Abstract
Loss of function of senataxin (SETX), a bona-fide RNA/DNA helicase, is associated with neuronal degeneration leading to Ataxia and Ocular Apraxia (AOA) in human patients. SETX is proposed to promote transcription termination, DNA replication, DNA repair, and to unwind deleterious RNA:DNA hybrids in the genome. In all the above-mentioned mechanisms, SETX unwinds transcription complex-associated nascent RNA which is then degraded by the RNA exosome complex. Here we have used B cells isolated from a SETX mutant mouse model and compared genomic instability and immunoglobulin heavy chain locus (IgH) class switch recombination (CSR) to evaluate aberrant and programmed genomic rearrangements, respectively. Similar to RNA exosome mutant primary B cells, SETX mutant primary B cells display genomic instability but a modest decrease in efficiency of CSR. Furthermore, knockdown of Setx mRNAs from CH12–F3 B-cell lines leads to a defect in IgA CSR and accumulation of aberrant patterns of mutations in IgH switch sequences. Given that SETX mutant mice do not recapitulate the AOA neurodegenerative phenotype, it is possible that some aspects of SETX biology are rescued by redundant helicases in mice. Overall, our study provides new insights into the role of the SETX/RNA exosome axis in suppressing genomic instability so that programmed DNA breaks are properly orchestrated.
Keywords: Biological sciences, Immunology, Genetics, Biochemistry, Molecular biology, RNA exosome, Senataxin, DNA/RNA hybrids, Class switch recombination
Biological sciences; Immunology; Genetics; Biochemistry; Molecular biology; RNA exosome, Senataxin, DNA/RNA hybrids, Class switch recombination
1. Introduction
Immunoglobulin (Ig) gene diversification plays an essential role in adaptive immunity. Faced with a continuous yet varied stream of self, non-self, and possibly harmful molecules, many organisms have mechanisms that have evolved to match the diversity of the encountered antigens. In humans and mice, pro- and pre-B and -T lymphocytes go through a first round of genomic alteration — V(D)J recombination — in the bone marrow and the thymus, respectively (Schatz and Swanson, 2011). B cells can subsequently undergo two additional Ig gene diversification processes in secondary lymphoid tissues. Through somatic hypermutation (SHM), Ig variable regions of stimulated germinal center (GC)-forming B cells are mutated and further diversified, enabling affinity maturation. During class-switch recombination (CSR) GC B cells recombine Ig constant regions, swapping the IgM-encoding locus for a downstream locus to allow for different effector functions. Both B cell-specific genomic alterations are initiated when the single-stranded DNA (ssDNA) mutator enzyme activation-induced cytidine deaminase (AID) (Chaudhuri et al., 2007; Muramatsu et al., 2000; Petersen-Mahrt et al., 2002) catalyzes the removal of the amino group from deoxycytidine residues, resulting in deoxyuridines and dU:dG mismatches. Low-fidelity cellular responses to the presence of dU, including the mismatch repair (MMR) and the base-excision repair (BER) pathways, then are thought to introduce mutations in SHM and CSR, as well as cause double-strand breaks (DSBs) repaired through non-homologous end-joining (NHEJ) and alternative end-joining (A-EJ) in CSR (Chaudhuri and Alt, 2004; Gearhart and Wood, 2001; Goodman et al., 2007).
Though necessary for proper physiological function, these lymphocytic diversification processes are rife with danger for B cells; strong selective pressure exists to orchestrate and target them carefully so as not to threaten the genomic integrity of the cells through breaks or other mutations at non-Ig loci (Casellas et al., 2016; Liu et al., 2008; Liu and Schatz, 2009). Yet these deleterious events can nevertheless occur, as demonstrated by the involvement of AID in the mutagenic events at Ig and non-Ig loci that result in oncogenic translocations. The mechanisms underlying AID mutagenic activity targeting to physiological deamination substrates accordingly have been the focus of several studies (Lim et al., 2017; Liu et al., 2008; Meng et al., 2014; Pefanis et al., 2014, 2015; Qian et al., 2014).
It has been reported that the elongation-competent RNA polymerase II complex, when transcriptionally stalled at the IgH switch sequence (IgS), recruits AID using the RNA processing activity of the 3′-5′ noncoding RNA exonuclease RNA exosome complex (Basu et al., 2011; Pavri et al., 2010; Pavri and Nussenzweig, 2011; Rajagopal et al., 2009; Wang et al., 2009). AID and RNA exosome directly interact with each other, providing evidence for ncRNA-processing-mediated targeting of AID to its physiological DNA substrates (Basu et al., 2011; Chandra et al., 2015; Laffleur et al., 2017). Another observation is the role of RNA exosome in providing AID access to the template DNA of the IgS (switch) regions. Based on published literature, it is likely that RNA exosome and RNaseH1 combine to strip away ncRNAs associated with the template DNA to provide single-strand DNA substrates to AID (Basu et al., 2011; Maul et al., 2017). The G-rich nature of the non-template strand is posited to help stabilize R-loop and G-quadruplex DNA structures, allowing the ssDNA mutator AID to use the exposed, non-template strand as a substrate. AID must then access the template strand. In this context, it has been shown previously that RNA exosome utilizes the function of the RNA helicase MTR4 to unwind and degrade nascent germline transcripts associated with the template strand of Sμ and Sx DNA (where “x” is any of the switch regions downstream of Sμ.). This generates a ssDNA template strand for AID to hypermutate and allows for the creation of DNA double-strand breaks at Sμ regions (Lim et al., 2017). While both RNA exosome are important for overall genomic integrity of B cells, and although MTR4 is the dominant RNA helicase that functions with RNA exosome in all the processes mentioned above, the RNA/DNA helicase SETX may provide some support. In MTR4 mutant and MTR4/SETX double mutant B-cell lines CSR is reduced, leading to the conclusion that in the absence of proper unwinding of RNA:DNA hybrids in switch sequences, coordinated DNA double-strand breaks are compromised, eventually leading to weak CSR.
SEN1 was identified as a tRNA splicing endonuclease-encoding gene in the yeast Saccharomyces cerevisiae (Winey and Culbertson, 1988). Mutations in its human ortholog senataxin (SETX) were first described in patients with ataxia-ocular apraxia 2 (AOA2) (Moreira et al., 2004). SETX mutations are also associated with amyotrophic lateral sclerosis 4 (ALS4), an autosomal dominant motor neuron disease (Chen et al., 2004). The function of SETX (or Sen1p) as an RNA/DNA helicase implicated in resolving R loops in mammalian (or yeast) cells has also been reported (Kim et al., 1999; Martin-Tumasz and Brow, 2015; Mischo et al., 2011; Chan et al., 2014; Skourti-Stathaki et al., 2011).
Given that the SETX mutation in this study was generated through a CRISPR/Cas9 knockout approach (Lim et al., 2017) and the decrease in CSR was mild in comparison to the MTR4 mutation, concerns exist that in a SETX knockout model system the effects on CSR could be due to off-targeting caused by the Cas9 enzyme. Here, we report that the knockdown of SETX with an shRNA-based approach likewise leads to reduced CSR. Although SETX expression is not completely down in SETX knockdown CH12–F3 B cells, the effect on CSR seems to be stronger than that seen in SETX knockout cells (both in CRISPR/Cas9 mutated CH12–F3 B cells and in a SETX mutant mouse (Becherel et al., 2013)), indicating that constitutive inactivation of SETX may promote other RNA helicases to function in the IgH locus. Finally, we formally demonstrate that deficiency in both SETX and RNA exosome leads to an increase in genomic instability in B cells. Our data suggest that chromosomal alterations accumulate in activated SETX mutant primary B cells, reminiscent of the accumulation seen in activated EXOSC3 mutant primary B cells. These results highlight the function of SETX in suppressing levels of DNA double-strand breaks in B cells either through preventing their occurrence or through promoting their repair, presumably through an RNA:DNA hybrid resolution-associated mechanism. Taken together, our results provide new insights regarding the role of the SETX/RNA exosome axis in programmed and aberrant DNA alterations in developing B cells and clarify some points regarding current SETX model systems that are utilized to study B cells.
2. Materials and methods
2.1. Splenic B-cell isolation
Animals 6–8 weeks in age were euthanized by CO2 inhalation as recommended by the 2000 Report of the American Veterinary Medical Association Panel on Euthanasia and approved by our institution's IACUC (Approval number AC-AAAF5450). In addition, cervical dislocation was used as a secondary physical method of euthanasia. Spleens were then dissected out, and the tissue was dissociated using a syringe plunger as mortar and a cell strainer as pestle, and B lymphocytes were isolated by negative selection using CD43 beads (Miltenyi Biotec).
2.2. Cell culture conditions
All cells were incubated at 37 °C in a 5% CO2 humidified incubator. Primary B cells were grown in RPMI 1640 medium supplemented with 15% Fetal Bovine Serum (FBS) and 1X Non-Essential Amino Acids, 1mM Sodium Pyruvate, 10mM (HEPES), Penicillin-Streptomycin (100 U/ml Penicillin and 100 μg/ml Streptomycin), 55 μM β-mercaptoethanol. CH12–F3 and Ramos cells were grown in similar medium with the addition of 2mM L-glutamine. HEK293T cells were cultured in DMEM supplemented with 1X Non-Essential Amino Acids, 1mM Sodium Pyruvate, 10mM HEPES, Penicillin-Streptomycin (100 U/ml Penicillin and 100 μg/ml Streptomycin), 55 μM β-mercaptoethanol. ES cells were grown in DMEM supplemented with 15% Fetal Bovine Serum (FBS), 4mM L-glutamine, 1X Non-Essential Amino Acids, 1mM Sodium Pyruvate, Penicillin-Streptomycin (100 U/ml Penicillin and 100 μg/ml Streptomycin), 110 μM β-mercaptoethanol, and 120 μL ESGRO (Leukemia Inhibitory Factor, LIF).
