Abstract
Objective
To build a knowledge base of dietary supplement (DS) information, called the integrated DIetary Supplement Knowledge base (iDISK), which integrates and standardizes DS-related information from 4 existing resources.
Materials and Methods
iDISK was built through an iterative process comprising 3 phases: 1) establishment of the content scope, 2) development of the data model, and 3) integration of existing resources. Four well-regarded DS resources were integrated into iDISK: The Natural Medicines Comprehensive Database, the “About Herbs” page on the Memorial Sloan Kettering Cancer Center website, the Dietary Supplement Label Database, and the Natural Health Products Database. We evaluated the iDISK build process by manually checking that the data elements associated with 50 randomly selected ingredients were correctly extracted and integrated from their respective sources.
Results
iDISK encompasses a terminology of 4208 DS ingredient concepts, which are linked via 6 relationship types to 495 drugs, 776 diseases, 985 symptoms, 605 therapeutic classes, 17 system organ classes, and 137 568 DS products. iDISK also contains 7 concept attribute types and 3 relationship attribute types. Evaluation of the data extraction and integration process showed average errors of 0.3%, 2.6%, and 0.4% for concepts, relationships and attributes, respectively.
Conclusion
We developed iDISK, a publicly available standardized DS knowledge base that can facilitate more efficient and meaningful dissemination of DS knowledge.
Keywords: dietary supplements, knowledge representation, terminology, RxNorm, unified medical language system
INTRODUCTION
The Dietary Supplement Health and Education Act (DSHEA) of 1994 defines dietary supplements (DS) in part as products ingested or administered to the body that contain a “dietary ingredient.” This includes vitamins, minerals, amino acids, and herbs or botanicals, as well as other substances that can be used to supplement the diet.1 The National Health and Nutrition Examination Survey, a nationally representative, cross-sectional survey, has reported that 49% of the total US population uses DS (males 44%, females 53%).2 DS are primarily considered as food, compared to prescription and over-the-counter drugs, and are regulated by the FDA under a different, less stringent set of rules. Additionally, the use of DS is often self-initiated rather than based on clinicians’ recommendations. This results in unique challenges pertaining to efficacy, safety, regulatory policies, and clinical practices for various stakeholders, such as researchers, clinicians, and consumers.3 For example, there are around 23 000 emergency department visits per year resulting from DS-related adverse events.4 These challenges underscore the need for accessible resources for consumers and prescribers to safely select DS if they are desired.
There are several commercially and publicly available resources covering DS ingredients and products. The Natural Medicines Comprehensive Database (NMCD)5 is a commercial ingredient-level database, built on evidence-based data and represented in free text monographs. The “About Herbs” page on the Memorial Sloan Kettering Cancer Center (MSKCC) website6 is a free resource for consumer and healthcare professionals to find information on using common herbs and other DS. The Dietary Supplement Label Database (DSLD)7 includes full product labels with detailed ingredient information for over 76 000 DS products marketed in the US. The products are further categorized using LanguaL codes, a thesaural system originally generated for describing data about food.8,9 The Natural Health Products Database (NHP), comprised of the Natural Health Product Ingredients Database10 and the Licensed Natural Health Products Database,11 contains information about natural health products that have been issued a product license by Health Canada, including data such as geographic area of origin, ingredient category, and dose forms.
Standardized biomedical terminologies and ontologies have facilitated cross-platform communicability and the reuse of knowledge, alleviating challenges associated with increasingly computerized clinical data. A few well-established and commonly employed terminology resources are the Unified Medical Language System (UMLS),12 RxNorm,13 the Medication Reference Terminology,14 the Medical Dictionary of Regulatory Activities (MedDRA),15 and the Anatomical, Therapeutic, and Chemical classification system/Defined Daily Dose.16 However, standardized knowledge representation is still lacking in the DS domain. According to our previous studies, none of the supplement databases or existing terminologies comprehensively covers supplement terms17,18 and the related information (eg, effectiveness, safety) is often incomplete.19 Furthermore, these resources are not built on standardized knowledge representation principles and are thus unable to communicate with other terminologies or across systems and healthcare organizations.20 A standardized terminology of DS would support informatics research related to DS, such as the mining of DS use status from clinical reports,21–23 the discovery DS adverse effects24–26 and drug interactions27 from the literature, and assess the effectiveness of DS for various conditions.28,29 Furthermore, a structured and searchable knowledge base of DS-related information, such as drug interactions and uses, would help clinicians and consumers make informed decisions regarding the usage of DS. It is thus necessary to develop a structured and standardized data store of DS-related information in order to facilitate the search and retrieval of DS information by a wide range of users.
There has been some previous work on the knowledge representation of DS and related substances. Sharma and Sarkar developed a thesaurus of DS terms for identifying DS mentions in adverse event reports, but their work did not address the integration of related data elements such as adverse effects and interactions.30 Similarly, the Normalized Chinese Clinical Drugs (NCCD) knowledge base published by Wang et al was built by integrating data from various resources and representing it following the RxNorm model in order to improve interoperability.31 Like Sharma and Sarkar’s work, however, NCCD is primarily a thesaurus, and its domain is Chinese clinical drugs, not DS. In other related domains, the WATRIMed knowledge graph compiles information on West-African herbal traditional medicine into a standardized data model32 and the Romedi dataset of French clinical drugs was created by integrating data from publicly available resources, standardizing it according to the RxNorm model, and linking it to existing terminologies.33
To fill the gap in DS knowledge representation, we present the first integrated DIetary Supplement Knowledge base (iDISK), which encompasses both a terminology of DS ingredients and a structured knowledge base of DS-related information. iDISK was built according to established terminology and ontology development guidelines and definitions34 by integrating knowledge from existing DS resources and representing it in a standardized and structured form. The iDISK data elements are further linked to existing controlled vocabularies thus increasing interoperability, a fundamental element for successful health information exchange.
