Abstract
Background: Uncertainty in the mechanism and directionality of observational associations between thyroid function and kidney function may be addressed by genetic analysis with an instrumental variable method termed bidirectional Mendelian randomization (MR).
Methods: In the Women's Genome Health Study (WGHS), observational associations between thyroid measures and kidney function were evaluated. Genetic instruments for MR were from recent genome-wide association studies (GWAS) of hypothyroidism, thyrotropin (TSH), and free thyroxine (fT4) concentrations within the reference range, thyroid peroxidase antibodies (TPOAb), estimated glomerular filtration rate from creatinine (eGFRcrea), eGFR from cystatin C (eGFRcys), and chronic kidney disease (CKD). In WGHS individual-level data, these instruments were used for bidirectional MR between thyroid (N = 3336) and kidney (N = 23,186) functions. To increase power, MR was also performed using GWAS summary statistics from the Chronic Kidney Disease Genetics Consortium (CKDGen) for eGFRcrea (N = 567,460), eGFRcys (N = 24,063), CKD [N(total) = 480,698, N(cases) = 41,395], and urinary albumin/creatinine ratio (UACR/N = 54,450).
Results: In the WGHS, hypothyroidism was observationally associated with decreased eGFRcrea [beta (standard error, SE): −0.024 (0.009) ln(mL/min/1.73 m2), p = 0.01]. By MR, hypothyroidism was associated with decreased eGFRcrea in the WGHS [beta (SE): −0.007 (0.002) per doubled odds hypothyroidism, p = 1.7 × 10−3] and in CKDGen [beta (SE): −0.004 (0.0005), p = 2.0 × 10−22], and robust to sensitivity analysis. Hypothyroidism was also associated by MR with increased CKD in CKDGen (odds ratio, OR [confidence interval, CI]: 1.05 [1.03–1.08], p = 3.3 × 10−5), but not in the WGHS (OR [CI]: 1.02 [0.95–1.10], p = 0.57). Increased TSH within the reference range had an MR association with increased eGFRcrea in the WGHS [beta (SE): −0.018 (0.007) ln(mL/min/1.73 m2)/standard deviation, SD, p = 6.5 × 10−3] and CKDGen [beta (SE): −0.008 (0.001) ln(mL/min/1.73 m2)/SD, p = 6.8 × 10−17], and with CKD in CKDGen (OR [CI]: 1.10 [1.04–1.15], p = 3.1 × 10−4). There were no MR associations of hypothyroidism or TSH with eGFRcys or UACR, and MR associations of fT4 in the reference range with kidney function were inconsistent in both the WGHS and CKDGen. However, by MR in CKDGen, TPOAb were robustly associated with decreased eGFRcrea [beta (SE): −0.041 (0.009), p = 6.2 × 10−6] and decreased eGFRcys [beta (SE): −0.294 (0.065), p = 6.2 × 10−6]. TPOAb were less robustly associated with CKD but not associated with UACR. In reverse MR in the WGHS, kidney function was not consistently associated with thyroid function.
Conclusions: Bidirectional MR supports a directional association from hypothyroidism, increased TSH, and TPOAb, but not fT4, to decreased eGFRcrea and increased CKD.
Keywords: hypothyroidism, kidney function tests, thyrotropin, creatinine, cystatin C
Introduction
Primary hypothyroidism is characterized by a high concentration of thyrotropin (TSH) concomitant with concentrations of thyroid hormones being low (overt) or within the reference range (subclinical). In conventional nongenetic observational studies, both overt and subclinical hypothyroidism are associated with increased plasma creatinine, decreased estimated glomerular filtration rate (eGFR), chronic kidney disease (CKD), and increased urinary albumin/creatinine ratio (UACR) (1–12). Overt hypothyroidism is most often an autoimmune disease in adults, (13) affecting predominantly middle-aged and older women. Thyroid hormones within the reference range may also affect kidney function through direct effects on glomerular and tubular functions and indirect prerenal effects on cardiovascular hemodynamics and renal blood flow (14). Increased TSH within the reference range is associated with reduced eGFR (1,15–22), but whether related triiodothyronine and thyroxine are also associated with kidney function is debated (3,4,15,17,21–23). Diagnosis of kidney disorders may also be related to thyroid dysfunction due to the depletion of TSH, free thyroxine (fT4), and relevant binding proteins from the circulation through leakage into the urine or alternatively to nonthyroidal illness (14,24–27).
Attempts to clarify the causal directionality between thyroid and kidney function from conventional nongenetic observational studies remain inconclusive. Observational designs or small randomized trials show that thyroid hormone replacement therapy improves kidney function in patients with subclinical or overt hypothyroidism (28–32); but no large-scale, long-term randomized trial has been undertaken to more convincingly determine the potential existence and directionality of a causal relationship between thyroid and kidney function.
Mendelian randomization (MR) analysis, which is conceptually analogous to a randomized controlled trial, can be used in a bidirectional design to evaluate effects of thyroid function on kidney function, and vice versa (33). In MR, genetic variation is used as an instrumental variable for a clinical trait, and its randomization at conception is leveraged to evaluate associations through methods that are less affected by reverse causation compared with conventional, nongenetic observational designs (33). Previous MR studies using genetic instruments from genome-wide association studies (GWAS) for TSH and fT4 investigated associations with cardiovascular disease (34,35), type 2 diabetes (36), and bone mineral density (37). A recent MR study using only three variants for TSH investigated the association with creatinine-based eGFR in a Chinese population, but did not find an association (38). No MR study has been performed using a bidirectional approach or using a full set of GWAS instruments for hypothyroidism, TSH, fT4, and thyroid peroxidase antibody (TPOAb) on creatinine- and cystatin C-based eGFR, UACR, and CKD in Europeans to infer the direction behind the conventional, nongenetic observational associations between thyroid and kidney dysfunction. Nevertheless, a comprehensive MR analysis of associations between thyroid and kidney functions has potential for clinical translation in the context of thyroid hormone treatment in patients with hypothyroidism.
We used bidirectional MR to assess the mechanisms and direction of effects between thyroid and kidney functions. We selected single-nucleotide polymorphism (SNP) GWAS instruments to address various measures of thyroid function, namely hypothyroidism (39–41), increased TSH within the reference range (42–44), increased fT4 within the reference range (42–44), and increased antibodies against thyroid peroxidase (TPOAb) (45,46), to predict kidney function [estimated glomerular filtration rate from creatinine [eGFRcrea] (47), eGFR from cystatin C [eGFRcys] (48), CKD (47), and UACR (49)] in the Women's Genome Health Study (WGHS) and through published GWAS summary statistics from the Chronic Kidney Disease Genetics Consortium (CKDGen). In addition, in the WGHS, we used GWAS-identified SNP instruments for kidney function as predictors of thyroid function, and thereby investigate reverse directionality.
Materials and Methods
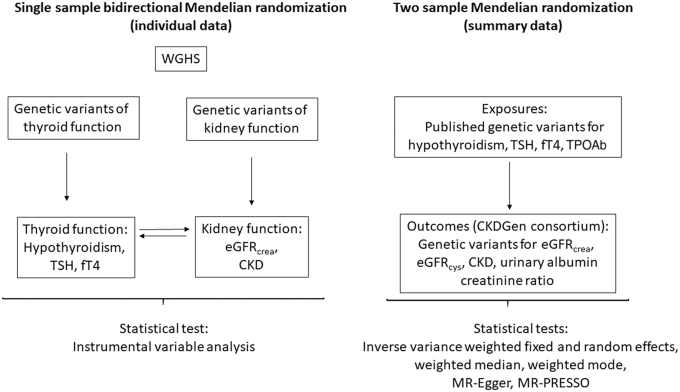
The analytic approach consisted of two parts. First, in a single-sample analysis using individual-level data from the WGHS, we performed MR with genetic risk scores (GRSs) to assess the relevance of hypothyroidism, TSH, and fT4, both within the reference range, to eGFRcrea and CKD (Fig. 1 and Table 1). Similarly, in the reverse direction, we tested the relevance of eGFRcrea and CKD to TSH, fT4, and prevalence of hypothyroidism. Second, to leverage the greater power implicit in the much larger sample sizes of GWASs for thyroid and kidney measures (the latter from the CKDGen), we performed two sample MR to assess the relevance of hypothyroidism, TSH, and fT4 within the reference range, and anti-TPO to eGFRcrea, eGFRcys, UACR, and CKD. For eGFRcrea and CKD, the WGHS and CKDGen were not fully independent due to overlapping samples (4%).
FIG. 1.
Flowchart representing the main design in the MR of thyroid and kidney function. CKDGen, Chronic Kidney Disease Genetics Consortium; eGFRcrea, estimated glomerular filtration rate from creatinine [in ln(mL/min/1.73 m2)]; eGFRcys, estimated glomerular filtration rate from cystatin C [in ln(mL/min/1.73 m2)]; fT4, free thyroxine (per SD); MR, Mendelian randomization; MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier; SD, standard deviation; TPOAb, thyroid peroxidase antibody (per SD); TSH, thyrotropin (per SD); WGHS, Women's Genome Health Study.
Table 1.
Description of Consortia and Cohorts for Each Phenotype of the Association of Thyroid Function with Kidney Function
| Phenotype | First author (year) | Consortium/cohort | Sample size | No. of studies |
|---|---|---|---|---|
| Single sample | ||||
| Hypothyroidism | — | WGHS | 2923 (676 hypothyroidism) | 1 |
| TSH | — | WGHS | 3321 | 1 |
| fT4 | — | WGHS | 3320 | 1 |
| eGFRcrea | — | WGHS | 23,186 | 1 |
| CKD | — | WGHS | 23,186 (2124 CKD) | 1 |
| Two sample | ||||
| Hypothyroidism | Pickrell (2016) | — | 134,641 (17,558 hypothyroidism) | 1 (23andMe, Inc.) |
| TSH | Teumer (2018) | — | 54,288 | 22 |
| fT4 | Teumer (2018) | — | 49,269 | 19 |
| TPOAb | Medici (2014) | — | 12,322 | 11 |
| TPOAb | Schultheiss (2015) | — | 12,214 | 3 |
| eGFRcrea | Wuttke (2019) | CKDGen | 567,460 | 85 |
| CKD | Wuttke (2019) | CKDGen | 480,698 (41,395 CKD) | NR |
| eGFRcys | Gorski (2017) | CKDGen | 24,063 | 11 |
| UACR | Teumer (2016) | CKDGen | 54,450 | 20 |
Individuals contributing to these analyses all had European ancestry.
