Abstract
The opportunistic pathogen Malassezia pachydermatis causes bloodstream infections in preterm infants or individuals with immunodeficiency disorders and has been associated with a broad spectrum of diseases in animals such as seborrheic dermatitis, external otitis and fungemia. The current approaches to treat these infections are failing as a consequence of their adverse effects, changes in susceptibility and antifungal resistance. Thus, the identification of novel therapeutic targets against M. pachydermatis infections are highly relevant. Here, Gene Essentiality Analysis and Flux Variability Analysis was applied to a previously reported M. pachydermatis metabolic network to identify enzymes that, when absent, negatively affect biomass production. Three novel therapeutic targets (i.e., homoserine dehydrogenase (MpHSD), homocitrate synthase (MpHCS) and saccharopine dehydrogenase (MpSDH)) were identified that are absent in humans. Notably, L-lysine was shown to be an inhibitor of the enzymatic activity of MpHCS and MpSDH at concentrations of 1 mM and 75 mM, respectively, while L-threonine (1 mM) inhibited MpHSD. Interestingly, L- lysine was also shown to inhibit M. pachydermatis growth during in vitro assays with reference strains and canine isolates, while it had a negligible cytotoxic activity on HEKa cells. Together, our findings form the bases for the development of novel treatments against M. pachydermatis infections.
Subject terms: Fungal infection, Antifungal agents
Introduction
The yeast M. pachydermatis is part of the skin microbiota of domestic and wild animals and behaves as an opportunistic pathogen causing external otitis and seborrheic dermatitis in dogs and cats. Particular conditions such as the presence of lipid-rich microenvironments, a local imbalance of the natural microbiota and altered immune states favor these infections1. Dermatologic infections caused by M. pachydermatis often exhibit a chronic (recurrent) course and their treatment can be complicated due to the ability of this yeast to form biofilms1. In addition, M. pachydermatis causes bloodstream infections in preterm infants or in individuals with immunodeficiency disorders. These infections are related to contamination of medical devices such as catheters, the transmission through medical staff and the administration of lipids through intravenous way2,3. Recently, several factors contributing to M. pachydermatis virulence have been determined, which include the production of proteinases, phospholipases, hyaluronidases, and chondroitin-sulfatases4.
Currently, five classes of antifungal agents are used orally, topically or intravenously for the treatment of fungal infections. The first class is formed by the azoles (ketoconazole, itraconazole, clotrimazole, miconazole, and voriconazole) that interfere with ergosterol synthesis by interacting with sterol-14α-demethylase. The second and third class, i.e. allylamines (terbinafine and naftifine) and polyenes (nystatin, natamycin, and amphotericin B) also target ergosterol by interfering with its synthesis by inhibiting squalene sterol-14α-demethylase and by producing pores in membranes by binding ergosterol, respectively. Echinocandins (caspofungin, micafungin, and anidulafungin) are the only available antifungal drugs targeting the cell wall, acting as noncompetitive inhibitors of the β-(1,3)-D-glucan synthase enzyme complex. The fifth class of anti-fungals are formed by the pyrimidine analogs like flucytosine that interfere with pyrimidine metabolism and RNA/DNA and protein synthesis2,3,5–8. Azoles and amphotericin B are mainly used to treat M. pachydermatis infections6,9. These infections have been classified as chronic, which may require prolonged treatment and thereby causing adverse effects1,3,6,8,10. The increase in incidence of azole-resistant strains and the number of therapeutic failures in animals2,11 also underline the importance to identify new therapeutic targets for the treatment of M. pachydermatis infections.
Searching therapeutic targets through metabolic network reconstructions has been proposed as a strategy to control the virulence of pathogens12,13. A frequently used approach is Gene Essentiality Analysis (GEA) that analysis the impact of in silico deletions to identify potentially essential genes for growth of an organism12,14. This approach provides useful information about the metabolism of target organisms, which can be used to nominate therapeutic candidates13,15,16. The aim of this study was to identify novel therapeutic targets for M. pachydermatis by GEA and to confirm their potential by assessing the inhibitory capacity of inhibitors. Results indicate that MpHSD, MpHCS, MpSDH are targets to treat M. pachydermatis infections.
Results
Novel potential therapeutic targets against M. pachydermatis
Curation of the metabolic network of M. pachydermatis17 identified 19 metabolites that cannot be produced and/or consumed by any of the reactions or imported/exported through any of the available uptake/secretion pathways in the model (Table S1). This was solved by adding 21 reactions (Table S2) and adjusting the upper and lower bounds of another 18 reactions (Table S3). The Flux Balance Analysis (FBA) of the curated network had a biomass flux of 3.13 and consisted of 45% active fluxes (reactions with a flux >0.01 mmol gDW−1h−1). Flux Variability Analysis (FVA) was performed to identify the flux range variability of each reaction. FVA revealed that 42.3% of the reactions of the M. pachydermatis metabolic network showed a difference between the maximum and minimum fluxes other than zero. These reactions represent the defined space of flux distributions of the optimal solution. This means that these reactions do not affect the overall flux of biomass as alternative pathways could be used to fulfill the objective function. This natural flexibility has been associated to the ability of organisms to face environmental changes (i.e. fitness of the cell)18. In contrast, reactions with a low range of plasticity (that is, reactions with a difference value between maximum and minimum fluxes equal to zero) or essential reactions related to the biomass reaction were around 16.4%. These essential reactions are catalyzed by 606 enzymes in the network.
GEA was applied to identify potential therapeutic targets for M. pachydermatis. In silico deletion of candidate genes should have a negative effect on the growth of the organism according to FBA performed in this study. The 606 enzymes catalyzing essential reactions in the network were grouped into 602 enzyme clusters by sequence similarity. A total of 67 enzyme clusters were identified as potential targets for growth inhibition based on the fact that their in-silico deletion resulted in a reduction of the flow of biomass ≥70%. This same procedure was also performed in M. furfur (data not shown) showing an overlap of 15 enzymes that are predicted to inhibit growth. Only three of these 15 enzymes (i.e. Imidazoleglycerol-phosphate dehydratase (IGPD), 6,7-dimethyl-8-ribitilumazine synthetase (RIBH) and riboflavin synthetase (RIB)) did not have a significant match with a human protein (Table 1). However, these enzymes were not selected for further studies as no commercial kits for their quantification are available. The M. pachydermatis enzymes homoserine dehydrogenase (MpHSD), homocitrate synthase (MpHCS) and saccharopine dehydrogenase (MpSDH) were selected for further studies as they showed a similarity <20% with a human protein (Table 1), there are no enzyme homologues reported in mammals and there are inhibitors of orthologues already reported in the literature for instance in Schizosaccharomyces pombe (HCS)19, Corynebacterium glutamicum (HSD)20, and S. cerevisiae (SDH)21.In fact, a literature review identified 58 inhibitors for HSD, 62 for HCS and 67 for SHD.
Table 1.
Potential therapeutic targets against M. pachydermatis.
| EC Number | Abbreviature | Enzyme | Human Match | % Protein similarity |
|---|---|---|---|---|
| 4.2.1.19 | IGPD | Imidazoleglycerol-phosphate dehydratase | NA | NA |
| 2.5.1.78 | RIBH | 6,7-Dimethyl-8-ribityllumazine synthase | NA | NA |
| 2.5.1.9 | RIB | Riboflavin synthase | NA | NA |
| 2.3.3.14 | HCS | Homocitrate synthase | HMGCL | 14.4 |
| 1.1.1.3 | HSD | homoserine dehydrogenase | PSB5 | 18.31 |
| 1.5.1.7; 1.5.1.10 | SDH | Saccharopine dehydrogenase | F8WE53 | 19.71 |
| 4.2.1.36 | HACN | Homoaconitate hydratase | ACON | 21.36 |
| 1.1.1.41; 1.1.1.42 | IDH | Isocitrate dehydrogenase | IDH3A | 32.72 |
| 1.14.13.70 | CYP51A1 | Lanosterol 14 alpha-demethylase | CP51A | 36.48 |
| 2.7.4.22 | UK | Uridylate kinase | KCY | 40.7 |
| 1.3.1.- | ERG4/ERG24 | Ergosterol biosynthesis sterol reductase ERG4/ERG24 | LBR | 41.18 |
| 5.4.2.8 | PMM | Phosphomannomutase | H3BV55 | 51.06 |
| 6.4.1.1 | PC | Pyruvate carboxylase | PC | 51.43 |
| 3.6.1.9 | ITPA | Inosine triphosphate pyrophosphatase | ITPA | 52.91 |
| 6.3.4.5 | AS | Argininosuccinate synthase | ASSY | 55.3 |
The in-silico deletion of these enzymes resulted in a decrease in the production of biomass of at least 30%. The percentage of similarity and human match to the human proteome is also shown. Prioritized enzymes are shown in bold and underline.