MATERIALS AND METHODS
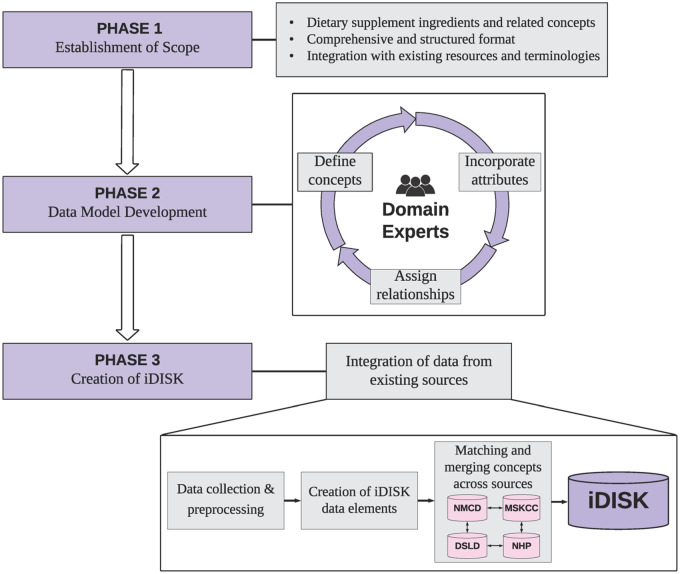
iDISK was developed by integrating essential DS information from multiple commonly used and well-trusted DS resources (ie, NMCD, MSKCC, DSLD, and NHP) into a common data model. NMCD is a commercial and subscription-based resource, and we have arranged an agreement with its copyright holder, Therapeutic Research Center (TRC), according to which we may publicly redistribute the NMCD information as represented in iDISK. iDISK was built in 3 phases, illustrated in Figure 1: 1) establishment of the scope of iDISK, 2) development of the data model by domain experts, and 3) creation of iDISK by integrating data from existing DS resources, including mapping to existing biomedical terminologies. In the rest of this paper, we use italics to denote instances of iDISK data elements and brackets are used to denote collections of data elements such as attributes [attribute: “value”] and relationships [subject, relationship, object].
Figure 1.
Overview of the design and creation of iDISK.
Phase 1: establishment of scope
To date, none of the available online resources fully represent DS knowledge in a complete and standardized form. To address this, we planned to create iDISK as a comprehensive and structured DS knowledge base by integrating related terms from different resources and mapping relevant terms to existing standardized terminologies such as the UMLS and MedDRA. The current iDISK version is primarily focused on DS ingredients, their attributes (eg, the type of the ingredient, the UMLS semantic type), and related concepts (eg, DS products, diseases, symptoms).
Phase 2: development of the data model
The iDISK data model was inspired by the RxNorm13 model of data representation with the addition of other relevant concepts related to DS ingredients. RxNorm is developed by the US National Library of Medicine as a part of the Unified Medical Language System (UMLS). It provides a normalized naming system for drugs which supports semantic interoperatability between 16 drug terminologies and pharmacy knowledge bases. As the normalization of DS ingredient names is a major contribution of iDISK, it is in this respect similar to RxNorm. We created the iDISK data model through a methodological and iterative process centered around the scope as described in Phase 1, according to the knowledge gained from our previous study on DS knowledge representation19 and the information available from the data sources. The development process entailed repeated discussions and consensus among a team of researchers, which included informaticists (RR, RZ, JV), physician informaticists (RR, TA, GM) and physicians/pharmacologists (TA, JB).
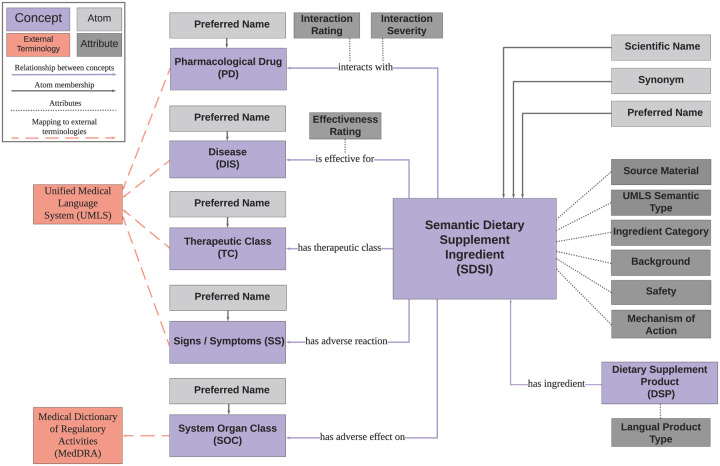
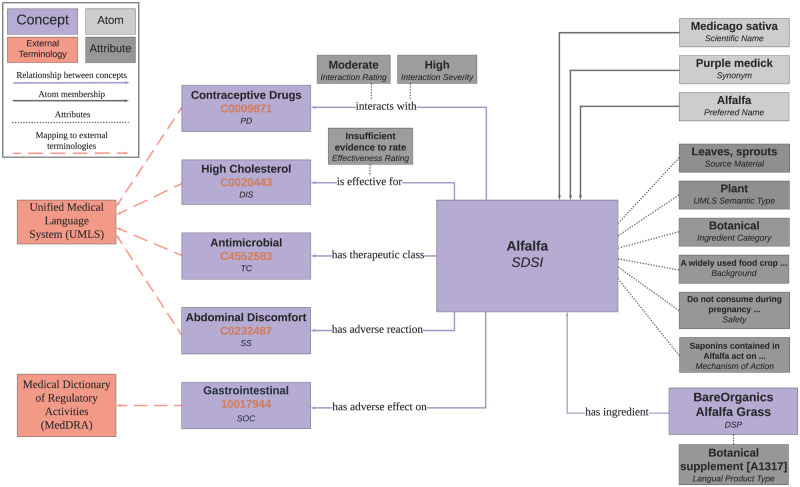
The final data model is given in Figure 2. iDISK is comprised of 4 data elements: concept, atom, relationship, and attribute, each of which is assigned a unique identifier. iDISK has 7 concept types, described in Table 1. A concept is a collection of atoms, which encode the synonymous names denoting that concept. Each atom is a unique combination of a term (eg, an ingredient name), a term type (the role of an atom in its source, eg, scientific name or common name), a data source (eg, DSLD), and a source code (the unique identifier which allows an atom to be tracked back to its source). Relationships connect concepts with the relationship type specifying the meaning and direction of the connection. A total of 6 unique relationship types are used to establish relationships between concepts: is_effective_for, has_therapeutic_class, has_adverse_effect_on, has_adverse_reaction, has_ingredient, and interacts_with. Concepts and relationships can have 1 or more attributes, whose value is free text. The attributes used in iDISK are described in Table 2. In Figure 3, we populate the data model with Alfalfa as a representative example of how iDISK represents DS information in a structured and consistent format.
