Abstract
Corynebacterium diphtheriae is a human pathogen that causes diphtheria. In response to immune system-induced oxidative stress, C. diphtheriae expresses antioxidant enzymes, among which are methionine sulfoxide reductase (Msr) enzymes, which are critical for bacterial survival in the face of oxidative stress. Although some aspects of the catalytic mechanism of the Msr enzymes have been reported, several details still await full elucidation. Here, we solved the solution structure of C. diphtheriae MsrB (Cd-MsrB) and unraveled its catalytic and oxidation-protection mechanisms. Cd-MsrB catalyzes methionine sulfoxide reduction involving three redox-active cysteines. Using NMR heteronuclear single-quantum coherence spectra, kinetics, biochemical assays, and MS analyses, we show that the conserved nucleophilic residue Cys-122 is S-sulfenylated after substrate reduction, which is then resolved by a conserved cysteine, Cys-66, or by the nonconserved residue Cys-127. We noted that the overall structural changes during the disulfide cascade expose the Cys-122–Cys-66 disulfide to recycling through thioredoxin. In the presence of hydrogen peroxide, Cd-MsrB formed reversible intra- and intermolecular disulfides without losing its Cys-coordinated Zn2+, and only the nonconserved Cys-127 reacted with the low-molecular-weight (LMW) thiol mycothiol, protecting it from overoxidation. In summary, our structure-function analyses reveal critical details of the Cd-MsrB catalytic mechanism, including a major structural rearrangement that primes the Cys-122–Cys-66 disulfide for thioredoxin reduction and a reversible protection against excessive oxidation of the catalytic cysteines in Cd-MsrB through intra- and intermolecular disulfide formation and S-mycothiolation.
Keywords: enzyme mechanism, enzyme structure, nuclear magnetic resonance (NMR), kinetics, redox regulation, biochemistry, hydrogen peroxide, methionine sulfoxide
Introduction
Corynebacterium diphtheriae is the causative agent of diphtheria. Once this pathogenic bacterium enters the host system, it encounters the immune system of the host (1, 2). There, it is exposed to oxidative stress of our immune system (3–7), and tries to survive using several antioxidant systems, such as catalase and the NADPH-dependent thioredoxin, GSH or mycothiol reduction pathways (8–11).
One of the antioxidant enzymes are the methionine sulfoxide reductases (Msr).3 Oxidation of methionine results in methionine-S-sulfoxide (Met-S-SO) and methionine-R-sulfoxide (Met-R-SO). MsrA reduces the S-epimeric form of MetSO, whereas MsrB reduces the R-epimeric form (12). Their active sites are mirrored with hydrophobic residues on one side and H-bond donating residues on the other side (13–15). This mirror design allows these enzymes to specifically reduce the stereoisomeric forms of MetSO.
Msr enzymes are important for the survival of pathogenic and nonpathogenic bacteria under oxidative stress conditions, as shown in several studies with msr deletion mutants. For example, msrA deletion mutants of Mycobacterium smegmatis showed increased sensitivity for hydroperoxides (16), and also the msrB deletion mutants of Enterococcus faecalis and Francisella tularensis showed increased sensitivity toward oxidative stress (17, 18).
Also structural studies gave insight into the catalytic mechanisms and the role of MsrB as an antioxidant regulatory enzyme and methionine sulfoxide scavenger (19, 20). In general, the catalytic mechanism of MsrB is similar to MsrA. Once a MetSO substrate is reduced by the MsrB nucleophilic cysteine, it forms a sulfenic acid (−SOH). This more electrophilic sulfenic acid is being attacked by the sulfur of a resolving cysteine to form a disulfide bond, and the disulfide is reduced by one of the reducing pathways, such as the thioredoxin pathway, or by a low-molecular-weight thiol (LMW-thiol) pathway. The most abundant LMW-thiol of Gram-negative bacteria and eukaryotes is GSH, whereas in Actinobacteria, it is mycothiol (21). Aside from the catalytic cysteines, some MsrB enzymes also have two CXXC motifs, which are distant in their primary amino acid sequence but in proximity in the three-dimensional structure. These cysteines are not involved in the catalytic mechanism, but they coordinate Zn2+ to maintain its overall structure.
We present detailed insights into the catalytic and oxidation protection mechanisms of C. diphtheriae MsrB (Cd-MsrB). We combined structural, biochemical, and kinetic approaches with MS and ICP-MS to show that Cd-MsrB catalyzes via a disulfide cascade mechanism. Furthermore, Cd-MsrB reversibly protects its catalytic cysteines from overoxidation by disulfide bond formation and S-mycothiolation.
Results
Solution structure of Cd-MsrB shows a nonconserved cysteine and cysteine-coordinated Zn2+
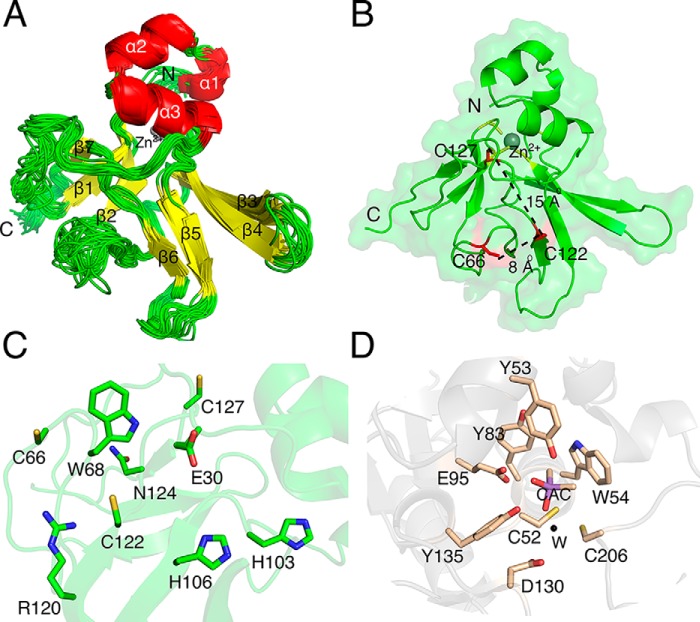
Cd-MsrB has seven cysteines (Fig. S1): two are conserved catalytic cysteines (Cys-122 and Cys-66, black asterisks) (22, 23), four belong to the conserved two CXXC motifs (Cys-48, Cys-51, Cys-97, and Cys-100, red asterisks) (24, 25), and one is a nonconserved cysteine located close to the C terminus (Cys-127, black dot). To determine the overall architecture of Cd-MsrB, and the location of its conserved and nonconserved cysteines, we determined the solution structure of reduced Cd-MsrB (Fig. 1 and Fig. S2).
Figure 1.
NMR solution structure of reduced Cd-MsrB and its active site. A, an overlay of the 20 lowest-energy structures of reduced Cd-MsrB. The secondary structural elements (yellow for β-sheets and red for α-helices) and the zinc (gray) are labeled (PDB code 6TR8). B, Cd-MsrB monomeric structure in a semi-transparent surface is presented in green cartoon. The location of the conserved Cys-66 and Cys-122 (thiols are surface exposed) and the nonconserved Cys-127 (buried thiol) are shown in red, and the cysteines involved in Zn2+ coordination are colored yellow. The distance between Cys-122 and Cys-66, and Cys-122 and Cys-127 are 8 and 15 Å, respectively. The N and C termini are indicated. C, the active site of Cd-MsrB has the hydrophobic pocket (Trp-68), and the H-bond donors (conserved His-103, His-106, and Asn-124). The amino acids are indicated in a green stick representation. D, the active site of C. diphtheriae MsrA (PDB 4D7L) is shown in wheat color. It has a mirror-like image of the active site of Cd-MsrB, with a hydrophobic pocket (Tyr-53 and Trp-54) and conserved H-bond donors (Tyr-83, Glu-95, and Tyr-135, and Asp-130 via a water molecule (W, black)). The cacodylate molecule mimicking the interactions a substrate would make with the active site residues is colored purple and labeled CAC.
The Cd-MsrB [1H,15N]-HSQC spectrum features a set of well-resolved peaks, typical for a folded globular protein (Fig. S2). Using triple-resonance NMR experiments, we assigned 89% of all backbone amides, but the NH resonances of Thr-2, Asn-3, Ser-18, Asn-39–Thr-41, Asn-63–Gly-67, Gly-77, His-88, and Ser-125 were not observed in the spectra. Observable aliphatic and aromatic protons of the protein side chains were also assigned. The structure calculations show a good convergence with the position of the protein backbone well-defined along most of the polypeptide chain. Several loop regions (residues 37–42 and 62–68) display high structural variability, most likely due to the paucity of the resonance assignments and/or the scarcity of inter-residue NOEs (Fig. 1A).
Similar to other MsrB enzymes, Cd-MsrB has two anti-parallel β-sheets, and two α-helices (Fig. 1, A and B) (23–25). At its N terminus, it has an additional short α-helix (α1) (Fig. 1). Cd-MsrB active site, which features a hydrophobic pocket (Trp-68) and H-bond donors (His-103, His-106, and Asn-124) (Fig. 1C), is a mirror image of the C. diphtheriae MsrA active site (Fig. 1D) (26). As Cd-MsrB contains two CXXC motifs, we used ICP-MS to confirm that Cd-MsrB contains a Zn2+, which has been shown to be coordinated by the cysteines of the CXXC motifs (24, 25). The conserved Cys-122 is located 8 Å away from Cys-66, and 15 Å away from Cys-127. Both Cys-122 and Cys-66 thiol groups are surface exposed, whereas the thiols of Cys-127 and the cysteines of the two CXXC motifs are more buried (Fig. 1B). As the exact role of the conserved and nonconserved cysteines of Cd-MsrB was not known, we decided to identify how they contribute to the catalytic mechanism of Cd-MsrB.
