Abstract
Background & Aims
Self-renewal and multipotent differentiation are cardinal properties of intestinal stem cells (ISCs), mediated in part by WNT and NOTCH signaling. Although these pathways are well characterized, the molecular mechanisms that control the ‘stemness’ of ISCs are still not well defined. Here, we investigated the role of Krüppel-like factor 5 (KLF5) in regulating ISC functions.
Methods
We performed studies in adult Lgr5EGFP-IRES-creERT2;Rosa26LSLtdTomato (Lgr5Ctrl) and Lgr5EGFP-IRES-creERT2;Klf5fl/fl;Rosa26LSLtdTomato (Lgr5ΔKlf5) mice. Mice were injected with tamoxifen to activate Cre recombinase, which deletes Klf5 from the intestinal epithelium in Lgr5ΔKlf5 but not Lgr5Crtl mice. In experiments involving irradiation, mice were subjected to 12 Gy total body irradiation (TBI). Tissues were collected for immunofluorescence (IF) analysis and next generation sequencing. Oganoids were derived from fluoresecence activated cell sorted- (FACS-) single cells from tamoxifen-treated Lgr5ΔKlf5 or Lgr5Crtl mice and examined by immunofluorescence stain.
Results
Lgr5+ ISCs lacking KLF5 proliferate faster than control ISCs but fail to self-renew, resulting in a depleted ISC compartment. Transcriptome analysis revealed that Klf5-null Lgr5+ cells lose ISC identity and prematurely differentiate. Following irradiation injury, which depletes Lgr5+ ISCs, reserve Klf5-null progenitor cells fail to dedifferentiate and regenerate the epithelium. Absence of KLF5 inactivates numerous selected enhancer elements and direct transcriptional targets including canonical WNT- and NOTCH-responsive genes. Analysis of human intestinal tissues showed increased levels of KLF5 in the regenerating epithelium as compared to those of healthy controls.
Conclusion
We conclude that ISC self-renewal, lineage specification, and precursor dedifferentiation require KLF5, by its ability to regulate epigenetic and transcriptional activities of ISC-specific gene sets. These findings have the potential for modulating ISC functions by targeting KLF5 in the intestinal epithelium.
Keywords: Intestinal Stem Cell, Multipotent Differentiation, Tissue Regeneration, Epigenetic Regulation
Abbreviations used in this paper: ASCL2, achaete-scute family bHLH transcription factor 2; ChIP-seq, chromatin immunoprecipitation assay with sequencing; EdU, 5-ethynyl-2′-deoxyuridine; EGFP, enhanced green fluorescent protein; GSEA, gene set enrichment analysis; H&E, hematoxylin and eosin; IGV, Integrative Genomics Viewer; ISC, intestinal stem cell; IRR, irradiation; KLF5, Krüppel-like factor 5; LGR5, leucine rich repeat containing G protein-coupled receptor 5; RFP, red fluorescent protein; RNA-seq, RNA sequencing; RT-qPCR, reverse transcriptase quantitative polymerase chain reaction; TA, transit amplifying; TF, transcription factor; TSS, transcription start site; TUNEL, terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling
Graphical abstract
Summary.
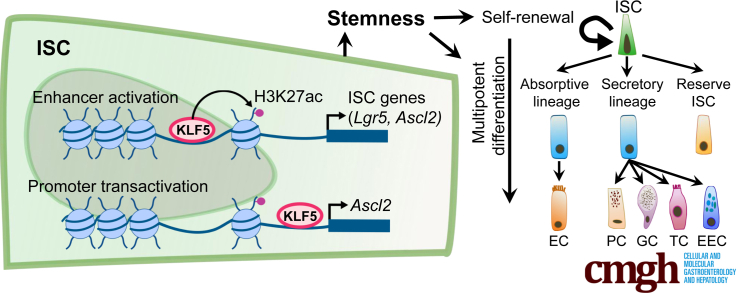
Intestinal stem cells are required for proliferation, differentiation, and regeneration of the intestinal epithelium. Krüppel-like factor 5 regulates intestinal stem cells in both physiologic and pathological conditions and may be a treatment target in certain diseases of the intestine.
The intestinal epithelium is replenished every 3–5 days and is driven by Lgr5+ intestinal stem cells (ISCs) at the crypt bottom.1 Stemness encompasses both self-renewal and multipotent differentiation, which must be carefully balanced to maintain a stable ISC pool while continuously supplying differentiated cells in the villi. ISCs divide stochastically and symmetrically, following “neutral drift” dynamics.2,3 When ISCs are depleted by gamma irradiation or other injuries, multiple crypt cell types dedifferentiate to replace them, revealing significant tissue plasticity.4, 5, 6, 7, 8 Both WNT and NOTCH signaling pathways are necessary for maintaining ISC stemness,9, 10, 11, 12 but the mechanisms by which they influence ISC division and differentiation are not well defined at this time.
Krüppel-like zinc-finger transcription factor (TF) KLF5 is expressed in both ISCs and transit-amplifying (TA)13, 14, 15 cells and regulates epithelial proliferation, differentiation, and development.13,16 Conditional Klf5 deletion from the entire mouse intestinal epithelium, using Villin-Cre as a driver, impairs epithelial cell proliferation.17, 18, 19 Previous studies in Lgr5EGFP-IRES-creERT2 mice indicated that KLF5 is important for crypt cell survival,15,20 but could not distinguish its requirement between ISCs and TA cells. Consequently, KLF5’s ability to regulate ISC stemness, its transcriptional targets, and possible links to WNT and NOTCH signaling remain undefined. In addition, whether epigenetic modifications such as covalent histone marks are regulated by KLF5 in ISCs has not been examined. In the current study, we hypothesize that KLF5 is critical for the maintenance of ISC identity and functions through transcriptional and epigenetic regulation.
To determine KLF5’s functions in ISCs, we investigated Lgr5EGFP-IRES-creERT2;Rosa26LSLtdTomato (Lgr5Ctrl) and Lgr5EGFP-IRES-creERT2;Klf5fl/fl;Rosa26LSLtdTomato (Lgr5ΔKlf5) mice following tamoxifen-induced activation of Cre recombinase. Surprisingly, absence of KLF5 increased ISC proliferation and induced premature enterocyte differentiation, with attendant loss of ISC identity. KLF5 is also required for the regeneration of the intestinal epithelium in response to radiation injury. Global gene analyses revealed a role of KLF5 in controlling both epigenetic and transcriptional activities of ISC-specific gene sets, including selected key elements related to WNT and NOTCH signaling. These findings identify a novel molecular mechanism by which a tissue-restricted TF maintains ISC identity and functions.
Results
KLF5 Deficiency Accelerates ISC Proliferation, Inhibits Self-Renewal, and Impairs Crypt Cell Dedifferentiation
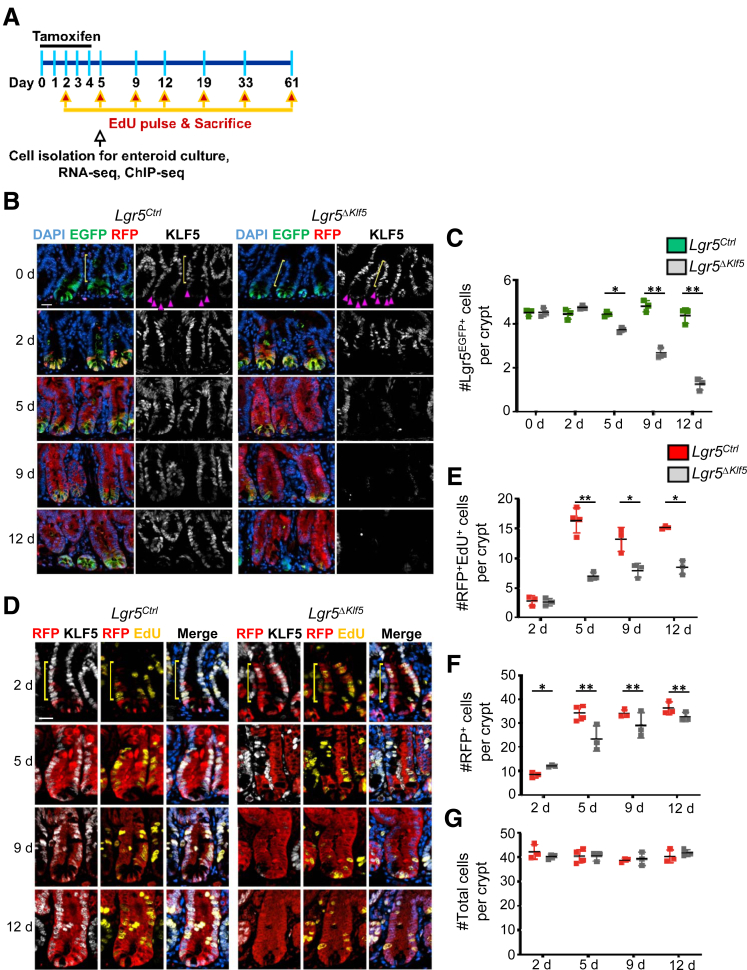
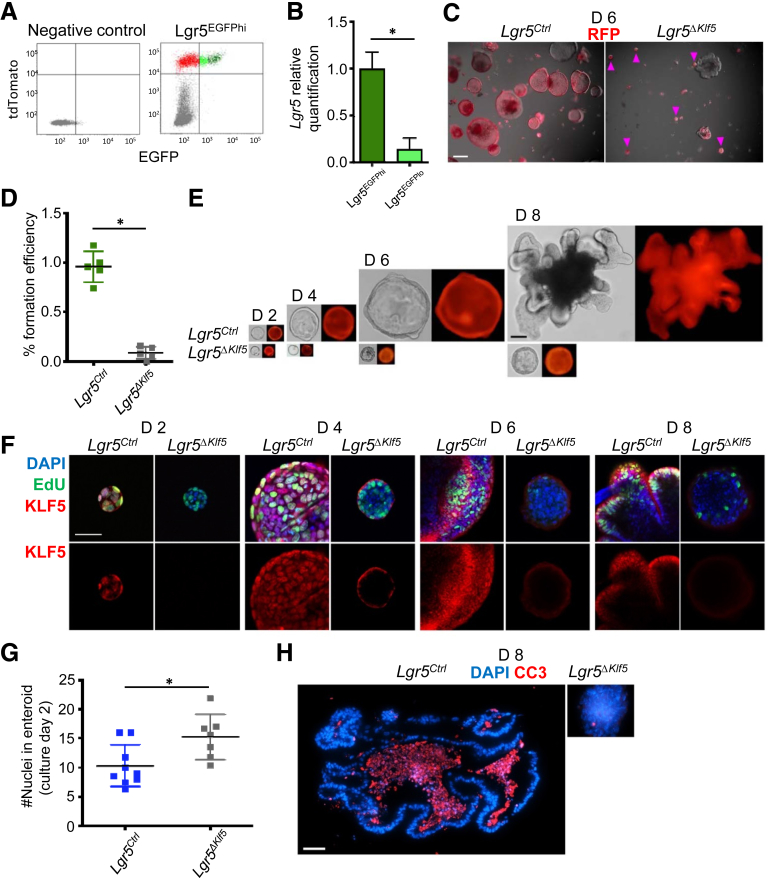
To investigate the role of KLF5 in regulating ISC self-renewal and maintenance, we injected Lgr5Ctrl and Lgr5ΔKlf5 mice with tamoxifen for 5 consecutive days to activate Cre recombinase and 5-ethynyl-2′-deoxyuridine (EdU) to selectively label cells in S-phase (Figure 1A). In Lgr5Ctrl mice or Lgr5ΔKlf5 mice before tamoxifen administration, KLF5 is expressed in both ISCs (Figure 1B, magenta arrowheads) and the TA zone of progenitor cells (Figure 1B, yellow brackets). Over a 12-day period following the initial tamoxifen treatment, the crypts of Lgr5ΔKlf5 mice showed a progressive loss of Lgr5EGFP+ ISCs (Figures 1B and 1C) and reduced expansion of EdU+RFP+ crypt cells (Figures 1D and 1E) when compared with control mice. Using a 3-hour EdU pulse treatment, we found at all studied time points that approximately 20% of Lgr5+ cells were in S-phase in Lgr5Ctrl mice (Figures 2A and 2B). In contrast, between 2 and 5 days after Klf5 deletion, up to 35% of Lgr5+ cells incorporated EdU (Figures 2A and 2B). This difference in cell proliferation between Lgr5Ctrl and Lgr5ΔKlf5 mice was no longer apparent after day 9, possibly because the number of Lgr5+ cells was significantly reduced (Figure 1C) and replaced by KLF5-expressing Lgr5EGFP+ cells that had escaped Cre recombination (Figure 2C).