Figure 2.
The iDISK data model.
Table 1.
The concept types present in iDISK, along with their descriptions and examples. Following the Unified Medical Language System (UMLS), concepts are collections of synonymous terms, called atoms, which are integrated from various sources. We therefore also provide the section in the data sources from which atoms for the corresponding concept were extracted
| iDISK Concept Type | Description | Example | Source | Corresponding Section in Source |
|---|---|---|---|---|
| Semantic dietary supplement ingredient (SDSI) | A non-branded, individual dietary supplement ingredient. | Ginkgo Biloba | DSLD | Synonym |
| NHP | Common name, Proper name | |||
| NMCD | Also known as, Synonym, Taxonomical synonym, Scientific name | |||
| Dietary supplement product (DSP) | A product that is marketed as a dietary supplement by its manufacturer. | Vitamer Laboratories Glucosamine Chondroitin Complete | DSLD | Product name, Brand name |
| NHP | Product name | |||
| Disease (DIS) | A disease or condition that may be treated by a given dietary supplement. | Emphysema | NMCD | Effectiveness |
| MSKCC | Purported uses | |||
| System organ class (SOC) | The broad biological or organ system in which the adverse effect manifests. | Gastrointestinal | NMCD | Adverse effects |
| Pharmacological drug (PD) | A prescription or over-the-counter drug, expressly intended to treat or prevent disease. | Aspirin | NMCD | Interactions with drugs |
| MSKCC | Herb-drug interactions | |||
| Therapeutic class (TC)a | A broad classification of the function of a dietary supplement. | Analgesic | NMCD | Mechanism of action |
| Signs/symptoms (SS) | The physical manifestation of an adverse effect. | Nausea | MSKCC | Adverse reactions |
Abbreviations: DSLD, Dietary Supplement Label Database; MSKCC, Memorial Sloan Kettering Cancer Center; NHP, Natural Health Products Database; NMCD, Natural Medicines Comprehensive Database.
The NMCD “Mechanism of Action” section, in fact, describes the therapeutic class of the DS (as opposed to a literal description of the pharmacologic mechanism), hence the name of the iDISK concept type.
Table 2.
The iDISK concept attributes and relationship attributes
| Attribute | Description | Associated Concept / Relationship | Source(s) |
|---|---|---|---|
| Concept Attributes(s) | |||
| Source Material | Source of the ingredient. | SDSI | MSKCC |
| UMLS Semantic Type | One of the broad categories described in the UMLS Semantic Network. | SDSI | UMLS |
| Ingredient category | Ingredient category classification by DSLD. | SDSI | DSLD |
| Background | A summary of information about this ingredient, including its origination, uses, constituent parts, etc. | SDSI | NMCD, MSKCC, NHP |
| Safety | A summary of the safety concerns in using this ingredient. | SDSI | NMCD, NHP |
| Mechanism of action | Mechanism by which an active substance produces an effect on a living organism or in a biochemical system. | SDSI | MSKCC |
| Product Type | LanguaL type classification by DSLD. | DSP | DSLD |
| Relationship Attributes(s) | |||
| Interaction Rating | Expert-reviewed, evidence-based likelihood of the occurrence of an interaction between a DS and a drug. Possible values are Likely, Probable, Possible, Unlikely.a | PD / interacts_with | NMCD |
| Interaction Severity | Expert-reviewed, evidence-based severity of the interaction, if it occurs. Possible values are High, Moderate, Mild, Insignificant.a | PD / interacts_with | NMCD |
| Effectiveness Rating | Expert-reviewed, evidence-based likelihood of effectiveness of a DS for a given disease or condition. Possible values are Likely, Probable, Possible, Unlikely.a | DIS / is_effective_for | NMCD |
Abbreviations: DIS, Disease; DSLD, Dietary Supplement Label Database; DSP, Dietary Supplement Product; MSKCC, Memorial Sloan Kettering Cancer Center; NHP, Natural Health Products Database; NMCD, Natural Medicines Comprehensive Database; PD, Pharmacological drug; SDSI, Semantic Dietary Supplement Ingredient; UMLS, Unified Medical Language System.
