Abstract
Management of the agricultural pathogen soybean cyst nematode (SCN) relies on the use of SCN-resistant soybean cultivars, a strategy that has been failing in recent years. An underutilized source of resistance in the soybean genotype Peking is linked to two polymorphisms in serine hydroxy-methyltransferase 8 (SHMT8). SHMT is a pyridoxal 5′-phosphate–dependent enzyme that converts l-serine and (6S)-tetrahydrofolate to glycine and 5,10-methylenetetrahydrofolate. Here, we determined five crystal structures of the 1884-residue SHMT8 tetramers from the SCN-susceptible cultivar (cv.) Essex and the SCN-resistant cv. Forrest (whose resistance is derived from the SHMT8 polymorphisms in Peking); the crystal structures were determined in complex with various ligands at 1.4–2.35 Å resolutions. We find that the two Forrest-specific polymorphic substitutions (P130R and N358Y) impact the mobility of a loop near the entrance of the (6S)-tetrahydrofolate–binding site. Ligand-binding and kinetic studies indicate severely reduced affinity for folate and dramatically impaired enzyme activity in Forrest SHMT8. These findings imply widespread effects on folate metabolism in soybean cv. Forrest that have implications for combating the widespread increase in virulent SCN.
Keywords: enzyme, X-ray crystallography, plant defense, folate, ligand-binding protein, host-pathogen interaction, disease resistance, Glycine max, quantitative trait loci (QTL), serine hydroxymethyltransferase 8 (SHMT8), soybean cyst nematode
Introduction
Soybean cyst nematode (SCN),4 a microscopic roundworm, is the most detrimental pathogen of soybean and a billion dollar problem annually in U.S. agriculture. Management of SCN infection relies almost exclusively on the use of SCN-resistant soybean cultivars (1). Unfortunately, this approach has become less effective over time (2) as nematodes have adapted to the current source of resistance derived from plant introduction (PI) 88788, which is present in more than 95% of commercially available resistant cultivars (cvs.). Because all but one soybean-producing state in the United States have reported SCN infestations, there is an urgent agricultural need for new sources of SCN resistance.
A promising candidate is the soybean cultivar known as “Forrest,” which derives SCN resistance from PI 548402 (or Peking), a genotype that differs mechanistically from PI 88788. Soybean cv. Forrest is well-known for saving the United States from billions of dollars in losses caused by SCN in the 1970s and 1980s (3, 4). The genes underlying two major quantitative trait loci associated with resistance to SCN infection, termed rhg (resistance to Heterodera glycines) on chromosomes 18 (rhg1) and 8 (Rhg4), have been identified (5). Soybean cv. Forrest was found to require both rhg1 and Rhg4, where Rhg4 is the dominant gene. Positional cloning of the Rhg4 gene identified mutations in the enzyme serine hydroxymethyltransferase 8 (SHMT8); mutation analysis, silencing, and transgenic complementation subsequently confirmed this result (6). The SCN-resistant allele of SHMT8 differs from the susceptible allele by only two amino acids: Forrest SHMT8 contains arginine and tyrosine at positions 130 and 358 of the polypeptide chain, respectively, compared with proline and asparagine in SHMT8 from the SCN-susceptible cv. Essex (6). Although the soybean genome encodes multiple SHMT isoforms (up to 18 predicted), SHMT8 is the only family member associated with resistance to SCN (7); none of the others can compensate in this regard for a loss of SHMT8 function (8).
SHMT is a pyridoxal 5′-phosphate (PLP)–dependent enzyme that catalyzes the reversible conversion of l-serine and (6S)-tetrahydrofolate (THF) to glycine and 5,10-methylenetetrahydrofolate (MTHF) (Fig. 1A). SHMT is a structurally conserved enzyme, typically a dimer or tetramer, and has been characterized from multiple organisms (currently 111 structures deposited in the PDB). SHMT is well-known for its role in cellular folate metabolism (Fig. 1B), making it a design target for antimalarial (9–15) and anti-cancer agents in humans (16–20). Folate is also a crucial nutrient for SCN to complete its life cycle; SCN does not itself produce folate and therefore must rely on host plants as a source (21). Perturbation of soybean folate metabolism linked to the two polymorphisms in Forrest SHMT8 could thus affect establishment and/or maintenance of the SCN feeding site. However, other possibilities, such as altered or novel enzymatic properties caused by the amino acid substitutions or impacts on protein–protein interactions in the cell, may also be relevant. To date, the role of soybean SHMT8 in SCN resistance has remained unclear.
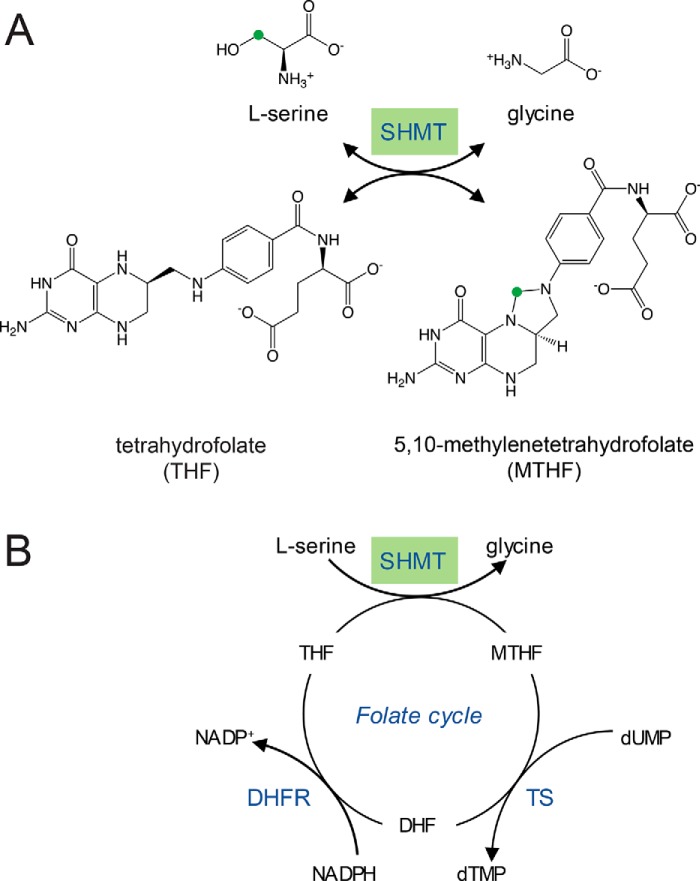
Figure 1.
An overview of the reaction catalyzed by SHMT and its context in the folate cycle. A, the conversion of serine and THF to glycine and MTHF. The green circle highlights the methylene group transferred from l-serine to THF. B, the cellular folate cycle involving SHMT, thymidylate synthase (TS), and dihydrofolate reductase (DHFR). This figure is adapted from Nonaka et al. (46).
Here we present a comprehensive structural and biochemical comparison of SHMT8 from soybean cvs. Essex and Forrest. Crystal structures show that the two Forrest polymorphic residues (P130R and N358Y) flank a mobile loop located near the entrance of the THF-binding site. In Essex SHMT8, this loop becomes ordered upon binding of 5-formyl-THF (FTHF), consistent with micromolar binding affinity and robust enzyme activity. In contrast, Forrest SHMT8 has dramatically impaired binding to FTHF and a corresponding defect in its folate-dependent enzyme activity. The different biochemical phenotypes of SHMT8 from cvs. Essex and Forrest have implications for understanding the environment of the nematode feeding cell during SCN infection and the mechanism of resistance to this agricultural pathogen.
