Abstract
The development of simple fluorescent and colorimetric assays that enable point-of-care DNA and RNA detection has been a topic of significant research because of the utility of such assays in resource limited settings. The most common motifs utilize hybridization to a complementary detection strand coupled with a sensitive reporter molecule. Here, a paper-based colorimetric assay for DNA detection based on pyrrolidinyl peptide nucleic acid (acpcPNA)-induced nanoparticle aggregation is reported as an alternative to traditional colorimetric approaches. PNA probes are an attractive alternative to DNA and RNA probes because they are chemically and biologically stable, easily synthesized, and hybridize efficiently with the complementary DNA strands. The acpcPNA probe contains a single positive charge from the lysine at C-terminus and causes aggregation of citrate anion-stabilized silver nanoparticles (AgNPs) in the absence of complementary DNA. In the presence of target DNA, formation of the anionic DNA-acpcPNA duplex results in dispersion of the AgNPs as a result of electrostatic repulsion, giving rise to a detectable color change. Factors affecting the sensitivity and selectivity of this assay were investigated, including ionic strength, AgNP concentration, PNA concentration, and DNA strand mismatches. The method was used for screening of synthetic Middle East respiratory syndrome coronavirus (MERS-CoV), Mycobacterium tuberculosis (MTB), and human papillomavirus (HPV) DNA based on a colorimetric paper-based analytical device developed using the aforementioned principle. The oligonucleotide targets were detected by measuring the color change of AgNPs, giving detection limits of 1.53 (MERS-CoV), 1.27 (MTB), and 1.03 nM (HPV). The acpcPNA probe exhibited high selectivity for the complementary oligonucleotides over single-base-mismatch, two-base-mismatch, and noncomplementary DNA targets. The proposed paper-based colorimetric DNA sensor has potential to be an alternative approach for simple, rapid, sensitive, and selective DNA detection.
Infectious diseases represent a major threat to human health in developed and developing countries alike. DNA alterations contribute to different types of diseases; therefore, the detection of specific DNA sequences plays a crucial role in the development method for early stage treatment and monitoring of genetic-related diseases. DNA diagnostics can provide sequence-specific detection, especially for single-nucleotide polymorphisms (SNPs),1 which critical for a range of applications including the diagnosis of human diseases and bacterial/viral infections.
Middle East respiratory syndrome (MERS), tuberculosis (TB), and cervical cancers related to human papilloma virus (HPV) are examples of infectious diseases caused by bacterial and viral infections that benefit greatly from DNA detection. TB is an infectious disease caused by mycobacteria, usually M. tuberculosis (MTB) in humans.2 HPV has been shown to be a major cause of cervical cancer.3 Middle East Respiratory Syndrome coronavirus (MERS-CoV) has recently emerged as an infectious disease with a high fatality rate in humans.4 Diagnostic methods developed for these infectious diseases include reverse transcription polymerase chain reaction (RT-PCR) for MERS-CoV,5 sputum smear microscopy, culture of bacilli, and molecular species diagnostics for MTB6−11 and Digene Hybrid Capture assay (HC2) and Pap smear test for HPV.12,13 While these techniques have been used for successful detection, they are difficult to implement in point-of-care clinical diagnostics particularly in developing countries lacking specialized medical facilities and skilled personnel. Therefore, simple, rapid, low-cost, and highly accurate on-site diagnostic platforms amenable to nucleic acid detection remain a challenge for early detection of infectious diseases for better patient management and infection control. Although DNA amplification is still needed with the current method to provide high sensitivity, we seek to further improve selectivity and assay simplicity to give immediate and quantitative responses in resource limited settings.
Paper-based analytical devices (PADs) are a point-of-use technology that recently received renewed interest because they are simple, inexpensive, portable, and disposable.14−16 To date, PADs have been extensively used for applications ranging from environmental analysis to clinical diagnostic assays.15,17,18 Colorimetric assays are particularly attractive when coupled with PADs due to their ease-of-use, lack of complicated external equipment and ability to provide semiquantitative results.19−21 Moreover, quantitative analysis of colorimetric assays can be accomplished using simple optical technologies such as digital cameras22−24 and office scanners20,25 combined with image processing software to carry out color, hue, and intensity measurements. In the field of clinical diagnostics, the advantages of simplicity, sensitivity, and low-cost are key reasons that make PADs coupled with colorimetric detection an effective diagnostic tool relative to traditional methods.
Colorimetric assays based on the aggregation of silver (AgNPs) and gold nanoparticles (AuNPs) have attracted increasing attention in biomedical applications. The optical properties of these nanomaterials depend on their size and shape.26−31 AgNPs are known to have a higher extinction coefficient compared to AuNPs,32−34 leading to improved optical sensitivity. Chemical reduction of silver salts is frequently used to synthesize AgNPs; while specific control of shape and size distribution is achieved by varying the reducing agents and stabilizers.35−37 Among stabilizing agents, negatively charged citrate has been widely used.38,39 Recently, colorimetric assays based on AgNPs aggregation for DNA detection has been reported.34 Colorimetric DNA detection using AgNPs usually involves modifying the particles with a DNA probe and mixing them with the DNA target containing the complementary sequence. When the hybridization of probe and target DNA occurs, the AgNPs aggregate and change color.33,34 The assay principal has been further adopted using charge-neutral peptide nucleic acids (PNA)40,41 as the hybridization agent. PNA causes aggregation of metal nanoparticles in solution without immobilization, thus, simplifying the assay.42,43 Finally, PNA-based nanoparticle aggregation assays also provide a high hybridization efficiency of PNA-DNA duplexes leading to a rapid color change.43
Recently, Vilaivan’s group proposed a new conformationally constrained pyrrolidinyl PNA system which possesses an α,β-peptide backbone derived from d-proline/2-aminocyclopentanecarboxylic acid (known as acpcPNA).44,45 Compared to Nielsen’s PNA,40 acpcPNA exhibits a stronger affinity and higher sequence specificity binding to DNA. acpcPNA exhibits the characteristic selectivity of antiparallel binding to the target DNA and low tendency to self-hybridize. Moreover, the nucleobases and backbone of acpcPNA can be modified to increase molecular functionality. These combined properties make acpcPNA an attractive candidate as a probe for biological applications.46−48
Here, the multiplex colorimetric PAD for DNA detection based on the aggregation of AgNPs induced by acpcPNA is reported. acpcPNA bearing a positively charged lysine modification at C-terminus was designed as the probe. The cationic PNA probe can interact with the negatively charged AgNPs leading to nanoparticle aggregation and a significant color change. This proposed sensor was used for simultaneous detection of MERS-CoV, MTB, and HPV. The developed paper-based DNA sensor has potential as an alternative diagnostic device for simple, rapid, sensitive, and selective DNA/RNA detection.