Possible values are adapted from NMCD.
Figure 3.
The iDISK data model populated with data about Alfalfa.
Phase 3: creation of iDISK
The iDISK build process is split into 3 steps, illustrated in the Phase 3 section of Figure 1: data collection and preprocessing, creation of iDISK data elements from the source data, and matching and merging synonymous data elements. These steps are described in detail below.
Data collection and preprocessing
The data were collected from each resource as follows. NMCD: We obtained data from the NMCD API with permission from the TRC. DSLD: Data were obtained from the DSLD data release (https://www.dsld.nlm.nih.gov/dsld/searchdownload.jsp#general). Product information was obtained via the DSLD API which provides a richer representation than the data release (https://www.dsld.nlm.nih.gov/dsld/faq.jsp#10). MSKCC: With permission, we developed a web scraper to obtain the ingredient monographs listed on the “About Herbs” page (https://www.mskcc.org/cancer-care/diagnosis-treatment/symptom-management/integrative-medicine/herbs/search). NHP: Ingredient and product information was obtained from the NHP data extract (https://www.canada.ca/en/health-canada/services/drugs-health-products/natural-non-prescription/applications-submissions/product-licensing/licensed-natural-health-product-database-data-extract.html).
While the ingredient information from NMCD and MSKCC could be used directly, that from DSLD and NHP required additional preprocessing. Many of the ingredient names in DSLD include extraneous information such as dosage (eg, “500 mg Aloe Vera”), product name, and preparation information (eg, “Dehydrated Barley Grass”). We therefore defined a set of regular expressions to remove dosage, product names, legal information (eg, ™, ®, ©), and unwanted punctuation. We further preprocessed the ingredient names by removing dose forms and plant preparations listed by the Australian Therapeutic Goods Administration (TGA).28 Some DSLD ingredient names contain additional synonyms in parentheses, for example, “African Mango (Irvingia gabonensis) extract.” We developed an additional regular expression system to extract the text in parentheses which we then treated as a separate synonym. We filtered the extracted parenthetical text using the TGA list of plant parts as well as the regular expressions for dosage and legal information so as to not extract parenthetical text as in “Acai (fruit) extract” and “infusion (1:6000) of Agrimonia eupatoria” which often appear in the DSLD data. NHP contains a variety of nonsensical ingredient names such as “8” or “%.” We therefore developed a set of patterns that removed any ingredients whose names were less than 2 characters, contained only numeric characters, or only punctuation.
Creation of the iDISK data elements
In order to facilitate downstream processing, such as mapping to existing terminologies and the merging of synonymous concepts, the data output by the previous step was converted to match the iDISK data model. This was achieved by creating an iDISK data element (atom, concept, attribute, or relationship) for each source data point.
Atoms and concepts: A concept was created for each ingredient and product listed in each data source by 1) creating an atom for each synonym listed in the data source for the ingredient or product and 2) collecting these atoms together. The locations in the data sources from which these synonyms were obtained are given in Table 1. An atom was designated “preferred” for a concept if it is the primary name for the corresponding entry in the source database (eg, the name in the header of the ingredient monograph).
Concept attributes: These were created by extracting the relevant free text from each concept’s source data. For example, the DSLD monograph for Alfalfa gives its ingredient category as “botanical.” This text was paired with the Alfalfa concept to form the attribute [ingredient category: “botanical”]. The UMLS semantic type attribute of the semantic dietary supplement ingredient (SDSI) concept is an exception to this process. We created these attributes by mapping the SDSI preferred name to the UMLS (described below) and extracting the semantic types of the matched UMLS entry.
Relationships and relationship attributes: Each data source contains 1 or more of the relationship types. These are contained, for example, in the columns in the data extract or the sections in the ingredient monograph. Thus for each concept we generated a set of candidate relationships. As relationships connect 2 concepts, we first create a concept for the object of the relationship from the value in the data source. This object concept contains only 1 atom and is assigned a concept type to fit the implied relationship. For example, “contraceptives” is listed as a possible drug interaction for Alfalfa in NMCD (Figure 3). As the object of the interacts_with relationship must be a drug, we created a pharmacological drug (PD) concept with a contraceptives atom. We then created a relationship between the subject and object concepts and assigned any attributes specified by the data source. Extending the above example, this results in the relationship [Alfalfa, interacts_with, contraceptives] with the relationship attributes [interaction_severity: high] and [interaction_rating: moderate].
After creating the iDISK data elements, we mapped each concept to either the UMLS or MedDRA as specified by the data model. We used QuickUMLS to map to the UMLS as it has been shown to outperform MetaMap on multiple tasks.35 System organ class (SOC) concepts were mapped to MedDRA and, there being only 17 unique values present in NMCD, a physician informaticist (RR) confirmed the mapping manually. Atoms were created for each of the resulting mappings and added to the corresponding concept. In addition to facilitating interoperability between iDISK and other systems, these mapped atoms serve as normalized terms for the concepts which facilitated the discovery of synonymous concepts discussed in the next section.
