborate the report of Hodges et al. [69], Ocejo et al. [70], Sulaiman et al. [71], and Vinueza-Burgos et al. [72]. In contrast, a low prevalence rate of C. fetus was detected, and this finding is also in agreement with the report of Sinulingga et al. [56]. C. coli and C. jejuni are known to cause infection in humans, but from the first report of Campylobacter infection to date, Campylobacter pathogenesis has not been clearly understood. Though, what has been clear about Campylobacter infections and has been proposed as virulence determinants includes Campylobacter’s invasive capability, adherence to intestinal mucosa, ability to produce toxins, and flagella-mediated motility [73]. Thus, the presence of these specific genes associated with Campylobacter invasion, adhesion, toxin production, and colonisation are all essential for the process of infection, and the mechanism by which they cause disease in humans is assumed to be multifactorial [74]. From the PCR results of the virulence gene assessed, most Campylobacter species were detected to harbour a high proportion of cadF, flaA, and iam genes responsible for colonisation, invasion, and adherence, and our results corroborate the report of Abu-Madi et al. [75]. The iam gene had the highest occurrence rate of 43.14% (Table 3) among the various virulence genes screened, and our results also corroborate the report of Redondo et al. [76].
Similarly, the high occurrence rate of the cadF (37.25%) gene and the lower prevalence of the flaA (1.96%) gene were detected in the Campylobacter isolates from meat samples, and these results are akin with the reports of Andrzejewska et al. [77] and Ripabelli et al. [78]. Our results also corroborated the report of Chukwu et al. [74], who also detected the cadF gene in Campylobacter strains isolated from water and paediatric stools. The ciaB gene was screened for but was not detected, and our findings are contrary to the reports of Melo et al. [79], Melo et al. [80], and Zhong et al. [81] who reported high detection rates of the ciaB gene in meats and in retailed food samples. Another virulence gene screened for was the cdtB gene, and the presence and expression of any of the cdt genes (cdtA, cdtB, and cdtC) are essential for the efficient activity of the CDT toxin. In this study, the cdtB gene was observed to be widespread among the Campylobacter species, with C. coli strains revealed to have a higher prevalence rate of cdtB than C. jejuni and C. fetus (Table 3), and our results are not in agreement with the reports of Wieczorek et al. [82] and Reddy and Zishiri [83], which reported a higher prevalence rate of the cdtB gene in C. jejuni than in the C. coli strain. Multiple virulence genes were also detected in both C. coli and C. jejuni with C. coli observed to harbour more virulence genes than C. jejuni, and these results are in contrast with the reports of Lim et al. [84] and Zhang et al. [85]. Campylobacter isolates have also been reported by Han et al. [86] to co-harbour more than three virulence genes, and our findings corroborate this report. Virulence genes in the Campylobacter genome are known to be implicated in human infection, and Campylobacter pathogenicity may be strengthened by the expression of a single virulence gene or multiple virulence genes that are enough to establish infection in the host [80].
The dissemination of virulence-associated genes in the identified Campylobacter strains isolated from meats showed a potential risk to humans and an impending risk of outbreak of Campylobacter infection if appropriate measures are not put into place. From the antimicrobial susceptibility testing result, a total of 78 antibiotic resistance profiles were generated among the 102 Campylobacter isolates characterised as C. coli, C. jejuni, and C. fetus, and 76.47% were resistant to more than two antimicrobial families and were classified as multi-drugs resistant strains (Table 5). The most common observed resistant profile was LEV-CRO-C-CIP-E-ATH-IMI-CD-T-GM-DXT-AP, which was common among C. coli and C. jejuni strains. The isolates displayed high phenotypic resistance to tetracycline (94.17%), erythromycin (87.03%), ampicillin (97.08%), and ciprofloxacin (76.25%). In Brazil, studies have shown that there is a high prevalence of quinolones (72.2%), tetracycline (43%), erythromycin (38.9%), and ampicillin (26.9%) resistance in Campylobacter in circulation [87,88]. In Bolivia, Argentina, Chile, and Peru, many Campylobacter strains are resistant to quinolones (47–78%) as well as tetracycline (40.8–65.9% in Argentina and Bolivia), erythromycin (58.6% to 61.4% in Bolivia and Chile), and ampicillin (47.2% in Argentina) [89]. The value of the MAR index is 0.2, and the MAR index is a good risk assessment tool used to distinguish high- and low-risk areas where antibiotics are overused [90]. The MAR indices of the isolates were calculated, and 77.83% were found to have a MAR index greater than 0.2, while 17 isolates had MAR indices of 1.0 (Table 5). A MAR index value of greater than 0.2 indicates a high-risk source of contamination where antibiotics are often used, and based on our results, most isolates had MAR indices of greater than 0.2, confirming that there is high selective pressure and high antibiotic use in these areas.