CKD, chronic kidney disease; CKDGen, Chronic Kidney Disease Genetics Consortium; eGFRcrea, estimated glomerular filtration rate from creatinine [in ln(mL/min/1.73 m2)]; eGFRcys, estimated glomerular filtration rate from cystatin C [in ln(mL/min/1.73 m2)]; fT4, free thyroxine (per SD); NR, not reported; SD, standard deviation; TPOAb, thyroid peroxidase antibody (per SD); TSH, thyrotropin (per SD); UACR, urinary albumin/creatinine ratio [ln(mg/g)]; WGHS, Women's Genome Health Study.
The Women's Genome Health Study
The WGHS includes 23,294 women with whole-genome genotype data and verified European ancestry who were 45 years or older and free of cardiovascular disease and cancer at the time of enrollment, that is, baseline (50). The cohort and the genotyping are described in Supplementary Data. Of the 23,186 women with serum creatinine at baseline, assessment of thyroid function was available in 3336 individuals through measures of TSH (mIU/mL, N = 3321), fT4 (ng/dL, N = 3320), and free triiodothyronine (fT3; pg/mL, N = 3321) (51). In this subsample, euthyroidism was defined as 0.27 ≤ TSH ≤4.2 mIU/L and 0.93 ≤ fT4 ≤ 1.7 ng/dL (N = 2247), and hypothyroidism (N = 676) was defined as TSH >4.2 mIU/L with either normal fT4 (i.e., subclinical, 0.93 ≤ fT4 ≤ 1.7 ng/dL) or decreased fT4 (i.e., overt hypothyroidism, fT4 < 0.93 ng/dL) (51). The study was approved by the Institutional Review Board at the Brigham and Women's Hospital (Boston, MA).
Genetic instruments
Thyroid function: Independent SNPs with minor allele frequency >1% have been identified in GWASs (p < 5 × 10−8) among Europeans for TSH (N SNPs = 42) and fT4 (N = 21) concentrations in the reference range (42–44), TPOAb concentration (N = 5) (45,46), and hypothyroidism (N = 30) (39–41) (Supplementary Data). In the study by Pickrell et al. using data from 23andMe, Inc., the diagnosis of hypothyroidism was self-reported (41). In the study by Teumer et al. using data from various cohorts, the reference range was 0.4–4 mIU/L and hypothyroidism (subclinical+overt) was defined as TSH above the upper reference limit (43,44). In the WGHS, genetic information was available for 14 SNPs for hypothyroidism (omitting SNPs that were either indels or had an ambiguous risk allele), 38 SNPs for TSH, and 21 SNPs for fT4. In the CKDGen GWAS, summary statistics were available for 15 SNPs for hypothyroidism, 39 SNPs for TSH, 21 SNPs for fT4, and 5 SNPs for TPOAb concentration. We included only SNPs in separate loci with minimal linkage disequilibrium (R2 ≤ 0.05).
Kidney function: Genetic instruments for kidney function were identified in summary statistics from the GWAS conducted by meta-analysis by the CKDGen (47,48,52). For use in the reverse MR analysis, the GWAS identified (a) 5 independent SNPs for CKD (52), 4 of which were available in the WGHS, and (b) 53 independent SNPs for eGFRcrea, of which we excluded 8 on the basis of their discovery only in the “no diabetes subgroup” (52), 7 SNPs associated with creatinine production and secretion (53), and 4 SNPs that were not available in the WGHS (49,52) (Supplementary Data).
Measurements of kidney function
In all studies in CKDGen, including the WGHS, serum creatinine was calibrated to the U.S. nationally representative National Health and Nutrition Examination Survey data, to account for between-laboratory variation (52) (Supplementary Data). Values of eGFRcrea were calculated from serum creatinine, using the equation for the Modification of Diet in Renal Disease (MDRD) Study and applied to define CKD (eGFRcrea <60 mL/min/1.73 m2) (52). eGFRcrea was age and sex adjusted using residuals, and then natural log transformed (52). In CKDGen, eGFRcys was estimated as 76.7 × (serum cystatin C)−1.19 (52), and UACR was calculated as urinary albumin/urinary creatinine (mg/g) to account for differences in urine concentration (49). The percentage overlap between CKDGen and WGHS was 4% for eGFRcrea and CKD.
Statistics
As we tested four primary but related traits by MR (hypothyroidism, TSH, fT4, and TPOAb), we considered a two-tailed p-value <0.013 (0.05/4) to be significant (Bonferroni correction). In published GWASs, TSH and fT4 were therefore scaled to study-specific standard deviation (SD) units (42,43). Traits in the WGHS were scaled using the WGHS SDs. All genetic instruments for MR had been reported as genome-wide significant (p < 5.0 × 10−8) associations. To detect a difference in eGFRcrea of 0.1 ln(mL/min/1.73 m2) at an alpha of 0.05 in the MR, we had 34% power for hypothyroidism and TSH in WGHS (N = 23,186) and 100% power in CKDGen (N = 567,460) (Supplementary Table S1 and Supplementary Fig. S1A, B) (54–56).
Single-sample GRS-based MR in the WGHS
Observational associations between thyroid measures (TSH, fT4, fT3, fT3/fT4-ratio) and kidney measures (eGFRcrea, CKD) were investigated at a cross-sectional level at baseline using multivariable adjusted linear regression for continuous outcomes and logistic regression for binary outcomes. Adjustments were made for the following covariates: age (years), smoking (current vs. no smoker), body mass index (kg/m2), HbA1c (%), alcohol (units/week), cholesterol (mg/dL), and systolic blood pressure (mm Hg). A GRS was created by summing the allele count of genotyped alleles or the maximum likelihood dose of imputed alleles at each of the variants, weighted by the published effect sizes (β coefficients) (44). For MR analyses in the WGHS, SNP-exposure and SNP-outcome associations were estimated by linear regression for continuous measures of thyroid function (TSH, fT4) and kidney function (eGFRcrea), while logistic regression was used for hypothyroidism and CKD (i.e., eGFRcrea < 60 mL/min/1.73 m2), adjusting for age at baseline, geographical location, and the first four principal components of population substrata for both types of models. F-statistics for instrument strength were obtained from linear or logistic regressions of “GRS-exposure” associations; and the GRSs for hypothyroidism, TSH, and fT4 all had sufficient instrument strength (F ≥ 10). MR instrumental effects were derived by the instrumental variable Wald estimator, that is, the ratio of the β coefficient from the GRS on the outcome association divided by the β coefficient from the GRS on exposure association, with standard errors (SEs) calculated using the delta method (57). As published GWAS results for TSH and fT4 were based on euthyroid individuals (42–44), we restricted the sample to euthyroid individuals for MR of TSH and fT4. For analyses of hypothyroidism (overt and subclinical combined), the comparison group was euthyroid individuals (Supplementary Data). We multiplied the MR estimate of a binary exposure (i.e., hypothyroidism or CKD) with 0.693 (i.e., loge2) to reflect the average change in the outcome per doubling (twofold increase) in the odds of the binary exposure (58,59).
Two-sample MR using CKDGen
For each SNP, we calculated the instrumental variable ratio as the quotient of the SNP-outcome to SNP-exposure effects, deriving the SE in this estimate with the delta method. We calculated the combined effect across all SNPs using five complementary methods with different assumptions about horizontal pleiotropy to assess robustness: inverse-variance weighted (IVW) fixed effects (IVW-FE) and random effects (IVW-RE), weighted median (WM), weighted mode, and MR-Egger regression (60–63). For the latter, we used simulation extrapolation (SIMEX) if the regression dilution statistic (I2GX) was below 0.90 indicating violation of the No Measurement Error (NOME) assumption (62). We used the Mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) to test for possible bias from horizontal pleiotropy (64). We calculated the proportion variance explained for the outcomes interrogated based on the instruments used (65). As for analysis using the GRS, we report instrumental MR estimates for binary exposures as the estimated average change in the outcome per doubling in the odds of the exposure. See Supplementary Data for details.
Results
Bidirectional single-sample MR using GRSs in the WGHS
Of the 3336 WGHS women with thyroid measures, 676 (20%) had subclinical or overt hypothyroidism and 2247 were euthyroid, while 2124 (9.2%) of 23,186 women with European ancestry had CKD (Table 1 and Supplementary Table S2). Compared with euthyroid women, hypothyroid women were older, had higher systolic blood pressure, and smoked less. Hypothyroidism was associated with decreased eGFRcrea [beta (SE): −0.024 (0.009) ln(mL/min/1.73 m2), p = 0.01], but nonsignificant for CKD (Supplementary Table S3). Increased TSH was also associated with a decreased eGFRcrea and an increased risk of CKD (Supplementary Table S3). However, in euthyroid participants, neither TSH nor fT4 was associated with either eGFRcrea or CKD (Supplementary Table S3).