L-lysine and L-threonine are potential inhibitors of MpHCS, MpHSD and MpSDH
The genes encoding MpHSD, MpHCS and MpSDH were heterologously expressed to assess their enzymatic activity in the presence of potential inhibitors. To this end, HIS-tag fusion genes were cloned into the expression vectors pET6xHN-N or pET6xHN-C (Supplementary Fig. S1) and introduced into E. coli BL21. SDS-PAGE revealed bands of 81, 51, and 43 kDa for MpHSD, MpHCS enzyme and MpSDH, respectively (Supplementary Figs. S1, S2). The additional bands likely correspond to proteins with histidine residues of E. coli BL21 that were co-purified with the histidine-tagged recombinant protein22,23.
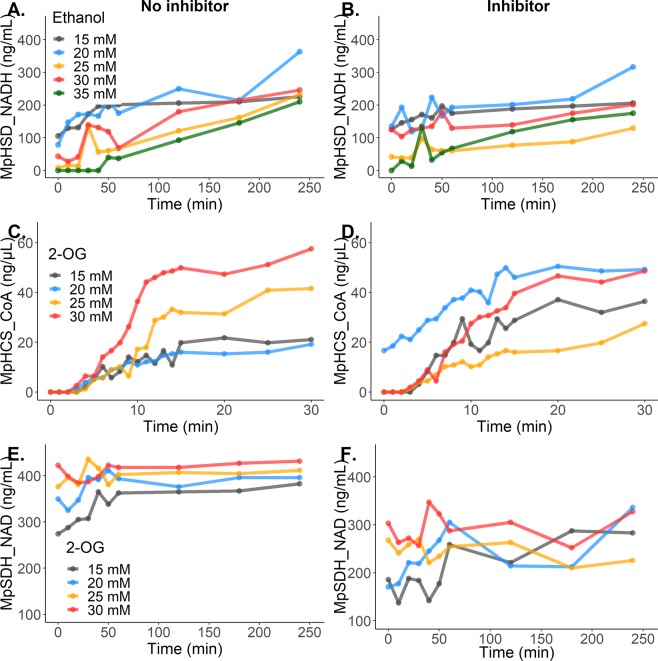
MpHSD, MpHCS and MpSDH activity was monitored in inhibition assays using NAD/NADH24 and coenzyme A25 quantification (see Material and Methods). The three enzymes carried out the expected catalytic activity in the absence of inhibitors (Fig. 1A,C,E; Supplementary Fig. S3A,C,E), while inhibitors reduced enzymatic activity (Fig. 1B,D,F; Supplementary Fig. 3B,D,F). The highest final concentrations of formed NADH were 363.3 ng/mL (Fig. 1A) and 316.42 ng/mL (Fig. 1B), respectively, in the first MpHSD activity test without and with 1 mM L-threonine as inhibitor. In the case of MpHCS activity, highest values of CoA were 57.56 ng/µL (Fig. 1C) in absence and 49.25 ng/µL in presence of the 1 mM L-lysine as inhibitor (Fig. 1D). Highest final concentrations of formed NAD were 431.2 ng/mL (Fig. 1E) without inhibitor and 336.1 ng/mL (Fig. 1F) with 75 mM L-lysine as inhibitor in the case of MpSDH. The second assays or biological replicates showed the same concentration differences in the absence and presence of inhibitor for each enzyme (Supplementary Fig. S3A–F), showing that threonine is an inhibitor of MpHSD and lysine of MpHCS and MpSDH. It should be noted that in the first trial MpHCS activity was higher in the presence than in the absence of inhibitor when 15 and 20 mM 2-oxoglutaric acid (2-OG) was used. This may have been caused by a higher actual 2-OG concentration or by a L-lysine concentration that was too low to inhibit (Fig. 1C,D). Note that these values were included in the Lineweaver-Burk diagrams (Supplementary Fig. S4C) still enabling the kinetic parameters to be determined (Table 2).
Figure 1.
Evaluation of the inhibitory capacity of amino acids upon candidates as therapeutic targets. (A) Enzymatic activity of HSD with ethanol as variable substrate (colored lines), NADH as reaction indicator and detected for four hours. (B) Enzymatic activity of HSD adding L-threonine 1 mM as an inhibitor. (C) Enzymatic activity of HCS with 2-OG as variable substrate (colored lines), CoA as reaction indicator and detected for thirty minutes. (D) Enzymatic activity of HCS adding L-lysine 1 mM as an inhibitor. (E) Enzymatic activity of SDH with 2-OG as variable substrate (colored lines), NAD as reaction indicator and detected for four hours. (F) Enzymatic activity of SDH adding L-lysine 75 mM as inhibitor. Representative results of two biological replicates.
Table 2.
Kinetic parameters for enzymatic activity of MpHSD, MpHCS and MpSDH in the presence and absence of the inhibitors L-threonine or L-lysine.
| Enzyme | Condition | Km (mM) | Vmax (mmol/min) for condition | Vmax for both assays (mmol/min) |
|---|---|---|---|---|
| MpHSD | Without inhibitor | 0.59 ± 2.1e-02 | 1.56e-06 ± 5.4e-08 (mM/min) | 1.98e-06 ± 5.9e-07 (mM/min) |
| With inhibitor | 1.41 ± 6.2e-02 | 2.40e-06 ± 1.2e-06 (mM/min) | ||
| MpHCS | Without inhibitor | 0.35 ± 2.1e-01 | 1.24e-06 ± 1.7e-06 | 2.11e-06 ± 2.9e-06 |
| With inhibitor | 1.36 ± 3.7e-02 | 2.98e-06 ± 2.1e-06 | ||
| MpSDH | Without inhibitor | 0.25 ± 7.1e-02 | 7.50e-09 ± 3.5e-09 | 7.80e-09 ± 4.0e-09 |
| With inhibitor | 0.83 ± 2.4e-01 | 8.06e-09 ± 4.3e-09 |
Data are shown with standard deviations.
Results of two biological replicates.
Kinetic parameters based on the Michaelis-Menten model26,27 and Lineweaver-Burk diagram28 showed competitive inhibition of MpHSD by 1 mM L-threonine, while in the case of MpHCS and MpSDH 1 mM and 75 mM L-lysine, respectively, showed competitive inhibition (Table 2). The Km of MpHSD with and without inhibitor was 1.41 ± 6.2e-02 and 0.59 ± 2.1e-02 mM, respectively, while Vmax was not affected with a value of 1.98e-06 ± 5.9e-07 (mM/min) (Table 2, Supplementary Fig. S4A,B). Km of MpHCS was 0.35 ± 2.1e-01 and 1.36 ± 3.7e-02 in absence and presence of inhibitor, respectively, while Vmax was not affected with a value of 2.11e-06 ± 2.9e-06 (Table 2, Supplementary Fig. S4B,C). Lastly, MpSDH had an increase in the Km of 0.25 ± 7.1e-02 without inhibitor to 0.83 ± 2.4e-01 with inhibitor, whereas also in this case Vmax was not affected by the inhibitor showing a value of 7.80e-09 ± 4.0e-09 (Table 3, Supplementary Fig. S4C,D). Together, these results indicate that L-threonine and L-lysine inhibit the enzymatic activity in a competitive way because these interfere with correct substrate-enzyme complex formation which is reflected in the increase of the Km value.
Table 3.
MICs (mg/mL) for Malassezia spp. evaluated with L-lysine and L-threonine and values for AB and Flz control (µg/mL).
| Strains | MIC L-lysine | MIC L-threonine | MIC to ABa | ||
|---|---|---|---|---|---|
| Microdilution assays | |||||
| M. pachydermatis CBS 1879b | 3.1 | >25 | 0.25 | ||
| M. pachydermatis C1c | 3.1 | >25 | — | ||
| M. pachydermatis M1c | 3.1 | >25 | — | ||
| M. pachydermatis N1c | 3.1 | >25 | — | ||
| Atypical M. furfurb | 3.1 | >25 | — | ||
| M. sympodialis CBS 7222b | 3.1 | >25 | — | ||
| M. furfur CBS 1878b | 1 | >25 | 4 | ||
| C. krusei ATCC 6258b | — | — | 2 | ||
| C. parapsilosis ATCC 22019b | — | — | 0.25 | ||
| E-test® method | |||||
| Strain | Flza | ABa | |||
| M. pachydermatis CBS 1879b | 48–64 | 1–2 | |||
| M. pachydermatis N1c | 64–128 | 3–6 | |||
| M. pachydermatis M1c | 8–16 | 3–4 | |||
| M. pachydermatis C1c | 4 | 8 | |||
| M. furfur CBS 1878b | 3–4 | >32 | |||
| M. sympodialis CBS 7222b | 0.75–1.5 | 2 | |||
aReference values (µg/mL) amphotericin B (AB) 0.125–8 and fluconazole (Flz) ≤64.
bReference strain.
cCanine isolate.