Matching and merging concepts across data sources
The result of the previous step is a set of concepts from each data source. However, there is significant overlap in the concepts across the source databases as well as duplicate concepts within each database. It was therefore necessary to discover synonymous concepts and merge them. Intuitively, 2 concepts would be synonymous if they share 1 or more synonyms. However, a preliminary review of the matches produced using this method revealed a large number of incorrect matches due to over-general or incorrect synonyms in the data sources. For example, DSLD contains “vitamin” as a synonym of both “vitamin D” and “vitamin A,” leading to an incorrect match using this method. We found the following more restrictive criteria effective according to a preliminary review of the matches. Two concepts were considered synonymous if 1) the preferred name of 1 concept occurs in the atoms of the other and 2) the concepts are mapped to the same UMLS or MedDRA entry. In the case where the mapping tool failed to map a concept, the system uses just the first criterion. For example, say the atoms of the “Açaí” concept in NMCD are (Açaí, Acai, Acai extract) (the preferred name in bold) and it is mapped to the UMLS concept C3850037 (Acai Berries), and the synonyms of the “Euterpe oleracea” concept in DSLD are (Acai, Açaí, Euterpe oleracea, Assai), and it is also mapped to C3850037. In this case the preferred name of the first (Açaí) appears in the synonyms of the second, satisfying criterion 1; and they are mapped to the same UMLS concept, satisfying criterion 2, so the 2 monographs match.
We performed the above check for each pair of concepts across each data source. The result of this step is a number of sets of synonymous concepts. Each of these sets was merged into a single concept by combining the atoms, attributes, and relationships of the individual concepts in that set. After merging, we updated the subject of each relationship to be the new concept and updated the object concept as it was itself merged with other concepts. After 2 or more concepts are merged, the resulting concept will have more than 1 atom that is preferred. In order to determine which preferred atom should be used as the default, we rank them according to their source. We use the following ranking, from most to least preferred: UMLS/MedDRA, NMCD, MSKCC, DSLD, NHP.
DS products were not matched in this version of iDISK. DSLD covers US products while NHP covers Canadian products. Because the US and Canada have very different DS labeling regulations, products of the same name across these 2 resources may have conflicting label information.
Evaluation
The iDISK build process was evaluated by manually checking that the data elements in the final database were correctly extracted and integrated from the source data. We randomly selected 50 out of 4208 DS ingredient concepts for manual review. The manual review of these 50 concepts involved checking their associated 3632 atoms, 2422 relationships, and 1645 attributes against the source from which they were extracted. Due to the size of the task, it was split between 4 health informaticists (RR, YW, SZ, and YR), who labeled each iDISK data element as either “correct” or “incorrect” according to whether it was correctly extracted from the associated source data. Accuracy was computed as the percentage of data elements with a “correct” label. We provide separate extraction accuracies for the atoms from each source database, as well as for each relationship and each attribute.
RESULTS
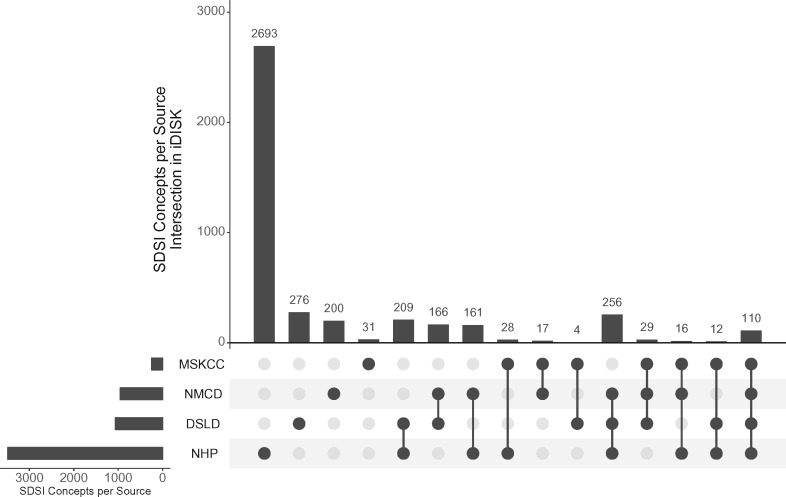
iDISK contains 144 654 unique concepts, including 4208 DS ingredient concepts and 137 568 DS product concepts, as well as 709 675 relationships and 84 674 attributes. Table 3 compares the number of concepts and attributes in iDISK to those extracted from the source databases. NHP provided the greatest number of ingredient concepts (3485) and product concepts (82 112) of all 4 data sources. NMCD, however, had the most comprehensive information, providing many of the relationships and attributes. The UpSet plot36 in Figure 4 shows the number of SDSI concepts containing information merged from each data source. This figure shows that while NHP provided the greatest number of ingredient concepts, over two-thirds of these were unmatched to any other concept from the other data sources.
Table 3.
The numbers of concepts, relationships and attributes in iDISK by data source
| NMCD | MSKCC | DSLD | NHP | iDISK | |
|---|---|---|---|---|---|
| Concepts(s) | |||||
| Semantic Dietary Supplement Ingredient (SDSI) | 955 | 247 | 1062 | 3485 | 4208 |
| Dietary Supplement Product (DSP) | – | – | 55 456 | 82 112 | 137 568 |
| Pharmacological Drug (PD) | 378 | 215 | – | – | 495 |
| Disease (DIS) | 722 | 201 | – | – | 776 |
| Therapeutic Class (TC) | 605 | – | – | – | 605 |
| System Organ Class (SOC) | 17 | – | – | – | 17 |
| Signs/Symptoms (SS) | – | 985 | – | – | 985 |
| Total concepts | 144 654 | ||||
| Relationships(s) | |||||
| is_effective_for | 4307 | 1056 | – | – | 5363 |
| has_therapeutic_class | 5454 | – | – | – | 5454 |
| has_adverse_effect_on | 3168 | – | – | – | 3168 |
| has_adverse_reaction | – | 2233 | – | – | 2233 |
| has_ingredient | – | – | 335 468 | 354 358 | 689 826 |
| interacts_with | 3076 | 555 | – | – | 3631 |
| Total relationships | 709 675 | ||||
| Attributes(s) | |||||
| Source Material | – | – | – | 5532 | 5532 |
| UMLS semantic type | – | – | – | – | 9230 |
| Ingredient Category | – | – | 1121 | – | 1121 |
| Background | 1140 | 259 | – | – | 1399 |
| Safety | 1150 | 69 | – | – | 1219 |
| Mechanism of action | – | 258 | – | – | 258 |
| LanguaL Product Type | – | – | 55 456 | – | 55 456 |
| Interaction_rating | 3076 | – | – | – | 3076 |
| Interaction_severity | 3076 | – | – | – | 3076 |
| Effectiveness_rating | 4307 | – | – | – | 4307 |
| Total attributes | 84 674 | ||||
The numbers in the columns for each data source represent the number of concepts extracted from that source, while the numbers in the iDISK column represent the number of concepts present in iDISK after matching and merging.
