Summary
Despite the immense growth of new anti-seizure drugs (ASDs), approximately one-third of epilepsy patients remain resistant to current treatment options. Advancements in whole genome sequencing technology continues to identify an increasing number of epilepsy-associated genes at a rate that is outpacing the development of in vivo animal models. Patient-derived induced pluripotent stem cells (iPSCs) show promise in providing a platform for modeling genetic epilepsies, high throughput drug screening, and personalized medicine. This is largely due to the ease of collecting donor cells for iPSC reprogramming, and their ability to be maintained in vitro, while preserving the patient’s genetic background. In this review, we summarize the current state of iPSC research in epilepsy and closely related syndromes, discuss the growing need for high-throughput drug screening (HTS), and review the use of stem cell technology for the purpose of autologous transplantation for epilepsy stem cell therapy. Although the use of iPSC technology, as it applies to ASD discovery, is in its infancy, we highlight the significant progress that has been made in phenotype and assay development to facilitate systematic HTS for personalized medicine.
Keywords: epilepsy, human induced pluripotent stem cells, anti-seizure drugs, personalized medicine, drug discovery
1. Generation and utility of patient-derived iPSCs
The development of anti-seizure drugs (ASDs) has seen tremendous growth since the 1930s (Loscher 2017), however, about one-third of epilepsy patients remain unresponsive to treatment (Gowers 1880; Shorvon 2009). It is estimated that ~75% of cases, previously identified as idiopathic, are thought to be caused by either a mono-genetic mutation or through complex inheritance (Thomas and Berkovic 2014). With the current advancements in genome wide sequencing, hundreds of epilepsy-associated genes have been identified (Noebels 2015; Wang, et al. 2017). In each of these cases, the effectiveness of current ASDs in seizure management varies (Balestrini and Sisodiya 2018). As the list of genes associated with epilepsy grows, the list of potential disease-causing genetic variants also grows. For example, in Dravet syndrome, 1,257 disease causing mutations within the SCN1A gene have been identified in patients (Meng, et al. 2015). Thus, it is becoming increasingly more difficult to develop models to study genetic epilepsies.
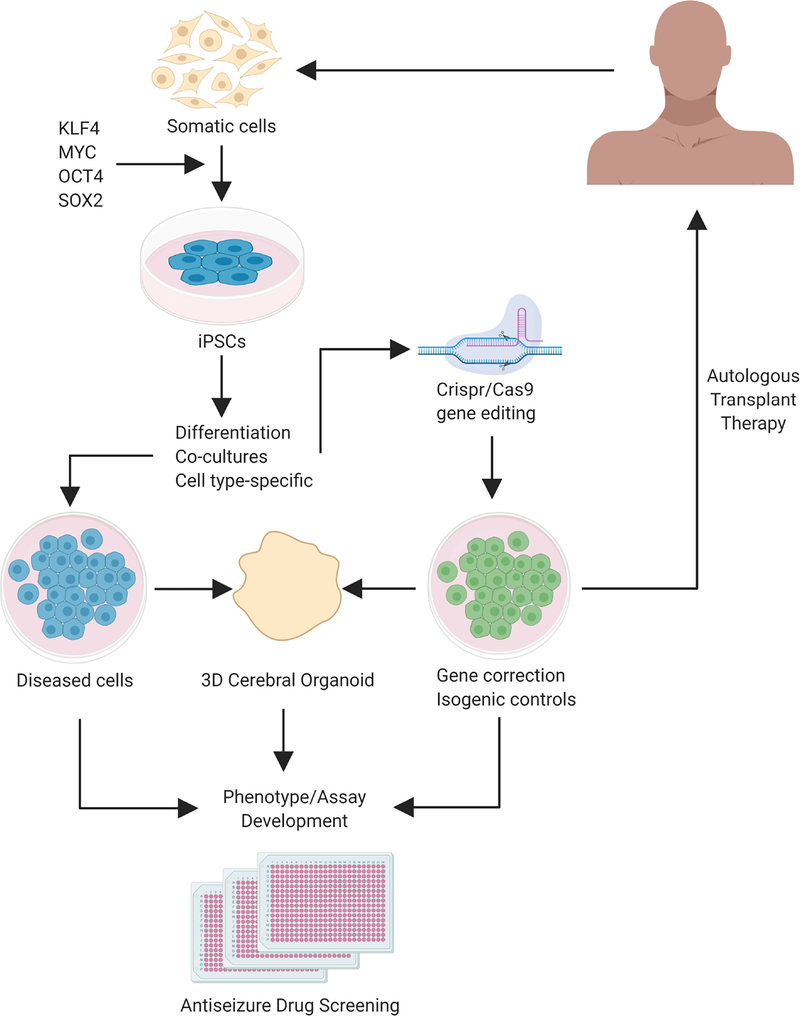
To keep up with the rapid discovery of genes and gene variants associated with epilepsy, scientists have developed stem cell-based technologies for patient-specific disease models. For example, induced pluripotent stem cells (iPSCs) are noninvasively derived from patient fibroblasts, peripheral blood mononuclear cells, dental pulp from deciduous teeth, and renal epithelial cells from urine (Staerk, et al. 2010; Xue, et al. 2013; Yan, et al. 2010). Researchers can reprogram these somatic cells into pluripotent stem cells by ectopically expressing transcription factors, and iPSCs can then be differentiated into a variety of post-mitotic cell lineages while maintaining the patient’s original genetic background (Takahashi, et al. 2007; Takahashi and Yamanaka 2006; Yu, et al. 2007) (Figure 1). Gene editing technology (e.g. CRISPR/Cas9) allows for gene correction and the generation of isogenic controls to study direct effects of gene mutations within the same genetic background (for review, Hockemeyer and Jaenisch 2016). Since the breakthrough of this technology, patient-derived iPSCs have been used to model a number of genetic neurological diseases including Down syndrome, schizophrenia, and Fragile X Syndrome (Baek, et al. 2009; Brennand, et al. 2011; Dimos, et al. 2008; Marchetto, et al. 2010; Pasca, et al. 2011; Urbach, et al. 2010).
Figure 1. Generation of human induced pluripotent stem cells for epilepsy therapies.
Somatic cells can be collected from patients, noninvasively. These cells are then reprogrammed by adding a combination of transcription factors that reprogram these cells to a pluripotent state. Differentiation of iPSCs can be directed towards specific neuronal subtypes, or mixed cultures that include glial cell types. iPSCs from patients can be genetically edited (i.e. CRISPR/Cas9, TALEN) to correct gene mutations and create isogenic controls. The patient’s cells and their gene edited equivalents are then subjected to a wide variety of differentiation protocols targeting specific cell types, of mixtures of cell types, in either two- or three-dimensional cultures. By comparing the diseased cell to isogenic repaired cells, multiple assays are employed to study disease phenotypes. Upon establishment of robust and reliable phenotypes and optimization/scaling-up of screening assays, libraries of candidate and existing antiseizure compounds can be tested at once to deliver personalized medicine. Alternatively, differentiated or repaired cells can be autologously transplanted into patients to restore epileptic circuits, however, this line of research will require extensive preclinical testing.
