Abstract
Microbial inoculation in drought challenged rice triggered multipronged steps at enzymatic, non-enzymatic and gene expression level. These multifarious modulations in plants were related to stress tolerance mechanisms. Drought suppressed growth of rice plants but inoculation with Trichoderma, Pseudomonas and their combination minimized the impact of watering regime. Induced PAL gene expression and enzyme activity due to microbial inoculation led to increased accumulation of polyphenolics in plants. Enhanced antioxidant concentration of polyphenolics from microbe inoculated and drought challenged plants showed substantially high values of DPPH, ABTS, Fe-ion reducing power and Fe-ion chelation activity, which established the role of polyphenolic extract as free radical scavengers. Activation of superoxide dismutase that catalyzes superoxide (O2−) and leads to the accumulation of H2O2 was linked with the hypersensitive cell death response in leaves. Microbial inoculation in plants enhanced activity of peroxidase, ascorbate peroxidase, glutathione peroxidase and glutathione reductase enzymes. This has further contributed in reducing ROS burden in plants. Genes of key metabolic pathways including phenylpropanoid (PAL), superoxide dismutation (SODs), H2O2 peroxidation (APX, PO) and oxidative defense response (CAT) were over-expressed due to microbial inoculation. Enhanced expression of OSPiP linked to less-water permeability, drought-adaptation gene DHN and dehydration related stress inducible DREB gene in rice inoculated with microbial inoculants after drought challenge was also reported. The impact of Pseudomonas on gene expression was consistently remained the most prominent. These findings suggested that microbial inoculation directly caused over-expression of genes linked with defense processes in plants challenged with drought stress. Enhanced enzymatic and non-enzymatic antioxidant reactions that helped in minimizing antioxidative load, were the repercussions of enhanced gene expression in microbe inoculated plants. These mechanisms contributed strongly towards stress mitigation. The study demonstrated that microbial inoculants were successful in improving intrinsic biochemical and molecular capabilities of rice plants under stress. Results encouraged us to advocate that the practice of growing plants with microbial inoculants may find strategic place in raising crops under abiotic stressed environments.
Subject terms: Plant signalling, Drought
Introduction
Under normal environmental conditions, crop plants maintain a delicate balance in optimum growth, development and productivity. However, under nutrient limiting conditions or environmental stresses, plants face physiological and biochemical challenges leading to growth disruption due to disturbed primary metabolism1. Cells further suffer from oxidative damage due to the accumulation of reactive oxygen species (ROS) (superoxide, OH− radical, H2O2 and singlet oxygen)2. ROS generated during aerobic metabolic processes usually impact cellular targets in concentration-dependent manner3. Normal ROS concentration in the cells regulates key cellular physiology and redox-sensitive metabolic mechanisms. However, increased level of ROS in plants growing under oxidative stresses becomes cytotoxic4. When exposed to abiotic stresses, normal pathways for photorespiration, photosynthesis and mitochondrial respiration lead to produce excessive ROS5 that disturbs intrinsic cellular homeostasis6. Environmental stresses also trigger activity of monoamine oxidase (MAO), xanthine oxidase (XOD) and NADPH oxidase that balance production and accumulation of ROS7. The consequences are observed in terms of negative cellular metabolic functions that damage nucleic acid, protein, lipid and carbohydrate metabolism2.
Plants are evolved with a sophisticated system to overcome ROS burden within the cells through prominent antioxidative defense mechanisms8. Enzymatic antioxidative mechanisms include regulation of the enzymes like superoxide dismutase, catalase, peroxidase, glutathione reductase, glutathione S-transferase and guaiacol peroxidase. These enzymes prevent or repair the oxidative damage caused due to disrupted cellular homoeostasis under stress conditions5. Cells also synthesize diverse antioxidant molecules that regulate signal pathways in redox mechanisms to overcome oxidative damage4. Increased production of antioxidative enzymes like SOD, POD, CAT, GPX and GST9 and the accumulation of antioxidant compounds such as carotenoids10 and phenylpropanoids11 successfully help plants reduce their load of ROS within the cells. These processes cumulatively help plants mitigate burden of oxidative mechanisms while maintaining their growth and development under stressful conditions.
Among various devastating environmental stresses for plants, drought conditions, either moderate to intense or short to prolonged, have remained a challenge for crop productivity12. Drought adaptation, avoidance and/or mitigation strategies in crop plants lie with their intrinsic metabolic and molecular mechanisms which, when triggered by environmental stimulus strengthen plant growth, development and productivity13. Beneficial microbial interactions with plants either under normal growth conditions or in stressful environment manifest diverse physiological, biochemical and molecular roles14–16. Microbial communities, the most natural inhabitants of the soils and the rhizosphere, the specific ecological niche associated with the root vicinity, tremendously influence plant growth and productivity17,18. Their interaction with the plant root system constitutes the most complex and intricate biological phenomenon that helps plant activate their adaptive capabilities against drought stress through induced defense mechanisms19,20. Plant growth promoting rhizobacteria (PGPR) colonize rhizosphere to promote growth and induce systemic drought tolerance21,22 through phytohormone, epoxypolysaccharides and ACC deaminase production23–26. Plant responses to Trichoderma inoculation as a biocontrol agent are manifested by early escape of abiotic stresses through activation of antioxidant machinery27,28. Inoculation of T. harzianum helped plants alleviate water deficit in tomato29 and rice28 through enhanced activation of ascorbate and glutathione-related defense enzymes30. Cumulatively, microbe-plant interaction and the resultant metabolic changes are being realized as a real time stress tolerance strategy in the plants for their survival and sustainable productivity31.
Rice (Oryza sativa L.) is the most important crop that feeds almost half of the world’s population32. Being a crop of tropical and subtropical origin, rice is usually sensitive to abiotic stresses, especially to drought conditions33. Water deficit is amongst the major limiting factors to produce rice in many parts of the world34. High sensitivity to drought and water deficit poses serious threat towards enhanced productivity of this crop35. Microbial communities are the dominant natural inhabitants of the plant rhizosphere36,37 including rice crop38,39. Their colonization and interaction with the rice roots impart beneficial plant growth promotion and abiotic stress mitigation impacts40,41. We demonstrated that the individual and combined inoculation of rice with Pseudomonas fluorescens and Trichoderma asperellum (T42) have contributed to strengthen intrinsic mechanisms in rice plants, thereby offering protective support against drought. Enzymatic and non-enzymatic antioxidant reactions in plants grown with microbial inoculation under non-drought and drought conditions were improved. The expression of defence-related genes that helped plants regulate ROS as key steps in microbe-mediated stress mitigation processes was explored. The study reveals that growing plants under microbe-inoculated conditions leads to modulate intrinsic biochemical and molecular mechanisms to help plants mitigate drought conditions. The observations warrant microbial inoculation as an efficient stress mitigation strategy for rice crop challenged with drought stress in the fields.
Materials and Methods
Seeds, microbial inoculants and experimental conditions
Seeds of rice variety Pusa Basmati (PB) 1612 were obtained from the seed bank of ICAR-Indian Institute of Seed Science, Mau, India. Rhizosphere compatible bioagents namely Pseudomonas fluorescens (Pf) (OKC; Genbank accession No. JN128891) and Trichoderma asperellum (Th) (T42; GenBank accession No. JN128894) were obtained from the Department of Mycology and Plant Pathology, Institute of Agricultural Sciences, Banaras Hindu University, Varanasi, India. Rice seeds were treated with both the cultures as described by Patel et al.42. For seed treatment, spore suspension of Th (spore count 1.3 × 108 ml−1) and cell suspension of Pf (1.2 × 108 cells ml−1, optical density equivalent to 0.39) was prepared in 0.5% sterilized carboxymethylcellulose (CMC). For combined application, equal proportion of fungal spores and bacterial cell suspension was mixed together and applied. Rice seeds (variety PB 1612) were surface sterilized with 0.1% HgCl2 solution for 2 min followed by washing thrice with sterilized distilled water. Dried seeds were coated with the inoculant suspension (individual and in combination) and kept for 3 h in air under sterilized conditions. Microbe-coated seeds were sown in earthen pots (10 inch diameter) containing 5.4 kg sterilized soil mixed with 20% vermicompost. Pots were kept in well-ventilated glasshouse throughout the Kharif season of 2017 from mid-June to November. Temperature ranged from 16.4 to 31.5 (min) to 30.1 to 39.2 °C (max) with gradual decrease as the plant development approached maturity. Regular watering was applied prior to flowering stage, before the onset of which, 7 days of continuous drought was given to one set of pots sown with the microbe-inoculated and non-inoculated (NI) rice seeds. All the plants were harvested after completion of drought period and leaves were collected for further experimentation.
Physicochemical determination
Plant growth parameters
Along with the protein concentration, dry shoot and root weight were recorded under inoculated, non-inoculated, stressed and non-stressed conditions. Protein concentration was estimated following Lowry et al. method43 in which bovine serum albumin (BSA) was used as standard. Protein concentration was expressed as mg protein per gram fresh wt. Plant shoot and root wt were recorded on dry wt basis by randomly uprooting 4 plants from 6 pots, each of which contained 2 plants. Shoot and roots were dried in an oven at 65 ± 2 °C for 72 h, the total dry matter (TDM) of shoot and root was weighed separately and recorded as g per plant.
Quantification of H2O2
Leaf samples (0.1 g) from each treatment were homogenized in 2.0 ml 0.1% (w/v) trichloroacetic acid (TCA) and centrifuged (12,000 g, 15 min). The supernatant (0.5 ml) was added with 10 mM phosphate buffer (pH 7.0). Afterwards, potassium iodide solution (1 M, 1 ml) was added following incubation for 5 min. The oxidation product formed was examined at 390 nm44. The concentration of H2O2 formed was determined as nMol H2O2 g−1 fresh weight (FW).
In situ examination of cell death
In situ cell death determination was carried out by treating plant leaves with 0.1% Evans blue solution. After 15 min, leaves were dipped in 95% boiling ethanol (30 min) for depigmentation. Necrotic spots were identified as indigo blue lesions at the leaf surface45.
Determination of total polyphenolic content (TPC)
TPC was determined following the method of Zheng and Shetty46 with modifications. Leaf tissues (0.1 g) were macerated in 5 ml water:methanol (1:1, v/v) at 4°C and extracted for 48 h. Homogenized samples were centrifuged at 15000 g (10 min). Polyphenolic content was quantified using Folin–Ciocalteau reagent. The extract (1 ml) was mixed with water:methanol (1:1, 1 ml, v/v), distilled water (3 ml) and Folin–Ciocalteau regent (0.5 ml) followed by thorough mixing. The reaction mixture containing 5% sodium carbonate (1 ml) was kept for 30 min and examined at 725 nm. TPC was calculated as mg gallic acid equivalents (GAE) per g FW.
Quantitative determination of enzymes
One g of fresh rice leaves were washed with the sterilized distilled water and macerated with 5 ml phosphate buffer (pH 7.8) in ice cooled pestle-mortar kept at 4 °C. The extract was centrifuged at 15,000 rpm for 15 min at 4 °C and used for enzymatic assays.
Superoxide dismutase (SOD)
SOD (EC 1.15.1.1) activity was determined by photochemical reduction method of nitrobluetetrazolium (NBT) chloride47. Reaction mixture containing methionine (200 mmol l−1), NBT (2.25 mmol l−1), EDTA (3 mmol l−1), phosphate buffer (100 mmol l−1; pH 7.8) and sodium carbonate (1.5 mol l−1) was mixed with the enzyme extract. In 3 ml final volume, 2µmol l−1 riboflavin (0.4 ml) was added following exposure to light (15 W fluorescent lamp, 15 min). The absorbance was taken at 560 nm after deactivating the enzyme activity in dark. One unit of SOD decreased the absorbance by 50% as compared to control, which lacked enzyme extract.