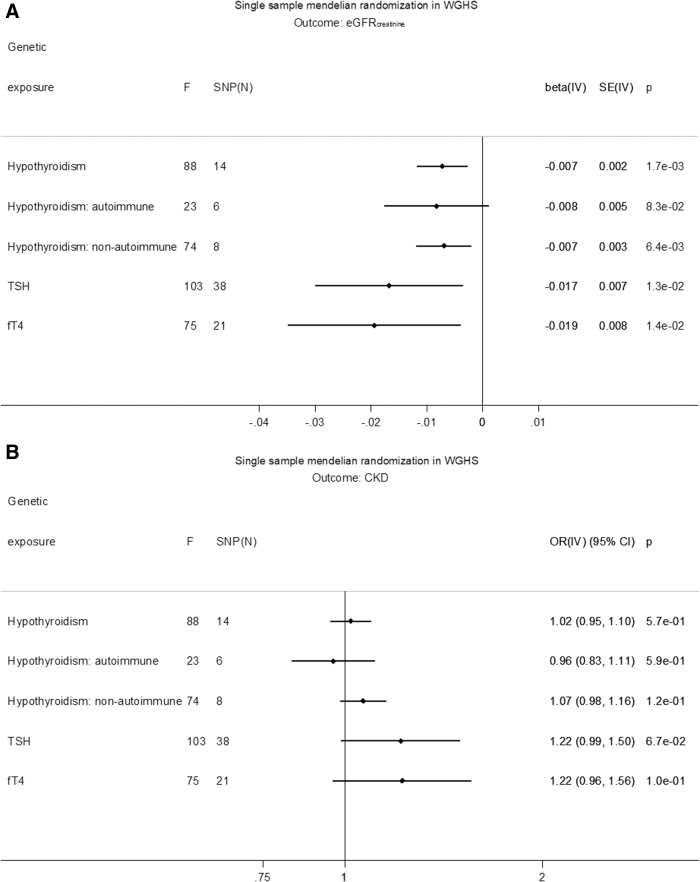
GRS associations with exposure, outcome, and covariates are shown in Supplementary Tables S4–S10. GRSs for hypothyroidism, TSH, fT4, eGFRcrea, and CKD were not associated with covariates, except for the GRS for eGFRcrea and total cholesterol, which was not significant after Bonferroni correction (Supplementary Table S4). In MR, hypothyroidism was associated with decreased eGFRcrea based on a GRS of all SNPs [beta (SE): −0.007 (0.002) ln(mL/min/1.73 m2), p = 1.7 × 10−3] (Fig. 2A). The association was similar for separate GRSs based on SNPs likely involved in either autoimmune [beta (SE): −0.008 (0.005), p = 8.3 × 10−2] or nonautoimmune functions [beta (SE): −0.007 (0.003), p = 6.4 × 10−3]. In MR, increased TSH in the reference range was associated with a decreased eGFRcrea [beta (SE): −0.018 (0.007) ln(mL/min/1.73 m2)/SD, p = 6.5 × 10−3], and increased fT4 in the reference range was associated with decreased eGFRcrea [beta (SE): −0.019 (0.008) ln(mL/min/1.73 m2)/SD, p = 0.01] (Fig. 2A). None of the thyroid measures associated with CKD in MR (Fig. 2B). Conversely, there was no MR association of eGFRcrea with hypothyroidism, TSH, or fT4 within the reference range (Supplementary Table S11). Similarly, there was no association of CKD with hypothyroidism [beta (SE): −0.31 (0.38), p = 0.41] or fT4 within the reference range [beta (SE): −0.21 (0.18), p = 0.25], and the association with TSH [beta (SE): −0.38 (0.18), p = 0.03] was not significant after Bonferroni correction (Supplementary Table S11).
FIG. 2.
(A) Single-sample MR in WGHS of eGFRcrea. (B) Single-sample MR in WGHS of CKD. The causal MR estimates of the various hypothyroidism exposures are multiplied with 0.693 (i.e., loge2) to reflect a doubling (twofold increase) in the odds of the binary exposure. CI, confidence interval; CKD, chronic kidney disease; F, instrument strength measured by F test; IV, instrumental variable analysis; OR, odds ratio. SNP, single-nucleotide polymorphism.
Two-sample MR using CKDGen
Hypothyroidism predicting kidney function
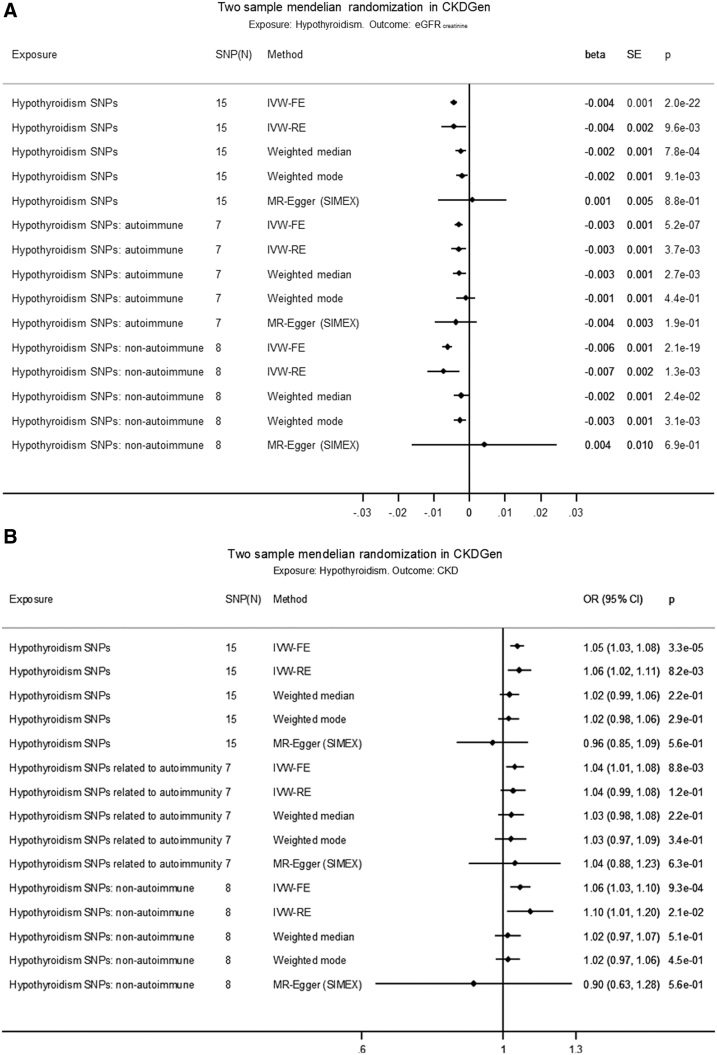
Using the fixed-effects approach (IVW-FE), hypothyroidism was associated with a decrease in eGFRcrea [beta (SE): −0.004 (0.001) ln(mL/min/1.73 m2), p = 2.0 × 10−22] based on all SNPs, with comparable estimates based on autoimmune SNPs and nonautoimmune SNPs [beta (SE), p: −0.003 (0.001), 5.2 × 10−7 d. −0.006 (0.001), 2.1 × 10−19] (Fig. 3A, Supplementary Tables S12 and S13, and Supplementary Fig. S2A, B). Using all SNPs, the IVW-RE, WM, and weighted mode estimates were similar and significant, but the regression dilution statistic (I2GX) in the MR-Egger regression was 0.85, indicating violation of NOME. However, using all SNPs, the heterogeneity in MR-Egger (I2 = 0.92) and IVW-FE (I2 = 0.93) was similar (Supplementary Table S14), indicating that MR-Egger was not a better fit to the data than IVW-FE. There was no evidence of horizontal pleiotropy influencing the estimates in that (a) the MR-Egger intercept was not significant (p = 0.34), (b) the MR-PRESSO outlier adjustment, which showed that even removing the instrument with the largest distorting effect, rs2396084 (at the VEGFA locus), did not result in a significantly different estimate [p(distortion test) = 0.14] (Supplementary Table S15), and (c) the funnel plot of individual SNP effects showed a symmetrical distribution around the overall IVW-FE effect estimate (Supplementary Fig. S2C).
FIG. 3.
(A) Two-sample MR in CKDGen of hypothyroidism on eGFRcrea. (B) Two-sample MR in CKDGen of hypothyroidism on CKD. The causal MR estimates of the various hypothyroidism exposures are multiplied with 0.693 (i.e., loge2) to reflect a doubling (twofold increase) in the odds of the binary exposure. IVW-FE, inverse-variance weighted fixed effects; IVW-RE, inverse-variance weighted random effects; SIMEX, simulation extrapolation algorithm for the MR-Egger model when the “No Measurement Error” assumption is violated, that is, I2GX below 0.9.
Regarding other measures of kidney function, hypothyroidism was associated with eGFRcys in IVW-FE [beta (SE): −0.008 (0.003) ln(mL/min/1.73 m2), p = 0.007], but other MR methods were nonsignificant (Supplementary Fig. S3A–D). The MR-PRESSO distortion test for eGFRcys was significant (p < 0.001), indicating outsized influences for both rs3184504 (SH2B3) and rs2396084 (VEGFA) (Supplementary Tables S14 and S15). However, hypothyroidism based on autoimmune SNPs was associated with eGFRcys in IVW-FE [beta (SE): −0.017 (0.004) ln(mL/min/1.73 m2), p = 3.8 × 10−5] with similar and significant results for IVW-RE and WM, and with similar but nonsignificant results for weighted mode and MR-Egger (Supplementary Fig. S3A). Hypothyroidism based on nonautoimmune SNPs was not associated with eGFRcys in any models (Supplementary Fig. S3A).
Hypothyroidism was associated with an increase in CKD in IVW-FE (odds ratio, OR [confidence interval, CI]: 1.05 [1.03–1.08], p = 3.3 × 10−5, I2 = 0.73) based on all SNPs (Fig. 3B and Supplementary Fig. S4A–C), with comparable estimates based separately on autoimmune SNPs (OR [CI]: 1.04 [1.01–1.08], p = 8.8 × 10−3) and nonautoimmune SNPs (OR [CI]: 1.06 [1.03–1.10], p = 9.3 × 10−4). Using all SNPs, the IVW-RE estimate was comparable and significant, whereas the WM or weighted mode was comparable but nonsignificant, and the MR-Egger not significant. There was no evidence (all p > 0.05) for an association between hypothyroidism and UACR (Supplementary Fig. S5A–D).
