Abstract
Fermented pastes are some of the most popular traditional products in China. Many studies reported a strong possibility that fermented pastes promote exposure to mycotoxins, including aflatoxins, ochratoxins, and cereulide, which were proven to be carcinogenic and neurotoxic to humans. The primary mechanism of pathogenicity is by inhibiting protein synthesis and inducing oxidative stress using cytochrome P450 (CYP) enzymes. The level of mycotoxin production is dependent on the pre-harvest or post-harvest stage. It is possible to implement methods to control mycotoxins by using appropriate antagonistic microorganisms, such as Aspergillus niger, Lactobacillus plantarum, and Saccharomyces cerevisiae isolated from ordinary foods. Also, drying products as soon as possible to avoid condensation or moisture absorption in order to reduce the water activity to lower than 0.82 during storage is also effective. Furthermore, organic acid treatment during the soaking process reduces toxins by more than 90%. Some novel detection technologies based on magnetic adsorption, aptamer probes, and molecular-based methods were applied to rapidly and accurately detect mycotoxins in fermented pastes.
Keywords: mycotoxins, fermented paste, pathogenicity, control measures, detection
1. Introduction
Fermented pastes are very popular in China and include soybean paste, thick broad-bean paste, flour paste, and chili paste. The unique flavor, taste, and nutritional components of the paste products mainly depend on the long-term fermentation by aroma-producing microorganisms [1]. The antioxidant activities of fermented pastes may be improved through prolonged fermentation [2]. However, several undesired mycotoxins may exist and can lead to human disease due to the high risk of paste contamination during the long-term fermentation process [3]. Mycotoxins produced by fungi play the most significant role in food contamination [4]. For instance, high levels of mycotoxins were detected in wheat, coffee, grape products, and even in animal feed [5]. Some fermented products were reported to contain an abundance of Aspergillus, Penicillium, and Fusarium spp., which are correlated with the presence of aflatoxins or ochratoxin A [6].
It is well known that mycotoxin contamination in food has a huge risk to human health by suppressing the immune system, leading to conditions such as nephrotoxicity and teratogenesis. Most mycotoxins are regarded as potent carcinogens by the International Agency for Research on Cancer (IARC) [7]. Previous researchers investigated the pathogenesis of mycotoxins, which are associated with alterations in the epigenetic state involving some primary enzymes [8]. In general, most mycotoxins possess properties such as stability, low molecular weight, and heat resistance, thereby making food contamination much more difficult to prevent [9].
The main objective of this review is to summarize the potential mycotoxins, including aflatoxins, ochratoxins, and cereulide, that can exist in fermented pastes. We also explore the significant risk to human health and the pathogenic mechanisms, including carcinogenicity, teratogenicity, nephropathy, and neurotoxicity. Furthermore, the most novel technologies to control and detect different toxins in fermented products are also discussed.
2. Mycotoxins in Fermented Pastes
2.1. Aflatoxins
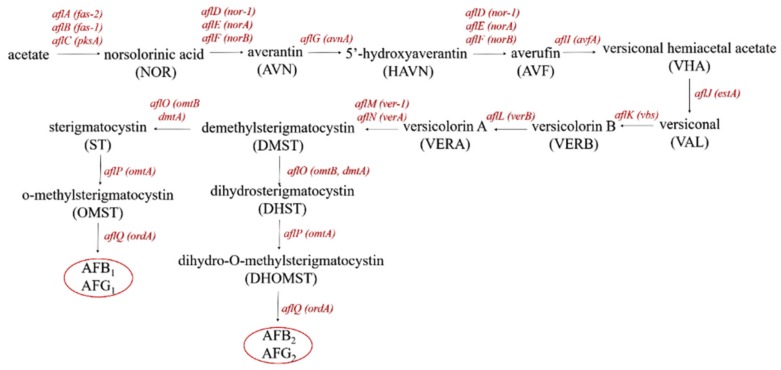
Aflatoxins are ubiquitously found in cereals, milk, tree nuts, and oilseeds and are considered to make up a group of extremely toxic metabolites produced by Aspergillus flavus, Aspergillus parasiticus, and Aspergillus nomius during almost all stages of paste production [10]. Previous research demonstrated that there more than 20 aflatoxins were present in pods and soybean seeds [11]. Aflatoxins slightly dissolve in water and are heat-stable [12]. The major aflatoxins include the B-types, i.e., aflatoxin B1 (AFB1) and aflatoxin B2 (AFB2), which are produced from the fusion of bifurano coumarins to cyclopentanone, and the G-types, i.e., aflatoxin G1 (AFG1) and aflatoxin G2 (AFG2), which are produced from the fusion of bifurano coumarins to lactone [13]. Efficient aflatoxin-producing species of the Aspergillus section Flavi were recently described (Table 1) [14]. Extensive investigations were performed, showing that aflatoxins are polyketide-derived secondary mebabolites produced via the conversion path of acetate polyketide to anthraquinones (norsolorinic acid, averantin, averufanin, averufin, and versicolorin A) to xanthones to aflatoxins (Figure 1) [15].
Table 1.
The list of the 18 identified aflatoxin-producing species.
| Species | Aflatoxins | Reference |
|---|---|---|
| Aspergillus aflatoxiformans | B1, B2, G1, G2 | [14] |
| Aspergillus arachidicola | B1, B2, G1, G2 | [16] |
| Aspergillus austwickii | B1, B2, G1, G2 | [14] |
| Aspergillus cerealis | B1, B2, G1, G2 | [14] |
| A. flavus | B1, B2 | [17] |
| Aspergillus luteovirescens | B1, B2, G1, G2 | [16] |
| Aspergillus minisclerotigenes | B1, B2, G1, G2 | [16] |
| Aspergillus mottae | B1, B2, G1, G2 | [18] |
| A. nomius | B1, B2, G1, G2 | [19] |
| Aspergillus novoparasiticus | B1, B2, G1, G2 | [20] |
| A. parasiticus | B1, B2, G1, G2 | [21] |
| Aspergillus pipericola | B1, B2, G1, G2 | [14] |
| Aspergillus pseudocaelatus | B1, B2, G1, G2 | [22] |
| Aspergillus pseudonomius | B1 | [22] |
| Aspergillus pseudotamarii | B1, B2 | [22] |
| Aspergillus sergii | B1, B2, G1, G2 | [18] |
| Aspergillus togoensis | B1 | [23] |
| Aspergillus transmontanensis | B1, B2, G1, G2 | [18] |
Figure 1.
The conversion pathway of aflatoxin production: aflA: fatty acid synthase α; aflB: fatty acid synthase β; alfC: polyketide synthase; aflD: reductase; aflE: NOR-reductase; aflF: dehydrogenase; aflG: P450 monooxygenase; aflH: alcohol dehydrogenase; aflI: oxidase; aflJ: esterase; aflK: VERB synthase; aflL: desaturase; aflM: dehydrogenase; aflN: monooxygenase; aflO: O-methyltransferase B; aflP: O-methyltransferase A; aflQ: oxidoreductase.
The production of aflatoxins during paste fermentation is affected by fermentation conditions, including the protein ratio of raw materials, starter cultures, and so on [24]. A. flavus and its close relatives are commensal with the crop and infect it during growth to produce aflatoxins before harvest. Some saprophytic species contaminate crops only after harvest [25]. Previous researchers showed that the growth conditions of these saprophytic species were above 37 °C and close to 0.80 water activity [26].
Crops contaminated by different Aspergillus species demonstrate various levels of risk regarding the presence of aflatoxins in final fermented pastes. For example, small grains like soybean and wheat do not have affinity with Aspergillus species, and aflatoxins can only be produced during untimely drying or storage at high- temperatures after harvest [27]. Crops such as peanuts and maize have higher risk levels than others, because A. flavus and A. parasiticus are commensal with these crops [28]. Contaminated soil further promotes mold on the plants if unharvested grains are not cleaned up in time [29]. Aflatoxins are not be degraded during the fermented process [30].
2.2. Ochratoxins
Ochratoxins are secondary metabolites produced by Penicillium or Aspergillus fungi under diverse conditions, with 18 related isomers, of which ochratoxin A (OTA) is the most toxic and considered to be teratogenic, neurotoxic, nephrotoxic, and carcinogenic in humans [31,32]. OTA contamination occurs in various foods, including soybeans, dried fruits, and coffee, and was also detected in fermented wines and grape juices [33]. Furthermore, the biosynthetic pathway of OTA in fungi was confirmed to be strongly related to polyketide synthase (otapksPN) and non-ribosomal peptide synthetase (npsPN), which are regarded as the primary enzymes [34]. Previous research demonstrated that there were three steps of OTA biosynthesis, namely, polyketide synthesis, acyl activation, and de-esterification by esterase [35].