Many neurological disorders, including epilepsy, affect multiple cell lineages that are not limited to neurons (e.g. glutamatergic and GABAergic) and glia (e.g. oligodendrocytes, astrocytes, microglia, and ependymal cells). Pluripotent stem cells can be differentiated into almost any cell type of interest to study mechanisms of disease comorbidity (Li, et al. 2018). For example, nearly 20% of Dravet’s patients die from sudden unexpected death in epilepsy (SUDEP) (Cooper, et al. 2016). iPSCs derived from these patients were differentiated into cardiac myocytes, and exhibited faster spontaneous contraction rate and increased sodium currents compared to control myocytes, which correlated to an abnormal electrocardiogram in the patient (Frasier, et al. 2018). Therefore, the versatility of iPSC technology enables the investigation of single gene mutation across multiple organ systems.
Stem cell technology to treat epilepsy uses in vitro and in vivo applications. In vitro assays aim (1) to establish patient-specific cellular and network phenotypes that confer a high degree of validity to clinical epilepsy, and (2) to enable high-throughput screening (HTS) of candidate ASDs for personalized medicine. Current in vivo applications explore stem cell replacement therapies to restore function in epileptic circuits. In this review, we first summarize the current state of “epilepsy-in-a-dish” models and discuss potential applications for HTS (Table 1). We also discuss the in vivo application of stem cell therapy as a treatment for epilepsy. Finally, we discuss future directions of using stem cell approaches in antiseizure drug discovery.
Table 1.
Studies describing epilepsy relevant phenotypes of patient-derived iPSCs
| Reference | Epilepsy Gene (mutation) | Encoded protein | Syndrome | Control | Epilepsy relevant phenotypes |
|---|---|---|---|---|---|
| Jiao et al., 2013 |
SCN1A (F1415I missense mutation) |
α subunit of Nav1.1 | Dravet syndrome | Unaffected iPSC line |
|
| Liu et al., 2013 |
SCN1A (c.2589+3A>T and c.975T>A nonsense mutation) |
α subunit of Nav1.1 | Dravet syndrome | Unaffected iPSC line |
|
| Higaroshi et al., 2013 |
SCN1A (c.4933C>T) |
α subunit of Nav1.1 | Dravet syndrome | Unaffected iPSC line |
|
| Liu et al., 2016 |
SCN1A (c. 5768A>G) |
α subunit of Nav1.1 | Dravet syndrome | Unaffected iPSC line & Isogenic control (by TALEN) |
|
| Sun et al., 2016 |
SCN1A (p.S328P) |
α subunit of Nav1.1 | Dravet syndrome | Unaffected iPSC line |
|
| Kim et al., 2018 |
SCN1A (c.4261G>T and c.3547_3551delATCAA) |
α subunit of Nav1.1 | Dravet syndrome | Unaffected iPSC line |
|
| Marchetto et al., 2010 |
MECP2 (Q244X and c.1155del32) |
X-linked methyl-CpG binding protein 2 | Rett syndrome | Unaffected iPSC line |
|
| Ananiev et al., 2011 |
MECP2 (T158M, V247X, R306C, and R294X) |
X-linked methyl-CpG binding protein 2 | Rett syndrome | Unaffected iPSC line & isogenic control (X chromosome inactivation from female lines) |
|
| Kim et al., 2011 |
MECP2 (c.473C>T, c.730C>T, c.705delG, c.916C>T, c1461A>G) |
X-linked methyl-CpG binding protein 2 | Rett syndrome | Embryonic stem cell line (H1) |
|
| Tang et al., 2016 |
MECP2 (Q83X) |
X-linked methyl-CpG binding protein 2 | Rett syndrome | Unaffected iPSC line from father |
|
| de Souza et al., 2017 |
MECP2 (Q83X) |
X-linked methyl-CpG binding protein 2 | Rett syndrome | Unaffected iPSC line |
|
| Ohashi et al., 2018 |
MECP2 (c.1461A>G, c.705delG) |
X-linked methyl-CpG binding protein 2 | Rett syndrome | Isogenic control (X chromosome inactivation from female lines) |
|
| Tang et al., 2019 |
MECP2 MECP2-null embryonic stem cells |
X-linked methyl-CpG binding protein 2 | Rett syndrome | Isogenic control embryonic stem cell line |
|
| Ricciardi et al., 2012 |
CDKL5 (R59X, L220P) |
Cyclin-dependent kinase-like 5 | Rett-like syndrome | Isogenic control (X chromosome inactivation from |
|
2. Patient-specific models for genetic epilepsies
2.1. Dravet Syndrome (DS)
Dravet syndrome (DS) represents one of the most severe genetic epileptic encephalopathies. Between 70–80% of DS patients carry a heterozygous loss-of-function mutation in the SCN1A gene, which encodes the voltage-gated sodium channel Nav1.1 (Marini, et al. 2011). Seizures present early in infancy and progress through childhood, resulting in developmental cognitive delays (Ceulemans, et al. 2004; Dravet 2011). Dravet syndrome is one of the most pharmacoresistant epilepsies, whereby some ASDs that block sodium channels, such as lamotrigine and carbamazepine, actually worsen seizure outcome (Chiron and Dulac 2011). Studies using DS patient-derived iPSCs have focused on neuronal hyperexcitability phenotypes. Jiao and colleagues found that SCN1A mutations increase sodium currents, in frequency and amplitude of evoked action potentials predominantly in excitatory neuron cultures (Jiao, et al. 2013). Moreover, phenytoin was shown to alleviate these hyperexcitability phenotypes, demonstrating predictive validity in using DS patient-derived iPSCs (Jiao, et al. 2013). These findings suggest that seizures from this patient may stem from excess activity of excitatory, glutamatergic neurons. However, other studies report predominantly GABAergic neurons in culture when differentiated from another DS iPSC line. Both GABAergic and glutamatergic cells exhibited increased sodium currents, along with more spontaneous bursts compared to neurons differentiated from control patient iPSCs (Liu, et al. 2013b). Therefore, whether SCN1A mutations preferentially alter excitatory or inhibitory cells is unclear. Possible explanations could lie in variations within differentiation protocols. Another possible explanation is that the SCN1A variant or the patient’s background might account for preferential differentiation of glutamatergic versus GABAergic cells.
In stark contrast to the above findings that suggest SCN1A mutations enhance sodium channel-mediated neuronal output, additional studies that focused on GABAergic cells have shown reduced action potential generation and a reduction in sodium currents, where the magnitude of change corresponded to the symptom severity of the patient from which the cells were derived (Higurashi, et al. 2013; Kim, et al. 2018; Liu, et al. 2016). This would suggest that reduced interneuron output could promote seizures in these patients. Confirming this interpretation, one study directly compared the effects of SCN1A mutations on GABAergic and glutamatergic cells derived from the same patient, and revealed a selective reduction in Nav1.1 channel function only in GABAergic cells (Sun, et al. 2016).