Results
Structural overview of soybean SHMT8
Crystal structures of soybean SHMT8 from cvs. Essex and Forrest were determined with various ligands at resolutions ranging from 1.4 to 2.35 Å. Three complexes of Essex SHMT8 were obtained: 1) one bound to the cofactor PLP, 2) one complexed with the PLP-glycine (PLP-Gly) external aldimine product, and 3) a ternary complex with PLP-Gly and the tightly binding inhibitor/alternative product FTHF. Complexes of SHMT8 from cv. Forrest were also obtained with PLP and PLP-Gly; a complex with FTHF was not obtained. See Tables S1 and S2 for a summary of data collection and refinement statistics.
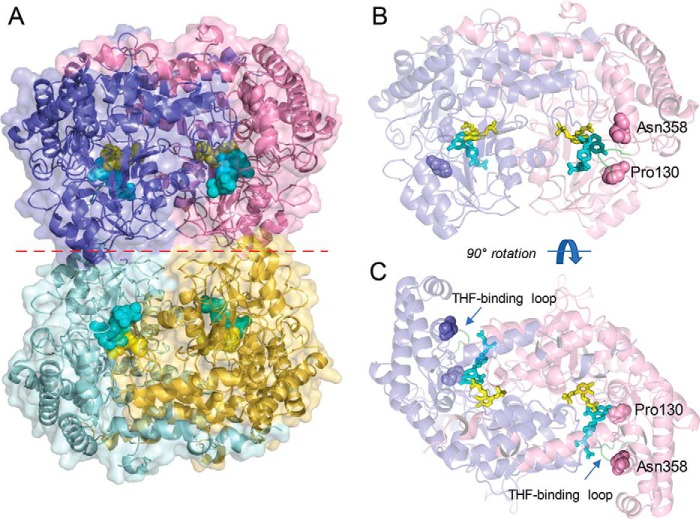
SHMT8 from cvs. Essex and Forrest adopts a tetrameric assembly in the crystal lattice (Fig. 2A). Two tightly intertwined dimers (hereafter called the “obligate dimer”) assemble to form a dimer-of-dimers tetramer. Within each obligate dimer (Fig. 2, B and C), the ligand-binding sites are found at the interface of two protomers, with residues from both chains required to create a complete, functional active site. Each protomer consists of a N-terminal arm (residues 1–32), a large domain (33–308) that binds PLP at Lys-244, and a small domain (residues 309–471) (Fig. S1). A survey of structural relatives in the PDB via the DALI server (22) confirms that soybean SHMT8 is closely related to SHMT from the legume Medicago truncatula (PDB code 6CCZ) (23), as well as the human cytosolic (PDB code 6FL5) and mitochondrial (PDB code 5V7I) isoenzymes (16, 17), with ∼60% sequence identity to all three enzymes.
Figure 2.
Overview of the structure of soybean SHMT8. The complex of Essex SHMT8 with PLP-Gly and FTHF is shown. A, structure of the tetramer with the four chains shown in different colors. The two obligate dimers are in purple/pink and cyan/yellow. PLP-Gly and FTHF are shown in the four active sites as spheres in yellow and cyan, respectively. The dimer–dimer interface is indicated by a dashed red line. B, side view of the obligate dimer in the same orientation as in A, showing the protomer–protomer interface, ligands as sticks, and the two residues affected by Forrest polymorphisms. The protein is shown in semitransparent mode to aid in visualization of the ligands. Residues 130 and 358 are shown as spheres. C, top down view of the obligate dimer (90° rotation from B), highlighting how the ligand-binding sites lie at the interface of the two polypeptide chains.
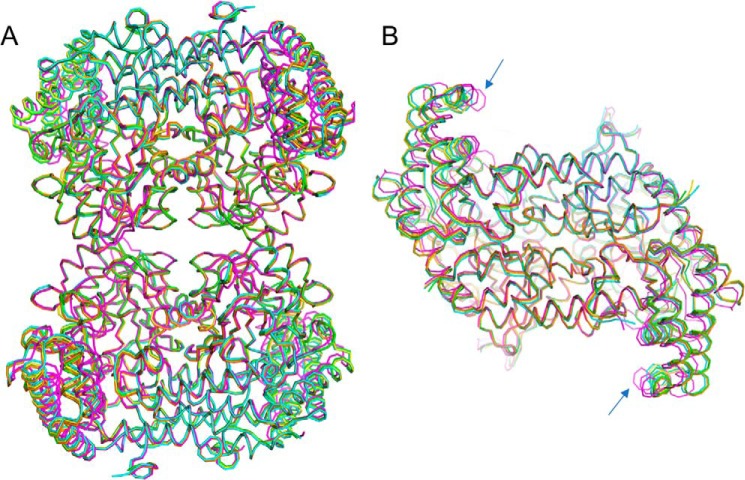
Structural superpositions of the various ligand complexes of SHMT8 from cvs. Essex and Forrest show a high degree of overall similarity (Fig. 3A). No major changes in the obligate dimer or tetrameric assemblies are observed. A minor difference in the Essex SHMT8 ternary complex entails a slight rotation of the small domain (Fig. 3B). Conserved quaternary structure in Essex and Forrest SHMT8 was confirmed in-solution by small-angle X-ray scattering (SAXS) and analytical ultracentrifugation, both of which indicated tetrameric assemblies (Fig. S2 and Table S3).
Figure 3.
Structural similarity of the SHMT8 structures in this study. Structures are colored as follows: Essex SHMT8 in complex with PLP (yellow), PLP-Gly (blue), PLP-Gly with THF (magenta), and Forrest SHMT8 in complex with PLP (green), and PLP-Gly (orange). A, a superposition of the polypeptide backbone the five tetramers. B, top-down view of the five obligate dimers, showing the slight difference (blue arrows) in the orientation of the small domain in the ternary complex with PLP-Gly and FTHF (magenta). A similar conformational change of the small domain was observed in the murine SHMT complex with FTHF (27).
The PLP-binding site of soybean SHMT8 is highly conserved with related enzymes
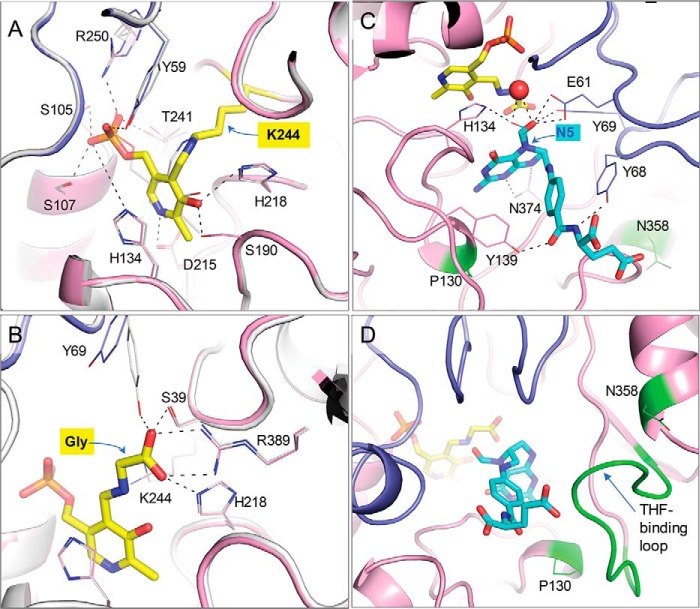
Crystal structures of Essex and Forrest SHMT8 complexed with PLP were determined to 1.85 and 2.1 Å, respectively. The SHMT8·PLP complexes represent the resting state of the active enzyme (Fig. 4A and Fig. S3). As observed in other SHMT structures, the PLP-binding pocket is located deep inside the obligate dimer (Fig. 2C). Lys-244 forms a covalent Schiff-base linkage (internal aldimine) with PLP. Nearby residues from both chains of the obligate dimer (Tyr-59′, Glu-61′, Ser-107, Asp-215, Thr-241, Arg-250, where prime indicates chain B) make conserved interactions with the phosphate, N1, and O3 hydroxyl of PLP, whereas the pyridine ring of PLP stacks against His-134. The PLP-binding pose and interactions of Forrest SHMT8 show no major perturbations compared with Essex (Fig. 4A). For reference, the Forrest polymorphic residues (positions 130 and 358) are >14 Å from PLP/PLP-Gly.