2.3. SETX-knockdown shRNAs and real-time quantitative PCR
An shRNA construct (MISSION®, Sigma) was introduced in the CH12–F3 cell line. The construct contains the following target sequence:
5′CCGGGCGGTTGATGAACTTATGAAACTCGAGTTTCATAAGTTCATCAACCGCTTTTTG3′
A randomized vector control was employed in the control CH12–F3 cells.
The polyethylenimine (PEI) method was used for transfection. The shRNA construct DNA was mixed with the intended DNA/packaging vectors (psPAX2 and pMD2.G) at a ratio of 4:3:1 (shRNA:psPAX2:pMD2.G) and PEI was added so as to have a DNA:PEI ratio of 3:4. Reduced serum medium Opti-MEM was added. The DNA-PEI-Optimem mixture was then added dropwise to HEK293T cells (passaged the night before so as to be ~75% confluent on the morning of the transfection and plated in 4 ml medium). The medium was changed once after 4–6 h and again after an overnight incubation. The virus-containing supernatant was collected 48 h later. In a 1:1 ratio, the virus-containing supernatant was added to CH12F3 cells plated at 2 × 105 cells/ml and containing a final polybrene concentration of 5–10 μg/ml. The cells were then centrifuged for 90 min at 1230xg at 30 °C. Subsequently 75% of the supernatant was removed and replaced with appropriate growth medium. After 48 h, the cells were exposed to the selection agent (puromycin at 0.5 μg/μl).
2.4. Class-switch recombination assay
For primary B cells, CSR was initiated by adding 20 μg/ml LPS (Sigma) and 20 ng/ml IL-4 (R&D Systems) to IgG1, 20 μg/ml LPS (Sigma) to IgG3, and 10 μg/ml LPS (Sigma), 2 ng/ml TGF-beta (R&D Systems) and 0.33 μg/ml anti-IgD dextran (Fina Biosolutions) to IgG2b for 72 h (or 48 h if the inversion-deletion reaction was taking place in the presence of LPS for 24 h). For CH12–F3 cells, CSR to IgA was initiated by adding 20 μg/ml LPS, 20 ng/ml IL-4, and 1 ng/ml TGF-beta. After culture, cells were placed in 2.5% FBS in 1X PBS, stained with the appropriate conjugated antibodies (FITC-conjugated anti-IgA (BD Bioscience), IgG1 (BD Bioscience), IgG3 (BD Bioscience), CD19 (Biolegend), B220 (Bioscience), and PE-conjugated anti-IgG1 (BD Bioscience) and IgG2b (Biolegend)). Data were acquired on a FACSAria cell-sorter and Accuri (BD Biosciences) and FlowJo software was used for analysis.
2.5. EXOSC3 allelic deletion
To allow for the inversion/deletion RNA exosome abrogation reaction to occur ROSACreERT2/+; EXOSC3COIN/COIN B lymphocytes and ES cells and their respective controls were exposed to 100 nM 4-hydroxytamoxifen (4-OHT, Sigma) in the appropriate cell culture conditions for 24 h, after which the 4-OHT was washed away, and CSR medium was added for B cells, or regular tissue culture growth medium was added for ES cells. B cells must receive a proliferation signal lest they die; therefore during the inversion-deletion reaction, LPS is added to the growth medium at 20 μg/mL.
2.6. Immunoprecipitation and immunoblotting assay
Cells are lysed by douncing in buffer A (20 mM Tris, pH 7.5, 1 mM DTT, 10 mM MgCl2) plus 10mM NaCl, 10 % glycerol and protease inhibitors, and an equal volume of buffer A plus 1 M NaCl,10 % glycerol and protease inhibitors was added. Nuclei are isolated by centrifugaion after douncing in buffer A plus 10 mM NaCl, 10 % glycerol and protease inhibitors, and lysed with buffer A plus 500 mM NaCl, 10% glycerol and protease inhibitors. The extract was dialyzed against buffer A plus 100 mM NaCl after removing debris by centrifugation. Lysates were pre-cleared with IgG-conjugated Pierce protein A/G magnetic beads (Life Technologies) for at least 2 h and then incubated with anti-human SETX antibody (Santa Cruz Biotechnology) plus protein A/G agarose beads for more than 6 h, followed by several washes with cell lysis buffer plus 10% glycerol. The immunoprecipitated beads were boiled in protein loading buffer for SDS-PAGE. Following gel electrophoresis and transfer to a PVDF membrane, immunoblotting was performed with the appropriate antibodies. Anti-RRP45 (Novusbio), anti-human SETX (Santa Cruz Biotechnology) were employed.
2.7. Electroporation of cells
An Amaxa Nucleofactor II electroporator device from Lonza was employed, following the manufacturer's instructions. Depending on the plasmid size, 2–4 x 106 cells to be electroporated were suspended in Lonza's electroporation buffer solution (100 μL) containing the desired plasmid and placed in a cuvette. After electroporation, 500 μl of 37˚C-medium were added to the cuvette, and the mixture was then added to 1 mL of growth medium pre-warmed to 37 °C.
2.8. Immunofluorescence assay
Cells were plated on glass coverslips pre-treated with poly-L-lysine for 1 h at room temperature and allowed to incubate for 60 min at room temperature or at 37 °C. The cells were then fixed using 3–4% paraformaldehyde (PFA) at room temperature for 20 min. To quench autofluorescence, the coverslips were placed in 50 mM NH4Cl. They were then permeabilized with 0.1 % Triton X-100 for no longer than 1 min, before undergoing blocking (with 0.25% fish skin gelatin and 0.01% saponin in PBS) for 30 min. The appropriate primary antibody was then used (anti-γH2AX or anti-H2AX (both from Abcam) with their appropriate fluorophore-tagged secondary antibodies). Images were captured with an EVOS digital inverted fluorescence microscope (Invitrogen), and analyzed with the ImageJ software from the United States National Institutes of Health.
2.9. Irradiation assay
Primary splenic B cells were harvested from mice and cultured for 72 h. Cell cultures were irradiated in a Gammacell 40 at 82 rad/min for a total of 0 Gy, 2 Gy (2.5 min), or 20 Gy (24.5 min). Cell cultures were removed from the Gammacell irradiator and immunofluorescence assay was performed.
2.10. RNA preparation and real-time quantitative PCR
Total RNA was isolated from cells using Trizol reagent (Life Technologies). RNA was resuspended in water and quantified using a Nanovue Plus spectrophotometer (GE Healthcare Life Sciences). RNA samples were treated with DNase I (Turbo DNA-free kit, Life Technologies), eluted in water, and re-quantified. 1.5 μg of RNA were then converted to cDNA using random hexamers and the Superscript III First-Strand Synthesis System for RT–PCR (Life Technologies). Two Setx primer pairs were employed for real-time quantitative PCR reactions.
Primers for the germline transcripts at switch sequences μ and α, Gapdh housekeeping control, and Aicda were as follows:
For μ: 5′-CTCTGGCCCTGCTTATTGTTG-3′
and 5′-GAAGACATTTGGGAAGGACTGACT-3′
For α: 5′-CCTGGCTGTTCCCCTATGAA-3′
and 5′-GAGCTGGTGGGAGTGTCAGTG-3′
For Gapdh: 5′-TGGCCTTCCGTGTTCCTAC-3′
and 5′-GAGTTGCTGTTGAAGTCGCA-3′
For Aicda: 5′-GGAACAGCAGAACTTCCAGACTTTG-3′
and 5′-CCTGAAAGTGAGCCTTAGAGGGAA-3′
Real-Time PCR was performed using the Light Cycler 480 II system by Roche Applied Science, using SYBR Green. Analysis was performed using the Δ-ΔCT method.
2.11. Statistical analysis
ANOVA and unpaired, two-tailed Student's t-tests were used when possible. The software program Prism (GraphPad Software) was employed for all statistical analysis calculations. Standard error of the mean was used whenever error bars are displayed.
2.12. Telomere fluorescence in-situ hybridization
Metaphase arrest was produced by cell exposure to colcemid at 200 ng/ml for 2–4 h. The cells were then centrifuged and resuspended in prewarmed hypotonic solution (75 mM KCl) and incubated at 37 °C for 15 min. Fixative solution (3:1 methanol:glacial acetic acid) was added drop-wise. The mixture was spun and the pellet resuspended in fixative solution, and incubated at 4 °C for 15 min. Centrifugation followed by resuspension and incubation was performed two additional times. Subsequently the pellet, resuspended in fixative solution, was dropped on glass slides that had been “hydrated” by being passed over water vapor emanating from a beaker containing water heated to 80–90 °C (with side on which the cells were to be dropped facing the vapor). Once the metaphases had been dropped using a Pasteur pipette and a bulb, the slide was passed over the water vapor again, on both sides, for 3–5 s at a time, then placed on a plate pre-heated to ~55–75 °C so as to dry the slide without burning the sample. The chromosomes can be fully visualized at this stage. The slides were left overnight, and the next day were washed with PBS, fixed in 4% formaldehyde/PBS, washed again in PBS, digested with acidified pepsin, washed and fixed again, and dehydrated with serial ethanol washes, before being allowed to air dry. The hybridization steps followed and consisted of adding the prepared probe to the slide and denaturing at 80 °C for 3 min. The probe mix contained 10 mM Tris at pH 7.2, buffer MgCl2, 70% deionized formamide, 0.25 % Boehringer Mannheim blocking regent, the hybridization probe (at 0.5 μg/ml) in water. After the denaturation step, the slide was allowed to incubate with the probe for 2 h at room temperature in a humified chamber, at the conclusion of which formamide washes were performed (70% formamide, 10mM Tris, 0.1% BSA in water), followed by PBS/Tween washes, serial dehydration ethanol washes, and air-drying. The slides were covered with Vectashield with DAPI mounting medium (Vector Laboratories), and sealed with nail polish, before being stored in the dark. Images were captured with an EVOS digital inverted fluorescence microscope, and analyzed with the ImageJ software from the United States National Institutes of Health. The telomere probes were cyanine PNA telomere probes purchased from Bio-synthesis (Lewisville, Texas) and were tagged with a Cy3 (green) or a Cy5 (red) fluorophore as follows: Cy3-OO-CCC TAA CCC TAA CCC TAA or Cy5-OO-CCC TAA CCC TAA CCC TAA.