(—) No tested.
>No inhibition, likely MIC higher.
Microdilution assays were performed in triplicate.
E-test® was done for duplicate and the results are showed among ranks.
L-lysine and L-threonine inhibit growth of M. pachydermatis
Microdilution and agar well diffusion assays were used to determine the effect of L-lysine and L-threonine on growth of reference strains of Malassezia spp. and canine isolates of M. pachydermatis (Table 4). The MIC of L-lysine was the same (i.e. 3.1 mg/mL) for all reference strains and canine isolates of M. pachydermatis, but not for the atypical M. furfur strain that was shown to be more susceptible with a MIC of 1 mg/mL of L-Lysine (Table 3). In contrast, L-threonine did not show inhibition for any strain at 25 mg/mL, which was the maximum concentration that could be evaluated due to the water solubility limit of the reagent used (Table 3). Agar diffusion assays did not show inhibited growth of any of the strains at levels of threonine up to 50 mg/mL (results not shown). In contrast, growth of M. pachydermatis strains was inhibited at ≥50 mg/mL lysine (Fig. 2A), while growth inhibition of M. sympodialis and M. furfur was observed above 100 mg/mL (Fig. 2A,B). More specifically, M. pachydermatis isolates CBS 1879 and M1 showed significant inhibition at 50 mg/mL L-lysine, whilst isolates C1, N1 were inhibited at 100 mg/mL. Growth of M. sympodialis strain CBS 7222 and the M. furfur strain CBS 1878 were only significantly inhibited at ≥150 mg/mL L-lysine (Fig. 2A). The atypical M. furfur strain was not evaluated by agar diffusion as grew poorly on culture media. As comparison, growth inhibition was evaluated with two antifungals. The E-test® indicated that M. pachydermatis strain CBS 1879 strain was most sensitive for amphotericin B (MIC 1–2 µg/mL) and the C1 strain for fluconazole (MIC 4 µg/mL) (Table 3). No inhibition of growth was observed at low concentrations of amphotericin B (4 or 16 µg/mL) and fluconazole (64 µg/mL), while growth was inhibited in all cases with higher concentrations used (400 or 1600 µg/mL) (Fig. 2). Together, results show that L-lysine inhibits growth of Malassezia spp.
Table 4.
Strains and plasmids used in this study.
| Strain | Description | Reference |
|---|---|---|
| Malassezia pachydermatis CBS 1879 | Reference strain. Genome Sequenced: NCBI: txid 77020. | 101,102 |
| Malassezia furfur CBS 1878 | Reference strain | 101 |
| Atypical Malassezia furfur 4DS | Reference strain | 103 |
| Malassezia sympodialis CBS 7222 | Reference strain | 101 |
| Candida krusei ATCC 6258 | Reference strain | 104 |
| Candida parapsilosis ATCC 22019 | Reference strain | 104 |
| Malassezia pachydermatis C1 | Canine isolate from the collection of the Cellular and Molecular research group of Microorganisms Pathogens (CeMoP, acronym in Spanish). From ears of a 2-years old female cocker spaniel dog. | 105 |
| Malassezia pachydermatis N1 | Canine isolation from the collection of the CeMoP. From ears of a 9-years old female cocker spaniel dog. | 105 |
| Malassezia pachydermatis M1 | Canine isolation from the collection of the CeMoP. From the ears of a male 1-year old Shih-Tzu dog. | 105 |
| DH5α-pUC57-mpsdh | Escherichia coli DH5α strain transformed with pUC57-mpsdh | This study |
| DH5α-pET6xHN-C-mpsdh | Escherichia coli DH5α strain transformed with pET6xHN-C-mpsdh | This study |
| BL21-pET6xHN-C-mpsdh | Strain Escherichia coli BL21(DE3) transformed with pET6xHN-C-mpsdh | This study |
| BL21-pET6xHN-N-mphcs | Strain Escherichia coli BL21(DE3) transformed with pET6xHN-N-mphcs | This study |
| BL21-pET6xHN-N-mphsd | Strain Escherichia coli BL21(DE3) transformed with pET6xHN-N-mphsd | This study |
| pET6xHN-C | Plasmid containing an IPTG inducible promoter system (T7/lac promoter) for high-level expression, a C-terminal 6xHN tag and conferring ampicillin resistance | 106 |
| pET6xHN-N | Plasmid containing an IPTG inducible promoter system (T7/lac promoter) for high-level expression, an N-terminal 6xHN tag and conferring ampicillin resistance | 106 |
| pUC57-mpsdh | Plasmid pUC57 containing a synthetic expression cassette encoding the enzyme saccharopine dehydrogenase (MpSDH) of M. pachydermatis flanked by NcoI and NotI restriction sites and conferring ampicillin resistance. | This study |
| pET6xHN-C-mpsdh | Plasmid pET6xHN-C containing a synthetic expression cassette encoding the enzyme saccharopine dehydrogenase (MpSDH) of M. pachydermatis flanked by NcoI and NotI restriction sites, conferring ampicillin resistance and with C-terminal 6xHN tag. | This study |
| pET6xHN-N-mphcs | Plasmid pET6xHN-N containing an amplified expression cassette encoding the enzyme homocitrate synthase (MpHCS) of M. pachydermatis flanked by HCSF and HCSR primers sites, conferring ampicillin resistance and with C-terminal 6xHN tag. | This study |
| pET6xHN-N-mphsd | Plasmid pET6xHN-N containing an amplified expression cassette encoding the enzyme homoserine dehydrogenase (MpHSD) of M. pachydermatis flanked by HSDF and HSDR primer sites, conferring ampicillin resistance and with C-terminal 6xHN tag. | This study |
Figure 2.
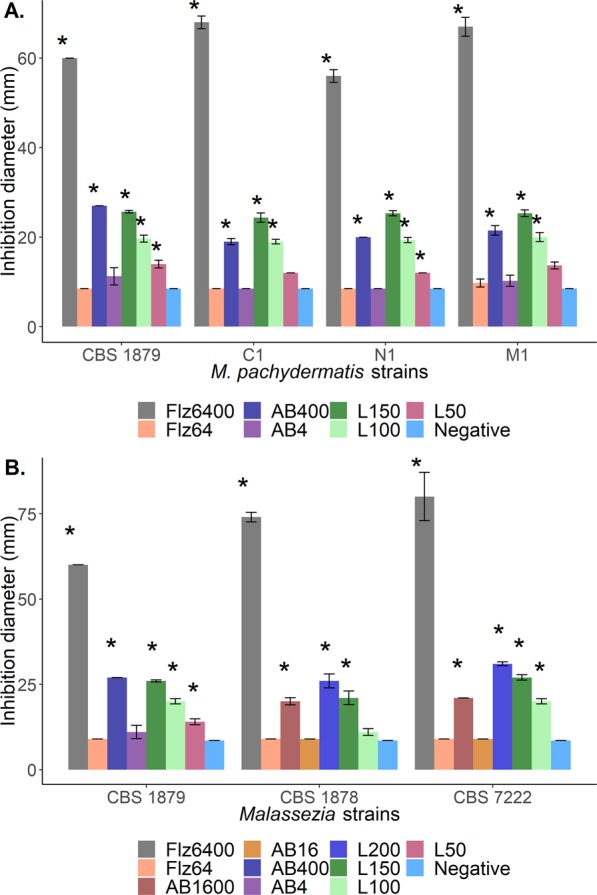
Agar diffusion assays to evaluate the inhibitory capacity of L-lysine upon the growth of M. pachydermatis and Malassezia strains. (A) The diameter of the inhibition zones of three different concentrations of L-lysine upon M. pachydermatis isolates. (B) Diameter of the inhibition zones measured for three different concentrations of L-lysine for M. pachydermatis, M. furfur and M. sympodialis, respectively. experiments were performed in triplicate and the results are shown with standard deviation. *Differences between the diameter of the zone of inhibition significantly different (p-value < 0.05) from the negative control. AB, amphotericin B; Flz, fluconazole; L, L-lysine. Concentrations in mg/mL for Lysine and µg/mL for AB and FLZ.
The amino acid L-lysine has a low cytotoxic effect on HEKa cells
A MTT viability assay was performed using HEKa29 (Primary Epidermal Keratinocytes) cells to determine if L-lysine is cytotoxic to human cells. L-lysine was shown to have a mild cytotoxic activity. Cell viability was 100% in the absence of lysine while 90.5% ± 0.06%, 79.3% ± 0.3 and 67.74% ± 0.08% of the cells survived in the presence of 100, 150, and 200 mg/mL L-lysine, respectively (Supplementary Fig. S5).