Peroxidase (PO)
PO (EC 1.11.1.7) was estimated in the reaction mixture containing 1.5 ml pyrogallol (0.05 mol), 0.05 ml enzyme extract and 0.5 ml H2O2 (1%; v⁄v)48. The change at 420 nm was determined at every 30 s intervals and the enzyme activity was recorded as U per min per g FW.
Ascorbate peroxidase (APX)
Plant leaves (100 mg) were suspended in 0.1 M sodium phosphate buffer (pH 6.8) containing 2 mM ascorbate, homogenized and centrifuged (15000 g, 20 min). The reaction mixture containing phosphate buffer (25 mM, pH 7.0), EDTA (0.1 mM), ascorbic acid (0.25 mM), H2O2 (1.0 mM) and enzyme extract (0.2 ml) was kept at room temp49. Reduction in absorbance was measured at 290 nm after 60 s and activity was expressed as U min−1 g−1 FW.
Catalase (CAT)
CAT (E.C. 1.11.1.6) was assayed by Aebi method50. Reaction mixture consisting of phosphate buffer (300 µM, pH 7.2) and H2O2 (100 µM) in enzyme extract (1 ml) was allowed to release O2 by enzymatic dissociation of H2O2 in the dark for 1 min. O2 produced due to enzyme reaction was determined at 240 nm (extinction coefficient of H2O2 is 0.036 mM−1 cm−1). The activity of the enzyme was expressed as µM H2O2 oxidized U min−1 g−1 FW.
Glutathione reductase (GR)
The method of Anderson51 was followed to determine GR (E.C. 1.6.4.2) activity. Reaction mixture contained Tris–HCl buffer (50 mM, pH 7.6), NADPH (0.15 mM, 10 ml), oxidized glutathione (1 mM GSSG, 100 µl), MgCl2 (3 mM) and enzyme extract (0.3 ml). GR was measured as gradual reduction in absorbance of NADPH at 340 nm. The activity of the enzyme was calculated in terms of U (nmol oxidized NADPH) min−1 mg−1 FW.
Guaiacol peroxidase (GPX)
GPX (E.C. 1.11.1.7) was measured by recording the increase in absorbance at 470 nm52. The reaction mixture consisting of sodium phosphate (10 mM; pH 6.0), H2O2 (0.3%, v/v), tetraguaiacol (1%, v/v) and enzyme extract (0.3 ml) was prepared. The enzyme activity was represented in terms of U min−1 mg−1 FW where one unit of enzyme catalyzes the oxidation of 1µmol of guaiacol min−1.
Phenylalanineammonia lyase (PAL)
Powdered leaf samples (0.5 g) were homogenized in 5 ml of ice-cold phosphate buffer (100 mM; pH 7.0 and 0.5 mM EDTA and mixed with 1.4 mmol l−1 β-mercaptoethanol53. The homogenate was centrifuged (15000 g, 15 min) and the supernatant was added with 0.1 mol l−1 l-phenylalanine (pH 8.7, 1 ml) along with the mixture of 0.5 ml 0.2 mol l−1 phosphate buffer (pH 8.7), 0.2 ml enzyme extract and 1.3 ml distilled water following incubation for 30 min. Trichloroacetic acid (TCA, 0.5 ml, 1 mol l−1) was added to terminate the reaction. The observations were recorded at 290 nm and activity was expressed in terms of µmol t-cinnamic acid g−1 FW.
Estimation of non-enzymatic antioxidative reactions
Free radical scavenging activity (FRSA)
The free radical scavenging activity was evaluated by 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging method using the stable radical DPPH54. Plant extract with TPC (100 μl) was mixed with 2.9 ml freshly prepared DPPH solution (60 μM in MeOH). The reduction in DPPH radical was determined at 515 nm till 1 h until stable values were obtained.
ABTS activity
The ABTS activity in TPC from the rice leaf was determined using the ABTS• + decolorization method55. The reaction mixture containing 10 ml ABTS• + radical (ABTS 9.5 mL, 7 mM) and potassium persulfate (245 μL, 100 mM) was kept in the dark for 18 h and then diluted with potassium phosphate buffer (0.1 M, pH 7.4) to an absorbance of 0.70 (±0.02) at 734 nm. TPC from rice leaves (50 μL) was mixed thoroughly with 2.95 mL ABTS radical solution. The absorbance was recorded at 734 nm and expressed as % inhibition of the activity.
Ferric reducing power antioxidant assay
The Fe-ion reducing power assay was performed with the leaf extracts taking quercetin as the standard compound56. To 200 and 500 μl aliquots, 1.0 ml MeOH, 2.5 ml phosphate buffer (pH 6.6) and 1% (w/v) potassium ferricyanide were added. Reaction mixture was incubated at 50 °C for 20 min and 2.5 ml TCA (10% w/v) was added to terminate the reaction. Absorbance was recorded at 700 nm and percent increase in Fe reducing activity was calculated.
Ferrous ion chelation activity
Ferrous ion chelation was determined by the method of Decker and Welch57. Rice leaf extract (200 μl) was maintained to 1.0 ml with MeOH and then added with 0.1 ml of ferrous chloride (2.0 mM), ferrozine [3-(2-pyridyl)-5,6-bis-(4-phenylsulfonic acid)-1,2,4-triazine] (0.2 ml of 5.0 mM) and methanol (3.7 ml). After 10 min, the absorbance was recorded at 562 nm where low absorbance indicated high ferrous ion chelating capacity.
Total RNA isolation and cDNA synthesis
Total RNA was isolated from 0.2 g fresh rice leaves using TRIzol™ LS reagent (Invitrogen; http://www.invitrogen.com). Three µg of the total RNA was digested with RNase-free DNase I (Thermo Scientific) to remove genomic DNA contamination. The poly(A)-RNA concentration was determined using NanoDrop 2000 spectrophotometer (Thermo Scientific). Samples with a 260/280 ratio of 1.9–2.1 and a 260/230 ratio ≥ 2.0 were chosen to determine the quality and purity of the RNA preparations. The integrity of the purified RNA was checked on 2% formamide denaturing gel. Subsequently, first-strand cDNA was synthesized in a 20 μL reaction mixture by using RevertAid H minus kit (Fermentas) following the manufacturer’s instructions and stored at −20 °C until use.
Quantitative qRT-PCR assay
Gene specific primer sequences for the defense-related genes as listed in Table 1 were obtained from TIGR Rice Genome Annotation Resource58 with the help of BLASTn and were synthesized from Helix Biosciences, India. qRT-PCR amplification was performed in 96-well plates with a iQ5 RT-PCR Detection System (BioRad Laboratories, Germany) using Green Supermix Kit Eva Green SYBR® (BioRad). Expression of the gene specific primers at a concentration of 0.1 µM was analyzed42. In brief the qPCR conditions were: denaturation at 95 °C for 2 min followed by 40 repeats at 95, 60 and 72 °C temp for 20, 30 and 25 s. The sense/antisense primer sequences for actin (5'-TCCATCTTGGCATCTCTCAG-3'/5'-GTACCCTCATCAGGCATCTG-3') and rRNA (5'- CTTCGGGATCGGAGTAATGA-3'/5'-AACTAAGAACGGCCATGCAC-3'), respectively were used as internal controls for normalizing relative gene expression levels in technically independent and triplicate biological experiments59. The threshold cycle (Ct) was measured automatically by the software.
Table 1.
Gene specific forward (F) and reverse (R) primers used in the study.
| Sl. No. | Gene Name | Primer 5′-3′ | |
|---|---|---|---|
| 1 | OsPIP1;1 | F | TACATGGGCAATGGCGGT |
| R | CAAGACCGTCACCCTTGGTG | ||
| 2 | DHN LOC_Os01g50700 | F | CAGCTCCAGCTCGGTAACTT |
| R | CTTCTGCTCCTCCTGCTTGT | ||
| 3 | DREB LOC_Os09g35030 | F | GGAGCAAGCAGAAACACACA |
| R | TCGTCTCCCTGAACTTGGTC | ||
| 4 | cCuZn-SOD1 | F | GAGATTCCAAACCAGCAGGA |
| R | TTGTAGTGTGGCCCAGTTGA | ||
| 5 | Fe-SOD | F | CTTGATGCCCTGGAACCTTA |
| R | GCCAGACCCCAAAAGTGATA | ||
| 6 | Mn-SOD1 | F | GGAGGCCATGTCAATCATTC |
| R | CACAAGGTCCAGAAGTGCAA | ||
| 7 | Chl_sAPX | F | CAATTGAGGAAGCTGGTGGT |
| R | ACTTCAGCGATCTGGCTCAT | ||
| 8 | CATa | F | CCACCACAACAACCACTACG |
| R | CCAACGACTCATCACACTGG | ||
| 9 | AU076282 | F | GCTACTACCGCAACCTCGTC |
| R | TCACTTTCCTGCAGTTGAGC | ||
| 10 | D14481 | F | CGCGATAAAGGAAGATCTCG |
| R | CGTCATAGTAAGGGCCTCCA | ||
| 11 | PAL 1 | F | CAGACACGGTCGTACCATTG |
| R | CCACCTCCTGCATTTGTTTT |
Statistical analysis
Data were subjected to Two Way ANOVA using PRISM version 5.0. Tests for normality of data and for homogeneity of variances were performed before running ANOVAs. PCA analysis was carried out using R-program. Except for the real time experiments using qRT-PCR, for which three replications were used, all the experiments were performed in complete randomized block design having six replications (n = 6). For the gene expression analyses, the expression values of the two housekeeping genes (actin and rRNA) were subjected to Two-way ANOVA using geometrical means of the internal controls and based on the mean values, the expression profile of all the genes was normalized. For all the experiments, the data were expressed as the mean value of the replicates. Standard error for each mean value was represented separately in the table and figures.
Results and Discussion
Plant responses to abiotic stresses are growth dependent and complex60. The underlying array of mechanisms for stress avoidance, tolerance and adaptation are conditional constraints involving multiple cellular physiological, metabolic and molecular alterations31. Stress induced antioxidative conditions within the cells generate reactive oxygen species (ROS) and lead to accumulation of free radicals that disrupt cellular homeostasis and adversely affect cell viability61. Stressed plants undergo multiple intrinsic equilibrations for early stress perception, signal channeling, gene expression and metabolic modifications to refrain from unfavorable conditions62. Microbial interactions with plants elicit modulation in molecular mechanisms to activate metabolic networks at gene, enzyme and metabolite level. This works in parallel to enhance plant’s intrinsic strength to support stress mitigation63. We inoculated rice with the strains of Trichoderma (Th) and Pseudomonas (Pf) as individual and combined inoculants (Th + Pf) and assessed whether microbial inoculation helped plants improve their metabolic capabilities to combat drought and if so, to what extent the biochemical and molecular level changes were linked with stress mitigation.