Taken together, it is unclear whether Nav1.1 channel dysfunction observed in DS-derived iPSCs is present in excitatory neurons, inhibitory neurons, or both. Increases in DS-derived excitatory neuron output or decreased inhibitory neuron output could both lead to hyperexcitable networks via distinct, but overlapping mechanisms, to promote seizures. Because of the high variability in SCN1A mutations is thought to be disease causing, possible differences in Nav1.1 expression may influence network excitability in a patient-specific manner. Additionally, since many of the current studies use healthy controls, phenotypic differences may be due to variation between genetic backgrounds of healthy control patients. For this reason, it is essential to compare between isogenic controls and DS patient iPSCs to better interpret these data. Thus, patient-derived iPSC-based studies highlight several robust phenotypes that can provide mechanistic insight to disease pathology on an individualized basis.
2.2. Rett syndrome
Rett syndrome is a severe autism spectrum developmental disorder characterized by an initial period of normal development followed by a rapid decline in acquired motor skills, language, and gait. An X-linked mutation in the MECP2 gene accounts for 95% of the typical Rett syndrome cases (Operto, et al. 2019), and 60–80% of these patients develop seizures (Glaze, et al. 2010; Operto, et al. 2019). These mutations occur de novo and currently about 800 different variants are attributed to causing Rett Syndrome (Ehrhart, et al. 2018). In addition, Rett-like syndromes result from mutations in genes that regulate MECP2, (e.g CDKL5 and FOXG1), which is characterized by early onset seizures (Operto, et al. 2019). Human and animal studies have observed reductions in cell size in postmortem Rett patient brain samples and MeCP2 knockout mouse models (Chahrour and Zoghbi 2007; Chen, et al. 2001). Some studies using Rett patient-derived iPSCs have also shown reductions in neuron size (Ananiev, et al. 2011; Marchetto, et al. 2010). Additional studies have suggested reductions in glutamatergic cell number due to neuronal maturation deficits (Kim, et al. 2011), reduced dendritic branching (Ohashi, et al. 2018), reduced dendritic spines (Marchetto, et al. 2010), and reduced putative excitatory synapses (Marchetto, et al. 2010; Ricciardi, et al. 2012). In some cases, these morphological alterations were accompanied by reduced calcium transients and spontaneous excitatory postsynaptic currents (Marchetto, et al. 2010). Consistent with rodent models of Rett syndrome, drastic dendritic reduction were observed in iPSC-derived neurons from MECP2 mutant cell lines. Pharmacological inhibition of p53 induction, a regulator of cellular senescence, with Pifithrin-α rescued the dendritic branching deficit in Rett iPSC-derived neurons (Ohashi, et al. 2018).
Similarly, in iPSCs derived from Rett syndrome patients, IGF-1 receptor expression was increased compared to a wild-type control line. When treated with IGF-1, neurons derived from Rett iPSCs were shown to recover their neurite length, suggesting targets for IGF-1 receptors can improve cell morphology in assays for neurite length (de Souza, et al. 2017). An additional study provided mechanistic insights linking deficits in potassium transporter (KCC2), a downstream target of MeCP2, in Rett iPSC-derived neurons to a delayed functional switch of GABA from excitation to inhibition (Tang, et al. 2016). Because of the major influence on maintaining the excitability/inhibitory (E/I) balance, reductions in KCC2 is thought to underlie reported dendritic deficits in spine morphogenesis and synapse development (Gauvain, et al. 2011; Li, et al. 2007; Puskarjov, et al. 2014). For iPSC models of Rett syndrome, dendritic changes appear the most robust. Additionally, since pharmacological targets can recover dendritic deficits, these models are apt for future drug screens to discover novel compounds.
More recently, this group used embryonic stem cells and CRISPR-Cas9 gene editing technology to generate Rett-like mutations in MECP2 and appropriate isogenic controls (Tang, et al. 2019). By screening 900 small molecules approved by the U.S. Food and Drug Administration they identified inhibitors of the fms-like tyrosine (FLT3) or glycogen synthase kinase 3β (GSK3β) pathways and activators of sirtuin 1 (SIRT1) and transient receptor potential cation channel subfamily V member 1 (TRPV1) pathways. These targets were sufficient to enhance KCC2 expression and rescue the E/I balance, previously shown to be disrupted in Rett-patient neurons. Further, they identified small molecules rescued dendritic branching deficits found in the MECP2 mutant neurons and ameliorated disease-related breathing and locomotor deficits in MECP2 mutant mice. While this study did not use patient-derived cells, it demonstrates the power of disease models-in-a-dish that can be translated to pre-clinical animal models.
It is currently unclear how these phenotypes could promote seizure phenotypes. One possibility is that cellular stress-induced injury could contribute to these morphological changes to indirectly cause epilepsy. Future studies linking changes to cell morphology to hypersynchronous networks will be valuable to identify phenotypic alterations that relevant to epileptogenesis. Together these studies using Rett patient-derived iPSCs, demonstrate the utility in studying cellular morphology-based assays.
2.3. Angelman syndrome (AS)
Angelman syndrome is characterized by microcephaly, intellectual impairments, speech deficits, paroxysms of laughter, and seizures occur in ~90% of AS patients (Pelc, et al. 2008) suggesting that neuronal hyperexcitability plays a prominent role in AS. These symptoms are linked to a loss-of-function mutation to the maternal imprinted UBE3A gene on chromosome 15 (Buiting, et al. 2016; Vu and Hoffman 1997). Around 75% of AS patients contain a 5–7Mb de novo interstitial deletion of the 15q11.2-q13 chromosome region. AS patient-derived iPSCs have successfully been differentiated into functional neurons (Chamberlain, et al. 2010; Fink, et al. 2017).
Fink and colleagues revealed several physiological phenotypes that manifested when iPSC-derived neurons spent 12–20 weeks in culture. Control neurons exhibited a developmental hyperpolarizing shift of the resting membrane potential (RMP) after 5 weeks in culture, which was absent in AS cells, and in neurons where the UBE3A gene was knocked out using CRISPR/Cas9. These investigators also observed a reduced incidence of “mature firing”, defined as spike amplitudes > 35mV and durations < 5.5 ms, in AS-derived neurons, and fewer spontaneous calcium transients. In line with alterations to active and passive membrane properties, decreases in the percent of synaptically-active cells were observed in AS-derived cells, as well as reduced frequency, but not amplitude of spontaneous excitatory synaptic currents. Additionally, employing a forskolin-induced long-term potentiation paradigm previously shown to be NMDA receptor dependent, these authors found that AS cells and UBE3A knockout cells failed to maintain LTP after its initial induction (Fink, et al. 2017). Due to neuron-specific genomic imprinting, activating the silenced, but present paternal copy of UBE3A may provide a therapeutic target. Indeed, the topoisomerase inhibitor, topotecan, partially restored UBE3A mRNA expression while rescuing the RMP, action potential, and synaptic phenotypes to control levels.