Cys-122 is essential for MetSO reduction
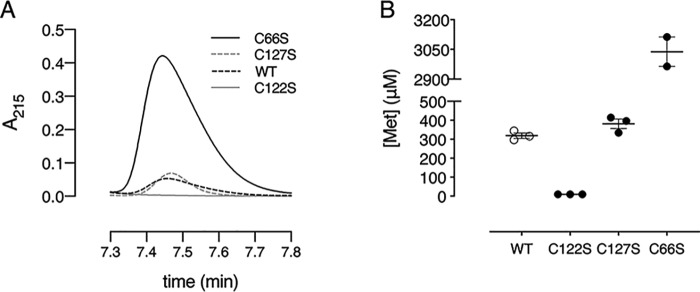
To determine which cysteines are involved in methionine sulfoxide reduction, three single Cys to Ser mutants were generated (C66S, C122S, and C127S). With reversed-phase chromatography, we monitored the formation of methionine after enzymatic reduction of methionine sulfoxide (MetSO) at 215 nm (Fig. 2A) (26). This experiment was performed in the presence of DTT to recycle the enzyme. Mutation of the nonconserved Cys-127 did not interfere with the production of methionine, as both Cd-MsrB wildtype (WT) (319 ± 13 μm) and MsrB C127S (382 ± 24 μm) produce almost similar concentrations of methionine. In the absence of the conserved nucleophilic Cys-122, no methionine is produced, which shows its role in MetSO reduction during the first step of the reaction. Mutation of the conserved resolving Cys-66 produced 10-fold more methionine (3038 ± 74 μm) compared with WT Cd-MsrB (Fig. 2B). With CD, we then found that mutation of Cys-66 (Cd-MsrB C66S) changes the overall secondary structure, which turned out to increase the MetSO reduction efficiency of Cd-MsrB (Fig. S3A). Structural changes were also observed for the C127S mutant; however, these overall changes had no effect on the reduction efficiency of Cd-MsrB (Fig. S3B).
Figure 2.
Cys-122 is required for substrate reduction. A, RPC-HPLC chromatograms of Cd-MsrB WT, C66S, C122S, and C127S mutants are shown, where Met peak formation is observed at a retention time 7.45 min. In the presence of MetSO and DTT, as a reducing agent, the C122S mutant does not convert MetSO to Met, whereas the WT and the other mutants (C66S and C127S) show Met production. B, the concentration of Met produced by the WT and Cys mutants are shown in a dot plot. WT and the C127S mutant show similar Met production, whereas the C66S mutant shows 10-fold excess Met production. The data are presented as a mean ± S.E. of at least two independent technical repeats and the graphs were generated using Prism8.
Formation of a sulfenic acid on Cys-122 after MetSO reduction
We first decided to confirm sulfenic acid formation on Cys-122 of Cd-MsrB, as this has already been shown for MsrB from other species (27, 28). Reduced WT Cd-MsrB, the C66S/C127S (only Cys-122 is present) mutant, and the C66S/C122S (only Cys-127 is present) mutant were incubated with dimedone, which specifically reacts with the sulfur of a cysteine once a sulfenic acid (−SOH) is formed (Fig. S4). On an anti-dimedone immunoblot, we clearly detected dimedone formation only when nucleophilic Cys-122 is present (Fig. S4, lanes 2 and 4).
MetSO reduction results in a Cys-122–Cys-66 and a Cys-122–Cys-127 disulfide
Next, we wanted to know whether a disulfide is formed after MetSO reduction. By using mass spectrometric analysis of Cd-MsrB WT in the presence and absence of MetSO, we detected Cys-122–Cys-127 to be the most predominant disulfide after substrate reduction, next to the Cys-66–Cys-122 disulfide. In addition, a minor population of inter- and intramolecular disulfides between the cysteines involved in zinc coordination was observed. This could be due to a population of Cd-MsrB, which might not contain zinc, making it possible for those cysteines of the CXXC motif to form a disulfide. Or, alternatively, these disulfides could have been generated during sample preparation for mass spectrometric analysis.
To confirm the formation of the two disulfide bonds after substrate reduction, we determined the free thiol (−SH) content of Cd-MsrB WT, C66S, C122S, and C127S mutants in the presence and absence of MetSO-substrate using the DTNB assay (29). In the presence of MetSO, both the C66S and C127S mutants showed loss of two free thiols (Table S2). This confirms that both the Cys-122–Cys-127 and the Cys-66–Cys-122 disulfides can be formed after substrate reduction (Table S2).
Methionine sulfoxide reduction induces structural changes
As the distances between the nucleophilic Cys-122 and the two resolving cysteines, Cys-66 and Cys-127, are more than 3 Å (Fig. 1B), we wanted to investigate whether structural changes occur upon disulfide bond formation. Both circular dichroism (CD) (Fig. S5) and NMR-HSQC spectral analysis were used (Figs. 3 and 4). Comparing the CD spectra of reduced and MetSO-treated MsrB, we observed a decrease in the molar ellipticity at 208 and 222 nm, which indicate an increase in the overall α helical content upon the addition of MetSO (Fig. S5). To further confirm this observation, we obtained NMR-HSQC spectra of MetSO-treated Cd-MsrB (oxidized sample) directly, and after 19 h of incubation at 25 °C.
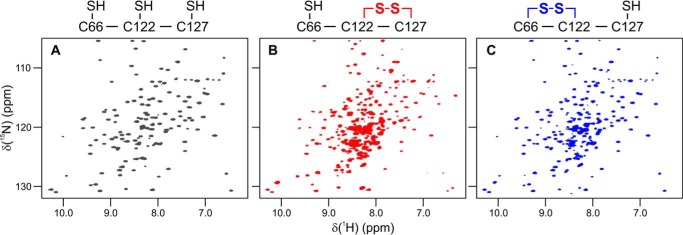
Figure 3.
MsrB oxidation with MetSO leads to pronounced changes in the NMR spectrum. The [1H,15N]-HSQC spectra of (A) reduced MsrB, (B) freshly prepared MetSO-oxidized MsrB, and (C) MetSO-oxidized MsrB incubated at 25 °C for 19 h are shown. The spectra can be attributed to distinct redox species with different Cys-66, Cys-122, and Cys-127 oxidation status (see text), schematically shown above the plots.
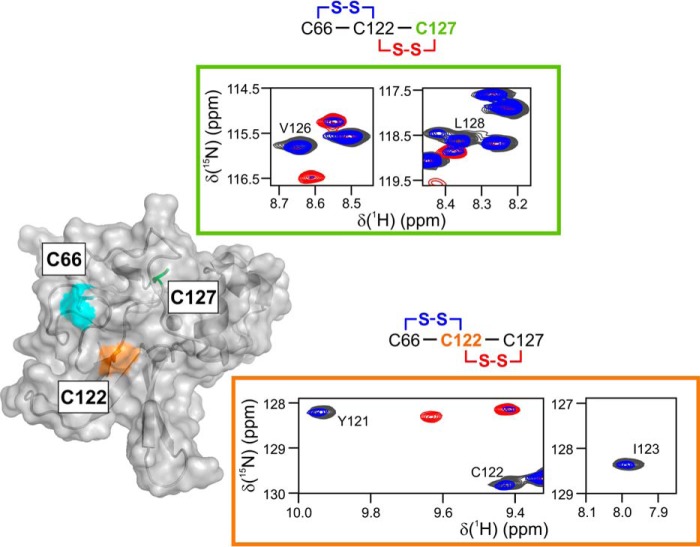
Figure 4.
Two disulfide forms of the oxidized MsrB can be discerned in the NMR spectra. The semi-transparent MsrB molecular surface shows solvent-exposed Cys-66 (cyan) and Cys-122 (orange), and buried Cys-127 (green sticks). The panels contoured in orange and green show HSQC spectral regions that report on the local chemical environment of Cys-122 and Cys-127, respectively. The spectra of reduced MsrB (black), freshly prepared MetSO-oxidized MsrB (red), and MetSO-oxidized MsrB incubated at 25 °C for 19 h (blue), correspond to those shown in Fig. 3. Note the absence of the red resonance for Ile-123 (orange panel), indicating a large shift to a new position lying outside of the plotted spectral region. Two possible disulfide forms are schematically shown above the plots, and the cysteine residue reported upon in a given panel is colored.
Also here we observed pronounced NMR spectral changes upon treatment of MsrB with MetSO (Fig. 3). The HSQC spectrum of the oxidized MsrB shows two sets of resonances: intense peaks of a major form and much weaker signals of a minor form (Fig. 3B). The presence of the two different forms agrees with the mass spectrometric analysis, where major (Cys-122–Cys-127) and minor (Cys-66–Cys-122) disulfide species are observed. Upon further incubation of the MetSO-treated MsrB at 25 °C for 19 h, the major HSQC peaks progressively diminish and eventually disappear, whereas the minor resonances increase in intensity. The resulting spectrum shows a single set of peaks after 19 h (Fig. 3C), which are very similar, but not identical, to those of the reduced MsrB (Fig. 3A). The observed spectral changes are characteristic for a slow chemical reaction and the associated structural reorganization process, which would be expected for the disulfide exchange. Unfortunately we were unable to obtain the NMR structure of oxidized MsrB, due to progressive protein aggregation, which leads to a dramatic degradation of the spectral quality.
Nevertheless, the detailed analysis of the NMR spectra of the oxidized MsrB allowed us to distinguish between the two disulfide forms. The large differences in structural properties of the two disulfides can be studied by following NMR-HSQC resonances of protein backbone amides, which are sensitive reporters of the local chemical environment. To probe the surroundings of Cys-127, we monitored the well-resolved NH resonances of its neighboring Val-126 and Leu-128 (Fig. 4, green panel). Important to note is that the HSQC peak of Cys-127 is excluded from the analysis as it strongly overlaps with another resonance. Compared with the spectrum of the reduced protein (Fig. 3A, black), the Val-126 and Leu-128 peaks shift to a different location in the initial form of the oxidized MsrB (Fig. 3B, red), yet return to the same spectral position in the final oxidized form (Fig. 3C, blue). This suggests that the chemical environment of Cys-127 is the same in the reduced and final oxidized forms, but different in the initial oxidized MsrB species, which indicates an initial Cys-122–Cys-127 and final Cys-66–Cys-122 disulfides. Similarly, we can focus on the Cys-122 region, by monitoring spectral changes of the Tyr-121, Cys-122, and Ile-123 NH resonances (Fig. 4, orange panel). The local environment of Cys-122 disulfide-bound to Cys-127 (red spectrum) is very different from that of Cys-122 coupled to Cys-66 (blue spectrum) or bearing a free thiol (black spectrum). A similar analysis of the Cys-66 surroundings in the two disulfides could not be performed because of the paucity of the resonance assignments in the Asn-63–Gly-67 region. Overall, MsrB oxidation by MetSO leads to the formation of an initial Cys-122–Cys-127 species, followed by a disulfide exchange reaction; yielding the final Cys-66–Cys-122 form.