Figure 4.
Details of the soybean SHMT8 ligand complexes. A, the enzyme–PLP complex as an internal aldimine with Lys-244 (yellow). Chain A of Essex SHMT8 is in pink, and chain B is purple. The Forrest SHMT8 (white) complex with PLP (gray) is superimposed. Side chains of key residues involved in ligand interactions are shown as sticks. B, The SHMT8 complexes with PLP-Gly complex as an external aldimine (yellow). The colors as described for A. Hydrogen bonds to the glycine adduct are shown. C, the 1.4 Å resolution Essex SHMT8 ternary complex with PLP-Gly and FTHF. FTHF is cyan, and the other colors are as in A; two conformers of FTHF are shown (major with thicker lines) with different positions of the formyl oxygen. Side chains of residues involved in ligand interactions are shown as sticks. The two Forrest polymorphic residues are highlighted in green. D, the THF-binding loop in the Essex SHTM8·PLP-Gly·FTHF complex showing how it packs against FTHF in its closed conformation.
Crystal structures of Essex and Forrest SHMT8 complexed with PLP-Gly were determined to 2.1 and 2.35 Å, respectively. The SHMT8·PLP-Gly complexes represent an intermediate step of the THF-dependent catalytic mechanism (Fig. S3). In the first step, l-Ser attacks the Schiff base linkage between Lys-244 and PLP to form a PLP-Ser external aldimine. Next, an active site general base deprotonates the hydroxyl side chain of l-Ser, liberating formaldehyde. Formaldehyde is attacked by THF N5, transferring the side chain of l-Ser to THF. The resulting external aldimine/quinonoid product is PLP-Gly (Fig. 4B and Fig. S3). We note that this PLP-Gly adduct can also be formed via a THF-independent retro-aldol mechanism (24).
The Essex SHMT8·PLP-Gly complex retains many of the enzyme–cofactor interactions as in the complex with PLP (Fig. 4B). Likewise, the Essex and Forrest SHMT8·PLP-Gly structures reveal nearly identical binding poses and active site interactions. Attachment of glycine as an external aldimine displaces the Lys-244 amino group, concomitant with rotation of the side chains of Thr-241 and/or Tyr-59. The pyridine ring shifts away from Lys-244, and the glycine moiety occupies space above Lys-244. A bidentate interaction is made between the glycine carboxylate group and Arg-389; additional interactions are made by Ser-39 and His-218. The Forrest SHMT8 PLP-Gly complex is similar to that of Essex except that the side chain of Tyr-69 has rearranged slightly to make an additional contact with the glycine carboxylate. Overall, the similarity in PLP and PLP-Gly binding poses and interactions between Essex and Forrest SHMT8 suggests no long-distance structural effects of the Forrest polymorphic residues.
Ternary complex of Essex SHMT8 with FTHF at atomic resolution
The crystal structure of the Essex SHMT8 ternary complex with PLP-Gly and FTHF was determined at 1.4 Å resolution (Fig. 4C). We note this exceeds the resolution of other SHMT·folate complexes in the PDB by 1.0 Å or more (PDB codes 1DFO (25), 1EQB (26), 1EJI (27), 1KL2 (28), 1LS3 (29), 2VMY (30), and 4OYT (9)). (A recent report of Arabidopsis thaliana SHMT with the folate analog permetrexed (PDB code 6SMW) has a resolution of 1.54 Å (31).) The THF-binding site in Essex SHMT8 comprises a channel that extends from the surface of the protein to the deeply buried PLP-binding site (Fig. 2). At the entrance of the channel, the FTHF glutamate moiety is exposed to solvent and has no specific interactions with the enzyme (Fig. 4C). This binding orientation suggests that the glutamate moiety of FTHF in the Essex ternary complex can accommodate the predominantly biologically relevant THF in plants, which is polyglutamylated (32, 33). The proximal section of the channel is lined with aliphatic and aromatic residues, including Tyr-68′, which stacks against the p-aminobenzyl ring of FTHF, and Tyr-139, which hydrogen bonds to a carbonyl group of the ligand. This Tyr–carbonyl interaction has not been seen in other SHMT complexes with folate derivatives, because Tyr is not conserved in these other members of the enzyme family.
The pteridine ring of FTHF lies at the base of the channel, with the pteridine ring positioned within 4–5 Å of PLP-Gly and parallel to the PLP-Gly pyridine ring (Fig. 4C). Multiple residues interact with the pteridine moiety, including Leu-129, Gly-133, Leu-135, and Asn-374. These contacts are highly conserved in other SHMT·folate complexes (25). The pteridine ring N5 is the site of condensation with l-Ser–derived formaldehyde during the SHMT reaction, making FTHF a product analog in the multistep reaction (Fig. S3). Our 1.4 Å maps reveal two orientations of the formyl O atom (occupancies ∼0.33 and 0.66). In one orientation, the oxygen participates in a hydrogen bond with the side chain of His-134 and a water molecule and is 3.2–3.4 Å from the nitrogen atom of the glycine aldimine. In the other orientation, the oxygen interacts with Glu-61, the same water molecule, and the side chain hydroxyl of Tyr-69. The proximity of the pteridine ring to the PLP-Gly adduct, as well as the presence of well-conserved pteridine-SHMT contacts, suggests that FTHF is present in the native binding pose required for catalysis.
The Forrest polymorphic residues flank the entrance of the THF-binding site
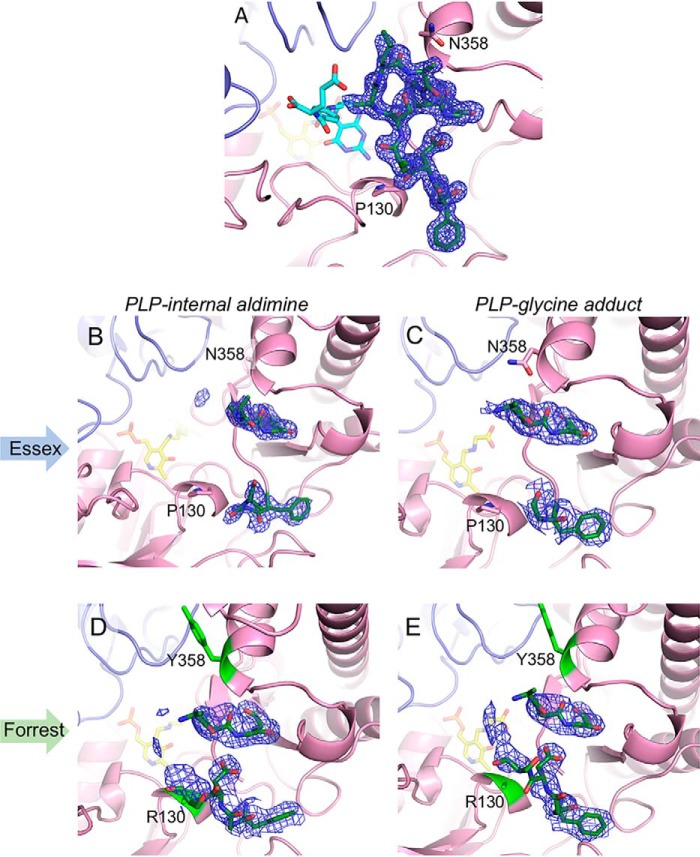
Initial comparison of the three Essex SHMT8 complexes revealed different conformers of a loop near the entrance of the THF-binding site, spanning residues 377–385 (hereafter: the THF-binding loop). In the PLP and PLP-Gly complexes, the THF-binding loop is partially disordered, with electron density maps suggesting a mostly open, flexible conformation (Fig. 5). In contrast, in the atomic-resolution ternary complex, the loop is highly ordered, with backbone and side chain atoms of all residues visible, and adopts a closed conformation (Figs. 4D and 5). Although no specific contacts are found between the THF-binding loop residues and FTHF, the loop packs closely against the ligand (e.g. Ala-382 is <4 Å from multiple FTHF atoms).