Much like the case of Rett syndrome, it is unclear how the reduced excitability phenotypes observed from AS-derived neurons could promote epilepsy, however, these studies represent a potential epileptogenic period of neuronal re-wiring that could ultimately lead to hypersynchronous neuronal networks. Future studies could test this possibility by measuring the activity patterns of neuronal networks generated by patient-derived iPSCs.
3. Potential for ASD discovery using epilepsy-in-a-dish models
A major goal for iPSC research is to develop assays for high-throughput screening (HTS). Use of patient-derived iPSCs has been successful for other neurological disorders including Amyotrophic Lateral Sclerosis (Bhinge, et al. 2017; Gendron, et al. 2017; Imamura, et al. 2017; Marrone, et al. 2018; Osborn, et al. 2018; Simone, et al. 2018), Autism Spectrum Disorder (Darville, et al. 2016), Alzheimer’s Disease (Kimura, et al. 2018; Kondo, et al. 2017; Wang, et al. 2018; Young, et al. 2018), Fragile X Syndrome (Kaufmann, et al. 2015; Kumari, et al. 2015), and Parkinson’s Disorder (Burbulla, et al. 2017; Chen, et al. 2017; Kouroupi, et al. 2017; Mittal, et al. 2017). These studies screened candidate drugs known or predicted to modulate a specific disease phenotype (Elitt, et al. 2018). Unfortunately, this has not yet occurred for epilepsy. Limiting factors include the large numbers of disease-causing gene variants, and the established epilepsy-like phenotypes that are robust, reproducible, and suited for HTS.
Despite these limiting factors, iPSC research has greatly advanced our understanding of disease mechanisms underlying genetic epilepsies, and the field is poised and ready for assay development. Current approaches include imaging-based screens, hyperexcitability assays, gene expression assays, and cell viability assays. Imaging-based screens are ideal to detect changes to cell morphology, soma size and shape, dendritic spine count, and dendritic branching can be quantified. For Rett syndrome, dendritic branching and spine changes were a robust phenotype characterized (Table 1). As previously discussed, HTS using embryonic stem cells with mutations in MECP2 via CRISPR-Cas9 found drug targets that rescued dendrite branching and these morphological changes translated to pre-clinical rodent models (Tang et al., 2019). Advancements in machine learning based image profiling enable morphology based assays to be used for HTS (Scheeder, et al. 2018).
Neuronal hyperexcitability assays that monitor real-time functional properties of neuronal populations would be a valuable approach for HTS. Standard procedures have relied on single-cell data obtained using whole-cell patch clamp techniques. Although this approach has provided much insight into cellular physiology underlying disease, it is labor-intensive and lower throughput compared to some newer assays better suited for HTT, including multielectrode arrays (MEA) and calcium imaging-based assays. MEA technology comes in multiwell platforms (e.g. 24, 48, 96 wells) where large numbers of iPSC-derived neurons are grown onto electrode arrays that detect local field potentials (LFPs) of neuronal populations. This allows for the direct comparison of neuronal activity between different genotype backgrounds, culturing conditions, or candidate compounds. In addition, live cell calcium imaging, using genetically encoded calcium indicators (e.g. GCaMP) or indicator dyes (e.g. fluo-4), can be used to monitor calcium flux from iPSC-derived neurons to allow for non-invasive, multi-neuronal activity to be monitored, and neural network level activity analyzed. An added compliment to this approach is the growing integration of machine learning and big data to identify disease characteristics in electrographic data. Signatures from MEA recordings could be used to classify healthy versus disease characteristics and future prospects to identify novel mutation specific electrographic biomarkers would greatly benefit patient care.
A third assay that can be used for HTS in epilepsy is gene expression assays. This assay depends on identifying targets to restore deficient gene function and can be done using gene microarrays, RNA sequencing, or fluorescent reporter-based assays. For example, in Fragile-X Syndrome, the Fmr1 gene is silenced on the X chromosome, and it is thought that reactivating the silenced Fmr1 gene rescues FXS phenotypes. FXS-derived iPSCs have successfully been screened to identify compounds that rescue lost FMRP protein expression and identified multiple compounds (Kaufmann, et al. 2015; Kumari, et al. 2015). For epilepsy, it remains unknown if rescuing gene expression is sufficient to reduce disease relevant phenotypes. However, recent studies using a heterozygous loss-of-function SCN1A mutant mouse model for DS demonstrated that Hm1a, a spider venom peptide, restores proper Nav1.1 activity in fast-spiking inhibitory interneurons. The rescue of mice from seizures and premature death presumably occurs by boosting wild type SCN1A expression (Richards, et al. 2018). This demonstrates that rescuing Nav1.1 function has the potential to ameliorate the DS phenotype, thus efforts to identify compounds that target Nav1.1 expression could rescue haploinsufficiency phenotypes in DS patients.
Epilepsy is often associated with either programmed or unprogrammed cell death. Cell viability assays can serve as valid phenotypes amenable to HTS. Possible outcome measures include readouts of cellular metabolism, combined with gene arrays to detect affected cell types, and cell counting assays. While the causative role of cell death in epileptogenesis is uncertain, drug development has been proposed to target the IL-1β, TNF-α, activated caspase, and inflammation pathways (Dingledine, et al. 2014).
iPSC technology is on the verge of new ASDs discoveries. A major limitation that should not be overlooked is the ability to predict antiseizure outcomes. As seizures are the primary symptom of epilepsy, further studies linking cellular phenotypes to seizure generation and propagation are required to ensure that iPSC technology can identify antiseizure drug targets. Validating salient phenotypes for morphology, hyperexcitability, gene expression, and cell viability to characterize “epilepsy-in-a-dish” models may come from complex phenotypes that utilize multiple assays.
4. Stem cell replacement therapies for epilepsy, in vivo
In addition to using iPSCs in vitro for mechanistic studies of genetic epilepsies and HTS of candidate ASDs, another goal is to use the patient’s own cells to heal themselves, in vivo. Examples of this approach can be found in cases of age-related macular degeneration (AMD). Initial clinical trials in Japan using iPSC transplants to treat age-related macular degeneration (AMD) have demonstrated iPSC transplant safety (Mandai, et al. 2017). However, initial concerns for possible oncogenic genes have caused some delay due to the need to optimize reprogramming protocols. A separate group has developed retinal patches from oncogenic mutation-free clinical grade iPSCs that rescued retinal degeneration in large animal models which demonstrated the safety and efficacy of iPSC-derived transplants (Sharma, et al. 2019).