The occurrence of OTA in fermented pastes is influenced by many factors, including the type of crop, the farming practice, the machining process, and environmental conditions. Previous studies showed that the prevalence of OTA in maize was very limited compared with other crops [36]. OTA was detected in 34.2% of wheat samples in six provinces of China [37]. Unlike aflatoxins, ochratoxin-producing pathogens are mostly regarded as post-harvest spoilage. For instance, Penicillium species were confirmed to be strongly correlated with stored cereals, such as soybeans, beans, and green coffee [38], therefore, the storage and process conditions were found to greatly influence the final products. Moisture is also a critical factor in the contamination of OTA during storage; the safe-storage water activity level is below 0.7 [39] and the risk of OTA occurrence is increased if the drying process is not rapid and moisture is absorbed on the surface of products in cold climates. Damage to the raw materials during harvest, processing, and storage is also a significant factor [40].
2.3. Cereulide
Bacillus cereus, a spore-forming, poisonous, Gram-positive bacteria, is especially relevant for immunocompromised patients, drug users, newborns, and post-surgical patients. Strains isolated from parenteral infections are able to synthesize necrotizing exotoxin-like phospholipase and hemolysin [41]. B. cereus can be isolated from human dentin plaques, but accounts for a small proportion of oral infections. Collegenase-producing Soc 67 and Ply 19 were isolated from plaque in a patient with gingivitis and a teenager with periodontitis [42].
Bacillus cereus can be isolated from a variety of fermented foods and environments, such as doenjang and soybean paste. B. cereus phages were isolated from 47 traditional Korean fermented foods, as well as 63.6% of doenjang samples, 100% of cheonggukjang samples, and 40% of gochujang samples [43]. Cereulide is considered to be an important foodborne toxin during paste fermentation due to its ability to cause diarrheal (heat-labile) and emetic (heat-stable) food poisoning after ingestion of more than 104 Colony-Forming Units (CFU) per gram of food [44,45]. Controlling cereulide in in industrial practices is difficult due to the wide range of temperature (5–55 °C) in which B. cereus can grow [46].
3. Pathogenicity Mechanism of Mycotoxins
3.1. Cancer Diseases
A large amount of research regarding the toxicological mechanisms of mycotoxins revealed that many foodborne mycotoxins inhibit protein synthesis and induce oxidative stress, thereby causing various cancers [47]. For instance, fermented pastes contaminated by AFB1 cause latent damage, leading to human hepatocellular carcinoma (HCC), a common liver cancer. Previous research confirmed that the metabolic activation of AFB1 had a strong correlation with cytochrome P450 (CYP) enzymes, including CYP1A2, and CYP3A4, which are mainly expressed in the human liver [48]. HCC is metabolized to an AFB1-epoxide, and then adducts into AFB1–DNA [49], induces the transversion of G to T within codon 249 of the tumor suppressor gene p53, which is associated with liver cancer [9].
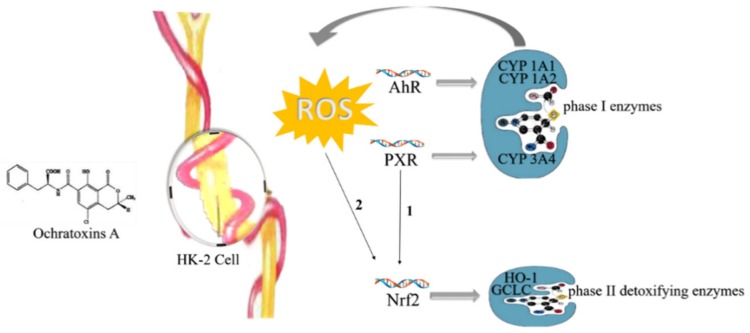
Furthermore, OTA was reported to be a group 2B human carcinogen by the International Agency for Research on Cancer (IARC) [32]. Due to the nephrotoxicity of OTA, ingesting products abundant in OTA increases the risk of suffering from Balkan Endemic Nephropathy (BEN) and Tunisian Nephropathy (TCIN) [50]. Recent research revealed that oxidative stress induced by OTA in HK-2 cells regulated the translocation of the transcription factors aryl hydrocarbon receptor (AhR) and pregnane X receptor (PXR) [51]. Activated AhR leads to immunosuppression and cancer by regulating CYP1A1 and CYP1A2 enzymes, and activated PXR has a relationship with CYP3A4, playing a role in phase I metabolism. Furthermore, activated Nrf2 inhibits cell death and decreases the expression of AhR and PXR in the presence of OTA, resulting in reduced renal damage [52]. Therefore, the regulation of transcription factors Nrf2, PXR, and AhR could help to treat and prevent renal injury by OTA (Figure 2) [53].
Figure 2.
Pathogenic mechanism of ochratoxin A in Human Kidney (HK)-2 cells: ROS: reactive oxygen species; AhR: aryl hydrocarbon receptor; PXR: pregnane X receptor; Nrf2: NF-E2-related factor 2; HO-1: heme oxygenase-1.
3.2. Neurodegenerative Diseases
Alongside carcinogenicity, OTA was demonstrated to impact migration, neuronal proliferation, and DNA content, thereby leading to neurodegenerative diseases and brain dysfunction of human such as parkinsonism and Alzheimer’s disease [54]. The neurotoxicity of the hippocampus, ventral mesencephalon, and striatum is more pronounced than in the cerebellum [55]. The mechanism of neurotoxicity involves OTA causing acute depletion of striatal dopamine and decreasing the immunoreactivity of striatum tyrosine hydroxylase, alongside a transient inhibition of DNA oxidative repair activity (oxyguanosine glycosylase, OGG1) [56]. Furthermore, OTA treatment inhibits the activation of SH-SY5Y, caspase-9, and caspase-3, which is accompanied by a decrease in mitochondrial membrane potential [57].
3.3. Gastrointestinal Diseases
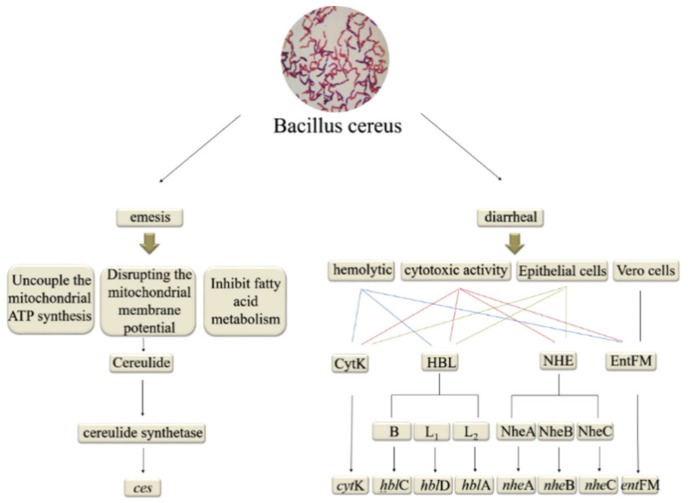
Diarrhea, a type of gastrointestinal syndrome induced by ingesting B. cereus-contaminated fermented pastes, is attributed to enterotoxins and virulence factors including cytotoxin K (CytK), hemolysin BL (HBL), nonhemolytic enterotoxin (NHE), and enterotoxin FM (EntFM) [58]. CytK, Nhe, and HBL cause osmotic lysis of intestinal epithelial cell membranes due to hemolytic and cytotoxic activity [59]. Both HBLand Nhe are pore-forming toxins that cause intestinal fluid secretion [60]. HBL is a three component enterotoxin encoded by the hblA, hblD, and hblC genes, respectively [61]. CytK, which is encoded by the cytK gene, is more prevalent than the Nhe and HBL complexes (Figure 3) [62].
Figure 3.
The enzymes and genes involved in Bacillus cereus toxin pathogenesis.
3.4. Emetic Illness
The emetic type of foodborne illness is induced by the small, cyclic, heat-stable toxin cereulide, which causes vomiting and nausea. [63]. The ces gene encodes cereulide synthetase, which is associated with the emetic toxin-producing strain of B. cereus [64]. Cereulide destroys the electrochemical gradient of membranes and inhibits fatty acid metabolism in mammalian cells by uncoupling mitochondrial ATP synthesis and disrupting the mitochondrial membrane potential [65]. Furthermore, the biological activity of the cereulide toxin impairs mitochondrial functionality (Figure 3) [66].
The pathogenicity mechanism of mycotoxins is briefly listed in Table 2.
Table 2.