Figure 1.
KLF5 is required for intestinal stem cell self-renewal. (A) Scheme of the experimental plan. Eight- to 12-week-old Lgr5Ctrl and Lgr5ΔKlf5 mice were injected with tamoxifen for 5 consecutive days and sacrificed at 0, 2, 5, 9, 12,19, 33, or 61 days after the first injection. Mice were injected with EdU 3 or 24 hours before sacrifice. Lgr5EGFPhi cells were FACS-isolated for 3-dimensional enteroid culture, RNA-seq, and ChIP-seq at day 5. (B) Representative immunofluorescence images of EGFP, RFP, KLF5, and DAPI in the PSI crypts of Lgr5Ctrl and Lgr5ΔKlf5 mice. KLF5 expression was observed in Lgr5EGFPhi cells at the base of the crypts (magenta arrowheads), as well as cells in the TA zone (yellow brackets). Scale bars represent 20 μm. (C) Quantification of average number of Lgr5EGFPhi cells per crypt. (D) Representative immunofluorescence images of RFP, KLF5, EdU, and DAPI of PSI crypts of Lgr5Ctrl and Lgr5ΔKlf5 mice. Mice were treated with 3 hours EdU pulse. Scale bar represents 20 μm. (E–G) Quantification of (E) EdU-incorporated RFP+, (F) an average number of RFP+, and (G) total cells per crypt. Data are expressed as mean ± SD, 20 crypts quantified per mouse (E–G), n = 3–5 mice per group. ∗P < .05, ∗∗P < .01 by linear mixed regression models.
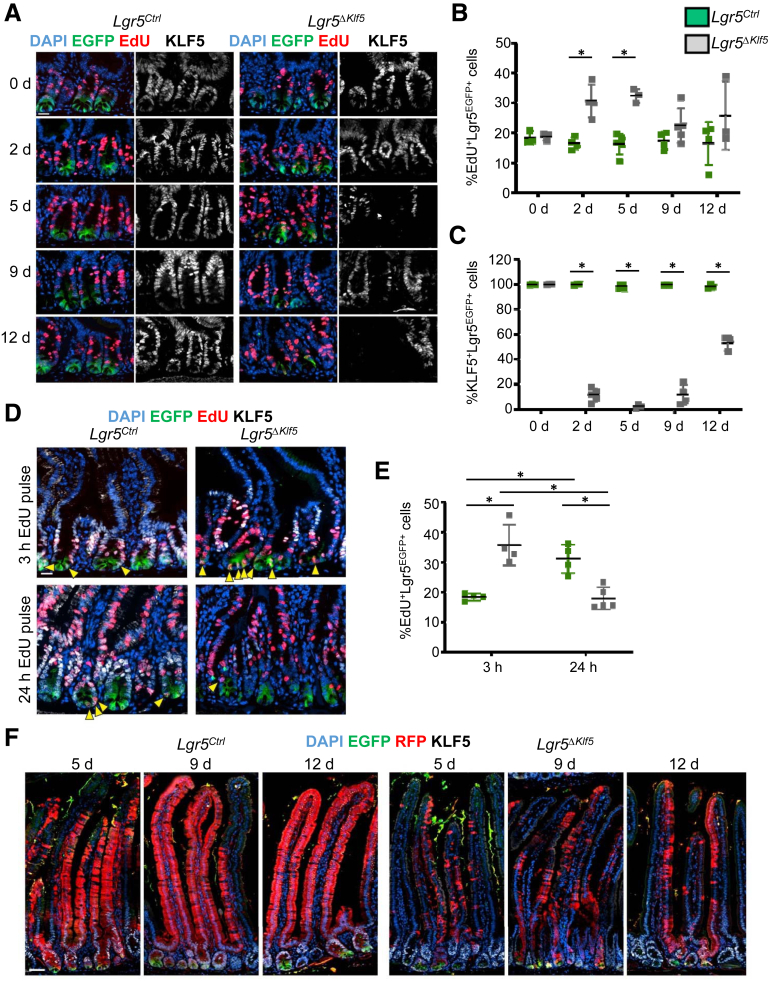
Figure 2.
KLF5 regulates proliferation of intestinal stem cells. (A) Representative immunofluorescence (IF) images of EGFP, EdU, KLF5, and DAPI of PSI crypts after 3 hours EdU pulse treatment at day 0, 2, 5, 9, and 12 after tamoxifen injections. Scale bar represents 20 μm. (B, C) Quantification of percent (B) EdU-incorporated or (C) KLF5-expressing Lgr5EGFP+ cells. (D) Representative IF images of EGFP, EdU, KLF5, and DAPI of the PSI crypts after 3 or 24 hours of pulse EdU treatment at day 5. EGFP+EdU+ cells are marked with yellow arrowheads. Scale bars represent 20 μm. (E) Quantification of percent of EdU-incorporated Lgr5EGFP+ cells. (F) Representative IF images of EGFP, RFP, KLF5, and DAPI of PSI crypt-villus axis of Lgr5Ctrl and Lgr5ΔKlf5 mice at 5, 9, and 12 days after the first tamoxifen injection. Scale bar represents 50 μm. Data are represented as mean ± SD, 250 cells quantified per mouse, n = 4–5 mice per group. ∗P < .05 by Mann-Whitney U test.
As increased EdU incorporation in Lgr5+ cells upon loss of KLF5 implies a faster rate of ISC proliferation, we traced the fate of ISC division after 3-hour and 24-hour EdU pulse treatments. In Lgr5Ctrl mice, the fraction of EdU-labeled Lgr5+ ISCs increased from 18.4 ± 0.6% at 3 hours to 31.1 ± 2.4% at 24 hours (Figures 2D and 2E, yellow arrowheads), providing evidence for self-renewal. This is confirmed by the significantly higher number of RFP+ progenitors within the crypts, from day 2 to 5, in Lgr5Ctrl mice (Figure 1F). In contrast, the proportion of EdU-labeled Lgr5+ cells in Lgr5ΔKlf5 mice decreased from 35.7 ± 3.4% at 3 hours to 17.9 ± 1.6% at 24 hours (Figures 2D and 2E), suggesting that self-renewal of ISCs is impaired, which leads to reduced numbers of Lgr5+ ISCs from the crypt base. These findings indicate that absence of KLF5 accelerates ISC division and reduces self-renewal, leading to ISC exhaustion. Importantly, these ISC functions contrast with those in crypts at large, where absence of KLF5 impairs cell replication.15,18,20
Although Klf5-deleted ISCs proliferate faster, generation of the lineage was stunted during the first 12 days, as evidenced by the scarcity of RFP+ cells within villi compared with Lgr5Ctrl mice (Figure 2F). Crypt cells, which predominantly drive tissue renewal, showed reduced EdU incorporation after Klf5 deletion (Figures 1D and 1E). Whereas RFP+ cells replaced most crypt cells in Lgr5Ctrl mice by day 5, lineage tracing by Klf5-deleted RFP+ cells was slower (Figures 1D and 1F). However, the total number of crypt cells from day 2 to 12 was similar between Lgr5Ctrl and Lgr5ΔKlf5 mice (Figure 1G).
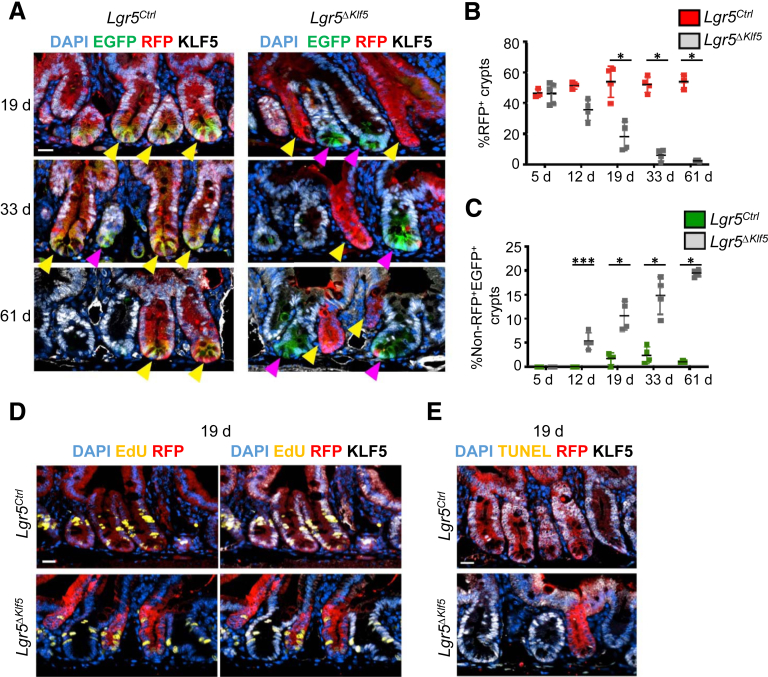
To determine long-term effects of KLF5 loss on the tissue lineage, we traced GFP+ and RFP+ cells for 19, 33, and 61 days following tamoxifen treatment. Klf5-null RFP+ crypts were rapidly depleted (Figures 3A and 3B) and the few residual crypts at day 61 were diminutive and devoid of Lgr5EGFP+ ISCs (Figure 3A, yellow arrowheads). KLF5-expressing RFP-Lgr5EGFP+ cells appeared in Lgr5ΔKlf5 mice starting at day 12 (Figure 3A, magenta arrowheads; and Figure 3C [such cells were infrequent in Lgr5Ctrl mice]). Furthermore, residual Klf5-null RFP+ crypts continued to incorporate EdU (Figure 3D) and terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) staining revealed absence of apoptosis (Figure 3E). Together, these findings imply that crypt loss resulted from impaired ISC self-renewal and impaired ability of Klf5-null progenitors to dedifferentiate in response to ISC attrition.
Figure 3.