Mesial temporal lobe epilepsy (mTLE) comprises a significant proportion of medically refractory epilepsy and commonly affects the hippocampus. Early models of mTLE revealed that while grafting hippocampal tissue from fetal rat brains into the adult epileptic animal was anticonvulsant, similar grafts into a non-epileptic hippocampus could be proconvulsant (Buzsaki, et al. 1988). The decades following this work has demonstrated that replenishing the GABAergic neuron pool, in particular, can improve seizure phenotypes in commonly used rodent epilepsy models (Baraban, et al. 2009; Calcagnotto, et al. 2010; Cunningham, et al. 2014; Hammad, et al. 2015; Handreck, et al. 2014; Hattiangady, et al. 2008; Henderson, et al. 2014; Hunt, et al. 2013; Loscher, et al. 1998; Maisano, et al. 2012). The medial ganglionic eminence (MGE) contains GABAergic neuron progenitors that populate the forebrain during development. To this end, cells derived from the MGE of E13.5 rodent embryos and dissociated into cell suspensions were transplanted into adult epileptic rodent hippocampi (Hunt, et al. 2013). After transplantation, surviving MGE progenitors exhibit similar migratory patterns to normal development and retain their neurochemical and physiological properties (Baraban, et al. 2009; Casalia, et al. 2017; Hunt, et al. 2013). Importantly, animals containing MGE transplants showed reduced seizure phenotypes for up to at least 6 months, but caudal ganglionic eminence (CGE) transplantation was ineffective at all-time points tested (Casalia, et al. 2017). Seizure rescue was also observed when H7 embryonic stem cells were differentiated into MGE-like cells prior to transplantation (Cunningham, et al. 2014). While most of these studies are promising, conflicting results were observed from another group, who despite observing behavioral rescue of cognitive function, found no effect on seizure activity (Anderson, et al. 2018). Possible sources of discrepancies include the duration and timing of seizure monitoring, differing culturing techniques, or differences in the extent of synaptic integration of the transplanted cells.
One conclusion from this research is that MGE transplants may ameliorate seizures by increasing levels of GABA in the host brain. However, increasing GABA levels is already a major mechanism of action of several current ASDs (e.g. barbiturates, phenobarbital, and valproic acid) and can lead to significant side effects in some patients (Cramer, et al. 2010). MGE transplants in mice lacking the α4 subunit of GABAA receptors that are associated with extrasynaptic GABA receptors, did not exhibit seizure reductions (for review, see (Brickley and Mody 2012)). Interestingly, extrasynaptic GABA receptors bind neurosteroids which have anticonvulsant properties (Biagini, et al. 2010; Yawno, et al. 2017). Therefore, the efficacy of transplantation assays could be combined with additional pharmacological treatment for the best possible patient outcome.
Given the ethical limitations of obtaining embryonic or fetal tissue, another focus of stem cell transplantation studies is to use iPSCs to provide seizure control using the patient’s own cells (Liu, et al. 2013a). Advantages include autologous transplantation, which bypasses graft rejection immune issues. A thorough investigation of human iPSC transplantation in a rodent mTLE model was performed by Upadhya and colleagues (Upadhya, et al. 2019). Human iPSCs were differentiated into MGE cells and transplanted into epileptic rats. Cells from these grafts survived, proliferated, and migrated after transplantation. Immunostaining revealed that these cells differentiated into mature, GABAergic interneurons, and expressed a number of associated peptides and calcium binding proteins. EEG recordings performed in animals receiving the MGE grafts revealed a reduction in seizure frequency. Finally, performance on a battery of behavioral tests demonstrated improvement in grafted rats. Together, these findings are promising in that the use of iPSCs can serve as a potential disease-modifying treatment option for refractory mTLE in animals. More work is needed to fully develop iPSC grafts into an applicable therapeutic. An open question that remains is how long do the anti-seizure effects of iPSCs grafts last. Many of the studies monitor seizures for <4 weeks after transplant, however it is important to determine long term effects of transplanted cells. Do they remain efficacious and are there long-term side effects (Baraban, et al. 2009; Hunt, et al. 2013; Upadhya, et al. 2019)?
Stem cells derived from other sources have also been investigated and include human adipose stem cells (Jahanbazi Jahan-Abad, et al. 2018), human fetal lung fibroblasts (Avaliani, et al. 2017), bone marrow mononuclear cells (DaCosta, et al. 2018), and ventral midbrain-derived cells (Backofen-Wehrhahn, et al. 2018). Furthermore, the administration of stem cells with a hydrogel matrix nanoscaffold was found to promote the survival of cell grafts and reductions in brain lesion volume, and ameliorate seizure phenotypes compared to stem cell grafts alone (Jahanbazi Jahan-Abad, et al. 2018). Since all of the approaches discussed so far require invasive surgical grafting procedures, additional studies are investigating the efficacy of intravenous delivery of neurospheres (de Gois da Silva, et al. 2018) and mesenchymal stem cells (Fukumura, et al. 2018), which also appear to reduce seizure phenotypes. However, more thorough analyses of these methods on seizure outcomes, cellular function, and cognition are necessary.
5. Future directions of stem cell approaches in antiseizure drug discovery
The two major goals of stem cell biology in ASD discovery are high throughput drug screening, and personalized medicine. Although each of these goals require very different strategies, they are not mutually exclusive. While HTS benefits from standardization of protocols and phenotypes, these protocols when applied to individual patient cell lines will allow personalized treatment plans even for the rarest epilepsies. This effort will help determine the contribution of individual genetic backgrounds in ASD responsiveness, possible side effects, and potential non-ictal biomarkers.
Epilepsy is often accompanied by abnormal EEG activity with or without structural abnormalities. However, depending on the type of epilepsy presented, these EEG signals are quite variable in both the spatial and temporal properties. They can be either generalized or focal, and can occur unpredictably as isolated events, or occur as closely spaced clusters followed by extensive seizure-free periods. Furthermore, diagnosis of epilepsy requires that epilepsy has already developed, precluding the ability to prevent the development of epilepsy in susceptible individuals. Therefore, biomarker discovery is an active area of investigation (Engel, et al. 2018; Hegde and Lowenstein 2014). Biomarkers found in the circulating blood or cerebrospinal fluid (CSF) might predict seizure recurrence, give insight to potential cause, or better our understanding of epileptogenesis. These biomarkers have relied mostly on animal models for validation but their translational significance is an area where iPSC technology can be applied to help elucidate. For example, superoxide dismutase 1 (SOD1) was found to be significantly decreased in the CSF of patients with epilepsy, specifically those with intractable epilepsy (Chen, et al. 2012). Because patient-derived iPSCs can be differentiated into multiple cell types, SOD1 can be studied in different neuronal and non-neuronal cell types to understand its role in epilepsy. This could lead to better prediction of a patient’s intractability, drug interactions, and develop possible drug targets for treatment.
