Abstract
Cytotoxic molecules can kill cancer cells by disrupting critical cellular processes or by inducing novel activities. 6-(4-(Diethylamino)-3-nitrophenyl)-5-methyl-4,5-dihydropyridazin-3(2H)-one (DNMDP) is a small molecule that kills cancer cells by generation of novel activity. DNMDP induces complex formation between phosphodiesterase 3A (PDE3A) and schlafen family member 12 (SLFN12) and specifically kills cancer cells expressing elevated levels of these two proteins. Here, we examined the characteristics and covariates of the cancer cell response to DNMDP. On average, the sensitivity of human cancer cell lines to DNMDP is correlated with PDE3A expression levels. However, DNMDP could also bind the related protein, PDE3B, and PDE3B supported DNMDP sensitivity in the absence of PDE3A expression. Although inhibition of PDE3A catalytic activity did not account for DNMDP sensitivity, we found that expression of the catalytic domain of PDE3A in cancer cells lacking PDE3A is sufficient to confer sensitivity to DNMDP, and substitutions in the PDE3A active site abolish compound binding. Moreover, a genome-wide CRISPR screen identified the aryl hydrocarbon receptor–interacting protein (AIP), a co-chaperone protein, as required for response to DNMDP. We determined that AIP is also required for PDE3A-SLFN12 complex formation. Our results provide mechanistic insights into how DNMDP induces PDE3A-SLFN12 complex formation, thereby killing cancer cells with high levels of PDE3A and SLFN12 expression.
Keywords: cancer therapy, cancer biology, cell death, phosphodiesterases, biomarker, AIP, DNMDP, PDE3A, PDE3B, SLFN12, protein-protein interaction, chaperone
Introduction
Despite advances in targeted therapies and immunotherapies, cancer remains a leading cause of human mortality (1), indicating a need for new therapeutic modalities. We recently discovered a compound, 6-(4-(diethylamino)-3-nitrophenyl)-5-methyl-4,5-dihydropyridazin-3(2H)-one (DNMDP),2 that selectively kills cancer cells using a unique mechanism of action (2).
DNMDP is active against a subset of cancer cell lines across cell lineages, with nanomolar potency and no obvious toxicity in a panel of nontumorigenic cell lines. Elevated expression of phosphodiesterase 3A (PDE3A) is the strongest genomic correlate of DNMDP sensitivity (2). DNMDP binds PDE3A and inhibits its enzymatic activity; however, enzyme inhibition alone is not sufficient for cell death. Most PDE3A inhibitors instead prevent cell killing by DNMDP when introduced in combination, possibly by competing away the binding of DNMDP to PDE3A (2). PDE3A knockout, moreover, abrogates the sensitivity of HeLa cells to DNMDP (2). Taken together, these findings suggest that DNMDP has a gain-of-function or neomorphic effect on PDE3A (2).
Consistent with the hypothesized gain or change of function, DNMDP induces formation of a novel complex between PDE3A and the poorly characterized protein, schlafen family member 12 (SLFN12) (2). Similar to PDE3A, elevated expression of SLFN12 correlates with, and is required for, DNMDP sensitivity (2). These observations suggest that DNMDP induces cancer cell death through a mechanism that is fundamentally different from the cancer cell dependencies exploited by typical targeted therapies. The cyclic nucleotide phosphodiesterase, PDE3A, is well-characterized with regard to its ability to hydrolyze the phosphodiester bonds of cAMP and cGMP to regulate and limit cellular responses to G protein–coupled receptor activation (3). More recently, evidence has also arisen for a role in hydrolysis of cUMP (4). Conversely, very little is known regarding SLFN12 function, although it may play a role in cell proliferation or differentiation (5–8). The molecular determinants of DNMDP response have not yet been explored.
Here, we define the determinants of cancer cell response to DNMDP. We characterize partial sensitivity at the single-cell level, investigate whether PDE3B can functionally substitute for PDE3A, and define the domains of PDE3A required for sensitivity. We furthermore use genome-wide CRISPR screening to identify additional genes required for DNMDP sensitivity. Results from these experiments indicate a central role for PDE3A protein expression levels in predicting the degree of DNMDP response and uncover AIP as a critical player in DNMDP-induced cancer cell killing.
Results
PDE3A- and SLFN12-expressing cell lines exhibit a gradient of sensitivity to DNMDP
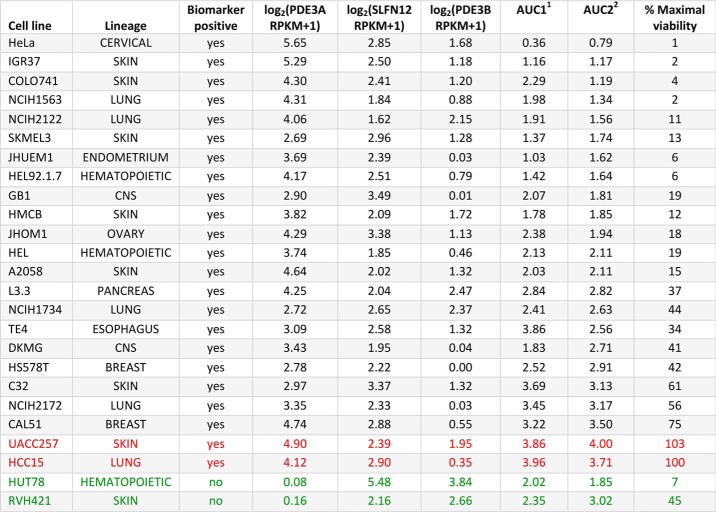
We have shown that PDE3A and SLFN12 expression levels together serve as a predictive biomarker for DNMDP sensitivity (2). Our previous analysis of sensitivity data from 766 cancer cell lines defined the positive predictive value (PPV) of this combined biomarker to be about 50%, with “sensitive” defined by an AUC equivalent to 1.6 on a scale of 0–4 (2). In other words, among biomarker-positive cell lines, about half are sensitive to DNMDP. We took two measures to further optimize PDE3A and SLFN12 expression as a predictive biomarker. First, we quantified gene expression using newly available RNA-Seq data from the Cancer Cell Line Encyclopedia (9), which provided greater resolution in the low expression range. Second, we more rigorously defined the optimal biomarker thresholds by maximizing the geometric mean of the sensitivity and the PPV over all possible biomarker thresholds (Fig. S1A). We found that the optimal expression thresholds for PDE3A and SLFN12 in this cell line panel were 2.65 and 1.47 log2(RPKM + 1), or 5.28 and 1.77 RPKM, respectively, resulting in a PPV of 62.5% and a sensitivity of 71.4% (Fig. S1B).
This PPV is comparable with the originally reported 50% objective response rate of HER2-positive metastatic breast cancer patients to the clinically approved targeted therapy, trastuzumab, in combination with chemotherapy (10). However, we sought to better understand the less-than-perfect prediction based on PDE3A and SLFN12 expression, which may be due to error in the high-throughput measurement of DNMDP response, or it may truly reflect the insufficient prediction power of these two expression markers alone, indicating the influence of additional factors. To distinguish between these two possibilities, we systematically assessed DNMDP response in 23 cell lines with PDE3A expression >5.28 RPKM and SLFN12 expression >1.77 RPKM with 18-point dose resolution, ranging from 0.26 nm to 3 μm (Table 1). We found good concordance between these results and AUCs from the published high-throughput data (2) (Fig. S1C). However, there was no correlation between AUC values and the expression levels of other phosphodiesterase family members or basal cAMP levels in these cell lines (Data set S1).
Table 1.
Biomarker expression and DNMDP response data
RNA-Seq expression values are from the Cancer Cell Line Encyclopedia (9). Maximal viability is defined as the bottom of the dose–response curve. Red, biomarker-positive but DNMDP-insensitive cell lines. Green, biomarker-negative but DNMDP-sensitive cell lines. CNS, central nervous system.
1 AUC1, DNMDP sensitivity data from high-throughput assay, normalized to an AUC scale of 0–4 (2). Originally reported DNMDP AUC values were normalized on a scale of 0-4 by capping values at 7 and multiplying by , to facilitate direct comparison with validation AUC values.