Cys-66–Cys-122 disulfide is recycled by the thioredoxin/thioredoxin reductase pathway
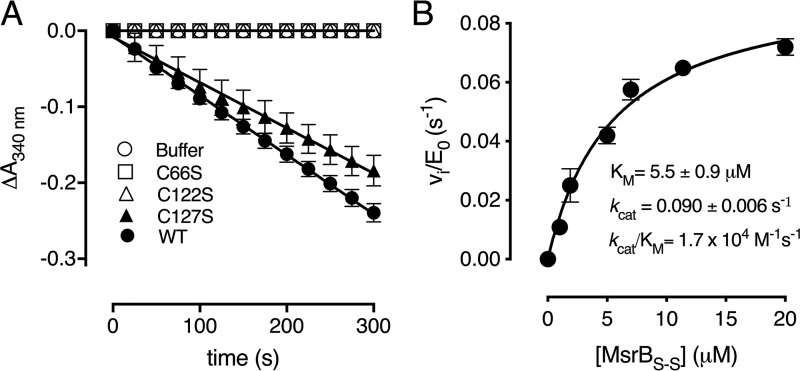
The thioredoxin/thioredoxin reductase (Trx/TrxR) pathway is used by most organisms to recycle protein disulfide bonds generated under oxidizing environment (30, 31). To determine whether the Cd-MsrB disulfide bonds formed after MetSO reduction could be recycled by Trx, a spectrophotometric coupled enzyme assay was performed (Fig. 5A). The consumption of NADPH was monitored at 340 nm in function of time for WT Cd-MsrB and the C66S, C122S, and C127S mutants. The C66S and C122S mutants showed no NADPH consumption, whereas mutation of Cys-127 (C127S) only marginally affected the coupling to the Trx/TrxR pathway (Fig. 5A). Based on these results, we showed that Trx only recognizes the disulfide between Cys-66 and Cys-122.
Figure 5.
Thioredoxin only reduces the Cys-66–Cys-122 disulfide. A, progress curves of WT Cd-MsrB and the cysteine mutants (C66S, C122S, and C127S) coupled to the Trx/TrxR pathway, following MetSO reduction, are shown. The C66S and C122S mutants do not consume NADPH, whereas mutating Cys-127 has almost no effect on the reaction rate. B, the reduction of Cys-66–Cys-122 disulfide bond by Trx follows the Michaelis-Menten steady-state kinetics. Increasing concentrations of Met-SO–oxidized Cd-MsrBS-S were used as substrate for thioredoxin. From the Michaelis-Menten curve, the rate of disulfide bond reduction was determined to be 1.7 × 104 m−1 s−1. The data are presented as a mean ± S.D. of three independent technical repeats and the graphs were generated using Prism8.
Next, we determined the steady-state kinetic parameters for MetSO reduction by WT Cd-MsrB, which resulted in a kcat/Km of 86 m−1 s−1 (Fig. S6). The rate of disulfide bond reduction by thioredoxin was determined by first incubating Cd-MsrB with 10 mm MetSO for 30 min at room temperature to allow for disulfide bond formation. MetSO was then removed, and increasing concentrations of oxidized Cd-MsrB (MsrBS-S) (0–20 μm) were used as substrate for Trx within the Trx/TrxR pathway. NADPH consumption at 340 nm was monitored in function of time, which indicated MsrBS-S disulfide bond reduction by Trx. The initial velocities obtained were divided by the concentration of Trx, and plotted against the concentration of MsrBS-S. The data were fitted with the Michaelis-Menten equation, and the rate of disulfide bond reduction by Trx was determined to be 1.7 × 104 m−1 s−1 (Fig. 5B).
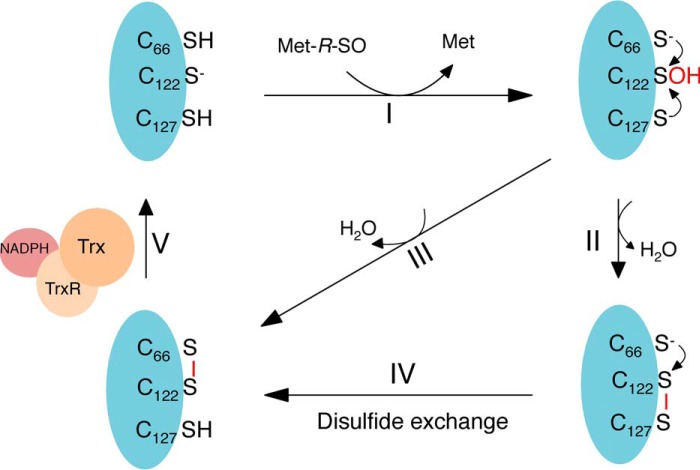
Based on these results, we propose the following mechanism (Fig. 6), where (step I), the nucleophilic Cys-122 attacks the sulfoxide of MetSO, which results in the release of Met, and the formation of a sulfenic acid on Cys-122. Then, either Cys-66 or Cys-127 performs a nucleophilic attack on the sulfur of the sulfenic acid on Cys-122, which results in the formation of Cys-122–Cys-127 (step II) or Cys-66–Cys-122 (step III) disulfides. After Cys-122–Cys-127 disulfide bond formation, a disulfide exchange occurs, where the thiolate of Cys-66 attacks the sulfur of Cys-122 and forms a Cys-122–Cys-66 disulfide (step IV), which is then recognized by thioredoxin and reduced (step V) (Fig. 6).
Figure 6.
Catalytic mechanism of Cd-MsrB coupled to the Trx/TrxR pathway. (I) Cd-MsrB Cys-122 performs a nucleophilic attack on the sulfoxide of MetSO, which results in the formation of a sulfenic acid on Cys-122 and the release of Met. Then, the catalytic cysteines, Cys-66 or Cys-127, attack the sulfur of the sulfenic acid, which results in the formation of the Cys-122–Cys-127 or Cys-122–Cys-66 disulfides (II or III). For Cys-122–Cys-127, a disulfide exchange occurs and Cys-66–Cys-122 is formed (IV), which is recognized by the Trx and reduced (V).
Cd-MsrB is not recycled through the mycothiol reducing pathway
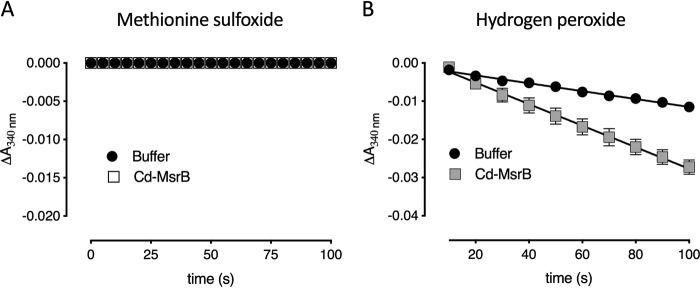
Aside from the Trx/TrxR pathway, most organisms use alternative reducing pathways such as the GSH pathway (32, 33). Actinobacteria, such as C. diphtheriae have an analogous GSH pathway, which is the mycothiol pathway (10, 26, 34). As has been reported for C. diphtheriae MsrA (26), we wanted to know whether Cd-MsrB could be recycled via the mycothiol pathway. In a spectrophotometric assay, the reduction of MetSO by Cd-MsrB was coupled to the mycothiol(MSH)/mycoredoxin1 (Mrx1)/mycothione reductase (Mtr) pathway (Fig. 7) (10). No NADPH consumption was observed, indicating that recycling of Cd-MsrB is not coupled to the mycothiol pathway (Fig. 7A). This result was also confirmed by MS, where Cd-MsrB incubated in the presence of mycothiol and MetSO did not show S-mycothiolation of its cysteines.
Figure 7.
Mycothiol protects Cys-127 in the presence of hydrogen peroxide. A, Cd-MsrB is not recycled via the mycothiol reduction pathway. Progress curves show no consumption of NADPH when Cd-MsrB is coupled to the MSH/Mrx1/Mtr pathway after adding MetSO as a substrate. B, in the presence of mycothiol and H2O2, Cd-MsrB gets S-mycothiolated and becomes substrate of the MSH/Mrx1/Mtr reduction pathway. The rate of demycothiolation is 17.0 ± 0.4 milli-absorbance units/min. Data are presented as a mean ± S.D. of three independent technical repeats and the graphs were generated using Prism8.
S-Mycothiolation reversibly protects the nonconserved Cys-127 from overoxidation
To test whether MSH is involved in the protection of Cd-MsrB cysteines during hydrogen peroxide (H2O2) stress, Cd-MsrB was incubated with mycothiol prior to the addition of H2O2. As a control, Cd-MsrB was incubated with MSH in the absence of H2O2. The samples were analyzed by MS and, from the three catalytic cysteines (Cys-66, Cys-122, and Cys-127), only the nonconserved Cys-127 was found S-mycothiolated (Fig. S7). Aside from Cys-127 S-mycothiolation, Cys-100 (involved in the coordination of zinc) was also observed to be S-mycothiolated. This could be due to the presence of a minor population of Cd-MsrB within the sample, where Cys-100 is not engaged in the coordination of Zn2+, and which could be S-mycothiolated after oxidation.
As S-mycothiolation of Cd-MsrB is used for the protection of the catalytic Cys-127, we next tested whether S-mycothiolated Cys-127 can be de-mycothiolated via coupling to the MSH/Mrx1/Mtr pathway (10, 34, 35). A spectrophotometric assay was performed, where the coupling of Cd-MsrB to the MSH/Mrx1/Mtr pathway in the presence of H2O2 was assessed (Fig. 7B). Upon addition of H2O2, we observed a decrease of the A340 nm in function of time, which indicates de-mycothiolation by Mrx1 (10). The control sample without Cd-MsrB showed only a minor decrease in the absorbance at 340 nm in function of time. This minor background of NADPH consumption is due to reduction of oxidized mycothiol present in the mycothiol batch.
