Abstract
The 31- and 32-nt 5′-fragment of Y4-RNA (Y4RNAfr) exists abundantly in human peripheral blood plasma. Although physiological roles of the plasma Y4RNAfr are not well established, its potential utility as a diagnostic/prognostic marker for acute coronary syndrome was suggested. In this paper, to establish a normal range of the Y4RNAfr level in plasma, we measured plasma Y4RNAfr levels of 40 healthy persons using the method we have developed, and compared them with other blood test data. From the obtained data, we tentatively regarded <0.1 fmol/ng as normal for the Y4RNAfr level in peripheral blood plasma. And the white blood cell count (WBC) and the C-reactive protein (CRP) level showed moderate positive correlations with the Y4RNAfr level, suggesting that Y4RNAfr could be a potential novel inflammatory marker. We also measured the Y4RNAfr level in peripheral blood plasma from four multiple myeloma patients. The plasma Y4RNAfr level was abnormal in all four myeloma patients, and the levels for two patients were far beyond the normal level. The WBC for each patient was normal and the CRP levels for two patients were normal. These observations together suggest that a high level of Y4RNAfr in peripheral blood plasma and a normal WBC could be indicative of multiple myeloma.
Keywords: Y4-RNA fragment, qRT-PCR, Plasma, Inflammatory marker, Multiple myeloma, White blood cell, C-reactive protein
1. Introduction
A biomarker is defined as a measurable indicator of normal biological processes, pathogenic processes, or pharmacologic responses to therapeutic interventions (https://www.fda.gov/drugs/cder-biomarker-qualification-program/more-about-biomarkers-qualification). A level of C-reactive protein (CRP) and a white blood cell count (WBC) are biomarkers of inflammation in a routine blood test, and M protein in serum is a typical diagnostic biomarker of multiple myeloma [1]. Nowadays, beyond proteins, small molecular metabolites, and cell type distribution in blood, potential utility of RNA molecules such as miRNA in plasma is investigated as biomarkers of cancers, non-alcoholic fatty liver disease, and neurodegenerative diseases [[2], [3], [4]].
We have found by chance that the 31-nt 5′-fragment of Y4-RNA exists abundantly in plasma from both healthy persons and multiple myeloma patients when investigating the gene regulatory network via tRNase ZL and small guide RNA (sgRNA) [[5], [6], [7]]. The cellular full-length Y4-RNA is a 94–96-nt non-coding RNA, and is thought to be involved in the initiation of DNA replication and RNA quality control [8,9]. The 31- and 32-nt 5′-fragments of Y4-RNA (Y4RNAfr) have been also shown to exist in saliva samples after cell removal [10]. Although physiological roles of the plasma Y4RNAfr are not well established [[11], [12], [13], [14]], its potential utility as a diagnostic/prognostic marker for acute coronary syndrome was suggested from the two observations: The Y4RNAfr exists in sera of coronary artery disease patients more abundantly than in sera of controls [15]. The plasma Y4RNAfr level correlates with platelet function in patients with acute coronary syndrome [16].
Recently we have established a method to quantitate an absolute amount of the Y4RNAfr using reverse transcription (RT) PCR [10]. This method should be more accurate than those in which only a relative level of the Y4RNAfr against that of an internal small RNA can be measured [15,16]. In this paper, to establish a normal range of the Y4RNAfr level in plasma, we measured plasma Y4RNAfr levels of 40 healthy persons using our method, and compared them with other blood test data. We also analyzed absolute amounts of the Y4RNAfr in peripheral blood plasma and bone marrow plasma from multiple myeloma patients. These analyses suggest that Y4RNAfr could be a potential novel inflammatory marker and might be used as a diagnostic/prognostic marker of multiple myeloma.
2. Materials and methods
2.1. Preparation of plasma RNA
Peripheral blood plasma or bone marrow plasma was filtrated with a sterile syringe filter with a 0.45-μm pore size membrane to remove cells. Total RNA was extracted from plasma with RNAiso Blood (Takara Bio, Shiga, Japan) according to manufacturer's instructions. The extracted RNA was quantitated with the fluorescent dye RiboGreen and a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, USA) according to manufacturer's protocol.
