Abstract
Cullin-RING E3 ligase (CRL) is the largest family of E3 ubiquitin ligase, responsible for ubiquitylation of ∼20% of cellular proteins. CRL plays an important role in many biological processes, particularly in cancers due to abnormal activation. CRL activation requires neddylation, an enzymatic cascade transferring small ubiquitin-like protein NEDD8 to a conserved lysine residue on cullin proteins. Recent studies have validated that neddylation is an attractive anticancer target. In this study, we report the establishment of an Alpha-Screen-based high throughput screen (HTS) assay for in vitro CUL5 neddylation, and screened a library of 17,000 compounds including FDA approved drugs, natural products and synthetic drug-like small-molecule compounds. Gossypol, a natural compound derived from cotton seed, was identified as an inhibitor of cullin neddylation. Biochemical studies showed that gossypol blocked neddylation of both CUL5 and CUL1 through direct binding to SAG-CUL5 or RBX1-CUL1 complex, and CUL5-H572 plays a key role for gossypol binding. On cellular level, gossypol inhibited cullin neddylation in a variety of cancer cell lines and selectively caused accumulation of NOXA and MCL1, the substrates of CUL5 and CUL1, respectively, in multiple cancer cell lines. Combination of gossypol with specific MCL1 inhibitor synergistically suppress growth of human cancer cells. Our study revealed a previously unknown anti-cancer mechanism of gossypol with potential to develop a new class of neddylation inhibitors.
Keywords: Neddylation, Ubiquitin–proteasome system, Small molecule inhibitors, Gossypol, Cullin-RING E3 ligase
Introduction
Protein ubiquitylation is one of the major post-translational modifications in eukaryotic cells that mainly regulates protein turnover [1]. By catalyzing the covalent attachment of a 76-amino-acid ubiquitin (Ub) to a lysine residue of a substrate protein, ubiquitylation controls the homeostasis of thousands of proteins and affects nearly every aspect of biological process in a living cell. A typical ubiquitylation modification is catalyzed by a three-step enzymatic cascade, consisting of an E1 activating enzyme, an E2 conjugating enzyme, and an E3 ligase [2]. Briefly, ubiquitin is first activated by E1 in an ATP-dependent manner, then transferred to an E2 via a thioester bond between E2 active site cysteine residue to the C-terminus of ubiquitin. Finally, E3 ligase interacts with both the E2 ∼ Ub and a specific substrate, transferring Ub to the ε-amino group of lysine residue on a substrate. Catalyzing the final step of ubiquitylation, E3 ligase is responsible for both reaction efficiency and substrate selectivity. Human genome encodes hundreds of E3 ligases, of which the cullin-RING E3 ligases (CRLs) constitute the largest family [3]. A cullin protein assembles with a RING domain protein to form the core subunit of a CRL [4]. The cullin family consists of eight canonical members (CUL1, 2, 3, 4A, 4B, 5, 7 and 9) [4]. And two specific RING domain proteins, RBX1 and RBX2 (also known as SAG), serve as the RING subunit of CRLs [3].
The activation of CRLs requires neddylation, another post-translational modification by transferring NEDD8 (neural precursor cell expressed developmentally downregulated protein 8) to a specific lysine residue on a cullin protein [5]. As one of the ubiquitin-like proteins (UBLs), NEDD8 shares 59% sequence identity with ubiquitin [6]. Similar to ubiquitylation, neddylation is also catalyzed by a three-step enzymatic cascade, including one NEDD8 E1 (APPBP1/UBA3) [7], and two NEDD8 E2s (UBE2M and UBE2F) [8], [9]. The two RING proteins, RBX1 and SAG serve as the NEDD8 E3s [5]. In addition, DCN1 (defective in cullin neddylation 1 protein) serves as a co-E3 ligase, which binds to acetylated N-terminus of UBE2M E2 [10]. Moreover, structural studies have revealed that UBE2M specifically pairs with RBX1 to neddylate CUL1, 2, 3, 4A, and 4B, while UBE2F specifically pairs with SAG/RBX2 to neddylate CUL5 [9], [11], [12].
CRLs are activated, upon neddylation modification, to ubiquitylate specific substrates, including a variety of tumor suppressors [13]. In human cancer cells, CRLs are abnormally activated due to overexpression of neddylation E1/E2/E3 enzymes, such as APPBP1/UBA3, UBE2M, UBE2F, RBX1, SAG and DCN1 with correlation of poor patient survival [14], [15], [16], [17], [18], [19], [20]. Thus, neddylation appears to be an attractive anti-cancer target. Indeed, several inhibitors have been discovered which target the activity of a specific enzyme or the protein–protein interactions (PPIs) in the neddylation cascade. MLN4924 (also known Pevonedistat), the first-in-class NAE inhibitor, exhibits potent inhibitory effect against neddylation of all cullins in vitro and in vivo [21], and has entered the Phase I/II clinical trials in treating myeloid leukemia, multiple myeloma, lymphoma, and some advanced solid tumors [22], [23], [24], [25]. Recently, high-throughput screening (HTS) and structure-based drug design have yielded a panel of inhibitors specifically targeting the DCN1-UBE2M interaction site [26], [27], [28], [29], [30]. Some of these potent DCN1 inhibitors selectively block the neddylation of CUL3 and accumulate CUL3-specific substrate (erythroid-derived 2)-like 2 (NRF2), an antioxidant transcription factor, showing the therapeutic potential of treating reactive oxygen species (ROS) related diseases [27], [28], [30].
In an effort to discover more selective and potent inhibitors targeting cullin neddylation, we developed an AlphaScreen-based in vitro CUL5 neddylation assay, and screened a library of 17,000 compounds, including all FDA approved drugs and ∼600 of natural products, leading to identification of gossypol as a potent inhibitor of cullin neddylation. Gossypol, a natural compound extracted from cotton seed, was initially used as a male contraceptive, and later developed as an antitumor agent against multiple types of human cancers [31]. An enantiomer of racemic gossypol, AT-101 [R-(−)-gossypol acetic acid; Ascenta Therapeutics, Inc.], has completed several Phase I/II clinical trials as a BCL-2 inhibitor [32], [33], [34], [35], [36].
In the present study, we repurposed gossypol as a potent inhibitor of cullin neddylation with evidence from both biochemical and cell-based studies involving multiple cancer cell lines. Specifically, our biochemical data showed that gossypol inhibits cullin neddylation by targeting the SAG-CUL5 and RBX1-CUL1 complex and caused accumulation of CRL substrates, such as pro-apoptotic protein NOXA and anti-apoptotic protein MCL1. Biologically, combination of gossypol and MCL-1 inhibitor showed synergistic effect in suppressing proliferation of cancer cells.
Materials and methods
Chemicals
MLN4924 was purchased from ApexBio (#B1036). Gossypol was purchased from Aladdin (#G133787). S63845 was purchased from Selleck (#S8383). Chlorhexidine (CHX) was purchased from Sigma-Aldrich (#C7698).
Constructs and protein purification
All constructs were generated by standard PCR/ligation molecular biology methods. The entire coding sequence for each construct was verified by automated sequencing. Plasmid of RBX1-CUL1CTD (CUL1 residues 411–776 with introduced mutations of L421E, V451E, V452K, and Y455K to increase solubility) was prepared as described previously [37]. Human UBE2F, UBE2M, APPBP1 and NEDD8 terminating at Gly76 were cloned into a home-made variant of pET-28b vector with a His6-SUMO tag fused at the N-terminus. CUL5CTD (CUL5 residues 401–780 with the mutations of L407E, L439K, V440K to increase solubility) was cloned into a home-made variant of pRSFDuet vector with a His6 tag [37]. UBA3 and SAG were cloned into the GST-fusion expression vector pGEX-6p-1 (GE Healthcare).
UBE2F, UBE2M, and NEDD8 were expressed in BL21 (DE3) Escherichia coli (TransGen Biotech) and purified by Ni-NTA agarose beads (QIAGEN). After Ulp1 digestion, the proteins were further purified by gel-filtration chromatography (GE Healthcare). His6-RBX1-CUL1 was purified by Ni-NTA agarose beads (QIAGEN) and gel-filtration chromatography. GST-UBA3 and His6-SUMO-APPBP1 were co-expressed in BL21 (DE3) E. coli (TransGen Biotech) and purified by Ni-NTA agarose beads (QIAGEN) and subsequent Glutathione Sepharose 4B beads (GE Healthcare) after treatment with Ulp1 to remove His6-SUMO tag. GST tag fused to UBA3 was retained with no influence on protein activity. His6-CUL5 and GST-SAG were co-expressed in BL21 (DE3) E. coli (TransGen Biotech) and purified by Ni-NTA agarose beads (QIAGEN) and Glutathione Sepharose 4B beads (GE Healthcare). GST tag fused to SAG was removed by 3C-Protease digestion to improve its purity. Finally, all proteins were purified by gel-filtration chromatography and stored in wash buffer (25 mM HEPEs pH 7.5 and 150 mM NaCl). Protein aliquots were rapidly frozen in liquid nitrogen and stored at − 80 °C.
Biochemical assays
In vitro E2 ∼ NEDD8 thioester assay
The reaction mixture contains 50 nM UBA3/APPBP1, 3 μM NEDD8, and 1 μM NEDD8 E2 (UBE2M or UBE2F) in a buffer containing 50 mM Tris-HCl (pH = 7.4), 5 mM MgCl2, 0.5 mM DTT, and 0.1 mg/ml BSA. The mixture was incubated with indicated compounds (final DMSO ≤1%) at 25 °C for 10 min, followed by addition of 20 μM ATP and incubation at 37 °C for 30 min. The reaction was quenched by adding 4× SDS loading buffer (without DTT). Final samples were separated by SDS–PAGE gel and detected by Coomassie-blue staining.
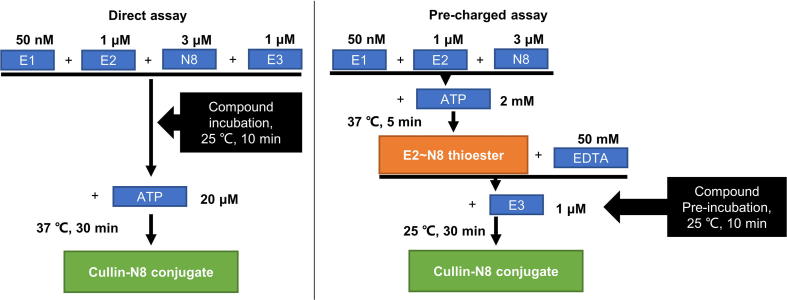
In vitro cullin neddylation assay (Direct)
The reaction mixture contains 50 nM UBA3/APPBP1, 3 μM NEDD8, 1 μM NEDD8 E2 (UBE2M or UBE2F), and 1 μM RING-Cullin E3 complex (RBX1-CUL1CTD or SAG-CUL5CTD) in a buffer containing 50 mM Tris-HCl (pH = 7.4), 5 mM MgCl2, 0.5 mM DTT, and 0.1 mg/ml BSA. The mixture was incubated with indicated compounds (final DMSO ≤1%) at 25 °C for 10 min, followed by addition of 20 μM ATP and incubation at 37 °C for 30 min. The reaction was quenched by adding 4× SDS loading buffer (without DTT). Final samples were separated by SDS–PAGE gel and detected by Coomassie-blue staining. For use of more sensitive detection method by western-blot (as shown in Fig. 2B), the above assay was done with an optimized reaction mixture containing 25 nM UBA3/APPBP1, 200 nM NEDD8, 300 nM NEDD8 E2, and 200 nM RING-Cullin E3 complex.