Microbial inoculation supports plant growth under drought stress
Protein concentration is one of the most prominent parameters to assess the impact of microbial inoculation on plants grown under drought or non-drought conditions. As indicated by two way ANOVA, the main effects of watering regime [F(1,40) = 281.8, p < 0.0001] and microbial inoculation [F(3,40) = 145.5, p < 0.0001] on protein concentration was significant. The impact of interaction of drought and microbial inoculation was also statistically significant [F(3,40) = 18.06, p < 0.0001]. In non-inoculated control plants, the concentration of protein (mg g−1) was significantly different (M = 8.96, SD = 0.51) than in the plants challenged with drought (M = 12.51, SD = 0.51). Maximum protein concentration was observed in plants challenged with drought and inoculated with Th + Pf (M = 16.90, SD = 0.5) followed by those inoculated with Th (M = 15.50, SD = 0.55) and Pf (M = 13.50, SD = 0.56) and grown under drought condition. Pair-wise analysis indicated significant differences between control and Th, Pf and Th + Pf inoculated non-drought plants. Further, the protein concentration in control plants was also significantly different than those grown under drought condition or in the plants challenged with the drought and given microbial inoculation (Table 2). Drought or desiccation tolerance in plants is known to promote accumulation of biomolecules including proteins64.
Table 2.
Impact of microbial inoculation on protein concentration and shoot and root dry weight of rice plants grown under non-drought and drought-challenged conditions.
| Treatments Parameters | Control (Non-Inoculation) | Trichoderma inoculation (Th) | Pseudomonas inoculation (Pf) | Combined inoculation of Trichoderma & Pseudomonas (Th + Pf) | Statistics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Protein Concentration (mg g−1) | Source | df | MS | F | P | ||||
| Non-Drought | 8.96 ± 0.51d | 12.91 ± 0.32c | 12.74 ± 0.41c | 13.30 ± 0.80c | Drought | 1 | 82.58 | 281.8 | <0.0001 |
| Inoculation | 3 | 42.64 | 145.5 | <0.0001 | |||||
| Drought | 12.51 ± 0.51c | 15.50 ± 0.55b | 13.50 ± 0.56c | 16.90 ± 0.50a | Drought*Inoculation | 3 | 5.29 | 18.06 | <0.0001 |
| Error | 40 | 0.2898 | |||||||
| Shoot dry weight (g per plant) | |||||||||
| Non-Drought | 5.48 ± 0.39cd | 6.66 ± 0.83bc | 7.78 ± 0.34ab | 8.21 ± 1.11a | Drought | 1 | 6.42 | 10.85 | 0.0021 |
| Inoculation | 3 | 16.92 | 28.58 | <0.0001 | |||||
| Drought | 4.81 ± 0.82d | 6.09 ± 1.06cd | 6.75 ± 0.59bc | 7.56 ± 0.58ab | Drought*Inoculation | 3 | 0.1215 | 0.205 | 0.892 |
| Error | 40 | 0.5899 | |||||||
| Root dry weight (g per plant) | |||||||||
| Non-Drought | 5.84 ± 0.21ef | 6.86 ± 0.77cd | 7.77 ± 0.65bc | 8.93 ± 0.40a | Drought | 1 | 4.23 | 16.67 | 0.0002 |
| Inoculation | 3 | 15.96 | 62.89 | <0.0001 | |||||
| Drought | 5.51 ± 0.44f | 6.62 ± 0.29de | 6.96 ± 0.38cd | 7.93 ± 0.60b | Drought*Inoculation | 3 | 0.4057 | 1.598 | 0.2049 |
| Error | 40 | 0.2507 | |||||||
p values in Bold are significantly different.
Drought reduced shoot and root dry weight although microbial inoculation substantially supported plant growth. Results of the two-way ANOVA for shoot dry weight showed significant effects of watering regime [F(1,40) = 10.85, p = 0.0021] and microbial inoculations [F(3,40) = 28.58, p < 0.0001], while the interaction effect was not significant [F (3,40) = 0.205, p 0.892] (Table 2). Shoot dry weight values of well-watered plants (M = 5.48, SD = 0.39) were significantly higher than those of drought-stressed plants (M = 4.81, SD = 0.82). On the other hand, Th + Pf inoculated plants had the highest shoot dry weight (M = 8.21, SD = 1.11), followed by Pf inoculated plants (M = 7.78, SD = 0.34), Th inoculated plants (M = 6.66, SD = 0.83) and uninoculated control plants (M = 5.48, SD = 0.39). Tukey’s pairwise tests indicated significant differences (p < 0.05) in between non-drought (control) plants and both Pf and Th + Pf inoculated plants, but no significant differences either between control and Th inoculated drought treated plants or between non-drought Pf and Pf + Th inoculated drought treated plants (Table 2). Similarly, on root dry wt, the impact of drought [F(1,40) = 16.67, p = 0.0002] and microbial inoculation [F(3,40) = 62.89, p < 0.0001] was statistically significant but the impact of interaction was non-significant [F(3,40) = 1.598, p < 0.2049]. Reduction in growth parameters in rice is the most obvious negative impact of drought and water deficit65. We reported that despite drought, microbial inoculation supported growth and development of shoot and root of rice plants in almost similar way as was evidenced under non-drought condition. Therefore, the negative impact of one factor (drought) is substantially being compensated by the other factor (microbial inoculation). Since growth promoting microorganisms enhance nutrient uptake by the plants, produce phytohormones and stimulate plant’s immune system14, the observed effect of microbial inoculation on developmental parameters, even in stress challenged plants, seems natural. These observations provided evidence that microbial inoculation may protect plants by bringing positive changes at physiological and morphological level under drought challenged condition.
Microbial inoculants help plants tolerate H2O2 impact and hypersensitive cell death
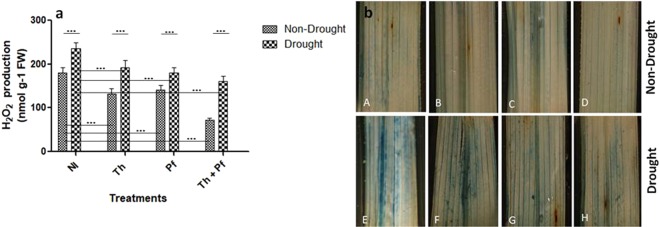
H2O2 level in the rice plants was reduced due to microbial inoculation. Compared to the non-inoculated control plants with high H2O2 level [M = 179.6 nmol g−1 FW, SD = 13.06], non-stressed plants inoculated with Th showed [M = 132.93, SD = 11.95], Pf [M = 141, SD = 10.07] and Th + Pf [M = 71.73, SD = 5.16] H2O2 concentration (Fig. 1a). Between control and the drought plants, the values of H2O2 content were significantly different. Pairwise tests also indicated significant differences in between non-inoculated drought challenged plants with those challenged with the drought and inoculated with the microbial species. Drought showed significant impact on H2O2 level in plants [F(1,40) = 296.9; p < 0.0001]. Microbial inoculation to plants also showed significant effects on H2O2 concentration in plants [F(3,40) = 112.1, p < 0.0001]. The interaction impact of drought and microbial inoculation was again significant [F(3,40) = 8.388, p = 0.0002) (Fig. 1a, Supplementary Table 1). We showed that although drought led to high H2O2 level, microbial inoculation lowered the magnitude of accumulation and thereby, lowered the toxic effect of H2O2 in the cells. This is further evidenced from the in situ hypersensitive reaction in the leaves of the rice plants (Fig. 1b). Leaves of non-stressed plants (Fig. 1b,A) grown with microbial inoculation (Fig. 1b,B,C,D) remained almost free from the lesions. Leaves of the plants grown under drought showed maximum stained lesions (Fig. 1b,E). However, microbial inoculation helped stressed plants minimize hypersensitive spots on the leaves (Fig. 1b,F,G) and minimum lesions were seen over the leaves of the plants inoculated with Th + Pf (Fig. 1b,H). Higher accumulation of H2O2 in plant cells is a toxic phenomenon leading to hypersensitive cell death. Microbial inoculation not only reduced the level of H2O2 in drought stressed plants, but it also minimized lesion development due to hypersensitive cell death in plant leaves. Drought as an unfavorable condition leads to the overproduction of H2O2 that eventually increased phytotoxicity leading to cell necrosis. Existing reports further confirm such processes in plants experiencing stressed conditions66–68.
Figure 1.
Generation of H2O2 in plants grown with microbial inoculation and post-drought stress (a) and in situ hypersensitive response in leaves (b). A & E: NI (non-inoculated); B & F: Trichoderma inoculation (Th); C & G: Pseudomonas inoculation (Pf); D & H: combined inoculation (Th + Pf). Level of significance was determined by two-way ANOVA. n = 6. Data are mean ± SEM.
Polyphenolics and PAL activity during drought stress
Accumulation of polyphenolics in plant leaves is shown to have protective role against stresses through anti-oxidation and ROS deactivation69,70. Polyphenolic metabolites play important role in plant defense against abiotic and biotic stresses71. Results of the two-way ANOVA for total polyphenol concentration showed significant effects of watering regime [F(1,40) = 549.2, p < 0.0001] and microbial inoculations [F(3,40) = 141.5, p < 0.0001]. The interaction effect was also significant [F(3,40) = 17.77, p < 0.0001] (Table S1). Drought-stressed plants had always significantly higher total polyphenol concentration than non-stressed plants (Fig. 2a). On the other hand, one-way ANOVAs and post hoc Tukey’s tests on both the drought stressed and non-stressed plant cohorts showed that significantly the lowest total polyphenol concentration was always in uninoculated plants. Among the three inoculation treatments in the cohort of drought-stressed plants, combined inoculation resulted in significantly high polyphenol concentration. Also, in the cohort of non-stressed plants, plants doubly inoculated with Trichoderma and Pseudomonas (Th + Pf) had significantly higher (p < 0.05) total polyphenol concentration than singly inoculated plants (Fig. 2a).
Figure 2.
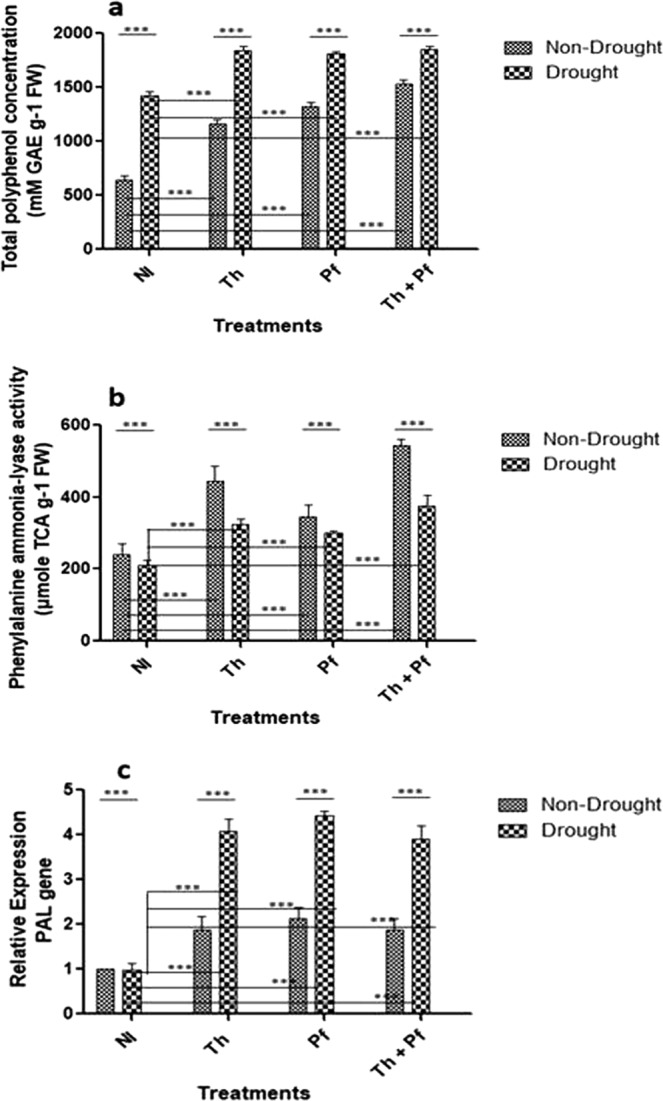
Accumulation of total polyphenol concentration (TPC) (a), PAL enzyme activity (b) and expression of PAL gene (c) in the leaves of microbe-inoculated and non-inoculated rice plants grown under non-drought and drought challenged conditions. GAE = gallic acid equivalents; TCA = trichloroacetic acid; Significance level was determined using two-way ANOVA. Data are mean ± SEM. n = 6 for TPC and enzyme assay, n = 3 for transcript analysis.