Blood samples from 40 healthy persons were obtained in Niitsu Medical Center Hospital, and blood and bone marrow samples from four myeloma patients were in Niigata Cancer Center Hospital. Written informed consent was obtained from each person, and this study was approved by the ethics committee of Niigata University of Pharmacy and Applied Life Sciences (Permit Numbers: H28–008 and H29-007) according to Ethical Guidelines for Medical and Health Research Involving Human Subjects (Public Notice of the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labour and Welfare, Japan, No. 3 of 2014).
2.2. Synthetic RNA/DNA
The 31-nt Y4RNAfr with a 5′-phosphate, 5′-pGGCUGGUCCGAUGGUAGUGGGUUAUCAGAAC-3′ was chemically synthesized by Nippon Bioservice (Saitama, Japan). The following DNA primers were obtained from Sigma-Aldrich (Tokyo, Japan): SL-forward_Y4_3, 5′-GGCGGCTGGTCCGATGGT-3′; Y4RNA_R4, 3′-CCCAATAGTCTTGGCG-5′; Y4RNA_R5, 3′-TTTAAACTGACCGGCG-5′.
2.3. RT-PCR
An RNA test sample was reverse transcribed using a reverse primer, Y4RNA_R4 or Y4RNA_R5, with a PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio, Shiga, Japan) according to manufacturer's protocol. Subsequently, a real time PCR for the obtained cDNA was carried out using the primer pair SL-forward_Y4_3/Y4RNA_R4 or SL-forward_Y4_3/Y4RNA_R5 and SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio, Shiga, Japan) with a Thermal Cycler Dice Real Time System (Takara Bio, Shiga, Japan) under the standard conditions according to manufacturer's protocol.
In case that the ~95-nt full-lengthY4-RNA co-exists in RNA samples and contributes to overestimation, we also performed RT-PCR for it [10]. Although the full-length Y4-RNA was detected in the samples examined, its amount was negligible compared with that of the Y4RNAfr.
2.4. Blood test