Because the effectiveness of antibiotic resistance might be compromised in the treatment of infections, antimicrobial resistance genes were screened. Multiple antibiotic resistance genes were highly detected in most of the isolates (Table 6), and this is in agreement with the multiple phenotypic resistance profiles displayed by the isolates (Table 5). Multiple resistance genes in the Campylobacter isolates were detected, and our results are in agreement with the reports of Chukwu et al. [74] and Wieczorek et al. [91]. There was a high rate of detection of resistance genes in chloramphenicol (catll (91.78%)), tetracycline (tetA (68.82%)), ciprofloxacin (gyra (61.76%)), ampicillin (ampC (55%)), gentamycin (aac(3)-IIa (aacC2)a (40.98%)), and tetracycline (tetM (38.71%) in the Campylobacter isolates. In this study, the high detection rate of resistance genes in tetracycline/ciprofloxacin-Campylobacter isolates was responsible for its phenotypic resistance, and this is in line with the report of Nguyen et al. [92]. A high rate of the gyra gene was also detected in the Campylobacter isolates recovered from meat carcasses, and our results are also in agreement with the report of Du et al. [93]. The results from the study of Rahimi et al. [94] show that Campylobacter isolates recovered from meat carcasses were all susceptible to chloramphenicol and gentamycin, and our results are contrary to their report. The implications in antimicrobial resistance show a strong connection between the use of antibiotics in animal production, veterinary medicine, and antibiotic-resistant Campylobacter isolates in humans [95], although the majority of the antibiotics used in the treatment of bacterial infections in humans are also used in animals. Nevertheless, a One Health approach to addressing the issue of antibiotic resistance and the spread of antibiotic-resistant bacteria includes plans to maintain and carry on with the efficiency of current antibiotics by abolishing their inappropriate use and by preventing the spread of infection [96]. Another One Health approach in the prevention of human campylobacteriosis and the solution to the spreading of antibiotic-resistant bacteria is to improve animal, environmental, and human health with key components such as access to safe food, clean water, and hygiene [97]. In addition, recent One Health approach in the prevention and spread of antimicrobial resistance have focused primarily on the reduction of the use of antibiotics in food animals [98].
5. Conclusions
We investigated the prevalence rate, characterisation, distribution patterns of virulence genes, antibiotic susceptibility patterns, and antibiotic resistance genes in Campylobacter species isolated from meat carcasses obtained from butcheries, open markets, and supermarkets. High prevalence rates of the genus Campylobacter and virulence markers were detected in meat samples obtained in Chris Hani and Amathole District Municipalities, Eastern Cape, South Africa, and PCR assay is one of the appropriate methods for the detection and characterisation of virulence genes and resistance genes in bacteria species. The majority of the isolates showed resistance to the test antibiotics, and multi-resistant isolates were also observed. In conclusion, there should be a continues surveillance of the presence of these pathogens and antibiotic resistance genes in Campylobacter isolates, and awareness of the impending risks associated with the consumption of undercooked, contaminated meats should be increased.
Acknowledgments
The authors are grateful to the South Africa Medical Research Council (SAMRC) for its financial support.
Supplementary Materials
The following are available online at https://www.mdpi.com/2304-8158/9/2/203/s1, Table S1: Primer sets used for the identification of different Campylobacter species; Table S2: Oligonucleotide sequences used for the detection of virulence genes; Table S3: Primer sequences used for the detection of resistance genes.