The pathogenicity mechanism of mycotoxins.
| Mycotoxins | Diseases | Aim Organ | Related Enzymes |
|---|---|---|---|
| AFB1 | Human hepatocellular carcinoma | Human liver | CYP1A2, CYP3A4 |
| Ochratoxin A | Balkan Endemic Nephropathy, Tunisian Nephropathy | HK-2 cells | CYP1A1, CYP1A2, CYP3A4 |
| Ochratoxin A | Parkinsonism, Alzheimer’s disease | Human brain | OGG1 |
| Cereulide | Gastrointestinal diseases | Intestinal epithelial cells | CytK, Hbl, Nhe, EntFM |
| Cereulide | Emetic illness | Mitochondria | Cereulide synthetase |
4. Methods to Control and Manage Mycotoxins
4.1. Biocontrol Method
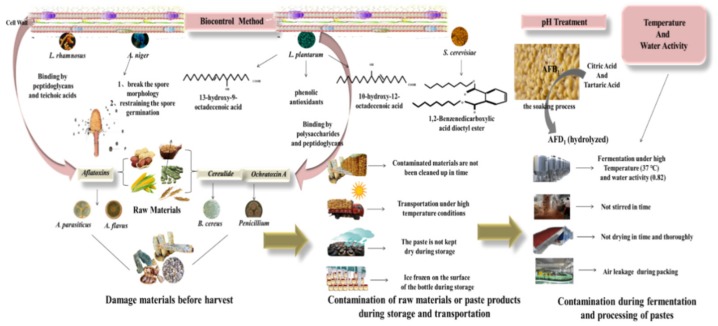
Biocontrol is one of the most effective techniques of controlling various mycotoxins; it uses safe microorganisms in fermented pastes to inhibit pathogen growth [67]. The advantage of the biocontrol method is that the growing conditions of related antagonistic microorganisms are simple enough, and they have strong abilities to multiply in foods for long periods of time [68]. In addition, unlike antagonists, they do not carry the risk of producing allergenic spores, which is harmful to the environment and human health [69].
Several works showed that many lactic acid bacteria (LAB) strains were able to restrict the growth of A. flavus. For instance, both L. plantarum and related low-molecular-weight substances showed significant antifungal activity [70]. Recent research revealed that various novel metabolites of LAB, including 13-hydroxy-9-octadecenoic acid, 10-hydroxy-12-octadecenoic acid [71], phenolic antioxidants [72], and spermine-like and short cyclic polylactates [73], showed lower minimum inhibitory concentrations of mycotoxins compared to the ordinary organic acids. Lactobacillus fermentum YML014 was also shown to reduce A. flavus by up to 50% after isolation from Nigerian fermented food (Cassava) [74].
The mechanism of LAB against fungal mycotoxins involves metabolic conversion by binding the related mycotoxin to the cell wall through adsorption and enzymatic degradation [75]. The coexistence of biocontrol agents and mycotoxins in the environment result in competition for nutrients and space, thereby directly impacting the growth of various mycotoxins. In addition, some biocontrol strains modify the external environment to inhibit mycotoxin production. For instance, LAB-induced decrease in pH led to a reduction in aflatoxin production by 91% by decreasing the stability of related mycotoxins [76]. Furthermore, oxidative stress induced by LAB was also responsible for the production of mycotoxins. It was reported that aflatoxins were produced under oxidative stress conditions, so antioxidants could play a restoration role [77]. The other widely accepted mechanism is that mycotoxins bind to LAB cell wall compounds through absorption. Polysaccharides and peptidoglycans are regarded as the most important components to bind OTA to L. plantarum and Lactobacillus sanfranciscensis, which could be enhanced through genetic modification [78]. Unlike OTA, aflatoxins were demonstrated to bind peptidoglycans and teichoic acids of the Lactobacillus rhamnosus cell wall; this binding ability was decreased by heat-treating [79].
Moreover, the mechanism of mycotoxin regulation by Aspergillus niger FS10 was also studied. The results confirmed that the culture filtrate of A. niger FS10 had a strong ability to disrupt spore morphology, deform the cell wall, and restrict spore germination [80]. Also, the degradation rate of AFB1 in the culture filtrate was 85.5%, which showed the feasibility of applying A. niger FS10 culture filtrate to control AFB1 contamination in fermented pastes.
Saccharomyces cerevisiae is one of the most important yeasts to produce various volatile aroma compounds, including acetate ester, which plays a significant role in the flavor formation of fermented pastes [81]. S. cerevisiae was revealed to be a potential biocontrol agent. For example, both S. cerevisiae EBF101 and S. cerevisiae 117 were proven to inhibit the growth of A. flavus Z103 by up to 85% and 83%, respectively [82]. Moreover, 1,2-benzenedicarboxylic acid dioctyl ester, which is one of the primary metabolites of S. cerevisiae, possesses antifungal and antibacterial properties (Figure 4).
Figure 4.
Mycotoxin production mechanism and control measures in fermented pastes.
4.2. Physicochemical Control Methods
Different grains make up fermented pastes. Various Aspergillus fungi strains generally exist after long-term grain storage. Hull-less crops have higher levels of AFB1 accumulation than hulled crops during storage [83]. Environmental conditions, such as temperature and water activity (AW), were confirmed to affect the production of mycotoxins by inhibiting the expression of related genes [84]. Regardless of temperature, AFB1 could not be generated at an AW of 0.82 [85]. Furthermore, the optimal temperatures of mycotoxin production ranged from 25 to 30 °C [86]. In general, temperature and AW are considered to be the most important factors in the management of crop storage. By controlling the AW to be stable and lower than 0.82 and the temperature to be lower than 37 °C, mycotoxins can be effectively inhibited at the source.
Most fermented pastes in China are made up of soybeans, which need to be soaked for 6–18 h in water. Adding organic acids (1 N citric acid and tartaric acid) during the soaking process reduced AFB1 by 94.1% and 95.1%, respectively [87]. It is well known that AFB1 is converted into β-keto acid and then to AFD1, which can be hydrolyzed [88]. Organic acids cannot impact the characteristics of various materials. Acidic or alkaline pH levels were shown to be effective in reducing the total amount of aflatoxins. Also the process of autoclaving significantly decreased the level of AFB1 at different pH levels compared to non-autoclaved samples [87] (Figure 4).
5. Detection Methods of Mycotoxins
At present, the main technologies regarding the determination of mycotoxins are based on chromatography, such as HPLC [89] and LC-MS/MS [90]. The methods of sample extraction include magnetic solid-phase extraction (MSPE) [91], solid-phase extraction (SPE) [92], and liquid–liquid extraction (LLE) [93]. Immunoassays are widely accepted to be able to detect mycotoxins sensitively in a variety of foods and feeds [94]. However, the techniques mentioned above include complicated and separated processes. Many novel methods exist which are simpler and more accurate.
5.1. Novel Magnetic Adsorbent Techniques
MSPE is a widely used method involving magnetic adsorption, whereby a product can be isolated from a sample solution using a magnet after the adsorption of analytes [95]. Recent studies focused on a new carbon-based nanomaterial named graphene as a magnetic adsorbent, which possesses unique properties such as strong adsorption capacity and excellent chemical stability. Furthermore, 3D graphene (3D-G), which has a low-density porous structure and versatile functionalization, overcomes the shortcomings of 2D graphene (2D-G) [96].
A new type of magnetic SPE, named graphene-coated magnetic-sheet SPE, was developed, in which magnetic 3D-G@Fe3O4 nanoparticles fixed on a perforated magnetic sheet were placed in a syringe filter holder and used as the adsorbent phase for the first time [97]. This technique was used to extract AFB1, B2, G1, and G2 rapidly, i.e., within 30 min, from fermented pastes. The relative recoveries of graphene-coated magnetic-sheet SPE were 83–102%, and the reusability was 20 times without significant loss of extraction recovery (< 4%).
Meanwhile, polydopamine-coated magnetic nanoparticles (PD-MNPs) were used as the adsorbent by preparation from amine-terminated magnetic nanoparticles (MNPs) and dopamine [98]. The extraction rates of AFB1 and AFG2 were 89.0% and 59.3%, and the limits of detection (LOD) were 0.0012 ng/mL for AFB1, AFB2, and AFG1, and 0.0031 ng/mL for AFG2.
Furthermore, the novel absorbent used an aptamer to functionalize magnetic agarosemicrospheres (MAMs) and bind to AFBs, then immobilized the AFB-aptamers on MAMs via a biotin–streptavidin interaction. The LOD values were 25 pg/mL for AFB1 and 10 pg/mL for AFB2 under reasonable conditions [99].