Experimental Section
Chemicals and Materials
Analytical grade reagents, including AgNO3, NaBH4, and sodium citrate from Sigma-Aldrich, KH2PO4 and KCl from Fisher Scientific, Na2HPO4 from Mallinckrodt, and NaCl from Macron, were used without further purification. A total of 18 M Ω·cm–1 resistance water was obtained from a Millipore Milli-Q water system. Synthetic DNA oligonucleotides were obtained from Biosearch Technologies. The sequences of DNA oligonucleotides are shown in Table 1.
Table 1. List of Oligonucleotide Used in This Study.
| oligonucleotide | sequence (5′-3′) |
|---|---|
| MERS-CoV | |
| complementary DNA | 5′-CGATTATGTGAAGAG-3′ |
| two-base-mismatch | 5′-CGATTATCTGAGGAG-3′ |
| noncomplementary DNA | 5′-TTCGCACAGTGGTCA-3′ |
| MTB | |
| complementary DNA | 5′-ATAACGTGTTTCTTG-3′ |
| single-base-mismatch | 5′-ATAACGTCTTTCTTG-3′ |
| noncomplementary DNA 1 | 5′-TGGCTAGCCGCTCCT-3′ |
| noncomplementary DNA 2 | 5′-CACTTGCCTACACCA-3′ |
| HPV | |
| complementary DNA (HPV type 16) | 5′-GCTGGAGGTGTATG-3′ |
| noncomplementary DNA 1 (HPV type 18) | 5′-GGATGCTGCACCGG-3′ |
| noncomplementary DNA 2 (HPV type 31) | 5′-CCAAAAGCCCAAGG-3′ |
| noncomplementary DNA 3 (HPV type 33) | 5′-CACATCCACCCGCA-3′ |
Synthesis of AgNPs
The AgNPs were synthesized using the citrate-stabilization method.49 Briefly, 4 mL of 12.6 mM sodium citrate and 50 mL of 0.3 mM AgNO3 were mixed together. Then, 1 mL of 37 mM NaBH4 was added to the mixture under vigorous stirring and the solution turned yellow. The formation of AgNPs and their size distribution were verified by dynamic light scattering measurement, and the average size of AgNPs was found to be 19 nm (Figure S1).
Synthesis of acpcPNA Probes
The acpcPNA probes were designed to detect the synthetic oligonucleotide targets with sequences corresponding to MERS-CoV, MTB, and HPV type 16. The sequences of acpcPNA probes are as follows:
MERS-CoV: CTCTTCACATAATCG-LysNH2
MTB: CAAGAAACACGTTAT-LysNH2
HPV type 16: CATACACCTCCAGC-LysNH2
*(written in the N → C direction)
The acpcPNA probe was synthesized by solid-phase peptide synthesis using Fmoc chemistry, as previously described.44 At the C-terminus, lysinamide was included as a positively charged group that could induce nanoparticle aggregation. All PNA were purified by reverse-phase HPLC (C18 column, 0.1% (v/v) trifluoroacetic acid (TFA) in H2O–MeOH gradient). The identity of the acpcPNA was verified by MALDI-TOF MS analysis (Figure S2), and the purity was confirmed to be >90% by reverse-phase HPLC.
Design and Operation of Paper-Based Multiplex DNA Sensor
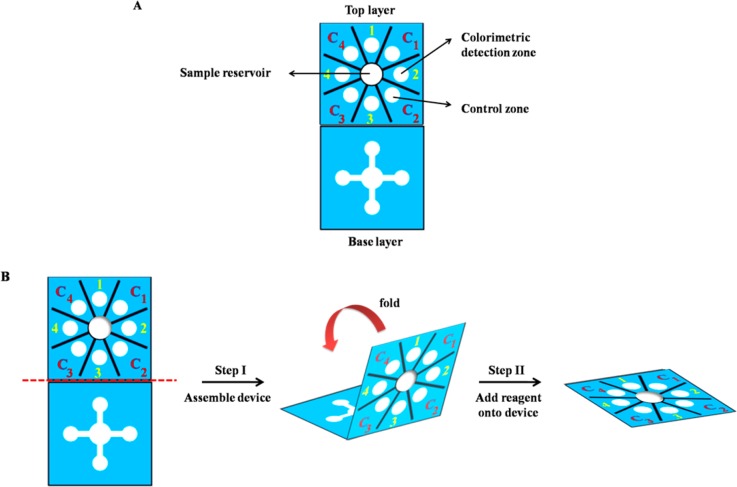
A wax-printing technique was used to create PADs.50 The sensor was designed using Adobe Illustrator. The wax colors were selected to be complementary to the colorimetric reactions to enhance visualization. For paper-based device fabrication, the wax design was printed onto Whatman grade 1 filter paper (VWR) using a wax printer (Xerox Phaser 8860). The wax pattern was subsequently melted at 175 °C for 50 s to generate the hydrophobic barriers and hydrophilic channels. The sensor was based on Origami concept consisting of two layers.51,52 As shown in Scheme 1A, the base layer contains four wax-defined channels extending outward from the sample reservoir (6 mm i.d.) and the top layer contains four detection and control zones (4 mm i.d.). Scheme 1B illustrates operation of the multiplex sensor. First, the sample reservoir of the top layer was punched to provide a solution connection directly from the top to the bottom layer, and then the device was assembled by folding the top layer over the base layer to create the three-dimension origami paper-based device. A polydimethylsiloxane (PDMS) lid was used for holding the two layers together. The lid consisting of one 6 mm diameter hole over the sample reservoir and eight 4 mm diameter holes over the colorimetric detection and control zones was aligned over the device to provide consistent pressure across the surface of the device. Next, the acpcPNA probe and AgNPs solution were added onto the detection and control zones. Finally, the sample solution was added onto the sample reservoir and flow through the channels to wet the colorimetric detection zones.