2 AUC2, manual validation DNMDP sensitivity data.
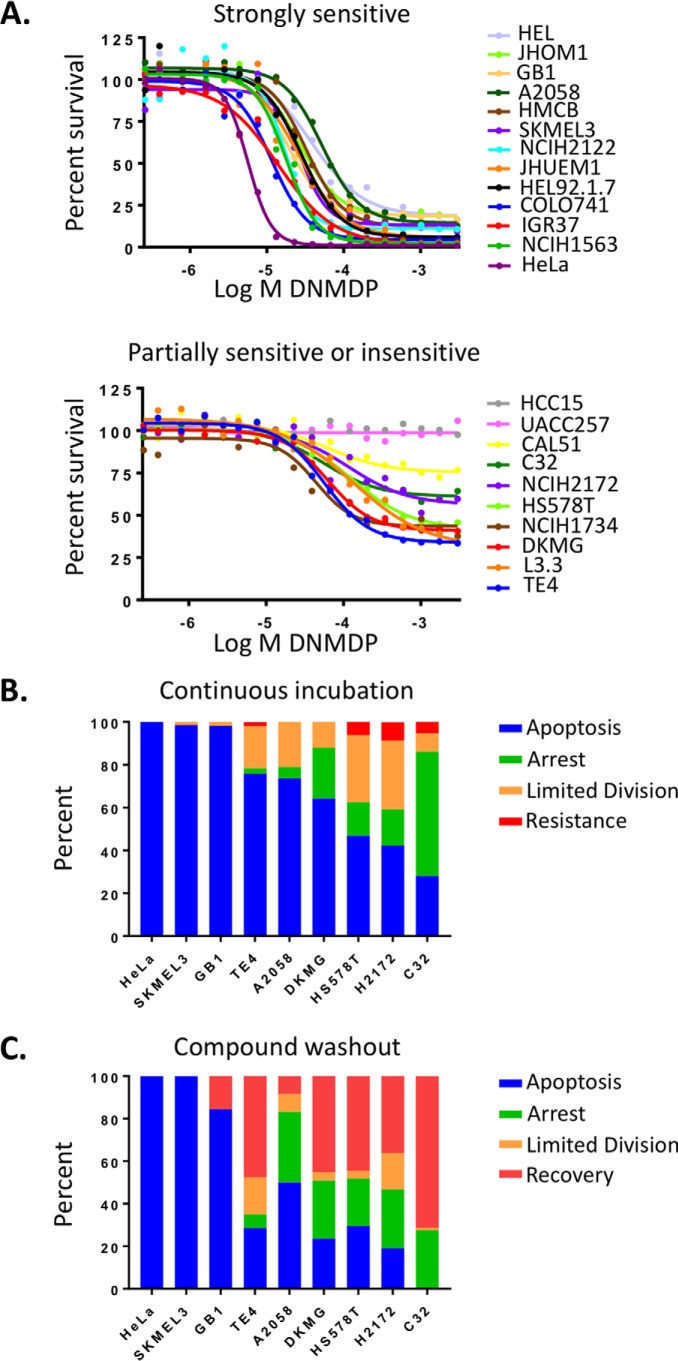
These experiments revealed that sensitivity is not binary (Fig. 1A); rather, dose-response curves showed a continuous gradient of inhibition response across the retested lines. Based on maximum viability values, the tested cell lines could be split into strongly sensitive cell lines (13 cell lines with <25% maximum viability), partially sensitive lines (8 cell lines with 25–75% maximum viability), and insensitive cell lines (2 cell lines with 100% maximal viability). Individual cell fate analysis revealed a mix of single-cell responses underlying this gradient of cellular response (Fig. 1B). For example, in the highly sensitive glioblastoma cell line, GB1, most individual cells underwent apoptosis within 72 h, and the remaining surviving cells apoptosed by about 96 h (Fig. S2). In the partially sensitive cell line, TE4, many cells underwent apoptosis by about 96 h, but several cells survived beyond 96 h, exhibiting a cytostatic phenotype (Fig. S2). In the more resistant cell line, NCI-H2172, many cells survived and continued to proliferate until the end of the experiment (Fig. S2). Washout experiments furthermore revealed that, whereas the most sensitive cell lines underwent full apoptosis even after compound removal at 72 h, many partially sensitive cells recovered and continued to proliferate (Fig. 1C).
Figure 1.
Cancer cell lines exhibit a range of sensitivities to DNMDP. A, gradient of DNMDP sensitivity among biomarker-positive cancer cell lines in a 72-h CellTiter-Glo assay. B, single-cell analysis of cell fates upon DNMDP treatment of cancer cell lines with a range of DNMDP sensitivities. Cells were treated continuously with 2 μm DNMDP, stained with an activated caspase substrate-conjugated dye, and imaged by Incucyte live-cell analysis. Cells were tracked starting at 24 h. Limited Division, cells divided at most twice between 24 h and the end of the observation period. C, cell fate tracking after 72-h treatment with 2 μm DNMDP, followed by a washout with control medium. Cells were tracked starting at 72 h. No HeLa cells survived beyond 72 h.
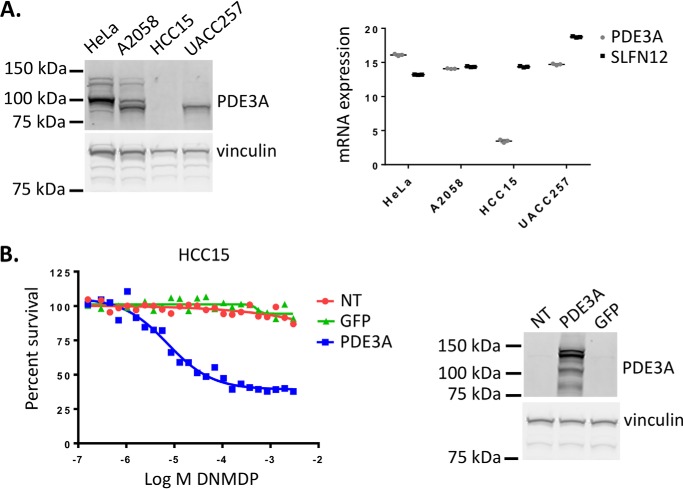
Two cell lines, HCC15 and UACC257, expressing high levels of PDE3A and SLFN12 mRNA, were curiously completely insensitive to DNMDP (Table 1 and Fig. 1A). We examined the expression of PDE3A in these two cell lines and found that the HCC15 cells expressed very little PDE3A mRNA and no detectable PDE3A protein despite high PDE3A RPKM values in the Cancer Cell Line Encyclopedia data set (9) (Fig. 2A). Ectopic expression of PDE3A in the HCC15 cells conferred response to DNMDP, confirming that the lack of DNMDP response was due to a lack of PDE3A expression (Fig. 2B). The UACC257 cells did express PDE3A protein (Fig. 2A), indicating that there must be another explanation for their lack of response (see Fig. 9).
Figure 2.
The insensitive cell line HCC15 does not express PDE3A protein. A (left), PDE3A protein expression was analyzed by immunoblotting in cancer cell lines. Right, PDE3A or GAPDH mRNA expression was analyzed by quantitative PCR. mRNA expression displayed as log2(relative gene expression) values. B, ectopic expression of PDE3A confers DNMDP sensitivity in the HCC15 cells, assayed by a 72-h CellTiter-Glo assay. Ectopic PDE3A expression was confirmed by immunoblotting. NT, no transduction.
Figure 9.
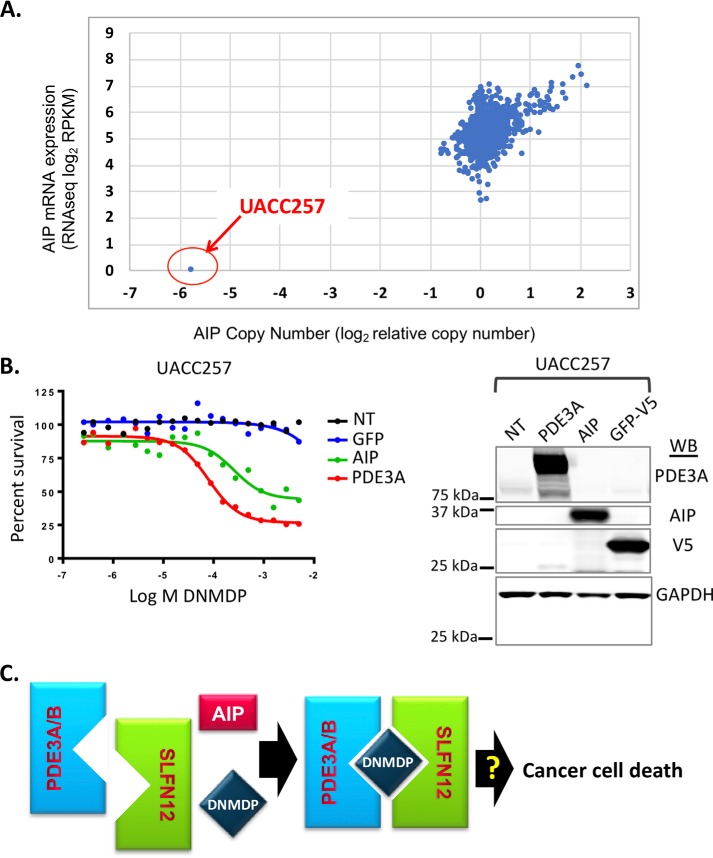
Melanoma cell line UACC257 does not respond to DNMDP due to lack of AIP expression. A, of 973 CCLE cancer cell lines, only UACC257 cells have an AIP deletion and express no AIP mRNA. B (left), ectopic expression of AIP in UACC257 cells confers DNMDP sensitivity in a 72-h CellTiter-Glo assay. Increased expression of PDE3A similarly confers DNMDP sensitivity. NT, no genes transduced. Right, immunoblots (WB) confirm expression of PDE3A, AIP, and V5-tagged GFP. C, model of DNMDP-induced complex formation between PDE3A and SLFN12. Complex formation is facilitated by AIP.