Discussion
In this study, a metabolic network of M. pachydermatis CBS 187917 was curated to identify potential novel therapeutic targets against M. pachydermatis infections. The improved network shows fluxes that are in agreement with those in other yeasts such as S. cerevisiae30 and Ustilago maydis31. FVA and GEA revealed several genes including MpHSD, MpHCS, and MpSDH whose in-silico deletions were predicted to result in a reduction in biomass formation.
HSD is involved in the aspartate route resulting in the L-amino acids lysine, threonine, cysteine and methionine. This enzyme catalyzes the third reversible reaction in this pathway producing L-homoserine20,32. Notably, this “sulfur assimilation pathway” is present in the fungal kingdom, but not in humans33,34. HCS and SDH catalyze the first and last step of the α-aminoadipate pathway that results in L-lysine biosynthesis. The former catalyzes the conversion of 2-OG into homocitrate35,36, while the latter catalyzes the reversible conversion of saccharopine to L-lysine and α-ketoglutarate (α-Kg) using NAD as an oxidant37. These two enzymes are also absent in mammals. In the latter, the bifunctional enzyme alpha-aminoadipic semialdehide synthase (EC 1.5.2.8) is present that has a saccharopine dehydrogenase domain, but it is not inhibited by L-lysine38. In contrast, HCS and SDH are regulated by feedback inhibition by an excessive amount of L-lysine, while HSD is similarly regulated by the end products of the aspartate route including L-threonine32,39,40.
The Km of MpHSD of 0.59 mM was similar to that of the HSDs of E. coli (0.68 mM), Staphylococcus aureus (0.69 mM) and Sulfolobus tokodaii (0.54 mM)32,41,42. Moreover, the kinetic parameters indicated that L-threonine is a competitive inhibitor for MpHSD activity (1.41 mM), as was shown in Brevibacterium flavum (at 3.3 mM)43, E. coli (at 0.08 mM or 0.5 mM)44,45 and Corynebacterium glutamicum (at 0.5 mM or 2 mM)20,46. The Km of HCS ranges from 0.044 to 1.3 mM for organisms like Thermus thermophiles, Candida albicans, Schizosaccharomyces pombe and S. cerevisiae19,35,39,47–50. Thus, the affinity constant of MpHCS of 0.35 mM is within this reported range. L-lysine was shown to compete with the substrate 2-OG increasing the Km to 1.36 mM. The inhibitory effect of L-lysine has been shown to be due to binding to free enzyme in S. cerevisiae, while the competitive inhibition of lysine versus α-KG (at a low concentration) can be explained by active-site and not allosteric inhibition51. The Km of SDH ranges between 0.11 and 0.66 mM in S. cerevisiae, Pichia guilliermondii and Candida maltosa21,40,52–55, while that of MpSDH again is in this range with 0.25 mM. The reductive condensation of α-ketoglutarate and lysine with SDH can be inhibited by concentrations of lysine >60 mM in S. cerevisiae21,56. In this study, the Michaelis-Menten constant changed but not the Vmax indicating that L-lysine at 75 mM was a competitive inhibitor for 2-OG.
We assessed whether the MpHSD, MpHCS, MpSDH inhibitors threonine and lysine can inhibit growth of the reference strains M. pachydermatis CBS 1879, M. furfur CBS 1878, M. sympodialis CBS 7222 and atypical M. furfur, and three M. pachydermatis canine isolates. L-lysine indeed reduced the growth of these strains most likely due to feedback inhibition and specific repression of the synthesizing enzymes. Similar observations have been reported in studies with S. cerevisiae57–59 and P. chrysogenum60, where increases on the concentration of lysine in the medium resulted in cell death. This was observed when lysine was used as the sole source of nitrogen and consequently it is not properly metabolized. This toxicity may also be due to negative regulation of SDH and HCS that can generate accumulation or absence of intermediate metabolites61. Moreover, growth repression may be due to phenomena like general control, which involves a simultaneous derepression of one or more enzymes of several unrelated amino acid biosynthetic pathways, in response to external imbalance61,62, and it can contribute to the internal imbalance of the amino acid pools even further, causing cell death63. Thus, the inhibition of M. pachydermatis growth, as well as other Malassezia strains by L-lysine could be by, at least, two mechanisms of regulation of the α-aminoadipate pathway.
It was not possible to determine the inhibitory capacity of L-threonine on growth of the Malassezia strains due to the limitations of water solubility64. Previous studies showed that feedback inhibition of HSD by 5 mm L-threonine reduced growth of Pseudomonas putida by 22%65. In contrast, no growth inhibition was observed in the case of Acetobacter aceti at 10 mM L-threonine despite inhibition of HSD66. There were differences in the concentrations of L-lysine, amphotericin B (AB), and fluconazole (Flz) that inhibited growth of the Malassezia strains between the microdilution and agar well diffusion assays. This might be due to differences in lipidic composition of the broth and agar cultivation mediums, which influences the antifungal sensitivity67,68, despite of this, the agar well diffusion method has been shown suitable to evaluate the inhibition of natural extracts on M. furfur69 and for both tests, L-lysine presented growth inhibition.
The MTT cell viability assay showed that L-lysine concentrations up to 150 mg/mL is not cytotoxic to Heka cells considering the definition in ISO 10993-570 that states that any solution or item that reduces the cell viability to 70% or less has a cytotoxic potential70. Nonetheless, a L-lysine concentration of 200 mg/mL did reduce the cell viability to 67.74%, exhibiting a cytotoxic potential activity. The cytotoxic activity of the most concentrated L-lysine solution could be explained by the cationic properties of L-lysine as has been reported71,72. Morgan et al., (1989) suggested that the cytotoxicity displayed by cationic macromolecules is strongly related to the high density of multivalent interactions or bindings with anionic groups at the surface of the cell72. Although these studies were performed with cationic polyaminoacids, it could be deduced that a high concentration of cationic amino acids, as L-lysine, could interact with the cell surface as well, exhibiting a similar cytotoxicity as shown in Choksakulnimitr’s research71. Overall, our results suggest that L-lysine could be used as a potential treatment against M. pachydermatis associated-diseases. It is important to mention that the concentrations required for growth inhibition could limit their clinical utility. Thus, L-lysine could be used to treat dermatological infections instead of sepsis where higher concentrations will be required. Additional cytotoxicity, pharmacokinetic and toxicodynamic studies will be needed to confirm its potential as a novel pharmaceutical product and possible treatment against M. pachydermatis infections.
Conclusions
In this study, in silico modeling together with in vitro analysis identified and nominated three essential genes as novel therapeutic targets against M. pachydermatis-associated infections. Their activity can be inhibited in vitro using L-threonine or lysine. Interestingly, L-lysine was shown to be able to reduce also the growth of Malassezia spp., and presented no cytotoxic activity in keratinocytes at a concentration ≤150 mg/mL. Further studies will be required to evaluate clinical application including analysis of pharmacokinetic/ADME-Tox (Absorption, Distribution, Metabolism, and Excretion – Toxicology), pharmacodynamics, and biopharmacy studies.
Materials and Methods
Strains, plasmids, and media composition
Strains and plasmids used in this study are listed in Table 4. Cells of E. coli were grown and maintained in Luria Bertani (LB) agar or broth containing 10 g/L Tryptone [Oxoid], 5 g/L yeast extract [Oxoid], 10 g/L NaCl [Merk], 14.8 g/L agar bacteriological [Scharlau] and 1 mL/L ampicillin [Sigma-Aldrich]. Growth of Malassezia yeasts as well as agar diffusion assays were carried out in modified Dixon agar medium (mDixon) containing 36 g/L Mycosel [DB], 36 g/L yeast extract [Oxoid], 20 g/L Oxgall [DB], 2 mL/L glycerol [Sigma-Aldrich], 2 mL/L oleic acid [Carlo Erbba], 10 mL/L Tween 40 [Sigma-Aldrich], and 0.5 g/L chloramphenicol [Colmed International, Sigma]. Microdilution tests were performed in modified Sabouraud broth (mSabouraud) containing 30 g/L Sabouraud broth [Scharlau], 5 ml/L Tween 40 [Sigma-Aldrich], 5 mL/L Tween 60 [Sigma-Aldrich] and 0.25 g/L chloramphenicol [Colmed International, Sigma]. Candida spp. were grown in 30 g/L Sabouraud agar [Scharlau]) containing 0.25 g/L chloramphenicol [Colmed International, Sigma].
