Abstract
Background
Simeoni and colleagues introduced a compartmental model for tumor growth that has proved quite successful in modeling experimental therapeutic regimens in oncology. The model is based on a system of ordinary differential equations (ODEs), and accommodates a lag in therapeutic action through delay compartments. There is some ambiguity in the appropriate number of delay compartments, which we examine in this note.
Methods
We devised an explicit delay differential equation model that reflects the main features of the Simeoni ODE model. We evaluated the original Simeoni model and this adaptation with a sample data set of mammary tumor growth in the FVB/N-Tg(MMTVneu)202Mul/J mouse model.
Results
The experimental data evinced tumor growth heterogeneity and inter-individual diversity in response, which could be accommodated statistically through mixed models. We found little difference in goodness of fit between the original Simeoni model and the delay differential equation model relative to the sample data set.
Conclusions
One should exercise caution if asserting a particular mathematical model uniquely characterizes tumor growth curve data. The Simeoni ODE model of tumor growth is not unique in that alternative models can provide equivalent representations of tumor growth.
Keywords: Tumor growth, Cancer chemotherapy, Mathematical model, Ordinary differential equations, Delay differential equations
Background
Simeoni and colleagues [1–3] introduced and developed a pharmacokinetic/pharmacodynamic (PK/PD) model of tumor growth from in vivo animal studies, which could be summarized mathematically by a system of ordinary differential equations (ODEs). The model, and adaptations, have proved quite successful in predicting or modeling tumor growth and efficacy of cancer treatments [4–9].
A distinguishing characteristic of the Simeoni tumor growth model is that under chemotherapy, a drug’s action is not instantaneous: rather, tumor cells pass through progressive stages of damage because of the drug’s mechanism of action. This delay in drug action is modeled through a series of delay compartments through which the cells transit before elimination. In this note, we propose an alternative mathematical formulation of the Simeoni model using delay differential equations, without recourse to a series of delay compartments. We describe the two models in the next section, and then compare their performances with experimental data relating to mammary tumor growth in a mouse model. We conclude that one should be cautious if asserting that a particular tumor growth model uniquely characterizes experimental data.
Methods
The Simeoni model
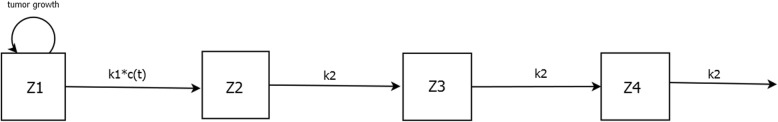
The Simeoni model is depicted in Fig. 1. The central compartment Z1 represents the actively growing tumor, which increases according to a growth function TGF. c(t) denotes the concentration of a chemotherapeutic or immunotherapeutic agent in the central compartment Z1 at time t. In turn, c(t) induces a fraction of tumor cells to commit to cell death with a killing constant k1. Tumor cells damaged by pharmacological treatment are shunted off successively into the peripheral compartments Z2, Z3, Z4 with a rate constant k2, followed by elimination representing cell death. The observed tumor volume is the sum of cells in compartments Z1, Z2, Z3, Z4. The system of differential equations prescribing the Simeoni model is as follows:
| 1 |
with initial conditions Z1(0) = V0, Z2(0) = Z3(0) = Z4(0) = 0. Total tumor volume is
| 2 |
and the tumor growth function TGF(t) is given by
| 3 |
Fig. 1.
Schematic representation of the Simeoni tumor growth model. The tumor resides in compartment Z1, with growth described by a tumor growth function. c(t) denotes the plasma concentration of an anticancer agent if present. The drug elicits its effect decreasing the tumor growth rate by a factor proportional to c(t)*Z1(t) through the constant parameter k1. Tumor cells cycle successively through transit compartments Z2, Z3, Z4 before cell death. k2 is a first-order rate constant of transit. The number of transit compartments is arbitrary. The system of ordinary differential equations describing this model is given in the text
Although we have depicted the Simeoni model with 3 peripheral compartments, we note that the number of peripheral compartments is in general arbitrary.
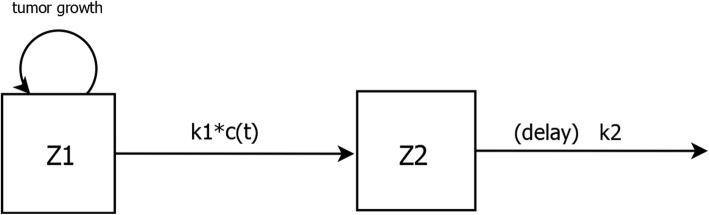
The model incorporating a delay differential equation is conceptually similar to the Simeoni model, the only difference being that the compartments Z2, Z3, and Z4 are replaced by a single compartment that incorporates a delay in elimination (Fig. 2). The system of differential equations describing this model is:
| 4 |
Fig. 2.