One criticism of using iPSC-derived neurons to model epilepsy is that the reduced niche limits the development of complex neural networks necessary for reproducing seizure-like activity. To overcome this hurdle, iPSC have been grown in three-dimensional (3D) organoid structures known to recapitulate early cortical development. Protocols for organoid development can be undirected, with the potential to grow complex neural structures similar to the retina, hindbrain, and cortical lamina (Lancaster, et al. 2013), or for specific brain regions like telencephalic (Mariani, et al. 2015), cerebral cortex (Pasca, et al. 2015), thalamic (Xiang, et al. 2019), forebrain, midbrain, or hypothalamus (Chen, et al. 2016). For epilepsy related diseases, iPSC-derived organoids have been used to study Rett syndrome (Mellios, et al. 2018), tuberous sclerosis (Blair, et al. 2018), and patients with CACNA1C mutations (Birey, et al. 2017). For in depth review of organoids used in other models of neurological diseases see (Amin and Pasca 2018). While these 3D organoids offer in vitro models of brain development they too come with limitations. Currently, cerebral organoids lack appropriate vascularization which has been argued to limit their growth, both in size and speed of maturation, by preventing proper gas exchange and nutrient availability. Overcoming these limitations would open up iPSC technology to model complex neuronal network function in healthy and disease states.
In summary, the historical contribution of animal models to ASD discovery has been highly successful in understanding epilepsy. As genome wide sequencing expands our understanding of the genetic contribution to epilepsy, modern medicine must move towards personalized medicine approaches that utilize patient-specific models (Figure 1). Stem cell technology, specifically iPSCs, offer the ability to investigate disease models and expand HTS for identifying novel compounds on a patient-specific level. The current status of the iPSC field is evaluating various differentiation protocols that can best model certain aspects of disease pathology and characterizing disease relevant phenotypes. While this foundation has been set and establishes the advantages of iPSCs to model genetic epilepsies and utilize them for ASD discovery, further testing to demonstrate their patient specific predictability of in vitro phenotypes must be accomplished. As a therapeutic themselves, transplanted stem cells have demonstrated to be safe and effective in reducing seizures in rodents. The potential is a non-pharmaceutical approach that one day may provide long-term or permanent management of seizures. Personalized therapy could transform the future of medicine and stem cell technology carves the path towards that potential.
Highlights:
The increase in number of identified epilepsy genes are out pacing the generation of in vivo models.
Induced pluripotent stem cells (iPSCs) offer patient-specific in vitro models to study disease mechanism for genetic epilepsies.
iPSCs are poised to use in high-throughput drug screens for personalized medicine.
Studies using in vivo stem cell transplantation in rodent models of epilepsy present a potential non-pharmaceutical therapeutic.
Acknowledgements
Figure illustration was constructed using BioRender. This work was supported by grants from the National Institute of Health (NIH) (R01NS093992, R01NS081203, R01NS089770, and R21NS090926 to J.H.), Department of Defense (W81XWH-15-1-0399 to J.H.). We are also grateful for support from the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation and the Semmes Foundation, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amin ND, and Pasca SP 2018. Building Models of Brain Disorders with Three-Dimensional Organoids. Neuron 100(2):389–405. [DOI] [PubMed] [Google Scholar]
- Ananiev G, et al. 2011. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS One 6(9):e25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NC, et al. 2018. Pluripotent stem cell-derived interneuron progenitors mature and restore memory deficits but do not suppress seizures in the epileptic mouse brain. Stem Cell Res 33:83–94. [DOI] [PubMed] [Google Scholar]
- Avaliani N, et al. 2017. Directly Converted Human Fibroblasts Mature to Neurons and Show Long-Term Survival in Adult Rodent Hippocampus. Stem Cells Int 2017:5718608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backofen-Wehrhahn B, et al. 2018. Anticonvulsant effects after grafting of rat, porcine, and human mesencephalic neural progenitor cells into the rat subthalamic nucleus. Exp Neurol 310:70–83. [DOI] [PubMed] [Google Scholar]
- Baek KH, et al. 2009. Down’s syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature 459(7250):1126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini S, and Sisodiya SM 2018. Pharmacogenomics in epilepsy. Neurosci Lett 667:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraban SC, et al. 2009. Reduction of seizures by transplantation of cortical GABAergic interneuron precursors into Kv1.1 mutant mice. Proc Natl Acad Sci U S A 106(36):15472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhinge A, et al. 2017. Genetic Correction of SOD1 Mutant iPSCs Reveals ERK and JNK Activated AP1 as a Driver of Neurodegeneration in Amyotrophic Lateral Sclerosis. Stem Cell Reports 8(4):856869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagini G, Panuccio G, and Avoli M 2010. Neurosteroids and epilepsy. Curr Opin Neurol 23(2):170–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F, et al. 2017. Assembly of functionally integrated human forebrain spheroids. Nature 545(7652):54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JD, Hockemeyer D, and Bateup HS 2018. Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat Med 24(10):1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, et al. 2011. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473(7346):221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickley SG, and Mody I 2012. Extrasynaptic GABA(A) receptors: their function in the CNS and implications for disease. Neuron 73(1):23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K, Williams C, and Horsthemke B 2016. Angelman syndrome - insights into a rare neurogenetic disorder. Nat Rev Neurol 12(10):584–93. [DOI] [PubMed] [Google Scholar]
- Burbulla LF, et al. 2017. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science 357(6357):1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G, et al. 1988. Restoration and deterioration of function by brain grafts in the septohippocampal system. Prog Brain Res 78:69–77. [DOI] [PubMed] [Google Scholar]
- Calcagnotto ME, et al. 2010. Grafting of GABAergic precursors rescues deficits in hippocampal inhibition. Epilepsia 51 Suppl 3:66–70. [DOI] [PubMed] [Google Scholar]
- Casalia ML, Howard MA, and Baraban SC 2017. Persistent seizure control in epileptic mice transplanted with gamma-aminobutyric acid progenitors. Ann Neurol 82(4):530–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans BP, Claes LR, and Lagae LG 2004. Clinical correlations of mutations in the SCN1A gene: from febrile seizures to severe myoclonic epilepsy in infancy. Pediatr Neurol 30(4):236–43. [DOI] [PubMed] [Google Scholar]
- Chahrour M, and Zoghbi HY 2007. The story of Rett syndrome: from clinic to neurobiology. Neuron 56(3):422–37. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, et al. 2010. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci U S A 107(41):17668–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, et al. 2012. Clinical value of decreased superoxide dismutase 1 in patients with epilepsy. Seizure 21(7):508–11. [DOI] [PubMed] [Google Scholar]
- Chen J, et al. 2016. Membrane fouling in a membrane bioreactor: High filtration resistance of gel layer and its underlying mechanism. Water Res 102:82–89. [DOI] [PubMed] [Google Scholar]