TSH and fT4 within the reference range predicting kidney function
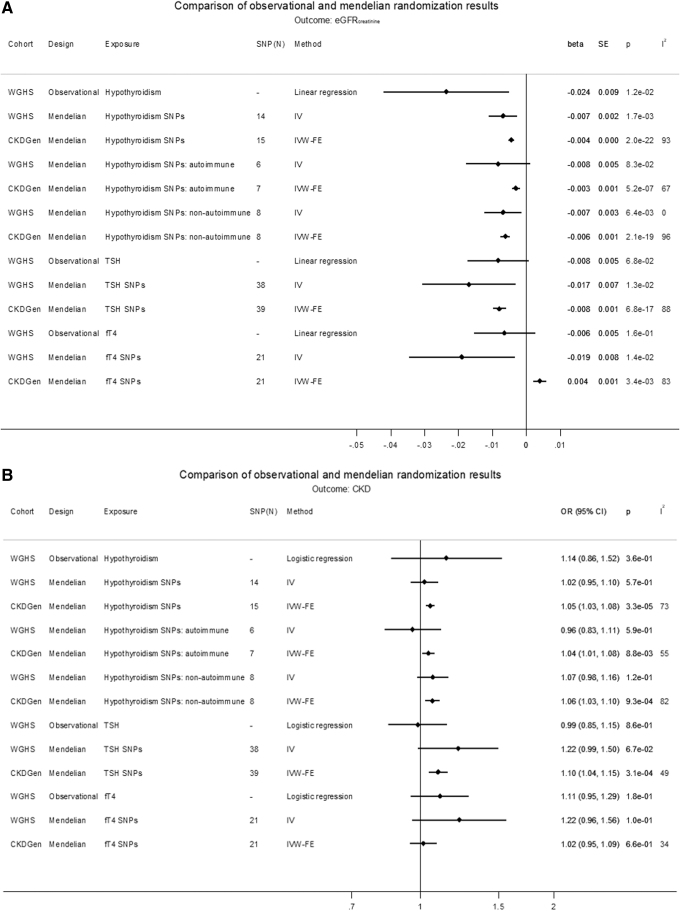
Increased TSH within the reference range was associated with a decrease in eGFRcrea [beta (SE): −0.008 (0.001) ln(mL/min/1.73 m2)/SD, p = 6.8 × 10−17], with comparable estimates for IVW-RE, WM, and weighted mode, and comparable but not significant for MR-Egger (Supplementary Fig. S6A−D). TSH also associated with an increase in CKD in IVW-FE (OR [CI]: 1.10 [1.04–1.15], p = 3.1 × 10−4) and IVW-RE, with comparable but nonsignificant estimates for WM, weighted mode, and MR-Egger. However, there was no MR association of TSH with eGFRcys or UACR (Fig. 4A, B, and Supplementary Figs. S6A–S9C). fT4 within the reference range associated with an increase in eGFRcrea in IVW-FE [beta (SE): 0.004 (0.001) ln(mL/min/1.73 m2)/SD, p = 3.4 × 10−3] (Fig. 4A), but with noncomparable estimates in other analyses, and did not associate with eGFRcys, CKD, or UACR (Supplementary Figs. S10A–S13C and Supplementary Tables S14–S17).
FIG. 4.
(A) Comparison of observational and MR results on eGFRcrea. (B) Comparison of observational and MR results on CKD. The causal MR estimates of the various hypothyroidism exposures are multiplied with 0.693 (i.e., loge2) to reflect a doubling (twofold increase) in the odds of the binary exposure.
TPOAb predicting kidney function
TPOAb were inversely associated with eGFRcrea with IVW-FE [beta (SE): −0.041 (0.009) ln(mL/min/1.73 m2)/SD, p = 6.2 × 10−6, I2 = 0.83], with similar and significant estimates in IVW-RE and WM, and with a similar but nonsignificant estimate in weighted mode but not MR-Egger (Supplementary Figs. S14, S15A–C and Supplementary Tables S14, S15, and S18). TPOAb were also inversely associated with eGFRcys in IVW-FE [beta (SE): −0.294 (0.065) ln(mL/min/1.73 m2), p = 6.2 × 10−6, I2 = 0.83], with a similar and significant estimate in IVW-RE (p = 0.02), and similar but nonsignificant estimates in WM and weighted mode (both p = 0.06), and nonsignificant for MR-Egger (Supplementary Figs. S14 and S16A–C). The MR-PRESSO distortion test was significant for TPOAb on eGFRcys (p < 0.001) for ATXN2 (rs653178) with an outlier-corrected estimate for eGFRcys of [beta (SE): −0.172 (0.036) ln(mL/min/1.73 m2)/SD, p = 4.0 × 10−2] (Supplementary Fig. S14 and Supplementary Tables S14 and S15). Increased TPOAb were associated with an increase in CKD (OR [CI]: 1.7 [1.06–2.74], p = 0.027) (Supplementary Fig. S17A−C), with comparable but nonsignificant estimates for IVW-RE, WM, weighted mode, and the MR-PRESSO outlier-corrected estimate, but nonsignificant for MR-Egger. TPOAb were not associated with UACR (Supplementary Fig. S18A−C).
Discussion
Using genetic analysis, we investigated the potential role of hypothyroidism in the development of decreased kidney function using the WGHS and summary statistics from the CKDGen. We found that genetically predicted hypothyroidism was associated with decreased eGFRcrea and an increased odds for CKD defined by eGFRcrea <60 mL/min/1.73 m2. Genetically predicted TSH within the reference range was associated with an increase in eGFRcrea in WGHS and CKDGen, and with an increase in CKD in CKDGen, with a similar but nonsignificant result in WGHS. There was no robust MR association of hypothyroidism or TSH with eGFRcys or UACR. In MR, fT4 in the reference range was inconsistently associated with kidney function in the WGHS and CKDGen. Using CKDGen summary statistics, genetically predicted TPOAb were associated with decreased eGFRcrea and eGFRcys but not with UACR. TPOAb were associated with increased CKD in some but not all analyses. In the reverse MR in the WGHS, eGFRcrea and CKD were not associated with hypothyroidism or either TSH or fT4 in the reference range.
For the association between hypothyroidism and decreased eGFRcrea, the observational analysis, the GRS-based MR in the WGHS, and the various MR methods in CKDGen (IVW, WM, and weighted mode) were all significant and in the same directions and with similar effects for autoimmune versus nonautoimmune instruments. These findings were strengthened by the MR association of TPOAb, a measure of autoimmune hypothyroidism, and TSH in the physiological range with kidney function. The finding is also consistent with the glomerular involvement seen in patients with Hashimoto's thyroiditis, an autoimmune thyroid disease (66). The power for fT4 should have been comparable with TSH, given the comparable samples contributing to the two GWASs. Given that both measures reflect thyroid function, it is therefore difficult to reconcile the TSH MR association with kidney function with the lack of an fT4 MR association. However, we note that the instruments for TSH and fT4 were different and that TSH may better represent thyroid function in as much as the TSH instruments are associated with TSH levels both outside and within the reference range, whereas the fT4 instruments are only associated with thyroid function within the reference range (44). Observational estimates in the WGHS also suggested an association between kidney and thyroid functions as in previous observational studies (14,24–26), but neither genetically predicted eGFRcrea nor CKD was consistently associated with thyroid function.
Bidirectional MR may clarify directionality of associations from observational studies. We performed bidirectional MR analyses in the WGHS, but we could only perform the forward direction with the thyroid genetic instruments in two-sample analysis using CKDGen, due to lack of available genome-wide summary statistics for thyroid measures, which therefore prohibited formal assignment of effect direction with Steiger filtering analysis (67).
The hypothalamic/pituitary/thyroid axis represents a system of hormones dependent on each other through tight regulation. This vertical pleiotropy risks also being accompanied by horizontal pleiotropy that may have contributed to the high heterogeneity, for example, I2 >0.50, in many analyses. However, the MR sensitivity analysis suggested that potential directional horizontal pleiotropy (i.e., consistent genetic effects on kidney not mediated by thyroid function) did not bias the findings, specifically due to SNPs at SH2B3 (rs3184504) and VEGFA (rs2396084) in the MR association of hypothyroidism with eGFRcys, and the SNP at ATXN2 (rs653178) for the MR association of TPOAb with eGFRcys. These SNPs are nevertheless interesting. Rrs3184504 at SH2B3 (encoding SH2B adaptor protein 3), neighboring and in high linkage disequilibrium (LD) (R2 > 0.9) with rs653178 at ATXN2, is highly pleiotropic and also associated with systolic and diastolic blood pressure, fibrinogen, red and white cell traits, platelet count, and cardiovascular disease, strongly suggesting the potential to affect kidney function through mechanisms other than thyroid function (68). Rs2396084 near VEGFA was strongly associated with both TSH in the reference range and hypothyroidism in previous GWAS (41,42), and was also GWAS significant for decreased eGFRcrea, decreased eGFRcys, risk of CKD, and decreased UACR after accounting for multiple testing (49,52,53), and therefore unsuitable as an MR instrument as revealed by the MR-PRESSO analysis. It is possible that variation at the VEGFA locus influences vascular development in a way that creates susceptibilities to dysfunction for both thyroid and kidney. For example, genetic variants in the VEGFA gene have been associated with vasculitis that in some settings may be exacerbated by an autoimmune response that may underlie hypothyroidism (69).
Associations between exposures and outcomes may not always be linear. Recent developments in MR statistical methodology include exploration of nonlinear associations by approximating associations as linear within piecewise ranges of a continuous exposure (70). Implicitly, this was also our approach by leveraging separate instruments from a separate GWAS for the reference ranges of fT4 and TSH (euthyroid individuals) and for hypothyroidism (elevated TSH). It was not possible to explore nonlinearity in the MR for anti-TPO GWAS because separate ranges of this biomarker are not available.
The power to detect small differences increases with a larger sample size. The CKDGen was 20 times larger than WGHS and thus had a greater power to detect smaller differences in kidney function explained by the genetic variants compared with WGHS. For hypothyroid, TSH, and fT4 SNPs, the power was above 80% and the precision better with narrower CIs for eGFRcrea and CKD in the larger CKDGen compared with WGHS. We also note that the significant MR point estimates for TPOAb predicting eGFRcrea and eGFRcys differed in the IVW-RE/FE analysis even though the two measures are estimating the same aspect of kidney function, that is, GFR. The estimates for eGFRcys generally had wider CIs compared with the eGFRcrea estimates because the GWAS sample for eGFRcys was substantially smaller than for eGFRcrea. In the sensitivity analysis, these CIs overlapped the estimates for eGFRcrea, possibly reconciling the different primary estimates of TPOAb effects.
Despite leveraging some of the best powered samples and data sets available, misclassification remains a potential limitation. The diagnosis of hypothyroidism in both the WGHS and for the GWAS summary statistics was either by self-reporting (41) or by TSH cutoff (44), but not ICD-coding or physician verified. Thus, with either method it was not possible to uniformly distinguish subclinical from overt hypothyroidism, and there is a risk of misclassification. It was also not possible to investigate Hashimoto's thyroiditis directly in this MR study, as only one GWAS article has addressed Hashimoto's disease (71), finding only one SNP in PTPN22 (rs2476601) that was also GWAS significant for Graves' disease and in complete LD (R2, and D2 = 1) with the SNP (rs6679677) we included for hypothyroidism. Larger GWASs need to be performed for these autoimmune diseases. However, we stratified into autoimmune (including the variant in PTPN22) versus nonautoimmune instruments, thereby potentially addressing hypothyroidism due to Hashimoto's thyroiditis somewhat. Similarly, in assessing CKD, we used the same definition in the WGHS as was used by CKDGen, namely eGFRcrea <60 mL/min/1.73 m2, but this definition was not based on kidney disease verified by a nephrologist or ICD-codes.