2.13. VPD450 proliferation assay
The cells were washed with PBS prior to being labeled through the addition of the VPD450 dye from BD Bioscience to a final concentration of 1 μM and incubation in a 37 °C water bath for 10–15 min. The cells were then thoroughly washed and resuspended in appropriate growth medium and placed in a tissue culture incubator to be analyzed at downstream time points. As with the CSR assay, the FlowJo software program was used to process the flow cytometry data.
2.14. 5' Sμ sequencing
CH12–F3 cells were stimulated with 20 μg/ml LPS, 20 ng/ml IL-4, and 1 ng/ml TGF-beta for 48h, then genomic DNA was extracted and purified. The 5′ subregion of Sμ was amplified by PCR with the following primers: Forward: 5′-AATGGATACCTCAGTGGTTTTTAATG, Reverse: 5′-GCGGCCCGGCTCATTCCAGTTCATTA. The PCR program was as follows: 28 cycles of 94 °C (30 s), 58 °C (30 s), and 72 °C (40 s). Turbo Pfu was the Taq polymerase employed in the reaction. The PCR products were then incubated with GoTaq DNA polymerase at 72 °C for 10 min and purified with a gel extraction kit (Qiagen) after electrophoresis in an agarose gel. Purified PCR products were ligated with pGEM-T Easy vector (Promega) and electroporated into XL-1 blue competent cells. White colonies were selected for minipreps and sequenced with the T7 primer. Sequences were aligned with ClustalX.
3. Results
3.1. Senataxin (SETX) associates with RNA exosome
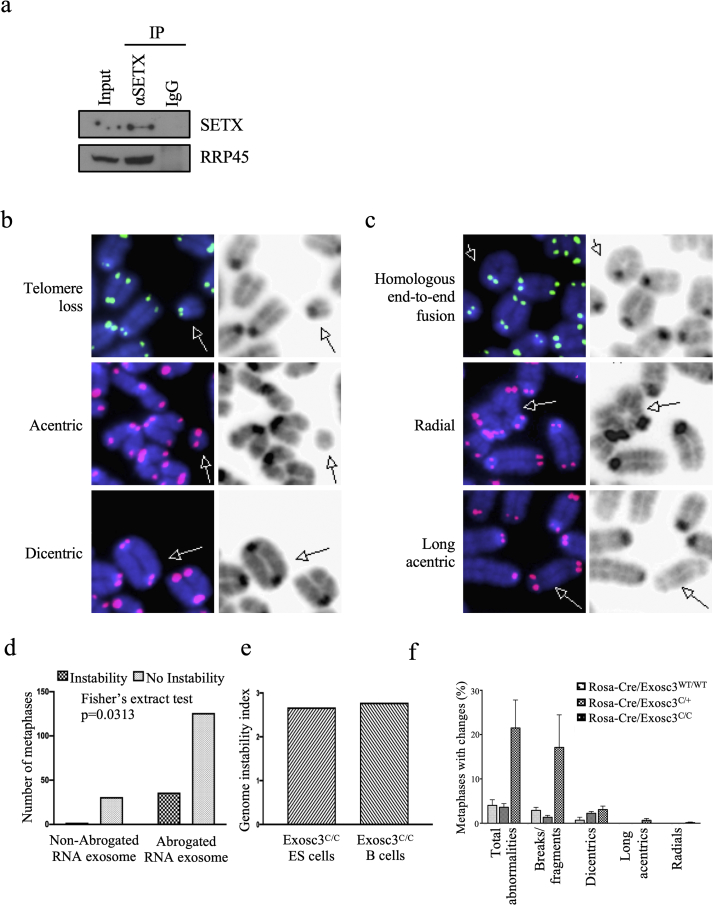
To investigate the possible involvement of SETX in antibody diversification mechanisms, we initially ensured that SETX interacts with an important component of the antibody diversification machinery pathway: RNA exosome. As observed previously, RNA exosome plays an important role in CSR (Basu et al., 2011; Pefanis et al., 2014). It has been shown that a complex formed by Sen1p (the yeast ortholog of SETX), Nrd1 and Nab3 (the NNS complex) physically interacts with the RNA exosome in Saccharomyces cerevisiae (Vasiljeva and Buratowski, 2006). More recently, a report using a yeast two-hybrid screen, a human brain cDNA library, and the amino terminal region of SETX provided evidence for an interaction between the RNA exosome subunit RRP45 and SETX (Richard et al., 2013). The RNA exosome is composed of a heterononameric core that can interact with catalytic cofactors to bring about its 3'→5' exo- or endoribonucleolytic activities. RRP45 is a core subunit of the RNA exosome complex. To observe an interaction between RRP45 and SETX in a B-cell background we employed protein co-immunoprecipitation assays using Ramos cells, a human B-cell line, as there are no efficient anti-murine SETX antibodies available. We observed that SETX interacts with RRP45 in Ramos cells, confirming reports utilizing other cell backgrounds (Figure 1a, Supplemental Fig. 1a). AID has been previously demonstrated to directly or indirectly interact with various proteins that are associated with the transcription complex, chromatin or DNA repair factories (Chaudhuri and Alt, 2004; Gearhart and Wood, 2001; Goodman et al., 2007). SETX could be a component of one of these protein complexes/pathways.
Figure 1.
Senataxin-interacting RNA exosome complex is important for maintenance of B-cell genomic integrity. (a) Human Burkitt's lymphoma Ramos cells were harvested, lysed, and prepared for SETX immunoprecipitation. Input (1/15 of lysate used for IP and normal mouse IgG1 controls are displayed, along with Western blotting using antibodies against RNA exosome core subunit RRP45. Full images are in the supplemental figure 1a. (b) Structural abnormalities in RNA exosome-deficient stimulated B cells. Examples are shown of telomeric telomere loss, acentric chromosomal fragments, and dicentric chromosomes. These abnormal chromosomal structures accrue at a higher degree in activated ROSA26Cre-ERT2/+; EXOSC3COIN/COIN B lymphocytes than in their RNA exosome-competent counterparts. Structures are shown with Cy3 (green) or Cy5 (red) fluorophore-tagged peptide nucleic acid (PNA) telomeric probes and 4′,6-diamidino-2-phenylindole (DAPI, blue) staining (on the left) and with transmitted light (on the right). Densities are indicative of centromeres. Chromosomal fragments without centromeres are acentric (middle row), while chromosomal structures with 2 centromeres are dicentric (bottom row). Fluorophore-labeled telomeric probes denote the presence or absence of telomeres (centromeric or telomeric telomeres; seen here is telomeric telomere loss in the top row). (c) Additional representations are shown for homologous end-to-end fusion, radial, and long acentric chromosomes. These abnormal chromosomal structures accrue at a higher degree in activated ROSA26Cre-ERT2/+; EXOSC3COIN/COIN B lymphocytes than in their RNA exosome-competent counterparts. Note the chromatid fusion and absence of telomeric telomeres in the top picture pair; the middle pair shows a quadriradial chromosome; the bottom pair shows a long acentric chromosome. Cognizant that in 2 mouse chromosomes lack of a centromere is expected, this chromosome is excessively long to be one of those. (d) Comparison of the percentage of fixed metaphases with chromosomal abnormalities before and after RNA exosome abrogation in embryonic stem (ES) cells. There is a statistically significant increase in the number of metaphases with genomic instability in the RNA exosome-abrogated background compared to the non-abrogated RNA exosome background (Fisher's exact test, p = 0.0313). (e) Genomic instability indices devised to compare the level of genomic instability observed when RNA exosome activity is impaired in both ESCs and B cells. Relative to their exosome-sufficient baselines, both ES and B cells have increased genomic instability upon abrogation of RNA exosome function. The genomic instability index [(D–S)/S] normalizes the change in instability seen in RNA exosome-deficient backgrounds (“D-S”) to baseline instability levels observed in RNA exosome-sufficient backgrounds (“S”) in our assay. (f) Breakdown of chromosomal instability events in activated B cells. A compilation of the different phenotypes is plotted for the three backgrounds. For all abnormalities, we observed a statistically significant difference between the RNA exosome-deficient B cells and the RNA exosome-competent backgrounds: ANOVA, p = 0.0409, R2 = 0.5989. 4 animals were used for the WT background, 2 for the heterozygous background, and 4 for the ROSA26Cre-ERT2/+; EXOSC3COIN/COIN background. The respective number of metaphases analyzed is indicated for each mouse background.