5.2. Aptamer Probes Techniques
Aptamers are specific affinity reagents with small sequences of oligonucleotides that can be screened for in vitro [100] and mimic antibodies by folding into complex 3D shapes that bind to specific targets [101]. Several works investigated aptamers in terms of designing them to construct various detection methods, such as the rapid detection of zearalenone in corn samples [102] and chloramphenicol determination [103]. Their best advantage is that they can be designed into numerous structures, including oligonucleotide probes to output signals directly.
An electrochemical aptasensor for aflatoxin B (AFB) detection using cyclic voltammetry (CV) and electochemical impedance spectroscopy (EIS) was developed with high sensitivity and selectivity. This aptasensor can be applied to detect AFB1 in white wine, peanuts, cashew nuts, and soy sauce, with a recovery of 85–100% [104].
In order to amplify the signal of aptamer probes, a novel AFB1 aptamer probe was studied using complementary DNA as a signal generator for amplification using real-time quantitative polymerase chain reaction (PCR) [105]. Many amplification strategies were adopted, involving complex enzyme catalysis and precise temperature control, which hindered the application of on-the-spot aflatoxin detection. Xia et al. proposed an aptamer probe design strategy to achieve AFB1 detection with enzyme-free amplification and homogeneous reactions [106], which was applied successfully in broad bean paste and peanut oil within one minute, which is the quickest detection of AFB1 so far.
Besides detecting AFBs, the aptamer was regarded as a high-affinity, specific method to detect OTA [107]. For example, the detection platform for OTA based on fluorescence aptamers was established, and exonuclease III (Exo III) was used to amplify signals and gold nanoparticles (AuNPs) to reduce fluorescence [108]. This aptasensor was confirmed to have excellent selectivity, with a limit of detection of 4.82 nM. Methods based on homogeneous electrochemical sensing protocol using a thionine–aptamer/graphene nanocomplex were also developed. The electronic signal starts off weak, but after making a connection with OTA, the thionine–aptamer/OTA generates an electrochemical signal using to detect OTA contamination in foods [109]. Moreover, biosensors using core–satellite nanostructures were applied to quantify mycotoxins, whereby the chiral intensity weakened by increasing OTA [110].
In recent years, a highly selective method integrating the electrochemistry and chemiluminescence of aptamers, electrochemiluminescence (ECL), has been widely used. Taking advantage of cadmium sulfide semiconductor quantum dots (CdS QDs) and DNA walkers, a novel technique based on the ECL aptasensor was confirmed to be a highly sensitive piece of equipment to quantify OTA during production, with a detection limit of 0.012 nM [111].
5.3. On-Site Test
Immunoassays are conventional techniques used AFB1 pretreatment in many foods. Due to this process being time-consuming and complex, many novel methods based on immunoassaying were developed. Immunomagnetism for pretreatment and enzyme-linked immunosorbent assay (ELISA) for quantification were established to detect AFB1 in soybean pastes [112]. These methods combined the active superparamagnetic beads and anti-AFB1 monoclonal antibodies to form immunomagnetic beads (IMBs). The magnetic beads coupled with the antibodies bind to a target material via an affinity reaction. The average recovery was between 0.5 and 7 µg/kg, or 83.6%~104%, and the relative standard deviation was 4.2%~11.7%.
A rapid on-site test for a wide range of aflatoxins in fermented soybean pastes is necessary. A novel strip test was developed based on lateral flow immunoassay, in which 3B6 was used to detect aflatoxins B1, M1, G1, G2, and B2, which belonged to monoclonal antibodies. The colloidal gold particles coated with 3B6 were considered to be the detector reagent, whereas the AFB1–BSA conjugate was regarded as a competitor reagent. This method was applied as a rapid and cost-effective screening tool, which had a limit of detection (0.5 µg/kg) within 10 min [113].
5.4. Molecular-Based Techniques
In recent years, molecular-based techniques such as PCR have been widely applied to detect B. cereus through the amplification of specific cereulide genes [114]. However, post-PCR analysis and gel electrophoresis assays increase the possibility of cross-contamination [115]. Furthermore, quantitative real-time PCR (qPCR), which uses intercalating dyes such as SYTO9 or dual-labeled fluorescence probes, offers simultaneous amplification and target-gene detection, which traditional PCR does not [116]. Many works were based on qPCR. For instance, the multiplex qPCR technique was developed to detect different pathogens simultaneously, including B. cereus in a variety of foods. Based on targeting various specific genes as indicators in a single reaction, the multiplex qPCR assay showed high efficiency and accuracy with a detection limit of 103 CFU/g for every pathogen [117].
Nevertheless, recent studies demonstrated the limitations of PCR-based methods, which cannot identify DNA from cells, regardless of viability. DNA amplification in nonviable cells leads to overestimation of bacterial counts [118]. Some researchers used propidium monoazide (PMA) as a nucleic acid to overcome this limitation by combining PMA with qRNA to detect and quantify B. cereus in ultra-high-temperature (UHT) milk samples, where the detection limit was 7.5 × 102 CFU/mL. Due to cell apoptosis during the UHT process, the detection percentage of PMA–qPCR was 32.6% compared with 57.8% of traditional qPCR, indicating that the application of PMA treatment prevented the overestimation of B. cereus in food [119].
However, the techniques mentioned above either require dual-labeled probes or a separate probe, which are very expensive. A novel pentaplex RT-PCR high-resolution melt-curve assay using DNA intercalating dye SYTO9 was developed [120], which has the ability to quantify four enterotoxin genes, namely nheA, entFM, cytK, and hblD, simultaneously, as well as ces, thereby allowing a wide range of detection of B. cereus. Furthermore, the detection limit in foods is 101 CFU/g with seven hours of enrichment. This investigation revealed an economical alternative method for melt-curve analysis and probe-based real-time detection technology.
Although PCR assays are sensitive enough and widely applied in cereulide detection, the fact that B. cereus strains have widely distributed genes presents a limitation [60]. A recent study demonstrated that expanding the number of genes though whole genome sequencing (WGS) was considered to be a more powerful technique than PCR-based methods [121]. Therefore, using WGS as pre-screening measure and BTyper as a data analysis tool would be a useful method to detect toxin genes in mixed cultures.
6. Conclusions
Three kinds of mycotoxins, namely, aflatoxins, ochratoxins, and cereulide, are often detected in fermented pastes. Different harvest stages, appropriate drying processes, relatively low temperatures, and organic acid pretreatments are the primary influential factors which will significantly help to guide fermentation industries to effectively avoid mycotoxin contamination. This review provided feasible suggestions for effective control, such as biocontrol agents and rapid detection methods throughout the whole production process of fermented pastes.
Author Contributions
Conceptualization, G.Z. and Y.Y.; software, Y.-F.W. and J.C.; investigation, Y.-F.W.; writing—original draft preparation, Y.-F.W.; writing—review and editing, G.Z. and Y.Y.; funding acquisition, G.Z. and J.C. All authors read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (No. 31972194 and 31601450) and Shanxi Province Key R&D Plan (No. 201703D211001–06–02).
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
Mycotoxins are always found in fermented pastes and can lead to serious illness. The purpose of this review is to summarize the sources, pathogenicity mechanisms, control methods, and novel detection technologies of mycotoxins.