Microbial inoculation resulted in enhanced activity of PAL enzyme in rice leaves. One way ANOVA and Tuckey’s test results on drought and non-drought plants indicated that significantly low PAL activity was always reflected in stressed plants (Fig. 2b). In the cohort of non-stressed plants that always showed higher PAL activity than stressed plants, those with combined inoculation of Th + Pf had significantly high PAL activity than any other single microbial inoculation. In non-stressed plants without inoculation, PAL activity was [M = 240.77 µM TCA g−1 FW, SD = 30.88]. In non-stressed plants inoculated with Th the activity was [M = 443.2, SD = 43.38], with Pf it was [M = 344.1, SD = 35.11] and Th + Pf it was [M = 543.7, SD = 16.01] (Fig. 2b). The impact of watering regime on PAL activity in plants was significant [F(1,40) = 135.0, p < 0.0001]. The impact of microbial inoculation was also significant [F(3,40) = 163.0, p < 0.0001] and so was the effect of interaction [F(3,40) = 17.04, p < 0.0001 (Fig. 2b). Microbial inoculation to plants under stressed condition influences accumulation of polyphenolics and activates PAL enzyme activity72–75. Since polyphenolics are strong antioxidants and PAL is a defense-related enzyme, high accumulation of polyphenolics and enhanced PAL enzyme activity in the leaves are supposed to strengthen plants under drought challenged condition. Having shown that the microbial inoculation enhanced polyphenolic accumulation and improved PAL enzyme activity, the expression of PAL gene was checked in plant leaves (Fig. 2c). Microbial inoculation enhanced PAL gene expression in the non-drought plants. In the cohort of plants grown under drought following microbial inoculation, expression of PAL gene was multi-fold enhanced (Fig. 2c). The impact of watering regime on PAL gene expression was statistically significant [F(1,16) = 102.5, p < 0.0001]. The effect of microbial inoculation showed statistical significance [F(3,16) = 42.08, p < 0.0001]. The interaction impact also had statistically significant values [F(3,16) = 11.79, p0.0003] (Supplementary Table 2). Stressed conditions usually activate phenylpropanoid pathway, in which PAL is a key gene to offer physiological and structural support to the plants76,77. A correlative activation pattern of the PAL gene, the enzyme activity and accumulation of polyphenolics in the leaves of rice plants grown with microbial inoculation was found under drought stress. Such biochemical and molecular strategies are presumed to confer cumulative support to rice to tolerate the adverse impact of stress.
Polyphenolics accumulation enhanced antioxidant profile in inoculated plants
Normal concentration of intracellular ROS regulates redox state in the cells and also acts as signals for defense against stresses78,79. Unfavourable conditions enhance production and prolonged accumulation of ROS in cellular compartments80, a condition that becomes phytotoxic with deleterious impact due to oxidative damage of cell membrane81,82. Small molecule metabolites like phenolics, tocopherol, carotenoids and proline maintain redox state in cells during oxidative damage as ROS scavengers2,83. This is why enhanced polyphenolics concentration usually favours ROS scavenging in the plants grown under stress conditions.
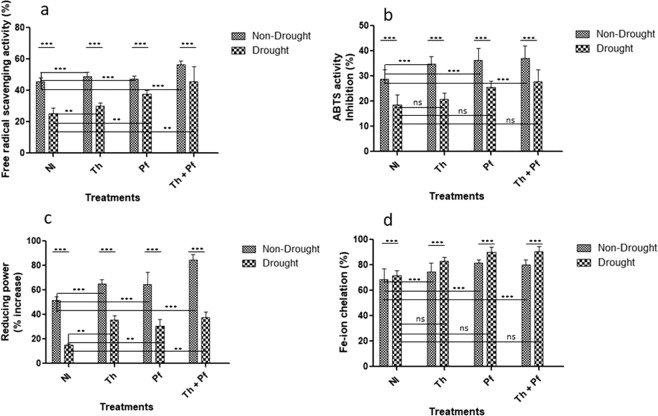
Two way ANOVA results showed that the effect of microbial inoculation on free radical scavenging activity (FRSA) of polyphenolic extract from rice plants had significant values [F(3,40) = 29.85, p < 0.0001] (Fig. 3a; Supplementary Table 3). Within the group of plants grown under non-stressed condition, the plants receiving combined inoculation of Th + Pf showed high FRSA activity [M = 56.29, SD = 2.66] as compared to control plants [M = 45.58, SD = 2.46] and single inoculations. The impact of drought on FRSA activity was significant [F(1,40) = 151.7, p < 0.0001], so was the impact of interaction [F(3,40) = 5.154, p0.0042]. Reduction in the radical cation as measured through decolorization of ABTS•+ was observed. Polyphenolic extract of rice leaves from the cohort of non-stressed plants inoculated with microbial inoculants showed high ABTS inhibition in comparison to stressed and microbe inoculated plants. Doubly inoculated plants showed high inhibition of ABTS activity [M = 37.08, SD = 5.05] than control (M = 28.79, SD = 3.84) and single Th [M = 34.86, SD = 2.98] or Pf [M = 36.31, SD = 4.65] inoculation (Fig. 3b). Two way ANOVA results showed that the effects of microbial inoculation on ABTS inhibition was significant [F(3,40) = 11.80, p < 0.0001]. The impact of watering regime was again found to be significant [F(1,40) = 96.15, p < 0.0001]. However, the impact of interaction of inoculation vs drought was statistically non-significant [F(3,40) = 0.8662, p0.4666]. Reduction of Fe3+-ferricyanide complex to Fe2+ by the plant extracts is an important assay to measure antioxidant activity in terms of reducing power (RP). Reduced RP activity was observed in the cohort of plants challenged with drought and inoculated with microbial species. Two way ANOVA results reflected that the impact of drought [F (1,40) = 639.8] and microbial inoculation [F ratio (3,40) = 61.13] was statistically significant at p < 0.0001 (Fig. 3c, Supplementary Table 3). The effect of interaction was also found significant [F(3,40) = 6.339, p 0.0013]. Within the set of non-stressed plants, doubly inoculation increased reducing power [M = 84.64, SD = 4.64] compared to single inoculation of Th [M = 64.94, SD = 3.69] and Ps [M = 64.74, SD = 9.78] and non-inoculated control [M = 51.81, SD = 2.81] (Fig. 3c).
Figure 3.
Impact of microbial inoculation on the antioxidant activity of the leaf extract of rice plants grown under non-drought and drought challenged condition. Free radical scavenging activity (a), ABTS activity (b), Reducing power assay (c) and Fe-iron chelation activity (d). The level of statistical significance was determined by two-way ANOVA; ns is non-statistical significance; n = 6; Data are mean values ± SEM; ns is non-significant.
The impact of microbial inoculation on Fe+2 chelation activity in plants was statistically significant [F(3,40) = 26.28, p < 0.0001]. The effects of drought was again found to be significant [F(1,40) = 27.63, p < 0.0001] (Fig. 3d, Supplementary Table 3). However, the impact of interaction was statistically non-significant [F(3,40) = 1.255, p0.3029]. Drought induced H2O2 production in plants has been obvious from the results (Fig. 1a) that could lead to high ROS accumulation. We presume that due to high concentration of polyphenolics in leaf extracts, rice plants show ROS scavenging strategy to neutralize the impact of oxidative toxicity. The results apparently describe that polyphenols in leaves of rice plants grown under microbial inoculation has profound non-enzymatic ROS scavenging impact. This strategy appears to be a promising stress tolerance mechanism in plants grown under drought6,9,84.
Microbial inoculation activate antioxidant defense enzymes in rice
Among the antioxidant machinery against oxidative damage, plants activate antioxidant enzymes as primary ROS scavengers. Antioxidative enzymes are ubiquitous in plant cells85 to perform defense related action under induced oxidative stress conditions86. We examined elicitation of SOD, PO, APX, catalase, GR and GPX as key inducible enzymes in drought stressed plants subsequently inoculated with microbial species.
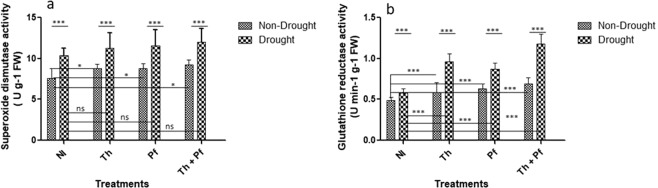
SOD with strong ROS scavenging functions catalyzes superoxide (O2−) in to O2 and H2O287. The group of enzyme copper-zinc-SOD (Cu/Zn-SOD), iron-SOD (Fe-SOD) and manganese-SOD (Mn-SOD) is compartmentalized into the cells to act against oxidative damage88. SOD helps in removing O−2 from the cells by forming O2 and H2O2 through dismutation89. Enhanced activity of the enzyme so as to discard as much O−2 formed due to oxidative condition as possible is a positive sign for cellular protection90. In the cohort of plants grown under non-drought and drought conditions separately, microbial inoculation enhanced SOD activity. Among the treatments, doubly inoculated plants showed high values of SOD activity under both the drought and non-drought plants than single inoculation (Fig. 4a). In non-drought plants with Th + Pf inoculation, the SOD activity was high [M = 9.2, SD = 0.57] than in Trichoderma [M = 8.8, SD = 0.73], Pseudomonas [M = 8.7, SD = 0.61] inoculated and non-inoculated rice leaves [M = 7.5, SD = 1.21]. The impact of drought on the SOD activity was statistically significant [F(1,40) = 52.30, p < 0.0001] (Fig. 4a, Supplementary Table 4). The effects of microbial inoculation was also significant [F(3,40) = 3.598, p0.0216]. The interaction impact was however, found to be non-significant [F(3,40) = 0.0541, p0.9832] (Supplementary Table 4). Results indicated that microbial inoculation to plants enhanced SOD activity even under drought challenged conditions. It is presumed that SOD is helpful in extending the first line of defense to the plants as they play vital role as ROS scavengers.
Figure 4.
Microbial inoculation leads to enhance superoxide dismutase (SOD) (a) and glutathione reductase (GR) (b) enzyme activity in rice leaves. n = 6; Data are mean ± SEM; The level of statistical significance was determined by two-way ANOVA; ns is non-significant.
Glutathione reductase (GR) is a potential enzyme in the antioxidative enzyme system of the plants. Two way ANOVA indicated that the effects of watering regime on GR activity in plants was significant [F(1,40) = 147.2, p < 0.0001] and so was the impact of microbial inoculation [F(3,40) = 44.07, p < 0.0001] and that of interaction [F(3,40) = 12.46, p < 0.0001) (Fig. 4b, Supplementary Table 4). In the cohort of plants challenged with drought and inoculated with the microbial inoculants, the value of GR activity was high in doubly inoculated plants [M = 1.18, SD = 0.12] than in plants with single inoculation of Th [M = 0.96, SD = 0.09] and Pf [M = 0.87, SD = 0.07]. Results indicated that microbial inoculation enhanced GR activity in drought challenged plants than in non-inoculated plants grown under drought. Glutathione, a tripeptide is abundant in cellular components and is widely involved in cell growth and regulation of gene expression linked with stress responses91. The enzyme replenishes cellular pool of glutathione that has a reductant role against detrimental ROS83. Multifold increase in GR activity in rice leaves following microbial inoculation, even under drought stress indicated for a defense support to the plants under microbial inoculation.