- Chen RZ, et al. 2001. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet 27(3):327–31. [DOI] [PubMed] [Google Scholar]
- Chen ZC, et al. 2017. Phosphorylation of amyloid precursor protein by mutant LRRK2 promotes AICD activity and neurotoxicity in Parkinson’s disease. Sci Signal 10(488). [DOI] [PubMed] [Google Scholar]
- Chiron C, and Dulac O 2011. The pharmacologic treatment of Dravet syndrome. Epilepsia 52 Suppl 2:72–5. [DOI] [PubMed] [Google Scholar]
- Cooper MS, et al. 2016. Mortality in Dravet syndrome. Epilepsy Res 128:43–47. [DOI] [PubMed] [Google Scholar]
- Cramer JA, et al. 2010. Adverse effects of antiepileptic drugs: a brief overview of important issues. Expert Rev Neurother 10(6):885–91. [DOI] [PubMed] [Google Scholar]
- Cunningham M, et al. 2014. hPSC-derived maturing GABAergic interneurons ameliorate seizures and abnormal behavior in epileptic mice. Cell Stem Cell 15(5):559–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaCosta JC, et al. 2018. Safety and seizure control in patients with mesial temporal lobe epilepsy treated with regional superselective intra-arterial injection of autologous bone marrow mononuclear cells. J Tissue Eng Regen Med 12(2):e648–e656. [DOI] [PubMed] [Google Scholar]
- Darville H, et al. 2016. Human Pluripotent Stem Cell-derived Cortical Neurons for High Throughput Medication Screening in Autism: A Proof of Concept Study in SHANK3 Haploinsufficiency Syndrome. EBioMedicine 9:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gois da Silva ML, et al. 2018. Neurochemical properties of neurospheres infusion in experimental-induced seizures. Tissue Cell 54:47–54. [DOI] [PubMed] [Google Scholar]
- de Souza JS, et al. 2017. IGF1 neuronal response in the absence of MECP2 is dependent on TRalpha 3. Hum Mol Genet 26(2):270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, et al. 2008. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321(5893):1218–21. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Varvel NH, and Dudek FE 2014. When and how do seizures kill neurons, and is cell death relevant to epileptogenesis? Adv Exp Med Biol 813:109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravet C 2011. The core Dravet syndrome phenotype. Epilepsia 52 Suppl 2:3–9. [DOI] [PubMed] [Google Scholar]
- Ehrhart F, Sangani NB, and Curfs LMG 2018. Current developments in the genetics of Rett and Rett-like syndrome. Curr Opin Psychiatry 31(2):103–108. [DOI] [PubMed] [Google Scholar]
- Elitt MS, Barbar L, and Tesar PJ 2018. Drug screening for human genetic diseases using iPSC models. Hum Mol Genet 27(R2):R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J Jr., Bragin A, and Staba R 2018. Nonictal EEG biomarkers for diagnosis and treatment. Epilepsia Open 3(Suppl Suppl 2):120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink JJ, et al. 2017. Disrupted neuronal maturation in Angelman syndrome-derived induced pluripotent stem cells. Nat Commun 8:15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasier CR, et al. 2018. Channelopathy as a SUDEP Biomarker in Dravet Syndrome Patient-Derived Cardiac Myocytes. Stem Cell Reports 11(3):626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura S, et al. 2018. Intravenous infusion of mesenchymal stem cells reduces epileptogenesis in a rat model of status epilepticus. Epilepsy Res 141:56–63. [DOI] [PubMed] [Google Scholar]
- Gauvain G, et al. 2011. The neuronal K-Cl cotransporter KCC2 influences postsynaptic AMPA receptor content and lateral diffusion in dendritic spines. Proc Natl Acad Sci U S A 108(37):15474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, et al. 2017. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72associated amyotrophic lateral sclerosis. Sci Transl Med 9(383). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaze DG, et al. 2010. Epilepsy and the natural history of Rett syndrome. Neurology 74(11):909–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers WR 1880. The Gulstonian Lectures on Epilepsy. Br Med J 1(1006):547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad M, et al. 2015. Transplantation of GABAergic Interneurons into the Neonatal Primary Visual Cortex Reduces Absence Seizures in Stargazer Mice. Cereb Cortex 25(9):2970–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handreck A, et al. 2014. Anticonvulsant effects by bilateral and unilateral transplantation of GABA-producing cells into the subthalamic nucleus in an acute seizure model. Cell Transplant 23(1):11132. [DOI] [PubMed] [Google Scholar]
- Hattiangady B, Rao MS, and Shetty AK 2008. Grafting of striatal precursor cells into hippocampus shortly after status epilepticus restrains chronic temporal lobe epilepsy. Exp Neurol 212(2):468–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde M, and Lowenstein DH 2014. The search for circulating epilepsy biomarkers. Biomark Med 8(3):413–27. [DOI] [PubMed] [Google Scholar]
- Henderson KW, et al. 2014. Long-term seizure suppression and optogenetic analyses of synaptic connectivity in epileptic mice with hippocampal grafts of GABAergic interneurons. J Neurosci 34(40):13492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi N, et al. 2013. A human Dravet syndrome model from patient induced pluripotent stem cells. Mol Brain 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, and Jaenisch R 2016. Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell 18(5):573–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt RF, et al. 2013. GABA progenitors grafted into the adult epileptic brain control seizures and abnormal behavior. Nat Neurosci 16(6):692–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, et al. 2017. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci Transl Med 9(391). [DOI] [PubMed] [Google Scholar]
- Jahanbazi Jahan-Abad A, et al. 2018. Human Neural Stem/Progenitor Cells Derived From Epileptic Human Brain in a Self-Assembling Peptide Nanoscaffold Improve Traumatic Brain Injury in Rats. Mol Neurobiol 55(12):9122–9138. [DOI] [PubMed] [Google Scholar]
- Jiao J, et al. 2013. Modeling Dravet syndrome using induced pluripotent stem cells (iPSCs) and directly converted neurons. Hum Mol Genet 22(21):4241–52. [DOI] [PubMed] [Google Scholar]
- Kaufmann M, et al. 2015. High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. J Biomol Screen 20(9):1101–11. [DOI] [PubMed] [Google Scholar]
- Kim HW, et al. 2018. Differential effects on sodium current impairments by distinct SCN1A mutations in GABAergic neurons derived from Dravet syndrome patients. Brain Dev 40(4):287–298. [DOI] [PubMed] [Google Scholar]
- Kim KY, Hysolli E, and Park IH 2011. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci U S A 108(34):14169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura J, et al. 2018. Nobiletin Reduces Intracellular and Extracellular beta-Amyloid in iPS Cell-Derived Alzheimer’s Disease Model Neurons. Biol Pharm Bull 41(4):451–457. [DOI] [PubMed] [Google Scholar]
- Kondo T, et al. 2017. iPSC-Based Compound Screening and In Vitro Trials Identify a Synergistic Anti-amyloid beta Combination for Alzheimer’s Disease. Cell Rep 21(8):2304–2312. [DOI] [PubMed] [Google Scholar]
- Kouroupi G, et al. 2017. Defective synaptic connectivity and axonal neuropathology in a human iPSC-based model of familial Parkinson’s disease. Proc Natl Acad Sci U S A 114(18):E3679–E3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari D, et al. 2015. High-Throughput Screening to Identify Compounds That Increase Fragile X Mental Retardation Protein Expression in Neural Stem Cells Differentiated From Fragile X Syndrome Patient-Derived Induced Pluripotent Stem Cells. Stem Cells Transl Med 4(7):800–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, et al. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501(7467):373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2007. KCC2 interacts with the dendritic cytoskeleton to promote spine development. Neuron 56(6):1019–33. [DOI] [PubMed] [Google Scholar]