References
- 1.Petra S., Peter S. Characterization of the key aroma compounds in soy sauce using approaches of molecular sensory science. J. Agric. Food Chem. 2007;55:6262–6269. doi: 10.1021/jf0709092. [DOI] [PubMed] [Google Scholar]
- 2.Durazzo A., Lucarini M. Current shot and re-thinking of antioxidant research strategy. Braz. J. Anal. Chem. 2018;5:9–11. doi: 10.30744/brjac.2179-3425.2018.5.20.9-11. [DOI] [Google Scholar]
- 3.Adebiyi J.A., Kayitesi E., Adebo O.A., Changwa R., Njobeh P.B. Food fermentation and mycotoxin detoxification: An African perspective. Food Control. 2019;106:8. doi: 10.1016/j.foodcont.2019.106731. [DOI] [Google Scholar]
- 4.Priyanka S., Sajid G.M., Kanade S.R. Aflatoxin B1 induced multiple epigenetic modulators in human epithelial cell lines. Toxicon. 2018;23:243–254. doi: 10.1016/j.toxicon.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Ahn J.S., Jang H.S., Song Y.J., Yang T.H., Jahng K.Y. Occurrence and biotransformation of ochratoxin A during pepper sauce fermentation. J. Korean Soc. Appl. Biol. Chem. 2011;54:972–977. doi: 10.1007/BF03253188. [DOI] [Google Scholar]
- 6.Trucksess M.W., Scott P.M. Mycotoxins in botanicals and dried fruits: A review. Food Addit. Contam. 2008;25:181–192. doi: 10.1080/02652030701567459. [DOI] [PubMed] [Google Scholar]
- 7.Schatzmayr G., Zehner F., Taubel M., Schatzmayr D., Klimitsch A., Loibner A.P., Binder E.M. Microbiologicals for deactivating mycotoxins. Mol. Nutr. Food Res. 2006;50:543–551. doi: 10.1002/mnfr.200500181. [DOI] [PubMed] [Google Scholar]
- 8.Dai Y., Huang K., Zhang B., Zhu L., Xu W. Aflatoxin B1-induced epigenetic alterations: An overview. Food Chem. Toxicol. 2017;109:586–591. doi: 10.1016/j.fct.2017.06.034. [DOI] [PubMed] [Google Scholar]
- 9.Guimarães F.R., Vieira C.M., Maria D.S.F.K., Monteiro E.K.M., Pereira A.R., Chagas M.M. Epigenetic alterations caused by aflatoxin B1: A public health risk in the induction of hepatocellular carcinoma. Transl. Res. 2018;22:32–40. doi: 10.1016/j.trsl.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Daohong Z., Peiwu L., Qi Z., Wen Z. Ultrasensitive nanogold probe-based immunochromatographic assay for simultaneous detection of total aflatoxins in peanuts. Biosens. Bioelectron. 2011;26:2877–2882. doi: 10.1016/j.bios.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 11.Roy K.W., Baird R.E., Abney T.S. A review of soybean (Glycine max) seed, pod, and flower mycofloras in north America, with methods and a key for identification of selected fungi. Mycopathologia. 2001;150:15–27. doi: 10.1023/A:1010805224993. [DOI] [PubMed] [Google Scholar]
- 12.Peng Z., Chen L., Zhu Y., Huang Y., Hu X., Wu Q., Nüssler A.K., Liu L., Yang W. Current major degradation methods for aflatoxins: A review. Trends Food Sci. Technol. 2018;80:155–166. doi: 10.1016/j.tifs.2018.08.009. [DOI] [Google Scholar]
- 13.Gourama H., Bullerman L.B. Aspergillus flavus and Aspergillus parasiticus: Aflatoxigenic fungi of concern in foods and feeds: A review. J. Food Prot. 1995;58:1395–1404. doi: 10.4315/0362-028X-58.12.1395. [DOI] [PubMed] [Google Scholar]
- 14.Frisvad J.C., Hubka V., Ezekiel C.N., Hong S.B., Nováková A., Chen A.J., Arzanlou M., Larsen T.O., Sklenář F., Mahakarnchanakul W. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019;93:1–10. doi: 10.1016/j.simyco.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiujiang Y., Perng-Kuang C., Ehrlich K.C., Cary J.W., Deepak B., Cleveland T.E., Payne G.A., Linz J.E., Woloshuk C.P., Bennett J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microb. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pildain M.B., Frisvad J.C., Vaamonde G., Cabral D., Varga J., Samson R.A. Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int. J. Syst. Evol. Microbiol. 2008;58:725–735. doi: 10.1099/ijs.0.65123-0. [DOI] [PubMed] [Google Scholar]
- 17.Rank C., Klejnstrup M.L., Petersen L.M., Kildgaard S., Frisvad J.C., Held G.C., Ostenfeld L.T. Comparative chemistry of Aspergillus oryzae (RIB40) and A. flavus (NRRL 3357) Metabolites. 2012;2:39–56. doi: 10.3390/metabo2010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soares C., Rodrigues P., Peterson S.W., Lima N. Three new species of Aspergillus section Flavi isolated from almonds and maize in Portugal. Mycologia. 2012;104:682–697. doi: 10.3852/11-088. [DOI] [PubMed] [Google Scholar]
- 19.Kurtzman C.P., Horn B.W., Hesseltine C.W. Aspergillus nomius: A new aflatoxin-producing species related to Aspergillus flavus and Aspergillus tamarii. Antonie Van Leeuwenhoek. 1987;53:147–158. doi: 10.1007/BF00393843. [DOI] [PubMed] [Google Scholar]
- 20.Gonçalves J.S., Ferracin L.M., Vieira M.L.C., Iamanaka B.T., Taniwaki M.H., Fungaro M.H.P. Molecular analysis of Aspergillus section Flavi isolated from Brazil nuts. World J. Microb. Biot. 2012;28:1817–1825. doi: 10.1007/s11274-011-0956-3. [DOI] [PubMed] [Google Scholar]
- 21.Basaran P., Demirbas R.M. Spectroscopic detection of pharmaceutical compounds from an aflatoxigenic strain of Aspergillus parasiticus. Microbiol. Res. 2010;165:516–522. doi: 10.1016/j.micres.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Varga J., Frisvad J.C., Samson R.A. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud. Mycol. 2011;69:57–80. doi: 10.3114/sim.2011.69.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rank C., Nielsen K.F., Larsen T.O., Varga J., Samson R.A., Frisvad J.C. Distribution of sterigmatocystin in filamentous fungi. Fungal Biol. 2011;115:406–420. doi: 10.1016/j.funbio.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Shukla S., Park H.K., Lee J.S., Kim J.K., Kim M. Reduction of biogenic amines and aflatoxins in Doenjang samples fermented with various Meju as starter cultures. Food Control. 2014;42:181–187. doi: 10.1016/j.foodcont.2014.02.006. [DOI] [Google Scholar]
- 25.Taniwaki M.H., Pitt J.I., Magan N. Aspergillus species and mycotoxins: Occurrence and importance in major food commodities. Curr. Opin. Food Sci. 2018;54:234–246. doi: 10.1016/j.cofs.2018.05.008. [DOI] [Google Scholar]
- 26.Pitt J.I., Hocking A.D. Aspergillus and related teleomorphs. Fungi Food Spoilage. 1985;36:217–221. [Google Scholar]
- 27.Katsurayama A.M., Martins L.M., Iamanaka B.T., Mhp F., Silva J.J., Frisvad J.C., Pitt J.I., Taniwaki M.H. Occurrence of Aspergillus section Flavi and aflatoxins in Brazilian rice: From field to market. Int. J. Food Microbiol. 2018;266:213–221. doi: 10.1016/j.ijfoodmicro.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Mohale S., Medina A., Rodríguez A., Sulyok M., Magan N. Mycotoxigenic fungi and mycotoxins associated with stored maize from different regions of Lesotho. Mycotoxin Res. 2013;29:209–219. doi: 10.1007/s12550-013-0176-9. [DOI] [PubMed] [Google Scholar]
- 29.Viaro H.P., Silva J.J.D., Ferranti L.D.S., Bordini J.G., Massi F.P., Fungaro M.H.P. The first report of A. novoparasiticus, A. arachidicola and A. pseudocaelatus in Brazilian corn kernels. Int. J. Food Microbiol. 2017;243:46–51. doi: 10.1016/j.ijfoodmicro.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Dong M.K., Chung S.H., Chun H.S. Multiplex PCR assay for the detection of aflatoxigenic and non-aflatoxigenic fungi in a Korean fermented soybean food starter. Food Microbiol. 2011;28:1402–1408. doi: 10.1016/j.fm.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 31.Cabanes F.J., Bragulat M.R., Castella G. Ochratoxin A producing species in the genus Penicillium. Toxins. 2010;2:1111–1120. doi: 10.3390/toxins2051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khoury A., Atoui A. Ochratoxin A: General overview and actual molecular status. Toxins. 2010;2:461–493. doi: 10.3390/toxins2040461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen M.T., Tozovanu M., Tran T.L., Pfohl-Leszkowicz A. Occurrence of aflatoxin B1, citrinin and ochratoxin A in rice in five provinces of the central region of Vietnam. Food Chem. 2007;105:42–47. doi: 10.1016/j.foodchem.2007.03.040. [DOI] [Google Scholar]