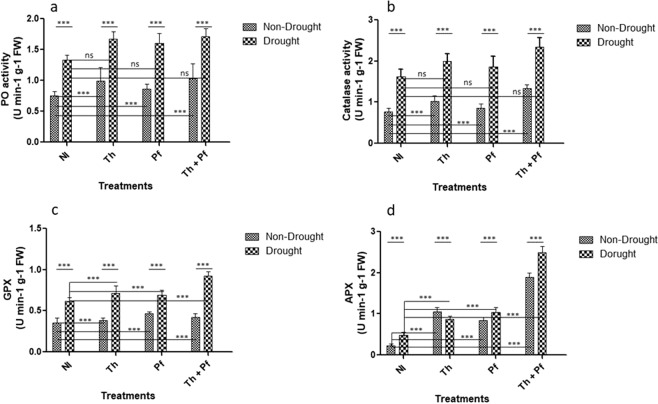
The enzyme activity of peroxidase (PO) indicates tolerance in plants against water stress92. We demonstrated that in the cohort of non-drought plants, microbial inoculation led to enhance peroxidase activity in rice leaves and maximum activity was found due to doubly inoculation of Th + Pf. Within the cohort of inoculated plants challenged with the drought, again doubly inoculation of Th + Pf showed maximum PO activity than single inoculation or drought plants alone (Fig. 5a). The effects of drought on PO activity was found to be significant [F(3,40) = 239.6, p < 0.0001]. The effects of microbial inoculation was also significant [F(3,40) = 11.96, p < 0.0001] but that of interaction was non-significant [F(3,40) = 0.6073, p0.6141] (Fig. 5a; Supplementary Table 5).
Figure 5.
Impact of microbial inoculation and drought condition on peroxidase (a), catalase (b), guaiacol peroxidase (c) and ascorbate peroxidase (d) enzyme activity in rice plants. Significance level was determined by two-way ANOVA; n = 6; Data are mean ± SEM; ns is non-significant.
Catalase possesses high affinity for H2O2 and catalyzes its dismutation into H2O and O27,92. In the cohort of stressed plants, plants doubly inoculated with Th + Pf had high catalase activity than single inoculations. Likewise, within the cohorts of inoculated non-stressed plants, double inoculation again led to high catalase activity (Fig. 5b). The impact of watering regime on catalase activity was significant [F(1,40) = 379.9, p < 0.0001] and so was the significant impact of microbial inoculation in plants [F(3,140) = 30.42, p < 0.0001]. However, the impact of interaction on catalase activity was found to be non-significant [F(3,40) = 0.6272, p0.6017] (Fig. 5b, Supplementary Table 5).
GPX reduces the level of H2O2 in the cells during stress conditions93,94. We showed that in the cohort of drought stressed plants, those inoculated with Th + Pf showed high GPX activity than those with single microbial inoculations (Fig. 5c). The impact of watering regime on GPX activity was significant [F(1,40)) = 423.7, p < 0.0001]. Similarly, microbial inoculation further showed significant effects on GPX activity [F(3,40) = 23.26, p < 0.0001] and so was the impact of interaction [F(3,40) = 15.40, p < 0.0001] (Supplementary Table 5).
The enzyme ascorbate peroxidase (APX) having great affinity for H2O2 reduces hydrogen peroxide to water molecules in chloroplasts, cytosol and mitochondria92. Under drought stress conditions, plants inoculated with Th + Pf showed maximum APX activity [M = 2.48, SD = 0.14] than the single inoculation of Th [M = 0.87, SD = 0.06] and Pf [M = 1.03, SD = 0.11] (Fig. 5d). The impact of inoculation of the inoculants on APX activity was statistically significant [F(3,40) = 776.9, p < 0.0001] in two way ANOVA. The effects of drought on APX activity was also significant [F(1,40) = 59.45, p < 0.0001] and so was the interaction impact on APX [F(3,40) = 33.47, p < 0.0001] (Supplementary Table 5).
Enhanced level of defense related enzymes is directly related to the degree of drought experienced by the plants95. Cell-bound peroxidases act as detoxifier of H2O2 produced as a byproduct of antioxidative mechanism9. The PO acts in H2O2-scaveginging and oxidize flavonoid and phenylpropanoids72. APX also performs H2O2 scavenging in the cytosol and chloroplast with the help of ascorbate as specific electron donor91. Thus, higher activity of both PO and APX is presumed to have a role in detoxification of enhanced H2O2 accumulation in the cells. Enhanced level of GPX and catalase is supposed to support plant’s biochemical strategy to mitigate drought under microbial inoculation. The enhanced activity of PO, APX, GPX and CAT enzymes in different cohorts of experiments led us to affirm the role of i) the enzyme activation and activity in imparting protection against stresses and ii) the microbial species in modulating enzyme activity in plants challenged with drought. Enhanced level of the defense related enzymes due to microbial inoculation go in parallel to different molecular mechanisms and strengthen the plant’s performance under stressed conditions.
PCA analysis showed the effect of drought (red colored) and non-drought (normal) (indigo colored) plants in two subgroups. The effect of drought was significant on CAT, PO, SOD, GR, GPX. The quantitative level of these biochemical products was found enhanced in inoculated plants growing under drought condition than in normal irrigated plants. Total polyphenolics concentration (TPC), protein, H2O2, Fe-Chelation, ABTS and DPPH were also high in drought challenged plants than in normal irrigated plants. Apart from PAL activity, APX and RP were high in normal irrigated plants than drought challenged plants. Co-inoculation of Trichoderma and Pseudomonas improved activity of PAL, APX and RP. The activity of CAT, SOD and PO were enhanced in Trichoderma inoculated drought challenged plants. The other antioxidant tests such as DPPH, ABTS, GR, and iron-chelation activity were high in co-inoculation of Trichoderma and Pseudomonas inoculated and drought challenged plants (Fig. 6).
Figure 6.
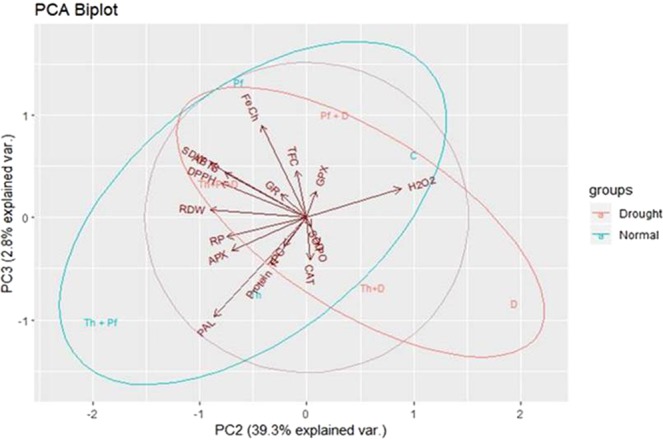
PCA analysis of enzymatic assays and plant biomass.
Microbial inoculation up-regulates the genes encoding dehydration tolerance
We analysed gene expression of OsPIP1;1, a prominent representative of rice plasma-membrane protein gene family that regulates aquaporin96. The impact of inoculation on the expression of OsPIP1;1 was statistically significant [F(3,16) = 12.34, p0.0002] but that of drought was non-significant [F(1,16) = 0.1953, p0.6644] (Fig. 7a, Supplementary Table 6). The interaction effect on the expression of this gene was statistically significant [F(3,16) = 6.054, p0.0059]. Microbial inoculation therefore, up-regulated OsPIP1;1 of the PIP gene family in both the cohorts of stressed and non-stressed plants. OsPIP1;1 is an important gene, the protein product of which is related to less water permeability in the plant cells97. We showed that microbial inoculation in plants growing normally (non-stress) led to up-regulation of OsPIP1;1 gene. Within the cohort of stressed plants, maximum upregulation was observed in plants inoculated with Pf alone (Fig. 7a). The results indicate positive role of microbial inoculation in the modulation of OsPIP1 gene, which regulates aquaporin, the water channel protein that mediates stress tolerance in rice plants.
Figure 7.
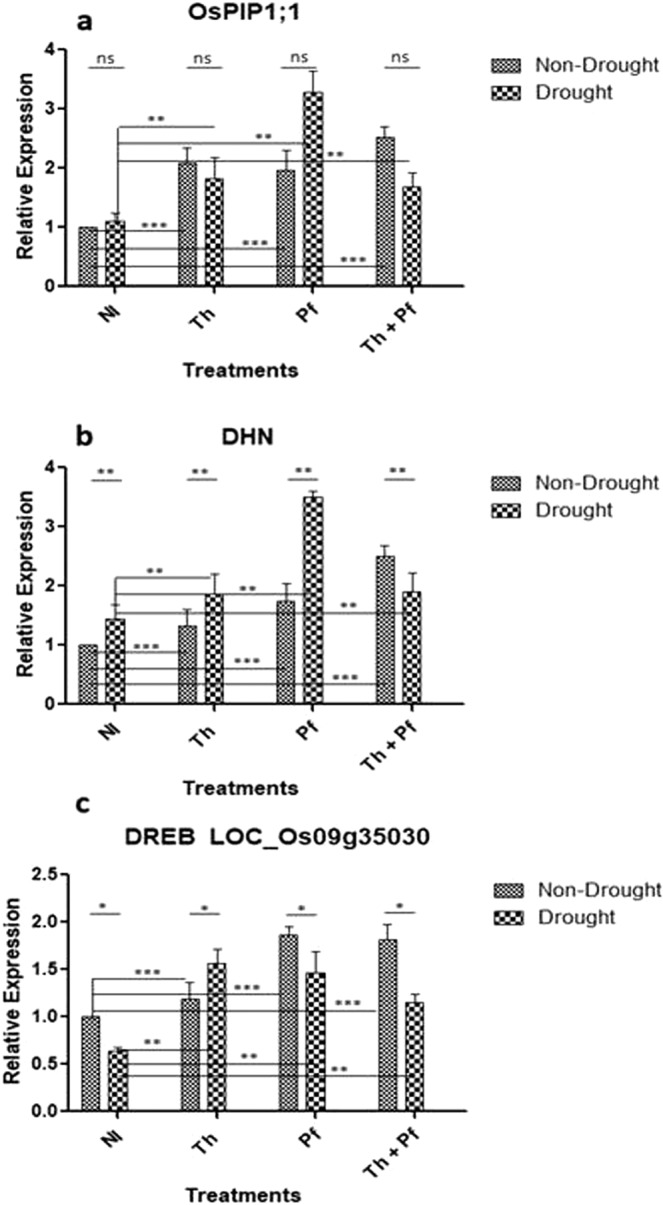
Effect of microbial inoculation and drought stress on the expression of OsPIP1(a), DHN (b) and DREB (c) genes related to less water permeability and dehydration tolerance in rice. Significance values were determined using two-way ANOVA. n = 3; Data are shown as mean ± SEM for each sample; ns is non-significant.