Metabolic network curation
The metabolic network of M. pachydermatis10 was manually curated using the biomass objective function as a starting point. First, metabolites with no production, accumulation and absorption problems were identified using standard protocols (network gap filling)15. Subsequently, a review and search of reactions in specialized literature and in databases such as KEGG73 and BioCyc74 was carried out, allowing the identification of missing reactions in the metabolic pathways. Those reactions were added in the network if the enzyme that catalyzed each reaction was present in the M. pachydermatis genome. Furthermore, FBA was run using GAMS software75 and CPLEX as the optimization solver76.
Gene essentiality analysis
The curated metabolic network of M. pachydermatis was used to perform the Genetic Essentiality Analysis (GEA). Initially, the flux of biomass was defined as objective function as we wanted to identify essential genes related to growth10. Then, the network was analyzed using Flux Balance Analysis (FBA) and Flux Variability Analysis (FVA). FBA was used to identify the core reactions, enzymes, and genes related to biomass production, while FVA was used to determine the plasticity77. Here, FVA was used to determine the feasible region of the FBA problem, which is the variability range and the plasticity of each flux to satisfy a fixed biomass value78. After that, the flux of each reaction was minimized and maximized, as follows:
| 1 |
Subject to
| 2 |
| 3 |
| 4 |
| 5 |
where z is the variable to optimize, is the flux of the reaction and is the stoichiometry matrix. The FVA was implemented in GAMS software75 and the resulting data was analyzed in R79. Furthermore, those reactions that had a minimum and maximum value of zero were filtered (blocked reactions) out and finally, the enzymes that catalyzed at least one non-blocked reaction were selected (non-zero).
Enzymes related to previously selected reactions were deleted one by one in the metabolic model and FBA was recalculated each time to determine their effect on the metabolic phenotype. Deletions were performed by setting upper and lower fluxes bounds to zero80. GEA was completed as previously described by Edwards and Palsson80.
Since genomes can present protein duplications and, therefore, enzymatic redundancy, deletions were also made in terms of clusters of enzymes based on sequence similarity. This clustering allows classifying each group into a specific enzyme. In this study, enzymatic groups were determined by CD-Hit75 with an identity threshold of 90%.
Nomination of therapeutic targets
Three nomination criteria were defined in order to identify potential therapeutic targets: 1. Target enzymes must not have counterpart versions within the human proteome, which was expected to reduce the effect of inhibitors on host’s metabolism. This was verified by comparing the sequences of potential enzymes with those reported in the human genome using BlastP81, where the candidate enzymes were used as a query and the human proteome (UniProt: UP000005640) as the database. 2. Target enzymes were selected to be easily quantified (i.e. there are commercially available quantification kits), and 3. Inhibitors must be reported for selected enzymes in specialized databases such as BindingDB82 and BRENDA83.
Heterologous expression of MpHCS, MpHSD, and MpSDH from M. pachydermatis in E. coli
Total RNA from M. pachydermatis CBS 1878 was isolated using TRIzol [Invitrogen] according to the manufacturer’s instructions84. Cells were homogenized with a TissueLyser [Qiagen]85 and total RNA was purified using RNeasy Mini Kit [Qiagen]86. 1 μg of purified RNA was reverse transcribed into cDNA using the SMARTer PCR cDNA Synthesis Kit [Clontech]87. The cDNA of HCS and HSD genes (mphcs and mphsd) were amplified by PCR Phusion® DNA polymerase [NEB]88. Primers for cloning (Supplementary Table S4) were designed with 15 bp overlaps to the vector pET6xHN-N (Table 4). PCR products were purified using QIAquick PCR Purification Kit [Qiagen]89, then fused in-frame into the pre-linearized vector and transformed into E. coli strain BL21 (Table 4) for expression.
A synthetic MpSDH gene (ID MN527521, NCBI) flanked by the restriction enzymes NcoI and NotI was designed using the reported MpSDH gene from M. pachydermatis CBS 1879 as a template and synthetized by Shangai ShineGene Biotech, Inc.90. This synthetic gene was initially placed into the pUC57-mpsdh plasmid and later introduced into the final vector pET6xHN-C for expression in E. coli DH5α (Table 4)90. Plasmids were purified through GeneJET Plasmid Miniprep Kit [ThermoScientific]91, and digested with NcoI and NotI following standard protocols92. The mpsdh gene and the open pET6xHN-C were separated by agarose-electrophoresis and purified by GeneJET Gel Extraction Kit [Thermo Scientific]93 for subsequently ligation with NEB94 protocol and the plasmid pET6xHN-C-mpsdh ligation product was inserted into E. coli DH5α and into E. coli BL21 (Table 4) by heat shock transformation95.
Once E. coli strains BL21 were constructed (Table 4), growth and biomass profiles were established for each strain in order to determine the time in which an optimal optical density (OD) of 0.6–0.8 is reached and the approximate volume to obtain sufficient biomass to perform protein purification.
Expression was induced by the addition of 2 mM (final concentration) of isopropyl-d-1-thiogalactopyranoside (IPTG [Sigma; Calbiochem]) to cultures of 1200 mL in LB medium with an OD600 of 0.6–0.8. The cultures were maintained at 37 °C and 5 h after initiation of induction the biomass was recovered by centrifugation. The cells were re-suspended in lysis buffer (6.89 g/L NaH2PO4.H2O [Merck], 17.55 g/L NaCl [Merck], 100 µL/L Tween 20 [Sigma-Aldrich], 10 mL/L Triton X-100 [Panreac Quimica], pH 8.0) in a ratio 6 mL per 1 g of pellet and the lysis was carried out using a VibraCellTM sonicator with an amplitude of 37% for 40 cycles of 40 seconds of dosing and 20 seconds of sonication. After, the protein purification was performed through immobilized metal affinity chromatography using ProfinityTM IMAC Resins [Bio-Rad]96 with Nickel charged resin (Nickel (II) chloride [Scharlau; Chemi]) following the instruction manual96. Finally, the protein concentration was quantified using a NanoDrop ThermoscientificTM spectrophotometer and the proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12.5% polyacrylamide gel under denaturing conditions and stained with Coomassie blue.
Enzymatic activity
For the evaluation of the enzymatic activity of MpHSD and MpSDH, NAD/NADH quantification Test Kit [Sigma-Aldrich] protocol24 was used. The reverse reaction of MpHSD enzyme was evaluated by NADH detection using15 mM, 20 mM, 25 mM, 30 mM, and 35 mM ethanol as substrate and 5.22 mg/mL total protein concentration. For inhibition condition, the same concentrations of substrate plus 1 mM L-threonine [Sigma-Aldrich] was used. Both assays were detected using a StatFax 2100 [Wiener lab Group] following standard protocols24; MpSDH activity was also evaluated with the reverse reaction, employing 4.32/4.65 mg/mL total protein, 15 mM, 20 mM, 25 mM, 30 mM 2-OG substrate, in the presence or absence of 75 mM L-lysine [Sigma-Aldrich]. The NAD cycling buffer and NADH developer were not added in the master reaction mix but NADH standard was added. The colorimetric detection was made in Multiskan GO [Thermo Scientific] at 259 nm (detect NAD) and 340 nm (detect NADH) keeping the read time.
For MpHCS the Coenzyme A Test Kit [Sigma-Aldrich] protocol25 was used with 15 mM, 20 mM, 25 mM, 30 mM 2-OG [MERK] as substrate and 3.45 mg/mL enzyme. For the condition with inhibitor, 1 mM L-lysine [Sigma-Aldrich] was employed and the final volume was the same, adjusting the amount of Coenzyme A Assay Buffer. Finally, colorimetric detection was made using a Multiskan GO [Thermo Scientific] at 535 nm (λex), using horseradish peroxidase (3.23 mg/mL) to identify CoA product following standard protocols25.
In vitro susceptibility tests
In-vitro susceptibility assays on broth microdilution and agar diffusion were performed to evaluate the inhibitory capacity of L-lysine and L-threonine on M. pachydermatis isolates (Table 3) and on the other reference strains, M. furfur, M. sympodialis and atypical M. furfur (Table 4). Broth microdilution methods were carried out according to the M27-A3 reference document of CLSI97,98. Inoculums were adjusted using Neubauer hemocytometer at 2 × 106 CFU/mL, the medium used was modified Sabouraud broth67 and the concentrations employed (mg/mL) for L-lysine was from 1.5 to 4 and for L-threonine was from 16 to 25.
For the agar well diffusion method99, first a microbial suspension of 2 × 106 CFU/mL was prepared for each strain and isolate followed by massive culture on a modified Dixon agar. Wells with a diameter of 8.5 mm was created in the medium and filled with 100 µL of the extract in concentrations (mg/mL) of 30, 40 and 50 of L-threonine or 50, 100, 150 and 200 of L-lysine and negative control with tween 80 (0.05%). Next, the plates were incubated at 33 °C for 3 days and results were recorded after 24, 48 and 72 hours. After that, the mean diameter of the inhibition zones was reported in millimeters and the inhibition was determined by comparing these to negative control (ultrapure water). The difference between the treatments was analyzed with ANOVA using the Tukey method100 (p-value < 0.05, confidence level 95%).