Blood samples were subjected to measurement of fasting blood glucose level (FBGL), WBC, red blood cell count (RBC), hemoglobin (HGB), hematocrit (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), platelets (PLT), neutrophil (Neu), eosinophil (Eos), basophil (Bas), monocyte (Mon), lymphocyte (Lym), red blood cell distribution width (RDW-SD), platelet distribution width (PDW), mean platelet volume (MPV), platelet-large cell ratio (P-LCR), aspartate aminotransferase (AST), alanine transaminase (ALT), alkaline phosphatase (ALP), lactate dehydrogenase (LDH), γ-guanosine triphosphate (γ-GTP), total bilirubin (T-BiL), total cholesterol (TCHO), triglyceride (TG), high density lipoprotein cholesterol (HDL-C), low density lipoprotein cholesterol (LDL-C), blood urea nitrogen (BUN), uric acid (UA), creatinine (Cre), serum amylase (AMY), total protein (TP), albumin/globulin ratio (A/G), sodium (Na), potassium (K), chloride (Cl), calcium (Ca), and CRP. The measurement was carried out by standard procedures.
2.5. Statistical analysis
Correlation coefficients and p-values in the Student's t-test were calculated with the software Microsoft Excel 2016.
3. Results and discussion
3.1. Measurement of the plasma Y4RNAfr level
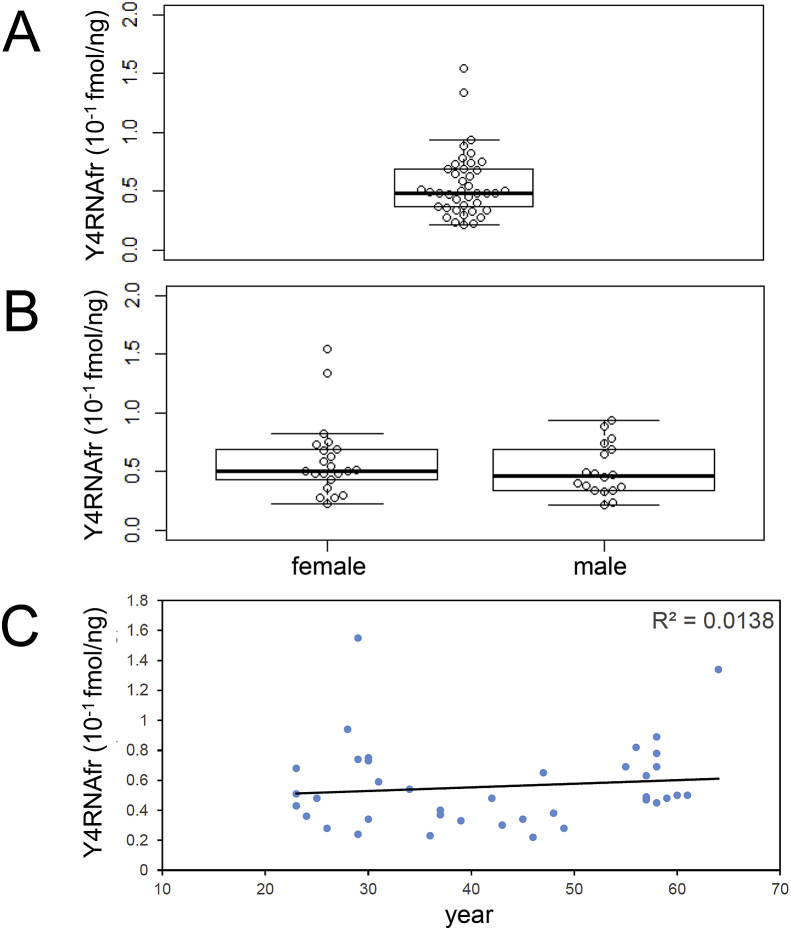
We measured the Y4RNAfr level in peripheral blood plasma from each of 40 healthy persons according to our standard protocol, and calculated a fmol amount per 1 ng of total plasma RNA. The mean, median, maximum, and minimum values of the Y4RNAfr level were 0.056, 0.049, 0.155, and 0.022 fmol/ng, respectively, and there were two outliers (Fig. 1A). The distribution pattern did not differ largely between females and males (Fig. 1B), and there was no correlation between the Y4RNAfr level and age (Fig. 1C). From these observations, we tentatively regarded <0.1 fmol/ng as normal for the Y4RNAfr level in peripheral blood plasma.
Fig. 1.
The Y4RNAfr levels in peripheral blood plasma from 40 healthy persons. Box-and-whisker plots for the whole data set (A) and for the female (n = 22) and male (n = 18) data sets (B) are shown. There was no significant difference in the distribution pattern between females and males (p-value = 0.33). (C) A plot of the Y4RNAfr level against age is shown. R, correlation coefficient.
3.2. Persons with abnormality in the WBC and the CRP level show an abnormal Y4RNAfr level
