Abstract
Food-producing animals may be a reservoir of vancomycin-resistant enterococci (VRE), potentially posing a threat to animal and public health. The aims of this study were to estimate the faecal carriage of VRE among healthy cattle (n = 362), pigs (n = 350), sheep (n = 218), and poultry (n = 102 flocks) in Switzerland, and to characterise phenotypic and genotypic traits of the isolates. VRE were isolated from caecum content of six bovine, and 12 porcine samples respectively, and from pooled faecal matter collected from 16 poultry flock samples. All isolates harboured vanA. Three different types of Tn1546-like elements carrying the vanA operon were identified. Conjugal transfer of vanA to human Enterococcus faecalis strain JH2-2 was observed for porcine isolates only. Resistance to tetracycline and erythromycin was frequent among the isolates. Our data show that VRE harbouring vanA are present in healthy food-producing animals. The vanA gene from porcine isolates was transferable to other enterococci and these isolates might play a role in the dissemination of VRE in the food production chain.
Keywords: Enterococcus faecium, E. faecalis, E. durans, vanA, whole genome sequencing, Tn1546, food chain
1. Introduction
Antimicrobial resistance has now become a permanent aspect of human medicine with vancomycin-resistant enterococci (VRE) gaining importance as nosocomial pathogens worldwide [1]. The World Health Organization (WHO) ranks vancomycin-resistant Enterococcus faecium (VREfm) as a pathogen of high priority in its global list of important antibiotic-resistant bacteria [2]. For European countries, the population-weighted mean percentage of resistance to vancomycin in invasive VREfm increased from 10.5% in 2015 to 17.3% in 2018 [3]. By contrast, in E. faecalis, vancomycin resistance remains infrequent in Europe [3].
Nosocomial VREfm may arise through independent events of introduction and subsequent dissemination within hospitals, but are also thought to generate within patients under antimicrobial therapy, most probably by the acquisition of resistance genes by means of horizontal gene transfer (HGT) [4,5,6,7]. One of the most important genetic determinants of vancomycin resistance is represented by the vanA gene cluster, which is organised as an operon consisting of vanRSHAXYZ, and is typically associated with transposons, such as Tn1546 [8,9]. Tn1546-type transposons play a key role in the acquisition and dissemination of vancomycin resistance among enterococci [9,10]. Tn1546 transposons vary structurally, because of point mutations, deletions, or the presence of insertion sequence (IS) elements [11,12]. These variations provide potential markers to type and trace the spread of vanA genes among enterococci isolated from different sources [13,14,15].
Most human clinical VREfm strains belong to the E. faecium lineage designated Clade A1 [16,17]. This clade contains the vast majority of strains isolated from clinical settings, including isolates belonging to clonal complex (CC)17 [17,18], and to the recently emerged sequence types (STs)203 and ST796 [4,19,20,21]. Clade A2 contains strains that are predominantly associated with sporadic human infections and with livestock [18].
The proliferation of VRE in livestock in Europe is attributed to the past use of avoparcin, which was introduced in 1975 as a growth promoter, but which confers cross-resistance to vancomycin [22]. The EU ban on antimicrobial growth promoters enforced in 2006 (EC no. 1831/2003) lead to a decline of the prevalence of VRE among farm animals [22]. Nevertheless, VRE continues to be readily detected in samples from livestock when using selective media, and its persistence is suggested to be maintained by co-selection, i.e., the use of macrolides, tetracycline, or copper, or by the presence of plasmid addiction systems [22].
Accordingly, food-producing animals may be considered a reservoir of VRE that affects veterinary and human medicine, either by horizontal transfer of vancomycin resistance genes between animal and human adapted enterococci, or by clonal dissemination of resistant strains [22].
This study aimed to assess the occurrence of VRE among healthy cattle, pigs, poultry, and sheep at slaughter and to characterise and compare phenotypic and genotypic traits of the isolates from these different sources.
2. Materials and Methods
2.1. Sampling and Bacterial Isolation
For slaughter cattle (n = 362), pigs (n = 350), and sheep (n = 218), swab samples were aseptically collected on 14 different days from caecal contents by cutting through the caecal wall with sterile scissors. Each caecum was swabbed once, avoiding an overload of faecal matter. Swabs were placed in sterile stomacher bags and transported to the laboratory. Animal and herd identification were collected along with each sample.
For poultry samples, faecal matter was collected on 9 different days at the entry of a poultry slaughterhouse from the crates of 102 poultry flocks (approximately 6000 chicken per flock). Pooled samples were placed in sterile bags and flock identification was noted for each sample.
In the case of caecum samples, the excess lengths of the swabs were broken off, and 20 mL brain heart infusion (BHI) broth with 6.5% NaCl (Oxoid, Pratteln, Switzerland) was added to each bag. For poultry samples, broth was added directly to each bag. Samples were homogenised for 30–60′′ using a stomacher and incubated for 18–24h at 37 °C. From the pre-enrichment broth, one loopful was streaked onto VRE select agar (BioRad, Cressier, Switzerland) and incubated for 48 h at 37 °C.
From samples with presumptive enterococci positive colonies, colonies of different colony morphology were selected and purified on sheep blood agar (BioRad, Cressier, Switzerland). In case of unclear identity, isolates were tested for catalase activity and catalase negative isolates were further analysed.
Species identification was performed by matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF-MS, Bruker Daltonics, Billerica, MA, USA) using Compass FlexControl version 3.4 software with the Compass database version 4.1.80. All isolates were stored in 20% glycerol at –20 °C for further analysis.