- 34.Bhatnagar D., Ehrlich K.C., Cleveland T.E. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2003;61:83–93. doi: 10.1007/s00253-002-1199-x. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Wang L., Liu F., Wang Q., Selvaraj J.N., Xing F., Zhao Y., Liu Y. Ochratoxin A producing fungi, biosynthetic pathway and regulatory mechanisms. Toxins. 2016;8:83. doi: 10.3390/toxins8030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi H., Li S., Bai Y., Prates L.L., Lei Y., Yu P. Mycotoxin contamination of food and feed in China: Occurrence, detection techniques, toxicological effects and advances in mitigation technologies. Food Control. 2018;7:43–51. doi: 10.1016/j.foodcont.2018.03.036. [DOI] [Google Scholar]
- 37.Lai X., Liu R., Ruan C., He Z., Liu C. Occurrence of aflatoxins and ochratoxin A in rice samples from six provinces in China. Food Control. 2015;50:401–404. doi: 10.1016/j.foodcont.2014.09.029. [DOI] [Google Scholar]
- 38.Varga J., Rigó K., Téren J., Mesterházy Á. Recent advances in ochratoxin research I. production, detection and occurrence of ochratoxins. Cereal Res. Commun. 2001;29:85–92. doi: 10.1007/BF03543646. [DOI] [Google Scholar]
- 39.Atanda S.A., Pessu P.O., Agoda S., Isong I.U., Adekalu O.A., Echendu M.A., Falade T.C. Fungi and mycotoxins in stored foods. Afr. J. Microbiol. Res. 2011;5:4373–4382. doi: 10.5897/AJMR11.487. [DOI] [Google Scholar]
- 40.Jafar M., Gisoo M. Effects of processing on mycotoxin stability in cereals. J. Sci Food Agric. 2014;94:2372–2375. doi: 10.1002/jsfa.6600. [DOI] [PubMed] [Google Scholar]
- 41.Kotiranta A., Lounatmaa K., Haapasalo M. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2000;2:189–198. doi: 10.1016/S1286-4579(00)00269-0. [DOI] [PubMed] [Google Scholar]
- 42.Tenovuo J., Makinen K.K., Sievers G. Antibacterial effect of lactoperoxidase and myeloperoxidase against Bacillus cereus. Antimicrob. Agents Chemother. 1985;27:96–101. doi: 10.1128/AAC.27.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin H., Bandara N., Shin E., Ryu S., Kim K.P. Prevalence of Bacillus cereus bacteriophages in fermented foods and characterization of phage JBP901. Res. Microbiol. 2011;162:791–797. doi: 10.1016/j.resmic.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Tewari A., Abdullah S. Bacillus cereus food poisoning: International and Indian perspective. J. Food Sci. 2014;52:1–12. doi: 10.1007/s13197-014-1344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartoszewiez M., Hansen B.M., Swiecicka I. The members of the Bacillus cereus group are commonly present contaminants of fresh and heat-treated milk. Food Microbiol. 2008;25:588–596. doi: 10.1016/j.fm.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 46.King N.J., Whyte R., Hudson J.A. Presence and significance of Bacillus cereus in dehydrated potato products. J. Food Prot. 2007;70:514–520. doi: 10.4315/0362-028X-70.2.514. [DOI] [PubMed] [Google Scholar]
- 47.Juil K., Seong-Hwan P., Hun D.K., Kim D., Moon Y. Interference with mutagenic aflatoxin B1-induced checkpoints through antagonistic action of Ochratoxin A in intestinal cancer cells: A molecular explanation on potential risk of crosstalk between carcinogens. Oncotarget. 2016;7:39627–39639. doi: 10.18632/oncotarget.8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding X., Kaminsky L.S. Human extrahepatic cytochromes P450: Function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu. Rev. Pharm. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 49.Bedard L.L., Massey T.E. Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 2006;241:174–183. doi: 10.1016/j.canlet.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 50.Pfohlleszkowicz A. Ochratoxin A and aristolochic acid involvement in nephropathies and associated urothelial tract tumours. Arh Hig Rada Toksikol. 2009;60:465–483. doi: 10.2478/10004-1254-60-2009-2000. [DOI] [PubMed] [Google Scholar]
- 51.Lu Y., Cederbaum A.I. CYP2E1 potentiation of LPS and TNFα-induced hepatotoxicity by mechanisms involving enhanced oxidative and nitrosative stress, activation of MAP kinases, and mitochondrial dysfunction. Genes Nutr. 2010;5:149–167. doi: 10.1007/s12263-009-0150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yim J.H., Kim K.Y., Chon J.W., Kim D.H., Kim H.S., Choi D.S., Choi I.S., Seo K.H. Incidence, Antibiotic susceptibility, and toxin profiles of Bacillus cereus sensu lato isolated from Korean fermented soybean products. J. Food Sci. 2015;80:1266–1270. doi: 10.1111/1750-3841.12872. [DOI] [PubMed] [Google Scholar]
- 53.Lee H.J., Pyo M.C., Shin H.S., Ryu D., Lee K.W. Renal toxicity through AhR, PXR, and Nrf2 signaling pathway activation of ochratoxin A-induced oxidative stress in kidney cells. Food Chem. Toxicol. 2018;45:325–331. doi: 10.1016/j.fct.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Doi K., Uetsuka K. Mechanisms of mycotoxin-induced neurotoxicity through oxidative stress-associated pathways. Int. J. Mo.l Sci. 2011;12:5213–5237. doi: 10.3390/ijms12085213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belmadani A., Betbeder A.M., Tramu G., Steyn P.S., Creppy E.E. P1E124–Regional selectivity to ochratoxin A, distribution and cytotoxicity in rat brain. Arch. Toxicol. 1998;72:656–662. doi: 10.1007/s002040050557. [DOI] [PubMed] [Google Scholar]
- 56.Sava V., Reunova O., Velasquez A., Harbison R., Sanchez-Ramos J. Acute neurotoxic effects of the fungal metabolite ochratoxin-A. Neurotoxicology. 2006;27:82–92. doi: 10.1016/j.neuro.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X., Boesch-Saadatmandi C., Lou Y., Wolffram S., Huebbe P., Rimbach G. Ochratoxin A induces apoptosis in neuronal cells. Genes Nutr. 2009;4:41–48. doi: 10.1007/s12263-008-0109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ceuppens S., Uyttendaele M., Drieskens K., Heyndrickx M., Rajkovic A., Boon N., Van de Wiele T. Survival and germination of Bacillus cereus spores without outgrowth or enterotoxin production during in vitro simulation of gastrointestinal transit. Appl. Environ. Microb. 2012;78:76–98. doi: 10.1128/AEM.02142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haug T.M., Sand S.L., Sand O., Phung D., Granum P.E., Hardy S.P. Formation of very large conductance channels by Bacillus cereus Nhe in vero and GH4 Cells identifies NheA. J. Membr. Biol. 2010;237:1–11. doi: 10.1007/s00232-010-9298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arnesen L.P.S., Fagerlund A., Granum P.E. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 2008;32:579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- 61.Chang H.C., Wei Y.F., Dijkshoorn L., Vaneechoutte M., Tang C.T., Chang T.C. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 2005;43:1632–1639. doi: 10.1128/JCM.43.4.1632-1639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wijnands L.M., Dufrenne J.B., Rombouts F.M., In’t Veld P.H., van Leusden F.M. Prevalence of potentially pathogenic Bacillus cereus in food commodities in The Netherlands. J. Food Prot. 2006;69:2587–2594. doi: 10.4315/0362-028X-69.11.2587. [DOI] [PubMed] [Google Scholar]