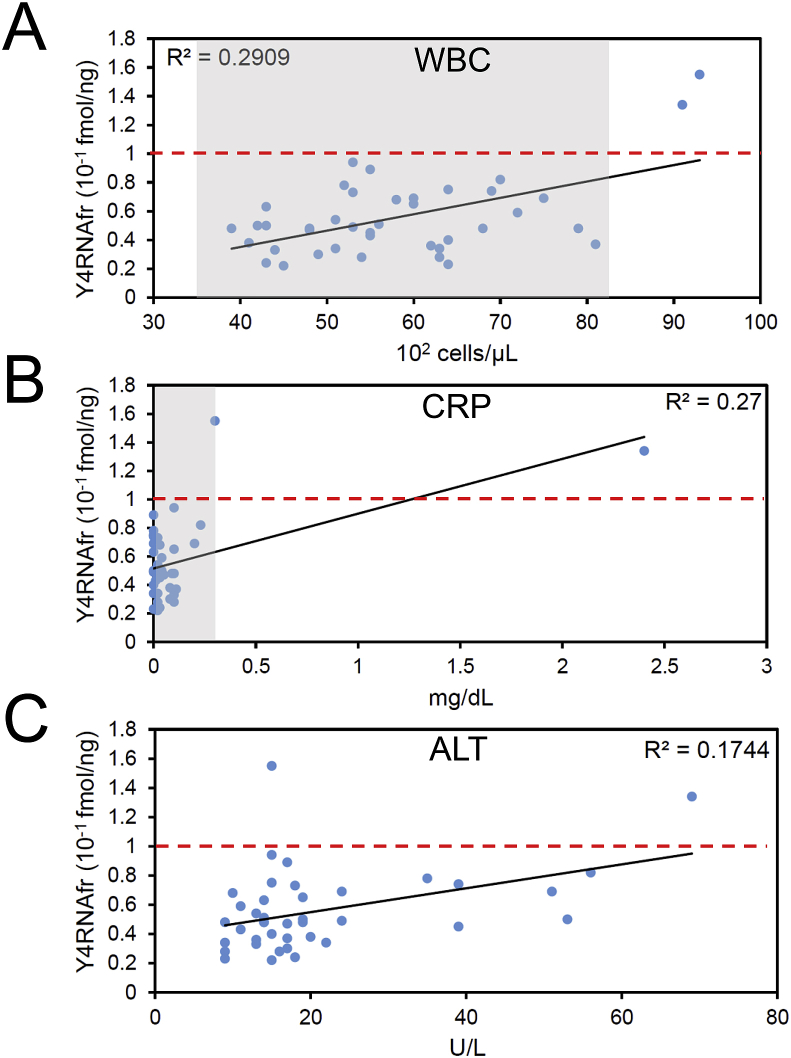
Next, we examined if the plasma Y4RNAfr level correlates with levels of 39 test items in blood analyzed by the routine blood test (Fig. 2 and Supplementary Fig. S1). The levels of WBC, CRP, and ALT showed moderate positive correlations (R2 = 0.16–0.49) with the Y4RNAfr level (Fig. 2). The levels of FBGL, RBC, HCT, AST, ALP, LDH, γ-GTP, TG, TP, and UA were positively correlated weakly (R2 = 0.04–0.16) with the Y4RNAfr level, and the levels of MCHC, Mon, HDL-C, and Cl were negatively correlated weakly (Supplementary Fig. S1A).
Fig. 2.
Correlations of the Y4RNAfr level in peripheral blood plasma with the levels of WBC (A), CRP (B), and ALT (C). A region for a normal range is shaded with respect to WBC (3500–8100 cells/μL) and CRP (<0.3 mg/dL). R denotes a correlation coefficient, and p-values in WBC, CRP, and ALT are 0.00033, 0.00059, and 0.0073, respectively.
The correlations of the Y4RNAfr level with the WBC and CRP levels were the strongest with the correlation coefficients R = + 0.54 and R = + 0.52, respectively. Two examinees who showed an abnormal WBC level and an abnormal or borderline CRP level showed an abnormal Y4RNAfr level (Fig. 2A and B). These observations suggest that Y4RNAfr could be a potential novel inflammatory marker.
3.3. Multiple myeloma patients show a high level of the plasma Y4RNAfr
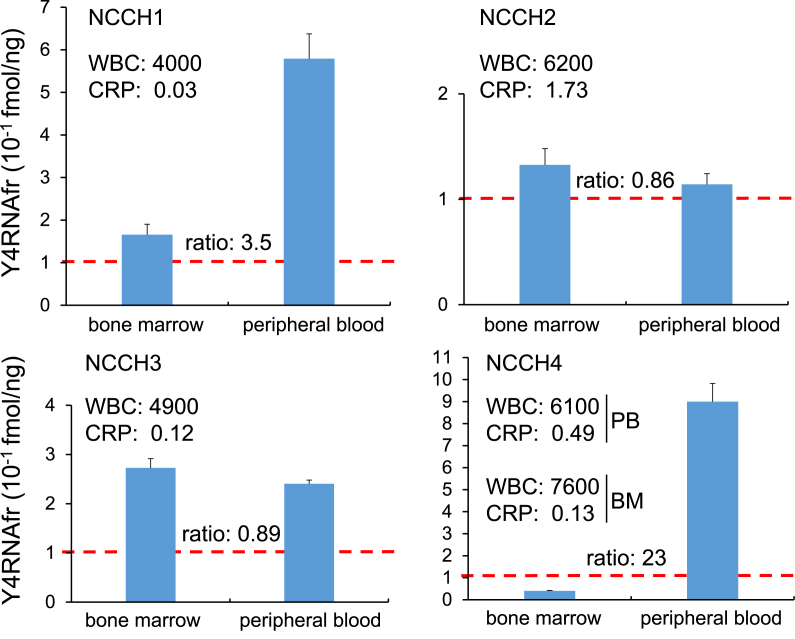
Furthermore, we measured the Y4RNAfr level in peripheral blood plasma and bone marrow plasma from each of the four multiple myeloma patients NCCH1–NCCH4, and calculated a fmol amount per 1 ng of total plasma RNA (Fig. 3). The Y4RNAfr level in peripheral blood plasma was abnormal in all four myeloma patients, and the levels for NCCH1 (0.58 fmol/ng) and NCCH4 (0.90 fmol/ng) were far beyond the normal level (<0.1 fmol/ng), although they showed no obvious inflammation symptoms. The WBC for each patient was normal and the CRP levels for NCCH1 and NCCH3 were normal. These observations together suggest that a high level of Y4RNAfr in peripheral blood plasma and a normal WBC could be indicative of multiple myeloma.