2.2. Multiplex PCR Detection of vanA, vanB, vanC-1, and vanC-2/3 Genes
Bacterial DNA was prepared by a standard lysis and boiling method [23]. Multiplex PCR targeting vanA, vanB, vanC-1, and vanC-2/3 genes was carried out on all isolates using custom synthesised primers (Microsynth, Balgach, Switzerland) and conditions published previously [24]. Amplicons were visualised under ultraviolet light after electrophoresis in 1% agarose gel stained with ethidium bromide. E. faecalis ATCC51299 (vanB) and Enterococcus casseliflavus (vanC) [25] were used as positive controls.
2.3. Antimicrobial Susceptibility Testing
Determination of the minimal inhibitory concentrations (MICs) of vancomycin was performed using a gradient strip (Etest, Biomérieux, Geneva, Switzerland) according to the manufacturer’s instructions. Antimicrobial susceptibility profiling was performed using the Micronaut-S livestock 4 susceptibility plate (Merlin diagnostics GmbH, Berlin, Germany) with H-broth (Merlin diagnostics GmbH, Berlin, Germany). The MIC values were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) susceptibility breakpoints, where available [26]. Antimicrobials with existing CLSI breakpoints included ampicillin (AMP), erythromycin (ERY), penicillin G (PEN), and tetracycline (TE).
2.4. Conjugation Experiments
Transfer experiments were performed using a modified solid mating protocol and E. faecalis JH2-2 (rifampicin resistant, vancomycin susceptible) as a recipient [27]. In brief, 40 µl volumes of overnight cultures of donor and recipient cells grown in BHI broth were mixed, concentrated by centrifugation, and resuspended in 20 µl of BHI broth. The mixture was dispensed onto BHI agar plates and incubated at 37 °C for 18–20 h. One loopful of cells were collected in 400 µl BHI broth. Serial dilutions were plated on BHI supplemented with 6 mg/L vancomycin and 50 mg/L rifampicin. The resulting transconjugants were purified and subject to identification by MALDI-TOF-MS.
Conjugation frequency was expressed as the number of transconjugants per recipient cell. Transconjugants were tested by singleplex PCR to confirm the presence of vanA using primers and conditions, described previously [24].
2.5. Whole Genome Sequencing
Bacterial cultures were grown overnight in 5 mL BHI with 6.5% NaCl. Chromosomal DNA was isolated from 1 mL overnight cultures using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Sequencing was done on an Illumina MiniSeq (Illumina, San Diego, CA, USA) and reads were assembled using Spades 3.12.0 [28] and Shovill [29], as described previously [30]. Species were identified using SpeciesFinder2.0 [31] available at https://cge.cbs.dtu.dk/services/SpeciesFinder/.
Draft genomes were annotated using Prokka and standard settings [32]. Resistance genes were identified using the resistance gene identifier (RGI) from the comprehensive antibiotic resistance database (CARD, available at https://card.mcmaster.ca/analyze/rgi) [33,34]. Searches were performed with standard setting and performed locally against the CARD database downloaded in September 2019.
Variations of the Tn1546 element were identified based on what is considered the Tn1546 prototype (GenBank M97297) [15,35]. Tn1546 sequences were compared by using average nucleotide identity (ANI). The ANI was calculated according to Richter et al. [36], using the python script PyANI.py [37].
An ANI-base tree was constructed using an in-house R script. In short, the Euclidean distance in the relative-identity matrix produced by PyANI.py was calculated using the function “dist” from the package cluster [38] und subsequently clustered using the function “hclust”.
Core-genome multilocus sequence typing (cgMLST) was performed in the software package SeqSphere 4.1.9 (Ridom, Münster, Germany), the Enterococcus faecium MLST database (https://pubmlst.org/efaecium/), and the Enterococcus faecalis MLST database (https://pubmlst.org/efaecalis/), respectively, to determine sequence types (STs).
Alignments of the vanA-operon were visualised using CLC Main Workbench 8.1.3.
2.6. Accession Numbers
The whole genome shotgun sequences have been deposited at GenBank numbers VYUB00000000 to VYUX00000000.
3. Results
3.1. Prevalence of VRE among Healthy Food-Producing Animals
To determine the occurrence of VRE in slaughter animals in Switzerland, faecal samples collected during a 14-day trial were analysed. Overall, VRE were isolated from six (2%) of the cattle samples (all from cattle belonging to the same herd), 12 (3%) of the swine samples, and 16 (16%) of the poultry flocks. No VRE were isolated from sheep samples. A multiplex PCR targeting vancomycin resistance genes revealed the presence of vanA in all isolates. No vanB, vanC-1, or vanC-2/3 genes were detected.
3.2. Antimicrobial Susceptibility Testing and Detection of Resistance Genes
A subset of 23 strains were selected for further analysis. To avoid sample clustering, one isolate from the positive cattle herd, and a selection of isolates from different pig herds were chosen. Strains included E. faecalis isolated from cattle (n = 1), E. faecium isolated from pigs (n = 6), E. faecium (n = 6) and Enterococcus durans (n = 10) isolated from poultry. Taken together, 12 E. faecalis, 10 E. durans, and one E. faecalis were analysed in this study.
All 23 strains exhibited MICs of vancomycin of > 32 µg/mL, confirming they were VRE (Table 1).
Table 1.