Reversible intra- and intermolecular disulfides protect cysteines from overoxidation
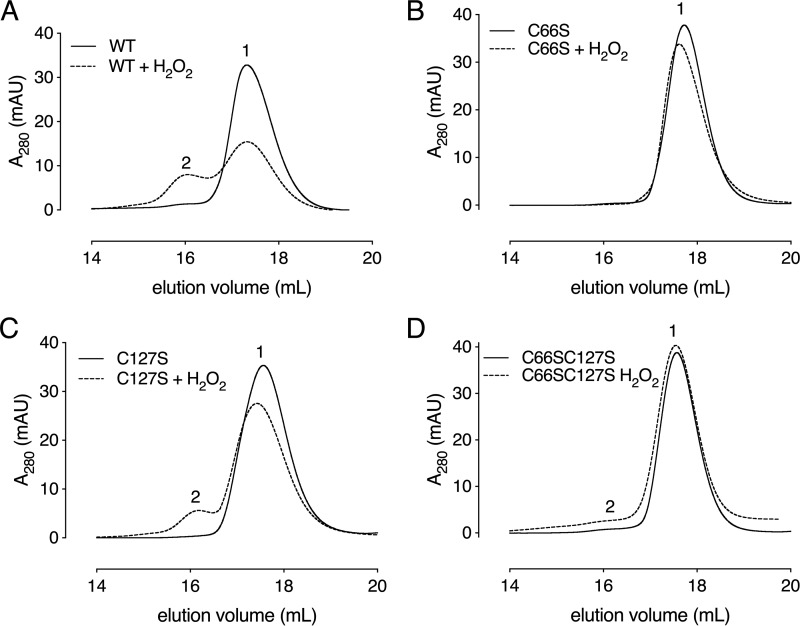
Next, we decided to study the impact of oxidation on the oligomerization state of Cd-MsrB by evaluating its elution position on a Superdex 200 size-exclusion column (Fig. 8). Incubation with a 5-fold excess of H2O2 resulted in a decrease of the monomeric form and an increase of the dimeric population of Cd-MsrB. To determine whether this shift is due to the presence of a disulfide bond, a gel-shift assay was used with increasing concentrations of H2O2 up to 2 mm (Fig. S8). We observed an increase in the dimerization and oligomerization state of Cd-MsrB with increasing H2O2 concentrations (Fig. S8). An additional monomeric Cd-MsrB band, which migrates slightly faster than the reduced band, was also observed (Fig. S8). Addition of DTT shifted both the dimeric and the additional monomeric band to the migration position of the reduced monomeric band (Fig. S8), indicating that inter- and intramolecular disulfides are being reduced. Overall, we showed that in the presence of H2O2, Cd-MsrB forms reversible disulfide bonds, which might be interpreted as a protection mechanism against overoxidation.
Figure 8.
Cys-66 and Cys-127 are involved in Cd-MsrB dimerization in the presence of H2O2. Size-exclusion chromatograms of Cd-MsrB (A) WT, and its Cys mutants, (B) C66S, (C) C127S, and (D) C66S/C127S), in the presence and absence of H2O2 are shown. Elution peaks 1 and 2 correspond to the elution position of the monomer (17.4 kDa) and the dimer (34.8 kDa), respectively. These results indicate that Cys-66 and Cys-127 are involved in the dimerization of Cd-MsrB. The data presents the union of two independent technical repeats.
To investigate which cysteines are involved in the dimerization, we repeated the size-exclusion chromatography (SEC) experiment with C66S, C127S, and C66S/C127S mutants. Oxidation of the C66S mutant did not result in a peak shift on SEC (Fig. 8B), indicating its role in the dimerization of Cd-MsrB. Oxidation of the C127S single mutant and the C66S/C127S double mutant resulted in only a minor increase of the dimeric population (Fig. 8, C and D). These results indicate that both Cys-127 and Cys-66 are engaged in the dimerization of Cd-MsrB in the presence of H2O2.
As Zn2+ is coordinated by cysteines in Cd-MsrB, we decided to investigate whether Zn2+ is released in the presence of H2O2. Therefore, we analyzed the dimeric and monomeric peaks of the SEC with ICP-MS. We found that oxidized WT Cd-MsrB (3000 μg/liter) has a similar zinc content as reduced WT Cd-MsrB (3300 μg/liter), which indicates that Zn2+ is still present after the H2O2-induced dimerization.
Discussion
Msr enzymes regulate protein function and downstream cellular signaling processes by reducing MetSO (36–39). In this study, we focus on the catalytic and oxidation protection mechanisms of C. diphtheriae MsrB (Cd-MsrB), which has three catalytic cysteines, two conserved and one nonconserved cysteine. Most MsrB enzymes have one or two catalytic cysteines (Fig. S1) that form a sulfenic acid or a disulfide after substrate reduction, which is then reduced by the Trx system (31, 40, 41). Unlike these enzymes, Cd-MsrB has an additional nonconserved catalytic cysteine (Cys-127) located on a loop between the β6 and β7 strands, located close to the C terminus (Fig. 1 and Fig. S1). At the position of this cysteine, we find a serine or alanine in the MsrB enzymes from other species (28) (Fig. S1). By generating three Cys to Ser mutants (C66S, C122S, and C127S), we showed that only mutating Cys-122 leads to complete loss of activity, which indicate its role as the nucleophilic cysteine essential for the first step in the catalytic mechanism, the reduction of MetSO (Fig. 6, I, and Fig. S4). Loss of activity upon mutation of this conserved cysteine has also been seen for both human MsrB3 (Cys-126) and Drosophila MsrB (Cys-124) (28, 42). The kcat/Km of methionine formation by Cd-MsrB (86 m−1 s−1) is in the same range as other MsrB enzymes, such as Saccharomyces cerevisiae (90 m−1 s−1), Arabidopsis thaliana (47 m−1 s−1), and Neisseria meningitidis (19 m−1 s−1) (27, 43, 44). Important to note is that MsrB enzymes are more efficient in reducing the protein MetSO compared with free MetSO. Examples include the catalytic efficiency of N. meningitidis MsrB (180 m−1 s−1) and S. cerevisiae MsrB (1180 m−1 s−1) measured with N-acetyl or dabsyl MetSO mimics of protein MetSO (27, 43). Comparing the kcat/Km of free MetSO reduction by Cd-MsrB (86 m−1 s−1) with Cd-MsrA (7.5 × 104 m−1 s−1) (26), more than 1000-fold difference was observed. This shows that free MetSO is the preferred substrate for MsrA, as described (27, 43, 45, 46).
Once MetSO is reduced, a sulfenic acid is formed on the nucleophilic Cys-122 of Cd-MsrB (Fig. 6, I, and Fig. S4), which is then attacked by one of the resolving cysteines (27, 28). Cd-MsrB is unique in having two resolving cysteines, which can form a disulfide bond with the nucleophilic Cys-122 (Fig. 6, II and III). Based on the MS analysis, DTNB assay, and the HSQC spectra, it becomes clear that two populations of disulfides are formed after MetSO reduction: Cys-122–Cys-127 (nonconserved disulfide), which is the major disulfide form, and Cys-122–Cys-66 (conserved disulfide), which is the minor disulfide form (Figs. 3 and 6, and Table S2). From the solution structure and the HSQC spectra of oxidized Cd-MsrB, we observed that major structural changes are required for the formation of Cys-122–Cys-127 disulfide bond, which is buried and inaccessible to Trx (Figs. 1B, 3, 4, and Fig. S5). On the other hand, the Cys-122–Cys-66 disulfide is exposed, and no major conformational changes are required to form this disulfide (Fig. 3C). This disulfide is accessible for reduction by Trx (Fig. 5A). Structural studies on Xanthomonas campestris MsrB (Xc-MsrB) also showed that drastic structural changes are required for the formation of a disulfide bond between the conserved nucleophilic and nonconserved resolving cysteines, which are located more than 3 Å apart (24). This observation for Xc-MsrB could fit with the overall structural changes that we observe for Cd-MsrB following MetSO reduction (Fig. 3 and Fig. S5).
Several enzymes with major structural changes during thiol-disulfide exchange have been reported. One example that we have studied was the reduction of arsenate by arsenate reductase from Staphylococcus aureus plasmid pI258 (47, 48). Here, the first resolving Cys-82 attacks the nucleophilic Cys-10, which results in the release of the reduced substrate, and the formation of a buried Cys-10–Cys-82 disulfide (47). Next, the second resolving Cys-89, which is located more than 10 Å away from Cys-82, moves out of its hydrophobic pocket to attack the first disulfide, which results in the formation of a surface-exposed Cys-82–Cys-89 disulfide, accessible for Trx reduction (47, 49). Another example is seen for Cd-MsrA after MetSO reduction (26). Here, a buried disulfide is formed between the nucleophilic Cys-52 and the first resolving Cys-206. Next, disulfide exchange results in the formation of a second disulfide between Cys-206–Cys-215, which becomes accessible to Trx (26).
Our findings suggest that after substrate reduction, Cd-MsrB undergoes a large structural reorganization, which results in the formation of a buried and Trx inaccessible Cys-122–Cys-127 disulfide (Figs. 1B, 5A, and 6). By comparing the HSQC spectra of the MetSO-oxidized Cd-MsrB, which we measured directly and after 19 h (Figs. 3 and 4), we observed a shift from a Cys-122–Cys-127 disulfide to a Cys-122–Cys-66 disulfide (Fig. 3, B and C). Our data suggest that in the absence of Trx, the Cys-122–Cys-127 disulfide form of Cd-MsrB accumulates, and over time slowly exchanges to form a minor population with a Cys-122–Cys-66 disulfide (Figs. 3 and 4). However, driven by thioredoxin, which only reduces the minor population (Cys-122–Cys-66 disulfide) with a catalytic efficiency of 1.7 × 104 m−1 s−1 (Fig. 5B), the exchange from the Cys-122–Cys-127 disulfide to Cys-122–Cys-66 becomes much faster (Fig. 5).
A recent study by Cao et al. (42) characterized human MsrB3 (hMsrB3) with three catalytic cysteines, but unlike Cd-MsrB, hMsrB3 has two nonconserved resolving cysteines (Cys-3 and Cys-9) located at the N terminus. Similar to Cd-MsrB, hMsrB3 resolving cysteines (Cys-3 and Cys-9) can form a disulfide with the conserved nucleophilic Cys-126. Here, the resolving cysteine that is not engaged in the first disulfide, then attacks the Cys-126–Cys-3 or the Cys-126–Cys-9 disulfide bond, resulting in a Cys-3–Cys-9 disulfide between two resolving cysteines. This disulfide is recognized and reduced by Trx. Although hMsrB3 has three catalytic cysteines, it has a different mechanism than the one we observed for Cd-MsrB, where the disulfide recognized by Trx is between a nucleophilic and only one specific resolving cysteine (Cys-122–Cys-66) (42).