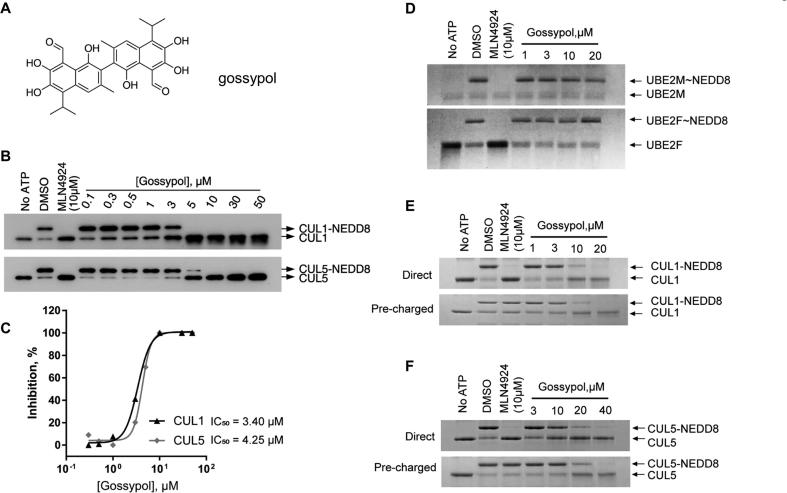
Fig. 2.
Gossypol inhibits in vitro Cullin neddylation. (A) Chemical structure of gossypol. (B-C) IC50 of gossypol inhibition of neddylation of cullins 1 and 5: various concentrations of gossypol, along with MLN4924 as a positive control was incubated in complete reaction mixture, followed by PAGE and western-blot detection using anti-CUL5 or anti-CUL1 antibodies (B). The band density was quantified and an IC50 curve plotted. The calculated IC50 against CUL1 and CUL5 neddylation was 3.4 μM and 4.25 μM, respectively (C). (D-F) Gossypol inhibits CUL1 and CUL5 neddylation, but not E2 conjugation: Gossypol at various concentrations was added into the reaction mixture, followed by detection of E2 conjugation via PAGE-Coomassie-blue staining (D). Gossypol at various concentrations was added into pre-charged reaction mixture (see M&M for details), followed by PAGE-Coomassie-blue staining (E-F).
In vitro cullin neddylation assay (Pre-charged)
Step 1: Preparation of reaction mixture A (Pre-charged E2 ∼ NEDD8 thioesters): The reaction mixture of 50 nM UBA3/APPBP1, 3 μM NEDD8, and 1 μM NEDD8 E2 (UBE2M or UBE2F) was mixed in the same reaction buffer with addition of 2 mM ATP, followed by incubation at 37 °C for 5 min. The reaction was quenched by addition of 50 mM EDTA and put on ice for 5 min.
Step 2: Preparation of reaction mixture B (E3 + Compound): The RING-Cullin E3 complex (RBX1-CUL1CTD or SAG-CUL5CTD, 1 μM) was mixed in a reaction buffer containing 50 mM Tris-HCl (pH = 7.4), 5 mM MgCl2, 0.5 mM DTT, 0.1 mg/ml BSA and 50 mM EDTA. The mixture was incubated with indicated compounds (final DMSO ≤1%) at 25 °C for 10 min.
Step 3: Reaction mixtures A and B were mixed and incubated at 25 °C for 30 min, followed by addition of 4× SDS loading buffer (without DTT). Final samples were separated by SDS–PAGE gel electrophoresis and detected by Coomassie-blue staining.
AlphaScreen assay
NEDD8 biotinylation: The purified NEDD8 was biotinylated using EZ-Link Sulfo-NHS-LC-Biotin Kit (Thermo Fisher Scientific Inc.) followed by the instructions from the manufacturer. The biotinylated NEDD8 was tested in the in vitro cullin neddylation assays shown above methods to validate its activity.
AlphaScreen-based cullin neddylation assay: The reaction mixture of 170 nM Biotin-NEDD8, 25 nM NEDD8 E1 (UBA3/APPBP1), 300 nM UBE2F and 170 nM SAG/CUL5CTD were incubated in a buffer containing 25 mM HEPES, 50 mM NaCl, 500 μM DTT, 0.01% BSA, and 2.5 mM MgCl2 in the presence of a compound or equal volume of DMSO (the “DMSO” control for a complete reaction) at 25 °C for 20 min in a 384-well Opti-plate (Perkin Elmer. Inc). The reaction was initiated by addition of 25 μM ATP or equal volume of reaction buffer (the “no-ATP” control). After incubation at 37 °C for 5 min, the reaction mixture was added with Nickel-chelated AlphaScreen Acceptor beads (Perkin Elmer. Inc) at a final concentration of 5 μg/ml and was incubated in dark at 22 °C for 1 h. Streptavidin-coated AlphaScreen Donor Beads were then added with a final concentration of 5 μg/ml. After incubation in dark at 22 °C for 1 h, the plate was read by Envision Multi-reader (Perkin Elmer. Inc) and the inhibition rate of an active compound was calculated using the captured signaling intensity of a given well by the following formula:
A: AlphaScreen Signal.
The HTS
Library
The library consists of a total of ∼17,000 compounds, consisting of 1700 FDA approved drugs (TargetMol. Inc), ∼600 natural compounds derived from plants, animals or microorganisms (TargetMol. Inc), ∼2300 biologically active and structurally diverse compounds (The Spectrum Collection) and >10,000 synthetic drug-like small-molecule compounds (MRCT Index library). The HTS in 384-well format was carried out as described above. Each compound was added to reach a final concentration of 20 μM. To get a homogeneous solution, the reaction mixture was mixed thoroughly upon the addition of AlphaScreen beads. On each 384-well screen plate, the left two columns were added with DMSO (DMSO control) while the right two columns were added with reaction buffer without ATP (no-ATP control). All plates were carefully sealed before reading to avoid light and evaporation. The compounds with 50% of inhibition of Alpha-signal were counted as primary hits.
The hit confirmation
The primary hits were cherry-picked by a Mosquito liquid station (TTP Labtech) to the reaction plates containing enzymes and reaction buffer as described above. The inhibitory effect of the primary hits was confirmed by re-testing at the final concentration of 20 μM. Meanwhile, a copy of these picked primary hits was further diluted and tested at a final concentration of 10 μM for evaluation of dose-dependency.
The in vitro thermal shift assay (TSA)
The purified E3 complex proteins (RBX1/CUL1CTD or SAG/CUL5CTD, 0.5 μg) were added to the regular PCR tubes with total volume adjusted to 50 μl by buffer containing 25 mM HEPES (pH = 7.4), 150 mM NaCl and 1 mM DTT. The reaction mixtures were first incubated with the compounds (or DMSO) at 25 °C for 10 min, then heated at indicated temperatures in a BIO-RAD T100TM Thermal Cycler for 3 min. Heated mixtures were centrifuged at 20,000×g for 20 min at 4 °C and the supernatant was collected, added with 4× SDS loading buffer, and subjected to SDS PAGE gel electrophoresis and western blotting.
The cellular thermal shift assay (CETSA)
The assay was performed according to the protocol described previously [38]. Briefly, the cells with indicated treatment (e.g. gossypol, MLN4924 or DMSO) were collected, washed with cold PBS, and suspended in 50 μl cold PBS containing complete protease inhibitor cocktail. The cell suspension was then heated at indicated temperatures on a PCR machine (described above) for 3 min and then lysed by 3 freeze-thawing cycles with liquid nitrogen. The lysates were centrifuged at 20,000×g for 20 min at 4 °C and the supernatant was collected and analyzed by western blotting as described above.
Docking study
The crystal structures of RBX2/SAG (PDB entry: 2ECL), CUL1/RBX1 (PDB entry: 1LDJ) [12], CUL4B-RBX1 (PDB entry: 4A0C) [39] and CUL5/RBX1 (PDB entry: 3DPL) [40] complexes were utilized and prepared by the Protein Preparation wizard module in Schrödinger 9.0 [41]. All water molecules were deleted from the structures, and the missing hydrogens were added to the proteins. The CUL1, CUL5 and RBX1 docking structures were generated by removing all the other binding partners in each protein complex. Then, a restrained partial minimization for each system was performed with the maximum root-mean-square deviation (RMSD) value set to 0.3 Å. The potential binding sites were detected by the SiteMap module and the receptor grid boxes were generated by the Receptor gird generation module. Gossypol was prepared by using the LigPrep module with protonated states generated at pH = 7.0 ± 2.0 and subsequently docked into the structures of CUL1, CUL5 RBX1 and RBX2/SAG using the Glide XP model.
Cell culture
Human lung cancer cell lines H1299, H358, and H2170, breast cancer cell lines MDA-MB-231, and BT-549, prostate cancer cell line PC3, and cervical cancer cell line HeLa, Acute promyelocytic leukemia cell line HL-60 was obtained from ATCC. Myeloma cell lines ARP1, JJN3 and 8226 were a kind gift from Dr. Zhen Cai. MDA-MB-231, H1299, and HeLa cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% (v/v) fetal bovine serum (FBS). H358, H2170, BT-549, PC3, HL-60, ARP1, JJN3 and 8226 were cultured in RPMI 1640 Medium with 10% FBS. All cells were incubated at 5% CO2, 37 °C, with 95% humidity.
Western blotting and antibodies
Cells after indicated treatment were collected, lysed and examined by standard western blot procedures. SAG monoclonal Ab was raised against the RING domain (AA44-113) [42]. The other antibodies were purchased from a variety of vendors as follows: CUL1CTD (Proteintech, 12895-1-AP), CUL1 (Santa Cruz, SC-11384), CUL5CTD (Sigma-Aldrich, AV35127), CUL5 (Santa Cruz, sc-373822), CUL3 (Cell signaling, 2759S), CUL2 (abcam, ab166917), CUL4A (Cell signaling, 2699S), CUL4B (Proteintech, 12916-1-AP), RBX1 (Cell signaling, 11922S), NEDD8 (abcam, ab81264), UBA3 (abcam, ab124728), NAE1 (Cell signaling, 14321S), UBE2F (Santa Cruz, sc-398668), UBE2M (Santa Cruz, sc-390064), p21 (Cell signaling, 2947S), p27 (Cell signaling, 2552S), MCL1 (Cell signaling, 5453S), NRF2 (Santa Cruz, sc-722), CDT1 (Santa Cruz, sc-28262), NOXA (EMD Millipore, OP180), and β-Actin (Sigma-Aldrich, A5441).