3.2. Genomic instability in RNA exosome subunit EXOSC3-deficient B cells
Given that RNA exosome and SETX appear to function together, it was necessary to determine if the RNA exosome has a role in preventing genomic instability as well as to discover whether the abrogation of RNA exosome function affects the genomic integrity of B cells lacking the complex. Numerous accounts have suggested that the accumulation of RNA:DNA species is detrimental to cellular health by generating genomic instability (Rondon and Aguilera, 2019; Skourti-Stathaki and Proudfoot, 2014). These RNA:DNA species may accumulate as a result of deficient RNA processing mechanisms that increase the relative amounts of RNA available for base-pairing with negatively supercoiled DNA, either through co-transcriptional defects that may result in increased RNA:DNA hybrids formed in cis, or through post-transcriptional defects that may result in increased RNA:DNA hybrids formed perhaps in trans at the overlapping locus or a different DNA locus sharing some homology. Given the implication in yeast of catalytic RNA exosome subunit Rrp6 in the genesis of genomic instability (Wahba et al., 2011) and the role of RNA exosome in maintenance of genomic stability in mouse embryonic stem (ES) cells (Domingo-Prim et al., 2019; Pefanis et al., 2015), we investigated the consequences of RNA exosome activity abrogation in conditional null mouse B cells using a telomeric fluorescence in situ hybridization (T-FISH) probe. A T-FISH assay permits the detection of general structural abnormalities in chromosomal integrity in the absence of the key RNA processing machinery, RNA exosome. We isolated splenic B lymphocytes from mice with different susceptibilities to the CreERT2-mediated inversion-deletion reaction (Pefanis et al., 2014), exposed the cells to 4-hydroxytamoxifen causing translocation of the Cre recombinase into the nucleus, induced the cells to undergo IgM-to-IgG1 class-switch recombination by stimulating them with a lipopolysaccharide (LPS)-interleukin 4 (IL-4) cocktail for 3 days, and provided them with colcemid (a less toxic relative of the microtubule-disruptive compound colchicine), arresting them in metaphase (Rieder and Palazzo, 1992). Cells were washed and fixed and the metaphase preparations were “dropped” on glass microscopy slides. A fluorophore-tagged telomeric probe was hybridized to the genomic material. When compared to RNA exosome-sufficient backgrounds, ex vivo CSR-activated RNA exosome-deficient B lymphocytes displayed a marked increase in the proportion of metaphases containing genomic instability. A number of changes were noted, including the loss of telomeres (both at centromeric and telomeric chromosomal ends), the presence of acentric fragments (which denotes the absence of a centromere in the DNA specimen), the fusion of two chromosomes into one dicentric chromosome, the end-to-end fusion of sister chromatids intra-chromosomally, and, rarely, the formation of so-called radials (Figure 1 b-c). Thought to occur at DNA polymerase stalling sites, radials are chromatid-type errors where a chromatid break is repaired by the insertion of another chromosome (which also bears a chromatid break in the case of quadriradials) (Savage, 1976; Scully et al., 2000). For both ES cells and B cells, more genomic instability catastrophic events occurred in the RNA exosome-deficient background than in the RNA exosome-sufficient background (Figure 1 d-e). In absence of RNA exosome activity, in ex vivo culture conditions ES cells and B cells survive well for a few cell cycles but eventually undergo cell death due to activation of the p53 pathway (Pefanis et al., 2015). Breaks and fragments, tallied from metaphases with telomeric loss (whether telomeric telomeres or centromeric telomeres) and from metaphases with acentric chromosomes, are mainly responsible for the difference observed in the proportion of metaphases with instability between RNA exosome-deficient and RNA exosome-sufficient backgrounds (Figure 1 d-f).
3.3. Genomic instability in IR-exposed stimulated RNA exosome-deficient B cells
The possibility exists that our assay restricted analysis only to those genomic instability instances resulting in chromosomal structural changes perceptible by T-FISH, namely, improperly repaired chromosomal breaks, dicentric or acentric chromosomes, and intra-chromosomal chromatid end-to-end fusion events. Were a break to occur and be successfully repaired, our assay could not recognize such an accidental occurrence; were a balanced, pathophysiological translocation event to take place, again our T-FISH assay would not be able to reveal that disease-predisposing rearrangement. We would be unable to detect any changes occurring more proximally to the presumably RNA exosome absence-related insult, which would have been repaired by the a priori intact cellular repair mechanism. Accordingly, the decision was made to increase the stress encountered by the B cells through exposure to ionizing radiation.
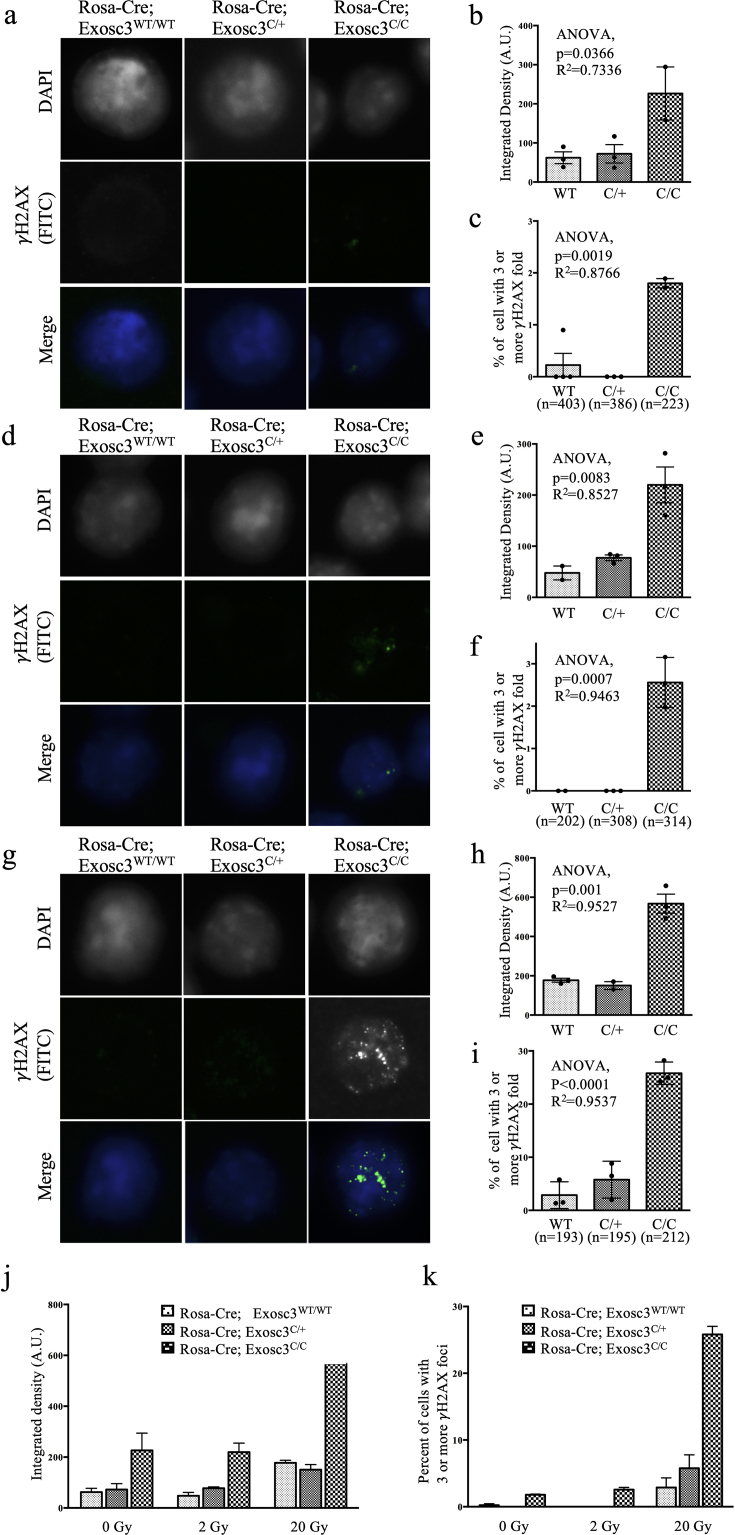
As performed previously, splenic B lymphocytes were isolated and expanded ex vivo in the presence of LPS and 4-OHT for the first 24 h to abrogate RNA exosome activity, followed by an LPS and IL-4 treatment for an additional 48 h to allow for CSR. The cells were then irradiated for 0 min, 2.5 min, or 24.5 min (corresponding to 0 Gy, 2 Gy, and 20 Gy.). Multiple attempts to perform T-FISH assays on these cells yielded no usable metaphase spreads, perhaps because the exposure to colcemid in addition to irradiation proved excessively toxic. However, we were able to perform immunofluorescence (IF) assays on these cells using antibodies recognizing proteins involved in the repair of double-stranded DNA breaks. The irradiated cells were allowed to recover for 45 min, then washed, fixed, and prepared for IF. 45 min was selected as longer recovery periods (5.5 h and 10.5 h) rendered DSB repair proteins undetectable by IF, and shorter recovery periods showed considerable variability. IF was performed using antibodies against γH2AX, the phosphorylated form of the histone variant protein H2A. It is thought that upon sensing of double-stranded DNA breaks by the complex formed by MRE11 (meiotic recombination homolog 11), RAD50 (radiation sensitive mutant 50), and NBS1 (Nijmegen breakage syndrome 1), phosphatidylinositol-3-kinase-like family of protein kinases (which include DNA-dependent protein kinase catalytic subunit (DNA-PKcs), ataxia telangiectasia mutated (ATM), and ATM- and RAD3-related (ATR)) will be activated and in turn will phosphorylate H2AX at its serine 139 residue to produce γH2AX, which will induce the recruitment of additional DNA repair proteins (Kinner et al., 2008). The aggregation of these phosphorylated γH2AX proteins into foci can therefore be used as a biomarker proxy for the presence of a double-stranded DNA break. At 0 Gy stimulated B cells, RNA exosome-sufficient and -deficient alike, have minimal amounts of γH2AX (Figure 2 a-c). The percentage of stimulated, irradiated, fixed cells that accumulate 3 or more γH2AX foci was evaluated. At baseline levels of 0 Gy (Figure 2 a-c) and 2 Gy (Figure 2 d-f) very few to no RNA exosome-competent B cells accumulate these foci, but some B cells with compromised RNA exosome exhibit a statistically significant increase (albeit still low level). Apparently, the increase in IR stress from 0 Gy to 2 Gy did not translate into a higher preponderance of DSBs in our assay as measured by γH2AX focus accumulation. Accordingly, IR stress was increased by exposing the stimulated B cells to 20 Gy, leading to a tenfold change in focus formation in B cells with intact RNA exosome. Overall, at 20 Gy, γH2AX detection levels increased in all three backgrounds compared to the previous IR exposures, with the RNA exosome-sufficient cells seeing an approximate twofold increase in γH2AX and RNA exosome-deficient cells exhibiting approximately triple the levels of γH2AX foci staining (Figure 2 g-i). Analyzing the subsets of cells that had 3 or more foci revealed that the increase in the RNA exosome-deficient cells was drastically larger, at about tenfold (Figure 2 j-k). A small but non-negligible portion of RNA exosome-sufficient cells also displayed an accumulation of γH2AX foci, though much less than the percentage seen in the deficient background. The differences observed at 20 Gy were statistically significant (Figure 2 h-i). To ensure that the changes in γH2AX were not due to higher levels of non-phosphorylated H2AX protein in the cells, the same IF assay was performed on the various RNA exosome backgrounds using an anti-H2AX antibody after irradiating the cells at 20 Gy (Supplemental Fig. 1b). No statistically significant difference among the three RNA exosome backgrounds was observed, indicating that the changes reported in γH2AX likely were not due to differences in H2AX protein levels. Taken together, these results indicate that RNA exosome deficiency causes genomic instability in activated B cells.