As most organisms could use an alternative reducing system, such as the LMW thiol glutathione (GSH)/GSH reductase system (21, 50), we decided to investigate whether Cd-MsrB could also be coupled to an alternative reducing pathway found in Actinobacteria, which is the MSH pathway (10, 26, 33–35, 51). MSH is involved in different processes during oxidative stress, including being a cofactor of important antioxidant enzymes (52). When thiol peroxidases, such as mycothiol peroxidase (Mpx), thiol peroxidase (Tpx), and AhpE, reduce hydroperoxides, they form a sulfenic acid on their peroxidatic cysteine (34, 51, 53, 54). Mycothiol then attacks this S-sulfenylated thiol to form a mixed disulfide (34, 51, 53, 54). Mrx1, which uses either a monothiol or a dithiol mechanism to reduce its target proteins (34, 51), de-mycothiolates the thiol peroxidases via the MSH pathway. In the case of Mpx, Tpx, and AhpE, Mrx1 attacks the mixed disulfide between the MSH and peroxidases, releasing the reduced form of the peroxidase and a mycothiolated form of Mrx1. Another molecule of MSH then attacks the sulfur of the MSH on the mixed disulfide, which releases reduced Mrx1 and oxidized mycothiol (MSSM). MSSM is then recycled by Mtr, which uses NADPH as electron donor (34, 51, 53, 54).
Using a spectrophotometric assay and MS, we showed that upon reduction of MetSO, Cd-MsrB does not couple to the MSH pathway (Fig. 7A). A similar study performed by Si et al. (55) showed that Corynebacterium glutamicum MsrB is also not coupled to the MSH pathway after substrate reduction. Although MSH is present in millimolar concentrations in the cell (33), it is not necessarily used in the catalytic mechanism of all enzymes. However, in this study, what was interesting is the S-mycothiolation of the nonconserved catalytic Cys-127 in the presence of H2O2, which indicates that Cd-MsrB gets S-mycothiolated to protect its Cys-127 from overoxidation (Fig. S7). We also showed that Cd-MsrB is de-mycothiolated via the MSH pathway (Fig. 7B). Therefore, MSH is used both for Cys-127 protection against overoxidation and deprotection, to restore the reduced form of the protein. In addition to S-mycothiolation, Cd-MsrB dimerizes and forms both inter- and intramolecular disulfides in the presence of H2O2. Mutation of the conserved Cys-66 and/or the nonconserved Cys-127 eliminates the dimerization in the presence of H2O2 (Fig. 8). This shows that both cysteines are involved in forming intermolecular disulfides in the presence of H2O2. With a gel-shift assay, we showed the reversibility of the intermolecular disulfides of Cd-MsrB (Fig. S8).
The solution structure of Cd-MsrB shows that a Zn2+ atom is coordinated by 4 cysteines (Fig. 1B), which seems to be important for its overall structural fold (24, 25). Studies on MsrB enzymes where the coordinating cysteine residues have been mutated show loss of the Zn2+ and as a consequence loss of MsrB activity (25, 28). In the presence of H2O2, Cd-MsrB does not lose the Zn2+. This implies that under H2O2 stress, Cd-MsrB maintains its structural fold and can be reactivated by the Trx and mycothiol reduction pathways. Overall, MsrB enzymes are flexible in employing different types of catalytic mechanisms depending on the number and location of catalytic cysteines involved in the reduction of MetSO. With structural and functional studies, we unraveled the detailed catalytic mechanism of Cd-MsrB, which involves a major structural rearrangement to expose the Cys-122–Cys-66 disulfide for Trx reduction. To maintain functionality after reduction, Cd-MsrB reversibly protects its catalytic cysteines from overoxidation by combining intra- and intermolecular disulfide formation with S-mycothiolation.
Experimental procedures
Cloning, expression, and purification of Cd-MsrB
Cloning, expression, and purification was performed as described by Tossounian et al. (20).
Site-directed mutagenesis to generate Cd-MsrB Cys to Ser mutants
Using the QuikChangeTM site-directed mutagenesis protocol (Stratagene), site-directed mutagenesis was performed on the Cd-msrB-pET-28b(+) vector. Forward primers 5′-ACTGAGAAATTTAATTCACATTCTGGGTGGCCGTCCTTCTTCTCG-3′, 5′-ACCCCTACGGATCTGCGCTATTCCATCAACAGCGTGTGCTTGACC-3′ and 5′-CGCTATTGCATCAACAGCGTGTCCTTGACCCTCATTCCGGCAGAA-3′, and reverse primers 5′-CGAGAAGAAGGACGGCCACCCAGAATGTGAATTAAATTTCTCAGT-3′, 5′-GGTCAAGCACACGCTGTTGATGGAATAGCGCAGATCGTAGGGGT-3′, and 5′-TTCTGCCGGAATGAGGGTCAAGGACACGCTGTGATGCAATAGCG-3′ were used to construct the Cd-msrB C66S, C122S, and C127S gene mutants, respectively. To construct the C66S/C122S and C66S/C127S double mutants, the Cd-msrB-C66S vector was used as a template for further site-directed mutagenesis, using the C122S or C127S forward and reverse primers, obtaining the double mutant constructions C66S/C122S and C66S/C127S, respectively.
HPLC reversed-phase chromatography analysis of methionine formation
The Met formation assay was performed as described (26). Briefly, prior to the start of the assay, Cd-MsrB was reduced with 20 mm DTT at room temperature for 30 min. Size-exclusion chromatography was used to remove DTT and to buffer exchange to 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl. The reduced Cd-MsrB or its Cys mutants (25 μm) were incubated with 10 mm DTT, as a recycling agent, and 10 mm MetSO substrate for 10 min at room temperature. The reaction was stopped by the addition of 1% TFA and the sample was then diluted 5-fold with 15% acetonitrile (ACN) and 0.1% TFA solution. The samples were centrifuged to remove precipitants and injected onto an ACE 5 C18 AR column (Achrom), equilibrated in 15% acetonitrile and 0.1% TFA and eluted isocratically at 0.5 ml/min. The methionine peak formation was followed at 215 nm in function of time. To determine the concentration of methionine, a methionine standard curve (0–2 mm) was made by plotting the peak area (μV·s) in function of injected methionine concentration. The experiments were performed with at least two independent replicates and the results were visualized using Prism8.
Kinetic parameter determination of MetSO reduction by Cd-MsrB
Cd-MsrB WT (2.5 μm) was incubated with DTT (15 mm) and varying substrate concentrations (0–4 mm). At several time points, the reaction was stopped and the samples were diluted 5-fold and injected onto the ACE 5 C18 AR column. The methionine peak formation was monitored at 215 nm in function of time. The results obtained were analyzed by linear regression. Product formation (μm) in function of time (min) was obtained from the slope (vi), which was used to plot vi/E0 in function of MetSO concentration. From the Michaelis-Menten curve, the Km, kcat, and kcat/Km values were calculated. Knowing that MsrB only reduces the R-stereoisomer of MetSO and assuming that the commercial MetSO contains a 1/1 ratio of R- and S-stereoisomer, the concentration of Met-R-SO was obtained by dividing the MetSO concentration by two. Data were obtained in triplicate and the results were analyzed with Prism8.
Anti-dimedone Western blotting
To determine the presence of Cd-MsrB sulfenic acid formation following MetSO reduction, anti-dimedone Western blotting was used. This experiment was performed under anaerobic conditions using the Whitely A35 anaerobic workstation. Cd-MsrB WT, C66S/C122S, and C66S/C127S mutants were reduced as described, and size-exclusion chromatography (Superdex 75 10/300 column) on an AKTA pure system (GE Healthcare Life Sciences) was used to remove excess of DTT and to buffer change to 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl. The reduced Cd-MsrB WT or double mutants (25 μm) were first incubated with 1 mm dimedone (Sigma) for 5 min at room temperature. Afterward, MetSO (10 mm) was added to the reaction mixture, which was incubated for 1 h in the dark at room temperature. To stop the reaction, the sample was incubated in the dark with 10 mm N-ethylmaleimide (NEM) at room temperature for 20 min. The samples were analyzed by Western blotting.
Western blotting was developed with rabbit anti-sulfenic acid antibody (1:10,000 dilution) at room temperature for 1 h, followed by 30 min incubation at room temperature with the secondary goat anti-rabbit antibody conjugated to alkaline phosphatase (1:10,000). Dimedone containing bands were visualized 20 min following the addition of 5-bromo-4-chloro-3-indoly phosphate/nitro-blue tetrazolium chloride (BCIP/NBT) premixed solution (Sigma-Aldrich) in the dark at room temperature.
Determining free thiol content with DTNB
The free thiol content of Cd-MsrB WT, and the C66S, C122S, and C127S mutants was monitored in the presence and absence of MetSO substrate using DTNB. The protein samples were reduced with 20 mm DTT for 30 min at room temperature, and Superdex 75 10/300 column on AKTA pure system (GE Healthcare Life Sciences) was used to remove excess DTT and to change buffer to 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl. Reduced Cd-MsrB WT or Cys mutant (1 μm) was incubated with DTNB (300 μm) for 30 min at room temperature. The sample (200 μl) was then transferred to a 96-well-microplate (Thermo ScientificTM NuncTM MicrowellTM), and the increase in absorbance was followed spectrophotometrically at A412 nm. To prepare the oxidized sample, Cd-MsrB WT or the mutants were incubated with MetSO (8 mm) for 40 min. Important to note, the free thiol content of the C66S mutant was determined in its denatured form. Therefore, the reduced and MetSO-treated C66S mutant was incubated with 6 m guanidinium hydrochloride for 30 min at room temperature, followed by dilution and incubation with DTNB for 30 min. Three independent replicates were analyzed. The number of free thiol was calculated based on Equation 1, where ϵ represents the extinction coefficient of TNB2−, which is 14,150 m−1 cm−1 for nondenaturing conditions and 13,700 m−1 cm−1 for denaturing conditions.
| (Eq. 1) |
Secondary structural changes monitored by CD
To monitor the secondary structure changes of Cd-MsrB in the reduced and MetSO substrate-treated samples, CD was used to record the far-UV spectra (190–260 nm). Prior to the CD experiment, Cd-MsrB was reduced with 20 mm DTT for 30 min at room temperature. Gel filtration using the AKTA pure system was used to remove excess DTT and change buffer in 20 mm sodium phosphate and 200 mm sodium fluoride, pH 7.5. To obtain MetSO-treated samples, reduced Cd-MsrB (50 μm) was incubated with a 10-fold excess of MetSO for 1 h at 25 °C. Excess MetSO was removed using Micro Bio-Spin® chromatography columns (Bio-Rad) equilibrated in 20 mm sodium phosphate and 200 mm sodium fluoride, pH 7.5. Following sample preparation, Cd-MsrB–reduced and MetSO–treated samples (4 μm) were analyzed by a Jasco J-810 spectropolarimeter at 25 °C in a quartz cuvette (1-mm path length). The same experimental conditions were used to compare the overall secondary structural differences between reduced Cd-MsrB WT, C66S, and C127S mutants. Far-UV CD spectra were recorded and the results were analyzed with Prism8.