Half-life analysis
Cells were treated with 20 μM gossypol, along with the DMSO control for 8 h, then incubated with 50 μg/ml CHX for indicated time points and collected for western blot. The quantification was done by analyzing the band density using ImageJ software (NIH).
Quantitative real-time reverse-transcription PCR (qRT-PCR)
Total RNA was extracted using TRIzol Reagent (Ambion, 15596–026, Life-Technologies, USA) and transcribed into first strand of DNA (cDNA) using SuperScript III reagents (Invitrogen) with an oligo(dT)20 primer. qRT-PCR was performed using SYBR green reagent on StepOnePlus Real-Time PCR system (Applied Biosystems). Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control.
The primers for NOXA are:
5′-GACTGTTCGTGTTCAGCTCG-3′ (forward);
5′-CACTCGACTTCCAGCTCTGCT-3′ (reverse).
The primers for MCL-1 are:
5′-CCAAGAAAGCTGCATCGAACCAT-3′ (forward);
5′-CAGCACATTCCTGATGCCACCT-3′ (reverse).
Transfection and siRNAs
Cells were transfected using the Transfection GenMuteTM siRNA Transfection Reagent (SignaGen® Laboratories) according to the manufacturer’s protocol. The siRNA sequence for NOXA was reported previously [43], and foe MCL1 was 5′-GGCAGTCGCTGGAGATTAT-3′.
Cell proliferation assays
Cell proliferation assays were performed using Cell Counting Kit-8 (CCK-8) (MedChem Express, HY-K0301-3000T) according to the manufacturer’s protocol. Briefly, cells were seeded in 96-well plates and incubated for 24 h, followed by addition of gossypol at indicated concentrations as a single agent or in combination of the MCL1 inhibitor S63845 (Selleck, #S8383). For the combination assays, S63845 was either added at the same concentration of gossypol (1:1 combination) or added at the concentration of IC20 which was determined in a pre-experiment. After 72-hours treatment, 10 μl CCK-8 solution was added to each well and incubated for 2 h. The optical density (OD) values were measured at 450 nm on a microplate reader (SpectraMax iD3, Molecular Devices). The data was analyzed and the IC50 was determined using a nonlinear regression model by GraphPad Prism software (La Jolla, CA, USA). The synergistic effect was analyzed by calculating the combination index (CI) using Compusyn software (Combosyn Inc., Paramus, NJ, USA) to generate isobolograms according to the algorithm described by Chou and Talalay [44].
Results
AlphaScreen-based in vitro CUL5 neddylation assay
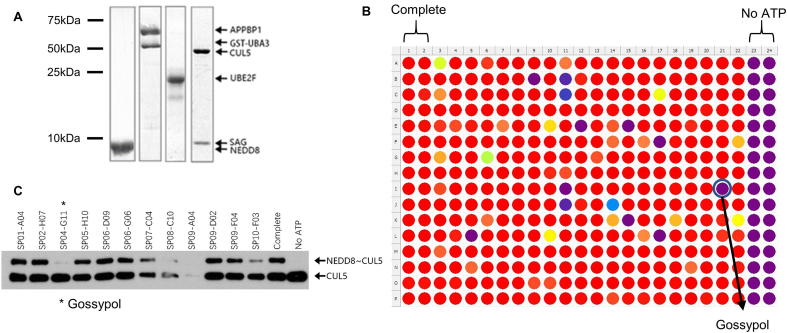
To identify small molecules that inhibit CUL5 neddylation catalyzed by NAE E1, UBE2F E2 and SAG E3, we developed a high-throughput AlphaScreen-based in vitro CUL5 neddylation assay. For this purpose, pure recombinant proteins were prepared, including NEDD8 E1 (UBA3/APPBP1), UBE2F E2, SAG-CUL5CTD (Cullin-5C-terminal domain) E3-substrate complex and NEDD8, with purity >95% (Fig. S1A). CUL5CTD containing 6x His tag and NEDD8 was labeled with biotin during protein preparation. Ni2+-chelated Alpha acceptor beads and Streptavidin-coated Alpha donor beads were successively added to the reaction mixture. The resulting NEDD8-CUL5CTD conjugates brought the donor and acceptor beads to a close proximity to generate energy transfer that produced the Alpha signals which could be detected by Multi-mode plate readers at 520–620 nm (Fig. 1A).
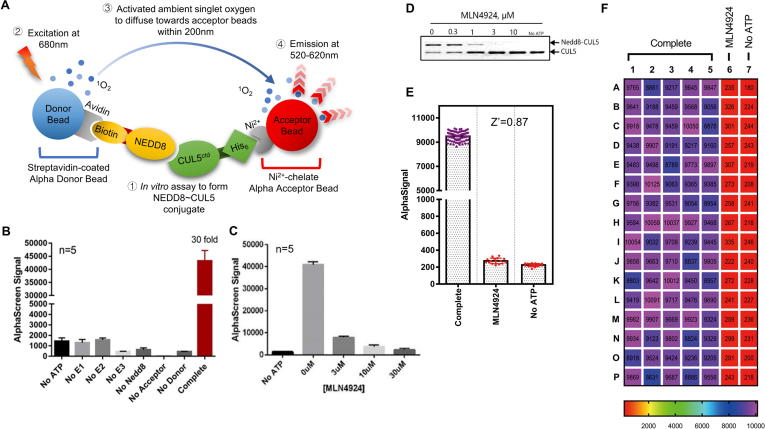
Fig. 1.
AlphaScreen based SAG-CUL5 neddylation assay. (A) Reaction schemes for AlphaScreen based SAG/CUL5 neddylation assay (see M&M for details). (B) Components omission test: The reaction mixture was placed in a 384-well plate, followed by addition of AlphaScreen Donor and Acceptor beads, and subjected to analysis with a Perkin-Elmer Envision reader. Only the complete reaction yielded high AlphaScreen signal (AlphaSignal). (C) MLN4924 titration: MLN4924 at various concentrations was added into the reaction mixture which generated a dose–response inhibition of AlphaSignal. (D) MLN4924 inhibition of in vitro CUL5 neddylation reaction: MLN4924 at various concentrations was added into the reaction mixture and subjected to PAGE and Coomassie-blue staining. (E-F) Heatmap and statistics for a High-throughput format assay: The AlphaScreen assay was carried out using automatic dispensers with CV <10% in Complete and No ATP groups, Signal/Noise ratio = 42.12, and Z′ factor = 0.87.
The Alpha signals are produced only when all components are present in the reaction mixture, and missing of any single component will abrogate the signal production (Fig. 1B), demonstrating that the formation of NEDD8-CUL5CTD conjugate is solely dependent on the presence of the E1/E2/E3 enzymes. The Signal/background ratio was about 30:1, providing a suitable HTS window for inhibitor detection. To validate, we used MLN4924 as a positive control in both the AlphaScreen assay (Fig. 1C) and gel-based assay (Fig. 1D). The results showed a dose-dependent inhibition of CUL5 neddylation (Fig. 1D), which were entirely consistent with diminished Alpha-signals in response to the inhibitor (Fig. 1C). Thus the observed Alpha-signals truly reflect the production of CUL5-NEDD8 conjugates in the enzymatic reaction.
To apply the AlphaScreen assay to a high throughput screening (HTS), we scaled down the assay reaction mixture to a 384-well format and optimized the procedures to fit into automation (Fig. 1F). We reached a calculated Z’ factor, an index for the quality of HTS screening [45] of 0.87 (0.5 is acceptable and 1 is the best in theory) in our optimized AlphaScreen-based SAG/CUL5 neddylation assay (Fig. 1E), indicating the assay is of excellent quality for the HTS.
Gossypol as a neddylation inhibitor of CUL1 and CUL5 identified by the HTS
We performed an HTS using this AlphaScreen assay with a pooled library containing about 17,000 compounds including natural products, FDA-approved drugs, and small-molecule compounds with diverse drug-like structures. Gossypol, a natural polyphenol (Fig. 2A) was identified as one of the positive hits (Fig. S1B). Its inhibition against in vitro CUL5CTD neddylation could be reproducibly detected in an independent gel-based assay using the library compound (Fig. S1C). Using freshly purposed gossypol, we determined that it inhibits neddylation of CUL1 or CUL5, with IC50 of 3.4 and 4.3 μM, respectively (Fig. 2B-C).
Unlike MLN4924, gossypol didn’t inhibit the formation of E2 ∼ NEDD8 thioesters (neither UBE2M nor UBE2F) (Fig. 2D). We then performed a pair of direct and pre-charged CUL1/CUL5 neddylation assays to further explore the target of gossypol. In the direct assay, gossypol was directly mixed with NEDD8, NAE/E1, UBE2M/E2 and RBX1/E3 for CUL1 neddylation or E1, UBE2F/E2, SAG/E3 for CUL5 neddylation before addition of ATP, while in the pre-charged assay, NEDD8, NAE and E2 (UBE2M or UBE2F) were pre-incubated to form E2 ∼ NEDD8 thioesters, followed by addition of E3 and gossypol to complete the final conjugation step (see assay scheme in Fig. S2). In these two assays, we used MLN4924, the specific NAE inhibitor as a control. MLN4924 forms an adduct with NEDD8 to prevent the transferring of NEDD8 from NAE to E2 and thus blocks all cullin neddylations [21]. As expected, MLN4924 effectively inhibited neddylation of both CUL1 and CUL5 in the direct assay, but had no effect in the Pre-charged assay in which E1-E2 transthiolation was pre-completed (Fig. 2E-F). Interestingly, unlike MLN4924, while showing no inhibition of the E2 ∼ NEDD8 thioester formation (Fig. 2D), gossypol effectively inhibited neddylation of CUL1 and CUL5 in both the direct and pre-charged assays (Fig. 2E-F). Taken together, our results showed that compare to MLN4924, an NEDD8 E1 inhibitor, gossypol had a totally different mode of action against cullin neddylation, and suggested that gossypol inhibitory effect was at the final E3 conjugation step in the neddylation cascade.