Dehydrins (DHNs) play key role in responding to adaptation against abiotic stresses98. Microbial inoculation up-regulated the expression of DHN gene in rice plants grown under stressed and non-stressed conditions both (Fig. 7b). In the cohort of stressed plants, inoculation of Pf showed maximum DHN expression. Results of the two-way ANOVA for DHN gene expression indicated significant effects of watering regime [F(1,16) = 9.408, p0.0074] and microbial inoculation [F(3,16) = 13, p0.0001]. Interaction also had significant impact on DHN gene expression [F(3,16) = 7.799, p0.002] (Fig. 7b, Supplementary Table 6). In the dehydrating plant cells due to prolonged drought, over-expression of dehydrin genes in the cytoplasm and vicinity of plasma membrane is an important phenomenon28,99,100. The up-regulation of the gene protects structural and functional enzymes, proteins and nucleic acids during oxidative damage101,102 and enhances efficiency of crop plants against abiotic stresses103. The microbial inoculation was shown to facilitate high expression of DHN gene in rice to offer protection to vegetative tissues against dehydration and desiccation under challenged osmotic stress.
Dehydration responsive element binding (DREB) transcription factors improve abiotic stress tolerance in plants through regulation of stress-inducible gene expression98,100,104. The impact of microbial inoculation in rice plants for the expression of DREB gene was significant [F(3,16) = 14.71, p < 0.0001]. The effects of drought [F(1,16) = 7.527, p0.0144) and interaction [F(3,16) = 5.383, p0.0094) was also significant (Fig. 7c, Supplementary Table 6). These observations, together with the enzyme activity provided evidences to confirm that microbial inoculation modulates expression of stress responsive genes linked with dehydration. This further makes a clearer understanding on the activation of strategic molecular mechanisms meant for avoidance or adaptation against stress damage in rice due to microbial inoculation.
Inoculants improved expression of genes encoding dismutation of superoxide radicals
In plants, SOD constitutes the first line of defense against ROS-induced damage82. To gain insight into the expression of SOD gene group CuZn-SOD (localized to chloroplasts and cytosol), Mn-SOD (bound to mitochondria) and Fe-SOD (localized to chloroplast), their expression in rice grown under drought following microbial inoculation was assessed. On CuZn-SOD, the impact of all the three experimental factors, viz. drought [F(1,16) = 35.57, p < 0.0001], microbial inoculation [F(3,16) = 62.73, p < 0.0001) and interaction [F(3,16) = 67.17, p < 0.0001] was significant (Fig. 8a, Supplementary Table 7). On Fe-SOD gene also, the impact of microbial inoculation [F(1,16) = 32.01, p < 0.0001], watering regime [F(3,16) = 31.35, p < 0.0001] and interaction [F(3,16) = 28.02., p < 0.0001] was significant (Fig. 8b, Supplementary Table 7). Like CuZn- and Fe-SOD, the effects of drought [F(1,16) = 61.24], microbial inoculation [F(3,16) = 59.83] and interaction factor [F(3,16) = 52.86] on the expression of Mn-SOD1 was also significant at p < 0.0001 (Fig. 8c). It was interesting that within the cohort of the three treatments of microbial inoculation in plants growing under stressed condition, inoculation of Pf bacteria showed high upregulation values for all the three genes (Fig. 8). Except for the DREB gene which showed maximum over expression in the cohort of non-stressed plants inoculated with Pf (Fig. 7c), inoculation of plants with the bacteria Pseudomonas alone showed consistently high expression values of OsPIP1;1, DHN and all the three isomorphs of SOD genes in the cohort of stressed plants (Figs. 7a,b and 8a–c). It was concluded that the over-expression of SOD gene isoforms leads to enhanced activity of SOD enzyme in rice plants grown under microbial inoculation and drought challenged condition. Presumably, the enhanced gene expression and subsequent enzyme activity level might have played an important role in reducing the deleterious impact of ROS in rice grown under stress.
Figure 8.
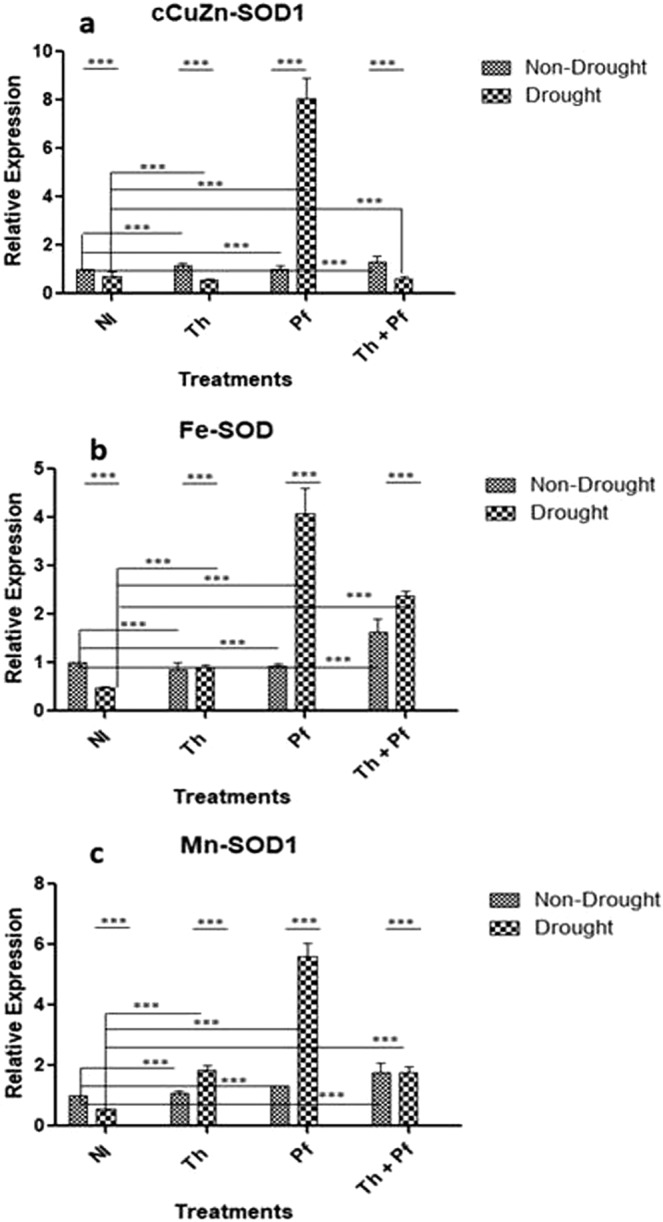
Effect of microbial inoculation on the expression of CuZn-SOD (a), Fe-SOD (b) and Mn-SOD (c) genes in the leaves of rice plants grown under drought and non-drought condition. Data are represented as mean ± SEM for each sample; Two-way ANOVA was performed to determine statistical significance; n = 3 for transcript analysis.
Microbial inoculation enhanced expression of genes encoding peroxidation of H2O2
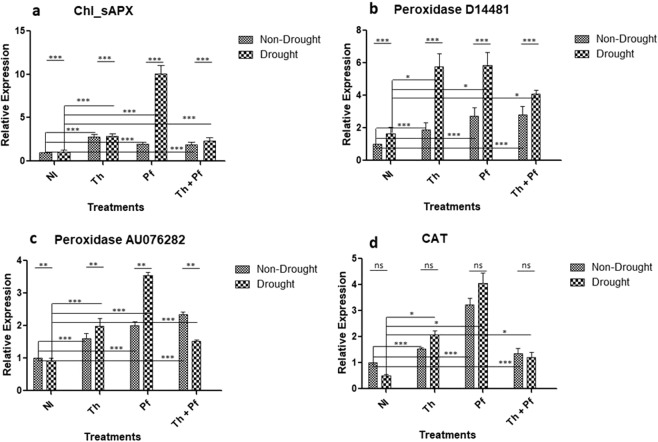
APX gene regulates ascorbate-glutathione (AsA-GSH) cycle that plays key role in the reduction of H2O2 to H2O105,106. Over-expression of APX gene in plants improves oxidative defense and offers tolerance to abiotic stress105. In the cohort of plants grown with stress and inoculated with Th, Pf and Th + Pf, single inoculation of Pf showed high overexpression of APX gene than Th or combined inoculation of Th + Pf (Fig. 9a). The impact of drought [F(1,16) = 46.30], microbial inoculation [F(3,16) = 45.21], interaction [F(3,16) = 38.55] was significant at p < 0.0001 (Fig. 9a, Supplementary Table 8). The bacterial inoculant Pf showed maximum over-expression of APX gene in plants under drought condition than Th or doubly inoculation of Th + Pf. Inoculating plants with microbial inoculants enhanced expression of the peroxidase (PO) genes (PO D14481 and PO AU076282) in rice. Within the cohort of stressed and non-stressed plants, inoculation resulted in enhanced over-expression than the control. Maximum over-expression was again recorded in plants grown under stressed conditions and inoculated with Pf (Fig. 9b,c). The impact of watering regime on the expression of the gene PO D14481 was significant [F(1,16) = 35.54, p < 0.0001] and similar was the effect of microbial inoculation [F(3,16) = 12.27, p0.0002]. The interaction impact on expression of this gene was also significant [F(3,16) = 4.150, p0.0236] (Fig. 9b; Supplementary Table 8). Likewise, the impact of drought [F(1,16) = 8.962, p0.0086], microbial inoculation [F(3,16) = 73.23, p < 0.0001] and interaction of drought and inoculation factor [F (3,16) = 32.28, p < 0.0001] on another peroxidase gene PO AU076282 was significant (Fig. 9c, Supplementary Table 8). The effect of the inoculation of bacterial inoculant Pf on the expression of PO AU076282 gene in plants grown under drought was maximum than Th or doubly inoculation of Th + Pf. Results indicated that microbial inoculation helped rice plants in over-expressing peroxidases and the inoculation of Pseudomonas was invariably instrumental in highest over-expression of these genes. Peroxidases are the key genes in regulating ROS scavenging and thus, their over-expression in rice can have protective role in plants exposed to drought.
Figure 9.
Impact of microbial inoculation on the expression of Chl_sAPX (a), peroxidase D14481 (b) peroxidase AU076282 (c) and Catalase (d) genes in rice plants grown under drought and non-drought conditions. Statistical significance was determined by two-way ANOVA; data are mean ± SEM; n = 3 for transcript analysis. ns is non-significant.
Over-expression of CAT gene enhances oxidative defense response in plants107. Inoculation of rice grown under drought condition with Pf resulted in highest level of expression of CAT gene in the cohort of drought stressed and inoculated plants (Fig. 9d). The impact of drought on the gene expression was non-significant [F(1,16) = 1.898, p0.1873]. The effects of microbial inoculation [F(3,16) = 81.48, p < 0.0001] and interaction (drought vs. inoculation) [F(3,16) = 4.739, p0.0150] were significant (Fig. 9d; Supplementary Table 8). The results strongly suggested that microbial inoculation had a positive role in the over-expression of the genes linked with the peroxidation of H2O2 in the plants challenged with the drought. Invariably, the effect of inoculation of Pseudomonas substantially enhanced APX, PO and CAT gene expression in plants grown under stressed condition. These modulations in gene expression may support improved drought tolerance in rice plants.