Author Contributions
A.I. carried out the experiment, analysed the results, and wrote the paper. A.I.O. applied for funding, designed the project, supervised the work, and corrected and proofread the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the South Africa Medical Research Council.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Harvala H., Rosendal T., Lahti E., Engvall E.O., Brytting M., Wallensten A., Lindberg A. Epidemiology of Campylobacter jejuni infections in Sweden, November 2011–October 2012: Is the severity of infection associated with C. jejuni sequence type? Infect. Ecol. Epidemiol. 2016;6:1–10. doi: 10.3402/iee.v6.31079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iglesias-Torrens Y., Miró E., Guirado P., Llovet T., Muñoz C., Cerdà-Cuéllar M., Madrid C., Balsalobre C., Navarro F. Population structure, antimicrobial resistance, and virulence-associated genes in Campylobacter jejuni isolated from three ecological niches: Gastroenteritis patients, broilers, and wild birds. Front. Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.01676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nisar M., Mushtaq M.H., Shehzad W., Hussain A., Nasar M., Nagaraja K.V., Goyal S.M. Occurrence of Campylobacter in retail meat in Lahore, Pakistan. Acta Trop. 2018;185:42–45. doi: 10.1016/j.actatropica.2018.04.030. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh Y.-H., Sulaiman I.M. Chapter 5—Campylobacteriosis: An Emerging Infectious Foodborne Disease. Foodborne Dis. 2018;1:119–155. [Google Scholar]
- 5.Havelaar A.H., Kirk M.D., Torgerson P.R., Gibb H.J., Hald T., Lake R.J., Praet N., Bellinger D.C., De Silva N.R., Gargouri N., et al. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 2015;12:1–23. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carron M., Chang Y.M., Momanyi K., Akoko J., Kiiru J., Bettridge J., Chaloner G., Rushton J., O’Brien S., Williams N., et al. Campylobacter, a zoonotic pathogen of global importance: Prevalence and risk factors in the fast-evolving chicken meat system of Nairobi, Kenya. PLoS Negl. Trop. Dis. 2018;12:1–18. doi: 10.1371/journal.pntd.0006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma K.P., Chattopadhyay U.K., Naskar K. Prevalence of’ Campylobacter species in raw meat samples sold in open markets of Kolkata city. Int. J. Agric. Environ. Biotechnol. 2016;9:535–539. doi: 10.5958/2230-732X.2016.00070.X. [DOI] [Google Scholar]
- 8.El-Aziz A.D., Abd-Allah S.M.S. Incidence of Campylobacter species in wholesale chicken carcasses and chicken meat products in Assiut city, Egypt. Int. Food Res. J. 2017;24:2660–2665. [Google Scholar]
- 9.Ndahi M.D., Kwaga J.K.P., Bello M., Kabir J., Umoh V.J., Yakubu S.E., Nok A.J. Prevalence and antimicrobial susceptibility of Listeria monocytogenes and methicillin-resistant Staphylococcus aureus strains from raw meat and meat products in Zaria, Nigeria. Lett. Appl. Microbiol. 2013;58:262–269. doi: 10.1111/lam.12183. [DOI] [PubMed] [Google Scholar]
- 10.Luo X., Xu X., Chen H., Bai R., Zhang Y., Hou X., Zhang F., Zhang Y., Sharma M., Zeng H., et al. Food safety related knowledge, attitudes, and practices (KAP) among the students from nursing, education and medical college in Chongqing, China. Food Control. 2019;95:181–188. doi: 10.1016/j.foodcont.2018.07.042. [DOI] [Google Scholar]
- 11.Guerra M.M.M., de Almeida A.M., Willingham A.L. An overview of food safety and bacterial foodborne zoonoses in food production animals in the Caribbean region. Trop. Anim. Health Prod. 2016;48:1095–1108. doi: 10.1007/s11250-016-1082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law J.W.F., Ab Mutalib N.S., Chan K.G., Lee L.H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015;5:1–19. doi: 10.3389/fmicb.2014.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newell D.G., Koopmans M., Verhoef L., Duizer E., Aidara-Kane A., Sprong H., Opsteegh M., Langelaar M., Threfall J., Scheutz F., et al. Food-borne diseases-the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010;139:3–15. doi: 10.1016/j.ijfoodmicro.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torgerson P.R., Devleesschauwer B., Praet N., Speybroeck N., Willingham A.L., Kasuga F., Rokni M.B., Zhou X.N., Fèvre E.M., Sripa B., et al. World Health Organization estimates of the global and regional disease burden of 11 foodborne parasitic diseases, 2010: A data synthesis. PLoS Med. 2015;12:1–22. doi: 10.1371/journal.pmed.1001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camino F.M.M., Arisseto-Bragotto A.P., Block J.M. Food quality, food-borne diseases, and food safety in the Brazilian food industry. Food Qual. Saf. 2017;1:13–27. doi: 10.1093/fqs/fyx003. [DOI] [Google Scholar]