Gossypol binds to the RBX1/CUL1 and SAG/CUL5 complex
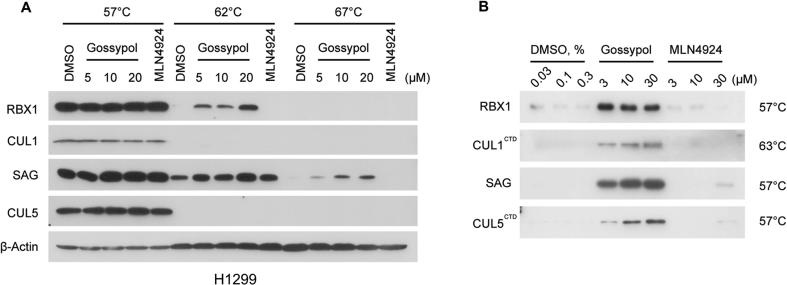
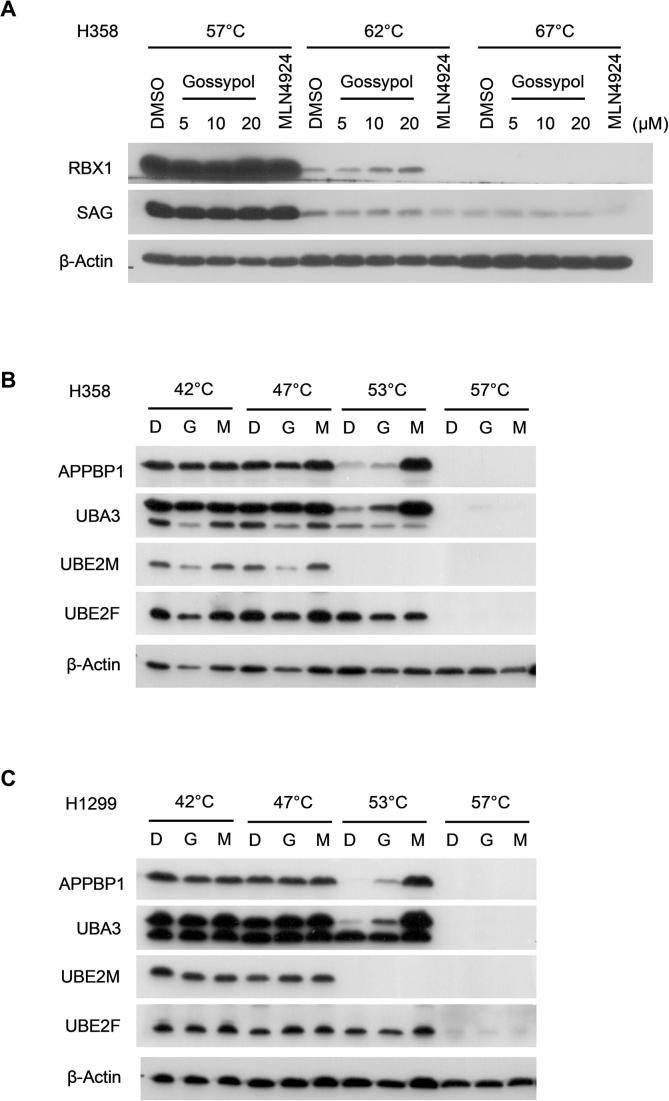
In the NEDD8 conjugation step, a RING E3 binds both NEDD8-loaded E2 and cullin substrate, and promotes the ligation of NEDD8 to the lysine residue on the cullin protein [12], [46]. We therefore hypothesized that gossypol may interrupt either the RING-E2 interaction or RING-cullin interaction. To this end, we investigated the target engagement of gossypol in cells using cellular thermal shift assay (CETSA) in two lung cancer cell lines, H1299 and H358. As shown, the thermal stability of cellular RBX1 and SAG protein was clearly enhanced by gossypol at 62 °C and 67 °C in a dose-dependent manner, whereas these proteins were largely degraded in the cells treated with DMSO or MLN4924 (Figs. 3A, S3A). In contrast, gossypol is unable to stabilize UBA3, APPBP1, UBE2F or UBC12 in cells (Fig. S3B-S3C). Of note, CUL1 or CUL5 was also not stabilized by gossypol in either H1299 (Fig. 3A) or H358 cells (data not shown). To further validate our findings, we performed in vitro thermal shift assay with purified RBX1-CUL1CTD and SAG-CUL5CTD protein complexes. In consistent with the results from CETSA, gossypol clearly stabilized purified RBX1-CUL1CTD and SAG-CUL5CTD in a dose-dependent manner (Fig. 3B).
Fig. 3.
Gossypol binds to the RBX1-CUL1 and SAG-CUL5 complexes. (A) Cellular thermal shift assay was performed in H1299 cells, which showed gossypol stabilized RBX1 and SAG, but not CUL1 or CUL5 in dose and temperature dependent manners. See also Fig. S3. (B) The in vitro thermal shift assay using purified RBX1/CUL1CTD and SAG/CUL5CTD complexes was performed, which showed that gossypol bond to RBX1-CUL1 and SAG-CUL5 complexes.
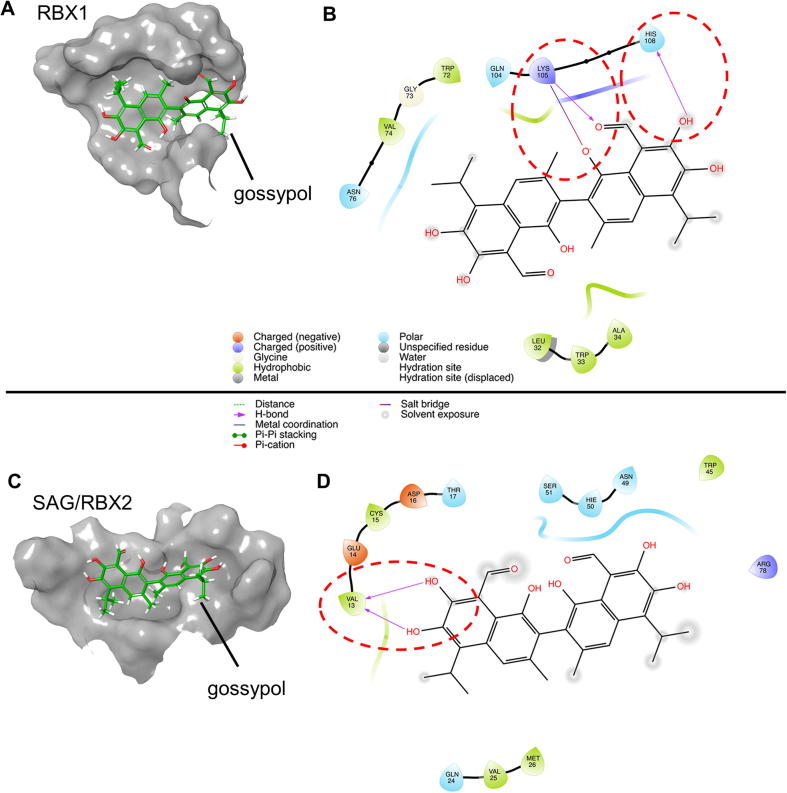
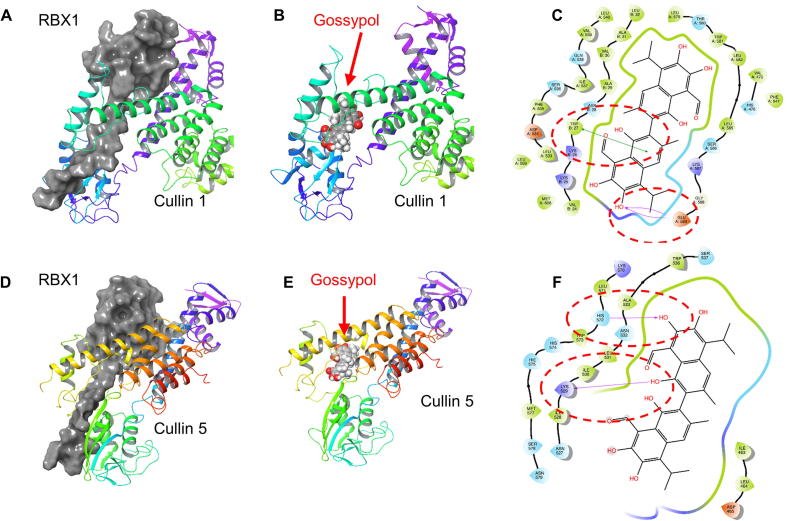
To find the structural basis for the binding of gossypol to RBX1-CUL1 and SAG-CUL5, we used computational modeling method to predict the binding mode. The unbound structures of RBX1, SAG/RBX2, CUL1 and CUL5 were individually generated and optimized based on the corresponding bounded protein–protein complexes. Gossypol was then docked into each protein structure to elucidate potential binding mechanisms. According to the calculation results, gossypol could bind to RBX1 and SAG/RBX2. The hydrogen bonds between gossypol and Lys105 and His108 play the vital roles in the binding of gossypol to RBX1 (Fig. 4A-B). For the binding with SAG/RBX2, two hydrogen bonds could be observed between gossypol and Val13 in SAG/RBX2 (Fig. 4C-D). Interestingly, our docking simulations also showed that gossypol could also strongly bind to both CUL1 and CUL5 with docking scores of −9.604 and −10.678, respectively. For the binding with CUL1, gossypol was found to form hydrogen binding interaction with Glu589 and hydrophobic interaction with Trp27 (Fig. S4A–S4C). While for the binding with CUL5, gossypol could form two hydrogen bonds with Lys529 and His572 (Fig. S4D-S4F), which might contribute to the high binding activity of gossypol to CUL5. For the binding with both CUL1 and CUL5, gossypol was located at the protein–protein interface between RBX1 and CUL1/CUL5, which likely disrupted the protein–protein interactions of the RBX1-CUL1/CUL5 complexes. Taken together, these biochemical data and computer modeling analysis suggested that gossypol could physically bind to the RBX1-CUL1 and SAG-CUL5 complex to disrupt cullin neddylation.
Fig. 4.