- Li L, Chao J, and Shi Y 2018. Modeling neurological diseases using iPSC-derived neural cells : iPSC modeling of neurological diseases. Cell Tissue Res 371(1):143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. 2016. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry 6:e703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. 2013a. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat Protoc 8(9):1670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. 2013b. Dravet syndrome patient-derived neurons suggest a novel epilepsy mechanism. Ann Neurol 74(1):128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher W 2017. Animal Models of Seizures and Epilepsy: Past, Present, and Future Role for the Discovery of Antiseizure Drugs. Neurochem Res 42(7):1873–1888. [DOI] [PubMed] [Google Scholar]
- Loscher W, et al. 1998. Seizure suppression in kindling epilepsy by grafts of fetal GABAergic neurons in rat substantia nigra. J Neurosci Res 51(2):196–209. [DOI] [PubMed] [Google Scholar]
- Maisano X, et al. 2012. Differentiation and functional incorporation of embryonic stem cell-derived GABAergic interneurons in the dentate gyrus of mice with temporal lobe epilepsy. J Neurosci 32(1):46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai M, et al. 2017. Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N Engl J Med 376(11):1038–1046. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, et al. 2010. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143(4):527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, et al. 2015. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162(2):375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini C, et al. 2011. The genetics of Dravet syndrome. Epilepsia 52 Suppl 2:24–9. [DOI] [PubMed] [Google Scholar]
- Marrone L, et al. 2018. Isogenic FUS-eGFP iPSC Reporter Lines Enable Quantification of FUS Stress Granule Pathology that Is Rescued by Drugs Inducing Autophagy. Stem Cell Reports 10(2):375389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, et al. 2018. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol Psychiatry 23(4):1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, et al. 2015. The SCN1A mutation database: updating information and analysis of the relationships among genotype, functional alteration, and phenotype. Hum Mutat 36(6):573–80. [DOI] [PubMed] [Google Scholar]
- Mittal S, et al. 2017. beta2-Adrenoreceptor is a regulator of the alpha-synuclein gene driving risk of Parkinson’s disease. Science 357(6354):891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noebels J 2015. Pathway-driven discovery of epilepsy genes. Nat Neurosci 18(3):344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi M, et al. 2018. Loss of MECP2 Leads to Activation of P53 and Neuronal Senescence. Stem Cell Reports 10(5):1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Operto FF, et al. 2019. Epilepsy and genetic in Rett syndrome: A review. Brain Behav:e01250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn TM, Beagan J, and Isacson O 2018. Increased motor neuron resilience by small molecule compounds that regulate IGF-II expression. Neurobiol Dis 110:218–230. [DOI] [PubMed] [Google Scholar]
- Pasca AM, et al. 2015. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12(7):671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca SP, et al. 2011. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med 17(12):1657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelc K, et al. 2008. Epilepsy in Angelman syndrome. Seizure 17(3):211–7. [DOI] [PubMed] [Google Scholar]
- Puskarjov M, et al. 2014. A variant of KCC2 from patients with febrile seizures impairs neuronal Cl- extrusion and dendritic spine formation. EMBO Rep 15(6):723–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi S, et al. 2012. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol 14(9):911–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KL, et al. 2018. Selective NaV1.1 activation rescues Dravet syndrome mice from seizures and premature death. Proc Natl Acad Sci U S A 115(34):E8077–E8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeder C, Heigwer F, and Boutros M 2018. Machine learning and image-based profiling in drug discovery. Curr Opin Syst Biol 10:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, et al. 2019. Clinical-grade stem cell-derived retinal pigment epithelium patch rescues retinal degeneration in rodents and pigs. Sci Transl Med 11(475). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorvon SD 2009. Drug treatment of epilepsy in the century of the ILAE: the first 50 years, 1909–1958. Epilepsia 50 Suppl 3:69–92. [DOI] [PubMed] [Google Scholar]
- Simone R, et al. 2018. G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol Med 10(1):22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staerk J, et al. 2010. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell 7(1):20–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, et al. 2016. A deleterious Nav1.1 mutation selectively impairs telencephalic inhibitory neurons derived from Dravet Syndrome patients. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, et al. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5):861–72. [DOI] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–76. [DOI] [PubMed] [Google Scholar]
- Tang X, et al. 2019. Pharmacological enhancement of KCC2 gene expression exerts therapeutic effects on human Rett syndrome neurons and Mecp2 mutant mice. Sci Transl Med 11(503). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, et al. 2016. KCC2 rescues functional deficits in human neurons derived from patients with Rett syndrome. Proc Natl Acad Sci U S A 113(3):751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RH, and Berkovic SF 2014. The hidden genetics of epilepsy-a clinically important new paradigm. Nat Rev Neurol 10(5):283–92. [DOI] [PubMed] [Google Scholar]
- Upadhya D, et al. 2019. Human induced pluripotent stem cell-derived MGE cell grafting after status epilepticus attenuates chronic epilepsy and comorbidities via synaptic integration. Proc Natl Acad Sci U S A 116(1):287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, et al. 2010. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell 6(5):407–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TH, and Hoffman AR 1997. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet 17(1):12–3. [DOI] [PubMed] [Google Scholar]
- Wang C, et al. 2018. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat Med 24(5):647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. 2017. Epilepsy-associated genes. Seizure 44:11–20. [DOI] [PubMed] [Google Scholar]
- Xiang Y, et al. 2019. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 24(3):487–497 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, et al. 2013. Generating a non-integrating human induced pluripotent stem cell bank from urine-derived cells. PLoS One 8(8):e70573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, et al. 2010. iPS cells reprogrammed from human mesenchymal-like stem/progenitor cells of dental tissue origin. Stem Cells Dev 19(4):469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawno T, et al. 2017. Ganaxolone: A New Treatment for Neonatal Seizures. Front Cell Neurosci 11:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JE, et al. 2018. Stabilizing the Retromer Complex in a Human Stem Cell Model of Alzheimer’s Disease Reduces TAU Phosphorylation Independently of Amyloid Precursor Protein. Stem Cell Reports 10(3):1046–1058. 318(5858):1917–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318(5858):1917–20. [DOI] [PubMed] [Google Scholar]