- 16.Epps S., Harvey R., Hume M., Phillips T., Anderson R., Nisbet D. Foodborne Campylobacter: Infections, metabolism, pathogenesis and reservoirs. Int. J. Environ. Res. Public Health. 2013;10:6292–6304. doi: 10.3390/ijerph10126292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gourley C.R., Negretti N.M., Konkel M.E. The food-borne pathogen Campylobacter jejuni depends on the AddAB DNA repair system to defend against bile in the intestinal environment. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-14646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Zhang S., He M., Zhang Y., Fu Y., Liang H., Jing H., Li Y., Ma H., Zhang M. Prevalence and molecular characterization of Campylobacter spp. isolated from patients with diarrhea in Shunyi, Beijing. Front. Microbiol. 2018;9:1–8. doi: 10.3389/fmicb.2018.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wainwright L.M., Elvers K.T., Park S.F., Poole R.K. A truncated haemoglobin implicated in oxygen metabolism by the microaerophilic food-borne pathogen Campylobacter jejuni. Microbiology. 2005;151:4079–4091. doi: 10.1099/mic.0.28266-0. [DOI] [PubMed] [Google Scholar]
- 20.Reich F., Valero A., Schill F., Bungenstock L., Klein G. Characterisation of Campylobacter contamination in broilers and assessment of microbiological criteria for the pathogen in broiler slaughterhouses. Food Control. 2018;87:60–69. doi: 10.1016/j.foodcont.2017.12.013. [DOI] [Google Scholar]
- 21.Rahimi E., Alipoor-Amroabadi M., Khamesipour F. Investigation of prevalence of thermotolerant Campylobacter spp. in livestock faeces. Can. J. Anim. Sci. 2017;97:207–213. doi: 10.1139/cjas-2015-0166. [DOI] [Google Scholar]
- 22.Man S.M. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011;8:669–685. doi: 10.1038/nrgastro.2011.191. [DOI] [PubMed] [Google Scholar]
- 23.Granić K., Krčar D., Uhitil S., Jakšić S. Determination of Campylobacter spp. in poultry slaughterhouses and poultry meat. Vet. Arhiv. 2009;79:79–491. [Google Scholar]
- 24.Modi S., Brahmbhatt M.N., Chatur Y.A., Nayak J.B. Prevalence of Campylobacter species in milk and milk products, their virulence gene profile and anti-biogram. Vet. World. 2015;8:1–8. doi: 10.14202/vetworld.2015.1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platts-Mills J.A., Kosek M. Update on the burden of Campylobacter in developing countries. Curr. Opin. Infect. Dis. 2014;27:444–450. doi: 10.1097/QCO.0000000000000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naravaneni R., Jamil K. Rapid detection of food-borne pathogens by using molecular techniques. J. Med. Microbiol. 2005;54:51–54. doi: 10.1099/jmm.0.45687-0. [DOI] [PubMed] [Google Scholar]
- 27.Katiyo W., de Kock H.L., Coorey R., Buys E.M. Assessment of safety risks associated with handling chicken as based on practices and knowledge of a group of South African consumers. Food Control. 2019;101:104–111. doi: 10.1016/j.foodcont.2019.02.027. [DOI] [Google Scholar]
- 28.NICD Communicable Diseases Communique. [(accessed on 1 April 2018)]; Available online: http://www.nicd.ac.za/wp-content/uploads/2018/08.
- 29.Jahan S. Epidemiology of Foodborne Illness. In: Valdez B., editor. Scientific, Health and Social Aspects of the Food Industry. Volume 1. IntechOpen; Qassim, Saudi Arabia: 2012. pp. 321–342. [Google Scholar]
- 30.Magana M., Chatzipanagiotou S., Burriel A.R., Ioannidis A. Inquiring into the Gaps of Campylobacter Surveillance Methods. Vet. Sci. 2017;4:36. doi: 10.3390/vetsci4030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei B., Kang M. In vitro activity of fosfomycin against Campylobacter isolates from poultry and wild birds. PLoS ONE. 2018;13:1–8. doi: 10.1371/journal.pone.0200853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Post A., Martiny D., van Waterschoot N., Hallin M., Maniewski U., Bottieau E., Van Esbroeck M., Vlieghe E., Ombelet S., Vandenberg O., et al. Antibiotic susceptibility profiles among Campylobacter isolates obtained from international travelers between 2007 and 2014. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:2101–2107. doi: 10.1007/s10096-017-3032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zollner-Schwetz I., Krause R. Therapy of acute gastroenteritis: Role of antibiotics. Clin. Microbiol. Infect. 2015;21:744–749. doi: 10.1016/j.cmi.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Szczepanska B., Andrzejewska M., Spica D., Klawe J.J. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. 