Conclusion
We have shown that although drought suppressed growth of rice plants, as is evident from reduced shoot and root weight, microbial inoculation managed to reduce the impact of drought. There have been multi-pronged mechanisms utilized and adopted by the plants to mitigate and/or minimize the impact of drought if the plants were inoculated with the microbial species. Generation of ROS is a common phenomenon in plant cells under stressed conditions. We reported hyperaccumulation of H2O2 in rice leaves and the resultant hypersensitive cell death responses thereafter. Induced accumulation of the PAL gene transcripts and resultant activation of PAL enzyme facilitated higher accumulation of the phenylpropanoids that have strong ROS scavenging activity and might have helped plants to overcome oxidative burden created due to drought stress.With the activation of the antioxidant enzymes SOD, PO, APX and CAT, rice plants were supposed to minimize tissue damaging impact of high H2O2 levels. Over expression of all the isoforms of SOD, Cu-Zn SOD, Mn-SOD and Fe-SOD genes suggested that microbial inoculation helped plants activate SOD activity as first line of defence at various levels of cellular compartments strongly to overcome ROS burden. Microbial inoculation in plants further improved the activity of the enzymes PO, APX, GPX and GR that have also contributed in reducing ROS burden in the plants following drought challenge. We also observed enhanced regulation of less-water permeability-linked gene, OSPiP1 that regulates aquaporin, drought-adaptation gene DHN and dehydration related DREB gene. Presumably, up-regulation of genes encoding phenylpropanoids, dismutation of superoxide radicals and peroxidation of H2O2 in microbe inoculated and drought challenged condition strongly contributed towards stress mitigation. Enhanced enzymatic and non-enzymatic antioxidant activities were thought to be the repercussions of the enhanced gene expression levels in microbial inoculated plants and have also helped in minimizing antioxidative load to overcome the oxidative stress. We further conclude that the physiological, biochemical and molecular mechanisms contributing to drought mitigation in rice following microbial interaction are multi-faceted, multi-channeled and interlinked. Results have shown that microbial inoculants have succeeded in improving intrinsic physiological and molecular capabilities of the plants mostly by reducing the damaging impact of the ROS, which was managed at multiple layers. Therefore, microbial inoculation could find an essential place in raising crops under abiotic stress conditions.
Supplementary information
Acknowledgements
DPS is thankful to Indian Council of Agricultural Research (ICAR), India for Institutional support and funding under Centre for Agricultural Bioinformatics (CABin) Network Project. Help rendered by Ms. Amrita Gupta, Dr. Pramod K. Sahu, ICAR-NBAIM, Mau and Dr. Abhishek Singh, Department of Agricultural Statistics, Institute of Agricultural Sciences, Banaras Hindu University, Varanasi is duly acknowledged.
Author contributions
D.P.S. designed the experiments and with R.P. and R.S. was involved in data analysis, interpretation and M.S. writing. V.S. and J.S.P. conducted the experiment. V.K.G. and B.K.S. reviewed the M.S. and helped in M.S. writing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-61140-w.
References
- 1.Law DR, Crafts-Brandner SJ, Salvucci ME. Heat stress induces the synthesis of a new form of ribulose-1, 5-bisphosphate carboxylase/oxygenase activase in cotton leaves. Planta. 2001;214:117–125. doi: 10.1007/s004250100592. [DOI] [PubMed] [Google Scholar]
- 2.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant. Physiol. Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Tripathy BC, Oelmüller R. Reactive oxygen species generation and signaling in plants. Plant. Signal. Behav. 2012;7:1621–1633. doi: 10.4161/psb.22455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weydert CJ, Cullen JJ. Measurement of superoxide dismutase, catalase, and glutathione peroxidase in cultured cells and tissues. Nat. Protoc. 2010;5:51–66. doi: 10.1038/nprot.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain S, Khan F, Cao W, Wu L, Geng M. Seed priming alters the production and detoxification of reactive oxygen intermediates in rice seedlings grown under sub-optimal temperature and nutrient supply. Front. Plant. Sci. 2016;7:439. doi: 10.3389/fpls.2016.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.You J, Chan Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant. Sci. 2015;6:1092. doi: 10.3389/fpls.2015.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant. Sci. 2002;7:405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 8.Mittler R. ROS are good. Trends Plant. Sci. 2017;22:11–19. doi: 10.1016/j.tplants.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Sharma P, Jha AB, Dubey RS, Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012;2012:26. [Google Scholar]
- 10.Nisar N, Li L, Lu S, Khin NC, Pogson BJ. Carotenoid metabolism in plants. Mol. Plants. 2015;8:68–82. doi: 10.1016/j.molp.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Korkina LG. Phenylpropanoids as naturally occurring antioxidants: from plant defense to human health. Cell Mol. Biol. 2007;53:15–25. [PubMed] [Google Scholar]
- 12.Lamaoui M, Jemo M, Datla R, Bekkaoui F. Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front. Chem. 2018;6:26. doi: 10.3389/fchem.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gairola S, Al Shaer KI, Al Harthi EK, Mosa KA. Strengthening desert plant biotechnology research in the United Arab Emirates: a viewpoint. Physiol. Mol. Biol. Plants. 2018;24:521–533. doi: 10.1007/s12298-018-0551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rolli E, et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 2014;17:316–331. doi: 10.1111/1462-2920.12439. [DOI] [PubMed] [Google Scholar]
- 15.Farrar K, Bryant D, Cope-Selby N. Understanding and engineering beneficial plant–microbe interactions: plant growth promotion in energy crops. Plant. Biotechnol. J. 2014;12:1193–206. doi: 10.1111/pbi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A, Verma JP. Does plant-microbe interaction confer stress tolerance in plants: A review? Microbiological Res. 2018;207:41–52. doi: 10.1016/j.micres.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt R, et al. Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 2014;5:64. doi: 10.3389/fmicb.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahu Pramod K., Singh Dhananjaya P., Prabha Ratna, Meena Kamlesh K., Abhilash P.C. Connecting microbial capabilities with the soil and plant health: Options for agricultural sustainability. Ecological Indicators. 2019;105:601–612. doi: 10.1016/j.ecolind.2018.05.084. [DOI] [Google Scholar]
- 19.Armada E, Roldan A, Azcon R. Differential activity of autochthonous bacteria in controlling drought stress in native Lavandula and Salvia plants species under drought conditions in natural arid soil. Microb. Ecol. 2014;67:410–420. doi: 10.1007/s00248-013-0326-9. [DOI] [PubMed] [Google Scholar]
- 20.Cherif H, et al. Oasis desert farming selects environment-specific date palm root endophytic communities and cultivable bacteria that promote resistance to drought. Environ. Microbiol. Rep. 2015;7:668–678. doi: 10.1111/1758-2229.12304. [DOI] [PubMed] [Google Scholar]
- 21.de Zelicourt A, Al-Yousif M, Hirt H. Rhizosphere microbes as essential partners for plant stress tolerance. Mol. Plant. 2013;6:242–245. doi: 10.1093/mp/sst028. [DOI] [PubMed] [Google Scholar]
- 22.Kakar KU, et al. A consortium of rhizobacterial strains and biochemical growth elicitors improve cold and drought stress tolerance in rice (Oryza sativa L.) Plant. Biol. 2016;18:471–83. doi: 10.1111/plb.12427. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Kloepper JW, Ryu CM. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant. Sci. 2009;14:1–4. doi: 10.1016/j.tplants.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Timmusk S, et al. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: Enhanced biomass production and reduced emissions of stress volatiles. PLoS One. 2014;9:e96086. doi: 10.1371/journal.pone.0096086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vurukonda SS, Vardharajula S, Shrivastava M, SkZ A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiological Res. 2016;184:13–24. doi: 10.1016/j.micres.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Ngumbi E, Kloepper J. Bacterial-mediated drought tolerance: Current and future prospects. Appl. Soil. Ecol. 2016;105:109–125. doi: 10.1016/j.apsoil.2016.04.009. [DOI] [Google Scholar]
- 27.Brotman Y, Kapuganti JG, Viterbo A. Trichoderma. Curr. Biol. 2010;20:R390–391. doi: 10.1016/j.cub.2010.02.042. [DOI] [PubMed] [Google Scholar]
- 28.Pandey V, et al. Dose-dependent response of Trichoderma harzianum in improving drought tolerance in rice genotypes. Planta. 2016;243:1251–64. doi: 10.1007/s00425-016-2482-x. [DOI] [PubMed] [Google Scholar]
- 29.Mastouri F, Bjorkman T, Harman GE. Trichoderma harzianum enhances antioxidant defense of tomato seedlings and resistance to water deficit. Mol. Plant. Microbe Interact. 2012;25:1264–1271. doi: 10.1094/MPMI-09-11-0240. [DOI] [PubMed] [Google Scholar]
- 30.Brotman Y, et al. Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLOS Pathog. 2013;9:e1003221. doi: 10.1371/journal.ppat.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meena KK, et al. Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front. Plant. Sci. 2017;8:172. doi: 10.3389/fpls.2017.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki T, Burr B. International Rice genome sequencing project: the effort to completely sequence the rice genome. Curr. Opin. Plant. Biol. 2002;3:138–142. doi: 10.1016/S1369-5266(99)00047-3. [DOI] [PubMed] [Google Scholar]
- 33.Zu X, et al. A new method for evaluating the drought tolerance of upland rice cultivars. Crop. J. 2017;5:488–498. doi: 10.1016/j.cj.2017.05.002. [DOI] [Google Scholar]
- 34.Hu HH, Xiong LZ. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant. Biol. 2014;65:715–741. doi: 10.1146/annurev-arplant-050213-040000. [DOI] [PubMed] [Google Scholar]
- 35.Vikram P, et al. Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Sci. Rep. 2015;2015:14799. doi: 10.1038/srep14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berendsen RL, Pieterse CMJ, Bakker P. The rhizosphere microbiome and plant health. Trends Plant. Sci. 2012;17:478–86. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 38.Breidenbach B, Pump J, Dumont MG. Microbial Community Structure in the Rhizosphere of Rice Plants. Front. Microbiol. 2016;6:1537. doi: 10.3389/fmicb.2015.01537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu T, et al. Investigation of Rhizospheric Microbial Communities in Wheat, Barley, and Two Rice Varieties at the Seedling Stage. J. Agric. Food Chem. 2018;66:2645–2653. doi: 10.1021/acs.jafc.7b06155. [DOI] [PubMed] [Google Scholar]
- 40.Nihorimbere V, Ongena M, Smargiassi M, Thonart P. Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol. Agron. Soc. Env. 2011;15:327–337. [Google Scholar]
- 41.Patel JS, Kharwar RN, Singh HB, Sarma BK. Trichoderma asperellum (T42) and Pseudomonas fluorescens (OKC) enhances resistance in pea against Erysiphe pisi through enhanced ROS generation and lignification. Front. Microbiology. 2017;8:306. doi: 10.3389/fmicb.2017.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowry OH, Rosbrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265. [PubMed] [Google Scholar]
- 43.Velikova V, Yordanov I, Edreva A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: protective role of exogenous polyamines. Plant. Sci. 2000;151:59–66. doi: 10.1016/S0168-9452(99)00197-1. [DOI] [Google Scholar]
- 44.NV P, PA V, Vemanna RS, MS S, Makarla U. Quantification of Membrane Damage/Cell Death Using Evan’s Blue Staining Technique. Bio-protocol. 2017;7:e2519. doi: 10.21769/BioProtoc.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Z, Shetty K. Solid-state bioconversion of phenolics from cranberry pomace and role of Lentinus edodes beta-glucosidase. J. Agric. Food Chem. 2000;48:895–900. doi: 10.1021/jf990972u. [DOI] [PubMed] [Google Scholar]
- 46.Fridovich I. Superoxide dismutases. Adv. Enzymol. Relat. Areas Mol. Biol. 1974;41:35–97. doi: 10.1002/9780470122860.ch2. [DOI] [PubMed] [Google Scholar]
- 47.Hammerschmidt R, Nuckles EM, Kuc J. Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiol. Plant. Pathol. 1982;20:73–82. doi: 10.1016/0048-4059(82)90025-X. [DOI] [Google Scholar]
- 48.Nakano Y, Asada K. Hydrogen Peroxide Is Scavenged by Ascorbate-Specific Peroxidase in Spinach Chloroplasts. Plant. Cell Physiol. 1981;22:867–880. [Google Scholar]
- 49.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–6. doi: 10.1016/S0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 50.Anderson, M. Glutathione in Free Radicals, A Practical Approach, ed. N.A. Punchard and F.J. Kelly Oxford University Press, New York, p 213 (1996).