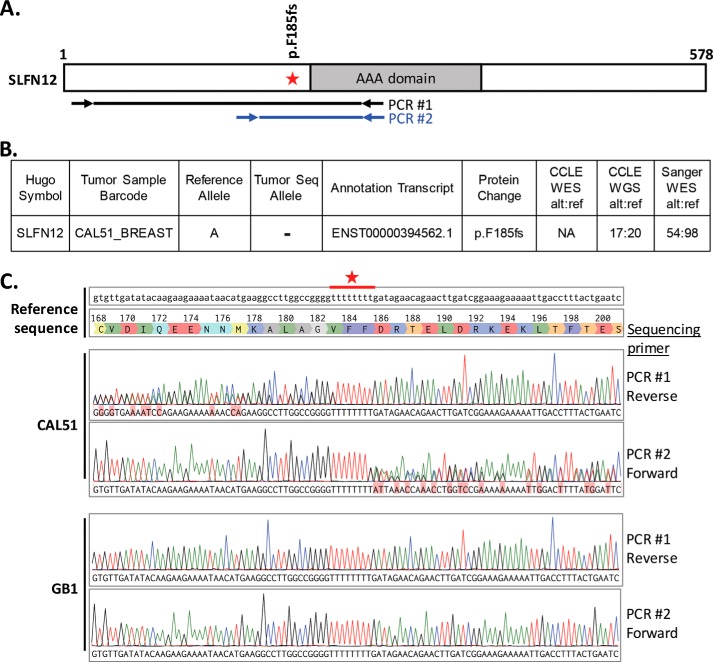
One additional cell line, CAL51, was similarly initially classified as biomarker-positive, based on mRNA expression, but was only slightly sensitive to DNMDP, with 75% viability at the bottom of the dose-response curve (Table 1 and Fig. 1A). Upon examination of genomic sequencing data from the Cancer Cell Line Encyclopedia (9), we found that the CAL51 cells harbor a heterozygous 1-bp deletion in SLFN12, shifting the reading frame after amino acid Phe-185 and causing an early translation stop at amino acid 196 (Fig. 3, A and B). We confirmed this finding by PCR-amplifying the relevant region from genomic DNA and Sanger sequencing the PCR products (Fig. 3, A and C).
Figure 3.
The CAL51 breast cancer cell line harbors a heterozygous SLFN12 Phe-185 frameshift mutation. A, SLFN12 gene diagram showing the position of the F185fs mutation. The locations of the primers, located within a single exon, used for genomic DNA PCR and sequencing are indicated below. B, heterozygous 1-bp deletion, as reported by CCLE (8). C, sequence alignment of PCR products to the reference sequence. Note that clean sequence peaks align perfectly to reference sequence before the stretch of Ts (marked by an asterisk), and overlapping peaks appear after the stretch of Ts. GB1 glioma cells, used as a WT control, showed perfect alignment across the entire region. fs, frameshift; WES, whole-exome sequencing; WGS, whole-genome sequencing; alt, alternate allele; ref, reference allele.
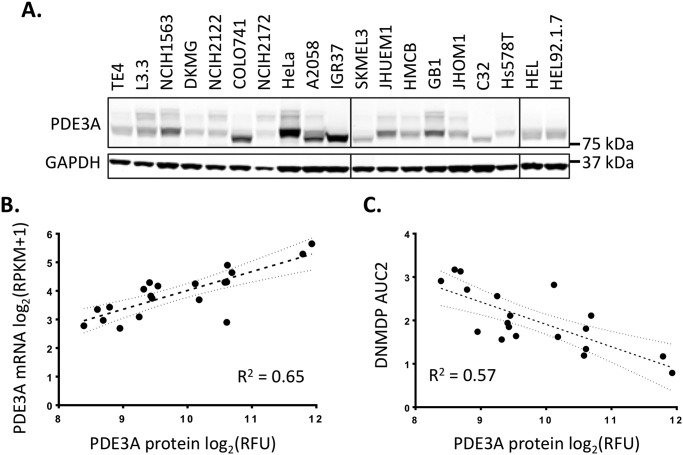
For the remaining biomarker-positive cells, the level of PDE3A protein expression (Fig. 4A) correlated with RNA expression (Fig. 4B) and DNMDP sensitivity, as demonstrated by the inverse correlation with DNMDP AUC (Fig. 4C). There is no antibody available that recognizes endogenous SLFN12 protein, so it was not possible to perform a similar analysis for SLFN12. However, no continuous correlation was observed between DNMDP AUC and SLFN12 mRNA expression (data not shown).
Figure 4.
A, immunoblot measuring PDE3A protein expression across biomarker-positive cell lines. B, correlation between PDE3A RNA expression, as measured by CCLE RNA-Seq, and PDE3A protein expression, as measured by LI-COR quantification of the immunoblot in A. The 95% confidence interval of the best-fit line is indicated. C, correlation between PDE3A protein expression and DNMDP response (AUC2 from Table S1). The 95% confidence interval of the best-fit line is indicated. RFU, relative fluorescence units.
PDE3B can also mediate DNMDP sensitivity
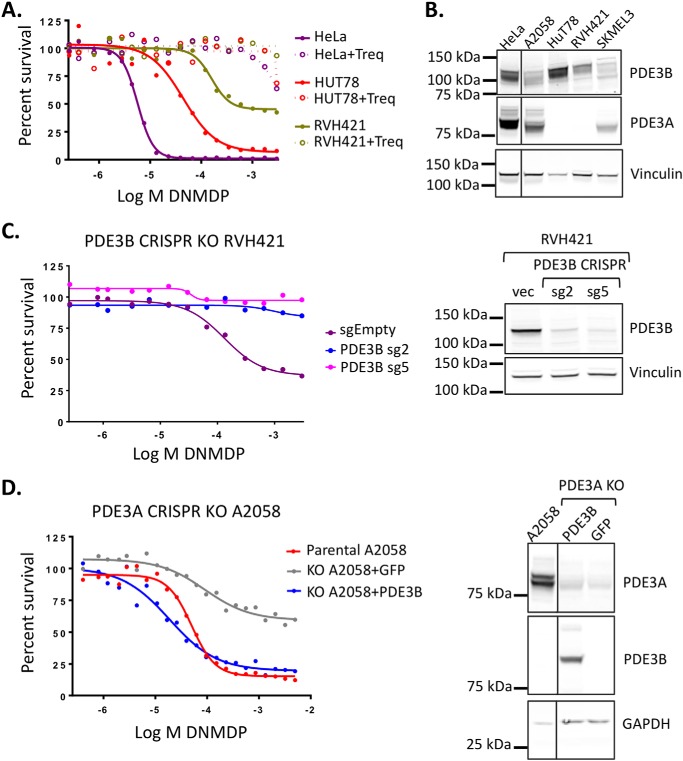
Two cancer cell lines, T-cell lymphoma HUT78 and melanoma cell line RVH421, were found to be sensitive to DNMDP in the absence of PDE3A expression (Table 1). We hypothesized that PDE3B, which is homologous to PDE3A in the catalytic domain, might substitute for PDE3A in these cells to support DNMDP cancer cell killing. Consistent with this idea, the cytotoxic response of HUT78 and RVH421 cells to DNMDP was competed away by trequinsin, suggesting a PDE3-mediated mechanism of response (Fig. 5A). RNA-Seq data indicated that these two cell lines express PDE3B mRNA (Table 1), and immunoblotting analysis confirmed that both express high levels of PDE3B but not PDE3A protein (Fig. 5B), suggesting that PDE3B could be the target for DNMDP in HUT78 and RVH421 cells.
Figure 5.
PDE3B can mediate DNMDP sensitivity. A, DNMDP sensitivity of HUT78 and RVH421, which lack PDE3A mRNA expression, can be competed away by co-treatment with 100 nm trequinsin (Treq) in a 72-h CellTiter-Glo assay. B, immunoblotting reveals that HUT78 and RVH421 do not express any PDE3A protein but do express high levels of PDE3B protein. C (left), CRISPR knockout of PDE3B in the partially sensitive cell line, RVH421, abolished DNMDP sensitivity in a 72-h CellTiter-Glo assay. Right, immunoblot analysis showing loss of PDE3B protein expression in knockout cells made with two PDE3B-specific CRISPR guide RNAs, sg2 and sg5. D (left), ectopic expression of PDE3B in PDE3A knockout A2058 cells restores sensitivity to DNMDP in a 72-h CellTiter-Glo assay. Right, immunoblot analysis showing loss of PDE3A protein and ectopic expression of PDE3B protein in the PDE3A knockout A2058 cells. Vinculin or GAPDH was used a loading control.
To functionally investigate the role of PDE3B in the response to DNMDP, we knocked out PDE3B in RVH421 cells. Knockout of PDE3B in RVH421 cells abolished DNMDP sensitivity, confirming that PDE3B is necessary for sensitivity in these cells (Fig. 5C). We then asked whether PDE3B can complement PDE3A function and restore sensitivity in PDE3A knockout cell lines. Ectopic expression of the PDE3B cDNA fully restored DNMDP sensitivity to the PDE3A-knockout melanoma cell line, A2058, and the PDE3A-knockout cervical cancer cell line, HeLa (Fig. 5D and Fig. S3A). Taken together, these results indicate that PDE3B can function in place of PDE3A to mediate DNMDP response. However, inclusion of PDE3B expression level in the optimized biomarker of DNMDP response improved biomarker sensitivity but not PPV (Fig. S3, B and C) when compared with PDE3A alone with SLFN12 (Fig. S1B). This indicates that there are cancer cell lines that express high levels of PDE3B and SLFN12 but do not respond to DNMDP (Fig. S3C). Nonetheless, the ability of PDE3B to replace PDE3A in DNMDP response suggests that the catalytic domain, which is highly conserved between the two family members, may be the determinant for DNMDP sensitivity.