Figure 5.
Polder-style electron density maps of the THF-binding loop in soybean SHMT8. Positions of residues 130 and 358 are noted. Maps are contoured at 3 σ, and density is shown spanning residues 377–387. A, the well-ordered loop in the Essex SHMT8 ternary complex with PLP-Gly and FTHF. B and C, show density for the loop in the Essex SHMT8 complexes with PLP and PLP-Gly, respectively. D and E, corresponding complexes with Forrest SHMT8, for comparison with those of Essex. Note the more extensive partial density for the loop in the Forrest complexes, suggesting a change in loop mobility caused by the substitutions at residues 130 and 358.
Amino acid positions 130 and 358, the two sites of the Forrest SHMT8 polymorphic residues, flank the THF-binding loop (Fig. 4C). In Essex SHMT8, Pro-130 is buried in a short helical region in the large domain of SHMT and is within 4 Å of the backbone of Asp-379 in the THF-binding loop. Likewise, Essex SHMT8 Asn-358 is positioned in a helix in the small domain and is also buried. It is also within 3.3 Å of the backbone of Leu-383, a residue in the THF-binding loop. Neither Pro-130 nor Asn-358 makes direct contact with FTHF (>8 Å away).
In the Forrest SHMT8 PLP and PLP-Gly complexes, the THF-binding loop is also partially disordered, as seen in the analogous Essex structures. However, some differences relative to Essex are apparent in the unmodeled electron density for the loop (Fig. 5), suggesting possible changes in loop mobility/position. Electron density maps are consistent with the Arg-130 and Tyr-358 substitutions, although the arginine side chain is not clearly visible past Cβ. The introduced tyrosine is solvent-exposed and makes no specific interactions with other residues. Overall, it appears that the proximity of the Forrest SHMT8 polymorphic residues to the THF-binding loop and the increased steric bulk of the polymorphic substitutions may have an effect on loop dynamics and/or conformation.
Forrest SHMT8 has severely impaired binding affinity for folate
To investigate potential effects on folate binding in Forrest SHMT8, we determined binding constants in a two-substrate system that exploits a spectrophotometrically observable change associated with folate binding (34). FTHF was used rather than THF because of increased stability and solubility. For Essex SHMT8, we observed saturable binding of FTHF, with a dissociation constant (Kd) of ∼17 μm (Table 1). We note that this Kd was glycine concentration-dependent (Fig. S4). Conversely, for Forrest SHMT8, although binding of FTHF could be observed, it was not saturable, and a Kd could not be determined under the conditions tested (Table 1 and Fig. S4). These results highlight a distinct biochemical difference in THF binding between Essex and Forrest SHMT8 and are consistent with our inability to obtain a ternary complex with Forrest SHMT8 for structural characterization.
Table 1.
Folate-binding and steady-state kinetic parameters
Reported uncertainties are standard errors as calculated from curve fitting in Origin 2019. ND, not determined.
| Essex SHMT8 | Forrest SHMT8 | |
|---|---|---|
| Spectrophotometric FTHF-binding assay | ||
| SHMT8·glycine + FTHF (μm) | 17 | ND |
| Steady-state kinetics: MTHFD-coupled reaction | ||
| kcat (s−1) | 14 ± 3 | ND |
| Km,THF (μm) | 30 ± 11 | ND |
| Km,Ser (μm) | 240 ± 30 | 1300 ± 200 |
| Ki,THF (μm) | 170 ± 70 | |
| kcat/Km,THF (mm−1 s−1) | 500 ± 190 | 0.22 ± 0.02a |
| Steady-state kinetic parameters: retro-aldol cleavage of l-phenylserine | ||
| kcat (s−1) | 1.3 ± 0.1 | 0.93 ± 0.04 |
| Km (mm) | 70 ± 10 | 63 ± 5 |
| kcat/Km (m−1 s−1) | 18 ± 3.3 | 15 ± 1.3 |
a Catalytic efficiency approximated from linear regression.
Forrest SHMT8 has a profound defect in MTHF production
The observed impairment in FTHF binding for Forrest SHMT8 suggested potential alterations in enzymatic activity. To test this, we utilized a coupled spectrophotometric assay with 5,10-methylenetetrahydrofolate dehydrogenase (MTHFD; Fig. S5) (14). Essex SHMT8 displayed THF-concentration dependent substrate inhibition at a fixed, saturating concentration of l-Ser. Fitting of initial velocity data to a substrate inhibition model returned a Km,THF of 30 ± 11 μm, a kcat of 14 ± 3 s−1, and an overall catalytic efficiency (kcat/Km,THF) of 500 ± 190 mm−1 s−1 (Table 1). The apparent Km,THF is similar in magnitude to the Kd for FTHF determined by the binding assay above (Table 1). Likewise, Essex SHMT8 displayed Michaelis–Menten behavior at varied concentrations of l-Ser at a fixed, saturating concentration of THF. Fitting of the initial velocity data to the Michaelis–Menten model returned a Km,l-Ser of 240 ± 30 μm (Table 1). These kinetic parameters are also similar to those of the human enzyme using the same assay (35).
When assayed under the same conditions, Forrest SHMT8 displayed only residual catalytic activity. With THF as the varied substrate at a fixed concentration of l-Ser, Forrest SHMT8 displayed catalytic efficiency of 0.22 ± 0.02 mm−1 s−1 (Table 1), a 2300-fold decrease compared with Essex. This is consistent with the FTHF-binding data, which indicated lack of saturation by folate. Residual activity when l-Ser was varied with THF at a fixed concentration was fit to a Michaelis–Menten model, returning a Km,l-Ser of 1300 ± 200 μm, a kcat of 0.36 ± 0.04 s−1, and a catalytic efficiency (kcat/Km,l-Ser) of 0.28 ± 0.06 mm−1 s−1 (Table 1). Surprisingly, Forrest SHMT8 displayed nearly identical kinetic parameters as Essex SHMT8 when assaying an alternative, THF-independent enzymatic reaction: retro-aldol cleavage of the β-hydroxy-amino acid l-phenylserine (Table 1). Overall, analysis of the catalytic properties of Essex and Forrest SHMT8 reveals a severe defect in MTHF production, but not glycine production, in Forrest SHMT8 compared with Essex SHMT8.
Discussion
We present herein a detailed biochemical and structural analysis of soybean SHMT8, one of only a few from the plant kingdom and the first from an agriculturally relevant species to be so characterized. More importantly, we show, for the first time, dramatic differences in molecular properties of SHMT8 from two soybean cultivars: the SCN-susceptible cv. Essex and SCN-resistant cv. Forrest. We find that Essex SHMT8 has robust catalytic activity (Table 1 and Fig. S5) comparable with SHMTs studied in other higher organisms (17, 35, 36). In stark contrast, Forrest SHMT8 is severely compromised in its primary, THF-dependent catalytic function (Table 1 and Fig. S5). We further demonstrate that this kinetic impairment is due to decreased binding affinity for folate and does not impact a THF-independent activity of the enzyme (Table 1 and Fig. S5).