Phenotypic and genotypic features of vancomycin-resistant Enterococcus spp. isolated from cattle, pigs, and poultry.
| Host/Species | No. of Strains | Resistance Phenotype | Resistance Genotype | MLST | ||
|---|---|---|---|---|---|---|
| MIC [µg/mL] Vancomycin |
Additional Resistances1 | Resistance Genes | Tn1546 Type | |||
| Cattle | ||||||
| E. faecalis | 1 | ≥128 | – | dfrE, emeA, efrA, efrB, lsaA, vanA | I | 29 |
| Pigs | ||||||
| E. faecium | 1 | ≥256 | PEN, ERY, TE | aac(6′)-Ii, eat(A)v, cadA, cadC, copZ, czrA, merA, merR, tetW/N/W, vanA, zosA | II | 133 |
| E. faecium | 5 | ≥256 | PEN, TE | aac(6′)-Ii, eat(A)v, cadA, cadC, copZ, czrA, merA, merR, tetW/N/W, vanA, zosA | II | 133 |
| Poultry | ||||||
| E. faecium | 1 | ≥256 | ERY | aac(6′)-Ii, aadK, eat(A)v, vanA | II | 13 |
| E. faecium | 1 | ≥256 | PEN | aac(6′)-Ii, aadK, eat(A)v, vanA | II | 157 |
| E. faecium | 1 | ≥256 | – | aac(6′)-Ii, aadK, eat(A)v, vanA | II | 157 |
| E. faecium | 3 | ≥256 | ERY | aac(6′)-Ii, aadK, eat(A)v, vanA | II | 310 |
| E. durans | 1 | ≥256 | TE | aac(6′)-Iid, tetW/N/W, vanA | I | – |
| E. durans | 2 | ≥256 | ERY, TE | aac(6′)-Iid, ermB, vanA | I | – |
| E. durans | 1 | 256 | ERY, TE | aac(6′)-Iid, ermB tetW/N/W, vanA | I | – |
| E. durans | 1 | ≥256 | TE | aac(6′)-Iid, ermB, tetW/N/W, vanA | II | – |
| E. durans | 1 | ≥256 | ERY, TE | aac(6′)-Iid, ermB tetW/N/W, vanA | II | – |
| E. durans | 3 | ≥256 | ERY, TE | aac(6′)-Iid, ermB tetW/N/W, vanA | III | – |
| E. durans | 1 | ≥256 | TE | aac(6′)-Iid, tetW/N/W, vanA | III | – |
Abbreviations: aac(6′)-Ii and aac(6′)-Iid: genes for aminoglycoside N-acetyltransferases; aadK, aminoglycoside 6-adenylyl-transferase; cadA, cadC, cadmium resistance genes; cop, copper resistance gene; czrA, metal transport repressor gene; dfrE, dihydrofolate reductase gene; eat(A)v, allelic variant of eat(A) gene for resistance to lincosamides, streptogramins A, and pleuromutilins (LSAP); emeA, enterococcal multidrug resistance efflux gene; efrA, efrB, ABC multidrug efflux pump genes; ermB, gene for 23S ribosomal RNA methyl-transferase; lsaA, active efflux ABC transporter gene for intrinsic LSAP resistance; merA, merR, mercury resistance genes; MIC, minimal inhibitory concentration; MLST, multilocus sequence type; tetW/N/W, mosaic tetracycline resistance gene and ribosomal protection protein; vanA, vancomycin resistance gene; zosA, zinc transporter gene.
Susceptibility profiling for additional antimicrobials with existing CLSI breakpoints showed that among the six E. faecium from pigs, the resistance phenotype PEN/TE, found in five isolates, represented the most common pattern (83%), followed by ERY/PEN/TE, detected in one isolate (17%).
Among the E. faecium from poultry, the ERY phenotype (determined in four isolates, 67%) predominated over the PEN phenotype (one isolate, 17%), and one isolate remained susceptible.
For E. durans, the resistance pattern ERY/TE was observed in seven (70%) of the isolates, while TE was found in three (30%) of the isolates, respectively.
The E. faecalis isolated from cattle did not exhibit an additional resistance phenotype.
Other antimicrobials included in the Micronaut-S livestock 4 susceptibility plate panel, to which enterococci are intrinsically resistant, or for which no breakpoints are available, had MIC values that are listed in Supplementary Table S1.
Regarding the resistance genotype, aminoglycoside resistance genes were detected among 22 (95.7%) of the 23 strains analysed, corresponding to all the E. faecium and E. durans strains and excluding the E. faecalis isolate. Tetracycline resistance genes were observed uniformly among E. faecium from pigs, in eight (80%) of the E. durans, but were absent in E. faecium for poultry and in the E. faecalis isolate (Table 1). Erythromycin genes were detected in eight (80%) of the E. durans strains. Notably, the lincosamide, streptogramin A, and pleuromutilin (LSAP) resistance gene eat(A)v was identified uniformly in E. faecium from pigs and from poultry (Table 1).
Genes potentially conferring metal-resistance including cadmium resistance genes cadA and cadC, the copper resistance gene copZ, the metal transport repressor gene czrA, mercury resistance genes merA and merR, and the putative zinc transporter gene zosA, were detected exclusively in isolates from pigs.
3.3. Mating Experiments
To determine the risk of VRE spread, transfer experiments were performed. A subset of 16 strains, including one E. faecalis from cattle, six E. faecium from pigs, six E. faecium from poultry, and three E. durans from poultry were selected as donors for conjugative transfer to the recipient E. faecalis JH2-2. Vancomycin-resistant transconjugants were obtained from five donors, all of which were E. faecium isolates from pigs. Vancomycin resistance was transferred with frequencies of 1.7 × 10−7 (donor Sw245), 4.3 × 10−7 (donor Sw253), 5 × 10−6 (donor Sw290), 2 × 10−6 (Sw292), and 1.5 × 10−5 (Sw342), per recipient, respectively.