The PDE3A catalytic domain is sufficient to confer DNMDP sensitivity
The full-length PDE3A protein consists of two membrane association regions at the N terminus followed by a catalytic domain that is conserved across phosphodiesterases (11–13). Interestingly, PDE3A undergoes alternative translation and exists as a set of protein isoforms that represent different N-terminal truncations. These naturally occurring truncations affect one or both of the membrane-associated regions and multiple kinase phosphorylation sites (12, 14, 15), resulting in different subcellular localizations and modes of regulation (14–19).
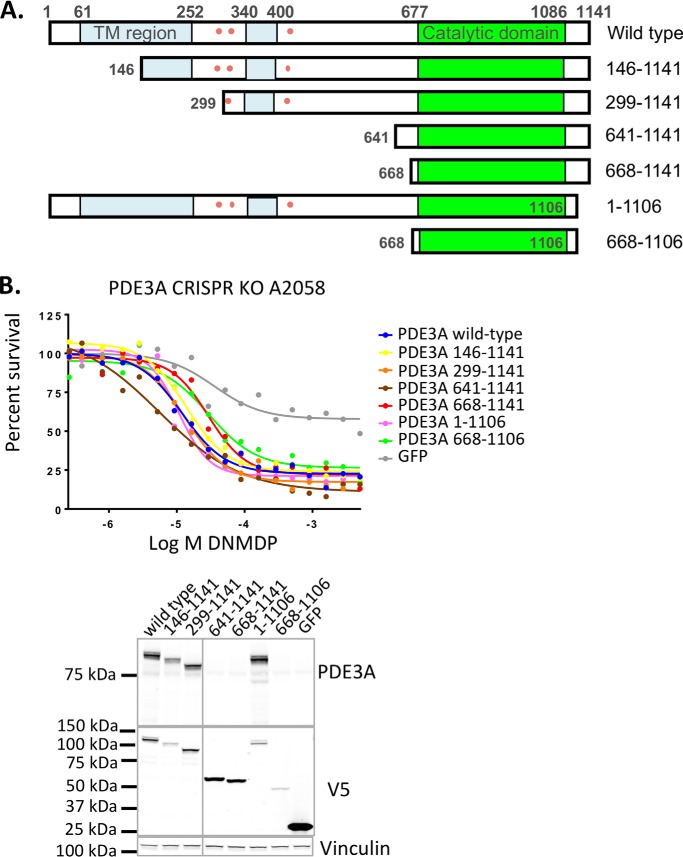
We generated a series of V5-tagged, N-terminally truncated cDNA expression constructs representing two of the shorter PDE3A isoforms as well as truncations that more closely isolate the catalytic domain (Fig. 6A). The ability of the PDE3A deletion constructs to restore DNMDP sensitivity of PDE3A-knockout A2058 cells was tested. All truncation constructs fully restored DNMDP sensitivity in these cells, including the shortest construct, comprised of amino acids 668–1106 (Fig. 6B). This suggests that the isolated catalytic domain of PDE3A is sufficient to mediate DNMDP sensitivity.
Figure 6.
The PDE3A catalytic domain is sufficient to confer DNMDP sensitivity. A, V5-tagged PDE3A deletion mutants tested for the ability to support DNMDP cancer cell killing. Amino acid numbers are shown. TM (blue), membrane-associated region. Pink dots, phosphorylation sites. B (top), DNMDP sensitivity of PDE3A CRISPR knockout A2058 cells can be restored by ectopic expression of full-length, WT PDE3A or any of the truncation constructs tested in a 72-h CellTiter-Glo assay. Bottom, truncation construct expression was confirmed by immunoblotting with anti-PDE3A (N-terminal epitope) or anti-V5. Note that constructs with larger N-terminal deletions are not recognized by anti-PDE3A.
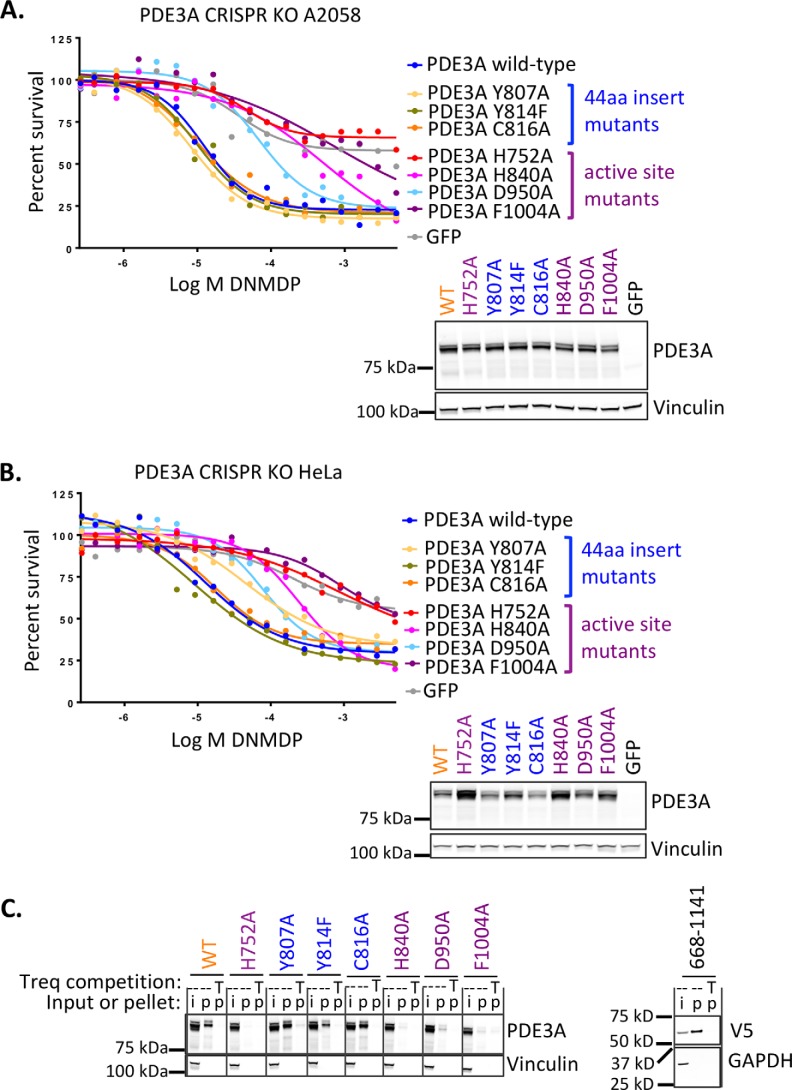
Because the cytotoxic effects of DNMDP can be competed away by trequinsin, a potent PDE3A/PDE3B inhibitor that does not have cancer cell killing activity (2), and because the catalytic domain is the only region of PDE3A required for the activity of DNMDP, we hypothesized that DNMDP binds to the active site of PDE3A, within the catalytic domain. To test this hypothesis, we generated PDE3A active-site mutants H840A, D950A, and F1004A, which are impaired for substrate binding, and H752A, affecting Mg2+ coordination, homologous to residues reported for PDE3B (20). We found that all four mutants exhibited impaired binding of and response to DNMDP (Fig. 7, A–C). In contrast, three point mutations, Y807A, Y814F, and C816A, in the 44-amino acid catalytic domain insert unique to the PDE3 family (21) had no effect on the ability of PDE3A to bind DNMDP and support cancer cell killing (Fig. 7, A–C). Consistent with the survival assay results, a nonmutated C-terminal construct of PDE3A encoding primarily the catalytic domain retained the ability to bind DNMDP (Fig. 7C).
Figure 7.
PDE3A active site mutants impair DNMDP sensitivity. A, A2058 cells. Top, 72-h CellTiter-Glo assay. Bottom, anti-PDE3A immunoblot analysis confirming construct expression. B, HeLa cells. Top, 72-h CellTiter-Glo assay. Bottom, anti-PDE3A immunoblot analysis confirming construct expression. C (left), active-site mutations (purple), but not 44-amino acid catalytic domain insert mutations (blue), abolish binding of PDE3A in 200 μg of cell lysate from transduced A2058 cells to resin-linked DNMDP. Right, the isolated V5-tagged PDE3A catalytic domain fragment (amino acids 668–1141) in 200 μg of cell lysate from transduced A2058 cells binds resin-linked DNMDP. Treq or T, trequinsin; i, input; p, pellet.
Taken together, these data indicate that the catalytic domain of PDE3A is necessary and sufficient for mediating DNMDP sensitivity, and the active site of the PDE3A catalytic domain is required for DNMDP binding, although inhibition of enzymatic activity is likely not the mechanism of cancer cell killing by DNMDP (2).