Figure 4.
UpSet plot36 depicting the number of SDSI concepts in iDISK matched and merged from each data source. Connected filled circles indicate the data sources, with the vertical bars showing the number of SDSI concepts in iDISK with atoms extracted from only those sources and not the others. For example, iDISK contains 110 SDSI concepts with atoms from all 4 data sources (MSKCC, NMCD, DSLD, NHP), 16 from MSKCC, NMCD, and NHP (not including DSLD), 28 from MSKCC and NHP (not including NMCD and DSLD), and 2693 SDSI concepts sourced only from NHP.
As illustrated in Table 4, accuracy across the DS data elements in iDISK demonstrates that the data extraction and integration methods used to create iDISK are effective, achieving accuracies in the range 89.6%–100%. Note that the number of data points for the Source material, Background, Safety, Mechanism of action, and LanguaL Product type attributes is low (< 100). However, since these attributes were extracted directly and without modification from the source databases, we do not expect much, if any, extraction error for these values.
Table 4.
Accuracy of the data elements for the 50 concepts evaluated against the relevant source databases
| Data element | N | Accuracy | Data element | N | Accuracy |
|---|---|---|---|---|---|
| SDSI Atoms | |||||
| NMCD | 1497 | 100.0% | Attributesa | ||
| MSKCC | 152 | 100.0% | Source material | 9 | 100.0% |
| DSLD | 1787 | 99.4% | Ingredient category | 141 | 100.0% |
| NHP | 195 | 100.0% | Background | 77 | 100.0% |
| Average Accuracy | 3632 | 99.7% | Safety | 58 | 100.0% |
| Relationships | Mechanism of action | 28 | 100.0% | ||
| is_effective_for | 874 | 99.3% | Langual Product type | 95 | 100.0% |
| has_therapeutic_class | 409 | 98.5% | Interaction rating | 252 | 99.7% |
| has_adverse_effect_on | 272 | 100.0% | Interaction severity | 252 | 99.7% |
| has_adverse_reaction | 240 | 89.6% | Effectiveness rating | 733 | 99.2% |
| ingredient_of | 277 | 99.3% | Average Accuracy | 1645 | 99.6% |
| interacts_with | 350 | 92.9% | |||
| Average Accuracy | 2422 | 97.4% | |||
Abbreviations: DSLD, Dietary Supplement Label Database; MSKCC, Memorial Sloan Kettering Cancer Center; NHP, Natural Health Products Database; NMCD, Natural Medicines Comprehensive Database; SDSI, semantic dietary supplement ingredient.
We do not include the UMLS semantic type attribute as an evaluation of the QuickUMLS tool used; to generate its values is outside the scope of this work.
DISCUSSION
iDISK integrates DS-related information from 4 well-regarded DS resources. As such, it contains more comprehensive information than any of the individual data sources. Furthermore, by standardizing this information according to a data model and linking it to existing controlled vocabularies, it renders this information more searchable and improves interoperability. iDISK’s terminology of DS ingredients can facilitate information retrieval of DS mentions from other resources, such as biomedical literature or electronic health records, and the inclusion of related information can assist clinicians and consumers find pertinent information about various supplements.
Error analysis
Figure 4 shows that over 2600 ingredient entries in NHP were not matched to entries in any other data source. A preliminary review of these ingredients revealed that many were unmatched because they were uncommon DS concepts that are not present in the other data sources, such as “Oryzin” (an enzyme of a type of mold) and “Partially hydrolyzed chicken eggshell membrane.” In some cases, synonymous concepts are present in 2 data sources, but unmatched due to nonoverlapping synonyms. For example, NHP and DSLD both contain entries corresponding to the DS ingredient Immortelle (a type of flowering plant). However, the closest synonyms are “Helichrysum italicum” in NHP and simply “Helichrysum” in DSLD, which were not matched using our method, which requires exact matches between synonym strings.
The imperfect accuracy for SDSI atoms sourced from DSLD (99.4%) was due to side-case errors during the preprocessing stage. For example, iDISK incorrectly contains “NITRO2GRANIT” as a synonym of pomegranate. This occurs because DSLD lists the product name “NITRO2GRANIT™” as a synonym of pomegranate. Due to our assumption that the data sources would only list ingredient names as synonyms, our preprocessing pipeline did not filter out product names, which means “NITRO2GRANIT” was added as a synonym after removing the “™”.