Scheme 1. (A) Design and (B) Operation of Multiplex Paper-Based Colorimetric Device.
Colorimetric Detection of MERS-CoV, MTB, and HPV DNA Target
According to the concept of PNA-induced AgNPs aggregation,42,43 acpcPNA was designed as a specific probe for quantitative detection of synthetic MERS-CoV, MTB, and HPV DNA targets. For colorimetric detection, the detection zone was prepared by adding 10 μL of AgNPs in 0.1 M phosphate buffer saline (PBS) pH 7.4 in a ratio of 5:1 (AgNPs: PBS), followed by 1 μL of specific acpcPNA probe. Control zones were prepared using the same conditions as the colorimetric detection zones. Next, 25 μL of DNA target was added to the open sample reservoir. Upon sample addition, solution moved outward through the channels to wet the colorimetric detection zone of the top layer. Finally, the AgNPs aggregation occurred and the color intensity was measured.
Image Processing
The detection images were recorded using a scanner (XEROX DocuMate 3220) and saved in JPEG format at 600 dpi. ImageJ software (National Institutes of Health) was used to analyze the mean intensity of the color for each colorimetric reaction zone by applying a color threshold window for removing the blue background. Images were then inverted, and the mean intensity was measured.20,53
Results and Discussion
acpcPNA-Induced AgNPs Aggregation
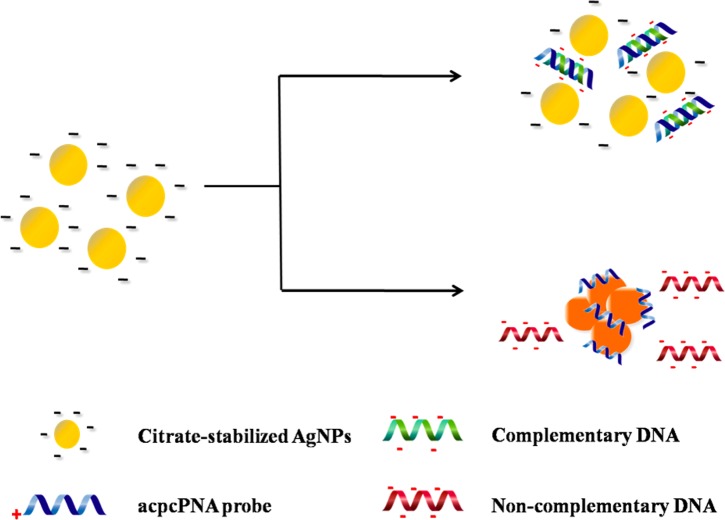
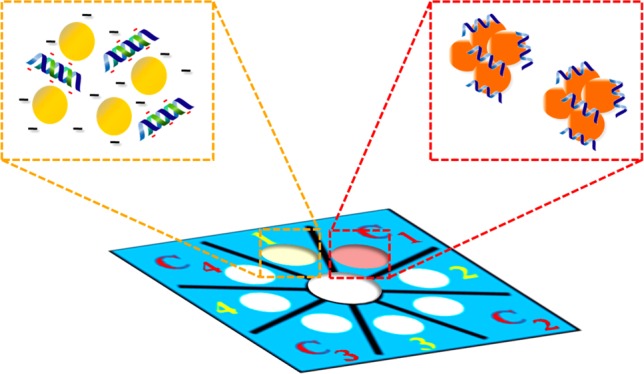
The process of acpcPNA-induced AgNPs aggregation is shown in Scheme 2. The anionic AgNPs are initially well dispersed due to electrostatic repulsion. On addition of the cationic acpcPNA, the electrostatic repulsion is shielded, resulting in nanoparticle aggregation. When complementary DNA (DNAcom) is present, the specific PNA–DNA interaction outcompetes the less specific PNA–AgNPs interaction, resulting in a negatively charged PNA–DNAcom duplex and deaggregation of the anionic nanoparticles. Upon addition of noncomplementary DNA (DNAnc), the acpcPNA should remain bound to the AgNPs and no color change occurs. To prove the concept, we designed and synthesized acpcPNA probes to detect synthetic oligonucleotide targets with sequences corresponding to MERS-CoV, MTB, and HPV type 16. The photographs of the results are shown in Figure 1. The yellow AgNPs turned red when the acpcPNA was added. When the solution contained of the acpcPNA and DNAnc, the color also changed to red due to aggregation of the AgNPs. On the other hand, the color changed from red (aggregated) to yellow (nonaggregated) in the presence of DNAcom, with the intensity dependent on the DNA concentration. Next, the sequence of adding the PNA probe and DNA target was investigated. As shown in Figure S3, when equimolar DNAcom was added either before or after the addition of acpcPNA probe into the AgNPs, the same color intensities were obtained indicating that the sequence of adding acpcPNA and DNAcom did not impact the final signal.
Scheme 2. Process of acpcPNA-Induced AgNP Aggregation in the Presence of DNAcom and DNAnc.
Figure 1.
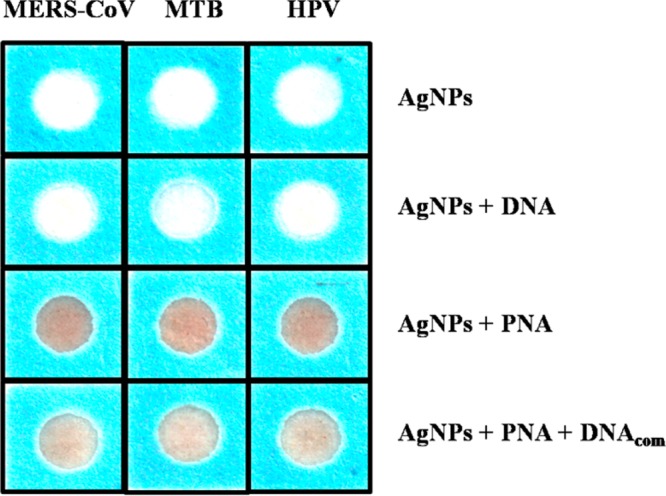
Photograph of visual color changes obtained from detection of MERS-CoV, MTB, and HPV in the presence of DNAcom.