3.4. Characterisation of the Tn1546 Structures
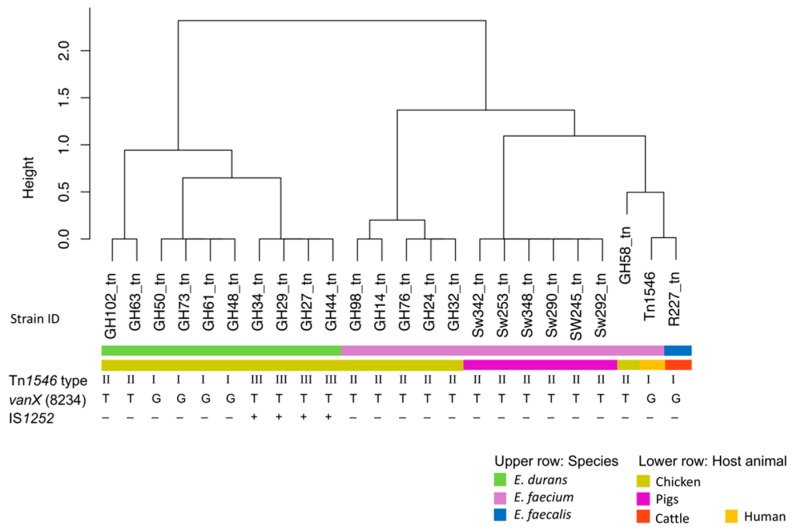
Analysis of the Tn1546 structures distinguished three different Tn1546-like types I-III, respectively (Table 1). A cluster dendrogram showing the Tn1546 types of all the isolates analyzed in this study and the prototype Tn1546 (M97297) is presented in Figure 1.
Figure 1.
Average nucleotide identity (ANI) based cluster dendrogram showing three types of Tn1546-like elements carrying vanA operons identified in 23 vanA- type vancomycin-resistant enterococci from healthy food-producing animals. Type I corresponds to the prototype Tn1546 (GenBank M97297) from human E. faecium B4147 [35]. Type II carries the G to T nucleotide point mutation at position 8234 in vanX. Tn1546-like element type III additionally carries an IS1252 in the orf2-vanR intergenic region.
The structure of the van operon in type I was identical to the van operon prototype described previously (GenBank M97297), and included four E. durans isolates from poultry and the E. faecalis isolate from cattle (Table 1). The Tn1546-like type I elements detected in E. durans contained a topoisomerase gene downstream of vanZ, placing them in a highly similar but distinct cluster to the E. faecalis Tn1546 (Figure 1).
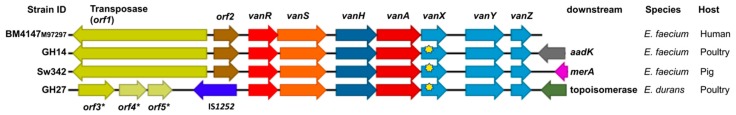
Type II Tn1546-like elements carried a G to T point mutation in vanX at position 8234 of the van operon and were identified in the six E. faecium from poultry, in the six E. faecium from pigs, and in two E. durans isolates from poultry (Table 1 and Figure 1). The Tn1546-like type II elements identified in E. faecium from poultry contained an aadK gene downstream of vanZ, whereas those found in pigs carried merA and those detected in E. durans contained a topoisomerase gene. Examples of type II Tn1546-like elements are shown in Figure 2.
Figure 2.
Linear maps of vanA encoding regions of the prototype Tn1546 (GenBank M97297) from human E. faecium B4147 [35], and of vancomycin-resistant enterococci from healthy food-producing animals. Yellow stars indicate the G to T nucleotide point mutation at position 8234 in vanX; aadK, aminoglycoside 6-adenylyltransferase; merA, mercury resistance gene; *, putative open reading frames.
Finally, type III Tn1546 was identical to the type II structure, but disrupted by IS1252 in the orf2-vanR intergenic region. Type III elements were detected in four E. durans isolates from poultry and carried a topoisomerase gene located downstream of vanZ (Figure 2).
3.5. Multilocus Sequence Typing
To determine the genetic diversity of the isolated, cgMLST was performed. Analysis of the 12 VREfm revealed the occurrence of four different sequence types (STs). The most frequent ST was ST133, which was found in the six isolates from pigs. ST310 was identified in three, ST157 in two, and ST13 in one isolate from poultry, respectively (Table 1). MLST further assigned the E. faecalis from cattle to ST29 (Table 1).
4. Discussion
The use of antimicrobials as growth promoters was banned by law in Switzerland in 1999 [39]. During the decades that followed, VRE were detected at very low levels in the context of resistance monitoring of livestock, with only two porcine VREfc isolated in 2009, and none from 2010 to 2012 [40]. However, between 2013 and 2016, one E. faecalis isolate from cattle, one E. faecalis from broilers, and two E. faecium isolates from fattening pigs were resistant to vancomycin, indicating that VRE are present in food animals in Switzerland [40,41].