KLF5 is required for long-term intestinal stem cell survival. (A) Representative immunofluorescence images of EGFP, RFP, KLF5, and DAPI of PSI crypts of Lgr5Ctrl and Lgr5ΔKlf5 mice scarified at day 19, 33, or 61. Yellow arrowheads mark RFP+ crypts. Magenta arrowheads mark crypts with non-RFP, Lgr5EGFPhi cells. Scale bar represents 20 μm. (B, C) Quantification of the percentage of (B) RFP+ crypts and (C) crypts with non-RFP, Lgr5EGFPhi cells of Lgr5Ctrl and Lgr5ΔKlf5 mice at 5, 12, 19, 33, and 61 days. Data are expressed as mean ± SD, n = 3–6 mice per group. ∗P < .05 by Mann-Whitney U test. (D, E) Representative immunofluorescence images of RFP, KLF5, DAPI, and (D) EdU or (E) TUNEL of PSI crypts of Lgr5Ctrl and Lgr5ΔKlf5 mice injected with 3 hours EdU pulse treatment at day 19. Scale bar represents 20 μm.
KLF5 Is Required for ISC Clonal Expansion
To test the ability of ISCs to expand clonally in 3D enteroid cultures in the absence of KLF5, we isolated Lgr5EGFPhi cells from Lgr5Ctrl and Lgr5ΔKlf5 mice (Figures 4A and 4B). We did so on day 5 after tamoxifen treatment, based on the high recombination efficiency and strong in vivo phenotypes evident at this time (Figures 1 and 2). Approximately 1% of control Lgr5EGFPhi cells formed enteroids by day 6, while Klf5-null cells expanded briefly but failed to form typical, mature enteroids (Figures 4C, 4D, and 4E). A majority of Klf5-deleted cells incorporated EdU on the second day of ex vivo culture, and the average number of nuclei was higher than in control cultures (Figures 4F and 4G), but EdU incorporation ceased by day 6 (Figure 4F). Moreover, staining with CC3 on day 8 of culture showed absence of apoptosis in the arrested enteroids derived from Lgr5ΔKlf5 cells (Figure 4H). These data confirm that Klf5 deletion initially accelerates ISC proliferation, but the cells subsequently fail at clonal expansion.
Figure 4.
Loss of KLF5 in Lgr5EGFP+cells impairs clonal expansion of ISCs in 3D enteroid culture. (A) FACS isolation of RFP-expressing Lgr5EGFPhi and Lgr5EGFPlo cells. Rosa26LSLtdTomato mice were used as negative control. (B) RT-qPCR analysis of Lgr5 in Lgr5EGFPhi or Lgr5EGFPlo populations of Lgr5Ctrl mice after the sorting. (C) Representative bright field and RFP images of enteroid culture at day 6. Magenta arrowheads mark Klf5-deleted cell clumps. Scale bar represents 200 μm. (D) Quantification of the percent enteroid formation. (E) Representative images of enteroids at culture day 2, 4, 6, and 8. (F) Representative confocal images of EdU, KLF5, and DAPI of enteroids treated with EdU 3 hours before formalin fixation. Scale bar represents 20 μm. (G) Quantification of the number of nuclei per enteroids at day 2 of culture. (H) Representative immunofluorescence images of CC3 and DAPI of enteroid at day 8. Scale bar represent 50 μm. Data are represented as mean ± SD, n = 3–6 mice per group, ∗P < .05, **P < .01 by (B, G) Mann-Whitney U test or (D) linear mixed regression models.
KLF5 Deficiency Results in Premature ISC Differentiation
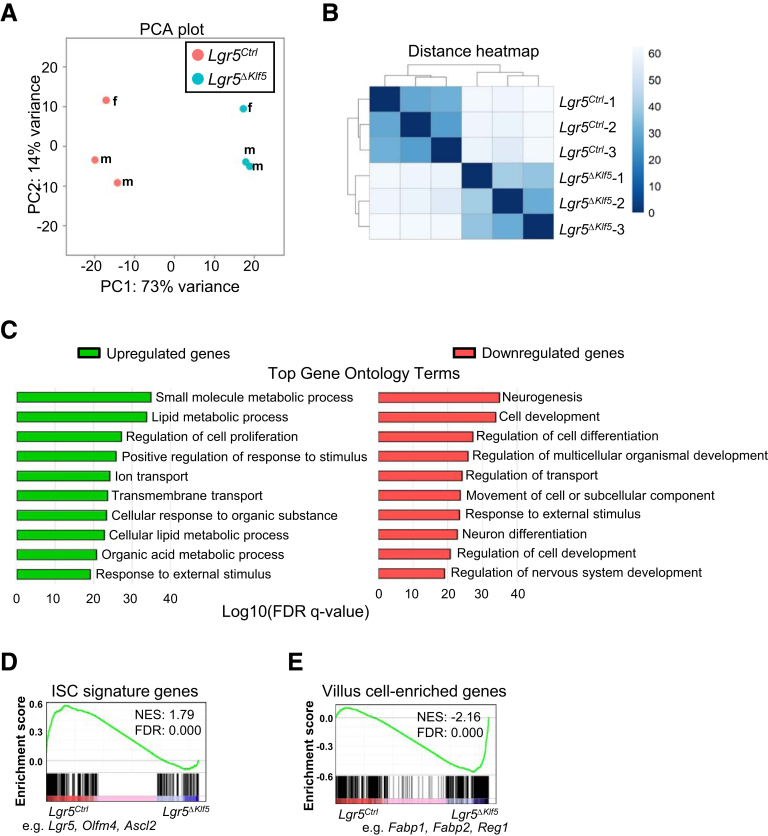
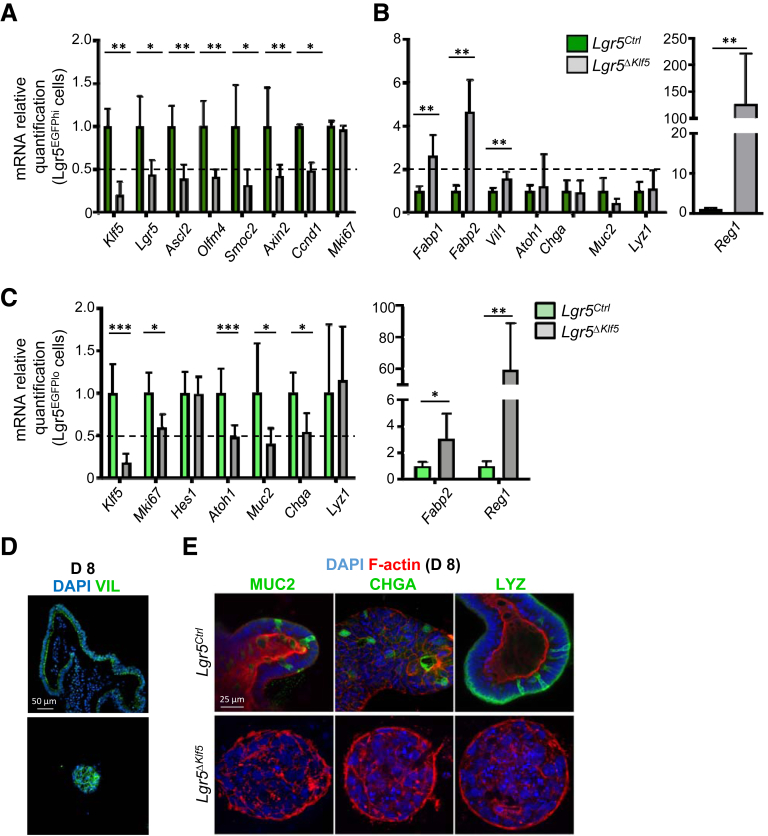
To define the transcriptional impact of Klf5 loss in Lgr5+ ISCs, we profiled the transcriptomes of Lgr5EGFPhi cells isolated from Lgr5Ctrl and Lgr5ΔKlf5 mice and observed differences in 2209 protein-coding genes (log2 fold-change >|1.5|; 1064 upregulated; 1145 downregulated) at a false discovery rate <.05 (Supplementary Table S1). Control and mutant cells clustered distinctly (Figures 5A and 5B), and by Gene Ontology analysis, genes upregulated in Klf5-null cells display metabolic functions associated with villus differentiation,21,22 whereas downregulated genes exhibit functions related to development and differentiation (Figure 5C). To characterize these changes further, we performed gene set enrichment analysis (GSEA) on the full dataset against ISC signature23 and villus-enriched24 genes. Klf5-null ISCs were depleted of ISC signature genes, such as Lgr5, Olfm4, Ascl2, and Smoc2 (Figure 5D), and enriched for genes that are highly expressed in villus cells, such as Fabp1, Fabp2, Reg1, and Krt20 (Figure 5E). Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) analysis of selected RNA sequencing (RNA-seq) findings confirmed elevation of enterocyte transcript levels in Klf5-null ISCs, such as Fabp1, Fabp2, and Reg1 (Figures 6A and 6B). RT-qPCR on Lgr5EGFPlo progenitors showed significant reduction of secretory lineage-specific genes Atoh1, Muc2, and Chga, whereas enterocyte markers Fabp2 and Reg1 were significantly increased over control cells (Figure 6C). Furthermore, the few Klf5-deleted enteroids contained VIL1-expressing, but lacked MUC2+, CHGA+, and LYZ+, cells (Figures 6D and 6E). Together, these findings indicate that loss of KLF5 in Lgr5EGFP+ cells results in loss of the ISC transcriptional signature, with premature enterocyte-biased differentiation, revealing a cardinal role for KLF5 in determining ISC “stemness.”
Figure 5.
Loss of KLF5 modifies transcriptome of Lgr5EGFPhiintestinal stem cells. (A) Principal component analysis (PCA) plot of RNA-seq analysis of FACS-isolated Lgr5EGFPhi cells from Lgr5Ctrl and Lgr5ΔKlf5 mice, n = 3. (B) Heatmap analysis of RNA-seq data. (C) Top 10 significant Gene Ontology terms of biological processes from differentially expressed genes. (D-E) GSEA. Intestinal stem cell signature genes23 are enriched in (D) control Lgr5EGFPhi cells, whereas genes differentially expressed in (E) villus cells (GSE71713) are enriched in Klf5-deleted Lgr5EGFPhi cells. FDR, false discovery rate; NES, normalized enrichment score.
Figure 6.
KLF5 is required to preserve the intestinal stem cell transcriptome of Lgr5EGFP+cells to prevent premature differentiation. (A, B) RT-qPCR analysis of genes in FACS-isolated Lgr5EGFPhi cells from Lgr5Ctrl and Lgr5ΔKlf5 mice. Data are represented as mean ± SD, n = 4–6, ∗P < .05, ∗∗P < .01 by Mann-Whitney U test. (C) RT-qPCR analysis of genes in FACS-isolated Lgr5EGFPlo cells (early progenitors) from Lgr5Ctrl and Lgr5ΔKlf5 mice. Data are represented as mean ± SD, n = 4–6, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 by Mann-Whitney U test. (D) Representative immunofluorescence images of Villin1 and DAPI of day 8 enteroids cultured from FACS-isolated Lgr5EGFPhi cells from Lgr5Ctrl and Lgr5ΔKlf5 mice. Scale bars represent 50 μm. (E) Representative confocal images of MUC2, CHGA, or LYZ with F-actin and DAPI of day 8 enteroids. Scale bars represent 25 μm.