Cd-MsrB NMR sample preparation
For the production of double labeled [U-13C,15N]Cd-MsrB, the minimal medium described by Volkov et al. (56) was used. Briefly, Rosetta (DE3) Escherichia coli cells containing Cd-msrB-pET28(+) plasmid were grown in minimal medium at 37 °C until A600 reached 1. The cells were then induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and further incubated overnight at 30 °C. The cells were then harvested, sonicated, and purified as described by Tossounian et al. (20). Following purification, the Cd-MsrB double-labeled samples were reduced with 20 mm DTT for 30 min at room temperature, and the buffer changed to 20 mm sodium phosphate, pH 7.0, 150 mm NaCl. Prior to NMR data collection, the reduced double-labeled Cd-MsrB sample (1 mm) was mixed with DTT (2 mm). To prepare the MetSO-treated oxidized sample, reduced Cd-MsrB was incubated with MetSO (1:2 ratio, respectively) for 10 min at 25 °C. To remove excess of MetSO, the sample was buffer changed to 20 mm sodium phosphate, pH 7.0, 150 mm NaCl, before determining its HSQC spectrum. After 19 h of incubation at 25 °C, another HSQC spectrum was determined.
NMR solution structure determination
All NMR experiments were performed at 25 °C on a Bruker Avance III HD 800 MHz spectrometer equipped with a TCI cryoprobe. For the protein structure determination, the sample contained 1 mm U-13C,15N-labeled MsrB and 2 mm DTT in 20 mm sodium phosphate, pH 7.0, 150 mm NaCl, and 10% D2O for the lock. All NMR data were processed in TopSpin 3.5 (Bruker) or NMRPipe (57) and analyzed in CCPN (58). Nearly complete, unambiguous 1H, 13C, and 15N resonance assignments of the protein nuclei were obtained from a suite of standard multidimensional NMR experiments: 2D [1H,15N]-HSQC, [1H,13C]-HSQC, and constant-time [1H,13C]-HSQC for the aromatic region; triple-resonance HNCACB, HN(CO)CACB, HNCO, HN(CA)CO, HBHA(CO)NH, (H)CCH-TOCSY, and H(C)CH-TOCSY experiments; 2D (HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE spectra for the aromatic resonances; and 3D 15N-edited NOESY-HSQC and 13C-edited NOESY-HSQC for aliphatics and aromatics. The resonance assignments were deposited in the BMRB data bank under accession number 28052.
The 3D 15N-edited NOESY-HSQC and 13C-edited NOESY-HSQC spectra for aliphatics and aromatics, all acquired with the mixing time of 120 ms, were subsequently used for the protein structure calculation. The NOE cross-peaks, determined with CCPN Analysis (58), were combined with the dihedral angle restraints, obtained with DANGLE (59), and used as an input for the automated NOE assignment and structure calculations in CYANA version 3 (60), followed by the explicit solvent and torsion angle refinement in CNS (61) and Xplor-NIH (62), respectively. The coordination geometry of the Zn-(Cys)4 cluster was obtained from the high-resolution X-ray structure of a homologous MsrB (PDB ID 3HCJ) and applied as a set of S-S distance restraints during the final refinement step. To complete the Td coordination polyhedron, the Zn atom was subsequently added to the molecular frame in Xplor-NIH (62). The 20 lowest-energy structures were retained and deposited in PDB under the accession code 6TR8. The NMR structure calculation and refinement statistics are presented in Table S1.
Coupled enzyme assay with the thioredoxin/thioredoxin reductase pathway
The MetSO reduction by Cd-MsrA coupled to the Trx/TrxR pathway described by Tossounian et al. (26) was adapted to include Cd-MsrB. Briefly, Cd-MsrB WT and Cys mutants (C66S, C122S, and C127S) were reduced with 20 mm DTT at room temperature for 30 min. The samples were gel filtered using a Superdex 75 HR 10/30 column equilibrated in 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl. A reaction mixture composed of 0.5 mm NADPH, 6 μm C. glutamicum TrxR, 3 μm C. glutamicum Trx, and 5 μm Cd-MsrB was incubated for 10 min at 37 °C in the same buffer solution. Following the incubation, 5 mm MetSO was added to start the reaction, and the decrease in NADPH was monitored at A340 nm using a SpectraMax 340PC spectrophotometer (Molecular Devices). Three independent replicates were obtained and the progress curves were analyzed using Prism8.
To determine the rate of Cys-66–Cys-122 disulfide bond reduction by thioredoxin, reduced Cd-MsrB was incubated with 10 mm MetSO at room temperature for 30 min. Excess MetSO was removed using a Micro Bio-Spin® chromatography column (Bio-Rad), equilibrated in the same buffer solution. A reaction mixture was composed of 1 mm NADPH, 6 μm C. glutamicum TrxR, and 3 μm C. glutamicum. Trx was incubated for 10 min at 37 °C. Increasing concentrations of MetSO-oxidized Cd-MsrBS-S (0–20 μm) were added to the reaction mixture and NADPH consumption was monitored at A340 nm. Three independent replicates were obtained and the progress curves were analyzed using Prism8. Trx and TrxR were cloned and purified as described elsewhere (35).
Coupled enzyme assay with the mycothiol/mycoredoxin1/mycothione reductase pathway
The MetSO reduction by Cd-MsrA coupled to the mycothiol/mycoredoxin1/mycothione reductase pathway (MSH pathway) as described by Tossounian et al. (26) was adapted. Briefly, Cd-MsrB WT was reduced with 20 mm DTT at room temperature for 30 min. The sample was then gel filtered using a Superdex 75 HR 10/30 column equilibrated in 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl. A reaction mixture composed of 0.5 mm NADPH, 5 μm C. glutamicum Mtr, 0.5 μm C. glutamicum Mrx1, 100 μm MSH, and 10 μm Cd-MsrB was incubated for 20 min at 37 °C in the same buffer solution. Following the incubation, 5 mm MetSO was added to start the reaction, and the decrease in NADPH was monitored at A340 nm in function of time, using a SpectraMax 340PC spectrophotometer (Molecular Devices). To determine whether the MSH pathway is used in the presence of hydrogen peroxide (H2O2), the same reaction composition was used, but the MetSO was replaced by 400 μm H2O2. Three independent replicates were obtained and the progress curves were analyzed using Prism8.
Determining Cd-MsrB oligomerization with size-exclusion chromatography
Using a standard of proteins with known molecular weights (gel filtration standard, Bio-Rad), Cd-MsrB oligomerization states were determined in the absence and presence of H2O2. Reduced Cd-MsrB (50 μm) was incubated with 250 μm H2O2 for 30 min at room temperature. The sample was injected on a Superdex 200 HR 10/30 column equilibrated in 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl. Based on the elution volume, the molecular weights of both the reduced and oxidized Cd-MsrB samples were determined. Two independent replicates were obtained, and the chromatogram is presented as the union of both replicates.
Determining Cd-MsrB oligomerization state with a gel-shift assay
Cd-MsrB was reduced and buffer exchanged to remove DTT, as described. Next, Cd-MsrB was incubated with increasing concentrations of H2O2 (0–2 mm) at room temperature for 20 min. The reaction was stopped by removing the oxidant with a microbiospin column, samples were incubated with 10 mm NEM in the dark for 10 min, and analyzed on SDS-PAGE gel. To determine whether DTT restores the reduced form of Cd-MsrB, the oxidized samples were incubated with 5 mm DTT, blocked with 10 mm NEM, and analyzed on SDS-PAGE gel.
Mass spectrometric analysis of disulfide bond formation and S-mycothiolation
Cd-MsrB was reduced and buffer exchanged to remove DTT. Reduced Cd-MsrB (30 μm) was incubated with excess MetSO (5 mm) at room temperature for 20 min, and excess of MetSO and Met were removed on a Micro Bio-Spin column. The remaining free thiols were blocked with NEM. As control, reduced Cd-MsrB was used. Both reduced and MetSO-incubated samples were trypsin digested.
The generated peptides were dissolved in solvent A (0.1% TFA in 2% ACN), directly loaded onto a reversed-phase pre-column (Acclaim PepMap 100, Thermo Scientific) and eluted in backflush mode. Peptide separation was performed on a reversed-phase analytical column (Acclaim PepMap RSLC, 0.075 × 250 mm, Thermo Scientific) developed with a linear gradient of 4–36% solvent B (0.1% FA in 98% ACN) for 36 min, 40–99% solvent B for 10 min, and holding at 99% for the last 5 min at a constant flow rate of 300 nl/min on an Ultimate 3000 RSLN nanoHPLC system (Thermo Fisher Scientific). The peptides were analyzed by an Orbitrap Fusion Lumos tribrid mass spectrometer (Thermo Fisher Scientific). The peptides were subjected to the NSI source followed by tandem mass spectrometry (MS/MS) in Fusion Lumos coupled online to the UPLC. Intact peptides were detected in the Orbitrap at a resolution of 120,000. Peptides were selected for MS/MS using HCD setting 35; ion fragments were detected in the Orbitrap at a resolution of 30,000. A data-dependent procedure of MS/MS scans was applied for the top precursor ions above a threshold ion count of 2.5E4 in the MS survey scan with 30.0-s dynamic exclusion. The total cycle time was set to 3 s. The electrospray voltage applied was 2.1 kV. MS1 spectra were obtained with an AGC target of 4E5 ions and a maximum injection time of 50 ms, and MS2 spectra were acquired with an AGC target of 5E4 ions and a maximum injection time of 100 ms. For MS scans, the m/z scan range was 350 to 1800. The resulting MS/MS data were processed using the Sequest HT search engine within Proteome Discoverer 2.2 against a custom database containing the C. diphtheriae MsrB sequence. Trypsin was specified as cleavage enzyme allowing up to 2 missed cleavages, 4 modifications per peptide, and up to 7 charges. Mass error was set to 10 ppm for precursor ions and 0.02 Da for fragment ions. Presence of NEM and Cys modifications were considered as variable modifications. The false discovery rate was assessed using a fixed value PSM validator and thresholds for protein, peptide, and modification site were specified at 1%. For disulfide bridge mapping, the MS raw files were analyzed with pLink 2.3 (63) with peptide masses set from 600 to 6000 Da and tolerance for precursor and fragment set to 20 ppm. False discovery rate was below 5% and MS/MS fragmentation was manually validated.