Schematic representation of the Simeoni tumor growth model, with transit compartments replaced by a single compartment in which tumor cell death is delayed relative to drug treatment. The delay is explicitly incorporated into the system of ordinary differential equations describing this model
where
TGF(t) and c(t) are as before, V(t) = Z1(t) + Z2(t), and initial conditions are Z1(0) = V0, Z2(0) = 0, and Z2(t) = 0 for t < 0. The variable t2 represents the time delay before elimination.
In the absence of chemotherapeutic or immunotherapeutic intervention (i.e., c(t) = 0), tumor growth in the Simeoni formulation reduces to the biphasic process TGF(t), characterized by initial exponential growth followed by linear growth. In particular, in contrast to more classic models of tumor growth, e.g. logistic, Gompertz, Von Bertalanffy, there is no plateau or upper limit of tumor size. One of these alternative models might credibly be more appropriate than TGF in certain experimental settings. The generalized logistic model is defined by
| 5 |
the Gompertz model is given by
| 6 |
and the Von Bertalanffy model is
| 7 |
These three models have analytic solutions [10], which might aid in nonlinear fitting to experimental data.
In vivo tumor growth experiments
A series of experiments was undertaken at PRISM, the goal being to establish a viable and reproducible mouse model for mammary tumors. The FVB/N-Tg(MMTVneu)202Mul/J model was used for this purpose (Jackson stock #002376). All mice were bred at PRISM, with female mice genotyped by polymerase chain reaction to confirm transgene expression. Focal mammary tumors appear in female mice at about 100 days, reset as day 0 to mark the onset of the experiment. Data from one particular experiment are considered in the present work. In this experiment, 40 mice were randomly assigned to either no treatment (n = 21) or a single dose of cisplatin (5 mg/kg, n = 19) on day 0; all animals were relatively healthy and functioning on day 0, with all tumors smaller than 700 mm3 at initiation. The primary experimental outcome consisted of the series of tumor measurements from each animal, which were taken daily (excepting weekends, holidays) by a single individual (TJF) using digital calipers, and tumor volumes were recorded using the approximating formula V = (Length x Width2)/2, where L > W. [In this approximating formula, tumor shape is taken as an ellipsoid generated from the rotation of a semi-ellipsis around its larger axis (length), and a multiplicative constant involving p is ignored.] Mice were euthanized when tumors reached a volume of ~ 2500 mm3 (or measured 17 mm in length + width + height), or for complications, e.g., interference, ulceration. The method of euthanization was CO2 narcosis followed by cervical dislocation, a method consistent with the 2013 recommendations of the American Veterinary Medical Association Panel on Euthanasia. The protocol for this study was approved by the Institutional Animal Care and Use Committee of Prism, confirmed by the National Institutes of Health Office of Laboratory Animal Welfare Assurance, #D16–00819. Animals were housed in Prism’s animal care facility.
All 40 animals initially randomized to the untreated and the cisplatintreated groups were included in the subsequent analyses. There were a total of 386 observations among the 21 control animals, and 545 observations among the 19 treated animals.
Statistical methods
A nonlinear mixed effects (NLME) model was used to fit growth curves to the longitudinal tumor size data. In our context, a general form for NLME models would be:
where N is the number of individuals, ni is the number of observations for individual i, t is the regression variable time, and y are the observations (tumor volumes). The term f is the structural model, expressed from the systems of ordinary differential equations shown earlier.
The residual error is g(tij, φi, ζ)εij, where εij ∼ N(0, σ2) In our modeling,
we chose the function g to be a linear combination of a constant term and a term proportional to the structural model f with the additional parameters ζ = (a, b), that is, y = f + (a + b ∗ f)e.
The individual parameters φi are defined as follows:
where m is a p-dimensional vector of fixed population parameters, li is a p-vector of random effects, W is the p x p variance-covariance matrix of the random effects, and h is a fixed transformation. We assume that all of the model parameters are log-normally distributed among the individuals in each cohort, so that h(x) = exp(x).
Parameter estimation of the typical population values and the interindividual variability of each model parameter were estimated using Monolix 2019R1 (Lixoft, Antony, France). The Monolix suite of programs implements a stochastic approximation expectation maximization (SAEM) algorithm [11, 12] for maximum likelihood estimation of the model parameters. With the treated animals, following a single bolus injection of cisplatin we took c(t) to represent first order elimination, that is, a constant proportion of the drug is eliminated per unit time.