Figure 2.
Accumulation of genomic instability in RNA exosome mutant B cells following IR radiation treatment. γH2AX immunofluorescence assay of (a–c) 0-Gy (d–f) 2-Gy and (g–i) 20-Gy irradiated stimulated B cells of different backgrounds. (a) Low levels of γH2AX are detected by (b) integrated density, though a small, significant difference is apparent among RNA exosome-non-deficient and RNA exosome-deficient backgrounds by ANOVA. (c) Cells with 3 or more γH2AX foci as detected in our assay. RNA exosome-deficient cells show a statistically significant increase in focus formation. Two tailed Student's t-test performed to calculate significance. Similar analyses as shown in (a–c) for 2-Gy IR (d-f) and 20-Gy (g-i) IR exposure. (j-k) Compilation of immunofluorescence/irradiation data. The different RNA exosome backgrounds are plotted for γH2AX accumulation after ionizing radiation at different levels. (j) The plot represents the detected γH2AX intensity in integrated density arbitrary units. (k) The plot shows the percentage of analyzed fixed cells with 3 or more γH2AX foci. Irrespective of the amount of IR, RNA exosome-deficient backgrounds accumulate more double-stranded breaks as assayed by γH2AX IF; at 20 Gy, in conditions of high mutagenic stress, RNA exosome-deficient cells show a dramatic increase in γH2AX accumulation, suggesting they are more sensitive to increases in mutagenic stress.
3.4. SETX-deficient CH12–F3 cells exhibit impaired IgA CSR
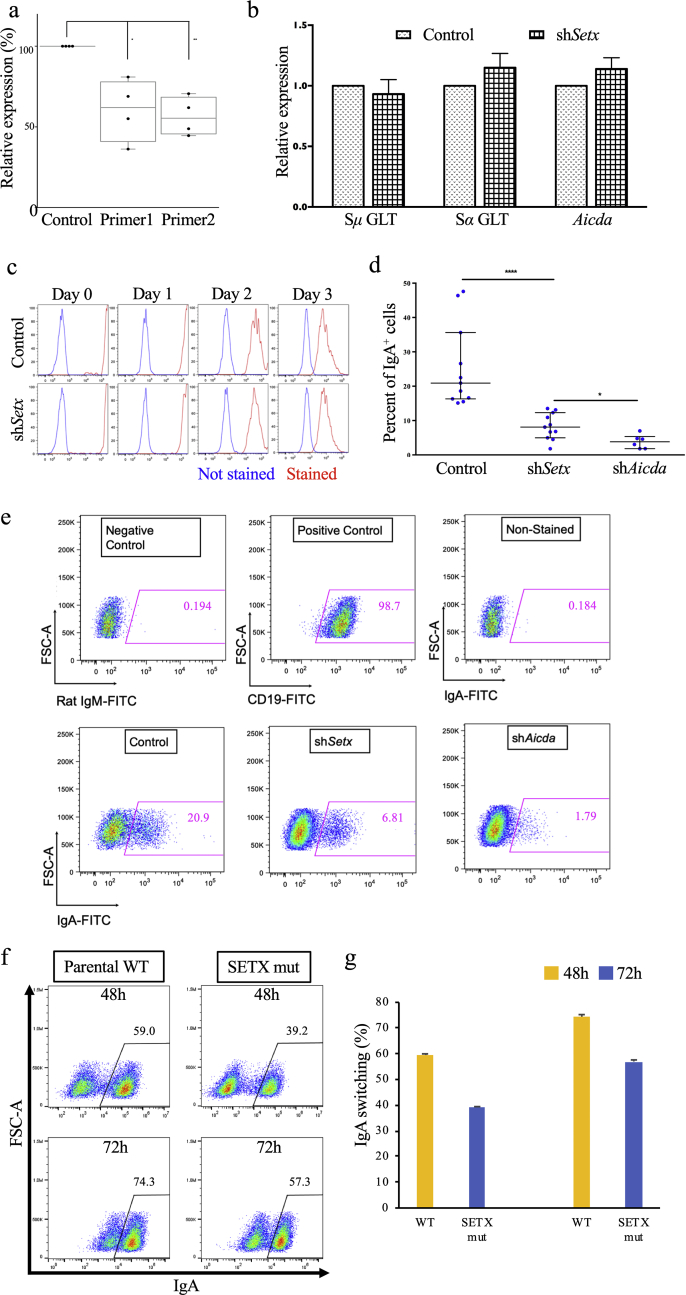
We next chose to determine if, in fact, SETX is important for the catalysis of programmed DNA rearrangements, such as CSR. The CH12–F3 cell line, a murine B-cell lymphoma line able to undergo isotype switching upon stimulation, was employed. Using shRNA, the expression of the Setx gene was knocked down (Figure 3a). Our knockdown approach, evaluated by RT-PCR using sets of primers that recognize two distinct regions of the gene, resulted in a statistically significant decrease in Setx mRNA of about 40 percentage points (Figure 3a). Transcription plays a crucial role for efficient, AID-initiated physiological CSR (Chaudhuri et al., 2003; Dickerson et al., 2003; Ramiro et al., 2003; Sohail et al., 2003). To evaluate whether germline transcription decreased as a result of knocking down SETX, μ and α switch sequence transcription was monitored in a SETX knockdown background and it was observed that germline transcription at switch sequences was not impaired (Figure 3b). AID is the critical mutagen in SHM and CSR (Muramatsu et al., 2000). Given its role in CSR, AID mRNA levels in the SETX knockdown background were obtained and no decrease in AID mRNA levels was observed (Figure 3b). Furthermore, Violet Proliferation Dye 450 (VPD 450) staining was employed to determine the proliferation profile of the knockdown background but no proliferative alteration between the knockdown and control backgrounds was noted (Figure 3c).
Figure 3.
SETX is important for robust CSR in CH12–F3 B cell lines. (a) Setx transcription levels in the knockdown background. Quantitative RT-PCR was performed on murine CH12–F3 cells containing a control vector or a SETX knockdown construct. Displayed here are results of an analysis using two primer sets. Results are shown relative to control vector levels. Student's t-test was used in either case, and primer pair 1 had p = 0.0061, while primer pair 2 had p = 0.0003. (b) Transcription at IgH switch sequences and Aicda in knockdown background. Sμ and Sα germ line transcripts (GLT) in a shSETX knockdown background are shown, as is the transcription at mouse Aicda, relative to control background. We observed no statistically appreciable defect in transcription at any of the three loci. (c) Proliferation of SETX knockdown CH12–F3 cells. Using the dilution dye VPD450, the proliferation of CH12–F3 cells containing control vector or the shSETX knockdown construct was tracked over 3 days. Unstained cells are shown in blue; red denotes stained cells. As the cells proliferate, the VPD450 intensity of stained cells is diluted and nears that of unstained cells. (d) Summary of multiple experiments for CSR efficiency of AID-deficient (shAID), SETX-deficient (shSETX) and vector control (control) backgrounds. CSR decrease in shSETX relative to control is statistically significant (unpaired Student's t-test, p = 0.0002), as is CSR decrease in shAID relative to shSETX (unpaired Student's t-test, p = 0.0182). (e) Representation of one experiment from (d). (f) FACS plots of IgA CSR following 48 h and 72 h of stimulation of parental CH12–F3 cells and SETX KO CH12–F3 cells, generated by the CRISPR-Cas9 genome editing approach. (g) The FACS plot shown in (f) is quantitated through three independent stimulations of the SETX knockout clones.