KLF5 Maintains H3K27ac at Genomic Loci Associated With ISC Gene Expression
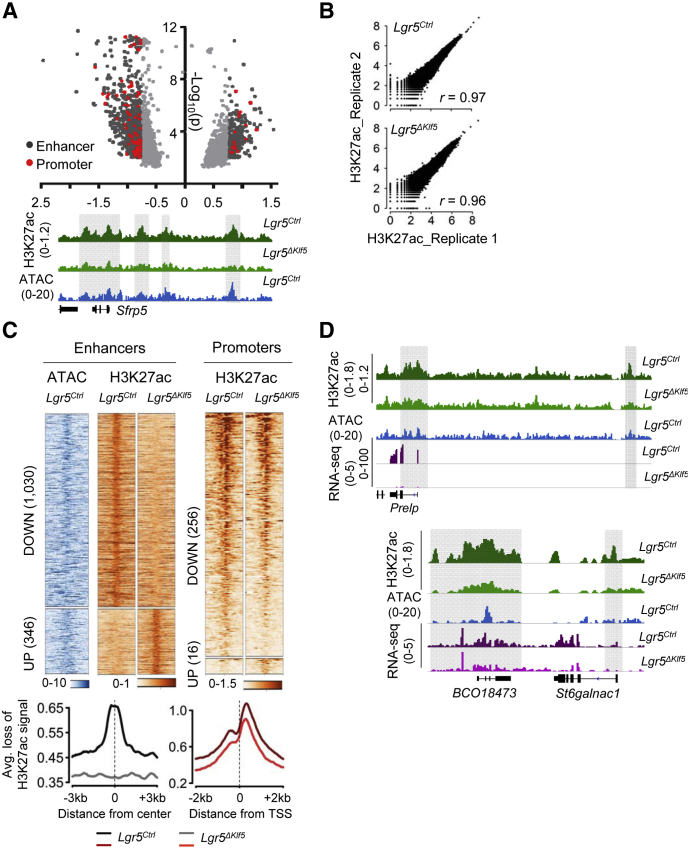
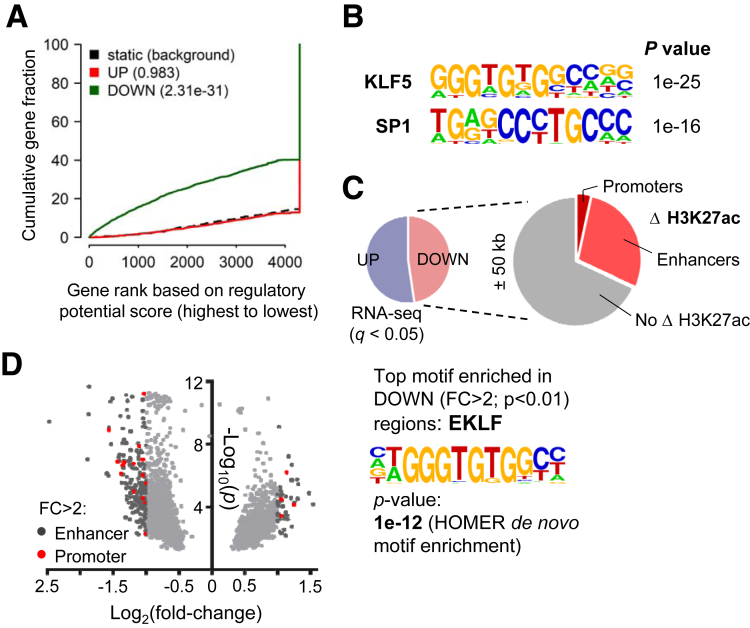
TFs occupy their target gene promoters and enhancers marked by active histone marks, such as H3K27ac.25,26 To identify KLF5-dependent cis-elements that may underlie the transcriptional response to KLF5 loss, we used chromatin immunoprecipitation followed by sequencing (ChIP-seq) to assess genome-wide H3K27ac distributions in Klf5-null ISCs. Compared with Lgr5Ctrl ISCs, duplicate samples of Lgr5ΔKlf5 ISCs showed 1,286 regions of reduced and 362 sites with increased H3K27ac (fold-change >1.7, P < .01, identified by diffReps) (Figures 7A and 7B).27 Compared with transcription start sites (TSSs) (promoters), many more distant regions lost H3K27ac in Lgr5ΔKlf5 ISCs (Figure 7C). Affected distant regions, such as those in the Sfrp5, Prelp, and St6galnac1 loci (Figures 7A and 7D), had accessible chromatin28,29 in control ISCs (Figure 7C) (GSE83394) and were significantly associated with genes having reduced expression in Lgr5ΔKlf5 ISCs (Figure 8A), indicating that they are bona fide cis-elements. Moreover, the TF-binding sequence most enriched in these regions corresponds to the KLF binding motif (Figure 8B), which implies that KLF5 occupancy at these sites underlies enhancer activity. Furthermore, 31.8% of downregulated genes (q < .05) had a H3K27ac-depleted site within 50 kb of the TSS (Figure 8C). In contrast, the many fewer sites (346 enhancers and 16 promoters) that acquired H3K27ac lacked association with upregulated genes (data not shown). Even by a more stringent cut-off (fold-change >2), enhancers with reduced H3K27ac in Lgr5ΔKlf5 ISCs were significantly enriched for the KLF motif (Figure 8D). Together, these data indicate that KLF5 primarily activates genes through distant enhancers and that increases in gene expression are likely secondary effects unrelated to KLF5 binding.
Figure 7.
Loss of Klf5 in intestinal stem cells leads to depletion of H3K27ac at genomic loci. (A) Genome-wide differential H3K27ac analysis27 reveals up- and downregulated regions in Lgr5ΔKlf5. Volcano plot shows all regions achieving P < .01 with fold-change (FC) depicted by shades of gray (light gray FC <1.7, dark gray FC ≥1.7); red dots mark promoters. Representative IGV tracks for H3K27ac28 and ATAC-seq (blue) at Sfrp5 in Lgr5Ctrl and Lgr5ΔKlf5 or Lgr5Ctrl ISCs, respectively. The shaded boxes mark regions of H3K27ac loss at promoter and putative enhancers. (B) Scatter plots show correlation between duplicate Lgr5Ctrl or Lgr5ΔKlf5 H3K27ac ChIP-seq samples. r is the Pearson correlation coefficient. (C) Heatmaps represent ATAC-seq (in Lgr5Ctrl ISCs; GSE83394) and H3K27ac at 1030 down and 346 upregulated enhancers in Lgr5ΔKlf5 compared with control ISCs (FC ≥1.7, P < .01). H3K27ac at 256 and 16 down and upregulated promoters, respectively, in Lgr5ΔKlf5 is depicted to the right. Aggregate plots show average signal intensities at enhancers (left) and promoters depleted for H3K27ac in Lgr5ΔKlf5. (D) Representative IGV tracks for H3K27ac ChIP-seq,28 ATAC-seq (blue), and RNA-seq (purple) at Prelp and St6galnac1 loci. The shaded regions depict loss of H3K27ac at promoters or enhancers.
Figure 8.
Depletion of H3K27ac associates with downregulated gene expression upon loss of Klf5. (A) BETA (binding and expression target analysis)31 reveals a strong association of DOWN promoters and enhancers with downregulated genes in Lgr5ΔKlf5 within ±50 kb of TSS. These regions do not associate with upregulated genes.24 Plot depicts the cumulative regulatory potential score, dashed line represents the background, and p-values denote the significance of UP or DOWN associations relative to the background. (B) Downregulated regions are significantly enriched for KLF5 motif as predicted by HOMER motif analysis. (C) Approximately 32% of all downregulated genes (q < .05) show diminished H3K27ac levels at promoters and enhancers as derived from BETA. Left pie chart shows the fractions of up- or downregulated genes (q < .05) in Lgr5ΔKlf5. The right pie chart represents the fractions of downregulated genes associated with a loss (shades of red) or no change (gray slice) in H3K27ac. (D) Genome-wide differential H3K27ac analysis by diffReps as in Figure 7A. Fold-change (FC) is depicted by shades of gray (light gray FC <2, dark gray FC ≥2), red dots mark promoters. Downregulated regions are most enriched for EKLF motif as predicted by HOMER motif analysis.
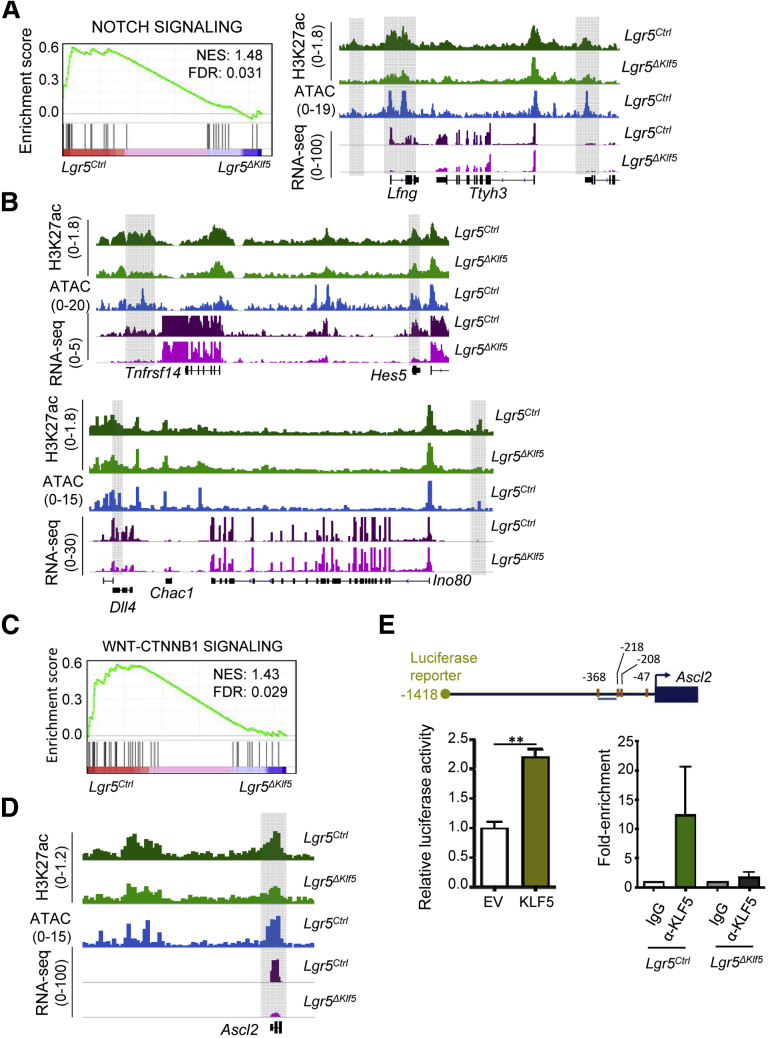
ISC self-renewal depends on WNT and NOTCH signaling,30 and integrative analysis of reduced messenger RNA and H3K27ac in Lgr5ΔKlf5 ISCs implicated KLF5 in regulation of selected genes in both pathways. Genes reduced in expression were enriched by GSEA for the NOTCH pathway (Figure 9A) and BETA analysis,31 independently revealed loss of H3K27ac within 50 kb of certain NOTCH pathway genes, including Lfng, Hes5, and Dll4, where coordinate losses of H3K27ac and messenger RNA were evident (Figures 9A and 9B). Similarly, genes activated by WNT signaling (β-catenin accumulation)32 were enriched in control Lgr5+ ISCs and reduced in Klf5-deleted ISCs (Figure 9C). Of note, H3K27ac losses occurred only at a subset of WNT target genes: diminished H3K27ac levels and significantly reduced messenger RNA were particularly apparent at Ascl2, a WNT target gene and ISC marker known to maintain the ISC compartment (Figure 9D).33 To verify the presumptive role of KLF5 in Ascl2 transcriptional control, we coexpressed a luciferase reporter construct carrying ∼1.4 kb of upstream Ascl2 sequence (Figure 9E) with a pMT3-KLF5 expression vector in RKO colorectal cancer cells, which express negligible KLF5 levels. Compared with an empty pMT3 vector, forced KLF5 expression increased luciferase activity significantly (Figure 9E), confirming the prediction that KLF5 activates Ascl2. Furthermore, JASPAR CORE 2018 vertebrate34 identified potential KLF5 binding sites in the 1.4-kb region upstream of Ascl2 (P < .001) (Figure 9E) and ChIP-qPCR with KLF5 antibody showed enrichment in control Lgr5+ ISCs but not in Klf5-null ISCs or in ChIP assays using an isotype control antibody (Figure 9E). Thus, the hundreds of KLF5-dependent cis-elements include the ISC-restricted Ascl2 promoter. As KLF5-dependent loss of H3K27ac associates particularly well with downregulated genes, including those related to NOTCH and WNT signaling, KLF5 activity upstream of selected genes in these pathways is likely responsible for maintaining ISC identity and self-renewal.