Hypothyroidism is more common in women than in men (72). While we did not investigate sex differences, the eGFR measure used to derive the CDKGen summary statistics is already sex adjusted, the WGHS included only women, and there were no sex differences for the TSH and fT4 GWAS SNPs published by Teumer et al. (44), all supporting generalizability of our findings in both men and women. However, the GWAS SNPs for hypothyroidism or TPOAb were not assessed for sex differences (41,46).
In summary, bidirectional MR supports the direction of the association of hypothyroidism and increased TSH with decreased eGFRcrea and increased CKD, but not vice versa.
Supplementary Material
Acknowledgments
The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication. We gratefully acknowledge Dr. Alexander Teumer for facilitating access to summary statistics for thyroid measures as well as for insights into the genetic analysis of both thyroid and kidney function.
Contributor Information
Collaborators: on behalf of the CKDGen Consortium
Authors' Contributions
D.I.C. and P.M.R. collected the data in the WGHS. S.M. collected information on thyroid function in the WGHS. C.E. and D.I.C. designed the present study. C.E. and D.I.C. analyzed the data. C.E. made the figures and tables. C.E. and D.I.C. drafted the article. All authors revised the article. All authors approved the final version of the article. Data from CKDGen were publicly available.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
The Women's Genome Health Study (WGHS) is supported by the National Heart, Lung, and Blood Institute (HL043851 and HL080467) and the National Cancer Institute (CA047988 and UM1CA182913), with collaborative scientific support and funding for genotyping provided by Amgen. Additional funding was also provided to Dr. Mora by an investigator-initiated grant from Atherotech Diagnostics (for thyroid measurements), the National Heart, Lung, and Blood Institute by R01HL134811 and K24 HL136852, and the National Institute of Diabetes and Digestive and Kidney Diseases (DK112940). The funding sources did not have any influence on the study design, analyses, interpretation of data, writing of the article, or the decision to submit the article for publication. S.M. received investigator-initiated institutional research grant from Atherotech Diagnostics to measure the thyroid function in the WGHS.
Supplementary Material
References
- 1. Asvold BO, Bjoro T, Vatten LJ. 2011. Association of thyroid function with estimated glomerular filtration rate in a population-based study: the HUNT study. Eur J Endocrinol 164:101–105 [DOI] [PubMed] [Google Scholar]
- 2. Zhou JB, Li HB, Zhu XR, Song HL, Zhao YY, Yang JK. 2017. Subclinical hypothyroidism and the risk of chronic kidney disease in T2D subjects: a case-control and dose-response analysis. Medicine (Baltimore) 96:e6519. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3. Meuwese CL, Gussekloo J, de Craen AJ, Dekker FW, den Elzen WP. 2014. Thyroid status and renal function in older persons in the general population. J Clin Endocrinol Metab 99:2689–2696 [DOI] [PubMed] [Google Scholar]
- 4. Abebe N, Kebede T, Wolde M. 2016. Assessment of renal function and electrolytes in patients with thyroid dysfunction in Addis Ababa, Ethiopia: a cross sectional study. Pan Afr Med J 24:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saini V, Yadav A, Arora MK, Arora S, Singh R, Bhattacharjee J. 2012. Correlation of creatinine with TSH levels in overt hypothyroidism - a requirement for monitoring of renal function in hypothyroid patients? Clin Biochem 45:212–214 [DOI] [PubMed] [Google Scholar]
- 6. Chuang MH, Liao KM, Hung YM, Wang PY, Chou YC, Chou P. 2016. Abnormal thyroid-stimulating hormone and chronic kidney disease in elderly adults in Taipei city. J Am Geriatr Soc 64:1267–1273 [DOI] [PubMed] [Google Scholar]
- 7. Zhang L, Yang G, Su Z, Yang J. 2017. Correlation between subclinical hypothyroidism and renal function in patients with diabetes mellitus. Nephrology (Carlton) 22:790–795 [DOI] [PubMed] [Google Scholar]
- 8. Chen HS, Wu TE, Jap TS, Lu RA, Wang ML, Chen RL, Lin HD. 2007. Subclinical hypothyroidism is a risk factor for nephropathy and cardiovascular diseases in type 2 diabetic patients. Diabet Med 24:1336–1344 [DOI] [PubMed] [Google Scholar]
- 9. El-Eshmawy MM, Abd El-Hafez HA, El Shabrawy WO, Abdel Aal IA. 2013. Subclinical hypothyroidism is independently associated with microalbuminuria in a cohort of prediabetic egyptian adults. Diabetes Metab J 37:450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Furukawa S, Yamamoto S, Todo Y, Maruyama K, Miyake T, Ueda T, Niiya T, Senba T, Torisu M, Kumagi T, Miyauchi S, Sakai T, Minami H, Miyaoka H, Matsuura B, Hiasa Y, Onji M, Tanigawa T. 2014. Association between subclinical hypothyroidism and diabetic nephropathy in patients with type 2 diabetes mellitus. Endocr J 61:1011–1018 [DOI] [PubMed] [Google Scholar]
- 11. Yasuda T, Kaneto H, Kuroda A, Yamamoto T, Takahara M, Naka T, Miyashita K, Fujisawa K, Sakamoto F, Katakami N, Matsuoka TA, Shimomura I. 2011. Subclinical hypothyroidism is independently associated with albuminuria in people with type 2 diabetes. Diabetes Res Clin Pract 94:e75–e77 [DOI] [PubMed] [Google Scholar]
- 12. Suher M, Koc E, Ata N, Ensari C. 2005. Relation of thyroid disfunction, thyroid autoantibodies, and renal function. Ren Fail 27:739–742 [DOI] [PubMed] [Google Scholar]
- 13. Roberts CG, Ladenson PW. 2004. Hypothyroidism. Lancet 363:793–803 [DOI] [PubMed] [Google Scholar]
- 14. Iglesias P, Bajo MA, Selgas R, Diez JJ. 2017. Thyroid dysfunction and kidney disease: an update. Rev Endocr Metab Disord 18:131–144 [DOI] [PubMed] [Google Scholar]
- 15. Lee DY, Jee JH, Jun JE, Kim TH, Jin SM, Hur KY, Kim SW, Chung JH, Lee MK, Kim JH. 2017. The effect of TSH change per year on the risk of incident chronic kidney disease in euthyroid subjects. Endocrine 55:503–512 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Y, Chang Y, Ryu S, Cho J, Lee WY, Rhee EJ, Kwon MJ, Pastor-Barriuso R, Rampal S, Han WK, Shin H, Guallar E. 2014. Thyroid hormone levels and incident chronic kidney disease in euthyroid individuals: the Kangbuk Samsung Health Study. Int J Epidemiol 43:1624–1632 [DOI] [PubMed] [Google Scholar]
- 17. Schultheiss UT, Daya N, Grams ME, Seufert J, Steffes M, Coresh J, Selvin E, Kottgen A. 2017. Thyroid function, reduced kidney function and incident chronic kidney disease in a community-based population: the Atherosclerosis Risk in Communities study. Nephrol Dial Transplant 32:1874–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaker L, Sedaghat S, Hoorn EJ, Elzen WP, Gussekloo J, Hofman A, Ikram MA, Franco OH, Dehghan A, Peeters RP. 2016. The association of thyroid function and the risk of kidney function decline: a population-based cohort study. Eur J Endocrinol 175:653–660 [DOI] [PubMed] [Google Scholar]
- 19. Sun MT, Hsiao FC, Su SC, Pei D, Hung YJ. 2012. Thyrotropin as an independent factor of renal function and chronic kidney disease in normoglycemic euthyroid adults. Endocr Res 37:110–116 [DOI] [PubMed] [Google Scholar]
- 20. Peixoto de Miranda EJF, Bittencourt MS, Goulart AC, Santos IS, de Oliveira Titan SM, Ladeira RM, Barreto SM, Lotufo PA, Bensenor IJ. 2017. Thyrotropin levels are associated with chronic kidney disease among healthy subjects in cross-sectional analysis of the Brazilian Longitudinal Study of Adult Health (ELSA-Brasil). Clin Exp Nephrol 21:1035–1043 [DOI] [PubMed] [Google Scholar]
- 21. Lippi G, Montagnana M, Targher G, Salvagno GL, Guidi GC. 2008. Relationship between thyroid status and renal function in a general population of unselected outpatients. Clin Biochem 41:625–627 [DOI] [PubMed] [Google Scholar]
- 22. Gopinath B, Harris DC, Wall JR, Kifley A, Mitchell P. 2013. Relationship between thyroid dysfunction and chronic kidney disease in community-dwelling older adults. Maturitas 75:159–164 [DOI] [PubMed] [Google Scholar]