We stimulated the CH12–F3 cells to undergo isotype switching to IgA. Knocking down SETX in CH12–F3 cells resulted in a substantial reduction in their ability to undergo CSR, as measured by the proportion of SETX-deficient CH12–F3 cells expressing IgA on their surface (Figure 3 d-e). In this regard, CSR in SETX-deficient CH12–F3 cells greatly resembled CSR in AID-deficient CH12–F3 cells (Figure 3 d-e). Repeated experiments in the shSetx background corroborated these findings, and demonstrated that the decrease in CSR in SETX-deficient CH12–F3 cells is statistically significant (Figure 3d). Finally, we confirmed our previous observations that CRISPR/Cas9-mediated deletion of the Setx gene also leads to a clear but modest deficit in CSR in CH12–F3 cells. As can be seen in Figure 3f (plotted in Figure 3g), the kinetics of IgM-to-IgA CSR at 48h and 72h post-stimulation are delayed in SETX KO cells when compared to the parental WT CH12–F3 cells. Taken together, these data suggest that SETX deficiency by RNA interference results in a relatively stronger CSR phenotype than SETX deficiency by frame-shift DNA mutation.
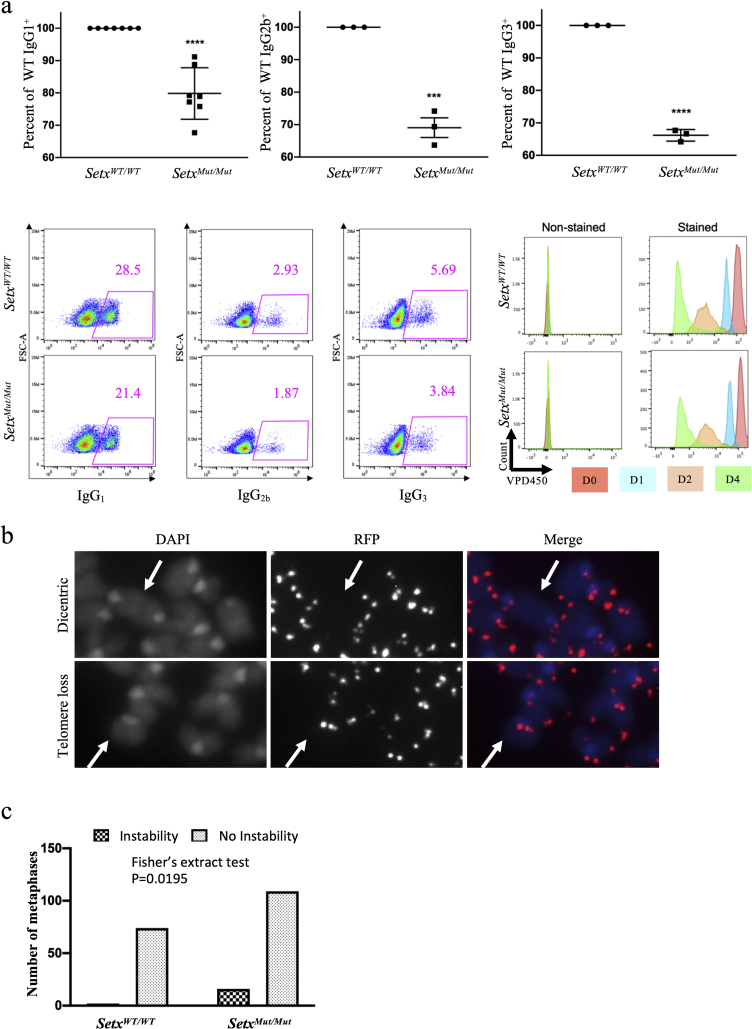
3.5. SETX mutant mouse model
We analyzed B cells from a SETX mutant mouse model that was generated previously (Becherel et al., 2013) for CSR. The Setx gene consists of 23 exons; the mutant mice have exon 4 of the Setx gene deleted. B cells obtained from Setx mutant allele mice (homozygous for the Setx Δexon 4 allele) exhibit relatively weaker CSR defects, when compared to SETX knockdown in CH12–F3 cells (Figure 4a, upper and lower left panels, Supplemental Fig. 2b). The SETX mutant B cells proliferated in a manner almost similar to wild-type B cells (Figure 4a, lower right panel). It is possible that in the absence of SETX, cells adapt to use other RNA helicases, namely MTR4, a component of the RNA exosome complex that unwinds RNA:RNA and RNA:DNA hybrids (Lim et al., 2017; Puno and Lima, 2018; Weick et al., 2018) to promote CSR. shRNA-mediated knockdown causes SETX deficiency at the RNA level and does not allow the cells to select alternative pathways/factors over a longer period of time. It is also possible that the SETX knockout mouse model or CRISPR/Cas9-edited SETX mutant cell lines express an alternatively spliced form of SETX, which is difficult to verify in the absence of a proper anti-murine SETX antibody.
Figure 4.
SETX mutant primary B cells have genomic instability and mild CSR defects. (a) SETX mutant primary B-cell isotype-switching (upper panel and lower left panel) and proliferation (lower right panel). Several assays suggest a small, statistically significant defect in CSR in SETX mutant primary B cells compared to their wild-type counterparts (unpaired Student's t-test, p < 0.0001 (IgG1), p = 0.0005 (IgG2b), p < 0.0001 (IgG3)). Proliferation assay using VPD450 shows no noticeable defect in proliferation in the mutant background. (b-c) SETX mutant primary B cell T-FISH. (b) Examples of telomere loss and dicentric chromosomes observed in the T-FISH performed on SETX-mutant cells. (c) Analysis of T-FISH assay results suggests a statistically significant increase in the number of metaphases with instability in the SETX mutant background compared to the wild-type background (Fisher's exact test, p = 0.0195).
3.6. SETX-deficient primary B cells have genomic instability
We determined whether splenic B cells from mutant SETX mice exhibit structural chromosomal alterations akin to those observed with the RNA exosome-deficient splenic B cells. An accumulation of RNA:DNA hybrids, as could be conjectured in the absence of properly active SETX, would be conducive to an increase in genomic instability by making R loops longer lasting. Accordingly, a T-FISH assay on splenic B cells isolated from SETX mutant and wild-type mice was performed, activating them for isotype switching for 72 h, metaphase-arresting them by colcemid exposure, and preparing them for telomeric probe hybridization. Activated SETX mutant B cells exhibit a statistically significant increase in genomic instability as measured by T-FISH when compared to activated wild-type B cells (Fisher's exact test, p = 0.0195, Figure 4 b-c). These observations and possible interpretations are discussed later in the report.
3.7. SETX-deficient CH12-F3 cells exhibit an altered mutational profile
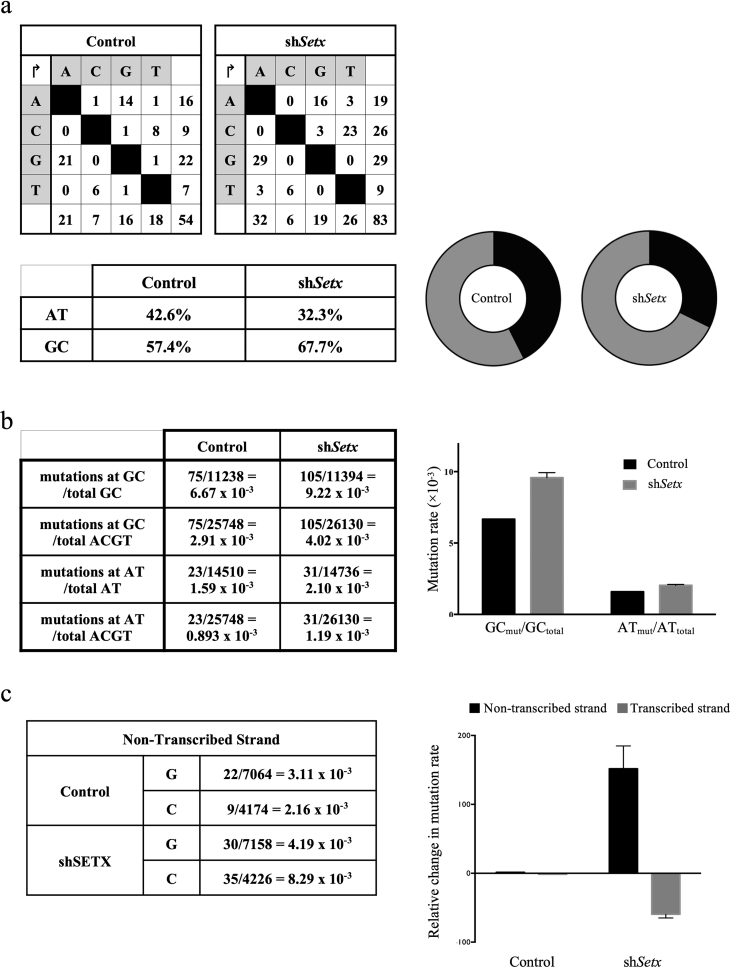
Accumulating RNA:DNA heteroduplexes would result in a different mutational profile at switch sequences, as the ssDNA mutagen AID would be unable to access the transcribed strand and use it as a substrate. To analyze this prediction the 5′-region of switch sequence μ (Sμ) was probed in SETX-sufficient and -deficient CH12–F3 cells stimulated to undergo isotype switching to IgA. The proportion of mutations that occur at GC residues is higher in SETX-deficient CH12–F3 cells than in control CH12–F3 cells; in parallel, the proportion of mutations that occur at AT residues is lower in SETX-deficient CH12–F3 cells than in SETX-sufficient CH12–F3 cells (Figure 5a). To know whether the proportional mutational increase at GC residues occurs at the expense of mutations at AT residues, an analysis shows that the mutation rate at GC residues in SETX-deficient CH12–F3 cells is higher than in SETX-sufficient CH12–F3 cells. Our analysis also shows that the mutation rate at AT residues is higher in SETX-deficient CH12–F3 cells than in SETX-sufficient CH12–F3 cells. However, the mutation rate increase from SETX-sufficient to SETX-deficient is more pronounced for GC residues than for AT residues (Figure 5b).