Fig. 3.
The Y4RNAfr levels in peripheral blood plasma and bone marrow plasma from the multiple myeloma patients NCCH1–NCCH4. The WBC, the CRP levels, and the ratio of the Y4RNAfr level in peripheral blood plasma to that in bone marrow plasma are also shown. Sampling dates for peripheral blood (PB) and bone marrow (BM) differed by 6 days with respect to NCCH4. Error bars indicate SD (n = 3).
The Y4RNAfr level in bone marrow plasma varied from 0.04 to 0.27 fmol/ng, and the ratio of the Y4RNAfr level in peripheral blood plasma to that in bone marrow plasma also varied largely from 0.86 to 23. These varied values might reflect different types of myeloma and might become prognostic indicators.
3.4. The plasma Y4RNAfr as a potential novel biomarker
This study suggests that Y4RNAfr could be a potential novel inflammatory marker and that its level might be a diagnostic/prognostic indicator of multiple myeloma. However, further intensive and extensive studies using large cohorts are needed to validate the current observations, and new studies could also discover further utility of Y4RNAfr as diagnostic/prognostic markers for other diseases.
Funding information
This work was supported by the Council for Science, Technology and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP) and ‘Technologies for creating next-generation agriculture, forestry and fisheries’ (funding agency: Bio-oriented Technology Research Advancement Institution, NARO). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interest
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2019.12.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Rajkumar S.V., Dimopoulos M.A., Palumbo A. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15:538–548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 2.Xi X., Li T., Huang Y. RNA biomarkers: frontier of precision medicine for cancer. Noncoding RNA. 2017;3:9. doi: 10.3390/ncrna3010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turchinovich A., Baranova A., Drapkina O. Cell-free circulating nucleic acids as early biomarkers for NAFLD and NAFLD-associated disorders. Front. Physiol. 2018;9:1256. doi: 10.3389/fphys.2018.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson C.N., Belli A., Di Pietro V. Small non-coding RNAs: new class of biomarkers and potential therapeutic targets in neurodegenerative disease. Front. Genet. 2019;10:364. doi: 10.3389/fgene.2019.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ninomiya S., Kawano M., Abe T. Potential small guide RNAs for tRNase ZL from human plasma, peripheral blood mononuclear cells, and cultured cell lines. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbarbary R.A., Takaku H., Uchiumi N. Modulation of gene expression by human cytosolic tRNase ZL through 5′-half-tRNA. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbarbary R.A., Takaku H., Uchiumi N. Human cytosolic tRNase ZL can downregulate gene expression through miRNA. FEBS Lett. 2009;583:3241–3246. doi: 10.1016/j.febslet.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Zhang A.T., Langley A.R., Christov C.P. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J. Cell Sci. 2011;124:2058–2069. doi: 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sim S., Wolin S.L. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscip. Rev. RNA. 2011;2:686–699. doi: 10.1002/wrna.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa T., Haino A., Seki M. The Y4-RNA fragment, a potential diagnostic marker, exists in saliva. Non-coding RNA Res. 2017;2:122–128. doi: 10.1016/j.ncrna.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hizir Z., Bottini S., Grandjean V. RNY (YRNA)-derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2016.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haderk F., Schulz R., Iskar M. Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aah5509. pii: eaah5509. [DOI] [PubMed] [Google Scholar]
- 13.Li C., Qin F., Hu F. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018;8:2. doi: 10.1186/s13578-018-0202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ninomiya S., Ishikawa T., Takahashi M. Potential physiological roles of the 31/32-nucleotide Y4-RNA fragment in human plasma. Non-Coding RNA Res. 2019 doi: 10.1016/j.ncrna.2019.11.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Repetto E., Lichtenstein L., Hizir Z. RNY-derived small RNAs as a signature of coronary artery disease. BMC Med. 2015;13:259. doi: 10.1186/s12916-015-0489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaudewitz D., Skroblin P., Bender L.H. Association of microRNAs and YRNAs with platelet function. Circ. Res. 2016;118:420–432. doi: 10.1161/CIRCRESAHA.114.305663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.