Molecular modeling of gossypol binding with RBX1 and SAG/RBX2. Crystal structures of RBX1 (PDB: 4A0C) and SAG/RBX2 (PDB: 2ECL) were utilized for docking simulations. Charged amino acids were colored orange (negative) and purple (positive). Hydrophobic environment was colored green, and polar environment was color blue. Hydrogen bonds were marked as purple arrows. (A) Predicted binding structures of gossypol and RBX1. (B) Detailed interaction patterns between gossypol and RBX1. (C) Predicted binding structures of gossypol and SAG/RBX2. (D) Detailed interaction patterns between gossypol and SAG/RBX2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
CUL5-H572 plays a key role for CUL5 ∼ NEDD8 conjugation and gossypol binding
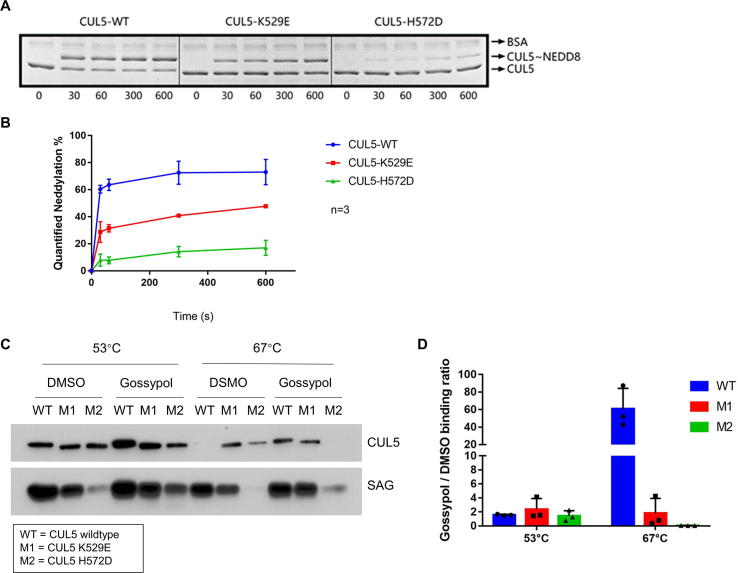
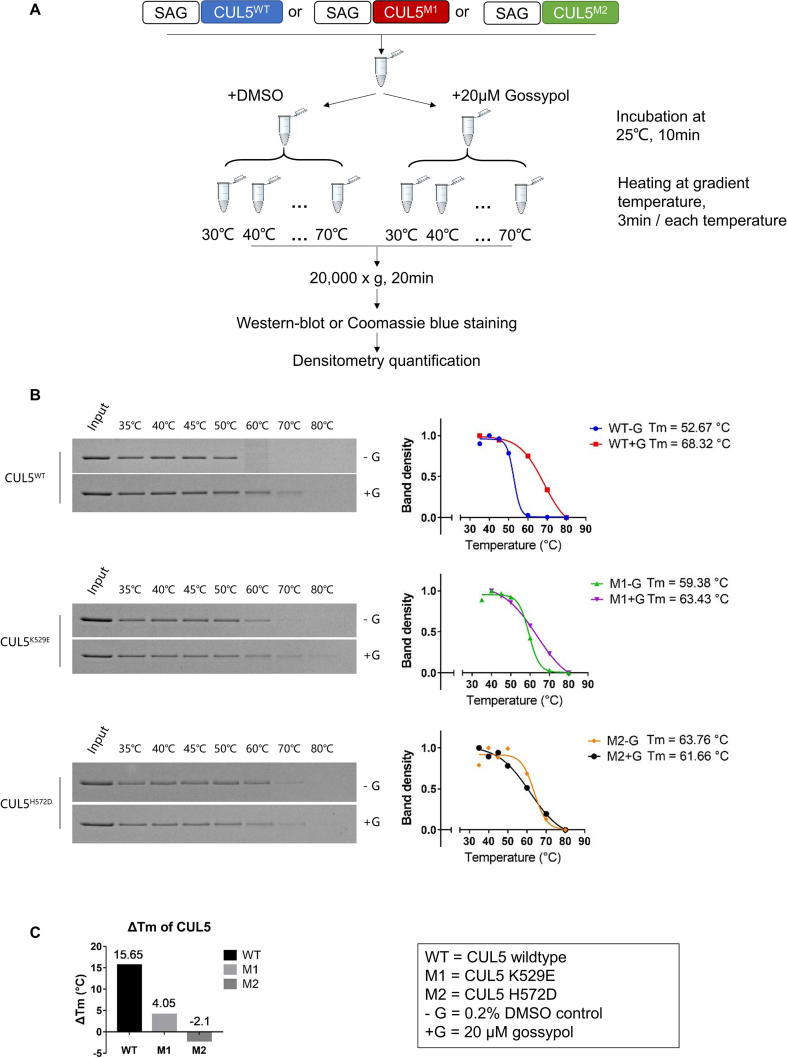
We next determined whether the residues K529 and H572 on CUL5, and G589 on CUL1 indeed play the role in gossypol binding, as predicted by our computer-modeling based in silico analysis. To this end, we first performed computational modeling to determine potential residue changes on K529 and H572 with a high likelihood to alter the CUL5-gossypol binding, and made K529E and H572D two mutants. We then purified these mutant CUL5-SAG E3 complexes and tested them in the pre-charged CUL5 neddylation enzyme reactions.
Indeed, we found that compared to wild-type CUL5, K529E mutant had decreased, whereas H572D mutant had nearly lost the ability in forming CUL5 ∼ NEDD8 conjugates (Fig. 5A&B). The results strongly suggest that these two sites, especially H572 on CUL5, play the critical role to facilitate the formation of CUL5 ∼ NEDD8 conjugation.
Fig. 5.
CUL5 mutations inhibit CUL5 ∼ NEDD8 conjugation, and interrupt the gossypol binding. (A) K529E and H572D mutants of CUL5CTD were cloned, co-expressed and purified with wild-type SAG. The recombinant CUL5-SAG E3 complexes were then tested in the in vitro pre-charged CUL5-neddylation assay. (B) The band density on the Coomassie blue staining SDS–PAGE gels in (A) were quantified by Image J software (NIH). The results were graphed by GraphPad Prism software (La Jolla, CA, USA). (C) The CUL5-SAG E3 complexes containing the wild-type (WT), K529E (M1) or H572D (M2) CUL5 mutants were treated with DMSO or 20 µM gossypol and were tested through the western-blot based in vitro thermal shift assay. (D) The band density in (C) were quantified by Image J software (NIH). The gossypol-treated CUL5 protein level was divided by the DMSO-treated CUL5 protein level in each group to get Gossypol/DMSO binding ratio. The results were graphed by GraphPad Prism software (La Jolla, CA, USA). See also Fig. S5.
We then tested the binding of the purified wild-type/mutant CUL5-SAG E3 complex with gossypol, using our established in vitro thermal shift assay (Fig. S5A). At the low temperature (53 °C), three forms of cullin had similar binding capacity with DMSO, but the highest binder with gossypol was wild type CUL5. At the high temperature (67 °C), wild-type lost DMSO binding, but maintained the highest binding capacity with gossypol. Reduced or lost gossypol binding was observed in K529E (M1) or H572D (M2) mutant, respectively (Fig. 5C). We then calculated the gossypol/DMSO binding ratio and found that a ∼30-fold higher binding ratio in wild-type CUL5 than M1 mutant, and a complete loss for M2 mutant (Fig. 5D). Interestingly, the M2 mutant had much reduced binding with SAG, consistent with its much reduced formation of CUL5 ∼ NEDD8 conjugation (Fig. 5C).
Furthermore, we generated the gossypol v.s DMSO-binding melting curves for each CUL5s by heating the CUL5 proteins at gradient temperatures with or without gossypol. Only the wild-type CUL5 (WT) exhibited significant shift of its melting temperature (Tm) in the presence of gossypol with ΔTm of 15.65 °C, in comparison of 4.05 °C for M1 or −2.1 °C in M2 mutants, respectively (Fig. S5B&C). Taken together, our study clearly demonstrated that the CUL5 residue K529 or H572 plays a minor or major role in mediating the CUL5-gossypol binding, respectively.
We also performed the same computational modeling analysis and made CUL1-E589A mutant as well as a nearby CUL1 (G588F) mutant with prediction to block gossypol binding. Unfortunately, after multiple attempts, both mutant constructs failed to express mutant CUL1 proteins. We reason that induction of either of these mutations likely changes the conformation of CUL1 protein, leading to a rapid degradation as the misfolded proteins. Nevertheless, although we were unable to measure the potential alterations in gossypol binding, the results indeed suggested that either of these two residues is important for the maintenance of proper CUL1 conformation.
Gossypol inhibits cullin neddylation and selectively causes accumulation of NOXA and MCL1 in multiple cancer cell lines
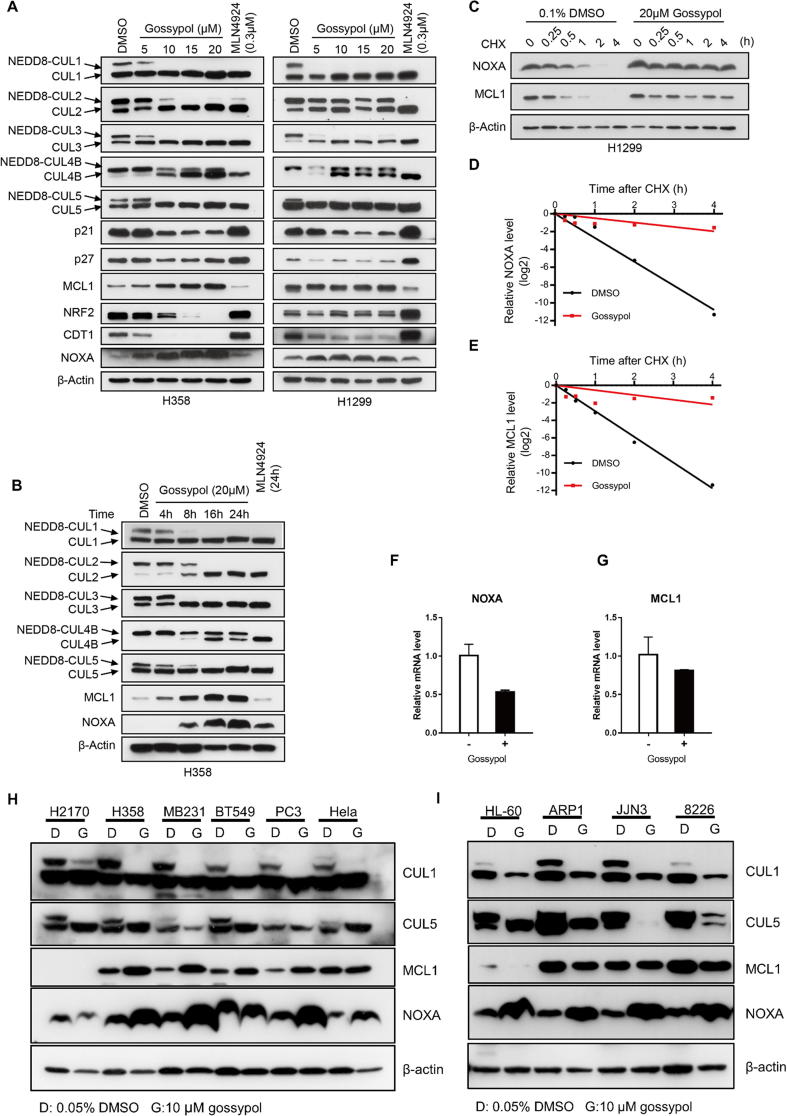
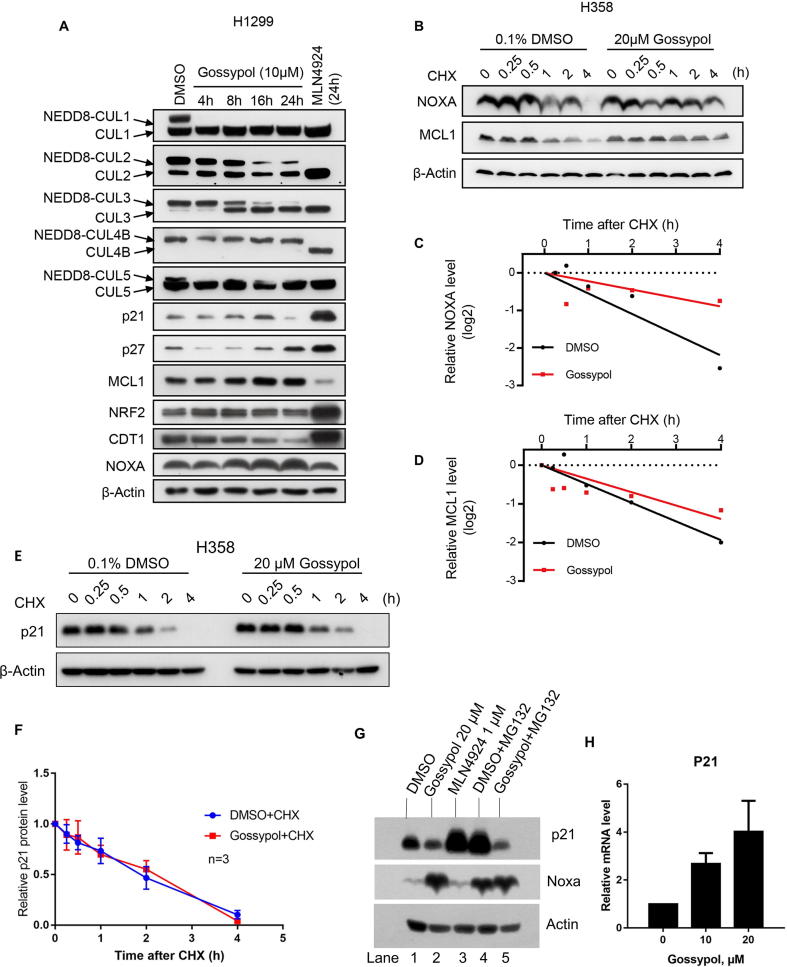
To investigate the cellular effect of gossypol on cullin neddylation, we treated two lung cancer cell lines, H358 and H1299 with gossypol and measured the neddylation levels of cullins 1–5 and the protein levels of typical cullin substrates. As shown in Fig. 6A, gossypol effectively inhibited the neddylation of CUL1, 2, 3, 4B, and 5 in H358 and H1299 cells in a dose-dependent manner. Inhibition kinetic assay showed that gossypol’s inhibitory effect against most cullin neddylation appeared within 8 h (Figs. 6B and S6A). Of note, gossypol selectively blocked the neddylation of CUL1 and CUL5 within 4 h in H1299 cells (Fig. S6A).