To determine whether Cd-MsrB can be S-mycothiolated in the presence of MetSO or H2O2, reduced Cd-MsrB (30 μm) was first incubated with MSH (200 μm) for 5 min at room temperature, followed by incubation with excess MetSO (5 mm) or H2O2 (300 μm) for 20 min at room temperature. Micro Bio-Spin columns were used to remove excess MetSO, Met, or H2O2, and samples were alkylated with NEM to block the remaining free thiols. As control, a sample of reduced Cd-MsrB and MSH in the absence of MetSO or H2O2 was used. The reduced and MetSO- or H2O2-treated samples were then trypsin digested. For the identification of S-mycothiolation sites, the LC-MS/MS data were acquired as described, except that the ion fragments were detected in the ion trap after CID fragmentation at 35%. Multistage fragmentation was enabled to promote richer fragmentation of daughter ions resulting from neutral loss of inositol from S-mycothiolated peptides.
High resolution ICP-MS
Both reduced and oxidized samples were analyzed for their zinc content using ICP-MS. Background zinc content was eliminated by analyzing the zinc content of the buffer solution. Zinc was determined by ICP-MS using a Thermo Finnigan Element II instrument (64). The extract was diluted 100-fold in 2% HNO3. 1 ppb of indium was used as an internal standard. Zinc was measured in medium resolution mode. Calibration standards (1–40 μg/liter) were prepared from a multielement stock solution (Merck XIII). The detection limit based on the standard deviation of the blank was 0.08 μg/liter for zinc and reproducibility at a concentration range of 10 μg/liter was 1.5%.
Author contributions
M.-A. T. and J. M. conceptualization; M.-A. T. formal analysis; M.-A. T., A.-C. K. T., L. B., K. W., A. M., M. L., D. V., A. N. V., and J. M. investigation; M.-A. T., A. N. V., and J. M. writing-original draft; M.-A. T., A. N. V., and J. M. writing-review and editing; L. B., D. V., and A. N. V. methodology; J. M. supervision; J. M. funding acquisition; L. M. M. critically read the manuscript.
Supplementary Material
This work was supported by the Vlaams Instituut voor Biotechnologie (VIB), a FWO Ph.D. fellowship grant (to M. A. T.), Hercules Foundation Grant HERC16, Anaerobic Workstation Grant 1508316N from FWO, and Strategic Research Programme Grant SRP34 of the Vrije Universiteit Brussel (to J. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Tables S1 and S2.
The atomic coordinates and structure factors (code 6TR8) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The resonance assignments reported in this paper have been submitted to the Biological Magnetic Resonance Data Bank database under accession no. 28052.
- Msr
- methionine sulfoxide reductase
- Cd-MsrB
- Corynebacterium diphtheriae methionine sulfoxide reductase B
- MetSO
- methionine sulfoxide
- Met-S-SO and Met-R-SO
- R and S epimeric forms of methionine sulfoxide
- ICP
- inductively coupled plasma
- ACN
- acetonitrile
- NEM
- N-ethylmaleimide
- DTNB
- 5,5-dithiobis-(2-nitrobenzoic acid)
- Trx
- thioredoxin
- TrxR
- thioredoxin reductase
- MSH
- mycothiol
- MSSM
- oxidized mycothiol (mycothione)
- Mrx1
- mycoredoxin1
- Mtr
- mycothione reductase
- NMR
- nuclear magnetic resonance
- HSQC
- heteronuclear single-quantum coherence
- SEC
- size-exclusion chromatography
- LMW
- low-molecular-weight.
References
- 1. Bell C. E., and Eisenberg D. (1996) Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry 35, 1137–1149 10.1021/bi9520848 [DOI] [PubMed] [Google Scholar]
- 2. Armstrong J. A., and Hart P. D. (1971) Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J. Exp. Med. 134, 713–740 10.1084/jem.134.3.713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosen G. M., Pou S., Ramos C. L., Cohen M. S., and Britigan B. E. (1995) Free radicals and phagocytic cells. FASEB J. 9, 200–209 10.1096/fasebj.9.2.7540156 [DOI] [PubMed] [Google Scholar]
- 4. Young I. S., and Woodside J. V. (2001) Antioxidants in health and disease. J. Clin. Pathol. 54, 176–186 10.1136/jcp.54.3.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park S., and Imlay J. A. (2003) High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 185, 1942–1950 10.1128/JB.185.6.1942-1950.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Winterbourn C. C., Hampton M. B., Livesey J. H., and Kettle A. J. (2006) Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J. Biol. Chem. 281, 39860–39869 10.1074/jbc.M605898200 [DOI] [PubMed] [Google Scholar]
- 7. Rosen H., Klebanoff S. J., Wang Y., Brot N., Heinecke J. W., and Fu X. (2009) Methionine oxidation contributes to bacterial killing by the myeloperoxidase system of neutrophils. Proc. Natl. Acad. Sci. U.S.A. 106, 18686–18691 10.1073/pnas.0909464106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Matés J. M., Pérez-Gomez C., and Nuñez de Castro I. (1999) Antioxidant enzymes and human diseases. Clin. Biochem. 32, 595–603 10.1016/S0009-9120(99)00075-2 [DOI] [PubMed] [Google Scholar]
- 9. Couturier J., Vignols F., Jacquot J. P., and Rouhier N. (2012) Glutathione- and glutaredoxin-dependent reduction of methionine sulfoxide reductase A. FEBS Lett. 586, 3894–3899 10.1016/j.febslet.2012.09.020 [DOI] [PubMed] [Google Scholar]
- 10. Van Laer K., Buts L., Foloppe N., Vertommen D., Van Belle K., Wahni K., Roos G., Nilsson L., Mateos L. M., Rawat M., van Nuland N. A., and Messens J. (2012) Mycoredoxin-1 is one of the missing links in the oxidative stress defence mechanism of Mycobacteria. Mol. Microbiol. 86, 787–804 10.1111/mmi.12030 [DOI] [PubMed] [Google Scholar]
- 11. Boschi-Muller S., Olry A., Antoine M., and Branlant G. (2005) The enzymology and biochemistry of methionine sulfoxide reductases. Biochim. Biophys. Acta 1703, 231–238 10.1016/j.bbapap.2004.09.016 [DOI] [PubMed] [Google Scholar]
- 12. Frelinger A. L. 3rd, and Zull J. E. (1986) The role of the methionine residues in the structure and function of parathyroid hormone. Arch. Biochem. Biophys. 244, 641–649 10.1016/0003-9861(86)90632-6 [DOI] [PubMed] [Google Scholar]
- 13. Grimaud R., Ezraty B., Mitchell J. K., Lafitte D., Briand C., Derrick P. J., and Barras F. (2001) Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276, 48915–48920 10.1074/jbc.M105509200 [DOI] [PubMed] [Google Scholar]
- 14. Lee B. C., and Gladyshev V. N. (2011) The biological significance of methionine sulfoxide stereochemistry. Free Radic. Biol. Med. 50, 221–227 10.1016/j.freeradbiomed.2010.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boschi-Muller S., Gand A., and Branlant G. (2008) The methionine sulfoxide reductases: catalysis and substrate specificities. Arch. Biochem. Biophys. 474, 266–273 10.1016/j.abb.2008.02.007 [DOI] [PubMed] [Google Scholar]
- 16. Dhandayuthapani S., Jagannath C., Nino C., Saikolappan S., and Sasindran S. J. (2009) Methionine sulfoxide reductase B (MsrB) of Mycobacterium smegmatis plays a limited role in resisting oxidative stress. Tuberculosis (Edinb) 89, S26–S32 10.1016/S1472-9792(09)70008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao C., Hartke A., La Sorda M., Posteraro B., Laplace J. M., Auffray Y., and Sanguinetti M. (2010) Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence. Infect. Immun. 78, 3889–3897 10.1128/IAI.00165-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saha S. S., Hashino M., Suzuki J., Uda A., Watanabe K., Shimizu T., and Watarai M. (2017) Contribution of methionine sulfoxide reductase B (MsrB) to Francisella tularensis infection in mice. FEMS Microbiol. Lett. 364, fnw260 10.1093/femsle/fnw260 [DOI] [PubMed] [Google Scholar]
- 19. Lee S. H., Li C. W., Koh K. W., Chuang H. Y., Chen Y. R., Lin C. S., and Chan M. T. (2014) MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress. J. Exp. Bot. 65, 5049–5062 10.1093/jxb/eru270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tossounian M. A., Van Molle I., Wahni K., Jacques S., Gevaert K., Van Breusegem F., Vertommen D., Young D., Rosado L. A., and Messens J. (2018) Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage. Biochim. Biophys. Acta Gen. Subj. 1862, 775–789 10.1016/j.bbagen.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 21. Van Laer K., Hamilton C. J., and Messens J. (2013) Low-molecular-weight thiols in thiol-disulfide exchange. Antioxid. Redox Signal. 18, 1642–1653 10.1089/ars.2012.4964 [DOI] [PubMed] [Google Scholar]
- 22. Neiers F., Sonkaria S., Olry A., Boschi-Muller S., and Branlant G. (2007) Characterization of the amino acids from Neisseria meningitidis methionine sulfoxide reductase B involved in the chemical catalysis and substrate specificity of the reductase step. J. Biol. Chem. 282, 32397–32405 10.1074/jbc.M704730200 [DOI] [PubMed] [Google Scholar]
- 23. Lowther W. T., Weissbach H., Etienne F., Brot N., and Matthews B. W. (2002) The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB. Nat. Struct. Biol. 9, 348–352 [DOI] [PubMed] [Google Scholar]
- 24. Ranaivoson F. M., Neiers F., Kauffmann B., Boschi-Muller S., Branlant G., and Favier F. (2009) Methionine sulfoxide reductase B displays a high level of flexibility. J. Mol. Biol. 394, 83–93 10.1016/j.jmb.2009.08.073 [DOI] [PubMed] [Google Scholar]
- 25. Olry A., Boschi-Muller S., Yu H., Burnel D., and Branlant G. (2005) Insights into the role of the metal binding site in methionine-R-sulfoxide reductases B. Protein Sci. 14, 2828–2837 10.1110/ps.051711105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tossounian M. A., Pedre B., Wahni K., Erdogan H., Vertommen D., Van Molle I., and Messens J. (2015) Corynebacterium diphtheriae methionine sulfoxide reductase a exploits a unique mycothiol redox relay mechanism. J. Biol. Chem. 290, 11365–11375 10.1074/jbc.M114.632596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olry A., Boschi-Muller S., Marraud M., Sanglier-Cianferani S., Van Dorsselear A., and Branlant G. (2002) Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis. J. Biol. Chem. 277, 12016–12022 10.1074/jbc.M112350200 [DOI] [PubMed] [Google Scholar]