- 63.Dommel M.K., Frenzel E., Strasser B., Blöchinger C., Scherer S., Ehling-Schulz M. Identification of the main promoter directing cereulide biosynthesis in emetic Bacillus cereus and its application for real-time monitoring of ces gene expression in foods. Immunol. Lett. 2010;56:12–32. doi: 10.1128/AEM.02317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dommel M.K., Genia L., Siegfried S., Monika E.S. Transcriptional kinetic analyses of cereulide synthetase genes with respect to growth, sporulation and emetic toxin production in Bacillus cereus. Food Microbiol. 2011;28:284–290. doi: 10.1016/j.fm.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Andersson M.A., Pasi H., Ulla H.H.M.L.I., Douwe H., Jean-Claude L., Jorma M.K.P., Martti S., Isabelle S., Anna-Laura S., Laura T. Toxicological profile of cereulide, the Bacillus cereus emetic toxin, in functional assays with human, animal and bacterial cells. Toxicon. 2007;49:351–367. doi: 10.1016/j.toxicon.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 66.Decleer M., Jovanovic J., Vakula A., Udovicki B., Rek A., Madder A., De Saeger S., Rajkovic A. Oxygen consumption rate analysis of mitochondrial dysfunction caused by Bacillus cereus cereulide in Caco-2 and HepG2 Cells. Toxins. 2018;10:266. doi: 10.3390/toxins10070266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meftah S., Abid S., Dias T., Rodrigues P. Effect of dry-sausage starter culture and endogenous yeasts on Aspergillus westerdijkiae and Penicillium nordicum growth and OTA production. Lwt-Food Sci. Technol. 2018;87:250–258. doi: 10.1016/j.lwt.2017.08.090. [DOI] [Google Scholar]
- 68.Chanchalchaovivat A., Ruenwongsa P., Panijpan B. Screening and identification of yeast strains from fruits and vegetables: Potential for biological control of postharvest chilli anthracnose (Colletotrichum capsici) Biol. Control. 2007;42:326–335. doi: 10.1016/j.biocontrol.2007.05.016. [DOI] [Google Scholar]
- 69.Nunes C.A. Biological control of postharvest diseases of fruit. Eur. J. Plant Pathol. 2012;133:181–196. doi: 10.1007/s10658-011-9919-7. [DOI] [Google Scholar]
- 70.Ahlberg S., Joutsjoki V., Laurikkala S., Varmanen P., Korhonen H. Aspergillus flavus growth inhibition by Lactobacillus strains isolated from traditional fermented Kenyan milk and maize products. Arch. Microbiol. 2017;75:89–95. doi: 10.1007/s00203-016-1316-3. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y.Y., Liang N.Y., Curtis J.M., Gänzle M.G. Characterization of linoleate 10-Hydratase of Lactobacillus plantarum and novel antifungal metabolites. Front. Microbiol. 2016;7:243–254. doi: 10.3389/fmicb.2016.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sellamani M., Kalagatur N.K., Siddaiah C., Mudili V., Krishna K., Natarajan G., Putcha V.L.R. Antifungal and zearalenone inhibitory activity of Pediococcus pentosaceus isolated from dairy products on Fusarium graminearum. Front. Microbiol. 2016;7:12–25. doi: 10.3389/fmicb.2016.00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mosbah A., Delavenne E., Souissi Y., Mahjoubi M., Jehan P., Le Yondre N., Cherif A., Bondon A., Mounier J., Baudy-Floc’h M., et al. Novel antifungal compounds, spermine-like and short cyclic polylactates, produced by Lactobacillus harbinensis K.V9.3.1Np in yogurt. Front. Microbiol. 2018;9:10–19. doi: 10.3389/fmicb.2018.02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adedokun E.O., Rather I.A., Bajpai V.K., Park Y.H. Biocontrol efficacy of Lactobacillus fermentum YML014 against food spoilage moulds using the tomato puree model. Front. Life Sci. 2016;9:64–68. doi: 10.1080/21553769.2015.1084951. [DOI] [Google Scholar]
- 75.Sadiq F.A., Yan B.W., Tian F.W., Zhao J.X., Zhang H., Chen W. Lactic acid bacteria as antifungal and anti-mycotoxigenic agents: A comprehensive review. Compr. Rev. Food. Sci. Food Saf. 2019;10:1541–4337. doi: 10.1111/1541-4337.12481. [DOI] [PubMed] [Google Scholar]
- 76.Guimarães A., Santiago A., Teixeira J.A., Venâncio A., Abrunhosa L. Anti-aflatoxigenic effect of organic acids produced by Lactobacillus plantarum. Int. J. Food Microbiol. 2017;264:31–42. doi: 10.1016/j.ijfoodmicro.2017.10.025. [DOI] [PubMed] [Google Scholar]
- 77.Furukawa T., Sakuda S. Inhibition of aflatoxin production by paraquat and external superoxide dismutase in Aspergillus flavus. Toxins. 2019;11:107. doi: 10.3390/toxins11020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khattab A.A., Ibrahim M.I.M., El-Kady A.A. Ochratoxin A biosorption onto genetically improved of Lactobacillus delbrueckii mutants. Int. Food Res. J. 2018;25:515–522. [Google Scholar]
- 79.Liew W.P., Mohdredzwan S. Mycotoxin: Its impact on gut health and microbiota. Front. Cell Infect. Microbiol. 2018;8:60–71. doi: 10.3389/fcimb.2018.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dan X., Wang H., Zhang Y., Yang Z., Sun X. Inhibition of non-toxigenic Aspergillus niger FS10 isolated from Chinese fermented soybean on growth and aflatoxin B1 production by Aspergillus flavus. Food Control. 2013;32:359–365. [Google Scholar]
- 81.Hong K.Q., Fu X.M., Dong S.S., Xiao D.G., Dong J. Modulating acetate ester and higher alcohol production in Saccharomyces cerevisiae through the cofactor engineering. J. Ind. Microbiol. Biol. 2019;5:261–272. doi: 10.1007/s10295-019-02176-4. [DOI] [PubMed] [Google Scholar]
- 82.Abdel-Kareem M.M., Rasmey A.M., Zohri A.A. The action mechanism and biocontrol potentiality of novel isolates of Saccharomyces cerevisiae against the aflatoxigenic Aspergillus flavus. Lett. Appl. Microbiol. 2019;68:104–111. doi: 10.1111/lam.13105. [DOI] [PubMed] [Google Scholar]
- 83.Krulj J., Markov S., Bocarov-Stancic A., Pezo L., Kojic J., Curcic N., Janic-Hajnal E., Bodroza-Solarov M. The effect of storage temperature and water activity on aflatoxin B-1 accumulation in hull-less and hulled spelt grains. J. Sci. Food Agric. 2019;99:3703–3710. doi: 10.1002/jsfa.9601. [DOI] [PubMed] [Google Scholar]
- 84.Gilbert M.K., Medina A., Mack B.M., Lebar M.D., Rodrã Guez A., Bhatnagar D., Magan N., Obrian G., Payne G. Carbon dioxide mediates the response to temperature and water activity levels in Aspergillus flavus during infection of maize kernels. Toxins. 2018;10:5. doi: 10.3390/toxins10010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Casquete R., Benito M.J., Córdoba M.D.G., Ruiz-Moyano S., Martín A. The growth and aflatoxin production of Aspergillus flavus strains on a cheese model system are influenced by physicochemical factors. J Dairy Sci. 2017;100:217–221. doi: 10.3168/jds.2017-12865. [DOI] [PubMed] [Google Scholar]
- 86.Mousa W., Ghazali F.M., Jinap S., Ghazali H.M., Radu S. Modelling the effect of water activity and temperature on growth rate and aflatoxin production by two isolates of Aspergillus flavus on paddy. J. Applmicrobiol. 2011;111:1262–1274. doi: 10.1111/j.1365-2672.2011.05134.x. [DOI] [PubMed] [Google Scholar]
- 87.Lee J., Her J.Y., Lee K.G. Reduction of aflatoxins (B 1, B 2, G 1, and G 2 ) in soybean-based model systems. Food Chem. 2015;189:45–51. doi: 10.1016/j.foodchem.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 88.Méndez-Albores A., Arambula-Villa G., Loarca-Piña M.G.F., Castano-Tostado E., Moreno-Martínez E. Safety and efficacy evaluation of aqueous citric acid to degrade B-aflatoxins in maize. Food Chem. Toxicol. 2005;43:233–238. doi: 10.1016/j.fct.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 89.Cao J., Zhou S., Kong W., Ma X., Yang M., Li W., Yang S. Simultaneous determination of aflatoxins B1, B2, G1, G2 in Fructus Bruceae by high-performance liquid chromatography with online post-column photochemical derivatization. J. Sep. Sci. 2015;37:2771–2778. doi: 10.1002/jssc.201400501. [DOI] [PubMed] [Google Scholar]
- 90.Fan S., Li Q., Zhang X., Cui X., Zhang D., Zhang Y. Simultaneous determination of aflatoxin B1, B2, G1, and G2 in corn powder, edible oil, peanut butter, and soy sauce by liquid chromatography with tandem mass spectrometry utilizing turbulent flow chromatography. J. Sep. Sci. 2015;38:1310–1317. doi: 10.1002/jssc.201401376. [DOI] [PubMed] [Google Scholar]
- 91.Zarrin E.H., Hamed Reza B., Javad F. Extraction of aflatoxins from food samples using graphene-based magnetic nanosorbents followed by high-performance liquid chromatography: A simple solution to overcome the problems of immunoaffinity columns. J. Sep. Sci. 2015;37:2566–2573. doi: 10.1002/jssc.201400260. [DOI] [PubMed] [Google Scholar]