- 23. Huang X, Ding L, Peng K, Lin L, Wang T, Zhao Z, Xu Y, Lu J, Chen Y, Wang W, Bi Y, Ning G, Xu M. 2016. Thyroid hormones associate with risk of incident chronic kidney disease and rapid decline in renal function: a prospective investigation. J Transl Med 14:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bajaj S, Purwar N, Gupta A, Gupta P, Srivastava A. 2016. Prevalence of hypothyroidism in diabetic kidney disease and effect of thyroid hormone replacement on estimate glomerular filtration rate. Indian J Endocrinol Metab 20:795–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chonchol M, Lippi G, Salvagno G, Zoppini G, Muggeo M, Targher G. 2008. Prevalence of subclinical hypothyroidism in patients with chronic kidney disease. Clin J Am Soc Nephrol 3:1296–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rhee CM, Kalantar-Zadeh K, Streja E, Carrero JJ, Ma JZ, Lu JL, Kovesdy CP. 2015. The relationship between thyroid function and estimated glomerular filtration rate in patients with chronic kidney disease. Nephrol Dial Transplant 30:282–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meuwese CL, van Diepen M, Cappola AR, Sarnak MJ, Shlipak MG, Bauer DC, Fried LP, Iacoviello M, Vaes B, Degryse J, Khaw KT, Luben RN, Asvold BO, Bjoro T, Vatten LJ, de Craen AJM, Trompet S, Iervasi G, Molinaro S, Ceresini G, Ferrucci L, Dullaart RPF, Bakker SJL, Jukema JW, Kearney PM, Stott DJ, Peeters RP, Franco OH, Volzke H, Walsh JP, Bremner A, Sgarbi JA, Maciel RMB, Imaizumi M, Ohishi W, Dekker FW, Rodondi N, Gussekloo J, den Elzen WPJ; Thyroid Studies Collaboration. 2019. Low thyroid function is not associated with an accelerated deterioration in renal function. Nephrol Dial Transplant 34:650–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adrees M, Gibney J, El-Saeity N, Boran G. 2009. Effects of 18 months of L-T4 replacement in women with subclinical hypothyroidism. Clin Endocrinol (Oxf) 71:298–303 [DOI] [PubMed] [Google Scholar]
- 29. Shin DH, Lee MJ, Lee HS, Oh HJ, Ko KI, Kim CH, Doh FM, Koo HM, Kim HR, Han JH, Park JT, Han SH, Yoo TH, Kang SW. 2013. Thyroid hormone replacement therapy attenuates the decline of renal function in chronic kidney disease patients with subclinical hypothyroidism. Thyroid 23:654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu Y, Guo H, Liu D, Zhao Z. 2016. Preservation of renal function by thyroid hormone replacement in elderly persons with subclinical hypothyroidism. Arch Med Sci 12:772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. den Hollander JG, Wulkan RW, Mantel MJ, Berghout A. 2005. Correlation between severity of thyroid dysfunction and renal function. Clin Endocrinol (Oxf) 62:423–427 [DOI] [PubMed] [Google Scholar]
- 32. Liu P, Liu R, Chen X, Chen Y, Wang D, Zhang F, Wang Y. 2015. Can levothyroxine treatment reduce urinary albumin excretion rate in patients with early type 2 diabetic nephropathy and subclinical hypothyroidism? A randomized double-blind and placebo-controlled study. Curr Med Res Opin 31:2233–2240 [DOI] [PubMed] [Google Scholar]
- 33. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. 2008. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 27:1133–1163 [DOI] [PubMed] [Google Scholar]
- 34. Ellervik C, Roselli C, Christophersen IE, Alonso A, Pietzner M, Sitlani CM, Trompet S, Arking DE, Geelhoed B, Guo X, Kleber ME, Lin HJ, Lin H, MacFarlane P, Selvin E, Shaffer C, Smith AV, Verweij N, Weiss S, Cappola AR, Dorr M, Gudnason V, Heckbert S, Mooijaart S, Marz W, Psaty BM, Ridker PM, Roden D, Stott DJ, Volzke H, Benjamin EJ, Delgado G, Ellinor P, Homuth G, Kottgen A, Jukema JW, Lubitz SA, Mora S, Rienstra M, Rotter JI, Shoemaker MB, Sotoodehnia N, Taylor KD, van der Harst P, Albert CM, Chasman DI. 2019. Assessment of the relationship between genetic determinants of thyroid function and atrial fibrillation: a Mendelian randomization study. JAMA Cardiol 4:144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao JV, Schooling CM. 2017. Thyroid function and ischemic heart disease: a Mendelian randomization study. Sci Rep 7:8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bos MM, Smit RAJ, Trompet S, van Heemst D, Noordam R. 2017. Thyroid signaling, insulin resistance, and 2 diabetes mellitus: a Mendelian randomization study. J Clin Endocrinol Metab 102:1960–1970 [DOI] [PubMed] [Google Scholar]
- 37. van Vliet NA, Noordam R, van Klinken JB, Westendorp RG, Bassett JD, Williams GR, van Heemst D. 2018. Thyroid stimulating hormone and bone mineral density: evidence from a two-sample Mendelian randomization study and a candidate gene association study. J Bone Miner Res 33:1318–1325 [DOI] [PubMed] [Google Scholar]
- 38. Chen C, Xia F, Chen Y, Zhang K, Cheng J, Li Q, Han B, Zhao L, Zhu C, Wang N, Lu Y. 2018. Association between thyroid-stimulating hormone and renal function: a Mendelian randomization study. Kidney Blood Press Res 43:1121–1130 [DOI] [PubMed] [Google Scholar]
- 39. Eriksson N, Tung JY, Kiefer AK, Hinds DA, Francke U, Mountain JL, Do CB. 2012. Novel associations for hypothyroidism include known autoimmune risk loci. PLoS One 7:e34442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, Chai HS, Bastarache L, Zuvich R, Peissig P, Carrell D, Ramirez AH, Pathak J, Wilke RA, Rasmussen L, Wang X, Pacheco JA, Kho AN, Hayes MG, Weston N, Matsumoto M, Kopp PA, Newton KM, Jarvik GP, Li R, Manolio TA, Kullo IJ, Chute CG, Chisholm RL, Larson EB, McCarty CA, Masys DR, Roden DM, de Andrade M. 2011. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 89:529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA. 2016. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 48:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taylor PN, Porcu E, Chew S, Campbell PJ, Traglia M, Brown SJ, Mullin BH, Shihab HA, Min J, Walter K, Memari Y, Huang J, Barnes MR, Beilby JP, Charoen P, Danecek P, Dudbridge F, Forgetta V, Greenwood C, Grundberg E, Johnson AD, Hui J, Lim EM, McCarthy S, Muddyman D, Panicker V, Perry JR, Bell JT, Yuan W, Relton C, Gaunt T, Schlessinger D, Abecasis G, Cucca F, Surdulescu GL, Woltersdorf W, Zeggini E, Zheng HF, Toniolo D, Dayan CM, Naitza S, Walsh JP, Spector T, Davey Smith G, Durbin R, Richards JB, Sanna S, Soranzo N, Timpson NJ, Wilson SG; UK0K Consortium. 2015. Whole-genome sequence-based analysis of thyroid function. Nat Commun 6:5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Porcu E, Medici M, Pistis G, Volpato CB, Wilson SG, Cappola AR, Bos SD, Deelen J, den Heijer M, Freathy RM, Lahti J, Liu C, Lopez LM, Nolte IM, O'Connell JR, Tanaka T, Trompet S, Arnold A, Bandinelli S, Beekman M, Bohringer S, Brown SJ, Buckley BM, Camaschella C, de Craen AJ, Davies G, de Visser MC, Ford I, Forsen T, Frayling TM, Fugazzola L, Gogele M, Hattersley AT, Hermus AR, Hofman A, Houwing-Duistermaat JJ, Jensen RA, Kajantie E, Kloppenburg M, Lim EM, Masciullo C, Mariotti S, Minelli C, Mitchell BD, Nagaraja R, Netea-Maier RT, Palotie A, Persani L, Piras MG, Psaty BM, Raikkonen K, Richards JB, Rivadeneira F, Sala C, Sabra MM, Sattar N, Shields BM, Soranzo N, Starr JM, Stott DJ, Sweep FC, Usala G, van der Klauw MM, van Heemst D, van Mullem A, Vermeulen SH, Visser WE, Walsh JP, Westendorp RG, Widen E, Zhai G, Cucca F, Deary IJ, Eriksson JG, Ferrucci L, Fox CS, Jukema JW, Kiemeney LA, Pramstaller PP, Schlessinger D, Shuldiner AR, Slagboom EP, Uitterlinden AG, Vaidya B, Visser TJ, Wolffenbuttel BH, Meulenbelt I, Rotter JI, Spector TD, Hicks AA, Toniolo D, Sanna S, Peeters RP, Naitza S. 2013. A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet 9:e1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teumer A, Chaker L, Groeneweg S, Li Y, Di Munno C, Barbieri C, Schultheiss UT, Traglia M, Ahluwalia TS, Akiyama M, Appel EVR, Arking DE, Arnold A, Astrup A, Beekman M, Beilby JP, Bekaert S, Boerwinkle E, Brown SJ, De Buyzere M, Campbell PJ, Ceresini G, Cerqueira C, Cucca F, Deary IJ, Deelen J, Eckardt KU, Ekici AB, Eriksson JG, Ferrrucci L, Fiers T, Fiorillo E, Ford I, Fox CS, Fuchsberger C, Galesloot TE, Gieger C, Gogele M, De Grandi A, Grarup N, Greiser KH, Haljas K, Hansen T, Harris SE, van Heemst D, den Heijer M, Hicks AA, den Hollander W, Homuth G, Hui J, Ikram MA, Ittermann T, Jensen RA, Jing J, Jukema JW, Kajantie E, Kamatani Y, Kasbohm E, Kaufman JM, Kiemeney LA, Kloppenburg M, Kronenberg F, Kubo M, Lahti J, Lapauw B, Li S, Liewald DCM; Lifelines Cohort Study, Lim EM, Linneberg A, Marina M, Mascalzoni D, Matsuda K, Medenwald D, Meisinger C, Meulenbelt I, De Meyer T, Meyer Zu Schwabedissen HE, Mikolajczyk R, Moed M, Netea-Maier RT, Nolte IM, Okada Y, Pala M, Pattaro C, Pedersen O, Petersmann A, Porcu E, Postmus I, Pramstaller PP, Psaty BM, Ramos YFM, Rawal R, Redmond P, Richards JB, Rietzschel ER, Rivadeneira F, Roef G, Rotter JI, Sala CF, Schlessinger D, Selvin E, Slagboom PE, Soranzo N, Sorensen TIA, Spector TD, Starr JM, Stott DJ, Taes Y, Taliun D, Tanaka T, Thuesen B, Tiller