Figure 9.
H3K27ac-depleted regions enrich for NOTCH and WNT signaling pathway genes. (A) GSEA shows enrichment of NOTCH pathway genes in control Lgr5-EGFPhi cells. Representative tracks for ChIP-seq,28 ATAC-seq (blue), and RNA-seq (purple) at Lfng locus show loss of H3K27ac and expression in Lgr5ΔKlf5. (B) Representative IGV tracks for H3K27ac ChIP-seq,28 ATAC-seq (blue) and RNA-seq (purple) at NOTCH pathway genes Hes5 and Dll4. The gray shaded box marks promoters and regions of H3K27ac loss at putative enhancers. (C) WNT-CTNNB1 signaling genes are enriched in Lgr5EGFPhi cells as shown by GSEA. (D) IGV tracks for H3K27ac ChIP-seq,28 ATAC-seq (blue) and RNA-seq (purple) at Ascl2 locus show loss of H3K27ac and expression in Lgr5ΔKlf5. (E) Schematic represents ∼1.4-kb region upstream of Ascl2 TSS marked with potential KLF5 binding sites (orange vertical lines), ChIP-qPCR primer locations (blue horizontal line), and the length of luciferase promoter assay constructs. Luciferase assay in RKO cells showed significant increase in the promoter activity with overexpression of KLF5. EV, empty vector. Data are represented as mean ± SD, n = 8, ** P < .01 by Mann-Whitney U test. ChIP-qPCR for KLF5 in Lgr5EGFPhi cells from Lgr5Ctrl mice shows enrichment of KLF5 at the promoter of Ascl2. Rabbit IgG and Lgr5EGFPhi cells from Lgr5ΔKlf5 mice were used as negative control.
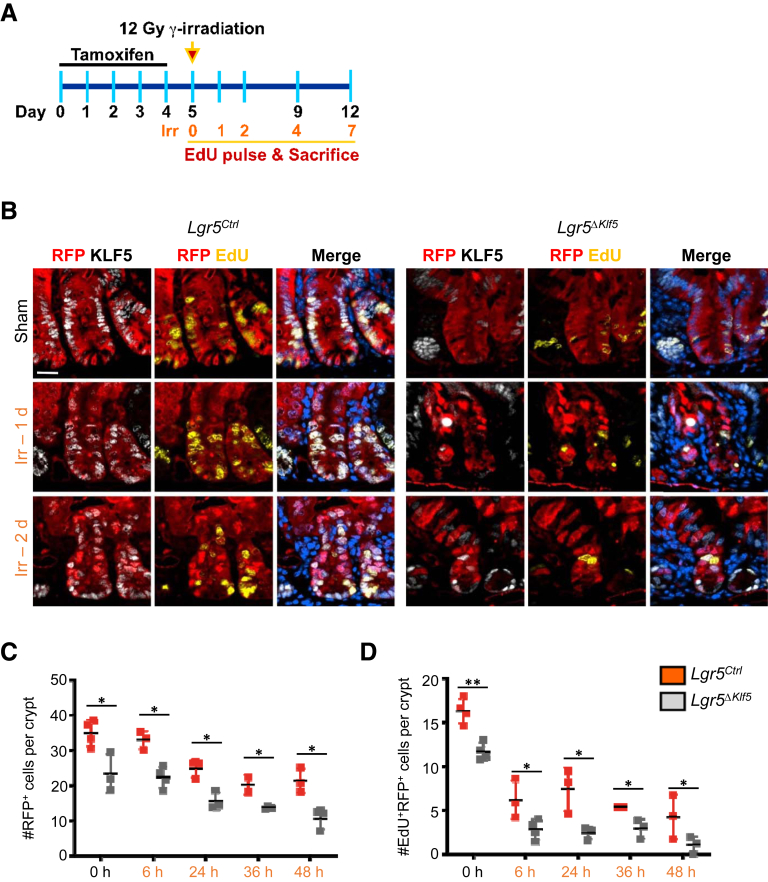
KLF5 Is Required for the Regenerative Response After Irradiation Injury
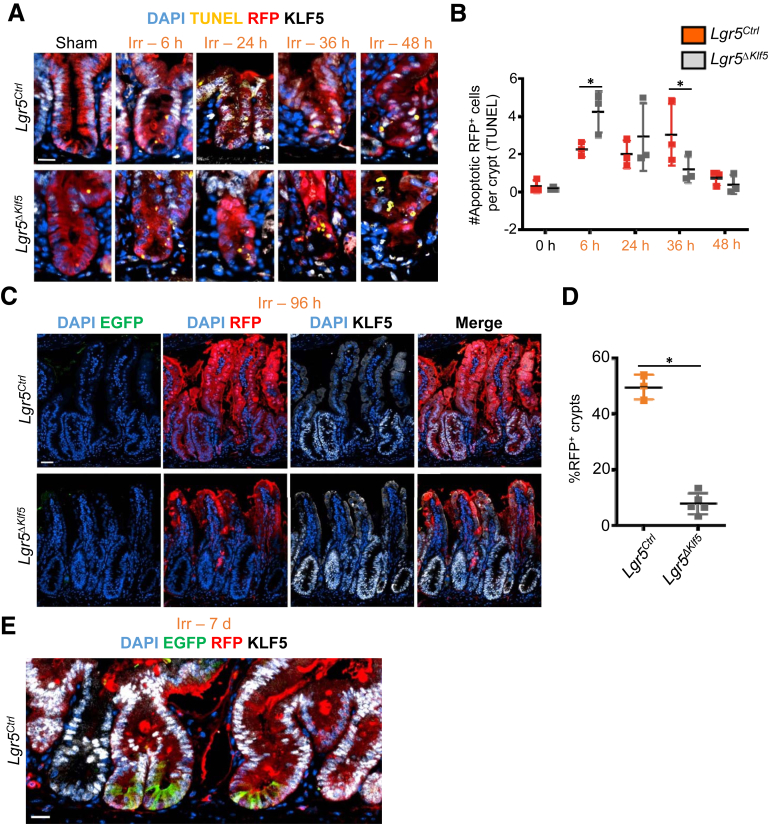
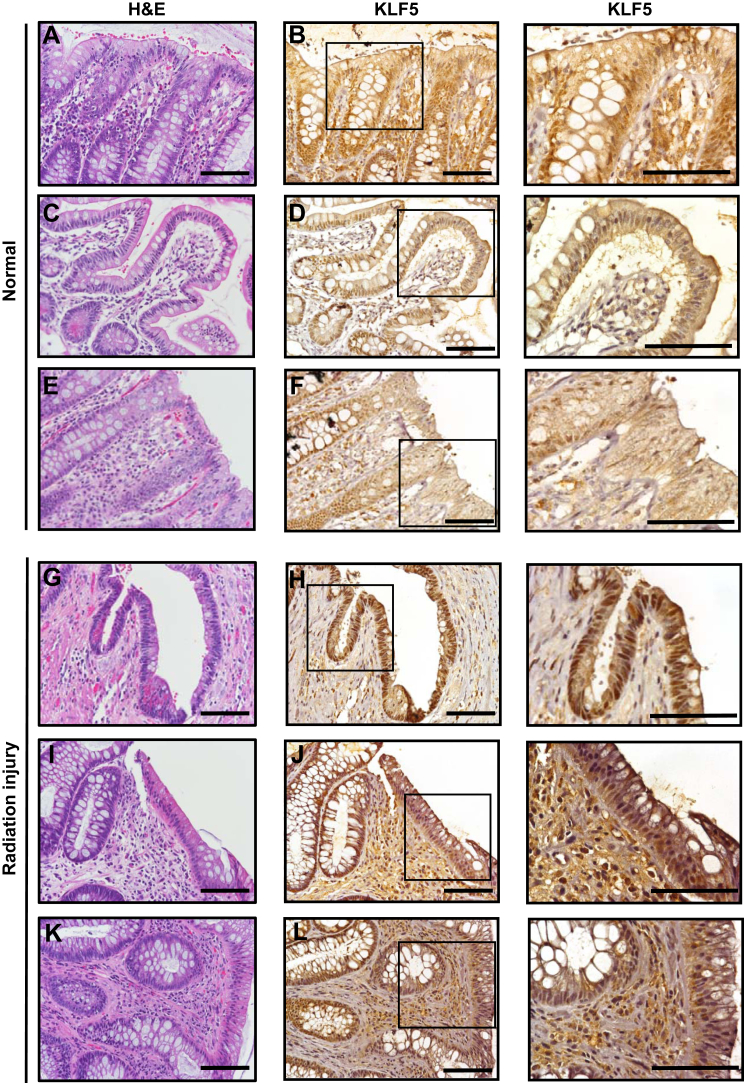
Intestinal epithelial regeneration following 12-Gy γ-irradiation injury in mice can be divided into 3 phases: apoptosis (0–48 hours), regeneration (72–96 hours), and normalization (after 96 hours).4 Multiple crypt cells populations are capable of dedifferentiating into ISC during the regenerative phase.4,5,35, 36, 37, 38, 39 To determine this capacity in Klf5-deleted crypt cells, we treated Lgr5Ctrl and Lgr5ΔKlf5 mice with 12-Gy whole-body γ-irradiation (Figure 10A). During the apoptotic phase, Klf5-deleted RFP+ crypt cells were consistently fewer in Lgr5ΔKlf5 mice compared with Lgr5Ctrl mice (Figures 10B and 10C), which may be due to lower EdU incorporation by Klf5-deleted RFP+ crypt cells (Figures 10B and 10D). Furthermore, Klf5-deleted RFP+ TA cells were more sensitive to apoptosis immediately after irradiation injury (Figures 11A and 11B). Ninety-six hours after irradiation, robust crypt regeneration was apparent in both Lgr5Ctrl and Lgr5ΔKlf5 mice, with the majority of cells expressing KLF5 (Figure 11C), but regenerating RFP+ crypts were markedly reduced in Lgr5ΔKlf5 mice in the setting of early apoptosis and decreased proliferation (Figures 11C and 11D). Lgr5+ ISCs appeared within regenerated RFP+ crypts 7 days after irradiation in Lgr5Ctrl mice, which demonstrates dedifferentiation of RFP+ precursors to ISCs (Figure 11E). Additionally, we performed hematoxylin and eosin (H&E) and KLF5 immunohistochemistry analysis of colonic or intestinal tissues obtained from control group (Figures 12A–F) and patients who underwent radiation treatments with a pathological diagnosis of radiation colitis or enteritis (Figures 12G–L). We observed that in control tissues KLF5 stain is limited to the 2/3 of the crypts while in tissues after irradiation KLF5 expression extends to the upper section of the crypts and to the surface of the colonic or intestinal epithelium. Collectively, these data indicate that KLF5 is required for crypt cells to dedifferentiate and regenerate the intestinal epithelium following radiation injury.