Critical Coagulation Concentration (CCC)
The influence of electrolyte solution on the aggregation behavior of citrate-stabilized AgNPs was investigated based on the CCC.54The CCC represents the electrolyte concentration required to cause aggregation of the nanoparticles in the absence of acpcPNA. In Figure S4, the color intensity of citrate-stabilized AgNPs in the absence of acpcPNA probe is shown as a function of NaCl concentration. The intensity and, therefore, the degree of aggregation, increased with the concentration of NaCl, indicating that increasing ionic strength led to enhanced aggregation.55 We believe that the ionic strength can decrease the electrostatic repulsion of citrate-stabilized AgNPs as a result of shielding, accelerating the AgNPs aggregation. The CCC was obtained when the degree of aggregation reached a maximum and became independent of NaCl concentration. In this experiment, the CCC of citrate-stabilized AgNPs was found to be 30 mM. Above this concentration, PNA-induced aggregation was not observed.
Optimization of Assay Parameters
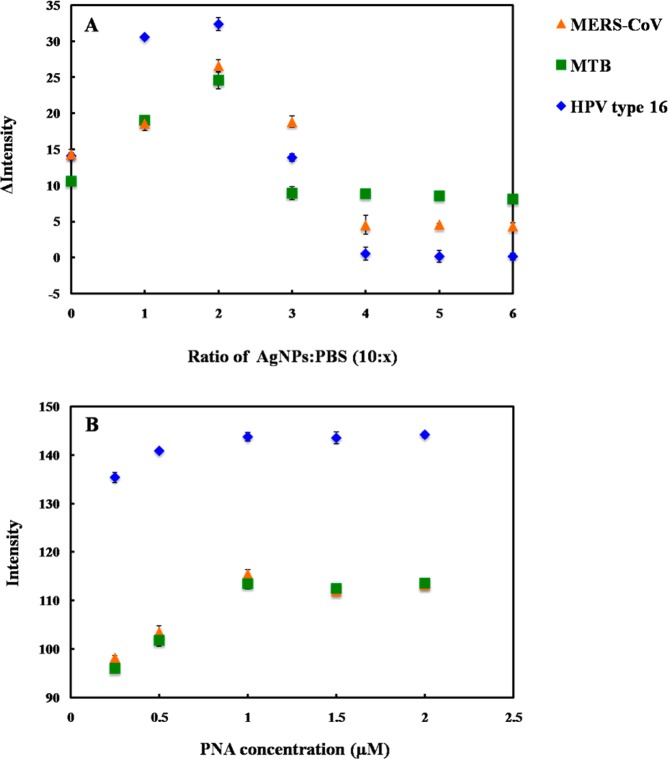
For a colorimetric assay based on acpcPNA-induced AgNPs aggregation, assay parameters including 0.1 M PBS (pH 7.4) ratio and acpcPNA concentration were optimized using a simple paper-based design. The degree of AgNPs aggregation was determined by measuring the color intensity of the resulting solution in the presence of acpcPNA without target DNA. First, the impact of the PBS concentration on AgNPs aggregation was measured. The differential color intensity (Δ intensity, ΔI) obtained before and after addition of acpcPNA as a function of AgNPs to PBS ratio is shown in Figure 2A. ΔI increased until the ratio of AgNPs/PBS reached 5:1 and then decreased until it plateaued at 5:2. Thus, the ratio of 5:1 AgNPs/PBS was selected as the optimal condition because it gave the largest ΔI. Another important aspect for the DNA assay is probe concentration. The influence of acpcPNA probe concentration on absolute intensity was studied. As shown in Figure 2B, the acpcPNA concentration was varied within a range of 0–2.5 μM, and the highest aggregation was obtained at the concentration of 1.0 μM. At this concentration, the aggregation became independent of acpcPNA concentration, which was desirable for simplifying the assay. Higher concentrations of AgNPs were not tested in order to minimize reagent consumption. As a result, the optimal conditions consisting of AgNPs/PBS ratio of 5:1 and acpcPNA concentration of 1.0 μM were selected for further experiments.
Figure 2.
Influence of (A) AgNPs/PBS ratio and (B) acpcPNA probe concentration on color intensity for MERS-CoV, MTB, and HPV detection. The error bars represent one standard deviation (SD) obtained from three independent measurements (n = 3).
Selectivity of MERS-CoV, MTB, and HPV Detection
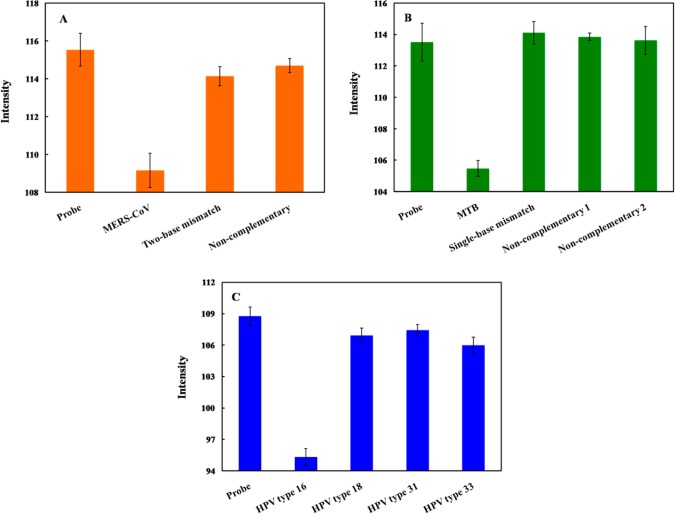
To investigate the selectivity of this system, the color intensity obtained from the DNAcom of MERS-CoV, MTB, and HPV was compared to that of single-base mismatch (DNAm1), two-base mismatch (DNAm2), and DNAnc sequences. The color intensity decreased significantly in the presence of DNAcom; whereas, the intensity did not change for the mismatched and noncomplementary targets (Figure 3). We believe the affinity of PNA–DNA hybridization was reduced due to the contribution of one- and two-base mismatches, leaving free PNA to aggregate the nanoparticles. PNA–DNAcom complex can retard the ability of PNA to induce AgNPs aggregation as discussed above and result in different color intensities. These results suggest that the fully complementary DNA selectively hybridized the acpcPNA probe and yielded measurable signals. In addition, bovine serum albumin (BSA), which is commonly used in cell culture protocols, was used to investigate the protein interference of the proposed system. The DNA target was prepared in the presence of 3% BSA solution. It was observed that the color intensities of the DNA targets for MERS-CoV, MTB, and HPV in 3% BSA solution were statistically identical to the ones without BSA (Figure S5). Hence, common proteins should not negatively affect the analysis of this system.