D, Toniolo D, Uitterlinden AG, Visser WE, Walsh JP, Wilson SG, Wolffenbuttel BHR, Yang Q, Zheng HF, Cappola A, Peeters RP, Naitza S, Volzke H, Sanna S, Kottgen A, Visser TJ, Medici M. 2018. Genome-wide analyses identify a role for SLC17A4 and AADAT in thyroid hormone regulation. Nat Commun 9:4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schultheiss UT, Teumer A, Medici M, Li Y, Daya N, Chaker L, Homuth G, Uitterlinden AG, Nauck M, Hofman A, Selvin E, Volzke H, Peeters RP, Kottgen A. 2015. A genetic risk score for thyroid peroxidase antibodies associates with clinical thyroid disease in community-based populations. J Clin Endocrinol Metab 100:E799–E807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Medici M, Porcu E, Pistis G, Teumer A, Brown SJ, Jensen RA, Rawal R, Roef GL, Plantinga TS, Vermeulen SH, Lahti J, Simmonds MJ, Husemoen LL, Freathy RM, Shields BM, Pietzner D, Nagy R, Broer L, Chaker L, Korevaar TI, Plia MG, Sala C, Volker U, Richards JB, Sweep FC, Gieger C, Corre T, Kajantie E, Thuesen B, Taes YE, Visser WE, Hattersley AT, Kratzsch J, Hamilton A, Li W, Homuth G, Lobina M, Mariotti S, Soranzo N, Cocca M, Nauck M, Spielhagen C, Ross A, Arnold A, van de Bunt M, Liyanarachchi S, Heier M, Grabe HJ, Masciullo C, Galesloot TE, Lim EM, Reischl E, Leedman PJ, Lai S, Delitala A, Bremner AP, Philips DI, Beilby JP, Mulas A, Vocale M, Abecasis G, Forsen T, James A, Widen E, Hui J, Prokisch H, Rietzschel EE, Palotie A, Feddema P, Fletcher SJ, Schramm K, Rotter JI, Kluttig A, Radke D, Traglia M, Surdulescu GL, He H, Franklyn JA, Tiller D, Vaidya B, de Meyer T, Jorgensen T, Eriksson JG, O'Leary PC, Wichmann E, Hermus AR, Psaty BM, Ittermann T, Hofman A, Bosi E, Schlessinger D, Wallaschofski H, Pirastu N, Aulchenko YS, de la Chapelle A, Netea-Maier RT, Gough SC, Meyer Zu Schwabedissen H, Frayling TM, Kaufman JM, Linneberg A, Raikkonen K, Smit JW, Kiemeney LA, Rivadeneira F, Uitterlinden AG, Walsh JP, Meisinger C, den Heijer M, Visser TJ, Spector TD, Wilson SG, Volzke H, Cappola A, Toniolo D, Sanna S, Naitza S, Peeters RP. 2014. Identification of novel genetic Loci associated with thyroid peroxidase antibodies and clinical thyroid disease. PLoS Genet 10:e1004123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, Tin A, Wang L, Chu AY, Hoppmann A, Kirsten H, Giri A, Chai JF, Sveinbjornsson G, Tayo BO, Nutile T, Fuchsberger C, Marten J, Cocca M, Ghasemi S, Xu Y, Horn K, Noce D, van der Most PJ, Sedaghat S, Yu Z, Akiyama M, Afaq S, Ahluwalia TS, Almgren P, Amin N, Arnlov J, Bakker SJL, Bansal N, Baptista D, Bergmann S, Biggs ML, Biino G, Boehnke M, Boerwinkle E, Boissel M, Bottinger EP, Boutin TS, Brenner H, Brumat M, Burkhardt R, Butterworth AS, Campana E, Campbell A, Campbell H, Canouil M, Carroll RJ, Catamo E, Chambers JC, Chee ML, Chee ML, Chen X, Cheng CY, Cheng Y, Christensen K, Cifkova R, Ciullo M, Concas MP, Cook JP, Coresh J, Corre T, Sala CF, Cusi D, Danesh J, Daw EW, de Borst MH, De Grandi A, de Mutsert R, de Vries APJ, Degenhardt F, Delgado G, Demirkan A, Di Angelantonio E, Dittrich K, Divers J, Dorajoo R, Eckardt KU, Ehret G, Elliott P, Endlich K, Evans MK, Felix JF, Foo VHX, Franco OH, Franke A, Freedman BI, Freitag-Wolf S, Friedlander Y, Froguel P, Gansevoort RT, Gao H, Gasparini P, Gaziano JM, Giedraitis V, Gieger C, Girotto G, Giulianini F, Gogele M, Gordon SD, Gudbjartsson DF, Gudnason V, Haller T, Hamet P, Harris TB, Hartman CA, Hayward C, Hellwege JN, Heng CK, Hicks AA, Hofer E, Huang W, Hutri-Kahonen N, Hwang SJ, Ikram MA, Indridason OS, Ingelsson E, Ising M, Jaddoe VWV, Jakobsdottir J, Jonas JB, Joshi PK, Josyula NS, Jung B, Kahonen M, Kamatani Y, Kammerer CM, Kanai M, Kastarinen M, Kerr SM, Khor CC, Kiess W, Kleber ME, Koenig W, Kooner JS, Korner A, Kovacs P, Kraja AT, Krajcoviechova A, Kramer H, Kramer BK, Kronenberg F, Kubo M, Kuhnel B, Kuokkanen M, Kuusisto J, La Bianca M, Laakso M, Lange LA, Langefeld CD, Lee JJ, Lehne B, Lehtimaki T, Lieb W; Lifelines Cohort Study, Lim SC, Lind L, Lindgren CM, Liu J, Liu J, Loeffler M, Loos RJF, Lucae S, Lukas MA, Lyytikainen LP, Magi R, Magnusson PKE, Mahajan A, Martin NG, Martins J, Marz W, Mascalzoni D, Matsuda K, Meisinger C, Meitinger T, Melander O, Metspalu A, Mikaelsdottir EK, Milaneschi Y, Miliku K, Mishra PP, Program VAMV, Mohlke KL, Mononen N, Montgomery GW, Mook-Kanamori DO, Mychaleckyj JC, Nadkarni GN, Nalls MA, Nauck M, Nikus K, Ning B, Nolte IM, Noordam R, O'Connell J, O'Donoghue ML, Olafsson I, Oldehinkel AJ, Orho-Melander M, Ouwehand WH, Padmanabhan S, Palmer ND, Palsson R, Penninx B, Perls T, Perola M, Pirastu M, Pirastu N, Pistis G, Podgornaia AI, Polasek O, Ponte B, Porteous DJ, Poulain T, Pramstaller PP, Preuss MH, Prins BP, Province MA, Rabelink TJ, Raffield LM, Raitakari OT, Reilly DF, Rettig R, Rheinberger M, Rice KM, Ridker PM, Rivadeneira F, Rizzi F, Roberts DJ, Robino A, Rossing P, Rudan I, Rueedi R, Ruggiero D, Ryan KA, Saba Y, Sabanayagam C, Salomaa V, Salvi E, Saum KU, Schmidt H, Schmidt R, Schottker B, Schulz CA, Schupf N, Shaffer CM, Shi Y, Smith AV, Smith BH, Soranzo N, Spracklen CN, Strauch K, Stringham HM, Stumvoll M, Svensson PO, Szymczak S, Tai ES, Tajuddin SM, Tan NYQ, Taylor KD, Teren A, Tham YC, Thiery J, Thio CHL, Thomsen H, Thorleifsson G, Toniolo D, Tonjes A, Tremblay J, Tzoulaki I, Uitterlinden AG, Vaccargiu S, van Dam RM, van der Harst P, van Duijn CM, Velez Edward DR, Verweij N, Vogelezang S, Volker U, Vollenweider P, Waeber G, Waldenberger M, Wallentin L, Wang YX, Wang C, Waterworth DM, Bin Wei W, White H, Whitfield JB, Wild SH, Wilson JF, Wojczynski MK, Wong C, Wong TY, Xu L, Yang Q, Yasuda M, Yerges-Armstrong LM, Zhang W, Zonderman AB, Rotter JI, Bochud M, Psaty BM, Vitart V, Wilson JG, Dehghan A, Parsa A, Chasman DI, Ho K, Morris AP, Devuyst O, Akilesh S, Pendergrass SA, Sim X, Boger CA, Okada Y, Edwards TL, Snieder H, Stefansson K, Hung AM, Heid IM, Scholz M, Teumer A, Kottgen A, Pattaro C. 2019. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 51:957–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gorski M, van der Most PJ, Teumer A, Chu AY, Li M, Mijatovic V, Nolte IM, Cocca M, Taliun D, Gomez F, Li Y, Tayo B, Tin A, Feitosa MF, Aspelund T, Attia J, Biffar R, Bochud M, Boerwinkle E, Borecki I, Bottinger EP, Chen MH, Chouraki V, Ciullo M, Coresh J, Cornelis MC, Curhan GC, d'Adamo AP, Dehghan A, Dengler L, Ding J, Eiriksdottir G, Endlich K, Enroth S, Esko T, Franco OH, Gasparini P, Gieger C, Girotto G, Gottesman O, Gudnason V, Gyllensten U, Hancock SJ, Harris TB, Helmer C, Hollerer S, Hofer E, Hofman A, Holliday EG, Homuth G, Hu FB, Huth C, Hutri-Kahonen N, Hwang SJ, Imboden M, Johansson A, Kahonen M, Konig W, Kramer H, Kramer BK, Kumar A, Kutalik Z, Lambert JC, Launer LJ, Lehtimaki T, de Borst M, Navis G, Swertz M, Liu Y, Lohman K, Loos RJF, Lu Y, Lyytikainen LP, McEvoy MA, Meisinger C, Meitinger T, Metspalu A, Metzger M, Mihailov E, Mitchell P, Nauck M, Oldehinkel AJ, Olden M, Wjh Penninx B, Pistis G, Pramstaller PP, Probst-Hensch N, Raitakari OT, Rettig R, Ridker PM, Rivadeneira F, Robino A, Rosas SE, Ruderfer D, Ruggiero D, Saba Y, Sala C, Schmidt H, Schmidt R, Scott RJ, Sedaghat S, Smith AV, Sorice R, Stengel B, Stracke S, Strauch K, Toniolo D, Uitterlinden AG, Ulivi S, Viikari JS, Volker U, Vollenweider P, Volzke H, Vuckovic D, Waldenberger M, Jin Wang J, Yang Q, Chasman DI, Tromp G, Snieder H, Heid IM, Fox CS, Kottgen A, Pattaro C, Boger CA, Fuchsberger C. 2017. 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep 7:45040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Teumer A, Tin A, Sorice R, Gorski M, Yeo NC, Chu AY, Li M, Li Y, Mijatovic V, Ko YA, Taliun D, Luciani A, Chen MH, Yang Q, Foster MC, Olden M, Hiraki LT, Tayo BO, Fuchsberger C, Dieffenbach AK, Shuldiner AR, Smith AV, Zappa AM, Lupo A, Kollerits B, Ponte B, Stengel B, Kramer BK, Paulweber B, Mitchell BD, Hayward C, Helmer C, Meisinger C, Gieger C, Shaffer CM, Muller C, Langenberg C, Ackermann D, Siscovick D; DCCT/EDIC, Boerwinkle E, Kronenberg F, Ehret GB, Homuth G, Waeber G, Navis G, Gambaro G, Malerba G, Eiriksdottir G, Li G, Wichmann HE, Grallert H, Wallaschofski H, Volzke H, Brenner H, Kramer H, Mateo Leach I, Rudan I, Hillege HL, Beckmann JS, Lambert JC, Luan J, Zhao JH, Chalmers J, Coresh J, Denny JC, Butterbach K, Launer LJ, Ferrucci L, Kedenko L, Haun M, Metzger M, Woodward M, Hoffman MJ, Nauck M, Waldenberger M, Pruijm M, Bochud M, Rheinberger M, Verweij N, Wareham NJ, Endlich N, Soranzo N, Polasek O, van der Harst P, Pramstaller PP, Vollenweider P, Wild PS, Gansevoort RT, Rettig R, Biffar R, Carroll RJ, Katz R, Loos RJ, Hwang SJ, Coassin S, Bergmann S, Rosas SE, Stracke S, Harris TB, Corre T, Zeller T, Illig T, Aspelund T, Tanaka T, Lendeckel U, Volker U, Gudnason V, Chouraki V, Koenig W, Kutalik Z, O'Connell JR, Parsa A, Heid IM, Paterson AD, de Boer IH, Devuyst O, Lazar J, Endlich K, Susztak K, Tremblay J, Hamet P, Jacob HJ, Boger CA, Fox CS, Pattaro C, Kottgen A. 2016. Genome-wide association studies identify genetic loci associated with albuminuria in diabetes. Diabetes 65:803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ridker PM, Chasman DI, Zee RY, Parker A, Rose L, Cook NR, Buring JE, Women's Genome Health Study Working Group. 2008. Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy American women. Clin Chem 54:249–255 [DOI] [PubMed] [Google Scholar]