The quantitative analyses herein differ somewhat from our results in an earlier study (6). Previously, we estimated the apparent activities of the Forrest and Essex SHMT8 enzymes using a cumbersome, discontinuous assay consisting of quenching the reaction with DTT, followed by a sodium borohydride reduction step, boiling the sample, centrifugation, and finally analysis by HPLC. The data from the HPLC assay could not be fit to any standard kinetic models, so kinetic parameters could not be estimated, preventing either a quantitative comparison between Essex and Forrest SHMT8, or a meaningful comparison with SHMTs from other organisms. Nevertheless, the qualitative HPLC assay data did suggest that Essex SHMT8 is more active than Forrest SHMT8, in general agreement with the current results. Here we used an enzyme-coupled assay that allowed us to continuously monitor product formation using simple absorbance spectroscopy. This method enabled us to study the enzyme activity in the physiologically meaningful THF concentration range of 0.003–0.4 mm, as compared with the very high THF concentration of 0.2–4 mm used in our previous work. Using the enzyme-coupled continuous assay, we obtained data that fit well to standard kinetic models (Fig. S5). Moreover, the kinetic parameters for Essex SHMT8 are in good quantitative agreement with those of other eukaryotic SHMTs (31, 35). Thus, we suggest that the current study provides a more accurate assessment of the enzymatic activities of Essex and Forrest SHMT8 and establishes that these two enzymes have distinct biochemical phenotypes with multiple potential implications for folate metabolism in soybean.
The results of our kinetic and ligand-binding studies on soybean SHMT8 are consistent with a wealth of data on enzyme structure presented here. SAXS and analytical ultracentrifugation studies show similar oligomeric states in solution for Essex and Forrest SHMT8 (Fig. S2). Detailed crystallographic analyses reveal nearly identical PLP-binding sites (Fig. 4) and an overall absence of significant protein structure perturbations caused by the P130R and N358Y amino acid polymorphic residue changes (Figs. 3 and 4). The only exception to this is an apparent change in mobility of the THF-binding loop (Fig. 5), which may help explain the dramatic discrepancy in folate-binding affinity between Essex and Forrest SHMT8. Although some conformational variability of this loop has been seen in other SHMT structures (25, 27, 28), its critical role in THF binding has not been previously shown. Thus, our study adds to a growing list of critical functional loops in SHMT, which include another recently characterized in human mitochondrial SHMT (35).
Our structural characterization of a high-resolution ternary complex of Essex SHMT8 with PLP-Gly and FTHF provides atomic accuracy to the active site changes that accompany folate binding in SHMT. The atomic resolution ternary complex reported here, combined with the additional Essex SHMT8 structures, presents a more comprehensive picture of the structural changes associated with binding of multiple ligands at the various steps of the catalytic cycle. Importantly, these structural data have implications beyond the study of soybean SHMT8 and its role in SCN resistance. In particular, details of these enzyme–ligand interactions have relevance to designing inhibitors for SHMTs from other organisms, which are known targets for chemotherapeutic (16, 18, 19) and antimalarial (10, 14) agents. Likewise, functional loops of SHMT, such as the THF-binding loop, may be targets for development of chemotherapeutics with an alternate mechanism of enzyme inhibition distal from the active site.
The urgent need for new sources of SCN resistance in soybeans (2) makes characterization of potential molecular differences between SHMT8 from resistant and susceptible soybean cultivars a topic of great interest in agriculture. Our data herein demonstrate that differences in SCN resistance related to Rhg4 are associated with impaired folate binding, resulting in a loss of normal enzymatic function of SHMT8, the extent of which was not previously appreciated. This finding opens the door for future studies to pinpoint the biological outcomes of reduced SHMT8 activity in the plant and may enable engineering of more durable resistance to SCN.
Materials and methods
Protein expression and purification
The Glycine max SHMT8 protein expression constructs used in this work were previously described (6). For protein expression, 1 liter of terrific broth supplemented with 50 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol was inoculated with 10 ml of starter culture. The cultures were shaken at 37 °C and 250 rpm. When A600 ≈ 0.8, the temperature was shifted to 18 °C, and protein expression was induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside. Cultures were shaken overnight at 180 rpm.
For purification, cell pellets were resuspended in a lysis buffer containing 50 mm HEPES, pH 7.5, 500 mm NaCl, 20 mm imidazole, 10% (v/v) glycerol, 1% Tween 20, and 1 mm PLP. The cells were lysed by sonication followed by centrifugation at 16,000 rpm for 1 h at 4 °C. The resulting supernatant was then filtered through a 0.45-μm syringe-driven filter and loaded onto a nickel–nitrilotriacetic acid column. After washing the column with 40 bed volumes of 50 mm HEPES, pH 7.5, 500 mm NaCl, 20 mm imidazole, and 10% (v/v) glycerol, the protein was eluted with 50 mm HEPES, pH 7.5, 500 mm NaCl, 250 mm imidazole, and 10% (v/v) glycerol.
The nickel–nitrilotriacetic acid eluate was further purified by size-exclusion chromatography (SEC) on a Superdex 200 10/300 column using a buffer containing 50 mm HEPES, pH 7.5, 10% (v/v) glycerol, and 0.5 mm Tris(carboxyethyl)phosphine (TCEP) as the mobile phase. Both Essex and Forrest SHMT8 eluted as a single uniform peak on the SEC chromatogram. Protein-containing fractions were pooled and concentrated to 24 mg ml−1. Protein was then flash frozen in liquid nitrogen and stored at −80 °C.
Crystallization of Essex and Forrest SHMT8
For crystallization experiments, SHMT8 samples were diluted to 12 mg ml−1 using a buffer containing 25 mm HEPES, pH 7.5, 50 mm NaCl, and 0.5 mm TCEP. All crystallization experiments were carried out using the sitting-drop vapor-diffusion format using a 1:1 protein:reservoir ratio. Initial crystals of SHMT8 were obtained in Hampton Index condition 62, which contains 0.2 m trimethylamine N-oxide hydrate, 0.1 m Tris (pH 8.5), 20% (w/v) PEG monomethyl ether 2000. These crystals were crushed and used as seeds for optimization screens. Optimization screens were carried out using an Oryx8 crystallization robot (Douglas Instruments). All crystals of SHMT8 that resulted in the reported structures were obtained in 0.175–0.24 m trimethylamine N-oxide hydrate, 0.1 m Tris (pH 8.5), and 19–23 (w/v) PEG monomethyl ether 2000.
For SHMT8 complexes with PLP, the crystals were cryoprotected in the reservoir solution supplemented with 15% (v/v) ethylene glycol and flash-cooled in liquid nitrogen. For the structure of Essex SHMT8 complexed with PLP-Gly, the crystals were obtained via cocrystallization experiments in the presence of 20 mm glycine in both the protein sample and the reservoir solution. Essex SHMT8·PLP-Gly crystals were then cryoprotected in the reservoir solution supplemented with 15% (v/v) ethylene glycol and flash-cooled in liquid nitrogen. For the structure of Forrest SHMT8 complexed with PLP-Gly, crystals were obtained by soaking Forrest SHMT8·PLP crystals in the reservoir solution supplemented with 20 mm l-serine and 15% (v/v) ethylene glycol for cryoprotection. After a short (∼30 s) soak, the crystals were flash-cooled in liquid nitrogen.
For the Essex SHMT8 ternary complex with PLP-Gly and FTHF, crystals were obtained via cocrystallization experiments in the presence of 20 mm glycine and 10 mm FTHF in both the protein sample and reservoir solution. We note that stock solutions of both glycine and FTHF were prepared in a buffer containing 25 mm HEPES, pH 7.5, 50 mm NaCl, and 0.5 mm TCEP. SHMT8·PLP-Gly·FTHF crystals were then cryoprotected in the reservoir solution supplemented with 15% (v/v) ethylene glycol and flash-cooled in liquid nitrogen.
X-ray diffraction data collection, phasing, and refinement
X-ray diffraction data were collected on Advanced Light Source Beamline 4.2.2 using a Taurus-1 CMOS detector and 5.0.2 using a Pilatus 3 6M detector in shutterless mode. The data sets consisted of 900 images spanning 180°. The data sets were integrated and scaled with XDS (37), and the intensities were merged and converted to amplitudes with Aimless (38). We note that for the Essex SHMT8·PLP and Forrest SHMT8·PLP structures, five and three data sets, respectively, collected on a single crystal were merged using XSCALE (39) and used for phasing and refinement.