A genome-wide CRISPR knockout screen for resistance to DNMDP uncovers AIP
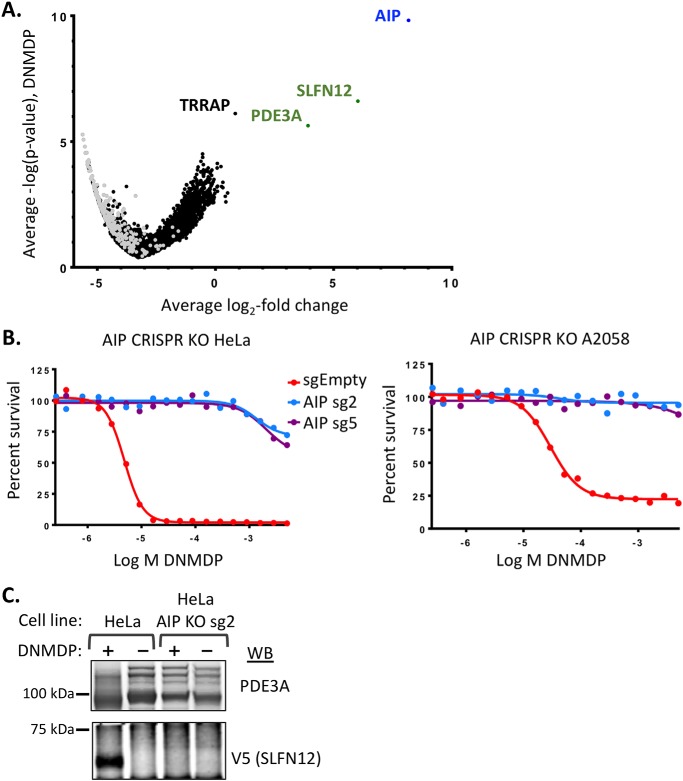
To identify additional genes required for DNMDP cancer cell killing, we performed a genome-wide CRISPR loss-of-function screen in HeLa cells treated with DNMDP or DMSO. We identified sgRNAs that were consistently enriched under DNMDP treatment, compared with DMSO treatment, suggesting cell survival in the presence of DNMDP. The sgRNAs best supporting survival in the presence of DNMDP were those targeting the aryl hydrocarbon receptor–interacting protein (AIP) (Fig. 8A and Data set S2). As expected, SLFN12 and PDE3A knockout also strongly supported HeLa cell survival in the presence of DNMDP, ranking second and third behind AIP, respectively. Knockout of the histone acetyltransferase complex protein, transformation/transcription domain–associated protein (TRRAP), exhibited a significant but much weaker resistance phenotype (Fig. 8A and Data set S2).
Figure 8.
AIP is required for DNMDP sensitivity. A, a genome-wide CRISPR knockout screen in HeLa cells reveals that AIP expression is required for DNMDP sensitivity. Shown is a volcano plot of average log2 -fold change (LFC) across guide RNAs and average −log p value across two replicates for each gene following 14-day exposure with 25 nm DNMDP. Only genes with 3–8 guide RNAs are plotted to eliminate guides with likely off-target effects. Gray dots, nontargeting control guide RNAs, randomly grouped into symbolic genes at 4 guides/gene. Full data are available in Data set S2. B (left), 72-h CellTiter-Glo assay with independent AIP CRISPR gRNAs (sg) confirming that AIP expression is required for sensitivity of HeLa cells to DNMDP. Right, 72-h CellTiter-Glo assay validating a requirement of AIP for DNMDP sensitivity in the melanoma cell line A2058. C, AIP knockout prevents DNMDP-induced complex formation. PDE3A immunoprecipitates from HeLa cells transiently transfected with V5-tagged SLFN12 and treated with 10 μm DNMDP were immunoblotted (WB) with anti-V5 to assess DNMDP-induced PDE3A-SLFN12 complex formation.
AIP is an HSP90 co-chaperone that is known to interact with several other proteins, regulating protein stability and subcellular localization (22). Interestingly, AIP is also a tumor suppressor gene in familial isolated pituitary adenoma (23). We confirmed that AIP knockout eliminates HeLa cell response to DNMDP by validating with independent sgRNAs (Fig. 8B). Similar results were observed upon knockout of AIP from the melanoma cell line A2058 (Fig. 8B).
To determine whether AIP knockout impacts DNMDP response upstream or downstream of PDE3A-SLFN12 complex formation, we tested the effects of AIP knockout directly on complex formation. We ectopically expressed V5-tagged SLFN12 in parental or AIP-knockout HeLa cells, immunoprecipitated endogenous PDE3A, and assessed whether we could detect V5-SLFN12 in the PDE3A immunoprecipitates. AIP knockout completely abolished PDE3A-SLFN12 complex formation in response to DNMDP (Fig. 8C), confirming that AIP functions upstream of DNMDP-induced complex formation.
Among the biomarker-positive cell lines, the melanoma cell line, UACC257, was conspicuously insensitive to DNMDP (Table 1). When we examined AIP expression across 973 cancer cell lines (9), we found that UACC257 was the only cell line that did not express AIP (Fig. 9A). Ectopic expression of AIP sensitized the UACC257 cells to DNMDP (Fig. 9B), indicating that loss of AIP was a major determinant of resistance of these cells. Interestingly, ectopic expression of PDE3A also increased sensitivity of the UACC257 cells to DNMDP (Fig. 9B), suggesting that loss of AIP can be compensated for by an increase in PDE3A levels. Taken together, our results demonstrate an essential role for DNMDP-induced and AIP-facilitated PDE3A-SLFN12 complex formation in cancer cell death in response to treatment of cancer cells with DNMDP (Fig. 9C).
Discussion
Predictive biomarkers have become an essential component of cancer treatment, enabling identification of the patients most likely to respond to a given therapy. We have identified a new method of cancer cell killing involving complex formation between PDE3A and SLFN12. Because sensitive tumor cells are found across cancer lineages, predictive biomarkers will play an especially important role in patient identification if DNMDP-like molecules that induce PDE3A-SLFN12 complex formation are evaluated in the clinic.
We previously showed that DNMDP response depends upon expression of PDE3A and SLFN12 (2). Optimization of the mRNA expression thresholds for these two proteins resulted in a PPV of 62.5% (Fig. S1), with expression level of PDE3A protein being a major determinant in degree of response (Fig. 4). It is likely that establishment of a protein-based biomarker assay would further improve PPV, as mRNA expression may not precisely reflect protein levels. For example, we used genomic techniques to uncover a frameshift mutation of one SLFN12 allele in the CAL51 cell line that is likely to result in decreased overall expression of SLFN12 protein, explaining the cryptic resistance in this seemingly biomarker-positive cell line (Fig. 3). However, a protein-based biomarker assay awaits development of a suitable antibody for SLFN12.
Other factors could also contribute to a biomarker with increased PPV. Although PDE3B was found to support DNMDP sensitivity in the absence of PDE3A expression (Fig. 5), and PDE3B expression was essential for response of two biomarker-negative but DNMDP-sensitive cell lines, inclusion of PDE3B did not improve biomarker PPV (Fig. S3). We identified another gene, AIP, more broadly required for DNMDP sensitivity (Fig. 8). AIP acts upstream of DNMDP-induced PDE3A-SLFN12 complex formation. A second biomarker-positive but DNMDP-insensitive cell line, UACC257, was found to express no AIP, and ectopic expression of AIP conferred sensitivity (Fig. 9). Thus, although it appears to be only rarely deleted, AIP expression could be considered as a component of a further improved biomarker.
We found that the catalytic domain of PDE3A was sufficient for complex formation and DNMDP sensitivity and that mutations in the active site of PDE3A abolish DNMDP response (Figs. 6 and 7). However, although DNMDP is a PDE3A inhibitor, inhibition of phosphodiesterase activity does not correlate with complex formation or cancer cell killing (2). It is possible that some early and more recent reports of PDE3A dependence in cancer cells may in fact also involve PDE3A-SLFN12 complex formation (24–28), as zardaverine and anagrelide phenocopy DNMDP (2, 29). Others have subsequently also observed sensitivity of cancer cells to DNMDP (30) and induction of PDE3A-SLFN12 complex formation (31).
Unlike traditional targeted inhibitors that leverage dependencies in cancer cells created by genomic alterations, DNMDP instead initiates induced cell death in cancer cells expressing elevated levels of PDE3A and SLFN12 by a likely gain-of-function or change-of-function mechanism. DNMDP initiates these processes by binding to PDE3A and inducing PDE3A-SLFN12 complex formation. Here, we defined the factors that influence PDE3A-SLFN12 complex formation and response to DNMDP; however, elucidation of the downstream mechanism of action of DNMDP requires further study. Despite this open question, DNMDP-like molecules may represent a new therapeutic opportunity for cancer patients with tumors expressing elevated levels of PDE3A and SLFN12.