Figure 3.
Color intensity of (A) MERS-CoV, (B) MTB, and (C) HPV detection after hybridization of DNAm1, DNAm2, and DNAnc. The error bars represent one standard deviation (SD) obtained from three independent measurements (n = 3).
Analytical Performance
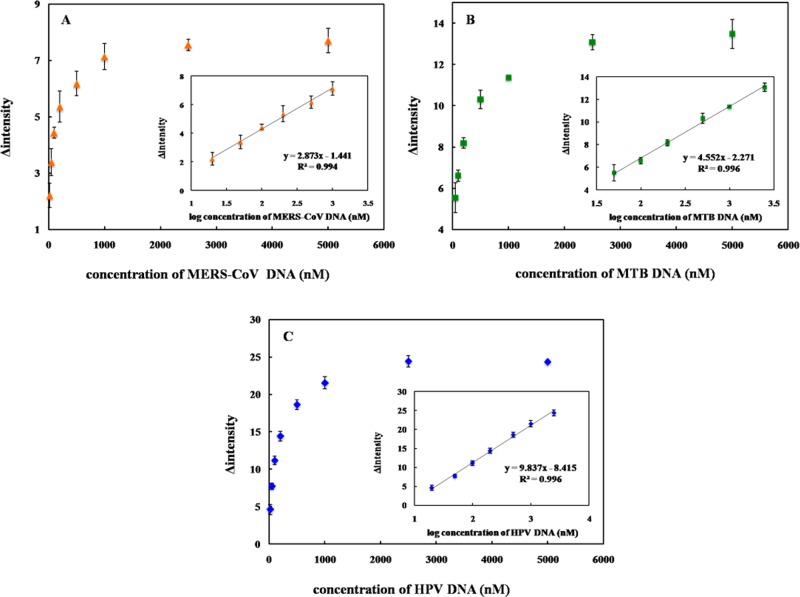
To assess the sensitivity of the proposed method for DNA quantification, the intensity as a function of the target DNA concentration was determined. The color intensity decreases with the target DNA concentration. The calibration curves for each species are shown in Figure 4A, B, and C for MERS-CoV, MTB, and HPV, respectively. The linear range for each DNA target using a logarithmic DNA concentration and color intensity (Figure 4, inset) was also obtained. The analytical performances for all three DNA targets are summarized in Table 2. It can be seen that a sufficiently low detection limit could be obtained for MERS-CoV, MTB, and HPV detection without the need for multiple PCR cycles. Moreover, this multiplex system can provide sensitive and selective detection for simultaneous analysis of multiple DNA targets in a single device, which simplifies the analysis compared to traditional diagnostics.9,56−59
Figure 4.
Change of probe color intensity vs DNA target concentration (ΔI) and calibration graph between ΔI and log DNA target concentration (inset) for (A) MERS-CoV, (B) MTB, and (C) HPV detection. The error bars represent standard deviation (SD) obtained from three independent measurement (n = 3).
Table 2. Summarized Analytical Performance of the Multiplexed 3DPAD for Colorimetric DNA Assay.
| DNA target | linearity (nM) | LOD (nM) | %RSD (n = 3) |
|---|---|---|---|
| MERS-CoV | 20–1000 | 1.53 | 0.17–0.50 |
| MTB | 50–2500 | 1.27 | 0.12–0.67 |
| HPV | 20–2500 | 1.03 | 0.43–0.93 |
Device Design
Next, a multiplex device (Scheme 1) was designed for simultaneous detection of MERS-CoV, MTB, and HPV. The top layer contained four detection zones and four control zones. Each zone contained AgNPs with a single acpcPNA probe to provide selectivity for DNA. The base layer contained four wax-defined channels extending outward from a sample inlet. After the device was folded and stacked together, the channels of the base layer were connected to four detection zones of the top layer. Upon sample addition, the solution moved outward through the channels of the base layer to wet the colorimetric detection zones of the top layer and lead to color change. Figure 5 illustrates the ability of the proposed sensor for detection of 100 nM MERS-CoV, MTB, and HPV. Only the colorimetric detection zones that contained the selective probes changed color compared to their control zones. This result indicated that the multiplex paper-based colorimetric sensor is promising for simultaneous determination of MERS-CoV, MTB, and HPV.
Figure 5.
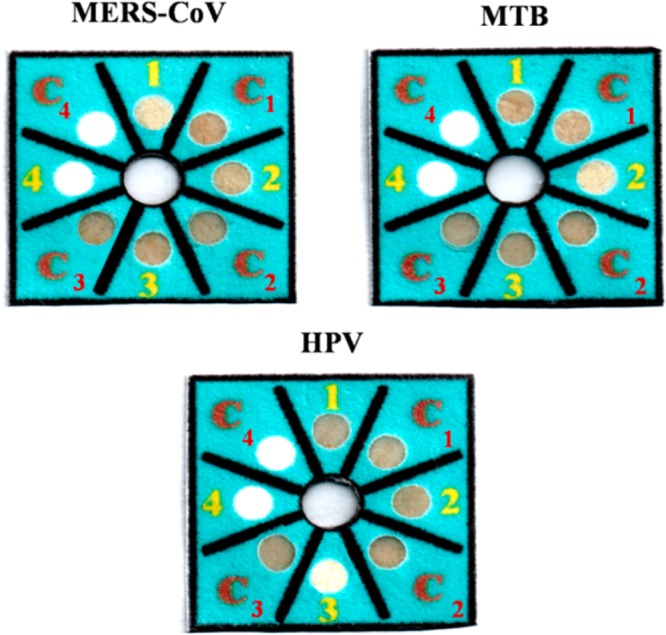
Selectivity of 100 nM MERS-CoV, MTB, and HPV detection using multiplex colorimetric PAD (1, C1= AgNPs + MERS-CoV acpcPNA probe; 2, C2 = AgNPs + MTB acpcPNA probe; 3, C3 = AgNPs + HPV acpcPNA probe).