Initial phases of the Essex SHMT8·PLP data set were determined by molecular replacement in Phaser (40) using the structure of rabbit SHMT (PDB code 1CJ0) as a template structure. The initial map, experimental data, and sequence were then input into phenix.autobuild (41) for phase improvement and automated ab initio model building. The resulting phenix.autobuild model was used for subsequent rounds of model building in COOT (42) and refinement in PHENIX (41). The refined Essex SHMT8·PLP structure was used to determine the phases for all other data sets.
Steady-state kinetics for the SHMT two-substrate reactions
Steady-state kinetic parameters for the SHMT reaction in which l-serine and THF are converted to glycine and 5,10-MTHF were determined using the previously reported coupled reaction utilizing the NADP+-dependent enzyme 5,10-methylenetetrahydrofolate dehydrogenase (MTHFD) (14). The Escherichia coli gene encoding MTHFD (EcMTHFD; FolD) was PCR-amplified from the E. coli genome using previously reported primers (43). The EcMTHFD gene was then cloned into pET24b for expression of the C-terminally His-tagged enzyme. Previous studies noted that the presence of the C-terminal His tag has no effect on the kinetic parameters of EcMTHFD (43). MTHFD-coupled assay studies were carried out in an Epoch II plate reader (BioTek) in a reaction buffer containing 50 mm HEPES, pH 7.5, and 0.5 mm EDTA. The reactions contained either 0.025 μm Essex SHMT8 or 2.5 μm Forrest SHMT8. NADPH production was continuously monitored at 375 nm. Assays to determine the apparent Michaelis constant of THF contained 1 mm NADP+, 5 μm EcMTHFD, 2 mm l-serine, and a concentration series of THF ranging from 0.003 to 0.4 mm. Reported kinetic parameters for Essex SHMT8 with THF as the variable substrate were determined by fitting a substrate inhibition model to the average of four biological replicates, each with three internal technical replicates, for a total of 12 individual replicates. Reported kinetic parameters for Forrest SHMT8 with THF as the variable substrate were determined by linear regression to the average of three biological replicates, each with three internal technical replicates, for a total of nine individual replicates. Assays to determine the apparent Michaelis constant of l-serine contained 1 mm NADP+, 5 μm EcMTHFD, 400 μm THF, and a concentration series of l-serine ranging from 0.016 to 2 mm. (The high concentration of THF in assays with l-serine as the variable substrate was used to compensate for the low THF-binding affinity of Forrest SHMT8.) Kinetic parameters with serine as the variable substrate were determined by fitting a Michaelis–Menten model to the average of three independent replicates. The initial slopes of the reactions were used to determine enzyme activity, and the calculation utilized a molar extinction coefficient of 1920 m−1 cm−1 for NADPH at 375 nm (10). The results were graphed and analyzed in Origin 2019. The data were fit to either a substrate inhibition or Michaelis–Menten model. The error bars in Fig. S5 are the S.D. values calculated from technical replicates. The uncertainties reported in the text and Table 1 are the standard errors from curve fitting with Origin 2019.
To minimize the effects of THF oxidation on the assay, the following precautions were taken. The reaction buffer was autoclaved the placed in a purge chamber during which the buffer was subjected to three cycles of vacuum and purging with either N2 or mixed gas (95% N2, 5% H2). Following this purge step, the buffer was immediately placed in an anaerobic glove bag. These steps reduced the presence of dissolved oxygen in the reaction buffer. Both the (6S)-THF (Cayman Chemical) and the DMSO used to resuspend the (6S)-THF were only opened in the anaerobic glove box, and the DMSO was stored desiccated in the anaerobic glove box. Additionally, all THF, l-serine and NADP+ stock solutions used in the assays were prepared in an anaerobic glove box.
Determination of specific activities for the SHMT retro-aldol cleavage reaction
To determine and compare the specific activities for the folate-independent retro-aldol cleavage reaction of Essex and Forrest SHMT8, dl-phenylserine (Millipore-Sigma) was used as a substrate. The reactions were carried out in a buffer containing 25 mm HEPES, pH 7.5, 50 mm NaCl, and 0.5 mm TCEP in an Epoch II plate reader (BioTek). Each reaction contained 1 μm SHMT8 and 1–250 mm dl-phenylserine. Benzaldehyde formation was monitored at 279 nm. The initial slopes of the reactions were used to determine enzyme activity, and the calculation utilized a molar extinction coefficient of 1400 m−1 cm−1 for benzaldehyde (44). The results were graphed and analyzed in Origin 2019.
Ligand-binding assays
Folate-binding assays were adapted from previously reported protocols (34, 45). Briefly, enzyme solutions containing 1 mg ml−1 enzyme were formulated in the presence of either FTHF or glycine. This solution was mixed 1:1 with a solution of glycine or FTHF, respectively, in a 96-well plate. The plate was analyzed by absorbance scan (450–550 nm for Essex SHMT8 and 450–700 nm for Forrest SHMT8) in a BioTek Epoch II plate reader following the formation of a peak at ∼500 nm, consistent with the formation of the SHMT8·folate complex, as previously reported (45). The absorbance at 500 nm was baseline subtracted from the absorbance at 540 or 700 nm for Essex and Forrest, respectively. The working concentrations of glycine tested were 0.5–20 mm for Essex and 0.5–100 mm for Forrest. The working concentrations of FTHF tested were 10–100 μm for Essex and 10–100,000 μm (100 mm) for Forrest.
The data were analyzed by plotting ΔAbsorbance versus substrate concentration. The data were globally fit to the Michaelis–Menten equation, assuming Km ∼ Kd. Determination of the dependence of the Kd of one ligand on the presence of the other was carried out graphically using double reciprocal plots as previously described (34). The data were plotted and analyzed in Origin 2019.
Data and software availability
The atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Bank under accession codes 6UXH, 6UXI, 6UXJ, 6UXK, and 6UXL. All other data are available from the corresponding author upon reasonable request.
Author contributions
D. A. K., J. J. T., M. G. M., and L. J. B. conceptualization; D. A. K., J. J. T., M. G. M., and L. J. B. formal analysis; D. A. K., J. J. T., and L. J. B. validation; D. A. K., J. J. T., M. G. M., and L. J. B. investigation; D. A. K. and L. J. B. methodology; D. A. K. and L. J. B. writing-original draft; D. A. K., P. K. K., J. J. T., M. G. M., and L. J. B. writing-review and editing; P. K. K. and M. G. M. resources; J. J. T., M. G. M., and L. J. B. supervision; J. J. T., M. G. M., D. A. K., and L. J. B. funding acquisition; D. A. K., M. G. M., and L. J. B. project administration.
Supplementary Material
Acknowledgments
We thank Jay Nix of the Advanced Light Source Beamline 4.2.2 for assistance with X-ray data collection and processing. We thank Srinivas Chakravarthy of Sector 18 (BioCAT) at the Advanced Photon Source at Argonne National Lab for assistance with SEC–multi-angle light scattering–SAXS data collection and processing. This research used resources of the Advanced Light Source, which is a Department of Energy Office of Science User Facility under contract no. DE-AC02–05CH11231. This research used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility operated for the Department of Energy Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Use of the Pilatus 3 1M detector was provided by grant 1S10OD018090-01 from NIGMS, National Institutes of Health.
This work was supported by Grant 9 P41 GM103622 from the NIGMS, National Institutes of Health. This work was also supported by National Institute of Food and Agriculture Award 2019-67012-29653 from the U.S. Department of Agriculture National Institute of Food and Agriculture (to D. A. K.), the Missouri Soybean Mercchanising Council Project 258 (to M. G. M.), and by a seed grant from the University of Missouri Bond Life Sciences Center (to M. G. M., J. J. T., and L. J. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supporting text, Tables S1–S3, and Figs. S1–S5.