Experimental procedures
Cell culture and viability assays
To measure cancer cell death in response to DNMDP treatment, cells were plated in 384w assay plates at the following cell density per well: 500 cells of HeLa (DMEM), A2058 (DMEM), HMCB (EMEM), IGR37 (DMEM), and NCIH1734 (RPMI); 750 cells of CAL51 (DMEM), COLO741 (RPMI), DKMG (RPMI), GB1 (EMEM), HEL (RPMI), HEL9217 (RPMI), JHUEM1 (DMEM + F-12), L3.3 (RPMI), TE4 (RPMI), HCC15 (RPMI), and UACC257 (RPMI); 1000 cells for HUT78 (IMDM), NCIH1563 (RPMI), NCIH2122 (RPMI), NCIH2172 (RPMI), RVH421 (RPMI), and SKMEL3 (McCoy's 5A); and 1500 cells for C32 (EMEM), HS578T (DMEM), and JHOM1 (DMEM + F-12). Cells were incubated at 37 °C overnight and then treated with a DNMDP dose dilution series using an HP D300 digital dispenser. For experiments involving trequinsin competition, 100 nm trequinsin was also added to each well at the same time as DNMDP addition. After 72 h, the viability of cells in each well was measured by Cell Titer Glo (Promega, G755B and G756B). Percent viability values were determined using the values from untreated wells, and AUC values were calculated using a 4-parameter fit. x axis values are reported in log10 M. IC50 and maximal killing values for all dose-response curves are listed in Table S2. DNMDP was purchased from Life Chemicals (F1638-0042), and trequinsin was purchased from Sigma–Aldrich (T2057).
Single-cell fate mapping
1400 cells/well were seeded in a 96 well plate in medium that had been centrifuged at 500 × g for 5 min to remove particulates. The next day, the red fluorescent DNA-staining dye, Incucyte Nuclite Rapid Red, and green fluorescent apoptosis dye, Incucyte Caspase-3/7 Green Apoptosis Reagent (Essen Biosciences), were added in 2 μl of fetal bovine serum to a final concentration of 1:1000 and 1:1500, respectively. Two hours later, 2 μm DNMDP + 0.2% DMSO or 0.2% DMSO alone was added. Because even sensitive cells sometimes divided before 24 h, cells were tracked starting at 24 h, although cells that apoptosed before 24 h were also counted. For the washout study, the medium was removed from DNMDP-treated cells at 72 h, the cells were rinsed with medium, and incubation was continued in the absence of DNMDP. Cells were tracked starting at 72 h. Images were taken every 1 h up to 96 h, and every 2 h thereafter, with an Incucyte S3 machine (Essen Biosciences). Three channels were recorded: phase contrast, red fluorescence (DNA), and green fluorescence (apoptosis). For cell tracking, a movie superimposing all three channels was analyzed. To avoid effects due to depletion of medium components over time, cells were followed up to the last hour before DMSO control cells started to show slowed division or increased apoptosis (136 h for HeLa, 194 h for SKMEL3, 160 h for GB1, 130 h for TE4, 130 h for A2058, 144 h for DKMG, 106 h for HS578T, 186 h for H2172, and 220 h for C32).
Cell lysis and immunoblotting
Cells were plated in 10-cm Petri dishes and collected at 50–90% confluence. For PDE3A immunoblotting in biomarker-positive cells and in AIP KO cells, cells were seeded in 15-cm plates at a similar density as in viability assays with a vessel scaling factor of 5000 (e.g. 500 cells/well was scaled to 106 cells/10-cm plate or 2.5 × 106 cells/15-cm plate) and then cultured for 72 h before collection. Cell pellets were lysed at 4 °C for 20 min in modified radioimmune precipitation assay buffer (150 mm NaCl, 10% glycerol, 50 mm Tris-Cl, pH 8.0, 50 mm MgCl2, 1% Nonidet P-40) supplemented with EDTA-free protease inhibitors (Sigma-Aldrich, 4693159001) and PhosSTOP phosphatase inhibitors (Sigma-Aldrich, 4906837001). Lysates were clarified by centrifugation at 13,000 rpm for 10 min at 4 °C and quantified using BCA protein assays (Thermo Fisher Scientific, 23225). Clarified lysates were resolved on 4–12% BisTris polyacrylamide gels, transferred to nitrocellulose membranes (Thermo Fisher Scientific, IB23001), and immunoblotted with primary antibodies against PDE3A (Bethyl, A302-740A; 1:2000), PDE3B (Bethyl, A302-743A), V5 (Life Technologies, Inc., R96205; 1:5000), AIP (Thermo Fisher Scientific, MA3-16515; 1:2000), and vinculin (Sigma–Aldrich, V9264; 1:5000), GAPDH (Cell Signaling Technology, 2118; 1:2000) and secondary antibodies from LI-COR Biosciences (92632210 and 926068021, each at 1:10,000). Blots were washed and imaged using a LI-COR Odyssey IR imager, and fluorescent signals were quantified using the Image Studio software provided by LI-COR. See Fig. S1D for full-length PDE3A and PDE3B immunoblots indicating sufficient antibody specificity.
RNA extraction and real-time quantitative RT-PCR
To measure PDE3A mRNA expression levels in cell lines, cells were collected, and total RNA was made using a Monarch total RNA miniprep kit (New England Biolabs, T2010S), reverse-transcribed using the SuperScript III First-Strand synthesis system (Thermo Fisher Scientific, 18080051). Parallel negative control reactions were carried out by leaving out the reverse transcriptase. TaqMan real-time PCR was performed on the cDNA products for PDE3A (Hs01012698_m1) and GAPDH (Hs02758991_g1), and relative gene expression levels were calculated as −ΔCT values between sample and respective no-reverse transcriptase control.
cDNA ectopic expression
ORF overexpression constructs were obtained from TRC consortium: PDE3A (ccsbBroad304_06701), PDE3B (ccsbBroad304_06702), GFP (ccsbBroad304_99997), and AIP (BRDN0000465180), for which the Lys-228 snp was corrected to Gln-228 to match the UniprotKB protein sequence (accession number O00170). For lentivirus-based gene delivery, 293T cells were transfected with ORF overexpression constructs and packaging plasmids psPAX2 and pMD2.G. Virus was collected 48 h after transfection and added to target cells for spin infection with 4 or 8 μg/ml of Polybrene. Transduced target cells were selected using 15 μg/ml blasticidin and then expanded for 3 days before plating into 384-well plates for DNMDP sensitivity testing.
Assay for SLFN12 Phe-185 frameshift mutation status in cell lines
Genomic DNA was isolated from cells using the QIAamp DNA mini kit (Qiagen 51304), and the SLFN12 genomic region was amplified by PCR using Q5 High-Fidelity 2X Master Mix (New England Biolabs, M0492) and primers SLFN12_2_F or SLFN12_428_F and SLFN12_858_R. PCR products were purified using the QIAquick PCR purification kit (Qiagen 28104) and sent for sequencing using forward or reverse primers used for PCR. Sequencing reads were aligned to reference sequence using Benchling alignment tools.
CRISPR knockout
PDE3A CRISPR KO cells (sgRNA#2) were generated according to de Waal et al. (2). CRISPR target sites for PDE3B and AIP were identified using the CHOPCHOP CRISPR Design Tool (http://chopchop.cbu.uib.no)3 (35, 36). Oligonucleotide sequences are listed in Table S1. For cloning of sgRNAs, forward and reverse oligonucleotides were annealed, phosphorylated, and ligated into a BsmBI-digested lentiCRISPRv2 vector. Lentivirus carrying each guide construct was packaged as described above and used to infect target cells. Transduced target cells were selected using 1 μg/ml puromycin and passaged for 7 days before use.
Site-directed mutagenesis
A pDONR-PDE3A plasmid from the TRC consortium (ccsbBroadEn_06701) was used to generate mutated PDE3A ORFs. Primers used for mutagenesis are listed in Table S1. PDE3A deletion mutant and H752A point mutant expression constructs were generated using the GeneArt Site-Directed Mutagenesis PLUS system (Thermo Fisher Scientific, A14604) whereas point mutants Y807A, C816A, Y814A, H840A, D950A, and F1004A were generated by recombination of PCR products (32). Mutated ORFs were then shuttled into the pLX304 lentiviral expression vector using Gateway LR Clonase (Thermo Fisher Scientific, 11791020). To generate the SNP-corrected AIP expression construct, the AIP ORF was shuttled into pDONR223 using Gateway BP Clonase (Thermo Fisher Scientific, 11789020), mutagenized using the GeneArt Site-Directed Mutagenesis PLUS system, and then shuttled back into pLX304 using Gateway LR Clonase. All plasmids were sequence-verified along the entire ORF length to ensure the presence of desired mutation and the absence of additional unwanted mutations.
Linker resin pulldown
To measure the binding of DNMDP to WT and mutant PDE3A proteins, A2058 PDE3A KO cells stably transduced with lentiviral ORF overexpression constructs were harvested. Linker resin pulldown experiments were performed as described previously (2). Briefly, cells were lysed in modified radioimmune precipitation assay lysis buffer (150 mm NaCl, 10% glycerol, 50 mm Tris-Cl, pH 8.0, 50 mm MgCl2, 1% Nonidet P-40) supplemented with EDTA-free protease inhibitors (Sigma–Aldrich, 4693159001) and PhosSTOP phosphatase inhibitors (Sigma–Aldrich, 4906837001). 200 μg of total protein (0.5 mg/ml, 400 μl) was incubated with 3 μl of DNMDP-2L affinity linker resin (2) with or without 10 μm trequinsin for 4 h at 4 °C. Beads were then washed three times with lysis buffer, and proteins bound were eluted with 50 μl of LDS sample-loading buffer with reducing agent added (Thermo Fisher Scientific, NP0007 and NP0009). 20 μl of each eluted sample and 50 μg of corresponding matched input lysate (50% input) were separated on 4–12% BisTris polyacrylamide gels (Thermo Fisher Scientific) and immunoblotted with anti-PDE3A antibody (1:1000; Bethyl, A302-740) as described above.