Figure 5.
Mutation profile of stimulated SETX-deficient CH12F3 cells. (a) Distribution of mutations in control and a SETX-deficient background. In the SETX-deficient background, compared to the control background, a decrease in the proportion of mutations at AT residues and an increase in the proportion of mutations at GC residues are noticeable. (b) Further analysis of the mutations in the SETX-deficient background shows that overall mutations at AT and GC both increase, but that this increase is more pronounced at GC residues, accounting for the opposite trends in panel a. (c) Mutation rate at C residues increases on the non-transcribed strand, and decreases on the transcribed strand, relative to the control vector.
We further probed the alterations to the mutational profile in SETX-deficient CH12–F3 cells. We compared the mutation rate at deoxycytidine residues on the non-transcribed strand compared to the transcribed strand. One would predict that dC residues would be mutated at higher rates on the non-transcribed strand than on the transcribed strand upon impairment of SETX activity. Indeed, compared to the control background, there is a marked increase in the mutation rate at dC residues found on the non-transcribed strand in SETX-deficient CH12–F3 cells. In parallel, there is a decrease in the mutation rate at dC residues found on the transcribed strand of SETX-sufficient CH12–F3 cells (Figure 5c). Taken together, these observations using a SETX knockdown approach reflect SETX's potential role in RNA:DNA hybrid unwinding and concomitant effects on AID-mediated somatic hypermutation in the IgH switch sequence.
4. Discussion
Realizing that RNA exosome is centrally involved in the regulation of processes like CSR, preventing chromatin associated noncoding RNA and RNA:DNA hybrid accumulation at various oncogenic regions of the B cell genome (Pefanis et al., 2014), we propose that RNA exosome activity is important for resolution of both programmed and aberrant DNA double strand breaks. RNA exosome also has been shown to process RNA to promote DNA double-strand break repair through its degradative activity on RNA:DNA hybrids (Domingo-Prim et al., 2019; Pefanis et al., 2015), thus it is likely that in the absence of RNA exosome activity mouse B cells will exhibit widespread genomic instability. Here we show that B lymphocytes experience a number of chromosomal abnormalities, as evidenced through the T-FISH assay, and are more sensitive to additional mutagenic stress, as evidenced by the ionizing radiation immunofluorescence assay following depletion of RNA exosome activity through deletion of RNA exosome core subunit-encoding Exosc3.
The chromosomal abnormalities witnessed in RNA exosome-deleted B cells included breaks and fragments (telomere loss, acentric chromosomal fragments), as well as rearrangements (dicentric chromosomes, homologous end-to-end fusions, radials). The rearrangement phenotypes can appear when breaks and fragments are repaired either intra-chromosomally through end-to-end homologous fusion or with a different chromosome to form a dicentric chromosome (rearranged chromosomes are only perceptible to a certain extent with the T-FISH assay; some balanced translocations or short intra-chromosomal deletions, for instance, would not be detected). These observations are reminiscent of Breakage-Fusion-Bridge cycles seen in some cancers, when chromosome breakage is repaired by a fusion event (between two sister chromatids or between two chromosomes), which is followed by the formation of a “bridge” during mitosis that is broken again as the spindle pulls on each centromere to re-create broken ends available for another cycle (Feijoo et al., 2014). As for the radials, they are thought of as a chromatid-type break (affecting only one chromatid, in contrast to both chromatids being affected in chromosome-type breaks) that arises at sites of stalled replication forks and that is repaired by the presumably non-physiological insertion of another chromosome (Savage, 1976; Scully et al., 2000). Our results would then hint at the possibility that chromosome-type breaks (involving both chromatids, typically at G1 in the cell cycle and therefore pre-replicative in origin) occur more frequently than chromatid-type breaks (involving one chromatid). Common fragile sites, where DNA polymerase stalling occurs, and which exhibit greater susceptibility to replication fork stalling and collapse, often are associated with these chromatid-type breaks that occur later in replication. These observations also highlight the possibility that instability events might be occurring at different times in the cell cycle (like the less frequent radials) or at additional sites sensitive to RNA exosome activity abrogation.
Attempting to understand the workings of the Ig RNA:DNA hybrid resolution reaction by investigating the role played by the RNA exosome complex led naturally to the study of the possible involvement of the putative RNA/DNA 5′ → 3′ helicase senataxin (SETX). The proposal is that SETX helps unwind RNA:DNA hybrid duplexes to facilitate the RNA exosome-mediated removal/degradation of RNA:DNA heteroduplex RNA moieties (Richard et al., 2013). We observed that in the human Burkitt's lymphoma cell line RA.1 (Ramos cells) SETX immunoprecipitates together with RNA exosome core subunit RRP45. A SETX-knockdown system in the murine CH12–F3 cell line was established, where class-switch recombination could be assayed, and it was found that CSR becomes impaired when SETX expression is decreased. We also found evidence that RNA:DNA hybrids accumulate in SETX-mutant (CRISPR/Cas9-edited) CH12–F3 cells (Lim et al., 2017). Our studies in this system were limited because no satisfactory commercial antibody against mouse SETX is available. Nevertheless, and importantly, we studied a 5′-most switch sequence Sμ segment for mutational analysis, and observed that in SETX-deficient backgrounds, the mutational profile at these AID target loci changes, with mutations accruing on the non-transcribed strand relative to the transcribed strand.
A mouse model was employed where Setx exon 4 has been deleted, abrogating the function of the protein constitutively (Becherel et al., 2013). Our findings using this model were less striking than in the knockdown CH12–F3 background. We saw a small though statistically significant decrease in IgG1+, IgG2b+ and IgG3+ activated B cells from mutant mice compared to activated B cells from wild-type mice, when assayed by ex vivo CSR experiments. Given that AOA patients have not been reported to be immunodeficient, it is unlikely that this mild level of CSR defect seen in Setx mutant mice affects overall circulating levels of antibodies or an adaptive immune response. T-FISH assay on these cells revealed a mild but statistically significant increase in genomic instability. These observations suggest the possible existence of redundant pathways in B cells that can partly substitute for SETX function. Several candidates could fall into this category. MTR4 (mRNA transport 4) is a member of the RNA helicase superfamily 2 that forms the TRAMP complex (Trf4/Trf5 poly(A) polymerases, Air1/Air2 Zn-knuckle RNA-binding proteins, and Mtr4 RNA helicase) known to interact with the RNA exosome complex (Houseley et al., 2006). Another potential candidate is Aquarius (AQR), which, unlike MTR4, but like SETX, is a putative RNA/DNA helicase member of the RNA helicase superfamily 1; AQR activity, like that of SETX, has also been associated with R-loop resolution (Sollier et al., 2014). Studies of the RNA:DNA hybrid resolution reaction in the absence or impairment of one or a combination of all these helicases surely would be insightful. Indeed, MTR4-deficiency shows a clear defect in RNA:DNA hybrid resolution and promotes AID-mediated asymmetric DNA mutagenesis as was demonstrated via genetic experiments (Lim et al., 2017) and modeled in biophysical cryo-EM structure analysis (Weick et al., 2018); other helicases also should be investigated. Another explanation for the mouse phenotype is that it may represent a hypomorph of some sort, or an alternatively-spliced gene product, given the absence of a pronounced neurological phenotype despite the known association of SETX with maladies such as ALS4 and AOA2 (Becherel et al., 2013). This may be worth exploring given reports of alternative splicing and transcription start sites (Fogel et al., 2009).
Finally, RNA exosome and SETX have important roles in early termination of elongating RNA polII (Lemay et al., 2014; Pefanis et al., 2014; Richard and Manley, 2009; Skourti-Stathaki et al., 2011). SETX, similar to Sen1p in the yeast NNS complex, can play a role in termination by unwinding RNA from DNA/RNA hybrids that are subsequently degraded by the RNA exosome complex. In absence of exosome activity, the termination complex function could be inhibited for termination function by accumulation of associated inhibitory RNAs that interfere with the function of the termination complexes in the cells. It is possible that lack of early termination of RNA polII in RNA exosome-deficient cells, particularly in noncoding RNA expressing regions that may not follow canonical transcription termination, creates secondary DNA structures that are deleterious to the cells. Alternatively, lack of termination of ncRNAs (e.g. antisense RNAs) may lead to RNA polII-RNA polII clashes or RNA polII-DNA polymerase clashes that lead to genomic instability (Rothschild and Basu, 2017). Although additional experimentation will be needed to further investigate the milder phenotype of SETX mutant primary B cells, it is clear that disruption of the SETX/RNA exosome axis leads to some level of genomic instability.
5. Conclusion
We aimed to examine the role played by the SETX/RNA exosome axis in programmed and aberrant genomic rearrangements in B cells. We found that stimulated RNA exosome-deficient B cells have increased genomic instability catastrophic events. Additionally, RNA exosome-deficient B cells are more sensitive to increases in IR mutagenic stress. We also found that CSR is impaired in stimulated SETX-deficient CH12–F3 cells and that stimulated SETX-deficient CH12–F3 cells exhibit an altered Sμ mutational profile. Activated SETX mutant primary B cells also have genomic instability and milder CSR defects.