For microdilution method, the quality control included an AB test on Candida krusei and Candida parapsilosis strains (Table 4), as described in the CLSI M27A-A reference method. For the method of agar diffusion AB and Flz in solution or E-tests® were used as a positive growth inhibition control.
HEKa cell culture viability using MTT colorimetric assay
HEKa cells were grown to 80% confluence in Dulbecco’s Modified Eagle Medium (DMEM) in a cell culturing incubator with 5% CO2. Then, these cells were trypsinizated following the recommended procedure by ATCC29. After the trypsinization, the cell density was determined using a Neubauer hemocytometer with the purpose of seeding 10e4 cells in each well of a 96-well flat bottom plate in 100 µL of DMEM and plates were subsequently incubated for 1 day at 37 °C in 5% CO2. Then, 100 µL of 3 L-lysine solution with different concentration were added to a well, experiments were performed in triplicate. The 3 tested L-lysine concentrations were the same that caused the growth inhibition in the agar well diffusion method (200,150,100 mg/mL). The negative control consisted of 100 µL of DMEM plus 100 µL of cell culture, and the blank of DMEM alone. Then, the cells in the 96-well plate were incubated for 5 days under 5% CO2. At the 5th day, 10 uL of MTT was added to each well. At the 6th day, the absorbance was read at 562 nm using a flat bottom plate-reader. The absorbance was normalized to 100%, where 100% indicated that all cells were viable (normalized with the absorbance of the negative control minus the absorbance of the blank).
Supplementary information
Acknowledgements
We thank Universidad de Los Andes (Faculty of Sciences & Department of Chemical Engineering) for financial support, and the researchers at Celular y Molecular de Microorganismos Patogenos (CeMoP) research group and at Grupo de Diseño de Productos y Procesos (GDPP) and for their constant contributions, correct observations, and great knowledge. Also, thanks to the researchers Carol Melo and Elizabeth Jimenez from Grupo de Investigación en Bioquímica Aplicada of the Chemistry Department for their assistance in the MTT viability assay. This work was supported by Colciencias grant No. 120465741393, the Netherlands fellowship program NFP – phd.14/99 and the Darwin Trust of Edinburgh PhD scholarship.
Author contributions
Conceptualization: S.T., S.R., A.F.G.B. and A.M.C.R. Data curation: A.S., S.T., L.S. and M.F.N. Formal analysis: A.S., S.T., K.E., L.S., M.F.N., A.F.G.B. and A.M.C.R. Funding acquisition: S.R., H.W., H.D.C., A.F.G.B. and A.M.C.R. Investigation: A.S., S.T., K.E. and L.S. Methodology: A.S., S.T., K.E., M.F.N., A.F.G.B. and A.M.C.R. Project administration: S.T., S.R., A.F.G.B. and A.M.C.R. Supervision & Validation: M.F.N., A.F.G.B. and A.M.C.R. Visualization: A.S., S.T., K.E. and L.S. Writing ± original draft: A.S., S.T., L.S., and M.F.N. Writing ± review & editing: A.S., K.E., L.S., S.R., H.W., H.D.C., M.F.N., A.F.G.B. and A.M.C.R. A.M.C.R. and A.F.G.B. contributed in the same way. All authors have approved the final version.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information Files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Angie Sastoque and Sergio Triana.
Contributor Information
Andrés Fernando González Barrios, Email: andgonza@uniandes.edu.co.
Adriana Marcela Celis Ramírez, Email: acelis@uniandes.edu.co.
Supplementary information
is available for this paper at 10.1038/s41598-020-61729-1.
References
- 1.Brilhante RS, et al. Malassezia pachydermatis from animals: Planktonic and biofilm antifungal susceptibility and its virulence arsenal. Vet. Microbiol. 2018;220:47–52. doi: 10.1016/j.vetmic.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Merkel S, Heidrich D, Danilevicz CK, Scroferneker ML, Zanette RA. Drosophila melanogaster as a model for the study of Malassezia pachydermatis infections. Vet. Microbiol. 2018;224:31–33. doi: 10.1016/j.vetmic.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Peano A, et al. Methodological issues in antifungal susceptibility testing of Malassezia pachydermatis. J. Fungi. 2017;37:1–15. doi: 10.3390/jof3030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ledbetter EC, Starr JK. Malassezia pachydermatis keratomycosis in a dog. Med. Mycol. Case Rep. 2015;10:24–26. doi: 10.1016/j.mmcr.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campoy Sonia, Adrio José L. Antifungals. Biochemical Pharmacology. 2017;133:86–96. doi: 10.1016/j.bcp.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Vuran E, Karaarslan A, Karasartova D, Turegun B, Sahin F. Identification of Malassezia species from pityriasis versicolor lesions with a new multiplex PCR method. Mycopathologia. 2014;177:41–49. doi: 10.1007/s11046-013-9704-6. [DOI] [PubMed] [Google Scholar]
- 7.Angileri M, Pasquetti M, De Lucia M, Peano A. Azole resistance of Malassezia pachydermatis causing treatment failure in a dog. Med. Mycol. Case Rep. 2019;23:58–61. doi: 10.1016/j.mmcr.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi E, Tan CL, Aw D. Malassezia: a case of coexisting pityriasis versicolor and Malassezia folliculitis. Singapore Med. J. 2000;14:353–353. doi: 10.11622/smedj.2018079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jesus FPK, et al. In vitro susceptibility of fluconazole-susceptible and -resistant isolates of Malassezia pachydermatis against azoles. Vet. Microbiol. 2011;152:161–164. doi: 10.1016/j.vetmic.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 10.Garau M, Pereiro M. In vitro susceptibilities of Malassezia species to a new triazole, Albaconazole (UR-9825), and other antifungal compounds. Antimicrob. Agents Chemother. 2003;47:2342–2344. doi: 10.1128/AAC.47.7.2342-2344.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cafarchia C, et al. In vitro evaluation of Malassezia pachydermatis susceptibility to azole compounds using E-test and CLSI microdilution methods. Med. Mycol. 2012;87:795–801. doi: 10.3109/13693786.2012.674219. [DOI] [PubMed] [Google Scholar]
- 12.Bazzani S, Hoppe A, Holzhütter H-G. Network-based assessment of the selectivity of metabolic drug targets in Plasmodium falciparum with respect to human liver metabolism. BMC Syst. Biol. 2012;6:118. doi: 10.1186/1752-0509-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uddin R, Masood F, Azam SS, Wadood A. Identification of putative non-host essential genes and novel drug targets against Acinetobacter baumannii by in silico comparative genome analysis. Microb. Pathog. 2019;128:28–35. doi: 10.1016/j.micpath.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Rawls KD, et al. A simplified metabolic network reconstruction to promote understanding and development of flux balance analysis tools. Comput. Biol. Med. 2019;105:64–71. doi: 10.1016/j.compbiomed.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Dreyfuss Jonathan M., Zucker Jeremy D., Hood Heather M., Ocasio Linda R., Sachs Matthew S., Galagan James E. Reconstruction and Validation of a Genome-Scale Metabolic Model for the Filamentous Fungus Neurospora crassa Using FARM. PLoS Computational Biology. 2013;9(7):e1003126. doi: 10.1371/journal.pcbi.1003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu N, et al. Reconstruction and analysis of the genome-scale metabolic network of Candida glabrata. Mol. Biosyst. 2013;9:205–216. doi: 10.1039/C2MB25311A. [DOI] [PubMed] [Google Scholar]
- 17.Triana S, et al. Lipid metabolic versatility in Malassezia spp. yeasts studied through metabolic modeling. Front. Microbiol. 2017;8:1–18. doi: 10.3389/fmicb.2017.01772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiele I, Fleming RMT, Bordbar A, Schellenberger J, Palsson B. Functional characterization of alternate optimal solutions of Escherichia coli’s transcriptional and translational machinery. Biophys. J. 2010;98:2072–2081. doi: 10.1016/j.bpj.2010.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulfer SL, Scott EM, Couture JF, Pillus L, Trievel RC. Crystal structure and functional analysis of homocitrate synthase, an essential enzyme in lysine biosynthesis. J. Biol. Chem. 2009;284:35769–35780. doi: 10.1074/jbc.M109.046821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong X, Zhao Y, Zhao J, Wang X. Characterization of aspartate kinase and homoserine dehydrogenase from Corynebacterium glutamicum IWJ001 and systematic investigation of L-isoleucine biosynthesis. J. Ind. Microbiol. Biotechnol. 2016;43:873–885. doi: 10.1007/s10295-016-1763-5. [DOI] [PubMed] [Google Scholar]
- 21.Xu H, West AH, Cook PF. Overall kinetic mechanism of saccharopine dehydrogenase from Saccharomyces cerevisiae. Am. Chem. Soc. 2006;45:12156–12166. doi: 10.1021/bi0610808. [DOI] [PubMed] [Google Scholar]
- 22.Gräslund S, et al. Protein production and purification. Nat. Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood DW. New trends and affinity tag designs for recombinant protein purification. Curr. Opin. Struct. Biol. 2014;26:54–61. doi: 10.1016/j.sbi.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Sigma-Aldrich®. NAD/NADH Quantification Kit. Sigma-Aldrich Co. MAK037 (2012).