Genome-wide CRISPR screen
The Brunello CRISPR KO library (33) was used for the DNMDP resistance screen. Lentiviral infection was carried out in duplicate and for each replicate with enough HeLa cells to achieve >1000 infected cells/library member (80,000 sgRNAs, >8 × 107 cells total) and at low multiplicity of infection to achieve transduction of a single sgRNA per cell. Infection efficiencies for the two replicates were 24 and 31%, respectively, corresponding to a multiplicity of infection of about 0.3, meaning about 85% of infected cells would be predicted to have single sgRNA integration (34). At the time of infection, HeLa cells were resuspended in medium and mixed with Brunello library virus in the presence of 8 μg/ml Polybrene (library lentivirus provided by the Genetic Perturbation Platform at the Broad Institute), plated in 12-well dishes at 3 × 106 cells/well, and spun at 931 × g for 2 h at 30 °C. 2 h after the spin infection, virus-containing medium was removed, and fresh medium was added for incubation overnight. The day after the infection, cells were trypsinized and pooled into T225 flasks at 50% confluence (1.6 × 107 cells/flask), and puromycin was added to 1 μg/ml to select for infected cells. At the same time, in-line infection efficiency assays were performed by comparing cell counts after puromycin selection with those without selection. After 4 days of puromycin selection, infected cells were collected and passaged in T225 flasks at 25% confluence (8 × 106 cells/flask) for three additional days to allow CRISPR KO to complete. Cells were collected at 8 days after infection, and 8 × 107 cells each were split into the DMSO control arm (plating at 8 × 106 cells/T225 flask) or the 25 nm DNMDP treatment arm (plating at 2 × 107 cells/T225 flask). Cells were passaged every 3–4 days at 25% confluence for the next 14 days. For the DMSO arm, 8 × 107 cells were maintained at every passage, whereas all surviving cells were passaged for the DNMDP arm. After 14 days of compound treatment, cells were harvested, washed with cold PBS, and flash-frozen at 2 × 107 cells (DMSO arm) or lesser portions for genomic DNA isolation. Genomic DNA was isolated using the Nucleospin Blood XL kit (DMSO samples, four preparations to cover 8 × 107 cells; Machere-Nagel, 740950.50) or the QIAamp DNA blood minikit (DNMDP-treated samples; Qiagen, 51104). PCR amplification of sgRNA tags and pooled library sequencing were carried out as described by Sanson et al. (33).
CRISPR screen data analysis was done largely as described by Sanson et al. (33). Briefly, deconvolution of sequencing reads yielded read counts for each sgRNA under each replicate treatment condition. Log2-normalized reads for each guide per condition were calculated using the formula, log2(guide/total × 1,000,000 + 1) and averaged across the two replicates. Subtracting DMSO values from those for 25 nm DNMDP generated log2-fold change values for each sgRNA, which were then averaged across all sgRNAs targeting the same gene to generate gene-level average log2-fold change score. To statistically evaluate gene-level enrichment in DNMDP treatment relative to DMSO, sgRNAs were rank-ordered based on average log2-fold change, and p values for each sgRNA relative to the rank order were determined by running a hypergeometric distribution without replacement, equivalent to a one-sided Fisher's exact test. The average of the negative log10 p values for each sgRNA targeting the same gene was calculated to generate the average negative log10 p value for each gene. A volcano plot was generated using the average log2 -fold change and the average negative log10 p value for all genes with 3–8 sgRNAs/gene to visualize gene enrichments after the positive selection of 25 nm DNMDP treatment.
Transient transfection and immunoprecipitation
HeLa or HeLa AIP KO cells were transfected with ORF overexpression constructs expressing V5-tagged SLFN12 (TRC consortium, TRCN0000476272) using the Fugene transfection reagent (VWR E2691). At 72 h post-transfection, cells were treated with 10 μm DNMDP or DMSO for 8 h and collected and snap-frozen until lysis. Cell pellets were lysed as described above. 3 mg of total protein was incubated with 4 μg of anti-PDE3A antibody (Bethyl 302-740A) at 4 °C overnight, followed by the addition of 20 μl each of Protein A– and Protein G–Dynabeads (Thermo Fisher Scientific, 10001D and 10003D) and additional incubation for 2 h. Beads were washed, and bound proteins were eluted with 20 μl of LDS sample-loading buffer (Thermo Fisher Scientific, NP0007) with reducing agent added (Thermo Fisher Scientific, NP0009) and subjected to SDS-PAGE separation and anti-PDE3A and anti-V5 immunoblotting as described above.
Author contributions
X. W., T. A. L., C. W. G., J. D., M. M., and H. G. conceptualization; X. W., G. F. G., L. W., S. H., H. S., A. D. C., and H. G. data curation; X. W., G. F. G., L. W., S. H., H. S., and A. D. C. formal analysis; X. W., G. R. S., B. D., A. R. B., B. K., K. W., S. L., T. A. L., M. L., C. K., and H. G. investigation; X. W., G. R. S., G. F. G., B. D., L. W., T. A. L., M. L., S. H., H. S., J. D., and A. D. C. methodology; X. W. writing-original draft; X. W. and H. G. writing-review and editing; C. W. G., J. D., A. D. C., C. K., M. M., and H. G. supervision; M. M. and H. G. funding acquisition.
Supplementary Material
Acknowledgments
We thank Alex Burgin and Jon Goldstein for critical reading of the manuscript. This work was supported by funding from Bayer AG.
This work was supported by a Broad Institute Next Generation Fund award (to H. G.). Several authors on this study received funding from Bayer AG (X. W., G. R. S., G. F. G., A. R. B., B. K., L. W., T. A. L., C. W. G., A. D. C., M. M., and H. G.) and are co-inventors on patent applications submitted with Bayer AG (X. W., T. A. L., M. M., and H. G.).
This article contains Tables S1 and S2, Figs. S1–S3, and Data sets S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- DNMDP
- 6-(4-(diethylamino)-3-nitrophenyl)-5-methyl-4,5-dihydropyridazin-3(2H)-one
- PPV
- positive predictive value
- RPKM
- reads per kilobase million
- AUC
- area under the dose-response curve
- sgRNA
- single guide RNA
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- KO
- knockout
- DMEM
- Dulbecco's modified Eagle's medium
- EMEM
- Eagle's minimal essential medium
- IMDM
- Iscove's modified Dulbecco's medium.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2018) Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2. de Waal L., Lewis T. A., Rees M. G., Tsherniak A., Wu X., Choi P. S., Gechijian L., Hartigan C., Faloon P. W., Hickey M. J., Tolliday N., Carr S. A., Clemons P. A., Munoz B., Wagner B. K., Shamji A. F., Koehler A. N., Schenone M., Burgin A. B., Schreiber S. L., Greulich H., and Meyerson M. (2016) Identification of cancer-cytotoxic modulators of PDE3A by predictive chemogenomics. Nat. Chem. Biol. 12, 102–108 10.1038/nchembio.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maurice D. H., Ke H., Ahmad F., Wang Y., Chung J., and Manganiello V. C. (2014) Advances in targeting cyclic nucleotide phosphodiesterases. Nat. Rev. Drug Discov. 13, 290–314 10.1038/nrd4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berrisch S., Ostermeyer J., Kaever V., Kälble S., Hilfiker-Kleiner D., Seifert R., and Schneider E. H. (2017) cUMP hydrolysis by PDE3A. Naunyn Schmiedebergs Arch. Pharmacol. 390, 269–280 10.1007/s00210-016-1328-1 [DOI] [PubMed] [Google Scholar]
- 5. Basson M. D., Wang Q., Chaturvedi L. S., More S., Vomhof-DeKrey E. E., Al-Marsoummi S., Sun K., Kuhn L. A., Kovalenko P., and Kiupel M. (2018) Schlafen 12 interaction with SerpinB12 and deubiquitylases drives human enterocyte differentiation. Cell Physiol. Biochem. 48, 1274–1290 10.1159/000492019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kovalenko P. L., and Basson M. D. (2014) Schlafen 12 expression modulates prostate cancer cell differentiation. J. Surg. Res. 190, 177–184 10.1016/j.jss.2014.03.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Puck A., Hopf S., Modak M., Majdic O., Cejka P., Blüml S., Schmetterer K., Arnold-Schrauf C., Gerwien J. G., Frederiksen K. S., Thell E., Leitner J., Steinberger P., Aigner R., Seyerl-Jiresch M., et al. (2017) The soluble cytoplasmic tail of CD45 (ct-CD45) in human plasma contributes to keep T cells in a quiescent state. Eur. J. Immunol. 47, 193–205 10.1002/eji.201646405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rhead B., Brorson I. S., Berge T., Adams C., Quach H., Moen S. M., Berg-Hansen P., Celius E. G., Sangurdekar D. P., Bronson P. G., Lea R. A., Burnard S., Maltby V. E., Scott R. J., Lechner-Scott J., Harbo H. F., Bos S. D., and Barcellos L. F. (2018) Increased DNA methylation of SLFN12 in CD4+ and CD8+ T cells from multiple sclerosis patients. PLoS ONE 13, e0206511 10.1371/journal.pone.0206511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slamon D. J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A., Fleming T., Eiermann W., Wolter J., Pegram M., Baselga J., and Norton L. (2001) Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 10.1056/NEJM200103153441101 [DOI] [PubMed] [Google Scholar]