2017;17:1–9. doi: 10.1186/s12866-017-0991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kashoma I.P., Mkomba F.D., Bunyaga A.S. Prevalence and antimicrobial resistance in Campylobacter from different stages of the chicken meat supply chain in Morogoro, Tanzania. Microb. Drug Res. 2016;22:40–52. doi: 10.1089/mdr.2015.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Giannatale E., Di Serafino G., Zilli K., Alessiani A., Sacchini L., Garofolo G., Aprea G., Marotta F. Characterization of antimicrobial resistance patterns and detection of virulence genes in Campylobacter isolates in Italy. Sensors. 2014;14:3308–3322. doi: 10.3390/s140203308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habib I., Berkvens D., De Zutter L., Dierick K., Van Huffel X., Speybroeck N., Geeraerd A.H., Uyttendaele M. Campylobacter contamination in broiler carcasses and correlation with slaughterhouses operational hygiene inspection. Food Microbiol. 2012;29:105–112. doi: 10.1016/j.fm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 38.ISO . Microbiology of Food and Animal Feeding Stuffs-Horizontal Method for Detection and Enumeration of campylobacter spp. International Organization for Standardization; Geneva, Switzerland: 2006. Part 1: Detection method. UNI EN ISO Norm 10272-1:2006. [Google Scholar]
- 39.Sierra-Arguello Y.M., Furian T.Q., Perdoncini G., Moraes H.L., Salle C.T., Rodrigues L.B., dos Santos L.R., Gomes M.J.P., do Nascimento V.P. Fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli from poultry and human samples assessed by PCR-restriction fragment length polymorphism assay. PLoS ONE. 2018;13:1–9. doi: 10.1371/journal.pone.0199974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno Y., Botella S., Luis Alonso J., Ferru’s M.A., Herna’ndez M., Herna’ndez J. Specific detection of Arcobacter and Campylobacter strains in Water and Sewage by PCR and Fluorescent in Situ Hybridization. Appl. Environ. Microbiol. 2003;69:1181–1186. doi: 10.1128/AEM.69.2.1181-1186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamazaki-Matsune W., Taguchi M., Seto K., Kawahara R., Kawatsu K., Kumeda Y., Kitazato M., Nukina M., Misawa N., Tsukamoto T. Development of a multiplex PCR assay for identification of Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis subsp. hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis. J. Med. Microbiol. 2007;56:1467–1473. doi: 10.1099/jmm.0.47363-0. [DOI] [PubMed] [Google Scholar]
- 42.Carvalho A.C., Ruiz-Palacios G.M., Ramos-Cervantes P., Cervantes L.E., Jiang X., Pickering L.K. Molecular characterization of invasive and noninvasive Campylobacter jejuni and Campylobacter coli isolates. J. Clin. Microbiol. 2001;39:1353–1359. doi: 10.1128/JCM.39.4.1353-1359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casabonne C., Gonzalez A., Aquili V., Subils T., Balague C. Prevalence of seven virulence genes of Campylobacter jejuni isolated from patients with diarrhea in Rosario, Argentina. Intern. J. Infect. 2016;3:1–6. doi: 10.17795/iji-37727. [DOI] [PubMed] [Google Scholar]
- 44.Wilson D.L., Rathinam V.A., Qi W., Wick L.M., Landgraf J., Bell J.A., Plovanich-Jones A., Parrish J., Finley R.L., Mansfield L.S., et al. Genetic diversity in Campylobacter jejuni is associated with differential colonization of broiler chickens and C57BL/6J IL10-deficient mice. Microbiology. 2010;156:2046–2057. doi: 10.1099/mic.0.035717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clinical and Laboratory Standards Institute (CLSI) Method for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. 3rd ed. CLSI; Wayne, NJ, USA: 2015. [DOI] [PubMed] [Google Scholar]
- 46.Krumperman P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46:165–170. doi: 10.1128/AEM.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng L.K., Martin I., Alfa M., Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes. 2001;15:209–215. doi: 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- 48.Strommenger B., Kettlitz C., Werner G., Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 2003;41:4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan S.S., Fox M.L., Holland S.M., Stock F., Gill V.J., Fedorko D.P. Resistance to multiple fluoroquinolones in a clinical isolate of Streptococcus pyogenes: Identification of gyrA and parC and specification of point mutations associated with resistance. Antimicrob. Agents Chemother. 2000;44:196–3198. doi: 10.1128/AAC.44.11.3196-3198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osode A.N., Okoh A.I. Impact of discharged wastewater final effluent on the physicochemical qualities of a receiving watershed in a suburban community of the Eastern Cape Province. Clean Soil Air Water. 2009;37:938–944. doi: 10.1002/clen.200900098. [DOI] [Google Scholar]