- 92.Alcaide-Molina M., Ruiz-Jiménez J., Mata-Granados J.M., Castro L.D. High through-put aflatoxin determination in plant material by automated solid-phase extraction on-line coupled to laser-induced fluorescence screening and determination by liquid chromatography–triple quadrupole mass spectrometry. J Chromatogr. A. 2009;1216:1115–1125. doi: 10.1016/j.chroma.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 93.Neda S.F., Jalal H., Yousefi S.R. Determination of aflatoxin B1 in cereals by homogeneous liquid-liquid extraction coupled to high performance liquid chromatography-fluorescence detection. J. Sep. Sci. 2015;34:1333–1337. doi: 10.1002/jssc.201000882. [DOI] [PubMed] [Google Scholar]
- 94.Duan N., Wu S., Dai S., Gu H., Hao L., Ye H., Wang Z. Advances in aptasensors for the detection of food contaminants. Analyst. 2016;141:3942–3961. doi: 10.1039/C6AN00952B. [DOI] [PubMed] [Google Scholar]
- 95.ŠafaříKová M., ŠafaříK I. Magnetic solid-phase extraction. J. Magn. Magn. Mater. 1999;194:108–112. doi: 10.1016/S0304-8853(98)00566-6. [DOI] [Google Scholar]
- 96.Wang M., Duan X., Xu Y., Duan X. Functional Three-Dimensional Graphene/Polymer Composites. ACS Nano. 2016;10:7231–7242. doi: 10.1021/acsnano.6b03349. [DOI] [PubMed] [Google Scholar]
- 97.Nouri N., Sereshti H., Farahani A. Graphene-coated magnetic-sheet solid-phase extraction followed by high-performance liquid chromatography with fluorescence detection for the determination of aflatoxins B-1, B-2, G(1), and G(2) in soy-based samples. J. Sep. Sci. 2018;41:10–21. doi: 10.1002/jssc.201800471. [DOI] [PubMed] [Google Scholar]
- 98.Cassandra M.C., Paul T., Li-Sheng D., Xun L., Yi-Ming L. Extraction of aflatoxins from liquid foodstuff samples with polydopamine-coated superparamagnetic nanoparticles for HPLC-MS/MS analysis. J. Agrfood Chem. 2014;62:4261–4267. doi: 10.1021/jf501659m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu H., Lu A., Fu H., Li B., Yang M., Wang J., Luan Y. Affinity capture of aflatoxin B1 and B2 by aptamer-functionalized magnetic agarose microspheres prior to their determination by HPLC. Microchim. Acta. 2018;185:326–337. doi: 10.1007/s00604-018-2849-8. [DOI] [PubMed] [Google Scholar]
- 100.Wang J., Yu J., Yang Q., Mcdermott J., Scott A., Vukovich M., Lagrois R., Gong Q., Greenleaf W., Eisenstein M. Multiparameter particle display (MPPD): A quantitative screening method for the discovery of highly specific aptamers. AngewChem. 2017;56:744–753. doi: 10.1002/anie.201608880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dunn M.R., Jimenez R.M., Chaput J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017;1:76–84. doi: 10.1038/s41570-017-0076. [DOI] [Google Scholar]
- 102.Wu S., Liu L., Duan N., Li Q., Zhou Y., Wang Z. An aptamer-based lateral flow test strip for rapid detection of zearalenone in corn samples. J. Agric. Food Chem. 2018;66:1949–1954. doi: 10.1021/acs.jafc.7b05326. [DOI] [PubMed] [Google Scholar]
- 103.Wu S.J., Zhang H., Shi Z., Duan N., Fang C.C., Dai S.L., Wang Z.P. Aptamer-based fluorescence biosensor for chloramphenicol determination using upconversion nanoparticles. Food Control. 2015;50:597–604. doi: 10.1016/j.foodcont.2014.10.003. [DOI] [Google Scholar]
- 104.Evtugyn G., Porfireva A., Stepanova V., Sitdikov R., Hianik T. Electrochemical aptasensor based on polycarboxylic macrocycle modified with neutral red for aflatoxin B1 detection. Electroanalysis. 2015;26:2100–2109. doi: 10.1002/elan.201400328. [DOI] [Google Scholar]
- 105.Guo X., Wen F., Zheng N., Luo Q., Wang H., Wang H., Li S., Wang J. Development of an ultrasensitive aptasensor for the detection of aflatoxin B1. BiosensBioelectron. 2014;56:340–344. doi: 10.1016/j.bios.2014.01.045. [DOI] [PubMed] [Google Scholar]
- 106.Xia X.H., Wang Y.X., Yang H., Dong Y., Zhang K.X., Lu Y.H., Deng R.J., He Q. Enzyme-free amplified and ultrafast detection of aflatoxin B-1 using dual-terminal proximity aptamer probes. Food Chem. 2019;283:32–38. doi: 10.1016/j.foodchem.2018.12.117. [DOI] [PubMed] [Google Scholar]
- 107.Cruz-Aguado J.A., Penner G. Determination of Ochratoxin A with a DNA aptamer. J. Agric. Food Chem. 2008;56:10456–10461. doi: 10.1021/jf801957h. [DOI] [PubMed] [Google Scholar]
- 108.Zhao Y.Y., Liu R.J., Sun W.Y., Lv L., Guo Z.J. Ochratoxin A detection platform based on signal amplification by Exonuclease III and fluorescence quenching by gold nanoparticles. Sens. Actuator B-Chem. 2018;255:1640–1645. doi: 10.1016/j.snb.2017.08.176. [DOI] [Google Scholar]
- 109.Sun A.L., Zhang Y.F., Sun G.P., Wang X.N., Tang D.P. Homogeneous electrochemical detection of ochratoxin A in foodstuff using aptamer-graphene oxide nanosheets and DNase I-based target recycling reaction. BiosensBioelectron. 2017;89:659–665. doi: 10.1016/j.bios.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 110.Cai J.R., Hao C.L., Sun M.Z., Ma W., Xu C.L., Kuang H. Chiral shell core-satellite nanostructures for ultrasensitive detection of mycotoxin. Small. 2018;14:8–16. doi: 10.1002/smll.201703931. [DOI] [PubMed] [Google Scholar]
- 111.Wei M., Wang C.L., Xu E.S., Chen J., Xu X.L., Wei W., Liu S.Q. A simple and sensitive electrochemiluminescence aptasensor for determination of ochratoxin A based on a nicking endonuclease-powered DNA walking machine. Food Chem. 2019;282:141–146. doi: 10.1016/j.foodchem.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 112.Xie F., Lai W.H., Saini J., Shan S., Cui X., Liu D.F. Rapid pretreatment and detection of trace aflatoxin B 1 in traditional soybean sauce. Food Chem. 2014;150:99–105. doi: 10.1016/j.foodchem.2013.10.147. [DOI] [PubMed] [Google Scholar]
- 113.Santos V.O., Pelegrini P.B., Mulinari F., Lacerda A.F., Moura R.S., Cardoso L.P.V., Buhrer-Sekula S., Miller R.N.G., Grossi-de-Sa M.F. Development and validation of a novel lateral flow immunoassay device for detection of aflatoxins in soy-based foods. Anal. Methods. 2017;9:2715–2722. doi: 10.1039/C7AY00601B. [DOI] [Google Scholar]
- 114.Law J.W., Ab Mutalib N.S., Chan K.G., Lee L.H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015;5:770–782. doi: 10.3389/fmicb.2014.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ding T., Suo Y., Zhang Z., Liu D., Ye X., Chen S., Zhao Y. A multiplex RT-PCR assay for S. aureus, L. monocytogenes, and Salmonella spp. detection in raw milk with pre-enrichment. Front. Microbiol. 2017;8:989–993. doi: 10.3389/fmicb.2017.00989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhao X., Lin C.W., Wang J., Oh D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbbiot. 2014;24:297–303. doi: 10.4014/jmb.1310.10013. [DOI] [PubMed] [Google Scholar]
- 117.Wei S., Daliri E.B.M., Chelliah R., Park B.J., Lim J.S., Baek M.A., Nam Y.S., Seo K.H., Jin Y.G., Oh D.H. Development of a multiplex real-time PCR for simultaneous detection of Bacillus cereus, Listeria monocytogenes, and Staphylococcus aureus in food samples. J. Food Saf. 2019;39:7–14. doi: 10.1111/jfs.12558. [DOI] [Google Scholar]
- 118.Knut R., Birgitte M., Signe Marie D.M., Holck A.L. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl Environ. Microbiol. 2005;71:1018–1024. doi: 10.1128/AEM.71.2.1018-1024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cattani V.C., Jr., Nasário J.S.R., Ferreira C.A.S., Oliveira S.D. Detection and quantification of viable Bacillus cereus group species in milk by propidium monoazide quantitative real-time PCR. J. Dairy Sci. 2016;99:2617–2624. doi: 10.3168/jds.2015-10019. [DOI] [PubMed] [Google Scholar]
- 120.Forghani F., Singh P., Seo K.H., Oh D.H. A novel pentaplex real time (RT)- PCR high resolution melt curve assay for simultaneous detection of emetic and enterotoxin producing Bacillus cereus in food. Food Control. 2016;60:560–568. doi: 10.1016/j.foodcont.2015.08.030. [DOI] [Google Scholar]
- 121.Nguyen A.T., Tallent S.M. Screening food for Bacillus cereus toxins using whole genome sequencing. Food Microbiol. 2019;78:164–170. doi: 10.1016/j.fm.2018.10.008. [DOI] [PubMed] [Google Scholar]