Model selection and comparison was based on the Akaike information criterion (AIC), the corrected Akaike information criterion (AICc), and the Bayesian information criterion (BIC). AICc is a modified version of the AIC, and includes a correction term for small sample sizes [13–15]:
| 8 |
where k denotes the number of free parameters, and n is the number of observations. In our setting, we also included variance components from the random effects in the parameter count.
Given a set of candidate models, each with a specific IC (AIC, AICc, BIC) value, we calculate IC model weights [15–17] for comparative purposes. We first compute for each model j the difference in IC relative to the IC of the best candidate model: Dj = ICj - min IC. We then obtain the model weights by transforming to the likelihood scale and normalizing:
| 9 |
These weights are measures of the strength of evidence. They sum to one, and reflect the probability of each model j given the data and the M candidate models [15, 17].
Results
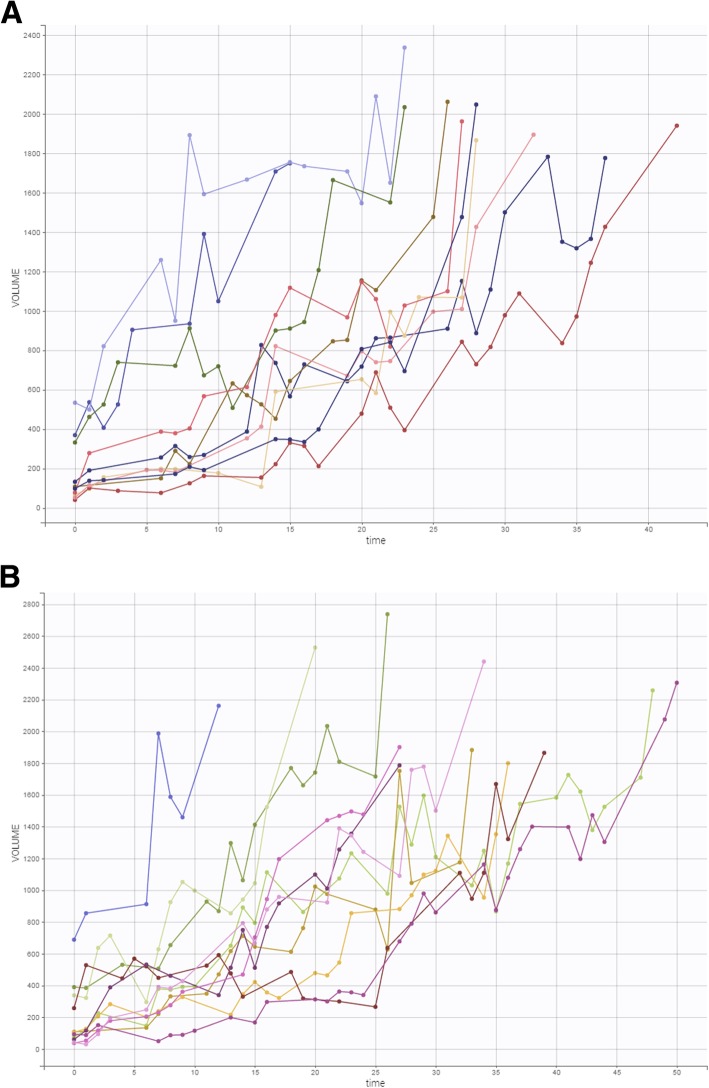
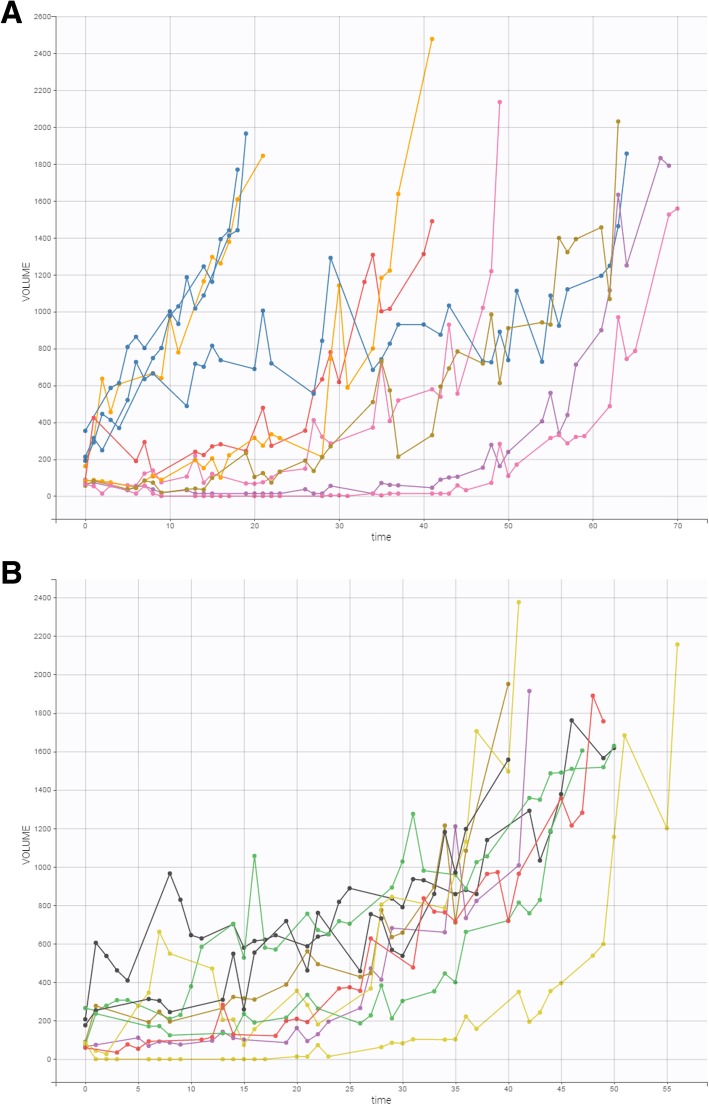
In the experiment outlined above, a total of 21 tumor-bearing mice were untreated. Tumor growth over time in this group of animals is shown in Fig. 3.