Figure 10.
Loss of KLF5 in intestinal stem cells and progenitors impairs the regenerative response of intestinal epithelial cells following γ-irradiation injury. (A) Experimental timeline. Lgr5Ctrl and Lgr5ΔKlf5 mice were injected with tamoxifen for 5 consecutive days and irradiated with 12-Gy γ-irradiation. (B) Representative immunofluorescence images of RFP, KLF5, EdU, and DAPI in the PSI crypts of Lgr5Ctrl and Lgr5ΔKlf5 mice treated with 3 hours EdU pulse. (C, D) Quantification of number of RFP+ cells (C) and EdU-incorporated RFP+ cells (D) per crypt. Scale bars represent 20 μm. Data are represented as mean ± SD, n = 4–5 mice per group, *P < .05, **P < .01 by (C, D) linear mixed regression models.
Figure 11.
KLF5 controls both short- and long-term regenerative response following radiation injury. (A) Representative immunofluorescence (IF) images of TUNEL, RFP, KLF5, and DAPI of the PSI crypts of Lgr5Ctrl and Lgr5ΔKlf5 mice 0, 6, 24, 36, or 48 hours following 12-Gy γ-irradiation. (B) Quantification of number of apoptotic RFP+ cells per crypt. Data are represented as mean ± SD, 20 crypts quantified per mouse, n = 3–5 mice per group. ∗P < .05 by linear mixed regression model. (C) Representative IF images of EGFP, RFP, KLF5, and DAPI in the PSI crypts of Lgr5Ctrl and Lgr5ΔKlf5 mice 4 days after γ-irradiation injury. (D) Quantification of percent regenerating RFP+ crypts in PSI of Lgr5Ctrl and Lgr5ΔKlf5 mice at day 4 after γ-irradiation injury. Data are represented as mean ± SD, n = 4–5 mice per group, *P < .05, **P < .01 by linear-mixed regression models Mann-Whitney U test. (E) Representative IF images of EGFP, RFP, KLF5, and DAPI of the PSI crypts of Lgr5Ctrl mice at day 7 postirradiation. Scale bars represent 20 μm.
Figure 12.
Increased KLF5 levels in the regions of human intestinal epithelium after radiation injury as compared with control. Representative images of (A, C, E) H&E and (B, D, F) and insets KLF5 immunohistochemistry stain from control human intestinal tissues. The control tissues have been characterized as benign/normal with no active disease (either Crohn’s disease, ulcerative colitis or colon cancer) by a pathologist. (G, I,K) H&E stain of regions of colonic or intestinal epithelium after radiation treatment, (H, J, L) with insets KLF5 immunohistochemistry stain of regions of colonic or intestinal epithelium after irradiation. The tissues shown were given the diagnosis of (G, H) radiation colitis, (I, J) radiation enteritis, and (K, L) radiation colitis. Scale bar = 100 μM.
Discussion
We report that KLF5 controls ISC proliferation and stemness, preventing their premature differentiation along the enterocyte lineage. Because previous studies implicate KLF5 in promoting intestinal epithelial cell proliferation,15,18,20 the increased proliferation of Klf5-null ISC is an unexpected finding. Accelerated proliferation results in upward migration of EdU-labelled Klf5-null progenitors and ultimately ISCs exhaustion from the crypt bottom. Interestingly, the initial burst of proliferation of Klf5-null cells is not maintained in the TA zone, supporting the notion that KLF5 has a precursor-specific function as a pro-proliferative factor. A recent study suggested that WNT signaling suppression induces conversion of ISCs into TA cells, resulting in accelerated proliferation.40 Based on transcriptome profile of Klf5-null ISCs that showed reduction of WNT target genes, we speculate that Klf5-null ISCs undergo premature differentiation to rapidly-cycling enterocyte precursors via suppression of WNT signaling pathway. However it remains possible that Klf5-null ISCs spontaneously lose Lgr5 expression, thus further contributing to loss of ISCs.
Moreover, KLF5 is expressed in the majority of crypt cells, which have shown to contribute to tissue regeneration postinjury,41 and we find that this regenerative capacity is abrogated in the absence of KLF5. Since KLF5 is required to maintain the proliferative capacity of these cells, it may also be required for proliferation and dedifferentiation during regeneration. Recent studies have suggested that activated NOTCH signaling stimulates Paneth cell plasticity during injury-induced regeneration.42,43 While KLF5 is not expressed in Paneth cells during homeostasis, the majority of cells within regenerative crypts express KLF5, indicating that KLF5 remains a critical player in the regenerative response. Furthermore, this response may be facilitated via KLF5-mediated NOTCH signaling regulation in other precursors.
Accessible chromatin and active histone modifications, such as H3K27ac, mark TF-bound cis-elements that control cell-specific genes. KLF5-dependent genes were strongly correlated with KLF5-dependent enhancers enriched for the cognate binding motif, indicating that at least part of KLF5’s mechanism is to maintain TF access and active histone marks at selected ISC enhancers. Among the panoply of bona fide target genes, KLF5-dependent enhancers control selected genes in the WNT and NOTCH pathways. KLF5 thus maintains stem cell homeostasis in part by preserving cis-regulatory elements upstream of these ISC signals, which may be also required in the dedifferentiation process post-injury. In contrast, genes that gain expression in Klf5-null ISCs are mature villus genes associated with enhancers that lack KLF5 motif enrichment, and these are likely not direct transcriptional targets. Furthermore, we observed that Klf5-null ISCs fail to produce secretory lineages in the context of reduction of NOTCH signaling and WNT target genes in ISCs. As we observed a reduction in Atoh1 expression while Hes1 expression did not change in Lgr5EGFPlo cells, it is possible that KLF5 has unique functions in precursors and would be of interest to explore its potential role in lineage determination of precursors.
In summary, our study has shown that KLF5 is required for ISC identity and functions through preserving cis-regulatory elements of ISC genes to regulate transcription, and is required in tissue regeneration postinjury and dedifferentiation of precursors into ISCs.
Materials and Methods
All authors had access to the study data and had reviewed and approved the final manuscript.
Mice
Klf5fl/fl,44 Lgr5EGFP-IRES-creERT2,45 and Rosa26LSLtdTomato46 mice were described previously and Lgr5EGFP-IRES-creERT2 and Rosa26LSLtdTomato mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Lgr5Ctrl and Lgr5ΔKlf5 mice were generated by cross-breeding. Animal studies were carried out in accordance with the Animal Research: Reporting In Vivo Experiments guidelines and were approved by the Stony Brook University Institutional Animal Care and Use Committee. Animals were kept on ad libitum normal chow and water. Female and male of mice at 8–12 weeks of age were used in this study. Animals were treated and sacrificed during the light cycle hours. To induce recombination, mice were injected intraperitoneally with tamoxifen dissolved in corn oil (10 mg/mL) (Sigma-Aldrich, St. Louis, MO) at 1 mg per injection for 2 or 5 days. Mice were sacrificed at 0, 2, 5, 9, 12, 19, 33, or 61 days after the initial tamoxifen injection, and small intestines were harvested for processing. Before euthanasia at 3 or 24 hours, all mice were injected with 100 μg of EdU (Santa Cruz Biotechnology, Dallas, TX) dissolved in 1:5 of DMSO and H2O. For γ-irradiation injury model, mice were exposed to total body γ-irradiation on day 5 after tamoxifen treatment with a dose rate of 0.8 Gy/min for total of 12 Gy. The mice were sacrificed at 0, 6, 24, 36, 48, 96 hours, and 7 days after γ-irradiation, and small intestines were harvested for processing.
Samples From Patients
Surgical specimens of resected colorectal cancer specimen obtained from Stony Brook University and SUNY Downstate were used in this study. A total of 17 specimens were processed for H&E and immunohistochemistry. The protocol for the sample collection has been originally approved by the Institutional Review Board by the State University of New York at Stony Brook on October 17, 2014 (CORIHS 2014-2821-F) and qualified for a waiver under the Federal Law of Department of Health and Human Services per article 45CFR46.116.d.
H&E Staining
Histology of sections was observed on stained 5-μm sections that were fixed, paraffin embedded, deparaffinized, and rehydrated, as mentioned previously. Then, they were stained with Hematoxylin Stain Solution, Gill 3 (Ricca Chemical Company, Pocomoke City, MD) and Eosin Y (Sigma-Aldrich). Sections were dehydrated in an increasing series of ethanol baths (70%, 95%, and 100%), cleared in xylene, and mounted with Cytoseal XYL xylene-based mounting media (Thermo Fisher Scientific, Waltham, MA). The H&E stains were used for histopathological assessment.
Immunofluorescence and Immunohistochemistry Staining
Tissue fixation and stain was done as described previously.15 The list of antibodies used in this study is presented in Table 1.
Table 1.
List of Antibodies Used in the Study
| Antibody | Source | Catalog # |
|---|---|---|
| Goat polyclonal anti-KLF5 (used at 1:300) | R&D Systems | Cat# AF3758 |
| Rabbit polyclonal anti-KLF5 (used at 1:300) | Abcam | Cat# Ab137676 |
| Chicken polyclonal anti-GFP (used at 1:500) | Aves Labs | Cat# GFP-1020 |
| Rabbit polyclonal anti-RFP (used at 1:300) | Rockland | Cat# 600-401-379 |
| Goat polyclonal anti-GFP (used at 1:300) | Rockland | Cat# 200-101-379 |
| Rabbit polyclonal anti-Cleaved caspase-3 (Used at 1:200) | Cell Signaling | Cat# 9661 |
| Rabbit polyclonal anti-E-cadherin (Used at 1:200) | Cell Signaling | Cat# 3195 |
| Rabbit polyclonal anti-Mucin 2 (Used at 1:100) | Santa Cruz Biotech. | Cat# SC-15334 |
| Rabbit polyclonal anti-Chromogranin A (Used at 1:200) | Abcam | Cat# 1773-1 |
| Rabbit polyclonal anti-Lysozyme (Used at 1:200) | Dako | Cat# A0099 |
| Mouse monoclonal anti-Vil1 | BD Bioscience | Cat# 610358 |
| AF647-conjugated goat polyclonal anti-bovine IgG (Used at 1:300) | Jackson ImmunoResearch | Cat# 101-605-003 |
| Bovine polyclonal anti-goat IgG (Used at 1:300) | Jackson ImmunoResearch | Cat# 805-005-180 |
| Cy3-conjugated donkey polyclonal anti-mouse IgG | Jackson ImmunoResearch | Cat# 715-165-150 |
| Mouse polyclonal anti-rabbit IgG | Jackson ImmunoResearch | Cat# 211-005-109 |
| AF488-conjugated donkey polyclonal anti-chicken | Jackson ImmunoResearch | Cat# 703-545-155 |
| Rabbit IgG, polyclonal – isotype control | Abcam | Cat# ab171870 |
EdU and TUNEL Staining
EdU-labeled cells were stained using the Click-IT Plus EdU Imaging kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. TUNEL staining was performed according to the manufacturer’s protocol (Sigma-Aldrich).
Cell and Crypt Counting
Countable crypts were selected based on the presence of 3 to 5 Paneth cells at the bottom of the crypt using red fluorescent protein (RFP) or enhanced green fluorescent protein (EGFP) immunofluorescent staining. The numbers were represented as average number of stained cells per crypt, or percent of stained cells of total number of cells. For average number of crypt cells, a minimum of 20 crypts were counted per mouse for n = 3–4. For percent of EdU-incorporated cells, a total of 250 cells were counted per mouse for n = 3.