- 51.Maynard C., Bekal S., Sanschagrin F., Levesque R.C., Brousseau R., Masson L., Lariviere S., Harel J. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J. Clin. Microbiol. 2004;42:5444–5452. doi: 10.1128/JCM.42.12.5444-5452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velusamy S., Barbara E.G., Mark J.L., Lien T.N., Susan I.H., Ynte H.S., Stephen P.O. Phenotypic and genotypic antimicrobial resistance patterns of Escherichia coli isolated from dairy cows with mastitis. Vet. Microbiol. 2007;124:319–328. doi: 10.1016/j.vetmic.2007.04.040. [DOI] [PubMed] [Google Scholar]
- 53.Dallenne C., Da Costa A., Decré D., Favier C., Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 54.Heredia N., García S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018;4:250–255. doi: 10.1016/j.aninu.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stella S., Soncini G., Ziino G., Panebianco A., Pedonese F., Nuvoloni R., Di Giannatale E., Colavita G., Alberghini L., Giaccone V. Prevalence and quantification of thermophilic Campylobacter spp. in Italian retail poultry meat: Analysis of influencing factors. Food Microbiol. 2007;62:232–238. doi: 10.1016/j.fm.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 56.Sinulingga T.S., Aziz S.A., Bitrus A.A., Zunita Z., Abu J. Occurrence of Campylobacter species from broiler chickens and chicken meat in Malaysia. Trop. Anim. Health Prod. 2019;1:1–7. doi: 10.1007/s11250-019-01995-y. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y., Dong Y., Deng F., Liu D., Yao H., Zhang Q., Shen J., Liu Z., Gao Y., Wu C., et al. Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008–2014. J. Antimicrob. Chemother. 2015;71:666–669. doi: 10.1093/jac/dkv382. [DOI] [PubMed] [Google Scholar]
- 58.Guyard-Nicodème M., Rivoal K., Houard E., Rose V., Quesne S., Mourand G., Rouxel S., Kempf I., Guillier L., Gauchard F., et al. Prevalence and characterization of Campylobacter jejuni from chicken meat sold in French retail outlets. Int. J. Food Microbiol. 2015;203:8–14. doi: 10.1016/j.ijfoodmicro.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Chon J.W., Lee S.K., Yoon Y., Yoon K.S., Kwak H.S., Joo I.S., Seo K.H. Quantitative prevalence and characterization of Campylobacter from chicken and duck carcasses from poultry slaughterhouses in South Korea. Poult. Sci. 2018;97:2909–2916. doi: 10.3382/ps/pey120. [DOI] [PubMed] [Google Scholar]
- 60.García-Sánchez L., Melero B., Diez A.M., Jaime I., Rovira J. Characterization of Campylobacter species in Spanish retail from different fresh chicken products and their antimicrobial resistance. Food Microbiol. 2018;76:457–465. doi: 10.1016/j.fm.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 61.Zou C., Zhang H., Yang X., Tang H., Xu X., Ren F., Huang J. Qualitative and quantitative epidemiological analysis of Campylobacter in chicken sold in Yangzhou. J. Food Saf. Qual. 2018;9:5040–5043. [Google Scholar]
- 62.Andrzejewska M., Klawe J., Szczepańska B., Śpica D. Occurrence of virulence genes among Campylobacter jejuni and Campylobacter coli isolates from domestic animals and children. Pol. J. Vet. Sci. 2011;4:207–211. doi: 10.2478/v10181-011-0031-x. [DOI] [PubMed] [Google Scholar]
- 63.Seliwiorstow T., Baré J., Berkvens D., Van Damme I., Uyttendaele M., De Zutter L. Identification of risk factors for Campylobacter contamination levels on broiler carcasses during the slaughter process. Int. J. Food Microbiol. 2016;226:26–32. doi: 10.1016/j.ijfoodmicro.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 64.Maktabi S., Ghorbanpoor M., Hossaini M., Motavalibashi A. Detection of multi-antibiotic resistant Campylobacter coli and Campylobacter jejuni in beef, mutton, chicken and water buffalo meat in Ahvaz, Iran. Vet. Res. Forum. 2019;10:37–42. doi: 10.30466/vrf.2019.34310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kashoma I.P., Kassem I.I., John J., Kessy B.M., Gebreyes W., Kazwala R.R., Rajashekara G. Prevalence and antimicrobial resistance of Campylobacter isolated from dressed beef carcasses and raw milk in Tanzania. Microb. Drug Resist. 2016;2:40–52. doi: 10.1089/mdr.2015.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Premarathne J.M., Anuar A.S., Thung T.Y., Satharasinghe D.A., Jambari N.N., Abdul-Mutalib N.A., Huat J.T.Y., Basri D.F., Rukayadi Y., Nakaguchi Y., et al. Prevalence and antibiotic resistance against tetracycline in Campylobacter jejuni and C. coli in cattle and beef meat from Selangor, Malaysia. Front. Microbiol. 2017;8:1–9. doi: 10.3389/fmicb.2017.02254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noormohamed A., Fakhr M.K. Prevalence and antimicrobial susceptibility of Campylobacter species in Oklahoma conventional and organic retail poultry. Open Microbiol. J. 2014;8:130–137. doi: 10.2174/1874285801408010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szosland-Fałtyn A.N.N.A., Bartodziejska B., Krolasik J., Paziak-Domańska B.E.A.T.A., Korsak D., Chmiela M. The Prevalence of Campylobacter spp. Pol. J. Microbiol. 2018;67:117–120. doi: 10.5604/01.3001.0011.6152. [DOI] [PubMed] [Google Scholar]