Conclusions
A multiplex colorimetric PAD was developed for simultaneous detection of DNA associated with viral and bacterial infectious diseases, including Middle East respiratory syndrome coronavirus (MERS-CoV), Mycobacterium tuberculosis (MTB), and human papillomavirus (HPV). AgNPs were used as a colorimetric reagent for DNA detection based on acpcPNA-induced nanoparticle aggregation. This colorimetric DNA sensor exhibited high selectivity against single-base mismatch, two-base mismatch and noncomplementary target DNA. Under the optimized condition, the limit of detection for MERS-CoV, MTB, and HPV were found to be 1.53, 1.27, and 1.03 nM, respectively. As a result, this developed multiplex colorimetric PAD could be a low-cost and disposable alternative tool for rapid screening and detecting in infectious diagnostics.
Acknowledgments
P.T. gratefully appreciates the financial supports from Thailand Graduate Institute of Science and Technology (TGIST 01-55-014) and The Thailand Research Fund (RTA5780005). C.S.H. acknowledges financial support from Colorado State University and the United States Department of Agriculture through the National Wildlife Research Center (1574000859CA). T.V. acknowledges technical assistance of Ms. Chotima Vilaivan (Organic Synthesis Research Unit, Chulalongkorn University) and the financial support from Thailand Research Fund (DPG5780002, to T.V.) for the PNA synthesis. The authors thank Dr. Yuanyuan Yang for assistance with manuscript editing. The authors also acknowledge important discussions with Dr. Christopher Ackerson surrounding the critical coagulation concentration.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.7b00255.
Supporting Figures S1–S5 (PDF).
The authors declare no competing financial interest.
Supplementary Material
References
- Wei F.; Lillehoj P. B.; Ho C.-M. DNA Diagnostics: Nanotechnology-Enhanced Electrochemical Detection of Nucleic Acids. Pediatr. Res. 2010, 67, 458–468. 10.1203/PDR.0b013e3181d361c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I. Mycobacterium tuberculosis Pathogenesis and Molecular Determinants of Virulence. Clin. Microbiol. Rev. 2003, 16, 463–496. 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd E. M. Human Papillomavirus and Cervical Cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E.; Rasmussen A. L.; Falzarano D.; Bushmaker T.; Feldmann F.; Brining D. L.; Fischer E. R.; Martellaro C.; Okumura A.; Chang J.; Scott D.; Benecke A. G.; Katze M. G.; Feldmann H.; Munster V. J. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 16598–16603. 10.1073/pnas.1310744110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S.; Jiang Y. S.; Kumar M. R.; Johnson R. F.; Hensley L. E.; Ellington A. D. Real-Time Sequence-Validated Loop-Mediated Isothermal Amplification Assays for Detection of Middle East Respiratory Syndrome Coronavirus (MERS-CoV). PLoS One 2015, 10, e0123126. 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies P. D. O.; Pai M. The diagnosis and misdiagnosis of tuberculosis. International Journal of Tuberculosis and Lung Disease 2008, 12, 1226–1234. [PubMed] [Google Scholar]
- Steingart K. R.; Ng V.; Henry M.; Hopewell P. C.; Ramsay A.; Cunningham J.; Urbanczik R.; Perkins M. D.; Aziz M. A.; Pai M. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect. Dis. 2006, 6, 664–674. 10.1016/S1473-3099(06)70602-8. [DOI] [PubMed] [Google Scholar]
- Lee J. J.; Suo J.; Lin C. B.; Wang J. D.; Lin T. Y.; Tsai Y. C. Comparative evaluation of the BACTEC MGIT 960 system with solid medium for isolation of mycobacteria. International Journal of Tuberculosis and Lung Disease 2003, 7, 569–574. [PubMed] [Google Scholar]
- Al-Zamel F. A. Detection and diagnosis of Mycobacterium tuberculosis. Expert Rev. Anti-Infect. Ther. 2009, 7, 1099–1108. 10.1586/eri.09.92. [DOI] [PubMed] [Google Scholar]
- Noordhoek G. T.; Kolk A. H.; Bjune G.; Catty D.; Dale J. W.; Fine P. E.; Godfrey-Faussett P.; Cho S. N.; Shinnick T.; Svenson S. B. Sensitivity and specificity of PCR for detection of Mycobacterium tuberculosis: a blind comparison study among seven laboratories. Journal of Clinical Microbiology 1994, 32, 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.-C.; Lu P.-L.; Huang S. C.; Jenh Y.-S.; Jou R.; Chang T. C. Evaluation of the Cobas TaqMan MTB Test for Direct Detection of Mycobacterium tuberculosis Complex in Respiratory Specimens. Journal of Clinical Microbiology 2011, 49, 797–801. 10.1128/JCM.01839-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lörincz A.; Anthony J. Advances in HPV detection by Hybrid Capture. Papillomavirus Report 2001, 12, 145–154. [Google Scholar]
- Gravitt P. E.; Jamshidi R. Diagnosis and Management of Oncogenic Cervical Human Papillomavirus Infection. Infectious Disease Clinics of North America 2005, 19, 439–458. 10.1016/j.idc.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Martinez A. W.; Phillips S. T.; Butte M. J.; Whitesides G. M. Patterned Paper as a Platform for Inexpensive, Low-Volume, Portable Bioassays. Angew. Chem., Int. Ed. 2007, 46, 1318–1320. 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate D. M.; Adkins J. A.; Mettakoonpitak J.; Henry C. S. Recent Developments in Paper-Based Microfluidic Devices. Anal. Chem. 2015, 87, 19–41. 10.1021/ac503968p. [DOI] [PubMed] [Google Scholar]