- 28. Kumar R. A., Koc A., Cerny R. L., and Gladyshev V. N. (2002) Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J. Biol. Chem. 277, 37527–37535 10.1074/jbc.M203496200 [DOI] [PubMed] [Google Scholar]
- 29. Riener C. K., Kada G., and Gruber H. J. (2002) Quick measurement of protein sulfhydryls with Ellman's reagent and with 4,4′-dithiodipyridine. Anal. Bioanal. Chem. 373, 266–276 10.1007/s00216-002-1347-2 [DOI] [PubMed] [Google Scholar]
- 30. Arnér E. S., and Holmgren A. (2000) Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109 10.1046/j.1432-1327.2000.01701.x [DOI] [PubMed] [Google Scholar]
- 31. Holmgren A. (1995) Thioredoxin structure and mechanism: conformational changes on oxidation of the active-site sulfhydryls to a disulfide. Structure 3, 239–243 10.1016/S0969-2126(01)00153-8 [DOI] [PubMed] [Google Scholar]
- 32. Fahey R. C., Brown W. C., Adams W. B., and Worsham M. B. (1978) Occurrence of glutathione in bacteria. J. Bacteriol. 133, 1126–1129 10.1128/JB.133.3.1126-1129.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Newton G. L., Arnold K., Price M. S., Sherrill C., Delcardayre S. B., Aharonowitz Y., Cohen G., Davies J., Fahey R. C., and Davis C. (1996) Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178, 1990–1995 10.1128/JB.178.7.1990-1995.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pedre B., Van Molle I., Villadangos A. F., Wahni K., Vertommen D., Turell L., Erdogan H., Mateos L. M., and Messens J. (2015) The Corynebacterium glutamicum mycothiol peroxidase is a reactive oxygen species-scavenging enzyme that shows promiscuity in thiol redox control. Mol. Microbiol. 96, 1176–1191 10.1111/mmi.12998 [DOI] [PubMed] [Google Scholar]
- 35. Ordoñez E., Van Belle K., Roos G., De Galan S., Letek M., Gil J. A., Wyns L., Mateos L. M., and Messens J. (2009) Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J. Biol. Chem. 284, 15107–15116 10.1074/jbc.M900877200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levine R. L., Mosoni L., Berlett B. S., and Stadtman E. R. (1996) Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 15036–15040 10.1073/pnas.93.26.15036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim G., Weiss S. J., and Levine R. L. (2014) Methionine oxidation and reduction in proteins. Biochim. Biophys. Acta 1840, 901–905 10.1016/j.bbagen.2013.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vogt W. (1995) Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Radic. Biol. Med. 18, 93–105 10.1016/0891-5849(94)00158-G [DOI] [PubMed] [Google Scholar]
- 39. Drazic A., and Winter J. (2014) The physiological role of reversible methionine oxidation. Biochim. Biophys. Acta 1844, 1367–1382 10.1016/j.bbapap.2014.01.001 [DOI] [PubMed] [Google Scholar]
- 40. Kim H. Y., and Gladyshev V. N. (2005) Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLos Biol. 3, e375 10.1371/journal.pbio.0030375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Collet J. F., and Messens J. (2010) Structure, function, and mechanism of thioredoxin proteins. Antioxid. Redox Signal. 13, 1205–1216 10.1089/ars.2010.3114 [DOI] [PubMed] [Google Scholar]
- 42. Cao Z., Mitchell L., Hsia O., Scarpa M., Caldwell S. T., Alfred A. D., Gennaris A., Collet J. F., Hartley R. C., and Bulleid N. J. (2018) Methionine sulfoxide reductase B3 requires resolving cysteine residues for full activity and can act as a stereospecific methionine oxidase. Biochem. J. 475, 827–838 10.1042/BCJ20170929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tarrago L., Kaya A., Weerapana E., Marino S. M., and Gladyshev V. N. (2012) Methionine sulfoxide reductases preferentially reduce unfolded oxidized proteins and protect cells from oxidative protein unfolding. J. Biol. Chem. 287, 24448–24459 10.1074/jbc.M112.374520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vieira Dos Santos C., Cuiné S., Rouhier N., and Rey P. (2005) The Arabidopsis plastidic methionine sulfoxide reductase B proteins: sequence and activity characteristics, comparison of the expression with plastidic methionine sulfoxide reductase A, and induction by photooxidative stress. Plant Physiol. 138, 909–922 10.1104/pp.105.062430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin Z., Johnson L. C., Weissbach H., Brot N., Lively M. O., and Lowther W. T. (2007) Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function. Proc. Natl. Acad. Sci. U.S.A. 104, 9597–9602 10.1073/pnas.0703774104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tarrago L., and Gladyshev V. N. (2012) Recharging oxidative protein repair: catalysis by methionine sulfoxide reductases towards their amino acid, protein, and model substrates. Biochemistry 77, 1097–1107 [DOI] [PubMed] [Google Scholar]
- 47. Messens J., Martins J. C., Van Belle K., Brosens E., Desmyter A., De Gieter M., Wieruszeski J. M., Willem R., Wyns L., and Zegers I. (2002) All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade. Proc. Natl. Acad. Sci. U.S.A. 99, 8506–8511 10.1073/pnas.132142799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zegers I., Martins J. C., Willem R., Wyns L., and Messens J. (2001) Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty. Nat. Struct. Biol. 8, 843–847 10.1038/nsb1001-843 [DOI] [PubMed] [Google Scholar]
- 49. Messens J., Van Molle I., Vanhaesebrouck P., Limbourg M., Van Belle K., Wahni K., Martins J. C., Loris R., and Wyns L. (2004) How thioredoxin can reduce a buried disulphide bond. J. Mol. Biol. 339, 527–537 10.1016/j.jmb.2004.04.016 [DOI] [PubMed] [Google Scholar]
- 50. Lillig C. H., and Berndt C. (2013) Cellular functions of glutathione. Biochim. Biophys. Acta 1830, 3137–3138 10.1016/j.bbagen.2013.02.019 [DOI] [PubMed] [Google Scholar]
- 51. Hugo M., Van Laer K., Reyes A. M., Vertommen D., Messens J., Radi R., and Trujillo M. (2014) Mycothiol/mycoredoxin 1-dependent reduction of the peroxiredoxin AhpE from Mycobacterium tuberculosis. J. Biol. Chem. 289, 5228–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reyes A. M., Pedre B., De Armas M. I., Tossounian M. A., Radi R., Messens J., and Trujillo M. (2018) Chemistry and redox biology of mycothiol. Antioxid. Redox Signal. 28, 487–504 10.1089/ars.2017.7074 [DOI] [PubMed] [Google Scholar]
- 53. Si M., Xu Y., Wang T., Long M., Ding W., Chen C., Guan X., Liu Y., Wang Y., Shen X., and Liu S. J. (2015) Functional characterization of a mycothiol peroxidase in Corynebacterium glutamicum that uses both mycoredoxin and thioredoxin reducing systems in the response to oxidative stress. Biochem. J. 469, 45–57 10.1042/BJ20141080 [DOI] [PubMed] [Google Scholar]
- 54. Chi B. K., Busche T., Van Laer K., Bäsell K., Becher D., Clermont L., Seibold G. M., Persicke M., Kalinowski J., Messens J., and Antelmann H. (2014) Protein S-mycothiolation functions as redox-switch and thiol protection mechanism in Corynebacterium glutamicum under hypochlorite stress. Antioxid. Redox Signal. 20, 589–605 10.1089/ars.2013.5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Si M., Feng Y., Chen K., Kang Y., Chen C., Wang Y., and Shen X. (2017) Functional comparison of methionine sulphoxide reductase A and B in Corynebacterium glutamicum. J. Gen. Appl. Microbiol. 63, 280–286 10.2323/jgam.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 56. Volkov A. N., Wohlkonig A., Soror S. H., and van Nuland N. A. (2013) Expression, purification, characterization, and solution nuclear magnetic resonance study of highly deuterated yeast cytochrome c peroxidase with enhanced solubility. Biochemistry 52, 2165–2175 10.1021/bi400220w [DOI] [PubMed] [Google Scholar]
- 57. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 58. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., and Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- 59. Cheung M. S., Maguire M. L., Stevens T. J., and Broadhurst R. W. (2010) DANGLE: a Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J. Magn. Reson. 202, 223–233 10.1016/j.jmr.2009.11.008 [DOI] [PubMed] [Google Scholar]
- 60. Güntert P., and Buchner L. (2015) Combined automated NOE assignment and structure calculation with CYANA. J. Biomol. NMR 62, 453–471 10.1007/s10858-015-9924-9 [DOI] [PubMed] [Google Scholar]
- 61. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 10.1107/S0907444998003254, 10.1107/S0108767398011465 [DOI] [PubMed] [Google Scholar]
- 62. Schwieters C. D., Kuszewski J. J., Tjandra N., and Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 10.1016/S1090-7807(02)00014-9 [DOI] [PubMed] [Google Scholar]
- 63. Yang B., Wu Y. J., Zhu M., Fan S. B., Lin J., Zhang K., Li S., Chi H., Li Y. X., Chen H. F., Luo S. K., Ding Y. H., Wang L. H., Hao Z., Xiu L. Y., Chen S., Ye K., He S. M., and Dong M. Q. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 10.1038/nmeth.2099 [DOI] [PubMed] [Google Scholar]
- 64. Leermakers M., Gao Y., Gabelle C., Lojen S., Ouddane B., Wartel M., and Baeyens W. (2005) Determination of high resolution pore water profiles of trace metals in sediments of the Rupel River (Belgium) using DET (diffusive equilibrium in thin films) and DGT (diffusive gradients in thin films) techniques. Water Air Soil Pollut. 166, 265–286 10.1007/s11270-005-6671-7 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.