- SCN
- soybean cyst nematode
- THF
- (6S)-tetrahydrofolate
- FTHF
- 5-formyl-THF
- MTHFD
- 5,10-methylenetetrahydrofolate dehydrogenase
- PDB
- Protein Data Bank
- SEC
- size-exclusion chromatography
- SAXS
- small-angle X-ray scattering
- SHMT
- serine hydroxy-methyltransferase
- PLP
- pyridoxal 5′-phosphate
- cv.
- cultivar
- PI
- plant introduction
- TCEP
- Tris(carboxyethyl)phosphine
- EcMTHFD
- Escherichia coli gene encoding MTHFD.
References
- 1. Allen T. W., Bradley C. A., Sisson A. J., Byamukama E., Chilvers M. I., Coker C. M., Collins A. A., Damicone J. P., Dorrance A. E., Dufault N. S., Esker P. E., Faske T. R., Giesler L. J., Grybauskas A. P., Hershman D. E., et al. (2017) Soybean yield loss estimates due to diseases in the United States and Ontario, Canada from 2010 to 2014. Plant Health Prog. 18, 19–27 10.1094/PHP-RS-16-0066 [DOI] [Google Scholar]
- 2. McCarville M. T., Marett C. C., Mullaney M. P., Gebhart G. D., and Tylka G. L. (2017) Increase in soybean cyst nematode virulence and reproduction on resistant soybean varieties in Iowa from 2001 to 2015 and the effects on soybean yields. Plant Health Prog. 18, 146–155 10.1094/PHP-RS-16-0062 [DOI] [Google Scholar]
- 3. Lightfoot D. A. (2008) Soybean genomics: developments through the use of cultivar “Forrest.” Int. J. Plant Genomics 2008, 793158 10.1155/2008/793158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hartwig E. E., and Epps J. M. (1973) Registration of “Forrest” soybeans. Crop Sci. 13, 287 10.2135/cropsci1973.0011183X001300020047x [DOI] [Google Scholar]
- 5. Cook D. E., Lee T. G., Guo X., Melito S., Wang K., Bayless A. M., Wang J., Hughes T. J., Willis D. K., Clemente T. E., Diers B. W., Jiang J., Hudson M. E., and Bent A. F. (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209 10.1126/science.1228746 [DOI] [PubMed] [Google Scholar]
- 6. Liu S., Kandoth P. K., Warren S. D., Yeckel G., Heinz R., Alden J., Yang C., Jamai A., El-Mellouki T., Juvale P. S., Hill J., Baum T. J., Cianzio S., Whitham S. A., Korkin D., et al. (2012) A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492, 256–260 10.1038/nature11651 [DOI] [PubMed] [Google Scholar]
- 7. Wu X. Y., Zhou G. C., Chen Y. X., Wu P., Liu L. W., Ma F. F., Wu M., Liu C. C., Zeng Y. J., Chu A. E., Hang Y. Y., Chen J. Q., and Wang B. (2016) Soybean cyst nematode resistance emerged via artificial selection of duplicated serine hydroxymethyltransferase genes. Front. Plant Sci. 7, 998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lakhssassi N., Patil G., Piya S., Zhou Z., Baharlouei A., Kassem M. A., Lightfoot D. A., Hewezi T., Barakat A., Nguyen H. T., and Meksem K. (2019) Genome reorganization of the GmSHMT gene family in soybean showed a lack of functional redundancy in resistance to soybean cyst nematode. Sci. Rep. 9, 1506 10.1038/s41598-018-37815-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chitnumsub P., Jaruwat A., Riangrungroj P., Ittarat W., Noytanom K., Oonanant W., Vanichthanankul J., Chuankhayan P., Maenpuen S., Chen C. J., Chaiyen P., Yuthavong Y., and Leartsakulpanich U. (2014) Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control. Acta Crystallogr. D Biol. Crystallogr. 70, 3177–3186 10.1107/S1399004714023128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pinthong C., Maenpuen S., Amornwatcharapong W., Yuthavong Y., Leartsakulpanich U., and Chaiyen P. (2014) Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: implications for selective inhibition. FEBS J. 281, 2570–2583 10.1111/febs.12803 [DOI] [PubMed] [Google Scholar]
- 11. Schwertz G., Frei M. S., Witschel M. C., Rottmann M., Leartsakulpanich U., Chitnumsub P., Jaruwat A., Ittarat W., Schäfer A., Aponte R. A., Trapp N., Mark K., Chaiyen P., and Diederich F. (2017) Conformational aspects in the design of inhibitors for serine hydroxymethyltransferase (SHMT): biphenyl, aryl sulfonamide, and aryl sulfone motifs. Chemistry 23, 14345–14357 10.1002/chem.201703244 [DOI] [PubMed] [Google Scholar]
- 12. Schwertz G., Witschel M. C., Rottmann M., Bonnert R., Leartsakulpanich U., Chitnumsub P., Jaruwat A., Ittarat W., Schäfer A., Aponte R. A., Charman S. A., White K. L., Kundu A., Sadhukhan S., Lloyd M., et al. (2017) Antimalarial inhibitors targeting serine hydroxymethyltransferase (SHMT) with in vivo efficacy and analysis of their binding mode based on X-ray cocrystal structures. J. Med. Chem. 60, 4840–4860 10.1021/acs.jmedchem.7b00008 [DOI] [PubMed] [Google Scholar]
- 13. Schwertz G., Witschel M. C., Rottmann M., Leartsakulpanich U., Chitnumsub P., Jaruwat A., Amornwatcharapong W., Ittarat W., Schäfer A., Aponte R. A., Trapp N., Chaiyen P., and Diederich F. (2018) Potent inhibitors of plasmodial serine hydroxymethyltransferase (SHMT) featuring a spirocyclic scaffold. Chem. Med. Chem. 13, 931–943 10.1002/cmdc.201800053 [DOI] [PubMed] [Google Scholar]
- 14. Sopitthummakhun K., Thongpanchang C., Vilaivan T., Yuthavong Y., Chaiyen P., and Leartsakulpanich U. (2012) Plasmodium serine hydroxymethyltransferase as a potential anti-malarial target: inhibition studies using improved methods for enzyme production and assay. Malar. J. 11, 194 10.1186/1475-2875-11-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Witschel M. C., Rottmann M., Schwab A., Leartsakulpanich U., Chitnumsub P., Seet M., Tonazzi S., Schwertz G., Stelzer F., Mietzner T., McNamara C., Thater F., Freymond C., Jaruwat A., Pinthong C., et al. (2015) Inhibitors of plasmodial serine hydroxymethyltransferase (SHMT): cocrystal structures of pyrazolopyrans with potent blood- and liver-stage activities. J. Med. Chem. 58, 3117–3130 10.1021/jm501987h [DOI] [PubMed] [Google Scholar]
- 16. Ducker G. S., Ghergurovich J. M., Mainolfi N., Suri V., Jeong S. K., Hsin-Jung Li S., Friedman A., Manfredi M. G., Gitai Z., Kim H., and Rabinowitz J. D. (2017) Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 114, 11404–11409 10.1073/pnas.1706617114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giardina G., Paone A., Tramonti A., Lucchi R., Marani M., Magnifico M. C., Bouzidi A., Pontecorvi V., Guiducci G., Zamparelli C., Rinaldo S., Paiardini A., Contestabile R., and Cutruzzolà F. (2018) The catalytic activity of serine hydroxymethyltransferase is essential for de novo nuclear dTMP synthesis in lung cancer cells. FEBS J. 285, 3238–3253 10.1111/febs.14610 [DOI] [PubMed] [Google Scholar]
- 18. Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A. L., Kafri R., Kirschner M. W., Clish C. B., and Mootha V. K. (2012) Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 10.1126/science.1218595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paone A., Marani M., Fiascarelli A., Rinaldo S., Giardina G., Contestabile R., Paiardini A., and Cutruzzolà F. (2014) SHMT1 knockdown induces apoptosis in lung cancer cells by causing uracil misincorporation. Cell Death Dis. 5, e1525 10.1038/cddis.2014.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patil G. B., Lakhssassi N., Wan J., Song L., Zhou Z., Klepadlo M., Vuong T. D., Stec A. O., Kahil S. S., Colantonio V., Valliyodan B., Rice J. H., Piya S., Hewezi T., Stupar R. M., et al. (2019) Whole-genome re-sequencing reveals the impact of the interaction of copy number variants of the rhg1 and Rhg4 genes on broad-based resistance to soybean cyst nematode. Plant Biotechnol. J. 17, 1595–1611 10.1111/pbi.13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goverse A., and Smant G. (2014) The activation and suppression of plant innate immunity by parasitic nematodes. Annu. Rev. Phytopathol. 52, 243–265 10.1146/annurev-phyto-102313-050118 [DOI] [PubMed] [Google Scholar]