Our results provide evidence that the SETX/RNA exosome axis is important for the resolution of both programmed and aberrant DNA double-strand breaks in the B-cell genome. The SETX/RNA exosome axis accordingly is important in promoting DNA double-strand breaks at AID target loci. Our results also suggest the involvement of redundant helicases in activated primary B-cell genomic rearrangements, and highlight the need for further investigation into these, as well as into Setx alternative splicing and transcription start sites.
Declarations
Author contribution statement
D. Kazadi and J. Lim: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
G. Rothschild and V. Grinstein: Performed the experiments.
B. Laffleur: Analyzed and interpreted the data.
O. Becherel and M. Lavin: Contributed reagents, materials, analysis tools or data.
U. Basu: Conceived and designed the experiments; Analyzed and interpreted the data.
Funding statement
This work was supported by grants to U.B. (NIAID (1R01AI099195 and Ro1AI134988), Leukemia & Lymphoma Society, and the Pershing Square Sohn Cancer Research Alliance) and to D.K. (NIAID, 1F31AI098411-01A1).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank Dr. Murty Vundavalli (Institute for Cancer Genetics, Columbia University, New York, NY, USA) for his help with the T-FISH assay, and Dr. Suprawee Tepsuporn (Harvard University, Cambridge, MA, USA). We also thank Dr. Matthew Miller (McMaster University, Hamilton, Ontario, Canada) and members of the Basu laboratory for valuable discussions.
Contributor Information
Junghyun Lim, Email: jl4045@cumc.columbia.edu.
Uttiya Basu, Email: ub2121@cumc.columbia.edu.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Basu U., Meng F.L., Keim C., Grinstein V., Pefanis E., Eccleston J., Zhang T., Myers D., Wasserman C.R., Wesemann D.R. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherel O.J., Yeo A.J., Stellati A., Heng E.Y., Luff J., Suraweera A.M., Woods R., Fleming J., Carrie D., McKinney K. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R., Basu U., Yewdell W.T., Chaudhuri J., Robbiani D.F., Di Noia J.M. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat. Rev. Immunol. 2016;16:164–176. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.A., Aristizabal M.J., Lu P.Y.T., Luo Z., Hamza A., Kobor M.S., Stirling P.C., Hieter P. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLOS Genet. 2014;10 doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V., Bortnick A., Murre C. AID targeting: old mysteries and new challenges. Trends Immunol. 2015;36:527–535. doi: 10.1016/j.it.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Alt F.W. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Basu U., Zarrin A., Yan C., Franco S., Perlot T., Vuong B., Wang J., Phan R.T., Datta A. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F.W. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Chen Y.-Z., Bennett C.L., Huynh H.M., Blair I.P., Puls I., Irobi J., Dierick I., Abel A., Kennerson M.L., Rabin B.A. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am. J. Hum. Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson S.K., Market E., Besmer E., Papavasiliou F.N. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Prim J., Endara-Coll M., Bonath F., Jimeno S., Prados-Carvajal R., Friedlander M.R., Huertas P., Visa N. EXOSC10 is required for RPA assembly and controlled DNA end resection at DNA double-strand breaks. Nat. Commun. 2019;10:2135. doi: 10.1038/s41467-019-10153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijoo P., Dominguez D., Tusell L., Genesca A. Telomere-dependent genomic integrity: evolution of the fusion-bridge-breakage cycle concept. Curr. Pharmaceut. Des. 2014;20:6375–6385. doi: 10.2174/1381612820666140630085416. [DOI] [PubMed] [Google Scholar]
- Fogel B.L., Lee J.Y., Perlman S. Aberrant splicing of the senataxin gene in a patient with ataxia with oculomotor apraxia type 2. Cerebellum. 2009;8:448–453. doi: 10.1007/s12311-009-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart P.J., Wood R.D. Emerging links between hypermutation of antibody genes and DNA polymerases. Nat. Rev. Immunol. 2001;1:187–192. doi: 10.1038/35105009. [DOI] [PubMed] [Google Scholar]
- Goodman M.F., Scharff M.D., Romesberg F.E. AID-initiated purposeful mutations in immunoglobulin genes. Adv. Immunol. 2007;94:127–155. doi: 10.1016/S0065-2776(06)94005-X. [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Kim H.-D., Choe J., Seo Y.-S. The sen1+ gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry. 1999;38:14697–14710. doi: 10.1021/bi991470c. [DOI] [PubMed] [Google Scholar]
- Kinner A., Wu W., Staudt C., Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffleur B., Basu U., Lim J. RNA exosome and non-coding RNA-coupled mechanisms in AID-mediated genomic alterations. J. Mol. Biol. 2017;429:3230–3241. doi: 10.1016/j.jmb.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J.F., Larochelle M., Marguerat S., Atkinson S., Bahler J., Bachand F. The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nat. Struct. Mol. Biol. 2014;21:919–926. doi: 10.1038/nsmb.2893. [DOI] [PubMed] [Google Scholar]
- Lim J., Giri P.K., Kazadi D., Laffleur B., Zhang W., Grinstein V., Pefanis E., Brown L.M., Ladewig E., Martin O. Nuclear proximity of Mtr4 to RNA exosome restricts DNA mutational asymmetry. Cell. 2017;169:523–537. doi: 10.1016/j.cell.2017.03.043. e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., Schatz D.G. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Liu M., Schatz D.G. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Martin-Tumasz S., Brow D.A. Saccharomyces cerevisiae sen1 helicase domain exhibits 5′- to 3′-helicase activity with a preference for translocation on DNA rather than RNA. J. Biol. Chem. 2015;290:22880–22889. doi: 10.1074/jbc.M115.674002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul R.W., Chon H., Sakhuja K., Cerritelli S.M., Gugliotti L.A., Gearhart P.J., Crouch R.J. R-loop depletion by over-expressed RNase H1 in mouse B cells increases activation-induced deaminase access to the transcribed strand without altering frequency of isotype switching. J. Mol. Biol. 2017;429:3255–3263. doi: 10.1016/j.jmb.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F.L., Du Z., Federation A., Hu J., Wang Q., Kieffer-Kwon K.R., Meyers R.M., Amor C., Wasserman C.R., Neuberg D. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo H.E., Gómez-González B., Grzechnik P., Rondón A.G., Wei W., Steinmetz L., Aguilera A., Proudfoot N.J. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira M.-C., Klur S., Watanabe M., Németh A.H., Le Ber I., Moniz J.-C., Tranchant C., Aubourg P., Tazir M., Schöls L. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermuation require Activation-Induced Cytindine Deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Nussenzweig M.C. AID targeting in antibody diversity. Adv. Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- Pefanis E., Wang J., Rothschild G., Lim J., Chao J., Rabadan R., Economides A.N., Basu U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E., Wang J., Rothschild G., Lim J., Kazadi D., Sun J., Federation A., Chao J., Elliott O., Liu Z.P. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Puno M.R., Lima C.D. Structural basis for MTR4-ZCCHC8 interactions that stimulate the MTR4 helicase in the nuclear exosome-targeting complex. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E5506–E5515. doi: 10.1073/pnas.1803530115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Wang Q., Dose M., Pruett N., Kieffer-Kwon K.R., Resch W., Liang G., Tang Z., Mathe E., Benner C. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal D., Maul R.W., Ghosh A., Chakraborty T., Khamlichi A.A., Sen R., Gearhart P.J. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J. Exp. Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro A.R., Stavropoulos P., Jankovic M., Nussenzweig M.C. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Richard P., Feng S., Manley J.L. A SUMO-dependent interaction between Senataxin and the exosome, disrupted in the neurodegenerative disease AOA2, targets the exosome to sites of transcription-induced DNA damage. Genes Dev. 2013;27:2227–2232. doi: 10.1101/gad.224923.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P., Manley J.L. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Palazzo R.E. Colcemid and the mitotic cycle. J. Cell Sci. 1992;102(Pt 3):387–392. doi: 10.1242/jcs.102.3.387. [DOI] [PubMed] [Google Scholar]
- Rondon A.G., Aguilera A. DNA repair; 2019. What Causes an RNA-DNA Hybrid to Compromise Genome Integrity? p. 102660. [DOI] [PubMed] [Google Scholar]
- Rothschild G., Basu U. Lingering questions about enhancer RNA and enhancer transcription-coupled genomic instability. Trends Genet. 2017;33:143–154. doi: 10.1016/j.tig.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage J.R. Classification and relationships of induced chromosomal structual changes. J. Med. Genet. 1976;13:103–122. doi: 10.1136/jmg.13.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., Swanson P.C. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- Scully R., Puget N., Vlasakova K. DNA polymerase stalling, sister chromatid recombination and the BRCA genes. Oncogene. 2000;19:6176–6183. doi: 10.1038/sj.onc.1203971. [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K., Proudfoot N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K., Proudfoot N.J., Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A., Klapacz J., Samaranayake M., Ullah A., Bhagwat A.S. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J., Stork C.T., Garcia-Rubio M.L., Paulsen R.D., Aguilera A., Cimprich K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L., Buratowski S. Nrd1 interacts with the nuclear exosome for 3' processing of RNA polymerase II transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Wahba L., Amon J.D., Koshland D., Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wuerffel R., Feldman S., Khamlichi A.A., Kenter A.L. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J. Exp. Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick E.M., Puno M.R., Januszyk K., Zinder J.C., DiMattia M.A., Lima C.D. Helicase-dependent RNA decay illuminated by a cryo-EM structure of a human nuclear RNA exosome-MTR4 complex. Cell. 2018;173:1663–1677 e1621. doi: 10.1016/j.cell.2018.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Culbertson M.R. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988;118:609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.