Finally, the lower accuracies for relationships (average 97.4%) compared to other data elements were largely due to errors in mapping the object concepts of the relationships to the UMLS. While QuickUMLS has been shown to outperform MetaMap,35 it is not without issues. For example, QuickUMLS fails to map the string “Antigout drugs” extracted from NMCD to the correct UMLS entry “Antigout Agents” (C4722035), instead mapping it to the general concept “Pharmaceutical Preparations” (C0013227) which does not accurately represent the information in the source. Such errors then propagate to the relationship attributes, which are incorrect if their associated relationship is incorrect.
Limitations and future work
The method for matching synonymous concepts is a limitation in the current version of iDISK. We developed our matching criteria according to a preliminary review of the matches produced, but a formal evaluation is needed in the future to assess the performance of this module fully. We also plan to address this limitation by investigating methods for matching concepts based on noisy sets of synonyms, such as those we obtain from our data sources.
As discussed in the error analysis, errors in concept mapping are another limitation in this version of iDISK. These errors affect both the creation of relationships, which are incorrect when their object concepts are mapped incorrectly, and the matching of concepts, in which false matches may occur if 2 nonsynonymous concepts are incorrectly mapped to the same UMLS entry. In the future, we plan to evaluate QuickUMLS, MetaMap, and other mapping tools to determine the best tools to use to minimize the mapping error in iDISK.
There are 2 limitations regarding the scope of iDISK. First, because the information in iDISK is collected from existing resources, it is necessarily limited to the information available in those resources. Thus, it is possible that iDISK does not include important information related to DS. However, it does provide a foundation for DS knowledge representation, which can be expanded to include new data elements and resources as they become available. Second, iDISK is primarily a DS ingredient knowledge base, and thus contains limited DS product information. We plan to include more product information (eg, dose, dose form, route, packaging, pharmacokinetics, licensing) in future iDISK versions, leveraging our preliminary work on the normalization of DS product names.29
Distribution and maintenance
The iDISK data files and associated code base are publicly available as described in the “Data Availability” section below. iDISK follows the semantic versioning system,37 which assigns each version 3 numbers of the format MAJOR.MINOR.PATCH. Major numbers correspond to changes incompatible with previous versions, minor numbers to backwards compatible changes, and patch numbers to bug fixes. NMCD, MSKCC, and DSLD provide rolling updates to their monographs while the NHP data extracts are released yearly. In light of this, we plan to release major iDISK updates when 1 or more of these data sources changes substantially or when we identify a new data source. We also plan to continuously improve iDISK via updates to the build process, such as the improvements to the concept mapping and matching modules discussed in the limitations section above.
CONCLUSION
We developed the first integrated DIetary Supplements Knowledge base (iDISK), where DS-related information is represented in a comprehensive and standardized form. We achieved this by integrating DS information from 4 existing and well-established DS resources. iDISK can serve as a one-stop DS information resource for a wide range of users, facilitating DS information extraction as well as interoperability across various DS systems and applications. We will continue to expand and improve iDISK as new resources become available and new techniques for data extraction and normalization are implemented.
DATA AVAILABILITY
iDISK is released in 2 formats: a Neo4j database and a set of UMLS-style pipe-delimited flat files. The current version of iDISK is publicly available for download at https://doi.org/10.13020/d6bm3v. The code used to build this release is publicly available at https://github.com/zhang-informatics/iDISK.
FUNDING
This work was supported by the National Center for Complementary & Integrative Health (NCCIH) and the Office of Dietary Supplements (ODS) grant number R01AT009457 (Zhang). The content is solely the responsibility of the authors and does not represent the official views of the NCCIH or ODS.
AUTHOR CONTRIBUTIONS
RZ, RR and JV conceived the study idea and design. RR and JV contributed equally to this project and the production of the manuscript. RR led the development of the knowledge base and was also lead annotator for the evaluation. JV implemented the code and generated the knowledge base data files and managed the evaluation infrastructure. RZ managed the project as a whole, providing guidance throughout. All authors contributed to the planning of the knowledge base, especially during the development of the data model.
ACKNOWLEDGMENTS
We would like to thank Changye Li for her efforts extracting the MSKCC data, and Yefeng Wang, Shuqin Zhou, and Yuanhao Ruan for their contribution to the evaluation.
Conflict of Interest statement
None to declare.
REFERENCES
- 1.Dietary Supplement Health and Education Act of 1994: Pub L. No. 103--417;1994.
- 2. Bailey RL, Gahche JJ, Lentino LC, et al. Dietary supplement use in the United States, 2003–2006. J Nutr 2011; 141 (2): 261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dwyer JT, Coates PM.. Why Americans need information on dietary supplements. J Nutr 2018; 148(suppl 2): 1401S–5S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geller AI, Shehab N, Weidle NJ, et al. Emergency department visits for adverse events related to dietary supplements. N Engl J Med 2015; 373 (16): 1531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natural Medicines Comprehensive Database (NMCD). https://naturalmedicines.therapeuticresearch.com/. Accessed October 2019.
- 6.Memorial Sloan Kettering Cancer Center: About Herbs, Botanicals, & Other Products. https://www.mskcc.org/cancer-care/diagnosis-treatment/symptom-management/integrative-medicine/herbs. Accessed October 2019.
- 7.Dietary Supplement Label Database (DSLD). https://www.dsld.nlm.nih.gov/dsld/index.jsp. Accessed October 2019.
- 8.LanguaL-The International Framework for Food Description. http://www.langual.org. Accessed October 2019.
- 9. Saldanha LG, Dwyer JT, Holden JM, et al. A structured vocabulary for indexing dietary supplements in databases in the United States. J Food Compost Anal 2012; 25 (2): 226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natural Health Products Ingredients Database (NHPID). http://webprod.hc-sc.gc.ca/nhpid-bdipsn/search-rechercheReq.do? lang=eng. Accessed October 2019.