Fig. 3.
a, b Time course of tumor growth in 21 untreated tumor-bearing mice over the course of the experiment. The X-axis (time) denotes days, and the Y-axis (volume) denotes mm3
We use this cohort of controls to investigate various tumor growth functions, namely, generalized logistic, Gompertz, Von Bertalanffny, and Simeoni. We present summary data relative to these fits in Table 1. It turns out that the Simeoni tumor growth function provides the best fit to these data, with minimal AIC, AICc, and BIC. The weights convey the overwhelming preference for the Simeoni tumor growth function among the candidate functions.
Table 1.
Summary statistics relating to growth curve models fit to the control data
| model | − 2*LL | AIC | w(AIC) | AICc | w(AICc) | BIC | w(BIC) |
|---|---|---|---|---|---|---|---|
| generalized logistic | 5159.69 | 5179.69 | .0037 | 5180.07 | .0038 | 5190.13 | .0038 |
| Gompertz | 5182.79 | 5198.79 | 2.66E-07 | 5199.01 | 2.88E-07 | 5207.15 | 7.72E-07 |
|
von Bertalanffy |
5189.97 | 5209.97 | 9.94E-10 | 5210.35 | 9.95E-10 | 5226.42 | 5.05E-11 |
| Simeoni | 5148.52 | 5168.52 | .9963 | 5168.90 | .9963 | 5179.01 | .9962 |
Notes
LL log likelihood, AIC Akaike information criterion, w(AIC) weights derived from candidate model AIC values, AICc corrected Akaike information criterion, w(AICc) weights derived from candidate model AICc values, BIC Bayesian information criteron, w(BIC) weights derived from candidate model BIC values
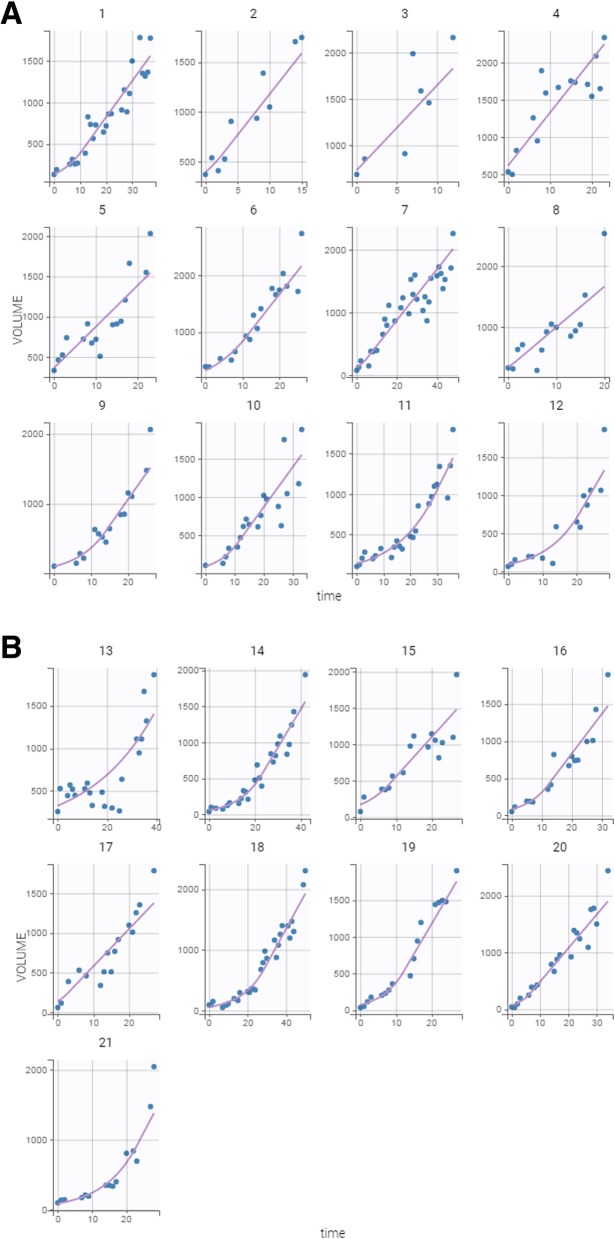
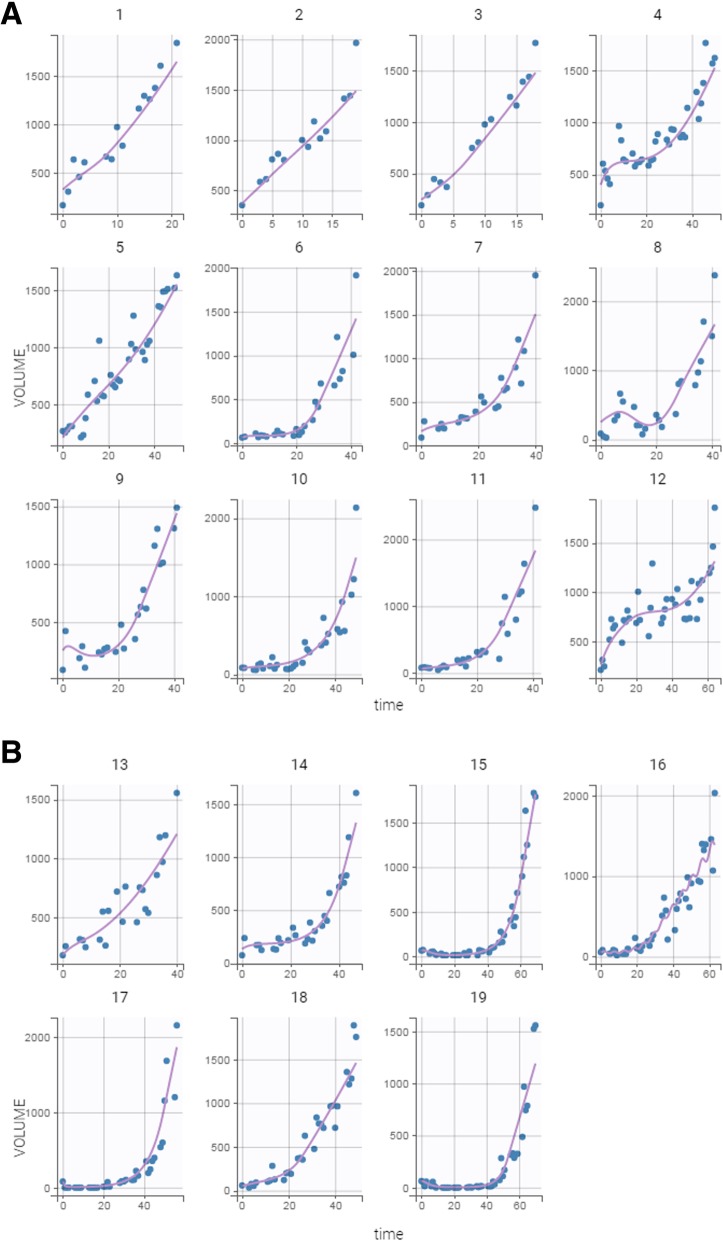