- 11. Kenan Y., Murata T., Shakur Y., Degerman E., and Manganiello V. C. (2000) Functions of the N-terminal region of cyclic nucleotide phosphodiesterase 3 (PDE 3) isoforms. J. Biol. Chem. 275, 12331–12338 10.1074/jbc.275.16.12331 [DOI] [PubMed] [Google Scholar]
- 12. Omori K., and Kotera J. (2007) Overview of PDEs and their regulation. Circ. Res. 100, 309–327 10.1161/01.RES.0000256354.95791.f1 [DOI] [PubMed] [Google Scholar]
- 13. Shakur Y., Takeda K., Kenan Y., Yu Z. X., Rena G., Brandt D., Houslay M. D., Degerman E., Ferrans V. J., and Manganiello V. C. (2000) Membrane localization of cyclic nucleotide phosphodiesterase 3 (PDE3). Two N-terminal domains are required for the efficient targeting to, and association of, PDE3 with endoplasmic reticulum. J. Biol. Chem. 275, 38749–38761 10.1074/jbc.M001734200 [DOI] [PubMed] [Google Scholar]
- 14. Hambleton R., Krall J., Tikishvili E., Honeggar M., Ahmad F., Manganiello V. C., and Movsesian M. A. (2005) Isoforms of cyclic nucleotide phosphodiesterase PDE3 and their contribution to cAMP hydrolytic activity in subcellular fractions of human myocardium. J. Biol. Chem. 280, 39168–39174 10.1074/jbc.M506760200 [DOI] [PubMed] [Google Scholar]
- 15. Hunter R. W., Mackintosh C., and Hers I. (2009) Protein kinase C-mediated phosphorylation and activation of PDE3A regulate cAMP levels in human platelets. J. Biol. Chem. 284, 12339–12348 10.1074/jbc.M807536200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahmad F., Shen W., Vandeput F., Szabo-Fresnais N., Krall J., Degerman E., Goetz F., Klussmann E., Movsesian M., and Manganiello V. (2015) Regulation of sarcoplasmic reticulum Ca2+ ATPase 2 (SERCA2) activity by phosphodiesterase 3A (PDE3A) in human myocardium: phosphorylation-dependent interaction of PDE3A1 with SERCA2. J. Biol. Chem. 290, 6763–6776 10.1074/jbc.M115.638585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vandeput F., Szabo-Fresnais N., Ahmad F., Kho C., Lee A., Krall J., Dunlop A., Hazel M. W., Wohlschlegel J. A., Hajjar R. J., Houslay M. D., Manganiello V. C., and Movsesian M. A. (2013) Selective regulation of cyclic nucleotide phosphodiesterase PDE3A isoforms. Proc. Natl. Acad. Sci. U.S.A. 110, 19778–19783 10.1073/pnas.1305427110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wechsler J., Choi Y. H., Krall J., Ahmad F., Manganiello V. C., and Movsesian M. A. (2002) Isoforms of cyclic nucleotide phosphodiesterase PDE3A in cardiac myocytes. J. Biol. Chem. 277, 38072–38078 10.1074/jbc.M203647200 [DOI] [PubMed] [Google Scholar]
- 19. Zhang W., and Colman R. W. (2007) Thrombin regulates intracellular cyclic AMP concentration in human platelets through phosphorylation/activation of phosphodiesterase 3A. Blood 110, 1475–1482 10.1182/blood-2006-10-052522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scapin G., Patel S. B., Chung C., Varnerin J. P., Edmondson S. D., Mastracchio A., Parmee E. R., Singh S. B., Becker J. W., Van der Ploeg L. H., and Tota M. R. (2004) Crystal structure of human phosphodiesterase 3B: atomic basis for substrate and inhibitor specificity. Biochemistry 43, 6091–6100 10.1021/bi049868i [DOI] [PubMed] [Google Scholar]
- 21. Hung S. H., Zhang W., Pixley R. A., Jameson B. A., Huang Y. C., Colman R. F., and Colman R. W. (2006) New insights from the structure-function analysis of the catalytic region of human platelet phosphodiesterase 3A: a role for the unique 44-amino acid insert. J. Biol. Chem. 281, 29236–29244 10.1074/jbc.M606558200 [DOI] [PubMed] [Google Scholar]
- 22. Trivellin G., and Korbonits M. (2011) AIP and its interacting partners. J. Endocrinol. 210, 137–155 10.1530/JOE-11-0054 [DOI] [PubMed] [Google Scholar]
- 23. Hernández-Ramirez L. C., Martucci F., Morgan R. M., Trivellin G., Tilley D., Ramos-Guajardo N., Iacovazzo D., D'Acquisto F., Prodromou C., and Korbonits M. (2016) Rapid proteasomal degradation of mutant proteins is the primary mechanism leading to tumorigenesis in patients with missense AIP mutations. J. Clin. Endocrinol. Metab. 101, 3144–3154 10.1210/jc.2016-1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cartledge D. M., Robbins K. M., Drake K. M., Sternberg R., Stabley D. L., Gripp K. W., Kolb E. A., Sol-Church K., and Napper A. D. (2017) Cytotoxicity of zardaverine in embryonal rhabdomyosarcoma from a Costello syndrome patient. Front. Oncol. 7, 42 10.3389/fonc.2017.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fryknäs M., Rickardson L., Wickström M., Dhar S., Lövborg H., Gullbo J., Nygren P., Gustafsson M. G., Isaksson A., and Larsson R. (2006) Phenotype-based screening of mechanistically annotated compounds in combination with gene expression and pathway analysis identifies candidate drug targets in a human squamous carcinoma cell model. J. Biomol. Screen. 11, 457–468 10.1177/1087057106288048 [DOI] [PubMed] [Google Scholar]
- 26. Nazir M., Senkowski W., Nyberg F., Blom K., Edqvist P. H., Jarvius M., Andersson C., Gustafsson M. G., Nygren P., Larsson R., and Fryknäs M. (2017) Targeting tumor cells based on phosphodiesterase 3A expression. Exp. Cell Res. 361, 308–315 10.1016/j.yexcr.2017.10.032 [DOI] [PubMed] [Google Scholar]
- 27. Pulkka O. P., Gebreyohannes Y. K., Wozniak A., Mpindi J. P., Tynninen O., Icay K., Cervera A., Keskitalo S., Murumägi A., Kulesskiy E., Laaksonen M., Wennerberg K., Varjosalo M., Laakkonen P., Lehtonen R., et al. (2018) Anagrelide for gastrointestinal stromal tumor. Clin. Cancer Res. 25, 1676–1687 [DOI] [PubMed] [Google Scholar]
- 28. Sun L., Quan H., Xie C., Wang L., Hu Y., and Lou L. (2014) Phosphodiesterase 3/4 inhibitor zardaverine exhibits potent and selective antitumor activity against hepatocellular carcinoma both in vitro and in vivo independently of phosphodiesterase inhibition. PLoS ONE 9, e90627 10.1371/journal.pone.0090627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. An R., Liu J., He J., Wang F., Zhang Q., and Yu Q. (2019) PDE3A inhibitor anagrelide activates death signaling pathway genes and synergizes with cell death-inducing cytokines to selectively inhibit cancer cell growth. Am. J. Cancer Res. 9, 1905–1921 [PMC free article] [PubMed] [Google Scholar]
- 30. Vandenberghe P., Hagué P., Hockman S. C., Manganiello V. C., Demetter P., Erneux C., and Vanderwinden J. M. (2017) Phosphodiesterase 3A: a new player in development of interstitial cells of Cajal and a prospective target in gastrointestinal stromal tumors (GIST). Oncotarget 8, 41026–41043 10.18632/oncotarget.17010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li D., Chen J., Ai Y., Gu X., Li L., Che D., Jiang Z., Li L., Chen S., Huang H., Wang J., Cai T., Cao Y., Qi X., and Wang X. (2019) Estrogen-related hormones induce apoptosis by stabilizing schlafen-12 protein turnover. Mol. Cell 75, 1103–1116.e9 10.1016/j.molcel.2019.06.040 [DOI] [PubMed] [Google Scholar]
- 32. Suzuki Y., Kagawa N., Fujino T., Sumiya T., Andoh T., Ishikawa K., Kimura R., Kemmochi K., Ohta T., and Tanaka S. (2005) A novel high-throughput (HTP) cloning strategy for site-directed designed chimeragenesis and mutation using the Gateway cloning system. Nucleic Acids Res. 33, e109 10.1093/nar/gni103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanson K. R., Hanna R. E., Hegde M., Donovan K. F., Strand C., Sullender M. E., Vaimberg E. W., Goodale A., Root D. E., Piccioni F., and Doench J. G. (2018) Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 5416 10.1038/s41467-018-07901-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ellis E. L., and Delbrück M. (1939) The growth of bacteriophage. J. Gen. Physiol. 22, 365–384 10.1085/jgp.22.3.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Labun K., Montague T. G., Gagnon J. A., Thyme S. B., and Valen E. (2016) CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276 10.1093/nar/gkw398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Montague T. G., Cruz J. M., Gagnon J. A., Church G. M., and Valen E. (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.