Cell Isolation for Enteroid Culture
Proximal small intestine was harvested from mice injected with tamoxifen for 5 consecutive days. Intestinal epithelial cells were dissociated as previously described.47 RFP+EGFPhi cells were sorted by flow cytometry (BD FACSARIA III; BD Biosciences, San Jose, CA) (Figure 4A) and embedded in Matrigel (Corning, Corning, NY). Enteroid culture medium was prepared using L-WRN cells as previously described,48 and supplemented with transforming growth factor β inhibitor A83-01 (500 nM) (Tocris Bioscience, Bristol, United Kingdom) and antibiotic cocktail Primocin (100 μg/mL) (Thermo Fisher Scientific). GSK3β inhibitor CHIR99021 (10 μM) (Tocris) and ROCK inhibitor Y-27632 (10 μM) (Sigma-Aldrich) were also added during the first 2 days of culture. The media were changed every 2 days. Live enteroids were imaged using Olympus (Center Valley, PA) phase contrast microscope. At day 6 of the enteroid culture, number of enteroids per well were quantified to measure enteroid-forming efficiency.
Enteroid Paraffin Section Preparation
Enteroids were washed with phosphate-buffered saline. Matrigel from multiple wells was gently scraped and dissolved with Cell Recovery Solution (Corning) on an orbital shaker (250 rpm) at 4°C for 30 minutes. Enteroids were centrifuged at 300 g for 10 minutes at 4°C, suspended in HistoGel (Thermo Fisher Scientific), moved to a disposable base mold, and placed on ice for 10 minutes. Hardened gel was fixed for 24 hours and processed for paraffin embedding.
Enteroid Whole-Mount Immunofluorescent Staining and Nuclei Quantification
Whole-mount immunofluorescent staining was performed as previously described.49 Three hours before fixation, enteroids were treated with 10-μM EdU (Thermo Fisher Scientific). Images were obtained using Leica Inverted Confocal Sp8 (Leica Microsystems Inc, Buffalo Grove, IL) equipped with a White Light Laser and a Leica HyD Detector. Number of nuclei of enteroids at day 2 were quantified using confocal images.
Cell Isolation for Total RNA Analysis
Mice were injected with tamoxifen for 5 consecutive days. Proximal small intestines were harvested, and cells were isolated and dissociated as previously described.24 Lgr5EGFPhi and Lgr5EGFPlo cells were sorted by flow cytometry (BD FACSARIA III).
RNA Isolation and Gene Expression Analysis by RT-qPCR
Total RNA was extracted using RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. RNase-Free DNase Set (Qiagen) was used to remove DNA. Total RNA was used for RT-qPCR and RNA-Sequencing. cDNA was synthesized using the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. RT-qPCR assay was performed using TaqMan Gene Expression Master Mix (Thermo Fisher Scientific) and QuantStudio 3 qPCR machine (Thermo Fisher Scientific). List of the primers used in this study is listed in Table 2.
Table 2.
List of TaqMan Primers and ChIP-qPCR Primers Used in the Study
| Primer | Sequence/TaqMan Gene Expression Primers Catalog Number |
|---|---|
| Klf5 | Cat. #: 4331182; Mm00438890_m1 |
| Lgr5 | Cat. #: 4331182; Mm00438905_m1 |
| Ascl2 | Cat. #: 4331182; Mm01268891_g1 |
| Olfm4 | Cat. #: 4331182; Mm01320260_m1 |
| Smoc2 | Cat. #: 4331182; Mm00491553_m1 |
| Msi1 | Cat. #: 4331182; Mm01203522_m1 |
| Axin2 | Cat. #: 4331182; Mm00443610_m1 |
| Ccnd1 | Cat. #: 4331182; Mm00432359_m1 |
| Mki67 | Cat. #: 4331182; Mm01278617_m1 |
| Fabp1 | Cat. #: 4453320; Mm00444340_m1 |
| Fabp2 | Cat. #: 4331182; Mm00433188_m1 |
| VIl1 | Cat. #: 4331182; Mm00494146_m1 |
| Atoh1 | Cat. #: 4448892; Mm00476035_s1 |
| Chga | Cat. #: 4448892; Mm00514341_m1 |
| Muc2 | Cat. #: 4448892; Mm01276696_m1 |
| Lyz1 | Cat. #: 4448892; Mm00657323_m1 |
| Reg1 | Cat. #: 4448892; Mm00485651_m1 |
| Reg3b | Cat. #: 4331182; Mm00440616_g1 |
| Hes1 | Cat. #: 4448892; Mm01342805_m1 |
| Alpi | Cat. #: 4448892; Mm00476035_s1 |
| Hprt | Cat. #: 4448490; Mm03024075_m1 |
| Ascl2 Promoter | F: CTGGGCACCTGTACCCATTTA |
| R: TCTCTCAGGTCAGGGCAACC |
ChIP-qPCR, chromatin immunoprecipitation assay with quantitative polymerase chain reaction.
RNA Library Preparation and Sequencing
RNA quality (RNA Integrity Number >7.0) was measured using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Total 500 ng of RNA was used to prepare RNA-seq libraries. The RNA library was prepared and sequenced as previously described.50 Quality of the sequencing data was assessed through multiple metrics, including number of pass filter reads per sample, base quality per cycle, percent base content per cycle, and the overall distribution of base quality scores.
RNA-seq Analysis
The reads were aligned with STAR (version 2.4.0c),51 and genes annotated in Gencode vM5 were quantified with featureCounts (v1.4.3-p1),52 and transcript abundance was quantified using kallisto.53 Normalization and differential expression was done with the Bioconductor package DESeq2.54 QC metrics were computed with a mix of RSeQC,55 picard (v1.83), and featureCounts. P values were adjusted for multiple comparisons using Bonferroni correction. Significant genes have a minimum 1.5-fold change and adjusted P value of .05. Gene Ontology terms of biological processes enriched in differentially expressed genes were identified using GSEA,56 Molecular Signatures Database (MsigDB). The villus-enriched gene list was obtained from a previously published RNA-seq data comparing transcriptomes of ISCs and villus cells, with differentially expressed genes with fold change ≥3.24
ChIP-seq
Lgr5EGFPhi cells were collected as for ChIP-PCR. ChIP-seq was performed as in.29 Briefly, cross-linked cells were lysed and sonicated in RIPA lysis buffer to obtain 200- to 800-bp chromatin fragments. Chromatin were incubated overnight at 4°C with H3K27ac antibody followed by Protein A and G Dynabeads (10002D and 10004D) (Thermo Fisher Scientific) at 2 hours. Chromatin-antibody complex bound beads were washed twice in the sonication buffer, once in high-salt buffer, once in LiCl buffer, and once in Tris-EDTA, pH 8. Cross-links were reversed overnight by incubation at 65°C followed by treatment with Proteinase K (25530049; Thermo Fisher Scientific) for 1 hour at 55°C. DNA was purified with MinElute PCR purification kit (28004; Qiagen). Libraries were prepared using ThruPLEX DNA-seq kit (R400427; Rubicon Genomics, Ann Arbor, MI), purified using Ampure XP beads (Beckman Coulter, A63881) and sequenced on Illumina HiSeq X (Illumina, San Diego, CA) to obtain 150-bp paired-end reads.
ChIP-seq Data Analysis
The first mates of paired-end reads were used as single-end reads for further analysis. Reads were aligned to the mouse reference genome mm9 (NCBI Build 37) or mm10 (GRCm38) using Bowtie2.57 diffReps27 was used for whole genome differential analysis of H3K27ac in ISCs isolated from Lgr5Ctrl and Lgr5ΔKlf5 mice. multiBamSummary module of deepTools258 was used to determine read coverage for duplicate bam files and correlation plots were created in R. HOMER v4.8.259 was used for motif analysis at differential regions detected by diffReps. For representation, ChIP and RNA-seq bigwigs were created using bamCoverage in deepTools2. For comparative visualization, experimental and control groups were quantile-normalized using Haystack.60 Heatmaps were plotted using deepTools2 and bigwig traces depicted on Integrative Genomics Viewer (IGV).61 BETA31was used to quantify promoter/enhancer gene associations for differential regions using ±50-kb distance limit from a TSS, a significance threshold of FDR-adjusted q < .05 for differential gene expression in Lgr5Ctrl and Lgr5ΔKlf5 ISCs, and other default parameters.
ChIP-PCR
Approximately 1 × 106 Lgr5EGFPhi cells pooled from 2–4 mice were used for ChIP-PCR. ChIP was performed as previously described,62 with a few modifications. Cells were cross-linked for 15 minutes with 1% formaldehyde, and cross-linking was stopped by adding glycine at a final concentration of 125 mM. Cells were washed once with cold phosphate-buffered saline. Chromatin digested with micrococcal nuclease was incubated with 1.5 μg of anti-KLF5 antibody (Abcam, Cambridge, United Kingdom) or rabbit IgG (Abcam), precipitated using Protein A– and Protein G–coated Dynabeads (Thermo Fisher Scientific). Beads were washed 6–8 times. Immunoprecipitated chromatin fragments were reverse cross-linked in elution buffer (0.1M NaHCO3, 1% sodium dodecyl sulfate) with NaCl and RNase A at 65°C for 4 hours. DNA was treated with Protease A for 1 hour at 60°C, extracted using UltraPure Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) (Thermo Fisher Scientific) in MaXtract High Density tubes (Qiagen), and purified using Agencourt Ampure XP DNA purification kit (Beckman Coulter, Brea, CA). Potential binding sites for KLF5 were identified using Eukaryotic Promoter Database63 and JASPAR CORE 2018 vertebrate.34 The list of the primers used in this study is provided in Table 2.
Luciferase Assay
RKO colorectal cancer cell line was purchased from American Type Culture Collection (CRL-2577) and cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. RKO cells were transfected with pEGFP-N1 plasmids (6085-1; Clontech). The signal from EGFP is used as a control. Cells were seeded in 96-well plate at 5 × 104 cells per well. Gaussia luciferase reporter construct bearing Ascl2 promoter (MPRM39895-PG02; GeneCopoeia, Rockville, MD) was transfected with pMT3 or pMT3-KLF5-HA using Lipofectamine 2000 (Thermo Fisher Scientific), according to manufacturer’s instructions. Vectors pMT3 and pMT3-KLF5-HA were previously described.64 Luciferase activities were determined at 72 hours after transfection using Secrete-Pair Gaussia Luciferase Assay Kit (GeneCopoeia).
Statistics
Mann-Whitney U test and linear mixed regression models were performed using GraphPad Prism version 7.0 for Windows (GraphPad Software, San Diego, CA) and SAS 9.4 (SAS Institute, Cary, NC), respectively. Log and square transformations were applied to the outcomes as needed to ensure the validity of assumptions of normal residuals for linear mixed regression models. All animal studies used tissues from at least 3 animals (n ≥ 3). To ensure quality and reproducibility of cell purification, all experiments involving FACS isolation of single Lgr5EGFP+ cells were done with at least 3 mice (n ≥ 3), with multiple individual experiments. ChIP-qPCR used approximately 1 × 106 Lgr5EGFPhi cells per sample pooled from 2–4 mice. Luciferase assay was performed with at least 7 wells per group, with multiple individual experiments. A P < .05 was considered significant.