- 51. Harada PHN, Buring JE, Cook NR, Cobble ME, Kulkarni KR, Mora S. 2017. Impact of subclinical hypothyroidism on cardiometabolic biomarkers in women. J Endocr Soc 1:113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, Garnaas M, Tin A, Sorice R, Li Y, Taliun D, Olden M, Foster M, Yang Q, Chen MH, Pers TH, Johnson AD, Ko YA, Fuchsberger C, Tayo B, Nalls M, Feitosa MF, Isaacs A, Dehghan A, d'Adamo P, Adeyemo A, Dieffenbach AK, Zonderman AB, Nolte IM, van der Most PJ, Wright AF, Shuldiner AR, Morrison AC, Hofman A, Smith AV, Dreisbach AW, Franke A, Uitterlinden AG, Metspalu A, Tonjes A, Lupo A, Robino A, Johansson A, Demirkan A, Kollerits B, Freedman BI, Ponte B, Oostra BA, Paulweber B, Kramer BK, Mitchell BD, Buckley BM, Peralta CA, Hayward C, Helmer C, Rotimi CN, Shaffer CM, Muller C, Sala C, van Duijn CM, Saint-Pierre A, Ackermann D, Shriner D, Ruggiero D, Toniolo D, Lu Y, Cusi D, Czamara D, Ellinghaus D, Siscovick DS, Ruderfer D, Gieger C, Grallert H, Rochtchina E, Atkinson EJ, Holliday EG, Boerwinkle E, Salvi E, Bottinger EP, Murgia F, Rivadeneira F, Ernst F, Kronenberg F, Hu FB, Navis GJ, Curhan GC, Ehret GB, Homuth G, Coassin S, Thun GA, Pistis G, Gambaro G, Malerba G, Montgomery GW, Eiriksdottir G, Jacobs G, Li G, Wichmann HE, Campbell H, Schmidt H, Wallaschofski H, Volzke H, Brenner H, Kroemer HK, Kramer H, Lin H, Leach IM, Ford I, Guessous I, Rudan I, Prokopenko I, Borecki I, Heid IM, Kolcic I, Persico I, Jukema JW, Wilson JF, Felix JF, Divers J, Lambert JC, Stafford JM, Gaspoz JM, Smith JA, Faul JD, Wang JJ, Ding J, Hirschhorn JN, Attia J, Whitfield JB, Chalmers J, Viikari J, Coresh J, Denny JC, Karjalainen J, Fernandes JK, Endlich K, Butterbach K, Keene KL, Lohman K, Portas L, Launer LJ, Lyytikainen LP, Yengo L, Franke L, Ferrucci L, Rose LM, Kedenko L, Rao M, Struchalin M, Kleber ME, Cavalieri M, Haun M, Cornelis MC, Ciullo M, Pirastu M, de Andrade M, McEvoy MA, Woodward M, Adam M, Cocca M, Nauck M, Imboden M, Waldenberger M, Pruijm M, Metzger M, Stumvoll M, Evans MK, Sale MM, Kahonen M, Boban M, Bochud M, Rheinberger M, Verweij N, Bouatia-Naji N, Martin NG, Hastie N, Probst-Hensch N, Soranzo N, Devuyst O, Raitakari O, Gottesman O, Franco OH, Polasek O, Gasparini P, Munroe PB, Ridker PM, Mitchell P, Muntner P, Meisinger C, Smit JH; ICBP Consortium; AGEN Consortium; CARDIOGRAM; CHARGe-Heart Failure Group; ECHOGen Consortium, Kovacs P, Wild PS, Froguel P, Rettig R, Magi R, Biffar R, Schmidt R, Middelberg RP, Carroll RJ, Penninx BW, Scott RJ, Katz R, Sedaghat S, Wild SH, Kardia SL, Ulivi S, Hwang SJ, Enroth S, Kloiber S, Trompet S, Stengel B, Hancock SJ, Turner ST, Rosas SE, Stracke S, Harris TB, Zeller T, Zemunik T, Lehtimaki T, Illig T, Aspelund T, Nikopensius T, Esko T, Tanaka T, Gyllensten U, Volker U, Emilsson V, Vitart V, Aalto V, Gudnason V, Chouraki V, Chen WM, Igl W, Marz W, Koenig W, Lieb W, Loos RJ, Liu Y, Snieder H, Pramstaller PP, Parsa A, O'Connell JR, Susztak K, Hamet P, Tremblay J, de Boer IH, Boger CA, Goessling W, Chasman DI, Kottgen A, Kao WH, Fox CS. 2016. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 7:10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O'Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Pare G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tonjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstatter A, Kollerits B, Kedenko L, Magi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Volzke H, Kroemer HK, Nauck M, Volker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Kramer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS. 2010. New loci associated with kidney function and chronic kidney disease. Nat Genet 42:376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brion MJ, Shakhbazov K, Visscher PM. 2013. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 42:1497–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Freeman G, Cowling BJ, Schooling CM. 2013. Power and sample size calculations for Mendelian randomization studies using one genetic instrument. Int J Epidemiol 42:1157–1163 [DOI] [PubMed] [Google Scholar]
- 56. Burgess S. 2014. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol 43:922–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Burgess S, Butterworth A, Thompson SG. 2013. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37:658–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Burgess S, Labrecque JA. 2018. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol 33:947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ohnhaus EE, Burgi H, Burger A, Studer H. 1981. The effect of antipyrine, phenobarbitol and rifampicin on thyroid hormone metabolism in man. Eur J Clin Invest 11:381–387 [DOI] [PubMed] [Google Scholar]
- 60. Bowden J, Davey Smith G, Burgess S. 2015. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44:512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bowden J, Davey Smith G, Haycock PC, Burgess S. 2016. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40:304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. 2016. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol 45:1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. 2017. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med 36:1783–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Verbanck M, Chen CY, Neale B, Do R. 2018. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50:693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shim H, Chasman DI, Smith JD, Mora S, Ridker PM, Nickerson DA, Krauss RM, Stephens M. 2015. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS One 10:e0120758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kocak G, Huddam B, Azak A, Ortabozkoyun L, Duranay M. 2012. Coexistent findings of renal glomerular disease with Hashimoto's thyroiditis. Clin Endocrinol (Oxf) 76:759–762 [DOI] [PubMed] [Google Scholar]
- 67. Hemani G, Tilling K, Davey Smith G. 2017. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 13:e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Olden M, Teumer A, Bochud M, Pattaro C, Kottgen A, Turner ST, Rettig R, Chen MH, Dehghan A, Bastardot F, Schmidt R, Vollenweider P, Schunkert H, Reilly MP, Fornage M, Launer LJ, Verwoert GC, Mitchell GF, Bis JC, O'Donnell CJ, Cheng CY, Sim X, Siscovick DS, Coresh J, Kao WH, Fox CS, O'Seaghdha CM; AortaGen CA RDIoGRAM CH ARGE Eye CH ARGE IMT IC BP NeuroCHARGE, and CKDGen Consortia. 2013. Overlap between common genetic polymorphisms underpinning kidney traits and cardiovascular disease phenotypes: the CKDGen consortium. Am J Kidney Dis 61:889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wei N, Chen Z, Xue Z, Zhu Y. 2015. Polymorphism of VEGF gene in susceptibility to chronic immune-mediated inflammatory diseases: a meta-analysis. Rheumatol Int 35:1351–1360 [DOI] [PubMed] [Google Scholar]
- 70. Staley JR, Burgess S. 2017. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet Epidemiol 41:341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cooper JD, Simmonds MJ, Walker NM, Burren O, Brand OJ, Guo H, Wallace C, Stevens H, Coleman G; Wellcome Trust Case Control Consortium, Franklyn JA, Todd JA, Gough SC. 2012. Seven newly identified loci for autoimmune thyroid disease. Hum Mol Genet 21:5202–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Giorda CB, Carna P, Romeo F, Costa G, Tartaglino B, Gnavi R. 2017. Prevalence, incidence and associated comorbidities of treated hypothyroidism: an update from a European population. Eur J Endocrinol 176:533–542 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.