Fig. 6.
Gossypol is a potent inhibitor of cullin neddylation in cells. (A-B) Gossypol inhibits cullin neddylation in lung cancer cells: H358 and H1299 lung cancer cells were treated with gossypol at indicated concentrations for 24 h (A) or H358 cells were treated with 20 μM gossypol for indicated time periods (B), along with MLN4924 as a positive control, followed by Western-blotting with indicated antibodies. (C) Gossypol extends the protein half-live of NOXA and MCL1. H1299 cells were treated with 20 μM gossypol, along with DMSO control for 8 h, followed by incubation with CHX for 0, 0.25, 0.5, 1, 2 and 4 h before harvesting. Cells were then lysed and the protein levels of NOXA and MCL1 were analyzed by western-blotting with indicated antibodies. (D-E) The band density in (C) was quantified using ImagJ software and normalized to β-Actin. The decay curves were plotted for NOXA (D), and MCL1 (D). (F-G) Gossypol has minimal effect on mRNA levels of NOXA and MCL1. H1299 cells were treated with 20 μM gossypol for 24 h, along with DMSO control, followed by total RNA isolation and qRT-PCR analysis. (H-I) Gossypol inhibits cullin neddylation in multiple human cancer cell lines. Indicated cancer cell lines were treated with 10 μM gossypol for 24 h, followed by Western blotting with indicated antibodies. G stands for gossypol and D stands for DMSO control. See also Fig. S6.
We then measured the protein levels of typical substrates of the cullin E3 ligases, including CUL1 substrates p21, p27, and MCL1, CUL3 substrate NRF2, CUL4 substrate CDT1 and CUL5 substrate NOXA (Figs. 6A, S6A). Surprisingly, while MLN4924 caused most of the substrates accumulated, gossypol only stabilized MCL1 and NOXA among the substrates tested (Figs. 6A&B, S6A), which indicates a strong association between gossypol and the two specific cullins, CUL1 and CUL5.
To investigate whether gossypol indeed disrupted the degradation of NOXA and MCL1, we measured the half-life of these two proteins. As shown, gossypol significantly increased the half-life of both NOXA and MCL1 in both H1299 and H358 cells (Figs. 6C-E and S6B-D). Moreover, the mRNA levels of NOXA and MCL1 was not increased, but moderately decreased by gossypol (Fig. 6F&G), indicating the accumulation of the two proteins was a consequence of post-translational modification.
We then asked whether gossypol’s effect on cullin neddylation and the substrates is cell-type specific. We treated gossypol in multiple cancer cell lines including lung cancer, breast cancer, prostate cancer, cervical cancer, and several hematologic tumors, and measured the levels of cullin neddylation and substrate accumulation. As shown, gossypol inhibited cullin neddylation and accumulated NOXA in a variety of cell lines. Interestingly, gossypol caused accumulation of MCL1 in most solid tumor cells, but not hematologic tumor cells (Fig. 6H&I). Collectively, our results shown that gossypol inhibits neddylation of all cullins, but selectively causes accumulation of CRL substrates.
Of note, we noticed that unlike MLN4924, which caused p21 accumulation, gossypol caused reduction of the protein level of the canonical CUL1 substrate, p21, in both H358 and H1299 cells (Figs. 6A&S6A). We found that gossypol did not change p21 protein half-life (Fig. S6E&F), suggesting that gossypol is not activating ubqiutinating complex. Furthermore, proteasome inhibitor MG132 could not reverse p21 decrease in gossypol-treated H358 cells (Fig. S6G), which also indicated that the gossypol-induced reduction of p21 protein level was not through enhancing ubiquitin-dependent protein degradation. Interestingly, gossypol increased p21 mRNA (Fig. S6H), suggesting that gossypol affects the rate of p21 mRNA translation. Elucidation of the mechanism by which gossypol caused p21 reduction is out of scope of this manuscript, but certainly an interesting topic for future investigation.
Combination of gossypol and MCL1 inhibitor synergistically suppresses growth of cancer cells
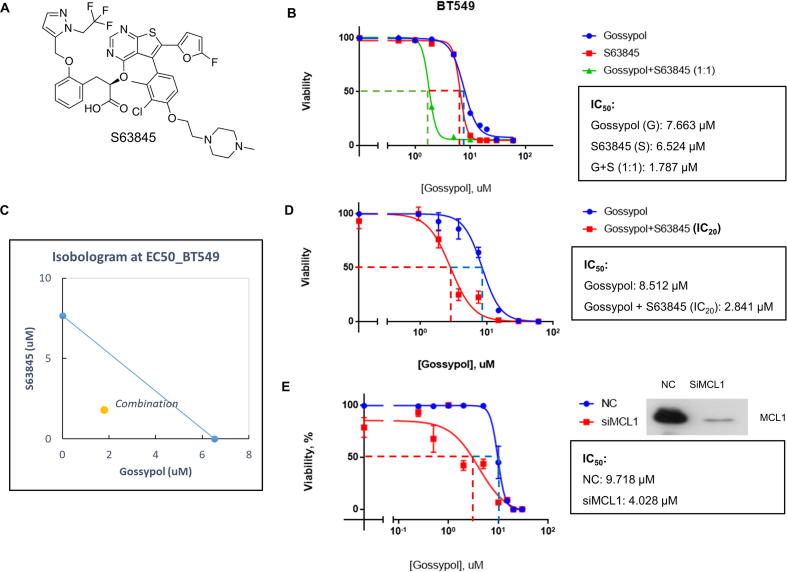
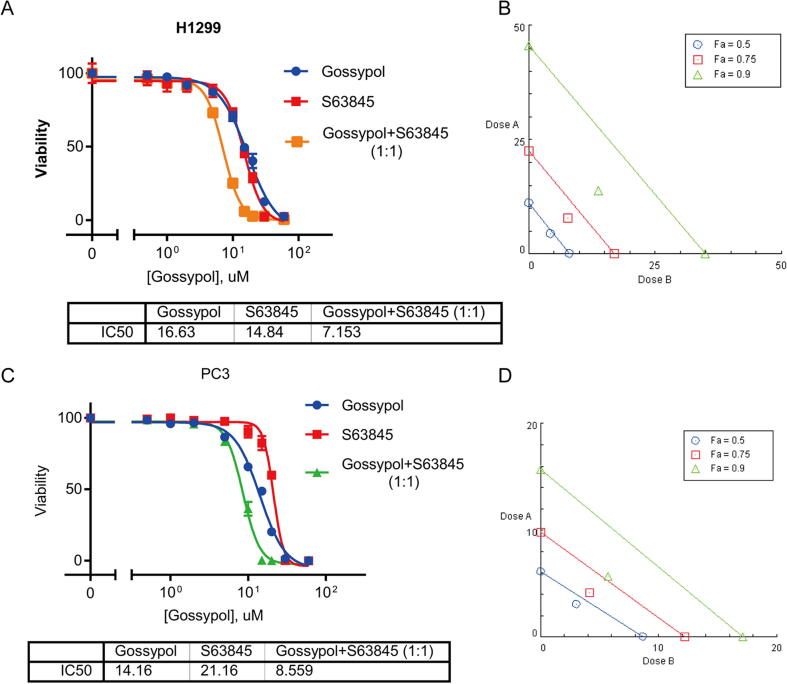
NOXA and MCL1 are both Bcl-2 family members [47]. While NOXA is a pro-apoptosis BH3 only protein, MCL1 acts as a pro-survival protein. These two proteins are tightly regulated to modulate cell death of survival under different conditions [47]. Since MCL1 is an anti-apoptotic protein, we hypothesized that the elevation of MCL1 may contribute to the less efficacy or drug resistance of gossypol in solid tumor cells. We therefore tested this hypothesis by determining if blockage of MCL1 by MCL1-specific inhibitor would sensitize cancer cells to gossypol. S63845 (Fig. 7A) is a potent small molecule inhibitor selectively binds to MCL1 with nano-molar affinity [48]. We combined gossypol with S63845 in a molar ratio of 1:1 to treat breast cancer BT549 cells, followed by cell proliferation assays using CCK-8. As shown, compared to sole treatment, the combinational treatment significantly lowered the IC50 (Fig. 7B). Isobologram analysis showed that the combination effect of gossypol and S63845 was synergistic (Fig. 7C). We then titrated gossypol against BT549 cells in combination with S63845 at IC20 level, and also observed a significant enhancement of growth suppression (Fig. 7D). Furthermore, MCL1 knockdown by siRNA in BT549 also sensitized cells to gossypol (Fig. 7E). Similar synergistic effect, although to a lesser extent, was also observed in H1299 and PC3 cells (Fig. S7A–D). Taken together, these results showed that gossypol in combination with MCL1 inhibition (via both pharmacological and genetic approaches) synergistically suppressed growth of certain solid tumor cells, providing a sound rationale for this previously unrealized combinational therapy.