- 69.Hodges L.M., Carrillo C.D., Upham J.P., Borza A., Eisebraun M., Kenwell R., Mutschall S.K., Haldane D., Schleihauf E., Taboada E.N. A strain comparison of Campylobacter isolated from retail poultry and human clinical cases in Atlantic Canada. PLoS ONE. 2019;14:1–16. doi: 10.1371/journal.pone.0215928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ocejo M., Oporto B., Hurtado A. Occurrence of Campylobacter jejuni and Campylobacter coli in Cattle and Sheep in Northern Spain and Changes in Antimicrobial Resistance in Two Studies 10-years Apart. Pathogens. 2019;8:98. doi: 10.3390/pathogens8030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sulaiman I.M., Hsieh Y.H., Simpson S. Species identification of Campylobacter jejuni and Campylobacter coli isolates from raw poultry products by MALDI–TOF MS and rRNA sequence analysis. J. AOAC Int. 2019;103:197–204. doi: 10.5740/jaoacint.19-0170. [DOI] [PubMed] [Google Scholar]
- 72.Vinueza-Burgos C., Wautier M., Martiny D., Cisneros M., Van Damme I., De Zutter L. Prevalence, antimicrobial resistance and genetic diversity of Campylobacter coli and Campylobacter jejuni in Ecuadorian broilers at slaughter age. Poult. Sci. 2017;96:2366–2374. doi: 10.3382/ps/pew487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silva J., Leite D., Fernandes M., Mena C., Gibbs P.A., Teixeira P. Campylobacter spp. as a foodborne pathogen: A review. Front. Microbiol. 2011;2:1–12. doi: 10.3389/fmicb.2011.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chukwu M.O., Luther King Abia A., Ubomba-Jaswa E., Obi L., Dewar J.B. Characterization and Phylogenetic Analysis of Campylobacter Species Isolated from Paediatric Stool and Water Samples in the Northwest Province, South Africa. Int. J. Environ. Res. Public Health. 2019;16:2205. doi: 10.3390/ijerph16122205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abu-Madi M., Behnke J.M., Sharma A., Bearden R., Al-Banna N. Prevalence of virulence/stress genes in Campylobacter jejuni from chicken meat sold in Qatari retail outlets. PLoS ONE. 2016;11:1–15. doi: 10.1371/journal.pone.0156938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Redondo N., Carroll A., McNamara E. Molecular characterization of Campylobacter causing human clinical infection using whole-genome sequencing: Virulence, antimicrobial resistance and phylogeny in Ireland. PLoS ONE. 2019;14:1–11. doi: 10.1371/journal.pone.0219088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andrzejewska M., Szczepańska B., Śpica D., Klawe J.J. Trends in the occurrence and characteristics of Campylobacter jejuni and Campylobacter coli isolates from poultry meat in Northern Poland. Food Control. 2015;51:190–194. doi: 10.1016/j.foodcont.2014.11.014. [DOI] [Google Scholar]
- 78.Ripabelli G., Tamburro M., Minelli F., Leone A., Sammarco M.L. Prevalence of virulence-associated genes and cytolethal distending toxin production in Campylobacter spp. isolated in Italy. Comp. Immunol. Microbiol. Infect. Dis. 2010;33:355–364. doi: 10.1016/j.cimid.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 79.Melo R.T., Nalevaiko P.C., Mendonça E.P., Borges L.W., Fonseca B.B., Beletti M.E., Rossi D.A. Campylobacter jejuni strains isolated from chicken meat harbour several virulence factors and represent a potential risk to humans. Food Control. 2013;33:227–231. doi: 10.1016/j.foodcont.2013.02.032. [DOI] [Google Scholar]
- 80.Melo R.T., Grazziotin A.L., Júnior E.C.V., Prado R.R., Mendonça E.P., Monteiro G.P., Peres P.A., Rossi D.A. Evolution of Campylobacter jejuni of poultry origin in Brazil. Food Microbiol. 2019;82:489–496. doi: 10.1016/j.fm.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 81.Zhong X., Wu Q., Zhang J., Shen S. Prevalence, genetic diversity and antimicrobial susceptibility of Campylobacter jejuni isolated from retail food in China. Food Control. 2016;62:10–15. doi: 10.1016/j.foodcont.2015.09.032. [DOI] [Google Scholar]