Plots of the individual fits with the Simeoni tumor growth function are shown in Fig. 4. Although the individual growth curves are quite heterogeneous (Fig. 3), the fitted curves quite nicely represent the observed data.
Fig. 4.
Observed tumor sizes and fitted values of the 21 untreated tumor-bearing mice over the course of the experiment. The Simeoni tumor growth function was fit to the tumor size data from the entire cohort of animals, and individual fits were then derived from the mixed model analysis undertaken in Monolix
Nineteen tumor-bearing mice were treated with cisplatin. Their individual growth patterns are shown in Fig. 5.
Fig. 5.
a, b Time course of tumor growth in 19 treated tumorbearing mice over the course of the experiment. Treatment consisted of a single dose of cisplatin (5 mg/kg) on day 0. The X-axis (time) denotes days, and the Y-axis (volume) denotes mm3
We used the Simeoni tumor growth function, and proceeded to fit the Simeoni model with varying numbers of peripheral delay compartments as well as the delay model to these data. Summary statistics are given in Table 2. Among the candidate models, the delay model evinces minimal AIC and AICc values, whereas the Simeoni model with 2 delay compartments minimizes the BIC. The weights reflect this model uncertainty: one might deem the delay model preferable to the Simeoni - 2 and Simeoni - 3 models relative to AIC or AICc, but this order is reversed with BIC. Regardless, the Simeoni - 1 model appears inferior to the other candidate models.