Fig. 7.
MCL1 inhibition sensitizes cancer cells to gossypol. (A) Chemical structure of MCL1 specific inhibitor S63845. (B) Combination of gossypol and S63845 synergistically inhibits growth of breast cancer cells: BT549 breast cancer cells were plated in 96-well plate and treated with gossypol or S63845 alone or in combination (1:1 ratio) at a variety of concentrations for 72 h, and cell viability was examined by CCK-8 kit. The relative viability was expressed as % growth inhibition, as compared to DMSO control, setting at 100%. The IC50 values were calculated by GraphPad software. (C) The data collected from (B) was analyzed by CompuSyn software to generate isobologram. The calculated inhibition rate at each concentration was used as Fa value (effective level). Note that the data points on the lower-left of the hypotenuse indicate synergism. The data points on the upper-right of the hypotenuse indicate antagonism. And data points on the hypotenuse indicate addictive effect. (D) S63845 at IC20 value sensitizes breast cancer cells to gossypol: BT549 breast cancer cells were plated in 96-well plate and treated with gossypol alone at a variety of concentrations or in combination with S63845 at IC20 value for 72 h, and cell viability was examined by CCK-8 kit. The relative viability was expressed as % growth inhibition, as compared to DMSO control, setting at 100%. The IC50 values were calculated by GraphPad software. (E) MCL1 knockdown sensitizes breast cancer to gossypol: BT549 cells were transfected with siRNA oligonucleotide targeting MCL1 (siMCL1), along with scramble control (NC). A portion of cells were then plated in 96-well plate and treated with various concentrations of gossypol for IC50 determination, and another portion was subjected to Western blotting for MCL1 levels. See also Fig. S7.
Discussion
The discovery of NAE inhibitor MLN4924 in 2009 [21] opens new eras in targeting the neddylation pathway for cancer therapy. Impressive anti-cancer activity in a numerous preclinical studies have advanced MLN4924 into multiple Phase I/II clinical trials [22], [23], [24], [25]. Given its universal inhibitory activity as an E1 inhibitor to block the entire neddylation modification, MLN4924 has shown unavoidable cytotoxicity [22], [23], [25] as well as some unexpected “off-target” effects [49], [50], [51]. Therefore, it is highly desirable in an effort to discover small molecule inhibitors targeting downstream E2s or E3s in the neddylation cascade for higher selectivity and lesser toxicity.
To this end, we developed the AlphaScreen-based in vitro neddylation assay and used it in an HTS format to identify small molecule inhibitor of SAG/RBX2 NEDD8 E3 ligase to block CUL5 neddylation. The AlphaScreen system has some prominent properties, including high sensitivity, accessible for assay modification, and readily to support high-throughput format [52]. As a result, gossypol was identified as one of the top hits and further validated as a potent inhibitor of cullin neddylation in vitro biochemical assays and in multiple cancer cell lines.
Gossypol as a natural compound has been studied for therapeutic purpose for a long time [31]. Previous studies have uncovered multiple biological activities of gossypol such as anti-trypanosome [53], anti-virus [54], and anti-tumors [55], [56]. Behind these biological activities are manifold mechanisms and multiple targets involving distinct cellular pathways [57]. In terms of the anti-cancer efficacy of gossypol, the most-accepted mechanism is that gossypol acts as a BH3 mimetic to bind anti-apoptotic proteins (e.g. Bcl-2 or MCL1) [57], which leads to the clinical trials of the gossypol derivative AT-101. However, AT-101 failed to meet the pre-specified endpoints or show benefit to the patients in many clinical trials due to low efficacy [58], [59], [32], [33], [34]. Associated side effects also limit its application in clinics [60], [61], [62]. The fact that exact anti-cancer mechanisms of gossypol remains elusive limits our understanding of side-effects or drug resistance associated with gossypol.
In this study, we demonstrated for the first time, to the best of our knowledge, that gossypol is a potent inhibitor of cullin neddylation. Our conclusion is supported by the following experimental results: (a) gossypol effectively inhibited cullin neddylation in in vitro enzyme assays; (b) gossypol showed the binding to the RING E3s in TSA and CETSA assays, and docking studies also supported gossypol binding to RBX1-CUL1/5 complex; and (c) gossypol potently inhibited cellular cullin neddylation in multiple cancer cell lines. It is worth noting, however, that while the in vitro thermal shift assay and the docking studies both supported the binding between gossypol and the two CRLs (i.e., RBX1-CUL1 and SAG-CUL5), the cellular thermal shift assays (CETSA) showed that gossypol only stabilizes RING proteins (RBX1 and SAG/RBX2) but not the cullins (CUL1 or CUL5). This contradictory observation maybe explained by the nature of CETSA, as described by Molina et al. [38] and Jafari et al. [63], that false negatives may occur as a result of (a) weak response by large proteins or protein complexes, (b) a nonfunctional form of target protein missing antibody recognition, or (c) unexpected global cellular effects in the cells in response to thermal changes. To address this potential issue, we attempted to perform alternative experiments including biolayer interferometry (BLI), isothermal titration calorimetry (ITC) and co-crystallization (gossypol co-crystallize with NEDD8 E1, E2s or E3s), but failed to obtain any convincing results likely due to the poor water solubility of gossypol. Future studies should be directly to using water-soluble derivatives of gossypol, if developed, to further investigate the true binding mode [64].
Unlike MLN4924, which targets NAE E1, gossypol has a totally different mode of action. The in vitro enzyme assays showed that gossypol could inhibit cullin neddylation without blocking the E1-E2 transthiolation, strongly suggesting that its targeting is likely the NEDD8 E3. Indeed, gossypol binds and targets both NEDD8 E3s (RBX1 and SAG/RBX2), thus acting as a novel class of compound to block universal cullin neddylation, leading to inactivation of CRL1-5. Interestingly and unexpectedly, at the cellular levels, again unlike MLN4924, gossypol failed cause accumulation of CRL substrates, such as p21, p27, CDT1 or NRF2. Rather, it selectively causes the accumulation of MCL1 and NOXA.
The selective accumulation by gossypol of MCL1 and NOXA, two BCL-2 family members, is unexpected, yet interesting. NOXA is known as a single domain BH3-only protein with pro-apoptotic function, whereas MCL1 functions as a pro-survival Bcl-2-like protein [47]. Our recent study showed that SAG/CUL5 E3 ligase promotes NOXA ubiquitylation via the K11 ubiquitin linkage [43]. Others have shown MCL2 is subjected to ubquitylation by four E3 ligases (SCFβ-TrCP, SCFFBXW7, MULE, and Trim17) [65], [66], [67], [68]. Thus, both NOXA and MCL1 are under post translational regulation by CRL1. Indeed, our findings reported here showed that gossypol selectively accumulated NOXA and MCL1 levels by at least in part blocking their degradation. The underlying mechanism for this selective accumulation of NOXA and MCL1, but not other tested CRL substrates, is unknown at the present time, but is an interesting subject for future investigation.
It was previously reported that NOXA binds MCL1 to promote MCL1 degradation [69]. However, we observed that gossypol increases the levels of both NOXA and MCL1. To demonstrate therapeutic application/significance of our finding, we performed combinational drug treatment of gossypol and MCL1 inhibitor, S63845 in multiple human cancer cell lines and observed a synergistic effect in suppression of cancer cell growth. We also showed an enhanced growth suppression of cancer cells when gossypol was combined with MCL1 knockdown. Our study, therefore, provides a sound rationale to combine two inhibitors of BCL2 family members for enhanced anti-cancer efficacy.
In conclusion, we identified gossypol, a natural product, as a new and potent inhibitor of cullin neddylation by targeting neddylation E3 ligases. Gossypol blocked cullin neddylation in multiple cancer cell lines and selectively accumulated CRL1 substrate MCL1 and CRL5 substrate NOXA. Finally, combinational treatment of gossypol with MCL1 inhibitor S63845 synergistically enhanced the anti-tumor efficacy.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgment
We gratefully acknowledged the financial support by the National Key R&D Program of China (2016YFA0501800 to YS).
Author Contributions
Y.S, Z.-Q.P., and J.H conceived and directed the project. Q.Y and Z.H prepared the proteins, established the AlphaScreen assays, conducted the high-throughput screen and performed the post-screen biochemical assays. Q.Y, Y.-W.S and Y.J conducted the cell-based assays. P.P and T.H performed the docking assays. Q.Y and Y.S wrote the paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.02.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S1.
Supplementary Fig. S2.
Supplementary Fig. S3.
Supplementary Fig. S4.
Supplementary Fig. S5.
Supplementary Fig. S6.
Supplementary Fig. S7.