- 11.Licensed Natural Health Products Database (LNHPD). https://www.canada.ca/en/health-canada/services/drugs-health-products/natural-non-prescription/applications-submissions/product-licensing/licensed-natural-health-products-database.html. Accessed October 2019.
- 12. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32(Database issue): D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.RxNorm Overview. https://www.nlm.nih.gov/research/umls/rxnorm/overview.html. Accessed October 2019.
- 14.Medication Reference Terminology (MED-RT) Documentation. https://evs.nci.nih.gov/ftp1/MED-RT/MED-RT%20Documentation.pdf. Accessed October 2019.
- 15.Medical Dictionary for Regulatory Activities (MedDRA). https://www.meddra.org/. Accessed October 2019.
- 16.The Anatomical Therapeutic Chemical (ATC) Classification System. https://www.who.int/medicines/regulation/medicines-safety/toolkit/en/ Accessed October 2019.
- 17. Manohar N, Adam TJ, Pakhomov S, et al. Evaluation of herbal and dietary supplement resource term coverage. Stud Health Technol Inform 2015; 216: 785–9. [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y, Adam T, Zhang R.. Term coverage of dietary supplements ingredients in product labels. In: proceedings AMIA Annual Symposium; 2016: 2053–61. [PMC free article] [PubMed]
- 19. Rizvi RF, Adam TJ, Lindemann EA, et al. Comparing existing resources to represent dietary supplements. AMIA Jt Summits Transl Sci Proc2018; 2017: 207–16. [PMC free article] [PubMed] [Google Scholar]
- 20. Boyce RD, Ryan PB, Norén GN, et al. Bridging islands of information to establish an integrated knowledge base of drugs and health outcomes of interest. Drug Saf 2014; 37 (8): 557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharma V, Sarkar IN.. Identifying supplement use within clinical notes: an application of natural language processing.AMIA Jt Summits Transl Sci Proc2018; 2018: 196–205. [PMC free article] [PubMed] [Google Scholar]
- 22. Fan Y, Zhang R.. Using natural language processing methods to classify use status of dietary supplements in clinical notes. BMC Med Inform Decis Mak 2018; 18(Suppl 2): 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan Y, Pakhomov S, McEwan R, et al. Using word embeddings to expand terminology of dietary supplements on clinical notes. J Am Med Inform Assoc Open 2019; 2 (2): 246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Friedman J, Birstler J, Love G, et al. Diagnoses associated with dietary supplement use in a national dataset. Complement Ther Med 2019; 43: 277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mazzanti G, Moro PA, Raschi E, et al. Adverse reactions to dietary supplements containing red yeast rice: assessment of cases from the Italian surveillance system. Br J Clin Pharmacol 2017; 83 (4): 894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sullivan R, Sarker A, O'Connor K, et al. Finding potentially unsafe nutritional supplements from user reviews with topic modeling. Pac Symp Biocomput2016; 21: 528–39. [PMC free article] [PubMed] [Google Scholar]
- 27. Trinh K, Pham D, Le L.. Semantic relation extraction for herb-drug interactions from the biomedical literature using an unsupervised learning approach. In: IEEE 18th International Conference on Bioinformatics and Bioengineering (BIBE); October 29–31, 2018; Taichung, Taiwan.
- 28. Meertens LJE, Scheepers HCJ, Willemse J, et al. Should women be advised to use calcium supplements during pregnancy? A decision analysis. Matern Child Nutr 2018; 14 (1): e12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan JW, Lussier YA.. Word-of-mouth innovation: hypothesis generation for supplement repurposing based on consumer reviews. AMIA Annual Symposium Proc 2018; 2017: 689–95. [PMC free article] [PubMed] [Google Scholar]
- 30. Sharma V, Sarkar IN.. Identifying natural health product and dietary supplement information within adverse event reporting systems. Pac Symp Biocomput2018; 23: 268–79. [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, Zhang Y, Jiang M, et al. Toward a normalized clinical drug knowledge base in China-applying the RxNorm model to Chinese clinical drugs. J Am Med Inform Assoc 2018; 25 (7): 809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Somé BMJ, Bordea G, Thiessard F, et al. Enabling West African herbal-based traditional medicine digitizing: the WATRIMed knowledge graph. Stud Health Technol Inform 2019; 264: 1548–9. [DOI] [PubMed] [Google Scholar]
- 33. Cossin S, Lebrun L, Lobre G, et al. Romedi: an open data source about French drugs on the semantic web. Stud Health Technol Inform 2019; 264: 79–82. [DOI] [PubMed] [Google Scholar]
- 34. Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998; 37 (4-5): 394–403. [PMC free article] [PubMed] [Google Scholar]
- 35. Soldaini L, Goharian N. QuickUMLS: a fast, unsupervised approach for medical concept extraction. In: MedIR Workshop, Special Interest Group on Information Retrieval (SIGIR); July 17–21, 2016; Pisa, Italy. https://github.com/Georgetown-IR-Lab/QuickUMLS. Accessed October 2019.
- 36. Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H.. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph 2014; 20 (12): 1983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Preston-Werner T. Semantic Versioning 2.0.0; 2013. https://semver.org/. Accessed October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
iDISK is released in 2 formats: a Neo4j database and a set of UMLS-style pipe-delimited flat files. The current version of iDISK is publicly available for download at https://doi.org/10.13020/d6bm3v. The code used to build this release is publicly available at https://github.com/zhang-informatics/iDISK.