The present study demonstrates the occurrence of vanA-type E. faecalis, E. faecium, and E. durans among Swiss cattle, pigs, and poultry flocks 20 years after the ban on avoparcin use. The persistence of VRE in the absence of an obvious selective pressure has been observed previously and is thought to be a consequence of co-selection through the therapeutic use of other antimicrobial agents, such as macrolides or tetracycline, and the use of metals, such as copper or zinc, as feed additives [42,43,44]. Accordingly, a high rate of phenotypic resistance to erythromycin and tetracycline was observed among the isolates in this study. Genetically, macrolide resistance encoded by ermB and tetracycline resistance encoded by tet genes, has been linked to the transposons of the Tn1546 family that contain the vanA gene [42,45,46]. Correspondingly, erm(B), and tet(W) were frequently detected among the isolates. Furthermore, the E. faecium isolated from pigs in this study contained zinc and copper resistance genes, which are typical adaptations to the porcine environment [47].
Other resistance genes identified among the E. faecium isolates included the aac(6′)-Ii gene, which is ubiquitous in E. faecium and thought to contribute to intrinsic aminoglycoside resistance [48]. Similarly, aac(6′)-Iid, which likewise is E. durans specific [49], was found uniformly among the E. durans isolates in this study. E. faecalis is intrinsically resistant to lincosamide, streptogramins A, and pleuromutilins (LSAP) due to the presence of linA, which was accordingly observed in the E. faecalis isolate R277 from cattle in this study. By contrast, in E. faecium, LSAP resistance is acquired by a C1349T point mutation in the Enterococcus ABC transporter gene eat(A) [50]. In human isolates, the mutated allelic variant eat(A)v has been reported in 23% of a collection of epidemiologically unrelated clinical isolates, including isolates corresponding to colonization or faecal carriage [50,51]. In the present study, eat(A)v was identified uniformly in E. faecium from poultry and from pigs. To our knowledge, eat(A)v has not been described previously among porcine and poultry associated E. faecium isolates.
VanA-type resistance is generally mediated by Tn1546-like transposons that are frequently carried by self-transferable plasmids [9]. However, under the experimental conditions applied in this study, transfer of vancomycin resistance was obtained only from porcine donors. Our data confirm the possibility of vanA transfer from porcine E. faecium to human E. faecalis, as demonstrated previously in vitro and in vivo in the intestines of mice [52].
The Tn1546 structures among the enterococci from this study were very similar. The majority contained the G8234T point mutation within vanX, which has been found in enterococcal isolates from healthy and hospitalised humans, in pig isolates, in food isolates, and in environmental enterococci [53,54,55,56]. This indicates that this transposon type is widely disseminated and shared between different enterococcal species and ecological niches.
Many vanA type Tn1546-like structures contain insertion sequence (IS) elements that likely play a role in the evolution of vancomycin resistance [9]. IS1216V is one of the main IS elements frequently observed in the orf2-vanR and the vanX–vanY intergenic regions within Tn1546 from different sources worldwide [14,53,57]. The absence of IS1216V within the Tn1546 elements of the isolates from this study is characteristic for VREfm identified previously in strains in Europe in the late 1990s and 2000s, indicating that the vanA-type resistance mechanism may be very conserved among livestock enterococci in Switzerland. The lack of the diversity observed in Tn1546 structures, which is typical for human clinical isolates, suggests a limited sharing of resistance genes between livestock and human VRE, as observed previously for livestock and human enterococci isolates analysed in the United Kingdom [58].
Using cgMLST indicated that the E. faecalis and the E. faecium isolates belonged to STs typically identified among livestock-associated strains [55]. E. faecium isolated from pigs in this study were represented by a distinct population belonging to ST133. ST133 clusters within subgroup complex-5 (ST5), which contains STs that has been identified in pig hosts in Europe, including pigs from Denmark, Portugal, Spain, and Switzerland [54]. Further, ST5 has been found previously in healthy and in hospitalised patients in Denmark, Germany, and Portugal, notably however, unrelated to nosocomial spread [17,54]. These findings suggest a limited potential for transmission of VRE between humans and pigs. Similarly, E. faecium ST310, detected in three poultry isolates, is a poultry-adapted ST that is prevalent among broilers in Sweden [59,60], and E. faecium ST13 and ST157 have been reported in poultry in Sweden, Denmark, and Korea [55]. Taken together, MLST analysis did not reveal any close relationship to typical nosocomial strains belonging to CC17, or to recently emerged endemic strains ST203 and ST796. Likewise, E. durans is detected frequently in healthy poultry, but is reported only sporadically in human clinical infections [55]. E. durans of human and animal origin have been found to contain similar genetic arrangements of the vanA gene cluster, and it has been shown in vitro that E. durans transfers vanA to human clinical E. faecium at a high frequency [61,62]. Conversely, in our study, the E. durans did not result in transferability to E. faecalis, at least under the given experimental circumstances.
5. Conclusions
Our study provides further evidence of the occurrence of vanA-type VRE in livestock, including healthy cattle, pigs, and poultry. Our results suggest that porcine E. faecium may be prone to transfer vanA genes to human related E. faecalis. Furthermore, our data confirm previous studies that show that there is limited sharing of livestock-associated VRE strains with strains associated with sporadic human disease, and we did not identify any clones related to hospital-related outbreak strains, such as CC17.
Acknowledgments
We gratefully acknowledge the support and assistance of the staff of the Veterinary Service at the slaughterhouse in Zürich, and the staff of the Bell company slaughterhouse in Zell, Switzerland. We thank Professor Christian Lacroix, Institute of Food Science and Nutrition, ETH Zürich, for providing the E. faecalis JH2-2 strain.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/2/261/s1, Table S1: Minimal inhibitory concentrations (MICs) of antimicrobial agents for 23 vancomycin-resistant Enterococcus spp. isolated from healthy cattle, pigs, and poultry.