- Yetisen A. K.; Akram M. S.; Lowe C. R. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 2013, 13, 2210–2251. 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- Adkins J.; Boehle K.; Henry C. Electrochemical paper-based microfluidic devices. Electrophoresis 2015, 36, 1811–1824. 10.1002/elps.201500084. [DOI] [PubMed] [Google Scholar]
- Mettakoonpitak J.; Boehle K.; Nantaphol S.; Teengam P.; Adkins J. A.; Srisa-Art M.; Henry C. S. Electrochemistry on Paper-based Analytical Devices: A Review. Electroanalysis 2016, 28, 1420–1436. 10.1002/elan.201501143. [DOI] [Google Scholar]
- Nery E. W.; Kubota L. T. Sensing approaches on paper-based devices: a review. Anal. Bioanal. Chem. 2013, 405, 7573–7595. 10.1007/s00216-013-6911-4. [DOI] [PubMed] [Google Scholar]
- Rattanarat P.; Dungchai W.; Cate D.; Volckens J.; Chailapakul O.; Henry C. S. Multilayer Paper-Based Device for Colorimetric and Electrochemical Quantification of Metals. Anal. Chem. 2014, 86, 3555–3562. 10.1021/ac5000224. [DOI] [PubMed] [Google Scholar]
- Liana D. D.; Raguse B.; Gooding J. J.; Chow E. Recent Advances in Paper-Based Sensors. Sensors 2012, 12, 11505. 10.3390/s120911505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apilux A.; Siangproh W.; Praphairaksit N.; Chailapakul O. Simple and rapid colorimetric detection of Hg(II) by a paper-based device using silver nanoplates. Talanta 2012, 97, 388–394. 10.1016/j.talanta.2012.04.050. [DOI] [PubMed] [Google Scholar]
- Apilux A.; Dungchai W.; Siangproh W.; Praphairaksit N.; Henry C. S.; Chailapakul O. Lab-on-Paper with Dual Electrochemical/Colorimetric Detection for Simultaneous Determination of Gold and Iron. Anal. Chem. 2010, 82, 1727–1732. 10.1021/ac9022555. [DOI] [PubMed] [Google Scholar]
- Chaiyo S.; Siangproh W.; Apilux A.; Chailapakul O. Highly selective and sensitive paper-based colorimetric sensor using thiosulfate catalytic etching of silver nanoplates for trace determination of copper ions. Anal. Chim. Acta 2015, 866, 75–83. 10.1016/j.aca.2015.01.042. [DOI] [PubMed] [Google Scholar]
- Cate D. M.; Nanthasurasak P.; Riwkulkajorn P.; L’Orange C.; Henry C. S.; Volckens J. Rapid Detection of Transition Metals in Welding Fumes Using Paper-Based Analytical Devices. Ann. Occup. Hyg. 2014, 58, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S.-Y.; Lim D.-K.; Nam J.-M. Ultrasensitive optical biodiagnostic methods using metallic nanoparticles. Nanomedicine 2008, 3, 215–232. 10.2217/17435889.3.2.215. [DOI] [PubMed] [Google Scholar]
- Baptista P.; Pereira E.; Eaton P.; Doria G.; Miranda A.; Gomes I.; Quaresma P.; Franco R. Gold nanoparticles for the development of clinical diagnosis methods. Anal. Bioanal. Chem. 2008, 391, 943–950. 10.1007/s00216-007-1768-z. [DOI] [PubMed] [Google Scholar]
- Zhao W.; Brook M. A.; Li Y. Design of Gold Nanoparticle-Based Colorimetric Biosensing Assays. ChemBioChem 2008, 9, 2363–2371. 10.1002/cbic.200800282. [DOI] [PubMed] [Google Scholar]
- Thaxton C. S.; Georganopoulou D. G.; Mirkin C. A. Gold nanoparticle probes for the detection of nucleic acid targets. Clin. Chim. Acta 2006, 363, 120–126. 10.1016/j.cccn.2005.05.042. [DOI] [PubMed] [Google Scholar]
- Li H.; Cui Z.; Han C. Glutathione-stabilized silver nanoparticles as colorimetric sensor for Ni2+ ion. Sens. Actuators, B 2009, 143, 87–92. 10.1016/j.snb.2009.09.013. [DOI] [Google Scholar]
- Vilela D.; González M. C.; Escarpa A. Sensing colorimetric approaches based on gold and silver nanoparticles aggregation: Chemical creativity behind the assay. A review. Anal. Chim. Acta 2012, 751, 24–43. 10.1016/j.aca.2012.08.043. [DOI] [PubMed] [Google Scholar]
- Wei H.; Chen C.; Han B.; Wang E. Enzyme Colorimetric Assay Using Unmodified Silver Nanoparticles. Anal. Chem. 2008, 80, 7051–7055. 10.1021/ac801144t. [DOI] [PubMed] [Google Scholar]
- Lee J.-S.; Lytton-Jean A. K. R.; Hurst S. J.; Mirkin C. A. Silver Nanoparticle–Oligonucleotide Conjugates Based on DNA with Triple Cyclic Disulfide Moieties. Nano Lett. 2007, 7, 2112–2115. 10.1021/nl071108g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. G.; Enright A.; Faulds K.; Smith W. E.; Graham D. Ultrasensitive DNA Detection Using Oligonucleotide–Silver Nanoparticle Conjugates. Anal. Chem. 2008, 80, 2805–2810. 10.1021/ac702403w. [DOI] [PubMed] [Google Scholar]
- Yeo S. Y.; Lee H. J.; Jeong S. H. Preparation of nanocomposite fibers for permanent antibacterial effect. J. Mater. Sci. 2003, 38, 2143–2147. 10.1023/A:1023767828656. [DOI] [Google Scholar]
- Chimentao R. J.; Kirm I.; Medina F.; Rodriguez X.; Cesteros Y.; Salagre P.; Sueiras J. E. Different morphologies of silver nanoparticles as catalysts for the selective oxidation of styrene in the gas phase. Chem. Commun. 2004, 846–847. 10.1039/B400762J. [DOI] [PubMed] [Google Scholar]
- He B.; Tan J. J.; Liew K. Y.; Liu H. Synthesis of size controlled Ag nanoparticles. J. Mol. Catal. A: Chem. 2004, 221, 121–126. 10.1016/j.molcata.2004.06.025. [DOI] [Google Scholar]
- Abou El-Nour K. M. M.; Eftaiha A. a.; Al-Warthan A.; Ammar R. A. A. Synthesis and applications of silver nanoparticles. Arabian J. Chem. 2010, 3, 135–140. 10.1016/j.arabjc.2010.04.008. [DOI] [Google Scholar]