- 82.Wieczorek K., Denis E., Lynch O., Osek J. Molecular characterization and antibiotic resistance profiling of Campylobacter isolated from cattle in Polish slaughterhouses. Food Microbiol. 2013;34:130–136. doi: 10.1016/j.fm.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 83.Reddy S., Zishiri O.T. Genetic characterisation of virulence genes associated with adherence, invasion and cytotoxicity in Campylobacter spp. isolated from commercial chickens and human clinical cases. Onderstepoort. J. Vet. Res. 2018;85:1–9. doi: 10.4102/ojvr.v85i1.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lim S.K., Moon D.C., Chae M.H., Kim H.J., Nam H.M., Kim S.R., Jang G.C., Lee K., Jung S.C., Lee H.S. Macrolide resistance mechanisms and virulence factors in erythromycin-resistant Campylobacter species isolated from chicken and swine feces and carcasses. J. Vet. Med. Sci. 2016;72:1791–1795. doi: 10.1292/jvms.16-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang T., Luo Q., Chen Y., Li T., Wen G., Zhang R., Luo L., Lu Q., Ai D., Wang H., et al. Molecular epidemiology, virulence determinants and antimicrobial resistance of Campylobacter spreading in retail chicken meat in Central China. Gut Pathogens. 2016;8:1–9. doi: 10.1186/s13099-016-0132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han X., Guan X., Zeng H., Li J., Huang X., Wen Y., Zhao Q., Huang X., Yan Q., Huang Y., et al. Prevalence, antimicrobial resistance profiles and virulence-associated genes of thermophilic Campylobacter spp. isolated from ducks in a Chinese slaughterhouse. Food Control. 2019;104:157–166. doi: 10.1016/j.foodcont.2019.04.038. [DOI] [Google Scholar]
- 87.Miranda K.L., Lage A.P. Antimicrobial susceptibility of Campylobacter sp strains isolated from calves with and without diarrhea in Minas Gerais state, Brazil. Braz. J. Microbiol. 2007;38:357–362. doi: 10.1590/S1517-83822007000200032. [DOI] [Google Scholar]
- 88.Kuana S.L., Santos L.R., Rodrigues L.B., Borsoi A., Moraes H.L.S., Salle C.T.P., Nascimento V.P. Occurrence and characterization of Campylobacter in the Brazilian production and processing of broilers. Avian Dis. 2008;52:680–684. doi: 10.1637/8296-032608-Reg.1. [DOI] [PubMed] [Google Scholar]
- 89.Giangaspero M. A Cosmopolitan One Health Issue: Campylobacteriosis. Clin. Microbiol. 2018;7:1–5. doi: 10.4172/2327-5073.1000308. [DOI] [Google Scholar]
- 90.Davis R., Brown P.D. Multiple antibiotic resistance index, fitness and virulence potential in respiratory Pseudomonas aeruginosa from Jamaica. J. Med. Microbiol. 2016;65:261–271. doi: 10.1099/jmm.0.000229. [DOI] [PubMed] [Google Scholar]
- 91.Wieczorek K., Wołkowicz T., Osek J. Antimicrobial resistance and virulence-associated traits of Campylobacter jejuni isolated from poultry food chain and humans with diarrhea. Front. Microbiol. 2018;9:1–11. doi: 10.3389/fmicb.2018.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nguyen T.N.M., Hotzel H., Njeru J., Mwituria J., El-Adawy H., Tomaso H., Neubauer H., Hafez H.M. Antimicrobial resistance of Campylobacter isolates from small scale and backyard chicken in Kenya. Gut Pathogens. 2016;8:1–9. doi: 10.1186/s13099-016-0121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Du Y., Ye Y., Liu Y., Wang A., Li Y., Zhou X., Pan H., Zhang J., Xu X. Molecular identification of multidrug-resistant Campylobacter species from diarrheal patients and poultry meat in Shanghai, China. Front. Microbiol. 2018;9:1–8. doi: 10.3389/fmicb.2018.01642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rahimi E., Ameri M., Kazemeini H.R. Prevalence and antimicrobial resistance of Campylobacter species isolated from raw camel, beef, lamb, and goat meat in Iran. Foodborne Path. Dis. 2010;7:443–447. doi: 10.1089/fpd.2009.0421. [DOI] [PubMed] [Google Scholar]
- 95.Wieczorek K., Osek J. Antimicrobial resistance mechanisms among Campylobacter. BioMed Res. Int. 2013:1–12. doi: 10.1155/2013/340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McEwen S.A., Collignon P.J. Antimicrobial Resistance in Bacteria from Livestock and Companion Animals. ASM Press; Washington, DC, USA: 2018. Antimicrobial resistance: A One Health perspective; pp. 521–547. [Google Scholar]
- 97.Garcia S.N., Osburn B.I., Cullor J.S. A one health perspective on dairy production and dairy food safety. One Health. 2019;7:1–9. doi: 10.1016/j.onehlt.2019.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.White A., Hughes J.M. Critical importance of a one health approach to antimicrobial resistance. EcoHealth. 2019;16:404–409. doi: 10.1007/s10393-019-01415-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.