References
- 1.Hershko A., Ciechanover A., Aaron C. The ubiquitin system. Ann Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Pickart C.M. Mechanisms Underlying Ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Deshaies R.J., Joazeiro C.A.P. RING domain E3 ubiquitin ligases. Ann Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 4.Petroski M.D., Deshaies R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 5.Bedford L., Lowe J., Dick L.R., Mayer R.J., Brownell J.E. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discovery. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitby F.G., Xia G., Pickart C.M., Hill C.P. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J Biol Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 7.Gong L., Yeh E.T. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 8.Liakopoulos D., Doenges G., Matuschewski K., Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang D.T., Ayrault O., Hunt H.W. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott D.C., Monda J.K., Bennett E.J., Harper J.W., Schulman B.A. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng N., Wang P., Jeffrey P.D., Pavletich N.P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 12.Zheng N., Schulman B.A., Song L. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L., Zhang W., Sun Y., Jia L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal. 2018;44:92–102. doi: 10.1016/j.cellsig.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., Wang M., Yu G. Overactivated neddylation pathway as a therapeutic target in lung cancer. J Natl Cancer Inst. 2014;106:15–21. doi: 10.1093/jnci/dju083. [DOI] [PubMed] [Google Scholar]
- 15.Xie P., Yang J.-P., Cao Y. Promoting tumorigenesis in nasopharyngeal carcinoma, NEDD8 serves as a potential theranostic target. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie P., Zhang M., He S. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat Commun. 2014;5:1–16. doi: 10.1038/ncomms4733. [DOI] [PubMed] [Google Scholar]
- 17.Hua W., Li C., Yang Z. Suppression of glioblastoma by targeting the overactivated protein neddylation pathway. Neuro-Oncology. 2015;17:1333–1343. doi: 10.1093/neuonc/nov066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia L., Soengas M.S., Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69:4974–4982. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen P., Hu T., Liang Y. Neddylation inhibition activates the extrinsic apoptosis pathway through ATF4-CHOP-DR5 axis in human esophageal cancer cells. Clin Cancer Res. 2016;22:4145–4157. doi: 10.1158/1078-0432.CCR-15-2254. [DOI] [PubMed] [Google Scholar]
- 20.Sarkaria I., O-Charoenrat P., Talbot S.G. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res. 2006;66:9437–9444. doi: 10.1158/0008-5472.CAN-06-2074. [DOI] [PubMed] [Google Scholar]
- 21.Soucy T.A., Smith P.G., Milhollen M.A. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 22.Swords R.T., Erba H.P., DeAngelo D.J. Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: a phase 1 study. Br J Haematol. 2015;169:534–543. doi: 10.1111/bjh.13323. [DOI] [PubMed] [Google Scholar]
- 23.Bhatia S., Pavlick A.C., Boasberg P. A phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with metastatic melanoma. Invest New Drugs. 2016;34:439–449. doi: 10.1007/s10637-016-0348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarantopoulos J., Shapiro G.I., Cohen R.B. Phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin Cancer Res. 2016;22:847–857. doi: 10.1158/1078-0432.CCR-15-1338. [DOI] [PubMed] [Google Scholar]
- 25.Shah J.J., Jakubowiak A.J., O'Connor O.A. Phase I study of the novel investigational NEDD8-activating enzyme inhibitor pevonedistat (MLN4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin Cancer Res. 2016;22:34–43. doi: 10.1158/1078-0432.CCR-15-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott D.C., Hammill J.T., Min J. Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase. Nat Chem Biol. 2017;13:850–857. doi: 10.1038/nchembio.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H., Lu J., Liu L. A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation. Nat Commun. 2017;8:1150. doi: 10.1038/s41467-017-01243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou H., Zhou W., Zhou B. High-Affinity Peptidomimetic Inhibitors of the DCN1-UBC12 Protein-Protein Interaction. J Med Chem. 2018;61:1934–1950. doi: 10.1021/acs.jmedchem.7b01455. [DOI] [PubMed] [Google Scholar]
- 29.Wang S., Zhao L., Shi X.-J. Development of highly potent, selective, and cellular active Triazolo[1,5-a]pyrimidine-based inhibitors targeting the DCN1-UBC12 protein-protein interaction. J Med Chem. 2019;62:2772–2797. doi: 10.1021/acs.jmedchem.9b00113. [DOI] [PubMed] [Google Scholar]
- 30.Zhou W., Ma L., Ding L. Potent 5-cyano-6-phenyl-pyrimidin-based derivatives targeting DCN1-UBE2M interaction. J Med Chem. 2019;62:5382–5403. doi: 10.1021/acs.jmedchem.9b00003. [DOI] [PubMed] [Google Scholar]
- 31.Zeng Y., Ma J., Xu L., Wu D. Natural product gossypol and its derivatives in precision cancer medicine. Curr Med Chem. 2019;26:1849–1873. doi: 10.2174/0929867324666170523123655. [DOI] [PubMed] [Google Scholar]
- 32.Xie H., Yin J., Shah M.H. A phase II study of the orally administered negative enantiomer of gossypol (AT-101), a BH3 mimetic, in patients with advanced adrenal cortical carcinoma. Invest New Drugs. 2019 doi: 10.1007/s10637-019-00797-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein M.N., Hussain M., Stadler W.M. A phase II study of AT-101 to overcome Bcl-2-mediated resistance to androgen deprivation therapy in patients with newly diagnosed castration-sensitive metastatic prostate cancer. Clin Genitourinary Cancer. 2016;14:22–27. doi: 10.1016/j.clgc.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baggstrom M.Q., Qi Y., Koczywas M. A phase II study of AT-101 (Gossypol) in chemotherapy-sensitive recurrent extensive-stage small cell lung cancer. J Thorac Oncol. 2011;6:1757–1760. doi: 10.1097/JTO.0b013e31822e2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suk Heist R., Fain J., Chinnasami B. Phase I/II study of AT-101 with topotecan in relapsed and refractory small cell lung cancer. J Thorac Oncol. 2010;5:1637–1643. doi: 10.1097/JTO.0b013e3181e8f4dc. [DOI] [PubMed] [Google Scholar]
- 36.Liu G., Kelly W.K., Wilding G., Leopold L., Brill K., Somer B. An open-label, multicenter, phase I/II study of single-agent AT-101 in men with castrate-resistant prostate cancer. Clin Cancer Res. 2009;15:3172–3176. doi: 10.1158/1078-0432.CCR-08-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu K., Chong R.A., Yu Q. Suramin inhibits cullin-RING E3 ubiquitin ligases. Proc Natl Acad Sci. 2016;113:E2011–E2018. doi: 10.1073/pnas.1601089113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molina D.M., Jafari R., Ignatushchenko M. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 39.Fischer E.S., Scrima A., Bohm K. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147(5):1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 40.Duda D.M., Borg L.A., Scott D.C., Hunt H.W., Hammel M., Schulman B.A. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LLC S. Schrödinger M. New York, NY: Schrödinger LLC; 2009
- 42.Jia L., Yang J., Hao X. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou W., Xu J., Li H. Neddylation E2 UBE2F promotes the survival of lung cancer cells by activating CRL5 to degrade NOXA via the K11 linkage. Clin Cancer Res. 2017;23:1104–1116. doi: 10.1158/1078-0432.CCR-16-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chou T.C., Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Chung, Oldenburg. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 46.Scott D.C., Sviderskiy V.O., Monda J.K. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157:1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guikema J.E., Amiot M., Eldering E. Exploiting the pro-apoptotic function of NOXA as a therapeutic modality in cancer. Expert Opin Therap Targets. 2017;21:767–779. doi: 10.1080/14728222.2017.1349754. [DOI] [PubMed] [Google Scholar]
- 48.Kotschy A., Szlavik Z., Murray J. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–482. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Q., Li H., Li Y. Inhibiting neddylation modification alters mitochondrial morphology and reprograms energy metabolism in cancer cells. JCI Insight. 2019;4 doi: 10.1172/jci.insight.121582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mao H., Tang Z., Li H. Neddylation inhibitor MLN4924 suppresses cilia formation by modulating AKT1. Protein Cell. 2019 doi: 10.1007/s13238-019-0614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X., Tan M., Nyati M.K., Zhao Y., Wang G., Sun Y. Blockage of neddylation modification stimulates tumor sphere formation in vitro and stem cell differentiation and wound healing in vivo. PNAS. 2016;113:E2935–E2944. doi: 10.1073/pnas.1522367113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Austin S., Taouji S., Chevet E., Wodrich H., Rayne F. Using AlphaScreen(®) to identify small-molecule inhibitors targeting a conserved host-pathogen interaction. Methods Mol Biol (Clifton, N.J.) 2016;1449:453–467. doi: 10.1007/978-1-4939-3756-1_30. [DOI] [PubMed] [Google Scholar]
- 53.Yin J., Jin L., Chen F., Wang X., Kitaygorodskiy A., Jiang Y. Novel O-glycosidic gossypol isomers and their bioactivities. Carbohydr Res. 2011;346:2070–2074. doi: 10.1016/j.carres.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 54.Li L., Li Z., Wang K., Liu Y., Li Y., Wang Q. Synthesis and antiviral, insecticidal, and fungicidal activities of gossypol derivatives containing alkylimine, oxime or hydrazine moiety. Bioorg Med Chem. 2016;24:474–483. doi: 10.1016/j.bmc.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 55.Xiong J., Li J., Yang Q., Wang J., Su T., Zhou S. Gossypol has anti-cancer effects by dual-targeting MDM2 and VEGF in human breast cancer. Breast Cancer Res. 2017;19:1–10. doi: 10.1186/s13058-017-0818-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lan L., Appelman C., Smith A.R. Natural product (−)-gossypol inhibits colon cancer cell growth by targeting RNA-binding protein Musashi-1. Mol Oncol. 2015;9:1406–1420. doi: 10.1016/j.molonc.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu Y., Li J., Dong C.-E., Huang J., Zhou H.-B., Wang W. Recent advances in gossypol derivatives and analogs: a chemistry and biology view. Fut Med Chem. 2017;9:1243–1275. doi: 10.4155/fmc-2017-0046. [DOI] [PubMed] [Google Scholar]
- 58.Sonpavde G., Matveev V., Burke J.M. Randomized phase II trial of docetaxel plus prednisone in combination with placebo or AT-101, an oral small molecule Bcl-2 family antagonist, as first-line therapy for metastatic castration-resistant prostate cancer. Ann Oncol. 2012;23:1803–1808. doi: 10.1093/annonc/mdr555. [DOI] [PubMed] [Google Scholar]
- 59.Van Poznak C., Seidman A.D., Reidenberg M.M. Oral gossypol in the treatment of patients with refractory metastatic breast cancer: a phase I/II clinical trial. Breast Cancer Res Treat. 2001;66:239–248. doi: 10.1023/a:1010686204736. [DOI] [PubMed] [Google Scholar]
- 60.Zbidah M., Lupescu A., Shaik N., Lang F. Gossypol-induced suicidal erythrocyte death. Toxicology. 2012;302:101–105. doi: 10.1016/j.tox.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 61.Aneja R., Dass S.K., Chandra R. Modulatory influence of tin-protoporphyrin on gossypol-induced alterations of heme oxygenase activity in male wistar rats. Eur J Drug Metab Pharmacokinet. 2003;28:237–243. doi: 10.1007/BF03190491. [DOI] [PubMed] [Google Scholar]
- 62.Akingbemi B.T., Ogwuegbu S.O., Onwuka S.K., Oke B.O., Aire T.A. The effects of protein malnutrition and experimental infection with Trypanosoma brucei on gossypol treatment in the rat: haematological and serum biochemical changes. J Comp Pathol. 1995;112:361–371. doi: 10.1016/s0021-9975(05)80017-9. [DOI] [PubMed] [Google Scholar]
- 63.Jafari R., Almqvist H., Axelsson H. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 64.Yan F., Cao X.X., Jiang H.X. A novel water-soluble gossypol derivative increases chemotherapeutic sensitivity and promotes growth inhibition in colon cancer. J Med Chem. 2010;53:5502–5510. doi: 10.1021/jm1001698. [DOI] [PubMed] [Google Scholar]
- 65.Magiera M.M., Mora S., Mojsa B., Robbins I., Lassot I., Desagher S. Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons. Cell Death Differentiation. 2013;20:281–292. doi: 10.1038/cdd.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Inuzuka H., Fukushima H., Shaik S., Liu P., Lau A.W., Wei W. Mcl-1 ubiquitination and destruction. Oncotarget. 2011;2:239–244. doi: 10.18632/oncotarget.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding Q., He X., Hsu J.-M. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong Q., Gao W., Du F., Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 69.Willis S.N., Chen L., Dewson G. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]