- Iravani S.; Korbekandi H.; Mirmohammadi S. V.; Zolfaghari B. Synthesis of silver nanoparticles: chemical, physical and biological methods. Research in Pharmaceutical Sciences 2014, 9, 385–406. [PMC free article] [PubMed] [Google Scholar]
- Nielsen P.; Egholm M.; Berg R.; Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science (Washington, DC, U. S.) 1991, 254, 1497–1500. 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- Egholm M.; Buchardt O.; Christensen L.; Behrens C.; Freier S. M.; Driver D. A.; Berg R. H.; Kim S. K.; Norden B.; Nielsen P. E. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature 1993, 365, 566–568. 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- Su X.; Kanjanawarut R. Control of Metal Nanoparticles Aggregation and Dispersion by PNA and PNA–DNA Complexes, and Its Application for Colorimetric DNA Detection. ACS Nano 2009, 3, 2751–2759. 10.1021/nn9005768. [DOI] [PubMed] [Google Scholar]
- Kanjanawarut R.; Su X. Colorimetric Detection of DNA Using Unmodified Metallic Nanoparticles and Peptide Nucleic Acid Probes. Anal. Chem. 2009, 81, 6122–6129. 10.1021/ac900525k. [DOI] [PubMed] [Google Scholar]
- Vilaivan T.; Srisuwannaket C. Hybridization of Pyrrolidinyl Peptide Nucleic Acids and DNA: Selectivity, Base-Pairing Specificity, and Direction of Binding. Org. Lett. 2006, 8, 1897–1900. 10.1021/ol060448q. [DOI] [PubMed] [Google Scholar]
- Vilaivan T. Pyrrolidinyl PNA with α/β-Dipeptide Backbone: From Development to Applications. Acc. Chem. Res. 2015, 48, 1645–1656. 10.1021/acs.accounts.5b00080. [DOI] [PubMed] [Google Scholar]
- Jampasa S.; Wonsawat W.; Rodthongkum N.; Siangproh W.; Yanatatsaneejit P.; Vilaivan T.; Chailapakul O. Electrochemical detection of human papillomavirus DNA type 16 using a pyrrolidinyl peptide nucleic acid probe immobilized on screen-printed carbon electrodes. Biosens. Bioelectron. 2014, 54, 428–434. 10.1016/j.bios.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Kongpeth J.; Jampasa S.; Chaumpluk P.; Chailapakul O.; Vilaivan T. Immobilization-free electrochemical DNA detection with anthraquinone-labeled pyrrolidinyl peptide nucleic acid probe. Talanta 2016, 146, 318–325. 10.1016/j.talanta.2015.08.059. [DOI] [PubMed] [Google Scholar]
- Jirakittiwut N.; Panyain N.; Nuanyai T.; Vilaivan T.; Praneenararat T. Pyrrolidinyl peptide nucleic acids immobilised on cellulose paper as a DNA sensor. RSC Adv. 2015, 5, 24110–24114. 10.1039/C4RA15287E. [DOI] [Google Scholar]
- Laliwala S. K.; Mehta V. N.; Rohit J. V.; Kailasa S. K. Citrate-modified silver nanoparticles as a colorimetric probe for simultaneous detection of four triptan-family drugs. Sens. Actuators, B 2014, 197, 254–263. 10.1016/j.snb.2014.02.087. [DOI] [Google Scholar]
- Carrilho E.; Martinez A. W.; Whitesides G. M. Understanding Wax Printing: A Simple Micropatterning Process for Paper-Based Microfluidics. Anal. Chem. 2009, 81, 7091–7095. 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- Liu H.; Crooks R. M. Three-Dimensional Paper Microfluidic Devices Assembled Using the Principles of Origami. J. Am. Chem. Soc. 2011, 133, 17564–17566. 10.1021/ja2071779. [DOI] [PubMed] [Google Scholar]
- Liu H.; Xiang Y.; Lu Y.; Crooks R. M. Aptamer-Based Origami Paper Analytical Device for Electrochemical Detection of Adenosine. Angew. Chem. 2012, 124, 7031–7034. 10.1002/ange.201202929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentele M. M.; Cunningham J.; Koehler K.; Volckens J.; Henry C. S. Microfluidic Paper-Based Analytical Device for Particulate Metals. Anal. Chem. 2012, 84, 4474–4480. 10.1021/ac300309c. [DOI] [PubMed] [Google Scholar]
- Huynh K. A.; Chen K. L. Aggregation Kinetics of Citrate and Polyvinylpyrrolidone Coated Silver Nanoparticles in Monovalent and Divalent Electrolyte Solutions. Environ. Sci. Technol. 2011, 45, 5564–5571. 10.1021/es200157h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Lenhart J. J.; Walker H. W. Dissolution-Accompanied Aggregation Kinetics of Silver Nanoparticles. Langmuir 2010, 26, 16690–16698. 10.1021/la101768n. [DOI] [PubMed] [Google Scholar]
- Shirato K.; Yano T.; Senba S.; Akachi S.; Kobayashi T.; Nishinaka T.; Notomi T.; Matsuyama S. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Virol. J. 2014, 11, 139–139. 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E. I.; Hashem A. M.; El-Kafrawy S. A.; Sohrab S. S.; Aburizaiza A. S.; Farraj S. A.; Hassan A. M.; Al-Saeed M. S.; Jamjoom G. A.; Madani T. A. Detection of the Middle East Respiratory Syndrome Coronavirus Genome in an Air Sample Originating from a Camel Barn Owned by an Infected Patient. mBio 2014, 5, e01450. 10.1128/mBio.01450-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu A. L. P.; Souza R. P.; Gimenes F.; Consolaro M. E. L. A review of methods for detect human Papillomavirus infection. Virol. J. 2012, 9, 262–262. 10.1186/1743-422X-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L. L.; Denny L. CHAPTER 7 Methods for detection of HPV infection and its clinical utility. Int. J. Gynecol. Obstet. 2006, 94, S71–S80. 10.1016/S0020-7292(07)60013-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.