Table 2.
Summary statistics relating to growth curves models fit to the treated data
| model | -2*LL | AIC | w(AIC) | AICc | w(AICc) | BIC | w(BIC) |
|---|---|---|---|---|---|---|---|
| Simeoni -1 | 6786.12 | 6818.12 | .015 | 6818.91 | .016 | 6833.23 | .020 |
| Simeoni - 2 | 6780.10 | 6812.10 | .310 | 6812.89 | .325 | 6827.21 | .410 |
| Simeoni - 3 | 6780.32 | 6812.32 | .278 | 6813.11 | .291 | 6827.44 | .366 |
| delay | 6775.61 | 6811.61 | .397 | 6812.64 | .368 | 6828.61 | .204 |
Notes
1, 2, and 3 in the Simeoni model designations refer to the number of delay compartments incorporated into these models (Fig. 1). The delay model has one peripheral compartment, with an explicit delay in elimination (Fig. 2)
LL log likelihood, AIC Akaike information criterion, w(AIC) weights derived from candidate model AIC values, AICc corrected Akaike information criterion, w(AICc) weights derived from candidate model AICc values, BIC Bayesian information criteron, w(BIC) weights derived from candidate model BIC values
Plots of the individual fits with the delay model are shown in Fig. 6. Again, inter-animal variability is quite high, especially in terms of tumor response to treatment, with the individual fits modeling the observed data appropriately.
Fig. 6.
Observed tumor sizes and fitted values of the 19 treated tumor-bearing mice over the course of the experiment. A system of delay differential equations incorporating the Simeoni tumor growth function was fit to the tumor size data from the entire cohort of animals, and individual fits were then derived from the mixed model analysis undertaken in Monolix
Discussion
The Simeoni tumor growth function switches between exponential and linear growth, but without a plateau phase, in contrast to the classic tumor growth functions. It works quite well in our experimental setting, in which tumor-bearing mice were euthanized for ethical reasons when tumor volume reached the maximum allowable tumor mass, and this generally occurred before a plateau phase was reached. We caution, however, that in other experimental or clinical settings, the Simeoni tumor growth function might be supplanted by a more biologically realistic growth law [10, 18]. And, as a reviewer has pointed out, even the classical growth curve models logistic, Gompertz, and von Bertalanffy (our eqs. 5–7) enjoy many variations, specializations, and extensions, which might be judiciously employed to good effect in experimental settings.
The original Simeoni model essentially is a transit compartment model: tumor cells pass through progressive stages of damage because of the drug’s mechanism of action. Dying tumor cells stop proliferating and pass through several stages of dying (depicted by n delay compartments) before eventual death. However, there is some ambiguity over the exact number of delay compartments. With our experimental data, the appropriate number of delay compartments remains somewhat uncertain, relative to goodness of fit. Moreover, we have found that the delay compartments can effectively be replaced by a model incorporating a single peripheral compartment with a delay in elimination; that is, explicit incorporation of a delay term in the Simeoni tumor growth model can operationally replace the multiple transit compartments originally proposed, and effectively model the duration of the death process subsequent to drug action. Delay differential equation models have often been successfully investigated in tumor growth studies [19–24], and seem especially applicable in settings where tumor volume decrease appears delayed with respect to observed drug concentrations. With agents such as cisplatin, the delay in cell kill relative to the time course of extracellular exposure likely reflects the kinetics of cellular drug uptake and binding to intracellular targets [25]. Alternatively, more physiologically based models can ameliorate the ambiguity inherent with an arbitrary number of transit compartments [25–28].
With regard to model selection, we are more interested in the relative performances of the models rather than their absolute AIC, AICc, or BIC values. In this regard, the weights derived from the information criterion values quantify the probabilities of each model being optimal, given the data and the set of candidate models, hence provide useful criteria for model comparisons. There is little uncertainty about the suitability of the Simeoni growth function in Table 1, whereas the identifiability of the best approximating model in Table 2 is not incontrovertible. Regardless, the delay model is certainly competitive with the Simeoni model, and does provide excellent agreement with the experimental data, even with substantial tumor growth heterogeneity and inter-animal diversity in response.
Conclusions
One should exercise caution if asserting a particular mathematical model uniquely characterizes tumor growth curve data. The Simeoni ODE model of tumor growth is not unique in that alternative models can provide equivalent representations of tumor growth.