Author Contributions
Conceptualization, R.S.; methodology, K.Z., M.M., M.S., N.C., and V.W.; software, M.J.A.S.; validation, R.S., and V.W.; formal analysis, M.N.-I., M.J.A.S., V.W., and R.S.; investigation, V.W.; data curation, M.J.A.S., V.W., and R.S.; writing—original draft preparation, M.N.-I.; writing—review and editing, K.Z., M.M., M.N.-I., M.J.A.S., M.S., R.S., and V.W.; supervision, R.S.; funding acquisition, R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially funded by the Federal Office for Public Health, Switzerland.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Puchter L., Chaberny I.F., Schwab F., Vonberg R.P., Bange F.C., Ebadi E. Economic burden of nosocomial infections caused by vancomycin-resistant enterococci. Antimicrob. Resist. Infect. Control. 2018;7:1. doi: 10.1186/s13756-017-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 3.ECDC . Surveillance of antimicrobial resistance in Europe 2018. European Centre for Disease Prevention and Control; Stockholm, Sweden: 2019. pp. 57–60. [Google Scholar]
- 4.Wassilew N., Seth-Smith H.M., Rolli E., Fietze Y., Casanova C., Führer U., Egli A., Marschall J., Buetti N. Outbreak of vancomycin-resistant Enterococcus faecium clone ST796, Switzerland, December 2017 to April 2018. Eurosurveillance. 2018;23:1800351. doi: 10.2807/1560-7917.ES.2018.23.29.1800351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howden B.P., Holt K.E., Lam M.M.C., Seemann T., Ballard S., Coombs G.W., Tong S.Y.C., Grayson M.L., Johnson P.D.R., Stinear T.P. Genomic insights to control the emergence of vancomycin-resistant enterococci. MBio. 2013;4:e00412-13. doi: 10.1128/mBio.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinholt M., Gumpert H., Bayliss S., Nielsen J.B., Vorobieva V., Pedersen M., Feil E., Worning P., Westh H. Genomic analysis of 495 vancomycin-resistant Enterococcus faecium reveals broad dissemination of a vanA plasmid in more than 19 clones from Copenhagen, Denmark. J. Antimicrob. Chemother. 2016;72:40–47. doi: 10.1093/jac/dkw360. [DOI] [PubMed] [Google Scholar]
- 7.Raven K.E., Gouliouris T., Brodrick H., Coll F., Brown N.M., Reynolds R., Reuter S., Török M.E., Parkhill J., Peacock S.J. Complex routes of nosocomial vancomycin-resistant Enterococcus faecium transmission revealed by genome sequencing. Clin. Infect. Dis. 2017;64:886–893. doi: 10.1093/cid/ciw872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed M.O., Baptiste K.E. Vancomycin-resistant enterococci: A review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb. Drug Resist. 2018;24:590–606. doi: 10.1089/mdr.2017.0147. [DOI] [PubMed] [Google Scholar]
- 9.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 2006;42:S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 10.Sletvold H., Johnsen P.J., Wikmark O.-G., Simonsen G.S., Sundsfjord A., Nielsen K.M. Tn 1546 is part of a larger plasmid-encoded genetic unit horizontally disseminated among clonal Enterococcus faecium lineages. J. Antimicrob. Chemother. 2010;65:1894–1906. doi: 10.1093/jac/dkq219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acar J., Casewell M., Freeman J., Friis C., Goossens H. Avoparcin and virginiamycin as animal growth promoters: A plea for science in decision-making. Clin. Microbiol. Infect. 2000;6:477–482. doi: 10.1046/j.1469-0691.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- 12.Yu H.S., Seol S.Y., Cho D.T. Diversity of Tn1546-like elements in vancomycin-resistant enterococci isolated from humans and poultry in Korea. J. Clin. Microbiol. 2003;41:2641–2643. doi: 10.1128/JCM.41.6.2641-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willems R.J.L., Top J., van den Braak N., van Belkum A., Mevius D.J., Hendriks G., van Santen-Verheuvel M., van Embden J.D.A. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 1999;43:483–491. doi: 10.1128/AAC.43.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wardal E., Kuch A., Gawryszewska I., Żabicka D., Hryniewicz W., Sadowy E. Diversity of plasmids and Tn1546-type transposons among vanA Enterococcus faecium in Poland. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:313–328. doi: 10.1007/s10096-016-2804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López M., Sáenz Y., Alvarez-Martínez M.J., Marco F., Robredo B., Rojo-Bezares B., Ruiz-Larrea F., Zarazaga M., Torres C. Tn1546 structures and multilocus sequence typing of vanA-containing enterococci of animal, human and food origin. J. Antimicrob. Chemother. 2010;65:1570–1575. doi: 10.1093/jac/dkq192. [DOI] [PubMed] [Google Scholar]
- 16.Bayjanov J.R., Baan J., Rogers M.R.C., Troelstra A., Willems R.J.L., van Schaik W. Enterococcus faecium genome dynamics during long-term asymptomatic patient gut colonization. Microb. Genom. 2019;5:e000277. doi: 10.1099/mgen.0.000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willems R.J.L., Top J., Marga van Santen D., Coque T.M., Baquero F., Grundmann H., Bonten M.J.M. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 2005;11:821. doi: 10.3201/1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebreton F., van Schaik W., McGuire A.M., Godfrey P., Griggs A., Mazumdar V., Corander J., Cheng L., Saif S., Young S. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio. 2013;4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammerum A.M., Baig S., Kamel Y., Roer L., Pinholt M., Gumpert H., Holzknecht B., Røder B., Justesen U.S., Samulioniené J., et al. Emergence of vanA Enterococcus faecium in Denmark, 2005–2015. J. Antimicrob. Chemother. 2017;72:2184–2190. doi: 10.1093/jac/dkx138. [DOI] [PubMed] [Google Scholar]