- 25.Sigma-Aldrich®. Coenzyme A Assay Kit. Sigma-Aldrich Co. MAK034 (2012).
- 26.Michaelis L, Menten ML, Goody RS, Johnson KA. Die kinetik der invertinwirkung/The kinetics of invertase action. Biochemistry. 1913;49:333–369. [Google Scholar]
- 27.Johnson K, Goody R. The original Michaelis constant. Biochemistry. 2012;50:8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lineweaver H, Burk D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934;56:658–666. doi: 10.1021/ja01318a036. [DOI] [Google Scholar]
- 29.American Type Culture Collection. Primary Epidermal Keratinocytes; Normal, Human, Adult (HEKa) (ATCC®PCS-200-011TM) (2018).
- 30.Zomorrodi AR, et al. Improving the iMM904 S. cerevisiae metabolic model using essentiality and synthetic lethality data. BMC Syst. Biol. 2010;4:178. doi: 10.1186/1752-0509-4-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitkänen Esa, Jouhten Paula, Hou Jian, Syed Muhammad Fahad, Blomberg Peter, Kludas Jana, Oja Merja, Holm Liisa, Penttilä Merja, Rousu Juho, Arvas Mikko. Comparative Genome-Scale Reconstruction of Gapless Metabolic Networks for Present and Ancestral Species. PLoS Computational Biology. 2014;10(2):e1003465. doi: 10.1371/journal.pcbi.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomonaga Y, Kaneko R, Goto M, Ohshima T, Yoshimune K. Structural insight into activation of homoserine dehydrogenase from the archaeon Sulfolobus tokodaii via reduction. Biochem. Biophys. Reports. 2015;3:14–17. doi: 10.1016/j.bbrep.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroeder AC, et al. Threonine-insensitive homoserine dehydrogenase from soybean: Genomic organization, kinetic mechanism, and in vivo activity. J. Biol. Chem. 2010;285:827–834. doi: 10.1074/jbc.M109.068882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai PW, et al. Candida albicans Hom6 is a homoserine dehydrogenase involved in protein synthesis and cell adhesion. J. Microbiol. Immunol. Infect. 2017;50:863–871. doi: 10.1016/j.jmii.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Bulfer SL, Mcquade TJ, Larsen MJ, Trievel RC. Application of a high-throughput fluorescent acetyltransferase assay to identify inhibitors of homocitrate synthase. Anal. Biochem. 2011;410:133–140. doi: 10.1016/j.ab.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bulfer SL, Scott EM, Pillus L, Trievel RC. Structural basis for L-lysine feedback inhibition of homocitrate synthase. J. Biol. Chem. 2010;285:10446–10453. doi: 10.1074/jbc.M109.094383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheng X, Gao J, Liu Y, Liu C. Theoretical study on the proton shuttle mechanism of saccharopine dehydrogenase. J. Mol. Graph. Model. 2013;44:17–25. doi: 10.1016/j.jmgm.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Pink DBS, et al. Lysine α-ketoglutarate reductase, but not saccharopine dehydrogenase, is subject to substrate inhibition in pig liver. Nutr. Res. 2011;31:544–554. doi: 10.1016/j.nutres.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Wulandari AP, et al. Characterization of bacterial homocitrate synthase involved in lysine biosynthesis. FEBS Lett. 2002;522:35–40. doi: 10.1016/S0014-5793(02)02877-6. [DOI] [PubMed] [Google Scholar]
- 40.Ekanayake DK, West AH, Cook PF. Contribution of K99 and D319 to substrate binding and catalysis in the saccharopine dehydrogenase reaction. Arch. Biochem. Biophys. 2011;514:8–15. doi: 10.1016/j.abb.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.James CL, Viola RE. Production and characterization of bifunctional enzymes. Domain swapping to produce new bifunctional enzymes in the aspartate pathway. Biochemistry. 2002;41:3720–3725. doi: 10.1021/bi015909o. [DOI] [PubMed] [Google Scholar]
- 42.Navratna V, Reddy G, Gopal B. Structural basis for the catalytic mechanism of homoserine dehydrogenase. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71:1216–1225. doi: 10.1107/S1399004715004617. [DOI] [PubMed] [Google Scholar]
- 43.Miyajima R, Otsuka SI, Shiio I. Regulation of aspartate family amino acid biosynthesis in Brevibacterium flavum. J. Biochem. 1968;63:139–148. doi: 10.1093/oxfordjournals.jbchem.a128754. [DOI] [PubMed] [Google Scholar]
- 44.Hama H, Kayahara T, Tsuda M, Tsuchiya T. Inhibition of homoserine dehydrogenase I by L-serine in Escherichia coli. J. Biochem. 1991;109:604–608. doi: 10.1093/oxfordjournals.jbchem.a123427. [DOI] [PubMed] [Google Scholar]
- 45.Wedler FC, Ley BW. Kinetic and regulatory mechanisms for (Escherichia coli) homoserine dehydrogenase-I. Equilibrium isotope exchange kinetics. J. Biol. Chem. 1993;268:4880–4888. [PubMed] [Google Scholar]
- 46.Chen Zhen, Rappert Sugima, Zeng An-Ping. Rational Design of Allosteric Regulation of Homoserine Dehydrogenase by a Nonnatural Inhibitor l-Lysine. ACS Synthetic Biology. 2014;4(2):126–131. doi: 10.1021/sb400133g. [DOI] [PubMed] [Google Scholar]
- 47.Andi B, West AH, Cook PF. Stabilization and characterization of histidine-tagged homocitrate synthase from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2004;421:243–254. doi: 10.1016/j.abb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 48.Gabriel I, Milewski S. Characterization of recombinant homocitrate synthase from Candida albicans. Protein Expr. Purif. 2016;125:7–18. doi: 10.1016/j.pep.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Okada T, Tomita T, Wulandari AP, Kuzuyama T, Nishiyama M. Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus. J. Biol. Chem. 2010;285:4195–4205. doi: 10.1074/jbc.M109.086330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian J, Khandogin J, West AH, Cook PF. Evidence for a catalytic dyad in the active site of homocitrate synthase from Saccharomyces cerevisiae. Biochemistry. 2008;47:6851–6858. doi: 10.1021/bi800087k. [DOI] [PubMed] [Google Scholar]
- 51.Andi B, West AH, Cook PF. Regulatory mechanism of histidine-tagged homocitrate synthase from Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:31624–31632. doi: 10.1074/jbc.M502846200. [DOI] [PubMed] [Google Scholar]
- 52.Fujioka M, Nakatani Y. A kinetic study of saccharopine dehydrogenase reaction. Eur. J. Biochem. 1970;16:180–186. doi: 10.1111/j.1432-1033.1970.tb01070.x. [DOI] [PubMed] [Google Scholar]
- 53.Saunders PP, Broquist HP. Saccharopine, an intermediate of the aminoadipic acid pathway of lysine biosynthesis. J. Biol. Chem. 1966;240:3435–3440. [PubMed] [Google Scholar]
- 54.Schmidt H, Bode R, Birnbaum D. Regulation of the lysine biosynthesis in Pichia guilliermondii. Antonie Van Leeuwenhoek. 1989;56:337–347. doi: 10.1007/BF00443747. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt H, Bode R, Lindner M, Birnbaum D. Lysine biosynthesis in the yeast Candida maltosa: properties of some enzymes and regulation of the biosynthetic pathway. J. Basic Microbiol. 1985;25:675–681. doi: 10.1002/jobm.3620251018. [DOI] [Google Scholar]
- 56.Fujioka M. Saccharopine dehydrogenase. Substrate inhibition studies. J. Biol. Chem. 1975;250:8986–8989. [PubMed] [Google Scholar]
- 57.Ye Z, Garrad R, Winston M, Bhattacharjee J. Use of a-aminoadipate and lysine as sole nitrogen source by Schizosaccharomyces pombe and selected pathogenic fungi. J. Basic Microbiol. 1991;31:149–156. doi: 10.1002/jobm.3620310215. [DOI] [PubMed] [Google Scholar]
- 58.Tucci AF, Ceci LN. Homocitrate synthase from yeast. Arch. Biochem. Biophys. 1972;153:742–750. doi: 10.1016/0003-9861(72)90393-1. [DOI] [PubMed] [Google Scholar]
- 59.Tucci A, Ceci L. Control of lysine biosynthesis in yeast. Arch. Biochem. Biophys. 1972;153:751–754. doi: 10.1016/0003-9861(72)90394-3. [DOI] [PubMed] [Google Scholar]
- 60.Bañuelos O, et al. Characterization and lysine control of expression of the lys1 gene of Penicillium chrysogenum encoding homocitrate synthase. Gene. 1999;226:51–59. doi: 10.1016/S0378-1119(98)00551-4. [DOI] [PubMed] [Google Scholar]
- 61.Bhattacharjee JK. α-aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes. Crit. Rev. Microbiol. 1985;12:131–151. doi: 10.3109/10408418509104427. [DOI] [PubMed] [Google Scholar]
- 62.Feller A, Ramos F, Piérard A, Dubois E. In Saccharomyces cerevisae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur. J. Biochem. 1999;261:163–170. doi: 10.1046/j.1432-1327.1999.00262.x. [DOI] [PubMed] [Google Scholar]
- 63.Niederberger P, Miozzari G, Hütter R. Biological role of the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2015;1:584–593. doi: 10.1128/MCB.1.7.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sigma-Aldrich®. Product specification, L-threonine. 1–2 (2016).