Acknowledgments
We than the reviewers for their insightful comments, which led to significant improvements in the manuscript.
Abbreviations
- AIC
Akaike information criterion
- AICc
Corrected Akaike information criterion
- BIC
Bayesian information criterion
- nlme
Nonlinear mixes effects
- ODEs
Ordinary differential equations
- pk/pd.
Pharmacokinetic/pharmacodynamic
- PRISM
Proteogenomics Research Institute for Systems Medicine
- SAEM
Stochastic approximation expectation maximization
Author contributions
JES conceived and designed the underlying animal studies, and directed the project. TJF assisted with planning the animal studies and carried out the implementation and data acquisition. JAK performed the mathematical analysis and drafted the manuscript. All authors read, edited, and approved the final manuscript.
Funding
This research is supported in part by grant 1PO1HL119165 from the National Institutes of Health. The funding agency played no role in the design of the study, collection, analysis, and interpretation of data, nor in writing the manuscript.
Availability of data and materials
The datasets analysed in this study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Institutional Animal Care and Use Committee of PRISM.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Simeoni Monica, Magni Paolo, Cammia Cristiano, De Nicolao Giuseppe, Croci Valter, Pesenti Enrico, Germani Massimiliano, Poggesi Italo, Rocchetti Maurizio. Predictive Pharmacokinetic-Pharmacodynamic Modeling of Tumor Growth Kinetics in Xenograft Models after Administration of Anticancer Agents. Cancer Research. 2004;64(3):1094–1101. doi: 10.1158/0008-5472.CAN-03-2524. [DOI] [PubMed] [Google Scholar]
- 2.Magni P., Simeoni M., Poggesi I., Rocchetti M., De Nicolao G. A mathematical model to study the effects of drugs administration on tumor growth dynamics. Mathematical Biosciences. 2006;200(2):127–151. doi: 10.1016/j.mbs.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 3.Simeoni Monica, De Nicolao Giuseppe, Magni Paolo, Rocchetti Maurizio, Poggesi Italo. Modeling of human tumor xenografts and dose rationale in oncology. Drug Discovery Today: Technologies. 2013;10(3):e365–e372. doi: 10.1016/j.ddtec.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Rocchetti M, Del Bene F, Germani M, Fiorentini F, Poggesi I, Pesenti E, Magni P, De Nicolao G. Testing additivity of anticancer agents in pre-clinical studies: a PK/PD modelling approach. Eur J Cancer. 2009;45:3336–3346. doi: 10.1016/j.ejca.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 5.Zhou R, Mazurchuk RV, Tamburlin JH, Harrold JM, Mager DE, Straubinger RM. Differential pharmacodynamic effects of paclitaxel formulations in an intracranial rat brain tumor model. J Pharmacol Exp Ther. 2010;332:479–488. doi: 10.1124/jpet.109.160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terranova Nadia, Germani Massimiliano, Del Bene Francesca, Magni Paolo. A predictive pharmacokinetic–pharmacodynamic model of tumor growth kinetics in xenograft mice after administration of anticancer agents given in combination. Cancer Chemotherapy and Pharmacology. 2013;72(2):471–482. doi: 10.1007/s00280-013-2208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li JY, Ren YP, Yuan Y, Ji SM, Zhou SP, Wang LJ, Mou ZZ, Li L, Lu W, Zhou TY. Preclinical PK/PD model for combined administration of erlotinib and sunitinib in the treatment of A549 human NSCLC xenograft mice. Acta Pharmacol Sin. 2016;37:930–940. doi: 10.1038/aps.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pigatto MC, Roman RM, Carrara L, Buffon A, Magni P, Dalla CT. Pharmacokinetic/pharmacodynamic modeling of etoposide tumor growth inhibitory effect in Walker−256 tumor-bearing rat model using free intratumoral drug concentrations. Eur J Pharm Sci. 2017;97:70–78. doi: 10.1016/j.ejps.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 9.Yates JWT, Holt SV, Armelle Logie A, et al. A pharmacokinetic–pharmacodynamic model predicting tumour growth inhibition after intermittent administration with the mTOR kinase inhibitor AZD8055. Br J Pharmacol. 2017;174:2652–2661. doi: 10.1111/bph.13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benzekry Sébastien, Lamont Clare, Beheshti Afshin, Tracz Amanda, Ebos John M. L., Hlatky Lynn, Hahnfeldt Philip. Classical Mathematical Models for Description and Prediction of Experimental Tumor Growth. PLoS Computational Biology. 2014;10(8):e1003800. doi: 10.1371/journal.pcbi.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delyon B, Lavielle M, Moulines E. Convergence of a stochastic approximation version of the EM algorithm. Ann Stat. 1999;27:94–128.