- 20.Mahony A.A., Buultjens A.H., Ballard S.A., Grabsch E.A., Xie S., Seemann T., Stuart R.L., Kotsanas D., Cheng A., Heffernan H., et al. Vancomycin-resistant Enterococcus faecium sequence type 796—Rapid international dissemination of a new epidemic clone. Antimicrob. Resist. Infect. Control. 2018;7:44. doi: 10.1186/s13756-018-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buultjens A.H., Lam M.M., Ballard S., Monk I.R., Mahony A.A., Grabsch E.A., Grayson M.L., Pang S., Coombs G.W., Robinson J.O., et al. Evolutionary origins of the emergent ST796 clone of vancomycin resistant Enterococcus faecium. Peer J. 2017;5:e2916. doi: 10.7717/peerj.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson O. Vancomycin resistant enterococci in farm animals—Occurrence and importance. Infect. Ecol. Epidemiol. 2012;2:16959. doi: 10.3402/iee.v2i0.16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual (3-Volume Set) 4th ed. Cold Spring Harbor Laboratory Press; Cold Spring Habor, NY, USA: 2001. pp. 11–28. [Google Scholar]
- 24.Dutka-Malen S., Evers S., Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 1995;33:1434. doi: 10.1128/JCM.33.5.1434-1434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wambui J., Tasara T., Njage P.M.K., Stephan R. Species distribution and antimicrobial profiles of Enterococcus spp. isolates from Kenyan small and medium enterprise slaughterhouses. J. Food Prot. 2018;81:1445–1449. doi: 10.4315/0362-028X.JFP-18-130. [DOI] [PubMed] [Google Scholar]
- 26.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Volume 38. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. pp. 1–198. CLSI supplement M100. [Google Scholar]
- 27.Jacob A.E., Hobbs S.J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 1974;117:360–372. doi: 10.1128/JB.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seemann T. Shovill. [(accessed on 15 January 2019)]; Available online: https://githubcom/tseemann/shovill.
- 30.Stevens M.J.A., Cernela N., Corti S., Stephan R. Draft genome sequence of Streptococcus parasuis 4253, the first available for the species. Microbiol. Res. Announc. 2019;8:18. doi: 10.1128/MRA.00203-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen M.V., Cosentino S., Lukjancenko O., Saputra D., Rasmussen S., Hasman H., Sicheritz-Pontén T., Aarestrup F.M., Ussery D.W., Lund O. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 2014;52:1529–1539. doi: 10.1128/JCM.02981-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 33.Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016:gkw1004. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J., Bhullar K., Canova M.J., De Pascale G., Ejim L. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arthur M., Depardieu F., Molinas C., Reynolds P., Courvalin P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene. 1995;154:87–92. doi: 10.1016/0378-1119(94)00851-I. [DOI] [PubMed] [Google Scholar]
- 36.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. PNAS. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard L., Glover R.H., Humphris S., Elphinstone J.G., Toth I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods. 2016;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 38.Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K. Cluster: Cluster Analysis Basics and Extensions. 2019. R package version 2.1.0.
- 39.Arnold S., Gassner B., Giger T., Zwahlen R. Banning antimicrobial growth promoters in feedstuffs does not result in increased therapeutic use of antibiotics in medicated feed in pig farming. Pharmacoepidemiol. Drug Saf. 2004;13:323–331. doi: 10.1002/pds.874. [DOI] [PubMed] [Google Scholar]
- 40.FOPH . Swiss Antibiotic Resistance report 2018. Usage of Antibiotics and Occurrence of Antibiotic Resistance in Bacteria from Humans and Animals in Switzerland. November 2018. Federal Office of Public Health and Federal Food Safety and Veterinary Office; Berne, Switzerland: 2018. pp. 98–102. Publication number:2018-OEG-87. [Google Scholar]
- 41.FOPH . Swiss Antibiotic Resistance Report 2016. Usage of Antibiotics and Occurrence of Antibiotic Resistance in Bacteria from Humans and Animals in Switzerland. November 2016. Federal Office of Public Health and Federal Food Safety and Veterinary Office; Berne, Switzerland: 2016. pp. 80–87. Publication number:2016-OEG-30. [Google Scholar]
- 42.Aarestrup F.M. Characterization of glycopeptide-resistant Enterococcus faecium (GRE) from broilers and pigs in Denmark: Genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J. Clin. Microbiol. 2000;38:2774–2777. doi: 10.1128/JCM.38.7.2774-2777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cattoir V., Leclercq R. Twenty-five years of shared life with vancomycin-resistant enterococci: Is it time to divorce? J. Antimicrob. Chemother. 2013;68:731–742. doi: 10.1093/jac/dks469. [DOI] [PubMed] [Google Scholar]
- 44.Hammerum A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 2012;18:619–625. doi: 10.1111/j.1469-0691.2012.03829.x. [DOI] [PubMed] [Google Scholar]