- 65.Robert-Gero M, Poiret M, Cohen GN. Homoserine dehydrogenase of Pseudomonas putida properties and regulation. Biochim. Biophys. Acta. 1970;6:31–37. doi: 10.1016/0005-2744(70)90078-1. [DOI] [PubMed] [Google Scholar]
- 66.O’Sullivan J. Growth inhibition of Acetobacter aceti by L-threonine and L-homoserine: the primary regulation of the biosynthesis of amino acids of the aspartate family. J. Gen. Microbiol. 1974;85:153–159. doi: 10.1099/00221287-85-1-153. [DOI] [PubMed] [Google Scholar]
- 67.Galvis-Marín JC, et al. Actividad antifúngica in vitro de azoles y anfotericina B frente a Malassezia furfur por el método de microdilución M27-A3 del CLSI y Etest®. Rev. Iberoam. Micol. 2017;34:89–93. doi: 10.1016/j.riam.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 68.Cafarchia C, et al. Assessment of the antifungal susceptibility of Malassezia pachydermatis in various media using a CLSI protocol. Vet. Microbiol. 2012;159:536–540. doi: 10.1016/j.vetmic.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 69.Valgas C, Machado de Souza S, Smânia EFA, Smânia AJ. Screening methods to determine antibacterial activity of natural products. Brazilian J. Microbiol. 2007;38:369–380. doi: 10.1590/S1517-83822007000200034. [DOI] [Google Scholar]
- 70.International Organization for Standarization. ISO 10993-5. Biological evaluation of medical devices. (2009).
- 71.Choksakulnimitr S, Masuda S, Tokuda H, Takakura Y, Hashida M. In vitro cytotoxicity of macromolecules in different cell culture systems. J. Control. Release. 1995;34:233–241. doi: 10.1016/0168-3659(95)00007-U. [DOI] [Google Scholar]
- 72.Morgan DML, Larvin VL, Pearson JD. Biochemical characterisation of polycation-induced cytotoxicity to human vascular endothelial cells. J. Cell Sci. 1989;94:553–559. doi: 10.1242/jcs.94.3.553. [DOI] [PubMed] [Google Scholar]
- 73.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:2730. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2015;44:471–480. doi: 10.1093/nar/gkv1164. [DOI] [Google Scholar]
- 75.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 76.Triana, S. et al. Metabolic reconstruction of five Malassezia genomes. (Universidad de los Andes, 2015).
- 77.Gudmundsson S, Thiele I. Computationally efficient flux variability analysis. BMC Bioinformatics. 2010;1:1–7. doi: 10.1186/1471-2105-11-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suthers PF, Zomorrodi A, Maranas CD. Genome-scale gene/reaction essentiality and synthetic lethality analysis. Mol. Syst. Biol. 2009;5:1–17. doi: 10.1038/msb.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing (2018).
- 80.Edwards, J. S. & Palsson, B. O. Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC Bioinformatics1 (2000). [DOI] [PMC free article] [PubMed]
- 81.Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: A web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Placzek S, et al. BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 2016;45:380–388. doi: 10.1093/nar/gkw952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Invitrogen®. TRIzolTMreagent user guide. ThermoFisher Scientific15596018 (2016).
- 85.Qiagen®. TissueLyser Handbook. Sample & Assay Technologies1064945, (Qiagen, 2010).
- 86.Chakrabarty Jagabandhu. Applied Plasticity, Second Edition. Boston, MA: Springer US; 2010. [Google Scholar]
- 87.Clontech. SMARTerTMPCR cDNA synthesis kit user manual. Clontech Laboratories, Inc. PT4097-1 (2014).
- 88.New England BioLabs® (NEB). PCR Protocol Phusion® DNA Polymerase. Available at: https://www.neb.com/protocols/1/01/01/pcr-protocol-m0530 (Accessed: 14th July 2019) (2018).
- 89. Qiagen®. QIAquick®Spin Handbook. Sample to Insight (Qiagen, 2018).
- 90.Shangai ShineGene Molecular Biotech, Inc. Specifications for Custom Gene Synthesis. Available at: http://www.shinegene.org.cn/english/genesynthesis.html. (Accessed: 20th October 2018) (2007).
- 91.Thermo Scientific. Manual GeneJET plasmid miniprep kit. Thermo Fisher Scientific, Inc. K0502 (2014).
- 92.New England BioLabs® (NEB). NEB cloner tool. Available at: http://nebcloner.neb.com/#!/redigest (Accessed: 7th October 2018) (2018).
- 93.Thermo Scientific. Manual GeneJET gel extraction kit. Thermo Fisher Scientific, Inc. (2015).
- 94.New England BioLabs® (NEB). Ligation protocol with T4 DNA Ligase. Available at: https://www.neb.com/protocols/1/01/01/dna-ligation-with-t4-dna-ligase-m0202. (Accessed: 7th January 2019) (2018).
- 95.New England BioLabs® (NEB). FAQ: How can I increase transformation efficiency? Available at: https://www.neb.com/faqs/0001/01/01/how-can-i-increase-transformation-efficiency (Accessed: 17th January 2019) (2019).
- 96.Bio-Rad Laboratories Inc. ProfinityTMIMAC Resins Instruction Manual. 10001677 (2000).
- 97.Cantón, E., Martín, E. & Espinel-Ingroff, A. Métodos estandarizados por el CLSI para el estudio de la sensibilidad a los antifúngicos (documentos M27-A3, M38-A y M44-A). Rev. Iberoam. Micol. 24 (2007).
- 98.Clinical and Laboratory Standards Institute (CLSI). Reference method for broth dilution. Antifungal Susceptibility Testing of Yeast; Approved Standard28, (2008).
- 99.Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016;6:71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tukey, J. W. & Cleveland, W. S. The collected works of John W. Tukey: Time series 1949-1964. I, (Chapman and Hall/CRC, 1984).
- 101.Westerdijk Fungal Biodiversity Institute. CBS-KNAW Collections. Available at: http://www.westerdijkinstitute.nl/Collections/BioloMICS.aspx?TableKey=14682616000000089&Rec=1406&Fields=All (Accessed: 7th December 2019) (2019).
- 102.Triana S, et al. Draft genome sequence of the animal and human pathogen Malassezia pachydermatis strain CBS 1879. Genome Announc. 2015;3:5–6. doi: 10.1128/genomeA.01197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.González A, et al. Physiological and molecular characterization of atypical isolates of Malassezia furfur. J. Clin. Microbiol. 2009;47:48–53. doi: 10.1128/JCM.01422-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.American Type Culture Collection. ATCC the Global Bioresource Center. 2–10 Available at: https://www.google.com.sa/url?sa=t&rct=j&q=&esrc=s&source=web&cd=2&ved=0CC0QFjABahUKEwiqg-vF7pLIAhWJvhQKHaDGAK4&url=http://www.ncbi.nlm.nih.gov/pmc/articles/PMC514607/&usg=AFQjCNG2yzD3FsdzmSNkZce_yX_a4cXFYw. (Accessed: 18th June 2019) (2016).
- 105.Celis Ramírez, A. M. CeMoP – Grupo de investigación celular y molecular de microorganismos patógenos. Universidad de los Andes Available at: https://gcemop.wordpress.com/ (Accessed: 7th August 2019) (2018).
- 106.Clontech. pET6xHN expression vector set. Certificate of analysis. Clontech Laboratories Inc. 631432 (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information Files).