- 12.Comets E, Lavenu A, Lavielle M. Parameter estimation in nonlinear mixed effect models using saemix,an R implementation of the SAEM algorithm. Journal of Statistical Software. 2017. 10.18637/jss.v080.i03.
- 13.Sugiura Nariaki. Further analysts of the data by akaike' s information criterion and the finite corrections. Communications in Statistics - Theory and Methods. 1978;7(1):13–26. doi: 10.1080/03610927808827599. [DOI] [Google Scholar]
- 14.Hurvich Clifford M., Tsai Chih-Ling. Model Selection for Extended Quasi-Likelihood Models in Small Samples. Biometrics. 1995;51(3):1077. doi: 10.2307/2533006. [DOI] [PubMed] [Google Scholar]
- 15.Burnham KP, Anderson DR. Model selection and multi model inference: A practical information-theoretic approach. New York: Springer-Verlag; 2002.
- 16.Akaike H. On the likelihood of a time series model. The Statistician. 1974;27:217–235. doi: 10.2307/2988185. [DOI] [Google Scholar]
- 17.Wagenmakers E-J, Farrell S. AIC model selection using Akaike weights. Psychon Bull Rev. 2004;11:192–196. doi: 10.3758/BF03206482. [DOI] [PubMed] [Google Scholar]
- 18.Mould DR, Walz A-C, Lave T, Gibbs JP, Frame B. Developing Exposure/Response Models for Anticancer Drug Treatment: Special Considerations. CPT: Pharmacometrics & Systems Pharmacology. 2015;4(1):12–27. doi: 10.1002/psp4.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villasana Minaya, Radunskaya Ami. A delay differential equation model for tumor growth. Journal of Mathematical Biology. 2003;47(3):270–294. doi: 10.1007/s00285-003-0211-0. [DOI] [PubMed] [Google Scholar]
- 20.Cui Shangbin, Xu Shihe. Analysis of mathematical models for the growth of tumors with time delays in cell proliferation. Journal of Mathematical Analysis and Applications. 2007;336(1):523–541. doi: 10.1016/j.jmaa.2007.02.047. [DOI] [Google Scholar]
- 21.Bodnar M, Piotrowska MJ, Forys U. Gompertz model with delays and treatment: mathematical analysis. Math Biosci Eng. 2013;10:551–63. [DOI] [PubMed]
- 22.Xu Shihe, Wei Xiangqing, Zhang Fangwei. A Time-Delayed Mathematical Model for Tumor Growth with the Effect of a Periodic Therapy. Computational and Mathematical Methods in Medicine. 2016;2016:1–8. doi: 10.1155/2016/3643019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazlan MSA, Rosli N, Azmi NS. Modelling the cancer growth process by stochastic delay differential equations under Verhults and Gompertz’s law. Jurnal Teknologi. 2016;78:77–82.
- 24.Kim Peter S., Lee Peter P. Modeling Protective Anti-Tumor Immunity via Preventative Cancer Vaccines Using a Hybrid Agent-based and Delay Differential Equation Approach. PLoS Computational Biology. 2012;8(10):e1002742. doi: 10.1371/journal.pcbi.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Kareh Ardith W., Secomb Timothy W. A Mathematical Model for Cisplatin Cellular Pharmacodynamics. Neoplasia. 2003;5(2):161–169. doi: 10.1016/S1476-5586(03)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribba Benjamin, Watkin Emmanuel, Tod Michel, Girard Pascal, Grenier Emmanuel, You Benoît, Giraudo Enrico, Freyer Gilles. A model of vascular tumour growth in mice combining longitudinal tumour size data with histological biomarkers. European Journal of Cancer. 2011;47(3):479–490. doi: 10.1016/j.ejca.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Ribba B., Kaloshi G., Peyre M., Ricard D., Calvez V., Tod M., Cajavec-Bernard B., Idbaih A., Psimaras D., Dainese L., Pallud J., Cartalat-Carel S., Delattre J.-Y., Honnorat J., Grenier E., Ducray F. A Tumor Growth Inhibition Model for Low-Grade Glioma Treated with Chemotherapy or Radiotherapy. Clinical Cancer Research. 2012;18(18):5071–5080. doi: 10.1158/1078-0432.CCR-12-0084. [DOI] [PubMed] [Google Scholar]
- 28.Ribba B, Holford N H, Magni P, Trocóniz I, Gueorguieva I, Girard P, Sarr C, Elishmereni M, Kloft C, Friberg L E. A Review of Mixed-Effects Models of Tumor Growth and Effects of Anticancer Drug Treatment Used in Population Analysis. CPT Pharmacometrics Syst. Pharmacol. 2014;3(5):e113. doi: 10.1038/psp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analysed in this study are available from the corresponding author on reasonable request.