Acknowledgments
The authors thank Jie Yang, Donglei Yin, and Lizhou Nie (Department of Applied Mathematics and Statistics, Stony Brook University) for assistance with RNA sequencing and statistical analyses, and Rebecca C. Connor and Todd P. Rueb (Research Flow Cytometry Core, Department of Pathology, Stony Brook University) for assistance with flow cytometry.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by National Institutes of Health grants R01DK052230 (to Vincent W. Yang), R01CA084197 (to Vincent W. Yang), R01DK081113 (to Ramesh A. Shivdasani), R01DK082889 (to Ramesh A. Shivdasani), and F32DK115080 (to Madhurima Saxena).
Supplementary Material
References
- 1.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 2.Snippert H.J., van der Flier L.G., Sato T., van Es J.H., van den Born M., Kroon-Veenboer C., Barker N., Klein A.M., van Rheenen J., Simons B.D., Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 3.Lopez-Garcia C., Klein A.M., Simons B.D., Winton D.J. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 4.Kim C.K., Yang V.W., Bialkowska A.B. The Role of Intestinal Stem Cells in Epithelial Regeneration Following Radiation-Induced Gut Injury. Curr Stem Cell Rep. 2017;3:320–332. doi: 10.1007/s40778-017-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan K.S., Chia L.A., Li X., Ootani A., Su J., Lee J.Y., Su N., Luo Y., Heilshorn S.C., Amieva M.R., Sangiorgi E., Capecchi M.R., Kuo C.J. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tetteh P.W., Basak O., Farin H.F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J.H., van Oudenaarden A., Clevers H. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong V.W., Stange D.E., Page M.E., Buczacki S., Wabik A., Itami S., van de Wetering M., Poulsom R., Wright N.A., Trotter M.W., Watt F.M., Winton D.J., Clevers H., Jensen K.B. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol. 2012;14:401–408. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhnert F., Davis C.R., Wang H.T., Chu P., Lee M., Yuan J., Nusse R., Kuo C.J. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan K.S., Janda C.Y., Chang J., Zheng G.X.Y., Larkin K.A., Luca V.C., Chia L.A., Mah A.T., Han A., Terry J.M., Ootani A., Roelf K., Lee M., Yuan J., Li X., Bolen C.R., Wilhelmy J., Davies P.S., Ueno H., von Furstenberg R.J., Belgrader P., Ziraldo S.B., Ordonez H., Henning S.J., Wong M.H., Snyder M.P., Weissman I.L., Hsueh A.J., Mikkelsen T.S., Garcia K.C., Kuo C.J. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi Z., Li Y., Zhao B., Xu C., Liu Y., Li H., Zhang B., Wang X., Yang X., Xie W., Li B., Han J.J., Chen Y.G. BMP restricts stemness of intestinal Lgr5(+) stem cells by directly suppressing their signature genes. Nat Commun. 2017;8:13824. doi: 10.1038/ncomms13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud A., Grosse A.S., Gumucio D.L., Ernst S.A., Tsai Y.H., Dempsey P.J., Samuelson L.C. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C.K., He P., Bialkowska A.B., Yang V.W. SP and KLF transcription factors in digestive physiology and diseases. Gastroenterology. 2017;152:1845–1875. doi: 10.1053/j.gastro.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nandan M.O., Yang V.W. The role of Kruppel-like factors in the reprogramming of somatic cells to induced pluripotent stem cells. Histol Histopathol. 2009;24:1343–1355. doi: 10.14670/hh-24.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nandan M.O., Ghaleb A.M., Bialkowska A.B., Yang V.W. Kruppel-like factor 5 is essential for proliferation and survival of mouse intestinal epithelial stem cells. Stem Cell Res. 2015;14:10–19. doi: 10.1016/j.scr.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McConnell B.B., Yang V.W. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConnell B.B., Kim S.S., Yu K., Ghaleb A.M., Takeda N., Manabe I., Nusrat A., Nagai R., Yang V.W. Kruppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology. 2011;141:1302–1313. doi: 10.1053/j.gastro.2011.06.086. 1313.e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nandan M.O., Ghaleb A.M., Liu Y., Bialkowska A.B., McConnell B.B., Shroyer K.R., Robine S., Yang V.W. Inducible intestine-specific deletion of Kruppel-like factor 5 is characterized by a regenerative response in adult mouse colon. Dev Biol. 2014;387:191–202. doi: 10.1016/j.ydbio.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Chidgey M., Yang V.W., Bialkowska A.B. Kruppel-like factor 5 is essential for maintenance of barrier function in mouse colon. Am J Physiol Gastrointest Liver Physiol. 2017;313:G478–G491. doi: 10.1152/ajpgi.00172.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakaya T., Ogawa S., Manabe I., Tanaka M., Sanada M., Sato T., Taketo M.M., Nakao K., Clevers H., Fukayama M., Kuroda M., Nagai R. KLF5 regulates the integrity and oncogenicity of intestinal stem cells. Cancer Res. 2014;74:2882–2891. doi: 10.1158/0008-5472.CAN-13-2574. [DOI] [PubMed] [Google Scholar]
- 21.Mariadason J.M., Nicholas C., L'Italien K.E., Zhuang M., Smartt H.J., Heerdt B.G., Yang W., Corner G.A., Wilson A.J., Klampfer L., Arango D., Augenlicht L.H. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 22.San Roman A.K., Tovaglieri A., Breault D.T., Shivdasani R.A. Distinct processes and transcriptional targets underlie CDX2 requirements in intestinal stem cells and differentiated villus cells. Stem Cell Rep. 2015;5:673–681. doi: 10.1016/j.stemcr.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S., Myant K., Versteeg R., Sansom O.J., van Es J.H., Barker N., van Oudenaarden A., Mohammed S., Heck A.J., Clevers H. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jadhav U., Nalapareddy K., Saxena M., O'Neill N.K., Pinello L., Yuan G.C., Orkin S.H., Shivdasani R.A. Acquired tissue-specific promoter bivalency is a basis for PRC2 necessity in adult cells. Cell. 2016;165:1389–1400. doi: 10.1016/j.cell.2016.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., Boyer L.A., Young R.A., Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen L., Shao N.Y., Liu X., Maze I., Feng J., Nestler E.J. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jadhav U., Saxena M., O'Neill N.K., Saadatpour A., Yuan G.C., Herbert Z., Murata K., Shivdasani R.A. Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell. 2017;21:65–77.e5. doi: 10.1016/j.stem.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian H., Biehs B., Chiu C., Siebel C.W., Wu Y., Costa M., de Sauvage F.J., Klein O.D. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 2015;11:33–42. doi: 10.1016/j.celrep.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C.A., Zhang Y., Liu X.S. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8:2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuijers J., Junker J.P., Mokry M., Hatzis P., Koo B.K., Sasselli V., van der Flier L.G., Cuppen E., van Oudenaarden A., Clevers H. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16:158–170. doi: 10.1016/j.stem.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R., Bessy A., Cheneby J., Kulkarni S.R., Tan G., Baranasic D., Arenillas D.J., Sandelin A., Vandepoele K., Lenhard B., Ballester B., Wasserman W.W., Parcy F., Mathelier A. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montgomery R.K., Carlone D.L., Richmond C.A., Farilla L., Kranendonk M.E., Henderson D.E., Baffour-Awuah N.Y., Ambruzs D.M., Fogli L.K., Algra S., Breault D.T. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., Guo Y., Shyr Y., Aronow B.J., Haigis K.M., Franklin J.L., Coffey R.J. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li N., Nakauka-Ddamba A., Tobias J., Jensen S.T., Lengner C.J. Mouse label-retaining cells are molecularly and functionally distinct from reserve intestinal stem cells. Gastroenterology. 2016;151:298–310.e7. doi: 10.1053/j.gastro.2016.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asfaha S., Hayakawa Y., Muley A., Stokes S., Graham T.A., Ericksen R.E., Westphalen C.B., von Burstin J., Mastracci T.L., Worthley D.L., Guha C., Quante M., Rustgi A.K., Wang T.C. Krt19(+)/Lgr5(-) Cells are radioresistant cancer-initiating stem cells in the colon and intestine. Cell Stem Cell. 2015;16:627–638. doi: 10.1016/j.stem.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barriga F.M., Montagni E., Mana M., Mendez-Lago M., Hernando-Momblona X., Sevillano M., Guillaumet-Adkins A., Rodriguez-Esteban G., Buczacki S.J.A., Gut M., Heyn H., Winton D.J., Yilmaz O.H., Attolini C.S., Gut I., Batlle E. Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell. 2017;20:801–816.e7. doi: 10.1016/j.stem.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabiri Z., Greicius G., Zaribafzadeh H., Hemmerich A., Counter C.M., Virshup D.M. Wnt signaling suppresses MAPK-driven proliferation of intestinal stem cells. J Clin Invest. 2018;128:3806–3812. doi: 10.1172/JCI99325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santos A.J.M., Lo Y.H., Mah A.T., Kuo C.J. The intestinal stem cell niche: homeostasis and adaptations. Trends Cell Biol. 2018;28:1062–1078. doi: 10.1016/j.tcb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones J.C., Brindley C.D., Elder N.H., Myers M.G., Jr., Rajala M.W., Dekaney C.M., McNamee E.N., Frey M.R., Shroyer N.F., Dempsey P.J. Cellular plasticity of Defa4(Cre)-expressing Paneth cells in response to notch activation and intestinal injury. Cell Mol Gastroenterol Hepatol. 2019;7:533–554. doi: 10.1016/j.jcmgh.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu S., Tong K., Zhao Y., Balasubramanian I., Yap G.S., Ferraris R.P., Bonder E.M., Verzi M.P., Gao N. Paneth cell multipotency induced by notch activation following injury. Cell Stem Cell. 2018;23:46–59.e5. doi: 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shindo T., Manabe I., Fukushima Y., Tobe K., Aizawa K., Miyamoto S., Kawai-Kowase K., Moriyama N., Imai Y., Kawakami H., Nishimatsu H., Ishikawa T., Suzuki T., Morita H., Maemura K., Sata M., Hirata Y., Komukai M., Kagechika H., Kadowaki T., Kurabayashi M., Nagai R. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8:856–863. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 45.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 46.Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., Lein E.S., Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 48.Miyoshi H., Stappenbeck T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Rourke K.P., Dow L.E., Lowe S.W. Immunofluorescent staining of mouse intestinal stem cells. Bio Protoc. 2016;6 doi: 10.21769/bioprotoc.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He P., Yang J.W., Yang V.W., Bialkowska A.B. Kruppel-like factor 5, increased in pancreatic ductal adenocarcinoma, promotes proliferation, acinar-to-ductal metaplasia, pancreatic intraepithelial neoplasia, and tumor growth in mice. Gastroenterology. 2018;154:1494–1508.e13. doi: 10.1053/j.gastro.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 53.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 54.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Wang S., Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 56.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinello L., Xu J., Orkin S.H., Yuan G.C. Analysis of chromatin-state plasticity identifies cell-type-specific regulators of H3K27me3 patterns. Proc Natl Acad Sci U S A. 2014;111:E344–E353. doi: 10.1073/pnas.1322570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brind'Amour J., Liu S., Hudson M., Chen C., Karimi M.M., Lorincz M.C. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- 63.Dreos R., Ambrosini G., Groux R., Cavin Perier R., Bucher P. The eukaryotic promoter database in its 30th year: focus on non-vertebrate organisms. Nucleic Acids Res. 2017;45:D51–D55. doi: 10.1093/nar/gkw1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nandan M.O., Yoon H.S., Zhao W., Ouko L.A., Chanchevalap S., Yang V.W. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23:3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.