- 51.Zheng X, Van Huystee. RB. Peroxidase-regulated elongation of segments from peanut hypocotyls. Plant. Sci. 1992;81:47–56. doi: 10.1016/0168-9452(92)90023-F. [DOI] [Google Scholar]
- 52.Bruske CH. Phenylalanine ammonia lyase activity in tomato roots infected and resistant to the root-knot nematode (Meloidogyne incognita) Physiol. Pl. Path. 1980;16:409–414. doi: 10.1016/S0048-4059(80)80012-9. [DOI] [Google Scholar]
- 53.Yen GC, Duh PD. Scavenging effect of methanolic extract of peanut hulls on free radical and active oxygen species. J. Agric. Food Chem. 1994;42:629–632. doi: 10.1021/jf00039a005. [DOI] [Google Scholar]
- 54.Re R, et al. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free. Radic. Biol. Med. 1999;26:1231–1237. doi: 10.1016/S0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- 55.Apati P, et al. Herbal remedies of Solidago, correlation of phytochemical characteristics and antioxidative properties. J. Pharm. Biomed. Anal. 2003;32:1045–1053. doi: 10.1016/S0731-7085(03)00207-3. [DOI] [PubMed] [Google Scholar]
- 56.Decker EA, Welch B. Role of ferritin as a lipid oxidation catalyst in muscle food. J. Agric. Food Chem. 1990;38:674–678. doi: 10.1021/jf00093a019. [DOI] [Google Scholar]
- 57.Quyang S, et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acid. Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J-H, Chung BY, Kim J-S, Wi SG. Construction of gene-specific primers for various antioxidant isoenzyme genes and their expressions in rice (Oryza sativa L.) seedlings obtained from gamma-irradiated seeds. J. Photoscience. 2004;11:115–120. [Google Scholar]
- 59.Lata C, Prasad M. Role of DREB in regulation of abiotic stress response in plants. J. Exp. Botany. 2011;62:4731–48. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- 60.Bartels D, Sunkar R. Drought and salt stress in plants. Crit. Rev. Plant. Sci. 2005;21:1–36. [Google Scholar]
- 61.Bray, E.A., Bailey-Serres, J., Weretilnyl, E. Responses to abiotic stresses: In: Buchman B.B., Gruissem, W. Jones R. L. eds. Biochemistry and molecular biology of plants. Rockville, M.D. Am Soc Pl Biologists, 1158–1203 (2000).
- 62.Rais A, Jabeen Z, Shair F, Hafeez FY, Hassan MN. Bacillus spp., a bio-control agent enhances the activity of antioxidant defense enzymes in rice against Pyricularia oryzae. PLoS ONE. 2017;12:e0187412. doi: 10.1371/journal.pone.0187412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sahebi Mahbod, Hanafi Mohamed M., Rafii M. Y., Mahmud T. M. M., Azizi Parisa, Osman Mohamad, Abiri Rambod, Taheri Sima, Kalhori Nahid, Shabanimofrad M., Miah Gous, Atabaki Narges. Improvement of Drought Tolerance in Rice (Oryza sativa L.): Genetics, Genomic Tools, and the WRKY Gene Family. BioMed Research International. 2018;2018:1–20. doi: 10.1155/2018/3158474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lafitte HR, Yongsheng G, Yan S, Lil ZK. Whole plant responses, key processes, and adaptation to drought stress: the case of rice. J. Exp. Bot. 2007;58:169–175. doi: 10.1093/jxb/erl101. [DOI] [PubMed] [Google Scholar]
- 65.Montillet J-L, et al. Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant. Physiol. 2005;138:1516–1526. doi: 10.1104/pp.105.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant. Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Breusegem F, Dat JF. Reactive Oxygen Species in Plant Cell Death. Plant. Physiol. 2006;141:384–390. doi: 10.1104/pp.106.078295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ali Q, Anwar F, Ashraf M, Saari N, Perveen R. Ameliorating effects of exogenously applied proline on seed composition, seed oil quality and oil antioxidant activity of maize (Zea mays L.) under drought stress. Int. J. Mol. Sci. 2013;14:818–835. doi: 10.3390/ijms14010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emam MM, Khattab HE, Helal NM, Deraz AE. Effect of selenium and silicon on yield quality of rice plant grown under drought stress. Aust. J. Crop. Sci. 2014;8:596–605. [Google Scholar]
- 70.Huang J, et al. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant. Physiol. 2010;153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ksouri R, et al. Salinity effects on polyphenol content and antioxidant activities in leaves of the halophyte Cakile maritima. Plant. Physiol. Biochem. 2007;45:244–249. doi: 10.1016/j.plaphy.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 72.Janas KM, Amarowicz R, Zielińska-Tomaszewska J, Kosińska A, Posmyk MM. Induction of phenolic compounds in two dark-grown lentil cultivars with different tolerance to copper ions. Acta Physiologiae Plant. 2009;31:587–595. doi: 10.1007/s11738-008-0268-x. [DOI] [Google Scholar]
- 73.Wada Kaede C., Mizuuchi Kaori, Koshio Aya, Kaneko Kentaro, Mitsui Toshiaki, Takeno Kiyotoshi. Stress enhances the gene expression and enzyme activity of phenylalanine ammonia-lyase and the endogenous content of salicylic acid to induce flowering in pharbitis. Journal of Plant Physiology. 2014;171(11):895–902. doi: 10.1016/j.jplph.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 74.Meena B, et al. Induction of pathogenesis-related proteins, phenolics and phenylalanine ammonia-lyase in groundnut by Pseudomonas fluorescens. JSTOR. 2000;107:514–527. [Google Scholar]
- 75.Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 76.Ramakrishna A, Ravishankar GA. Influence of abiotic stress signals on secondary metabolites in plants. Plant. Signal. Behav. 2011;6:1720–1731. doi: 10.4161/psb.6.11.17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dat J, et al. Dual action of the active oxygen species during plant stress responses. Cel. Mol. Life Sci. 2000;57:779–795. doi: 10.1007/s000180050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noctor G, Mhamdi A, Foyer CH. The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant. Physiol. 2014;164:1636–48. doi: 10.1104/pp.113.233478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Carvalho MHC. Drought stress and reactive oxygen species Production, scavenging and signaling. Plant. Sig Behav. 2008;3:156–165. doi: 10.4161/psb.3.3.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vranova E, Inzé D, Van Breusegem F. Signal transduction during oxidative stress. J. Exp. Bot. 2002;53:1227–1236. doi: 10.1093/jxb/53.372.1227. [DOI] [PubMed] [Google Scholar]
- 81.Kar RK. Plant responses to water stress: role of reactive oxygen species. Plant. Signal. Behav. 2011;6:1741–5. doi: 10.4161/psb.6.11.17729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Das K, Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Env. Sci. 2014;2:53. doi: 10.3389/fenvs.2014.00053. [DOI] [Google Scholar]
- 83.Roychoudhury A, Basu S, Sengupta DN. Antioxidants and stress-related metabolites in the seedlings of two indica rice varieties exposed to cadmium chloride toxicity. Acta Physiol. Plant. 2012;34:835–847. doi: 10.1007/s11738-011-0881-y. [DOI] [Google Scholar]
- 84.Iba K. Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu. Rev. Plant. Biol. 2002;53:225–45. doi: 10.1146/annurev.arplant.53.100201.160729. [DOI] [PubMed] [Google Scholar]
- 85.Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: an overview. Environ. Exp. Botany. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- 86.Perry JJ, Shin DS, Getzoff ED, Tainer JA. The structural biochemistry of the superoxide dismutases. Biochimica Biophysica Acta. 2010;1804:245–62. doi: 10.1016/j.bbapap.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Botany. 2002;53:1331–41. doi: 10.1093/jexbot/53.372.1331. [DOI] [PubMed] [Google Scholar]
- 88.Apel, K. & Hirt, H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol.55, 373–399 (2004). [DOI] [PubMed]
- 89.Boguszewska D, Grudkowska M, Zagdańska B. Drought-responsive antioxidant enzymes in potato (Solanum tuberosum L.) Potato Res. 2010;53:373–382. doi: 10.1007/s11540-010-9178-6. [DOI] [Google Scholar]
- 90.Mullineaux PM, Rausch T. Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth. Res. 2005;86:459–74. doi: 10.1007/s11120-005-8811-8. [DOI] [PubMed] [Google Scholar]
- 91.Sofo A, Scopa A, Nuzzaci M, Vitti A. Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int. J. mol. Sci. 2015;16:13561–13578. doi: 10.3390/ijms160613561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Caverzan A, et al. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012;35:1011–1019. doi: 10.1590/S1415-47572012000600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Asada K. The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- 94.Basu S, Roychoudhury A, Saha PP, Sengupta DN. Differential antioxidative responses of indica rice cultivars to drought stress. Plant. Growth Regul. 2010;60:51–59. doi: 10.1007/s10725-009-9418-4. [DOI] [Google Scholar]
- 95.Pinheiro C, Chaves MM. Photosynthesis and drought: Can we make metabolic connections from available data? J. Exp. Bot. 2011;62:869–882. doi: 10.1093/jxb/erq340. [DOI] [PubMed] [Google Scholar]
- 96.Guo L, et al. Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res. 2006;16:277–286. doi: 10.1038/sj.cr.7310035. [DOI] [PubMed] [Google Scholar]
- 97.Matsumoto T, et al. Role of the aquaporin PIP1 subfamily in the chilling tolerance of rice. Plant. Cell Physiol. 2009;50:216–29. doi: 10.1093/pcp/pcn190. [DOI] [PubMed] [Google Scholar]
- 98.Hanin M, et al. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant. Sign Behav. 2011;6:1503–1509. doi: 10.4161/psb.6.10.17088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamaguchi-Shinozaki Kazuko, Shinozaki Kazuo. Molecular Breeding of Forage and Turf. New York, NY: Springer New York; 2009. DREB Regulons in Abiotic-Stress-Responsive Gene Expression in Plants; pp. 15–28. [Google Scholar]
- 100.Kumar M, et al. Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.) J. Pl. Biol. 2014;57:383–393. doi: 10.1007/s12374-014-0487-1. [DOI] [Google Scholar]
- 101.Guo P, et al. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J. Exp.Botany. 2009;60:3531–44. doi: 10.1093/jxb/erp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J. Exp. Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 103.Zhang Z, et al. Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE. 2013;8:e57472. doi: 10.1371/journal.pone.0057472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Refli SM, Dewi K, Rachmawati D. Expression analysis of antioxidant genes in response to drought stress in the flag leaf of two Indonesian rice cultivars. Indonesian J. Biotechnol. 2014;19:43–55. doi: 10.22146/ijbiotech.8633. [DOI] [Google Scholar]
- 105.Sato Y, Masuta Y, Saito K, Murayama S, Ozawa K. Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant. Cell Rep. 2011;30:399–406. doi: 10.1007/s00299-010-0985-7. [DOI] [PubMed] [Google Scholar]
- 106.Tseng MJ, Liu CW, Yiu JC. Enhanced tolerance to sulfur dioxide and salt stress of transgenic Chinese cabbage plants expressing both superoxide dismutase and catalase in chloroplasts. Plant Physiology Biochem. 2007;45:822–833. doi: 10.1016/j.plaphy.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 107.Mhamdi Amna, Queval Guillaume, Chaouch Sejir, Vanderauwera Sandy, Van Breusegem Frank, Noctor Graham. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. Journal of Experimental Botany. 2010;61(15):4197–4220. doi: 10.1093/jxb/erq282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.