- 45.Silva V., Igrejas G., Carvalho I., Peixoto F., Cardoso L., Pereira J.E., del Campo R., Poeta P. Genetic characterization of vanA Enterococcus faecium isolates from wild red-legged partridges in Portugal. Microb. Drug Resist. 2018;24:89–94. doi: 10.1089/mdr.2017.0040. [DOI] [PubMed] [Google Scholar]
- 46.Lozano C., Gonzalez-Barrio D., Camacho M.C., Lima-Barbero J.F., de la Puente J., Höfle U., Torres C. Characterization of faecal vancomycin-resistant enterococci with acquired and intrinsic resistance mechanisms in wild animals, Spain. Microb. Ecol. 2016;72:813–820. doi: 10.1007/s00248-015-0648-x. [DOI] [PubMed] [Google Scholar]
- 47.Yazdankhah S., Rudi K., Bernhoft A. Zinc and copper in animal feed—Development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb. Ecol. Health Dis. 2014;25:25862. doi: 10.3402/mehd.v25.25862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright G.D., Ladak P. Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob. Agents Chemother. 1997;41:956–960. doi: 10.1128/AAC.41.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Del Campo R., Galán J.C., Tenorio C., Ruiz-Garbajosa P., Zarazaga M., Torres C., Baquero F. New aac (6′)-I genes in Enterococcus hirae and Enterococcus durans: Effect on β-lactam/aminoglycoside synergy. J. Antimicrob. Chemother. 2005;55:1053–1055. doi: 10.1093/jac/dki138. [DOI] [PubMed] [Google Scholar]
- 50.Isnard C., Malbruny B., Leclercq R., Cattoir V. Genetic basis for in vitro and in vivo resistance to lincosamides, streptogramins A, and pleuromutilins (LSAP phenotype) in Enterococcus faecium. Antimicrob. Agents Chemother. 2013;57:4463–4469. doi: 10.1128/AAC.01030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bérenger R., Bourdon N., Auzou M., Leclercq R., Cattoir V. In vitro activity of new antimicrobial agents against glycopeptide-resistant Enterococcus faecium clinical isolates from France between 2006 and 2008. Med. Mal. Infect. 2011;41:405–409. doi: 10.1016/j.medmal.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 52.Bourgeois-Nicolaos N., Moubareck C., Mangeney N., Butel M., Doucet-Populaire F. Comparative study of vanA gene transfer from Enterococcus faecium to Enterococcus faecalis and to Enterococcus faecium in the intestine of mice. FEMS Microbiol. Lett. 2006;254:27–33. doi: 10.1111/j.1574-6968.2005.00004.x. [DOI] [PubMed] [Google Scholar]
- 53.Novais C., Freitas A.R., Sousa J.C., Baquero F., Coque T.M., Peixe L.V. Diversity of Tn1546 and its role in the dissemination of vancomycin-resistant enterococci in Portugal. Antimicrob. Agents Chemother. 2008;52:1001–1008. doi: 10.1128/AAC.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freitas A.R., Coque T.M., Novais C., Hammerum A.M., Lester C.H., Zervos M.J., Donabedian S., Jensen L.B., Francia M.V., Baquero F. Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. J. Clin. Microbiol. 2011;49:925–931. doi: 10.1128/JCM.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres C., Alonso C.A., Ruiz-Ripa L., León-Sampedro R., Del Campo R., Coque T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018;6:4. doi: 10.1128/microbiolspec.ARBA-0032-2018. [DOI] [PubMed] [Google Scholar]
- 56.Guardabassi L. Dalsgaard, A. Occurrence, structure, and mobility of Tn1546-like elements in environmental isolates of vancomycin-resistant enterococci. Appl. Environ. Microbiol. 2004;70:984–990. doi: 10.1128/AEM.70.2.984-990.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cha J.O., Yoo J.I., Kim H.K., Kim H.S., Yoo J.S., Lee Y.S., Jung Y.H. Diversity of Tn1546 in vanA-positive Enterococcus faecium clinical isolates with VanA, VanB, and VanD phenotypes and susceptibility to vancomycin. J. Appl. Microbiol. 2013;115:969–976. doi: 10.1111/jam.12300. [DOI] [PubMed] [Google Scholar]
- 58.Gouliouris T., Raven K.E., Ludden C., Blane B., Corander J., Horner C.S., Hernandez-Garcia J., Wood P., Hadjirin N.F., Radakovic M. Genomic surveillance of Enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. MBio. 2018;9:e01780-18. doi: 10.1128/mBio.01780-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nilsson O., Greko C., Top J., Franklin A., Bengtsson B. Spread without known selective pressure of a vancomycin-resistant clone of Enterococcus faecium among broilers. J. Antimicrob. Chemother. 2009;63:868–872. doi: 10.1093/jac/dkp045. [DOI] [PubMed] [Google Scholar]
- 60.Nilsson O., Alm E., Greko C., Bengtsson B. The rise and fall of a vancomycin-resistant clone of Enterococcus faecium among broilers in Sweden. J. Glob. Antimicrob. Resist. 2019;17:233–235. doi: 10.1016/j.jgar.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 61.Robredo B., Torres C., Singh K.V., Murray B. Molecular analysis of Tn1546 in vanA-containing Enterococcus spp. isolated from humans and poultry. Antimicrob. Agents Chemother. 2000;44:2588–2589. doi: 10.1128/AAC.44.9.2588-2589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vignaroli C., Zandri G., Aquilanti L., Pasquaroli S., Biavasco F. Multidrug-resistant enterococci in animal meat and faeces and co-transfer of resistance from an Enterococcus durans to a human Enterococcus faecium. Curr. Microbiol. 2011;62:1438–1447. doi: 10.1007/s00284-011-9880-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.