- 22. Dietmann S., Park J., Notredame C., Heger A., Lappe M., and Holm L. (2001) A fully automatic evolutionary classification of protein folds: DALI domain dictionary version 3. Nucleic Acids Res. 29, 55–57 10.1093/nar/29.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruszkowski M., Sekula B., Ruszkowska A., and Dauter Z. (2018) Chloroplastic serine hydroxymethyltransferase from Medicago truncatula: a structural characterization. Front. Plant Sci. 9, 584 10.3389/fpls.2018.00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ulevitch R. J., and Kallen R. G. (1977) Purification and characterization of pyridoxal 5′-phosphate dependent serine hydroxymethylase from lamb liver and its action upon beta-phenylserines. Biochemistry 16, 5342–5350 10.1021/bi00643a027 [DOI] [PubMed] [Google Scholar]
- 25. Scarsdale J. N., Radaev S., Kazanina G., Schirch V., and Wright H. T. (2000) Crystal structure at 2.4 A resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate. J. Mol. Biol. 296, 155–168 10.1006/jmbi.1999.3453 [DOI] [PubMed] [Google Scholar]
- 26. Contestabile R., Angelaccio S., Bossa F., Wright H. T., Scarsdale N., Kazanina G., and Schirch V. (2000) Role of tyrosine 65 in the mechanism of serine hydroxymethyltransferase. Biochemistry 39, 7492–7500 10.1021/bi000032z [DOI] [PubMed] [Google Scholar]
- 27. Szebenyi D. M., Liu X., Kriksunov I. A., Stover P. J., and Thiel D. J. (2000) Structure of a murine cytoplasmic serine hydroxymethyltransferase quinonoid ternary complex: evidence for asymmetric obligate dimers. Biochemistry 39, 13313–13323 10.1021/bi000635a [DOI] [PubMed] [Google Scholar]
- 28. Trivedi V., Gupta A., Jala V. R., Saravanan P., Rao G. S., Rao N. A., Savithri H. S., and Subramanya H. S. (2002) Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism. J. Biol. Chem. 277, 17161–17169 10.1074/jbc.M111976200 [DOI] [PubMed] [Google Scholar]
- 29. Fu T. F., Scarsdale J. N., Kazanina G., Schirch V., and Wright H. T. (2003) Location of the pteroylpolyglutamate-binding site on rabbit cytosolic serine hydroxymethyltransferase. J. Biol. Chem. 278, 2645–2653 10.1074/jbc.M210649200 [DOI] [PubMed] [Google Scholar]
- 30. Pai V. R., Rajaram V., Bisht S., Bhavani B. S., Rao N. A., Murthy M. R., and Savithri H. S. (2009) Structural and functional studies of Bacillus stearothermophilus serine hydroxymethyltransferase: the role of Asn341, Tyr60 and Phe351 in tetrahydrofolate binding. Biochem. J. 418, 635–642 10.1042/BJ20081739 [DOI] [PubMed] [Google Scholar]
- 31. Ruszkowski M., Sekula B., Ruszkowska A., Contestabile R., Nogues I., Angelaccio S., Szczepaniak A., and Dauter Z. (2019) Structural basis of methotrexate and pemetrexed action on serine hydroxymethyltransferases revealed using plant models. Sci. Rep. 9, 19614 10.1038/s41598-019-56043-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mehrshahi P., Gonzalez-Jorge S., Akhtar T. A., Ward J. L., Santoyo-Castelazo A., Marcus S. E., Lara-Núñez A., Ravanel S., Hawkins N. D., Beale M. H., Barrett D. A., Knox J. P., Gregory J. F. 3rd, Hanson A. D., Bennett M. J., et al. (2010) Functional analysis of folate polyglutamylation and its essential role in plant metabolism and development. Plant J. 64, 267–279 10.1111/j.1365-313X.2010.04336.x [DOI] [PubMed] [Google Scholar]
- 33. Wei Z., Sun K., Sandoval F. J., Cross J. M., Gordon C., Kang C., and Roje S. (2013) Folate polyglutamylation eliminates dependence of activity on enzyme concentration in mitochondrial serine hydroxymethyltransferases from Arabidopsis thaliana. Arch. Biochem. Biophys. 536, 87–96 10.1016/j.abb.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 34. Florini J. R., and Vestling C. S. (1957) Graphical determination of the dissociation constants for two-substrate enzyme systems. Biochim. Biophys. Acta 25, 575–578 10.1016/0006-3002(57)90529-2 [DOI] [PubMed] [Google Scholar]
- 35. Ubonprasert S., Jaroensuk J., Pornthanakasem W., Kamonsutthipaijit N., Wongpituk P., Mee-Udorn P., Rungrotmongkol T., Ketchart O., Chitnumsub P., Leartsakulpanich U., Chaiyen P., and Maenpuen S. (2019) A flap motif in human serine hydroxymethyltransferase is important for structural stabilization, ligand binding, and control of product release. J. Biol. Chem. 294, 10490–10502 10.1074/jbc.RA119.007454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tramonti A., Nardella C., di Salvo M. L., Barile A., Cutruzzolà F., and Contestabile R. (2018) Human cytosolic and mitochondrial serine hydroxymethyltransferase isoforms in comparison: full kinetic characterization and substrate inhibition properties. Biochemistry 57, 6984–6996 10.1021/acs.biochem.8b01074 [DOI] [PubMed] [Google Scholar]
- 37. Kabsch W. (2010) XDS. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol. Crystallogr. 66, 133–144 10.1107/S0907444909047374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., and Adams P. D. (2008) Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 10.1007/978-1-60327-058-8_28 [DOI] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., yet al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Cryst. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 43. Sah S., and Varshney U. (2015) Impact of mutating the key residues of a bifunctional 5,10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase from Escherichia coli on its activities. Biochemistry 54, 3504–3513 10.1021/acs.biochem.5b00400 [DOI] [PubMed] [Google Scholar]
- 44. Chiba Y., Terada T., Kameya M., Shimizu K., Arai H., Ishii M., and Igarashi Y. (2012) Mechanism for folate-independent aldolase reaction catalyzed by serine hydroxymethyltransferase. FEBS J. 279, 504–514 10.1111/j.1742-4658.2011.08443.x [DOI] [PubMed] [Google Scholar]
- 45. Schirch L., and Ropp M. (1967) Serine transhydroxymethylase: affinity of tetrahydrofolate compounds for the enzyme and enzyme-glycine complex. Biochemistry 6, 253–257 10.1021/bi00853a039 [DOI] [PubMed] [Google Scholar]
- 46. Nonaka H., Nakanishi Y., Kuno S., Ota T., Mochidome K., Saito Y., Sugihara F., Takakusagi Y., Aoki I., Nagatoishi S., Sumoto K. and Sando S. (2019) Design strategy for serine hydroxymethyltransferase probes based on retro-aldol-type reaction. Nat. Commun. 10, 876 10.1038/s41467-019-08833-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Bank under accession codes 6UXH, 6UXI, 6UXJ, 6UXK, and 6UXL. All other data are available from the corresponding author upon reasonable request.